Abstract
Background
Drugs that work based on the mechanism of RNA interference have shown strong potential in cancer gene therapy. Although significant progress has been made in small interfering RNA (siRNA) design and manufacturing, ideal delivery system remains a limitation for the development of siRNA-based drugs. Particularly, it is necessary to focus on parameters including delivery efficiency, stability, and safety when developing siRNA formulations for cancer therapy.
Methods
In this work, a novel degradable siRNA delivery system cRGD-R9-PEG-PEI-Cholesterol (rrPPC) was synthesized based on low molecular weight polyethyleneimine (PEI). Functional groups including cholesterol, cell penetrating peptides (CPPs), and poly(ethylene oxide) were introduced to PEI backbone to attain enhanced transfection efficiency and biocompatibility.
Results
The synthesized rrPPC was dispersed as nanoparticles in water with an average size of 195 nm and 41.9 mV in potential. rrPPC nanoparticles could efficiently deliver siRNA into C26 clone cancer cells and trigger caveolae-mediated pathway during transmembrane transportation. By loading the signal transducer and activator of transcription 3 (STAT3) targeting siRNA, rrPPC/STAT3 siRNA (rrPPC/siSTAT3) complex demonstrated strong anti-cancer effects in multiple colon cancer models following local delivery. In addition, intravenous (IV) injection of rrPPC/siSTAT3 complex efficiently suppressed lung metastasis tumor progression with ideal in vivo safety.
Conclusion
Our results provide evidence that rrPPC nanoparticles constitute a potential candidate vector for siRNA-based colon cancer gene therapy.
Keywords: RNA interfering, nanoparticle, STAT3, colon cancer
Introduction
Colon cancer is a common malignant tumor in the gastrointestinal tract, and its morbidity and mortality rates rank third and second among global cancer morbidity and mortality data, respectively.1 Currently, the treatment of colon cancer is mainly encompasses conventional methods including chemotherapy, radiotherapy and surgery and the recurrence rate and adverse reaction rate are both still high.2 Identifying novel and effective treatments has become a focus in the field of colon cancer treatment.
RNA interference (RNAi) has been widely applied as a posttranscriptional gene regulation technology for a variety of pathological conditions including viral infections, cancer, genetic disorders and autoimmune disorders.3 As such, great progress has been made in the development of RNAi drugs in past decades. For example, a lipid nano-formulated siRNA modality from Alnylam Pharmaceuticals (Cambridge, MA, USA), ONPATTRO™ (patisiran) was recently approved by the FDA for the first time commercially therapeutic siRNA drug.4,5 Meanwhile, at least 30 siRNA drug candidates are undergoing different phases of clinical trials.6,7 Among them, cancer therapy with siRNA nano-formulations is currently being actively tested. Suppression of the transcription activities of several important gene targets concerning proliferation, migration, and immune escape has already been proven to be effective in cancer gene therapy.8–13 However, despite the outstanding progress made in siRNA design, chemical modification, and manufacturing, ideal delivery system remains a limitation for the development of siRNA-based drugs.14,15 “Naked” siRNA is highly unstable and rapidly degraded by nucleases in vivo.16,17 In addition, without the help of an appropriate vector, it is difficult to accomplish the tissue-targeted transportation as well as the cellular uptake tasks of therapeutic siRNA.16 Thus, the development of a safe and effective siRNA delivery system is required.
As a typical cationic agent, polyethyleneimine (PEI) possesses great gene condensation potential owing to its rich amino groups. High molecular weight PEI (PEI25K) has long attracted significant attention due to its strong capacity for gene delivery against DNA and RNA molecules. However, the widespread application of PEI25K is attenuated by its serious cytotoxicity on cells. Conversely, although low molecular weight PEI (LMW PEI, such as PEI2K) with much fewer amino groups has exhibited acceptable toxicity, lacking an adequate gene condensation ability makes it less attractive than PEI25K.18 As such, researchers have tried to crosslink LMW PEI onto backbone materials to simulate high molecular PEIs and, hence, enhance its gene delivery capacity.19–22 However, in these studies, LMW PEI was only used as a functional group rather than a backbone core, which underestimated its value. In fact, owning to its safety, low molecular weight property, and amino group-functionalized structure, LMW PEI is an ideal backbone material in developing vectors for small nucleic acids including siRNA.
Unlike genetic diseases, cancer is usually multi-organ metabolized in the body. In addition, the gene uptake by cancer cells is comparatively more difficult. Therefore, significant attention should be paid to the parameters of delivery efficiency, stability, and safety when developing siRNA formulations for cancer therapy. When adopting LMW PEI as a backbone, these requirements can be met by introducing different functional moieties to it through amino group “harbors”. With regard to the delivery efficiency, cell penetrating peptides (CPPs) are a group of natural or artificial short peptides that able to facilitate cell intake.23,24 These small peptides can be conjugated onto nanoparticles and chemical compounds, thus providing additional nonspecific delivery capacity.25 In particular, the CPPs of the RGD family have been intensively studied and their integrin receptors have already been indicated.26 On the other hand, three main internalization pathways including the caveolae-mediated pathway, clathrin-mediated pathway, and micropinocytosis are reported to be involved in the uptake process of nano-sized vectors.27,28 These pathways can be triggered by certain biological structures and thus can promote substantial transmembrane delivery. As an essential structural component of animal tissues, cholesterol plays an important role in membrane stabilization. When being applied in drug delivery, cholesterol not only enhances tissue biocompatibility and in vivo stability but may also triggers the caveolae-mediated pathway, which can further reinforce transmembrane delivery efficiency.29,30 In addition to endogenous molecules, artificial products such as poly(ethylene oxide) (PEG) could also provide extra biocompatibility and serum stability for delivery systems. Owning to its hair-like linear structure, PEGylation may alter the vehicle’s pharmacokinetic properties, providing long circulation property.31,32 Furthermore, PEG’s flexible dynamic structure can avoid potential steric hindrance caused by other functional modifications, thus helping to better retain their performance. These safety-enhancing functions is particularly necessary for intravenous administration.
In this study, we intended to develop a novel siRNA delivery system based on LMW PEI backbone, where multifunctional modifications including CPPs, cholesterol, and PEG were going to be introduced through amino group “harbors” to form a nano-sized rrPPC non-viral siRNA vector. We assumed that the fusing of these function moieties would result in an efficient delivery system with high degrees of stability, safety and siRNA protection ability. To further evaluate its capacity, a siRNA targeting stat3 gene was selected to complex with the rrPPC nano-vector. As a transcription factor, STAT3 can promote oncogenesis by being constitutively active through various pathways.33,34 Although the silencing of stat3 has been proved to be an effective strategy for cancer treatment, related drug development remains obstructed by delivery technology.35 In this work, the capacity of the rrPPC vector for siRNA delivery and protection ability of rrPPC vector will be characterized in detail. Meanwhile, the anti-cancer effects of rrPPC/siSTAT3 complex on different C26 colon cancer models will be evaluated.
Methods
Synthesizing of rrPPC
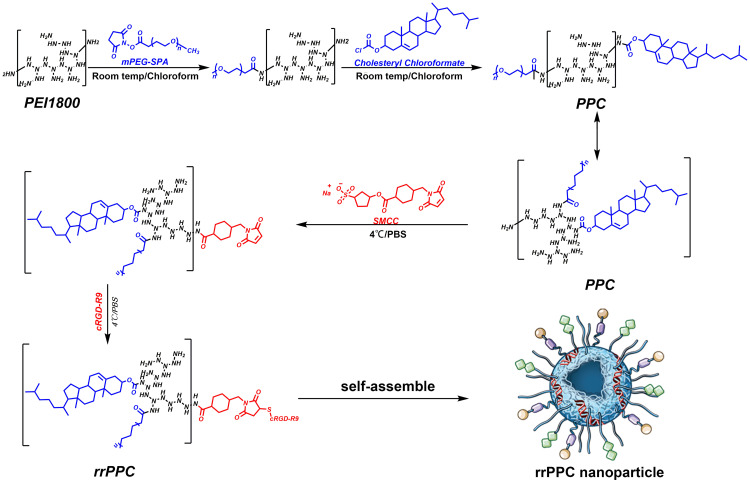
Branched PEI1.8K was dissolved in chloroform and stirred for 20 minutes at room temperature. Cholesteryl chloroformate and PEG (Mw 550Da) was dissolved in chloroform and transferred to an addition funnel. The mixture of Cholesteryl chloroformate and PEG was then slowly dropped to the PEI solution under stirring. After rotary evaporation, the remaining material was dissolved in ethyl acetate. The product was then precipitated by n-Hexane and rehydrated. The aqueous solution was then lyophilized to obtain the precursor product (PEG-PEI-Cholesterol, PPC). To synthesis rrPPC, PPC was then react with cRGD-R9 peptide with crosslinker SMCC (Thermo, USA). The mixture of PPC and SMCC solution were continually reacted for 24 hours before further to react with cRGD-R9. The final product was dialyzed and collected as rrPPC. Proton nuclear magnetic resonance (1H NMR) spectra of rrPPC was recorded on a Varian Mercury 400 NMR spectrometer, 1H NMR data is reported in parts per million (ppm) downfield from tetramethylsilane as an internal standard. Fourier transform infrared spectroscopy (FT-IR) analysis was performed using a Spectrum Two FT-IR spectrometer.
