Abstract
Nitrate is an essential macronutrient and a signal molecule that regulates the expression of multiple genes involved in plant growth and development. Here, we describe the participation of Arabidopsis DNA binding with one finger (DOF) transcription factor CDF3 in nitrate responses and shows that CDF3 gene is induced under nitrate starvation. Moreover, knockout cdf3 mutant plants exhibit nitrate-dependent lateral and primary root modifications, whereas CDF3 overexpression plants show increased biomass and enhanced root development under both nitrogen poor and rich conditions. Expression analyses of 35S::CDF3 lines reveled that CDF3 regulates the expression of an important set of nitrate responsive genes including, glutamine synthetase-1, glutamate synthase-2, nitrate reductase-1, and nitrate transporters NRT2.1, NRT2.4, and NRT2.5 as well as carbon assimilation genes like PK1 and PEPC1 in response to N availability. Consistently, metabolite profiling disclosed that the total amount of key N metabolites like glutamate, glutamine, and asparagine were higher in CDF3-overexpressing plants, but lower in cdf3-1 in N limiting conditions. Moreover, overexpression of CDF3 in tomato increased N accumulation and yield efficiency under both optimum and limiting N supply. These results highlight CDF3 as an important regulatory factor for the nitrate response, and its potential for improving N use efficiency in crops.
Keywords: CDF, nitrate, tomato, photosynthesis, crop yield, C/N metabolism, transcriptome
Introduction
Nitrogen (N) is an essential macronutrient and its availability in soil is a crucial factor for plant growth, distribution, and crop productivity. Nitrate (NO3−) is the main source of inorganic nitrogen for land plants (Krapp et al., 2014; Vidal et al., 2014). In addition, NO3− plays a key function as a signaling molecule in many aspects of plant metabolism and developmental processes like the ones involved in seed germination, root, and shoot development and senescence (Scheible et al., 1997; Stitt, 1999; Crawford and Forde, 2002; Little et al., 2005; Vidal and Gutierrez, 2008). Global expression analyses of Arabidopsis plants under different N treatments revealed changes in expression levels of a large set of genes, including those involved in N transport and assimilation. Moreover, N supply promotes changes in the expression of genes involved in abiotic stress responses, carbon (C) metabolism, regulation of C/N balance, and signaling transduction, like transcription factors (TFs), kinases, and phosphatases (Gutierrez et al., 2007). Thus, several TFs implicated in the regulation of gene expression and signaling by NO3− have been identified so far, including NIN Like protein 7, NLP7 (Castaings et al., 2009; Alvarez et al., 2020), NLP8 (Yan et al., 2016); TGA1 and TGA4 (Alvarez et al., 2014, 2019), SPL9 (Krouk et al., 2010), LBD37, LBD38, and LBD39 (Rubin et al., 2009), bZIP1 (Obertello et al., 2010), TCP20 (Guan et al., 2014), and ANR1 (Zhang and Forde, 1998). However, our understanding of the diverse regulatory pathways and the molecular mechanisms by which the different TFs modulate NO3− responses is still limited.
DNA binding with one finger (DOF) TFs are a group of plant specific proteins that contain a highly conserved DNA binding domain of 52 amino acids, with a C2-C2 structure that binds to a 5-T/AAAAG-3 DNA sequence motif (Yanagisawa and Schmidt, 1999). Different reports showed that DOF proteins are involved in a wide range of developmental processes such as root growth, seed development, and flowering time (Yanagisawa, 2002; Noguero et al., 2013; Rueda-Lopez et al., 2017). In addition, maize ZmDOF1 and ZmDOF2 have been also implicated in nitrogen assimilation and C/N balance (Yanagisawa and Sheen, 1998; Yanagisawa, 2004; Peña et al., 2017). The overexpression of ZmDOF1 in Arabidopsis and rice enhanced the expression of genes involved in N assimilation like glutamine synthetase (GS) and glutamate synthase and genes encoding enzymes for carbon skeleton production like C4-phosphoenol pyruvate carboxylase (PEPC) and pyruvate kinase1 (PK1). Moreover, ZmDOF1 overexpressing lines also showed increased amounts of amino acids, especially glutamine and an elevation in the nitrogen content (Yanagisawa, 2004). Most notably, Arabidopsis and rice plants expressing Dof1 showed better growth under low-nitrogen conditions (Yanagisawa, 2004; Kurai et al., 2011; Peña et al., 2017). Besides, a group of DOF factors, whose transcripts oscillate under constant light conditions named Cycling Dof Factors (CDF1-5; Imaizumi et al., 2005; Fornara et al., 2009), play a central role in the photoperiodic pathway controlling flowering-time in Arabidopsis. Moreover, Arabidopsis and tomato CDFs play additional functions in abiotic stress responses (Corrales et al., 2017; Renau-Morata et al., 2017, 2020a). In fact, both Arabidopsis and tomato (AtCDFs and SlCDFs) are differentially regulated by abiotic stress conditions like dehydration, osmotic, salt, heat stress, and cold stress (Corrales et al., 2014, 2017). Previously, we reported that the Arabidopsis KO mutant cdf3-1 is more sensitive to drought and low temperature stress, whereas CDF3 overexpression enhances the tolerance of transgenic plants to drought, cold, and osmotic stress and promotes late flowering (Corrales et al., 2017). Similar results have been also reported for tomato SlCDFs. In fact, overexpression of tomato SlCDF1 and SlCDF3 genes in Arabidopsis (Corrales et al., 2017) and AtCDF3 and SlCDF3 in tomato (Renau-Morata et al., 2017) increased drought and salt tolerance, and salt stress resistance, respectively. In addition, recent studies point to an involvement of CDFs in the responses to nitrogen. Network analyses using a time-based machine learning method applied to 2,174 dynamic N-responsive genes identified 155 regulators, including among them TFs previously validated in the N response (e.g., NLP7/8, TGA1/4, NAC4, HRS1, SNZ, and LBD37). This study also showed that those TFs are connected with additional second layer of TFs, which includes CDF1 (Varala et al., 2018). In addition, it is reported that master factor NLP7 targets multiple TFs including LBD37, LBD38, TGA4, HAP2C, NAC096, and CDF1 using the TARGET system (Alvarez et al., 2020), suggesting that CDF1 is a component of the regulatory network involved in N responses. In addition, we previously reported that tomato and Arabidopsis plants overexpressing the AtCDF3 gene, the closest homolog of CDF1, exhibited changes in primary metabolism, with increased amounts of amino acids like glutamine, asparagine and GABA, and higher sucrose contents (Corrales et al., 2017; Renau-Morata et al., 2017). Moreover, the tomato CDF3 overexpressor lines also showed higher yield and modified fruit amino acid and sugar content (Renau-Morata et al., 2017). These results suggested that CDF3 might play a role in nitrogen assimilation.
In the present work, we explored the function of CDF3 in N nutrition in Arabidopsis and evaluated the impact of its overexpression in tomato on N accumulation and use efficiency. We provide functional evidence that CDF3 is involved in NO3− assimilation. In addition, our data support that CDFs are potential candidate genes for improving nutrient use efficiency in tomato.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana
The Arabidopsis thaliana ecotype Columbia (Col-0) was used as wild type (WT). The 35S::CDF3 lines were previously described (Corrales et al., 2017). The cdf3-1 and cdf3-2 T-DNA insertion knockout mutants (GK-808605 and SAIL_434_09, respectively) were obtained from NASC. For studies on seedlings, plants were grown on MS-modified basal salt media without N (M531, Phytotechnology Laboratories) containing 1% (w/v) sucrose and 0.8% (w/v) plant agar at 22°C under long day 16-h light/8-h dark photoperiod for the time indicated in the figure legends. The full N treatment plate contained 10 mM KNO3 (Kiba et al., 2012). For N-limiting conditions (1 mM KNO3), the ion equilibrium of the medium was ensured by replacing KNO3 by KCl. For the phenotypic analyses plant images were acquired and biomass measurements were obtained after 12 days of treatment. Furthermore, roots and shoots were dried at 72°C for 48 h to determine their dry weights.
To analyze the role of CDFs in nitrogen signaling, we measure CDFs mRNA level in time-course experiments after nitrate treatments. First, we analyzed the early response to nitrate addition. To do so, WT plants were grown for 7 days in 10 mM KNO3, starved for 3 days (0 mM KNO3), and at the beginning of the next light cycle re-supplied with 5 mM KNO3, or 5 mM KCl as a control, for 0, 20 min, 2.5 h, or 8 h. We applied 5 mM KNO3 treatment, because it has been reported that this treatment elicit fast and robust responses to NO3− in Arabidopsis (Krouk et al., 2010; Alvarez et al., 2014; Canales et al., 2014). Second, we assayed CDF response for N starvation. To do so, WT plants were grown for 7 days in 10 mM KNO3 and at the start of the light period transferred to a nutrient solution with 0 or 10 mM KNO3 for 0, 1, 3, and 6 days. Then, plants were harvested and immediately frozen in liquid N2.
Solanum lycopersicum
Tomato (Solanum lycopersicum L. cv. Moneymaker) plants overexpressing the Arabidopsis CDF3 gene were previously as described in Renau-Morata et al. (2017). Two homozygous lines (L2 and L10) for the gene were selected for the present work and non-transformed Moneymaker plants were used as WT controls (C).
