Abstract
The aim of the present study was to evaluate the occurrence of Salmonella spp., Verotoxigenic E. Coli (VTEC) and enumerate E. coli in shellfish (Mytilus galloprovincialis and Ruditapes decussatus) collected before and after depuration from two class B harvesting areas located in Sardinia (Italy). All the samples were analyzed for Salmonella spp. detection according to European Commission Regulation (EC) 2073/2005 and examined using the five tube Most-Probable-Number (MPN) method for enumeration of E. coli in accordance with the European Union reference method ISO 16649-3:2015. E. coli VTEC was investigated following a direct multiplex Polymerase Chain Reaction (PCR) screening test. The enumeration of E. coli met the European law limit for Class A areas of 230 MPN/100g. The averaged enumeration of E. coli in samples of M. galloprovincialis and R. decussatus collected at the harvesting time was 39 and 37 MPN/100 g respectively. The average contamination levels in samples collected after purification were 58 MPN/100g (M. galloprovincialis) and 32 MPN/100 g (R. decussatus). E. coli VTEC was not detected, on the contrary, Salmonella ser. Typhimurium was detected in one sample of M. galloprovincialis and in one sample of R. decussatus collected at the harvesting time. No significant associations were observed between E. coli levels in shellfish and environmental parameters of water or with the detection of Salmonella ser. Typhimurium in M. galloprovincialis and R. decussatus samples. Nevertheless, the occurrence of Salmonella ser. Typhimurium, involved in human infection outbreaks, should be considered a potential risk for consumers.
Key words: Mytilus galloprovincialis, Ruditapes decussatus; Purification, E. coli, Salmonella ser. Typhimurium.
Introduction
The evaluation of shellfish safety is based entirely on the use of food safety criteria laid down in Commission Regulation (EC) 2073/2005 and Commission Regulation (EU) 2285/2015: absence of Salmonella spp. in 25g of flesh and intervalvular liquid and an upper limit of 230 MPN Escherichia coli/100 g of flesh and intervalvular liquid in 80% of the samples. The 20% of the samples may contain E. coli between 230 and 700 MPN/100g. It is well known that shellfish contamination occurs because bivalve molluscs are filter-feeding animals that selectively filter and accumulate small particles of phytoplankton, zooplankton, viruses, bacteria and inorganic matter from the environment (Carella et al., 2010; Leoni et al., 2017). This highlights the role of shellfish as vehicle for several hazards that could result in products that are unsuitable to guarantee the safety of consumers, particularly if live bivalve molluscs are consumed raw or lightly cooked (Rubini et al., 2018; Sferlazzo et al., 2018). Bivalve molluscs transmitted illness can be either due to indigenous bacteria as Vibrio species (Smaldone et al., 2014; Leoni et al., 2016) or from non-indigenous bacteria, usually enteric bacteria, derived from fecal contamination (mostly Salmonella spp., E. coli VTEC, Shigella spp.) (Marceddu et al., 2017). In marine environments, Salmonella has been detected in coastal waters, molluscs, as well as other seafood products (Bazzoni et al., 2019; Catalao Dionisio et al., 2000). The microorganisms of the genus Salmonella spp. are introduced into the aquatic environment via inappropriate disposal of human wastes, agricultural runoffs or sewage discharges (Malham et al., 2014) as well as wildlife (Obiri-Danso and Jones, 2000). Several authors reported a prevalence of Salmonella spp. in shellfish samples collected in Italy, ranging from 0 to 3.1% (Bazzoni et al., 2019; Carraro et al., 2015; Marceddu et al., 2017; Prato et al., 2013; Rubini et al., 2018; Sferlazzo et al., 2018;). Although Salmonella spp. is considered one of the most common causes of human gastroenteritis (EFSA and ECDC, 2016) and in spite