Abstract
Exploration of the chemical space of known influenza polymerase PB2 inhibitor Pimodivir, was performed by our group. We synthesized and identified compounds 16a and 16b, two novel thienopyrimidine derivatives displaying anti-influenza A activity in the single digit nanomolar range in cell culture. Binding of these unique compounds in the influenza polymerase PB2 pocket was also determined using molecular modeling.
Keywords: Flu, Influenza A, Antiviral, Virus
Introduction:
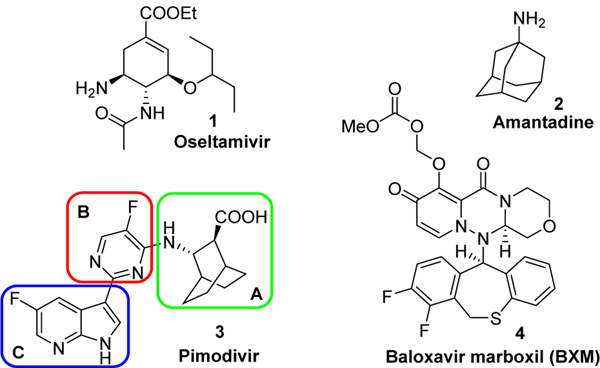
Seasonal influenza outbreaks are estimated to impact more than 3.5 million people every year and be responsible for over 290,000 to 650,000 death worldwide. Although influenza vaccines remain the most effective means to prevent seasonal influenza, they typically provide suboptimal protection. Moreover, they may be completely ineffective in the event of an antigenic mismatch between viruses in the seasonal vaccine and those circulating in the community, and their utility in response to rapid and pandemic spreads remain uncertain.1 Only three classes of antiviral agents are currently approved in the US for the treatment of influenza: Adamantanes (amantidine, rimantadine), neuramidinase (NA) inhibitors (Oseltamivir, Peramivir, Zanamivir) and baloxavir marboxil (BXM), which is the prodrug of baloxavir acid (BXA), an endonuclease (PA) inhibitor. However, adamantanes, which target the viral M2 protein involved in the uncoating of the virus during replication, are not part of the standard of care for seasonal influenza A viruses due to widespread viral resistance.2 The use of NA inhibitors, which impair the release of virus from infected cells, is also limited since their antiviral potency is relatively modest. Importantly, oseltamivir, the most prescribed NA drug, was completely ineffective during the 2008–2009 H1N1 flu outbreak.3, 4 BXM is the only endonuclease inhibitor approved for the treatment of both influenza A and B. Even though a single dose of BXM proved to positively improve outcomes, treatment-emergent monitoring in the phase 2 study identified PA I38T/F substitutions in A/H3N2 viruses conferring more than 10-fold reduction in BXM susceptibility.5 Furthermore, this loss of susceptibility occurred in 10% of the persons treated with BXM in phase III trials.6 Drugs targeting viral proteins are currently in the influenza drug pipeline, including Pimodivir, an influenza polymerase PB2 inhibitor which is now in phase III clinic trials.7,8 The PB2 subunit specifically plays a role in generating 5’-capped RNA fragments from cellular pre-mRNA molecules that are used as primers for viral transcription. Known inhibitors bind to active site of the PB2 cap-binding domain preventing the binding of the natural ligand, 7-methyl GTP, thereby preventing viral RNA synthesis.9 It is worth noting that because of major sequence difference between influenza A and B PB2 domains, PB2 inhibitor remains only active against influenza A.10 As part of our influenza research program, we divided Pimodivir into 3 key components (Parts A, B and C, Figure 1) and evaluated the influence of novel structural modifications for each part. Herein, we wish to report the synthesis of a small library of influenza polymerase PB2 inhibitor along with their antiviral evaluation.
Figure 1.
Structure of approved drugs oseltamivir, amantadine, baloxavir marboxil and experimental drug Pimodivir.
