Abstract
Blood biomarkers of multiple sclerosis (MS) can provide a better understanding of pathophysiology and enable disease monitoring. Here, we performed quantitative shotgun lipidomics on the plasma of a unique cohort of 73 monozygotic twins discordant for MS. We analyzed 243 lipid species, evaluated lipid features such as fatty acyl chain length and number of acyl chain double bonds, and detected phospholipids that were significantly altered in the plasma of co‐twins with MS compared to their non‐affected siblings. Strikingly, changes were most prominent in ether phosphatidylethanolamines and ether phosphatidylcholines, suggesting a role for altered lipid signaling in the disease.
Introduction
Multiple sclerosis (MS) exhibits high interpersonal variance and our capability to predict individual disease courses is limited. 1 Thus, there is an unmet need for easily accessible biomarkers to support the diagnosis and monitor disease activity as well as treatment response. 2 Single‐molecule arrays have uncovered serum neurofilament light chain as a protein‐marker for axonal damage in MS and other diseases. 3 However, in MS the inflammatory process also results in extensive damage to myelin sheaths, which are multilayered membrane stacks, highly enriched in lipids such as galactosylceramides, sulfogalactosylceramides, and ether‐linked phospholipids. 4 To date, the lipidome of MS‐patients is not well defined and unbiased studies with an open, untargeted approach to lipidomics are scarce. 5 , 6 , 7 While so far the reproducible quantification of entire lipid profiles in multiple samples has been challenging, the recent development of high throughput and quantitative shotgun lipidomics has overcome this limitation. 8 , 9 We used this technology to obtain a detailed and quantitative assessment of the plasma lipidome of MS patients including features such as fatty acyl chain length and number of acyl chain double bonds. To eliminate genetic heterogeneity and reduce environmental influences, we focused our analysis on 73 monozygotic twin pairs, discordant for MS.
Subjects/Materials And Methods
Study Cohort
Twins were recruited nation‐wide in Germany. Inclusion criteria for study participation were met for monozygotic twins with an MS diagnosis according to the 2010 revised McDonald criteria or clinically isolated syndrome in one twin only. 10 In total, we included 73 monozygotic twin pairs (see Table 1 for cohort description) as previously described. 11 Blood sampling was always performed in the morning in fasting condition. The local ethics committee of the Ludwig Maximilians University of Munich approved the study and all participants gave written informed consent.
Table 1.
Cohort description.
| Healthy | Multiple Sclerosis | Wilcoxon matched‐pairs signed rank test | |
|---|---|---|---|
| Number of twins | 73 | ||
| Age, yrs. [mean ± SD | 40.8 ± 12.1 | ||
| Female [%] | 75.3 % | ||
| BMI [mean ± SD] | 24.7 ± 4.8 | 24.3 ± 4.9 | P = 0.23 |
| Disease course [%] | |||
| RRMS | 75 % | ||
| SPMS | 19 % | ||
| PPMS | 3 % | ||
| CIS | 3 % | ||
| Patients without treatment, no. (%) | 20 (27%) | ||
| Disease duration, yrs. [mean ± SD] | ‐ | 11.4 ± 9.6 | |
| EDSS | 0.3 ± 0.5 | 2.8 ± 2.0 | P < 0.0001 |
BMI body mass index defined as body mass divided by the square of the body height, expressed in kg/m2. RRMS relapsing‐remitting multiple sclerosis (MS), SPMS secondary progressive MS, PPMS primary progressive MS, CIS clinically isolated syndrome. EDSS Expanded Disability Status Scale is a method of quantifying disability in multiple sclerosis only. Scale range from 0 to 10, where 0 comprises a normal neurological exam and 10 is defined as death due to MS. In the healthy co‐twins abnormal neurological signs were documented using the EDSS as well for better comparison, even though the etiology of these slight abnormal neurological findings remains unknown.
Lipidomics
Lipid species and subspecies are annotated according to their molecular composition as described previously and lipid identifiers are provided 12 (Table S1, Table 1).