of its presence in marine environments (Catalao Dionisio et al., 2000), the risk of foodborne diseases associated with shellfish consumption is very low (Iwamoto et al., 2010). According to Reg. (EU) 2285/2015, the enumeration of E. coli as an indicator of fecal contamination, is the standard way to estimate the associated potential risk to human health from all waterborne enteric pathogens (Balière et al., 2015). Moreover, in addition to being considered as a fecal indicator, E. coli includes strains that can be pathogenic to humans (Touchon et al., 2009). Pathogenic E. coli are distributed into diarrheagenic pathotypes including Enterotoxigenic E. coli (ETEC), Enteropathogenic E. coli (EPEC), Enterohemorrhagic E. coli (EHEC), Enteroinvasive E. coli (EIEC) and Enteroaggregative E. coli (EAEC) (Baliere et al., 2015). Among EHEC, verotoxigenic E. coli (VTEC) are strains that produce Shiga-like toxin (verotoxin). VTEC are not commonly found in seafood, but it is well known that bivalve molluscs harvested in areas contaminated by landfill can carry verotoxigenic E. coli (Marceddu et al., 2017). Moreover, VTEC have been isolated in breeding environments from different sites such as soil, manure, sewage, drinking water, irrigation water, crops, and various equipment (Avery et al., 2008). The gastrointestinal tract of ruminants is the main reservoir of VTEC (Caprioli et al., 2005): from agricultural environments, they can be transferred to watercourses, especially during periods of high rainfall, and subsequently spread to coastal areas (Williams et al., 2008). Shellfish harvested in these areas can consequently concentrate the pathogen and thereby pose a risk to the health of consumers (Marceddu et al., 2017). Sardinia is one of the nationally relevant Italian shellfish production areas: the annual production accounts for 83% of the regional aquaculture, and it almost exclusively rests on M. galloprovincialis (Meloni et al., 2015; Sferlazzo et al., 2018). Specific literature in the Sardinian shellfish harvesting areas is limited. Therefore, in this study, we aimed to i) enumerate E. coli and detect E. coli VTEC and Salmonella spp. in two shellfish species (Mytilus galloprovincialis and Ruditapes decussatus); ii) study the effects of environmental changes on the E. coli seasonal distribution considering the interrelations existing among those parameters.
Materials and Methods
Sampling
The study was performed from April 2011 to May 2012 on shellfish samples (n. 68) collected from two class-B harvesting areas (named A and O) located in Sardinia (Italy). Samples of two shellfish species were included in this study: M. galloprovincialis (n. 34) and R. decussatus (n. 34). Forty-eight total samples were collected at the harvesting time from batches (n. 12). To evaluate the effects of purification on the safety of shellfish, samples (n. 20) from selected batches (n. 5) were collected after purification. The purification centers annexed to both production areas, applied short ‘‘recirculating’’ purification protocols (8h) previously described in Sferlazzo et al. (2018). The shellfish harvesting areas (Figure 1) were on the central-western coast (production area A) and on the North- Eastern coast of Sardinia (production area O). Sampling was performed by the authors. Environmental conditions (temperature, pH, and salinity) of the water were recorded. The samples were shipped refrigerated to the laboratories of the Department of Veterinary Medicine at the University of Sassari (Italy) and were analyzed within 24h of harvesting. Depending on the size of the M. galloprovincialis or R. decussatus, 15 to 30 bivalves of each sample were randomly selected for microbiological analysis. Samples were processed for microbiological detection, enumeration and graduated dilutions according to the Reg. (EC) 2073/2005, Reg. (EU) 2285/2015 and ISO 6887-3 method (ISO, 2003).