Part A:
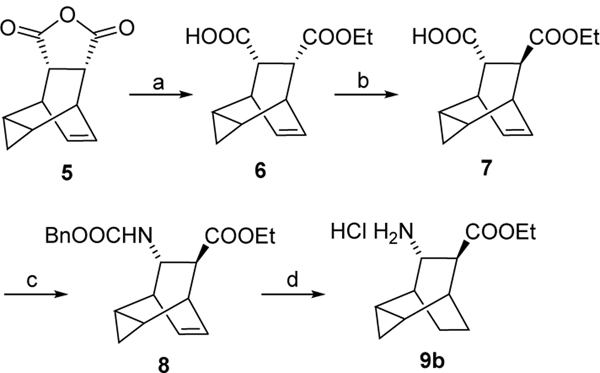
Key amine 9d was purchased while amines 9a and 9c were prepared by following reported procedures11. 9b was synthesized according to the chemistry described in scheme 112.
Scheme 1. Reagents and conditions:
a) quinine, toluene, EtOH, −15 °C, two days; b) i) 6N HCl, toluene, rt, 1 h; ii) KOtAmyl, toluene, −20 °C to −10 °C, 2.5 h; iii) HOAc, HCl, 45 min, 41 % over 2 steps; c) Et3N, DPPA, BnOH, toluene, 96 °C, overnight, 71 %; d) Pd(OH)2 /C, H2, HCl/EtOH, overnight, 80 %.
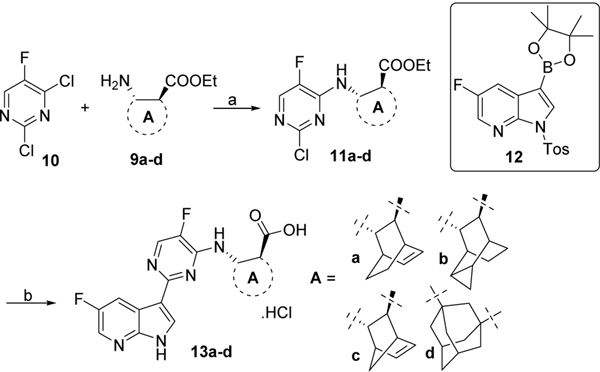
Dichloropyrimidine 10 was treated with amines 9a-d and DIPEA to give intermediates 11a-d which were then reacted with fluoro-3-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine 1213 in presence of Pd2(dba)3, X-Phos and K3PO4. Final saponification and deprotection with LiOH afforded the desired derivatives 13a-d (Scheme 2).
Scheme 2. Reagents and conditions:
a) DIPEA, dichloroethane, 70 °C, overnight, for 11a, 67%; for 11b, 74%; for 11c, 56%; for 11d, 45%; b) i) 12, Pd2(dba)3, X-Phos, K3PO4, 2-methylTHF/H2O, 110 °C, overnight, 45–70%; ii) LiOH, THF, 70 °C, 48 h, for 13a, 50%; for 13b, 65%; for 13c, 35%; for 13d, 67%.
Part B:
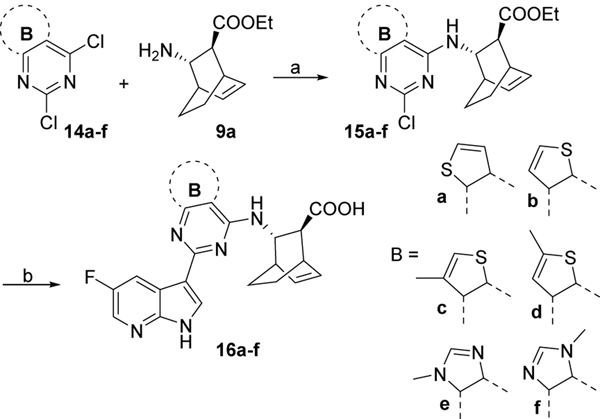
Compounds 16a-f were synthesized from dichloro heteromatic moieties 14a-f14 following the same coupling, saponification, deprotection sequence (Scheme 3)
Scheme 3. Reagents and conditions:
a) DIPEA, dichloroethane, 70 °C, overnight, for 15a, 55%; for 15b, 62%; for 15c, 54%; for 15d, 50%; for 15e, 36%; for 15f, 40%; b) i) 12, Pd2(dba)3, X-Phos, K3PO4, 2-methylTHF/H2O, 110 °C, overnight, 25–67%; ii) LiOH, THF, 70 °C, 2 days, for 16a, 65%; for 16b, 64%; for 16c, 58%; for 16d, 60%; for 16e, 21%; for 16f, 18%.
