Brahma is functionally involved in Piwi-piRISC–mediated transposon silencing in Drosophila ovarian somatic cells.
Abstract
Drosophila Piwi associates with PIWI-interacting RNAs (piRNAs) and represses transposons transcriptionally through heterochromatinization; however, this process is poorly understood. Here, we identify Brahma (Brm), the core adenosine triphosphatase of the SWI/SNF chromatin remodeling complex, as a new Piwi interactor, and show Brm involvement in activating transcription of Piwi-targeted transposons before silencing. Bioinformatic analyses indicated that Piwi, once bound to target RNAs, reduced the occupancies of SWI/SNF and RNA polymerase II (Pol II) on target loci, abrogating transcription. Artificial piRNA-driven targeting of Piwi to RNA transcripts enhanced repression of Brm-dependent reporters compared with Brm-independent reporters. This was dependent on Piwi cofactors, Gtsf1/Asterix (Gtsf1), Panoramix/Silencio (Panx), and Maelstrom (Mael), but not Eggless/dSetdb (Egg)–mediated H3K9me3 deposition. The λN-box B–mediated tethering of Mael to reporters repressed Brm-dependent genes in the absence of Piwi, Panx, and Gtsf1. We propose that Piwi, via Mael, can rapidly suppress transcription of Brm-dependent genes to facilitate heterochromatin formation.
INTRODUCTION
PIWI proteins and PIWI-interacting RNAs (piRNAs) bind with each other and assemble into piRNA-induced silencing complexes (piRISCs) to control transposons, which protects the germline genome from invasive elements (1–4). The loss of piRISC function desilences transposons, leading to DNA damage, defective gonadal development, and infertility (5, 6).
Drosophila expresses three PIWI members: Piwi, Aubergine (Aub), and Argonaute3 (Ago3) (1–3). While Aub and Ago3 repress transposons posttranscriptionally in the cytoplasm, Piwi is localized in the nucleus and represses transposons transcriptionally through heterochromatinization. A number of Piwi cofactors, including Gtsf1 and Mael, have been identified (7–18). Panx, Nxf2, and p15 associate with each other for their mutual stabilization and reinforce Piwi–target RNA association to facilitate heterochromatinization through Egg-mediated repressive histone mark (H3K9me3) deposition (7, 8, 14–18). Uniquely, loss of Mael has little impact on H3K9me3 accumulation, although transposons are desilenced (11). This suggests that the role of Mael is different from that of other Piwi cofactors. A recent study showed that Mael represses canonical transcription in the germ line by piRNA-dependent and piRNA-independent processes (19). However, the functions of Mael in these pathways remain unclear.
RESULTS
Brm associates with the Piwi complex in OSC nuclei
We immunopurified the Piwi complex from nuclear extracts of cultured fly ovarian somatic cells (OSCs) (Fig. 1A) (20). The presence of Mael, Panx, and Gtsf1 in the immunoprecipitates was verified by Western blotting (Fig. 1B). The protein contents were also visualized by silver staining (Fig. 1C). Immunoprecipitation was also performed using anti-Mael antibodies (Fig. 1C). Both Piwi and Mael immunoprecipitations were performed three times before and after Piwi and Mael depletion, respectively (fig. S1A), and each sample was subjected to mass spectrometric analysis.
Fig. 1. Brm associates with the Piwi complex in OSC nuclei.
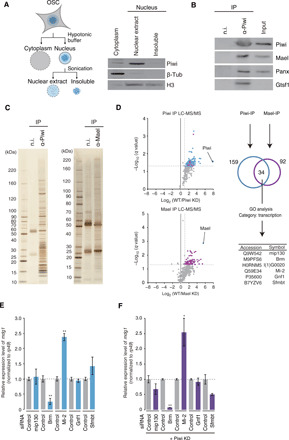
(A) Left: Scheme for OSC nuclear extract preparation. Right: Western blotting showing the protein levels of Piwi, β-tubulin (β-Tub), and histone H3 (H3) in the cytoplasmic fraction, nuclear extract, and insoluble fraction from OSCs. (B) Western blotting showing that Piwi co-immunoprecipitates with Mael, Panx, and Gtsf1 from the nuclear extract in (A). IP, immunoprecipitation; n.i., non-immune IgG. (C) Silver staining showing proteins co-immunoprecipitated with Piwi (~90 kDa) and Mael (~50 kDa) from the nuclear extract in (A). (D) Left: Volcano plots showing enrichment rates and significance levels of each protein in Piwi (top) and Mael (bottom) immunoprecipitates (n = 3). Blue and purple dots represent Piwi and Mael interactors, respectively. Magenta dots represent six proteins appearing in both Piwi and Mael interactors. Upper right: Scheme for bioinformatic analysis of Piwi and Mael interacting proteins. Lower right: Gene ontology (GO) analyses showed that seven proteins common to the Piwi and Mael complexes are related to transcription. LC-MS/MS, liquid chromatography–tandem mass spectrometry. (E) Changes in mdg1 expression levels in OSCs before (Control) and after depletion of mip130, Brm, Mi-2, Gnf1, or Sfmbt. n = 3. *P < 0.05, **P < 0.01. siRNA, small interfering RNA. (F) Changes in mdg1 expression levels in Piwi-depleted OSCs (Piwi KD) before (Control) and after depletion of mip130, Brm, Mi-2, Gnf1, or Sfmbt. n = 3. *P < 0.05, **P < 0.01.
The Sum posterior error probability (PEP) Score of each protein was normalized against the mean score of bovine serum albumin (BSA) spiked into the samples before mass spectrometry, and then PIWI and Mael datasets were analyzed individually using an enrichment score (twofold over background) and a q value (<0.05) (Fig. 1D and table S1). This analysis identified 159 and 92 proteins in Piwi and Mael datasets, respectively. Of these, 34 proteins overlapped between the two datasets. Gene ontology analysis revealed that 6 of 34 proteins were related to transcription (magenta in Fig. 1D and table S2). Proteins in other categories are listed in table S2.
We knocked down these six proteins individually in OSCs (fig. S1B) and examined the level of mdg1 by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 1E). mdg1 is one of the transposons repressed by Piwi in OSCs (21). L(1)G0020 was only partially depleted despite our best efforts (fig. S1B) and was, therefore, not subjected to further analyses. The loss of Mi-2 derepressed mdg1, while loss of mip130, Gnf1, or Sfmbt had little or weak impact on the level of mdg1 (Fig. 1E). Mi-2 is a nucleosome remodeler (22). We recently reported that Mi-2 acts in Piwi-mediated transcriptional repression, along with its partner, MEP-1, by accelerating local epigenetic modifications by Rpd3, Egg, and Su(var)2-10 (23). Su(var)2-10 acts in the piRNA pathway by associating with Egg upon its own sumoylation (24).
