Abstract
Purpose of the review:
In this review, we will summarize the recent progress made in generating stem cells-based organoid and enteroid models of the gastrointestinal tract and their importance in understanding the role of microbes in intestinal epithelial homeostasis and disease.
Recent finding:
Intestinal stem-cell derived culture systems are self-organizing three-dimensional organotypic cultures that recapitulate many cellular, architectural and functional aspects of the human intestine. Progress has been made in the development of methods to incorporate additional cell lineages and physiological cues to better mimic the complexity of the intestine. Current model systems have facilitated both the study of gastrointestinal infections and interactions with normally non-pathogenic microbial residents of the GI tract. These studies have illustrated how live microbes, or their metabolites, ligands and virulence factors influence epithelial cell differentiation, maintenance, repair, function and intestine development.
Summary:
Organotypic models are invaluable tools for studying host-microbe interactions that complement in vivo experimental model systems. These models have evolved in terms of complexity and fidelity. The stem-cell based models are already at forefront for studying host-microbe interactions and with continued development, the future looks even more promising.
Keywords: organoid, enteroid, host-microbe interaction, bacteria, epithelium
Introduction
The mucosal lining of the human gastrointestinal tract is an important site of direct contact between the host cells and microbes. These epithelial-microbe interactions play an important role in gut homeostasis. Multiple systems exist to study the molecular and physiologic interactions between the gut mucosa and resident microbes. Previously host-microbiota interactions were modeled with either in vitro transformed cell lines or in vivo animal models. Models employing cell lines are characterized by a homogenous cellular population and limited relevance to the in vivo environment (1). Animal model systems can recapitulate the complexity of the human gastrointestinal tract with the caveat of species-specific differences in intestinal physiology, anatomy, diet and resident microbiota (2). Stem-cell-derived in vitro model systems can bridge the gap between these models. Stem cell-based intestinal models originating from either pluripotent or adult human stem cells recapitulate the cellular, architectural and functional aspects of the human intestine. Here we highlight recent advances in the use of microbiota-associated stem cell-based model systems to study gastrointestinal health and disease.
Stem cell derived model systems
Several versions of stem-cell derived organotypic models have been developed. Organoids are three dimensional structures derived from either human embryonic stem cells (ESC) or induced pluripotent stem cells (iPSCs) (Figure 1). Organoids have been generated for multiple gastrointestinal tissues including liver, pancreas, stomach, small intestine and colon. Directed differentiation of stem cells along particular gastrointestinal lineages is driven by specific growth factors (3, 4). However, to date only organoids resembling duodenum and colon have been developed using this approach. In general, organoids consist of an inner lumen enclosed by an organ specific heterocellular polarized epithelium and an outer mesenchymal layer. However lately attempts have been done to generate mesenchyme free small intestinal and colonic organoids (5*). This emergent model will open possibilities for investigating epithelial specific functions and interactions of the iPSC derived organoids. Moreover, the intestinal organoids resemble fetal epithelial cells with ability for further differentiation and maturation upon in vivo transplantation (6).
Figure 1:
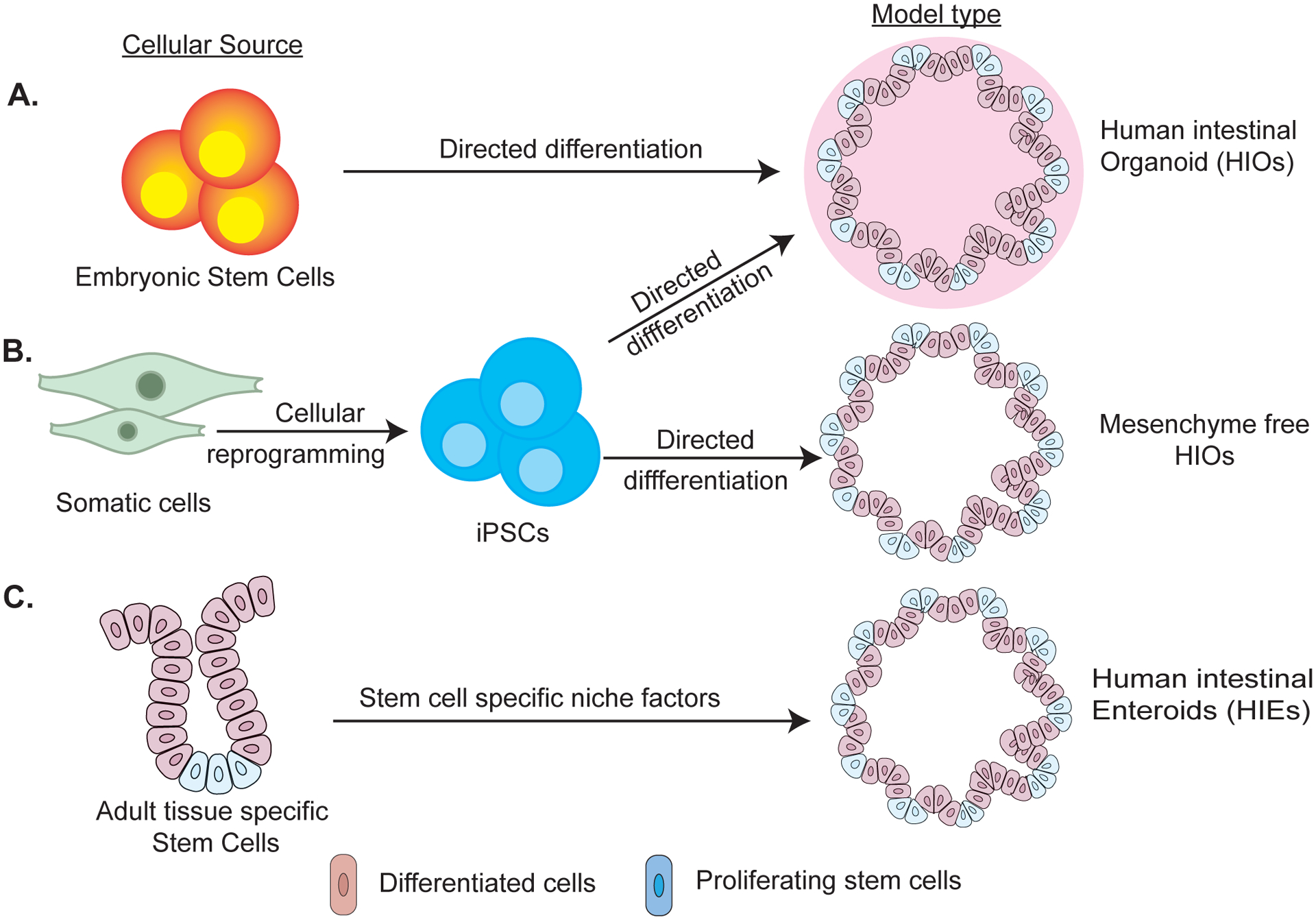