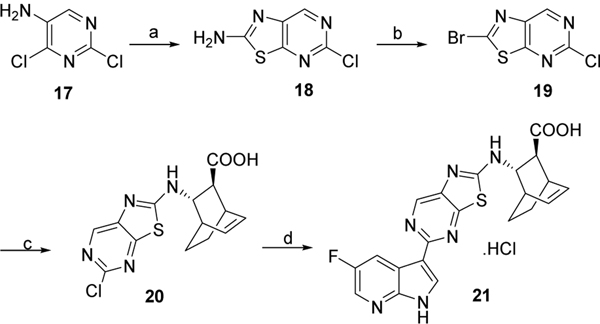
Compound 21 was prepared by following the chemistry described in Scheme 4. Dichloro pyrimidine 17 was treated with KSCN under acidic condition to form 18. Compound 18 was further brominated and then coupled with amine 9a in presence of DIPEA. Finally, Suzuki type coupling with 5-fluoro-3-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine 12 in presence of X-Phos and K3PO4, followed by treatment with LiOH gave compound 21.
Scheme 4. Reagents and conditions:
a) KSCN, acetic acid, reflux, 3 h, 89%;b) t-Butyl nitrite, CHBr3, 60 °C for 1 h then 90 °C for 1 h, 50%; c) 9a, DIPEA, dichloroethane, 70 °C, overnight, 65%; d) i) 12, Pd2(dba)3, X-Phos, K3PO4, 2-methylTHF/H2O, 110 °C, overnight, 43%; ii) LiOH, THF, 70 °C, 2 days, 42%.
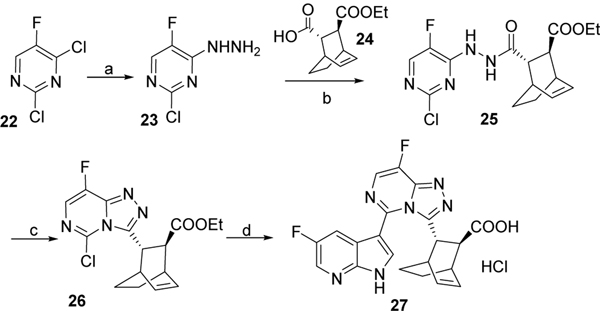
Triazolopyrimidine derivative 27 was synthesized by first reacting dichlorofluoro pyrimidine 22 with hydrazine followed by coupling of intermediate 23 with carboxylic acid derivative 2413 in presence of EDCI and DMAP. Subsequent reaction with POCl3 gave chlorinated intermediate 26 which was reacted with 5-fluoro-3-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine 12 in presence of X-Phos and K3PO4. Final treatment with LiOH afforded the desired compound 27 (Scheme 5).
Scheme 5. Reagents and conditions:
a) NH2NH2, ethanol, 0°C to rt, overnight, 68%; b) EDCI, DMAP, dichloromethane, overnight, 51%; c) POCl3, 80 °C, overnight, 80%; d) i) 12, Pd2(dba)3, X-Phos, K3PO4, 2-methylTHF/H2O, 110 °C, overnight, 25%; ii) LiOH, THF, 70°C, 2 days, 40%.
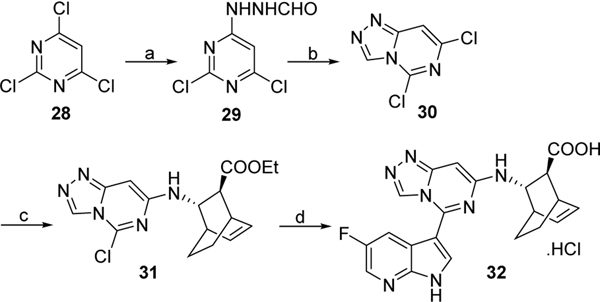
Compound 32 was prepared by first reacting trichloropyrimidine 28 with formic hydrazide and cyclization of intermediate 29 in presence of POCl3. Subsequent coupling of 30 with 9a in presence of DIPEA, followed by palladium mediated coupling with 5-fluoro-3-(4,4,5,5tetramethyl-1,3,2-dioxaborolan-2-yl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine 12 and treatment with LiOH gave targeted compound 32 (Scheme 6).