For phenotypic analyses in growth chamber (25/18°C and 16/8 h photoperiod), seeds were germinated in Petri dishes. After germination, seedlings were cultivated for 15 days in trays filled with vermiculite and fertilized with 1/2 strength Hoagland no. 2 (Hoagland and Arnon, 1950). Thirty-day-old plantlets were transferred to 1 L pots containing expanded clay balls (2–3 mm diameter; Arlita™, Spain). Plants were fertilized with 1/2 Hoagland no. 2 nutrient solution without N and supplemented with 8 or 4 mM NO3−, as N non-limiting and limiting, respectively (Wahle and Masiunas, 2003). The amount of other essential elements was maintained unaltered as described by Hoagland and Arnon (1950). Ten different plants for each genotype and experimental condition were used and physiological and biomass determinations were performed after 25 days.
For the greenhouse experiments, imbibed seeds were germinated on a moistened mixture of peat moss and sand in growth chambers at 25/18°C and a 16/8 h photoperiod. Thirty-days-old plantlets were transferred to 15 L pots that contained coconut coir fiber and grown for 6 months. Differential levels of N supply (8 and 4 mM nitrate) were applied as described previously for growth chamber experiments. Thirty plants were used per genotype and treatment.
In vitro Root Morphology Analysis
For root morphology analyses, Arabidopsis seeds were surface sterilized, stratified at 4°C for 2 days and grown for 12 days on vertical plates in a chamber at 22°C/18°C under long day (16 /8 h, light/dark) conditions. MS-modified basal salt media without N (M531, Phytotechnology Laboratories) containing 1% (w/v) sucrose, 0.8% (w/v) plant agar, and supplemented with 10 or 0.1 mM KNO3, as described by Alvarez et al. (2019). To compensate the potassium balance in the N-limiting media, KCl in its appropriate molarity was added. Three replicates per genotype and condition were performed and six seeds were used in every replicate. Plates were photographed and plant fresh weight was measured. Root length and number of lateral root (LR) were estimated using Image J software. Three replicates per genotype and condition were performed and six seeds were used in every replicate. Biomass and root morphology parameters were measure as previously mentioned.
Histochemical GUS Staining
For histochemical analyses, 4-day-old Arabidopsis pCDF3::GUS transgenic plants harboring a 1-kb promoter region fused to the uidA coding sequence (Corrales et al., 2017) were grown in N-free MS-modified basal salt media (M531, Phytotechnology Laboratories), containing 1% (w/v) sucrose, 0.8% (w/v) plant agar, and supplemented with 10 or 0 mM KNO3. GUS staining was performed as described by Jefferson et al. (1987).
Gene Expression Analyses
For qRT-PCR expression analyses, total RNA was isolated from 35S::CDF3, cdf3-1 and WT seedlings that were grown in plates under 10 or 1 mM KNO3 supply as N-non-limiting and limiting conditions for 12 days. In the case of tomato, qRT-PCR expression analyses the total RNA was isolated from leaves of 55-day-old Moneymaker cv. and 35S::CDF3 tomato plants grown in growth chamber conditions under 8 and 4 mM N nitrogen supply. Total RNA was isolated by phenol/chloroform method, following Oñate-Sánchez and Vicente-Carbajosa (2008), and its quality and quantification were assayed using a NanoDrop 2000 (Thermo Scientific). cDNAs were obtained from 2 μg of RNA using oligo(dT)23 primers (Promega) and the Avian Myeloblastosis Virus Reverse Transcriptase (AMV RT; Promega) according to the manufacturer’s instructions. The primers used for PCR amplification in Arabidopsis CDF3 (At3G47500) GLU1 (AT5G04140), GS1.1 (AT5G37600), GS1.4 (AT5G16570), GS2 (AT5G35630), ASN1 (AT3G47340), NIA1 (AT1G77760), NRT2.1 (AT1G08090), NRT2.4 (AT5G60770), NRT2.5 (AT1G12940), PK1 (AT3G08730), PEPC1 (AT3G08730), and tomato NR (Solyc011g01381), GAD2 (Solyc11g011920), and GS2 (Solyc04g014510) genes are described in Supplementary Table S1. UBIQUITIN21 (Czechowski et al., 2005) and UBIQUITIN3 (Hoffman et al., 1991) from A. thaliana and S. lycopersicum, respectively, were used as reference genes. A LightCycler®480 System (Roche) was used for real-time PCR (5 min at 95°C, and 45 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 30 s) using LightCycler®480 SYBR Green I Master (Roche). In order to analyze the melting dynamic of the amplified products, a final dissociation step was added (5 s at 95°C, 1 min at 65°C, continuous 97°C and 30 s at 40°C). Three independent samples were used and each reaction was performed in triplicate. The relative expression levels of target genes were calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001), where ΔCt is the difference in threshold cycle number (Ct) for target gene and references genes.
Amino Acid Quantification
To determine individual amino acids, 12-day-old control plants (Col-0) and the KO-mutant cdf3-1, and two independent 35S::CDF3 lines were grown in agar plates with the medium previously described, supplemented with 1 or 10 mM KNO3 as N limiting and non-limiting conditions, respectively, for 12 days. Extraction, manipulation, and mass spectrometric analysis of samples followed an adapted protocol described in Corrales et al. (2014). Protein content was determined as described by Sarasketa et al. (2014).
Physiological and Metabolic Characterization of Tomato Lines
Net CO2 photosynthetic rate and total plant biomass were determined in WT (Moneymaker cv.) and CDF3 overexpressing (lines L2 and L10) tomato plants after 25 days of differential N supply. Photosynthesis was measured using an LI-6400 infrared gas analyzer (LICOR Biosciences, Lincoln, USA) as described in Renau-Morata et al. (2017). Total soluble sugars, total α-amino acids, and starch content quantifications were performed as described in Monerri et al. (2011). The agronomic performance of the transgenic lines was assessed at the end of the experiment by measuring total yield (g fruits/plant) in the greenhouse as described in Renau-Morata et al. (2017). Total C and N content in reproductive and vegetative organs was measured with an elemental analyzer at the Ionomic service of CEBAS-CSIC (Murcia, Spain).
Enzyme Activity Assays
For Arabidopsis, protein was extracted from 12 day-old seedlings, and the activity of glutamine synthetase (GS), nitrate reductase (NR), and glutamate dehydrogenase (GDH) enzymes were determined as described by Sarasketa et al. (2014). For tomato plants, protein extracts were obtained from leaves of 55-day-old plants. NR activity was determined as described by Calatayud et al. (2008).
Determination of N Accumulation Efficiency and Its Components in Tomato
The N accumulation efficiency (NAE) and its components were calculated in tomato (Moneymaker cv) and CDF3 overexpressing lines according to the method described previously (Weih, 2014; Weih et al., 2018). See Appendix S1 for details.
Results
Expression of CDF3 Gene in Response to N Availability
In a previous work, we identified a set of five DOF TFs from group D (CDFs) in Arabidopsis and tomato that are differentially expressed in vegetative tissues in response to diverse environmental conditions like drought, salinity, or extreme temperatures (Corrales et al., 2014, 2017). In addition, we reported that the overexpression of AtCDF3 and SlCDF3 in both Arabidopsis and tomato, altered metabolism since several amino acids like GABA, proline, glutamine, and asparagine are accumulated among others, therefore suggesting that CDFs might play important functions in the control of N metabolism (Corrales et al., 2017; Renau-Morata et al., 2017). In addition, recent studies point to an involvement of CDF1, the closest CDF3 homolog, in the responses to nitrogen (Varala et al., 2018). Moreover, it is reported that NLP7, a master regulator of the nitrogen signaling pathway, targets multiple TFs including CDF1 (Alvarez et al., 2020), suggesting that CDF1 plays a role in the regulatory network involved in N responses. However, there is still a lack of information regarding the specific roles of CDF1 and CDF3 in N responses. In this context, the objective of this work was to assess the potential involvement of CDF3 in nitrogen signaling and plant responses to nitrogen availability.
To investigate the possible role of CDF3 in nitrate responses, we first evaluated whether its expression is regulated by N availability. We studied the expression patterns of CDF3 under N starvation or after a resupply of NO3−. In N starvation experiments, we performed qRT-PCR expression analyses using RNA isolated from 7-day-old WT plants that were initially grown with 10 mM NO3− and then transferred to a nutrient solution without N. Expression levels were measured at 0, 1, 3, and 6 days. Under these conditions, the expression levels of CDF3 increased showing the highest levels at 1 day after N starvation and decreased over the time (Figure 1A). We further analyzed the expression CDF3 in response to N resupply treatment in a time course experiment. Plants (WT) were grown with 10 mM nitrate for 7 days, and then transferred to N depleted medium for 3 additional days. Afterward, plants were moved again to medium, containing 5 mM KNO3 or 5 mM KCl as a negative control for 0, 20 min, 2.5 h, and 8 h. We used 5 mM KNO3 treatment, because it has been shown by different research groups that this treatment elicit fast and robust responses to NO3− in Arabidopsis (Krouk et al., 2010; Alvarez et al., 2014; Canales et al., 2014). As shown in Figure 1B, transcript levels of CDF3 changed significantly in response to NO3−. The expression of CDF3 was slight and transiently induced by NO3− reaching a maximum level at 20 min, but repressed at longer times (8 h). These results indicate that the expression of CDF3 is regulated by NO3− availability.
Figure 1.
