Abstract
Host cytochrome P450s (P450s) play important roles in the bioactivation and detoxification of numerous therapeutic drugs, environmental toxicants, dietary factors, as well as endogenous compounds. Gut microbiome is increasingly recognized as our “second genome” that contributes to the xenobiotic biotransformation of the host, and the first pass metabolism of many orally exposed chemicals is a joint effort between host drug metabolizing enzymes including P450s and gut microbiome. Gut microbiome contributes to the drug metabolism via two distinct mechanisms: direct mechanism refers to the metabolism of drugs by microbial enzymes, among which reduction and hydrolysis (or deconjugation) are among the most important reactions; whereas indirect mechanism refers to the influence of host receptors and signaling pathways by microbial metabolites. Many types of microbial metabolites, such as secondary bile acids (BAs), short chain fatty acids (SCFAs), and tryptophan metabolites, are known regulators of human diseases through modulating host xenobiotic-sensing receptors. To study the roles of gut microbiome in regulating host drug metabolism including P450s, several models including germ free mice, antibiotics or probiotics treatments, have been widely used. The present review summarized the current information regarding the interactions between microbiome and the host P450s in xenobiotic biotransformation organs such as liver, intestine, and kidney, highlighting the remote sensing mechanisms underlying gut microbiome mediated regulation of host xenobiotic biotransformation. In addition, the roles of bacterial, fungal, and other microbiome kingdom P450s, which is an understudied area of research in pharmacology and toxicology, are discussed.
I. Introduction and History
Biotransformation of xenobiotics, such as drugs, dietary factors, and environmental chemicals, often mitigates potential toxicities to the body, preventing the occurrence of xenobiotic-induced injury. Liver, intestine, and kidney are the major organs for the detoxification of various xenobiotics using two distinct classes of drug-metabolizing enzymes. Phase-I drug-metabolizing enzymes catalyze oxidation, reduction, and hydrolysis reactions, which generally produce a small functional group on the substrate for further reactivity. Phase-II drug-metabolizing enzymes catalyze various conjugation reactions, including glucuronidation, sulfonation, glutathione conjugation, acetylation, methylation, amino acid conjugation, and fatty acid conjugation. In general, phase-I and -II metabolism make the initial substrate more water soluble for excretion [102].
Cytochrome P450s (P450s or CYP) are a superfamily of enzymes in which a subset comprises a major portion of phase-I oxidation enzymes. Beyond drug metabolism, P450s are also important for the bio-synthesis of endogenous biomolecules. For example, CYP7A1 is the rate-limiting enzyme for the synthesis of bile acids from cholesterol in liver; CYP27A1, CYP7B1, and CYP8B1 are also involved in bile acid production [1]. P450s are present across all seven biological kingdoms with over 41,000 different naturally occurring P450 sequences [2]. The functions, genetic variability, tissue distribution, and species differences of P450s have been extensively studied and reviewed [3–9]. Briefly, P450s are expressed in the liver as well as extrahepatically with varying expression levels leading to differential metabolism [6]. Other organs expressing xenobiotic metabolizing P450s include the intestine, lung, trachea, nasal respiratory and olfactory mucosa, esophagus, stomach, kidney, etc.[3, 7–9]. Many orally exposed xenobiotics are subject to first-pass metabolism in the gastrointestinal tract and liver. Intestinal P450s can metabolize a major portion of orally exposed xenobiotics before these chemicals enter the blood circulation, whereas liver further contributes to the first-pass metabolism of xenobiotics that are delivered through the portal blood [8, 10]. Together the P450-mediated first-pass metabolism within the gut-liver axis limits the entry of many xenobiotics into systemic circulation. There are 57 functional P450s in humans and 12 of them are from the CYP1, 2, and 3 subfamilies, which biotransform 70-80% of clinical drugs [6]. Genetic polymorphisms in P450 genes may affect the pharmacokinetics of their substrates, accounting for a significant portion of the inter-individual variability in drug metabolism and xenobiotic toxicity among individuals [11–13]. In addition to host genetic factors that influence the P450s, the gut microbiome also affects the expression and activity of P450s related to the metabolism of xenobiotics [14, 15].
The gut microbiome is an increasingly recognized critical component of the first-pass metabolism of xenobiotics and is a mediator for “indirect xenobiotic metabolism” through modulation of hepatic signaling via remote-sensing mechanisms [15, 16]. The gut microbiome varies in composition and function along the gastrointestinal tract that is reflective of a chemical, nutritional, antimicrobial gradient [17–22]. Although the ratio of bacterial to human cells is approximately 1:1, there are nearly 10 million genes in the human microbiome [23] with 3.3 million genes in the human gut microbiome [24]. Within the vast pool of microbial genes, a subset are capable of directly metabolizing drugs, dietary factors, environmental chemicals, and other xenobiotics independent from the host, often through reduction or hydrolysis reactions. This was initially shown in the 1960s by the deacylation of N-acetyl derivatives [25, 26] as well as O-dealkylation by microflora [27–33]. Intestinal microflora were later shown to metabolize many drugs [34]. This initial metabolism can occur before first pass metabolism redefining pharmacokinetics outside of host genetic polymorphisms, xenobiotic partitioning, distribution, and other factors of inter-individual variability. Indirectly, the gut microbiome produces metabolites that can interact with host receptors, acting as signaling molecules to regulate the expression and activity of xenobiotic metabolizing enzymes. For example, primary bile acids are produced in the liver from cholesterol and are excreted into the intestines through the bile duct wherein the microbiota convert them to secondary bile acids that can be reabsorbed to the liver [1]. The secondary bile acid lithocholic acid (made from the primary bile acid chenodeoxycholic acid) can activate the major xenobiotic sensing nuclear receptor pregnane X receptor (PXR/NR1I2), which is responsible for regulating the expression of many xenobiotic metabolizing enzymes [35].
The indirect relationship between gut microbiota and hepatic drug metabolism was not deeply investigated until the 2000s. In rats, the administration of the antibiotic ciprofloxacin decreased the hepatic expression of CYP2C11 and CYP3A1 and increased the area under the curve (AUC) (decreased the parent compound to metabolite ratio by almost 50%) when the chemotherapeutic cyclophosphamide was co-administered [36]. This indicated that a change in gut microbiome induced by antibiotics could change the pharmacokinetics of drugs by altering the key xenobiotic metabolizing enzymes. A study in mice comparing the expression and activity of Cyp3a11 between conventional (CV; i.e. mice with a microbiome) and germ-free (GF; i.e. mice without a microbiome) mice treated with ciprofloxacin was dependent on microbiota sensitive to ciprofloxacin [37]. Dr. Kiyoshi Sugiyama’s group showed that the expression of many Cyp1, 2, and 3 enzymes as well as the major xenobiotic sensing transcription factors PXR, constitutive androstane receptor (CAR/NR1I3), and aryl hydrocarbon receptor (AHR) were decreased in the livers of GF compared to CV mice [38]. Dr. Sven Pettersson’s group used microarrays to show that the expression of 112 targeted genes were differentially expressed between CV and GF mice, and a major pathway for these genes was xenobiotic metabolism by P450s [39]. These studies were the impetus for characterizing and understanding the interactions of the gut microbiome and host P450s in xenobiotic metabolism. The purpose of this review is to highlight the known interactions between gut microbiome and P450s regarding the metabolism of xenobiotics and examine unknown areas that may lead to increased drug efficacy and therapeutic protection from harmful xenobiotic exposures.
