Abstract
Background
As the important components in polycomb repressive complexes 1 (PRC1) and heterochromatin protein 1 (HP1), Chromobox (CBX) family members are involved in epigenetic regulatory function, transcriptional repression, and other cellular metabolisms. Increasing studies have indicated significant associations between CBX and tumorigenesis, which is a progression in different types of cancers. However, the information about the roles of each CBX in gastric cancer is extremely limited.
Methods
We explored CBX mRNA expression, corrections with clinicopathological parameters, protein expression, prognostic values, enrichment analysis with several databases including Oncomine, Human Protein Atlas, UALCAN, Kaplan-Meier plotter, cBioPortal, GeneMANIA, and Enrichr.
Results
In our study, comparing to the normal tissues, higher mRNA expression of CBX1/2/3/4/5/8 and lower mRNA expression of CBX7 were found in GC tissues while upregulations of CBX1/2/3/4/5/8 and downregulations of CBX7 were indicated to be significantly correlated to the nodal metastasis status and individual cancer stages in GC patients. As for protein level, the expression of CBX2/3/4/5/6 was higher and the expression of CBX7 was lower in the GC tissues than those in the normal. What is more, higher mRNA expression of CBX1/5/6/8 and lower mRNA expression of CBX7 were markedly correlated to poor outcomes of OS and FP in GC patients. Besides, high mutation rate of CBXs (42%) was observed in GC patients.
Conclusions
We suggest that CBX5/7 may serve as potential therapeutic targets for GC while CBX1/8 may serve as potential prognostic indicators for GC.
1. Introduction
According to the Global Cancer Statistics 2018 report, gastric cancer (GC) ranks sixth among the most common malignant tumors worldwide [1], with close to 1.03 million new GC cases and 0.78 million cancer-related deaths in 2018. The GC incidence rates are much higher in the low/middle income countries with highest rates in Eastern Asia, Eastern Europe, and South America while they are much lower in the higher income countries [2–4]. Because a large number of patients are diagnosed with advanced GC at their initial diagnosis, the 5-year survival rate of the disease remains below 30% even with comprehensive treatment modalities [5]. Stomach adenocarcinoma (STAD) is the most common type of GC, which occurs in the gastric glandular cells and accounts for 95% of the total incidence of GC [6, 7]. Recently, researchers have made immense progress in understanding the mechanisms of occurrence, development, and metastasis in the disease. However, some of the molecular mechanisms of GC remain unclear. Early diagnosis can significantly improve the therapeutic outcome of GC. However, due to the lack of effective methods for early diagnosis, many patients are diagnosed in the advanced stages or with distant metastasis, which significantly decrease the therapeutic outcome. Thus, identifying effective biomarkers and potential targets are extremely important for the early detection and treatment of GC.
Eight members of the Chromobox (CBX) family of proteins have been identified in mammalian cells thus far, including the polycomb (CBX2/4/6/7/8) and heterochromatin protein 1 (HP1) (CBX1/3/5, also known as HP1β/γ/α) [8–10]. All of them contain a single N-terminal chromodomain [8]. The polycomb-related CBX proteins interact with H3K27me3 by their chromodomains and help in the recruitment and stabilization of PRC1 to specific regions of the chromatin [11, 12], while the HP1-related proteins also have the terminal chromoshadow domain. Moreover, the CBX family of proteins is reported to be involved in transcriptional repression, cell cycle regulation, tumor initiation, progression, development, and chromatin [9, 13]. Increasing evidence suggests that CBX family of proteins plays vital roles in various cancers. CBX5 was demonstrated to serve as an oncogenic role in GC and be targeted by microRNA-758-3p in the proliferation, migration, and invasion of GC [14]. CBX7 was overexpressed both in GC cell lines and GC tissues, and its overexpression was correlated with patient's age, clinical stage, and lymph node metastasis [15]. However, the precise functions of the different CBXs in the development and progression of GC remain unclear. Using several large public databases, we analyzed the expression and mutations in the different CBX family members in STAD patients to determine their expression levels, potential functions, and prognostic values in GC.
1.1. Method Oncomine
Oncomine (https://www.oncomine.org) is a publicly accessible online cancer database, containing gene expression array data [16, 17]. In our study, Oncomine was used to analyze the mRNA expression level of the CBX members in different cancers. We compared the mRNA expression of CBX members in cancer and normal tissues using Student's t-test after set the thresholds as follows: P value: 0.001 and fold − change : 1.5.
1.2. UALCAN
An interactive online public platform, UALCAN (http://ualcan.path.uab.edu/), can be used to estimate gene expression and correlation, methylation, and survival analysis [18] based on clinical data and level 3 RNA-seq data from TCGA database. In our study, we used UALCAN to analyze the expression of CBX family members, nodal metastasis status, and individual cancer stage in STAD patients based on mRNA levels with 34 normal and 415 STAD tissues. We compared the mRNA expression using Student's t-test with P < 0.05 considered as statically significant. The nodal metastasis status criterion of UALCAN was as follows: no regional lymph node metastasis was considered as N0, and metastases in 1 to 3, 4 to 9, and 10 or more axillary lymph nodes were considered to be N1, N2, and N3, respectively.
1.3. Human Protein Atlas
The Human Protein Atlas (https://www.proteinatlas.org) is a public database, which can be used to analyze mRNA and protein expression data and survival information of patients for nearly 20 common kinds of cancers [19]. In our study, we used this database to compare the protein expression level of different CBX members between normal and GC tissues using immunohistochemistry images.
1.4. Kaplan-Meier Plotter
Kaplan-Meier plotter (http://www.kmplot.com) is an interactive database to obtain mRNA and miRNA expression data and survival-related information for different cancers from the GEO, EGA, and TCGA databases [20–22]. In our study, we used the KM Plotter to explore the overall survival (OS) and first progression (FP) as the prognostic values for CBX members, according to the datasets (excluding GSE62254, based on KM Plotter suggestion) of GC having divided the 592 patient samples into a high and low group based on the median mRNA expression level. Moreover, we set the hazard ratio with 95% confidence intervals. A log rank P value <0.05 was considered statically significant difference.
1.5. cBioPortal
cBioPortal (http://www.cbioportal.org) is a publicly accessible tool which can be used to analyze and visualize different cancer genomics data [23, 24]. We selected the STAD (TCGA, provisional) dataset, including 369 cases with pathology reports, for the analysis of CBXs in cBioPortal. Then, we assessed the gene mutations, putative copy number alterations from GISTIC, and mRNA expression z-scores (RNA Seq V2 RSEM, z-score threshold ±2.0). Using cBioPortal, we analyzed the genetic alterations in the CBXs and their correlation with each other. For the correlation of CBXs with each other, Spearman′s correlation coefficient > 0.3 and P < 0.05 were considered to be significant.