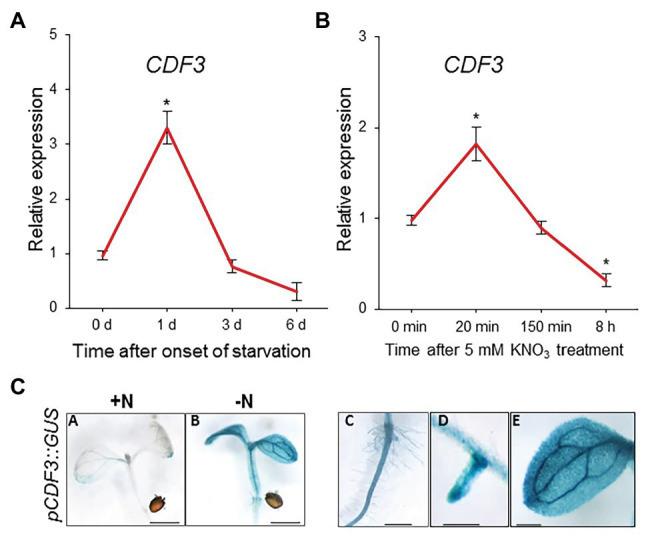
Expression patterns of Arabidopsis CDF3 gene in response to nitrogen availability. qRT-PCR expression analyses of Arabidopsis CDF3 gene in response to nitrogen availability. (A) To analyze the effect of starvation, the total RNA was extracted from plants grown in non-limiting N (10 mM KNO3) and thereafter transferred to 0 or 10 mM KNO3 for the indicated periods of time. Data are normalized to non-limiting N conditions. (B) To analyze the response to nitrate, 7-day-old plants were transferred, after 3 days of starvation, to MS medium supplemented with 5 mM KNO3 or KCl for the indicated periods of time. UBIQUITIN21 gene was used as a reference gene. Data are normalized to KCl conditions. Data are means ± SE (n = 3). Asterisks indicate significant differences from control (p < 0.05); analysis of variance, followed by a Student-Newman-Keuls test (C) Histochemical localization of GUS activity of 7-day-old pCDF3::GUS Arabidopsis plants grown on plates supplemented with 10 mM (+N) or 0 mM KNO3 (−N) for 1 day (A–E). Bars indicate 300 μm. GUS staining of 7-day-old pCDF3::GUS Arabidopsis plants grown on N-depleted (−N) conditions (1d) showing expression of CDF3 in (C) root hair zone in primary root, (D) emerging lateral root, and (E) close-up view of young leafs. Scale bars indicate 200 μm (C,D) and 1 mm (E). Photographs are representative of at least five independent experiments.
In order to perform more detailed analyses of the spatial expression patterns of CDF3 in response to NO3−, we analyzed 7-day-old pCDF3::GUS plants grown under N-non-limiting conditions (10 mM N) and then transferred to a nutrient solution without N (1 day). Figure 1C shows that under N non-limiting conditions GUS staining was mainly detected in the vascular systems of roots, stems, and cotyledons (A). When pCDF3::GUS seedlings were grown under N depleted conditions (B–E), GUS staining was increased in emerged LR, root hairs, stems, and cotyledons, being especially strong in the vascular tissues (C–E). All these results indicate that CDF3 gene is clearly expressed under N starvation conditions in tissues or cells that are involved in plant responses to N availability.
Overexpression of CDF3 Increases Plant Biomass Under Both N Limiting and Non-limiting Conditions
The results of the expression analyses were performed, suggested that CDF3 might play an important role in plant responses to NO3−. To further investigate this possibility, a phenotypic analysis of CDF3 gain- and loss-of-function plants was done by analyzing their growth under both limiting and non-limiting N conditions (Figure 2). We analyzed a previously identified T-DNA insertion mutant cdf3-1 (Fornara et al., 2009; Corrales et al., 2017) and a new mutant allele (cdf3-2; SAIL 434_G09) that we have identified with the T-DNA insertion site located at position 651 from the ATG (Supplementary Figure S1A). The disruption was verified by almost the absence of CDF3 expression (Supplementary Figure S1B). In addition, two CDF3 overexpressor Arabidopsis lines were included in the study (L2.1 and L5.4; Corrales et al., 2017). Plants were grown on MS medium supplemented with a range of N conditions including 1 and 10 mM KNO3, and plant biomass was evaluated after 12 days of growth. Under N limiting conditions (1 mM KNO3), CDF3 overexpressor lines showed better performance compared to WT and cdf3-1, keeping healthy greener leaves and showing higher values of root biomass (Figures 2A,C). In contrast, cdf3-1 and cdf3-2 displayed lower size and values of shoot and root biomass than WT plants on medium containing 1 mM nitrate. On the other hand, under N non-limiting conditions (10 mM KNO3), cdf3-1 and cdf3-2 lines showed similar values of shoot and root biomass and water content (about 81%) than the WT. But, CDF3 overexpressor plants exhibited significant higher values of shoot biomass than WT and cdf3 lines. Consequently, the shoot to root DW weight ratio, an important parameter influenced by nutrient availability (Lawlor et al., 2001), was significantly higher for CDF3-overexpressing plants at 10 mM NO3− conditions but slightly lower at limiting N supply (Figure 2C). However, cdf3-1 and cdf3-2 lines showed similar ratios than the WT at both N limiting and non-limiting conditions. All these data suggest that CDF3 might be involved in the adjustment of root and shoot growth in response to N availability.
Figure 2.
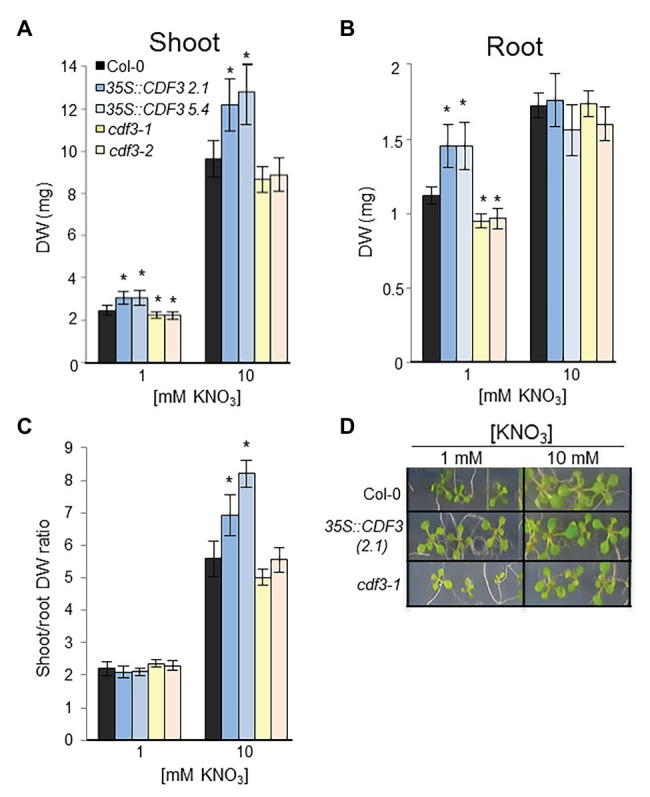
CDF3 effects on Arabidopsis plants growth under different N conditions. Phenotypes and biomass measurements of Col-0, cdf3-1, cdf3-2, and CDF3 overexpressor plants (Lines 2.1 and 5.4) on MS medium containing different concentrations of potassium nitrate as sole nitrogen source. Biomass was measured as dry weight per plant. The photographs and biomass measurements were obtained after 12 days after treatment. (A) Shoot and (B) root dry weight (DW) of 12-day-old plants WT (Col-0), and CDF3 gain- and loss-of-function lines grown under different nitrate conditions (1 and 10 mM KNO3). Values are the mean ± SE of three independent replications each containing 20 plants per genotype. (C) Shoot-root DW ratio. Asterisks indicate significant differences compared with wild-type (Col-0; p < 0.05); analysis of variance Student-Newman-Keuls tests. (D) Representative pictures of the analyzed plants.
CDF3 Impact on Root Morphology
Nitrogen availability modulates gene expression affecting primary and LR growth and development (Little et al., 2005; Remans et al., 2006; Li et al., 2007; Canales et al., 2017). To further investigate the role of CDF3 in root morphology in relation to N availability, we analyzed the root system by estimating primary root (PR) and LR length in gain- and loss-of-function plants grown in 0.1 and 10 mM NO3−. These conditions have been previously shown to promote significant changes in Arabidopsis root morphology in root length assays in vertical plates in short-term experiments (Zhang and Forde, 1998; Varala et al., 2018; Alvarez et al., 2020). As shown in Figure 3A, under 10 mM NO3− no significant differences in the main root length were found between gain- and loss-of-function lines and WT plants. In contrast, under 0.1 mM NO3− supply, 35S::CDF3 plants showed moderate but significant higher values of relative PR length growth than WT, whereas cdf3 plants exhibited lower values of PR relative growth. Notably, cdf3 lines also showed lower values of LR length compared to WT plants (Figures 3B,C). In contrast, CDF3 overexpression lines showed higher values of LR length compared to WT plants. More detailed analysis of the root system showed that both CDF3-overexpressing and cdf3 plants showed no significant differences in LR density compared to the WT under both N limiting and non-limiting conditions, suggesting that CDF3 might play a more important role in PR and LR elongation than in branching (Supplementary Figure S2). Overall, these data indicate that CDF3 is involved in NO3− modulation of root growth and development and overexpression of CDF3 promotes root elongation.
Figure 3.