Characterization of rrPPC Nanoparticles
The particle distribution and electric charge properties of rrPPC were studied by Malvern Sizer (Malvern, UK). The morphology of rrPPC cationic nanoparticles were observed in detail with a transmission electron microscope (TEM) (H-6009IV, Hitachi, Japan), and stored in 4°C for further use. The agarose gel retarding assay was used to study the loading ability of rrPPC nanoparticles to siRNA. Briefly, siRNA/rrPPC mixtures with different complexing ratios (1:0, 1:2, 1:4, 1:6, 1:8, 1:10) were electrophoresed on a 1% (w/v) agarose gel. One μg of siRNA (1 mg/mL) was mixed with different amounts of rrPPC (1 mg/mL). Electrophoresis was performed under 120V for 15 mins. The gel was then stained with ethidium bromide and illuminated by Bio-Rad ChemiDox XRS. To assess the protection effect of rrPPC against RNase degradation, RNaseA (1 μg, Sigma, USA) was incubated with naked scramble siRNA (0.5 μg) or rrPPC/siRNA complex at 37 °C. At the indicated times, sodium dodecyl sulfate (final concentration = 0.5%) was added and the mixture was denatured for 10 min. Released siRNA was analyzed by electrophoresis on an agarose gel (1%). siRNA band was visualized under UV light.
MTT Assay
The cell cytotoxicity of rrPPC was studied by MTT assay. 293t cells (5×103cells/well) were seeded into 96-well plates and incubated with various concentrations of rrPPC or PEI25K (18.75, 25, 37.5, 50, 75, 100, 150, and 200 μg/mL). 72 hours post transfection, 20 μL MTT solution was added to each well and incubated for 4 hours. 200 μL DMSO was then added to dissolve formed formazan crystals. The density of living cells was calculated by measuring the solution at 570 nm. The surviving rate of untreated cells was considered as 100%.
In vitro Transfection
C26 mouse colon carcinoma cells (catalog No. CRL-2638, purchased from ATCC, USA. 1×104cells/well) were pre-seeded into a 24-well plate and incubated for 24 hours. FAM-labeled scramble siRNA was used as a reporter gene. The rrPPC/FAM siRNA complex (FAM siRNA=1 μg/well) was added to each well and incubated for 4 hours. PEI25K, PEI1.8K, and PPC were used as controls. The mass ratio of rrPPC/PPC/PEI1.8K to siRNA was 10:1 while PEI25K to siRNA was 5:1. 24 hours later, fluorescence in each well was observed using a microscope and accordant transfection efficiency was measured by flow cytometry.
Internalization Study of rrPPC/siRNA Complex
To study the transmembrane internalization mechanism of rrPPC/siRNA complex, C26 cells in 24-well plate were pretreated with different inhibitors of known internalization pathways for 30 mins including amiloride (3 mM), genistein (150 μM), filipin ш (4 μg/mL). 24 hours post-transfection (FAM siRNA=1 μg/well), the transfection efficiency in each group was determined by flow cytometry. For fluorescent staining, cells grown on a cover glass were stained with DAPI and Alexa Fluor 594 for the labeling of nucleus and plasma membrane. The accordant fluorescence was observed and photographed by confocal microscopy (Olympus) and fluorescent microscope (Olympus, Japan).
Real-Time PCR
The silencing effect against stat3 gene in C26 cells was studied by real-time quantitative RT-PCR. Briefly, C26 cells seeded into 24-well plate were treated with rrPPC/siRNA complex (siRNA=1 μg/well, rrPPC=10 μg/well) for 4 hours. 24 hours later, total RNAs from transfected C26 cells were isolated using TRIzol Reagent and cDNAs were synthesized with a SuperScript II reverse transcriptase kit (Takara Bio, USA). Real-time quantitative PCR was performed with a SYBR Green ER quantitative PCR SuperMix Universal kit (Thermo Fisher, USA). Reactions were carried out through the standard cycle program on an AB7500 real-time PCR system. The sequence of siRNA targeting mouse stat3 is UUA GCC CAU GUG AUC UGA CAC CC UGA A (sense). The sequence of scramble siRNA is UUC UCC GAA CGU GUC ACG UTT (sense). The PCR primers to detect stat3 mRNA level (forward: TTC TCG TCC ACC ACC AAG, reverse: GAT ATT GTC TAG CCA GAC CC) and GAPDH (forward: 5ʹ-ATG GGG AAG GTG AAG GTC G-3ʹ, reverse: 5ʹ-TAA AAG CAG CCC TGG TGA CC-3ʹ) were synthesized and purified by TSINGKE Biological Technology (Chengdu, China).
Anti-Proliferation Assay
C26 cells were seeded into a 96-well plate at the density of 1.0×104 cells per well. Cells were then treated with rrPPC/siSTAT3 and rrPPC/scramble siRNA (rrPPC/siScr) complex separately (siRNA=0.2 μg, siRNA:rrPPC=1:10). Forty-eight hours post-transfection, cells were subjected to MTT cell proliferation assay to measure cell viability. This assay was repeated three times independently.
Clonogenic Assay
C26 cells pre-seeded in 6-well plate (1×103 cells/well) were exposed to different groups including rrPPC/siSTAT3 complex (siRNA=2 μg/well, rrPPC=10 μg/well) for 4 hours. The medium was refreshed with complete DMEM medium. Cells were continued to culture at 37 °C in 5% CO2 to form colonies. Two weeks later, after washing each well with PBS, colonies were stained with 10% crystal violet blue for 15 minutes. The number of clones in each well was counted and the suppression rate was measured.
In vitro Apoptosis Assay
The apoptosis inducing capacity of rrPPC/siSTAT3 complex was studied by flow cytometry. C26 cells were pre-seeded into 6-well culture plates (5×104 per well) and exposed to rrPPC/siSTAT3 complex and rrPPC/siScr complex (siRNA=5 μg/well, rrPPC=10 μg/well), respectively. Four hours later, the medium was replaced with complete medium. Seventy-two hours post treatment, all cells were stained with Propidium Iodide (PI) and Annexin V-FITC. The apoptotic cells were measured by flow cytometry (NovoCyte Flow Cytometer, ACEA Biosciences, USA).
Western Blot
C26 cells seeded into 6-well plate were treated with rrPPC/siRNA complex (siRNA=2 μg/well, rrPPC=10 μg/well) for 4 hours.72 hours later, total proteins were extracted from tumor cells and protein concentrations were quantified with Bicinchoninicacid (BCA) protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). 30 μg of protein was then separated by 12% SDS-PAGE gel electrophoresis and incubated with antibodies against mouse STAT3 and β-actin (Abcam, USA) at 4°C overnight. Protein bands were then further incubated with Horseradish peroxidase (HRP)-conjugated corresponding secondary antibody and detected with an enhanced chemiluminescence detection kit (EMD Millipore, Billerica, MA, USA).
Animal Study
All animal procedures were approved and controlled by the Institutional Animal Care and Treatment Committee of Sichuan University and carried out according to the Animal Care and Use Guidelines of Sichuan University. The abdominal cavity metastatic model was established by injecting C26 cells (1 × 105 cells/mouse) into the abdominal cavity of 6–8 weeks old female BALB/c mice on Day 0. On Day 3, mice were randomly divided into 3 groups (3 mice per group) and numbered separately. rrPPC/siSTAT3 complex (siRNA=5 μg/mouse, siRNA:rrPPC=1:10) was injected intraperitoneally every day for 7 treatments. The control groups received equivalent amounts of normal saline or rrPPC/siScr complex. On Day 12, all animals were sacrificed, and tumors were isolated for further analysis. Meanwhile, the volumes of ascites in each mouse also be collected and measured.
