Abstract
In this review, we outline the growing role that molecular dynamics simulation is able to play as a design tool in drug delivery. We cover both the pharmaceutical and computational backgrounds, in a pedagogical fashion, as this review is designed to be equally accessible to pharmaceutical researchers interested in what this new computational tool is capable of and experts in molecular modeling who wish to pursue pharmaceutical applications as a context for their research. The field has become too broad for us to concisely describe all work that has been carried out; many comprehensive reviews on subtopics of this area are cited. We discuss the insight molecular dynamics modeling has provided in dissolution and solubility, however, the majority of the discussion is focused on nanomedicine: the development of nanoscale drug delivery vehicles. Here we focus on three areas where molecular dynamics modeling has had a particularly strong impact: (1) behavior in the bloodstream and protective polymer corona, (2) Drug loading and controlled release, and (3) Nanoparticle interaction with both model and biological membranes. We conclude with some thoughts on the role that molecular dynamics simulation can grow to play in the development of new drug delivery systems.
Keywords: pharmaceutics, nanomedicine, molecular dynamics, drug delivery, nanoparticle
Introduction
The exponential advance of the computational power available to us has led to related approaches attaining a prominent, one can argue now dominant, position within pharmaceutical research. The majority of this toolkit, as we will elaborate below, are methodologies that fit experimental data to a mathematical model that provides a numerical answer, for example a specific drug molecule structure or delivery system formulation. A subset of computational methodologies provide something further: mechanistic understanding; in place of just an answer, i.e., an optimum value or set of values, mechanistic understanding means an elucidation of what is actually occurring in the system that produces the results: in simple terms, a model of the system, expressed as a cartoon in three dimensions, of what is happening. Such an output, often referred to as a simulation, has power far beyond that provided by a mere result of what is optimal for the specific application sought; it can lead to an informed design process that is more efficient, allows for broader intuitive leaps from its interpretation and provides insight that transcends the specific application studied.
An extremely successful computational scheme for attaining mechanistic understanding is molecular dynamics simulation (MD), a methodology that models the system as a set of particles that interact through classical mechanics. An intuitive choice for these particles, particularly for those with a background in chemistry, is for them to represent atoms, with interactions between the atoms producing the intramolecular forces that govern the structure of molecules and the intermolecular forces that govern interactions between molecules. This is, however, not the only choice that can be made, as particles can be chosen to represent larger structures than single atoms; they can represent groups of atoms, whole molecules, or even groups of molecules. Such models can obtain insight into the system on a larger length and time scale than can be achieved through a model with atomistic resolution and are known as coarse grained (Ingólfsson et al., 2014).
In this review we will highlight the growing role that MD has played and will continue to play in drug delivery, what has been referred to as computational pharmaceutics by Ouyang and Smith (2015), using computational methods to address issues related to drug delivery including dissolution, solubility, protection from the bodies defense mechanisms, controlled release and targeted delivery. The development of advanced mechanisms for drug delivery based on nanoscale drug delivery vehicles, a field known as nanomedicine (Riehemann et al., 2009; Lammers and Ferrari, 2020; Moghimi et al., 2020), is a particular area where MD methods have borne fruit. This review paper has two target audiences: (1) pharmaceutical researchers, intrigued by the rapid rise of computational methods applicable to their research, who are interested in learning what kind of insight MD can provide and (2) computational physicists and chemists, with a background in MD methods, atomistic and coarse grained, who, for reasons I most probably do not need to inform the reader of, realize that at this point in history pharmaceutical applications are an extremely desirable context for their research. Both of the target audiences will find certain elements of this review to be trivially basic and may even bristle at some oversimplifications; one should keep in mind the dual audience focused nature of this review. As the subject matter is extremely broad, with several areas covered by comprehensive reviews themselves, this publication can, to some extent, be seen as a meta-review, to be used as an initial jumping off point leading to many further review papers, in addition to original work.
At its core, pharmaceutical science is roughly (1) the search for small molecules that, over the scale of the entire organism, do more good than harm under certain conditions: drug design and (2) development of the means by which these molecules can enter the body and reach their target tissue intact: drug delivery or pharmaceutics. Pharmaceutical science begins with Paracelsus, the man who is to pharmacy what Isaac Newton is to physics and his maxim “the dose makes the poison” (Rozman and Doull, 2001); substances exist that, at too high a dose are a poison that will kill you, but when taken at a certain dose can actually help you. The substance enters the body, dissolution occurs and the drug molecules within the substance are freed and diffuse through the body and enough reaches, intact, the desired location in sufficient quantity to induce the desired effect. Any drug molecule will reach other parts of the body and have different effects which are undesirable: the toxicity, i.e., side effects, of the drug. The conventional drug design paradigm is thus a balancing act between efficacy, toxicity and solubility. A very efficacious drug can be found that either has intolerable toxicity or too poor solubility to be carried through the bloodstream or, due to the nature of the target tissue, insufficient quantities of the drug reach it to have the desired effect.