Notably, the loss of Brm reduced the level of mdg1, regardless of the presence or absence of Piwi (Fig. 1, E and F), indicating that Brm functions in activating transposon expression, most likely upstream of Piwi. These results also indicate that transposon transcription is being kept in an active, albeit minor, state even under Piwi regulation, which is consistent with previous observations showing that a constant supply of Piwi is required for the maintenance of transposon silencing (11, 25, 26).
Brm is the core adenosine triphosphatase (ATPase) of the SWI/SNF chromatin remodeling complex (27, 28). The association of Brm with the Piwi complex was verified by Western blotting (fig. S1C). The essentiality of Brm ATPase activity (29) in transposon regulation was also verified by rescue experiments, where an ATPase-deficient K804A Brm mutant failed to repress mdg1, unlike wild-type (WT) Brm, in endogenous Brm-depleted OSCs (fig. S1D).
Brm plays a role in the transcriptional activation of Piwi-targeted transposons
Genome-wide RNA sequencing (RNA-seq) confirmed that loss of Piwi increased the levels of a number of transposons, including mdg1 (fig. S2A) (21). Comparison of the RNA-seq data with our previous piRNA sequencing data (30) revealed that some transposons (e.g., Bari1 and mariner2) only had a few piRNAs against them, although they were up-regulated by Piwi loss (fig. S2A). We therefore eliminated transposons whose piRNA frequency was lower than 0.3% of the total transposon-targeting piRNA reads, no matter how much the expression levels were altered by Piwi loss, and selected from the remainder those whose RNA levels were increased at least twofold by Piwi depletion (i.e., log2 > 1). Ten transposons were obtained (fig. S2B), which we hereinafter designate as Piwi-dependent transposons.
Piwi-dependent transposons were up-regulated similarly by Mael or Piwi loss (Fig. 2A). Opposite effects were observed upon Brm depletion (Fig. 2B). The expression levels of other transposons were only weakly affected by Piwi, Mael, or Brm depletion (Fig. 2C). These results indicate that Piwi-dependent transposons in OSCs are mostly under the control of Brm. The abundances of Brm-dependent protein-coding transcripts were hardly affected by Piwi loss (fig. S2C). Thus, the inverse correlation effect observed between Piwi/Mael and Brm depletions is specific for Piwi-dependent transposons.
Fig. 2. Brm plays a role in the transcriptional activation of Piwi-targeted transposons.
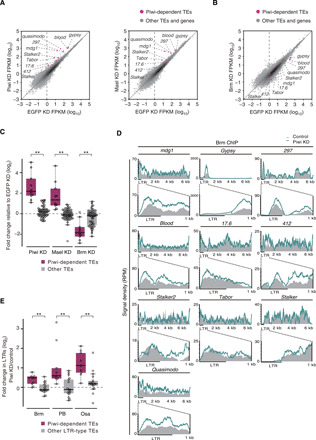
(A) Scatter plots showing the expression levels of Piwi-dependent transposons relative to EGFP KD OSCs in Piwi (left) and Mael (right) KD OSCs. Magenta plots represent Piwi-dependent transposons (TEs) (fig. S2B), and gray plots represent other TEs and genes (see Materials and Methods). (B) Scatter plots showing the expression levels of Piwi-dependent TEs relative to EGFP KD OSCs in Brm KD OSCs. Magenta plots represent Piwi-dependent TEs, and gray plots represent other TEs and genes. (C) Box plots showing fold changes in the expression levels of Piwi-dependent TEs (10 TEs) and other TEs (49 TEs) in Piwi, Mael, or Brm KD OSCs. **P < 0.01. (D) Density plots for normalized Brm ChIP-seq signals over Piwi-dependent TEs in control and Piwi KD OSCs (gray infill and colored lines, respectively). RPM, reads per million. (E) Box plots showing the fold changes in ChIP-seq signals of Brm, Osa, and PB in LTRs of Piwi-dependent LTR-type TEs except stalker (9 TEs) and other LTR-type TEs (17 TEs) upon Piwi depletion in OSCs. **P < 0.01.
Chromatin immunoprecipitation sequencing (ChIP-seq) showed that Brm tends to be enriched at promoter-transcriptional start sites (TSSs) of protein-coding genes (51.0%) (fig. S2D). Transposon mapping showed that Brm is accumulated more around the long terminal repeats (LTRs) of genes compared with other regions (Fig. 2D). The levels of Brm in the vicinity of LTRs were notably increased upon Piwi loss (Fig. 2D). These findings support the idea that Piwi down-regulates the occupancy of Brm around the LTR regions, leading to the abrogation of Pol II–mediated transcription.
SWI/SNF is also known as Brm-associated protein (BAP) and Polybromo (PB)–containing BAP (PBAP) complexes (27, 31, 32). BAP and PBAP share seven proteins including Brm, while Osa and PB/Bap170/SAYP are specific to BAP and PBAP, respectively (fig. S2E) (27, 31, 32). ChIP-seq showed that Osa was enriched at intron regions (53.1%) (fig. S2F), whereas PB was enriched at promoter-TSSs (72.8%) (fig. S2G), similarly to Brm (fig. S2D). Transposon mapping revealed that both Osa and PB patterns were similar to that of Brm (Fig. 2D and fig. S2H); the abundance of Osa and PB around the LTR regions was significantly increased upon Piwi loss. The abundance of Osa and PB at the LTRs of other LTR-type transposons was relatively weakly affected by Piwi loss (Fig. 2E). Depletion of Osa or PB in Piwi-lacking OSCs resulted in a strong reduction in mdg1 levels, similarly to Brm loss (fig. S2, I and J). Depletion of Snr1 (33), another common factor in BAP and PBAP (fig. S2E), also repressed mdg1 (fig. S2K). It is likely that both BAP and PBAP have similar, albeit not identical, effects on activating the transcription of Piwi-dependent transposons in OSCs. We also performed ChIP-seq using WT and mael mutant ovaries. Binding of Brm to the transposon loci, whose expression was affected by the loss of mael (19), was significantly increased by Mael loss as in OSCs (fig. S2, L to N). Thus, the phenomenon observed in OSCs was not unique to the cell culture system.
Artificial piRNA-driven Piwi induces repression of Brm-dependent genes
We targeted Piwi to the RNA transcripts of two protein-coding genes CG14072 and CG34330 by expressing artificial piRNAs (30) against them (Fig. 3A). The expression of CG14072 and CG34330 was sensitive to Brm depletion but insensitive to Piwi loss (fig. S3, A and B); thus, we considered them as Brm-dependent and Piwi-independent genes. The induction of artificial piRNA expression significantly reduced the mRNA levels of CG14072 and CG34330 (Fig. 3B and fig. S3C). Under the same conditions, expression of the Brm-independent genes, CG44194 and CG5119 (fig. S3, A and B), was little changed by the expression of artificial piRNAs against them (Fig. 3B and fig. S3C). Brm-independent genes were eventually silenced by the artificial piRNAs over a longer time period (fig. S3D). It seems that Piwi induces the repression of Brm-dependent genes more rapidly than Brm-independent genes.