Scheme 6. Reagents and conditions:
a) NH2NHCHO, ethanol, rt, overnight, 54 %; b) POCl3, 80 °C, 10 h, 78%; c) 9a, DIPEA, dichloroethane, 70 °C, overnight, 50 %. d) i) 12, Pd2(dba)3, X-Phos, K3PO4, 2-methylTHF/H2O, 110 °C, overnight, 56%; ii) LiOH, THF, 70°C, 2 days, 42%.
Part C:
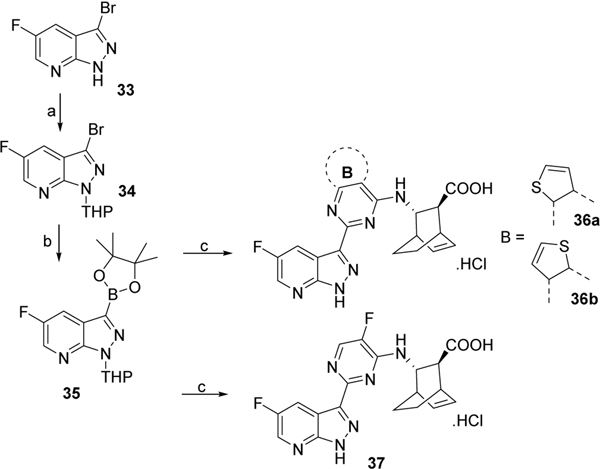
Compound 3315 was first protected with a THP group then reacted with bis(pinacolato) diboron in presence of potassium acetic and Pd(dppf)2Cl2 to form boronic ester derivative 35. Compound 35 was then coupled with chlorinated scaffolds 11a or 15a-b to give, after deprotection under acid condition and saponification of the ester group with LiOH, the desired compounds 36a-b and 37 (Scheme 7).
Scheme 7. Reagents and conditions:
a) dihydropyran (DHP), TsOH, dichloromethane, rt, overnight, 65%; b) bis(pinacolato)diboron, potassium acetic, Pd(dppf)2Cl2, 80 °C, overnight, 70%; c) i) 11a or 15a-b, Pd2(dba)3, X-Phos, K3PO4, 2-methylTHF/H2O, 110°C, overnight, 30–40%; ii) trifluoroacetic acid, dichloromethane, rt, overnight, 63–72%; iii) LiOH, THF, 70°C, 2 days, for 36a, 35%; for 36b, 45%; for 37, 40%.
The synthesized compounds were evaluated against influenza A virus PR8-GLuc (H1N1) strain. Briefly, A549 cells were seeded at a density of 40,000 cells/well and incubated at 37 °C and 5% CO2 overnight followed by inoculating with 0.1 MOI of the virus for 1 h and then were washed with 2 acidic washes (PBS, pH 2.2) and 3 neutral washes (PBS, pH 7.4) to remove unbound virus. Infected cells then were treated with test compound in a single concentration lower than maximum non-toxic concentration. (MNTC is defined as the maximum concentration at which at least 90% of the treated cells are viable and is determined by treating cells with increasing concentrations of tested compound). Those which showed 50% inhibitory activity against virus replication were then retested in a dose response manner to determine EC50 and EC90 values. In addition, cytotoxicity was assessed against a panel of cell lines including A459 cells, primary human peripheral blood mononuclear (PBM) cells, human lymphoblastoid cells (CEM), and African Green monkey Vero cells to determine therapeutic indexes. The results are summarized in Table 1.
Table 1.
Anti-influenza A activity in A549 cells and cytotoxicity of compounds 13a-d, 16a-f, 21, 27, 32, 36a-b and 37.
