Abstract
Background
T cell immunity plays a central role in the body's defense system, including maintaining homeostasis and preventing tumorigenesis and viral infection. Immune system functions degenerate with age, leading to immune senescence. Physiologically, immune senescence is characterized by a decrease in T cell receptor diversity, naive T cell deficiency, and alterations in T cell immune-related miRNAs. However, little is known about the characteristics of T cell immunosenescence in Chinese individuals.
Results
A significant decrease in the miR-17, miR-92a, and miR-181a levels in PBMCs was detected with age. The miR-92a and miR-181a levels were upregulated in CBMCs when comparing healthy individuals to group I (0~9 years), whereas miR-17 was downregulated. The sjTREC level in PBMCs was negatively correlated with age, and a sharp decrease in sjTRECs was found between groups I and II (10~19 years). Twenty-four TCR Vβ subfamilies could be detected in most samples, and most displayed polyclonality, while skewed expression of the Vβ subfamilies as well as an increased oligoclonal tendency was found with age. Similarly, the frequencies of the TCR Vγ and Vδ subfamilies decreased with age, and the alteration in clonality appeared to be stable at different ages.
Conclusion
We made the novel observation of T cell immunosenescence with age in Chinese individuals, which may provide information for immune targets to enhance the T cell immune response in immunotherapy settings for elderly patients.
1. Introduction
Aging leads to numerous alterations in physiological systems, and the immune system is one of the most influential systems resulting in immunosenescence. In particular, T cell subfamilies are associated with a reduced capacity for immune surveillance and increased cancer incidence [1, 2]. Physiologically, T cell immunosenescence is characterized by reduced T cell regeneration, lower differentiation ability [3], decreased naive T cell numbers [4], lesser T cell receptor (TCR) diversity [5], low activation, failure to respond to vaccines and neoantigens [6], and increased T cell terminal differentiation [7, 8].
Recently, investigators have been seeking biomarkers for immunosenescence with the hopes of enhancing the T cell response to vaccines and improving immunotherapy for cancer [9]. Several characteristics of T cell aging have been reported, such as loss of the costimulatory molecule CD28 [10]; high expression of Fas/Fas ligand on T cells [11]; decreased IL-2 and IFN-γ; increased IL-4, IL-6, IL-10, and TNF-α secretion [12]; decreased naive T cells; and increased memory T cells [13]. Based on the immunocompetence of T cells toward antigens and maintaining immune homeostasis, thymic recent output function and TCR repertoire diversity are the essential indices for evaluating T cell immunity. For example, low naive T cells and a skewed TCR repertoire result in poor T cell immune recovery after chemotherapy and hematopoietic stem cell transplantation [14–16].
Evaluation of TCR diversity and T cell clonality is based on analysis of the length and sequence of the complementarity-determining region 3 (CDR3) resulting from TCR V(D)J gene rearrangement [14, 17, 18]. Typical methods for evaluating thymic output function are based on quantitative analysis of signal joint T cell receptor excision circles (sjTRECs). sjTRECs are circular excision products formed by TCR gene segment deletion during TCR rearrangement steps. These molecules are stable, cannot amplify, are exported from the thymus to the periphery with thymic recent emigrants, and are ultimately diluted during T cell proliferation. Since 1998, sjTRECs have been used as markers for T cell neogenesis to represent the number of naive T cells as well as thymic output function [19, 20].
Recently, increasing data have shown that epigenetic regulation is an important factor in aging. It has been reported that microRNAs (miRNAs), such as the miR-17-92 cluster, miR-24, miR-92a, miR-103, miR-107, miR-128, miR-130a, miR-132, miR-142-3p/5p, miR-146a/b, miR-155, miR-221, miR-223, miR-496, and miR-1538 may be involved in cellular senescence and aging [21, 22]. miR-17-92 and miR-181a have been reported to play a critical role in T cell differentiation and proliferation [23, 24]. Thus, in this study, we analyzed the expression of miR-17, miR-92a, and miR-181a with aging combined with changes in TCR diversity and sjTRECs to attempt to characterize the potential role of the immune profile as biomarkers for immunosenescence in the Chinese population.
2. Materials and Methods
2.1. Samples
Cord blood was obtained from 20 full-term healthy babies at delivery. Peripheral blood samples were collected from 160 healthy donors including 85 males and 75 females with a median age of 40 years (range: 0 to 70 years) (Table 1). The healthy donors had a healthy status without any type of cancer, type 2 diabetes, or autoimmune disease. All experiments were conducted with the understanding and consent of each participant, and ethical approval was obtained from the Ethics Committee of the First Affiliated Hospital, School of Medicine, Jinan University.
Table 1.
Sample demographic information.
| Groups (years) | Male (n) | Female (n) | Median age (years) |
|---|---|---|---|
| 0~9 | 9 | 11 | 6 |
| 10~19 | 9 | 11 | 17 |
| 20~29 | 6 | 14 | 25 |
| 30~39 | 9 | 11 | 34.5 |
| 40~49 | 8 | 12 | 45 |
| 50~59 | 12 | 8 | 53 |
| 60~69 | 10 | 10 | 64 |
| ≥70 | 12 | 8 | 75.5 |
| CMBCs | 10 | 10 | 0 |
Cord and peripheral blood mononuclear cells (CBMCs and PBMCs) were isolated, DNA and total RNA were extracted, and cDNA for miR-17, miR-92a, and miR-181a was reverse-transcribed using the miScript II RT Kit (QIAGEN, Duesseldorf, Germany). First-strand cDNA for target gene assays was reverse-transcribed using random hexamer primers and the High Capacity cDNA Reverse Transcription kit (ABI, Carlsbad, CA, USA) according to the manufacturer's instructions.