Fig. 3. Artificial piRNA-driven Piwi induces repression of Brm-dependent genes.
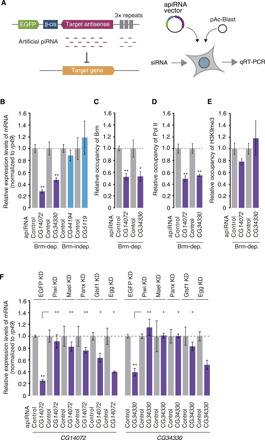
(A) Left: Schematic representation of the artificial piRNA–mediated Piwi tethering system. Right: Plasmid transfection procedure. qRT-PCR, quantitative reverse transcription polymerase chain reaction. (B) qRT-PCR showing changes in the RNA levels of Brm-dependent genes, CG14072 and CG34330, and Brm-independent genes, CG44194 and CG5119, 48 hours after the indicated artificial piRNA (apiRNA) expression. n = 3. *P < 0.05, **P < 0.01. (C) qRT-PCR showing changes in the abundance of Brm at the promoter regions of CG14072 and CG34330 48 hours after artificial piRNA expression. n = 3. *P < 0.05, **P < 0.01. (D) qRT-PCR showing changes in the abundance of Pol II at the promoter regions of CG14072 and CG34330 48 hours after artificial piRNA expression. n = 3. *P < 0.05, **P < 0.01. (E) qRT-PCR showing changes in the abundance of H3K9me3 at the promoter regions of CG14072 and CG34330 48 hours after artificial piRNA expression. n = 3. *P < 0.05, **P < 0.01. (F) qRT-PCR showing changes in the RNA levels of CG14072 and CG34330 48 hours after artificial piRNA expression. Piwi, Mael, Panx, Gtsf1, and Egg were individually down-regulated (fig. S3D). n = 3.
The occupancy of Brm at the 5′ untranslated regions (5′UTRs) of CG14072 and CG34330 was significantly reduced upon artificial piRNA expression (Fig. 3C). Pol II occupancy at these regions was also reduced upon artificial piRNA expression (Fig. 3D). In contrast, the levels of H3K9me3 at the target genes were little changed even at the time point when they were effectively silenced by the Piwi complex (Fig. 3E). These findings indicate that Piwi ceases Brm-dependent transcription before Egg-dependent H3K9me3 deposition. In agreement with this, Egg depletion had little impact on Piwi-artificial piRNA-mediated silencing (Fig. 3F and fig. S3E). In contrast, loss of Piwi, Mael, Panx, and Gtsf1 desilenced both genes. Thus, Mael, Panx, and Gtsf1, but not Egg, are essential for the initiation of Piwi-artificial piRNA-driven gene silencing.
Artificial tethering of Mael induces repression of Brm-dependent genes
The tethering of Panx, Nxf2, and p15 by the λN-box B system (34) to luciferase (luc) reporter RNAs induces transcription repression, but this silencing effect was little changed by Mael depletion (7, 8, 14–17). Here, we used the CG14072 promoter as a Brm-dependent promoter; a 1-kb DNA fragment directly upstream from the TSS of CG14072 was integrated into the luc reporter (Fig. 4A). As a control, the Hsp70 promoter was used, because it is often used as a Brm-independent promoter (35). The luc activity in OSCs that stably expressed the Hsp70-luc reporter was not down-regulated by Brm depletion, while that of the CG14072-luc reporter was significantly down-regulated (Fig. 4B and fig. S4A).
Fig. 4. Artificial tethering of Mael induces repression of Brm-dependent genes.
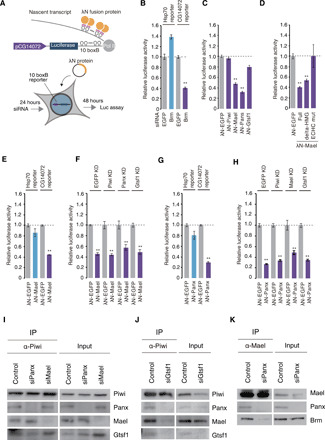
(A) Upper: Schematic representation of the λN-box B–mediated tethering system. Lower: Procedure for plasmid transfection and luciferase assays. (B) Bar graph showing relative luciferase activities of Brm KD OSC lysates and EGFP KD OSC lysates. The CG14072-luc and the Hsp70-luc reporters were used. n = 4. **P < 0.01. (C) Bar graph showing relative luciferase activities of OSC lysates upon transfection of each tethering construct. The CG14072-luc reporter was used. n = 3. **P < 0.01. (D) Bar graph showing relative luciferase activities of OSC lysates upon transfection of each tethering construct. The CG14072-luc reporter was used. n = 3. **P < 0.01. (E) Bar graph showing relative luciferase activities of OSC lysates, 48 hours after transfection of each tethering construct. The CG14072-luc and the Hsp70-luc reporters were used. n = 4. **P < 0.01. (F) Bar graph showing relative luciferase activities of OSC lysates upon KD of each factor and transfection of each tethering construct. The CG14072-luc reporter was used. **P < 0.01. (G) Bar graph showing relative luciferase activities of OSC lysates upon transfection of each construct. The CG14072-luc and the Hsp70-luc reporters were used. n = 3. **P < 0.01. (H) Bar graph showing relative luciferase activities of OSC lysates upon KD of each factor and transfection of each construct. The CG14072-luc reporter was used. n = 3. **P < 0.01. (I) Western blotting showing the levels of Panx, Mael, and Gtsf1 in the Piwi complex before and after Panx or Mael depletion. (J) Western blotting showing the levels of Panx, Mael, and Gtsf1 in the Piwi complex before and after Gtsf1 depletion. (K) Western blotting showing that Mael-Brm association was weakened by loss of Panx.
We tethered Piwi, Gtsf1, Panx, and Mael individually to CG14072-luc reporter transcripts by λN-box B. Tethering Piwi or Gtsf1 rarely repressed expression of the Brm-dependent reporter, but tethering Panx or Mael produced strong repression (Fig. 4C and fig. S4, B and C). Tethering Mael lacking the N-terminal HMG-box repressed the Brm-dependent reporter similarly to WT Mael (Fig. 4D and fig. S4D). However, when the core Glu-Cys-His-Cys (ECHC) motif of Mael was mutated to four alanines (36), repression was prevented. These results were consistent with our previous observations where the ECHC motif, but not the HMG-box, was important for Mael function in the piRNA pathway (36). WT Mael and the Mael ECHC mutant bound with Piwi and Brm (fig. S4E). It seems that the ECHC motif is the key in repressing the transposon loci, although the molecular details of how this occurs remain unknown.