One μL of plasma was analyzed using Shotgun Lipidomics platform by Lipotype GmbH (Dresden, Germany), as described previously. 8 Lipidomic data were analyzed as described, 8 with criteria on lipid identification, batch‐ and drift correction as applied earlier 13 using R version 3.6.1 (2019‐07‐05) and tidyverse packages (version 1.2.1). A 66% occupational threshold was applied to the data, that is, we only considered lipids, which occur in at least 66% of the samples in either healthy co‐twins or in co‐twins with MS, for further analysis. Data were mostly normally distributed and no bias of missing data was detectable before thresholding (Table S2, Figure S1). Threshold application leads to a reduction from 722 measured lipids to 243 lipids for further comparison. If not stated otherwise lipids are measured in molar amounts normalized by the total lipid amount (mol%) per person.
Statistics
If not stated otherwise, features between healthy co‐twins (H) and co‐twins with MS are analyzed by matched comparison using a paired ratio t‐test with significant results defined as P < 0.05. Adjustment for multiple testing is performed with the method of Benjamini and Hochberg (BH) (*P < 0.05, **P < 0.01, ***P < 0.001.).
Results
We performed shotgun lipidomics in plasma samples of 73 monozygotic twins discordant for MS (see Table 1 for cohort description) and analyzed a total of 243 lipid species after application of an occupational threshold of 66% (Table S1, Table S2). Total lipid amount did not differ between healthy co‐twins and MS co‐twins and principal component analysis revealed no separation between groups (Figure S1A and Figure S2A). Surprisingly, we found a significant reduction in ether phosphatidylcholines (PC O‐), ether phosphatidylethanolamines (PE O‐) and phosphatidylcholines (PC) in MS co‐twins, of which the first two persist after correction for multiple comparison (Fig 1A; Table S3, Table S1). Of note, both PC O‐ and PE O‐ represent ether lipids, in which an ether bond attaches the hydrocarbon chain, instead of an ester bond as in the more common diacyl phospholipids. Detailed analysis of these lipid classes revealed that the reduction in PC O‐ species is accompanied by a relative reduction in certain acyl chains (i.e., bound fatty acids) within this lipid class. Namely, docosapentaenoic acid (DPA, C22:5), other polyunsaturated fatty acids (C22:4, C20:3, C20:4) and ether bound saturated/ monounsaturated alkyls (O‐16:0;0, O‐16:1;0 and O‐18:1;0) were significantly reduced and the reduction in DPA maintained significance after correction for multiple testing (P = 0.00047, q = 0.011, mean reduction (MS/H) =0.838).
Figure 1.
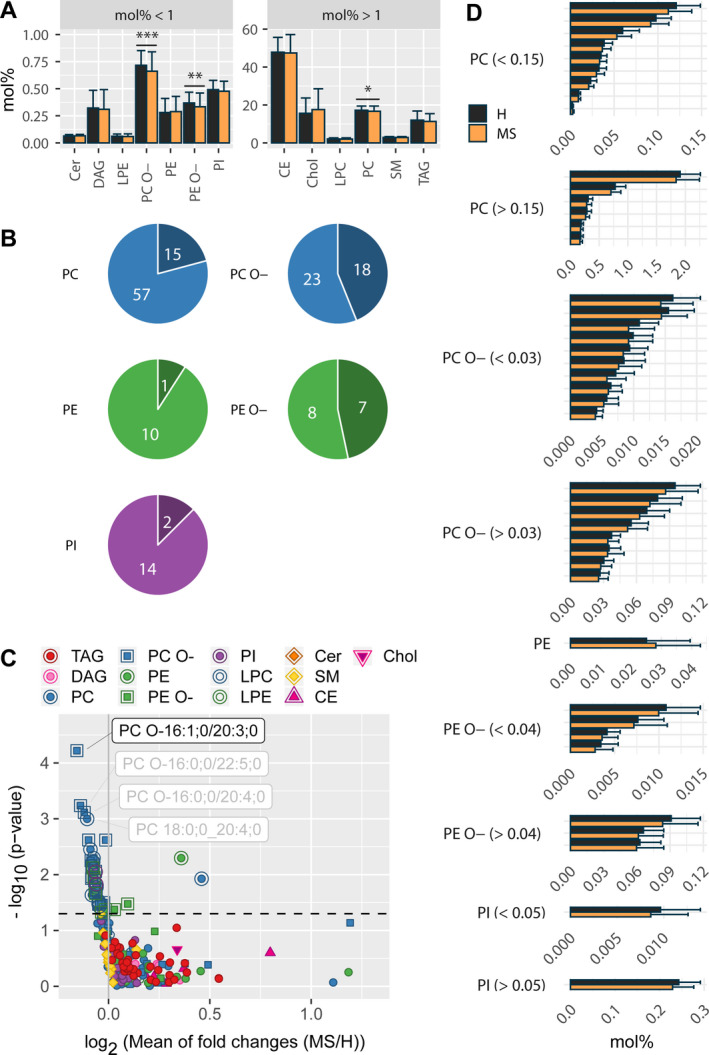