2.2. Quantitative Real-Time RT-PCR (qRT-PCR) for miRNA Detection
The expression levels of miR-17, miR-92a, and miR-181 were determined using the primers in Table 2 and the miScript SYBR Green PCR kit (QIAGEN, Duesseldorf, Germany). The miRNA expression levels were normalized to RNU6B snRNA [25].
Table 2.
miRNA primer sequences.
| Primer | Sequence (5′→3′) |
|---|---|
| miR-17-3p | ACUGCAGUGAAGGCACUUGUAG |
| miR-92a | UAUUGCACUUGUCCCGGCCUGU |
| miR-181a-5p | AACAUUCAACGCUGUCGGUGAGU |
| RNU6-2 | ACGCAAATTCGTGAAGCGTT |
2.3. Real-Time Quantitative PCR for sjTREC Detection
Quantitative detection of δRec-ψJα sjTRECs in genomic DNA from CBMCs and PBMCs was performed by TaqMan real-time PCR as described previously [26].
2.4. RT-PCR and GeneScan Analysis for the Expression and Clonality of the TCR Vβ, TCR Vγ, and TCR Vδ Subfamilies
The expression of the TCR Vβ, TCR Vγ, and TCR Vδ subfamilies was detected by RT-PCR using 24 Vβ, 3 Vγ, and 8 Vδ gene primers and a single Cβ, Cγ, or Cδ primer, respectively, for the first run (unlabeled PCR for amplification). Subsequently, runoff PCR was performed using fluorophore-labeled primers (Cβ-FAM, Cγ-FAM, or Cδ-FAM), and the products were the analyzed for clonality by GeneScan analysis. PCR and GeneScan analyses were performed as previously described [27–29].
2.5. Statistical Analyses
Pearson correlation and linear regression analysis was used to estimate the correlation between the expression level of miR-17, miR-92a, miR-181a, and TRECs and age from all the groups. The differences in miR-17, miR-92a, miR-181a, and TRECs among different age groups were analyzed by one-way ANOVA. Results are expressed as mean ± SEM. All data analyses, including statistical calculations and graphical displays, were performed using SPSS 13.0 and GraphPad software. The readxl and circlize packages in R, version 3.6.1 (http://www.r-project.org/), were used for constructing chord diagrams.
3. Results
3.1. Downregulation of miR-17, miR-92a, and miR-181a with Aging
The expression levels of miR-17, miR-92a, and miR-181a were quantitatively detected in peripheral blood mononuclear cells (PBMCs) from 160 healthy individuals (HIs) of different ages (0 to 70 years, groups I to VIII) and cord blood mononuclear cells (CBMCs) from 20 cases. The expression of three miRNAs in PBMCs was found to be significantly downregulated with age. Moreover, compared to group I (0~9 years), we found a lower expression of miR-17 (p < 0.0001) in group V (40~49 years), group VI (50~59 years), group VII (60~69 years), and group VIII (over 70 years), and the sharpest declines were found for those between the ages of 30 and 40 followed by those between 10 and 20. However, there was no statistically significant difference between neighboring age groups. The miR-17 level in the CBMC group was lower than that in group I, group II (10~19 years), and group III (20~29 years) and higher than that in groups IV (30~39 years) to VIII. Statistical significance was found when comparing CBMCs in groups VII and VIII (p < 0.05) (Figure 1(c)). Because miR-17 and miR-92a belong to the same gene cluster (Figure 1(a)), we found that they had similar expression characteristics. For example, compared with group I, low miR-92a expression was found in groups VI through VIII (p < 0.01), and the sharpest decline in the miR-92a level was also found for those between 30 and 40 years old. However, unlike miR-17, the expression of miR-92a also had its own characteristics. miR-92a was upregulated in CBMCs when compared with all groups of PBMCs, and statistical significance was found for CBMCs when comparing groups V to VIII (p < 0.05) (Figure 1(d)). For miR-181a (Figure 1(b)), when compared with group I, a significant lower expression was found in group VIII (p < 0.01). The sharpest decline was found between 60 and 70 years followed by 0 and 10 years. Moreover, miR-181a was highly expressed in CBMCs in comparison with all PBMCs (Figure 1(e)). Furthermore, the expression levels of miR-17, miR-92a, and miR-181a had a significant negative correlation with aging (r = −0.59465, -0.55174, and -0.47312, all p < 0.0001) (Figures 1(f)–1(h)).
Figure 1.