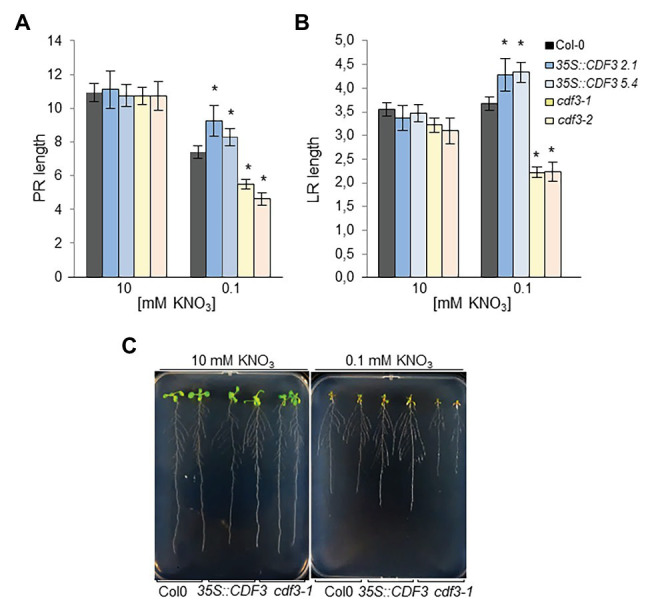
Root morphology of CDF3 overexpressing, cdf3 mutants, and WT plants under different N supply. Plants were grown on vertical plates with 10 or 0.1 mM KNO3 as sole N source, for 12 days. (A) Primary root (PR) length and (B) Lateral root (LR) length (cm) was estimated under different nitrate conditions. Data are means ± SE of three independent experiments with at least 20 plants each. Asterisks indicate significant differences compared with wild-type (Col-0; p < 0.05); analysis of variance, followed by Student-Newman-Keuls tests. (C) Representative pictures of the analyzed plants.
CDF3 Regulates the Expression of Genes Involved in Nitrogen Metabolism
To further gain insight into the molecular mechanisms underlying the observed NO3− responses in CDF3 gain- and loss-of-function lines, we performed detailed expression analyses of selected NO3− responsive genes involved in N assimilation: NIA1 encoding nitrate reductase 1, glutamine synthetase 1.1 (GLN1.1), GLN1.4 and GLN2, asparagine synthetase 1 (ASN1), and glutamate synthase 1 (GLU1). A set of genes encoding nitrate transporters NRT2.1, NRT2.4, and NRT2.5 were also surveyed. We performed qRT-PCR using RNA isolated of 35S::CDF3, cdf3-1 and WT seedlings that were grown under 10 or 1 mM KNO3 supply as N non-limiting and limiting conditions for 12 days. As shown in Figure 4, in N non-limiting conditions, CDF3 overexpressor lines showed higher levels of expression of genes involved in N assimilation: GLU1, GLN1.1, GLN2, ASN1, and NIA1 compared to WT and cdf3-1. Remarkably, we observed that expression levels of all the nitrate transporters analyzed are higher in 35S::CDF3 lines compared with the WT (Figures 4A,B). In contrast, in the case of cdf3-1 mutant, the genes analyzed display different expression patterns in N non-limiting conditions. While GLN1.4, GLU1, NIA1, NRT2.4, and NRT2.5 genes exhibited lower expression levels than the WT, the rest of genes were analyzed and displayed similar levels of expression compared to the WT. Under N limiting conditions, most of the genes analyzed in CDF3 overexpressor plants exhibited higher levels of expression than WT and cdf3-1. In the case of cdf3-1 mutant, the majority of the genes was analyzed and exhibited similar transcript levels to the ones in the WT. However, the transcript levels of GLN2, GLN1.4, NRT2.4, and NRT2.5 genes, which are involved in glutamine biosynthesis, nitrogen remobilization, and nitrate transport, respectively, especially in senescing organs (Masclaux-Daubresse et al., 2010; Kiba et al., 2012; Moison et al., 2018), were lower in cdf3-1 than in control plants. This suggests that CDF3 might be involved not only in NO3− assimilation but also in remobilization and transport in response to N availability.
Figure 4.
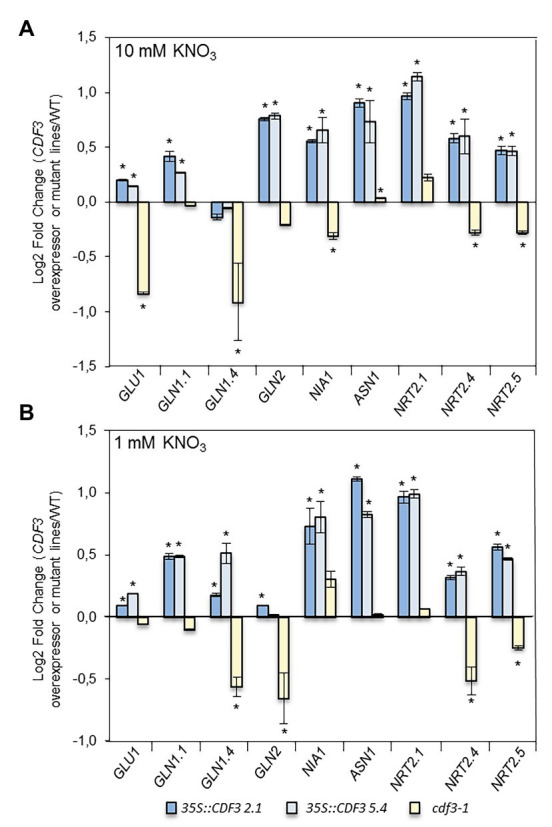
CDF3 regulates a set of genes related to nitrogen assimilation and transport. Expression analyses of N assimilation (GLU1, GLN1.1, GLN1.4, GLN2, ASN1, and NIA1) and nitrate transporter (NRT2.1, NRT2.4, and NRT2.5) genes by qRT-PCR in Col-0, cdf3-1, and 35S::CDF3 (L2.1 and L5.4) transgenic lines. Total RNA was extracted from 12-day-old plants grown on (A) MS medium containing 1 or 10 mM (B) KNO3 as sole nitrogen source. Log2 Fold Change (Log FC) values were generated by comparing the expression of genes at each N treatment of each line vs. the control (Col0) using the 2−ΔΔCt method. Arabidopsis UBIQUITIN21 gene was used as a reference gene. Data are means ± SE (n = 3). Asterisks indicate significant differences compared with Col-0; (p < 0.05) analysis of variance, followed by a Student-Newman-Keuls test.
N assimilation requires both a source of inorganic nitrogen and a carbon skeleton for its incorporation, mainly 2-oxoglutarate (2-OG) that is produced through sequential reactions from photoassimilated carbohydrates. In addition, it has been shown that the maize TF DOF1 plays an important role in nitrogen assimilation (Yanagisawa, 2004) by regulating the expression of two genes encoding key enzymes involved in this process PEPC1 and PK1. To further investigate the role of the Arabidopsis CDF3, we analyzed the expression of PEPC1 and PK1 genes in CDF3 overexpressor, cdf3-1 and WT plants. Under N non-limiting conditions, the expression levels of PK1 and PEPC1 are lower in the cdf3-1 mutant and higher in both CDF3 overexpressor lines compared with the WT control (Figure 5). In contrast, under N limiting conditions the expression of PK1 and PEPC1 are induced in the cdf3-1 mutant line compared to the WT, and in the case of PK1 slightly repressed in the CDF3 overexpressor lines compared to the control. These results indicate that CDF3 can modulate the expression of PK1 and PEPC1 genes involved in the production of C skeletons for amino acid biosynthesis, depending on N availability.
Figure 5.
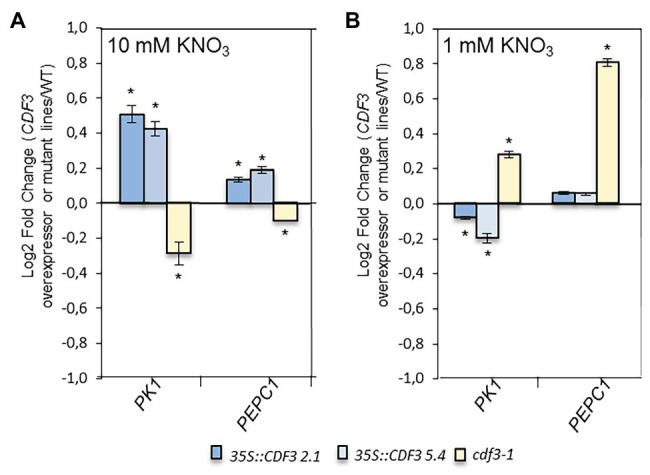
CDF3 regulates genes involved in carbon skeleton formation for N assimilation. Expression analyses by qRT-PCR of PK1 and PEPC1 genes in Col-0, cdf3-1, and 35S::CDF3 (L2.1 and L5.4) transgenic lines. Total RNA was extracted from 12-day-old plants grown on (A) MS medium containing 1 or 10 mM (B) KNO3 as sole nitrogen source. Log2 Fold Change (LogFC) values were generated by comparing the expression of genes at each N treatment of each line vs. the control (Col-0) using the 2−ΔΔCt method. Arabidopsis UBIQUITIN21 gene was used as a reference gene. Data are means ± SE (n = 3). Asterisks indicate significant differences compared with Col-0 (p < 0.05), analysis of variance, followed by a Student-Newman-Keuls test.