Schematic illustration summarizing generation of human intestinal stem-cell derived models from A) embryonic stem cells (ESCs) B) induced pluripotent stem cells and (iPSCs) C) adult stem cells. Both ESC and iPSC are differentiated into definitive endoderm followed by generation of CDX2 expressing spheroids. Spheroids are grown in 3D matrix to generate HIOs with mesenchyme. Mesenchyme free HIOs are generated from iPSCs by inhibiting BMP/TGF-β signaling pathway followed by sorting for gut progenitor cells that are differentiated into small or large intestine in specific growth media. HIEs are generated from stem cells present in the biopsies or tissue sections obtained from region of interest followed by their growth in 3D matrix in undifferentiated or differentiated state using specific growth factors.
Enteroids, conversely, are derived from adult tissue-specific stem cells and maintain structural and functional similarity to the tissue of origin (Figure 1). Enteroids are composed only of a mature epithelial layer without mesenchyme (7). The epithelial only nature of enteroids facilitate formation of two-dimensional monolayers which are heterocellular and demonstrate epithelial polarity mimicking in vivo conditions (8). Enteroids maintain genetic diversity and epigenetic signatures of their cellular origin over time (9*, 10).
Both enteroids and organoids express all major cellular lineages of the intestinal epithelium including enterocytes, goblet, enteroendocrine, Paneth cells, and stem cells. Additionally, they can be chemically and genetically manipulated and can be expanded and cryopreserved indefinitely (4). The various organotypic models offer specific advantages and limitations (Table 1). The specific research question addressed will dictate the use of one model system over another.
Table 1:
Key features of human intestinal stem-cell derived models
| Organotypic Model | Cellular Components | Key Features | Limitations |
|---|---|---|---|
| iPSC and ESC derived Organoids | Epithelium and mesenchyme |
|
|
| iPSC derived Mesenchyme free Organoids | Epithelium |
|
|
| Adult stem cell derived Enteroids | Epithelium |
|
|
| Adult stem cell derived inside out Enteroids | Epithelium |
|
|
Organotypic models to study host-epithelial interactions in the gut
For exploring the role of microbes in intestinal homeostasis, several different methods have been developed to expose microbes to organoids and enteroids (Figure 2). Live microorganisms or their products can be microinjected into the lumen of 3D organoids and enteroids. However, this is resource intensive, technically challenging and accumulation of debris inside the lumen interferes with direct and uniform interaction between microbes and epithelium (11). Recently developed apical-out enteroids overcome this limitation by providing free access to the apical surface. This adaptation, however, is not useful for studying microbes that preferentially interact with the basolateral surface of the epithelium (12*). Another approach used for co-culturing microbes with organoids, is spontaneous infection of fragmented organoids with microbes followed by resealing of organoids into 3D structures (13). Several host-microbe co-culture studies have also used human intestinal enteroids (HIEs) in two dimensional forms. These monolayers when grown on cell culture inserts with permeable membrane, provide access to both apical and basolateral sides of the epithelial layer (14). With several possibilities available, the choice of microbial exposure method should be determined by the accessibility to the physiological route of exposure, stem cell model used and the availability of resources. Another refinement of monolayer culture involves establishing an air-liquid interface wherein apical surface of the monolayer is devoid of any media (15**). This approach was successfully used for co-culturing Cryptosporidium with murine intestinal enteroids and did not affect epithelial cell viability or function. This technological advancement will thus enable investigation of epithelium derived factors that affect microbial growth and colonization within the intestine without interference from media derived nutrients from apical side.
