Abstract
INTRODUCTION:
In contrast to most colorectal carcinomas arising from pedunculated or sessile protruded adenomas, submucosal-invasive (pT1) colorectal carcinoma exhibiting a depressed surface (hereinafter, “depressed colorectal carcinoma,” identified by means of high-definition endoscopy) is considered to be derived from depressed precursors. We hypothesized that depressed colorectal neoplasms have unique clinicopathological features different that are different from those of protruded and flat colorectal neoplasms.
METHODS:
We classified 27,129 colorectal neoplasms (909 pT1 carcinomas and 26,220 adenomas) resected between 2001 and 2017 into depressed (211 carcinomas and 109 adenomas), flat (304 carcinomas and 11,246 adenomas), and protruded subtypes (394 carcinomas and 14,865 adenomas) and compared their clinicopathological features. As exploratory analyses of pT1 carcinomas, we conducted whole-exome sequencing for 19 depressed and 8 protruded subtypes and RNA sequencing for 8 depressed and 8 protruded subtypes.
RESULTS:
pT1 carcinomas were more common in depressed lesions (66%) than in protruded (2.6%) and flat lesions (2.6%) (P < 0.001). Compared with nondepressed pT1 carcinomas, depressed pT1 carcinomas were positively correlated with lymphovascular invasion, tumor budding, and massive submucosal invasion and inversely correlated with the presence of an adenoma component (all P < 0.001). Depressed adenomas were more likely to contain high-grade dysplasia than nondepressed adenomas (49% vs 11%, P < 0.001). A KRAS mutation was observed only in one of the 19 depressed pT1 carcinomas. Relative to protruded carcinomas, depressed carcinomas generally exhibited higher expression of genes related to angiogenesis and epithelial-mesenchymal transition.
DISCUSSION:
Depressed colorectal neoplasms may harbor a unique combination of malignant histopathological phenotypes and molecular features.
INTRODUCTION
With the advent of high-definition endoscopy and multiple imaging techniques, colorectal neoplasms with depressed morphology are increasingly being identified in clinical practice (1). The carcinogenesis of this depressed subset may differ from that used during the production of predominant protruded colorectal neoplasms that evolve through the traditional adenoma-carcinoma sequence or serrated pathway (2). Colorectal carcinomas are a heterogeneous collection of neoplasms arising from multiple carcinogenic pathways that result in distinct phenotypes based on intertumor variations regarding genetic and epigenetic alterations (3–5). A better understanding of the distinct pathways underlying colorectal carcinogenesis will help us refine the strategies of diagnosis, prevention, and treatment (6). However, given the difficulties in early endoscopic detection and the relative rarity of depressed colorectal tumors, the clinical, pathological, and molecular features of this subset have remained largely unexplored.
Accumulating evidence points to the distinct macroscopic and microscopic characteristics of depressed colorectal tumors (7,8). According to the Paris classification, there are 3 primary types of colorectal neoplasms, namely, protruded, flat, and depressed (9). To date, clinical efforts have focused on identifying and resecting polypoid-type colorectal tumors to manage colorectal cancer-related mortality (10–12). Compared with the nondepressed subtype, depressed colorectal neoplasms have been difficult to identify by colonoscopy and are associated with more invasive phenotypes, potentially contributing to the high incidence of metastasis of colorectal cancer (7,8,13,14). In a previous clinical study involving 18 depressed colorectal neoplasms among a total of 1,535 neoplasms, the nonpolypoid (depressed or flat) subtype was more likely to include the carcinoma component than the polypoid subtype; however, this study was limited by its small sample size (8). In addition, the molecular profiles of depressed colorectal tumors have not been examined. We hypothesized that depressed colorectal neoplasms might be distinctively different from the protruded and flat neoplasms regarding clinical, pathological, and molecular characteristics.
Using clinical and pathological data from consecutive colorectal neoplasms resected over 16 years, we examined the clinicopathological characteristics of depressed pT1 carcinomas and adenomas and compared them with the characteristics of the protruded and flat subtypes. In addition, we analyzed the mutation and gene expression in select pT1 carcinomas.
METHODS
Patients and colorectal neoplasms
We began by considering 27,129 colorectal neoplasms (adenomas and pT1 carcinomas) that had been resected endoscopically or surgically among 12,788 patients at Showa University Northern Yokohama Hospital between April 2001 and December 2017. Treatment selection was based on the guidelines proposed by the Japan Gastroenterological Endoscopy Society and the Japanese Society for Cancer of the Colon and Rectum (15,16). We excluded pT2 and more invasive carcinomas and neoplasms identified in individuals with a history of inflammatory bowel disease or familial adenomatous polyposis. None of the patients had received preoperative radiotherapy or neoadjuvant chemotherapy.
This study was approved by the institutional review boards at Showa University and Kyushu University (protocol numbers 213 and 629-00, respectively). The study was conducted according to the Declaration of Helsinki. Written informed consent was obtained from all patients included in this study. There were no animal experiments.