Altered lipid classes and species in monozygotic co‐twins discordant for Multiple Sclerosis. (A) On lipid class level, PC O‐, PE O‐, and PC are significantly decreased in MS co‐twins (P = 0.00081, P = 0.0015, P = 0.017, and q = 0.0095, q = 0.0095, q = 0.074, respectively). Y axis depicts lipids in molar amounts normalized by the total lipid amount per participant [mol%].) Paired ratio t‐test, *P < 0.05, **P < 0.01, ***P < 0.001. (B) Number of significantly altered lipid species (darker color) as parts of whole of the corresponding lipid class. (C) A Volcano plot of all measured lipid species shows that most significantly altered lipid species are less concentrated in participants with MS and that species with the smallest p‐values consist mainly of PC and PC O‐ species. X axis shows log2 of fold changes (MS/H), gray vertical line depicts x = 0 (MS = H), Y axis shows –log10 of P‐values (paired ratio t‐test), dotted horizontal line depicts P = 0.05. Significantly changed species (P < 0.05) are marked with an additional outline. (D) Significantly altered lipid species between healthy (black) and MS (orange) co‐twins grouped per lipid class. X axis depicts lipids in molar amounts normalized by the total lipid amount per participant [mol%]. Only significant results are illustrated. (B), (C) Lipids are colored according to lipid class. TAG Triacylglycerol, DAG Diacylglycerol, PC Phosphatidylcholine, LPC Lysophosphatidylcholines, PC O‐ ether‐phosphatidylcholine, PE Phosphatidylethanolamine, LPE Lysophosphatidylethanolamine, PE O‐ ether‐phosphatidylethanolamine, PI Phosphatidylinositol, Cer Ceramide, SM Sphingomyelin, Chol Cholesterol, CE Cholesteryl ester. H healthy, MS Multiple Sclerosis.
On lipid species level, we detected 43 differentially concentrated lipid species between healthy and MS co‐twins, thereby constituting 18% of all measured lipid species (Fig. 1B and C; Table S3, Table S2). In detail, 47% of PE O‐, 44% of PC O‐, 21% of PC, 13% of PI, and 9% of phosphatidylethanolamine (PEs) species were altered (7 PE O‐, 18 PC O‐, 15 PC, 2 PI, 1 PE species in absolute numbers). After correction for multiple comparison 1 lipid subspecies, namely PC O‐16:1;0/20:3;0 remained as true discovery (0.0162 ± 0.00437 mol% (H) vs. 0.0143 ± 0.00511 mol% (MS), P = 0.00006, q = 0.015, mean of fold change (MS/H) 0.898). Of note, top hits at the species level are dominantly PC O‐ species with polyunsaturated fatty acids and the vast majority of significantly changed species, which we define by a nominal P‐value < 0.05 for this report, are slightly decreased in MS co‐twins (Figure 1C and D). Of note, paired multivariate analyses reveal a small separation between healthy co‐twins and co‐twins with MS and discriminating features comprise the PC O‐ species from the top hits from univariate testing (Figure S2). To explore if the observed alterations are rather due to MS pathology or to a side effect of disease‐modifying treatment we analyzed the significantly altered lipid species in the subgroup of untreated twin pairs. Reassuringly, this led to an increase in changes for most species indicating a genuine link to MS pathology (median of fold change MS/H all = 0.96 vs MS/H untreated = 0.93, P = 0.0005, Wilcoxon matched‐pairs signed‐rank test).