Enhanced N Assimilation in CDF3-Overexpressing Arabidopsis Plants
To further investigate the observed differences in growth and gene expression of CDF3 gain- and loss-of-function mutant lines under the different nitrate conditions, we analyzed the levels of major amino acids like glutamine, glutamate, proline, and asparagine, which are well-known markers of N assimilation. The metabolites were evaluated in 12-day-old 35S::CDF3, cdf3-1, and WT plants that were grown under 1 or 10 mM nitrate supply, (N limiting and non-limiting conditions, respectively) as performed in growth and gene expression assays. As shown in Figure 6, under non-limiting N conditions, we found higher values of asparagine, glutamate, glutamine, and proline in CDF3 overexpresor lines than in WT plants, whereas cdf3-1 mutants showed similar values of proline, asparagine, and glutamate. On the other hand, under N limiting conditions, cdf3-1 mutant showed lower levels of glutamate, glutamine, and proline amino acids compared to the WT. In contrast, 35S::CDF3 plants exhibited higher levels of asparagine and proline than control plants, but similar levels of glutamine and glutamate. Consistently, the activity of GS, NR, and GDH enzymes were higher in both CDF3 overexpressor lines compared to the WT (Supplementary Figure S3), but lower in the cdf3-1, which might partly supported the glutamate and glutamine levels in the plants. Total protein content increased in the CDF3-overexpressing plants under 1 mM N but not under 10 mM N condition compared with WT plants, while in the case of cdf3 mutant, the total protein content was significantly lower under N limiting conditions respect to WT plants but no change was observed at 10mM N (Supplementary Figure S4). Altogether, these observations might explain the amino acid contents detected in the different genotypes and suggest that CDF3 plays an important role in nitrate assimilation.
Figure 6.
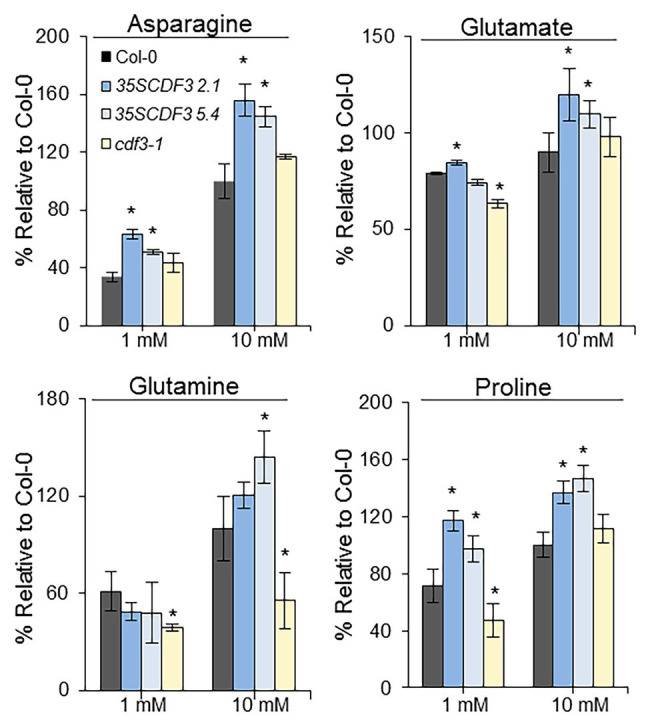
Effect of CDF3 expression on individual amino acid levels. Relative quantities (% relative to wild type) of selected metabolites analyzed by gas chromatography-selected ion monitoring-mass of 12-day-old control plants (Col-0) and cdf3-1, and 35S::CDF3 (lines L2.1 and L5.4) grown in agar plates supplemented with 1 or 10 mM KNO3 (N limiting and non-limiting, respectively). Results are shown as means ± SE (n = 15). Similar results were obtained in five independent experiments; Asterisks indicate significant differences compared with control (p < 0.01); analysis of variance, followed by a Student-Newman-Keuls test.
CDF3 Enhances Biomass Production and Crop Yield in Tomato Under Different N Supply
Since the data obtained in Arabidopsis suggested that CDF3 participates in the control N metabolism, we decided to assess whether CDF3 can be used to improve N assimilation in an economically important crop plant like tomato. For this purpose, phenotypic and physiological analysis of the tomato (Moneymaker) plants overexpressing CDF3 gene (lines L2 and L10) were studied when grown under different N supply. Tomato plantlets at the three-leaf-stage were grown for 25 days under contrasting 8 and 4 mM N supply, which in tomato have been shown as N non-limiting and limiting conditions, respectively (Wahle and Masiunas, 2003). Figure 7 shows that CDF3 overexpressing plants exhibited improved growth with increased fresh weight compared to control plants under different N conditions. In addition, CDF3 overexpressors displayed higher photosynthetic rates and biomass than control plants under both N conditions (Figures 7A–C; Supplementary Table S2). Consistently, CDF3 overexpressing plants exhibited higher total sugar and total free amino acids content in leaves compared to WT (Supplementary Table S2). Moreover, expression analyses of the tomato genes NR, GS2, and glutamate decarboxylase (GAD2) involved in N assimilation and metabolism showed significant higher transcript levels in CDF3 overexpressing plants than in the WT (Supplementary Figure S5). Besides, NR activity was higher in 35S::CDF3 plants than in control plants, both under N non-limiting (22.9 vs. 16.2 μmol NO2/m2s) and limiting (17.6 vs. 13.1 μmol NO2/m2s) conditions. These, results show that the overexpression of CDF3 gene in tomato increases biomass production under both N treatments, and that this higher growth is sustained by a higher N and C assimilation.
Figure 7.
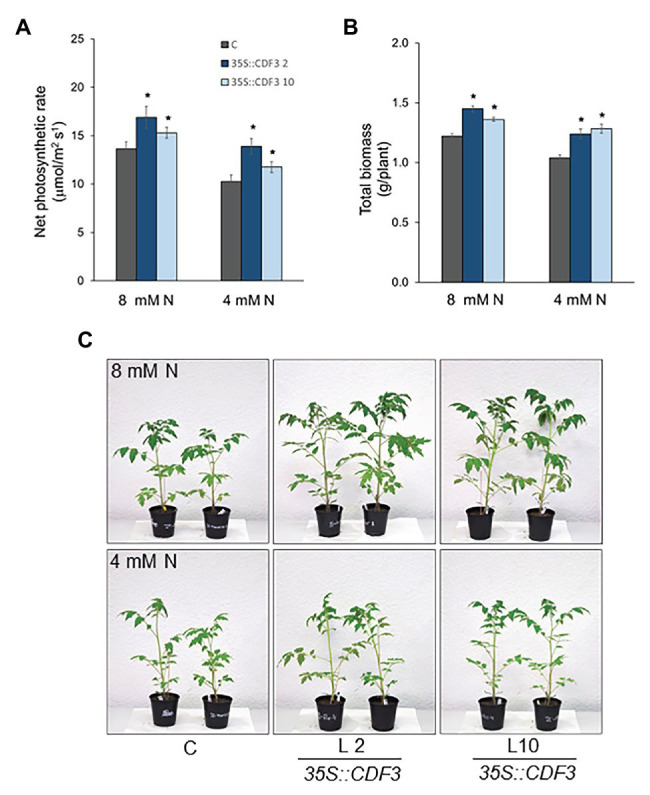
CDF3 overexpression in tomato enhanced photosynthesis and biomass production under different N supply. Net photosynthetic rate (A) and total dry biomass (B) in 35S::CDF3 (L2 and L10; blue bars) and control (C; gray bars) Moneymaker tomato plants grown in hydroponic conditions under N non-limiting (8 mM N) and limiting (4 mM N) nitrogen supply for 25 days. Each value is the mean ± SE of 10 different determinations in different plants. Asterisks indicate significant differences compared with control (p < 0.05), analysis of variance, followed by a Student-Newman-Keuls test. (C) Representative pictures of the plants analyzed.
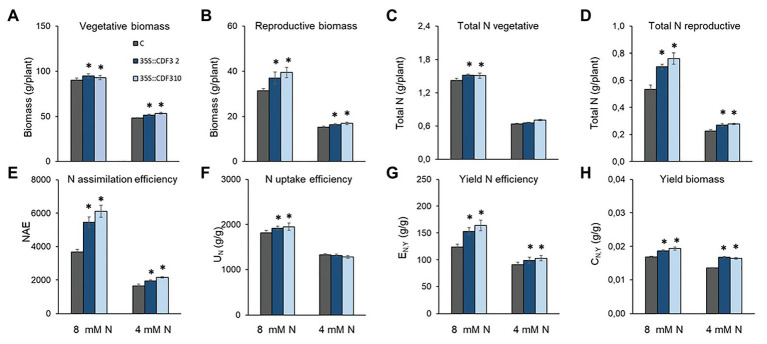
Further, we investigated if the higher biomass production of the 35S::CDF3 tomato plants resulted in increased fruit production by measuring the number of fruits and fruit weight in CDF3 overexpressor plants (L2 and L10 lines) and control plants. To do so, tomato plants were cultured in the greenhouse during 6 months under the different N treatments (8 and 4 mM N). Tomato CDF3 overexpressing plants showed improved fruit yield under both N conditions (Figure 8B; Supplementary Figure S6). Specially, under 8 mM N fertilization, the increase in yield was related to both larger fruit size and number of fruits, whereas under low N conditions resulted from only larger fruit size, since fruit number remained similar between WT and 35S::CDF3 plants (Supplementary Figure S6). Remarkably, 35S::CDF3 plants showed higher values of dry biomass in fruits and vegetative tissues under both N conditions at the end of the culture (Figures 8A,B). The higher values of biomass under high N supply conditions are connected to higher N content in both vegetative and reproductive organs of 35S::CDF3 plants (Figures 8C,D). Under limited N supply, the N content was higher in the fruits, but similar in the vegetative organs of 35S::CDF3 and control plants. Notably, under 8 mM N supply, the values of percentage of total fruit biomass were higher in 35S::CDF3 than control plants (29.8 vs. 25.8% of control fruits, respectively), which might suggest an increased partition to the fruits in this genotype. Overall, these results suggest that CDF3 influenced N assimilation and biomass partition to the fruits in tomato, leading to increased yield both under optimal and limited N supply.
