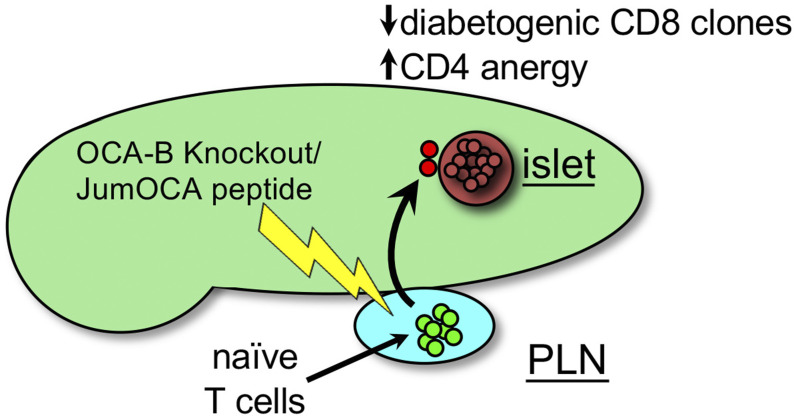
Kim and colleagues show that OCA-B in T cells is essential for the generation of type 1 diabetes. OCA-B loss leaves the pancreatic lymph nodes largely undisturbed but associates autoreactive CD4+ T cells in the pancreas with anergy while deleting potentially autoreactive CD8+ T cells.
Abstract
The transcriptional coregulator OCA-B promotes expression of T cell target genes in cases of repeated antigen exposure, a necessary feature of autoimmunity. We hypothesized that T cell–specific OCA-B deletion and pharmacologic OCA-B inhibition would protect mice from autoimmune diabetes. We developed an Ocab conditional allele and backcrossed it onto a diabetes-prone NOD/ShiLtJ strain background. T cell–specific OCA-B loss protected mice from spontaneous disease. Protection was associated with large reductions in islet CD8+ T cell receptor specificities associated with diabetes pathogenesis. CD4+ clones associated with diabetes were present but associated with anergic phenotypes. The protective effect of OCA-B loss was recapitulated using autoantigen-specific NY8.3 mice but diminished in monoclonal models specific to artificial or neoantigens. Rationally designed membrane-penetrating OCA-B peptide inhibitors normalized glucose levels and reduced T cell infiltration and proinflammatory cytokine expression in newly diabetic NOD mice. Together, the results indicate that OCA-B is a potent autoimmune regulator and a promising target for pharmacologic inhibition.
Graphical Abstract
Introduction
Type 1 diabetes (T1D) is an autoimmune disease in which the host immune system is directed toward antigens associated with pancreatic β cells (Cooke and Plotnick, 2008; Lernmark and Larsson, 2013). Pathologically, T1D is characterized by insulitis, β cell destruction, and inability to produce insulin. The main treatment for T1D, lifelong insulin therapy, treats symptoms but not cause. The development of new T1D treatments is limited by an incomplete understanding of disease mechanisms (Staeva et al., 2013). β cell regeneration is a promising line of therapy, but still requires methods to specifically block T1D autoimmunity. An ideal therapy would block autoimmunity early in the disease course to spare remaining β cell function while preserving normal immune function.
B cell–specific Oct1/2 coactivator (OCA-B), also known as Bob.1/OBF-1 (gene symbol Pou2af1), is a transcriptional coregulatory protein named for its strong expression in the B cell lineage, where it is dispensable until after B cell activation. Following B cell activation, it is essential for the generation of germinal centers (Shi et al., 2015). In CD4+ T cells, OCA-B docks with the transcription factor Oct1 to regulate a set of ∼150 target genes, including Il2, Ifng, and Csf2 (Gmcsf; Shakya et al., 2015). Upon T cell activation, many of these targets are activated by pathways that converge on transcription factors such as NFAT, AP-1, and NF-κB. Factors like NFAT can be thought of as the primary on/off switches for these genes, and drugs that block their activity effectively block target gene expression. Such drugs have utility in many contexts, but also have drawbacks including global dampening of immune function and side effects due to expression in other tissues. In contrast, Oct1 and OCA-B insulate silent but previously activated target genes against stable repression. Loss of either protein does not affect CD4+ T cell responses to stimulation in vitro or primary infection in mice (Kim et al., 2019; Shakya et al., 2015), but causes target gene expression defects upon secondary stimulation in vitro (Shakya et al., 2011, 2015). Loss of either protein also results in defective CD4+ memory T cell formation and recall responses in vivo (Shakya et al., 2015). In addition, OCA-B expression is largely confined to lymphocytes. OCA-B is undetectable in thymocytes and naive CD4+ T cells but becomes stably expressed after antigen stimulation (Shakya et al., 2015). Once expressed, OCA-B recruits the histone lysine demethylase Jmjd1a/Jhdm2a/Kdm3a to Oct1 at target loci, where it locally removes inhibitory H3K9me2 chromatin marks that would otherwise promote repressive chromatin environments (Shakya et al., 2015). OCA-B also potentiates polarization of activated helper T cells toward the Th17 phenotype, among the most pro-inflammatory helper T cell subsets (Yosef et al., 2013).
Persistent antigen exposure is a common property of autoimmune responses. Among profiled CD4+ T cell subsets, OCA-B levels are elevated in both central memory cells and pancreas-infiltrating, islet-reactive CD4+ T cells (Heng et al., 2008). Both Oct1 and OCA-B are highly conserved from mice to humans, and genome-wide association studies identify Oct1/OCA-B binding site polymorphisms that confer predisposition to a variety of human autoimmune diseases including T1D, multiple sclerosis, lupus, inflammatory bowel disease, and rheumatoid arthritis (Cunninghame Graham et al., 2006; Farh et al., 2015; Kiesler et al., 2009; Leon Rodriguez et al., 2016; Maurano et al., 2012; van Heel et al., 2002; Vince et al., 2016). Other studies implicate polymorphisms at the Ocab (Pou2af1) locus itself in different forms of autoimmunity (Ban et al., 2006; Nakamura et al., 2012). We therefore hypothesized that targeting OCA-B would inhibit autoreactive, diabetogenic T cell responses.
Here, we generate a T cell conditional mouse model of OCA-B deficiency. We show that OCA-B ablation protects animals from spontaneous, polyclonal T1D. Protection was associated with reduced autoreactivity of autoantigen-specific CD8+ T cells in the pancreatic LNs (PLNs), reduced representation of these same TCR clones in the pancreatic islets, and the induction of anergy in potentially autoreactive islet CD4+ T cell clones. The protection associated with OCA-B loss varied among different TCR transgenic and monoclonal antigen models and was weakest using synthetic antigens and monoclonal transplant models. Protection was stronger using a spontaneous autoantigen-specific TCR transgenic model. Using a rational-design approach, we developed membrane-penetrating OCA-B peptide inhibitors that are efficacious in vitro and in mice. Collectively, the results show that OCA-B is a potent and pharmacologically accessible autoimmune regulator.
Results
Generation of an Ocab conditional allele and nonobese diabetic (NOD) backcrossing
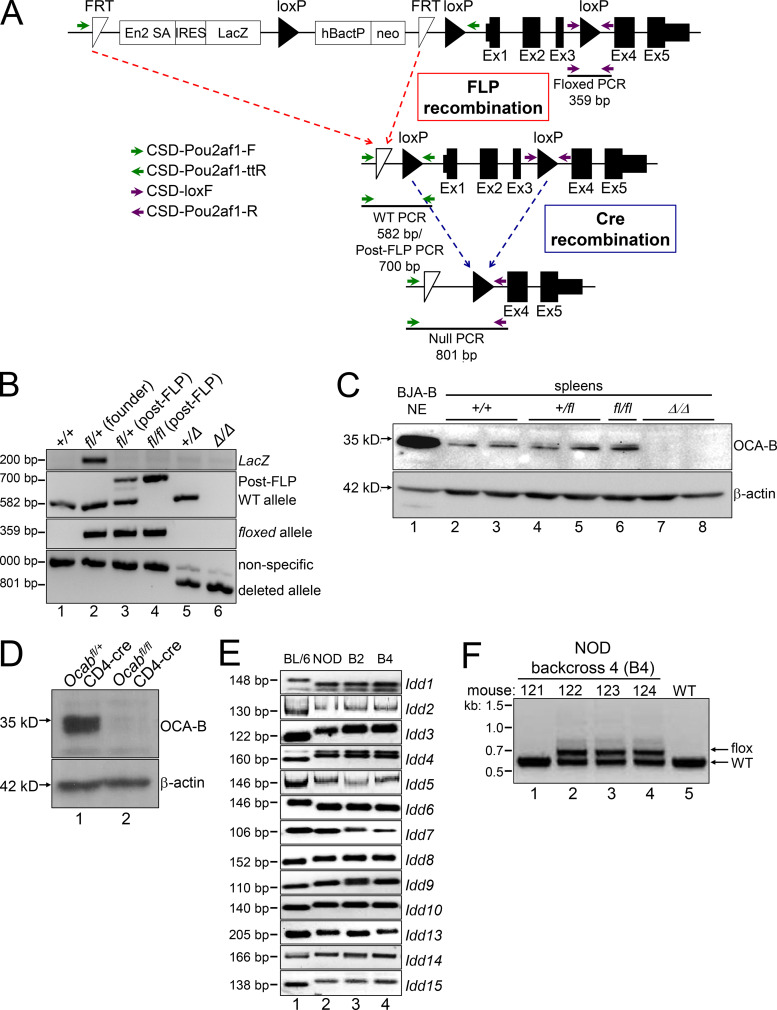
We generated a conditional Ocab (Pou2af1) mouse allele using embryonic stem cells provided by the Knockout Mouse Project (KOMP) repository. Deletion of the LoxP sites creates a null allele by removing the first three exons (Fig. S1 A). Breeding chimeric mice resulted in germline transmission and a founder mouse (Fig. S1 B, lane 2) that was crossed to a flippase (FLP) deleter mouse strain (FLPRosa26) to remove the reporter cassette and create a floxed conditional allele (Fig. S1, A and B, lane 3). Intercrossing these mice generated fl/fl homozygotes (Fig. S1 B, lane 4), which were crossed to a germline Cre deleter strain (CreRosa26) to generate germline null (Δ) alleles (lanes 5 and 6). As expected, homozygous fl/fl mice produce normal amounts of both p34 and p35 OCA-B protein isoforms (Fig. S1 C, lane 6), while no protein of either isoform is present in Δ/Δ spleens (lanes 7 and 8). These results indicate that the fl allele is OCA-B sufficient and the Cre-deleted allele is a null. Crossing the fl allele onto a CD4-cre driver, which deletes in both CD4 and CD8 compartments, resulted in efficient deletion in splenic T cells (Fig. S1 D). This allele therefore represents a robust system in which to study OCA-B function in T cells.
Figure S1.
Ocab (Pou2af1) conditional allele. (A) Targeting event. Crossing with FLPRosa26 results in conditional (fl) allele. Primer pairs used for genotyping are depicted with arrows. FRT, flippase recognition target. (B) Example genotyping of the targeted allele and recombination events. The founder animal is in lane 2. The primer pairs shown in A were used. (C) OCA-B immunoblots from spleens of control, fl/fl, and Δ/Δ animals. BJA-B B cell nuclear extract is shown as a positive control (lane 1). (D) The Ocab (Pou2af1) conditional allele was crossed to CD4-cre. Total splenic CD4+ T cells were isolated from an Ocabfl/flCD4-cre animal or a Ocab+/flCD4-cre littermate control, and stimulated for 2 d in vitro using plate-bound CD3ε and soluble CD28 antibodies. An OCA-B immunoblot of stimulated total T cells is shown. β-Actin is shown as a loading control. (E) To generate NOD.Ocab conditional mice by speed congenic backcross, microsatellite repeat polymorphisms at the Idd loci were tested by PCR using the primers in Table S1. Backcross generations 2 and 4 (B2 and B4) are shown (lanes 3 and 4), together with parent C57BL/6 (lane 1) and target NOD (lane 2) genomic DNA as controls. Images are of different PCR products resolved using agarose or PAGE. (F) Example genotyping of Ocab WT and conditional alleles in B4 backcrossed mice. An agarose gel image is shown. Mouse 122 corresponds to the B4 animal shown in B and was used as the founder animal. This animal was crossed with NOD.CD4-cre for subsequent experiments with the conditional allele.
The conditional allele was generated on a C57BL/6 background. To test the role of OCA-B expression in T cells in T1D emergence, we conducted congenic backcrosses to the NOD strain background. This method allows spontaneous diabetes to be produced rapidly by screening and selectively breeding mice with 13 microsatellite and single-nucleotide polymorphism markers associated with autoimmunity (Serreze et al., 1996). Ocab is located on mouse chromosome 9 and distant from any of the Idd loci. Following these markers with specific primer pairs (Table S1), we produced backcrossed animals with all 13 markers (Fig. S1 E) that recapitulate spontaneous autoimmunity (not shown). For example, Idd1, which maps to the MHC region (Hattori et al., 1986), converted to the homozygous NOD allele after two backcross generations (Fig. S1 E, lane 3), whereas Idd13, which maps near β-2-microglobulin (Serreze et al., 1998), became NOD homozygous after four generations (lane 4). Genotyping identified this fourth-generation backcrossed animal as having all 13 microsatellite markers and the Ocab conditional allele (Fig. S1 F, 122). This founder animal was crossed to NOD.CD4-cre in order to delete OCA-B in T cells. Similar NOD backcrosses conducted using Ocab null mice (Kim et al., 1996) resulted in OCA-B germline-deficient NOD mice (not shown). As with the original C57BL/6 OCA-B germline knockout (Kim et al., 1996), all alleles were viable and fertile with no obvious health issues.