The subcutaneous xenograft tumor model was established by subcutaneously injection of C26 cells (5×106 cells/mouse) on the right hind limb of 6–8 weeks old female BALB/c mice. When the average tumor volume reached approximately 50 mm3, mice were randomly divided into 3 groups (4 mice per group). rrPPC/siSTAT3 complex (siRNA=5 μg/mouse, siRNA:rrPPC=1:10) was intratumorally injected every day for 7 treatments. The control groups received equivalent amounts of normal saline or rrPPC/siScr complex. After all animals were sacrificed, tumors were isolated for further analysis.
For pulmonary metastatic model, mice were IV injected with 100 μL of cell suspension containing 3×105 C26 cells on day 0. On day 5, mice were randomized into 3 groups (4 mice per group) and IV injected with normal saline, rrPPC/siScr, and rrPPC/siSTAT3 every day for 10 treatments (siRNA=5 μg/mouse, siRNA:rrPPC=1:10). On day 19, mice were sacrificed, and lungs were harvested for further analysis.
For histological analysis, wax-embedded tissue sections were dewaxed and rehydrated before staining with Mayer’s H&E. Apoptotic levels in tumor cells were determined using a DeadEnd™ Fluorometric TUNEL System kit (Promega, USA) according to the manufacturer’s instructions. The fluorescent image of each group was taken through a fluorescence microscope (Olympus, Japan). For CD31 staining assay, tumor sections were blocked and subsequently incubated with rabbit anti-mouse CD31 antibody (Abcam, USA) at 4°C overnight. Appropriate horseradish peroxidase-conjugated secondary antibody was then applied. The density of newly formed micro-vessels were observed through a fluorescence microscope (Olympus, Japan).
Blood Test
Blood tests were applied to evaluate the safety of IV administrated rrPPC/siSTAT3 complex (siRNA=5 μg/mouse, siRNA:rrPPC=1:10) for a single treatment. 24 hours after injection, circulating red blood cell count as well as serum-based clinical chemistry evaluation of systemic toxicity were analyzed.
Statistical Analysis
Data were expressed as the means with 95% confidence intervals. All data were statistically analyzed using Prism 5.0c software (GraphPad Software, La Jolla, CA) by two trailing ttests or one-way analysis of variance (ANOVA). Statistical significance was assigned at value of P<0.05 (95% confidence level).
Results
Preparation and Characterization of rrPPC Nanoparticles
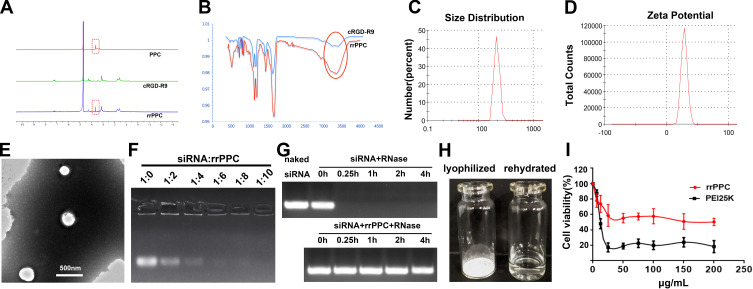
In this study, the cRGD-R9-PEG-PEI-Cholesterol carrier (rrPPC) was prepared by covalently modifying LMW PEI with PEG, cholesterol and cRGD-R9 peptide through nucleophilic substitution and Michael addition reaction (Scheme 1). In its structure, cholesterol, PEG, and cRGD-R9 peptide were all conjugated to PEI1.8K backbone through amido linkages, rendering it biodegradable when exposed to enzymes in cytoplasm. The chemical structure of rrPPC was characterized by 1H NMR and FT-IR spectra. As shown in Figure 1A, in the 1H NMR spectrum of rrPPC, the peaks corresponding to PPC and cRGD-R9 could be easily found. Obviously, it showed that cRGD-R9 has successfully connected with PPC and formed a new material rrPPC. In addition, by comparing the infrared spectra of rrPPC and cRGD-R9, it was not difficult to find that the absorption peak at 3300 cm−1 in the FT-IR spectrum of rrPPC is significantly higher (Figure 1B). This change also proved that cRGD-R9 is successfully connected to PPC since the structure of PPC contains a large number of amino groups. Therefore, we then characterized rrPPC solution in detail. As shown in Figure 1C, the dynamic diameter of cationic nanoparticles was 195.4 ± 3.5 nm with a polydispersity index of 0.22. Meanwhile, its zeta potential was 41.9 ± 1.2 mV (Figure 1D). In addition, rrPPC exhibited a share-like morphology when viewed under a TEM with an estimated average size of 205 ± 42 nm (Figure 1E). These results suggested that rrPPC could self-assemble into cationic nanoparticles in water solution, offering potential capacity for siRNA delivery. A siRNA retarding assay was then used to indicate the loading potential of rrPPC to siRNA. As shown in Figure 1F, when the ratio of siRNA and rrPPC reached 1:10 (w/w), no siRNA band could be observed. It suggested that under this ratio, siRNA could be fully loaded by positively charged rrPPC nanoparticles through electronic complexing. Hence, subsequent experiments were conducted with this indicated ratio. Protection of siRNA from enzymic degradation is a important property for delivery vector. Thus, a RNase degradation assay was performed. As shown in Figure 1G, siRNAs complexed with rrPPC were efficiently protected from enzymic degradation as long as 4 hours. The extracted siRNA bands from rrPPC/siRNA complexes showed equivalent brightness to that of naked siRNA, suggesting strong protection effect. The lyophilized rrPPC powder could be easily rehydrated in water, suggesting good solubility and distribution properties (Figure 1H). The cytotoxicity of rrPPC nanoparticles in vitro was evaluated by MTT assay. As shown in Figure 1I, as compared to PEI25K (the “gold standard” transfection agent), rrPPC exhibited much lower toxicity on 293t cells with an IC50 above 50 μg/mL, showing high safety as a potential siRNA vector.
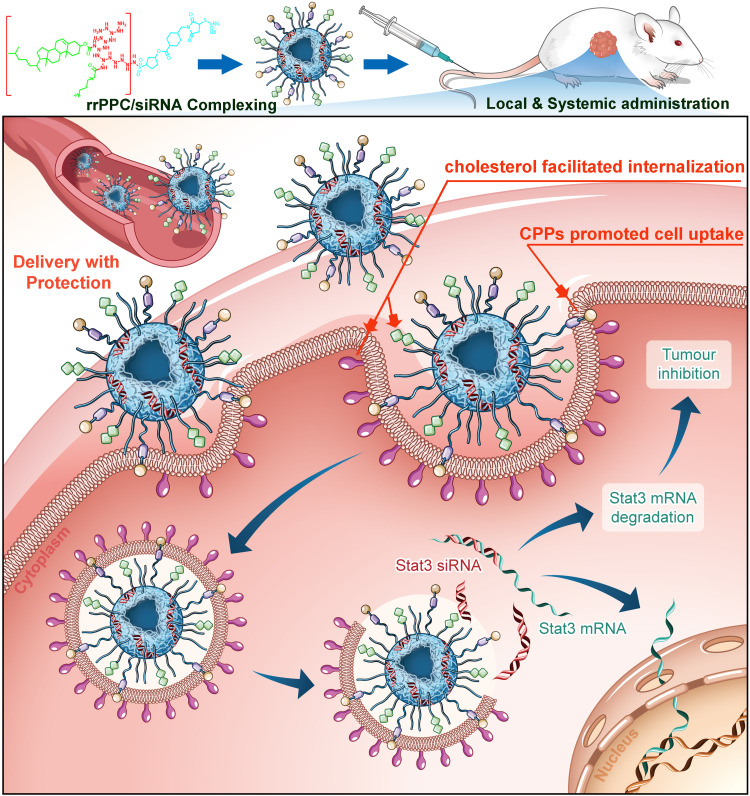
Scheme 1.
The synthesis process of rrPPC cationic nanoparticles.
Figure 1.