Figure 8.
CDF3 enhances nitrogen accumulation efficiency in tomato under contrasting N supply. Total biomass (g, DW/plant) in vegetative organs (A) and fruits (B); total nitrogen content (g/plant) in vegetative organs (C) and fruits (D) of 35S::CDF3 (lines L2 and L10) and control Moneymaker plants (C). Values are mean (±SE) of 10 different determinations in different 220-day-old plants grown under 4 mM (white bars) and 8 mM N (black bars) conditions. Values of (E) nitrogen accumulation efficiency (NAE) and its components: (F) N uptake efficiency (UN; g g−1), (G) yield-specific N efficiency (EN,y; g g−1) and (H) yield biomass (CN,y; g g−1). Asterisks indicate significant differences compared with control (p < 0.05); analysis of variance, followed by a Student-Newman-Keuls test.
The Overexpression of CDF3 Increased NAE in Tomato
In order to determine the possible impact of CDF3 in NAE, we analyzed CDF3 overexpressor (L2 and L10) and WT tomato plants, using the method proposed by Weih (2014). The results shown in Figure 8 indicated that the reported changes in C and N metabolism of plants overexpressing the CDF3 gene were clearly reflected by corresponding changes in NAE and its components N uptake efficiency (UN), yield-specific N efficiency (EN,y), and fruit N content (CN,y; Appendix S1). Thus, under N non-limiting conditions 35S::CDF3 lines showed higher UN than controls, indicating an increased capacity to take up and/or assimilate N during growth. Furthermore, the EN,y and CN,y were enhanced in CDF3 overexpressing plants compared to the control (Figure 8) and contributed significantly to the increased NAE. The results are consistent with the increased biomass partitioning to the fruits and yield observed in CDF3 overexpressor plants (Figures 8A–D). Under N limiting conditions, NAE was considerably lower compared to non-limiting N treatment in both genotypes, although 35S::CDF3 plants showed higher NAE than control plants due to higher values of EN, Y and CN,Y (Figure 8). Altogether, results indicate that the increased NAE of 35S::CDF3 plants is related to a greater efficiency of converting accumulated N into fruit biomass, allowing an increased partitioning of C and N compounds to these organs in both N treatments and increased N uptake efficiency only under optimum N supply.
Discussion
In recent years, different reports showed that CDF TFs not only participate in the control of plant growth and development but also in the responses to different abiotic stresses (Corrales et al., 2014, 2017; Fornara et al., 2015; Renau-Morata et al., 2017, 2020a). In this study, we have identified that Arabidopsis CDF3 expression is modulated by N availability. The presented data support that CDF3 plays a relevant role in relation to N supply, especially in nitrogen metabolism. CDF3 controls the expression of different nitrate-regulated genes involved in the acquisition, transport, and assimilation of N. Besides, this study also showed that the expression of CDF3 gene in tomato enhances growth under both N-limiting and non-limiting conditions, resulting in higher yield and increased NAE and partition of C and N compounds to the fruits of 35S::CDF3 plants.
New Function of CDF Factors in Nitrogen Responses
Previous works have shown that members of DOF TF family are implicated in N-regulated processes (Yanagisawa, 2004; Rueda-López et al., 2008; Renau-Morata et al., 2017, 2020a,b; Varala et al., 2018) motivating in this work a further comprehensive analysis of CDF3 role in plant response to N availability. Detailed expression analyses by RT-qPCR showed that CDF3 expression is transiently induced after 1 day of N starvation, indicating that it might participate in the assimilation/mobilization pathways. Further analyses of gain- and loss-of-function lines allowed us to confirm the role of CDF3 as an important factor in N-regulated processes. The overexpression of CDF3 in Arabidopsis led to significant higher values of shoot and root biomass under N non-limiting and limiting conditions, respectively, together with a notable rise in the content of major amino acids and total protein and concurrently with an increase in enzyme activities of N metabolism (Figure 6; Supplementary Figures S3, S4). On the contrary, cdf3 KO mutant plants had impaired N use ability and showed more severe N-deficient phenotypes including reduced growth, lower size and shoot, and root biomass reduction compared to WT, specially under N limiting conditions (Figures 2, 3). In sharp contrast to cdf3-1 mutant, the overexpression of CDF3 significantly enhanced the expression of important N-regulated genes involved N assimilation like NIA1, ASN1, GLU, GLN2, GLN1.1, and transport such as NRT2.1, NRT2.4, and NTR2.5, both under N limiting and non-limiting conditions (Figure 4). The increased transcript levels of this group of genes would be related to enhanced N assimilation as revealed in Figure 6 and Supplementary Figures S3, S4. Notably, the transcript levels of GLN1.4, GLN2, NRT2.5, and NRT2.4 were found to be downregulated in the cdf3 mutant under N limiting conditions. Interestingly, these groups of missregulated genes are involved in glutamine biosynthesis and N remobilization and NO3− transport, mainly in senescing organs (Masclaux-Daubresse et al., 2010; Kiba et al., 2012; Moison et al., 2018). Using growth analyses of multiple mutant lines, it was established that NRT2.4 and NRT2.5 are required to support growth of N-starved adult plants by ensuring the efficient uptake of NO3− collectively with NRT2.1 and NRT2.2 and by taking part in NO3− loading into the phloem during N remobilization (Kiba et al., 2012; Lezhneva et al., 2014). All these observations support a multifaceted role of CDF3 in response to N limitation, modulating the expression of genes involved N assimilation and a set of NO3− transporters, likely in different organs, and consequently would lead to enhanced metabolism and probably the transport/uptake of NO3−, to rapidly adapt to N availability and maintain plant N homeostasis. Supporting this hypothesis in silico expression analyses of CDF3 using publicly available expression data (eFP browser),1 indicated that CDF3 showed significant levels of expression in both root and shoot at the seedling stage and in senescing leaves, stems, and seeds in adult plants (Supplementary Figure S8). In Figure 9, a general scheme of the different group of genes regulated by CDF3 in Arabidopsis is proposed. In addition, the ectopic expression of Arabidopsis CDF3 in tomato also promoted similar effects as in Arabidopsis (Figures 7, 8). We confirmed that tomato CDF3 overexpressing plants exhibited improved growth and biomass under different N conditions together with higher levels of total free amino acids and N content compared to WT. In addition, the expression of CDF3 in tomato also promoted enhanced expression of tomato genes involved in N metabolism like NR, GS2 and GAD2 (Supplementary Figure S5). Remarkably, the activity of NR enzyme was higher in both tomato and Arabidopsis CDF3 overexpressing lines compared to WT plants (Supplementary Figure S3). All these results evidence a similar function of CDF3 in tomato and Arabidopsis, suggesting that this might be a conserved mechanism in plants. Overall, these data indicate that overexpression of CDF3 can improve plant N use ability by regulating N metabolism and transport pathways depending on N supply.
Figure 9.
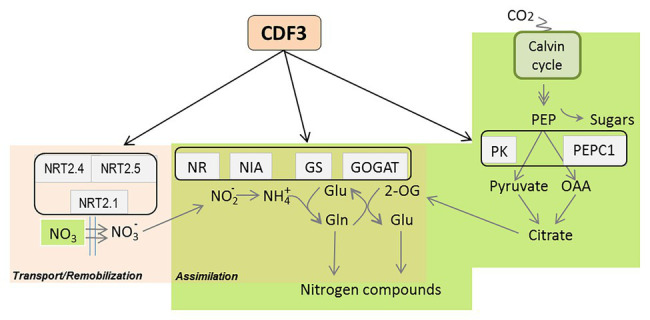
General scheme of CDF3 functions in nitrogen metabolism. CDF3 modulates the expression of the gene-modules involved in nitrogen assimilation and transport. PEP, phosphoenolpyruvate; PK, piruvate kinase; OAA, oxaloacetate; GOGAT, glutamate synthase; GS, glutamine synthetase; NIA/NR, nitrate/nitrite reductases; NRT2, nitrate transporter.
Implication of CDF3 in Root Morphology
Root and root hairs are organs that first sense N availability and display crucial morphological adaptations to N supply (Walch-Liu et al., 2005; Alvarez et al., 2014). Our results provide evidence of a new function for this transcription factor in nitrate regulation of gene expression and root developmental responses in Arabidopsis. CDF3 expression analyses using promoter-GUS fusions showed that CDF3 is expressed in roots and root hairs, and in the perivascular tissues of root/stem and leaves in response to N starvation treatments (Figure 1), implying a possible function of this gene in root development. In fact, our results reveal that in contrast to the cdf3-1 mutant, the overexpression of CDF3 promotes increased LR and PR length under low nitrate conditions in vertical grow assays (Figure 3). The expression of genes involved in root development in response to N availability, such as NRT2.1 (Little et al., 2005), changed significantly in the CDF3-overexpressing and knockout plants (Figure 4). However, we found no significant differences in LR density compared to the WT under both N conditions, which suggest that CDF3 could play a significant role in root elongation (Supplementary Figure S2). Overall, these data might indicate that CDF3 is involved in NO3− modulation of root growth and development and overexpression of CDF3 promotes root development.