Normal baseline T cell status in NOD mice lacking OCA-B
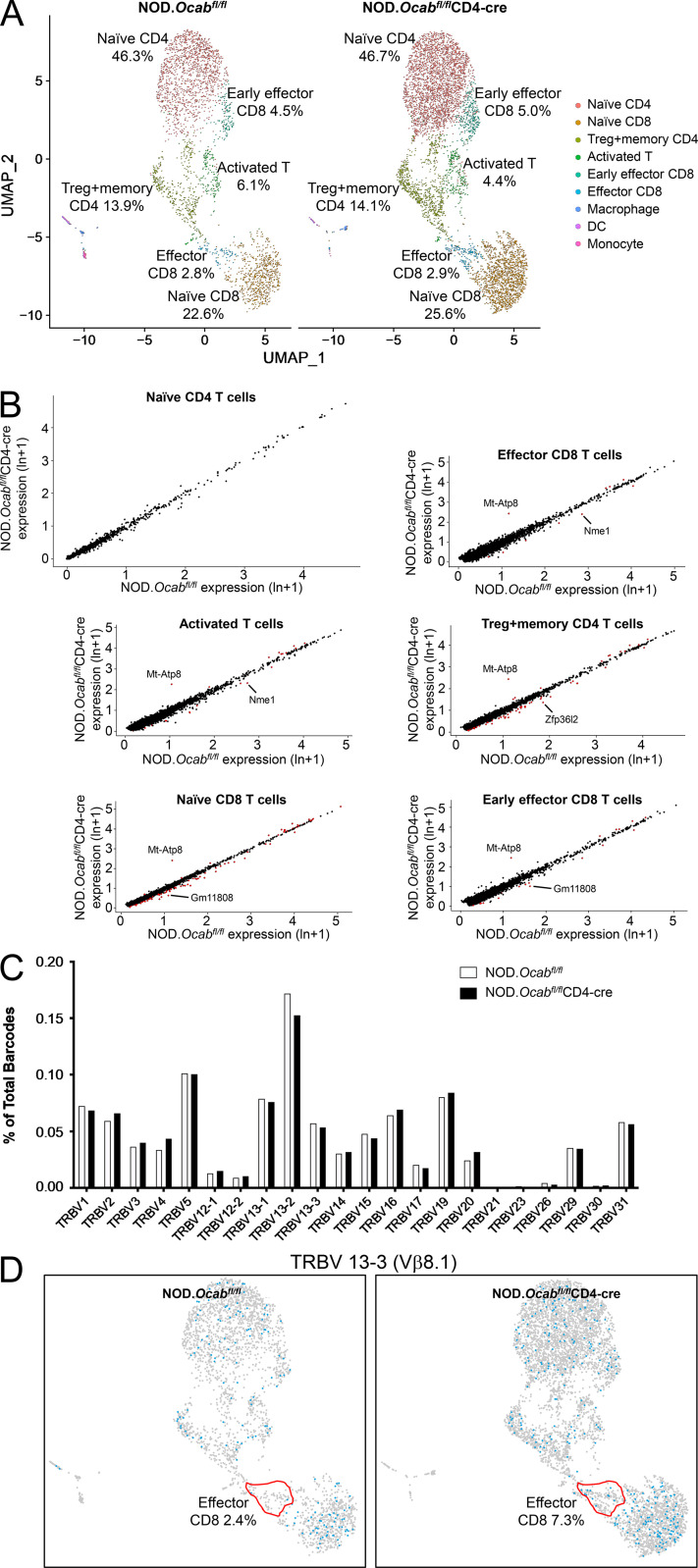
To study the effects of T cell–specific OCA-B loss on T cell function, we performed single-cell RNA sequencing (scRNAseq) and TCR clonotype analysis using CD3+ T cells isolated from PLNs of prediabetic 8-wk-old female NOD.Ocabfl/flCD4-cre or NOD.Ocabfl/fl littermate controls. For each group, cells from three animals were combined for microfluidics, sequencing, and analysis. Approximately 6,000 knockout and ∼3,000 control cells were identified in this analysis, most of which comprise naive CD4+ and CD8+ cells (Fig. S2 A, pink and orange) and regulatory T (T reg) cells (green). There was also a small population of activated CD4+ and CD8+ cells (blue-green), presumably in the process of exiting the lymph nodes. Other than a small decrease in the percentage of activated T cells (4.4% vs. 6.1%), few differences in relative abundance of the populations were observed. None of the populations expressed measurable amounts of Ocab (Pou2af1) mRNA (not shown); however, Pou2af1 is expressed at low levels in T cells, making it difficult capture by scRNAseq. In addition, few changes in gene expression were identified (Fig. S2 B and Table S2). We also determined TCR repertoires, identifying few changes across all populations (Fig. S2 C). However, specific subpopulations did show changes in TCR utilization. For example, >90% of NOD CD8+ T cells that display reactivity to islet-specific glucose-6-phosphatase catalytic subunit–related protein (IGRP) residues 206–214 express Trbv13-3 (Vβ8.1; Wong et al., 2006). Although this clone was unchanged across the entire population of T cells (Fig. S2 C), it was over-represented in the effector CD8+ population (Fig. S2 D). In contrast, other TCR clonotypes such as Trbv13-1, Trbv13-2, Trbv1, and Trbv15 were unchanged (not shown).
Figure S2.
Unaltered T cell populations and TCR representation in prediabetic NOD.Ocabfl/flCD4-cre PLNs compared with NOD.Ocabfl/fl littermate controls. (A) CD3+ T cells were isolated and combined from the PLNs of three 8-wk-old NOD.Ocabfl/flCD4-cre mice or three NOD.Ocabfl/fl littermate controls. scRNAseq was performed. Clusters were overlaid in UMAP plots. DC, dendritic cell. (B) Six different populations from A were analyzed for differential gene expression. Identified genes are shown as scatter plots. Significantly differentially expressed genes (adjusted P value <0.05) are shown in red. ln, natural logarithm. (C) Percentage contribution of individual variable β chains is shown. (D) Trbv13-3–expressing cells are shown in blue superimposed on the same UMAP scRNAseq data as in A. The effector CD8+ T cell cluster is highlighted in red.
NOD mice lacking OCA-B are protected from T1D
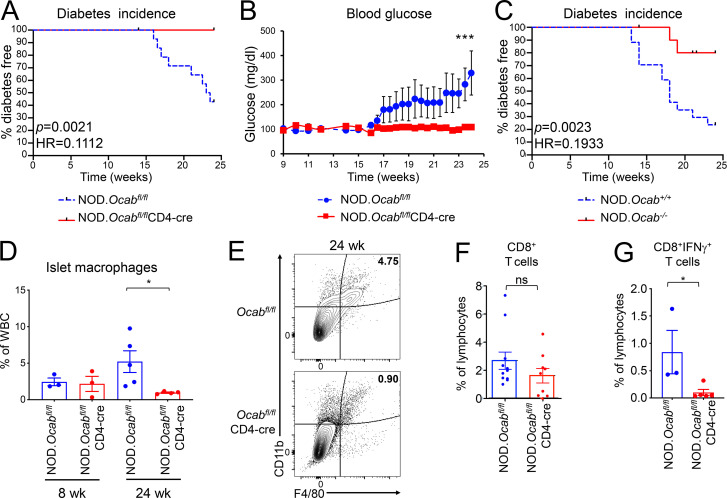
T1D onset in NOD mice is spontaneous and acute (Makino et al., 1985). To test the impact of OCA-B loss in T cells on spontaneous T1D, we compared incidence in female NOD.Ocabfl/flCD4-cre mice and littermate controls lacking CD4-cre. Glucose was monitored two or three times per week. Approximately 60% of Ocabfl/fl control mice manifested spontaneous diabetes by 24 wk of age, reaching an average of >300 mg/dl by 24 wk, while no Ocabfl/flCD4-cre mice became diabetic (Fig. 1, A and B). Similar results were observed using female whole-body knockout NOD.Ocab−/− compared with Ocab+/+ littermate controls. While ∼80% of control females developed T1D within 24 wk after birth, only ∼20% of NOD.OcabΔ/Δ females did so (Fig. 1 C). The differences in T1D penetration are likely attributable to differences in the NOD backcrosses, as the 13 Idd NOD determinants largely but not completely dictate the autoimmune phenotype. Cumulatively, the results show that OCA-B expression in T cells is critical for T1D pathogenesis in NOD mice.
Figure 1.
Loss of OCA-B protects NOD mice from T1D. (A) Kaplan–Meier plot of diabetes-free survival in littermate female NOD.Ocabfl/flCD4-cre (n = 12) and NOD.Ocabfl/fl (n = 14) mice. (B) Average blood glucose levels from mice shown in A. Student’s t test P values: day 23, 0.0458; day 24, 0.0403; and day 25, 0.0494. (C) Percentage of diabetes-free survival in germline knockout NOD.Ocab−/− (n = 16) and control NOD.Ocab+/+ (n = 17) mice was plotted. (D) Pancreatic islet leukocytes were isolated from 8-wk-old or 24-wk-old littermate female NOD.Ocabfl/flCD4-cre (8 wk, n = 3; 24 wk, n = 5) or NOD.Ocabfl/fl (8 wk, n = 3; 24 wk, n = 4) mice and analyzed by flow cytometry. Mean CD45+CD11b+F4/80+ cell frequencies are depicted. Student’s t test P value = 0.0397. (E) Frequencies of CD45+CD11b+F4/80+ cells from representative animals in D are shown. Plots were gated on CD45. (F) Mean percentages of total pancreatic-infiltrating CD8+ T cells in 9 experimental NOD.Ocabfl/flCD4-cre or 11 littermate control NOD.Ocabfl/fl 12-wk-old mice are plotted. Cells were gated on CD45. (G) IFN-γ–expressing CD8+ T cell percentages in 16-wk-old NOD.Ocabfl/flCD4-cre (n = 3) and littermate control islets (n = 5) are shown. Student’s t test P value = 0.0477. All error bars denote ±SEM. *, P ≤ 0.05; ***, P ≤ 0.001. ns, not significant; HR, hazard ratio.
We sacrificed Ocabfl/flCD4-cre female mice and controls at 24 wk and determined immune cell frequencies in the pancreata. We observed more T cells in the pancreata of OCA-B–deficient mice (not shown). A relative increase in infiltrating T cells in knockouts at endpoint is understandable because the controls have annihilated their pool of β cells and associated antigens. We also studied pancreatic macrophages in these mice. Islet infiltration of F4/80+CD11b+ macrophages is critical for NOD T1D pathogenesis in both spontaneous and adoptive transfer T1D models (Peterson et al., 1998; Rosmalen et al., 2000). CCL1 expressed in diabetogenic CD4+ T cells recruits CCR8-expressing macrophages (Cantor and Haskins, 2007). OCA-B deletion decreases Ccl1 mRNA expression in stimulated, rested, and restimulated CD4+ T cells compared with controls (Table S3). Despite the presence of antigen and greater numbers of T cells, we observed fewer macrophages in 24-wk NOD.Ocabfl/flCD4-cre islets (Fig. 1, D and E). In contrast, macrophages were unaffected by T cell–specific OCA-B loss in prediabetic (8 wk) mice (Fig. 1 D).
IFN-γ is a direct OCA-B target gene (Shakya et al., 2015). Serum and pancreatic IFN-γ levels gradually rise in young NOD mice, reaching maximum at diabetes onset (Schloot et al., 2002). Diabetogenic CD4+ and CD8+ T cells express IFN-γ in both humans and NOD mice (Krishnamurthy et al., 2008; Newby et al., 2017; Wang et al., 1997). We therefore predicted that OCA-B loss would reduce IFN-γ expression in islet CD4+ and CD8+ T cells. We compared IFN-γ–expressing pancreatic CD4+ and CD8+ T cells in female NOD.Ocabfl/flCD4-cre to littermate control mice lacking CD4-cre. In 12-wk prediabetic mice, CD8+ T cell frequencies were only slightly decreased (Fig. 1 F). CD4+ T cells at the same time point were also unchanged (not shown). At the time of diabetes onset (16 wk), fewer IFN-γ–expressing CD8+ T cells were present in Ocab conditional-deleted mice compared with controls (Fig. 1 G).
T cell–specific OCA-B deletion alters the islet-infiltrating TCR repertoire
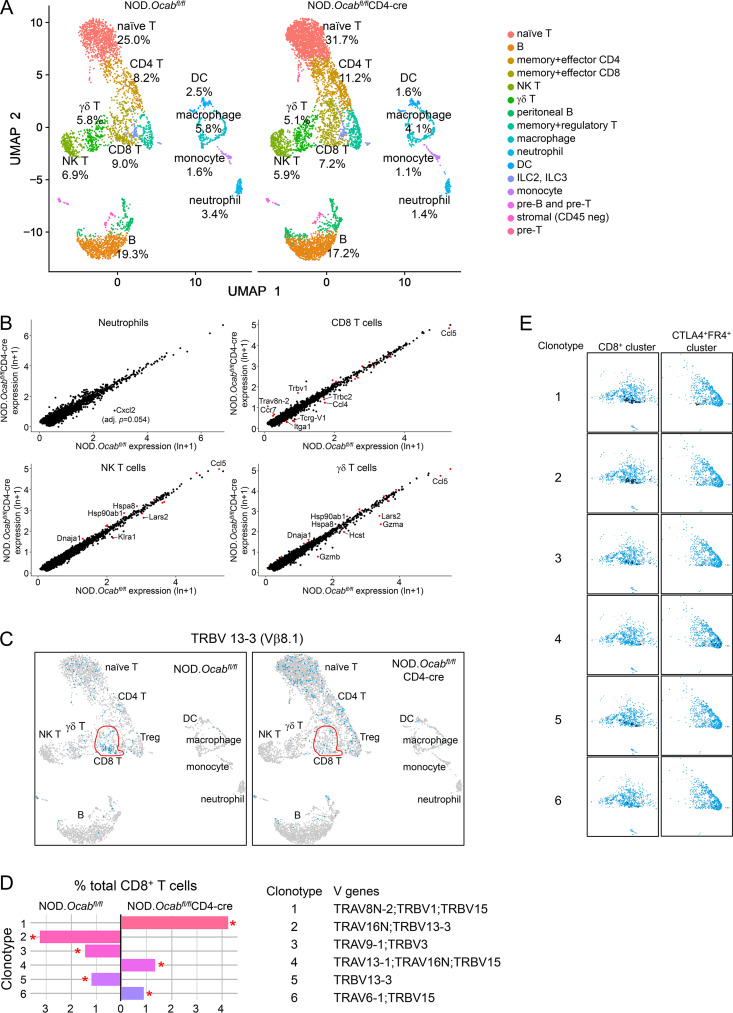
We performed scRNAseq and TCR clonotype analysis similar to Fig. S2 but using older (12 wk) prediabetic NOD.Ocabfl/flCD4-cre or NOD.Ocabfl/fl littermate control female mice and using total CD45+ cells isolated from pancreatic islets. Cells were isolated from four animals per group and combined for microfluidics and sequencing. After filtering, ∼6,200 knockout and ∼3,800 control cells were identified. Overlaying cells from both genotypes, we used their gene expression profiles to definitively identify cell lineages, including naive T cells, memory/effector CD4+ T cells, memory/effector CD8+ T cells, B cells, macrophages, and neutrophils (Fig. 2 A). Among the different populations, the biggest relative change was neutrophils, which decreased from 3.4% to 1.4% in conditional knockout islets (Fig. 2 A). B cells were also decreased, albeit more slightly. These changes were accompanied by increased percentages of naive T cells (32% vs. 25%). Prediabetic islets also showed significant changes in gene expression between the two groups (Fig. 2 B and Table S4), the strongest of which occurred in neutrophils (e.g., Cxcl2, down), memory/effector CD8+ T cells (Ccl4, Tcrg-v1, Itga, down; Tcrbv1, Trav8n-2, up), natural killer T cells (Klra2, Lars2, down), and γδT cells (Gzma, Gzmb, Ccl5, down).
Figure 2.