II. Interplay between host P450 and gut microbiota for xenobiotic metabolism
A first step in understanding the influential capacity of the gut microbiome on drug metabolism and, in particular, P450s in host liver is to investigate how the lack of gut microbiome alters the expression of drug-metabolizing enzymes. For these studies, researchers often rely on GF animal models, in particular GF mice. It is important to note that under basal conditions GF animals have different host physiology, such as enlarged cecum, as compared to CV animals. Therefore, with a GF animal model, cautions should be made while interpreting the data generated from GF studies, as a GF mouse is not merely a CV mouse minus the microbiome. Ingenuity Pathway Analysis of differentially expressed genes between livers of CV and GF mice showed that xenobiotic metabolism ranked among the most enriched network [40]. As previously stated, the lack of gut microbiome decreased the expression of P450s when comparing CV and GF adult male mouse livers by GeneChip or qPCR methods [38, 39]. A third study [41] using RNA-Seq examined the global differences in the hepatic expression of drug-metabolizing enzymes between CV and GF adult C57BL/6 male mice. Overall, the most differentially regulated drug-metabolizing enzymes in livers of GF mice were carboxylesterases and P450s with 20 upregulated and seven down-regulated P450s. A summary of the general changes in P450s is shown in Table 1 and selected nuclear receptors and transcription factors is shown in Table 2. Two P450s with increased expression (Cyp1a2 increased 51% and Cyp2a5 increased 143%) are regulated by the transcription factor AHR, whereas the Cyp3a and Cyp2b cluster controlled by both PXR and CAR were down regulated, respectively. An exception is Cyp2b9 which had increased expression (7454% increase with a fragments per kilobase of exon model per million reads mapped [FPKM] of 9.1) in GF mice. The omega fatty acid hydroxylase subfamily Cyp4a members, controlled by peroxisome proliferator-activated receptor α (PPARα), were also upregulated by 31-200% [41]. Comparing the mRNA expression of P450s in various sections of intestinal tissues (duodenum, jejunum, ileum, and large intestine) between CV and GF male, 46 P450s were differentially regulated by the lack of microbiome [42]. However, generally the same P450 transcript was not differentially regulated throughout all experimental tissues (intestines and liver), suggesting that there is a tissue-specific sensitivity in its regulation by absence of the microbiome. In lieu of using GF mice, poorly absorbed antibiotics with limited to no effects on the host can be used to deplete the microbiome to ascertain the necessity of the microbial affects. Kuno et al. (2016) treated ten-week-old C57BL/6NJcl male mice with vancomycin hydrochloride (500 mg/L) and polymyxin B sulfate (100 mg/L) in drinking water for 5 days and investigated hepatic and renal protein expression by liquid chromatography with tandem mass spectrometry (LC-MS/MS). Antibiotics decreased hepatic Cyp3a11 and Cyp2b10 proteins in liver, but did not alter the P450 proteins in kidney [43].
Table 1.
General changes in RNA expression of P450s in adult liver of GF mice compared to CV mice.
| Family and individual P450s | RNA expression change in GF mice compared to CV mice | Reference |
|---|---|---|
| Cyp1a and 2a | Increased 51-143% | [41] |
| Cyp2b | Decreased 57% | [41, 43] |
| Cyp2c | Increased minimally or no change | [40] |
| Cyp2e1 | Increased about 100% or no change | [40, 41] |
| Cyp3a | Decreased up to 87% | [40, 41, 43] |
| Cyp4a | Increased up to 200% | [40, 41] |
Table 2.
General changes in RNA expression of transcription factors and nuclear receptors that regulate P450s in adult liver of GF mice compared to CV mice.
Differentially expressed P450s between CV and GF mice are generally observed in adult mice, but not in developing mice. Specifically, the hepatic expression of Cyp3a11 and 3a44 between CV and GF mice is similar between 1- and 30-day-old mice, but the expression is decreased in livers of 90-day-old GF mice [40]. The protein expression and enzyme activity of Cyp3a were also decreased at adult age. Conversely, the mRNA expression of Cyp1a2, 2b9, 2c40, 2c54, 2c65, 2c67, 2c69, and 2e1 was increased up to about 100% in livers of GF mice in adult age only. In GF mice, subfamily Cyp4a (4a10, 4a14, 4a31, and 4a32) had increased expression up to about 200% compared to CV mice at day 1, 15, 30, and 90, but had decreased expression at day 5; Cyp4a10 protein was increased at day 15, 30, and 90, but Cyp4a14 protein was only increased (over 200%) at 90 days of age [40]. Expression of the transcription factors that are known to regulate drug-processing genes including P450s, namely AHR, CAR, PXR, and PPARα, were increased up to 100% at adult age in GF mouse livers (see Table 2). The reader is encouraged to read the references for a greater appreciation of the changes in gene expression due to lack of gut microbiome [40, 41]. Because the PXR-target genes were decreased in liver of GF mice in spite of the increase in the PXR mRNA, these data suggest that gut microbiome mainly modulates the PXR pathway at the receptor activity level.
Conventionalization or restoration of the microbiome to create exGF mice abrogates some of the differentially regulated drug metabolizing enzymes. Using C3H/Orl female mice at 8 weeks of age, Claus et al. (2011) placed the GF mice in cages previously habituated by CV mice and collected samples every 5 days for 20 days. Hepatic RNA expression of many P450s was normalized in exGF mice as compared to CV mice, including Cyp3a11 [44]. Cyp2e1 expression was increased after conventionalization of GF mice [44], whereas in Selwyn et al. (2015), Cyp2e1 was increased (about 100%) by lack of microbiome. Selwyn et al. (2016) compared exposing GF mice to CV feces versus exposing GF mice to the probiotic VSL3, which contains eight live bacterial strains (Bifidobacterium breve, Bifidobacterium longum, Bifidobacterium infantis, Lactobacillus acidophilus, Lactobacillus plantarum, Lactobacillus paracasei, Lactobacillus bulgaricus, and Streptococcus thermophilus). Conventionalization by exposing the GF mice to CV feces normalized the expression and activities of the Cyp3a (about a 7-fold increase in conventionalized mice compared to GF mice) and Cyp4a (decreased down to 1% of germ-free expression levels in conventionalized mice) genes [45]. Specifically, conventionalization increased the PXR-target genes Cyp3a cluster and decreased the PPARα-target genes Cyp4a cluster, corresponding to changes in PXR and PPARα binding. In contrast, VSL3 did not have a major impact on hepatic P450 gene expression, except an increase in Cyp4v3 mRNA [45]. Because the bacteria in VSL3 did not conventionalize the expression of PXR or PPARα-target genes whereas expression was restored by bacteria in a mouse habituate environment, distinct commensal bacteria in the gut other than the VSL3 composition are responsible for the production of PXR activators (increasing PXR-target gene expression from GF to CV mice) and the degradation of PPARα activators (decreasing PPARα from GF to CV mice.