Microbiological analysis
Enumeration of E. coli
All the bivalve samples were examined using the five-tube Most Probable Number (MPN) method in accordance with the EU reference method ISO 16649–3 (ISO, 2015). In briefly, 75-100g of flesh and intervalvular liquid were added to 2 parts of Peptone water (BioMérieux, France) and homogenised using a Stomacher for 2.5min. Ten ml of the liquid part of the 1+2 suspension were added to a flask containing 90 ml of Peptone water (BioMérieux, France) resulting in a final 1+9 dilution. Aliquots of 10 ml of the initial suspension (1+2) were transferred to each of five tubes of doublestrength Mineral Modified Glutamate Medium (MMGB) (Oxoid, UK). Aliquots of 1.0 ml of the 1+9 dilution were transferred to each of five tubes of singlestrength MMGB. Further dilutions were prepared in the same way. All the double and single-strength MMGB were incubated aerobically at 37°C for 24h. Subcultures from positive MMGB tubes that changed colour from purple to yellow, were plated on chromogenic Tryptone Bile Glucuronide Agar (TBX) plates (Oxoid, UK) incubated aerobically at 44°C for 20h. At the end of incubation, the number of positive tubes of double or single-strength MMGB tubes were counted in order to estimate the level of E. coli/100g using the MPN table, generated with the MPN calculator referenced in ISO 7218 (ISO, 2007). The enumeration of E. coli in bivalve molluscs by the MPN technique has limits of detection of fewer than 18 E. coli cells.
Figure 1.
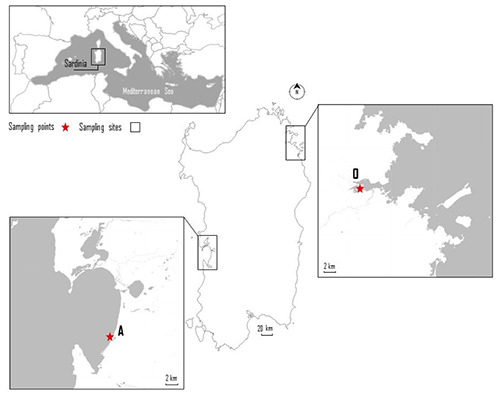
Shellfish harvesting areas located in Sardinia (Italy) included in the study.
Detection of E. coli VTEC
For the direct detection of E. coli VTEC, 10 g of each sample were subjected to selective enrichment in 90 ml of modified- Tryptone Soya Broth (m-TSB) containing novobiocin (20 mg/L) and incubated at 37°C for 18–20h. Subsequently, an aliquot was frozen at -20°C (Angelantoni Industrie Spa, Massa Martana, Italy) for Immuno-Magnetic Separation (IMS) screening test by using the protocol for the E. coli O157, O26, O103, O111, and O145 Dynabeads capture (Dynal, Oslo, Norway), as described by the manufacturer. Another aliquot equal to 1.0mL was used for DNA extraction using the Chelex 100 (BioRad, Hercules, CA, USA) resins. VTEC detection was carried out by a Polymerase Chain Reaction (PCR) one-step method for the detection of stx1 and stx2 genes using the primer sets MK1/MK2 (Karch and Meyer, 1989). A negative control (NCTC 12900) and a positive control (ATCC 35150) were included at each PCR test. All PCR amplifications were performed by using a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). All PCR-positive samples (VTEC presence) were subjected to qualitative detection of E. coli O157, O26, O103, O145, and O111 serogroups by IMS using Dynabeads anti-E. coli (Invitrogen, Carlsbad, CA, USA). Each Dynabeads-microorganism complex was streaked on CT-SMAC (MacConkey agar cefixime tellurite sorbitol, Thermo Fisher Scientific Oxoid Ltd., Basingstoke, UK) for the detection of serogroup O157, CTRMAC (MacConkey agar cefixime tellurite rhamnose, Thermo Fisher Scientific Oxoid Ltd, Basingstoke, UK) for O26, and EHLY (Enterohemolysin agar, Thermo Fisher Scientific Oxoid Ltd, Basingstoke, UK) for serogroups O103, O111, and O145. All the plates were incubated at 37°C for 24 h. Isolates with typical morphological characteristics were subjected to biochemical identification by the API 20E identification system (bioMérieux, Marcy l’Etoile, France). All the isolated E. coli were subjected to a multiplex PCR for the detection of stx1, stx2, hlyA, and eae genes (Paton and Paton, 1998).
Detection of Salmonella spp.