We then tethered Mael to the Hsp70-reporter: Repression was not remarkable at 48 or 96 hours after transfection (Fig. 4E and fig. S4, F and G). We assume that this promoter selectivity may explain, at least in part, why previous Mael tethering was not successful in target gene silencing (8). The difference may also be attributed to the expression levels of the tethering proteins, which may not have been consistent across studies.
The absence of Piwi, Panx or Gtsf1 had little effect on the repression (Fig. 4F and fig. S4, H and I). Thus, Mael-mediated repression of Brm-dependent transcription functions independently of other factors.
Panx tethering repressed Brm-dependent but not Brm-independent transcription (Fig. 4G and fig. S4J). This indicates that the repressive effect of Panx is not global but shows some, albeit minor, preference in target gene selection. Panx, but not Mael, tethering successfully repressed an α-tubulin 84D promoter-driven reporter (8). It seems that Panx handles a wider range of target genes than Mael in the piRNA pathway. Enforced tethering of Panx repressed transcription in a manner independent of Mael, Piwi, and Gtsf1 (Fig. 4H and fig. S4K). It seems that Mael and Panx function independently and additively in the silencing of transposons.
Mael and Panx maintained their interaction with Piwi even when the other was depleted (Fig. 4I), but loss of Gtsf1 caused a reduction in Piwi-Mael and Piwi-Panx interactions (Fig. 4J). These results suggest that Piwi requires Gtsf1 for assembling other factors and that Mael and Panx independently interact with Piwi. These findings agreed with our observations in the tethering assays that Mael and Panx function independently of each other. Our previous study showed that the loss of Gtsf1 derepressed transposons to an extent similar to the loss of Piwi (10). At that time, we could not explain the outcome. However, we now know the reason for this; namely, Gtsf1 bridges Piwi with Mael and Panx, the facilitators of Piwi-driven gene silencing. The Mael-Brm interaction was significantly weakened by Panx loss (Fig. 4K). This agreed with our earlier observation that Mael failed to repress fully Brm-dependent transcription without Panx in the artificial piRNA assays (Fig. 3F).
DISCUSSION
These findings support new concepts for Mael/Brm-dependent and Mael/Brm-independent mechanisms of Piwi-mediated transcriptional repression in OSCs (fig. S4L). In both cases, the silencing is initiated by Piwi-piRISC targeting nascent transcripts and subsequent participation of the ternary complex composed of Panx, Nxf2, and p15 (7, 8, 14–17). Piwi-piRISC might be pre-accompanied with Gtsf1 while translocating from the cytoplasm to the nucleus because loss of Piwi in the ovary caused Gtsf1 to be left in the cytoplasm (9). Mael then joins the complex and quickly ceases Pol II transcription by reducing Brm (SWI/SNF) occupancy of transposon promoter regions if the target genes are Brm (SWI/SNF) dependent. Panx caused a similar phenomenon. When the target genes are Brm (SWI/SNF) independent, Mael is ineffective. However, it seems that Panx has a higher probability of ceasing transcription and that this action is also effective on Brm-dependent genes. Last, Egg deposits H3K9me3 at the target loci, and heterochromatin formation is completed with help from linker histone H1, HP1, Lsd1, Su(var)2-10, and Mi-2 (fig. S4L). Unlike Mael, which is dedicated to transcription inactivation, Panx has an additional function to transcription inactivation, i.e., reinforcement of Piwi-target RNA association by binding to both Piwi and target RNAs to facilitate heterochromatinization. From this point of view, it makes sense that the effect of Panx loss resembled that of Piwi loss more than that of Mael; namely, Panx loss reduced the level of H3K9me3 at target loci similarly to Piwi loss, but Mael loss had little effect on H3K9me3 accumulation (11).
Conceptually, the cessation of Pol II–mediated transcription is key in piRNA-mediated transcriptional silencing. Without this cessation and as long as Pol II–mediated transcription continues, even though the Piwi-initiated silencing complex is successfully assembled on nascent RNAs, the complex and RNAs are freed from the target loci, resulting in failure of heterochromatin formation.
Most Piwi-dependent transposons in OSCs are LTR-type transposons (11). We also noticed this bias in the current study. Retroviruses (e.g., HIV-1, human T cell leukemia virus–1, and murine leukemia virus), which are considered as the origin of retrotransposons, “hijack” the host SWI/SNF to activate transcription (37–39). Therefore, SWI/SNF-dependent activation of LTR-type retrotransposons may be an inherited feature from retroviruses. In this regard, the duality of the piRISC-mediated transcriptional silencing mechanism of transposons, which relies on Panx and Mael, is considered to be the remnant of the arms race between piRNA and transposons.
MATERIALS AND METHODS
Cell culture and RNAi
OSCs were grown at 26°C in culture medium prepared from Shields and Sang M3 Insect Medium (Sigma-Aldrich) supplemented with glutathione (0.6 mg/ml), 10% fetal bovine serum, insulin (10 mU/ml), and 10% fly extract (40). For RNA interference (RNAi), trypsinized OSCs (3 × 106 cells) were suspended in 20 μl of Solution SF of the Cell Line Nucleofector Kit SF (Amaxa Biosystems) together with 200 pmol of small interfering RNA (siRNA) duplex. Transfection was conducted in a 96-well electroporation plate using a Nucleofector device 96-well Shuttle (Amaxa Biosystems). Transfected cells were transferred to fresh OSC medium and incubated at 26°C for 2 to 4 days for further experiments. The siRNA sequences used are shown in table S3.
Plasmid rescue assay
To construct Myc-Brm and Myc-Brm mutant plasmids, a full-length brm complementary DNA (cDNA) was amplified by RT-PCR and subcloned into pAcM under control of the actin 5C promoter. Trypsinized OSCs (3 × 106 cells) were suspended in 100 μl of Solution V of the Cell Line Nucleofector Kit V (Amaxa Biosystems) together with 200 to 400 pmol siRNA duplex and 4 μg of plasmid. Transfection was conducted in electroporation cuvettes using a Nucleofector device 2b (Amaxa Biosystems). The transfected cells were transferred to fresh OSC medium and incubated at 26°C for 2 to 4 days for further experiments. The primer sets used are shown in table S3.