T cell conditional OCA-B loss reduces the numbers of activated, autoreactive pancreatic islet T cells. (A) scRNAseq using total islet CD45+ cells from prediabetic NOD.Ocabfl/flCD4-cre or littermate control NOD.Ocabfl/fl mice (n = 4 for each group). Cell populations were plotted using UMAP (Seurat R package), and percentages in each cluster are shown for each genotype. Clusters were identified using the Seurat R package function FindMarkers. (B) Four clusters from A were analyzed for differential gene expression. Identified genes are shown as a scatter plot. Significantly differentially expressed genes (adjusted P value <0.05) are shown in red. For Cxcl2 in neutrophils, P = 3.75 × 10−6, adjusted P = 0.055. (C) UMAP plots similar to A, except cells expressing TCR clonotype 13–3 are shown. The nonnaive (activated + memory) CD8+ cell population identified in A is shown in red. (D) Percent contribution of the top six identified TCR clonotypes to total activated + memory CD8+ cells is shown. V genes comprising the clonotypes are shown below. (E) Cells positive for clonotypes 1–5 are shown for two clusters, nonnaive (activated + memory) CD8+ T cells and CTLA4+FR4+ anergic cells (consisting of mostly CD4+ cells). For each cluster, positive cells are shown in dark blue. An overlay of control and OCA-B–deficient cell populations is shown. Each clonotype is only observed in a single genotype (D), allowing all cells to be mapped back entirely to OCA-B–deficient (clonotypes 1 and 4) or control (clonotypes 2, 3, and 5).
We also profiled TCR utilization in pancreatic T cells. Across total T cells, few changes were observed (Fig. S3); however, this was largely due to similar representation in the naive T cell pool. Isolating antigen-experienced CD4+ and CD8+ effector + memory subpopulations revealed sharp differences. For example, >90% of diabetogenic CD8+ T cells displaying reactivity to IGRP206-214 express Trbv13-3 (Vβ8.1; Wong et al., 2006). Relative to controls, this clonotype was under-represented in OCA-B–deficient effector + memory CD8+ cells (Fig. 2 C; 20.9% vs. 8.6%). Based on this information, we repeated the clonotype analysis, but only using the effector + memory CD8+ T cell cluster. Within this population, dominant TCR clonotypes were strikingly different (Fig. 2 D). Clonotype 1, for example, is the most abundant, and corresponds to a TCRα V8N-2 chain paired with TCRβ V1 or V15. It has not been associated with T1D. This clonotype was absent in control islets but represented >4% of OCA-B–deficient islet CD8+ effector + memory T cells (Fig. 2 D). Clonotype 1 cells overlaid with both genotypes are shown in Fig. 2 E. Clonotype 2, by contrast, expresses the T1D-associated Vβ8.1 TCR β chain. This clonotype was strongly represented in controls (>3%) but absent from OCA-B–deficient effector + memory CD8+ cells (Fig. 2 D). In contrast to CD8, CD4-dominant TCR clonotypes associated with T1D were still present in knockouts but associated with markers of anergy. For example, clonotype 4 (Trav13-1/16N+Trbv15) is associated with pro-diabetogenic, BDC2.5 Ag-reactive CD4+ T cells (Li et al., 2009). This clonotype was enriched in the OCA-B–deficient effector + memory CD4+ cells, but only in a CTLA4- and FR4-expressing subpopulation (Fig. 2 E). Cumulatively, these results show that OCA-B loss from T cells significantly skews the TCR landscape of islet effector/memory-like T cells to an anti-diabetogenic phenotype.
Figure S3.
TCR β-chain representation across the entire population of prediabetic NOD.Ocabfl/flCD4-cre pancreatic islets compared with NOD.Ocabfl/fl littermate controls. Four 12-wk-old mice in each group were combined for scRNAseq TCR analysis.
Autoantigen-specific CD8+ T cells are present in PLNs of OCA-B–deficient mice but show reduced reactivity and islet penetration
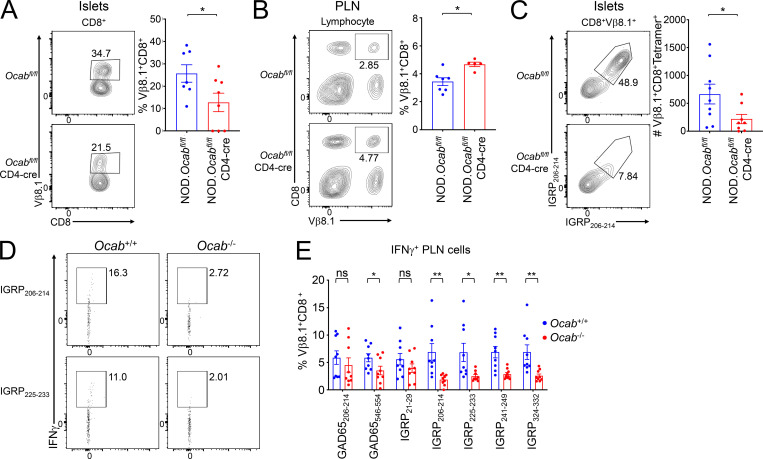
To understand the basis for the reduction in autoantigen-specific CD8+ T cell islet clones, we studied Vβ8.1 expression and IGRP206-214 reactivity in matched pancreatic islets and PLNs from 12-wk prediabetic female Ocabfl/flCD4-cre mice or littermate Ocabfl/fl controls. Islet CD8+ T cells showed decreased Vβ8.1 usage (Fig. 3 A), while PLNs from the same animals showed a corresponding increase (Fig. 3 B). To directly test diabetogenic, antigen-specific islet CD8+ T cells, we used IGRP206-214 H-2Kd class I tetramers, which recognize TCRs expressing Vβ8.1. Islet CD8+Vβ8.1+ T cells showed a strong decrease in IGRP206-214 specificity (Fig. 3 C).
Figure 3.
Autoantigen-specific CD8+ cells become nonreactive and fail to infiltrate islets in the OCA-B–deficient condition. (A) Pancreatic leukocytes were isolated from prediabetic 12-wk-old littermate female NOD.Ocabfl/flCD4-cre (n = 9) or NOD.Ocabfl/fl (n = 10) mice and analyzed by flow cytometry. Representative plots (left) and total CD8+Vβ8.1+ T cell numbers (right) are shown. Student’s t test P value = 0.0416. (B) PLN leukocytes were isolated from the same mice (NOD.Ocabfl/flCD4-cre, n = 7 or NOD.Ocabfl/fl, n = 5) as in A. Representative plots (left) and total CD8+Vβ8.1+ T cell frequencies (right) are shown. Student’s t test P value = 0.0355. (C) Representative plots (left) and total cell numbers (right) of CD8+Vβ8.1+ H-2Kd IGRP206-214 tetramer-positive islet T cells are depicted from NOD.Ocabfl/flCD4-cre (n = 9) or NOD.Ocabfl/fl (n = 10). Student’s t test P value = 0.0449. (D) Total PLN WBCs were isolated from prediabetic 12-wk-old littermate NOD.Ocab−/− or NOD.Ocab+/+ mice and restimulated with IGRP peptides (IGRP206-214 or IGRP225-233). Cells were analyzed for IFN-γ expression by flow cytometry. (E) Mean percentages of cells expressing IFN-γ from nine experiments (three replicates from three Ocab+/+ and three Ocab−/− mice) for the peptides in D, as well as two GAD65 peptides (206–214 and 546–554) and three additional IGRP peptides (21–29, 241–249, and 324–332). Significant Student’s t test P values were as follows: GAD65546-554, 0.046; IGRP206-214, 0.005; IGRP225-233, 0.0197; IGRP241-249, 0.0018; IGRP324-332, 0.0063. All error bars denote ±SEM. *, P ≤ 0.05; **, P ≤ 0.01. ns, not significant.
The decrease in autoantigen-specific islet CD8+ T cells and corresponding increase in the PLNs in OCA-B T cell–deficient mice raised two possibilities: cells in the PLNs could fail to become activated or could become activated but fail to migrate to the pancreas. In the first case, more antigen-reactive cells would be observed, while in the second, there would be fewer. To distinguish between these possibilities, we performed peptide stimulation experiments using seven different known diabetogenic self-peptides. We used five IGRP peptides, including IGRP206-214, which corresponds to the tetramer used above and the NY8.3 TCR transgenic mice used below. We also used two GAD65 epitopes, 206–214 and 546–554. Five of the seven peptides showed significant reductions in autoreactivity, including IGRP206-214 (Fig. 3, D and E). These results indicate that potentially autoreactive cells fail to become activated in the PLNs of OCA-B–deficient mice.
Protection conferred by OCA-B loss is model specific
T1D in NOD mice originates from defective negative selection in developing T cells, resulting in T cell autoreactivity. OCA-B is not expressed in thymocytes, and OCA-B loss does not appear to affect T cell development (Shakya et al., 2015). Therefore, the protective effect of OCA-B loss in T cells likely arises from blunting preexisting autoreactivity in the periphery. To test the hypothesis that OCA-B loss confers protection in simplified systems, we crossed OCA-B–deficient mouse models using TCR transgenes and model antigens.
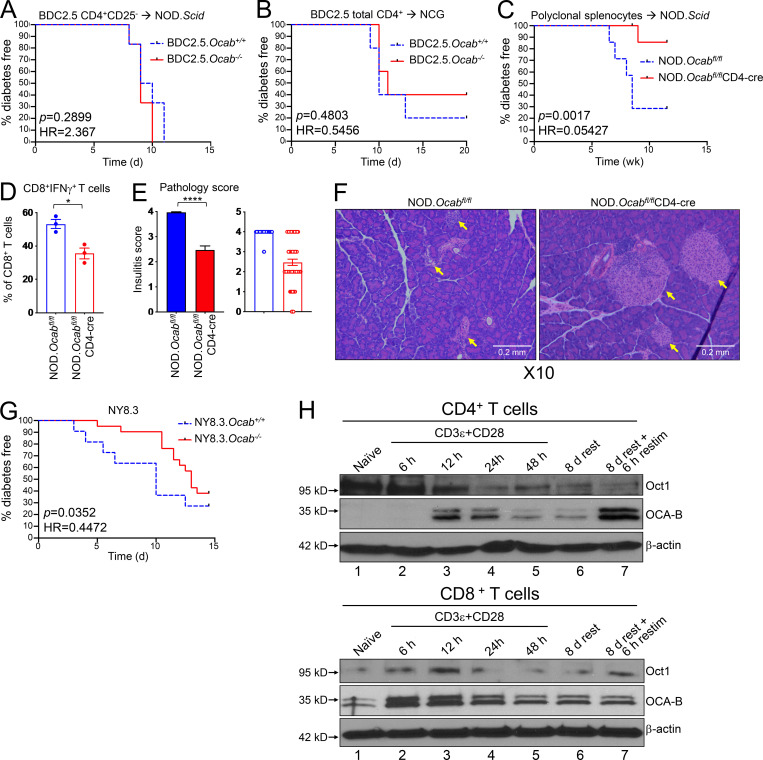
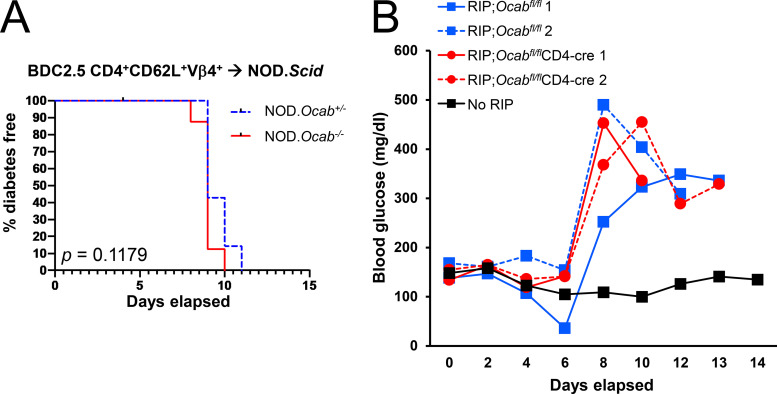
We crossed the germline null Ocab allele to BDC2.5 mice (Katz et al., 1993), which express a CD4-restricted TCR specific for a hybrid insulin–chromogranin A peptide (Delong et al., 2016). Transferring T reg cell–depleted T cells from BDC2.5 TCR transgenic mice into NOD.Scid mice results in disease after ∼10 d (Berry and Waldner, 2013; Presa et al., 2015). We transferred CD4+CD25− T cells from germline Ocab−/− or Ocab+/+ BDC2.5 mice into NOD.Scid recipients, monitoring diabetes emergence. Transplants from both Ocab−/− and control NOD.BDC2.5 mice resulted in equivalent disease kinetics (Fig. 4 A) and severity (not shown). Transferring Ocab−/− or control NOD.BDC2.5 CD4+CD62L+Vβ4+ T cells into NOD.Scid recipients, or total CD4+ T cells into NCG recipients, also resulted in equivalent disease (Fig. S4 A and Fig. 4 B).
Figure 4.
OCA-B loss protects NOD mice from T1D in spontaneous and polyclonal splenocyte transfer models but not monoclonal transfer models. (A) 2 × 105 purified CD4+CD25− splenic T cells from NOD.BDC2.5.Ocab−/− or control NOD.BDC2.5.Ocab+/+ donors were injected retro-orbitally into NOD.Scid (n = 6 for each group) mice. Diabetes-free survival is shown. (B) 1.5 × 106 purified splenic total CD4+ T cells from NOD.BDC2.5.Ocab−/− or control donors were transferred into NCG mice (n = 5 for each group). Mice were monitored for diabetes development. (C) Total NOD splenocytes (5 × 106) from prediabetic 6–8-wk-old NOD.Ocabfl/flCD4-cre or control NOD.Ocabfl/fl donors were adoptively transferred into sex-matched NOD.Scid (n = 7 for each group) recipients. T1D-free survival is shown. (D) 13 wk after transfer, the proportion of IFN-γ–expressing islet CD8+ T cells was assessed by flow cytometry from mice in C. Student’s t test P value = 0.0136. (E) Pancreata from the same mice as in C were fixed, sectioned, and H&E stained. Pathological scores are shown based on six or seven islets per slide, three slides per mouse, and three mice per group (>60 islets/group). Student’s t test P value = 1.46 × 10−15. (F) Example pancreatic images from endpoint animals. Yellow arrows indicate islet positions. Images were collected at 10× magnification. (G) The Ocab null allele was crossed to NY8.3 TCR transgenic mice. Spontaneous T1D was measured in female Ocab−/− (n = 20) or control Ocab+/+ (n = 12) littermates. (H) Naive CD4+ or CD8+ T cells (CD8aneg or CD4neg,CD11bneg,CD45Rneg,DX5neg,Ter-119neg,CD44lo) were isolated from C57BL/6 spleens and stimulated for up to 2 d in vitro using anti-CD3ε and CD28 antibodies. Cells were then washed and replated in the presence of exogenous IL-2. After 8 d rest in culture, cells were restimulated for 6 h. Lysates were prepared from each step and subjected to OCA-B immunoblotting to assess changes in expression. Oct1 and β-actin are shown as controls. All error bars denote ±SEM. *, P ≤ 0.05; ****, P ≤ 0.0001. HR, hazard ratio; restim, restimulated.
Figure S4.
Germline and T cell conditional OCA-B loss has no effect in two different T1D transplant model systems. (A) 5 × 105 purified CD4+CD62L+Vb4+ T cells from NOD.BDC2.5.Ocab−/− or heterozygous control NOD.BDC2.5.Ocab+/− donors were injected retro-orbitally into NOD.SCID recipients (n = 7 or 8 per group). T1D-free survival following transplant is shown. (B) 10,000 OT-I T cells were transferred retro-orbitally into C57BL6 RIP-mOVA;Ocabfl/flCD4-cre mice or littermate RIP-mOVA;Ocabfl/fl controls. Mice were infected with 2,000 colony-forming units L. monocytogenes expressing chicken OVA (Lm-OVA) later the same day. Glucose was measured every 2 d and plotted. Two mice in each group were used.