Figure 2:
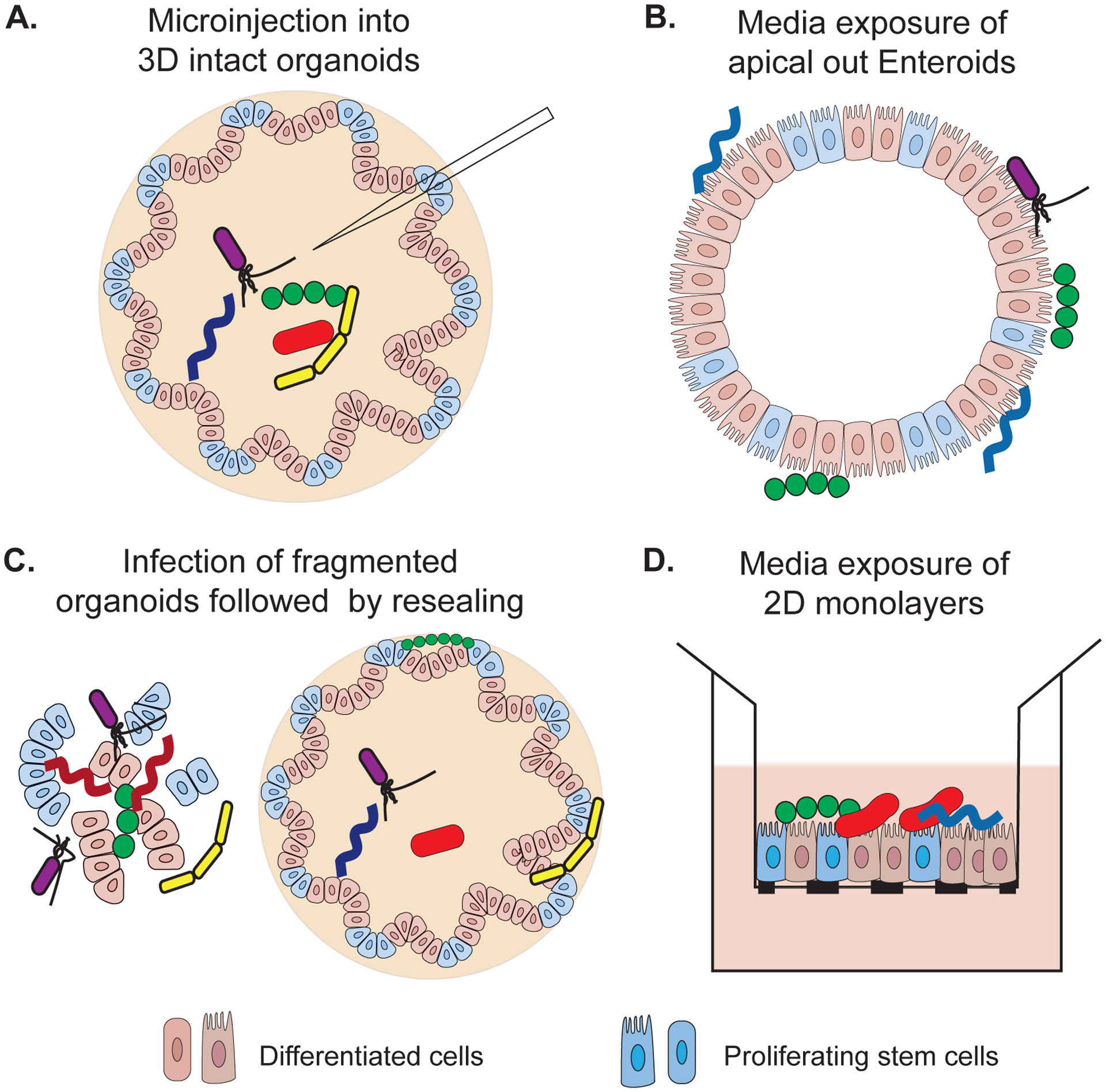
Methods used for exposing human intestinal stem-cell derived models with gut microbes. A) Microinjection is used for introducing microbes into the lumen of 3D organotypic models enabling microbe-epithelium interaction at the apical side. B) Polarity of enteroids in apical out HIEs is reversed by removing surrounding matrix and microbes are added in the outer media providing apical access. C) Dissociated 3D organotypic cultures are mixed with microbial species of interest. Microbes may get incorporated into the lumen or left outside in the matrix of reformed 3D structures. D) Organotypic models can be enzymatically dissociated to form 2D polarized monolayers and microbes can be added either on the apical or basolateral side.
Studying host-microbe interactions in vitro is challenging because most microbes residing in the GI tract are anaerobic whereas epithelial cells require oxygen for viability. The lower oxygen concentration inside 3D organoids allows survival of anaerobic bacteria, however, it is not clear if it permits the growth of strict anaerobes (6, 16). Two separate research groups have developed simple, cost-effective methods to develop oxygen gradients across the epithelial monolayer and successfully co-cultured strict anaerobic gut bacteria with the oxygen requiring enteroid monolayers (17**, 18). With these innovations, stem-cell based models have overcome the limitations of conventional aerobic in vitro models in studying host-gut microbe interactions.