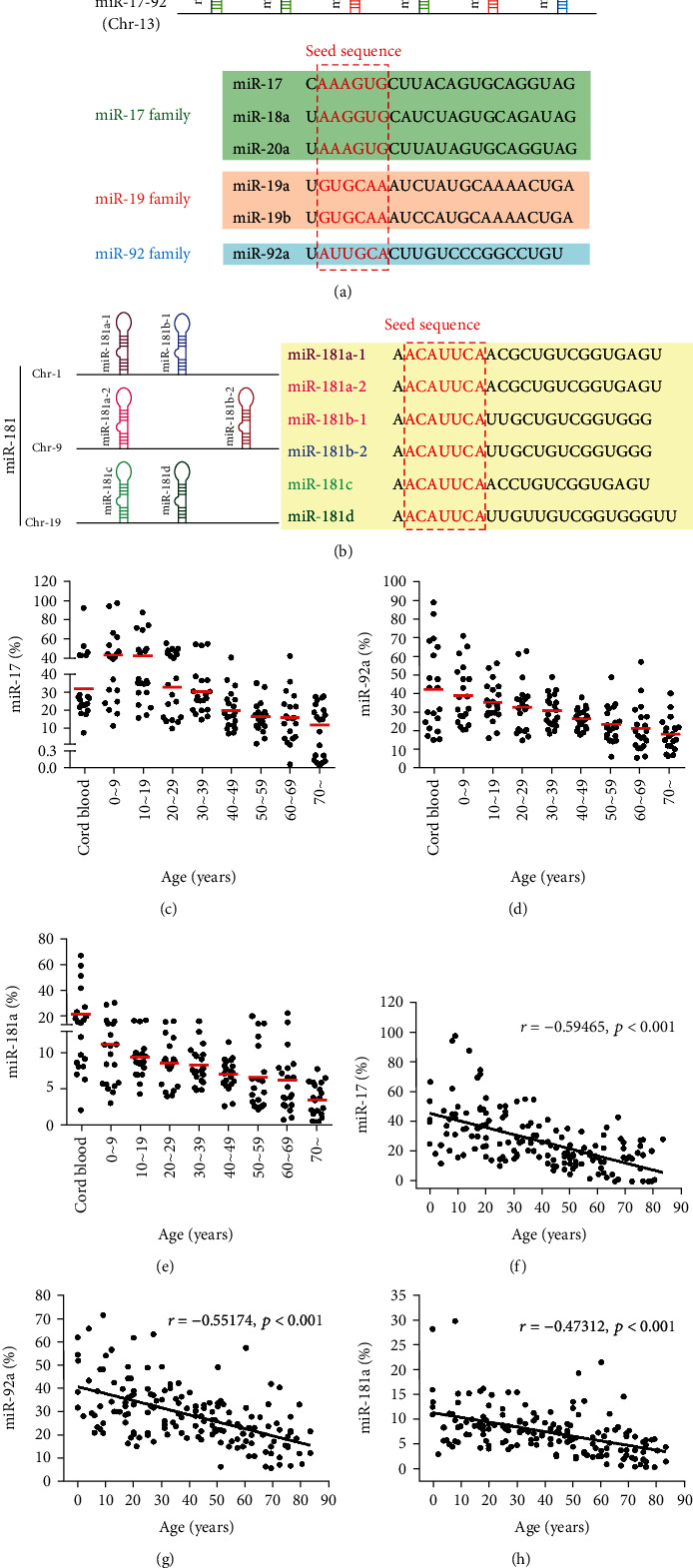
The expression levels of miR-17, miR-92a, and miR-181a in PBMCs are downregulated with age. (a) The location and members of the miR-17-92 cluster and the sequences of the six members in three families. Green: members of the miR-17 family; red: members of the miR-19 family; blue: members of the miR-92 family. (b) miR-181 family members and their genomic locations. (c–e) Quantification of the data revealed that the expression levels of miR-17 (c), miR-92a (d), and miR-181a (e) were downregulated with age (n = 20/each age group). (f–h) A negative correlation between age and miR-17 (f), miR-92a (g), and miR-181a (h) is shown.
3.2. Thymic Output Function Progressively Declines during Aging
To characterize alterations in thymic output function over the lifespan, absolute quantitative analysis of the sjTREC (TREC) level in DNA samples from PBMCs and CBMCs was performed with 160 healthy individuals (HI) of different ages (0 to 70 years old) and 20 cord blood samples (Figure 2(a)) [20]. Significantly, the number of TRECs progressively declined with age, and the biggest drop was between the ages of 0 and 9 (Figure 2(b)). A strong negative correlation between the number of TRECs in PBMCs and age was found (r = −0.64951; p < 0.0001) (Figure 2(c)). Interestingly, there were no obvious alterations in the TREC level in the 50 to 59 age group (group VI), and a relatively lower number of TRECs was found for those ≥ 60 years. However, minimal thymic output function remained in elder individuals.
Figure 2.
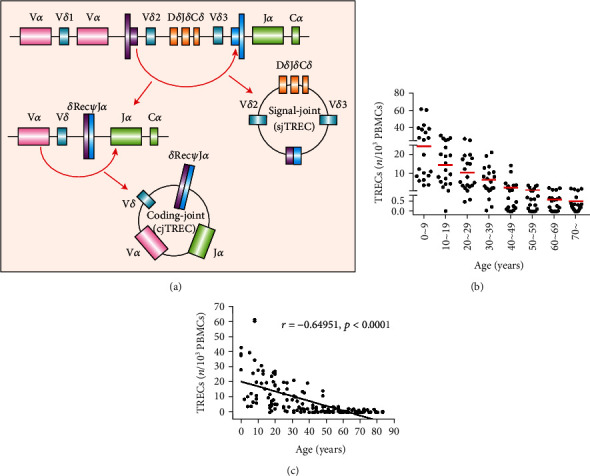
sjTREC are decreased in PBMCs with age. (a) Schematic diagram of T cell receptor excision circle (TREC) formation. TRECs are pieces of DNA fragments generated during TCR gene rearrangement in the thymus that are exported from the thymus to the periphery in T cell episomes. TRECs are not duplicated during mitosis and remain stable for a long time. However, as T cells divide, the level of TRECs is gradually diluted. Generation of TRECs occur in >70% of all new (naive) T cells, and they can be detected by PCR. (b) TREC levels in PBMCs from 160 healthy individuals at different ages (0 to 70 years old). (c) An inverse correlation between the TREC count and donor age was found.