1.6. GeneMANIA
GeneMANIA (http://www.genemania.org), an online system for network analysis, can be used for predicting and visualizing the protein-protein interaction (PPI) network and gene functional assays [25] and features several bioinformatics methods: physical interaction, gene coexpression, gene colocation, gene enrichment analysis, and website prediction. In our study, GeneMANIA was used to construct the gene networks and predict the functions of CBXs.
1.7. Enrichr Database
The online public database [26, 27], Enrichr (http://amp.pharm.mssm.edu/ Enrichr/), was used to obtain the data pertaining to Gene Ontology (GO) functional annotations and Reactome pathway enrichment analysis for CBXs. Finally, we used the R (ggplot2 package) to create plots and visualize the data from our study.
2. Results
2.1. mRNA Expression of CBXs in Different Cancers
We compared the mRNA expression of the CBXs in cancer and normal samples (Figure 1) using the Oncomine database and found that the transcriptional levels of CBX1/2//3/4/5/8 were upregulated while that of CBX6/7 were downregulated in majority of the cancers. In the GC samples (Table 1), CBX1 was markedly higher in GC patients in five datasets. In Cho's gastric dataset [28], CBX1 was overexpressed in gastric adenocarcinoma (fold − change = 2.415 and P = 4.52E − 6). In Chen's gastric dataset [29], CBX1 was found to be upregulated in diffuse gastric adenocarcinoma (fold − change = 1.516 and P = 2.83E − 08), gastric mixed adenocarcinoma (fold − change = 1.66 and P = 2.25E − 6), gastric intestinal-type adenocarcinoma (fold − change = 1.613 and P = 2.38E − 13). In D'Errico's gastric dataset [30], CBX1 was found to be upregulated in gastric intestinal-type adenocarcinoma (fold − change = 2.116 and P = 2.21E − 13). The mRNA expression of CBX2 was higher in GC patients in three datasets. In Cho's gastric dataset [28], CBX2 was overexpressed in diffuse gastric adenocarcinoma (fold − change = 2.29 and P = 6.01E − 09) and gastric mixed adenocarcinoma (fold − change = 2.077 and P = 3.75E − 04). D'Errico's gastric dataset [30] showed CBX2 to be upregulated in gastric intestinal-type adenocarcinoma (fold − change = 4.485 and P = 1.7E − 09). The mRNA expression of CBX3 was higher in GC patients in four datasets. Chen's gastric dataset [29] revealed that CBX3 was overexpressed in gastric mixed adenocarcinoma (fold − change = 1.998 and P = 1.62E − 07) and gastric intestinal-type adenocarcinoma (fold − change = 1.878 and P = 1.13E − 16). Moreover, CBX3 was overexpressed in gastric intestinal-type adenocarcinoma (fold − change = 3.014 and P = 6.64E − 14) in D'Errico's gastric dataset [30] and GC (fold − change = 1.736 and P = 6.79E − 04) of Wang's gastric dataset [31]. The mRNA expression of CBX4 was higher in GC patients in six datasets. In Chen's gastric dataset, CBX4 was overexpressed in gastric intestinal-type adenocarcinoma (fold − change = 1.783 and P = 2.55E − 17), gastric mixed adenocarcinoma (fold − change = 1.955 and P = 3.03E − 06), and diffuse gastric adenocarcinoma (fold − change = 1.730 and P = 4.23E − 04). D'Errico's gastric dataset [30] showed that CBX4 was upregulated in diffuse gastric adenocarcinoma (fold − change = 2.466 and P = 2.45E − 05) and gastric mixed adenocarcinoma (fold − change = 3.314 and P = 2.29E − 06). Cho's gastric dataset [28] indicated that CBX4 was higher in gastric mixed adenocarcinoma (fold − change = 1.625 and P = 7.18E − 04). The mRNA expression of CBX6 was higher in GC patients in one dataset. Chen's gastric dataset [29] revealed that CBX6 was upregulated in diffuse gastric adenocarcinoma (fold − change = 1.758 and P = 8.38E − 05). The mRNA expression of CBX7 was lower in GC patients in one dataset. In Cho's gastric dataset [28], CBX7 was downregulated in diffuse gastric adenocarcinoma (fold − change = −1.656 and P = 9.09E − 05).
Figure 1.
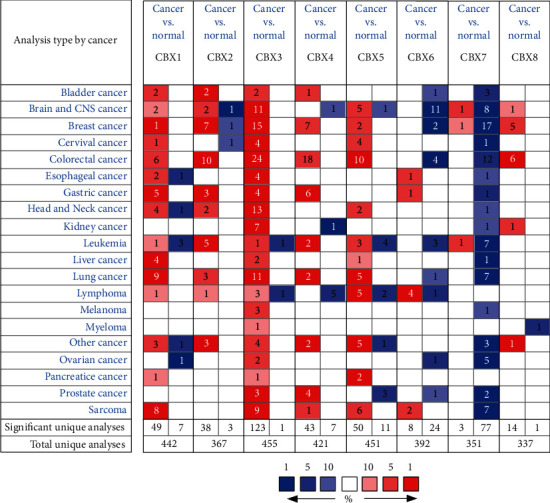
Transcriptional levels of the CBX family members in different cancers (Oncomine, P value: 0.001 and fold − change : 1.5).
Table 1.
The transcription levels of CBX family members between different types of gastric cancers and normal gastric tissues (Oncomine).