The Role of CDF3 in the Supply of C Skeletons for N Assimilation and Plant Growth
Nitrogen assimilation is closely interconnected to C metabolism, and plant growth relies on this interaction. Photosynthesis generates reducing power and C skeletons for the assimilation of nitrogen. It is well-known that PEPC1 and PK are key elements to fuel N assimilation machinery with carbon skeletons (Foyer et al., 2003). On this regard, plants have evolved complex mechanisms to sense and coordinate N assimilation with C metabolism and meet the demands required by growth and development (Stitt and Krapp, 1999; Sato et al., 2011; Sulpice et al., 2013). Previous results showed that maize DOF1 TF play a role in nitrogen assimilation through the control of the expression of PEPC1 and PK genes (Yanagisawa, 2004). In order to investigate the role of the Dof transcription factor CDF3 in the control of the supply of C skeletons for the synthesis of N compounds, we analyzed the expression of PEPC1 and PK1 genes in CDF3 overexpressor, cdf3-1 and WT plants. Interestingly, we found that CDF3 behaves in a dual manner by controlling the expression of both genes depending on the N supply. In fact, we observed that under N non-limiting conditions, CDF3 behaves as a transcriptional activator of both PEPC1 and PK1 genes, facilitating N assimilation by feeding C skeletons and thus increased biomass production. In contrast, it plays a role as a repressor of both genes under N limiting conditions, likely to avoid the metabolic unbalance and accumulation C compounds like 2-OG, which could then not be used in N assimilation. Altogether, our results support an integrative dual regulatory role of CDF3 in C/N balance through the control of both N assimilation and C skeleton production genes depending on nitrogen availability (Figure 9). These results are in agreement with previous studies suggesting that ZmDOF1 also controls C biosynthesis pathway depending on plant N status (Yanagisawa, 2004). However, protein sequence analyses show limited sequence similarity between both proteins, ZmDOF1 and CDF3, except in the DOF DNA binding domain (Supplementary Figure S7), suggesting that CDFs in dicot plant species could play similar functions in C/N metabolism as the ones displayed by DOF1-like genes in monocot species.
CDF3 Overexpression Enhances Tomato Biomass Production and N Accumulation Efficiency
The results obtained in this work support an important role of Arabidopsis CDF3 in N assimilation during plant growth and in the response to changes in N availability. This led us to explore whether the functionality and effects observed in Arabidopsis are conserved in other plant species and it could be used to improve N use efficiency in a crop plant like tomato. We confirmed that 35S::CDF3 tomato plants exhibited higher CO2 fixation capacity and sugar content under optimal N fertilization. In addition, we showed that the overexpression of CDF3 increased N assimilation, as inferred from the higher total N and amino acid contents (Figures 6, 8; Supplementary Table S2). Consistently, the CDF3 overexpressing tomato plants showed increased biomass production, and notably, increased fruit yield and NAE.
In general, NUE can be divided into the components of N acquisition, assimilation, distribution, and utilization (Plett et al., 2017). The methodology used in this work (Weih, 2014; Weih et al., 2018) allowed a robust and reliable determination of the equivalent NAE parameter and the components related to the N acquisition and assimilation (N uptake efficiency), and utilization and distribution (yield-specific N efficiency and fruit yield N concentration). The higher overall N accumulation efficiency of the plants overexpressing CDF3 gene under optimal N supply was due to increases in all three NAE components. Our data indicate that CDF3 improves N use efficiency under optimal N supply by enhancing the amounts of photoassimilates partitioned to the fruits, resulting from increased C and N metabolism, transport and sink strength of the tomato fruits. Moreover, we observed that under limited N condition, photosynthetic rate, biomass accumulation, and fruit yield were also enhanced in the 35S::CDF3 plants compared to controls (Figures 7, 8). Although a reduction was provoked by the limitation in N supply in control and 35S::CDF3 plants, photosynthetic rates, biomass accumulation, and yield were higher in the transgenic plants. Interestingly, under N limited supply, the N uptake efficiency did not differ between both genotypes. However, 35S::CDF3 plants maintained higher C/N supply to the fruits and in accordance higher yield under N limiting supply. These data support a key role of CDF3 in the regulation of photoassimilate partitioning to the fruits under both optimal and limiting N supply.
In conclusion, the present work, we identified CDF3 as a new important regulatory factor of the nitrogen responses in Arabidopsis and tomato, linking key aspects of C and N metabolism, root development, and plant growth. Altogether, our results highlight CDF3 as a new potential target to improve crop production in the context of sustainable agriculture that aims to improve crop production, while at the same time being environmentally sustainable, and ensuring healthy and productive soils for the future.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
R-VM, SN, JV-C, and JM designed, planned, and organized the experiments. JD-F, LC, BR-M, LY, MW, and DM performed the research. R-VM, SN, JC, DM, and JM wrote the article. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mar Gonzalez for technical assistance.
Funding. This work was supported by grants from the National Institute for Agriculture and Food Research and Technology (INIA; RTA2015-00014-c02-01 to JM) as well as from the National Commission for Scientific and Technological Research (CONICYT; REDI170024 to JM, JC), ANID – Millennium Science Initiative Program – ICN17_022 to JC, Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 1190812 and 1170926 to JC), Basque and Spanish Government (IT-932-16 and BIO2017-84035-R to DM), UE Prima (PCI2019-103610 to JM) MINECO-Spain (BIO2017-82873-R to JV-C) and INIA pre-doctoral fellowship to JD-F. We also want to acknowledge the “Severo Ochoa Program for Centers of Excellence in R&D” from the Agencia Estatal de Investigación of Spain (grant SEV-2016-0672) for supporting the scientific services used in this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.601558/full#supplementary-material
References
- Alvarez J. M., Moyano T. C., Zhang T., Gras D. E., Herrera F. J., Araus V., et al. (2019). Local changes in chromatin 648 accessibility and transcriptional networks underlying the nitrate response in Arabidopsis 649 roots. Mol. Plant 12, 1545–1560. 10.1016/j.molp.2019.09.002, PMID: [DOI] [PubMed] [Google Scholar]
- Alvarez J. M., Riveras E., Vidal E. A. (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80, 1–13. 10.1111/tpj.12618, PMID: [DOI] [PubMed] [Google Scholar]
- Alvarez J. M., Schinke A. L., Brooks M. D., Pasquino A., Leonelli L., Varala K., et al. (2020). Transient genome-wide interactions of the master transcription factor NLP7 initiate a rapid nitrogen-response cascade. Nat. Commun. 11:1157. 10.1038/s41467-020-14979-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calatayud A., Gorbe E., Roca D., Martinez P. F. (2008). Effect of two nutrient solution temperatures on nitrate uptake, nitrate reductase activity, NH4+concentration and chlorophyll a fluorescence in rose plants. Environ. Exp. Bot. 64, 65–74. 10.1016/j.envexpbot.2008.02.003 [DOI] [Google Scholar]
- Canales J., Contreras-Lopez O., Alvarez J. M., Gutierrez R. A. (2017). Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. Plant J. 92, 305–316. 10.1111/tpj.13656, PMID: [DOI] [PubMed] [Google Scholar]
- Canales J., Moyano T. C., Villarroel E., Gutiérrez R. A. (2014). Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front. Plant Sci. 5:22. 10.3389/fpls.2014.00022, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., et al. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57, 426–435. 10.1111/j.1365-313X.2008.03695.x, PMID: [DOI] [PubMed] [Google Scholar]
- Corrales A. R., Carrillo L., Lasierra P., Nebauer S. G., Domínguez-Figueroa J., Renau-Morata B., et al. (2017). Multifaceted role of cycling Dof Factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 40, 748–764. 10.1111/pce.12894, PMID: [DOI] [PubMed] [Google Scholar]
- Corrales A. R., Nebauer S. G., Carrillo L., Fernández-Nohales P., Marqués J., Renau-Morata B., et al. (2014). Characterization of tomato cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65, 995–1012. 10.1093/jxb/ert451, PMID: [DOI] [PubMed] [Google Scholar]
- Crawford N. M., Forde B. G. (2002). Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1:e0011. 10.1199/tab.0011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. 10.1104/pp.105.063743, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Montaigu A., Sánchez-Villarreal A., Takahashi Y., van Termaat E. V. L., Huettel B., et al. (2015). The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 81, 695–706. 10.1111/tpj.12759, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K. C. S., Gissot L., Sauerbrunn N., Rühl M., Jarilo J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. 10.1016/j.devcel.2009.06.015, PMID: [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Parry M., Noctor G. (2003). Markers and signals associated with nitrogen assimilation in higher plants. J. Exp. Bot. 54, 585–593. 10.1093/jxb/erg053, PMID: [DOI] [PubMed] [Google Scholar]
- Guan P., Wang R., Nacry P., Breton G., Kay S. A., Pruneda-Paz J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 111, 15267–15272. 10.1073/pnas.1411375111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R. A., Lejay L. V., Dean A., Chiaromonte F., Shasha D. E., Coruzzi G. M. (2007). Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 8:R7. 10.1186/gb-2007-8-1-r7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D. R., Arnon D. I. (1950). The water-culture method for growing plants without soil. Vol. 347. Berkeley, Calif: University of California, College of Agriculture, Agricultural Experiment Station. [Google Scholar]