Characterization of rrPPC/siRNA complex. (A) 1H NMR spectra of rrPPC; (B) FT-IR spectra of rrPPC; (C) size distribution of rrPPC nanoparticles; (D) zeta potential distribution of rrPPC nanoparticles; (E) transmission electron microscopy images; (F) gel retarding assay of rrPPC/siRNA complex; (G) RNase degradation assay; (H) rehydration of lyophilized rrPPC powder; and (I) cytotoxicity as evaluated by MTT assay.
rrPPC Could Efficiently Deliver siRNA with High Stability
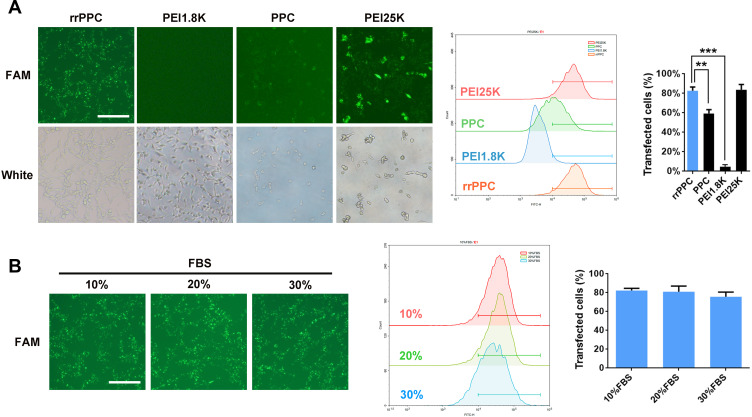
The ability of prepared rrPPC nanoparticles to deliver siRNA was next tested in C26 colon cancer cells in vitro. As shown in Figure 2A, rrPPC nanoparticles could efficiently deliver FAM reporter siRNA into C26 cells with good safety, exhibiting an average transfection rate up to 82.6% (FAM siRNA=1 μg/well, siRNA: rrPPC/PPC/PEI1.8K=1:10, siRNA:PEI25K=1:5). Although a similar degree of efficiency was also achieved by PEI25K (83.7%), obvious cytotoxicity was observed in the morphology change, and that safety difference was also spotted between rrPPC and PPC group. Furthermore, PEI1.8K (4.6%) and unmodified PPC (59.4%) demonstrated a much lower transfection ability than rrPPC, indicating the promoting effect functional modification on siRNA delivery. In addition, the delivery capacity of rrPPC under serum environment was further studied. As shown in Figure 2B, no obvious reduction in the transfection efficiency was observed under a serum concentration of up to 30%. This finding suggest that rrPPC could effectively protect siRNA in serum conditions and thus predicted a strong potential for in vivo delivery.
Figure 2.
rrPPC nanoparticles could efficiently deliver siRNA into C26 cells in vitro. (A) The evaluation and comparison of rrPPC with other gene vectors (**P < 0.01; ***P < 0.001). (B) Evaluation of the siRNA delivery ability of rrPPC under the existence of FBS.
Examination of the Mechanism of Cell Uptake
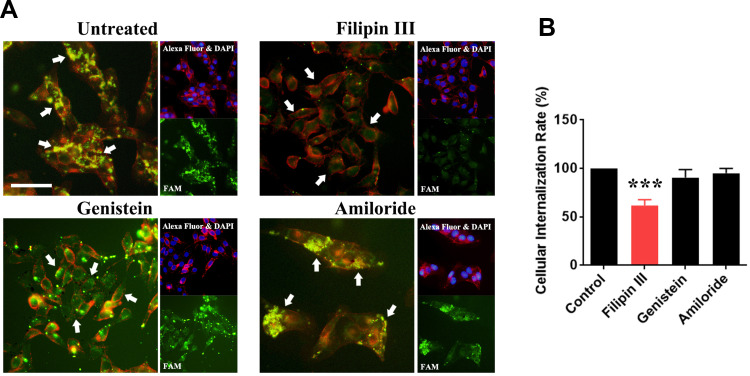
The cell uptake mechanism of rrPPC-delivered siRNA was then studied. Conventionally, three pathways including the caveolae-mediated pathway, clathrin-mediated pathway and macrocytosis are involved in the internalization process of nano-sized vehicles. We thus hypothesized that certain pathways would be triggered during the uptake of the rrPPC/siRNA complex. To validate that, inhibitors for each pathway were pre applied to seed C26 cells before being exposed to rrPPC/FAM siRNA complex (FAM siRNA=1 μg/well, siRNA:rrPPC=1:10). It could be found that the siRNA delivery efficiency in the group treated with Filipin III treated group was obviously reduced when compared with that in the other two inhibitor groups (Figure 3A). This effect was also detected by flow cytometry with an inhibition rate of 38.3% (Figure 3B). This result suggest that rrPPC/siRNA complex was specifically uptake through the caveolae-mediated pathway. Considering the cholesterol moiety of rrPCC nanoparticles, it is highly possible that this structure triggered caveolae, a cholesterol-rich invagination in plasma membrane, thus promoting cell uptake. These results also provide an explanation to the high gene delivery ability of rrPPC.
Figure 3.
Study of the cell-uptake mechanism of the rrPPC/siRNA complex. (A) Fluorescent images of transfected C26 cells pre-treated with various inhibitors. Different uptake levels of FAM siRNAs (white arrow) could be observed. Cells were co-stained with DAPI for nucleus and Alexa Fluor 594 for cell membranes, Bar=50 μm; (B) the influence of the cell uptake rate calculated by flow cytometry (***P < 0.001).
rrPPC/STAT3 siRNA Complex Suppressed the Growth of Cancer Cell in vitro
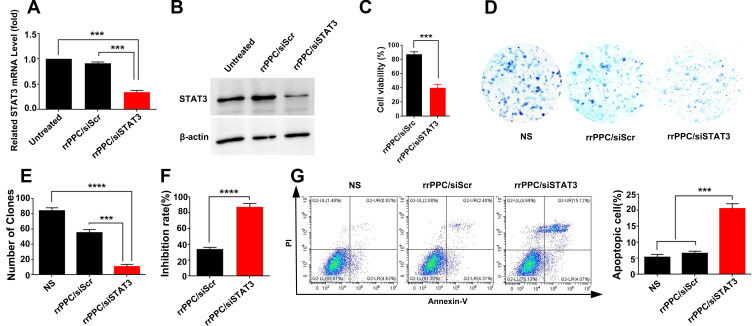
The tumor inhibition potential of rrPPC-delivered STAT3 siRNA was studied in vitro. At first, the rrPPC/siSTAT3 complex (siRNA=1 μg/well, rrPPC=10 μg/well) was evaluated for its ability to silence stat3 expression by RT-PCR. At 24 hours after treatment, the STAT3 mRNA level in rrPPC/siSTAT3 group was five-fold lower (P < 0.001) than that in the untreated group (Figure 4A), showing a high level of siRNA delivery efficiency. The siRNA delivery outcome was also evaluated by Western Blot. As seen in Figure 4B, compared to untreated group and scramble siRNA group (rrPPC/siScr), the protein level of mouse STAT3 was obviously down regulated by treating with rrPPC/siSTAT3 complex (siRNA=2 μg/well, siRNA:rrPPC=1:10), which is consistent with PCR results. MTT assay results demonstrate that rrPPC/siSTAT3 complex (siRNA=0.2 μg/well, siRNA:rrPPC=1:10) could efficiently inhibit the growth of C26 cells after 48 hours’ treatment (Figure 4C). The anti-proliferative effect of rrPPC/siSTAT3 complex was also studied by clonogenic assay. As compared with that in the rrPPC/siScr group and control group, rrPPC/siSTAT3 significantly reduced the clonogenicity of seeded C26 cells (siRNA=2 μg/well, siRNA:rrPPC=1:10, Figure 4D). A decrease in both the number and size of colonies could be directly observed. More specifically, the number of clones in rrPPC/siSTAT3 group was 10 ± 4 clones, while those of NS group and rrPPC/siScr group were 84 ± 4 clones and 55 ± 5 clones, respectively (Figure 4E). These results demonstrate that silencing the expression of stat3 by the rrPPC/siSTAT3 complex could significantly inhibit the growth of C26 cells with an inhibition rate of nearly 88% (P < 0.0001, Figure 4F).
Figure 4.
rrPPC/siSTAT3 complex suppressed C26 cells in vitro. (A) STAT3 mRNA levels in C26 cells after transfection (***P < 0.001); (B) STAT3 protein level of each treatment group; (C) anti-proliferation effect of rrPPC/siSTAT3 complex (***P < 0.001); (D) clonogenic assay used to detect the inhibitory effect of rrPPC/siSTAT3 complex; (E) average number of clones in each well (***P < 0.001; ****P < 0.0001); (F) inhibition rate calculated by clonogenic assay (****P < 0.0001); (G) rrPPC/siSTAT3 complex efficiently induced apoptosis in C26 cells (***P < 0.001).