We also tested the effect of T cell conditional OCA-B deletion in a different strain background, C57BL/6J, using the artificial membranous chicken OVA antigen expressed from the rat insulin promoter–membranous OVA (RIP-mOVA; Van Belle et al., 2009). WT OVA-reactive CD8+ OT-I T cells were transferred into either Ocabfl/flCD4-cre or control Ocabfl/fl RIP-mOVA mice. In this model, host CD4+ T cells help transferred OT-I T cells promote T1D pathogenesis (Kurts et al., 1997). Mice were infected with Listeria monocytogenes–expressing chicken OVA to induce strong and synchronous β cell destruction and loss of glucose control (>400 mg/dl). No differences were observed using host mice with or without OCA-B T cell deletion (Fig. S4 B). These results indicate that OCA-B does not act analogously to transcription factors that regulate many of the same genes, such as NFAT. Knockouts of these factors would result in broad immunosuppression and protection from T1D across models. Rather, our results suggest that OCA-B loss selectively blunts immune activation in contexts such as autoantigen recognition. To test this supposition and rule out the possibility that T cell transfer itself does not blunt the effect of OCA-B loss, we transferred total splenocytes from polyclonal NOD.Ocabfl/flCD4-cre mice or controls. This model, which takes weeks rather than days to develop T1D, recapitulates robust T1D protection with T cell–specific OCA-B loss (Fig. 4 C). Protection was associated with fewer IFN-γ–expressing islet cytotoxic T cells (Fig. 4 D) and reduced insulitis in pancreatic histological sections (Fig. 4, E and F).
Because the Vβ8.1 clonotype and IGRP206-214-reactive cells were significantly attenuated in the islets of T cell conditional OCA-B–deficient mice (Fig. 2, C–E; and Fig. 3, A–C), we crossed the Ocab germline allele to the NY8.3 TCR transgenic line, which expresses a CD8-restricted TCR directed toward IGRP206-214 and manifests spontaneous T1D (Verdaguer et al., 1997). We passively monitored Ocab−/− or control Ocab+/+ NY8.3 mice for T1D development. Significant protection was observed (Fig. 4 G; hazard ratio [HR] = 0.4472, P = 0.0352), though the degree of protection was less than in OCA-B germline knockout polyclonal models. Together with the decreased IGRP reactivity in OCA-B–deficient PLNs, this result indicates that OCA-B promotes T1D in peripheral auto-reactive CD8+ T cells. Using immunoblotting with OCA-B antibodies, we confirmed that OCA-B is expressed in primary splenic C57BL/6 CD8+ T cells. Unlike CD4+ cells, where OCA-B is undetectable in naive cells but becomes induced with prolonged activation (Fig. 4 H, top panels), OCA-B could be detected in naive CD8+ cells and was rapidly augmented upon activation (Fig. 4 H, lanes 1 and 2). Similar to CD4+ cells, expression was stably maintained when the stimulus was removed and cells were rested with IL-2 (lane 6).
OCA-B deficiency promotes CD4+ T cell anergy in vitro
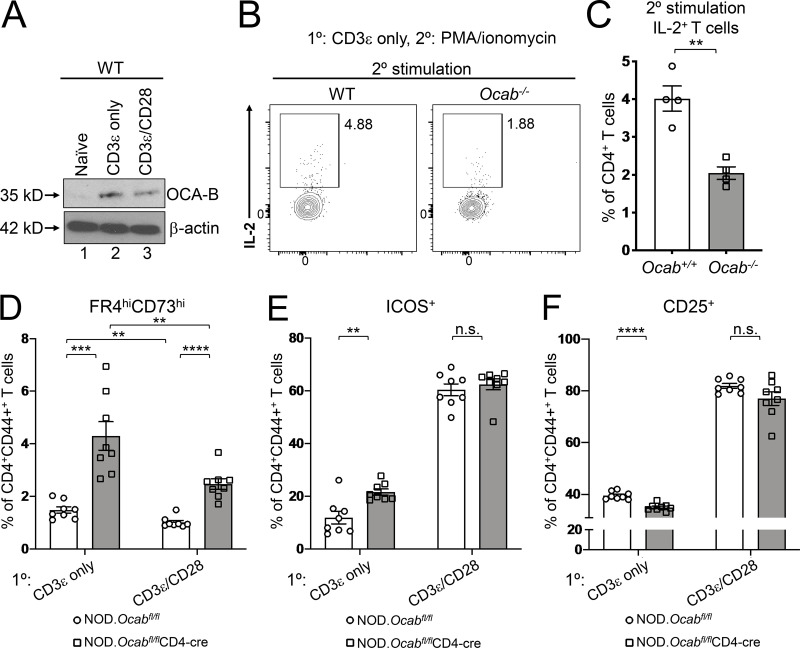
CD8-restricted TCR clones associated with T1D are depleted from the islets of OCA-B T cell conditional mice. In contrast, CD4-restricted clones are increased but associated with an anergic phenotype (Fig. 2 E). Anergy is a peripheral tolerance mechanism in which T cells that encounter antigen without costimulation become nonfunctional (Kalekar et al., 2016; Kearney et al., 1994; Vanasek et al., 2001). Normal naive primary CD4+ T cells can be activated in culture by TCR and costimulatory receptor activation, e.g., with anti-CD3ε/CD28 antibodies. Providing TCR signals without CD28 antibodies generates anergic responses (Chai and Lechler, 1997). Interestingly, OCA-B is induced to the same extent with or without costimulation (Fig. 5 A). To determine if OCA-B loss promotes anergic CD4+ T cell responses, we stimulated naive splenic CD4+ T cells from germline knockout Ocab−/− and control Ocab+/+ animals ex vivo using plate-bound anti-CD3ε antibodies without costimulation and restimulated the cells with PMA/ionomycin. IL-2 responses were weak in restimulated cells, as expected given the absence of initial costimulation. CD4+ T cells lacking OCA-B generated ∼2.5-fold less IL-2 (Fig. 5, B and C), indicating that OCA-B provides a barrier against anergic responses.
Figure 5.
OCA-B loss in CD4+ T cells increases anergy in vitro. (A) Naive CD4+ C57BL/6 T cells were stimulated in vitro for 24 h with anti-CD3ε antibodies ± costimulation with CD28 antibodies. Lysates were prepared and subjected to immunoblotting using OCA-B antibodies. β-Actin is shown as a loading control. (B) Naive OCA-B–deficient and control CD4+ T cells were stimulated in vitro with anti-CD3ε antibodies. 48 h later, the cells were restimulated with PMA and ionomycin for 6 h in the presence of brefeldin A, stained for intracellular IL-2, and analyzed by flow cytometry. Cells were gated on CD4 and CD44. (C) Quantitation using independently purified cells from the spleens of four mice treated similar to those in B. Student’s t test P value = 0.0019. (D) Naive OCA-B–deficient and littermate control splenic CD4+ T cells from 6-wk-old NOD mice were stimulated for 2 d in vitro with indicated antibodies, replated, and rested in the absence of antibody for 2 d, and analyzed by flow cytometry. Mean FR4hi CD73hiCD4+CD44+ cell frequencies are shown using independently purified cells from the spleens of two mice, with four independent culture replicates performed for each mouse (n = 8). Student’s t test P values: CD3ε only, 0.0002; CD3ε/CD28, 1.04 × 10−5; control CD3ε vs. CD3ε/CD28, 0.0053; OCA-B–deficient CD3ε vs. CD3ε/CD28, 0.0068. (E) Similar to D, except frequencies of CD4+CD44+ICOS+ cells are plotted. CD3ε-only Student’s t test P value = 0.0026. (F) Similar to D, except average percentages of CD4+CD44+CD25+ cells are plotted. CD3ε-only Student’s t test P value = 1.80 × 10−5. All error bars denote ±SEM. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. n.s., not significant.
In CD4+ cells, inducible T cell costimulator (ICOS) promotes the induction of T cell anergy (Dong et al., 2001; Dong and Nurieva, 2003; Tuettenberg et al., 2009) and is up-regulated by Oct1 loss in naive T cells subjected to stimulation under anergic conditions (Kim et al., 2019). Oct1 loss also elevates expression of FR4 and CD73 (Kim et al., 2019), the coexpression of which is a marker for anergy (Kalekar et al., 2016; Martinez et al., 2012). To determine whether OCA-B loss similarly affects expression of these proteins, we stimulated naive splenic CD4+ T cells (CD8anegCD11bnegCD45RnegDX5negTer-119negCD44lo) from 6-wk-old Ocabfl/flCD4-cre and littermate control CD4-cre male NOD mice for 2 d ex vivo using anti-CD3ε antibodies ± costimulation. Cells were then rested for an additional 2 d. Across all conditions, CD4+CD44+FR4hiCD73hi anergic cells were present at greater than twofold higher frequency in the OCA-B–deficient condition (Fig. 5 D). The fraction of CD44+ cells that express ICOS was similarly elevated in cells lacking OCA-B, though only without costimulation (Fig. 5 E). Additionally, the expression of the activation marker CD25 was slightly decreased (Fig. 5 F). Cumulatively, these results suggest that OCA-B loss in CD4+ T cells institutes higher immune thresholds by augmenting anergic responses.
Generation of an OCA-B peptide inhibitor
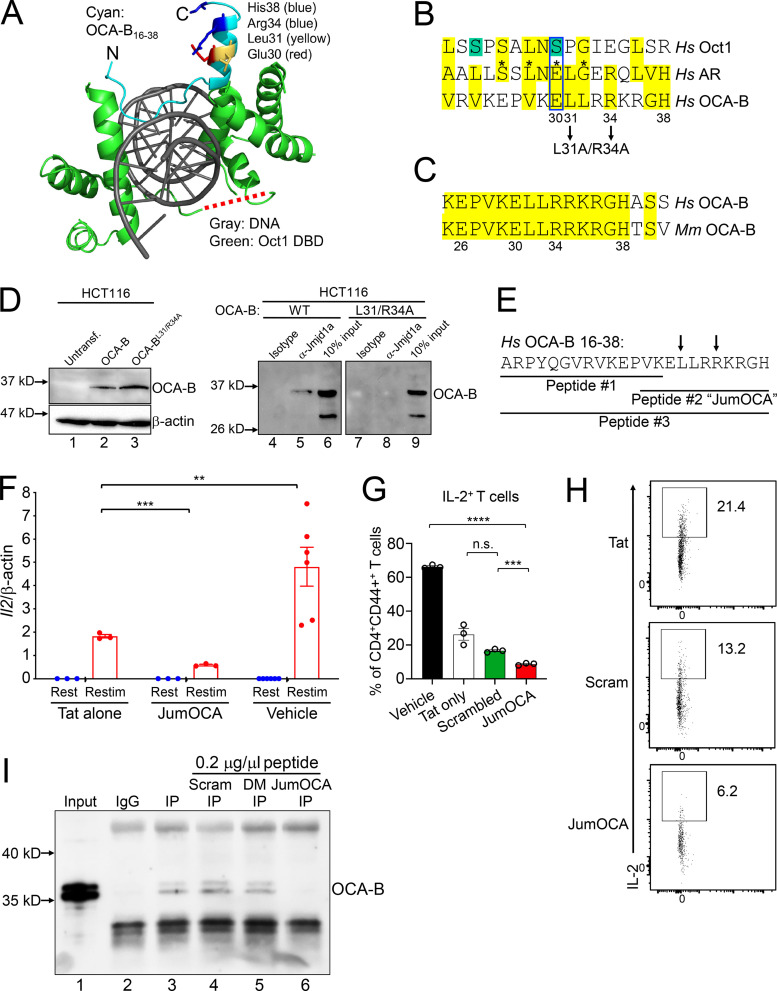
The normal T cell developmental and primary immune response phenotypes observed with OCA-B genetic deletion suggested the possibility of a “therapeutic window” in which targeting OCA-B pharmacologically would blunt autoimmunity while minimally affecting baseline immune function. Based on available OCA-B structural information, we generated a membrane-permeable peptide inhibitor of OCA-B’s downstream effector functions. Oct1 interacts with Jmjd1a in a mitogen-activated protein kinase (MEK)/ERK-dependent manner (Shakya et al., 2011, 2015) and contains two potential ERK phospho-acceptor serines. The structure of the Oct1 DNA binding domain complexed with consensus octamer binding site DNA (Klemm et al., 1994) reveals that these Oct1 serines are located in the flexible, solvent-exposed linker domain, which connects the two DNA binding subdomains (Fig. 6 A, red dashed line). In contrast with Oct1, OCA-B constitutively interacts with Jmjd1a (Shakya et al., 2015). The OCA-B N terminus has been solved in complex with the Oct1 DNA binding domain and consensus octamer DNA (Chasman et al., 1999). This region of OCA-B is critical for both Oct1 binding and transcription activity (Gstaiger et al., 1996). To identify potential Jmjd1a-interacting OCA-B regions, we aligned the full-length Oct1 and OCA-B amino acid sequences to a cofactor interaction surface of androgen receptor (AR), which also interacts with Jmjd1a (Yamane et al., 2006). Human AR mutations that cause androgen insensitivity have been mapped to residues 698–721 (Thin et al., 2002). Aligning this span to Oct1 identifies the linker domain as the top hit (Fig. 6 B). The alignment shows conservation of three out of four deleterious AR mutations (asterisks). The mutation site that is not conserved is a potential ERK target serine residue in Oct1, and a phospho-mimetic glutamic acid residue in AR (blue box). These findings suggest that the Oct1 linker domain constitutes a surface that, when phosphorylated, interacts with Jmjd1a.
Figure 6.