3.3. Skewed Distribution and Clonality of TCR Vβ, Vγ, and Vδ T Cells with Aging
TCR diversity and clonality are indices for T cell generation, proliferation potential, and the ability of specific amplifications to respond to antigen stimulation. In this study, we detected the distribution and clonality of the TCR Vβ, Vγ, and Vδ subfamilies in 80 cases in groups I to VIII using RT-PCR and GeneScan. First, the expression of TCR Vβ1, Vβ2, Vβ4~Vβ11, Vβ14~Vβ16, Vβ18, Vβ20, Vβ21, and Vβ23 could be detected in all samples (100%). The expression of Vβ3 (98.75%), Vβ12 (97.5%), Vβ17 (95%), Vβ19 (98.75%), and Vβ22 (98.75%) was absent in some cases, and the expression of Vβ13 and Vβ24 was lowest (91.25%) (Figure 3(a)). Based on the different CDR3 TCR rearrangement lengths, the clonality of the T cells was characterized as multipeaks, oligopeaks, and monopeaks corresponding to polyclonality, oligoclonality, and monoclonality, respectively, by GeneScan analysis (Figure 4(a)) [30]. Vβ polyclonality was identified in most samples, and oligoclonality was found for Vβ6 and Vβ23 in 23.75% of the samples. There was decreasing Vβ subfamily expression frequency with an increased oligoclonal tendency for Vβ T cells with age (Figures 3(c) and 4(c)). Second, for the Vγ subfamily, VγII was the main subfamily detected at 78.75% followed by VγIII at 70% and VγI at 61.25% in 80 total PBMC samples (Figure 3(b)). Similarly, the Vγ frequency decreased with age. Interestingly, an oligoclonal tendency was mainly found in group I, but there was no significant correlation with age (Figures 3(c), 3(d), 4(b), and 4(c)).
Figure 3.
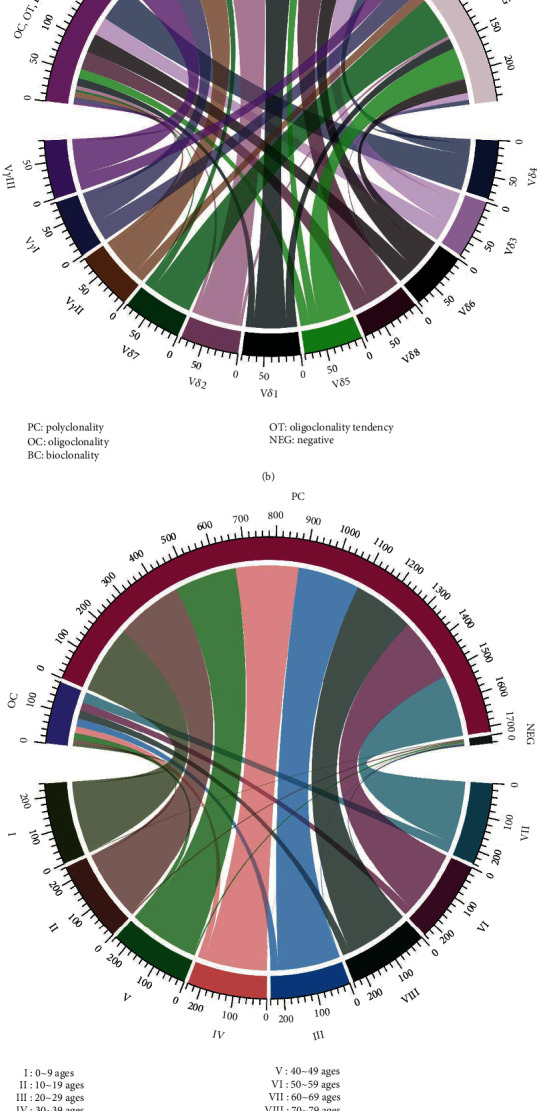
Chord diagram displaying the frequencies and clonalities of the TCR Vβ, Vγ, and Vδ subfamilies in different age groups. (a, b) Each sector of the circle represents one node of the TCR subfamilies, and its width indicates the expression frequencies of TCR Vβ (a), Vγ, and Vδ (b) that connect the clonalities of the TCR subfamilies. (c, d) Circularly arranged sectors represent the different age groups and the clonalities of the TCR Vβ (c), Vγ, and Vδ (d), and the scale bars indicate their relative expression frequencies (n = 10 in each age group).
Figure 4.