Gene silencing by artificial piRNAs
Gene silencing in OSCs by expressing artificial piRNAs was performed essentially as previously described (30). In brief, genomic DNA of target genes was amplified by PCR and subcloned into plasmids downstream of a tj-cis element in a reverse complementary orientation. A 300–base pair fragment was then amplified from the first methionine of each gene. The above derived plasmids (3.6 μg) and pAcBlast (0.4 μg), which expresses the blasticidin resistance gene under control of the actin 5C promoter, were transfected into OSCs (1 × 107 cells) as previously described (41). OSCs and plasmids were suspended in 100 μl of buffer [180 mM sodium phosphate buffer for Church and Gilbert hybridization (pH 7.2), 5 mM KCl, 15 mM MgCl2, 50 mM d-mannitol], and electroporation was performed in an electroporation cuvette with a 2-mm gap using program N-020 of a Nucleofector 2b device (Lonza Bioscience). After incubation for 24 hours, blasticidin was added. Cells were further incubated for 24 hours and then harvested. For long-term (96 hours) expression of artificial piRNAs, plasmids (3.6 μg) and pAcBlast (0.4 μg) were added using Xfect Transfection Reagent (Clontech) after incubation for 48 hours. After incubation for 24 hours, blasticidin was added. Cells were incubated for a further 24 hours and then harvested. Target gene expression levels were quantitatively measured by real-time PCR. The production of artificial piRNAs was confirmed by northern blotting using a DNA oligo probe against the piRNAs (30). Target genes were selected on the basis of RNA-seq data and ChIP-seq data. Target genes were selected from genes with FPKM (fragments per kilobase of exon per million mapped reads) values in control OSCs higher than 100. The Brm-dependent genes were selected under the following conditions: 0.8 < FPKM [Piwi KD (knock-down)]/FPKM (EGFP KD) < 1.2, FPKM (Brm KD)/FPKM (EGFP KD) < 0.5, and 1 < RPKM (Brm-, PB-, Osa-ChIP)/RPKM (input). The Brm-independent genes were selected under the following conditions: 0.8 < FPKM (Piwi KD)/FPKM (EGFP KD) < 1.2, 0.8 < FPKM (Brm KD)/FPKM (EGFP KD) < 1.2, and 1 > RPKM (Brm-, PB-, Osa-ChIP)/RPKM (input). The primer sets and DNA oligos used are shown in table S3.
Tethering assay
A tethering assay system was established in OSCs as previously described (15). For luciferase-reporter plasmids, we replaced the actin 5C promoter of pAc-Fluc-10boxB with a Hsp70 promoter or a CG14072 promoter at Bgl II and Kpn I sites. The Hsp70 promoter sequence was obtained from pBS-Hsp70-Cas9 (Addgene plasmid number 46294). The CG14072 genomic region from its 5′UTR to 1 kb upstream of its TSS was defined as the CG14072 promoter in this study. The CG14072 promoter sequence was obtained from OSC genomic DNA. These plasmids (4.0 μg) and pAcBlast (0.04 μg) were transfected into OSCs (3.0 × 106 cells) using Xfect Transfection Reagent (Clontech). After incubation for 24 hours, blasticidin was added (final concentration, 50 ng/μl), and incubation continued for another week. A small number of cells were transferred to new OSC medium with blasticidin, and the incubation continued for another week for single colony formation. Single colonies were individually transferred to new OSC medium without blasticidin, and incubation continued until the cells proliferated to sufficient quantities for subsequent use. Cell lines expressing luciferase and lacking blasticidin resistance were selected for tethering analysis using these cell lines. To yield λN-HA–fused Piwi and Gtsf1-expressing plasmids, we replaced the panx cDNA sequence of pAc-λN-HA-Panx (15) with cDNAs of respective genes using NEBuilder HiFi DNA Assembly (NEB). To yield λN-HA-Mael–expressing plasmid, a λN-HA cDNA was inserted into Nhe I site of pAcM-Mael (36). For tethering assays, luciferase-expressing OSCs (3 × 106 cells) were transfected with λN-HA fusion protein expression plasmids (3.6 μg) and pAcBlast (0.4 μg) as previously described in the artificial piRNA assay. After incubation for 24 hours, blasticidin was added. After incubation for 24 hours, cells were harvested and lysed in 150 μl of Glo Lysis Buffer (Promega). For long-term (96 hours) tethering of λN-HA fusion proteins, the same procedure was repeated after incubation for 48 hours. Firefly luciferase activities of cell lysates were measured using the ONE-Glo Ex Luciferase Assay System (Promega) and the GloMax-Multi Detection System (Promega). The luciferase activities were normalized against total protein concentration of cell lysates measured by the Bradford assay (Bio-Rad). The primer sets and DNA oligos used are shown in table S3.
Cell fractionation
OSCs were suspended in hypotonic buffer [10 mM Hepes KOH (pH 7.3), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol (DTT), pepstatin A (2 μg/ml), leupeptin (2 μg/ml), and 0.5% aprotinin] by pipetting gently five to six times and then passing through a 25-gauge needle three times. Nuclei were collected by centrifugation at 400g for 10 min at 4°C. The nuclear fraction was washed three to four times with hypotonic buffer and resuspended in chromatin co-immunoprecipitation (co-IP) buffer [50 mM Hepes KOH (pH 7.3), 200 mM KCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, pepstatin A (2 μg/ml), leupeptin (2 μg/ml), and 0.5% aprotinin]. The nuclear fraction was then sonicated on ice using a Branson Digital Sonifier and then centrifuged at 20,000g for 20 min at 4°C. The supernatant was collected as nuclear extract for further experiments.
Immunoprecipitation
Immunoprecipitation of Piwi and Mael nuclear complexes from OSC nuclear extracts was performed using anti-Piwi (40) and anti-Mael (42) antibodies in chromatin co-IP buffer. In brief, nuclear extract prepared from 2 × 108 OSCs was incubated with 5 μg of purified anti-Piwi or anti-Mael antibody in chromatin co-IP buffer (protein concentration was approximately 2 mg/ml) for 2 hours at 4°C with rotation. Then, Dynabeads Protein G (Thermo Fisher Scientific) was added, and incubation continued at 4°C for 1 hour with rotation. The bead fractions were then washed five times using the same buffer, and the protein complexes were eluted from the beads with sample buffer containing SDS by heating for 10 min at 70°C and then loaded onto SDS–polyacrylamide gel electrophoresis gels. After electrophoresis, proteins were visualized by silver staining using SilverQuest (Invitrogen) or processed for Western blot analysis.
Shotgun mass spectrometric analysis
Co-IP of Piwi and Mael was performed with antibodies cross-linked to beads by dimethyl pimelimidate (Thermo Fisher Scientific). Immunoprecipitation using Piwi and Mael KD OSCs was also performed to provide negative controls. The immunoprecipitants were eluted in elution buffer containing 125 mM tris-HCl (pH 6.8), 4% SDS, and 0.01% bromophenol blue by heating for 10 min at 70°C. The eluted immunoprecipitants were precipitated in acetone containing 20% trichloroacetic acid and 20 mM DTT. The same procedure was repeated twice more for each immunoprecipitation. Before digestion, BSA was added in an equal amount to each immunoprecipitant. After alkylation in iodoacetamide solution for 45 min at room temperature with shielding from light, the proteins were concentrated by chloroform/methanol precipitation and then digested using Trypsin Gold (Promega) at 37°C overnight. An LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with a nanoLC interface (AMR) was used for peptide separation and identification. The data were compared against the UniProt protein sequence database of Drosophila melanogaster using protein identification in the search program Proteome Discoverer 1.4 (Thermo Fisher Scientific). The Sum PEP Score for each protein was normalized to the mean value of BSA spiked into all experimental samples. The P value of the normalized Sum PEP Scores relative to negative controls was calculated using the Student’s t test, and then the q value was calculated by the Benjamini-Hochberg procedure. Note that proteins with score 0 were omitted, restating that only proteins detected in all three experiments were used. The fold change was calculated by dividing the mean value of the normalized Sum PEP Score +1 by the value of the negative control Sum PEP Score +1. To screen candidates for Piwi/Mael interactors, proteins with more than twofold changes and q values <0.05 were listed (Fig. 1D and table S1).