| Types of gastric cancer vs. normal | Fold-change | t-test | P value | Ref | PMID | |
|---|---|---|---|---|---|---|
| CBX1 | Gastric adenocarcinoma vs. normal | 2.415 | 6.913 | 4.52E-06 | Cho gastric | 21447720 |
| Diffuse gastric adenocarcinoma vs. normal | 1.516 | 6.996 | 2.83E-08 | Chen gastric | 12925757 | |
| Gastric mixed adenocarcinoma vs. normal | 1.66 | 7.026 | 2.25E-06 | Chen gastric | 12925757 | |
| Gastric intestinal-type adenocarcinoma vs. normal | 1.613 | 9.531 | 2.38E-13 | Chen gastric | 12925757 | |
| Gastric intestinal-type adenocarcinoma vs. normal | 2.116 | 10.157 | 2.21E-13 | D'Errico gastric | 19081245 | |
| CBX2 | Diffuse gastric adenocarcinoma vs. normal | 2.29 | 6.862 | 6.01E-09 | Cho gastric | 21447720 |
| Gastric mixed adenocarcinoma vs. normal | 2.077 | 4.349 | 3.75E-04 | Cho gastric | 21447720 | |
| Gastric intestinal type adenocarcinoma vs. normal | 4.485 | 7.31 | 1.7E-09 | D'Errico gastric | 19081245 | |
| CBX3 | Gastric mixed adenocarcinoma vs. normal | 1.998 | 8.875 | 1.62E-07 | Chen gastric | 1292575 |
| Gastric intestinal-type adenocarcinoma vs. normal | 1.878 | 11.061 | 1.13E-16 | Chen gastric | 12925757 | |
| Gastric intestinal-type adenocarcinoma vs. normal | 3.014 | 9.795 | 6.64E-14 | D'Errico gastric | 19081245 | |
| Gastric cancer vs. normal | 1.736 | 3.719 | 6.79E-04 | Wang gastric | 21132402 | |
| CBX4 | Gastric intestinal-type adenocarcinoma vs. normal | 1.783 | 10.753 | 2.55E-17 | Chen gastric | 12925757 |
| Gastric mixed adenocarcinoma vs. normal | 1.955 | 8.08 | 3.03E-06 | Chen gastric | 12925757 | |
| Diffuse gastric adenocarcinoma vs. normal | 1.73 | 4.295 | 4.23E-04 | Chen gastric | 12925757 | |
| Diffuse gastric adenocarcinoma vs. normal | 2.466 | 4.862 | 2.45E-05 | D'Errico gastric | 19081245 | |
| Gastric mixed adenocarcinoma vs. normal | 3.314 | 6.444 | 2.29E-06 | D'Errico gastric | 19081245 | |
| Gastric mixed adenocarcinoma vs. normal | 1.625 | 3.633 | 7.18E-04 | Cho gastric | 21447720 | |
| CBX5 | NA | NA | NA | NA | NA | NA |
| CBX6 | Diffuse gastric adenocarcinoma vs. normal | 1.758 | 4.643 | 8.38E-05 | Chen gastric | 12925757 |
| CBX7 | Diffuse gastric adenocarcinoma vs. normal | -1.656 | -4.072 | 9.09E-05 | Cho gastric | 21447720 |
| CBX8 | NA | NA | NA | NA | NA | NA |
2.2. mRNA and Protein Expression of CBXs in GC and Normal Samples
The mRNA expression data from the UALCAN database revealed that the expression of CBX1/2/3/4/5/8 was significantly upregulated in the STAD tissues compared to the normal tissues, while the expression of CBX6/7 was markedly downregulated (Figure 2(a); please see the Supplementary 1 for primary data). Then, using the Human Protein Atlas, we evaluated the immunohistochemistry (IHC) data pertaining to the protein expression of CBXs in GC and normal tissues. We found that the protein expression of CBX2/3/4/5/6 was higher in GC tissues than normal tissues while that of CBX7 was lower in GC tissues than in the normal tissues (Figure 2(b)). The protein expression of CBX1/8 did not show much difference.
Figure 2.
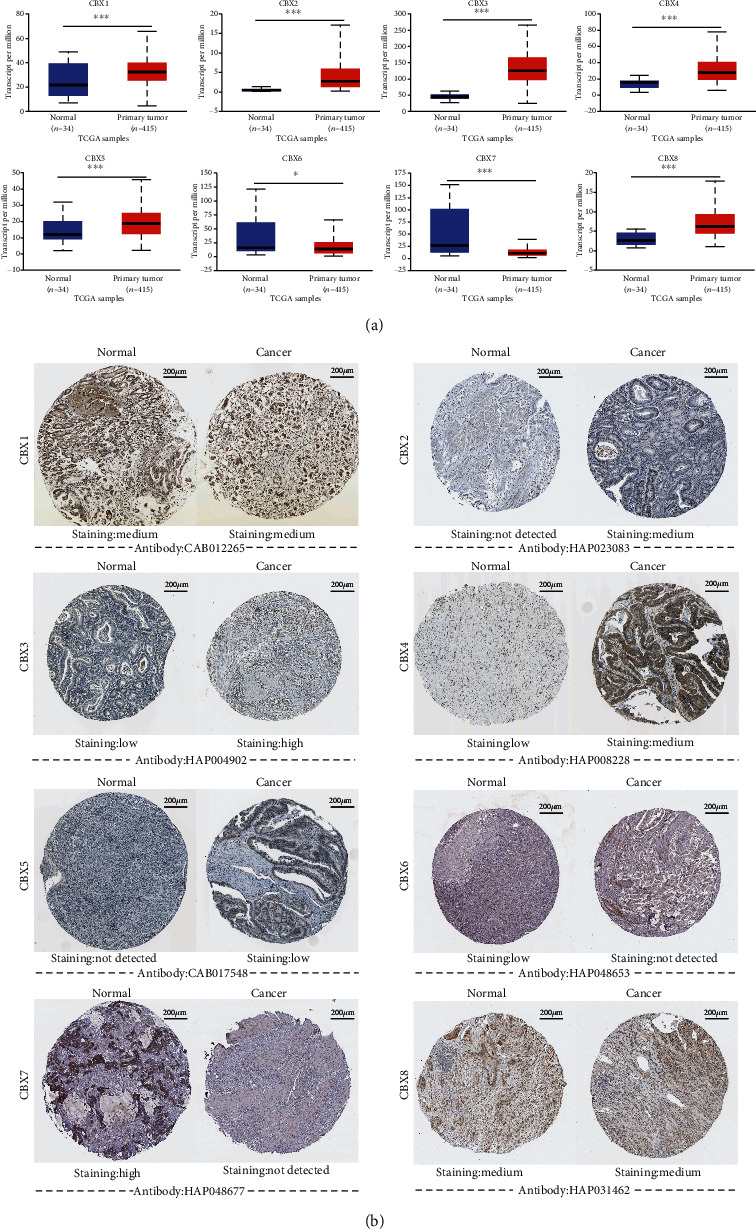
mRNA and protein expression of CBXs (UALCAN and Human Protein Atlas): (a) mRNA expression of different CBXs in STAD and normal tissues (UALCAN, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001); (b) protein expression of different CBXs in GC and the normal tissues (Human Protein Atlas).
2.3. Clinicopathological Parameters of CBXs in GC Patients
Using the STAD database in UALCAN, we evaluated the correlation of CBX mRNA expression with the nodal metastatic status (Figure 3(a); please see the Supplementary 2 for primary data) and cancer stage of individual patients (Figure 3(b); please see the Supplementary 3 for primary data). As shown in Figure 3(a), the expression of CBX1/2/3/4/5/7/8 was all correlated with the nodal metastatic status while the expression of CBX6 had no correlation with the nodal metastatic status. Compared to normal tissues, the mRNA expression of CBX2/3/4/8 was significantly higher in the cancer stages 1, 2, 3, and 4, while the mRNA expression of CBX1 and CBX5 was significantly upregulated in the cancer stages 2, 3, and 4. Moreover, the mRNA expression of CBX7 was found to be downregulated in all cancer stages. The mRNA expression of CBX6 was lower in the cancer stages 1 and 4 compared to the normal tissues.