III. Microbial metabolites and host receptors
III-1. Microbial metabolites as host receptor modulators
Microbial metabolites are key to indirect regulation of host xenobiotic metabolism in proximal and remote organs [16]. The metabolites produced by bacteria can be absorbed by the host and distributed to sites of action (e.g. microbial metabolites activating xenobiotic transcription factors in the liver). Metabolites produced from the gut microbiome are absorbed through the intestine and under-go first-pass metabolism in the liver before becoming systemic, with the potential to be modified by the host if the site of action is a peripheral organ. In addition, bacteria can modify host-derived metabolites, rendering them either active or inactive. Gram-negative bacteria produce lipopolysaccharides (LPS), which is an essential component in the outer cell membrane, and LPS induce immune and oxidative stress responses in the host. LPS, also referred to endotoxin, can reach 10-50 grams in a healthy human gut, but only up to 5 pg/mL of blood without any side effects [46]. Intravenous injection of LPS is rapidly cleared in the liver, although 1-2 μg of LPS can be lethal [47]. Mice are resistant to LPS exposure relative to humans likely due to specialized immunoglobulins [48]. For example, in a diet study of soy protein isolate with grape polyphenols LPS concentration in mouse serum was between 1-2 ng/mL [49]. An exposure of 100 μg of LPS in C57BL/6 and NMRI mice was 40% lethal [50]. Intermittent intraperitoneal injection of LPS (0.2 mg/kg of LPS once a week for three weeks) decreased the mRNA expression of Cyp3a11 (0.5 fold decrease) in mouse liver, and LPS also decreased CYP3A4 (the Cyp3a11 ortholog; 0.5 fold decrease) in human liver HepG2 cells [51]. The decreased expression of Cyp3a/CYP3A in mouse and human liver cells occurred through suppression of PXR. In rats, LPS injected into the lateral ventricle of the brain (25 μg) decreased the protein expression of several P450s in liver (CYP2D decreased 31%, and CYP2E1 decreased 36%) likely through a cytokine signaling cascade, whereas CYP1A remained unchanged [52]. Systemic LPS from gram-negative bacteria could influence the expression of PXR target genes and potentially decrease the metabolic capacity of the liver. Alternatively, host metabolites can be modified by microbial enzymes and reabsorbed, such as host primary bile acids modified to microbial secondary bile acids [1]. As mentioned previously, the secondary bile acid LCA can activate PXR and induce expression of Cyp3a11 in mice [35]. In addition to LPS and LCA, PXR can be activated by the microbial tryptophan metabolite indole-3-propionic acid (IPA) in intestine, but not in liver [53, 54]. IPA, known to be produced by Clostridium sporogenes from tryptophan, up-regulated the PXR target genes Cyp3a11, ATP-binding cassette, sub-family B, member 1B (Mdr1), and uridine diphosphate glucuronosyltransferase family 1 member A1 (Ugt1a1) in small intestine of mice [54]. PXR activation by IPA was shown to interact with Toll-like receptor 4 (Tlr4) to maintain mucosal integrity in the intestine.
III-1. Pharmacological activation of host receptors as regulators of microbiome and microbial metabolites
Activation of host receptors by xenobiotics may alter gut microbiome and host/microbial metabolite production and may at least partly contribute to the regulation of host P450 gene expression. For example, pharmacological activation of mouse PXR and CAR by oral exposure to their ligands—(pregnenolone-16α-carbonitrile [PCN] for PXR and 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-tetrachloro-1,4 bis(pyridyloxy)benzene [TCPOBOP] for CAR)—decreased two taxa in the Bifidobacterium genus, which corresponded with decreased gene abundance of the BA-deconjugating enzyme bile salt hydrolase. The absence of gut microbiome potentiated CAR ligand mediated increase in total, primary and conjugated bile acids corresponding with increased Cyp7a1 mRNA [55]. Statin therapy, which is used to lower serum cholesterol and to reduce the risk of heart disease, increased total bile acids and altered the gut microbiota in a PXR dependent manner [56]. This suggested that deleterious metabolic effects—increased risk of type 2 diabetes mellitus—of statin therapy may be related to the changes in the gut microbiome. Overall, activation of host drug receptors by bacteria pharmacologically alters the gut bacteria landscape, changing signaling patters and potentially the metabolic capacity of the liver.
Another major xenobiotic-sensing transcription factor that can interact with microbial metabolites is AHR. The P450s CYP1A1, 1B1, 1A2, and 2S1 are all regulated by AHR, and genetic variation in AHR may alter response to xenobiotics. AHR is general activated by 3-5 ring planar molecules, which includes polycyclic aromatic hydrocarbons (PAHs), benzimidazoles, flavonoids, polychlorinated biphenyls, and dioxins, and can interact with the nuclear hormone receptors estrogen and androgen receptors [57, 58]. In recent years, Drs. Gary Perdew and Andrew Patterson have explored host-microbe interactions in relation to AHR. Under basal conditions, the microbiome of wild-type mice compared to AHR-null mice is a stronger determinant of microbiome compositional differences than exposure to the persistent organic contaminant 2,3,7,8-tetrachlorodibenzofuran (TCDF) [59]. Increased expression of Cyp1a2 (about 125%) was also shown to be AHR dependent. Furthermore, TCDF altered the gut microbiome composition, and this was associated with altered bile acid metabolism and farnesoid X receptor (FXR) signaling in an AHR dependent manner. However, it is unclear if the activation of AHR by TCDF indirectly or if TCDF directly affect microbiome compositional change. Similar to PXR, AHR is also activated by microbial tryptophan metabolites. Indole, 3-methyl indole and 2-oxindole were shown to activate AHR in human and mouse liver cell lines. In silico analysis showed that two indole residues may be needed to effectively activate AHR such that two indole-like compounds resemble large planar molecules (e.g. indirubin) [60]. In addition, bacteria can produce quorum-sensing molecules in the form of quinoline derivatives. Nanomolar expression of 2,8-dihydroxyquinoline, which is a microbiome-dependent metabolite, activated human AHR and induced the expression of CYP1A1. Mouse AHR was modestly activated by 2,8-dihydroxyquinoline [61]. The short-chain fatty acid butyrate, produced from the fermentation of soluble fibers, was also shown to activate AHR in human intestinal cells [62]. These studies demonstrate that several microbial metabolites can activate AHR and further demonstrates that AHR is an important moderator of host-microbiota communication [63].
