3D high-resolution live imaging reveals the importance of ceramide chain length for protein sorting in selective export sites.
Abstract
Protein sorting in the secretory pathway is crucial to maintain cellular compartmentalization and homeostasis. In addition to coat-mediated sorting, the role of lipids in driving protein sorting during secretory transport is a longstanding fundamental question that still remains unanswered. Here, we conduct 3D simultaneous multicolor high-resolution live imaging to demonstrate in vivo that newly synthesized glycosylphosphatidylinositol-anchored proteins having a very long chain ceramide lipid moiety are clustered and sorted into specialized endoplasmic reticulum exit sites that are distinct from those used by transmembrane proteins. Furthermore, we show that the chain length of ceramide in the endoplasmic reticulum membrane is critical for this sorting selectivity. Our study provides the first direct in vivo evidence for lipid chain length–based protein cargo sorting into selective export sites of the secretory pathway.
INTRODUCTION
In eukaryotic cells, proteins synthesized in the endoplasmic reticulum (ER) are subsequently sorted during transport through the secretory pathway for delivery to their proper cellular destinations (1). In addition to coat-mediated sorting, it has long been postulated that certain lipids could also sort specific proteins by clustering them into specialized membrane domains that act as selective export sites (2–5). However, direct in vivo evidence for this possible lipid-based mechanism is still lacking. To address this fundamental issue, we have investigated in yeast how glycosylphosphatidylinositol (GPI)–anchored proteins (GPI-APs), a diverse class of lipid-linked cell surface proteins, are differentially exported from the ER (6, 7). GPI-APs are secretory proteins attached by a glycolipid moiety (GPI anchor) to the external leaflet of the plasma membrane. They receive the GPI anchor as a conserved posttranslational modification in the ER lumen (8). After attachment, GPI-APs travel from the ER to the plasma membrane via the Golgi apparatus (5, 9). The presence of the GPI anchor leads GPI-APs to be trafficked separately from transmembrane secretory proteins, including other plasma membrane proteins, along the secretory pathway (5, 9, 10). In yeast cells, GPI-APs are separated from other secretory proteins in the ER to be subsequently packaged into distinct coat protein complex II (COPII)–coated vesicles (6, 7). The determinants of this sorting process during ER export are unknown, but it has been speculated that this mechanism might require lipids and, specifically, the structural remodeling of the lipid moiety of the GPI anchor (5, 8). In yeast, GPI-lipid remodeling begins immediately after GPI attachment and, in many cases, leads to the incorporation of a ceramide with a very long-chain saturated fatty acid of 26 carbons (C26:0) (11, 12). C26 ceramide, which is by far the major ceramide produced by yeast cells, is synthesized in the ER and mostly exported to the Golgi by COPII vesicles (13). The ongoing ceramide synthesis is specifically required for the ER exit of GPI-APs (14, 15), and, in turn, ceramide conversion to inositolphosphoceramide (IPC) at the Golgi depends on the GPI anchor synthesis (16). Biophysical studies with artificial membranes suggest that very long acyl chain ceramides can coalesce to form ordered domains with unique physical properties (17, 18). These data have led to the hypothesis that C26 ceramides and GPI-APs with C26 ceramides take advantage of their physical properties to coalesce into ordered domains or zones in the relatively disordered lipid environment of the ER membrane, which is composed of mostly shorter and unsaturated glycerolipids (C16:1 and C18:1) (19, 20). These zones would be selectively concentrated at specific ER exit sites (ERES) from where ceramides and ceramide-based GPI-APs can be cotransported to the Golgi in the same specialized COPII vesicles (5).
In this study, we have directly tested this hypothetical lipid-based mechanism by using superresolution confocal live imaging microscopy (SCLIM), a cutting-edge microscopy technology that allows simultaneous three-color and three-dimensional (3D) observation of fluorescent-tagged proteins with extremely high resolution and high speed in living cells (21, 22).
RESULTS
Newly synthesized C26 ceramide–based GPI-APs segregate from transmembrane cargos by forming clusters associated with specific ERES
We first applied the SCLIM technology to further define how normal GPI-APs having C26 ceramide moieties are sorted from transmembrane secretory proteins upon exit from the ER in the yeast Saccharomyces cerevisiae. To examine ER sorting, we used a genetic system that allows direct in vivo visualization of the entry of newly synthesized cargos into ERES (7, 23). As cargos, we chose the C26 ceramide–based GPI-AP Gas1 tagged with green fluorescent protein (GFP) and the transmembrane secretory protein Mid2 tagged with near infra-red fluorescent protein (iRFP), both of which are targeted to the plasma membrane (24–26). These two cargos were expressed under a galactose-inducible promoter, together with a constitutive ERES marker in the sec31-1 temperature-sensitive mutant strain. At the restrictive temperature (37°C), the newly synthesized cargos accumulate at the ER because sec31-1 mutation affects the function of the COPII outer coat component Sec31 inhibiting COPII budding and ER export (23). Upon shift-down to low temperature (24°C), sec31-1 mutant cells are restored from the secretory block, and the accumulated newly synthesized cargo starts export from the ER. CLIM visualization showed that after incubation at 37°C and subsequent release at 24°C for 5 min, most of newly synthesized Gas1-GFP and Mid2-iRFP still accumulated in the ER of sec31-1 mutant cells (Fig. 1). Their distribution patterns were quite different since Mid2-iRFP was distributed throughout the ER membrane, and Gas1-GFP was concentrated and clustered into discrete ER membrane zones (Fig. 1, A to C, and movie S1). Furthermore, as shown in Fig. 1D, Gas1-GFP clusters were devoid of Mid2-iRFP. These results indicate that GPI-APs and transmembrane proteins are segregated early into different ER membrane zones. Gas1-GFP clusters were adjacent to specific ERES labeled with the COPII outer coat protein Sec13 tagged with mCherry (Fig. 1, E and F, and movie S1) (23).
Fig. 1. Newly synthesized C26 ceramide–based GPI-AP cargos form clusters in the ER membrane adjacent to specific ERES.
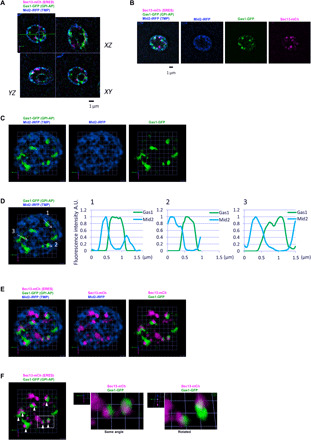
sec31-1 cells expressing galactose-inducible secretory cargos, the very long acyl chain (C26) ceramide GPI-AP Gas1-GFP (GPI-AP, green) and the transmembrane protein Mid2-iRFP (TMP, blue), and constitutive ERES marker Sec13-mCherry (ERES, magenta) were incubated at 37°C for 30 min, shifted down to 24°C and imaged by SCLIM after 5 min. (A to C) Representative merged or individual 2D images of one plane (A), 2D projection images of 10 z-sections (B), or 3D cell hemisphere images (C) of cargo and ERES markers are shown. Scale bar, 1 μm (A and B). Scale unit, 0.551 μm (C). Gas1-GFP was detected in discrete ER zones or clusters, whereas Mid2-iRFP was detected and distributed throughout the ER membrane (C). (D) Graphs show relative fluorescence intensities of Gas1-GFP and Mid2-iRFP along the white arrow lines in the Gas1-GFP clusters (left). A.U., arbitrary units. (E and F) Representative merged 3D images of cargo and ERES markers. Gas1-GFP clusters were detected adjacent to specific ERES. Scale unit, 0.551 μm. (F) The white filled arrowheads mark Gas1-GFP clusters associated with ERES. Middle and right panels show a merged enlarged 3D image and rotated view of selected Gas1-GFP clusters.