Figure 3.
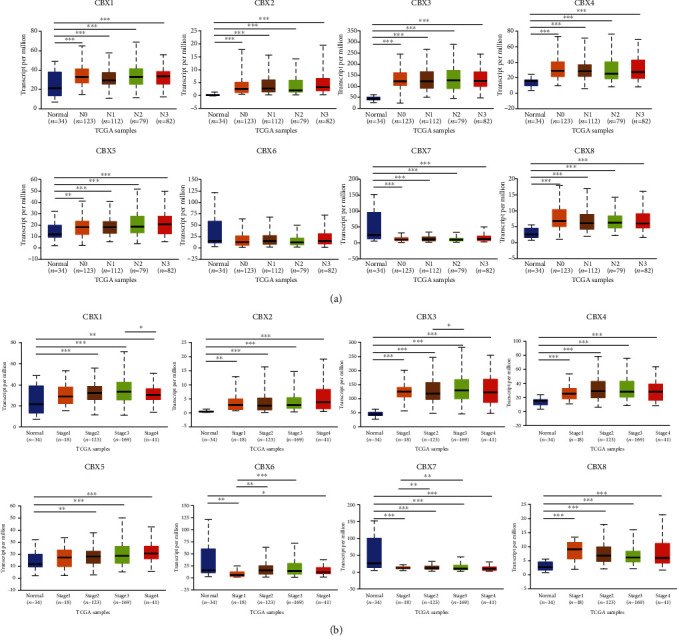
Clinicopathological parameters and CBX mRNA levels in STAD patients (UALCAN): (a) correlation between the expression level of CBXs and nodal metastatic status in STAD patients (UALCAN, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001); (b) correlation between expression of CBXs and individual cancer stage in STAD patients (UALCAN, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001).
2.4. Prognostic Value of mRNA Level of CBXs in GC Patients
Using the KM Plotter, we explored the prognostic value of mRNA expression of CBXs according to the OS and FP of GC patients. The survival curve indicated that higher mRNA expression of CBX1/5/6/8 and lower mRNA expression of CBX7 predicted poor OS and FP (Figure 4).
Figure 4.
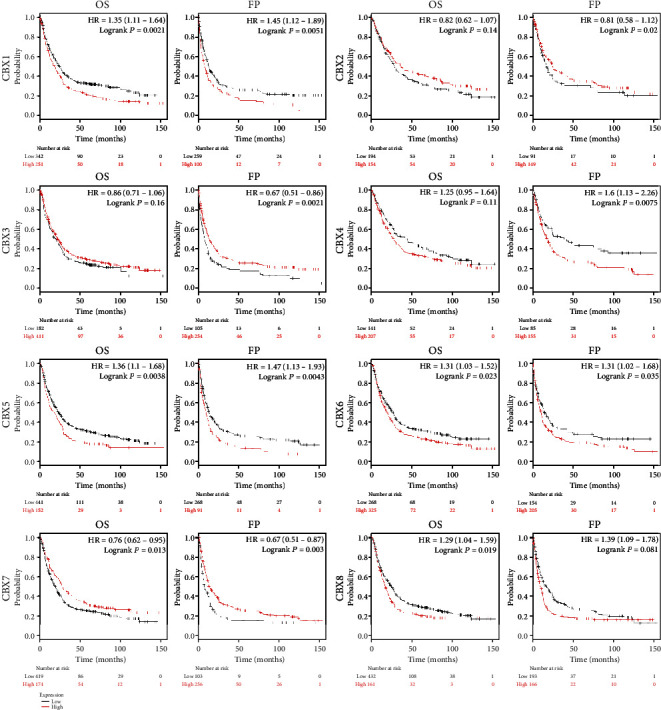
Prognostic value of mRNA level of CBX family members in GC patients (Kaplan-Meier plotter, P < 0.05 was considered statistically significant).
2.5. Genetic Alterations and Interaction Analysis of CBXs in GC
Using cBioPortal, we analyzed the alterations in CBXs and their correlations with each other in STAD. The results showed that the CBXs were altered in 155 out of 369 patients with STAD (42%) (Figure 5(a)). Based on TCGA provisional dataset, the percentage of genetic alterations in CBX1/2/3/4/5/6/7/8 was 9, 9, 15, 9, 8, 8, 5, and 11%, respectively, in STAD (Figure 5(b)). Further, by analyzing the mRNA expression of the CBXs, we calculated the correlation of the CBXs with each other using Spearman's correlation (Figure 5(c); please see the Supplementary 4 for primary data). The results revealed significant positive correlations between in the following CBX pairs: CBX2 with CBX4 (r = 0.58 and P = 3.53E − 04); with CBX8 (r = 0.56 and P = 5.45E − 04); CBX4 with CBX8 (r = 0.62 and P = 9.63E − 05); CBX6 with CBX7 (r = 0.56 and P = 5.27E − 04); while significant negative correlations were found between the following pairs of CBXs; CBX2 with CBX7 (r = −0.54 and P = 8.88E − 04); CBX3 with CBX6 (r = −0.40 and P = 1.81E − 02).
Figure 5.