Since it has been reported elsewhere that STAT3 targeted siRNA has apoptosis-inducing properties on cancer cells, such an effect was verified herein by flow cytometry. As shown in Figure 4G, rrPPC/siSTAT3 complex could efficiently induce apoptosis in C26 cells. After being exposed to rrPPC/siSTAT3 complex (5 μg siRNA) for 72 hours, in both the early and late apoptosis quadrants, a total of 22.7% ± 1.8% of apoptotic cells were detected (P < 0.001), while other groups did not exhibit equivalent ability. This result demonstrate that rrPPC nanoparticles could effectively deliver STAT3-siRNA into C26 cells as expected. In other words, the silencing of the stat3 gene resulted in a strong anti-cancer effect through apoptosis.
rrPPC/siSTAT3 Complex Efficiently Suppressed Colon Cancer in vivo
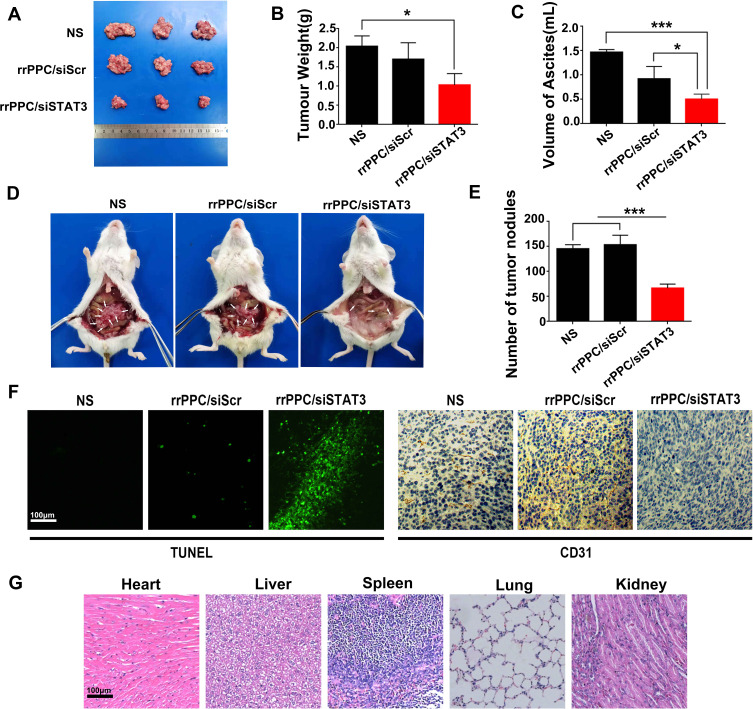
We first treated C26 abdominal cavity metastasis model by intraperitoneal injection (siRNA=5 μg/mouse, siRNA:rrPPC=1:10). The therapeutic effect of rrPPC/siSTAT3 complex can be obviously observed in Figure 5A and D. Much fewer tumor nodules were collected from mice in rrPPC/siSTAT3 complex group than in the other groups. In addition, a large amount of blood-like ascites could be observed in the control groups. The average weight of tumor nodules in rrPPC/siSTAT3 complex group was significantly lower than other groups (P < 0.05) (Figure 5B), with an average weight of 1.0 ± 0.2 g than that of NS group (2.1 ± 0.4 g) and rrPPC/siScr group (1.7 ± 0.3 g). Compared to other groups, the producing of ascites in rrPPC/siSTAT3 complex group was also significantly suppressed with 0.5 ± 0.1 mL in rrPPC/siSTAT3 complex group, while that of NS group and rrPPC/siScr group was 1.5 ± 0.4 mL and 0.92 ± 0.2 mL, respectively (Figure 5C). These of ascites collected from mouse abdominal cavities suggest severe tumor cell infiltration and local inflammation. In addition, as shown in Figure 5E, significantly fewer metastatic tumor nodules were collected from rrPPC/siSTAT3 complex group with an average number of only 66. In contrast, those numbers in the rrPPC/siScr and control groups were 145 and 153 nodules, respectively. These results suggest that rrPPC/siSTAT3 complex could efficiently suppress C26 tumor growth by intraperitoneal injection.
Figure 5.
rrPPC/siSTAT3 complex inhibited abdominal metastasis tumor growth in vivo. (A) Tumor nodules of each group; (B) average tumor weights in each group (*P < 0.05); (C) average volume of ascites in each group (*P < 0.05; ***P < 0.001); (D) representative images of abdominal cavity metastasis of C26 colon carcinoma; (E) number of tumor nodules in each group (***P < 0.001); (F) detection of vessel formation and apoptosis by CD31 staining and TUNEL assay; (G) H&E analysis.
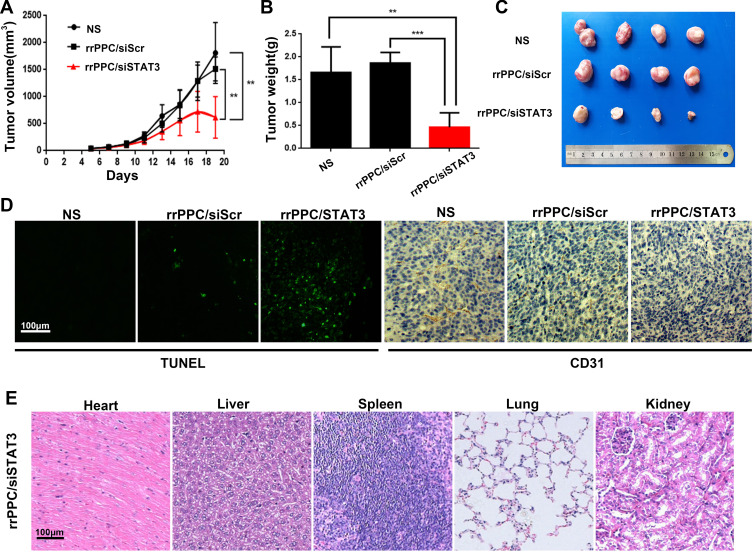
A subcutaneous xenograft tumor model was then established to evaluate the anti-tumor effect of rrPPC/siSTAT3 complex (siRNA=5 μg/mouse, siRNA:rrPPC=1:10). The growth curves and images of collected tumor tissues were shown in Figure 6A and C, respectively. It can be observed that a strong level of inhibition of tumor growth was achieved in rrPPC/siSTAT3 complex group. The tumor weight from each mouse was then isolated and weighted. As compared with the NS group (1.50 ± 0.2 g) and rrPPC/siScr group (1.64 ± 0.1 g), the average tumor weight in rrPPC/siSTAT3 complex group was only 0.3 6± 0.07 g (Figure 6B). These results indicate that rrPPC/siSTAT3 complex may strongly suppress colon cancer when administrated locally.
Figure 6.
rrPPC/siSTAT3 complex inhibited subcutaneous xenograft tumor growth in vivo. (A) Tumor growth curves of each group (**P < 0.01); (B) average tumor weight of each group; (C) representative images of subcutaneous tumor tissues (**P < 0.01; ***P < 0.001); (D) detection of vessel formation and apoptosis by CD31 staining and TUNEL assay; (E) H&E analysis.
The anti-tumor mechanisms that involved were studied by TUNEL assay and CD31 staining on tumor tissue sections. As shown in Figures 5F and 6D, much more positive dots with green fluorescence were detected by TUNEL assay in tissues from rrPPC/siSTAT3 complex group, suggesting an obvious apoptosis-inducing effect. Moreover, the formation of micro-vessels was detected by immunofluorescence staining with CD31 antibody. As seen in Figures 5F and 6D, as compared with in the other two groups, the density of micro-vessels in tumor tissues treated with rrPPC/siSTAT3 complex was obviously lower, suggesting strong anti-angiogenesis effects in both models. These results demonstrate that treatment with rrPPC/siSTAT3 complex resulted in obvious cell apoptosis and neovascular inhibition. In addition, the side effects of rrPPC/siSTAT3 complex on main organ tissues were further studied by H&E stain. As shown in Figures 5G and 6E, no obvious pathological changes were found in the heart, liver, spleen, lung, or kidney tissues collected from the rrPPC/siSTAT3 complex groups of both animal models, conforming the safety of rrPPC/siSTAT3 complex when delivered locally.