Segregated C26 ceramide–based GPI-APs and transmembrane cargos are sorted into different ERES
The close spatial association between Gas1-GFP clusters and specific ERES suggests that Gas1-GFP could enter into selective ERES different from the ones used by Mid2-iRFP to exit the ER. To address this possibility, we quantified the proportion of ERES in which only one type of cargo or both are present (Fig. 2, A to C). We found that most of the ERES (70%) exclusively contained only one type of cargo. Lower panels of Fig. 2C show two typical examples of ERES with only Gas1-GFP (panel 1) or only Mid2-iRFP (panel 2). By contrast, approximately 20% of ERES contained both cargos overlapping in the same area. Some ERES (10%) were found containing both cargos, but they segregated in clearly distinct zones. Therefore, this statistical analysis shows that the GPI-AP Gas1-GFP is sorted into different ERES from the transmembrane cargo Mid2-iRFP upon ER exit (Fig. 2D). This sorting efficiency agrees well with previous biochemical (6) and morphological determinations (7). We could also observe the behavior of segregated cargos during their entry in ERES (Fig. 2E and movie S2). Figure 2E shows that only a small portion of Gas1-GFP (panel 3) or Mid2-iRFP (panel 4) enters the ERES from one side and is confined in a discrete zone. Panel 5 of Fig. 2E shows that Gas1-GFP and Mid2-iRFP were occasionally found in the same ERES but entered from different sides and concentrated in separated zones that might represent distinct COPII vesicles. We also confirmed that the observed segregation and sorting of the C26 ceramide–based GPI-AP Gas1 into selective ERES are specific since another transmembrane secretory cargo, the plasma membrane protein Axl2 tagged with GFP (27), showed a similar behavior to Mid2-iRFP (fig. S1 and movie S3). Newly synthesized Axl2-GFP distributed through the ER membrane like Mid2-iRFP (fig. S1, A and B) and colocalized with Mid2-iRFP in the majority of ERES (fig. S1, B to D). Panels 1 and 2 of fig. S1C show two typical examples of ERES with both transmembrane cargos overlapping. In these cases, both cargos entered together into the ERES (fig. S1E, panel 3, and movie S3).
Fig. 2. Newly synthesized C26 ceramide–based GPI-APs and transmembrane cargos are recruited to different ERES.
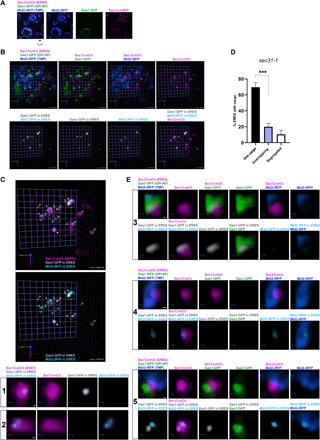
sec31-1 cells expressing galactose-inducible secretory cargos, Gas1-GFP (GPI-AP, green) and Mid2-iRFP (TMP, blue), and constitutive ERES marker Sec13-mCherry (ERES, magenta) were incubated at 37°C for 30 min, shifted down to 24°C for releasing secretion block, and imaged by SCLIM after 20 min. (A to C) Representative 2D projection images of 10 z-sections (A; scale bar, 1 μm) or 3D cell hemisphere images (B and C; scale unit, 0.456 μm) of cargo and ERES markers. Lower panels in (B) and panels in (C) show processed images to display only the cargo [Gas1-GFP (gray) and Mid2-iRFP (light blue)] present in the ERES (magenta). (C) Open arrowheads: ERES with just one cargo (1 to 4). Gray filled arrowheads: ERES containing segregated cargos (5). White filled arrowhead: ERES containing colocalizing cargos. Lower panels: selected individual ERES containing only Gas1-GFP (1) or Mid2-iRFP (2). Scale bar, 100 nm. (D) Quantification of micrographs described in (C). Average percentage of ERES containing only one cargo (Gas1-GFP or Mid2-iRFP), segregated cargos, and overlapping cargos. n = 432 in 54 cells in three independent experiments. Error bars = SD. Two-tailed, unpaired t test. ***P = 0.0002. (E) 3D images of selected ERES with segregated cargos marked in (C). Either Gas1-GFP (green) (3) or Mid2-iRFP (blue) (4) enters ERES (magenta) from one side and is confined in a small zone within the ERES. Occasionally, both cargos approach the same ERES (5) but from different sides and are restricted to segregated zones within the ERES. Scale bar, 100 nm.
C26 ceramide–based GPI-AP cargos are not clustered and enter the same ERES as transmembrane cargo in the ER membrane with shorter C18-C16 ceramide
We next tested the hypothesis that the very long acyl chain ceramide (C26) present in the ER membrane drives the specific clustering and sorting of Gas1 into selective ERES. For this purpose, we used a modified yeast strain GhLag1, in which the two endogenous ceramide synthases Lag1 and Lac1 were replaced by GhLag1, the Lag1 homolog from cotton, resulting in a yeast strain that produces cellular membranes with shorter ceramides than the wild-type strain (Fig. 3A) (28). The mass spectrometry (MS) analysis showed that while in wild-type strain, 95% of total ceramide is very long (C26) chain ceramide, in GhLag1, 85% is long (C18 and C16) and only 2% is very long (C26) chain ceramide. Although C18 and C16 ceramides are, by far, the major ceramides detected in GhLag1 membranes, the MS analysis also determined that the GPI anchor of Gas1-GFP expressed in GhLag1 strain contains C26 ceramide, the same lipid as in wild-type (Fig. 3A) (26). Therefore, this implies that the ceramide remodelase enzyme Cwh43 is highly selective for C26 ceramide, which is preferentially incorporated into the GPI anchors from the very small amount of C26 ceramide in GhLag1 strain, as shown in fig. S2 (29). Nevertheless, the fact that cellular membranes of GhLag1 contain essentially only C18-C16 ceramide whereas Gas1-GFP still has C26 ceramide makes this strain an ideal tool to specifically address the hypothetical role of the acyl chain length of membrane ceramides in ER clustering and sorting. We then investigated first by conventional fluorescence microscopy the capacity of C26 Gas1-GFP to aggregate into clusters in GhLag1 bearing the sec31-1 temperature-sensitive mutant allele in which only long (C18-C16) chain ceramides exists in the ER membrane (Fig. 3). We observed that whereas in sec31-1, most of Gas1-GFP was concentrated in clusters, Gas1-GFP in sec31-1 GhLag1 with long (C18-C16) chain ceramide ER membrane was mainly unclustered and distributed throughout the ER membrane. Precisely, because C26 ceramide–based clustering occurs intimately associated with specific ERES (Fig. 1), we next investigated whether this process might also involve the function of the ER export protein machinery. GPI-APs use a specialized COPII system for ER export, which is actively regulated by the structural remodeling of the glycan part of the GPI anchor by Ted1 (30, 31). The remodeled GPI-glycan is then recognized by the transmembrane cargo receptor p24 complex, which, in turn, selectively recruits Lst1, a specific isoform of the major COPII cargo binding subunit Sec24 required to form COPII vesicles enriched in GPI-APs (31–33). Thus, we constructed double mutants combining deletions of these individual proteins (the p24 complex component Emp24, the GPI-glycan remodelase Ted1, and the specific COPII subunit Lst1) with the sec31-1 mutant strain and examined whether they can form clusters of Gas1-GFP (Fig. 3). We observed that in both sec31-1 emp24Δ and sec31-1 ted1Δ, Gas1-GFP was mainly unclustered and distributed throughout the ER membrane as seen previously for sec31-1 GhLag1, whereas in sec31-1 lst1Δ, Gas1-GFP was clustered like in sec31-1. These results indicate that, in addition to the presence of C26 ceramide in the ER membrane, clustering of Gas1-GFP requires binding with the p24 complex, but not specific Lst1 recruitment. We then addressed the possibility that the chain length of the ceramide in the ER membrane could regulate the p24 binding of Gas1-GFP. However, we found that the presence of C18-C16 ceramide in the membrane did not affect the ability of the p24 complex to bind and export GPI-APs through their remodeled GPI-glycan (figs. S3 and S4, A and B) or to recruit the COPII subtype Lst1 (fig. S4C). Therefore, C26 ceramide–dependent clustering does not entail differential protein interactions with the ER export protein machinery, supporting instead the alternative, lipid length–driven, sorting mechanism. We then analyzed whether the ceramide acyl chain length in the ER membrane is important for the effective sorting of Gas1-GFP into selective ERES. Since Gas1 in GhLag1 strain with shorter chain ceramide exited the ER toward the plasma membrane (fig. S5), we reasoned that if sorting is driven by ceramide acyl chain length, then Gas1 in GhLag1 strain could then be rerouted toward the same ERES as transmembrane cargo.