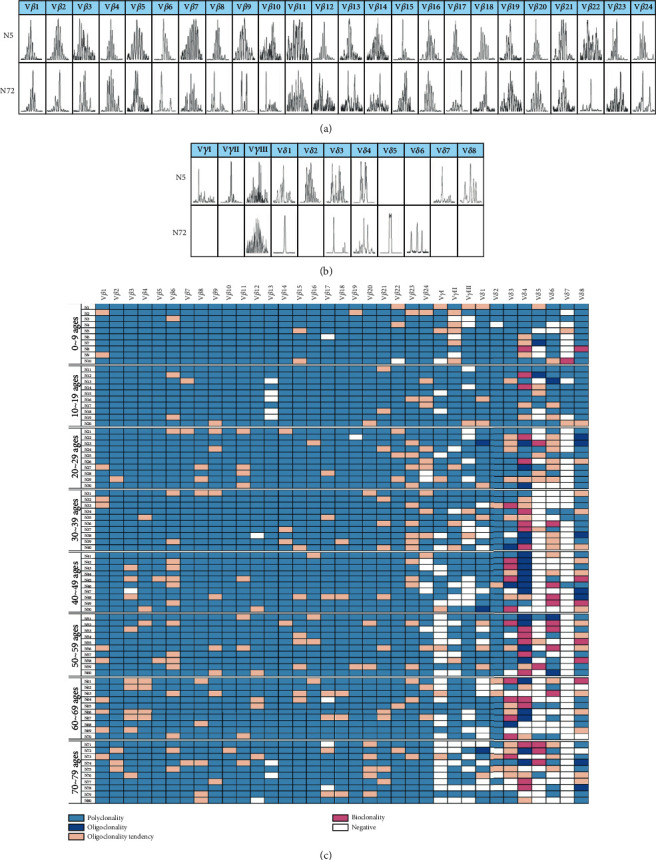
Distribution and clonality of the TCR Vβ, Vγ, and Vδ subfamilies in different age groups. (a) GeneScan analysis results from the TCR Vβ, Vγ, and Vδ subfamilies in PBMCs from two cases, group I (N5) and group VIII (N72). (b) The characteristic distribution and clonality of the TCR Vβ, Vγ, and Vδ subfamilies in PBMCs from 80 cases of healthy adult individuals.
Finally, unlike the high expression frequency of the Vβ subfamilies in healthy individuals, the expression frequency of the Vδ subfamilies was relatively different. The highest expression frequency was found for Vδ8 at 97.5% followed by Vδ2 at 93.75% and Vδ4 at 91.25%. Vδ5 and Vδ7 expression was detected in only 37.5% and 18.75% of the samples, and greater than 20% of the Vδ subfamilies was absent in groups IV to VIII, particularly in group VII (up to 36%) (Figures 3(b) and 3(c)). In addition, oligoclonality was relatively high for Vδ4 (70%), and oligoclonal expansion increased significantly beginning with group III (Figures 3(c), 4(b), and 4(c)).
4. Discussion
Understanding T cell immune alterations in healthy individuals during aging may help us evaluate the immune status of patients with different diseases, particularly cancer patients who may receive immunotherapy. There are limited basic data regarding the T cell immune status in the Chinese population [7, 8]. In this study, we examined T cell receptor diversity, thymic recent output function, and expression of the T cell immune-related miRNAs miR-17, miR-92a, and miR-181a in healthy individuals of different ages ranging from 0 to over 70.
Several studies have reported a number of miRNAs mediating cellular senescence [31]; however, how these miRNAs contribute to the aging process remains unclear. miR-17 and miR-92a downregulation was found in human CD8+ T cells, while miR-181a declined with age in CD4+ T cells [32, 33]. In this study, we detected a change in miR-17, miR-92a, and miR-181a in healthy individuals ranging in age from 0 to over 70 years. Significantly, the expression of these miRNAs decreased with age, and we also characterized a declining tendency with age for the different miRNAs. Healthy individuals were divided into eight different age groups in intervals of ten years according to calendar age. In this manner, we could identify more detailed miRNA expression patterns with obvious variation trends. Interestingly, the miR-17 level was high in groups I to III (0 to 29 years) compared with that in the CBMC groups, while the expression levels of miR-92a and miR-181a were highest in CBMCs compared with all of the PBMC groups. The sharpest decline in the expression of miR-17 and miR-92a was between 30 and 40 years, and the sharpest decline in the miR-181a level was delayed to over 60 years. Overall, these novel results are in agreement with other reports demonstrating that miR-17, miR-92a, and miR-181a are downregulated in various tissues with age [34, 35]. Moreover, we found that the regulation of miR-17, miR-92a, and miR-181a may have different patterns during aging. Thus, we assume that ages 40 and 70 may be cutoff points for miR-17~92 and miR-181a aging, respectively. It has been reported that miR-17~92 targets the control of BIM and PTEN, and the decrease in miR-17 and miR-92a may result in inhibition of AKT-mTOR by increasing PTEN. mTOR is a well-known regulator of cell aging and the cell cycle [36]. Additionally, the mechanism underlying miR-181a regulation of aging is thought to be via inhibition of Bcl-2 and an increase in the proapoptosis genes caspase 9 and caspase 3 and cell death [37]. Further investigation of the relationship between miR-17 and miR-92a and the AKT-mTOR pathway and that between miR-181a and the Bcl-2 pathway may confirm their role in aging.
In this study, we also examined the expression profile of miR-17, miR-92a, and miR-181a in cord blood as it is known that cord blood contains greater numbers of naive T cells [38]. As expected, the miR-92a and miR-181a levels were higher than that in all groups of healthy individuals, and only the miR-17 level was lower than that in group I (0~9 years), suggesting that the expression of these miRNAs may be out of sync in cord blood, and the underlying explanation for this phenomenon remains to be investigated.