Collection of clinicopathological data
According to the Paris classification of endoscopic findings of superficial colorectal neoplasms (7), the tumors were morphologically classified into depressed, flat (including laterally spreading tumors), or protruded types (9,17). Clinical information was collected by reviewing electronic medical charts. Resected specimens of colorectal neoplasms were examined histopathologically in accordance with World Health Organization criteria (15), the Vienna classification system and Japan Gastroenterological Endoscopy Society, and the Japanese Society for Cancer of the Colon and Rectum guidelines (16,18,19). High-grade adenomas are consistent with the carcinoma in situ in the Vienna classification system (19). For all carcinoma cases included in this study, carcinoma components invading beyond the muscularis mucosa were pathologically confirmed. For pT1 carcinomas, histological grade (well to moderately differentiated vs poorly differentiated or mucinous), presence of adenoma component, depth of submucosal invasion, and grade of tumor budding (1–3) were assessed using hematoxylin and eosin (H&E)-stained tissue slides (20). Immunohistochemistry was performed to evaluate lymphatic and vascular invasion. Lymphatic invasion was evaluated by H&E staining and immunohistochemistry using the D2-40 antibody (Dako North America, Carpinteria, CA). Vascular invasion was evaluated using double staining with H&E and Victoria blue (Muto Pure Chemicals, Tokyo, Japan). To assess the reliability of the pathological findings, we measured their inter-rater concordance. Two gastrointestinal pathologists reviewed the pathological findings of 30 depressed lesions and calculated the kappa value of each pathological characteristic.
Extraction of DNA and RNA
For mutation and expression analyses, we obtained tissue samples from pT1 colorectal carcinoma and adjacent normal mucosa during endoscopic or surgical resection in 2015 or 2016 from patients with depressed and protruded subtypes (n = 8, each), those we could obtain the written informed consent. DNA and RNA were extracted from fresh-frozen tumor samples and matched normal mucosa using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany). We extracted DNA from formalin-fixed paraffin-embedded samples of 11 depressed pT1 colorectal carcinomas resected in 2013 or 2014 using the AllPrep DNA/RNA formalin-fixed paraffin-embedded Kit (Qiagen).
Whole-exome sequencing
Whole-exome capture of the extracted DNA samples was performed using the SureSelect Human All Exon V5 kit (Agilent Technologies, Tokyo, Japan). Captured targets were sequenced on a HiSeq 2500 (Illumina, San Diego, CA) with the pair-end 100-bp read option. The sequencing data were processed using the in-house pipeline Genomon 2.5.0 (http://genomon.hgc.jp) (21). Briefly, the sequencing reads were aligned to the National Center for Biotechnology Information Human Reference Genome Build 37 hg19 using BWA version 0.7.8 with default parameters (http://bio-bwa.sourceforge.net/). Polymerase chain reaction duplicate reads were filtered out using Picard (http://www.picard.sourceforge.net). Mutation calling was performed using the EBcall algorithm with the following settings: (i) mapping quality score ≥20, (ii) base quality score ≥15, (iii) both tumor and normal depths ≥8, (iv) variant reads in tumor ≥4, (v) variant allele frequency in tumor ≥0.05, (vi) variant allele frequency in paired normal samples ≤0.1, (vii) minus logarithm of P value for the Fisher exact test ≥1.3, and (viii) minus logarithm of P value for EBcall ≥5. The filtered mutations were annotated using ANNOVAR version 2015Dec14 (http://www.openbioinformatics.org/annovar/).
Copy number alterations (CNAs) in the whole-exome sequencing (WES) data were detected using EXCAVATOR47 software (http://sourceforge.net/projects/excavatortool/), which not only reports chromosomal segments subjected to CNAs but also outputs the log-transformed ratio of copy number intensities between tumor and normal samples (log R ratio) for each locus. If the length was >50% of the chromosomal arm, CNAs were classified as chromosomal arm-level CNAs; otherwise, they were classified as focal CNAs. A CNA was considered to be present if the absolute value of the averaged log R ratio was >0.15.
RNA sequencing
RNA sequencing was performed on HiSeq 2500, and the sequence data were processed using Genomon 2.5.0. The sequencing reads were analyzed using TopHat-Fusion (for gene fusion), Cufflinks, and HTSeq (for expression) (22–24). Inconsistent read pairs (in SAM format) and reads with a mapping quality score ≤20 were filtered out. For each specific exon and RefSeq gene, aligned bases were calculated. For each gene symbol, the associated RefSeq genes with maximum mapped bases divided by region size were obtained. Fragments per kilobase of exon per million reads mapped (FPKM) values for each gene symbol were calculated. Gene expression was measured and compared between depressed and protruded carcinomas using edgeR software (25). We assessed normalized expression data using gene set enrichment analysis (GSEA) software and the Molecular Signature Database (http://www.broad.mit.edu/gsea/), as previously described (26). We compared the FPKM values between depressed and protruded carcinomas using GSEA software. We used hallmark gene sets (50 gene sets). For genes related to angiogenesis, epithelial-mesenchymal transition (EMT), and inflammatory response, we generated a receiver operating characteristic (ROC) curve and calculated the area under the ROC curve. Immunohistochemical analysis of genes related to angiogenesis, EMT, and inflammatory response was performed for 10 depressed and 10 protruded T1 carcinomas. Anti-CD31 (PECAM1) antibody (PA0414; Leica Biosystems, Wetzlar, Germany), anti-VIM antibody (Nichirei biosciences, Tokyo, Japan), and anti-CD14 antibody (ab183322; Abcam, Cambridge, United Kingdom) were used.
Analysis of The Cancer Genome Atlas data
Using the Firehose pipeline at Broad Institute (Cambridge, MA; https://gdac.broadinstitute.org/), we obtained clinical and genetic data associated with colorectal adenocarcinoma cases included in The Cancer Genome Atlas (TCGA). We excluded hypermutated colorectal carcinomas because they are generally associated with a low frequency of CNAs. We compared CNAs between KRAS-mutant and -wild type cases.
Statistical analysis
Demographic data are presented as number of patients (%) or median (interquartile range). Continuous variables, including FPKM values, were compared using the Wilcoxon rank-sum test, and categorical variables were compared using the Fisher exact test.