Initially drugs were found through trial and error, however, the search space is gigantic: the number of different small organic molecules that are theoretically possible to synthesize is ∼1063 (Bohacek et al., 1996; Hoffmann and Gastreich, 2019) a number that dwarfs such quantities as Avogadro’s number and the number of stars in the universe; drug design can be seen as searching this discrete “drug structure space.” The latter half of the twentieth century saw the onset of a systematic approach to searching this space based on the “lock and key” paradigm: drug molecules were designed to fit a certain active site on a certain protein to either inhibit or activate them. This was propelled by advances in three areas (1) robotics to enable massive simultaneous parallel screening experiments, (2) increasing numbers of high resolution protein structures, determined first through X-ray crystallography, but now increasingly through cryo-EM, and (3) the computational power and advanced algorithms to analyze the massive data sets produced. The computational component of this, computational drug design, can be divided into two methodologies: (1) ligand-based (Acharya et al., 2011) where the target protein structure is not known and (2) structure based (Alonso et al., 2006; Sousa et al., 2006; Sliwakosky et al., 2014; Ferreira et al., 2015), where the binding free energy of potential drug molecules is calculated, using the experimentally determined high resolution protein structure, a calculation known as “ligand docking and scoring.” Ligand based methods use pattern recognition, now trendily referred to as “machine learning,” algorithms where elements of the structural properties are mapped to either (1) high throughput screening results for activity, i.e., efficacy and other desirable properties, e.g., solubility parameters: Quantitative Structure Activity Relationship/Quantitative Structure Property Relationship (QSAR/QSPR) (Liu and Long, 2009; Nantasenamat et al., 2009; Ghasemi et al., 2018; Toporov and Toporova, 2020) or (2) elements of three dimensional structure of the molecule: pharmacophore modeling (Acharya et al., 2011).
Apart from the pharmacological research to determine appropriate target protein active sites, the above mentioned methodologies for drug discovery together are a fixed, simplified, purely empirical, paradigm: fitting data without insight. As is the case with research carried out using a fixed paradigm, metaphorically speaking continuing to turn the crank on the same machine, one reaches a point of diminishing returns; this is exactly what has occurred for the case of pharmaceutical research: as the resources spent globally on pharmaceutical research increase exponentially, the number of new drugs approved each year remains constant, a phenomenon referred to as “Eroom’s law” (Scanell et al., 2012) the reverse of the famous Moore’s law regarding the exponential increase in computational technology we have witnessed over the past half century: pharmaceutical research is slowing down exponentially; moving forward requires moving beyond this oversimplified model.
The situation for drug delivery, i.e., pharmaceutics, is similar. When a given molecule is designed, using the above methodology, a set of rules of thumb are applied regarding its properties, known as “Lipinski’s rule of 5” (Lipinski et al., 2001; Lipinski, 2004). This determines whether the molecule is “drug-like,” i.e., a molecular structure likely to have a sufficiently optimal solubility profile, or not. Behavior of the drug in the body apart from its drug action, known as its Absorption, Distribution, Metabolism, and Excretion (ADME) properties, is a critical aspect that partially determines both efficacy and toxicity. This is modeled using numerical solutions to complex sets of coupled differential equations that represent the interactions of drugs and drug metabolites, as their distribution varies in time in the different tissues of the organism; this form of numerical computational modeling is known as pharmacokinetic/pharmacodynamics modeling (Craig, 1998; Ruiz-Garcia et al., 2008; Belfo and Lemos, 2021). While this form of modeling is not entirely empirical, as it is dependent on known metabolic relations, it still remains a method to calculate a quantitative result from experimentally measured parameters.
Given that the global pharmaceutical industry is estimated to have a turnover in excess of 1 trillion USD, there is obviously a substantial continuing effort to break out of the rut of diminishing returns it finds itself in. Regarding pharmaceutics, the last 30 years has seen the development of increasingly sophisticated mechanisms for enhancing the solubility profile, carrying/protecting drugs in the bloodstream and targeting them to the desired tissue (Zhang W. et al., 2017): the aforementioned nanomedicine (Moghimi et al., 2020). These involve either covalently bonding the drug to a molecule or packaging the drug into a nanoscale (diameter 100nm or less) vehicle that performs this function. As this field has developed, these means have become increasingly complex and intricate and, as a result, this avenue has also become stuck (Park, 2016): while increasingly complex devices make for engaging narratives leading to well cited publications, the greater the complexity the more that can go wrong, resulting in a field of research that is far better at producing publications than real approved therapies; as Venditto and Szoka have put it “so many papers and so few drugs!” (Venditto and Szoka, 2013); the resulting system, coupled to the human physiological environment, is far too complex to be developed through the above described limited, mostly empirical, paradigm.
