Abstract
Objective
Image evaluation strategy for lung cancer patients has difficulty obtaining the appropriate quantity of diffuse lung nodules and bone metastases. The study was to demonstrate whether early variations in the levels of serum 4-tumor markers (4-TMs)(carcinoembryonic antigen [CEA], cancer antigen [CA]125, CA19-9, and CA15-3) after TKI targeted therapy were associated with treatment response in patients with lung adenocarcinoma.
Methods
Patients with stage IIIB-IV lung adenocarcinoma taking epidermal growth factor receptor (EGFR) TKIs or anaplastic lymphoma kinase (ALK) inhibitors were enrolled prospectively from June 2012 to February 2015. According to the variations of the percentage of change in 4-TM levels (4-TMpc), we divided patients into ascending (increases in 4-TMpc over the 7th- 14th day) and descending (decreases in 4-TMpc over the 7th- 14th day) groups.
Results
184 patients were enrolled, and 89% had at least one of the pre-treatment evaluable TMs and were further analyzed. An excellent response to the TKI targeted therapy was accurately predicted in the descending group, as determined using receiver operating characteristic curve analysis (an area under the curve, 0.83). Multivariate Cox hazards model analyses demonstrated that the type of 4-TMpc and mutation status were the strongest predictors of progression-free survival (PFS)(descending versus ascending, hazard ratios [HR] 0.30, 95% confidence interval [CI], 0.19–0.47; sensitive mutation versus wide type, HR 0.30, 95% CI, 0.19–0.48).
Conclusions
Type of 4-TMpc 14 days after TKI targeted therapy is associated with an image response and PFS, without regarding mutation status, in patients with advanced lung adenocarcinoma.
Introduction
The prognosis of advanced lung adenocarcinoma patients with genotype-driven mutations has improved due to targeted therapy [1–5]. Epidermal growth factor receptor (EGFR) mutation and anaplastic lymphoma kinase (ALK) rearrangement are two major oncogenic alterations that are targeted with available tyrosine kinase inhibitors (TKIs). EGFR TKIs, gefitinib [1], erlotinib [2], and afatinib [3], and ALK inhibitors, crizotinib [4] and ceritinib [5], have prolonged progression-free survival (PFS) rates in advanced lung adenocarcinoma patients with sensitive EGFR mutations and ALK rearrangement, respectively.
However, although sensitive EGFR mutations and ALK rearrangement are strong predictors of good response to TKIs targeted therapy, not all patients (about 60–70%) respond to the therapy [1–5], although a portion of patients with EGFR wild type mutations and ALK-negative have shown a response [6,7].
Morphologic imaging studies using the Response Evaluation Criteria in Solid Tumors (RECIST) remain the standard tool for evaluating treatment response [8]. However, this image evaluation strategy has several limitations, such as difficulty in obtaining the appropriate quantity of diffuse lung nodules, pleural effusions, and bone metastases [9].
Serum tumor marker (TM) concentration is a reflection of the synthesis potential of the tumor [10]. Assessment of TMs for evaluating treatment responses is clinically objective. Elevated carcinoembryonic antigen (CEA) levels have been observed in 40–80% of patients with non-small cell lung cancer (NSCLC) [11,12]. Nevertheless, a single assessment of CEA levels to evaluate lung cancer treatment response is not sensitive [9].
Thus far, not much is known about TM levels' changes and their genuine relationship to predict prognosis in adenocarcinoma patients receiving TKI targeted therapy [9,13,14]. We assessed 4-TMs, CEA, carbohydrate antigen (CA) 125 (CA125), CA19-9, and CA15-3. The selection of these tumor markers was based on reports [9–14] and our previous pilot study results (data not shown). There 4-TMs were also easy to assess in clinical practice and provided the most cost-effective coverage in patients with advanced lung adenocarcinoma. The study aimed to demonstrate whether early variations in serum 4-TMs after TKI targeted therapy were associated with treatment response and PFS in advanced lung adenocarcinoma patients.