Fig. 3. ER clustering of C26 ceramide–based GPI-APs requires the presence of C26 ceramides in the membrane and interaction with the p24 complex receptor through the GPI-glycan.
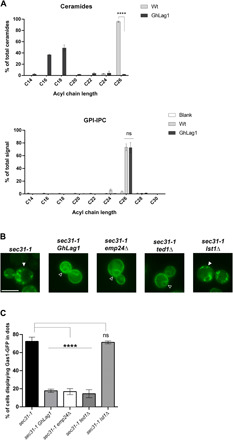
(A) The cellular membranes of GhLag1 mainly contain shorter C18-C16 ceramide, while the GPI anchor of Gas1-GFP still has C26 IPC as in wild-type cells. Upper graph: acyl chain length analysis by mass spectrometry (MS) of ceramides from cellular membranes of wild-type (Wt) and GhLag1p strains. Data represent the percentage of total ceramides. Mean of three independent experiments. Error bars = SD. Two-tailed, unpaired t test. ****P < 0.0001. Lower graph: acyl chain length analysis by MS of IPCs present in the GPI anchor of Gas1-GFP (GPI-IPC) expressed in wild-type and GhLag1p strains. Data represent the percentage of total IPC signals. Mean of five independent experiments. Error bars = SD. Two-tailed, unpaired t test. ns, not significant. P = 0.9134. (B) Fluorescent micrographs of sec31-1, sec31-1 GhLag1, sec31-1 emp24Δ, sec31-1 ted1Δ, and sec31-1 lst1Δ cells expressing galactose-inducible Gas1-GFP were incubated at 37°C for 30 min and visualized by conventional fluorescence microscopy after shifting down to 24°C. White arrowheads: ER Gas1-GFP clusters. Open arrowheads: unclustered Gas1-GFP distributed throughout the ER membrane showing the ER-characteristic nuclear ring staining. Scale bar, 5 μm. (C) Quantification of micrographs described in (B). Average percentage of the cells with dot-like Gas1-GFP structures. n ≥ 300 cells in three independent experiments. Error bars = SD. Two-tailed, unpaired t test. ****P < 0.0001.
To directly address this, we conducted SCLIM visualization of Gas1-GFP and Mid2-iRFP in GhLag1 bearing the sec31-1 temperature-sensitive mutant allele (Fig. 4 and movie S4). After ER retention at 37°C and subsequent release at 24°C, most of newly synthesized Gas1-GFP was unclustered and distributed throughout the ER membrane as observed by conventional microscopy (Fig. 4, A and B). Furthermore, a large percentage of ERES (67%) included both cargos colocalizing within them (Fig. 4D). Panels 1 and 2 of Fig. 4C show two typical examples of ERES with overlapping Gas1-GFP and Mid2-GFP. Moreover, both cargos were recruited into the same ERES (Fig. 4E, panel 3, and movie S4). Therefore, our findings demonstrate that ceramide acyl chain length in the ER membrane is an essential determinant for ER protein clustering and sorting.
Fig. 4. C26 ceramide–based GPI-AP cargos are rerouted and enter the same ERES as transmembrane cargo in the ER membrane with C18-C16 ceramides.
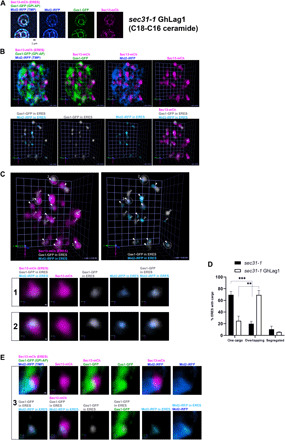
sec31-1 GhLag1 cells expressing galactose-inducible secretory cargos, Gas1-GFP (GPI-AP, green) and Mid2-iRFP (TMP, blue), and constitutive ERES marker Sec13-mCherry (ERES, magenta) were incubated at 37°C for 30 min, shifted down to 24°C for releasing secretion block, and imaged by SCLIM after 20 min. (A to C) Representative 2D projection images of 10 z-sections (A; scale bar, 1 μm) or 3D cell hemisphere images (B and C; scale unit, 0.45 μm) of cargo and ERES markers. Lower panels in (B) and panels in (C) show processed images to display only the cargo [Gas1-GFP (gray) and Mid2-iRFP (light blue)] present in the ERES (magenta). (C) White filled arrowheads: ERES with overlapping cargo. Open arrowheads: ERES containing only one cargo. Lower panels: selected ERES with overlapping cargos (1 and 2) marked in (C). Scale bar, 100 nm. (D) Quantification of micrographs described in (C). Average percentage of ERES containing only one cargo (Gas1-GFP or Mid2-iRFP), segregated cargos, and overlapping cargos in sec31-1 and sec31-1 GhLag1 cells. n = 432 in 54 cells (sec31-1) and n = 430 in 47 cells (sec31-1 GhLag1) in three independent experiments. Error bars = SD. Two-tailed, unpaired t test. ***P = 0.0002 (sec31-1) and **P = 0.0031 (sec31-1 GhLag1). (E) 3D images of a selected ERES with overlapping cargos (3) marked in (C). Gas1-GFP (green) and Mid2-iRFP (blue) approach the ERES (magenta) from the same side and stay in the same ERES restricted zone. Scale bar, 100 nm.