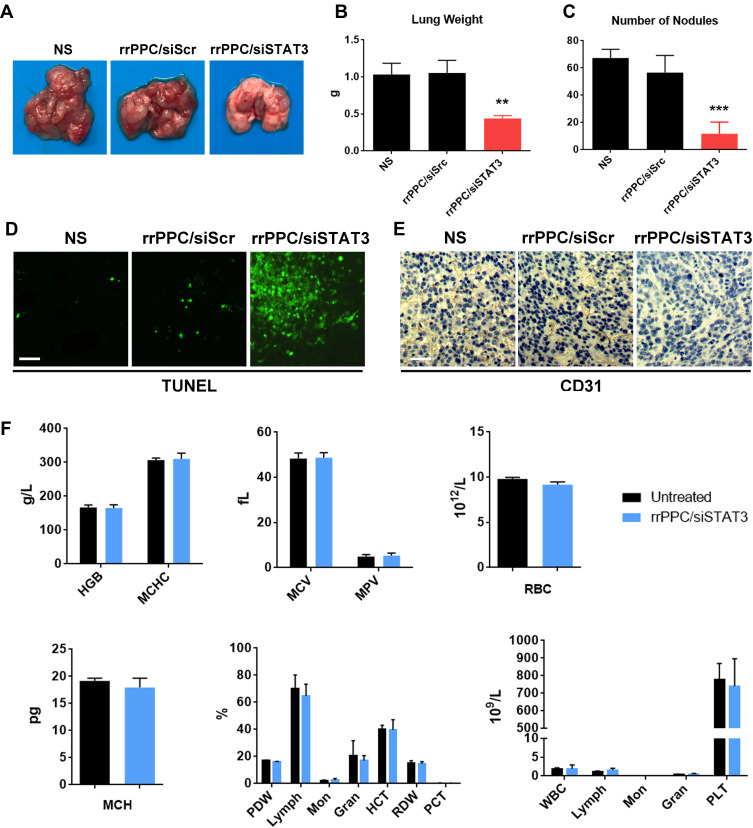
Next, the therapeutic efficacy and safety of rrPPC/siSTAT3 complex in lung metastasis tumor model were studied by intravenous administration (siRNA=5 μg/mouse, siRNA:rrPPC=1:10). As shown in Figure 7A, mice treated with rrPPC/siSTAT3 complex showed reduced tumor burdens as compared with in the other two groups. Obvious difference could be observed when lung tissues from each group were harvested and weighted. The average weight in rrPPC/siSTAT3 complex groups was 0.44 g while those in NS group and rrPPC/siScr complex groups were 1.03 g and 1.06 g, respectively (Figure 7B). The nodules in lung tissues were also counted to comprehend the therapeutic outcome. As seen in Figure 7C, as compared with the control groups, obviously fewer nodules were identified in rrPPC/siSTAT3 treatment group (11 on average), while averages of 67 and 56 nodules were collected in NS group and rrPPC/siScr treatment group, respectively. Meanwhile, the strong apoptosis-inducing and anti-angiogenesis effects of IV injected rrPPC/siSTAT3 complex were indicated by TUNEL assay and immunohistochemistry staining (Figure 7D and E). In addition, a blood routine and biochemical examinations were performed to evaluate the in vivo safety of rrPPC/siSTAT3 complex. As shown in Figure 7F, no significant impairment of liver or kidney function was detected following intravenous administration of this siRNA formulation. Different parameters of red blood cell level were also maintained within normal ranges. These results suggest that rrPPC delivered STAT3 siRNA is efficient and safe enough for the treatment of lung metastasis tumor by intravenous (IV) injection.
Figure 7.
rrPPC/siSTAT3 complex inhibited lung metastasis tumor growth by IV injection. (A) Representative images of lung tissues from each group; (B) average lung weight of each group (**P < 0.01); (C) average number of metastasis nodules (***P < 0.001); (D and E) vessel-formation and apoptosis-inducing in tumor tissues were detected by CD31 staining and TUNEL assay, respectively; (F) blood routine and biochemical examination after intravenous administration of the rrPPC/siSTAT3 complex.
Discussion
In recent years, RNAi-based cancer treatment technology has received much attention. The developing of new and suitable siRNA delivery vector is still highly demanded. In this report, a new siRNA delivery system named rrPPC nanoparticle was constructed and exhibited strong potential and high safety. By loading the STAT3 targeting siRNAs, rrPPC/siSTAT3 complex could significantly inhibit tumor growth both in vitro and in vivo (Scheme 2).
Scheme 2.
rrPPC/siSTAT3 complex efficiently inhibits colon cancer progression. rrPPC nanoparticles efficiently facilitated the loading, delivery, protection, and transmembrane transportation processes of STAT3 siRNA.
In recent years, significant progress has been made in developing gene delivery systems. In particular, large numbers of non-viral vectors derived from artificial or natural materials with novel structures have been designed especially for cancer therapy, and some candidates have reached the clinical stage.36–38 The efficiency and safety of gene delivery vectors play crucial roles in the success of gene therapy, while nanotechnology has contributed tremendously to this area as well.39–43 In the area of siRNA-based gene therapy, different non-viral nanoparticle carrier systems for siRNA delivery have been investigated extensively recently.44,45 The nanoparticle systems with the most interest include biodegradable polymeric nanoparticles, polyplex, lipid nanoparticles, lipoplex, liposomes, and dendrimers. For example, Kenny et al used a cationic liposome to deliver the survivin gene to effectively inhibit the growth of human ovarian cancer cells.43 Alshamsan et al using lipid-linked PEI polyplexes for the delivery of STAT3 siRNA, showing an effective inhibitory effect on the development of melanoma.46 James et al reported using a modified poly(amidoamine) (PAMAM) dendrimer to deliver a TWIST1 siRNA for metastatic breast cancer therapy.47 Jere et al discovered that PCL-PEI nanoparticles could efficiently deliver Akt1-siRNA into lung cancer cells with a good safety profile. Silencing Akt1 protein greatly reduced the cancer cell survival, proliferation, malignancy, and metastasis.48 In addition, several siRNA nanoparticle products are currently in preclinical or clinical phase studies including ALN-VSP02, EZN-2968, and TKM080301.16 Although RNAi-based gene suppression represents a promising direction of gene therapy, the lack of siRNA delivery vector with high efficacy, safety, and targeting properties persists as a limitation.
As a transcription factor, STAT3 is closely correlated with the pathogenesis, development, invasion and metastasis of many malignant tumors. Previous studies have shown that the delivery of STAT3 siRNA by nanomaterials could be applicable and potent in the treatment of malignant tumors such as melanoma, gastric cancer, glioblastoma. A few studies focusing on the use of STAT3 siRNA for the treatment of colon cancer have also been reported. For example, Yu Fan and Shi et al used liposomes to deliver STAT3 siRNA into colon cancer cells and evaluate their anti-tumor effects.49,50 However, limited attempts have been made to develop new delivery vectors for STAT3 siRNA and, thus, its therapeutic effects remain restricted in many aspects. For example, most of the cationic vectors are too toxic and thus unsuitable for intravenous administration, while efforts to reduce this cytotoxicity are usually complicated by simultaneously attenuated siRNA delivery efficiency. Meanwhile, although siRNA is comparatively smaller than other nucleic acid materials such as plasmid or messenger RNA, a high level of delivery efficiency cannot be easily achieved by every vector. The development of extra design strategies or special vector backbones is essential for successful siRNA delivery. Thus, developing new siRNA vector for STAT3-based cancer gene therapy is necessary.