Materials and methods
Patient selection and treatment
Patients with stage IIIB-IV lung adenosquamous cancer or adenocarcinoma taking EGFR TKIs (gefitinib, erlotinib, or afatinib) or ALK inhibitors (crizotinib or ceritinib) in different lines of therapy were enrolled in a prospective, single-center at the China Medical University Hospital from June 2012 to February 2015. We calculated the sample size needed for the kappa analysis by PASS software (version 20.0.1, NCSS, LLC. Kaysville, Utah, USA) via assuming that the proportion of good response to being around 50% in patients with advanced lung adenocarcinoma treated with targeted therapy. A sample of 161 patients achieves 90% power to detect a true Kappa value of 0.60 in a test of H0: Kappa = 0.40 vs. H1: Kappa>0.40, at a significance level of 0.05. Furthermore, considering that 10–15% of patients cannot be adapted to TM due to biochemical non-accessibility, we increased the sample size to around 190 patients. The study was approved by the China Medical University Hospital Institutional Review Board, Taichung, Taiwan (CMUH DMR 101-IRB1-087 and CMUH 104-REC1-108). Written informed consent was obtained from all patients.
EGFR mutation and ALK immunohistochemistry analysis
The tumor DNA sequences of exons 18 to 21 of EGFR were determined using direct forward and reverse sequencing via the polymerase chain reaction (PCR) product from nested PCR reactions [15]. Sensitizing mutations are defined as G719X in exon 18, in-frame deletions or insertion of exon 19, A763_Y764 insFQEA mutation, and S768I in exon 20 and L858R or L861Q in exon 21 [16–18].
ALK immunohistochemistry (IHC) was performed using the Ventana anti-ALK (D5F3) CDx assay. The staining results were evaluated using a binary scoring system: positive or negative following the manufacturer’s instructions [19].
Serum CEA, CA125, CA19-9, and CA15-3 level detection and analysis
TM levels obtained from peripheral blood samples were measured before TKI targeted therapy and after 7 and 14 days of treatment. To reduce TM levels' influence with inherent intra-individual biological variation and within-laboratory coefficients of variation (TMv) [20–22], we defined a cutoff level for each individual using pre-treatment TM levels of 2-fold over the standard upper limit. Therefore, enrollment criteria included CEA, CA125, CA19-9, and CA15-3 levels at 10.0 ng/mL, 70 units/mL, 70 units/mL, and 76 units/mL, respectively. Patients who did not show an elevation in TMs above this level were regarded as biochemically non-assessable and were excluded from further follow-up.
To evaluate changes in TM levels after TKIs targeted therapy and to account for patients having more than one evaluable TM, we created a formula “percentage of change of 4 TMs (4-TMpc)”. Assuming a distinct sub-clone released each TM within the tumor bulk (1-marker(later)/marker(previous)), a reasonable estimate of the proportion of tumor treatment for this sub-clone was made. Our Eq 1 represents the weighted average of the proportion of tumor treatment across different sub-clones.
| Eq-1 |
[note: subscript “p” = previous; subscript “l” = later; *2-fold over the standard upper limit was regarded as evaluable.]
For example, if on Day 0, a patient had the following serum TM values (CEA 2 ng/mL, CA125 225 units/mL, CA19-9 5 units/mL, and CA15-3 197 units/mL), then this patient had only 2 evaluable TMs (CA125 and CA15-3). If on Day 7, the TM levels were CA125 175 units/mL and CA15-3 132 units/mL, then the 4-TMpc over the 0th-7th day was 27.6%, as shown in Eq 2.
| Eq-2 |
According to variations in 4-TMpc on days 0, 7, and 14, we divided patients into four groups. Type 1. Ascending: patients who sustained an increase in 4-TMpc. Type 2. Descending-ascending: patients who showed a decreasing trend on the 7th day, and then showed an increasing trend on the 14th day. Type 3. Ascending-descending: patients who showed an increasing trend on the 7th day and then showed a decreasing trend at subsequent time points. Type 4. Descending: patients who showed a persistently decreased 4-TMpc. For minimizing TMv interference, the following definitions were created [20–22]: when 4-TMpc was <5% over the 7th-14th day, and it was defined as “type uncertain.” Confirmed decreases in 4-TMpc over the 7th- 14th day (types 3/4) were consistent with tumor response. Similarly, increases in 4-TMpc over the 7th-14th day (types 1/2) were regarded as tumor progression.
Imaging-based response
Tumor response was assessed on chest radiographs (CXR) and computed tomography (CT) scans, using the RECIST version 1.1 in an independent radiologic review by assessors who did not know the results of 4-TMpc studies and confirmed at least two scans obtained 28 days apart. Long-term follow-up was performed until July 31, 2015.