In addition to dioxin-like AhR activators, other persistent organic pollutants, such as the formerly used flame retardants polybrominated diphenyl ethers (PBDEs), can also interact with the gut microbiome to modulate the host P450s and other host drug-processing genes. In mice orally exposed to the non-coplanar PBDEs, namely BDE-47 and BDE-99, the lack of gut microbiota augmented PBDE-mediated up-regulation of many drug-processing genes, including Cyp1a2 and Cyp3a11 proteins in liver [64]. The lack of gut microbiome also augmented the Cyp3a enzyme activity in livers of GF mice [64]. In colon, the lack of gut microbiota also augmented the PBDE-mediated up-regulation of Cyp1a1 mRNA [64]. Regarding the BA-synthetic P450s in liver, PBDE-mediated down-regulation of Cyp8b1 and Cyp27a1 mRNAs was gut microbiota-dependent, whereas BDE-47 mediated up-regulation of Cyp7a1 mRNA was also gut microbiota-dependent [65]. Together these data indicate that gut microbiome is an important modifier for PBDE-mediated regulation of host P450s.
IV. Microbial P450s
The Cytochrome P450 Engineering Database (CYPED) has 18,851 bacterial CYP sequences [2]. Among these, 2,979 bacterial sequences are named in 602 prokaryotic CYP families. Yet, it is hypothesized that most new bacterial sequences will fit into existing families, which is extraordinarily high compared to only 57 P450s in humans [2]. Although the majority of the sequences in CYPED are from water and land samples, bacteria present in the human gut microbiome are present. For example, the species Enterococcus faecium, which can affect host response to pathogens and antibiotics [66–69], has 21 different P450 sequences in CYPED; however, the function of these P450s is not characterized. Recently, there has been a focus on the evolution of P450s in bacteria [70, 71]. It has been shown that bacterial P450s are highly conserved, suggesting that these P450s are essential to the generation of steroids, fatty acids, and terpenoids [71]. In particular, mycobacteria have the highest P450 diversity and have a high coverage of P450s in their genomes compared to other bacteria [71]. A comparison of P450s in Streptomyces spp. and Mycobacterium spp., which are both genera in the phylum Actinobacteria, revealed that there are more P450s in Streptomyces spp. that contribute to the antibiotic diversity, whereas P450s in mycobacteria were predicted to be important for the synthesis of lipids [72]. The high diversity in P450s suggests the evolution of endogenous and exogenous roles to secure competitive niches in an evolutionary pattern for bacteria.
The bacterial P450 CYP102 is known to weakly oxidize low molecular PAHs, such as phenanthrene, fluoranthene, and pyrene, to phenols and quinones [73]. Bacillus megaterium found in human ileum can express CYP102 and may affect stress pathways and cell cycle in epithelial cells [74], and protein engineering of Bacillus megaterium CYP102 increased the metabolism of PAHs [73]. CYP102 in B. megaterium can be induced by barbiturates, including the indirect CAR activator phenobarbital [75], peroxisome proliferators, and nonsteroidal anti-inflammatory drugs, whereas isoflavones (genistein, biochanin A, coumestrol, and equol) and green tea flavonoid epicatechin inhibit phenobarbital induction of CYP102 [76]. In addition, oral ibuprofen could induce an oxidative stress response in B. megaterium and deplete glutathione, making the cells more susceptible to oxidative insult [77]. Taken regularly, ibuprofen and other drugs have the potential to alter a healthy gut microbiome and potentially create an environment that is advantageous to pathogens.
Several progesterone-like PAHs can be oxidized by microbial P450s to compounds that have estrogenic activity; these compounds include methoxychlor, trans-stilbene, diphenyl, diphenylmethane, 2,2-diphenylpropane, benzo[a]pyrene, benzophenone, 2-nitrofluorene, chalcone, trans-4-phenyl-3-buten-2-one, and styrene oligomers [78]. In vitro assays of naphthalene, phenanthrene, pyrene, and benzo(a)pyrene display no estrogenic activity [79]. Upon colon digestion, but not stomach or small intestine, PAHs exhibit estrogenic activity similar to 17α-ethynylestradiol [79]. As shown by microbial isolates of skin microbiota, benzo[a]pyrene oxidation and degradation serve as a carbon source for bacteria, giving bacteria in a PAH contaminated site—gastrointestinal tract, contaminated soil, or other environment—a competitive advantage over other bacteria [80]. Overall, oxidation of PAHs by P450s in the colon and skin alter the toxicity from carcinogenic to endocrine disrupting due to their estrogenic activity.
In addition to bacterial P450s, fungal P450s also present a new area for xenobiotic biotransformation [81, 82]. Fungal P450s are capable of metabolizing anti-inflammatories, β-blockers, and antibiotics and may be useful in bioremediation processes [81]. Filamentous fungi have also been shown to have multidrug resistant properties and may overexpress P450s, which could confound detoxification as we all antifungal drugs [82]. This opens the research are to new possibilities of P450s in the microbiome beyond bacteria fungi to include Archea [83], protissts [84, 85], and viruses [86].
Because the function of microbiome P450s are affected by drugs and other xenobiotics, it is important to understand how microbial P450s may influence host health and drug efficacy. The functions of microbial P450s can be used in synthetic biology and biotechnology [87–89]. This can include drug development [89–94] and secondary metabolite discovery, bioremediation of pesticide contaminated soils [95], degradation of environmental chemicals such as bisphenol A or polybrominated diphenyl ethers [96, 97], oxidation of alkanes [98, 99] and wastewater treatment [103]. Some P450s have already been artificially mutated to perform catalytic reactions not previously observed in nature to improve known chemical reaction or create new molecular structures; this includes the modification C400S in cytochrome P450 BM3 (also referred to as P411) [100–102]. Similarly, modifying, regulating, or inhibiting microbial P450s may be important for personalized medicine, drug therapy, and detoxificiation of toxic environmental chemicals. Although not a P450, microbial β-glucuronidases can cause severe diarrhea by reactivation of the chemotherapeutic irinotecan due to metabolism of the glucuronide metabolite back to the active form of the drug [103]. Inhibition of β-glucuronidases in Escherichia coli prevented irinotecan-induced toxicity and established a new method of inhibiting bacterial enzymes to increase drug efficacy [103, 104]. Therefore, inhibiting or inducing microbial P450s may be a novel method to alter the pharmacokinates of drugs, to improve drug efficacy, or protect the host from toxic xenobiotic exposures.