In this research, rrPPC cationic nanoparticles were constructed and used as vehicles to deliver STAT3 siRNA into tumor cells and their anti-tumor ability was subsequently investigated. The synthesized rrPPC was composed of LMW PEI, cholesterol, CPPs, and PEG. PEI has always been considered as the “gold standard” for transfection agents.51,52 However, most of PEI-base gene vector development studies were focused on high molecular weight PEIs, typically PEI25K. PEI25K posses high density of positive charges with complex branched structure. It is rich of primary and secondary amino groups and thus provide tremendous gene delivery ability as well as strong toxicity. Comparing to PEI25K, low molecular weight PEIs such as PEI1.8K turns to be an alternative choice if certain structural modification is applied to gather the positive groups. Although various modifications have been made to PEI1.8K to enhance different aspects of gene delivery,53–55 PEI1.8K was seldom treated as a core. The potential of PEI1.8K has thus been greatly omitted. However, to become a qualified gene vector, structural modification is required to achieve additional abilities of protecting and delivering genes. In this study, we intend to build up a novel siRNA vector based on PEI1.8K, rather than just utilize it as a functional group. To this goal, cholesterol, PEG and CPPs were involved into its structure. PEG could inhibit the phagocytosis of monocyte phagocytes and reduce the uptake of carrier material by the reticuloendothelial system.56,57 According to our results, rrPPC efficiently protected siRNA oligos from the degradation of RNase for at least 4 hours (Figure 1G). This result demonstrated the stabilization capacity of rrPPC on delivered siRNA, thus further contributed to the systemic administration. Cholesterol had strong rigidity, biodegradability and membrane fusion ability.58 When being applied in drug delivery, it may also trigger caveolae-mediated pathway, which can further reinforce transmembrane delivery efficiency. According to our results, this unique property has been successfully achieved. Our results demonstrated that rrPPC/siRNA complex was primarily internalized through caveolae-mediated pathway (Figure 3). Alone with PEG, modification of PEI with cholesterol rendered rrPPC a highly biocompatible vector in vivo with high safety. As shown in Figure 7F, no obvious damage was detected to the main blood parameters, suggested high safety in circulation. Meanwhile, by cooperating with CPPs, cholesterol also contribute to the high siRNA delivery efficiency. A specialized uptake mechanism would promote fast transmembrane transportation of siRNA oligo. Efficiency is the ultimate goal of a delivery vector. Although the combination of PEG-PEI-Cholesterol (PPC) has been applied in gene therapy studies, its application was limited in delivering plasmid DNA. It exhibited weak capacity when handling small nucleic acid materials. As shown in Figure 2A, PPC could hardly enhance the transfection efficiency of siRNA on C26 cells. However, after further modifying it with R9-cRGD peptide, the transfection efficiency reached the same level with PEI25K. When complexed with siRNAs, the rrPPC/siSTAT3 complex demonstrated strong anti-cancer ability in multiple C26 colon cancer models. In particular, rrPPC/siSTAT3 complex demonstrated high therapeutic capacity as well as safety when administrated intravenously (Figures 5–7). Therefore, these results demonstrate that the design of rrPPC with multiple function group modifications have been successfully proved. It also suggest that with proper functional modification, PEI1.8K could be used as an idea core of gene vector. Our work not only introduces a novel PEI1.8K-based siRNA delivery system but also provides theoretical foundation for developing gene vector with low molecular PEIs.
Conclusion
In this article, a novel siRNA delivery system rrPPC was synthesized by modifying LMW PEI with multiple functional groups. The prepared rrPPC nanoparticles could efficiently deliver siRNA with high in vivo safety. By loading the STAT3 targeting siRNA, rrPPC/siSTAT3 complex demonstrated strong anti-cancer effects on multiple colon cancer models by local or systemic administration. These results suggest that rrPPC nanoparticle is a potential candidate vector for siRNA-based cancer gene therapy.
Acknowledgments
This work was supported by the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (No. 2018ZX09733001-006-008), Key Research and Development Program of Science and Technology Department of Sichuan Province (2020YFS0200), the Scientific Research Program of Chinese Medical Association Wu Jieping Medical Foundation (LCYX-Q015) and the Science Foundation of Chengdu (2020-YF05-00069-SN). We would like to thank Guangzhou Sagene Biotech Co.,Ltd. for providing illustrations assistance during the preparation of this manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Freddie B, Jacques F, Isabelle S. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. [DOI] [PubMed] [Google Scholar]
- 3.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2(12):711–719. doi: 10.1038/nchembio839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garber K. Alnylam Launches Era of RNAi Drugs. Nature Publishing Group; 2018. [DOI] [PubMed] [Google Scholar]
- 5.Adams D, Gonzalez-Duarte A, O’Riordan WD, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379(1):11–21. doi: 10.1056/NEJMoa1716153 [DOI] [PubMed] [Google Scholar]
- 6.Lee K, Jang B, Lee Y-R, et al. The cutting-edge technologies of siRNA delivery and their application in clinical trials. Arch Pharm Res. 2018;41(9):867–874. [DOI] [PubMed] [Google Scholar]
- 7.Hu B, Weng Y, Xia XH, Liang X, Huang Y. Clinical advances of siRNA therapeutics. J Gene Med. 2019;21(7):e3097. doi: 10.1002/jgm.3097 [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Zhou Z-G, Zheng X-L, et al. RNA interference against peroxisome proliferator-activated receptor δ gene promotes proliferation of human colorectal cancer cells. Dis Colon Rectum. 2008;51(3):318–328. [DOI] [PubMed] [Google Scholar]
- 9.Xia L, Xiaowen T. Research progress on siRNA interference technique in drug-resistant ovarian cancer. Surg Res N Tech. 2018. [Google Scholar]
- 10.Xu X, Wu J, Liu S, et al. Redox‐responsive nanoparticle‐mediated systemic RNAi for effective cancer therapy. Small. 2018;14(41):1802565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X, Saw PE, Tao W, et al. Tumor microenvironment-responsive multistaged nanoplatform for systemic RNAi and cancer therapy. Nano Lett. 2017;17(7):4427–4435. doi: 10.1021/acs.nanolett.7b01571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin Y, Huang M, Guo WW, Huang Q, Zhen Zhang L, Jiang G. Nano-based delivery of RNAi in cancer therapy. Mol Cancer. 2017;16(1):1–9. doi: 10.1186/s12943-017-0683-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansoori B, Shotorbani SS, Baradaran B. RNA interference and its role in cancer therapy. Adv Pharm Bull. 2014;4(4):313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reischl D, Zimmer A. Drug delivery of siRNA therapeutics: potentials and limits of nanosystems. Nanomed Nanotechnol Biol Med. 2009;5(1):8–20. doi: 10.1016/j.nano.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 15.Cullis PR, Hope MJ. Lipid nanoparticle systems for enabling gene therapies. Mol Ther. 2017;25(7):1467–1475. doi: 10.1016/j.ymthe.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A, Trivedi P, Jain NK. Advances in siRNA delivery in cancer therapy. Artif Cells, Nanomed Biotechnol. 2018;46(2):274–283. doi: 10.1080/21691401.2017.1307210 [DOI] [PubMed] [Google Scholar]
- 17.Scherman D, Rousseau A, Bigey P, Escriou V. Genetic pharmacology: progresses in siRNA delivery and therapeutic applications. Gene Ther. 2017;24(3):151–156. doi: 10.1038/gt.2017.6 [DOI] [PubMed] [Google Scholar]
- 18.Kunath K, von Harpe A, Fischer D, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89(1):113–125. doi: 10.1016/S0168-3659(03)00076-2 [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Yu M, Wang J, et al. Low molecular weight PEI‐based vectors via acid‐labile ortho ester linkage for improved gene delivery. Macromol Biosci. 2016;16(8):1175–1187. doi: 10.1002/mabi.201600071 [DOI] [PubMed] [Google Scholar]
- 20.Giron-Gonzalez MD, Salto-Gonzalez R, Lopez-Jaramillo FJ, et al. Polyelectrolyte complexes of low molecular weight PEI and citric acid as efficient and nontoxic vectors for in vitro and in vivo gene delivery. Bioconjug Chem. 2016;27(3):549–561. doi: 10.1021/acs.bioconjchem.5b00576 [DOI] [PubMed] [Google Scholar]