DISCUSSION
This study provides direct in vivo evidence for lipid-based protein cargo sorting into selective export sites in the secretory pathway and reveals the critical importance of the acyl chain length for sorting selectivity. We took advantage of a powerful and cutting-edge microscopy technology called SCLIM to demonstrate in yeast that newly synthesized Gas1-GFP, a major plasma membrane GPI-AP having a very long acyl chain (C26) ceramide lipid moiety, is clustered into discrete ER zones associated with specific ERES, whereas transmembrane secretory proteins distribute throughout the ER membrane (Fig. 1). Moreover, these two types of cargos selectively entered into different ERES (Fig. 2). Decreasing the acyl chain length of cellular ceramide in the membrane from C26 to C18-C16 disrupted Gas1-GFP clustering into the discrete ER zones, and Gas1-GFP was rerouted to exit the ER with transmembrane proteins via the same ERES (Figs. 3 and 4).
Although GPI-APs use a specialized protein machinery to exit the ER, we found that C26 ceramide–dependent segregation does not rely on differential protein interactions that could lead to ERES specialization (figs. S4 and S5). Instead, our findings support an alternative sorting mechanism driven by lipid-based protein clustering and consequent exclusion of other cargos. Our observation that Gas1-GFP zones or clusters associated with specific ERES are devoid of the transmembrane secretory protein Mid2-iRFP indicates that C26 ceramide–dependent clustering of GPI-APs would facilitate their entrance into the associated ERES and, at the same time, would exclude the transmembrane secretory cargo from entering into this specific ERES (Figs. 1 and 2). By contrast, the presence of C18-C16 ceramides in the ER membrane does not cause GPI-APs to form zones or clusters and, thus, they would not exclude or displace transmembrane secretory proteins from accessing the same ERES (Figs. 3 and 4). Therefore, we propose that C26 ceramide drives segregation and sorting by promoting protein clustering linked to specific ERES.
How could this C26 ceramide–dependent clustering into specific ER zones be achieved? The propensity of membrane ceramides to segregate laterally might lead GPI-APs with C26 ceramide to form small and transient-ordered lipid clusters in the less-ordered lipid environment of the ER membrane containing shorter and unsaturated glycerolipids (17, 18). These small and transient clusters could further coalesce into larger and more stable clusters upon binding with the p24 complex (34). Consistent with this, we show that C26 Gas1-GFP needs to interact with the p24 complex to form large visible clusters (Fig. 3). The p24 complex is a hetero-oligomer consisting of four different p24 transmembrane proteins in yeast (35), providing multivalent binding that could lead to the cross-linking of small GPI-AP clusters generating larger stabilized clusters (34). Interactions between the protein ectodomains of GPI-APs could also contribute to their clustering as shown during their Golgi transport in mammalian polarized epithelial cells (36). Nevertheless, larger segregated clusters are not formed when the p24 complex binds to Gas1-GFP in the presence of C18-C16 ceramides in the ER membrane. The underlying mechanism could rely on a specific physicochemical property of the very long acyl chain ceramides. Biophysical studies with artificial membranes have shown that although both very long (C24) and shorter (C18-C16) acyl chain ceramides induce phase separation, only very long acyl chain ceramides (C24) are able to remodel membranes by promoting high curvature and membrane bending by interdigitation (17, 37, 38). The transmembrane helix of the Emp24 human homolog TMED2 has been shown to selectively interact with the C18 ceramide–based sphingolipid sphingomyelin in the cytosolic leaflet (39). Using molecular dynamics (MD) simulations, we found that both C18 and C26 ceramides accumulate around the cytosolic leaflet of the transmembrane helix of Emp24 and that they do so with similar preferences (fig. S6). Noteworthy, this suggests that the transmembrane helix of Emp24 can cause an asymmetric distribution of lipids in the membrane, as it has been also shown recently for ether lipids in similar MD simulations based on results in mammalian cells (40). Therefore, we speculate that local enrichment of C26 ceramides in both leaflets of the ER membrane, upon direct multivalent p24 binding of GPI-APs in the luminal leaflet and C26 ceramide accumulation around p24 in the cytosolic leaflet, could promote concomitant protein clustering and membrane curvature generation by interdigitation (41), leading to GPI-AP segregation into discrete zones next to ERES, which also prefer highly curved zones of the ER membrane (42). This proposed mechanism is supported by previous reports (43, 44). Multivalent binding of oligomeric lectins, pathogens, or antibodies to ceramide-based glycosphingolipids (GSLs) at the plasma membrane triggers large GSL clustering, enhancing phase separation and resulting in membrane deformation and internalization (44). Iwabuchi et al. (43) found that multivalent ligand binding to the GSL lactosylceramide induced large cluster formation and membrane invagination in the presence of very long (C24) but not shorter long (C16) acyl chain and that lyn-mediated signaling at the cytosolic leaflet was coupled by acyl chain interdigitation in human neutrophils.
In mammalian polarized epithelial cells, clustering governs the segregation and sorting of GPI-APs at the level of the trans-Golgi network (TGN) to the apical plasma membrane (10, 45). This clustering is driven by GPI-AP oligomerization (36), but it might also depend on the ceramide chain length as we found in yeast. Although mammalian GPI-APs have an ether lipid–based anchor with a very different chemical structure from very long acyl chain ceramides, a recent study found that both lipids evolutionary share some similar physicochemical properties and functions (40). Thus, the ether lipid moiety in mammalian cells might act similarly to the C26 ceramide in yeast, by associating with very long chain ceramides in the membrane to facilitate GPI-AP clustering and sorting. Although this possibility still requires direct testing, it is supported by the previous findings that the transport to the Golgi of very long acyl chain ceramides is not carried out by cytosolic transfer proteins and depends on GPI anchor synthesis like in yeast. Therefore, an evolutionary conserved mechanism appears to operate to selectively cotransport very long acyl chain ceramides and GPI-APs in the same transport vesicles (13, 16, 20, 46, 47).
In both yeast and mammalian polarized epithelial cell systems, GPI-AP clustering and segregation from other plasma membrane proteins occur before arrival to the cell surface. Paladino et al. (48) found that at the TGN of mammalian polarized epithelial cells, GPI-AP clustering is not only required for the selective sorting of GPI-APs to the apical plasma membrane but also regulates clustered organization of GPI-APs and their biological activities at the cell surface. In yeast, the C26 ceramide–dependent clustering of GPI-APs at the ER shown in this study could regulate then the clustered organization and functional activity of GPI-APs at the plasma membrane (24, 49). Consistent with this model, hypersensitivity of GhLag1 cells to a GPI inhibitor or to drugs affecting cell wall integrity (28) and ceramide requirement for functional Gas1-GFP clustering at the tip of the mating projection of yeast cells (49) indicate possible physiological consequences of GPI-AP missorting. Nevertheless, further testing of whether the functional organization of the cell surface is already programmed from the ER by a lipid length–based sorting will be subject of our future research.