The sjTREC number in PBMCs or T cell subsets was used as a specific index for thymic recent emigrants and naive T cell number, which is important for evaluating T cell immunity [20]; however, little is known about how the numbers change with aging in the Chinese population. Thus, in this study, we quantified sjTRECs in PBMCs and CBMCs to determine baseline level data and plotted the changes in sjTREC numbers with age. We found that in 160 healthy volunteers of different ages, TRECs ranged from 0.004 to 95.124 copies/103 PBMCs, and linear correlation analysis showed that the level of TRECs was negatively correlated with the ages. The thymus produces significantly less TRECs with increasing age, which is consistent with the results of foreign reports [39, 40].TREC levels were high up to age 10, then gradually declined with age, and the levels of TRECs decreased significantly after age 50, but they were still detectable above age 70 with limited amounts. Furthermore, the study by Pido-Lopez et al. pointed out that the TREC content in PBMCs is quadratic with age, and the best fit curve is y = 0.002x2 − 0.2139x + 7.96181, R2 = 0.601. The authors noted that TREC levels decrease with age at a rate of approximately 2% per year up to age 40 and at a rate of approximately 1.5% per year after age 40 [40]. In addition, the study by Naylor et al. also suggests that TREC levels decrease significantly after age 65. Before age 65, only a small fraction of new thymocytes are required to maintain diversity, but after age 65, this diversity is maintained through increased spontaneous T cell activation and the conversion of naive cells to memory cells. Additional studies have also confirmed that the age of 40-50 years may be a watershed in thymic output function. The two biggest drops in number were between 10 and 60 years [41]. Since 1998, sjTRECs have been used to evaluate the recovery of the thymus after treatment in HIV-infected and AIDS patients [19]. Decreased immune function and imbalance in the elderly increases susceptibility to infection and ultimately reduces their quality of life. Therefore, restoration of immune function is beneficial in maintaining the health of the elderly. We hypothesized that greater thymic output would lead to a larger pool of naive T cells to reproduce a more competent immune response and reduced mortality.
The TCR includes α, β, γ, and δ chains, which are functionally expressed on the T cell surface as either α/β or γ/δ heterodimers corresponding to αβ+ or γδ+ T cells [42]. Each functional TCR gene is encoded by TCR variable (V), diversity (D, only for β and δ chain), and joining (J) segments in which the length and sequence of the complementarity-determining region 3 (CDR3) is specific for each TCR rearrangement, which could be used as a biomarker for evaluating the TCR repertoire diversity [43, 44]. In this study, we analyzed the CDR3 of the TCR Vβ, Vγ, and Vδ subfamilies to characterize the diversity and clonality of the αβ+ and γδ+ T cells with aging. For the TCR Vβ subfamilies, the frequency appeared to decrease slightly with age, while an increasing oligoclonality of TCR Vβ T cells with age also indicated reduced TCR diversity in the same subfamilies. Thus, evaluation of TCR diversity may include changes in both TCR Vβ frequency and clonality. Moreover, we recently found that T cell exhaustion displays a different distribution of TCR Vβ subfamily T cells [17]. Thus, it may be worth combining the detection of T cell exhaustion and senescence with aging. The expression pattern of the TCR Vγ subfamilies appeared to be similar to that of Vβ, while for the TCR Vδ subfamilies, the distribution and clonality were different from the TCR Vβ subfamilies with aging. High expression was only found for Vδ2, Vδ4, and Vδ8. Oligoclonality was relatively high, particularly for Vδ4, and significantly, the expression of the Vδ subfamilies was reduced over age 30. There are reports that Vγ9/Vδ2 (VγII/Vδ2) T cells, which have cytotoxic effects on tumor or infected cells, are the major population of γδ T cells in peripheral blood in healthy individuals [45]. In this study, we also found that VγII/Vδ2 had the highest subfamily expression, but we also found that the VγII/Vδ8 subfamilies also had a high frequency of expression in this group of samples. In a previous study, our group demonstrated that the TCR Vδ gene is mainly comprised of Vδ1, Vδ2, Vδ3, and Vδ8 chains. In addition, Vδ8 was the most frequently clonally expanded T cell subfamily member in patients with refractory anemia with an excess of blasts and acute myeloid leukemia [30, 46]. However, the function of Vδ8+ T cells is not clear. For this phenomenon, further analysis is needed to determine whether it is a characteristic of the TCR lineage in the Chinese population.
We know that most common aging-associated diseases are cardiovascular disease, cancer, arthritis, dementia, cataract, osteoporosis, diabetes, hypertension, and neurodegenerative diseases such as Alzheimer's disease, Huntington's disease, Parkinson's disease, and amyotrophic lateral sclerosis. Research on immunosenescence and testing methods can provide a basis for diagnosis and further understanding of immunodeficiency-related diseases and provide a basis for seeking specific therapies.
5. Conclusions
In conclusion, in this study, we reported for the first time the characteristics of T cell immunity based on thymic recent output function, TCR diversity, and T cell-related miRNAs in the healthy Chinese population; we made the novel observation of T cell immunosenescence with age. The biggest drops of sjTREC number clearly point to the age-related timing change of thymic function in Chinese individuals; combining with the dynamic change of miR-92a, miR-181a, and miR-17 levels, it may indicate the regulating time points in T cell proliferation with age. Finally, the findings of the distribution pattern of reduced TCR diversity with increasing oligoclonality of TCR subfamily T cells with age may help to build a foundation for immunosenescence evaluation indices and provide information for immune targets for enhancing the T cell immune response in the immunotherapy setting in elderly patients.