All the samples were analyzed for Salmonella spp. detection according to the ISO 6579 method (ISO, 2002). Briefly, 25 g of each sample were added to 225 ml of Buffered Peptone Water (BPW) and incubated at 37°C for 18 h (Thermo Fisher Scientific, Waltham, MA, USA). One-hundred μl of the BPW enrichment were inoculated in 10 ml of Rappaport-Vassiliadis Soya enrichment broth (RVS) and incubated at 42°C for 24 h while 100 μL of the BPW enrichment were streaked over the surface of a Modified Semi-Solid Rappaport-Vassiliadis agar plate (MSRV) and incubated at 42°C for 24 h. Finally, 1 ml of the BPW enrichment was transferred to 10 ml of Muller-Kaufmann tetrathionatenovobiocin broth (MKTTn) and incubated at 37°C for 24h. RVS, MKTTn, and MSRV were sub-cultured onto the surface of one Xylose-Lysine-Desoxycholate (XLD) agar plates and incubated at 37°C for 24h. Presumptive Salmonella-like colonies were submitted to phenotypic identification with the API ID 32E identification system (bioMérieux, Marcy l’Etoile, France). One colony from each positive sample was selected and sent to the laboratories of the “Centro Nazionale di Referenza per le Salmonellosi” in Legnaro (Padua, Italy), serotyped by agglutination tests with specific O and H antisera (Staten Serum Institute, Copenhagen, Denmark) and classified according to the White-Kauffmann-Le Minor scheme (Grimont and Weill, 2007).
Statistical analysis
Statistical analysis was performed with R software (version 1.0.153, R Foundation for Statistical Computing, Vienna, Austria). The enumeration of E. coli in M. galloprovincialis and R. decussatus collected in the two harvesting areas in relation to season, temperature, pH, and salinity were compared and analyzed with a linear regression model. The model was defined as follows: y= site + species + period + pH + temperature + salinity + e, where y is the contamination levels of E. coli expressed as MPN/100g; site is the effect of the harvesting area; species is the effect of the shellfish; period is the effect of the month and year of sampling; temperature (°C), pH, and salinity (%) express the effects of environmental parameters; and e is the error term. The mean values were compared with Tukey’s honestly significant difference test. The results were considered statistically significant when p<0.05 for all the tests performed.
Results and Discussion
Environmental parameters of water
The environmental parameters of the two harvesting areas were averaged and were as follows (mean ± standard deviation): temperature, 17 ± 4.20°C; salinity, 36 ± 1.50 %; pH, 8.21 ± 0.26. Data are summarized in Table 1.
Table 1.
Effects of purification on the levels of contamination by E. coli in shellfish (MPN/100g). Data from the two harvesting areas were averaged.
| Month | Mytilus galloprovincialis | Ruditapes decussatus | ||
|---|---|---|---|---|
| Before purification | After purification | Before purification | After purification | |
| May 2011 | <18 | 40 | <18 | <18 |
| July 2011 | 40 | 40 | 40 | 40 |
| November 2011 | 40 | 40 | 61 | 40 |
| March 2012 | 130 | 130 | 40 | 40 |
| May 2012 | 61 | 40 | 40 | 40 |
| Total | 54 | 58 | 36 | 32 |
Microbiological analysis
Enumeration of E. coli and detection of E. coli VTEC
The results of enumeration of E. coli conducted in shellfish samples are summarized in Table 2. E. coli was found in 100% of the samples. As reported in Table 2, the averaged values of E. coli load were respectively 39 and 37 MPN/100g in samples of M. galloprovincialis and R. decussatus collected at the harvesting time. The average contamination levels in samples collected after purification were 58 MPN/100g (M. galloprovincialis) and 32 MPN/100g (R. decussatus). The purification treatment allowed a slight reduction in E. coli loads (p<0.01) only in the latter species. On the contrary, in Sferlazzo et al. (2018), purification allowed a progressive and significant reduction in E. coli loads (p<0.01) in 10 batches of M. galloprovincialis. However, the results of E. coli load were consistently in agreement with the legislation limits for class A harvesting areas (EU 2285/2015). As previously reported by Sferlazzo et al. (2018), this evidence, with a moderate initial contamination, allowed the Food Business Operator to effectively reduce the purification times (~8h). The results of the present study have been used by the Food Business Operators to support requests for re-classification of the harvesting areas.