Selection of candidate proteins using gene ontology
We identified 159 and 92 proteins as potential Piwi and Mael interactors, respectively (Fig. 1D). Of these, 34 proteins overlapped, on which functional annotation clustering was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (classification stringency: high). The analysis revealed that 6 of the 34 proteins were related to transcription (Transcription, Transcription regulation, GO:0006355 ~ regulation of transcription, DNA-templated, GO:0006351 ~ transcription, DNA-templated).
ChIP for OSCs
To cross-link protein and DNA complexes, OSCs (1 × 108 cells) were incubated with formaldehyde directly added to the OSC medium to a final concentration of 0.75% for 10 or 15 min at room temperature (10 min for Brm, PB, H3K9me3, and Pol II and 15 min for Osa). Glycine was then added to a final concentration of 125 mM to stop cross-linking. Cells were rinsed with phosphate-buffered saline (PBS) twice and then harvested by thorough scraping. Cells were suspended in a swelling buffer containing 25 mM Hepes KOH (pH 7.3), 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 0.1% NP-40, pepstatin A (2 μg/ml), leupeptin (2 μg/ml), and 0.5% aprotinin by gently pipetting up and down five to six times. After inculcation on ice for 10 min, OSC nuclei were obtained by centrifugation at 1000g for 5 min at 4°C. For H3K9me3-, Brm-, and PB-ChIP, the nuclei were lysed in sonication buffer containing 50 mM Hepes KOH (pH 7.3), 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, pepstatin A (2 μg/ml), leupeptin (2 μg/ml), and 0.5% aprotinin for 20 min at 4°C, and then EDTA was added to a final concentration of 10 mM. The nuclei were lysed using a Covaris S220 focused-ultrasonicator for 10 min at 4°C with peak power 140, duty factor 5.0, and 200 cycle/burst. After centrifugation at 20,000g for 20 min at 4°C, the supernatant was collected. For Osa-ChIP, the nuclei were lysed with micrococcal nuclease (a final concentration of 2000 gel units/μl) in digestion buffer containing 50 mM Hepes KOH (pH 7.3), 140 mM NaCl, 40 mM MgCl2, 10 mM CaCl2, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, pepstatin A (2 μg/ml), leupeptin (2 μg/ml), and 0.5% aprotinin for 20 min at 4°C, and then EDTA was added to a final concentration of 10 mM. Next, the lysate was sonicated with a Branson Digital Sonifier (10% amplitude, pulse width 10 s ON and 50 s OFF, six times on ice). After centrifugation at 20,000g for 20 min at 4°C, the supernatant was collected. For immunoprecipitation, the supernatant was mixed by rotation with antibodies at 4°C overnight and then mixed with Dynabeads Protein G by rotation at 4°C for 1 hour. An anti–Pol II RPB1 antibody (BioLegend 664906) and an anti-H3K9me3 antibody (Active motif 61013) were used. The anti-Brm and anti-PB antibodies were a gift from S. Hirose (Division of Gene Expression, Department of Developmental Genetics, National Institute of Genetics) (43). The anti-Osa antibody was obtained from the Developmental Studies Hybridoma Bank. The beads were washed sequentially with low-salt wash buffer [20 mM tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, 0.1% SDS, and 1 mM phenylmethylsulfonyl fluoride (PMSF)], high-salt wash buffer [20 mM tris-HCl (pH 8.0), 500 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, 0.1% SDS, and 1 mM PMSF], LiCl wash buffer [20 mM tris-HCl (pH 8.0), 200 mM LiCl, 2 mM EDTA, 1% NP-40, 1% sodium deoxycholate, and 1 mM PMSF], and lastly with TE wash buffer [20 mM tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM PMSF] twice. DNA was eluted by adding 200 μl of elution buffer [50 mM tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS] to the beads. After incubation for 30 min at 65°C, the supernatant was collected, mixed with 4.8 μl of 5 M NaCl and 2 μl of ribonuclease (RNase) A (10 mg/ml), and incubated with gentle agitation at 65°C overnight. The next day, it was further mixed with 2 μl of proteinase K (20 mg/ml) and incubated with shaking at 60°C for 1 hour. The DNA was purified by phenol:chloroform extraction. The DNA levels were quantified by real-time PCR or sequencing. In Fig. 3 (C to E), the occupancy of Pol II, Brm, and H3K9me3 at the CG14072 and CG34330 regions was normalized by their occupancy at the CG34330 and CG14072 regions, respectively. ChIP-seq libraries were prepared using a NEBNext Ultra II FS DNA Library Prep Kit for Illumina (NEB) and an SMARTer Thruplex DNA-seq Kit (Clontech) according to the manufacturers’ instructions. The primer sets and the DNA oligos used are shown in table S3.