| cpd | Anti-Influenza A activity in A549 cells (μM ± SD) | Toxicity (μM) in | |||||
|---|---|---|---|---|---|---|---|
| PBM | CEM | Vero | Huh7 | A549 | |||
| EC50 | EC90 | CC50 | CC50 | CC50 | CC50 | CC50 | |
| pimodivir | 0.004 ± 0.002 | 0.027 ± 0.01 | > 100 | 48.9 | > 100 | 95.8 | 20.0 |
| 13a | 0.007 ± 0.005 | 0.027 ± 0.01 | > 100 | 91.1 | > 100 | > 100 | ≥ 100 |
| 13b | 0.05 ± 0.04 | 0.21 ± 0.07 | > 100 | 62.9 | > 100 | ND | 94.6 |
| 13c | 0.05 ± 0.03 | 0.28 ± 0.05 | > 100 | > 100 | > 100 | > 100 | > 100 |
| 13d | 0.95 ± 0.14 | 5.1 ± 2.1 | 23.1 | 10.1 | 30.3 | 14.3 | 16.2 |
| 16a | 0.0065 ± 0.0011 | 0.047 ± 0.008 | 9.3 | 4.2 | 65.9 | 24.0 | 14.5 |
| 16b | 0.017 ± 0.01 | 0.10 ± 0.06 | 64.6 | 31.6 | 74.9 | 68.7 | 14.7 |
| 16c | 0.029 ± 0.006 | 0.082 ± 0.01 | > 100 | > 100 | > 100 | > 100 | > 100 |
| 16d | 0.027 ± 0.014 | 0.14 ± 0.11 | 78.4 | 12.9 | 90.3 | 31.2 | 41.7 |
| 16e | 0.4 ± 0.23 | 2 ± 0.8 | >100 | 29.1 | > 100 | > 100 | ≥ 100 |
| 16f | >10 | ND | >100 | > 100 | > 100 | > 100 | > 100 |
| 21 | >100 | ND | 84.8 | 17.3 | 86.1 | 70.2 | 31.3 |
| 27 | >40 | ND | ≥ 100 | 41.7 | ≥ 100 | ≥ 100 | 65.4 |
| 32 | 0.68 ± 0.17 | 2.5 ± 0.79 | > 100 | 30.8 | > 100 | 71.0 | 51.9 |
| 36a | 0.012 ± 0.001 | 0.036 ± 0.007 | > 100 | 13.7 | > 100 | 62.7 | > 100 |
| 36b | 0.042 ± 0.007 | 0.14 ± 0.04 | > 100 | 67.0 | 52.5 | 70.7 | 32.5 |
| 37 | 0.016 ± 0.003 | 0.044 ± 0.011 | > 100 | 48.9 | > 100 | 83.0 | 30.6 |
ND: Not determined
Increasing the size of Pimodivir’s bicyclic part A (Figure 1) by adding another ring led to a 12–230 times decrease of potency (compounds 13b and 13d). A similar pattern was observed when the bicyclooctane ring of Pimodivir was changed to a smaller bicycloheptane ring (compound 13c, EC50 = 0.05 μM). On the other hand, introduction of an unsaturated bicylooctane ring led to the discovery of compound 13a which displayed an activity profile similar to Pimodivir (EC90 = 0.027 μM for both compounds). However, unlike Pimodivir, which displays a CC50 of 20 μM in A549 cells, 13a did not display toxicity in A549 cells to 100 μM. Further optimization of compound 13a was performed by evaluating the replacement of the central pyrimidine moiety (Figure 1 - Part B) by various bicylic heterocyclic rings. Introduction of a triazolopyrimidine ring in compounds 27 and 32, an imidazolopyrimidine in compound 16e and 16f or a thiazolopyrimidine ring in compound 21 was counterproductive and led to a significant loss of activity. However introduction of a thienopyrimidine (compounds 16a16 and 16b17) or a methylated thienopyrimidine (compounds 16c and 16d) was well tolerated and compounds 16a and 16b displayed activities similar to that of Pimodivir (EC50 = 0.006 and 0.017 μM respectively). It is worth noting that despite their high potencies, compounds 16a and 16b exhibited toxicity in the micromolar range in our panel of cell lines. Because oxidation of the 2-position of the 7-azaindole ring via aldehyde oxidase has been previously established in that series of compounds,18 introduction of a nitrogen at this position, to eventually block the formation of metabolites, was also evaluated (Figure 1 - Part C). Unfortunately, this modification led to significant loss of potency as seen in compounds 36a-b and 37.