The urinary metabolite ratio of acetaminophen metabolism was also investigated with regard to the microbiome. Specifically, patients who had high urinary levels of p-cresol sulfate before exposure to acetaminophen had a low ratio of acetaminophen to acetaminophen-glucuronide [105]. This was attributed to O-sulfonation competition between p-cresol versus acetaminophen, which may have the capacity to alter the pharmacokinetics of acetaminophen. In a separate study, probiotic exposure of Lactobacillus reuteri KCTC3679 increased metabolism of acetaminophen, decreased the AUC of oral acetaminophen, and increased the bacterial load of L. reuteri as well as cyanobacteria [106] . The spleen tyrosine kinase inhibitor R406 (produced from intestinal activation of the prodrug fostamatinib) is de-methylated in liver by hepatic CYP3A4 and is subsequently metabolized by microbial mono-oxygenases [107]. The experimental chemotherapeutic epacadostat that inhibits indoleamine 2,3-dioxygenase 1 (ID01) was shown to be metabolized by microbial P450s by pre-exposing mice to ciprofloxacin or 1-aminobenzotriazole to inhibit bacteria or all P450s, respectively [108]. Bacterial metabolism of drugs is a well-known confounder of drug therapy with over 30 drugs known to be co-metabolized by microbial enzymes [109–111]; however, characterization of these enzymes, and in particular, microbial P450s is less understood. Understanding the specificity of these enzymes and the particular bacteria containing these genes may be essential to providing personalized medicine health care [112].
V. Conclusion
Taken together, the present review has systematically summarized the current information regarding the interactions between microbiome (mainly gut microbiome) and the host P450s in drug-processing organs such as liver, intestine and kidney, highlighting the importance of the “remote-sensing” mechanisms underlying gut microbiome mediated regulation of host xenobiotic biotransformation. Important microbial metabolites from cholesterol and amino acid metabolism pathways have been shown to modulate the host transcription factors that subsequently regulate the host P450 gene expression. In addition, the roles of bacterial, fungal, and other microbiome kingdom P450s, which is an understudied area of research in pharmacology and toxicology, are discussed. Looking forward, through integrating microbiome sequencing and metabolomics, as well as improved resolution of microbial functions at single species/strain levels future investigations in the area of “functional metagenome” and P450s are expected markedly expand our understanding of physiologically based pharmacokinetic modeling for greater accuracy in personalized medicine.
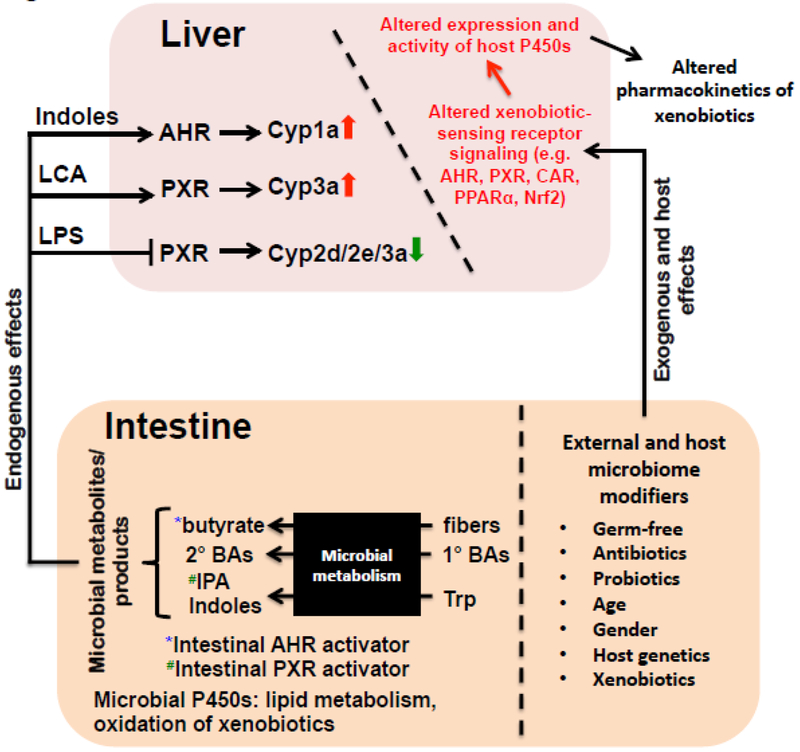
Figure 1.
Summary of the regulation of P450s within the gut-liver axis. Distinct microbial metabolites produced from the intestine, such as butyrate (from fermentation of fibers), secondary bile acids [BAs] (from primary bile acids), as well as indoles and indole-3-propionic acid (IPA) (from tryptophan), are known activators of certain host receptors, such as AHR (butyrate and indoles) and PXR (IPA). Certain microbial metabolites may enter the circulation and reach the liver to modulate the host receptor signaling and targeted P450 gene expression. Microbiome modifiers, such as germ free condition, the use of antibiotics/probiotics, age, gender, host genetics, and exposure to other drugs and environmental chemicals, may shift the composition and functions of gut microbiome, leading to altered host receptor signaling and target gene expression. Microbial P450s as a separate entity in the intestine have been shown to contribute to lipid metabolism as well as oxidation of xenobiotics. Trp: tryptophan; 1°: primary; 2°: secondary.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
The authors declared no conflict of interest.