Therapeutic efficacy was classified as partial response (PR), stable disease (SD) with tumor reduction <30% (SD-30), SD with the increase in tumor size <20% (SD+20), or progressive disease (PD). Patients who died due to cancer between these CT/CXR procedures were classified as having PD. While analyzing the correlation between 4-TMpc after 14 days TKI targeted therapy and RECIST assessed response, and we combined categories of PR and SD-30 into the “good response group.” In contrast, the “poor response group” included cases with PD and SD+20. This classification's rationales were that: (1) It was the straight forward approach to classify responders vs. non-responders. (2) It was also difficult for PR, SD, or PD to correlate with the ascending or descending of the 4-TMpc from a statistical point of view. The endpoint was PFS. PFS was assessed from the date of the beginning of TKI targeted therapy to the date of PD or death due to cancer. If a patient was lost to follow-up or had no event, time to progression was censored as the date of the last contact date.
Statistical analysis
To obtain a descriptive analysis, we resumed each continuous variable as median and 25–75, and categorical variables as proportion. We performed receiver operating characteristic (ROC) curve analysis for 4-TMs to predict TKI targeted therapy's response. The agreement between the 4-TMs and the image-based morphologic response was evaluated using the kappa statistic. PFS was analyzed according to the Kaplan-Meier method and was compared with the log-rank test. Cox proportional hazards model was used to evaluate independent predictive factors associated with PFS. Data were analyzed using SPSS-17 (IBM SPSS Statistics. Inc. Chicago, IL, USA). For all analyses, two-sided P<0.05 was taken as statistically significant.
Results
In all, 195 patients with a diagnosis of stage IIIb-IV adenocarcinoma (one with adenosquamous carcinoma) were screened for 4-TM levels before the start of TKI targeted therapy, and 11 were excluded: 1 because the patient had severe interstitial lung disease after taking erlotinib and ten because the standard protocol was not followed. Seven patients were recruited more than once because patients accepted re-challenge TKI targeted therapy. Therefore, 184 patients and 191 patient-times were enrolled in this study, including 29 accepted the diagnostic procedure of computed tomography-guided core needle biopsy, 68 accepted transbronchial biopsy, 20 accepted ultrasound-guided biopsy, 27 accepted thoracentesis or pericardiocentesis, and 40 accepted operative procedures. Detailed baseline characteristics are summarized in Table 1. Activating mutations were documented in 73% cases, and 3% had an unknown mutation status because no sufficient pathological material was available. Further, 98% of patient-times received EGFR TKIs, and 2% received ALK-inhibitors
Table 1. Baseline characteristics of patients.
| Variables | All patients | CEA/CA125/CA153/CA199 Elevation | CEA/CA125/CA153/CA199 No elevation |
|---|---|---|---|
| All patient times* | 191 | 170 | 21 |
| All patients | 184 | 163 | 21 |
| Age, yr | 62.5 (55.0–73.0) | 62.0 (55.0–72.0) | 68.0 (54.5–78.5) |
| Sex | |||
| Male | 76 (41.3) | 68 (41.7) | 8 (38.1) |
| Female | 108 (58.7) | 95 (58.3) | 13 (61.9) |
| Smoking | |||
| Never | 131 (71.2) | 118 (72.4) | 13 (61.9) |
| Former/current | 53 (28.8) | 45 (27.6) | 8 (38.1) |
| Stage | |||
| IIIb | 5 (2.7) | 5 (3.1) | 0 |
| Iva | 65 (35.3) | 54 (33.1) | 11 (52.4) |
| IVb | 114 (62.0) | 104 (63.8) | 10 (47.6) |
| Performance status* | |||
| 0–1 | 108 (56.6) | 95 (55.8) | 13 (61.9) |
| 2 | 26 (13.6) | 21 (12.4) | 5 (23.8) |