Design and validation of OCA-B peptide inhibitors. (A) OCA-B N terminus (residues 16 and 38)/Oct1 DNA binding domain/octamer binding DNA cocrystal structure (Protein Data Bank identification 1CQT; Green, 2016). Gray: DNA. Green: Oct1 DNA binding domain. Cyan: OCA-B. Red dashed line shows position of the Oct1 linker domain. N, number. DBD, DNA binding domain. Untransf., untransfected. (B) Top alignment of the human AR isoform transcript variant 1 (sequence identification ADD26780.1) coactivator interaction domain (residues 698–721) with full-length human Oct1 and OCA-B. The aligned regions are from the Oct1 linker domain and OCA-B N terminus. Green serines: putative ERK phospho-acceptor sites. Yellow: similar or identical amino acids. Asterisks: known human point mutations that block coactivator binding and cause androgen insensitivity syndrome in humans. Pairwise alignments were performed using the FASTA algorithm (https://embnet.vital-it.ch/software/LALIGN_form.html) and trimmed for three-way overlap. (C) Alignment of human and mouse primary OCA-B peptide sequences. (D) coIP of Jmjd1a with L31A/R34A double-point mutant OCA-B and WT control. HCT116 cells transfected with WT or mutant OCA-B constructs were used. Protein expression was checked 48 h after transfection by Western blotting. 50 µg input protein was loaded in lanes 1–3. (E) Indicated peptide sequences were synthesized as C-terminal Tat fusions for membrane permeability. Arrows indicate position of mutant in B and D. (F) Il2 mRNA expression in primary naive CD4+ T cells treated with 50 µM JumOCA peptide was measured relative to β-actin internal standard by RT quantitative PCR. Cells were stimulated with CD3ε/CD28 antibodies for 2 d, rested for a further 8 d in the presence of exogenous recombinant IL-2, and restimulated for 6 h. Peptide was included during resting and restimulation only and replaced every other day with media changes. Three independent biological replicates were used for each condition. Tat vs. JumOCA restimulated (Restim) Student’s t test P value = 0.0007. Tat vs. vehicle Student’s t test P value = 0.0022. (G) IL-2 cytokine expression in primary naive CD4+ T cells cultured with 50 µM peptide at initial treatment and with 25 µM peptide from the secondary treatment was measured by flow cytometry. Cells were treated similarly to those in F except collection, brefeldin A treatment, and processing for flow cytometry occurred 24 h after restimulation. Three independent biological replicates were used for each condition. Vehicle vs. JumOCA Student’s t test P value = 9.62 × 10−7. Scrambled peptide vs. JumOCA Student’s t test P value = 0.0006. (H) CD4+CD44+IL2+ T cell frequencies from representative samples in G. Plots were gated on CD4 and CD44. (I) Peptide effects on the interaction between OCA-B and Jmjd1a were measured by coIP. M12 B cells were used. After incubation with protein-G beads, 0.2 µg/µl control scrambled (Scram), double-point mutant (DM) peptide, or JumOCA peptide were added for a further 3 h before precipitation and washing. 1% input (lane 1) is shown as a control. All error bars denote ±SEM. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. n.s., not significant.
As with Oct1, alignment with OCA-B identifies a potential Jmjd1a interacting surface (Fig. 6 B). Unlike Oct1, a glutamic acid residue (Glu30) aligns with AR. Furthermore, the OCA-B residues aligning to AR lie on one side of an α-helix (Fig. 6 A). Glu30 (red), Leu31 (yellow), Arg34 (blue), and His38 (blue) may therefore constitute a Jmjd1a docking surface. These residues are conserved between mouse and human (Fig. 6 C). We mutated OCA-B L31 and R34 to alanine in a transient expression plasmid, and transfected the double-point mutant or parent plasmid control into HCT116 cells, which do not express endogenous OCA-B. Control coimmunoprecipitation (coIP) using Jmjd1a antibodies and lysates from cells expressing WT OCA-B confirms the interaction (Fig. 6 D, lane 5), while the mutant OCA-B protein failed to interact (lane 8). The mutant was expressed equivalently to WT (lanes 2 and 3). These results show the importance of OCA-B residues 30–38 for cofactor interactions.
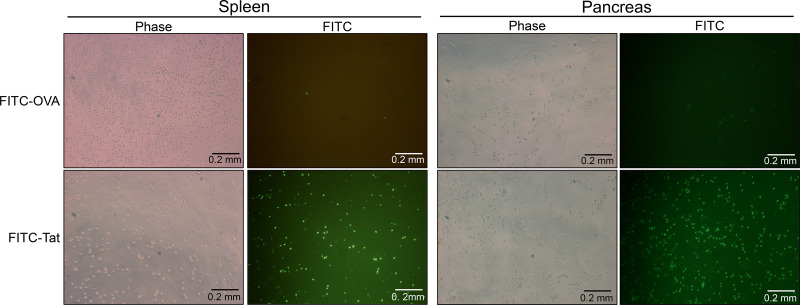
We synthesized three overlapping peptides corresponding to the OCA-B N terminus (Fig. 6 E, peptides #1, #2 [hereafter called “JumOCA”], and #3) as C-terminal fusions to the HIV trans-activator of transcription (Tat) protein for membrane permeability (Schwarze et al., 2000). We also conjugated HIV Tat to FITC to enable peptide tracking. Incubating total splenic or pancreatic CD3+ T cells in ex vivo culture with 45 µM Tat-fused peptide for 15 min significantly concentrated the peptide within cells. Control OVA-fused peptide lacking membrane-penetrating properties showed no effect (Fig. S5). We then treated splenic CD4+ T cells with the Tat-fused JumOCA peptides. Prior work has shown that 6 h restimulation of resting but previously activated OCA-B–deficient CD4+ T cells results in decreased expression of target genes such as Il2 (Shakya et al., 2015). Using this assay with 50 µM JumOCA peptide (every other day with media changes during rest and restimulation) inhibited Il2 mRNA expression relative to β-actin by ∼10-fold (Fig. 6 F, compare Tat peptide to JumOCA). Although treated cells showed no obvious changes in viability, morphology, or expansion during the course of the assay (not shown), there was a two- to threefold nonspecific diminution of activity associated with unfused Tat peptide (Fig. 6 F, compare Tat peptide with no peptide), which has been associated with toxicity (Caron et al., 2004; Krautwald et al., 2004; Polo et al., 2004). Similar results were obtained with the larger peptide, #3, but not with peptide #1 (not shown). Peptide #1 is missing residues important for the Jmjd1a interaction based on mutagenesis (Fig. 6 B). For all further experiments, we used the JumOCA peptide.
Figure S5.
Tat- but not OVA-conjugated peptides concentrate in cultured T cells. CD3+ T cells were isolated from the spleens or pancreata of NOD mice and were incubated with 40 µM FITC-conjugated OVA control peptide, or FITC-conjugated to the TAT membrane–penetrating peptide for 15 min. Cells with concentrated FITC fluorescence were observed by epifluorescence microscopy. Images were taken at 40× magnification.
We then synthesized a Tat-conjugated peptide using a scrambled JumOCA sequence. This peptide has the same amino acid composition, mass, and pI, but should not be able to efficiently compete for Jmjd1a. We used the Tat-only, scrambled, and JumOCA peptides with flow cytometry to assess IL-2 production in restimulated CD4+ T cells. JumOCA significantly inhibited IL-2 production relative to the scrambled control; however, there were significant toxicities associated with both the scrambled and Tat-only peptides relative to vehicle (Fig. 6 G). An example mouse from this experiment is shown in Fig. 6 H.
The above results are consistent with the JumOCA peptide operating via Jmjd1a competitive inhibition. To test this directly, we used scrambled, double-point mutant and WT (JumOCA) peptides (lacking the Tat membrane–penetrating domain) in a Jmjd1a coIP assay. We subjected lysates from mouse M12 B cells, which express endogenous OCA-B, to IP with Jmjd1a antibodies, resulting in OCA-B coIP (Fig. 6 I, lane 3). Incubation of precipitated material with scrambled or double-point mutant peptides had no effect on OCA-B coIP (lanes 4 and 5), while the same concentration of JumOCA peptide efficiently blocked OCA-B recovery (lane 6).
JumOCA protects NOD mice from newly arisen T1D
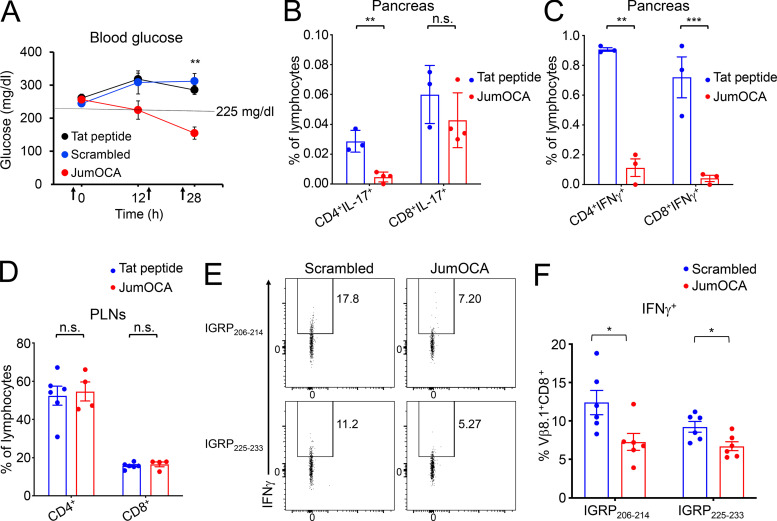
By the time symptoms arise in NOD mice, 90–95% of β cells have been destroyed. Remaining β cells are rendered nonfunctional due to insulitis. Alleviating this inflammation provides a “honeymoon period” during which glucose homeostasis can be restored. We treated 12–18-wk littermate female NOD mice whose glucose levels were newly risen above 225 mg/dl but still below 275 mg/dl with three intravenous injections of 10 mg/kg JumOCA peptide or Tat-only peptide control. Injections were spaced 12 h apart. Immediately after the second injection and 4 h after the final injection, blood glucose was measured. Strikingly, JumOCA but not Tat-only or scrambled peptide reversed elevated blood glucose (Fig. 7 A). Islet T cell pro-inflammatory IFN-γ and IL-17A cytokine production were reduced by JumOCA peptide treatment compared with Tat-only controls (Fig. 7, B and C). In contrast to the pancreas, PLNs showed no change in T cell numbers or percentages (Fig. 7 D) but similar decreases in pro-inflammatory cytokine production (not shown).
Figure 7.
OCA-B inhibitor peptide reduces blood glucose and inflammatory cytokine levels in the pancreas in NOD mice with newly arisen diabetes. (A) Three doses of 10 mg/kg JumOCA (n = 4), Tat only (n = 6), or scrambled peptide (n = 3) were injected retro-orbitally every 12 h into 12–18-wk-old NOD female littermate mice whose glucose levels were newly risen above 225 mg/dl but still below 275 mg/dl. Glucose levels are shown immediately before the first injection, after the second injection, and 4 h after the final injection. Data were collected on a rolling basis as mice became spontaneously diabetic. JumOCA vs. Tat peptide Student’s t test P value = 0.0004. JumOCA vs. scrambled peptide Student’s t test P value = 0.0064. (B) 12 h after the last injection of peptides in A, PLN CD4+ and CD8+ T cell percentages were analyzed by flow cytometry. Mean of islet IL-17–expressing T cells was analyzed by flow cytometry and plotted. Results are from three independent mice in the case of Tat and four independent mice in the case of JumOCA. CD4+IL-17+ Student’s t test P value = 0.002. (C) Similar analysis as B except for IFN-γ. CD4+ Student’s t test P value = 0.0062. CD8+ Student’s t test P value = 0.0002. (D) Percentages of PLN CD4+ and CD8+ T cells from mice in A. (E) NOD mice were treated with three intravenous peptide injections as before, except that 12-wk prediabetic mice were used, and scrambled peptide was additionally included as a control. 4 h after the final injection, PLN WBCs were stimulated with IGRP peptides and brefeldin A for 4 h, stained for IFN-γ, and analyzed by flow cytometry as in Fig. 3 D. Representative animals are shown. (F) Mean percentages of cells expressing IFN-γ from six experiments. IGRP206-214 Student’s t test P value = 0.0244. IGRP225-233 Student’s t test P value = 0.0201. All error bars denote ±SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. n.s., not significant.
Autoantigen-specific CD8+ T cells in the PLNs of prediabetic, OCA-B–deficient animals are present but poorly activated by self-peptide (Fig. 3, D and E). We treated animals with three injections of JumOCA or scrambled peptide control, and stimulated PLN white blood cells (WBCs) with IGRP self-peptides as in Fig. 3 D. Significant reductions in IFN expression were observed in JumOCA-treated animals compared with controls (Fig. 7 E). Quantification from multiple mice is shown in Fig. 7 F. These data show that JumOCA peptide treatment decreases CD8+ T cell autoreactivity within the PLNs, similar to genetic OCA-B deletion. Cumulatively, these data provide evidence that targeting OCA-B is a valid strategy to treat emerging T1D and identify a first-generation inhibitor that is efficacious in vivo.
Discussion
Autoimmune therapies aim to inhibit autoreactivity while preserving normal immune function. Here, we show that genetic and pharmaceutical targeting of OCA-B blocks T1D in mouse models. Although potentially autoreactive CD8+ T cell specificities are still generated and appear in the PLNs of OCA-B T cell–deficient mice, they fail to become activated and do not accumulate in the pancreas. In contrast, potentially autoreactive CD4+ T cells are present but associated with anergic cell populations. OCA-B is not expressed during thymic development or in naive CD4+ T cells but is stably induced upon activation. In contrast, OCA-B is expressed in naive CD8+ T cells and rapidly increased upon activation.
In CD4+ T cells, OCA-B associates with the POU (Pit-1, Oct 1/2, UNC-86)-domain transcription factor Oct1 at ∼150 immunomodulatory genes, among them Il2, Ifng, and Csf2 (Gmcsf). Unlike transcription factors such as NFAT, AP-1, and NF-κB that act as primary “on” switches, OCA-B removes inhibitory chromatin modifications to establish permissive chromatin environments that allow for silent but previously activated targets to be robustly expressed upon antigen reencounter (Shakya et al., 2011, 2015). More specifically, OCA-B interacts with Jmjd1a, a histone lysine demethylase that removes inhibitory histone H3 lysine 9 methyl marks. In vivo, OCA-B loss preserves T cell development and pathogen response but impairs the establishment of new central memory CD4+ T cells. The few cells that are formed respond poorly to antigen reencounter (Shakya et al., 2015). Repeated antigen encounters that drive high levels of proinflammatory cytokines are a key feature of autoimmunity. In both mice and humans, memory or memory-like cells can underlie autoimmunity, including T1D (Chee et al., 2014; Kawakami et al., 2005; Yeo et al., 2018).
The NOD model of T1D is spontaneous but only partially penetrant, much like the human disease (Parker et al., 2009). 60–80% of female NOD mice develop T1D in standard environments (Makino et al., 1985). We find that prediabetic OCA-B T cell–deficient NOD mice harbor normal T cell numbers and TCR specificities in their PLNs, consistent with prior observations that OCA-B–deficient T cells are largely immunocompetent (Shakya et al., 2015). Nevertheless, T cell conditional OCA-B knockouts are protected from spontaneous T1D. Whole-body NOD Ocab knockouts are also protected from T1D. Prior work showed that OCA-B whole-body knockout protects mice both from lupus-like disease mediated by loss of Izkf3/Aiolos (Sun et al., 2003) and in a MOG/EAE (myelin oligodendrocyte glycoprotein/experimental autoimmune encephalomyelitis) model of multiple sclerosis (Ikegami et al., 2019). In contrast, a third study showed exacerbation of systemic, antibody-driven autoimmunity in Sanroque mice (Chevrier et al., 2014). The protective vs. exacerbating effects in whole-body knockouts may be due to effects on class switching and affinity maturation in germinal centers vs. effects on BCR repertoire (Casellas et al., 2002). A whole-body OCA-B knockout also reduces fat accumulation and insulin resistance in aged mice, likely through ablation of B cell–mediated inflammation in white adipose tissue (Carter et al., 2018). T cell–specific knockout of Oct1, a transcription factor with which OCA-B docks, blocks EAE but preserves responses to infection with neurotropic viruses (Kim et al., 2019). Together, these data show that in T cells, Oct1 and OCA-B promote autoimmunity including T1D, but that their role in B cells may be more complex. OCA-B is typically expressed 50–100-fold higher in B cells compared with T cells. A correctly calibrated dose of competitive inhibitor could therefore be used to selectively blunt T cell–mediated autoimmunity.
scRNAseq experiments reveal decreased pancreatic neutrophil infiltration and an increased percentages of T cells with naive phenotypes in prediabetic OCA-B conditional knockouts. Gene expression changes were identified consistent with antidiabetogenic effects of OCA-B T cell knockout. For example, the remaining islet neutrophils in knockout mice show reductions in Cxcl2 expression. Cxcl2 is a potent neutrophil chemoattractant that promotes T1D in vivo (Citro et al., 2015; Diana and Lehuen, 2014). Strikingly, OCA-B T cell–deficient mice also lack T1D-associated TCR clones in their activated/memory CD8 pool. Flow cytometry using class I tetramers shows a significant reduction in CD8+ T cells specific for the IGRP206-214 autoantigen. PLN CD8+ T cells reactive to this and other autoantigens show reduced reactivity to peptide stimulation ex vivo, suggesting that these autoreactive CD8+ cells are produced in the thymus but fail to become activated in the PLNs. In contrast, CD4 clonotypes associated with T1D are present in the pancreata but associated with anergic gene expression profiles. Consistently, OCA-B loss promotes CD4+ T cell anergy in vitro. Loss of Oct1, the transcription factor with which OCA-B docks, also augments anergic responses (Kim et al., 2019).