All statistical analyses were performed using R software (version, 3.5.0.; http://www.r-project.org). All P values were 2 sided. Statistical significance was set at P ≤ 0.05. Research materials supporting this publication can be accessed by contacting the corresponding author.
RESULTS
Clinicopathological characteristics of depressed colorectal neoplasms
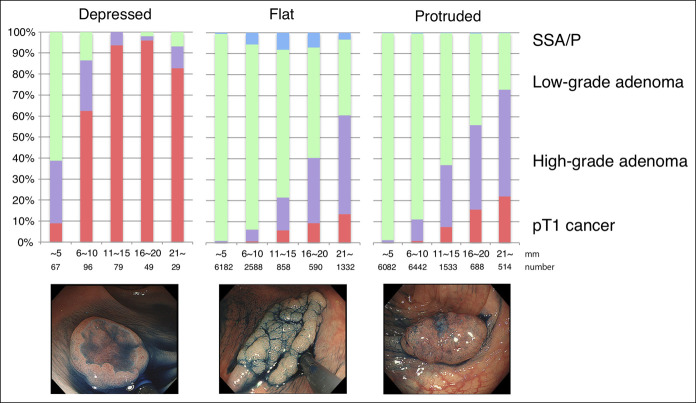
Among the 27,129 colorectal neoplasms resected, 320 (1.2%) were depressed, 11,550 (43%) were flat, and 15,259 (56%) were protruded. Histopathological diagnoses revealed a sessile serrated adenoma/polyp in 394 (1.5%), low-grade adenoma in 22,910 (84%), high-grade adenoma in 2,916 (11%), and pT1 carcinoma in 909 (3.4%). Among the 320 depressed neoplasms, 211 (66%) cases were pT1 carcinomas. The depressed subtype was more frequently observed in pT1 carcinomas than in adenomas (23 vs 0.4%, P < 0.005). Stage pT1 carcinoma was more frequently observed in depressed subtypes than in flat and protruded subtypes, regardless of the tumor size (Figure 1).
Figure 1.
Pathological characteristics of colorectal neoplasms according to tumor size and morphological subtypes. Pathological diagnoses of colorectal neoplasms according to the tumor size in each morphological subtype. Frequencies of pT1 carcinomas were compared between depressed and other types in each stratum of the tumor size. Values for P were based on the Wilcoxon rank-sum test for comparisons of rates of pT1 carcinomas between depressed and other subtypes. SSA/P, sessile serrated adenoma/polyp.
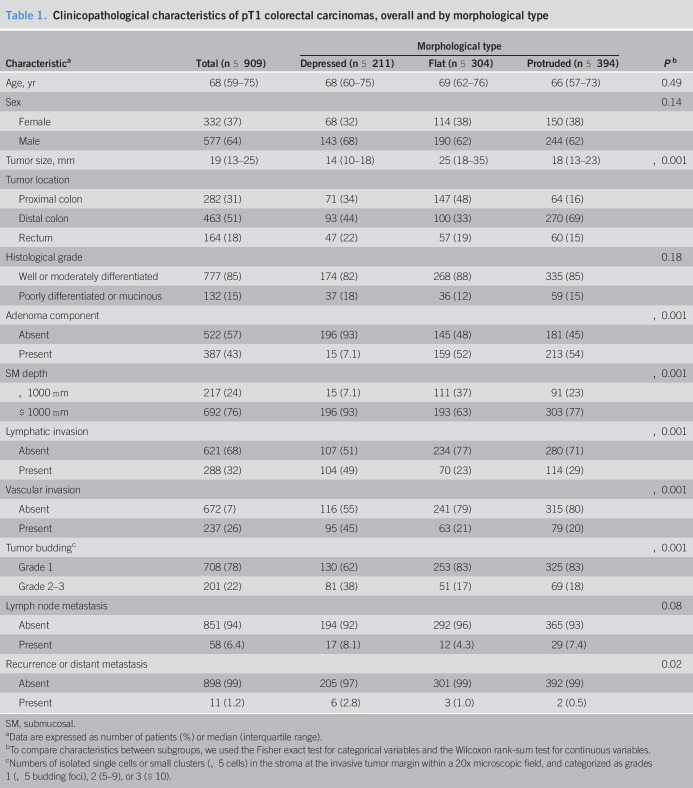
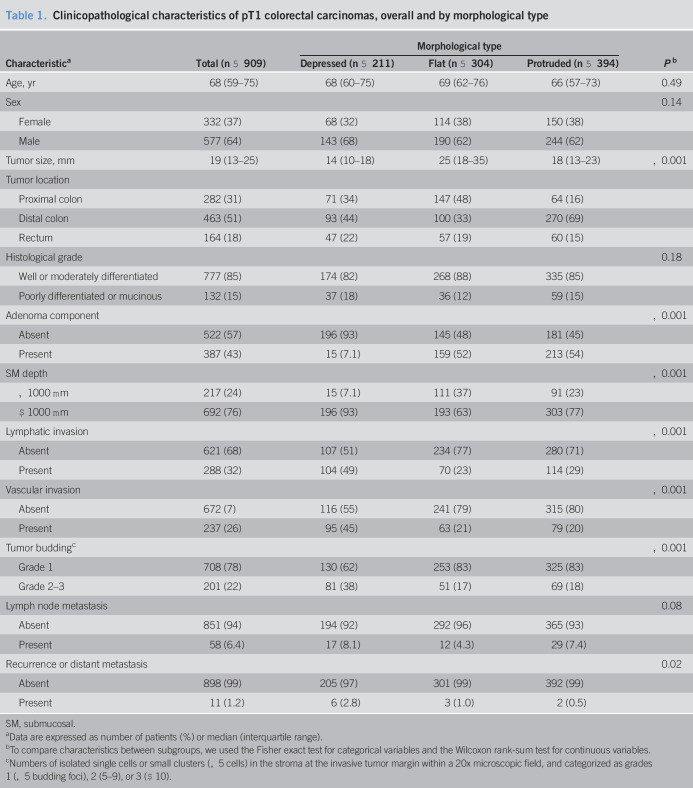
Relative to the protruded and flat pT1 carcinomas, depressed carcinomas were more strongly associated with lymphatic and vascular invasion, tumor budding, and massive submucosal invasion (all P < 0.005, Table 1). Adenoma components were less often observed in the depressed subtype than in the protruded and flat subtypes. The kappa values were 0.65 for histological grade, 0.74 for SM depth, 0.55 for lymphatic invasion, 0.81 for vascular invasion, and 0.64 for tumor budding. Supplementary Table 1 (see Supplementary Digital Content, http://links.lww.com/CTG/A452) summarizes the characteristics of 11 patients who underwent recurrence or distant metastasis after resection of pT1 colorectal carcinoma. Six patients (55%) had the depressed subtype. The median depth of submucosal invasion was 3,700 μm (range, 2,750–4,875 μm). Most of these tumors were derived from the rectum and had lymphatic and vascular invasion.