Acknowledgments
We would also like to thank the volunteers who donated cord blood or peripheral blood for this project. This study was supported by grants from the National Key R&D Program of China (2017YFE0131600), the National Natural Science Foundation of China (Nos. 81770152 and 82070152), and the Guangzhou Science and Technology Project Fund (Nos. 201807010004 and 201803040017).
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Larbi A., Franceschi C., Mazzatti D., Solana R., Wikby A., Pawelec G. Aging of the immune system as a prognostic factor for human longevity. Physiology. 2008;23(2):64–74. doi: 10.1152/physiol.00040.2007. [DOI] [PubMed] [Google Scholar]
- 2.Beirne C., Delahay R., Hares M., Young A. Age-related declines and disease-associated variation in immune cell telomere length in a wild mammal. PLoS One. 2014;9(9, article e108964) doi: 10.1371/journal.pone.0108964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linton P. J., Dorshkind K. Age-related changes in lymphocyte development and function. Nature Immunology. 2004;5(2):133–139. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- 4.Kline K. A., Bowdish D. M. E. Infection in an aging population. Current Opinion in Microbiology. 2016;29:63–67. doi: 10.1016/j.mib.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Goronzy J. J., Lee W. W., Weyand C. M. Aging and T-cell diversity. Experimental Gerontology. 2007;42(5):400–406. doi: 10.1016/j.exger.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodwin K., Viboud C., Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24(8):1159–1169. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 7.Xu L., Yao D., Tan J., et al. Memory T cells skew toward terminal differentiation in the CD8+T cell population in patients with acute myeloid leukemia. Journal of Hematology & Oncology. 2018;11(1):p. 93. doi: 10.1186/s13045-018-0636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M., Yao D., Zeng X., et al. Age related human T cell subset evolution and senescence. Immunity & Ageing. 2019;16(1):p. 24. doi: 10.1186/s12979-019-0165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasakovski D., Xu L., Li Y. Q. T cell senescence and CAR-T cell exhaustion in hematological malignancies. Journal of Hematology & Oncology. 2018;11(1):p. 91. doi: 10.1186/s13045-018-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broux B., Markovic-Plese S., Stinissen P., Hellings N. Pathogenic features of CD4(+)CD28(-) T cells in immune disorders. Trends in Molecular Medicine. 2012;18(8):446–453. doi: 10.1016/j.molmed.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Aggarwal S., Gupta S. Increased apoptosis of T cell subsets in aging humans: altered expression of Fas (CD95), Fas ligand, Bcl-2, and Bax. Journal of Immunology. 1998;160(4):1627–1637. [PubMed] [Google Scholar]
- 12.Koelman L., Pivovarova-Ramich O., Pfeiffer A. F. H., Grune T., Aleksandrova K. Cytokines for evaluation of chronic inflammatory status in ageing research: reliability and phenotypic characterisation. Immunity & Ageing. 2019;16(1) doi: 10.1186/s12979-019-0151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon J. J., Chu H. H., Pepper M., et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27(2):203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Li Y. T cell receptor-engineered T cells for leukemia immunotherapy. Cancer Cell International. 2019;19(1):p. 2. doi: 10.1186/s12935-018-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida K., Cologne J. B., Cordova K., et al. Aging-related changes in human T-cell repertoire over 20years delineated by deep sequencing of peripheral T-cell receptors. Experimental Gerontology. 2017;96:29–37. doi: 10.1016/j.exger.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Britanova O. V., Putintseva E. V., Shugay M., et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. Journal of Immunology. 2014;192(6):2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 17.Huang J., Tan J., Chen Y., et al. A skewed distribution and increased PD‐1+Vβ+CD4+/CD8+ T cells in patients with acute myeloid leukemia. Journal of Leukocyte Biology. 2019;106(3):725–732. doi: 10.1002/JLB.MA0119-021R. [DOI] [PubMed] [Google Scholar]
- 18.Lian J. Y., Liu J. L., Yue Y., et al. The repertoire features of T cell receptor β-chain of different age and gender groups in healthy Chinese individuals. Immunology Letters. 2019;208:44–51. doi: 10.1016/j.imlet.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Douek D. C., McFarland R. D., Keiser P. H., et al. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396(6712):690–695. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- 20.Li Y. Recent thymic output function in patients with hematological malignancy. Hematology. 2005;10(4):297–305. doi: 10.1080/10245330500093591. [DOI] [PubMed] [Google Scholar]
- 21.Park S., Kang S., Min K. H., Hwang K. W., Min H. Age-associated changes in micro RNA expression in bone marrow derived dendritic cells. Immunological Investigations. 2013;42(3):179–190. doi: 10.3109/08820139.2012.717328. [DOI] [PubMed] [Google Scholar]
- 22.Hooten N. N., Abdelmohsen K., Gorospe M., Ejiogu N., Zonderman A. B., Evans M. K. MicroRNA expression patterns reveal differential expression of target genes with age. PLoS One. 2010;5(5, article e10724) doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q. J., Chau J., Ebert P. J. R., et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Liao R. X., Sun J. G., Zhang L., et al. MicroRNAs play a role in the development of human hematopoietic stem cells. Journal of Cellular Biochemistry. 2008;104(3):805–817. doi: 10.1002/jcb.21668. [DOI] [PubMed] [Google Scholar]