| 3–4 | 57 (29.8) | 54 (31.8) | 3 (14.3) |
| Mutation | |||
| EGFR sensitizing mutation | |||
| Exon 19 deletion | 65 (35.3) | 54 (33.1) | 11 (52.4) |
| L858R | 55 (29.9) | 47 (28.8) | 8 (38.1) |
| Others+ | 10 (5.4) | 10 (6.1) | 0 |
| ALK rearrangement | 5 (2.7) | 5 (3.1) | 0 |
| EGFR and ALK-negative | 44 (23.9) | 42 (25.8) | 2 (9.5) |
| Unknown | 5 (2.7) | 5 (3.1) | 0 |
| Targeted therapy* | |||
| Gefitinib | 80 (41.9) | 70 (41.2) | 10 (77.7) |
| Erlotinib | 88 (46.1) | 78 (45.9) | 10 (47.6) |
| Afatinib | 19 (9.9) | 18 (10.6) | 1 (4.8) |
| Crizotinib | 2 (1.0) | 2 (1.2) | 0 |
| Ceritinib | 2 (1.0) | 2 (1.2) | 0 |
| Treatment-line* | |||
| First-line | 167 (87.4) | 150 (88.2) | 17 (81.0) |
| Second-line | 23 (12.0) | 19 (11.2) | 4 (19.0) |
| Third-line | 1 (0.5) | 1 (0.6) | 0 |
*All patient times
+One patient had L858R + T790M. One patient had exon 19 deletion + T790M. One patient had exon 19 deletion + L858R. One patient had exon 19 deletion + S768I. One patient had S768I. Three patients had G719X. Two patients had L861Q.
EGFR epidermal growth factor receptor, ALK anaplastic lymphoma kinase
CEA carcinoembryonic antigen, CA125 carbohydrate antigen 125, CA19-9 carbohydrate antigen 19–9, CA15-3 carbohydrate antigen 15–3
Of the 184 enrolled patients, baseline serum CEA levels, CA125, CA19-9, and CA15-3 were 2-fold over the standard upper limit in 71%, 62%, 27%, and 23% patients, respectively. In all, 89% of patients with at least one of the pre-treatment evaluable TM were further analyzed. Of these, 7% had all 4 TM levels elevated, 26% had 3, 33% had 2, and 34% had 1 TM elevated. The summarized data is shown in Table 2.
Table 2. a. Characteristics of carcinoembryonic antigen (CEA), cancer antigen (CA) 125, CA19-9, and CA15-3 in 184 patients with advanced lung adenocarcinoma. b. Items of 163 patients with advanced lung adenocarcinoma with evaluable tumor markers.
| a | ||
| Variables | N (%) | Median (25–75) |
| CEA ng/mL | 130 (70.7) | 121.2 (32.1–345.6) |
| CA125 units/mL | 114 (62.0) | 167.1 (112.8–360.2) |
| CA19-9 units/mL | 50 (27.2) | 334.6 (159.5–1423.5) |
| CA15-3 units/mL | 42 (22.8) | 155.3 (90.6–253.2) |
| b | ||
| Variables | N (%) | |
| One tumor marker | CEA | 38 (23.3) |
| CA125 | 17 (10.4) | |
| CA19-9 | 0 | |
| CA15-3 | 1 (0.6) | |
| Two tumor markers | CEA + CA125 | 32 (19.6) |
| CEA + CA15-3 | 5 (3.1) | |
| CEA + CA19-9 | 3 (1.8) | |
| CA125 + CA15-3 | 5 (3.1) | |
| CA125 + CA19-9 | 7 (4.3) | |
| CA15-3 + CA19-9 | 1 (0.6) | |
| Three tumor markers | CEA + CA125 + CA15-3 | 15 (9.2) |
| CEA + CA125 + CA19-9 | 24 (14.7) | |
| CEA + CA15-3 + CA19-9 | 1 (0.6) | |
| CA125 + CA15-3 + CA19-9 | 2 (1.2) | |
| Four tumor markers | CEA + CA125 + CA15-3 + CA19-9 | 12 (7.4) |
CEA carcinoembryonic antigen, CA125 carbohydrate antigen 125, CA19-9 carbohydrate antigen 19–9, CA15-3 carbohydrate antigen 15–3
Of 170 patient-times after TKI targeted therapy with evaluable TMs, PR, SD-30, SD+20, and PD were 55%, 23%, 1% and 21%, respectively. We divided the patients into 4 groups by 14 days 4-TMpc: type 1, 22%; type 2, 4%; type 3, 25%; and type 4, 42%. Further, 8% of patients were classified as “type uncertain.” Types 1/2 were observed in 32 of 44 patients (73%) who showed a poor response. On the contrary, types 3/4 were observed in 111 of 113 patients (98%), who showed a good response (Fig 1). The presence of types 3/4 could accurately predict “good response” by using ROC curve analysis, with an area under the curve (AUC) 0.83 (95% confidence interval [CI] 0.73 to 0.93). The Kappa value between 157 cases with the type of 4-TMpc and measurable radiographic lesions was 0.762 (P<0.001) (Fig 1).