Attempts have also been made to incorporate additional physiologic conditions such as peristalsis and intestinal luminal flow into these models using bioengineering approaches. Microfluidic-based gut organoid flow chip devices have been developed for three-dimensional HIOs to maintain steady state flow conditions through the organoid. This device enables removal of debris from the organoid lumen with a simultaneous exchange of nutrients (19). Similarly, an intestine-on-chip device was developed for 2D models to simulates the mechanical and physiological features of the human intestine (20*, 21). This in vitro platform consists of differentiated enteroid monolayer as well as endothelial cells on the opposite sides of a porous membrane, an intact mucus layer, oxygen gradient and a mechanism capable of generating luminal flow and peristalsis. Furthermore, this device can sustain complex human stool microbiota for a number of days without compromising epithelial function (20*). This success in recapitulating complex cross talk between multiple tissue lineages and anaerobic bacteria will potentially provide new insights on the mechanisms involved in gut microbiota associated gastrointestinal homeostasis and disease.
Use of microbe incorporated stem-cell derived models for studying gastrointestinal homeostasis and diseases
The technological advances in organotypic models have led to multiple studies that investigate the role of gastrointestinal microbes in epithelial maintenance, repair, regeneration, and barrier function. Here we highlight some of these studies that focus on gastrointestinal homeostasis and disease.
Epithelium regeneration, repair and maintenance
Intestinal stem cells (ISCs) play an important role in epithelium homeostasis by balancing between self-renewal and epithelial differentiation, regeneration and maintenance. ISCs were considered more protected from pathogens because of their deep location within the intestinal crypts. A recent study using murine and human enteroids provided the first direct evidence that the toxins produced by C. difficile can damage colonic stem cells. Colonic tissue from mice infected with toxigenic C.difficile had decreased expression of stem cell markers and were less efficient at generating enteroids, suggesting an effect on stem cells. This was associated with the production of the toxin TcdB by C. difficile. The study was further able to demonstrate that the stem cell damage delays epithelial repair and regeneration (22**). Organoids are also being explored to understand the protective effect of non-pathogenic bacteria on ISCs (23, 24*). A recent work unraveled the mechanism by which resident gut microbiota protects ISCs and promotes epithelial regeneration after oxidative stress (23). Muramyl dipeptide (MDP), a common bacterial motif recognized by the NOD pattern recognition receptor, reduced cell death due to apoptosis in enteroids following irradiation. Furthermore, an increase in NOD2 expression, expansion of Lgr5+ stem cells and decreased ROS generation were observed in MDP-stimulated irradiated enteroids. Another study explored how gut microbes restores homeostasis in the setting of inflammation. The authors stimulated murine enteroids with Lactobacillus reuteri and observed that under physiological conditions, L. reuteri maintained proliferation of ISCs by activating Wnt-β catenin signaling pathway (24*). Under pathological conditions caused by inflammatory mediator TNF, L.reuteri was able to reverse the epithelial damage by maintaining the number of ISCs and stimulating epithelial proliferation. Together these studies show that the protective or pathologic effect of individual microbial species on ISCs can be well investigated using stem-cell based microbial co-culture models and may provide clue to targets that can promote epithelial regeneration following injury, infection or cancer therapy.
Epithelial lineage differentiation and function
Recent studies have used organotypic models to show the effect of microbes on the differentiation of individual cellular lineages of the intestinal epithelium (25–27). Using Clostridium ramosum, Mandic et al demonstrated the role of gut microbes in programming differentiation of colonic ISCs towards enterochromaffin (EC) lineage of the epithelium (28**). This effect of C. ramosum on murine ISCs was indirect since a bacterial lysate stimulated release of serotonin from enteroids. Increased serotonin in turn programed differentiation of stem cells towards serotonin secreting EC cells by changing the expression of genes involved in maintaining a balance between secretory and absorptive lineages of the intestine. Organotypic models have been employed to investigate the role played by different cellular lineages in microbial recognition and stimulating their growth within the gut. It has been demonstrated that sparse cellular lineages in human intestinal organoids/enteroids can be enriched (29–31). Leveraging on this property of stem-cell models, tuft cells were expanded in mouse intestinal enteroids using IL-13. The increased population of tuft cells helped in investigating the response of tuft cell towards parasitic helminth Trichinella spiralis infection (32). In conjunction with animal work, enteroids were used in this research to identify the key components such as parasite receptors, intracellular transduction proteins and other molecules involved in the signaling cascade induced in tuft cells after parasite infection. The activation of signaling cascade resulted in release of IL-25 cytokine by the enteroids indicating activation of type 2 immunity. Similarly, the role of M cells in uptake of Shigella flexneri was confirmed by inducing expression of M cells in enteroids (31, 33*). The study showed a significant increase in intracellular bacteria in enteroids with increased M cells. The enhanced accessibility to these rare cellular populations via stem cell models demonstrates they are an ideal platform for understanding the role played by the scarce intestinal cell types in recognizing microbes and mediating reciprocal responses (32, 33*).