C57BL/6.RIP-mOVA and NOD.BDC2.5 transgenic mice are simplified monoclonal systems that allow tracking of uniform T cell responses to antigens in vivo (Haskins, 2005). RIP-mOVA expresses synthetic membranous chicken OVA in thymic epithelial and pancreatic β cells (Kurts et al., 1996; Van Belle et al., 2009). Disease can be induced in this model in 6–10 d by the addition of OT-I T cells and infection with OVA-expressing L. monocytogenes. OCA-B loss in host T cells has no effect in this model. BDC2.5 autoreactive T cells migrate to the islets and cause insulitis beginning at 3–4 wk (Katz et al., 1993). Due in part to T reg cells that have escaped allelic exclusion, in many animals insulitis is limited and does not progress to T1D. However, transferring CD4+CD62L+Vβ+ or CD4+CD25− T cells from BDC2.5 donor mice into NOD.SCID recipients also results in rapid (7–10 d) disease onset (Berry and Waldner, 2013). Transplants from both control and OCA-B null NOD/BDC2.5 mice results in disease with similar kinetics. Identical results were generated by depleting CD25+ T reg cells, or by transplanting total CD4+ T cells into immunodeficient NCG recipient mice.
NOD.NY8.3 TCR transgenic mice carry a monoclonal CD8-restricted TCR directed toward IGRP206-214 and manifest spontaneous T1D (Verdaguer et al., 1997). Unlike transplantation models, OCA-B T cell conditional NY8.3 mice show significant protection from spontaneous T1D. Cumulatively, the results support a model in which OCA-B loss preserves T cell functionality while desensitizing autoreactive T cells. This results in graded protection that drops off using models in which protection is inseparable from immunodeficiency. These properties make OCA-B a promising target for pharmaceutical inhibition.
Recent work indicates that the prevailing idea that transcription regulators and protein–protein interactions are “undruggable” is erroneous (Antony-Debré et al., 2017; Green, 2016; Skwarczynska and Ottmann, 2015; Zuber et al., 2011). Pre-clinical membrane-penetrating peptides have also been developed that successfully target transcription factors such as BCL6 (Polo et al., 2004). We applied rational-design principles to generate a membrane-permeable competitive peptide inhibitor of the interaction between OCA-B and a downstream effector, Jmjd1a/Kdm3a. The sequence of the inhibitor, termed JumOCA, does not coincide with other sequences in the human or mouse proteome and is not predicted to be strongly immunogenic. JumOCA administration blunts newly arisen T1D in NOD mice and significantly reduces islet T cell cytokine production. Total T cell numbers were unaffected; however, PLN CD8+ T cell autoreactivity to IGRP peptides was blunted, similar to T cell knockouts. OCA-B levels in B cells are at least 50-fold higher than in T cells (Heng et al., 2008; Zwilling et al., 1997), making it likely that the observed effects are due to inhibition in T cells. Consistently, three JumOCA injections have no effect of splenic germinal center architecture (not shown), which is known to depend on OCA-B expression in B cells (Qin et al., 1998). More than three peptide injections results in toxicity (not shown), complicating experiments testing T1D prevention and durable response. Tat-conjugated peptides are known to be associated with toxicity (Caron et al., 2004; Krautwald et al., 2004; Polo et al., 2004). While these peptides are unlikely to be used in a clinical setting, they offer proof of principle for OCA-B as a therapeutic target and can be used as tools for the further development of therapeutics, with the caveat that targeting this pathway would likely inhibit the generation of new memory CD4+ T cells.
Materials and methods
Mice
All animal experiments were approved by the University of Utah Institutional Animal Care and Use Committee (IACU approval 17–05008). NOD/ShiLtJ, NOD.Scid (NOD.Emv302/2.CB17-Prkdcscid), BDC-2.5 TCR transgenic, NY8.3 TCR transgenic, and all breeder mice were maintained in a specific pathogen-free research colony. NCG (NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl) mice were purchased from Charles River Laboratories. NOD.CD4-cre mice were a gift from Alexander Chervonsky (University of Chicago, Chicago, IL). Pou2af1tm1a(KOMP)Wtsi mice were provided by the KOMP repository (University of California, Davis, Davis, CA). Mice were derived from embryonic stem cell clone EPD0761_3_B03, generated by the Wellcome Trust Sanger Institute. These cells were derived from C57BL/6N mice, but subsequent mouse crosses were to C57BL/6J (>20 generations) for the RIP-mOVA model and NOD (four congenic backcrosses plus an additional backcross to NOD.CD4-cre). The embryonic stem cells contain a CSD-targeted allele (Testa et al., 2004). The presence of WT (582 bp) and post-FLP (700 bp) alleles was determined by PCR using CSD-Pou2af1-F and CSD-Pou2af1-ttR primers. The presence of the null (801 bp) allele was determined using CSD-Pou2af1-F and CSD-Pou2af1-R primers. The presence of the floxed (359 bp) allele was determined using CSD-Lox-F and CSD-Pou2af1-R primers. Primer sequences were as follows: CSD-Pou2af1-F, 5′-TACAGAGAGACTAGACACGGTCTGC-3′; CSD-Pou2af1-R, 5′-GATGAGGACTCTGGGTTCAGAGAGG-3′; CSD-loxF, 5′-GAGATGGCGCAACGCAATTAATG-3′; and CSD-Pou2af1-ttR, 5′-AGAAGGCCTCGTTACACTCCTATGC-3′.
Backcrossing Ocab germline null and conditional mice to the NOD background
To generate Ocab−/− and Ocab conditional mice on the NOD background, the previously described C57BL/6 Ocab germline null allele (Kim et al., 1996) and the newly generated Ocab conditional allele were backcrossed to NOD ShiLt/J mice using congenic markers based on Mouse Genome Informatics (http://www.informatics.jax.org; Table S1).
Diabetes development and assessment
Diabetes was monitored using blood glucose test strips (Contour; Bayer). Mice with blood glucose levels >250 mg/dl were considered diabetic.
Leukocyte isolation and flow cytometry
PLNs were disaggregated and passed through a nylon strainer. Pancreatic leukocytes were isolated as described previously (Sitrin et al., 2013). Briefly, pancreata were chopped and digested using collagenase IV (1 mg/ml; Gibco) in DMEM containing 1% FBS (Rocky Mountain Biologicals) and 10 U of DNase I for 15 min at 37°C. The digested tissues were passed through a 70-µm strainer. Red blood cells were lysed by ammonium-chloride-potassium lysis buffer (150 mM NH4Cl, 10 mM KHCO3, and 0.1 mM Na2EDTA). For intracellular cytokine staining, cell suspensions in RPMI medium supplemented with 10% FBS were restimulated for 4 h with PMA (50 ng/ml; Sigma-Aldrich) and ionomycin (1 µg/ml; Sigma-Aldrich) in the presence of brefeldin A (1 µl/ml; GolgiPlug, BD Bioscience) and fixed by cell fixation/permeabilization solution (BD Cytofix/Cytoperm) according to the manufacturer’s protocol. Antibodies used for flow cytometry were as follows: FITC-conjugated anti-mouse CD4, PE-conjugated anti-mouse CD45, PE-conjugated anti-mouse FR4 (BioLegend), PerCP-conjugated anti-mouse CD8a, APC-conjugated anti-mouse IFN-γ, PE-conjugated anti-mouse IL-17, PerCP-conjugated anti-mouse CD11b, APC-conjugated anti-mouse F4/80, PE-conjugated anti-mouse Gr-1, PE-conjugated anti-mouse ICOS (eBioscience), PerCP-conjugated anti-mouse FoxP3, V450-conjugated anti-mouse CD73 (BD Bioscience), and FITC-conjugated anti-mouse Vβ8.1/8.2 (Invitrogen). A FACS Canto II (BD Biosciences) was used for flow cytometry, and FlowJo software was used for analyses.
T cell adoptive transfer
6–8-wk-old NOD.Scid recipient mice were injected retro-orbitally with 2 × 105 of splenic CD4+CD25− T cells from prediabetic Ocab−/− or Ocab+/+ NOD.BDC2.5 donors (6–8 wk old, sex matched). T cells were purified using a CD4+CD25+ T cell isolation kit (Miltenyi Biotec). For CD4+CD25+ T cells, 1.5 × 106 purified splenic CD4+ T cells from prediabetic, BDC2.5 transgenic NOD.Ocab−/− or NOD.Ocab+/+ mice were transferred to sex-matched 6–8-wk-old NCG recipients (University of Utah Preclinical Resource Core) as previously described (Presa et al., 2015). For total splenic transfer experiments, 5 × 106 splenocytes from prediabetic NOD.Ocabfl/fl or NOD.Ocabfl/flCD4-cre mice were transferred into sex-matched 6–8-wk-old NOD.Scid recipient mice as described previously (Presa et al., 2015).
In vitro T cell culture
For anergic cell induction, spleens were harvested from NOD.Ocabfl/flCD4-cre or control NOD.Ocabfl/fl animals. Single-cell suspensions were generated by grinding and passage through 70-µm strainers. Cells were isolated using a mouse CD4+ T cell isolation kit (Miltenyi Biotec) and stimulated with 5 µg/ml plate-bound anti-CD3ε (BD Bioscience) ± 2 µg/ml anti-CD28 antibody (eBioscience) as described previously (Kim et al., 2019). For OCA-B inhibitor peptide treatments, cells were isolated from WT C57BL/6 spleens using a mouse naive CD4+ T cell isolation kit (Miltenyi Biotec) and cultured as described previously (Shakya et al., 2011). Indicated concentrations of peptides were incubated with cells, with media changes every 2 d after primary stimulation. Activation of C57BL/6 CD4+ cells and subsequent profiling of anergic responses were performed identically to the procedure by (Kim et al.,2019) For peptide restimulation, total WBCs from PLNs were incubated with 10 µg/ml specific peptide for 4 h in the presence of brefeldin A. The peptides used were as follows: GAD65206-214, TYEIAPVFV; GAD65546-554, SYQPLGDKV; IGRP21-29, TYYGFLNFM; IGRP206-214, VYLKTNVFL; IGRP225-233, LRLFGIDLL; IGRP241-249, KWCANPDWI; and IGRP324-332, SFCKSASIP. Cells were then fixed and stained for flow cytometry.
scRNAseq (GEO Series record GSE145228)
CD3+ T cells were isolated from 8-wk NOD.Ocabfl/fl or NOD.Ocabfl/flCD4-cre females using a pan T cell isolation kit (Miltenyi Biotec). CD45+ pancreatic leukocytes were isolated from 12-wk NOD.Ocabfl/fl or NOD.Ocabfl/flCD4-cre females by flow cytometry using a FACS Aria (Becton Dickinson). For each condition, cells were isolated from three mice and combined. Cells were processed using the 10× Genomics Chromium platform according the manufacturer’s instructions. Paired-end high-throughput sequencing (125 cycles) was performed via an Illumina NovaSeq instrument. Sequencing reads were processed by using the 10X Genomics CellRanger pipeline and further analyzed using the Seurat R package. Analysis of cells used a standard filtering protocol removing cells with unique feature counts of >4,000 or <500, as well as cells with >5% mitochondrial counts (indicative of dead cells). No more than 15% of total cells were removed by this process. Cells were subjected to unsupervised hierarchical clustering followed by uniform manifold approximation and projection (UMAP) to visualize clusters with similar gene expression and their relative positions.
Tetramer staining
H-2Kd mouse IGRP206-214 tetramers (mouse NRP-V7 mimotope KYNKANVFL) conjugated to APC or PE were synthesized by the National Institutes of Health tetramer core facility and were a gift from Maria Bettini (University of Utah, Salt Lake City, UT).
OCA-B mutagenesis and coIP
For generation of the OCA-B double-point mutant, a plasmid transiently expressing human OCA-B, pCATCH-Bob.1 (Gstaiger et al., 1995), was sequentially mutagenized using QuickChange (Thermo Fisher Scientific). Correct mutagenesis was confirmed by resequencing. HCT116 lysate preparation, coIP of OCA-B and endogenous Jmjd1a, and immunoblot detection were performed using the same conditions as described in Shakya et al. (2015). For coIP using M12 cells, lysis buffer consisted of 50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, 1 mM EGTA, and protease/phosphatase inhibitors (PhosSTOP; Roche). Lysates were incubated with 2.5 µg Jmjd1A antibody (Bethyl) and protein-G Dynabeads (Thermo Fisher Scientific) in lysis buffer containing 20% glycerol overnight at 4°C. After overnight incubation, the indicated concentrations of peptide or PBS vehicle were added and incubated for a further 3 h at 4°C before bead precipitation and washing 3× in lysis buffer plus 20% glycerol. Co-precipitated OCA-B was analyzed by SDS-PAGE and immunoblot.