Statistical analysis showed no significant difference (p>0.05) between E. coli levels and environmental parameters of water, seasonality, site and shellfish species. As reported by Bazzardi et al. (2014), Sardinian seawaters had stable salinity, temperature, dissolved oxygen, and pH during the year, without significant variations. However, recent studies showed a positive correlation between E. coli levels and seasonality: Bazzoni et al. (2019) found E. coli in three of the four seasonal mollusc samples, with the highest counts in autumn and winter (270 and 330 MPN/100 g, respectively). E. coli was not found in the summer. The same results were previously obtained by Sferlazzo et al. (2018), who found significantly lower (p<0.05) levels of E. coli contamination during the summer period. E. coli VTEC was not detected in samples of M. galloprovincialis and R. decussatus collected before as well after purification. In a recent study carried out in Sardinia, the prevalence of E. coli VTEC was 6.6% in samples of Cerastoderma spp. and R. decussatus (Marceddu et al., 2017). The ten E. coli VTEC strains belonged to the serogroups O157, O26, O103, O145 and O11 and showed a complete pathogenicity profile (stx1 +, stx2 +, eae +, hlyA +). As previously reported by Rodriguez-Manzano et al. (2014), the prevalence of E. coli VTEC in bivalve molluscs is low and could be related to the presence of competitive bacterial flora, and to the decreased vitality of these microorganisms in vitro.
Detection of Salmonella spp.
Salmonella spp. were detected only in one sample of R. decussatus (April 2011) and in one sample of M. galloprovincialis (November 2011) collected at the harvesting time. In a recent study carried out in Sardinia, Salmonella spp. were only present in samples of M. galloprovincialis collected in spring (Bazzoni et al., 2019). On the contrary, in Marceddu et al. (2017) Salmonella spp. were not detected in any of Cerastoderma spp. and R.decussatus samples. As previously reported by Rubini et al. (2018), these results were not surprising: the presence of Salmonella spp. is strongly related to the shellfish species, the production areas and the sampling period. All the Salmonella spp. strains were identified as S. enterica subsp. enterica and Salmonella ser. Typhimurium resulted the only serovar (100%). In a recent Italian study (Rubini et al., 2018), Salmonella ser. Typhimurium resulted the dominant serovar (26.9%). No significant associations (p>0.05) were observed between the detection of Salmonella ser. Typhimurium and the E. coli levels in M. galloprovincialis and R. decussatus samples.
Conclusions
In our study, we applied a combined microbiological and biomolecular approach to enumerate E. coli and detect E. coli VTEC and Salmonella spp. in two shellfish species harvested in Sardinia (Italy). Our results provided useful information regarding the health risks associated with the consumption of M. galloprovincialis and R. decussatus. Although the enumeration of E. coli was consistently within the legal limits (EU Regulation 2285/2015) in 100 % of the samples, this study has reported the presence of Salmonella ser. Typhimurium in M. galloprovincialis and R. decussatus samples. As it happens in the rest of South Italy, in Sardinia these shellfish species are usually consumed raw or slightly cooked (Sferlazzo et al., 2018). Salmonella ser. Typhimurium is involved in most of human infection outbreaks and its recovering could lead to products that are unsuitable for guaranteeing the safety of Sardinian consumers. Recent studies reporting collateral surveys carried out in M. galloprovincialis and R. decussatus harvested in the same period from the same production areas, highlighted the presence of potentially pathogenic V. parahaemolyticus isolates (tdh + or trh +) too (Lamon et al., 2019a; Lamon et al., 2019b).
Table 2.