Intestinal development
Alterations in gut microbiota structure and function impacts intestinal health and physiology in newborns and infants. The use of stem cell based models have shown an association between early life microbiota and intestinal maturation (34, 35*). Abo et al; used enteroids generated from conventional, germ-free and co-housed mice to demonstrate that the microbial exposure in early life positively influences epithelial regeneration, proliferation and repair of ISCs by controlling expression of erythroid differentiation regulator-1 in ISCs (36). Another group introduced human neonatal microbiota into mouse enteroids to understand the impact of early life microbiota on intestine development. In the study, microbiota from preterm infants, as opposed to microbiota from term neonates, induced epithelial proliferation marked by increased expression of stem cell gene markers and formation of large budding enteroids (35*). Although it is still under debate if microbial colonization occurs before or at birth; it is important to know how initial colonization of the gut affects intestinal development and function. Organoids, due to their resemblance to immature fetal epithelium can also be used to address these questions, thus providing a leverage in investigating the role of microbes in early life intestinal development. In fact, an earlier study found that introduction of E.coli, an early gut colonizer, into the immature organoids resulted in epithelial maturation as indicated by increased differentiation of enterocytes, mucus production and improved epithelial barrier (6). The microbe-stem cell-based co-culture models therefore may prove instrumental in advancing our understanding of the role played by microbes in intestinal development, physiology and function during postnatal adaptation.
Modeling gastrointestinal infections and diseases
Organotypic models have served as reliable model system for studying the pathophysiology of various infectious agents involved in gastrointestinal diseases including bacteria, viruses and parasites. This has been comprehensively reviewed previously (37–42*). Traditional in vitro and in vivo models of GI tract were lacking in their ability to study certain infections either because of lack of pathogen infectivity or poor disease manifestations (1, 2). It is beyond the scope of this review to provide details of each pathogen individually; we have compiled a list of key steps in the pathogenesis of several gastrointestinal infections that have been recently studied using these models (Table 2). Receptors for the recently emerged coronavirus, SARS-CoV2, were found on apical side of human small intestinal and colon derived enteroids permitting the study of gut infection by this virus (43, 44). It was also shown that the virus primarily infects and replicates in mature enterocytes and to a lesser extent in undifferentiated enteroids. Moreover, the infected epithelium elicited expression of cytokines and genes associated with interferon I and III responses (45**). The rapidity with which organotypic models can be used to study a new viral pathogen modeling demonstrates the power and flexibility of these systems.
Table 2:
Main pathogenesis steps recently studied using stem-cell derived models
| Aspects of intestinal infections studied | Pathogen studied | Main findings | Reference |
|---|---|---|---|
| Host susceptibility | Human astrovirus | Pathogen replication rates in human enteroids can be donor dependent | (53, 54) |
| Shigella flexineri | |||
| Pathogen receptors involved in epithelial adherence | Salmonella enterica serovar Typhi | Serovar specific role of phospholipid transporter (YrbE) in epithelial adherence | (55, 56) |
| Enteroaggregative E. coli | Aggregative adherence fimbriae II were involved in adherence and pathogenesis to human enteroids | ||
| Preferential route of pathogen entry | S. Typhimurium | Preferentially invades apical surface | (12*, 33*) |
| Shigella flexineri, Listeria monocytogenes | Attaches to basolateral surface | ||
| Cells involved in pathogen replication | Human astrovirus SARS-CoV-2 | Astrovirus shows tropism for multiple cell types whereas SARS-CoV-2 was mainly found in enterocytes | (44, 53) |
| Epithelial response | Human norovirus | RNA-seq analysis showed activation of JAK-STAT pathway | (57) |
| Effect on epithelial barrier | C. difficile | Effect on epithelial barrier function is strain and toxin specific | (58, 59) |
| Enterovirus | Not all enteric viruses affect epithelial barrier integrity | ||
| Activation of innate immune response | Enterovirus71 | Induction of type III interferon immune response upon infection | (54, 58) |
| Shigella flexineri | Release of proinflammatory cytokines and chemokines by HIE | ||
| Treatment strategies | Shigella flexneri | A S. flexneri targeting phage prevented its adhesion and invasion to HIEs | (60*, 61) |
| C.difficile | Bacitricin inhibits translocation of toxin B into the cytosol of epithelial cells thereby preventing their disruption |
With the ability to generate enteroids from diseased gut epithelium and successful use of CRISPR/CAS technology in creating organoids from modified pluripotent stem cells (46), stem cell based models have been used to study mechanistic aspects of the role of microbes in various multifactorial diseases of the gut including celiac disease (9*), Barrett’s esophagus (47), inflammatory bowel diseases (48), necrotizing enterocolitis (49) and cancer (50**). For example, repeated exposures to genotoxic E. coli over multiple passages of human pediatric enteroids induced unique mutational signatures in the enteroid epithelium (50**). Interestingly, similar mutational signatures were found in colorectal cancer patients providing a direct evidence for the role played by microbes in colorectal cancer development or progression. Although still in its infancy to accurately mimic such complex diseases, human enteroids have the potential to address the questions relevant to long term effect of presence or absence of common gut residents on individual human health and disease.
Conclusions
Role of microbes in host health and disease is well established but the mechanistic details are not well understood. Organotypic models offer an invaluable tool for studying host-microbe interactions and have filled the gap between other in vitro and in vivo experimental model systems. Till date, most studies have concentrated on studying individual microbial strains specifically pathogens and limited attempts have been made to co-culture the complex microbial community of the human gastrointestinal tract with the organoids or enteroids. Moving forward, for a clear understanding of the host-microbe interactions of the gut it will be necessary to include complex microbiota into these models. Significant new advances are made in these models with the introduction of other physiological and cellular components of the intestine (51, 52). These components play a critical role in modulating both microbes and epithelium. Thus, it will be important to incorporate them in microbe-stem cell-based co-culture models as well. Nonetheless, organotypic models are already at forefront for studying host-microbe interactions and with the continuous evolvement, the potential of these models to recreate complex host-microbiota interactions of the GI tract looks promising.