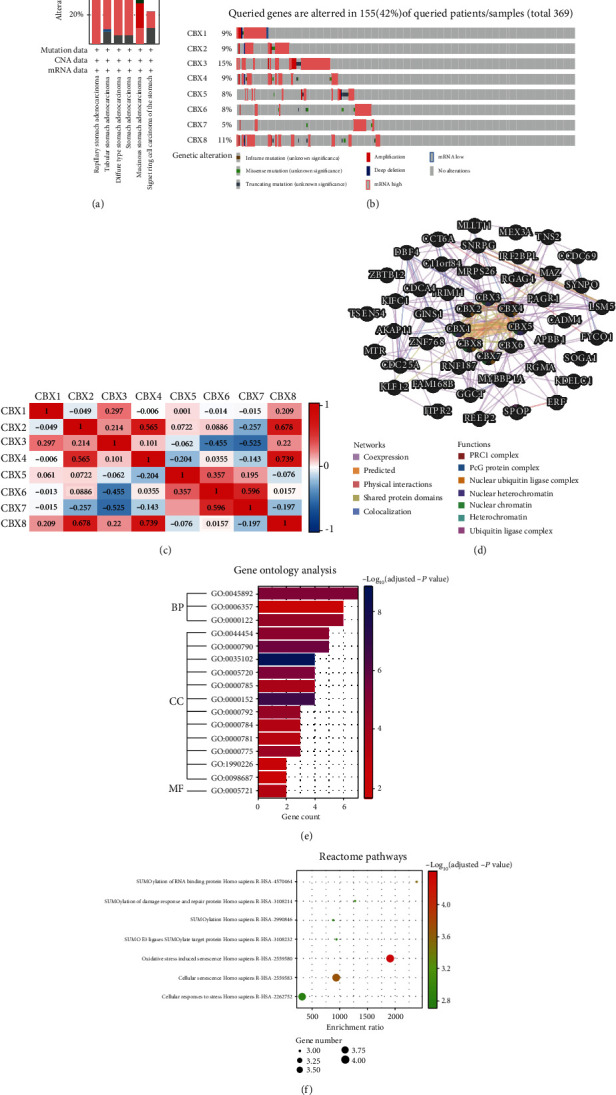
Genetic alterations, interactions, and enrichment analysis of CBXs in GC: (a) summary of CBX alteration in STAD; (b) alterations in CBXs in GC; (c) correction of CBXs with each other (cBioPortal); (d) interaction analysis of CBXs (GeneMANIA). Summary of alterations in CBXs in HCC: (e) GO enrichment analysis; (f) Reactome pathway prediction. BP: biological processes; CC: cellular components; MF: molecular functions.
The GeneMANIA (Figure 5(d); please see the Supplementary 5 for primary data) results revealed that the CBXs shared protein domains with each other. Interactions were predicted between CBX1 and CBX3, CBX1 and CBX5, CBX3 and CBX5, and CBX4 and CBX8. Moreover, CBX1 and CBX2, CBX1 and CBX3, CBX1 and CBX5, CBX2 and CBX4, CBX3 and CBX5, CBX4 and CBX6, CBX4 and CBX8, and CBX7 and CBX8 shared physical interactions. Relationships of coexpression were predicted between CBX1 and CBX8, CBX2 and CBX4, CBX3 and CBX5, and CBX6 and CBX7.
2.6. Enrichment Analysis of CBXs
To identify the potential signaling pathways of CBXs in GC, we used the Enrichr online database to analyze GO functional annotation (based on biological processes (BPs), cellular components (CCs), and molecular functions (MPs) and the Reactome pathways; please see the Supplementary 6 for primary data). According to the results (Figure 5(e)), CBXs were mainly involved in negative regulation of transcription, DNA templated (GO:0045892), negative regulation of transcription from RNA polymerase II promoter (GO:0000122), and regulation of transcription from RNA polymerase II promoter (GO:0006357) in BPs. As for CCs, the CBXs were mainly involved in PRC1 complex (GO:0035102), nuclear ubiquitin ligase complex (GO:0000152), nuclear heterochromatin (GO:0005720), heterochromatin (GO:0000792), and chromatin (GO:0000785). Moreover, CBXs influenced MFs through histone methyltransferase binding (GO:1990226).
In Reactome analysis (Figure 5(f)), 7 pathways were of significance, including oxidative stress-induced senescence Homo sapiens R-HSA-2559580, cellular senescence Homo sapiens R-HSA-2559583, SUMOylation of RNA binding protein Homo sapiens R-HSA4570464, SUMOylation of DNA damage response and repair protein Homo sapiens R-HSA-3108214, SUMO E3 ligases SUMOylate target protein Homo sapiens R-HSA3108232, SUMOylation Homo sapiens R-HSA-2990846, and cellular responses to stress Homo sapiens R-HSA-2262752.
3. Discussion
To date, some of CBXs have been reported to be involved in tumorigenesis and progression of several cancers, but the function of CBXs in GC is limited. This is the first study to systematically analyze their mRNA and protein expression, as well as evaluate their correlation with the clinicopathological parameters, prognostic values, genetic alterations, and potential functions in GC. We believe that our study will contribute toward improved early diagnosis, treatment outcome, and prognosis for patients with GC.
Increasing number of studies have shown that CBX1, also known as HP1-β, is overexpressed in different type of tumors, including prostate cancer (PRCA), breast cancer (BRCA), and hepatocellular carcinoma (HCC) [13, 32, 33]. Overexpression of CBX1 was associated with poor recurrence-free survival (RFS) in BRCA patients [13]. In addition, high expression of CBX1 was markedly correlated with larger tumor size, poor tumor differentiation, and tumor vascular invasion in hepatocellular carcinoma [33]. However, Tretiakova et al. [34] reported that the expression of CBX1 was lower in thyroid carcinoma, and the decrease in CBX1 followed by a reduction in CBX5 contributed to the pathogenesis of thyroid carcinoma. Our study indicated that CBX1 mRNA expression was upregulated in GC patients, which was in accordance with the data in other tumors. However, its protein expression did not show much difference between GC and normal tissues. Further, its mRNA expression was significantly correlated with nodal metastatic status, individual cancer stage, and poor OS and FP in GC patients. Taken together, our data indicate that CBX1 may serve as a potential prognostic marker in GC.
Recent studies have confirmed that CBX2 acts as an oncogene in several cancers, including BRCA, HCC, PRCA, and ovarian cancer (OVCA) [35–38]. Piqué et al. found that CBX2 promotes BRCA cell growth, and overexpression of CBX2 was related to poorer 5-year survival [37]. In HCC, Mao et al. confirmed that CBX2 was highly expressed both in HCC cell lines and tissues and was associated with poor prognosis in patients [36]. Clermont et al. revealed that elevated CBX2 expression was associated with poor clinical outcome in PRCA [35]. Our study indicated that mRNA and protein expression of CBX2 were significantly overexpressed in GC compared to normal tissues, while it was also correlated with nodal metastatic status and individual cancer stage in GC patients. However, lower CBX2 expression was associated with lower OS and PF in the patients with GC, but the difference was not significant.
CBX3 is highly expressed in lung adenocarcinoma (LUAD), pancreatic cancer (PACA), colorectal cancer (CRC), PRCA, non-small-cell lung cancer (NSCLC), and tongue squamous cell carcinoma (TSCC) [39–44]. Alam et al. found that CBX3 enhanced the expression of protumorigenic genes in LUAD by downregulating NCOR2 and ZBTB7A [39]. Chen et al. reported that CBX3 promoted aerobic glycolysis by suppressing FBP1 in PACA cell [42]. Liu et al. demonstrated that CBX3 was correlated with poor prognosis in CRC and promoted the proliferation and tumorigenesis of CRC, while miR-30a targeted CBX3 to specifically suppress the growth of CRC in a xenograft mouse model [43]. In our report, we confirmed that both mRNA and protein expression of CBX3 were upregulated in GC tissues compared to normal tissues. The mRNA expression was also significantly associated with nodal metastatic status and individual cancer stage. Lower mRNA expression of CBX3 was associated with poor FP in GC, but the OS result was not significant.