OCA-B inhibitor peptide synthesis
Unique chemically synthesized peptide sequences were as follows: Peptide#1, ARPYQGVRVKEPVK; Peptide#2/JumOCA, VKELLRRKRGH; Peptide#3, ARPYQGVRVKEPVKELLRRKRGH; and Scrambled peptide, VLREKGKRHLR. Peptides were synthesized with and without a covalent C-terminal linker and Tat membrane–penetrating peptide (GSG-GRKKRRQRRRGY). Fluorenylmethoxycarbonyl (Fmoc)-protected amino acids were obtained from Protein Technologies Inc. 1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU) was purchased from Chemimpex Inc. H-Rink-Amide-ChemMatrix was purchased from Biotage. N,N-diisopropylethylamine (DIEA), dichloromethane (DCM), and triisopropylsilane were purchased from Sigma-Aldrich. Dimethylformamide (DMF), TFA, acetonitrile, and ethyl ether were purchased from Thermo Fisher Scientific. Peptides were synthesized using automated Fmoc SPPS chemistry (Syro I). Briefly, 220 mg of H-Rink-Amide-ChemMatrix resin (loading = 0.45 mmol/g) was swelled in DCM for 15 min and followed by adding a solution of specific Fmoc-protected aa (aa = 0.1 mmol, DIEA = 0.2 mmol in DCM = 4 ml) and incubated at room temperature for 1.5 h. The resin was washed with DMF and DCM and incubated with 5 ml of DCM containing 16% vol/vol MeOH and 8% vol/vol DIEA for 5 min. This action was repeated five times before thoroughly washing with DCM and DMF. Coupling reactions were performed using HATU (5 equivalents [eq.]), di-isopropylethylamine (10 eq.), and amino acid (5 eq.) in DMF with 15 min heating to 70°C (50°C for Cys). Deprotection was performed using 20% (vol/vol) piperidine in DMF, 5 min at room temperature for two rounds. Peptides were cleaved from the resin by treatment with a cocktail buffer (3 ml/0.1 mmol, TFA:H2O:TIS:EDT = 95:2:2:1) for 2.5 h. Peptide-TFA solution was then filtered and precipitated in cold ether, centrifuged, washed with ether twice, and vacuum dried. The crude product was then purified by reverse phase HPLC. Peptide characterization was performed by liquid chromatography/mass spectrometry (LC/MS) on an Xbridge C18 5 µm (50 × 2.1 mm) column at 0.4 ml/min with a water/acetonitrile gradient in 0.1% formic acid on an Agilent 6120 Quadrupole LC/MS system. Fractions collected from HPLC runs were also analyzed by LC/MS. The purified fractions containing the targeted product were collected and lyophilized using a Labconco Freeze Dryer. All samples were analyzed by the following conditions: preparative reverse-phase HPLC of crude peptides was performed on a Jupiter 5 µ C18 300 Å (250 × 10 mm) column at 3 ml/min with a water/acetonitrile gradient in 0.1% TFA on an Agilent 1260 HPLC system. Purity, isomer coinjection, and stability checks were performed on HPLC on a Phenomenex Gemini C18 3 µm (110 Å 150 × 3 mm) column.
OCA-B inhibitor peptide treatment
Anesthetized prediabetic WT NOD females with glucose levels newly risen to between 225 and 275 mg/dl were treated with 10 mg/kg inhibitor or control peptides by intravenous (retro-orbital) injection three times every 12 h. Blood glucose levels were measured after the second and third injection. 4 h after the last injection, pancreata and PLNs were collected, and cell populations were analyzed by flow cytometry.
Histology
Formalin-fixed pancreatic tissues were embedded in paraffin. H&E-stained sections were scored for islet inflammation based on published precedents (Zhang et al., 2007): 0, sparse surrounding sentinel leukocytes, no insulitis; 1, peri-islet leukocytes; 2, some islet leukocytes, <50% of islet area; 3, islet insulitis with >50% of islet area occupied by leukocytes; and 4, islets destroyed with fibrotic remnants.
Statistical analyses
All error bars denote ±SEM. Two-tailed Student t tests were used to ascribe statistical significance. For all figures, *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ****, P ≤ 0.0001.
Online supplemental material
Fig. S1 outlines the construction and validation of the Ocab (Pou2af1) conditional mouse allele. Fig. S2 shows results from scRNAseq using PLNs from 8-wk-old prediabetic female NOD.Ocabfl/flCD4-cre or littermate control NOD.Ocabfl/fl mice. Fig. S3 shows TCR representation across all populations based on scRNAseq from islet CD45+ cells. Fig. S4 shows results from a BDC2.5 T cell transfer experiment using OCA-B–deficient and –sufficient NOD mice and results from RIP-mOVA T cell transfer experiments using C57BL/6 mice. Fig. S5 shows the concentration of Tat-conjugated peptides in cultured primary CD3+ T cells. Table S1 shows loci for T1D susceptibility and resistance in NOD/ShiLtJ mice and oligonucleotide primer pairs used for PCR amplification of Idd loci microsatellite markers. Table S2 shows gene expression comparisons in T cells taken from PLNs of 8-wk-old NOD.Ocabfl/flCD4-cre and NOD.Ocabfl/fl littermate controls. Table S3 shows expression levels of Ccl1 mRNA in naive, stimulated, rested, and re-stimulated OCA-B/Pou2af1-deficient T cells relative to controls. Table S4 shows gene expression comparisons in total CD45+ cells taken from pancreatic islets of 12-wk-old prediabetic NOD.Ocabfl/flCD4-cre and NOD.Ocabfl/fl littermate controls.
Supplementary Material
is related to Fig. S1 and shows loci for T1D susceptibility and resistance in NOD/ShiLtJ mice and oligonucleotide primer pairs used for PCR amplification of Idd loci microsatellite markers.
is related to Fig. S2 and shows gene expression comparisons in T cells taken from PLNs of 8-wk-old NOD.Ocabfl/flCD4-cre and NOD.Ocabfl/fl littermate controls.
is related to Fig. 1 and shows expression levels of Ccl1 mRNA in naive, stimulated, rested, and re-stimulated OCA-B/Pou2af1–deficient T cells relative to controls.
is related to Fig. 2 and shows gene expression comparisons in total CD45+ cells taken from pancreatic islets of 12-wk-old prediabetic NOD.Ocabfl/flCD4-cre and NOD.Ocabfl/fl littermate controls.
Acknowledgments
We thank P. Santamaria, M. Williams, M. Bettini, and F. Gounari for critical reading of the manuscript. We thank A. Chervonsky (University of Chicago) for the gift of NOD.CD4-cre mice. We thank J. Marvin and the University of Utah Health Sciences Center Flow Cytometry Core facility for assistance with flow cytometry, and D. Lum and the Preclinical Resource Core for immunodeficient mice. We thank M. Hanson and the University of Utah Health Sciences DNA/peptide synthesis core.
This work was supported by grants from the Praespero Foundation, Juvenile Diabetes Research Foundation United States of America (1-INO-2018-647-A-N), and National Institutes of Health (R01-AI100873) to D. Tantin.
Author contributions: D. Tantin conceived the study and designed experiments, supervised the study, and provided administrative and material support. H. Kim, A. Shakya, Z. Shen, A. Ibarra, and J.L. Jafek acquired and interpreted the data. J. Perovanovic performed analysis of scRNAseq and TCR clonotype data. C.N. German and N.-P. Lin helped generate critical reagents. B.D. Evavold, D.H.-C. Chou, X. He, and P.E. Jensen provided material and intellectual support. All authors were involved in writing, reviewing, and revising the manuscript.
References
- Antony-Debré, I., Paul A., Leite J., Mitchell K., Kim H.M., Carvajal L.A., Todorova T.I., Huang K., Kumar A., Farahat A.A., et al. 2017. Pharmacological inhibition of the transcription factor PU.1 in leukemia. J. Clin. Invest. 127:4297–4313. 10.1172/JCI92504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, M., Booth D., Heard R., Stewart G., Goris A., Vandenbroeck K., Dubois B., Laaksonen M., Ilonen J., Alizadeh M., et al. Games Collaborative Group . 2006. Linkage disequilibrium screening for multiple sclerosis implicates JAG1 and POU2AF1 as susceptibility genes in Europeans. J. Neuroimmunol. 179:108–116. 10.1016/j.jneuroim.2006.06.003 [DOI] [PubMed] [Google Scholar]
- Berry, G., and Waldner H.. 2013. Accelerated type 1 diabetes induction in mice by adoptive transfer of diabetogenic CD4+ T cells. J. Vis. Exp. 6:e50389. 10.3791/50389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor, J., and Haskins K.. 2007. Recruitment and activation of macrophages by pathogenic CD4 T cells in type 1 diabetes: evidence for involvement of CCR8 and CCL1. J. Immunol. 179:5760–5767. 10.4049/jimmunol.179.9.5760 [DOI] [PubMed] [Google Scholar]
- Caron, N.J., Quenneville S.P., and Tremblay J.P.. 2004. Endosome disruption enhances the functional nuclear delivery of Tat-fusion proteins. Biochem. Biophys. Res. Commun. 319:12–20. 10.1016/j.bbrc.2004.04.180 [DOI] [PubMed] [Google Scholar]
- Carter, S., Miard S., Caron A., Sallé-Lefort S., St-Pierre P., Anhê F.F., Lavoie-Charland E., Blais-Lecours P., Drolet M.C., Lefebvre J.S., et al. 2018. Loss of OcaB Prevents Age-Induced Fat Accretion and Insulin Resistance by Altering B-Lymphocyte Transition and Promoting Energy Expenditure. Diabetes. 67:1285–1296. 10.2337/db17-0558 [DOI] [PubMed] [Google Scholar]
- Casellas, R., Jankovic M., Meyer G., Gazumyan A., Luo Y., Roeder R., and Nussenzweig M.. 2002. OcaB is required for normal transcription and V(D)J recombination of a subset of immunoglobulin kappa genes. Cell. 110:575–585. 10.1016/S0092-8674(02)00911-X [DOI] [PubMed] [Google Scholar]
- Chai, J.G., and Lechler R.I.. 1997. Immobilized anti-CD3 mAb induces anergy in murine naive and memory CD4+ T cells in vitro. Int. Immunol. 9:935–944. 10.1093/intimm/9.7.935 [DOI] [PubMed] [Google Scholar]
- Chasman, D., Cepek K., Sharp P.A., and Pabo C.O.. 1999. Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface. Genes Dev. 13:2650–2657. 10.1101/gad.13.20.2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee, J., Ko H.J., Skowera A., Jhala G., Catterall T., Graham K.L., Sutherland R.M., Thomas H.E., Lew A.M., Peakman M., et al. 2014. Effector-memory T cells develop in islets and report islet pathology in type 1 diabetes. J. Immunol. 192:572–580. 10.4049/jimmunol.1302100 [DOI] [PubMed] [Google Scholar]
- Chevrier, S., Kratina T., Emslie D., Karnowski A., and Corcoran L.M.. 2014. Germinal center-independent, IgM-mediated autoimmunity in sanroque mice lacking Obf1. Immunol. Cell Biol. 92:12–19. 10.1038/icb.2013.71 [DOI] [PubMed] [Google Scholar]
- Citro, A., Valle A., Cantarelli E., Mercalli A., Pellegrini S., Liberati D., Daffonchio L., Kastsiuchenka O., Ruffini P.A., Battaglia M., et al. 2015. CXCR1/2 inhibition blocks and reverses type 1 diabetes in mice. Diabetes. 64:1329–1340. 10.2337/db14-0443 [DOI] [PubMed] [Google Scholar]
- Cooke, D.W., and Plotnick L.. 2008. Type 1 diabetes mellitus in pediatrics. Pediatr. Rev. 29:374–384, quiz:385. 10.1542/pir.29-11-374 [DOI] [PubMed] [Google Scholar]
- Cunninghame Graham, D.S., Wong A.K., McHugh N.J., Whittaker J.C., and Vyse T.J.. 2006. Evidence for unique association signals in SLE at the CD28-CTLA4-ICOS locus in a family-based study. Hum. Mol. Genet. 15:3195–3205. 10.1093/hmg/ddl395 [DOI] [PubMed] [Google Scholar]
- Delong, T., Wiles T.A., Baker R.L., Bradley B., Barbour G., Reisdorph R., Armstrong M., Powell R.L., Reisdorph N., Kumar N., et al. 2016. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 351:711–714. 10.1126/science.aad2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diana, J., and Lehuen A.. 2014. Macrophages and β-cells are responsible for CXCR2-mediated neutrophil infiltration of the pancreas during autoimmune diabetes. EMBO Mol. Med. 6:1090–1104. 10.15252/emmm.201404144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C., and Nurieva R.I.. 2003. Regulation of immune and autoimmune responses by ICOS. J. Autoimmun. 21:255–260. 10.1016/S0896-8411(03)00119-7 [DOI] [PubMed] [Google Scholar]
- Dong, C., Juedes A.E., Temann U.A., Shresta S., Allison J.P., Ruddle N.H., and Flavell R.A.. 2001. ICOS co-stimulatory receptor is essential for T-cell activation and function. Nature. 409:97–101. 10.1038/35051100 [DOI] [PubMed] [Google Scholar]
- Farh, K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A., et al. 2015. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 518:337–343. 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.R. 2016. A BH3 Mimetic for Killing Cancer Cells. Cell. 165:1560. 10.1016/j.cell.2016.05.080 [DOI] [PubMed] [Google Scholar]
- Gstaiger, M., Knoepfel L., Georgiev O., Schaffner W., and Hovens C.M.. 1995. A B-cell coactivator of octamer-binding transcription factors. Nature. 373:360–362. 10.1038/373360a0 [DOI] [PubMed] [Google Scholar]
- Gstaiger, M., Georgiev O., van Leeuwen H., van der Vliet P., and Schaffner W.. 1996. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 15:2781–2790. 10.1002/j.1460-2075.1996.tb00638.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins, K. 2005. Pathogenic T-cell clones in autoimmune diabetes: more lessons from the NOD mouse. Adv. Immunol. 87:123–162. 10.1016/S0065-2776(05)87004-X [DOI] [PubMed] [Google Scholar]
- Hattori, M., Buse J.B., Jackson R.A., Glimcher L., Dorf M.E., Minami M., Makino S., Moriwaki K., Kuzuya H., Imura H., et al. 1986. The NOD mouse: recessive diabetogenic gene in the major histocompatibility complex. Science. 231:733–735. 10.1126/science.3003909 [DOI] [PubMed] [Google Scholar]
- Heng, T.S., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Ikegami, I., Takaki H., Kamiya S., Kamekura R., and Ichimiya S.. 2019. Bob1 enhances RORγt-mediated IL-17A expression in Th17 cells through interaction with RORγt. Biochem. Biophys. Res. Commun. 514:1167–1171. 10.1016/j.bbrc.2019.05.057 [DOI] [PubMed] [Google Scholar]
- Kalekar, L.A., Schmiel S.E., Nandiwada S.L., Lam W.Y., Barsness L.O., Zhang N., Stritesky G.L., Malhotra D., Pauken K.E., Linehan J.L., et al. 2016. CD4(+) T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat. Immunol. 17:304–314. 10.1038/ni.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, J.D., Wang B., Haskins K., Benoist C., and Mathis D.. 1993. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 74:1089–1100. 10.1016/0092-8674(93)90730-E [DOI] [PubMed] [Google Scholar]
- Kawakami, N., Odoardi F., Ziemssen T., Bradl M., Ritter T., Neuhaus O., Lassmann H., Wekerle H., and Flügel A.. 2005. Autoimmune CD4+ T cell memory: lifelong persistence of encephalitogenic T cell clones in healthy immune repertoires. J. Immunol. 175:69–81. 10.4049/jimmunol.175.1.69 [DOI] [PubMed] [Google Scholar]