MATERIALS AND METHODS
Yeast strains, plasmids, and culture conditions
Strains of S. cerevisiae used for this work are listed in table S1. MMY1583 and MMY1635 strains used for live-cell imaging by SCLIM were constructed in the W303 background. These strains expressing fluorescent protein–tagged Sec13-mCherry were constructed by a polymerase chain reaction (PCR)–based method using pFA6a plasmids as a template (23). Strains expressing fluorescent protein–tagged Mid2-iRFP under the control of the GAL1 promoter were constructed as follows. The iRFP-KanMx sequence was PCR amplified from the pKTiRFP-KAN vector (a gift from E. O’Shea, Addgene plasmid no. 64687; http://n2t.net/addgene:64687; Research Resource identifier (RRID): Addgene_64687) and inserted at the C terminus of the endogenous Mid2. The Mid2-iRFP genomic sequence was amplified and cloned behind the GAL1 promoter into the Not I–Sac I sites of the integrative plasmid pRS306. The resulting plasmid pRGS7 was linearized with Pst I for integration at the URA3 locus.
Gas1-GFP fusion gene was expressed under the control of the GAL1 promoter in a centromeric (CEN) plasmid, which was constructed as follows. The Gas1-GFP sequence was amplified by PCR from pRS416-GAS1-GFP plasmid (24) (a gift from L. Popolo) and cloned into Xma I–Xho I sites of the CEN plasmid pBEVY-GL LEU2 (a gift from C. Miller; Addgene plasmid no. 51225; http://n2t.net/addgene:51225; RRID: Addgene_51225). The resulting plasmid was named pRGS6. Axl2-GFP fusion gene was also expressed under the control of the GAL1 promoter in the pBEVY-GL LEU2 vector, which was constructed as follows. The Axl2-GFP sequence was amplified by PCR from the pRS304-p2HSE-Axl2-GFP plasmid (23) and cloned into Bam HI–Pst I sites of the pBEVY-GL LEU2 vector. The resulting plasmid was named pRGS12. The sequences of oligonucleotides used in this study are listed in table S2.
Strains were grown either in rich yeast extract-peptone (YP) medium (1% yeast extract and 2% peptone) supplemented with 0.2% adenine and containing 2% glucose [YP-dextrose (YPD)], 2% raffinose [YP-raffinose (YPR)], or 2% galactose [YP-galactose (YPG)] as carbon source or in synthetic minimal medium (0.15% yeast nitrogen base and 0.5% ammonium sulfate) supplemented with the appropriate amino acids and bases as nutritional requirements, and containing either 2% glucose (synthetic dextrose minimal medium) or 2% galactose (synthetic galactose minimal medium) as carbon source.
Live-cell imaging by SCLIM
For live imaging, temperature-sensitive sec31-1 mutant cells expressing constructs under GAL1 promoter were grown to mid-log phase overnight at 24°C in YPR medium. After induction for 1 hour at 24°C in YPG, cells were incubated for 30 min at 37°C in SG and subsequently released from secretory block by shifting to 24°C. Cells were immobilized on glass slides using concanavalin A and imaged by SCLIM. SCLIM was developed by combining an Olympus model IX-71 inverted fluorescence microscope with a UPlanSApo 100× 1.4–numerical aperture oil objective lens (Olympus), a high-speed and high signal-to-noise ratio spinning-disk confocal scanner (Yokogawa Electric), a custom-made spectroscopic unit, image intensifiers (Hamamatsu Photonics) equipped with a custom-made cooling system, magnification lens system for giving ×266.7 final magnification, and electron-multiplying charge-coupled device cameras (Hamamatsu Photonics) (21). Image acquisition was executed by custom-made software (Yokogawa Electric). For 3D images, we collected optical sections spaced 100 nm apart in stacks by oscillating the objective lens vertically with a custom-made piezo actuator. Z-stack images were converted to 3D voxel data and processed by deconvolution with Volocity software (PerkinElmer) using the theoretical point-spread function for spinning-disk confocal microscopy. Measurement of ERES including cargo was performed by colocalization analysis with automatic thresholding of Volocity software. MetaMorph software (Molecular Devices) was used for presenting line-scan analysis.
Statistical analysis
Statistical significance was determined using GraphPad Prism software. For the two-tailed Student’s t test and ordinary one-way analysis of variance (ANOVA) test, differences among groups were considered significant for P < 0.05 (*).
Light microscopy
For fluorescence microscopy of Gas1-GFP, log-phase cells were grown overnight in YPD and collected by centrifugation, washed twice with phosphate-buffered saline, and incubated at least 15 min on ice before being examined under the microscope as described previously (24). Acquisition was performed using a Leica DMi8 microscope equipped with an objective lens (HCX PL APO 1003/1.40 Oil PH3 CS), L5 (GFP) filter, a Hamamatsu camera, and Application Suite X (LAS X) software following the instructions of the manufacturer.
Immunoblotting
Samples were denatured with SDS sample buffer for 10 min at 65°C and then separated by SDS–polyacrylamide gel electrophoresis (PAGE). For immunoblot analysis, 10 μl of sample was loaded in each lane. Primary antibodies: Rabbit polyclonal anti-Gas1 was used at 1:3000 dilution, rabbit polyclonal anti-Emp24 was used at 1:500 dilution, and rabbit polyclonal anti-GFP was used at 1:3000 dilution (gifts from H. Riezman). Mouse monoclonal anti-Pgk1 was used at 1:5000 dilution (gift from J. de la Cruz). Secondary antibody: Horseradish peroxidase (HRP)–conjugated goat anti-rabbit immunoglobulin G (IgG) was used at 1:3000 dilution (Pierce). HRP-conjugated goat anti-mouse IgG was used at 1:3000 dilution (Pierce). Immunoreactive bands were visualized by chemiluminescence with SuperSignal West Pico reagents (Thermo Fisher Scientific).
Native coimmunoprecipitation
The native coimmunoprecipitation experiment was performed on enriched ER fractions as described (31). Briefly, 100 optical density at 600 nm (OD600) units of yeast cells were washed twice with TNE buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonylfluoride, and protease inhibitor cocktail; Roche Diagnostics] and disrupted with glass beads, after which cell debris and glass beads were removed by centrifugation. The supernatant was then centrifuged at 17,000g for 15 min at 4°C. The pellet was resuspended in TNE, and digitonin was added to a final concentration of 1%. The suspension was incubated for 1 hour at 4°C with rotation, after which insoluble components were removed by centrifugation at 13,000g for 60 min at 4°C. For immunoprecipitation of Gas1-GFP, the sample was first preincubated with empty agarose beads (ChromoTek) at 4°C for 1 hour and subsequently incubated with GFP-Trap_A (ChromoTek) at 4°C for 3 hours. The immunoprecipitated beads were washed five times with TNE containing 0.2% digitonin, eluted with SDS sample buffer, resolved on SDS-PAGE, and analyzed by immunoblot.
Cross-linking assay
The cross-linking assay was performed on enriched ER fractions as described (31). Briefly, enriched ER fractions were incubated with 0.5 mM dithiobis(succinimidylpropionate) (Pierce, Thermo Fisher Scientific, Rockford, IL, USA; 20°C, 20 min). The cross-linking reaction was quenched by addition of glycine (50 mM final concentration, 5 min, 20°C).