Reference
- [1].Chiang JYL, Ferrell JM. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu Rev Nutr. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nelson DR. Cytochrome P450 diversity in the tree of life. Biochim Biophys Acta Proteins Proteom. 2018;1866:141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–73. [DOI] [PubMed] [Google Scholar]
- [4].Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC. The human intestinal cytochrome P450 “pie”. Drug Metab Dispos. 2006;34:880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Renaud HJ, Cui JY, Khan M, Klaassen CD. Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci. 2011;124:261–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013; 138:103–41. [DOI] [PubMed] [Google Scholar]
- [7].Knights KM, Rowland A, Miners JO. Renal drug metabolism in humans: the potential for drug-endobiotic interactions involving cytochrome P450 (CYP) and UDP-glucuronosyltransferase (UGT). Br J Clin Pharmacol. 2013;76:587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xie F, Ding X, Zhang QY. An update on the role of intestinal cytochrome P450 enzymes in drug disposition. Acta Pharm Sin B. 2016;6:374–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Miners JO, Yang X, Knights KM, Zhang L. The Role of the Kidney in Drug Elimination: Transport, Metabolism, and the Impact of Kidney Disease on Drug Clearance. Clin Pharmacol Ther. 2017;102:436–49. [DOI] [PubMed] [Google Scholar]
- [10].Zhang QY, Dunbar D, Ostrowska A, Zeisloft S, Yang J, Kaminsky LS. Characterization of human small intestinal cytochromes P-450. Drug Metab Dispos. 1999;27:804–9. [PubMed] [Google Scholar]
- [11].Hart SN, Zhong XB. P450 oxidoreductase: genetic polymorphisms and implications for drug metabolism and toxicity. Expert Opin Drug Metab Toxicol. 2008;4:439–52. [DOI] [PubMed] [Google Scholar]
- [12].Tracy TS, Chaudhry AS, Prasad B, Thummel KE, Schuetz EG, Zhong XB, et al. Interindividual Variability in Cytochrome P450-Mediated Drug Metabolism. Drug Metab Dispos. 2016;44:343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yu AM, Zhong XB. Advanced knowledge in drug metabolism and pharmacokinetics. Acta Pharm Sin B. 2016;6:361–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang J, Zhang J, Wang R. Gut microbiota modulates drug pharmacokinetics. Drug Metab Rev. 2018;50:357–68. [DOI] [PubMed] [Google Scholar]
- [15].Fu ZD, Cui JY. Remote Sensing between Liver and Intestine: Importance of Microbial Metabolites. Curr Pharmacol Rep. 2017;3:101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol. 2016;14:273–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol. 2005;54:1093–101. [DOI] [PubMed] [Google Scholar]
- [18].Wang M, Ahrne S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219–31. [DOI] [PubMed] [Google Scholar]
- [19].Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017;32:300–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Crespo-Piazuelo D, Estelle J, Revilla M, Criado-Mesas L, Ramayo-Caldas Y, Ovilo C, et al. Characterization of bacterial microbiota compositions along the intestinal tract in pigs and their interactions and functions. Sci Rep. 2018;8:12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Seekatz AM, Schnizlein MK, Koenigsknecht MJ, Baker JR, Hasler WL, Bleske BE, et al. Spatial and Temporal Analysis of the Stomach and Small-Intestinal Microbiota in Fasted Healthy Humans. mSphere. 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Uno T, Kono M. [Studies on the metabolism of sulfisoxazole. V. On the deacetylation of N-acetylsulfisoxazole by intestinal bacteria]. Yakugaku Zasshi. 1961;81:1434–6. [DOI] [PubMed] [Google Scholar]
- [26].Free Sjaastad O. and conjugated histamine in faeces from healthy individuals. Scand J Gastroenterol. 1966;1:1–8. [DOI] [PubMed] [Google Scholar]
- [27].Scheline RR. Decarboxylation and demethylation of some phenolic benzoic acid derivatives by rat caecal contents. J Pharm Pharmacol. 1966;18:664–9. [DOI] [PubMed] [Google Scholar]
- [28].Scheline RR. The decarboxylation of some phenolic acids by the rat. Acta Pharmacol Toxicol (Copenh). 1966;24:275–85. [DOI] [PubMed] [Google Scholar]
- [29].Scheline RR. Metabolism of phenolic acids by the rat intestinal microflora. Acta Pharmacol Toxicol (Copenh). 1968;26:189–205. [DOI] [PubMed] [Google Scholar]
- [30].Scheline RR. The metabolism of drugs and other organic compounds by the intestinal microflora. Acta Pharmacol Toxicol (Copenh). 1968;26:332–42. [DOI] [PubMed] [Google Scholar]
- [31].Griffiths LA, Smith GE. Metabolism of apigenin and related compounds in the rat. Metabolite formation in vivo and by the intestinal microflora in vitro. Biochem J. 1972;128:901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Griffiths LA, Smith GE. Metabolism of myricetin and related compounds in the rat. Metabolite formation in vivo and by the intestinal microflora in vitro. Biochem J. 1972;130:141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Meyer T, Scheline RR. 3,4,5-trimethoxycinnamic acid and related compounds. I. Metabolism by the rat intestinal microflora. Xenobiotica. 1972;2:383–90. [DOI] [PubMed] [Google Scholar]
- [34].Smith GE, Griffiths LA. Metabolism of N-acylated and O-alkylated drugs by the intestinal microflora during anaerobic incubation in vitro. Xenobiotica. 1974;4:477–87. [DOI] [PubMed] [Google Scholar]
- [35].Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xie HJ, Broberg U, Griskevicius L, Lundgren S, Carlens S, Meurling L, et al. Alteration of pharmacokinetics of cyclophosphamide and suppression of the cytochrome p450 genes by ciprofloxacin. Bone Marrow Transplant. 2003;31:197–203. [DOI] [PubMed] [Google Scholar]
- [37].Toda T, Ohi K, Kudo T, Yoshida T, Ikarashi N, Ito K, et al. Ciprofloxacin suppresses Cyp3a in mouse liver by reducing lithocholic acid-producing intestinal flora. Drug Metab Pharmacokinet. 2009;24:201–8. [DOI] [PubMed] [Google Scholar]
- [38].Toda T, Saito N, Ikarashi N, Ito K, Yamamoto M, Ishige A, et al. Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica. 2009;39:323–34. [DOI] [PubMed] [Google Scholar]
- [39].Bjorkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One. 2009;4:e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Selwyn FP, Cheng SL, Bammler TK, Prasad B, Vrana M, Klaassen C, et al. Developmental Regulation of Drug-Processing Genes in Livers of Germ-Free Mice. Toxicol Sci. 2015;147:84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Selwyn FP, Cui JY, Klaassen CD. RNA-Seq Quantification of Hepatic Drug Processing Genes in Germ-Free Mice. Drug Metab Dispos. 2015;43:1572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fu ZD, Selwyn FP, Cui JY, Klaassen CD. RNA-Seq Profiling of Intestinal Expression of Xenobiotic Processing Genes in Germ-Free Mice. Drug Metab Dispos. 2017;45:1225–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kuno T, Hirayama-Kurogi M, Ito S, Ohtsuki S. Effect of Intestinal Flora on Protein Expression of Drug-Metabolizing Enzymes and Transporters in the Liver and Kidney of Germ-Free and Antibiotics-Treated Mice. Mol Pharm. 2016;13:2691–701. [DOI] [PubMed] [Google Scholar]