Table 1.
Clinicopathological characteristics of pT1 colorectal carcinomas, overall and by morphological type
| Characteristica | Total (n = 909) | Morphological type | Pb | ||
| Depressed (n = 211) | Flat (n = 304) | Protruded (n = 394) | |||
| Age, yr | 68 (59–75) | 68 (60–75) | 69 (62–76) | 66 (57–73) | 0.49 |
| Sex | 0.14 | ||||
| Female | 332 (37) | 68 (32) | 114 (38) | 150 (38) | |
| Male | 577 (64) | 143 (68) | 190 (62) | 244 (62) | |
| Tumor size, mm | 19 (13–25) | 14 (10–18) | 25 (18–35) | 18 (13–23) | <0.001 |
| Tumor location | |||||
| Proximal colon | 282 (31) | 71 (34) | 147 (48) | 64 (16) | |
| Distal colon | 463 (51) | 93 (44) | 100 (33) | 270 (69) | |
| Rectum | 164 (18) | 47 (22) | 57 (19) | 60 (15) | |
| Histological grade | 0.18 | ||||
| Well or moderately differentiated | 777 (85) | 174 (82) | 268 (88) | 335 (85) | |
| Poorly differentiated or mucinous | 132 (15) | 37 (18) | 36 (12) | 59 (15) | |
| Adenoma component | <0.001 | ||||
| Absent | 522 (57) | 196 (93) | 145 (48) | 181 (45) | |
| Present | 387 (43) | 15 (7.1) | 159 (52) | 213 (54) | |
| SM depth | <0.001 | ||||
| <1000 μm | 217 (24) | 15 (7.1) | 111 (37) | 91 (23) | |
| ≥1000 μm | 692 (76) | 196 (93) | 193 (63) | 303 (77) | |
| Lymphatic invasion | <0.001 | ||||
| Absent | 621 (68) | 107 (51) | 234 (77) | 280 (71) | |
| Present | 288 (32) | 104 (49) | 70 (23) | 114 (29) | |
| Vascular invasion | <0.001 | ||||
| Absent | 672 (7) | 116 (55) | 241 (79) | 315 (80) | |
| Present | 237 (26) | 95 (45) | 63 (21) | 79 (20) | |
| Tumor buddingc | <0.001 | ||||
| Grade 1 | 708 (78) | 130 (62) | 253 (83) | 325 (83) | |
| Grade 2–3 | 201 (22) | 81 (38) | 51 (17) | 69 (18) | |
| Lymph node metastasis | 0.08 | ||||
| Absent | 851 (94) | 194 (92) | 292 (96) | 365 (93) | |
| Present | 58 (6.4) | 17 (8.1) | 12 (4.3) | 29 (7.4) | |
| Recurrence or distant metastasis | 0.02 | ||||
| Absent | 898 (99) | 205 (97) | 301 (99) | 392 (99) | |
| Present | 11 (1.2) | 6 (2.8) | 3 (1.0) | 2 (0.5) | |
SM, submucosal.
Data are expressed as number of patients (%) or median (interquartile range).
To compare characteristics between subgroups, we used the Fisher exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables.
Numbers of isolated single cells or small clusters (<5 cells) in the stroma at the invasive tumor margin within a 20x microscopic field, and categorized as grades 1 (<5 budding foci), 2 (5–9), or 3 (≥10).