ChIP for ovaries
Ovaries with the mutant allelic combination maelM391/Df(3L)BSC554 were used (described as maelM391/Df). maelM391/TM3 ovaries were used as a control (described as WT). To cross-link protein and DNA complexes, 40 to 30 ovaries were incubated with 0.75% formaldehyde in PBS for 10 min at room temperature. Cells were rinsed with PBS twice. Cells were suspended in a swelling buffer containing 25 mM Hepes KOH (pH 7.3), 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 0.1% NP-40, pepstatin A (2 μg/ml), leupeptin (2 μg/ml), and 0.5% aprotinin by gently pipetting up and down 10 to 15 times and were then passed through a 25-gauge needle three times. After inculcation on ice for 10 min, ovarian nuclei were obtained by centrifugation at 1000g for 5 min at 4°C. The nuclei were lysed using a Covaris S220 focused ultrasonicator with peak power 140, duty factor 5.0, and 200 cycle/burst for 20 min at 4°C in sonication buffer containing 50 mM Hepes KOH (pH 7.3), 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, pepstatin A (2 μg/ml), leupeptin (2 μg/ml), and 0.5% aprotinin. EDTA was then added to a final concentration of 10 mM. After centrifugation at 20,000g for 20 min at 4°C, the supernatant was collected. For immunoprecipitation, the supernatant was mixed by rotation with antibodies at 4°C overnight and then mixed with Dynabeads Protein G by rotation at 4°C for 1 hour. The anti-Brm antibody was kindly provided by S. Hirose (Division of Gene Expression, Department of Developmental Genetics, National Institute of Genetics) (43). The beads were washed sequentially with low-salt wash buffer [20 mM tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, 0.1% SDS, and 1 mM PMSF], high-salt wash buffer [20 mM tris-HCl (pH 8.0), 500 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, 0.1% SDS, and 1 mM PMSF], LiCl wash buffer [20 mM tris-HCl (pH 8.0), 200 mM LiCl, 2 mM EDTA, 1% NP-40, 1% sodium deoxycholate, and 1 mM PMSF], and lastly twice with TE wash buffer [20 mM tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM PMSF]. DNA was eluted by adding 200 μl of elution buffer [50 mM tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS] to the beads. After incubation for 30 min at 65°C, the supernatant was collected, mixed with 4.8 μl of 5 M NaCl and 2 μl of RNase A (10 mg/ml), and incubated with gentle agitation at 65°C overnight. The next day, it was further mixed with 2 μl of proteinase K (20 mg/ml) and incubated with shaking at 60°C for 1 hour. The DNA was purified by phenol:chloroform extraction. ChIP-seq libraries were prepared using a NEBNext Ultra II FS DNA Library Prep Kit for Illumina (NEB) according to the manufacturer’s instructions.
Western blotting
Western blotting was performed essentially as described previously (44). Production of anti-Piwi, anti-Mael, anti-Gtsf1, anti-Panx, and anti-Egg antibodies was described previously (10, 15, 40, 42). The anti-Brm antibody was a gift from L. Zhang (State Key Laboratory of Cell Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences). Anti-Myc (Sigma-Aldrich, C3956) and Histone H3 (Abcam) antibodies were used. Purification of antibodies from the culture supernatant of hybridoma cells was performed using Thiophilic-Superflow Resin (BD Biosciences). The anti–β-tubulin antibody was obtained from the Developmental Studies Hybridoma Bank (DSHB Hybridoma Product E7). Anti-mouse immunoglobulin G (IgG), horseradish peroxidase (HRP)–linked antibody (MP Biomedicals, 55558), and anti-rabbit IgG, HRP-linked antibody (Cell Signaling Technology) were purchased from the manufacturers.
RNA isolation and real-time PCR
Total RNAs were isolated using ISOGEN II (Nippon Gene) according to the manufacturer’s instructions. cDNAs were prepared using ReverTra Ace (Toyobo) according to the manufacturer’s instructions. Real-time PCR was performed as previously described (44). In brief, cDNAs or DNA fragments were amplified with StepOnePlus (Applied Biosystems) using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). The primer sets used are shown in table S3. The amplification efficiency of a quantitative PCR was calculated on the basis of the slope of the standard curve. After confirming amplification efficiency values (between 95 and 105%), relative steady-state RNA levels were determined from the threshold cycle for amplification.
Bioinformatic analysis of small RNA-seq, RNA-seq, and ChIP-seq
Adapter-trimmed sequences were mapped to the D. melanogaster genome assembly release 6 (dm6) by bowtie2 (ver. 2.2.4) using default parameters. Mapped reads were further mapped to the transcriptome, which consisted of gene and transposon sequences, including the LTR sequences, by bowtie2, and then FPKM values were calculated. The dm6 genome and transcriptome sets were downloaded through piPipes (45). Read counts corresponding to each genomic and genic position were obtained by generating bedgraph files from BAM files (binary version of SAM files) using BEDTools genomecov. All samples were normalized to have equivalent reads per million using the “-scale” option. Genes and transposons under detection were excluded in subsequent analyses. A small RNA-seq library for Piwi-bound piRNAs (30) in OSCs was used. RNA-seq libraries for control and mael mutant ovaries (SRA: PRJNA448445) were used (19). All transposons selected for statistical analysis have FPKM values in control OSCs higher than 1.0. The top 50 Brm-dependent genes were selected from genes whose FPKM values in control OSCs were higher than 1.0. For statistical analysis and data visualization, R packages implemented in R 3.2.1 were used. P values were calculated using the Wilcoxon rank sum test. For bar graphs, P values were calculated using the t test.
Accession numbers
Deep sequencing datasets have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database and are available under accession number GEO: GSE108329.
Supplementary Material
Acknowledgments
We thank A. Takahashi, K. Osumi, M. Horikoshi, and M. Ariura for technical assistance; T. Sumiyoshi, Y.W. Iwasaki, S. Hirakata, and H. Yoshitane for advice on bioinformatic analyses; and members of the Siomi laboratories, particularly Y. W. Iwasaki and S. Yamanaka, for insightful comments on the manuscript. We also thank L. Zhang and S. Hirose for sharing antibodies and K. Shirahige for advice on ChIP. Funding: This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K.S. (20K06596), K.M. (20H03439), H.S. (25221003), and M.C.S. (19H05466). Author contributions: R.O., K.S., K.M., H.S., and M.C.S. conceived the project, designed the experiments, and wrote the manuscript. R.O., K.S., and K.M. performed experiments and bioinformatic analyses. L.N. performed shotgun mass spectrometric analysis. All authors analyzed data and contributed to the preparation of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Correspondence and requests for materials should be addressed to M.C.S.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eaaz7420/DC1
REFERENCES AND NOTES
- 1.Ozata D. M., Gainetdinov I., Zoch A., O’Carroll D., Zamore P. D., PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 20, 89–108 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki Y. W., Siomi M. C., Siomi H., Piwi-interacting RNA: Its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Yamashiro H., Siomi M. C., PIWI-interacting RNA in Drosophila: Biogenesis, transposon regulation, and beyond. Chem. Rev. 118, 4404–4421 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Czech B., Hannon G. J., One loop to rule them all: The ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 41, 324–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schüpbach T., Wieschaus E., Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129, 1119–1136 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klattenhoff C., Bratu D. P., McGinnis-Schultz N., Koppetsch B. S., Cook H. A., Theurkauf W. E., Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45–55 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Yu Y., Gu J., Jin Y., Luo Y., Preall J. B., Ma J., Czech B., Hannon G. J., Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350, 339–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sienski G., Batki J., Senti K.