Enumeration of E. coli (MPN/100g) in Mytilus galloprovincialis and Ruditapes decussatus. Data from the two harvesting areas were averaged.
| Month | Mytilus galloprovincialis | Ruditapes decussatus | Temperature (°C) | Salinity (‰) | pH |
|---|---|---|---|---|---|
| April 2011 | <18 | <18 | 16.45 | 36.45 | 8.19 |
| May 2011 | <18 | <18 | 20.00 | 35.40 | 8.41 |
| June 2011 | <18 | 78 | 22.00 | 36.61 | 8.27 |
| July 2011 | 40 | 40 | 23.65 | 36.60 | 8.11 |
| September 2011 | 40 | 40 | 24.10 | 37.90 | 7.81 |
| October 2011 | 40 | 40 | 20.64 | 39.05 | 8.26 |
| November 2011 | 40 | 61 | 16.90 | 33.25 | 7.94 |
| December 2011 | 40 | 40 | 12.92 | 37.65 | 8.91 |
| January 2012 | 40 | 40 | 11.07 | 35.70 | 8,17 |
| March 2012 | 130 | 40 | 13.83 | 35.50 | 8.11 |
| April 2012 | 40 | 40 | 16.20 | 36.00 | 8.15 |
| May 2012 | 61 | 40 | 18.20 | 35.10 | 8.18 |
| Total | 39 | 37 | 17±4.20* | 36±1.50* | 8.21±0.26* |
*mean±sd.
Funding Statement
Funding: This work was funded by Regione Autonoma della Sardegna, L.R. 7 Agosto 2007, N. 7 “Promozione della Ricerca Scientifica e dell'Innovazione Tecnologica in Sardegna”. Progetto di Ricerca fondamentale o di base: "Sviluppo di tecnologie biomolecolari innovative per la sorveglianza epidemiologica di contaminanti batterici e virali ai fini della valorizzazione della produzione dei molluschi bivalvi della Sardegna". project ID: CRP2_470.
References
- Avery LM, Williams AP, Killham K, Jones DL, 2008. Survival of Escherichia coli O157:H7 in waters from lakes, rivers, puddles and animal-drinking troughs. Sci Total Environ 389:378-85. [DOI] [PubMed] [Google Scholar]
- Balière C, Rincé A, Thevenot D, Gourmelon M, 2015. Successful detection of pathogenic Shiga-toxin-producing Escherichia coli in shellfish, environmental waters and sediment using the ISO/TS-13136 method. Lett Appl Microbiol 60:315–20. [DOI] [PubMed] [Google Scholar]
- Bazzardi R, Fattaccio MC, Salza S, Canu A, Marongiu E, Pisanu M, 2014. Preliminary study on norovirus, hepatitis A virus, Escherichia coli and their potential seasonality in shellfish from different growing and harvesting areas in Sardinia region. Ital J Food Safety 3:125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni AM, Mudadu AG, Esposito G, Urru R, Ortu S, Mara L, Uda MT, Arras I, Lorenzoni G, Sanna G, Bazzardi R, Marongiu A, Virgilio S, Meloni D, 2019. Bacteriological and viral investigation combined with determination of phytoplankton and algal biotoxins in mussels and water from a Mediterranean coastal lagoon. J Food Prot 82, 9: 1501-11. [DOI] [PubMed] [Google Scholar]
- Caprioli A, Morabito S, Brugere H, Oswald E, 2005. Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Vet Res 36:289–311. [DOI] [PubMed] [Google Scholar]
- Carella F, Aceto S, Marrone R, Maiolino P, De Vico G, 2010. Marteilia refringens infection in cultured and natural beds of mussels (Mytilus galloprovincialis) along the Campania coast (Tirrenian sea, South of Italy). Bull Eur Ass Fish Pathol 30:189. [Google Scholar]
- Carraro V, Sanna C, Brandas V, Sanna A, Pinna A, Coroneo V, 2015. Hygiene and health risks associated with the consumption of edible lamellibranch molluscs. Int J Food Microbiol 201:52–7. [DOI] [PubMed] [Google Scholar]
- Catalao Dionisio LP, Ferreiro JP, Fidalgo ML, Garcia Rosado ME, Borrego JJ, 2000. Occurrence of Salmonella spp. in estuarine and coastal waters of Portugal. Antonie Van Leeuwenhoek 78:99-106. [DOI] [PubMed] [Google Scholar]
- European Commission, 2005. Regulation No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Official Journal, L 338:1–26. [Google Scholar]