As an important member in the CBX family, CBX4 was found to promote several cancers, including HCC, lung cancer, and BRCA [45–47]. Li et al. revealed that the expression of CBX4 was markedly related to VEGF expression, angiogenesis, and OS in HCC and that CBX4 was important for tumor angiogenesis by governing HIF-1α protein [46]. In lung cancer, CBX4 regulated the expression of BMI-1 to promote proliferation and metastasis in lung cancer cells, and its expression was positively correlated with tumor size [45]. Surprisingly, Wang et al. indicated that CBX4 suppressed metastasis through recruitment of HDAC3 to the runx2 promoter in CRC [48]. In our study, the mRNA and protein expression of CBX4 were higher in GC tissues compared to normal tissues, and the mRNA expression was significantly correlated with nodal metastatic status and individual cancer stage in patients with GC. Moreover, higher CBX4 mRNA expression was correlated with worse OS and FP in GC, although the differences were not significant.
CBX5, also known as HP1α, was reported to be associated with several cancers, including GC, PRCA, BRCA, and NSCLC [14, 49–51]. A previous study showed that overexpression of CBX5 promoted cell proliferation in NSCLC cell lines [51]. Lieberthal et al. revealed that the expression of CBX5 was reduced in BRCA cell lines, while YY1 expression was detected to be lower in the invasive BRCA cell line [50]. In our report, both the mRNA and protein expression of CBX5 were upregulated in GC tissues than that in normal tissues. The mRNA expression was significantly correlated with nodal metastatic status and individual cancer stage in patients with GC. Moreover, its higher mRNA expression was significantly correlated with poor OS and FP. Thus, the results revealed that CBX5 may serve an oncogenic role in GC.
The mechanism of CBX6 is complex in different types of cancers. Zheng et al. found that higher CBX6 expression in HCC patients was associated more frequently with larger tumor sizes and multiple tumors [52]. Deng et al. revealed that CBX6 was downregulated in BRCA and negatively regulated by EZH2 [53]. In our study, the function of CBX6 was ambiguous. The mRNA and protein expression of CBX6 were significantly increased in GC tissues than normal tissues. However, in the STAD databases, the mRNA expression of CBX6 was significantly decreased, and the downregulation of the mRNA was associated with individual cancer stages 1 and 4 but of no significant correction with nodal metastatic status. Higher mRNA expression of CBX6 was significantly correlated with poor OS. Taken together, our results seem inconsistent with the predicted role of CBX6 as a gene of prognostic value, and further studies are needed to determine the precise function of CBX6 in GC in the future.
Among the CBXs, CBX7 is most studied in cancers. CBX7 was reported to be a tumor suppressor gene. The loss or downregulation of CBX7 gene expression was associated with several cancer, including PACA, thyroid cancer (THCA), CRC, NSCLC, bladder carcinoma (BLCA), and HCC [54–59]. Pallante et al. [56] reported that compared to the normal colonic mucosa, CBX7 expression was decreased or missing in a significant number of CRC samples. In THCA, the loss of CBX7 expression was correlated with larger tumor size in THCA patients while CBX7 expression progressively decreased with malignancy grade and neoplasia stage [55]. Moreover, Karamitopoulou et al. [54] showed that loss of CBX7 expression was related to increased tumor grade in PACA.
Surprisingly, Zhang et al. [15] found that CBX7 was overexpressed in GC cell lines and tumor tissues while its overexpression was also correlated to patient's age, lymph node metastasis, and clinical stage. Our study demonstrated that the mRNA and protein expression of CBX7 were markedly decreased in GC tissues compared to normal tissues. Moreover, the downregulation of CBX7 mRNA expression was markedly correlated with nodal metastatic status and individual cancer stage and poor OS and FP in GC patients. These results indicate that CBX7 may serve as a tumor suppressor in GC.
Recent studies show that CBX8 promotes tumorigenesis in several cancers, including BRCA, HCC, and CRC [60–62]. Overexpression of CBX8 in HCC patients was positively associated with distant metastasis and inversely correlated with OS. CBX8 promoted HCC cell proliferation capacity [61]. Yang et al. [62] revealed that silencing CBX8 induced apoptosis in CRC cell lines. Our report confirmed that the mRNA expression of CBX8 was higher in GC tissues than in normal tissues while its protein expression did not show much difference. Moreover, the mRNA expression was significantly correlated with nodal metastatic status and individual cancer stage in patients with GC. High CBX8 mRNA expression was associated with poor OS and FP in GC. These results indicate that CBX8 may of prognostic value in GC.
However, our study has some limitations. Firstly, the data in our study were from public databases, and further studies are needed to validate our results. Secondly, we did not explore the molecular mechanisms of different CBXs in GC, and future studies are needed to investigate their detailed mechanisms.
4. Conclusion
In conclusion, compared to normal tissues, higher mRNA expression of CBX1/2/3/4/5/8 and lower mRNA expression of CBX7 were found in GC, while the results for CBX6 were ambiguous. The protein levels of CBX2/3/4/5/6 were higher while that of CBX7 was lower in the GC tissues compared to normal tissues. The upregulation of the mRNA of CBX1/2/3/4/5/8 and downregulation of CBX7 were found to be significantly correlated to the nodal metastatic status and individual cancer stage in GC patients. Further, higher mRNA expression of CBX1/5/6/8 and lower mRNA expression of CBX7 were markedly associated with poor OS and FP in GC patients. High mutation rate of CBXs (42%) was observed in GC patients and to varying degrees. In summary, CBX5/7 may serve as a potential therapeutic target, while CBX1/8 may serve as potential prognostic factor in GC.
Acknowledgments
We would like to give our sincere Ms. Yan-Si Yang for her support during the study. This study is supported by Hunan Provincial Natural Science Foundation of China (No. 2020JJ5331).
Abbreviations
- CBXs:
Chromobox family members
- GC:
Gastric cancer
- STAD:
Stomach adenocarcinoma
- GISTC:
Genomic Identification of Significant Targets in Cancer
- PPI:
Protein-protein interaction
- PcG:
Polycomb group
- HCC:
Hepatocellular carcinoma
- BRCA:
Breast cancer
- PRCA:
Prostate cancer
- LUAD:
Lung adenocarcinoma
- CRC:
Colorectal Cancer
- PACA:
Pancreatic cancer
- NSCLC:
Non-small-cell lung cancer
- THCA:
Thyroid cancer
- BLCA:
Bladder carcinoma
- CECA:
Cervical cancer
- OS:
Over survival
- FP:
First progression
- RFS:
Recurrence-free survival
- TCGA:
The Cancer Genome Atlas
- GO:
Gene Ontology
- BP:
Biological processes
- CC:
Cellular components
- MF:
Molecular functions.