Discussion
We performed comprehensive, untargeted lipidomics in plasma samples of a unique cohort of 73 monozygotic twins discordant for MS. We measured 243 lipid species, and found that PC O‐ containing polyunsaturated fatty acids are decreased in MS. This observation was mirrored on a class level by a decrease in the ether lipid classes PC O‐ and PE O‐ and was accompanied by a reduction in specific polyunsaturated acyls and ether‐bound saturated/monounsaturated alkyls.
One of the unique features of our study cohort is increased test power due to matched comparison among genetically identical twins. Thus, unmatched comparison results in a reduction in significant hits and maintains a total of seven differentially concentrated lipids, which all belong to the PC O‐ and PC class and include PC O‐16:1;0/20:3;0. On class level, reduction in PC O‐ persists, if an unmatched test is performed.
Ethers, as PC O‐ and PE O‐, do not only effect biophysical properties of membranes (e.g., lipid packing), but do also play an important role in lipid signaling and inflammation. 14 Ether lipids are enriched in polyunsaturated fatty acids as arachidonic acid (AA) and docosahexaenoic acid (DHA), which can be released upon activation of phospholipase A2. Eicosanoids, their oxygenated products, are major signaling compounds in inflammation. 15 Furthermore, ethers are thought to be protective against oxidative stress by acting as sacrificial antioxidants and are potential ligands of peroxisome proliferator‐activated receptor gamma, a transcription factor with metabolic and anti‐inflammatory functions. 14 , 16 , 17 Ether lipids and polyunsaturated fatty acids are also decreased in autoimmune diseases like systemic lupus erythematosus. 18 Therefore, it seems more likely that the observed reduction in ethers is a general consequence of prolonged inflammation, even if a MS‐specific pathophysiological role cannot be excluded. In this study, we used monozygotic twin pairs with discordant health status as a genetically matched case‐control cohort for a quantitative plasma lipidome analysis. As alterations increase in the untreated subgroup of twin pairs, the observed lipidome variations seem to be genuinely linked to disease rather than to treatment effects. Further studies are required to validate our results in larger, genetically heterogenous MS cohorts, to understand the specificity and pathophysiological role of lipidome alterations in MS and to explore if lipid profiles may function as biomarkers as recently suggested 19.
Conflicts of Interest
Christian Klose is shareholder of Lipotype GmbH. Mathias Gerl, Chris Lauber, and Markus Damm are employees of Lipotype GmbH. All other authors declare no conflict of interest.
Authors’ Contributions
H.P., L.G., and M.S. conceived the project and designed experiments. H.P., C.K., M.G. C.L., C.K., and D.F. carried out experiments, H.P., C.K., M.G. C.L., C.K., M.D., D.F, R.H., L.G., and M.S. analyzed the data or supervised data acquisition. H.P., M.G., C.L., and M.D. visualized the data. L.G., A.F.H., and T.K. recruited patients. M.K. and R.H. supervised the MS twin cohort. H.P. and M.S. supervised the project and wrote the manuscript.
Supporting information
Figure S1. Lipid distribution.
Figure S2. Paired multivariate analyses show a discrimination between healthy co‐twins and co‐twins with MS.
Table S1. Annotation of lipid species and subspecies and mol% raw data.
Table S2. Data distribution and test for normality.
Table S3. Quantification of lipid classes and species.
Acknowledgments
We are grateful to all subjects who participated in this study.
Funding Information
The work was supported by grants from the German Research Foundation (TRR274, TRR128, SyNergy Excellence Cluster, EXC2145, Projekt ID390857198), the Human Frontier Science Program (HFSP), the European Research Counsil (ERC, Consolidator Grant to M.S.), the Klaus Tschira Foundation, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Else Kröner Fresenius Foundation, the Non‐profit Hertie Foundation, the national and bavarian division of the German MS society (DMSG and DMSG‐Landesverband Bayern), and the association "Verein zur Therapieforschung für MS Kranke e.V."