Key points.
Intestinal organoids and enteroids recapitulate various structural and functional features of human intestinal epithelium.
Intestinal organoids or enteroids have served as valuable models for several studies focused on understanding the effect of microbes on intestinal homeostasis and disease.
Incorporation of more than one microbial species in these models should be considered to create more physiologic relevance.
Current strategies used for developing stem cell based engineered models of the intestine need further improvement to more precisely recapitulate in vivo conditions of the GI tract.
These model systems offer exciting opportunity for understanding the complex interactions between the intestine and its microbiota.
Acknowledgement
Financial Support and sponsorship
This work was supported by NIH-NIAID grants U19AI116482 and U01AI124255 to VBY.
Footnotes
Conflict of interest
Vincent Young has consulted for Bio-K+ International, Inc., Pantheryx, Exarca Pharmaceuticals, and Vedanta Biosciences.
References and recommended readings
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Pearce SC, Coia HG, Karl JP, Pantoja-Feliciano IG, Zachos NC, Racicot K. Intestinal in vitro and ex vivo Models to Study Host-Microbiome Interactions and Acute Stressors. Front Physiol. 2018;9:1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson AC, Greenwood-Van Meerveld B. Critical Evaluation of Animal Models of Gastrointestinal Disorders. Handb Exp Pharmacol. 2017;239:289–317. [DOI] [PubMed] [Google Scholar]
- 3.Daoud A, Munera JO. Insights Into Human Development and Disease From Human Pluripotent Stem Cell Derived Intestinal Organoids. Front Med. 2019;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh A, Poling HM, Spence JR, Wells JM, Helmrath MA. Gastrointestinal Organoids: A Next Generation Tool for Modeling Human Development. Am J Physiol Gastrointest Liver Physiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mithal A, Capilla A, Heinze D, Berical A, Villacorta-Martin C, Vedaie M, et al. Generation of mesenchyme free intestinal organoids from human induced pluripotent stem cells. Nat Commun 2020;11(1):215. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Differentiation of iPSCs towards mesenchyme free human intestinal organoids was demonstrated in this study. This novel approach will exclusively allow study of epithelial cell development.
- 6.Hill DR, Huang S, Nagy MS, Yadagiri VK, Fields C, Mukherjee D, et al. Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. eLife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schutgens F, Clevers H. Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu Rev Pathol. 2020;15:211–34. [DOI] [PubMed] [Google Scholar]
- 8.Altay G, Larranaga E, Tosi S, Barriga FM, Batlle E, Fernandez-Majada V, et al. Self-organized intestinal epithelial monolayers in crypt and villus-like domains show effective barrier function. Sci Rep. 2019;9(1):10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freire R, Ingano L, Serena G, Cetinbas M, Anselmo A, Sapone A, et al. Human gut derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci Rep. 2019;9(1):7029. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study explored the complex interaction between microbial metabolites, diet and genetic variation of celiac disease (CD) by deriving enteroids from CD and non-CD patients.
- 10.Kraiczy J, Nayak KM, Howell KJ, Ross A, Forbester J, Salvestrini C, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68(1):49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbester JL, Hannan N, Vallier L, Dougan G. Derivation of Intestinal Organoids from Human Induced Pluripotent Stem Cells for Use as an Infection System. Methods Mol Biol. 2019;1576:157–69. [DOI] [PubMed] [Google Scholar]
- 12.Co JY, Margalef-Catala M, Li X, Mah AT, Kuo CJ, Monack DM, et al. Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions. Cell Rep. 2019;26(9):2509–20 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The plasticity of three dimensional structures was further reported in this study. Contrary to the previous apical inside entroids, this study generated 3D entroids with apical surface on the outside.
- 13.Zhang YG, Sun J. Study Bacteria-Host Interactions Using Intestinal Organoids. Methods Mol Biol. 2019;1576:249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.In JG, Foulke-Abel J, Clarke E, Kovbasnjuk O. Human Colonoid Monolayers to Study Interactions Between Pathogens, Commensals, and Host Intestinal Epithelium. J Vis Exp. 2019(146). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilke G, Funkhouser-Jones LJ, Wang Y, Ravindran S, Wang Q, Beatty WL, et al. A Stem-Cell-Derived Platform Enables Complete Cryptosporidium Development In Vitro and Genetic Tractability. Cell host & microbe. 2019;26(1):123–34 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study showed that intestinal enteroid monolayers can be maintained at air-liquid interface without losing viability or function.
- 16.Williamson IA, Arnold JW, Samsa LA, Gaynor L, DiSalvo M, Cocchiaro JL, et al. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cell Mol Gastroenterol Hepatol. 2018;6(3):301–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fofanova T, Stewart C, Auchtung J, Wilson R, Britton R, Grande-Allen K, et al. A novel human enteroid-anaerobe co-culture system to study microbial-host interaction under physiological hypoxia. 2019:555755. [Google Scholar]; **This study is the first to demonstrate co-culture of intestinal enteroids and strict anaerobic bacteria for upto 24 hrs at aerboic-anaerobic interphase.