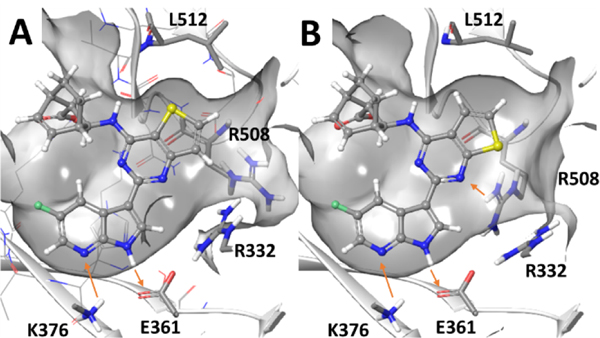
Based on the high potency and unusual structures of the thienopyrimidine derivatives 16a and 16b, we used molecular modeling to establish how these compounds bind to the influenza A PB2 pocket. The crystal structure of the PB2 subunit complexed with Pimodivir served as a locus for induced fit docking (PDBID 6EUV).19 The results of induced fit docking yielded poses that maintain key interactions observed for Pimodivir (Figure 2). The azaindole ring of 16a/16b acted as a hydrogen bond donor to Glu361 and a hydrogen bond acceptor from Lys376 while the bicyclic carboxylate formed an electrostatic interaction with Arg355. Interestingly, to accommodate the thienopyrimidine ring in both compounds, Leu512 rotated away from the agent thereby enlarging the pocket. The model also suggested that in the case of 16b, Arg508 and Arg332 position closer to the inhibitor than observed in the crystal structure.
Figure 2.
Molecular model of (A) 16a or (B) 16b bound to the influenza PB2 pocket obtained from induced fit into Pimodivir complexed crystal structure (PDBID 6EUV).
A small library of influenza polymerase PB2 inhibitors were designed by evaluating novel structural modifications of the 3 key portions of reference compound Pimodivir. Antiviral activity along with cytotoxicity of each compounds was assessed. Among them, thienopyrimidine derivatives 16a and 16b showed antiviral activities in the low nanomolar range, similar to that of Pimodovir. Further modifications of compounds 16a and 16b are currently being investigated in order to further improve potency and decrease observed toxicities.
Acknowledgments
This work was supported in part by NIH Grant 1-R01-AI-132833, and 5P30-AI-50409 (CFAR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harding AT; Heaton NS Efforts to Improve the Seasonal Influenza Vaccine. Vaccines (Basel). 2018, 6(2),19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bright RA; Medina MJ; Xu X; Perez-Oronoz G; Wallis TR; Davis XM; Povinelli L; Cox NJ; Klimov AI Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005, 366 (9492), 1175–81. [DOI] [PubMed] [Google Scholar]
- 3.Hurt AC; Ernest J; Deng YM; Iannello P; Besselaar TG; Birch C; Buchy P; Chittaganpitch M; Chiu SC; Dwyer D; Guigon A; Harrower B; Kei IP; Kok T; Lin C; McPhie K; Mohd A; Olveda R; Panayotou T; Rawlinson W; Scott L; Smith D; D’Souza H; Komadina N; Shaw R; Kelso A; Barr IG Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 2009, 83(1), 90–3. [DOI] [PubMed] [Google Scholar]
- 4.Hurt AC; Hardie K; Wilson NJ; Deng YM; Osbourn M; Gehrig N; Kelso A. Community transmission of oseltamivir-resistant A(H1N1)pdm09 influenza. N. Engl. J. Med 2011, 365(26), 2541–2. [DOI] [PubMed] [Google Scholar]
- 5.Omoto S; Speranzini V; Hashimoto T; Noshi T; Yamaguchi H; Kawai M; Kawaguchi K; Uehara T; Shishido T; Naito A; and Cusack S. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep 2018, 8(1), 9633–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayden FG; Sugaya N; Hirotsu N; Lee N; de Jong MD; Hurt AC; Ishida T; Sekino H; Yamada K; Portsmouth S,; Kawaguchi K; Shishido T; Arai M; Tsuchiya K; Uehara T; Watanabe A; Baloxavir Marboxil Investigators Group. Baloxavir Marboxil for Uncomplicated Influenza in Adults and Adolescents. N. Engl. J. Med 2018, 379(10), 913–23. [DOI] [PubMed] [Google Scholar]