- [44].Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, et al. Colonization-induced host-gut microbial metabolic interaction. MBio. 2011;2:e00271–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Selwyn FP, Cheng SL, Klaassen CD, Cui JY. Regulation of Hepatic Drug-Metabolizing Enzymes in Germ-Free Mice by Conventionalization and Probiotics. Drug Metab Dispos. 2016;44:262–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Salden HJ, Bas BM. Endotoxin binding to platelets in blood from patients with a sepsis syndrome. Clin Chem. 1994;40:1575–9. [PubMed] [Google Scholar]
- [47].Sauter C, Wolfensberger C. Interferon in human serum after injection of endotoxin. Lancet. 1980;2:852–3. [DOI] [PubMed] [Google Scholar]
- [48].Boes M, Prodeus AP, Schmidt T, Carroll MC, Chen J. A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection. J Exp Med. 1998;188:2381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Roopchand DE, Carmody RN, Kuhn P, Moskal K, Rojas-Silva P, Turnbaugh PJ, et al. Dietary Polyphenols Promote Growth of the Gut Bacterium Akkermansia muciniphila and Attenuate High-Fat Diet-Induced Metabolic Syndrome. Diabetes. 2015;64:2847–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Galanos C, Freudenberg MA, Reutter W. Galactosamine-induced sensitization to the lethal effects of endotoxin. Proc Natl Acad Sci U S A. 1979;76:5939–43.293694 [Google Scholar]
- [51].Sun HY, Yan YJ, Li YH, Lv L. Reversing effects of ginsenosides on LPS-induced hepatic CYP3A11/3A4 dysfunction through the pregnane X receptor. J Ethnopharmacol. 2019;229:246–55. [DOI] [PubMed] [Google Scholar]
- [52].Renton KW, Nicholson TE. Hepatic and central nervous system cytochrome P450 are down-regulated during lipopolysaccharide-evoked localized inflammation in brain. J Pharmacol Exp Ther. 2000;294:524–30. [PubMed] [Google Scholar]
- [53].Morgan ET, Dempsey JL, Mimche SM, Lamb TJ, Kulkarni S, Cui JY, et al. Physiological Regulation of Drug Metabolism and Transport: Pregnancy, Microbiome, Inflammation, Infection, and Fasting. Drug Metab Dispos. 2018;46:503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dempsey JL, Wang D, Siginir G, Fei Q, Raftery D, Gu H, et al. Pharmacological Activation of PXR and CAR Downregulates Distinct Bile Acid-Metabolizing Intestinal Bacteria and Alters Bile Acid Homeostasis. Toxicol Sci. 2019;168:40–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Caparros-Martin JA, Lareu RR, Ramsay JP, Peplies J, Reen FJ, Headlam HA, et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome. 2017;5:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ramadoss P, Marcus C, Perdew GH. Role of the aryl hydrocarbon receptor in drug metabolism. Expert Opin Drug Metab Toxicol. 2005;1:9–21. [DOI] [PubMed] [Google Scholar]
- [58].Hubbard TD, Murray IA, Perdew GH. Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab Dispos. 2015;43:1522–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang L, Nichols RG, Correll J, Murray IA, Tanaka N, Smith PB, et al. Persistent Organic Pollutants Modify Gut Microbiota-Host Metabolic Homeostasis in Mice Through Aryl Hydrocarbon Receptor Activation. Environ Health Perspect. 2015;123:679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hubbard TD, Liu Q, Murray IA, Dong F, Miller C 3rd, Smith PB, et al. Microbiota Metabolism Promotes Synthesis of the Human Ah Receptor Agonist 2,8-Dihydroxyquinoline. J Proteome Res. 2019;18:1715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Marinelli L, Martin-Gallausiaux C, Bourhis JM, Beguet-Crespel F, Blottiere HM, Lapaque N. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci Rep. 2019;9:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang L, Nichols RG, Patterson AD. The aryl hydrocarbon receptor as a moderator of host-microbiota communication. Curr Opin Toxicol. 2017;2:30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li CY, Lee S, Cade S, Kuo LJ, Schultz IR, Bhatt DK, et al. Novel Interactions between Gut Microbiome and Host Drug-Processing Genes Modify the Hepatic Metabolism of the Environmental Chemicals Polybrominated Diphenyl Ethers. Drug Metab Dispos. 2017;45:1197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li CY, Dempsey JL, Wang D, Lee S, Weigel KM, Fei Q, et al. PBDEs Altered Gut Microbiome and Bile Acid Homeostasis in Male C57BL/6 Mice. Drug Metab Dispos. 2018;46:1226–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Scharek-Tedin L, Kreuzer-Redmer S, Twardziok SO, Siepert B, Klopfleisch R, Tedin K, et al. Probiotic Treatment Decreases the Number of CD14-Expressing Cells in Porcine Milk Which Correlates with Several Intestinal Immune Parameters in the Piglets. Front Immunol. 2015;6:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Popovic N, Djokic J, Brdaric E, Dinic M, Terzic-Vidojevic A, Golic N, et al. The Influence of Heat-Killed Enterococcus faecium BGPAS1–3 on the Tight Junction Protein Expression and Immune Function in Differentiated Caco-2 Cells Infected With Listeria monocytogenes ATCC 19111. Front Microbiol. 2019;10:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kim B, Wang YC, Hespen CW, Espinosa J, Salje J, Rangan KJ, et al. Enterococcus faecium secreted antigen A generates muropeptides to enhance host immunity and limit bacterial pathogenesis. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].McKenney PT, Yan J, Vaubourgeix J, Becattini S, Lampen N, Motzer A, et al. Intestinal Bile Acids Induce a Morphotype Switch in Vancomycin-Resistant Enterococcus that Facilitates Intestinal Colonization. Cell Host Microbe. 2019;25:695–705 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Nelson DR. Progress in tracing the evolutionary paths of cytochrome P450. Biochim Biophys Acta. 2011;1814:14–8. [DOI] [PubMed] [Google Scholar]
- [71].Parvez M, Qhanya LB, Mthakathi NT, Kgosiemang IK, Bamal HD, Pagadala NS, et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci Rep. 2016;6:33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Senate LM, Tjatji MP, Pillay K, Chen W, Zondo NM, Syed PR, et al. Similarities, variations, and evolution of cytochrome P450s in Streptomyces versus Mycobacterium. Sci Rep. 2019;9:3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Carmichael AB, Wong LL. Protein engineering of Bacillus megaterium CYP102. The oxidation of polycyclic aromatic hydrocarbons. Eur J Biochem. 2001;268:3117–25. [DOI] [PubMed] [Google Scholar]
- [74].Di Luccia B, D’Apuzzo E, Varriale F, Baccigalupi L, Ricca E, Pollice A. Bacillus megaterium SF185 induces stress pathways and affects the cell cycle distribution of human intestinal epithelial cells. Benef Microbes. 2016;7:609–20. [DOI] [PubMed] [Google Scholar]
- [75].Kim BH, Fulco AJ. Induction by barbiturates of a cytochrome P-450-dependent fatty acid monooxygenase in Bacillus megaterium: relationship between barbiturate structure and inducer activity. Biochem Biophys Res Commun. 1983;116:843–50. [DOI] [PubMed] [Google Scholar]
- [76].Rajnarayanan RV, Rowley CW, Hopkins NE, Alworth WL. Regulation of phenobarbital-mediated induction of CYP102 (cytochrome P450(BM-3)) in Bacillus megaterium by phytochemicals from soy and green tea. J Agric Food Chem. 2001;49:4930–6. [DOI] [PubMed] [Google Scholar]