- 18.Sasaki N, Miyamoto K, Maslowski KM, Ohno H, Kanai T, Sato T. Development of a scalable co-culture system for gut anaerobes and human colon epithelium. Gastroenterology. 2020. [DOI] [PubMed] [Google Scholar]
- 19.Sidar B, Jenkins BR, Huang S, Spence JR, Walk ST, Wilking JN. Long-term flow through human intestinal organoids with the gut organoid flow chip (GOFlowChip). Lab Chip. 2019;19(20):3552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng. 2019;3(7):520–31. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This article developed and used a complex and physiologically relevant model of small intestine ‘Intestine-on-chip’ for co-culturing stool microbiota with host epithelium.
- 21.Tovaglieri A, Sontheimer-Phelps A, Geirnaert A, Prantil-Baun R, Camacho DM, Chou DB, et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome. 2019;7(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mileto SJ, Jarde T, Childress KO, Jensen JL, Rogers AP, Kerr G, et al. Clostridioides difficile infection damages colonic stem cells via TcdB, impairing epithelial repair and recovery from disease. Proc Natl Acad Sci U S A. 2020;117(14):8064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** By using enteorid models, this study provided the direct evidence that a bacterial infection can prevent intestinal stem cells to performs its basic function of regenrating intestinal epithelium.
- 23.Levy A, Stedman A, Deutsch E, Donnadieu F, Virgin HW, Sansonetti PJ, et al. Innate immune receptor NOD2 mediates LGR5(+) intestinal stem cell protection against ROS cytotoxicity via mitophagy stimulation. Proc Natl Acad Sci U S A. 2020;117(4):1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Xie S, Miao J, Li Y, Wang Z, Wang M, et al. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut Microbes. 2020:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]; * In this study, the dual role of non-pathogenic bacteria during intestinal homeostasis and inflammation was investigated using enteroids.
- 25.Liu R, Moriggl R, Zhang D, Li H, Karns R, Ruan HB, et al. Constitutive STAT5 activation regulates Paneth and Paneth-like cells to control Clostridium difficile colitis. Life Sci Alliance. 2019;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoi Y, Nakamura K, Yoneda T, Kikuchi M, Sugimoto R, Shimizu Y, et al. Paneth cell granule dynamics on secretory responses to bacterial stimuli in enteroids. Sci Rep. 2019;9(1):2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Zhang T, Guo C, Geng M, Gai S, Qi W, et al. Bacillus subtilis RZ001 improves intestinal integrity and alleviates colitis by inhibiting the Notch signalling pathway and activating ATOH-1. Pathogens and disease. 2020;78(2). [DOI] [PubMed] [Google Scholar]
- 28.Mandic AD, Woting A, Jaenicke T, Sander A, Sabrowski W, Rolle-Kampcyk U, et al. Clostridium ramosum regulates enterochromaffin cell development and serotonin release. Scientific Reports. 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** An intersting study demonstrating mechanistic details of how a gut bacteria modulates epithelial cell differentiation using HIEs.
- 29.Fujii M, Matano M, Toshimitsu K, Takano A, Mikami Y, Nishikori S, et al. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell. 2018;23(6):787–93 e6. [DOI] [PubMed] [Google Scholar]
- 30.Chang-Graham AL, Danhof HA, Engevik MA, Tomaro-Duchesneau C, Karandikar UC, Estes MK, et al. Human Intestinal Enteroids With Inducible Neurogenin-3 Expression as a Novel Model of Gut Hormone Secretion. Cell Mol Gastroenterol Hepatol. 2019;8(2):209–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fasciano AC, Blutt SE, Estes MK, Mecsas J. Induced Differentiation of M Cell-like Cells in Human Stem Cell-derived Ileal Enteroid Monolayers. J Vis Exp. 2019(149). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo XC, Chen ZH, Xue JB, Zhao DX, Lu C, Li YH, et al. Infection by the parasitic helminth Trichinella spiralis activates a Tas2r-mediated signaling pathway in intestinal tuft cells. Proc Natl Acad Sci U S A. 2019;116(12):5564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranganathan S, Doucet M, Grassel CL, Delaine-Elias B, Zachos NC, Barry EM. Evaluating Shigella flexneri Pathogenesis in the Human Enteroid Model. Infect Immun. 2019;87(4). [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study demonstrated the use of enteroids with enhanced M cells to study Shigella flexneri pathogenesis.
- 34.Beaumont M, Paes C, Mussard E, Knudsen C, Cauquil L, Aymard P, et al. Gut microbiota derived metabolites contribute to intestinal barrier maturation at the suckling-to-weaning transition. Gut Microbes. 2020:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dougherty MW, Kudin O, Muhlbauer M, Neu J, Gharaibeh RZ, Jobin C. Gut microbiota maturation during early human life induces enterocyte proliferation via microbial metabolites. BMC Microbiol. 2020;20(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]; * An interesting study that used murine enteroids to show differential effect of microbiota obtained from term and pre-term neonates on intestinal stem cell proliferation and regeneration.