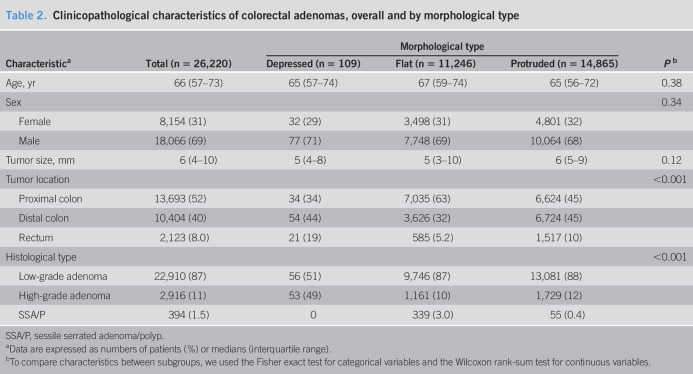
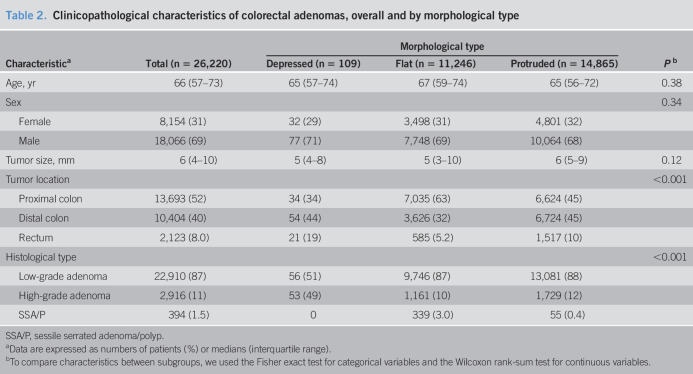
In an analysis limited to adenomas (Table 2), the depressed subtype was more likely to represent high-grade dysplasia than the protruded and flat subtypes. As noted for colorectal neoplasms overall, the depressed subtype was the most frequently observed subtype in rectal tumors.
Table 2.
Clinicopathological characteristics of colorectal adenomas, overall and by morphological type
| Characteristica | Total (n = 26,220) | Morphological type | Pb | ||
| Depressed (n = 109) | Flat (n = 11,246) | Protruded (n = 14,865) | |||
| Age, yr | 66 (57–73) | 65 (57–74) | 67 (59–74) | 65 (56–72) | 0.38 |
| Sex | 0.34 | ||||
| Female | 8,154 (31) | 32 (29) | 3,498 (31) | 4,801 (32) | |
| Male | 18,066 (69) | 77 (71) | 7,748 (69) | 10,064 (68) | |
| Tumor size, mm | 6 (4–10) | 5 (4–8) | 5 (3–10) | 6 (5–9) | 0.12 |
| Tumor location | <0.001 | ||||
| Proximal colon | 13,693 (52) | 34 (34) | 7,035 (63) | 6,624 (45) | |
| Distal colon | 10,404 (40) | 54 (44) | 3,626 (32) | 6,724 (45) | |
| Rectum | 2,123 (8.0) | 21 (19) | 585 (5.2) | 1,517 (10) | |
| Histological type | <0.001 | ||||
| Low-grade adenoma | 22,910 (87) | 56 (51) | 9,746 (87) | 13,081 (88) | |
| High-grade adenoma | 2,916 (11) | 53 (49) | 1,161 (10) | 1,729 (12) | |
| SSA/P | 394 (1.5) | 0 | 339 (3.0) | 55 (0.4) | |
SSA/P, sessile serrated adenoma/polyp.
Data are expressed as numbers of patients (%) or medians (interquartile range).
To compare characteristics between subgroups, we used the Fisher exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables.
WES for depressed pT1 carcinomas
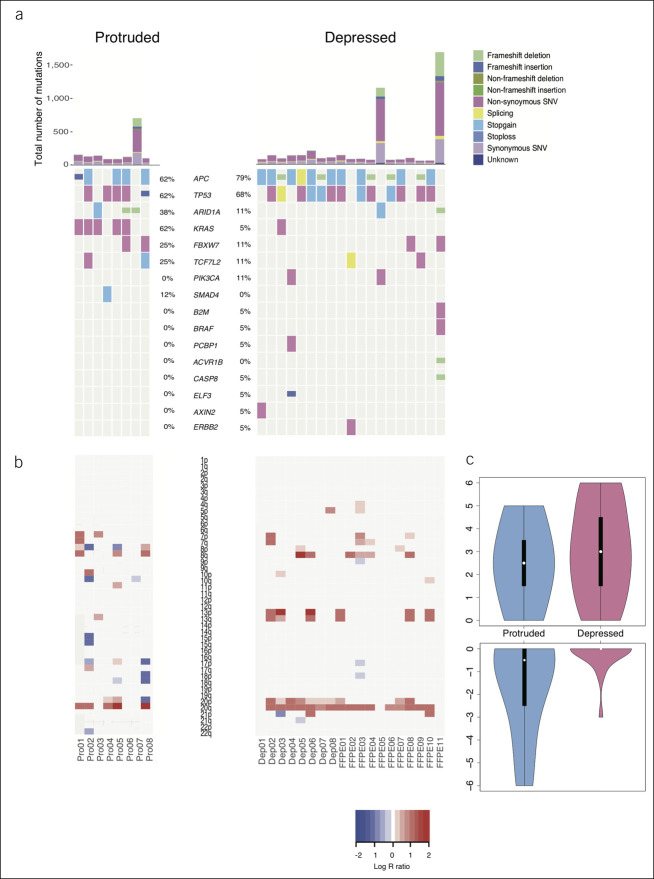
To define the mutational spectrum of depressed carcinomas, we performed WES for pT1 carcinomas (depressed for 19 and protruded for 8; detailed information provided in Supplementary Table 2, see Supplementary Digital Content, http://links.lww.com/CTG/A452). The mean target coverage was 121.3 (SD, 41.9) for >80% of the captured exons. The mean number of mutations was 276 ± 378 in depressed carcinomas and 235 ± 388 in protruded carcinomas, with 2 (11%) and 1 (13%), respectively, harboring >100 mutations per 106 base pairs.