Fig 1. Relevance between image-based response and the type of 4 tumor marker levels in 157 patients with advanced lung adenocarcinoma.
RECIST Response Evaluation Criteria in Solid Tumors, PR partial response, SD-30 stable disease with tumor reduction <30%, SD+20 stable disease with tumor increasing <20%, PD progressive disease.
However, 24% of patients had baseline CEA< 10 ng/mL. This group of patients could not be biochemically assessed using CEA levels alone. The AUC was only 0.51 (95% CI, 038–0.63) for predicting TKI targeted therapy response with the type of CEA. The Kappa value in 117 cases was 0.449 (Fig 2).
Fig 2. Relevance between image-based response and type of CEA percentage of change over the 0th-14th day in 117 patients with advanced lung adenocarcinoma.
CEA carcinoembryonic antigen, RECIST Response Evaluation Criteria in Solid Tumors, PR partial response, SD-30 stable disease with tumor reduction <30%, SD+20 stable disease with tumor increasing <20%, PD progressive disease.
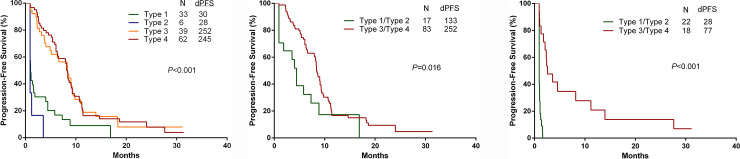
One hundred and forty patients in whom first-time TKI targeted therapy with the type of 4-TMpc was used, were further analyzed using Kaplan-Meier curves for PFS. As shown in Fig 3A, the median PFS had no significant difference between types 1 and 2 (30 and 28 days), nearly the same as types 3 and 4 (252 and 245 days). However, PFS was significantly longer in types 3/4 than in types 1/2 (P<0.001). Analysis for subgroups stratified according to mutation status found that PFS was still longer in patients with types 3/4 than in patients with types 1/2 (activated mutation group, P = 0.016 (Fig 3B); EGFR and ALK-negative/unknown group, P<0.001 (Fig 3C)).
Fig 3.
Kaplan-Meier curves for progression-free survival (A) in the entire cohort, (B) in activated mutation, (C) in EGFR and ALK-negative/unknown groups, respectively. EGFR epidermal growth factor receptor, ALK anaplastic lymphoma kinase, d-PFS progression-free days.
In the univariate analysis using the Cox hazards model, types 3/4 and sensitive mutation were the only two PFS predictive factors (P<0.001). Multivariate Cox hazard model analyses shows the same result (types 3/4 versus types 1/2, P<0.001, hazard ratio (HR) 0.30, 95% CI, 0.19–0.47; sensitive mutation versus EGFR and ALK-negative/unknown, P< 0.001, HR 0.30, 95% CI, 0.19–0.48) (Table 3).
Table 3. Univariate and multivariate prediction of progression-free survival.