- European Commission, 2015. Regulation No 2015/2285 of 8 December 2015 amending annex II to Regulation (EC) No 854/ 2004 of the European Parliament and of the Council laying down specific rules for the organisation of official controls on products of animal origin intended for human consumption as regards certain requirements for live bivalve molluscs, echinoderms, tunicates and marine gastropods and annex I to Regulation (EC) No 2073/2005 on microbiological criteria for foodstuffs. Official Journal, L 323:2–4. [Google Scholar]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC), 2016. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J 14:4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont PA, Weill FX, 2007. Antigenic Formulae of the Salmonella Serovars. 9th Paris, France: WHO Collaborating Center for Reference and Research on Salmonella, Institut Pasteur; 2007. [Google Scholar]
- ISO, 2002. ISO 6579:2002: Microbiology of food and animal feeding stuffs — Horizontal method for the detection of Salmonella spp. Geneva, Switzerland: International Organization for Standardization (ISO). [Google Scholar]
- ISO, 2003. ISO 6887-3:2003: Microbiology of food and animal feeding stuffs – Preparation of test samples, initial suspension and decimal dilutions for microbiological examination - Part 3: Specific rules for the preparation of fish and fishery products. Geneva, Switzerland: International Organization for Standardization (ISO). [Google Scholar]
- ISO, 2007. ISO 7218:2007. Microbiology of food and animal feeding stuffs – Generalrequirements and guidance for microbiological examinations. Geneva, Switzerland: International Organization for Standardization (ISO) [Google Scholar]
- ISO, 2015. ISO/FDIS 16649-3 Microbiology of the food chain - Horizontal method for the enumeration of beta-glucuronidase-positive Escherichia coli -Part 3: Detection and most probable number technique using 5-bromo-4-chloro-3-indolyl - ß - Dglucuronide. Geneva, Switzerland: International Organization for Standardization (ISO). [Google Scholar]
- Iwamoto M, Ayers T, Mahon BE, Swerdlow DL, 2010. Epidemiology of seafoodassociated infections in the United States. Clin Microbiol Rev 23:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch H, Meyer T, 1989. Single primer pair for amplifying segments of distinct Shiga-like-toxin genes by Polymerase Chain-Reaction. J Clin Microbiol 27:2751–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamon S, Bastardo A, Meloni D, Consolati SG, Fois F, Porcheddu G, Agus V, Pes M, Cambula MG, Mureddu A, Romalde J, 2019a. Clonal relationship among Vibrio parahaemolyticus isolated from Mediterranean mussels (Mytilus galloprovincialis) and Grooved carpet shells (Ruditapes decussatus) harvested in Sardinia (Italy). Food Microbiol 84:103258. [DOI] [PubMed] [Google Scholar]
- Lamon S, Consolati SG, Fois F, Cambula MG, Pes M, Porcheddu G, Agus V, Esposito G, Mureddu A, Meloni D, 2019b. Occurrence, seasonal distribution and molecular characterization of V. vulnificus, V. cholerae and V. parahaemolyticus in shellfish (Mytilus galloprovincialis and Ruditapes decussatus) collected in Sardinia (Italy). J Food Prot 82,11:1851-6. [DOI] [PubMed] [Google Scholar]
- Leoni F, Chierichetti S, Santarelli S, Talevi G, Masini L, Bartolini C, Rocchegiani E, Naceur Haouet M, Ottaviani D, 2017. Occurrence of Arcobacter spp. and correlation with the bacterial indicator of faecal contamination Escherichia coli in bivalve molluscs from the Central Adriatic, Italy. Int J Food Microbiol 245:6–12. [DOI] [PubMed] [Google Scholar]