MS analysis of cellular membrane ceramides
MS analysis of ceramides in wild-type and GhLag1 strains was performed as described previously (50). Briefly, cells were grown in YPD at 30°C to exponential phase (3 to 4 OD600 units/ml), and 25 × 107 cells were harvested. Their metabolism was quenched with trichloroacetic acid. Lipids were extracted with the extraction solvent [ethanol, water, diethyl ether, pyridine, and 4.2 N ammonium hydroxide (15:15:5:1:0.018 v/v)] and with 1.2 nmol of the internal standard C17 ceramide (860517, Avanti Polar Lipids). The extract was subjected to mild alkaline-hydrolysis using monomethylamine reagent [methanol, water, n-butanol, and methylamine solution (4:3:1:5 v/v)] followed by salt removal using water-saturated n-butanol. Last, the extract was resuspended in positive mode solvent [chloroform/methanol/water (2:7:1) + 5 mM ammonium acetate] and infused onto the mass spectrometer. Multiple reaction monitoring (MRM) was performed for the identification and quantification of sphingolipid molecular species. A TSQ Vantage Triple Stage Quadrupole Mass Spectrometer (Thermo Fisher Scientific) equipped with a robotic nanoflow ion source Nanomate HD (Advion Biosciences, Ithaca, NY) was used for the lipid analysis. The collision energy was optimized for each ceramide class. MS data were obtained in positive ion mode. For each biological replicate, the signal of the lipid is the median of three independent measurements.
MS analysis of the GPI-lipid of Gas1-GFP
Cells (800 × 107) expressing Gas1-GFP were subjected to native immunoprecipitation as described (31). The purified Gas1-GFP was separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membrane. The protein was visualized by staining the PVDF with amido black. The band of Gas1-GFP was excised from the PVDF and washed with methanol five times and one wash with liquid chromatography–MS (LC-MS) grade water. Lipid moieties were released from Gas1-GFP by incubating the membrane strips with a mixture of 500 μl of 0.3 M NaOAc (pH 4.0), buffer, and 500 μl of freshly dissolved 1 M sodium nitrite for 3 hours at 37°C, which cleaved between the glucosamine and inositol to release inositophosphorylceramide (51). Afterward, the membrane strips were washed four times with LC-MS grade water, dried at room temperature, and preserved in a nitrogen atmosphere at −80°C until the analysis. As control, a blank sample piece of the PVDF membrane was used for each experiment. The extracted lipids from Gas1-GFP were subsequently analyzed by MS as described (50). Briefly, PVDF strips containing the GPI-lipids were resuspended in 75 μl of negative mode solvent [chloroform/methanol (1:2) + 5 mM ammonium acetate], and sphingolipid species were analyzed with electrospray ionization (ESI)–MRM/MS (TSQ Vantage) as mentioned above. In this case, MS data were obtained in negative ion mode.
Analysis of GPI-lipid remodeling
Lipid moieties of GPI anchor were isolated from GPI-APs labeled with [3H] myo-inositol as described previously (16). The lipids were separated by thin-layer chromatography using a solvent system (55:45:10 chloroform-methanol–0.25% KCl) and visualized using FLA-7000 (Fujifilm).
MS analysis of the GPI-glycan of Gas1-GFP
Cells (600 × 107) expressing Gas1-GFP were washed twice with TNE buffer and disrupted with glass beads, after which cell debris and glass beads were removed by centrifugation. The supernatant was then centrifuged at 17,000g for 1 hour at 4°C. The pellet was washed in TNE and incubated with 1U PI-PLC (Invitrogen) in TNE containing 0.2% digitonin for 1 hour at 37°C. After the enzymatic treatment, membranes were removed by centrifugation at 17,000g for 1 hour at 4°C. For immunoprecipitation of Gas1-GFP, the supernatant was incubated with GFP-Trap_A (ChromoTek) overnight at 4°C. The purified Gas1-GFP separated by SDS-PAGE was stained with Coomassie brilliant blue. The stained band of Gas1-GFP was excised from the periaqueductal gray and then was subjected to in-gel digestion with trypsin after alkylation with iodoacetamide and reduction with dithiothreitol. Tryptic peptides and the peptides carrying GPI-glycan were extracted and dried. The dried peptides were dissolved in 20 μl of water. A portion (8 μl) was injected into LC. The peptides were separated using an octadecylsilane (ODS) column (Develosil 300ODS-HG-5; 150 mm × 1.0 mm inside diameter; Nomura Chemicals, Aichi, Japan) under specific gradient conditions. The mobile phases were solvent A (0.08% formic acid) and solvent B (0.15% formic acid in 80% acetonitrile). The column was eluted with solvent A for 5 min, at which point the concentration of solvent B was increased to 40% over 55 min at the flow rate of 50 μl min−1 using an Accela HPLC system (Thermo Fisher Scientific, Boston, MA, USA). The eluate was introduced continuously into an ESI source, and tryptic peptides and the peptides carrying GPI-glycan were analyzed by LTQ Orbitrap XL (hybrid linear ion trap-orbitrap mass spectrometer; Thermo Fisher Scientific). In the MS setting, the voltage of the capillary source was set at 4.5 kV, and the temperature of the transfer capillary was maintained at 300°C. The capillary voltage and tube lens voltage were set at 15 and 50 V, respectively. MS data were obtained in positive ion mode over the mass range mass/charge ratio (m/z) 300 to m/z 3000 (resolution, 60,000; mass accuracy, 10 parts per million). MS/MS data were obtained by ion trap in LTQ Orbitrap XL [data-dependent top 3, Collision-induced dissociation (CID)].
MD simulation
MD simulations were run with the GROMACS (52) software using the MARTINI 2 force field (53–55). The CHARMM GUI Membrane Builder (56, 57) was then used to build bilayers containing dioleoylphosphatidylcholine (DOPC) and Cer C18 or DOPC and Cer C26. The topology and coordinates for Cer C26 were derived from those of DXCE by removing extra beads in the sphingosine tail. The bilayers were equilibrated and run using the procedure described below, and the last coordinates of the systems were used to build systems containing Emp24. The yeast Emp24 transmembrane domain (residues 173 to 193) was built as an α-helix using the visual MD (VMD) tool molefacture (58). The protein was then coarse grained using CHARMM GUI and inserted in the bilayer, after removing overlapping lipids. The final systems contain 1202 DOPC and 302 Cer C26 or 1197 DOPC and 295 Cer C18 and Emp24. The systems were ionized to a concentration of 0.150 M. Four independent replicas for both bilayer compositions were run.
Lipid bilayers were equilibrated using the CHARMM GUI procedure that involves minimization followed by an equilibration for 405,000 steps in which positional restraints are gradually lowered and removed, while the time step increases from 0.005 to 0.02 ps. The equilibration was followed by a 6-μs production, with a time step of 0.02 ps. After insertion of Emp24, the same CHARMM GUI procedure was used for minimization and equilibration of the systems, and then production was run for 8 μs.
For all systems, during equilibration, pressure was controlled by a Berendsen barostat (59), while during production, pressure was controlled by a Parrinello-Rahman barostat (60). In all the cases, average pressure was 1 bar, and a semi-isotropic pressure coupling scheme was used. A velocity-rescale thermostat (61), with separate temperature coupling for the protein, lipids, and solvent particles, was used in both equilibration and production. Target temperature was 310 K during the entire run. Nonbonded interactions were calculated by generating a pair list using the Verlet scheme with a buffer tolerance of 0.005. Coulombic terms were calculated using reaction field and a cutoff distance of 1.1 nm. A cutoff scheme was used for the van der Waals terms, with a cutoff distance of 1.1 nm and the Verlet cutoff scheme for the potential shift (62).