- Hoffman N. E., Ko K., Milkowski D., Pichersky E. (1991). Isolation and characterization of tomato cDNA and genomic clones encoding the ubiquitin gene ubi3. Plant Mol. Biol. 17, 1189–1201. 10.1007/BF00028735, PMID: [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T. F., Harmon F. G., Ho L. A., Kay S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. 10.1126/science.1110586, PMID: [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta- glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. 10.1002/j.1460-2075.1987.tb02730.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Feria-Bourrellier A. B., Lafouge F., Lezhneva L., Boutet-Mercey S., Orsel M., et al. (2012). The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24, 245–258. 10.1105/tpc.111.092221, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A., David L. C., Chardin C., Girin T., Marmagne A., Leprince A. S., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65, 789–798. 10.1093/jxb/eru001, PMID: [DOI] [PubMed] [Google Scholar]
- Krouk G., Mirowski P., Lecun Y., Shasha D. E., Coruzzi G. M. (2010). Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11:R123. 10.1186/gb-2010-11-12-r123, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurai T., Wakayama M., Abiko T., Yanagisawa S., Aoki N., Ohsugi R. (2011). Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 9, 826–837. 10.1111/j.1467-7652.2011.00592.x, PMID: [DOI] [PubMed] [Google Scholar]
- Lawlor D. W., Lemaire G., Gastal F. (2001). “Nitrogen, plant growth and crop yield” in Plant nitrogen. eds. Lea P. J., Morot-Gaudry J. F. (Berlin, Heidelberg: Springer-Verlag; ), 343–367. [Google Scholar]
- Lezhneva L., Kiba T., Feria-Bourrellier A. B., Lafouge F., Boutet-Mercey S., Zoufan P., et al. (2014). The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80, 230–241. 10.1111/tpj.12626, PMID: [DOI] [PubMed] [Google Scholar]
- Li W., Wang Y., Okamoto M., Crawford N. M., Siddiqi M. Y., Glass A. D. M. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433. 10.1104/pp.106.091223, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little Y. D., Rao H., Oliva S., Daniel-Vedele F., Krapp A., Malamy J. E. (2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. U. S. A. 102, 13693–13698. 10.1073/pnas.0504219102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262, PMID: [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Daniel-Vedele F., Dechorgnat J., Chardon F., Gaufichon L., Suzuki A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. 10.1093/aob/mcq028, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moison M., Marmagne A., Dinant S., Soulay F., Azzopardi M., Lothier J., et al. (2018). Three cytosolic glutamine synthetase isoforms localized in different-order veins act together for N remobilization and seed filling in Arabidopsis. J. Exp. Bot. 69, 4379–4393. 10.1093/jxb/ery217, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monerri C., Fortunato-Almeida A., Molina R. V., Nebauer S. G., García-Luis A., Guardiola J. L. (2011). Relation of carbohydrate reserves with the forthcoming crop, flower formation and photosynthetic rate, in the alternate bearing “Salustiana” sweet orange (Citrus sinensis L.). Sci. Hortic. 129, 71–78. 10.1016/j.scienta.2011.03.009 [DOI] [Google Scholar]
- Noguero M., Atif R. M., Ochatt S., Thompson R. D. (2013). The role of the DNA- binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. 10.1016/j.plantsci.2013.03.016, PMID: [DOI] [PubMed] [Google Scholar]
- Obertello M., Krouk G., Katari M. S., Runko S. J., Coruzzi G. M. (2010). Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Syst. Biol. 4:111. 10.1186/1752-0509-4-111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L., Vicente-Carbajosa J. (2008). DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC. Res. Notes 1:93. 10.1186/1756-0500-1-93, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña P. A., Quach T., Sato S., Ge Z., Nersesian N., Changa T., et al. (2017). Expression of the maize Dof1 transcription factor in wheat and sorghum. Front. Plant Sci. 8:434. 10.3389/fpls.2017.00434, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett D., Garnett T., Okamoto M. (2017). “Molecular genetics to discover and improve nitrogen use efficiency in crop plants” in Plant macronutrient use efficiency. eds. Anwar M., Kamiya T., Burrit D. J., Phan L. -S., Fujiwara T. (London, UK: Elsevier; ), 93–122. [Google Scholar]
- Remans T., Nacry P., Pervent M., Girin T., Tillard P., Lepetit M., et al. (2006). A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 140, 909–921. 10.1104/pp.105.075721, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renau-Morata B., Carrillo L., Cebolla-Cornejo J., Molina R. V., Marti R., Dominguez-Figueroa J., et al. (2020b). The targeted overexpression of SlCDF4 in the fruit enhances tomato size and yield involving gibberellin signalling. Sci. Rep. 10:10645. 10.1038/s41598-020-67537-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renau-Morata B., Carrillo L., Dominguez-Figueroa J., Vicente-Carbajosa J., Molina R. V., Nebauer S. G., et al. (2020a). CDF transcription factors: plant regulators to deal with extreme environmental conditions. J. Exp. Bot. 71, 3803–3815. 10.1093/jxb/eraa088, PMID: [DOI] [PubMed] [Google Scholar]
- Renau-Morata B., Molina R. V., Carrillo L., Cebolla-Cornejo J., Sánchez-Perales M., Pollmann S., et al. (2017). Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front. Plant Sci. 8:660. 10.3389/fpls.2017.00660, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W. R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584. 10.1105/tpc.109.067041, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda-López M., Crespillo R., Cánovas F. M., Avila C. (2008). Differential regulation of two glutamine synthetase genes by a single Dof transcription factor. Plant J. 56, 73–85. 10.1111/j.1365-313X.2008.03573.x, PMID: [DOI] [PubMed] [Google Scholar]
- Rueda-Lopez M., Pascual M. B., Pallero M., Henao L. M., Lasa B., Jauregui I., et al. (2017). Overexpression of a pine Dof transcription factor in hybrid poplars: a comparative study in trees growing under controlled and natural conditions. PLoS One 12:e0174748. 10.1371/journal.pone.0174748, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasketa A., González-Moro M. B., González-Murua C., Marino D. (2014). Exploring ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. J. Exp. Bot. 65, 6023–6033. 10.1093/jxb/eru342, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Maekawa S., Yasuda S., Domeki Y., Sueyoshi K., Fujiwara M., et al. (2011). Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J. 68, 137–146. 10.1111/j.1365-313X.2011.04673.x, PMID: [DOI] [PubMed] [Google Scholar]
- Scheible W. R., Lauerer M., Schulze E. D., Caboche M., Stitt M. (1997). Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J. 11, 671–691. 10.1046/j.1365-313X.1997.11040671.x [DOI] [Google Scholar]
- Stitt M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2, 178–186. 10.1016/S1369-5266(99)80033-8, PMID: [DOI] [PubMed] [Google Scholar]
- Stitt M., Krapp A. (1999). The molecular physiological basis for the interaction between elevated carbon dioxide and nutrients. Plant Cell Environ. 22:58. [Google Scholar]
- Sulpice R., Nikoloski Z., Tschoep H., Antonio C., Kleessen S., Larhlimi A., et al. (2013). Impact of the carbon and nitrogen supply on relationships and connectivity between metabolism and biomass in a broad panel of Arabidopsis accessions. Plant Physiol. 162, 347–363. 10.1104/pp.112.210104, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varala K., Marshall-Colón A., Cirrone J., Brooks M. D., Pasquino A. V., Léran S., et al. (2018). Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. U. S. A. 115, 6494–6499. 10.1073/pnas.1721487115, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E. A., Gutierrez R. A. (2008). A system view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 11, 521–529. 10.1016/j.pbi.2008.07.003, PMID: [DOI] [PubMed] [Google Scholar]
- Vidal E. A., Moyano T. C., Canales J., Gutiérrez R. A. (2014). Nitrogen control of developmental phase transitions in Arabidopsis thaliana. J. Exp. Bot. 65, 5611–5656. 10.1093/jxb/eru326, PMID: [DOI] [PubMed] [Google Scholar]
- Wahle E. A., Masiunas J. B. (2003). Population density and nitrogen fertility effects on tomato growth and yield. HortScience 38, 367–372. 10.21273/HORTSCI.38.3.367 [DOI] [Google Scholar]
- Walch-Liu P., Filleur S., Gan Y., Forde B. G. (2005). Signaling mechanisms integrating root and shoot responses to changes in the nitrogen supply. Photosynth. Res. 83, 239–250. 10.1007/s11120-004-2080-9, PMID: [DOI] [PubMed] [Google Scholar]
- Weih M. (2014). A calculation tool for analyzing nitrogen use efficiency in annual and perennial crops. Agronomy 4, 470–477. 10.3390/agronomy4040470 [DOI] [Google Scholar]
- Weih M., Hamnér K., Pourazari F. (2018). Analyzing plant nutrient uptake and utilization efficiencies: comparison between crops and approaches. Plant Soil 430, 7–21. 10.1007/s11104-018-3738-y [DOI] [Google Scholar]
- Yan D., Easwaran V., Chau V., Okamoto M., Lerullo M., Kimura M., et al. (2016). NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 7:13179. 10.1038/ncomms13179, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560. 10.1016/S1360-1385(02)02362-2, PMID: [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2004). Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 45, 386–391. 10.1093/pcp/pch055, PMID: [DOI] [PubMed] [Google Scholar]
- Yanagisawa S., Schmidt R. J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214. 10.1046/j.1365-313X.1999.00363.x, PMID: [DOI] [PubMed] [Google Scholar]
- Yanagisawa S., Sheen J. (1998). Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10, 75–89. 10.1105/tpc.10.1.75, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Forde B. G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. 10.1126/science.279.5349.407, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.