- Leoni F, Talevi G, Masini L, Ottaviani D, Rocchegiani E. 2016. Trh (tdh−/trh+) gene analysis of clinical, environmental and food isolates of Vibrio parahaemolyticus as a tool for investigating pathogenicity. Int J Food Microbiol 225:43–53. [DOI] [PubMed] [Google Scholar]
- Malham SK, Rajko-Nenow P, Howlett E, Tuson KE, Perkins TL, Pallett DW, Wang H, Jago JF, Jones DL, McDonald JE, 2014. The interaction of human microbial pathogens, particulate material and nutrients in estuarine environments and their impacts on recreational and shellfish waters. Environ Sci Process Impact 16:2145-55. [DOI] [PubMed] [Google Scholar]
- Marceddu M, Lamon S, Consolati SG, Ciulli S, Mazza R, Mureddu A, Meloni D, 2017. Determination of Salmonella spp., E. coli VTEC, Vibrio spp., and Norovirus GI-GII in Bivalve Molluscs Collected from Growing Natural Beds in Sardinia (Italy). Foods, Special Issue Seafood Products: Safety and Quality 6:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni D. 2015. Survey on labelling and marketing of bivalve and gastropod mollusks retailed in Sardinia (Italy) between 2009 and 2013. It J Food Safety 4:104-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiri-Danso K, Jones K, 2000. Intertidal sediments as reservoirs for Hippurate negative campylobacters, salmonellae and faecal indicators in three EU recognised bathing waters in North West England. Water Res 34:519-27. [Google Scholar]
- Paton JC, Paton AW, 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. J Clin Microbiol 11:450–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prato R, Martinelli D, Tafuri S, Barbuti G, Quarto M, Germinario CA, Chironna M, 2013. Safety of shellfish and epidemiological pattern of enterically transmitted diseases in Italy. Int J Food Microbiol 162:125–8. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzano J, Hundesa A, Calgua B, Carratala A, Maluquer de Motes C, Rusinol M, Moresco V, Ramos AP, Martinez-Marca F, Calvo M, Monte Barardi CR, Girones R, Bofill-Mas Adenovirus and Norovirus contaminants commercially distributed shellfish. Food Environ Virol 6:31–41. [DOI] [PubMed] [Google Scholar]
- Rubini S, Galletti G, D’Incau M, Govoni G, Boschetti L, Berardelli C, Barbieri S, Merialdi G, Formaglio A, Guidi E, Bergamini M, Piva S, Serraino A, Giacometti S, 2018. Occurrence of Salmonella enterica subsp. enterica in bivalve molluscs and associations with Escherichia coli in molluscs and faecal coliforms in seawater. Food Control 84:429-35. [Google Scholar]
- Sferlazzo G, Meloni D, Lamon S, Marceddu M, Mureddu M, Consolati SG, Pisanu M, Virgilio S, 2018. Evaluation of short purification cycles in naturally contaminated Mediterranean mussels (Mytilus galloprovincialis) harvested in Sardinia (Italy). Food Microbiol 74:86-91. [DOI] [PubMed] [Google Scholar]
- Smaldone G, Marrone R, Cappiello S, Martin GA, Oliva G, Cortesi ML, Anastasio A, 2014. Occurrence of antibiotic resistance in bacteria isolated from seawater organisms caught in Campania Region: preliminary study. BMC Vet Res 10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Medigue C, Rocha EP, Denamur E, 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genetics 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AP, Gordon H, Jones DL, Strachan NJC, Avery LM, Killham K, 2008. Leaching of bioluminescent Escherichia coli O157:H7 from sheep and cattle faeces during simulated rainstorm events. J Appl Microbiol 105:1452–60. [DOI] [PubMed] [Google Scholar]