The number of lipids interacting with the protein was calculated using VMD, using a cutoff of 0.7 nm between the phosphate bead of DOPC or the AM1 bead of ceramides and the protein. The depletion-enrichment (D-E) factors were calculated, as in (63) according to the following formula
The values reported are obtained as average and the error bars as SE over four independent replicas. The statistical significance of the D-E factor was calculated from t test [(averageDE-factor-1)/SE]. P values were calculated from a one-tailed distribution.
2D lateral density maps for the systems containing Emp24 were calculated using GROMACS tools, over the last 250 ns of trajectory. To obtain an enrichment/depletion map for ceramides, the density maps of Cer was divided by the sum of the maps of Cer and DOPC and then divided by the concentration of Cer in bulk. The same colormap scale has been used.
All the images were rendered using VMD and the graphs created using MATLAB or python scripts.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers for critical comments and suggestions, L. Silva for discussion, L.M. Escudero for helpful technical comments, Biology and Microscopy Services of the University of Seville (SGI CITIUS) for technical support, and I. Riezman for technical support in MS. Funding: This research was funded by the Spanish Ministry of Economy and Competitiveness (MINECO; grant number BFU2017-89700-P to M.M.), “VI Own Research Plan” of the University of Seville (VIPPIT-2020-I.5 to M.M.), and Grants-in-Aid for Scientific Research from Japan Society for the Promotion of Science (JP25221103, JP17H06420, and JP18H05275 to A.N. and K. K.; and JP19H02922 to M.N. and K.F.). S.V. and V.Z. acknowledge support from the Swiss National Science Foundation (grant no. 163966) and from the Swiss National Supercomputing Centre (CSCS) under project IDs s726 and s842. H.R. is supported by the Swiss National Science Foundation and the NCCR Chemical Biology (grants 184949 and 185898). This research was funded by the “VI Own Research Plan “of the University of Seville (VIPPIT-2020-I.5); “V Own Research Plan” of the University of Seville (VPPI-US) contract (cofounded by the European Social Fund) to S.L.; University of Seville fellowship to S.R.-G.; Ministry of Education, Culture, and Sport (MECD) fellowship to S.S.-B.; and contract from the University of Seville by the Youth Employment Initiative to A.C.-G. Author contributions: S.R.-G. and K.K. established and performed SCLIM microscopy experiments. S.R.-G. designed and constructed strains and plasmids, with help from S.S.-B., A.M.P.-L., A.A.-R., S.L., A.C.-G., and M.W. S.R.-G., S.S.-B., A.M.P.-L., A.A.-R, S.L., A.C.-G. A.I., M.A., M.N., and K.F. performed biochemical studies and data analysis. S.S.-B. prepared samples for glycan analysis and M.N. performed the glycan analysis, with help from K.F. S.S.-B., A.C.-G., and A.A.-R prepared samples for lipid analysis, and H.R. performed the lipid analysis of yeast strains. V.Z. and S.V. designed, performed, and interpreted MD simulation experiments. M.M., K.K., and A.N. designed the experiments and supervised the project. M.M. wrote the manuscript, with input from all other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eaba8237/DC1
REFERENCES AND NOTES
- 1.Gomez-Navarro N., Miller E., Protein sorting at the ER-Golgi interface. J. Cell Biol. 215, 769–778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons K., Van Meer G., Lipid sorting in epithelial cells. Biochemistry 27, 6197–6202 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Simons K., Ikonen E., Functional rafts in cell membranes. Nature 387, 569–572 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Surma M. A., Klose C., Simons K., Lipid-dependent protein sorting at the trans-Golgi network. Biochim. Biophys. Acta 1821, 1059–1067 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Muñiz M., Riezman H., Trafficking of glycosylphosphatidylinositol anchored proteins from the endoplasmic reticulum to the cell surface. J. Lipid Res. 57, 352–360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muñiz M., Morsomme P., Riezman H., Protein sorting upon exit from the endoplasmic reticulum. Cell 104, 313–320 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Castillon G. A., Watanabe R., Taylor M., Schwabe T. M. E., Riezman H., Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic 10, 186–200 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita T., Fujita M., Biosynthesis of GPI-anchored proteins: Special emphasis on GPI lipid remodeling. J. Lipid Res. 57, 6–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muñiz M., Zurzolo C., Sorting of GPI-anchored proteins from yeast to mammals–Common pathways at different sites? J. Cell Sci. 127 ( Pt. 13), 2793–2801 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Zurzolo C., Simons K., Glycosylphosphatidylinositol-anchored proteins: Membrane organization and transport. Biochim. Biophys. Acta 1858, 632–639 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Umemura M., Fujita M., Yoko-o T., Fukamizu A., Jigami Y., Saccharomyces cerevisiae CWH43 is involved in the remodeling of the lipid moiety of GPI anchors to ceramides. Mol. Biol. Cell 18, 4304–4316 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghugtyal V., Vionnet C., Roubaty C., Conzelmann A., CWH43 is required for the introduction of ceramides into GPI anchors in Saccharomyces cerevisiae. Mol. Microbiol. 65, 1493–1502 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Funato K., Riezman H., Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell Biol. 155, 949–959 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath A., Sütterlin C., Manning-Krieg U., Movva N. R., Riezman H., Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 13, 3687–3695 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sütterlin C., Doering T. L., Schimmöller F., Schröder S., Riezman H., Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 110 ( Pt. 21), 2703–2714 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Kajiwara K., Watanabe R., Pichler H., Ihara K., Murakami S., Riezman H., Funato K., Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol. Biol. Cell 19, 2069–2082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto S. N., Silva L. C., Futerman A. H., Prieto M., Effect of ceramide structure on membrane biophysical properties: The role of acyl chain length and unsaturation. Biochim. Biophys. Acta 1808, 2753–2760 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Silva L., de Almeida R. F. M., Fedorov A., Matos A. P. A., Prieto M., Ceramide-platform formation and -induced biophysical changes in a fluid phospholipid membrane. Mol. Membr. Biol. 23, 137–148 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Melero A., Chiaruttini N., Karashima T., Riezman I., Funato K., Barlowe C., Riezman H., Roux A., Lysophospholipids facilitate COPII vesicle formation. Curr. Biol. 28, 1950–1958.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funato K., Riezman H., Muñiz M., Vesicular and non-vesicular lipid export from the ER to the secretory pathway. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158453 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Kurokawa K., Ishii M., Suda Y., Ichihara A., Nakano A., Live cell visualization of Golgi membrane dynamics by super-resolution confocal live imaging microscopy. Methods Cell Biol. 118, 235–242 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa K., Osakada H., Kojidani T., Waga M., Suda Y., Asakawa H., Haraguchi T., Nakano A., Visualization of secretory cargo transport within the Golgi apparatus. J. Cell Biol. 218, 1602–1618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurokawa K., Okamoto M., Nakano A., Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat. Commun. 