- 25.He Z., Liao Z., Chen S., et al. Downregulated miR-17, miR-29c, miR-92a and miR-214 may be related to BCL11B overexpression in T cell acute lymphoblastic leukemia. Asia-Pacific Journal of Clinical Oncology. 2018;14(5):e259–e265. doi: 10.1111/ajco.12979. [DOI] [PubMed] [Google Scholar]
- 26.Li Y., Geng S., Yin Q., et al. Decreased level of recent thymic emigrants in CD4+ and CD8+T cells from CML patients. Journal of Translational Medicine. 2010;8(1):p. 47. doi: 10.1186/1479-5876-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Z., Wu X., Chen S., Yang L., Liu Q., Li Y. Distribution and clonality of the Vα and Vβ T-cell receptor repertoire of regulatory T cells in leukemia patients with and without graft versus host disease. DNA and Cell Biology. 2014;33(3):182–188. doi: 10.1089/dna.2013.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Chen S., Yang L., et al. TRAV and TRBV repertoire, clonality and the proliferative history of umbilical cord blood T-cells. Transplant Immunology. 2007;18(2):151–158. doi: 10.1016/j.trim.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Chen S., Yang L., Zhou Y., Wu X., Huang M. Clonal expanded TCR Vbeta T cells in patients with APL. Hematology. 2005;10(2):135–139. doi: 10.1080/10245330400026196. [DOI] [PubMed] [Google Scholar]
- 30.Jin Z., Luo Q., Lu S., et al. Oligoclonal expansion of TCR Vδ T cells may be a potential immune biomarker for clinical outcome of acute myeloid leukemia. Journal of Hematology & Oncology. 2016;9(1):p. 126. doi: 10.1186/s13045-016-0353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hackl M., Brunner S., Fortschegger K., et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9(2):291–296. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohyashiki M., Ohyashiki J. H., Hirota A., Kobayashi C., Ohyashiki K. Age-related decrease of miRNA-92a levels in human CD8+ T-cells correlates with a reduction of naïve T lymphocytes. Immunity & Ageing. 2011;8(1):p. 11. doi: 10.1186/1742-4933-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G. J., Yu M. C., Lee W. W., et al. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nature Medicine. 2012;18(10):1518–1524. doi: 10.1038/nm.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gombar S., Jung H. J., Dong F., et al. Comprehensive microRNA profiling in B-cells of human centenarians by massively parallel sequencing. BMC Genomics. 2012;13(1):p. 353. doi: 10.1186/1471-2164-13-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serna E., Gambini J., Borras C., et al. Centenarians, but not octogenarians, up-regulate the expression of microRNAs. Scientific Reports. 2012;2(1):p. 961. doi: 10.1038/srep00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perluigi M., Di Domenico F., Butterfield D. A. mTOR signaling in aging and neurodegeneration: at the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiology of Disease. 2015;84:39–49. doi: 10.1016/j.nbd.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 37.Khanna A., Muthusamy S., Liang R. Q., Sarojini H., Wang E. Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging (Albany NY) 2011;3(3):223–236. doi: 10.18632/aging.100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szabolcs P., Park K. D., Reese M., Marti L., Broadwater G., Kurtzberg J. Coexistent naïve phenotype and higher cycling rate of cord blood T cells as compared to adult peripheral blood. Experimental Hematology. 2003;31(8):708–714. doi: 10.1016/S0301-472X(03)00160-7. [DOI] [PubMed] [Google Scholar]
- 39.Wu C. J., Chillemi A., Alyea E. P., et al. Reconstitution of T-cell receptor repertoire diversity following T-cell depleted allogeneic bone marrow transplantation is related to hematopoietic chimerism. Blood. 2000;95(1):352–359. doi: 10.1182/blood.V95.1.352. [DOI] [PubMed] [Google Scholar]
- 40.Pido-Lopez J., Imami N., Aspinall R. Both age and gender affect thymic output: more recent thymic migrants in females than males as they age. Clinical and Experimental Immunology. 2001;125(3):409–413. doi: 10.1046/j.1365-2249.2001.01640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naylor K., Li G. J., Vallejo A. N., et al. The influence of age on T cell generation and TCR diversity. Journal of Immunology. 2005;174(11):7446–7452. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Chen S., Yang L., Li B., Chan J. Y., Cai D. TRGV and TRDV repertoire distribution and clonality of T cells from umbilical cord blood. Transplant Immunology. 2009;20(3):155–162. doi: 10.1016/j.trim.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 43.Xu L., Lu Y., Lai J., et al. Characteristics of the TCR Vβ repertoire in imatinib-resistant chronic myeloid leukemia patients with ABL mutations. Science China. Life Sciences. 2015;58(12):1276–1281. doi: 10.1007/s11427-015-4930-4. [DOI] [PubMed] [Google Scholar]
- 44.Li Y., Geng S., Du X., et al. Restricted TRBV repertoire in CD4+ and CD8+ T-cell subsets from CML patients. Hematology. 2013;16(1):43–49. doi: 10.1179/102453311X12902908411634. [DOI] [PubMed] [Google Scholar]
- 45.Parker C. M., Groh V., Band H., et al. Evidence for extrathymic changes in the T cell receptor gamma/delta repertoire. The Journal of Experimental Medicine. 1990;171(5):1597–1612. doi: 10.1084/jem.171.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geng S., Weng J., du X., et al. Comparison of the distribution and clonal expansion features of the T-cell γδ repertoire in myelodysplastic syndrome-RAEB and RAEB with progression to AML. DNA and Cell Biology. 2012;31(10):1563–1570. doi: 10.1089/dna.2012.1769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