- 36.Abo H, Chassaing B, Harusato A, Quiros M, Brazil JC, Ngo VL, et al. Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration. Nat Commun. 2020;11(1):513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min S, Kim S, Cho SW. Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp Mol Med. 2020;52(2):227–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verma S, Senger S, Cherayil BJ, Faherty CS. Spheres of Influence: Insights into Salmonella Pathogenesis from Intestinal Organoids. Microorganisms. 2020;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolawole AO, Wobus CE. Gastrointestinal organoid technology advances studies of enteric virus biology. PLoS Pathog. 2020;16(1):e1008212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunasekera S, Zahedi A, O’Dea M, King B, Monis P, Thierry B, et al. Organoids and Bioengineered Intestinal Models: Potential Solutions to the Cryptosporidium Culturing Dilemma. Microorganisms. 2020;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estes MK, Ettayebi K, Tenge VR, Murakami K, Karandikar U, Lin SC, et al. Human Norovirus Cultivation in Nontransformed Stem Cell-Derived Human Intestinal Enteroid Cultures: Success and Challenges. Viruses. 2019;11(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duque-Correa MA, Maizels RM, Grencis RK, Berriman M. Organoids - New Models for Host-Helminth Interactions. Trends Parasitol. 2020;36(2):170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]; * A comprehensive review providing upto date information regarding the use of organoids as models for host-parasite interactions.
- 43.Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5(47). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nature medicine. 2020. [DOI] [PubMed] [Google Scholar]
- 45.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The timely publication of this study demonstrates how rapidly organotypic models can be developed and used to know key aspects of newly identified infectious agents.
- 46.Fujii M, Clevers H, Sato T. Modeling Human Digestive Diseases With CRISPR-Cas9-Modified Organoids. Gastroenterology. 2019;156(3):562–76. [DOI] [PubMed] [Google Scholar]
- 47.Munch NS, Fang HY, Ingermann J, Maurer HC, Anand A, Kellner V, et al. High-Fat Diet Accelerates Carcinogenesis in a Mouse Model of Barrett’s Esophagus via Interleukin 8 and Alterations to the Gut Microbiome. Gastroenterology. 2019;157(2):492–506 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angus HCK, Butt AG, Schultz M, Kemp RA. Intestinal Organoids as a Tool for Inflammatory Bowel Disease Research. Front Med. 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, et al. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. 2020;580(7802):269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study was first to use HIEs to demonstrate the development of distint mutational signatures in intestinal epithelium in response to exposure to bacteria carrying colibactin producing pathogenecity island.
- 51.Workman MJ, Mahe MM, Trisno S, Poling HM, Watson CL, Sundaram N, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nature medicine. 2017;23(1):49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jose SS, De Zuani M, Tidu F, Hortova Kohoutkova M, Pazzagli L, Forte G, et al. Comparison of two human organoid models of lung and intestinal inflammation reveals Toll-like receptor signalling activation and monocyte recruitment. Clinical & translational immunology. 2020;9(5):e1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolawole AO, Mirabelli C, Hill DR, Svoboda SA, Janowski AB, Passalacqua KD, et al. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog. 2019;15(10):e1008057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koestler BJ, Ward CM, Fisher CR, Rajan A, Maresso AW, Payne SM. Human Intestinal Enteroids as a Model System of Shigella Pathogenesis. Infect Immun. 2019;87(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma S, Prescott RA, Ingano L, Nickerson KP, Hill E, Faherty CS, et al. The YrbE phospholipid transporter of Salmonella enterica serovar Typhi regulates the expression of flagellin and influences motility, adhesion and induction of epithelial inflammatory responses. Gut Microbes. 2019:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonyar LA, Smith RM, Giron JA, Zachos NC, Ruiz-Perez F, Nataro JP. Aggregative adherence fimbriae II of enteroaggregative Escherichia coli are required for adherence and barrier disruption during infection of human colonoids. Infect Immun. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hosmillo M, Chaudhry Y, Nayak K, Sorgeloos F, Koo BK, Merenda A, et al. Norovirus Replication in Human Intestinal Epithelial Cells Is Restricted by the Interferon-Induced JAK/STAT Signaling Pathway and RNA Polymerase II-Mediated Transcriptional Responses. mBio. 2020;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Good C, Wells AI, Coyne CB. Type III interferon signaling restricts enterovirus 71 infection of goblet cells. Sci Adv. 2019;5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engevik MA, Danhof HA, Chang-Graham AL, Spinler JK, Engevik KA, Herrmann B, et al. Human intestinal enteroids as a model of Clostridioides difficile-induced enteritis. Am J Physiol Gastrointest Liver Physiol. 2020;318(5):G870–G88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Z, Schnell L, Muller B, Muller M, Papatheodorou P, Barth H. The Antibiotic Bacitracin Protects Human Intestinal Epithelial Cells and Stem Cell-Derived Intestinal Organoids from Clostridium difficile Toxin TcdB. Stem Cells Int. 2019;2019:4149762. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study demonstrated that in addition to direct pathogen killing, antibiotics (bacitracin) can also control infections by inhibiting virulence factor (toxin B) released by C.difficile.
- 61.Llanos-Chea A, Citorik RJ, Nickerson KP, Ingano L, Serena G, Senger S, et al. Bacteriophage Therapy Testing Against Shigella flexneri in a Novel Human Intestinal Organoid-Derived Infection Model. J Pediatr Gastroenterol Nutr. 2019;68(4):509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