When we examined mutations in driver genes of colorectal cancer reported in a previous TCGA study (27), KRAS mutations seemed to be less frequent in depressed carcinomas than in protruded carcinomas (P = 0.006, Figure 2a). No statistically significant difference was observed for other driver genes, including APC, TP53, and PIK3CA. In a CNA analysis (Figure 2b,c), arm-level copy number losses were less frequently observed in depressed carcinomas than in protruded carcinomas (P = 0.002). In the TCGA data set containing 306 colorectal carcinoma cases, KRAS-wild type tumors were less likely to harbor CNAs than were KRAS-mutant tumors, especially in chromosomes 7p, 13pq, and 20pq (see Supplementary Figure 1, Supplementary Digital Content, http://links.lww.com/CTG/A452).
Figure 2.
Genetic alterations in depressed pT1 colorectal carcinomas compared with those in protruded carcinomas. (a) Somatic mutations in driver genes of colorectal cancer. (b) Copy number alterations. (c) Arm-level gene gain and loss in depressed and protruded types. SNV, single-nucleotide variant.
RNA sequencing for depressed pT1 carcinomas
To investigate the transcriptomic profile of depressed colorectal carcinomas, we performed RNA sequencing of 8 depressed and 8 protruded pT1 carcinomas (Figure 3). In GSEA, depressed carcinomas were more likely to exhibit enrichment of gene sets related to angiogenesis, epithelial mesenchymal transition, and inflammatory response than the protruded carcinomas (Figure 3a). In addition, the gene set related to KRAS signaling was more enriched in depressed carcinomas than in protruded carcinomas (FDR q value = 0.17). Figure 3b compares the expression statuses of the 18 genes with the most variable expression, suggesting high expression of growth factor-related genes (FGF14 and IGF2) in the depressed subtype (P < 0.005 and FDR q value < 0.05). Figure 3c shows the ROC curves for the genes related to angiogenesis, EMT, and inflammatory response genes. The area under the ROC curve was 0.75–0.86 suggesting moderately predictive abilities of these genes. For some genes, we performed immunostaining, and angiogenesis (PECAM1), EMT (VIM), and inflammatory response-related genes (CD14) were more likely to be overexpressed in depressed lesions than in protruded lesions (Figure 3d).
Figure 3.
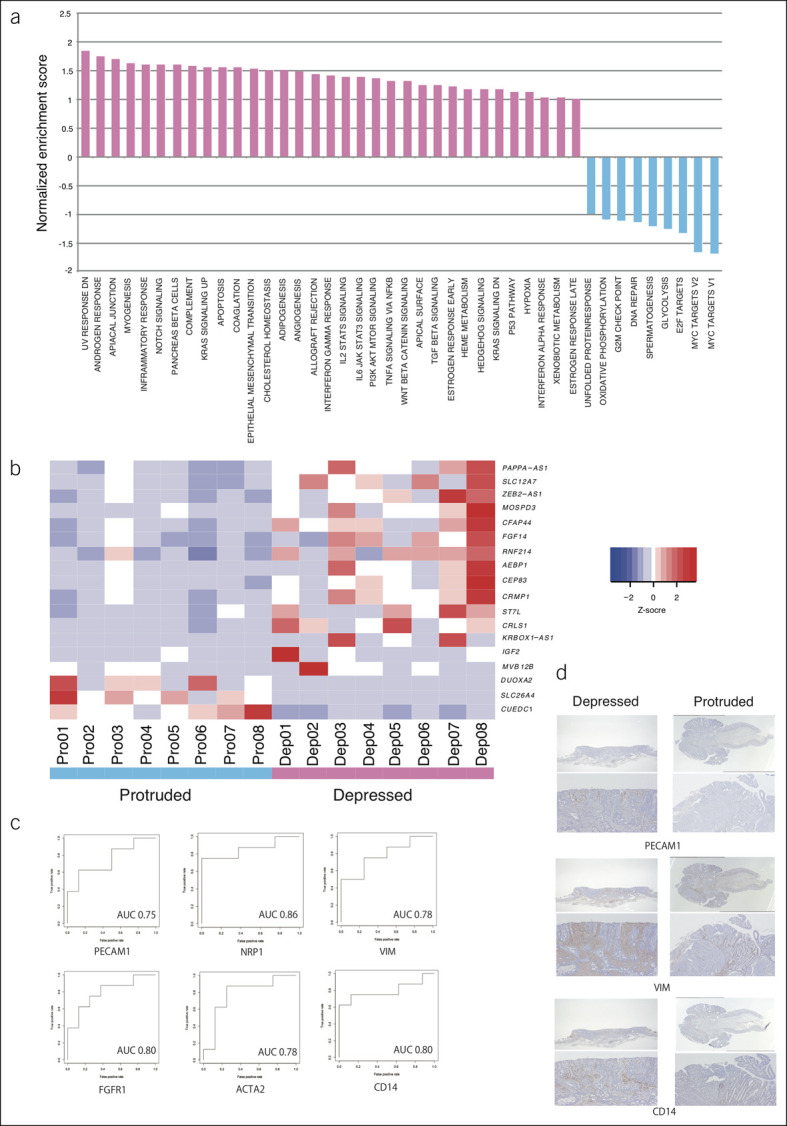
RNA sequencing of depressed pT1 colorectal carcinomas. (a) Gene set enrichment analysis. (b) Expression of genes that were most differentially expressed between depressed and protruded carcinomas. (c) ROC curves of RNA expression of angiogenesis, EMT, and inflammatory related genes. (d) Immunostaining for angiogenesis (PECAM1), EMT (VIM) and inflammatory response related gene (CD14). AUC, area under the ROC curve; EMT, epithelial-mesenchymal transition; ROC, receiver operating characteristic.