- [77].English NT, Rankin LC. Antioxidant-mediated attenuation of the induction of cytochrome P450BM-3(CYP102) by ibuprofen in Bacillus megaterium ATCC 14581. Biochem Pharmacol. 1997;54:443–50. [DOI] [PubMed] [Google Scholar]
- [78].Kitamura S, Sugihara K, Sanoh S, Fujimoto N, Ohta S. Metabolic activation of proestrogens in the environment by cytochrome P450 system. J Health Sci. 2008;54:343–55. [Google Scholar]
- [79].Van de Wiele T, Vanhaecke L, Boeckaert C, Peru K, Headley J, Verstraete W, et al. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environ Health Perspect. 2005;113:6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sowada J, Schmalenberger A, Ebner I, Luch A, Tralau T. Degradation of benzo[a]pyrene by bacterial isolates from human skin. FEMS Microbiol Ecol. 2014;88:129–39. [DOI] [PubMed] [Google Scholar]
- [81].Olicon-Hernandez DR, Gonzalez-Lopez J, Aranda E. Overview on the Biochemical Potential of Filamentous Fungi to Degrade Pharmaceutical Compounds. Front Microbiol. 2017;8:1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sang H, Hulvey JP, Green R, Xu H, Im J, Chang T, et al. A Xenobiotic Detoxification Pathway through Transcriptional Regulation in Filamentous Fungi. MBio. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].McLean MA, Maves SA, Weiss KE, Krepich S, Sligar SG. Characterization of a cytochrome P450 from the acidothermophilic archaea Sulfolobus solfataricus. Biochem Biophys Res Commun. 1998;252:166–72. [DOI] [PubMed] [Google Scholar]
- [84].Ndifor AM, Ward SA, Howells RE. Cytochrome P-450 activity in malarial parasites and its possible relationship to chloroquine resistance. Mol Biochem Parasitol. 1990;41:251–7. [DOI] [PubMed] [Google Scholar]
- [85].Barrett J Cytochrome P450 in parasitic protozoa and helminths. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;121:181–3. [DOI] [PubMed] [Google Scholar]
- [86].Lamb DC, Lei L, Warrilow AG, Lepesheva GI, Mullins JG, Waterman MR, et al. The first virally encoded cytochrome p450. J Virol. 2009;83:8266–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Girvan HM, Munro AW. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr Opin Chem Biol. 2016;31:136–45. [DOI] [PubMed] [Google Scholar]
- [88].Bhattacharya SS, Yadav JS. Microbial P450 Enzymes in Bioremediation and Drug Discovery: Emerging Potentials and Challenges. Curr Protein Pept Sci. 2018;19:75–86. [DOI] [PubMed] [Google Scholar]
- [89].Xu LH, Du YL. Rational and semi-rational engineering of cytochrome P450s for biotechnological applications. Synth Syst Biotechnol. 2018;3:283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–3. [DOI] [PubMed] [Google Scholar]
- [91].Bernhardt R, Urlacher VB. Cytochromes P450 as promising catalysts for biotechnological application: chances and limitations. Appl Microbiol Biotechnol. 2014;98:6185–203. [DOI] [PubMed] [Google Scholar]
- [92].McLean KJ, Hans M, Meijrink B, van Scheppingen WB, Vollebregt A, Tee KL, et al. Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum. Proc Natl Acad Sci U S A. 2015;112:2847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, et al. Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 2016;113:3209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zuo R, Zhang Y, Jiang C, Hackett JC, Loria R, Bruner SD, et al. Engineered P450 biocatalysts show improved activity and regio-promiscuity in aromatic nitration. Sci Rep. 2017;7:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Doolotkeldieva T, Konurbaeva M, Bobusheva S. Microbial communities in pesticide-contaminated soils in Kyrgyzstan and bioremediation possibilities. Environ Sci Pollut Res Int. 2018;25:31848–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Han J, Kim DH, Seo JS, Kim IC, Nelson DR, Puthumana J, et al. Assessing the identity and expression level of the cytochrome P450 20A1 (CYP20A1) gene in the BPA-, BDE-47, and WAF-exposed copepods Tigriopus japonicus and Paracyclopina nana. Comp Biochem Physiol C Toxicol Pharmacol. 2017;193:42–9. [DOI] [PubMed] [Google Scholar]
- [97].Mtibaa R, Olicon-Hernandez DR, Pozo C, Nasri M, Mechichi T, Gonzalez J, et al. Degradation of bisphenol A and acute toxicity reduction by different thermo-tolerant ascomycete strains isolated from arid soils. Ecotoxicol Environ Saf. 2018;156:87–96. [DOI] [PubMed] [Google Scholar]
- [98].Liang JL, JiangYang JH, Nie Y, Wu XL. Regulation of the Alkane Hydroxylase CYP153 Gene in a Gram-Positive Alkane-Degrading Bacterium, Dietzia sp. Strain DQ12–45-1b. Appl Environ Microbiol. 2016;82:608–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].McCarl V, Somerville MV, Ly MA, Henry R, Liew EF, Wilson NL, et al. Heterologous Expression of Mycobacterium Alkene Monooxygenases in Gram-Positive and Gram-Negative Bacterial Hosts. Appl Environ Microbiol. 2018;84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Coelho PS, Brustad EM, Kannan A, Arnold FH. Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome P450 enzymes. Science. 2013;339:307–10. [DOI] [PubMed] [Google Scholar]
- [101].Coelho PS, Wang ZJ, Ener ME, Baril SA, Kannan A, Arnold FH, et al. A serine-substituted P450 catalyzes highly efficient carbene transfer to olefins in vivo. Nat Chem Biol. 2013;9:485–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhang RK, Chen K, Huang X, Wohlschlager L, Renata H, Arnold FH. Enzymatic assembly of carbon-carbon bonds via iron-catalysed sp(3) C-H functionalization. Nature. 2019;565:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Biernat KA, Pellock SJ, Bhatt AP, Bivins MM, Walton WG, Tran BNT, et al. Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases. Sci Rep. 2019;9:825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kim JK, Choi MS, Jeong JJ, Lim SM, Kim IS, Yoo HH, et al. Effect of Probiotics on Pharmacokinetics of Orally Administered Acetaminophen in Mice. Drug Metab Dispos. 2018;46:122–30. [DOI] [PubMed] [Google Scholar]
- [107].Sweeny DJ, Li W, Clough J, Bhamidipati S, Singh R, Park G, et al. Metabolism of fostamatinib, the oral methylene phosphate prodrug of the spleen tyrosine kinase inhibitor R406 in humans: contribution of hepatic and gut bacterial processes to the overall biotransformation. Drug Metab Dispos. 2010;38:1166–76. [DOI] [PubMed] [Google Scholar]
- [108].Boer J, Young-Sciame R, Lee F, Bowman KJ, Yang X, Shi JG, et al. Roles of UGT, P450, and Gut Microbiota in the Metabolism of Epacadostat in Humans. Drug Metab Dispos. 2016;44:1668–74. [DOI] [PubMed] [Google Scholar]
- [109].Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. [DOI] [PubMed] [Google Scholar]
- [110].Li H, Jia W. Cometabolism of microbes and host: implications for drug metabolism and drug-induced toxicity. Clin Pharmacol Ther. 2013;94:574–81. [DOI] [PubMed] [Google Scholar]
- [111].Taneva E, Sinclair S, Mesquita PM, Weinrick B, Cameron SA, Cheshenko N, et al. Vaginal microbiome modulates topical antiretroviral drug pharmacokinetics. JCI Insight. 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 2017;179:204–22. [DOI] [PMC free article] [PubMed] [Google Scholar]