Data Availability
All the data of this article is derived from public database in methods.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
ZYC and JB planned the study; ZYC wrote the manuscript and prepared all the figures. SXS and SXZ conducted the statistical analysis; JB revised the manuscript. ZYC and SXS prepared the supplementary information.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: a Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Pellino A., Riello E., Nappo F., et al. Targeted therapies in metastatic gastric cancer: current knowledge and future perspectives. World Journal of Gastroenterology. 2019;25(38):5773–5788. doi: 10.3748/wjg.v25.i38.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre L. A., Siegel R. L., Ward E. M., Jemal A. Global cancer incidence and mortality rates and trends--an update. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2016;25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 5.Ajani J. A., Bentrem D. J., Besh S., et al. Gastric cancer, version 2.2013: featured updates to the NCCN guidelines. Journal of the National Comprehensive Cancer Network. 2013;11(5):531–546. doi: 10.6004/jnccn.2013.0070. [DOI] [PubMed] [Google Scholar]
- 6.Blank S., Rachakonda S., Keller G., et al. A retrospective comparative exploratory study on two methylentetrahydrofolate reductase (MTHFR) polymorphisms in esophagogastric cancer: the A1298C MTHFR polymorphism is an independent prognostic factor only in neoadjuvantly treated gastric cancer patients. BMC Cancer. 2014;14(1):p. 58. doi: 10.1186/1471-2407-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon M., Mahar A. L., Helyer L. K., Vasilevska-Ristovska J., Law C., Coburn N. G. Prognostic factors in metastatic gastric cancer: results of a population-based, retrospective cohort study in Ontario. Gastric cancer : official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2016;19(1):150–159. doi: 10.1007/s10120-014-0442-3. [DOI] [PubMed] [Google Scholar]
- 8.Ma R. G., Zhang Y., Sun T. T., Cheng B. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. Journal of Zhejiang University. Science. B. 2014;15(5):412–428. doi: 10.1631/jzus.B1400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ning G., Huang Y. L., Zhen L. M., et al. Transcriptional expressions of Chromobox 1/2/3/6/8 as independent indicators for survivals in hepatocellular carcinoma patients. Aging. 2018;10(11):3450–3473. doi: 10.18632/aging.101658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruddock-D'Cruz N. T., Prashadkumar S., Wilson K. J., et al. Dynamic changes in localization of Chromobox (Cbx) family members during the maternal to embryonic transition. Molecular Reproduction and Development. 2008;75(3):477–488. doi: 10.1002/mrd.20752. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein B. E., Mikkelsen T. S., Xie X., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 12.Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. The EMBO Journal. 2006;25(11):2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Y.-K., Lin H.-Y., Chen C.-F., Zeng D. Prognostic values of distinct CBX family members in breast cancer identification of miR-758-3p as potential modulator of CBX5 expression in gastric cancer. Oncotarget. 2017;8(54):92375–92387. doi: 10.18632/oncotarget.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo J., Zhang Z., Pan L., Zhou Y. Identification of miR-758-3p as potential modulator of CBX5 expression in gastric cancer. Technology in Cancer Research & Treatment. 2018;17:p. 153303381881606. doi: 10.1177/1533033818816061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Zhang X. W., Zhang L., Qin W., et al. Oncogenic role of the chromobox protein CBX7 in gastric cancer. Journal of experimental & clinical cancer research. 2010;29(1):p. 114. doi: 10.1186/1756-9966-29-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes D. R., Kalyana-Sundaram S., Mahavisno V., et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes D. R., Yu J., Shanker K., et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrashekar D. S., Bashel B., Balasubramanya S. A. H., et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asplund A., Edqvist P. H., Schwenk J. M., Pontén F. Antibodies for profiling the human proteome-the Human Protein Atlas as a resource for cancer research. Proteomics. 2012;12(13):2067–2077. doi: 10.1002/pmic.201100504. [DOI] [PubMed] [Google Scholar]
- 20.Győrffy B., Surowiak P., Budczies J., Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12, article e82241) doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy Á., Lánczky A., Menyhárt O., Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific Reports. 2018;8(1):p. 9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha S. K., Biswas P. K., Gil M., Cho S. G., Cho S.-G. High expression of TTYH3 is related to poor clinical outcomes in human gastric cancer. Journal of Clinical Medicine. 2019;8(11):p. 1762. doi: 10.3390/jcm8111762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E., Gao J., Dogrusoz U., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J., Aksoy B. A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6(269):p. pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warde-Farley D., Donaldson S. L., Comes O., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research. 2010;38(Supplement_2):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen E. Y., Tan C. M., Kou Y., et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):p. 128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuleshov M. V., Jones M. R., Rouillard A. D., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho J. Y., Lim J. Y., Cheong J. H., et al. Gene expression signature-based prognostic risk score in gastric cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(7):1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Leung S. Y., Yuen S. T., et al. Variation in gene expression patterns in human gastric cancers. Molecular Biology of the Cell. 2003;14(8):3208–3215. doi: 10.1091/mbc.e02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Errico M., de Rinaldis E., Blasi M. F., et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. European Journal of Cancer. 2009;45(3):461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q., Wen Y.-G., Li D.-P., et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Medical Oncology. 2012;29(1):77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 32.Ngollo M., Lebert A., Daures M., et al. Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression. BMC Cancer. 2017;17(1):p. 261. doi: 10.1186/s12885-017-3256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y. F., Pan Y. H., Tian Q. H., Wu D. C., Su S. G. CBX1 indicates poor outcomes and exerts oncogenic activity in hepatocellular carcinoma. Translational Oncology. 2018;11(5):1110–1118. doi: 10.1016/j.tranon.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tretiakova M. S., Bond S. D., Wheeler D., et al. Heterochromatin protein 1 expression is reduced in human thyroid malignancy. Laboratory Investigation. 2014;94(7):788–795. doi: 10.1038/labinvest.2014.68. [DOI] [PubMed] [Google Scholar]