-A., Dönertas D., Tirian L., Meixner K., Brennecke J., Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev. 29, 2258–2271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dönertas D., Sienski G., Brennecke J., Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 27, 1693–1705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtani H., Iwasaki Y. W., Shibuya A., Siomi H., Siomi M. C., Saito K., DmGTSF1 is necessary for Piwi–piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 27, 1656–1661 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sienski G., Dönertas D., Brennecke J., Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki Y. W., Murano K., Ishizu H., Shibuya A., Iyoda Y., Siomi M. C., Siomi H., Saito K., Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol. Cell 63, 408–419 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Huang X. A., Yin H., Sweeney S., Raha D., Snyder M., Lin H., A major epigenetic programming mechanism guided by piRNAs. Dev. Cell 24, 502–516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabry M. H., Ciabrelli F., Munafò M., Eastwood E. L., Kneuss E., Falciatori I., Falconio F. A., Hannon G. J., Czech B., piRNA-guided co-transcriptional silencing coopts nuclear export factors. eLife 8, e47999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murano K., Iwasaki Y. W., Ishizu H., Mashiko A., Shibuya A., Kondo S., Adachi S., Suzuki S., Saito K., Natsume T., Siomi M. C., Siomi H., Nuclear RNA export factor variant initiates piRNA-guided co-transcriptional silencing. EMBO J. 38, e102870 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batki J., Schnabl J., Wang J., Handler D., Andreev V. I., Stieger C. E., Novatchkova M., Lampersberger L., Kauneckaite K., Xie W., Mechtler K., Patel D. J., Brennecke J., The nascent RNA binding complex SFiNX licenses piRNA-guided heterochromatin formation. Nat. Struct. Mol. Biol. 26, 720–731 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao K., Cheng S., Miao N., Xu P., Lu X., Zhang Y., Wang M., Ouyang X., Yuan X., Liu W., Lu X., Zhou P., Gu J., Zhang Y., Qiu D., Jin Z., Su C., Peng C., Wang J.-H., Dong M.-Q., Wan Y., Ma J., Cheng H., Huang Y., Yu Y., A Pandas complex adapted for piRNA-guided transcriptional silencing and heterochromatin formation. Nat. Cell Biol. 21, 1261–1272 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Osumi K., Sato K., Murano K., Siomi H., Siomi M. C., Essential roles of Windei and nuclear monoubiquitination of Eggless/SETDB1 in transposon silencing. EMBO Rep. 20, e48296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang T. H., Mattei E., Gainetdinov I., Colpan C., Weng Z., Zamore P. D., Maelstrom represses canonical polymerase II transcription within bi-directional piRNA clusters in Drosophila melanogaster. Mol. Cell 73, 291–303.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., Siomi M. C., A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296–1299 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Saito K., Ishizu H., Komai M., Kotani H., Kawamura Y., Nishida K. M., Siomi H., Siomi M. C., Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24, 2493–2498 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brehm A., Längst G., Kehle J., Clapier C. R., Imhof A., Eberharter A., Müller J., Becker P. B., dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19, 4332–4341 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mugat B., Nicot S., Varela-Chavez C., Jourdan C., Sato K., Basyuk E., Juge F., Siomi M. C., Pélisson A., Chambeyron S., The Mi-2 nucleosome remodeler and the Rpd3 Histone deacetylase are involved in piRNA-guided heterochromatin formation. Nat. Commun. 11, 2818 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ninova M., Chen Y.-C. A., Godneeva B., Rogers A. K., Luo Y., Fejes Tóth K., Aravin A. A., Su(var)2-10 and the SUMO pathway link piRNA-guided target recognition to chromatin silencing. Mol. Cell 77, 556–570.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarot E., Payen-Groschêne G., Bucheton A., Pélisson A. E., Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166, 313–321 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pélisson A., Payen-Groschêne G., Terzian C., Bucheton A., Restrictive flamenco alleles are maintained in Drosophila melanogaster population cages, despite the absence of their endogenous gypsy retroviral targets. Mol. Biol. Evol. 24, 498–504 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Schuettengruber B., Martinez A.-M., Iovino N., Cavalli G., Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Fry C. J., Peterson C. L., Chromatin remodeling enzymes: Who’s on first? Curr. Biol. 11, 185–197 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Elfring L. K., Danie L. C., Papoulas O., Deuring R., Sarte M., Moseley S., Beek S. J., Waldrip W. R., Daubresse G., DePace A., Kennison J. A., Tamkun J. W., Genetic analysis of brahma: The Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148, 251–265 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishizu H., Iwasaki Y. W., Hirakata S., Ozaki H., Iwasaki W., Siomi H., Siomi M. C., Somatic primary piRNA biogenesis driven by cis-acting RNA elements and trans-acting Yb. Cell Rep. 12, 429–440 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Papoulas O., Beek S. J., Moseley S. L., McCallum C. M., Sarte M., Shearn A., Tamkun J. W., The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125, 3955–3966 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Moshkin Y. M., Mohrmann L., van Ijcken W. F., Verrijzer C. P., Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol. Cell. Biol. 27, 651–661 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dingwall A. K., Beek S. J., McCallum C. M., Tamkun J. W., Kalpana G. V., Goff S. P., Scott M. P., The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell 6, 77–91 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baron-Benhamou J., Gehring N. H., Kulozik A. E., Hentze M. W., Using the lambdaN peptide to tether proteins to RNAs. Methods Mol. Biol. 257, 135–154 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Armstrong J. A., Papoulas O., Daubresse G., Sperling A. S., Lis J. T., Scott M. P., Tamkun J. W., The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 21, 5245–5254 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto N., Sato K., Nishimasu H., Namba Y., Miyakubi K., Dohmae N., Ishitani R., Siomi H., Siomi M. C., Nureki O., Crystal structure and activity of the endoribonuclease domain of the piRNA pathway factor Maelstrom. Cell Rep. 11, 366–375 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Tréand C., du Chéné I., Brès V., Kiernan R., Benarous R., Benkirane M., Emiliani S., Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 25, 690–1699 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemasson I., Polakowski N. J., Laybourn P. J., Nyborg J. K., Tax-dependent displacement of nucleosomes during transcriptional activation of human T-cell Leukemia virus type 1. J. Biol. Chem. 281, 3075–3082 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Iba H., Mizutani T., Ito T., SWI/SNF chromatin remodelling complex and retroviral gene silencing. Rev. Med. Virol. 13, 99–110 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Saito K., Nishida K. M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M. C., Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirakata S., Ishizu H., Fujita A., Tomoe Y., Siomi M. C., Requirements for multivalent Yb body assembly in transposon silencing in Drosophila. EMBO Rep. 2019, e47708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato K., Nishida K. M., Shibuya A., Siomi M. C., Siomi H., Maelstrom coordinates microtubule organization during Drosophila oogenesis through interaction with components of the MTOC. Genes Dev. 25, 2361–2373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama T., Shimojima T., Hirose S., The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development 139, 4582–4590 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Sato K., Iwasaki Y. W., Siomi H., Siomi M. C., Tudor-domain containing proteins act to make the piRNA pathways more robust in Drosophila. Fly 9, 86–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han B. W., Wang W., Zamore P. D., Weng Z., piPipes: A set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq and genomic DNA sequencing. Bioinformatics 31, 593–595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eaaz7420/DC1