DISCUSSION
Using a large endoscopy database, we have characterized the depressed subtypes of colorectal cancer and its precursors. Our data suggest a potential novel model of colorectal carcinogenesis characterized by depressed morphology, distinct from the adenoma-carcinoma sequence model. We found that depressed colorectal neoplasms represented only a small fraction of colorectal tumors resected (approximately 1.5%) but exhibited a unique spectrum of clinicopathological and molecular characteristics that have been associated with tumor aggressiveness. Relative to the protruded subtype, the depressed subtype showed distinct patterns of KRAS mutation and gene expression related to tumor promotion. Given that depressed tumors are difficult to identify at an early stage and show malignant pathological findings, much effort should be focused on the development and utilization of advanced endoscopic screening technologies to increase their detection rate.
We observed more malignant phenotypes with depressed colorectal tumors. They were more likely to contain carcinomatous lesions. The depressed subtype was more strongly associated with lymphovascular invasion and massive submucosal invasion in carcinomas and high-grade dysplasia in adenomas, suggesting the high invasive potential of colorectal tumors with this morphology. The depressed subtype occurs most frequently in the rectum, which might explain the higher risk of recurrence after resection of rectal tumors (28). Of note, an adenoma component was not commonly observed within depressed carcinomas, suggesting that the depressed carcinogenic process might be distinct from the traditional adenoma-carcinoma sequence. More than half of recurrent pT1 carcinomas were associated with depressed morphology, and therefore, a meticulous investigation of local and metastatic recurrences is required after their resection. Very tiny depressed lesions are difficult to detect. We focus on the tiny reddish area in normal mucosa and spray indigo carmine (29,30).
Improvement of endoscopic devices such as high definition image quality, image enhanced system, and the evolution of AI endoscopy would improve the detection rate of depressed lesions (31).
This study demonstrated the unique molecular profile of depressed colorectal carcinomas. Activating KRAS mutations has been associated with dysregulated cell proliferation and the resultant initiation of polypoid-type colorectal tumors (32). KRAS mutations have been recognized as an important genetic alteration in the adenoma-carcinoma sequence, characterized by chromosomal instability, as validated in our analysis of the TCGA data. In our study, KRAS mutation was observed in only a small fraction of depressed colorectal carcinomas, although they have been reported to be present in up to 50% of all colorectal carcinomas (3). Genes related to angiogenesis, epithelial-mesenchymal transition, and inflammatory responses were overexpressed in the depressed subtype. These responses might not only help in the establishment of a tumor-promoting microenvironment but also confer migratory and invasive properties to tumor cells (33).
Our study has some notable strengths; the primary strength is that to our knowledge, this is the largest clinical study of depressed colorectal tumors. We used a large long-term cohort of patients with colorectal tumors that allowed us to analyze a larger number of depressed colorectal neoplasms. Integrated analysis combining clinical, pathological, and molecular data is another strength. Nevertheless, there are certain limitations to our study. First, it was conducted at a single referral center; therefore, there might be selection and referral biases. However, we included consecutive patients who underwent endoscopic or surgical resection of colorectal tumor(s); moreover, our endoscopic and pathological examinations were carried out in a consistent manner. Second, the cross-sectional study design might suggest a reverse causation; however, our study hypothesis was based on several previous studies that have already suggested the malignant potentials of depressed colorectal tumors. Third, exome and RNA sequencing were conducted only for depressed and protruded lesions. In the near future, we intend to clarify the molecular characteristics of flat lesions. Finally, the sample size was small in our molecular analyses. Therefore, our findings on the molecular characteristics of depressed colorectal carcinomas still need to be validated in larger independent cohorts.
In conclusion, we elucidated the genetic and phenotypic variations associated with morphologic patterns of colorectal neoplasms. The depressed morphology was associated with malignant potentials from the early stages of carcinogenesis. Our findings support a new paradigm for our understanding of the development of colorectal cancer and indicate a necessity for surveillance and prevention programs designed specifically for the depressed colorectal cancer subtype.
CONFLICTS OF INTEREST
Guarantor of the article: Koshi Mimori, MD, PhD.
Specific author contributions: S.K., Y.K., Y.O., K. Kato, and K.M.: conceived and designed the study. K.I., M.M., Y. Mori, T.K., T. Hayashi, K.W., H.M., N.S., and F.I.: provided the study materials and conducted clinical analyses. S.H. and T.N.: conducted pathological diagnosis and analyses. Y.K., T. Hamada, K. Kudo, T.M., H.O., T. Sato, T. Shibata, A.N., S.M., M.O., S.O., and K.M.: analyzed and interpreted the data. All authors approved the final version of the manuscript.
Financial support: This work was supported in part by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Science Research (grant numbers: JP16K07177, JP16K10543, JP16K19197, JP17K15670, JP17K16454, JP17K16521, JP17K10593, and JP17K19608); Daiwa Securities Health Foundation (grant number: 15H0912); Priority Issue on Post-K Computer (grant numbers: hp170227 and hp160219); USA National Institutes of Health (grant numbers: R35 CA197735 and R21 CA230873).
Potential competing interests: None to report.
Registration number: This study was approved by the institutional review boards at Showa University and Kyushu University (protocol numbers 213 and 629-00, respectively).
Study Highlights.
WHAT IS KNOWN
✓ Colorectal neoplasms with depressed morphology are increasingly being identified in clinical practice.