- 35.Clermont P. L., Crea F., Chiang Y. T., et al. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clinical Epigenetics. 2016;8(1):p. 16. doi: 10.1186/s13148-016-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao J., Tian Y., Wang C., et al. CBX2 regulates proliferation and apoptosis via the phosphorylation of YAP in hepatocellular carcinoma. Journal of Cancer. 2019;10(12):2706–2719. doi: 10.7150/jca.31845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piqué D. G., Montagna C., Greally J. M., Mar J. C. A novel approach to modelling transcriptional heterogeneity identifies the oncogene candidate CBX2 in invasive breast carcinoma. British Journal of Cancer. 2019;120(7):746–753. doi: 10.1038/s41416-019-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler L. J., Watson Z. L., Qamar L., et al. CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogene. 2018;7(11):p. 92. doi: 10.1038/s41389-018-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alam H., Li N., Dhar S. S., et al. HP1γ promotes lung adenocarcinoma by downregulating the transcription-repressive regulators NCOR2 and ZBTB7A. Cancer Research. 2018;78(14):3834–3848. doi: 10.1158/0008-5472.CAN-17-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang C., Liu J., He W., et al. A regulatory circuit HP1γ/miR-451a/c-Myc promotes prostate cancer progression. Oncogene. 2018;37(4):415–426. doi: 10.1038/onc.2017.332. [DOI] [PubMed] [Google Scholar]
- 41.Chang S. C., Lai Y. C., Chen Y. C., Wang N. K., Wang W. S., Lai J. I. CBX3/heterochromatin protein 1 gamma is significantly upregulated in patients with non-small cell lung cancer. Asia-Pacific Journal of Clinical Oncology. 2018;14(5):e283–e288. doi: 10.1111/ajco.12820. [DOI] [PubMed] [Google Scholar]
- 42.Chen L. Y., Cheng C. S., Qu C., et al. CBX3 promotes proliferation and regulates glycolysis via suppressing FBP1 in pancreatic cancer. Biochemical and Biophysical Research Communications. 2018;500(3):691–697. doi: 10.1016/j.bbrc.2018.04.137. [DOI] [PubMed] [Google Scholar]
- 43.Liu M., Huang F., Zhang D., et al. Heterochromatin protein HP1γ promotes colorectal cancer progression and is regulated by miR-30a. Cancer Research. 2015;75(21):4593–4604. doi: 10.1158/0008-5472.CAN-14-3735. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H., Fu X., Su X., Yang A. CBX3/HP1γ is upregulated in tongue squamous cell carcinoma and is associated with an unfavorable prognosis. Experimental and Therapeutic Medicine. 2018;15(5):4271–4276. doi: 10.3892/etm.2018.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu C., Zhang Q., Tang Q., et al. CBX4 promotes the proliferation and metastasis via regulating BMI-1 in lung cancer. Journal of Cellular and Molecular Medicine. 2020;24(1):618–631. doi: 10.1111/jcmm.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Xu Y., Long X. D., et al. Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25(1):118–131. doi: 10.1016/j.ccr.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Zeng J. S., Zhang Z. D., Pei L., et al. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. The International Journal of Biochemistry & Cell Biology. 2018;95:1–8. doi: 10.1016/j.biocel.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Li L., Wu Y., et al. CBX4 suppresses metastasis via recruitment of HDAC3 to the Runx2 promoter in colorectal carcinoma. Cancer Research. 2016;76(24):7277–7289. doi: 10.1158/0008-5472.CAN-16-2100. [DOI] [PubMed] [Google Scholar]
- 49.Kim J.-M., Shin Y., Lee S., et al. MacroH2A1.2 inhibits prostate cancer-induced osteoclastogenesis through cooperation with HP1α and H1.2. Oncogene. 2018;37(43):5749–5765. doi: 10.1038/s41388-018-0356-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lieberthal J. G., Kaminsky M., Parkhurst C. N., Tanese N. The role of YY1 in reduced HP1alpha gene expression in invasive human breast cancer cells. Breast Cancer Research. 2009;11(3):p. R42. doi: 10.1186/bcr2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K., Wang J., Yang L., et al. Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1α and APC2 gene expression in non-small cell lung cancer. Molecular Cancer. 2018;17(1):p. 153. doi: 10.1186/s12943-018-0896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng H., Jiang W. H., Tian T., et al. CBX6 overexpression contributes to tumor progression and is predictive of a poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8(12):18872–18884. doi: 10.18632/oncotarget.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng H., Guan X., Gong L., et al. CBX6 is negatively regulated by EZH2 and plays a potential tumor suppressor role in breast cancer. Scientific Reports. 2019;9(1):p. 197. doi: 10.1038/s41598-018-36560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karamitopoulou E., Pallante P., Zlobec I., et al. Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreatic cancer. European journal of cancer. 2010;46(8):1438–1444. doi: 10.1016/j.ejca.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 55.Pallante P., Federico A., Berlingieri M. T., et al. Loss of the CBX7 gene expression correlates with a highly malignant phenotype in thyroid cancer. Cancer Research. 2008;68(16):6770–6778. doi: 10.1158/0008-5472.CAN-08-0695. [DOI] [PubMed] [Google Scholar]
- 56.Pallante P., Terracciano L., Carafa V., et al. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. European Journal of Cancer. 2010;46(12):2304–2313. doi: 10.1016/j.ejca.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 57.Peng X., Guan L., Gao B. miRNA-19 promotes non-small-cell lung cancer cell proliferation via inhibiting CBX7 expression. Oncotargets and Therapy. 2018;11:8865–8874. doi: 10.2147/ott.s181433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie D., Shang C., Zhang H., Guo Y., Tong X. Up-regulation of miR-9 target CBX7 to regulate invasion ability of bladder transitional cell carcinoma. Medical Science Monitor. 2015;21:225–230. doi: 10.12659/MSM.893232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yongyu Z., Lewei Y., Jian L., Yuqin S. MicroRNA-18a targets IRF2 and CBX7 to promote cell proliferation in hepatocellular carcinoma. Oncology Research. 2018;26 doi: 10.3727/096504018X15165493852990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung C. Y., Sun Z., Mullokandov G., et al. Cbx8 acts non-canonically with Wdr5 to promote mammary tumorigenesis. Cell Reports. 2016;16(2):472–486. doi: 10.1016/j.celrep.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang B., Tian Y., Liao Y., et al. CBX8 exhibits oncogenic properties and serves as a prognostic factor in hepatocellular carcinoma. Cell Death & Disease. 2019;10(2):p. 52. doi: 10.1038/s41419-018-1288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang S., Liu W., Li M., Wen J., Zhu M., Xu S. Insulin-like growth Factor-1 modulates polycomb Cbx8 expression and inhibits colon cancer cell apoptosis. Cell Biochemistry and Biophysics. 2015;71(3):1503–1507. doi: 10.1007/s12013-014-0373-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data of this article is derived from public database in methods.