5, 3653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rolli E., Ragni E., Calderon J., Porello S., Fascio U., Popolo L., Immobilization of the glycosylphosphatidylinositol-anchored Gas1 protein into the chitin ring and septum is required for proper morphogenesis in yeast. Mol. Biol. Cell 20, 4856–4870 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philip B., Levin D. E., Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21, 271–280 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoko-O T., Ichikawa D., Miyagishi Y., Kato A., Umemura M., Takase K., Ra M., Ikeda K., Taguchi R., Jigami Y., Determination and physiological roles of the glycosylphosphatidylinositol lipid remodelling pathway in yeast. Mol. Microbiol. 88, 140–155 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Roemer T., Madden K., Chang J., Snyder M., Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 10, 777–793 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Epstein S., Castillon G. A., Qin Y., Riezman H., An essential function of sphingolipids in yeast cell division. Mol. Microbiol. 84, 1018–1032 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Bosson R., Guillas I., Vionnet C., Roubaty C., Conzelmann A., Incorporation of ceramides into Saccharomyces cerevisiae glycosylphosphatidylinositol-anchored proteins can be monitored in vitro. Eukaryot. Cell 8, 306–314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., Kinoshita T., GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell 139, 352–365 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Manzano-Lopez J., Perez-Linero A. M., Aguilera-Romero A., Martin M. E., Okano T., Silva D. V., Seeberger P. H., Riezman H., Funato K., Goder V., Wellinger R. E., Muñiz M., COPII coat composition is actively regulated by luminal cargo maturation. Curr. Biol. 25, 152–162 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Castillon G. A., Aguilera-Romero A., Manzano-Lopez J., Epstein S., Kajiwara K., Funato K., Watanabe R., Riezman H., Muñiz M., The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol. Biol. Cell 22, 2924–2936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C. S., Hamamoto S., Orci L., Schekman R., Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Kusumi A., Koyama-Honda I., Suzuki K., Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic 5, 213–230 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Marzioch M., Henthorn D. C., Herrmann J. M., Wilson R., Thomas D. Y., Bergeron J. J. M., Solari R. C. E., Rowley A., Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell 10, 1923–1938 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paladino S., Sarnataro D., Pillich R., Tivodar S., Nitsch L., Zurzolo C., Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J. Cell Biol. 167, 699–709 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ventura A. E., Varela A. R. P., Dingjan T., Santos T. C. B., Fedorov A., Futerman A. H., Prieto M., Silva L. C., Lipid domain formation and membrane shaping by C24-ceramide. Biochim. Biophys. Acta Biomembr. 1862, 183400 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Zelnik I. D., Ventura A. E., Kim J. L., Silva L. C., Futerman A. H., The role of ceramide in regulating endoplasmic reticulum function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158489 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Contreras F. X., Ernst A. M., Haberkant P., Björkholm P., Lindahl E., Gönen B., Tischer C., Elofsson A., von Heijne G., Thiele C., Pepperkok R., Wieland F., Brügger B., Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature 481, 525–529 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Jiménez-Rojo N., Leonetti M. D., Zoni V., Colom A., Feng S., Iyengar N. R., Matile S., Roux A., Vanni S., Weissman J. S., Riezman H., Conserved functions of ether lipids and sphingolipids in the early secretory pathway. Curr. Biol. 30, 3775–3787.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Sorre B., Callan-Jones A., Manneville J.-B., Nassoy P., Joanny J.-F., Prost J., Goud B., Bassereau P., Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 5622–5626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto M., Kurokawa K., Matsuura-Tokita K., Saito C., Hirata R., Nakano A., High-curvature domains of the ER are important for the organization of ER exit sites in Saccharomyces cerevisiae. J. Cell Sci. 125 ( Pt. 14), 3412–3420 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Iwabuchi K., Prinetti A., Sonnino S., Mauri L., Kobayashi T., Ishii K., Kaga N., Murayama K., Kurihara H., Nakayama H., Yoshizaki F., Takamori K., Ogawa H., Nagaoka I., Involvement of very long fatty acid-containing lactosylceramide in lactosylceramide-mediated superoxide generation and migration in neutrophils. Glycoconj. J. 25, 357–374 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Sych T., Mély Y., Römer W., Lipid self-assembly and lectin-induced reorganization of the plasma membrane. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lebreton S., Paladino S., Zurzolo C., Clustering in the Golgi apparatus governs sorting and function of GPI-APs in polarized epithelial cells. FEBS Lett. 593, 2351–2365 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Loizides-Mangold U., David F. P. A., Nesatyy V. J., Kinoshita T., Riezman H., Glycosylphosphatidylinositol anchors regulate glycosphingolipid levels. J. Lipid Res. 53, 1522–1534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaji T., Horie A., Tachida Y., Sakuma C., Suzuki Y., Kushi Y., Hanada K., Role of intracellular lipid logistics in the preferential usage of very long chain-ceramides in glucosylceramide. Int. J. Mol. Sci. 17, 1761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paladino S., Lebreton S., Tivodar S., Formiggini F., Ossato G., Gratton E., Tramier M., Coppey-Moisan M., Zurzolo C., Golgi sorting regulates organization and activity of GPI proteins at apical membranes. Nat. Chem. Biol. 10, 350–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bagnat M., Simons K., Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. U.S.A. 99, 14183–14188 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.da Silveira Dos Santos A. X., Riezman I., Aguilera-Romero M.-A., David F., Piccolis M., Loewith R., Schaad O., Riezman H., Systematic lipidomic analysis of yeast protein kinase and phosphatase mutants reveals novel insights into regulation of lipid homeostasis. Mol. Biol. Cell 25, 3234–3246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehlert A., Ferguson M. A. J., Proteomic scale high-sensitivity analyses of GPI membrane anchors. Glycoconj. J. 26, 915–921 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. J. Abraham, D. van der Spoel, E. Lindahl, B. Hess, GROOMACS Team, GROMACS User Manual version 2016.3 (Royal Institute of Technology and Uppsala University, Sweden, 2017).
- 53.Monticelli L., Kandasamy S. K., Periole X., Larson R. G., Tieleman D. P., Marrink S.-J., The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theory Comput. 4, 819–834 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Marrink S. J., Risselada H. J., Yefimov S., Tieleman D. P., de Vries A. H., The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Marrink S. J., De Vries A. H., Mark A. E., Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B 108, 750–760 (2004). [Google Scholar]
- 56.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Qi Y., Ingólfsson H. I., Cheng X., Lee J., Marrink S. J., Im W., CHARMM-GUI Martini Maker for coarse-grained simulations with the Martini force field. J. Chem. Theory Comput. 11, 4486–4494 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Di Nola A., Haak J. R., Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984). [Google Scholar]
- 60.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 61.Bussi G., Donadio D., Parrinello M., Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 62.de Jong D. H., Baoukina S., Ingólfsson H. I., Marrink S. J., Martini straight: Boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 199, 1–7 (2016). [Google Scholar]
- 63.Corradi V., Mendez-Villuendas E., Ingólfsson H. I., Gu R.-X., Siuda I., Melo M. N., Moussatova A., DeGagné L. J., Sejdiu B. I., Singh G., Wassenaar T. A., Delgado Magnero K., Marrink S. J., Tieleman D. P., Lipid-protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 4, 709–717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eaba8237/DC1