Funding Statement
This work was funded by ERC grant ERC‐CoG.
Contributor Information
Lisa A. Gerdes, Email: lgerdes@med.lmu.de.
Mikael Simons, Email: mikael.simons@dzne.de.
REFERENCES
- 1. Chung KK, Altmann D, Barkhof F, et al. A 30‐Year Clinical and Magnetic Resonance imaging observational study of multiple sclerosis and clinically isolated syndromes. Ann Neurol 2020;87:63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ziemssen T, Akgün K, Brück W. Molecular biomarkers in multiple sclerosis. J Neuroinflammation 2019;16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Disanto G, Barro C, Benkert P, et al. Serum Neurofilament light: A biomarker of neuronal damage in multiple sclerosis. Ann Neurol 2017;81(6):857–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stadelmann C, Timmler S, Barrantes‐Freer A, Simons M. Myelin in the central nervous system: structure, function, and pathology. Physiol Rev 2019;99:1381–1431. [DOI] [PubMed] [Google Scholar]
- 5. De Oliveira EML, Montani DA, Oliveira‐Silva D, et al. Multiple sclerosis has a distinct lipid signature in plasma and cerebrospinal fluid. Arq Neuropsiquiatr 2019;77:696–704. [DOI] [PubMed] [Google Scholar]
- 6. Senanayake VK, Jin W, Mochizuki A, et al. Metabolic dysfunctions in multiple sclerosis: implications as to causation, early detection, and treatment, a case control study [Internet]. BMC Neurol. 2015;15(1):154 10.1186/s12883-015-0411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Del Boccio P, Pieragostino D, Di Ioia M, et al. Lipidomic investigations for the characterization of circulating serum lipids in multiple sclerosis [Internet]. J Proteomics 2011;74:2826–2836. 10.1016/j.jprot.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 8. Surma MA, Herzog R, Vasilj A, et al. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur J Lipid Sci Technol 2015;117(10):1540–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gross RW. The evolution of lipidomics through space and time. Biochim Biophys Acta Mol Cell Biol Lipids 2017;1862:731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 2011;69:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Souren NY, Gerdes LA, Lutsik P, et al. DNA methylation signatures of monozygotic twins clinically discordant for multiple sclerosis [Internet]. Nat Commun 2019;10:1–12. 10.1038/s41467-019-09984-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aimo L, Liechti R, Hyka‐Nouspikel N, et al. The SwissLipids knowledgebase for lipid biology [Internet]. Bioinformatics 2015;31:2860–2866. 10.1093/bioinformatics/btv285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerl MJ, Klose C, Surma MA, et al. Machine learning of human plasma lipidomes for obesity estimation in a large population cohort. PLoS Biol. 2019;17:e3000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dean JM, Lodhi IJ. Structural and functional roles of ether lipids [Internet]. Protein. Cell 2018;9:196–206. http://link.springer.com/10.1007/s13238‐017‐0423‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation [Internet]. Nat Rev Immunol 2015;15:511–523. 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation [Internet]. Trends Endocrinol Metab 2012;23:351–363. 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 17. Villapol S. Roles of peroxisome proliferator‐activated receptor gamma on brain and peripheral inflammation. Cell Mol Neurobiol 2018;38:121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferreira HB, Pereira AM, Melo T, et al. Lipidomics in autoimmune diseases with main focus on systemic lupus erythematosus [Internet]. J Pharm Biomed Anal 2019;174:386–395. http://www.sciencedirect.com/science/article/pii/S0731708519307186 [DOI] [PubMed] [Google Scholar]
- 19. Lötsch J, Schiffmann S, Schmitz K, et al. Machine‐learning based lipid mediator serum concentration patterns allow identification of multiple sclerosis patients with high accuracy. Sci Rep 2018;8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Lipid distribution.
Figure S2. Paired multivariate analyses show a discrimination between healthy co‐twins and co‐twins with MS.
Table S1. Annotation of lipid species and subspecies and mol% raw data.
Table S2. Data distribution and test for normality.
Table S3. Quantification of lipid classes and species.