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | |
| Age (years) | ||||||
| <65 vs. ≥65 | 1.25 | 0.87–1.80 | 0.24 | 1.56 | 1.05–2.34 | .030 |
| Gender | ||||||
| Female vs. Male | 0.85 | 0.59–1.23 | 0.39 | 1.17 | 0.73–1.88 | .516 |
| Smoking habit | ||||||
| Never vs. Current/former | 0.70 | 0.47–1.06 | 0.09 | 0.82 | 0.48–1.40 | .469 |
| Performance status | ||||||
| 0–1 vs. ≥2 | 0.89 | 0.61–1.30 | 0.55 | 1.03 | 0.70–1.52 | .881 |
| Stage | ||||||
| IIIb/IVa vs. IVb | 0.79 | 0.54–1.16 | 0.22 | 0.89 | 0.60–1.33 | .572 |
| Mutation | ||||||
| MUT vs. WT/UNK | 0.34 | 0.23–0.52 | <0.001 | 0.30 | 0.19–0.48 | <0.001 |
| CEA/CA125/19-9/15-3 | ||||||
| Type 3/4 vs. Type 1/2 | 0.29 | 0.19–0.44 | <0.001 | 0.30 | 0.19–0.47 | <0.001 |
CI confidence interval, HR hazard ratio, MUT mutated patients, WT/UNK wild-type/unknown patients, CEA carcinoembryonic antigen, CA125 carbohydrate antigen 125, CA19-9 carbohydrate antigen 19–9, CA15-3 carbohydrate antigen 15–3
Discussion
This prospective study was the first to provide CEA, CA125, CA19-9, and CA15-3 permutation and combination in patients with advanced lung adenocarcinoma. We compared the AUC of CEA and 4-TMs to predict clinical responses of TKI targeted therapy. Our study demonstrated that 14 days of 4-TMpc type could be an early predictor of TKI targeted therapy efficacy. The descending type of 4-TMpc over the 7th- 14th days had longer PFS in mutated or non-mutated adenocarcinoma patients.
Although squamous cell lung cancer and adenocarcinoma are a subset of NSCLC, they have different driver mutations and treatment [23]. CEA is more frequently reported in patients with adenocarcinoma than squamous lung cancer [11,12]. Therefore, we focused on lung adenocarcinoma. Furthermore, not all lung adenocarcinoma patients had elevated CEA levels [9,13,14]. Advanced lung adenocarcinoma has other potentially valuable TMs in addition to CEA. In our series, CA125, CA 19–9, and CA15-3 levels reached evaluable criteria in 62%, 27%, and 23% patients, respectively. While combined with 4-TMs, only 11% of patients had 4-TMs below the evaluable levels (Table 2).
There is insufficient evidence to support a conclusion concerning the standardized combination of TMs to evaluate tumor status. Different intra-tumor sub-clones may release different TMs. We presumed “one TM, one evaluable clone” and combined TMs as “4-TMpc”.
Zhang et al. indicated that the descending type of CEA within one month correlated with PR and SD of EGFR-TKI in patients with lung adenocarcinoma [13]. However, the CEA type can be affected by TMv and can influence the clinician to make a wrong decision [20,22]. In our 170 patient-times with evaluable TM cohort, “type uncertain” was classified when 4-TMpc over the 7th- 14th day was <5%. We divided the others into four groups. Among treatment, patients showing effectiveness may have an ascending 4-TMpc pattern before the descending pattern (type 3). This transient increase in TM levels is known as surges [13,24]. Otherwise, type 2 with fluctuation in 4-TMpc and then an ascending pattern the over 7th-14th day is not logical if TKI targeted therapy is considered effective (Fig 1).
We evaluated the type of 4-TMpc within 14 days of TKI targeted therapy and the relationship with imaging-based response and PFS. The ROC curve analysis showed that using 4-TMs for predicting the efficacy of TKI targeted therapy response had a higher AUC (0.83) than that using CEA (0.51). The Kappa value for the agreement analysis between 157 cases with the type of 4-TMpc and radiographic results was "good" (0.762) (Fig 1). However, using CEA levels, 24% of patients were biochemically non-assessable in this 170 patient-time series. The kappa value was 0.449 only (Fig 2). These findings showed that using 4-TMs to predict TKI targeted therapy response was more accurate than using CEA alone.
The results of sensitizing mutations cannot guarantee a clinical response to TKI targeted therapy [6,7,25]. It is essential to develop a new strategy for early prediction of the effect of TKI targeted therapy. In our series, patients with types 3/4, 4-TMpc had a longer PFS than those with types 1/2. Regarding the ascending (type 3) or descending pattern (type 4) over the 0th-7th day, patients' outcomes were similar. On the contrary, similar PFS was observed in patients with types 1 and 2 (Fig 3A). Regarding activated mutation (Fig 3B) and EGFR and ALK-negative/unknown group (Fig 3C), PFS was also significantly longer in patients with types 3/4 than in patients with types 1/2. Similarly, in multivariate models, our results demonstrate that 4-TMpc and mutation status continues to be the strongest predictors of PFS (Table 3).