- 21.Zhupanyn P, Ewe A, Büch T, et al. Extracellular vesicle (ECV)-modified polyethylenimine (PEI) complexes for enhanced siRNA delivery in vitro and in vivo. J Control Release. 2020;319:63–76. doi: 10.1016/j.jconrel.2019.12.032 [DOI] [PubMed] [Google Scholar]
- 22.Yao W, Fu S, Yang G, Wang J, Wang X, Tang R. Low molecular weight PEI-grafted carboxyl-modified soybean protein as gene carriers with reduced cytotoxicity and greatly improved transfection in vitro. Int J Polym Mater. 2019;68(11):617–627. doi: 10.1080/00914037.2018.1482462 [DOI] [Google Scholar]
- 23.Habault J, Poyet J-L. Recent advances in cell penetrating peptide-based anticancer therapies. Molecules. 2019;24(5):927. doi: 10.3390/molecules24050927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Khan AR, Fu M, Wang R, Ji J, Zhai G. Cell-penetrating peptide: a means of breaking through the physiological barriers of different tissues and organs. J Control Release. 2019;309:106–124. doi: 10.1016/j.jconrel.2019.07.020 [DOI] [PubMed] [Google Scholar]
- 25.Osman G, Rodriguez J, Chan SY, et al. PEGylated enhanced cell penetrating peptide nanoparticles for lung gene therapy. J Control Release. 2018;285:35–45. doi: 10.1016/j.jconrel.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gajbhiye K, Gajbhiye V, Siddiqui IA, Gajbhiye J. cRGD functionalised nanocarriers for targeted delivery of bioactives. J Drug Target. 2019;27(2):111–124. doi: 10.1080/1061186X.2018.1473409 [DOI] [PubMed] [Google Scholar]
- 27.Lacerda L, Russier J, Pastorin G, et al. Translocation mechanisms of chemically functionalised carbon nanotubes across plasma membranes. Biomaterials. 2012;33(11):3334–3343. doi: 10.1016/j.biomaterials.2012.01.024 [DOI] [PubMed] [Google Scholar]
- 28.Xiao L, Xiong X, Sun X, et al. Role of cellular uptake in the reversal of multidrug resistance by PEG-b-PLA polymeric micelles. Biomaterials. 2011;32(22):5148–5157. doi: 10.1016/j.biomaterials.2011.03.071 [DOI] [PubMed] [Google Scholar]
- 29.Gratton J-P, Bernatchez P, Sessa WC. Caveolae and caveolins in the cardiovascular system. Circ Res. 2004;94(11):1408–1417. doi: 10.1161/01.RES.0000129178.56294.17 [DOI] [PubMed] [Google Scholar]
- 30.Qin L, Zhu N, Ao B-X, et al. Caveolae and caveolin-1 integrate reverse cholesterol transport and inflammation in atherosclerosis. Int J Mol Sci. 2016;17(3):429. doi: 10.3390/ijms17030429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nunes SS, Fernandes RS, Cavalcante CH, et al. Influence of PEG coating on the biodistribution and tumor accumulation of pH-sensitive liposomes. Drug Deliv Transl Res. 2019;9(1):123–130. doi: 10.1007/s13346-018-0583-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turecek PL, Bossard MJ, Schoetens F, Ivens IA. PEGylation of biopharmaceuticals: a review of chemistry and nonclinical safety information of approved drugs. J Pharm Sci. 2016;105(2):460–475. doi: 10.1016/j.xphs.2015.11.015 [DOI] [PubMed] [Google Scholar]
- 33.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston PA, Grandis JR. STAT3 signaling: anticancer strategies and challenges. Mol Interv. 2011;11(1):18. doi: 10.1124/mi.11.1.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huynh J, Chand A, Gough D, Ernst M. Therapeutically exploiting STAT3 activity in cancer—using tissue repair as a road map. Nat Rev Cancer. 2019;19(2):82–96. [DOI] [PubMed] [Google Scholar]
- 36.Brooks PJ, Yang NN, Austin CP. Gene therapy: the view from NCATS. Hum Gene Ther. 2016;27(1):7–13. doi: 10.1089/hum.2016.29018.pjb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Q, Lin D, Lei L, Li X, Shi S. Engineered Non-viral gene vectors for combination cancer therapy: a review. J Biomed Nanotechnol. 2017;13(12):1565–1580. doi: 10.1166/jbn.2017.2489 [DOI] [PubMed] [Google Scholar]
- 38.Ren E, Wang J, Liu G. Cell-surface cascaded landing location for nanotheranostics. Chin Chem Lett. 2017;28(9):1799–1800. [Google Scholar]
- 39.Shang S, Monfregola L, Caruthers MH. Peptide-substituted oligonucleotide synthesis and non-toxic, passive cell delivery. Signal Transduct Target Ther. 2016;1(1):16019. doi: 10.1038/sigtrans.2016.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khatri A, Mishra A, Chauhan VS. Characterization of DNA condensation by conformationally restricted dipeptides and gene delivery. J Biomed Nanotechnol. 2017;13(1):35–53. doi: 10.1166/jbn.2017.2325 [DOI] [PubMed] [Google Scholar]
- 41.Srivastava A, Babu A, Filant J, Moxley KM, Ramesh R. Exploitation of exosomes as nanocarriers for gene-, chemo-, and immune-therapy of cancer. J Biomed Nanotechnol. 2016;12(6):1159–1173. doi: 10.1166/jbn.2016.2205 [DOI] [PubMed] [Google Scholar]
- 42.Oliveira C, Ribeiro AJ, Veiga F, Silveira I. Recent advances in nucleic acid-based delivery: from bench to clinical trials in genetic diseases. J Biomed Nanotechnol. 2016;12(5):841–862. doi: 10.1166/jbn.2016.2245 [DOI] [PubMed] [Google Scholar]
- 43.Kenny GD, Kamaly N, Kalber TL, et al. Novel multifunctional nanoparticle mediates siRNA tumour delivery, visualisation and therapeutic tumour reduction in vivo. J Control Release. 2011;149(2):111–116. [DOI] [PubMed] [Google Scholar]
- 44.Ma J, Zhang J, Chi L, Liu C, Li Y, Tian H. Preparation of poly (glutamic acid) shielding micelles self-assembled from polylysine-b-polyphenylalanine for gene and drug codelivery. Chin Chem Lett. 2020. [Google Scholar]
- 45.Chen Y, Li B, Chen X, et al. A supramolecular co-delivery strategy for combined breast cancer treatment and metastasis prevention. Chin Chem Lett. 2020;31(5):1153–1158. doi: 10.1016/j.cclet.2019.06.022 [DOI] [Google Scholar]
- 46.Alshamsan A, Hamdy S, Samuel J, El-Kadi AO, Lavasanifar A, Uluda H. The induction of tumor apoptosis in B16 melanoma following STAT3 siRNA delivery with a lipid-substituted polyethylenimine. Biomaterials. 2011;31(6):1420–1428. [DOI] [PubMed] [Google Scholar]
- 47.James F. RNA-based TWIST1 inhibition via dendrimer complex to reduce breast cancer cell metastasis. Biomed Res Int. 2015;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jere D, Arote R, Jiang H-L, Kim Y-K, Cho M-H, Cho C-S. Biodegradable nano-polymeric system for efficient Akt1 siRNA delivery. J Nanosci Nanotechnol. 2010;10(5):3366–3369. doi: 10.1166/jnn.2010.2228 [DOI] [PubMed] [Google Scholar]
- 49.Shi N, Chen LJ, Jin QY. The interfering effect of STAT3-siRNA in human colorectal cancer cell. Progress Modern Biomed. 2010. [Google Scholar]
- 50.Fan Y, Zhang YL, Wu Y, Zhang W, Li H. Inhibition of signal transducer and activator of transcription 3 expression by RNA interference suppresses invasion through inducing anoikis in human colon cancer cells. World J Gastroenterol. 2008;14(3):428–434. doi: 10.3748/wjg.14.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J, Park J, Kim H, Singha K, Kim WJ. Transfection and intracellular trafficking properties of carbon dot-gold nanoparticle molecular assembly conjugated with PEI-pDNA. Biomaterials. 2013;34(29):7168–7180. doi: 10.1016/j.biomaterials.2013.05.072 [DOI] [PubMed] [Google Scholar]
- 52.Zwiorek K, Kloeckner J, Wagner E, Coester C. Gelatin nanoparticles as a new and simple gene delivery system. J Pharm Pharm Sci. 2004;7(4):22–28. [PubMed] [Google Scholar]
- 53.Brownlie A, Uchegbu I, Schätzlein A. PEI-based vesicle-polymer hybrid gene delivery system with improved biocompatibility. Int J Pharm. 2004;274(1–2):41–52. doi: 10.1016/j.ijpharm.2003.12.029 [DOI] [PubMed] [Google Scholar]
- 54.Thomas M, Klibanov AM. Enhancing polyethylenimine’s delivery of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A. 2002;99(23):14640–14645. doi: 10.1073/pnas.192581499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong Z, Feijen J, Lok MC, et al. Low molecular weight linear polyethylenimine-b-poly(ethylene glycol)-b-polyethylenimine triblock copolymers: synthesis, characterization, and in vitro gene transfer properties. Biomacromolecules. 2005;6(6):3440–3448. doi: 10.1021/bm050505n [DOI] [PubMed] [Google Scholar]
- 56.Bazile D, Prud’homme C, Bassoullet MT, Marlard M, Spenlehauer G, Veillard M. Stealth me. PEG‐PLA nanoparticles avoid uptake by the mononuclear phagocytes system. J Pharm Sci. 1995;84(4):493–498. doi: 10.1002/jps.2600840420 [DOI] [PubMed] [Google Scholar]
- 57.Gabizon A, Papahadjopoulos D. Liposome formulations with prolonged circulation time in blood and enhanced uptake by tumors. Proc Natl Acad Sci U S A. 1988;85(18):6949–6953. doi: 10.1073/pnas.85.18.6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu L, Wang Q, Li W, Hou Y. Stability analysis and stabilisation of full-envelope networked flight control systems: switched system approach. Iet Control Theory Appl. 2012;6(2):286–296. doi: 10.1049/iet-cta.2010.0654 [DOI] [Google Scholar]