- 7.a) Shaw ML The Next Wave of Influenza Drugs. ACS Infect. Dis 2017, 3 (10), 691–4. [DOI] [PubMed] [Google Scholar]; b) Trevejo JM; Asmal M; Vingerhoets J; Polo R; Robertson S; Jiang Y; Kieffer TL; Leopold L. Pimodivir treatment in adult volunteers experimentally inoculated with live influenza virus: a Phase IIa, randomized, double-blind, placebo-controlled study. Antivir. Ther 2018, 23(4), 335–44. [DOI] [PubMed] [Google Scholar]; c) Finberg RW; Lanno R; Anderson D; Fleischhackl R; van Duijnhoven W; Kauffman RS; Kosoglou T; Vingerhoets J; Leopold L. Phase 2b study of pimodivir (JNJ-63623872) as monotherapy or in combination with oseltamivir for treatment of acute uncomplicated seasonal influenza A: TOPAZ trial. J. Infect. Dis 2019, 219(7),1026–34. [DOI] [PubMed] [Google Scholar]
- 8.a) Clark MP; Ledeboer MW; Davies I; Byrn RA; Jones SM; Perola E; Tsai A; Jacobs M; Nti-Addae K; Bandarage UK; Boyd MJ; Bethiel RS; Court JJ; Deng H-B; Duffy JP; Dorsch WA; Farmer LJ; Gao H; Gu W-X; Jackson K; Jacobs DH; Kennedy JM; Ledford B; Liang J-L; Maltais F; Murcko M; Wang T-S; Wannamaker MW; Bennett HB; Leeman JR; McNeil C; Taylor WP; Memmott C; Jiang M; Rijnbrand R; Bral C; Germann U; Nezami A; Zhang Y-G; Salituro FG; Bennani YL; Charifson PS Discovery of a Novel, First-in-Class, Orally Bioavailable Azaindole Inhibitor (VX-787) of Influenza PB2. J. Med. Chem 2014, 57 (15), 6668–78. [DOI] [PubMed] [Google Scholar]; b) Farmer LJ; Clark MP; Boyd MJ; Perola E; Jones SM; Tsai A; Jacobs MD;Bandarage UK; Ledeboer MW; Wang T-S; Deng H-B; Ledford B; Gu W-X; Duffy JP; Bethiel RS; Shannon D; Byrn RA; Leeman JR; Rijnbrand R; Bennett HB; O’Brien C; Memmott C; Nti-Addae K; Bennani YL; Charifson PS Discovery of Novel, Orally Bioavailable β‑Amino Acid Azaindole Inhibitors of Influenza PB2. ACS Med. Chem. Lett 2017, 8, 256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palese P; Shaw ML Orthomyxoviridae: The Viruses and Their Replication In Fields Virology: Knipe DM, Howley PM, Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, 2007; pp 1647–89. [Google Scholar]
- 10.Yuan S-F; Wen L; Zhou J. Inhibitors of influenza A virus polymerase. ACS infect. Dis 2018, 4, 218–23. [DOI] [PubMed] [Google Scholar]
- 11.Tanoury GJ; Nugent WA; Dvornikovs V; Rose PJ Preparation of pyrimidinylpyrrolopyridine derivatives for use as influenza viruses replication inhibitors. WO2015073481, 2015, 165pp. [Google Scholar]
- 12.Jordan R; Bailey TR; Rippin SR; Dai D-C Preparation of isoindole derivatives for treatment and prevention of orthopoxvirus infections. WO2008130348, 2008, 82 pp. [Google Scholar]
- 13.Nti-Addae KW; Waldo M; O’Neil SA; Van Alsten JG; Macikenas D; Mudunuri P; Shi Y; Ledeboer MW; Jurkauskas V; Medek A; Jones S; Byrn R; Asmal M; Robertson SM; Tsai W-J Preparation of pyrimidinylpyrrolopyridine derivatives for use as influenza viruses replication inhibitors. WO2015073476, 2015, 126pp. [Google Scholar]