Our study's strengths include its prospective design, and a large number of patients included compared to previous TM studies in NSCLC patients under TKI targeted therapy [9,13,14]. However, certain drawbacks should be considered. Firstly, it is a single-center study. Secondly, there is insufficient evidence to define the best cutoff level for minimizing the interference of TMv. Thirdly, we did not extend our study after Feb 2015 because the enrolled cases and the follow-up time are sufficient to reflect the study results in which we did find high-level agreement [kappa = 0.762] by planned accrual [around 190 cases]. Furthermore, the investigated scenario (gefitinib, erlotinib, afatinib, crizotinib or ceritinib for lung adenocarcinoma patients) was still valid in Taiwan in 2020, although whether our finding was applicable for some new inhibitors (such as osimertinib [26] or alectinib [27]) deserved to be studied.
Conclusion
In conclusion, image evaluation strategy with RECIST for patients with lung cancer has difficulty in obtaining the appropriate quantity of diffuse lung nodules, pleural effusions, and bone metastases. The type of 4-TMpc after 14 days TKI targeted therapy is associated with image response and PFS without accounting for mutation status in advanced lung adenocarcinoma patients. Our study results could help early therapeutic decision making by identifying patients who may benefit from gefitinib, erlotinib, afatinib, crizotinib, or ceritinib 14 days after TKI targeted therapy.
Supporting information
(XLS)
Acknowledgments
The authors wish to thank Chia-Ing Li for her technical help with statistical analysis.
Abbreviations and acronyms
- EGFR
Epidermal growth factor receptor
- ALK
anaplastic lymphoma kinase
- TKI
tyrosine kinase inhibitor
- PFS
progression-free survival
- RECIST
Response Evaluation Criteria in Solid Tumors
- TM
tumor marker
- CEA
carcinoembryonic antigen
- NSCLC
non-small cell lung cancer
- CA125
carbohydrate antigen125
- CA19-9
carbohydrate antigen 19–9
- CA15-3
carbohydrate antigen153
- PCR
polymerase chain reaction
- IHC
Immunohistochemistry
- TMv
tumor markers with inherent intra-individual biological variation and within-laboratory coefficients of variation
- 4-TMpc
percent of change of 4 tumor markers
- TMp
previous TM
- TMl
later TM
- CXR
chest radiograph
- CT
computed tomography
- PR
partial response
- SD
stable disease
- SD-30
stable disease with tumor reduction less than 30%
- SD+20
stable disease with tumor increasing less than 20%
- PD
progressive disease
- ROC
receiver operating characteristic
- AUC
area under the curve
- CI
confidence interval
- HR
hazard ratio
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, et al. (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361: 947–957. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, et al. (2012) Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 13: 239–246. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 3.Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, et al. (2013) Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 31: 3327–3334. 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, et al. (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703. 10.1056/NEJMoa1006448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, et al. (2014) Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 370: 1189–1197. 10.1056/NEJMoa1311107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John T, Liu G, Tsao MS (2009) Overview of molecular testing in non-small-cell lung cancer: mutational analysis, gene copy number, protein expression and other biomarkers of EGFR for the prediction of response to tyrosine kinase inhibitors. Oncogene 28 Suppl 1: S14–23. 10.1038/onc.2009.197 [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, et al. (2014) Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 371: 1963–1971. 10.1056/NEJMoa1406766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, et al. (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45: 228–247. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 9.Chiu C-H, Shih Y-N, Tsai C-M, Liou J-L, Chen Y-M, et al. (2007) Serum tumor markers as predictors for survival in advanced non-small cell lung cancer patients treated with gefitinib. Lung Cancer 57: 213–221. 10.1016/j.lungcan.2007.02.016 [DOI] [PubMed] [Google Scholar]