✓ Compared with protruded and flat-type neoplasms, the clinicopathological or molecular features of depressed type colorectal neoplasms are not fully elucidated because of their rarity and difficulty in detection.
WHAT IS NEW HERE
✓ The depressed morphology is associated with malignant potential in the early stage.
✓ KRAS mutation is rarely observed in depressed colorectal carcinoma.
✓ Depressed carcinomas generally exhibited high-level expression of genes related to angiogenesis and epithelial-mesenchymal transition.
TRANSLATIONAL IMPACT
✓ This study shows the clinical importance and the molecular features of depressed type colorectal neoplasms.
Supplementary Material
ACKNOWLEDGMENT
We thank M. Oshiumi, M. Uto, K. Oda, M. Kasagi, S. Sakuma, N. Mishima, and T. Kawano for technical assistance and Editage for English proofreading.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A452
REFERENCES
- 1.Nakamura H, Ikematsu H, Osera S, et al. Visual assessment of colorectal flat and depressed lesions by using narrow band imaging. Endosc Int Open 2017;5:E1284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kudo S, Kashida H, Tamura T. Early colorectal cancer: Flat or depressed type. J Gastroenterol Hepatol 2000;15(Suppl):D66–70. [DOI] [PubMed] [Google Scholar]
- 3.Muzny D, Bainbridge M, Chang K, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: An emerging transdisciplinary and interdisciplinary field. Gut 2011;60:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dienstmann R, Vermeulen L, Guinney J, et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017;17:79–92. [DOI] [PubMed] [Google Scholar]
- 7.Kudo S, Lambert R, Allen JI, et al. Nonpolypoid neoplastic lesions of the colorectal mucosa. Gastrointest Endosc 2008;68:S3–S47. [DOI] [PubMed] [Google Scholar]
- 8.Soetikno R, Kaltenbach T, Rouse RV, et al. Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA 2008;299:1027–35. [DOI] [PubMed] [Google Scholar]
- 9.Lambert R. The Paris endoscopic classification of superficial neoplastic lesions: Esophagus, stomach, and colon: November 30 to December 1, 2002. Gastrointest Endosc 2003;58:S3–43. [DOI] [PubMed] [Google Scholar]
- 10.Kudo S, Tamure S, Nakajima T, et al. Depressed type of colorectal cancer. Endoscopy 1995;27:54–7. [DOI] [PubMed] [Google Scholar]
- 11.Winawer SJ, Zauber AG, Ho MN, et al. Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N Engl J Med 1993;329:1977–81. [DOI] [PubMed] [Google Scholar]
- 12.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 2013;369:1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soetikno R, Friedland S, Kaltenbach T, et al. Nonpolypoid (flat and depressed) colorectal neoplasms. Gastroenterology 2006;130:566–76. [DOI] [PubMed] [Google Scholar]
- 14.Burgess NG, Hourigan LF, Zanati SA, et al. Risk stratification for covert invasive cancer among patients referred for colonic endoscopic mucosal resection: A large multicenter cohort. Gastroenterology 2017;153:732–42. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka S, Kashida H, Saito Y, et al. JGES guidelines for colorectal endoscopic submucosal dissection/endoscopic mucosal resection. Dig Endosc 2015;27:417–34. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe T, Muro K, Ajioka Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2016 for the treatment of colorectal cancer. Int J Clin Oncol 2018;23:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kudo S. Endoscopic mucosal resection of flat and depressed types of early colorectal cancer. Endoscopy 1993;25:455–61. [DOI] [PubMed] [Google Scholar]
- 18.Flejou JF. WHO classification of digestive tumors: The fourth edition. Ann Pathol 2011;31:S27–31. [DOI] [PubMed] [Google Scholar]
- 19.Schlemper RJ, Riddell RH, Kato Y, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut 2000;47:251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueno H, Mochizuki H, Hashiguchi Y, et al. Risk factors for an adverse outcome in early invasive colorectal carcinoma. Gastroenterology 2004;127:385–94. [DOI] [PubMed] [Google Scholar]
- 21.Shiraishi Y, Kataoka K, Chiba K, et al. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res 2018;28:1111–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Salzberg SL. TopHat-fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol 2011;12:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolayeva O, Robinson MD. edgeR for differential RNA-seq and ChIP-seq analysis: An application to stem cell biology. Methods Mol Biol 2014;1150:45–79. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumor types. Nature 2014;505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouyama Y, Kudo SE, Miyachi H, et al. Risk factors of recurrence in T1 colorectal cancers treated by endoscopic resection alone or surgical resection with lymph node dissection. Int J Colorectal Dis 2018;33:1029–38. [DOI] [PubMed] [Google Scholar]
- 29.Kudo S, Kashida H. Flat and depressed lesions of the colorectum. Clin Gastroenterol Hepatol 2005;3:S33–6. [DOI] [PubMed] [Google Scholar]
- 30.Kashida H. Non-polypoid colorectal neoplasms are no longer unique to Japan, but do not mix up flat and depressed lesions. Dig Endosc 2015;27:300–2. [DOI] [PubMed] [Google Scholar]
- 31.Misawa M, Kudo SE, Wada Y, et al. Magnifying narrow-band imaging of surface patterns for diagnosing colorectal cancer. Oncol Rep 2013;30:350–6. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi M, Watanabe H, Ajioka Y, et al. Effect of K-ras mutation on morphogenesis of colorectal adenomas and early cancers: Relationship to distribution of proliferating cells. Hum Pathol 1996;27:1042–9. [DOI] [PubMed] [Google Scholar]
- 33.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer 2009;9:265–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.