- 14.14a-d are commercially available; 14e-f are synthesized by following the chemistry described in : Janssen PAJ; Lewi PJ; De Jonge MR; Koymans LMH; Daeyaert FFD; Heeres J; Vinkers HM; Leenders RGG; Vandenput DAL Preparation of purine derivatives for use in pharmaceutical compositions as HIV replication inhibitors. WO2005028479, 2005, 108pp. [Google Scholar]
- 15.Charifson PS; Clark MP; Bandarage UK; Bethiel RS; Boyd MJ; Davies I; Deng H-B; Duffy JP; Farmer LJ; Gao H; Gu W-X; Kennedy JM; Ledford B; Ledeboer MW; Maltais F; Perola E; Wang T-S Preparation of pyrrolopyridinylpyrimidinylamine deratives and analogs for use as influenza viruses replication inhibitors. WO2013019828, 2013, 174pp. [Google Scholar]
- 16.Compound 16a: 1H NMR (400MHz, DMSO-d6): δ 12.24 (s, 1H), 8.71 (dd, J = 2.8 Hz, J = 10.0 Hz, 1H), 8.35 (s, 1H), 8.29 (s, 1H), 8.06 (d, J = 5.2 Hz, 1H), 7.39 (d, J = 5.6 Hz, 1H), 6.55 (t, J = 7.2 Hz, 1H), 6.25 (t, J = 7.2 Hz, 1H), 4.84 (m, 1H), 3.10 (brs, 1H), 2.93 (brs, 1H), 2.65 (brs, 1H), 1.81 (m, 1H), 1.61 (m, 1H), 1.34 (m, 1H), 1.17 (m, 1H); 13C NMR (100 MHz, DMSO-d6): δ 175.2, 159.0, 157.2, 156.5, 154.8, 146.4, 135.2, 132.6, 131.8, 131.1, 128.5, 125.9, 124.6, 119.0,115.9, 114.8, 72.7, 60.7, 52.8, 50.5, 23.42, 20.26; 19F NMR (376 MHz,DMSO-d6): δ −138.42; MS: m/z = 436.4 [M+ H]+
- 17.Compound 16b: 1H NMR (400MHz, DMSO-d6): δ 12.29 (s, 1H), 8.67 (dd, J = 2.8 Hz, J = 10.0 Hz, 1H), 8.35 (s, 1H), 8.29 (m, 1H), 7.65 (d, J = 6.0 Hz, 1H), 7.40 (d, J = 6.0 Hz, 1H), 6.56 (t, J = 7.6 Hz, 1H), 6.25 (t, J = 7.2 Hz, 1H), 4.83 (s, 1H), 3.11 (m, 1H), 2.94 (m, 1H), 2.56 (m, 1H), 1.83 (m, 1H), 1.65 (m, 1H), 1.36 (m, 1H), 1.20 (m, 1H); 13C NMR (100 MHz, DMSO-d6): δ 167.1, 158.3, 157.2, 156.5, 154.8, 146.4, 135.4, 132.5, 131.8, 131.4, 121.0, 120.3, 119.0, 115.8, 114.7, 113.9, 52.8, 51.0, 34.0, 33.2, 23.4, 20.2; 19F NMR (376 MHz, DMSO-d6): δ −138.32; MS: m/z = 436.5 [M+ H]+
- 18.Bandarage UK; Clark MP; Perola E; Gao H; Jacobs MD; Tsai A; Gillespie J; Kennedy JM; Maltais F; Ledeboer MW; Davies I; Gu W; Byrn RA; Nti Addae K; Bennett H; Leeman JR; Jones SM; O’Brien C; Memmott C; Bennani Y; Charifson PS Novel 2-Substituted 7-Azaindole and 7-Azaindazole Analogues as Potential Antiviral Agents for the Treatment of Influenza. ACS Med. Chem. Lett 2017, 8, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Pflug A, Gaudon S, Resa-Infante P, Lethier M, Reich S, Schulze WM, & Cusack S. (2017). Capped RNA primer binding to influenza polymerase and implications for the mechanism of cap-binding inhibitors. Nucleic acids res, 46(2), 956–971. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sherman W, Beard HS, Farid R. Use of an induced fit receptor structure in virtual screening. Chemical biology & drug design, 2006, 67(1), 83–84. [DOI] [PubMed] [Google Scholar]; c) Sherman W; Day T; Jacobson MP; Friesner RA; Farid R, “Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects,” J. Med. Chem, 2006, 49, 534–553. [DOI] [PubMed] [Google Scholar]