- 10.Fritsche HA (1993) Serum tumor markers for patient monitoring: a case-oriented approach illustrated with carcinoembryonic antigen. Clin Chem 39: 2431–2434. [PubMed] [Google Scholar]
- 11.Ferrigno D, Buccheri G (1995) Clinical applications of serum markers for lung cancer. Respir Med 89: 587–597. 10.1016/0954-6111(95)90225-2 [DOI] [PubMed] [Google Scholar]
- 12.Grunnet M, Sorensen J (2012) Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer 76: 138–143. 10.1016/j.lungcan.2011.11.012 [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Jin B, Shao M, Dong Y, Lou Y, et al. (2014) Monitoring of carcinoembryonic antigen levels is predictive of EGFR mutations and efficacy of EGFR-TKI in patients with lung adenocarcinoma. Tumor Biology 35: 4921–4928. 10.1007/s13277-014-1646-1 [DOI] [PubMed] [Google Scholar]
- 14.Facchinetti F, Aldigeri R, Aloe R, Bortesi B, Ardizzoni A, et al. (2015) CEA serum level as early predictive marker of outcome during EGFR-TKI therapy in advanced NSCLC patients. Tumour Biol 36: 5943–5951. 10.1007/s13277-015-3269-6 [DOI] [PubMed] [Google Scholar]
- 15.Huang SF, Liu HP, Li LH, Ku YC, Fu YN, et al. (2004) High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 10: 8195–8203. 10.1158/1078-0432.CCR-04-1245 [DOI] [PubMed] [Google Scholar]
- 16.Joshi M, Rizvi SM, Belani CP (2015) Afatinib for the treatment of metastatic non-small cell lung cancer. Cancer Manag Res 7: 75–82. 10.2147/CMAR.S51808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu CH, Yang CT, Shih JY, Huang MS, Su WC, et al. (2015) Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas with G719X/L861Q/S768I Mutations. J Thorac Oncol 10: 793–799. 10.1097/JTO.0000000000000504 [DOI] [PubMed] [Google Scholar]
- 18.Shen Y-C, Tseng G-C, Tu C-Y, Chen W-C, Liao W-C, et al. (2017) Comparing the effects of afatinib with gefitinib or Erlotinib in patients with advanced-stage lung adenocarcinoma harboring non-classical epidermal growth factor receptor mutations. Lung Cancer 110: 56–62. 10.1016/j.lungcan.2017.06.007 [DOI] [PubMed] [Google Scholar]
- 19.Marchetti A, Di Lorito A, Pace MV, Iezzi M, Felicioni L, et al. (2016) ALK protein analysis by IHC staining after recent regulatory changes: a comparison of two widely used approaches, revision of the literature, and a new testing algorithm. Journal of Thoracic Oncology 11: 487–495. 10.1016/j.jtho.2015.12.111 [DOI] [PubMed] [Google Scholar]
- 20.Tuxen MK, Soletormos G, Petersen PH, Schioler V, Dombernowsky P (1999) Assessment of biological variation and analytical imprecision of CA 125, CEA, and TPA in relation to monitoring of ovarian cancer. Gynecol Oncol 74: 12–22. 10.1006/gyno.1999.5414 [DOI] [PubMed] [Google Scholar]
- 21.Tso E, Elson P, Vanlente F, Markman M (2006) The "real-life" variability of CA-125 in ovarian cancer patients. Gynecol Oncol 103: 141–144. 10.1016/j.ygyno.2006.02.010 [DOI] [PubMed] [Google Scholar]
- 22.Sturgeon CM (2001) Tumor markers in the laboratory: closing the guideline-practice gap. Clin Biochem 34: 353–359. 10.1016/s0009-9120(01)00199-0 [DOI] [PubMed] [Google Scholar]
- 23.Li T, Kung HJ, Mack PC, Gandara DR (2013) Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 31: 1039–1049. 10.1200/JCO.2012.45.3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HS, Park YH, Park MJ, Chang MH, Jun HJ, et al. (2009) Clinical significance of a serum CA15-3 surge and the usefulness of CA15-3 kinetics in monitoring chemotherapy response in patients with metastatic breast cancer. Breast Cancer Res Treat 118: 89–97. 10.1007/s10549-009-0377-2 [DOI] [PubMed] [Google Scholar]
- 25.Cappuzzo F, Ciuleanu T, Stelmakh L, Cicenas S, Szczésna A, et al. (2010) Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. The lancet oncology 11: 521–529. 10.1016/S1470-2045(10)70112-1 [DOI] [PubMed] [Google Scholar]
- 26.Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, et al. (2018) Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. New England Journal of Medicine 378: 113–125. 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 27.Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, et al. (2017) Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. New England Journal of Medicine 377: 829–838. 10.1056/NEJMoa1704795 [DOI] [PubMed] [Google Scholar]