- Kearney, E.R., Pape K.A., Loh D.Y., and Jenkins M.K.. 1994. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity. 1:327–339. 10.1016/1074-7613(94)90084-1 [DOI] [PubMed] [Google Scholar]
- Kiesler, P., Shakya A., Tantin D., and Vercelli D.. 2009. An allergy-associated polymorphism in a novel regulatory element enhances IL13 expression. Hum. Mol. Genet. 18:4513–4520. 10.1093/hmg/ddp411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, U., Qin X.F., Gong S., Stevens S., Luo Y., Nussenzweig M., and Roeder R.G.. 1996. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 383:542–547. 10.1038/383542a0 [DOI] [PubMed] [Google Scholar]
- Kim, H., Dickey L., Stone C., Jafek J.L., Lane T.E., and Tantin D.. 2019. T cell-selective deletion of Oct1 protects animals from autoimmune neuroinflammation while maintaining neurotropic pathogen response. J. Neuroinflammation. 16:133. 10.1186/s12974-019-1523-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm, J.D., Rould M.A., Aurora R., Herr W., and Pabo C.O.. 1994. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 77:21–32. 10.1016/0092-8674(94)90231-3 [DOI] [PubMed] [Google Scholar]
- Krautwald, S., Ziegler E., Tiede K., Pust R., and Kunzendorf U.. 2004. Transduction of the TAT-FLIP fusion protein results in transient resistance to Fas-induced apoptosis in vivo. J. Biol. Chem. 279:44005–44011. 10.1074/jbc.M401327200 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy, B., Mariana L., Gellert S.A., Colman P.G., Harrison L.C., Lew A.M., Santamaria P., Thomas H.E., and Kay T.W.. 2008. Autoimmunity to both proinsulin and IGRP is required for diabetes in nonobese diabetic 8.3 TCR transgenic mice. J. Immunol. 180:4458–4464. 10.4049/jimmunol.180.7.4458 [DOI] [PubMed] [Google Scholar]
- Kurts, C., Heath W.R., Carbone F.R., Allison J., Miller J.F., and Kosaka H.. 1996. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184:923–930. 10.1084/jem.184.3.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurts, C., Carbone F.R., Barnden M., Blanas E., Allison J., Heath W.R., and Miller J.F.. 1997. CD4+ T cell help impairs CD8+ T cell deletion induced by cross-presentation of self-antigens and favors autoimmunity. J. Exp. Med. 186:2057–2062. 10.1084/jem.186.12.2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon Rodriguez, D.A., Carmona F.D., Echeverría L.E., González C.I., and Martin J.. 2016. IL18 Gene Variants Influence the Susceptibility to Chagas Disease. PLoS Negl. Trop. Dis. 10:e0004583. 10.1371/journal.pntd.0004583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lernmark, A., and Larsson H.E.. 2013. Immune therapy in type 1 diabetes mellitus. Nat. Rev. Endocrinol. 9:92–103. 10.1038/nrendo.2012.237 [DOI] [PubMed] [Google Scholar]
- Li, L., He Q., Garland A., Yi Z., Aybar L.T., Kepler T.B., Frelinger J.A., Wang B., and Tisch R.. 2009. beta cell-specific CD4+ T cell clonotypes in peripheral blood and the pancreatic islets are distinct. J. Immunol. 183:7585–7591. 10.4049/jimmunol.0901587 [DOI] [PubMed] [Google Scholar]
- Makino, S., Muraoka Y., Kishimoto Y., and Hayashi Y.. 1985. Genetic analysis for insulitis in NOD mice. Jikken Dobutsu. 34:425–431. 10.1538/expanim1978.34.4_425 [DOI] [PubMed] [Google Scholar]
- Martinez, R.J., Zhang N., Thomas S.R., Nandiwada S.L., Jenkins M.K., Binstadt B.A., and Mueller D.L.. 2012. Arthritogenic self-reactive CD4+ T cells acquire an FR4hiCD73hi anergic state in the presence of Foxp3+ regulatory T cells. J. Immunol. 188:170–181. 10.4049/jimmunol.1101311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano, M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science. 337:1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, M., Nishida N., Kawashima M., Aiba Y., Tanaka A., Yasunami M., Nakamura H., Komori A., Nakamuta M., Zeniya M., et al. 2012. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am. J. Hum. Genet. 91:721–728. 10.1016/j.ajhg.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby, B.N., Brusko T.M., Zou B., Atkinson M.A., Clare-Salzler M., and Mathews C.E.. 2017. Type 1 Interferons Potentiate Human CD8+ T-Cell Cytotoxicity Through a STAT4- and Granzyme B-Dependent Pathway. Diabetes. 66:3061–3071. 10.2337/db17-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, M.J., Xue S., Alexander J.J., Wasserfall C.H., Campbell-Thompson M.L., Battaglia M., Gregori S., Mathews C.E., Song S., Troutt M., et al. 2009. Immune depletion with cellular mobilization imparts immunoregulation and reverses autoimmune diabetes in nonobese diabetic mice. Diabetes. 58:2277–2284. 10.2337/db09-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, J.D., Berg R., Piganelli J.D., Poulin M., and Haskins K.. 1998. Analysis of leukocytes recruited to the pancreas by diabetogenic T cell clones. Cell. Immunol. 189:92–98. 10.1006/cimm.1998.1377 [DOI] [PubMed] [Google Scholar]
- Polo, J.M., Dell’Oso T., Ranuncolo S.M., Cerchietti L., Beck D., Da Silva G.F., Prive G.G., Licht J.D., and Melnick A.. 2004. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 10:1329–1335. 10.1038/nm1134 [DOI] [PubMed] [Google Scholar]
- Presa, M., Chen Y.G., Grier A.E., Leiter E.H., Brehm M.A., Greiner D.L., Shultz L.D., and Serreze D.V.. 2015. The Presence and Preferential Activation of Regulatory T Cells Diminish Adoptive Transfer of Autoimmune Diabetes by Polyclonal Nonobese Diabetic (NOD) T Cell Effectors into NSG versus NOD-scid Mice. J. Immunol. 195:3011–3019. 10.4049/jimmunol.1402446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, X.F., Reichlin A., Luo Y., Roeder R.G., and Nussenzweig M.C.. 1998. OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J. 17:5066–5075. 10.1093/emboj/17.17.5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosmalen, J.G., Martin T., Dobbs C., Voerman J.S., Drexhage H.A., Haskins K., and Leenen P.J.. 2000. Subsets of macrophages and dendritic cells in nonobese diabetic mouse pancreatic inflammatory infiltrates: correlation with the development of diabetes. Lab. Invest. 80:23–30. 10.1038/labinvest.3780004 [DOI] [PubMed] [Google Scholar]
- Schloot, N.C., Hanifi-Moghaddam P., Goebel C., Shatavi S.V., Flohé S., Kolb H., and Rothe H.. 2002. Serum IFN-gamma and IL-10 levels are associated with disease progression in non-obese diabetic mice. Diabetes Metab. Res. Rev. 18:64–70. 10.1002/dmrr.256 [DOI] [PubMed] [Google Scholar]
- Schwarze, S.R., Hruska K.A., and Dowdy S.F.. 2000. Protein transduction: unrestricted delivery into all cells? Trends Cell Biol. 10:290–295. 10.1016/S0962-8924(00)01771-2 [DOI] [PubMed] [Google Scholar]
- Serreze, D.V., Chapman H.D., Varnum D.S., Hanson M.S., Reifsnyder P.C., Richard S.D., Fleming S.A., Leiter E.H., and Shultz L.D.. 1996. B lymphocytes are essential for the initiation of T cell-mediated autoimmune diabetes: analysis of a new “speed congenic” stock of NOD.Ig mu null mice. J. Exp. Med. 184:2049–2053. 10.1084/jem.184.5.2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serreze, D.V., Bridgett M., Chapman H.D., Chen E., Richard S.D., and Leiter E.H.. 1998. Subcongenic analysis of the Idd13 locus in NOD/Lt mice: evidence for several susceptibility genes including a possible diabetogenic role for beta 2-microglobulin. J. Immunol. 160:1472–1478. [PubMed] [Google Scholar]
- Shakya, A., Kang J., Chumley J., Williams M.A., and Tantin D.. 2011. Oct1 is a switchable, bipotential stabilizer of repressed and inducible transcriptional states. J. Biol. Chem. 286:450–459. 10.1074/jbc.M110.174045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya, A., Goren A., Shalek A., German C.N., Snook J., Kuchroo V.K., Yosef N., Chan R.C., Regev A., Williams M.A., and Tantin D.. 2015. Oct1 and OCA-B are selectively required for CD4 memory T cell function. J. Exp. Med. 212:2115–2131. 10.1084/jem.20150363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, W., Liao Y., Willis S.N., Taubenheim N., Inouye M., Tarlinton D.M., Smyth G.K., Hodgkin P.D., Nutt S.L., and Corcoran L.M.. 2015. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 16:663–673. 10.1038/ni.3154 [DOI] [PubMed] [Google Scholar]
- Sitrin, J., Ring A., Garcia K.C., Benoist C., and Mathis D.. 2013. Regulatory T cells control NK cells in an insulitic lesion by depriving them of IL-2. J. Exp. Med. 210:1153–1165. 10.1084/jem.20122248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skwarczynska, M., and Ottmann C.. 2015. Protein-protein interactions as drug targets. Future Med. Chem. 7:2195–2219. 10.4155/fmc.15.138 [DOI] [PubMed] [Google Scholar]
- Staeva, T.P., Chatenoud L., Insel R., and Atkinson M.A.. 2013. Recent lessons learned from prevention and recent-onset type 1 diabetes immunotherapy trials. Diabetes. 62:9–17. 10.2337/db12-0562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Matthias G., Mihatsch M.J., Georgopoulos K., and Matthias P.. 2003. Lack of the transcriptional coactivator OBF-1 prevents the development of systemic lupus erythematosus-like phenotypes in Aiolos mutant mice. J. Immunol. 170:1699–1706. 10.4049/jimmunol.170.4.1699 [DOI] [PubMed] [Google Scholar]
- Testa, G., Schaft J., van der Hoeven F., Glaser S., Anastassiadis K., Zhang Y., Hermann T., Stremmel W., and Stewart A.F.. 2004. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 38:151–158. 10.1002/gene.20012 [DOI] [PubMed] [Google Scholar]
- Thin, T.H., Kim E., Yeh S., Sampson E.R., Chen Y.T., Collins L.L., Basavappa R., and Chang C.. 2002. Mutations in the helix 3 region of the androgen receptor abrogate ARA70 promotion of 17beta-estradiol-induced androgen receptor transactivation. J. Biol. Chem. 277:36499–36508. 10.1074/jbc.M202824200 [DOI] [PubMed] [Google Scholar]
- Tuettenberg, A., Huter E., Hubo M., Horn J., Knop J., Grimbacher B., Kroczek R.A., Stoll S., and Jonuleit H.. 2009. The role of ICOS in directing T cell responses: ICOS-dependent induction of T cell anergy by tolerogenic dendritic cells. J. Immunol. 182:3349–3356. 10.4049/jimmunol.0802733 [DOI] [PubMed] [Google Scholar]
- Van Belle, T.L., Taylor P., and von Herrath M.G.. 2009. Mouse Models for Type 1 Diabetes. Drug Discov. Today Dis. Models. 6:41–45. 10.1016/j.ddmod.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel, D.A., Udalova I.A., De Silva A.P., McGovern D.P., Kinouchi Y., Hull J., Lench N.J., Cardon L.R., Carey A.H., Jewell D.P., and Kwiatkowski D.. 2002. Inflammatory bowel disease is associated with a TNF polymorphism that affects an interaction between the OCT1 and NF(-kappa)B transcription factors. Hum. Mol. Genet. 11:1281–1289. 10.1093/hmg/11.11.1281 [DOI] [PubMed] [Google Scholar]
- Vanasek, T.L., Khoruts A., Zell T., and Mueller D.L.. 2001. Antagonistic roles for CTLA-4 and the mammalian target of rapamycin in the regulation of clonal anergy: enhanced cell cycle progression promotes recall antigen responsiveness. J. Immunol. 167:5636–5644. 10.4049/jimmunol.167.10.5636 [DOI] [PubMed] [Google Scholar]
- Verdaguer, J., Schmidt D., Amrani A., Anderson B., Averill N., and Santamaria P.. 1997. Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J. Exp. Med. 186:1663–1676. 10.1084/jem.186.10.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince, N., Li H., Ramsuran V., Naranbhai V., Duh F.M., Fairfax B.P., Saleh B., Knight J.C., Anderson S.K., and Carrington M.. 2016. HLA-C Level Is Regulated by a Polymorphic Oct1 Binding Site in the HLA-C Promoter Region. Am. J. Hum. Genet. 99:1353–1358. 10.1016/j.ajhg.2016.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., André I., Gonzalez A., Katz J.D., Aguet M., Benoist C., and Mathis D.. 1997. Interferon-gamma impacts at multiple points during the progression of autoimmune diabetes. Proc. Natl. Acad. Sci. USA. 94:13844–13849. 10.1073/pnas.94.25.13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, C.P., Li L., Frelinger J.A., and Tisch R.. 2006. Early autoimmune destruction of islet grafts is associated with a restricted repertoire of IGRP-specific CD8+ T cells in diabetic nonobese diabetic mice. J. Immunol. 176:1637–1644. 10.4049/jimmunol.176.3.1637 [DOI] [PubMed] [Google Scholar]
- Yamane, K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., and Zhang Y.. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 125:483–495. 10.1016/j.cell.2006.03.027 [DOI] [PubMed] [Google Scholar]
- Yeo, L., Woodwyk A., Sood S., Lorenc A., Eichmann M., Pujol-Autonell I., Melchiotti R., Skowera A., Fidanis E., Dolton G.M., et al. 2018. Autoreactive T effector memory differentiation mirrors β cell function in type 1 diabetes. J. Clin. Invest. 128:3460–3474. 10.1172/JCI120555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef, N., Shalek A.K., Gaublomme J.T., Jin H., Lee Y., Awasthi A., Wu C., Karwacz K., Xiao S., Jorgolli M., et al. 2013. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 496:461–468. 10.1038/nature11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Todorov I., Lin C.L., Atkinson M., Kandeel F., Forman S., and Zeng D.. 2007. Elimination of insulitis and augmentation of islet beta cell regeneration via induction of chimerism in overtly diabetic NOD mice. Proc. Natl. Acad. Sci. USA. 104:2337–2342. 10.1073/pnas.0611101104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber, J., Shi J., Wang E., Rappaport A.R., Herrmann H., Sison E.A., Magoon D., Qi J., Blatt K., Wunderlich M., et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 478:524–528. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling, S., Dieckmann A., Pfisterer P., Angel P., and Wirth T.. 1997. Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science. 277:221–225. 10.1126/science.277.5323.221 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is related to Fig. S1 and shows loci for T1D susceptibility and resistance in NOD/ShiLtJ mice and oligonucleotide primer pairs used for PCR amplification of Idd loci microsatellite markers.
is related to Fig. S2 and shows gene expression comparisons in T cells taken from PLNs of 8-wk-old NOD.Ocabfl/flCD4-cre and NOD.Ocabfl/fl littermate controls.
is related to Fig. 1 and shows expression levels of Ccl1 mRNA in naive, stimulated, rested, and re-stimulated OCA-B/Pou2af1–deficient T cells relative to controls.
is related to Fig. 2 and shows gene expression comparisons in total CD45+ cells taken from pancreatic islets of 12-wk-old prediabetic NOD.Ocabfl/flCD4-cre and NOD.Ocabfl/fl littermate controls.