It can be argued that what is missing is mechanistic understanding: insight into what is actually physically happening, i.e., what are the molecules actually doing? The above described computational methods do not provide this; what they provide is a numerical answer. Mechanistic understanding is obtained by a computational method that can, given knowledge of the structure of molecules, provide insight into how the molecules interact, i.e., what structures they form and how they move with respect to each other with time: a three dimensional movie of what is happening on the molecular length scale. A molecule, or system of molecules, is a set of nuclei and electrons interacting in a specific way. How this interaction affects the motion of the atoms, i.e., the physics of the system, is quantum mechanics. Exact calculation is impossible, however, the discipline of theoretical quantum chemistry has developed many methods for approximating the behavior of molecules governed by quantum mechanics (Cramer, 2002). While these calculations can be simplified through the use of semi-empirical methods (Thiel, 2014), we are still left with a calculation that is too computationally intensive to simulate the length and time scales that are of interest to us. Making a set of approximations and accepting certain limitations of the variety of phenomena that can be observed, we arrive at the molecular mechanics paradigm: the molecule modeled as a set of particles with their interactions governed by classical mechanics.
The Molecular Mechanics Paradigm and Molecular Dynamics Simulation
The molecular mechanics paradigm is based on a combination of insight from the quantum mechanical interactions of atoms and empirical physical chemistry. The resulting model, illustrated in Figure 1, can be intuitively pictured as a set of sticky rubber balls (the short range attractive van der Waals (Israelachvili, 1985) and repulsive Pauli exclusion forces modeled through what is known as the Lennard-Jones potential term) that are charged (electronegativity of atoms and H-bonding behavior modeled through partial charges) connected by springs (the bond forces) with hinges (angular interactions), axels (proper dihedral potentials) and other 4-body interactions to produce correct structure (improper dihedral potentials); the atoms and molecules follow Newton’s equations of motions, knocking into each other and rattling about in response to these forces; the result is a three dimensional movie of the system with atomistic resolution: molecular dynamics simulation (Allen and Tildesley, 1989; Frenkel and Smit, 2001). This has been referred to as a “computational microscope” by Lee E. H. et al. (2009), however, we feel this analogy is misleading as this is not a visualization of a piece of a real system but rather the isolation and study of a specific aspect of the system that we have assembled the appropriate set of models of molecules to study. Discussion of the methods used to determine the parameters of this model can be found elsewhere (Plimpton, 1995; Karplus and McCammon, 2002; Case et al., 2005; Phillips et al., 2005; Hess et al., 2008; van Gunsteren et al., 2008; Brooks et al., 2009; Abraham et al., 2015).
FIGURE 1.
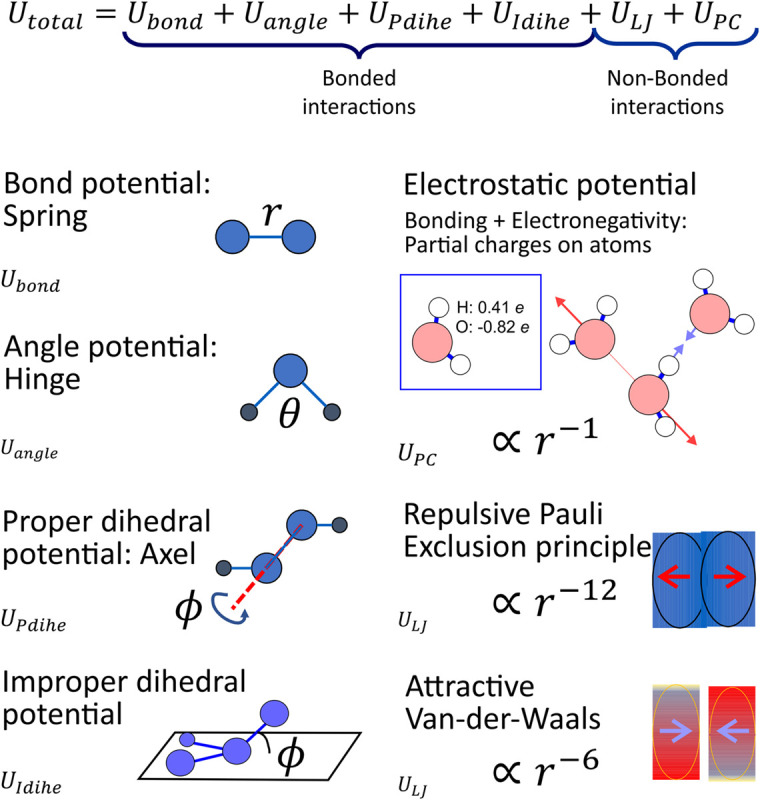
The set of interactions that are the molecular mechanics paradigm that defines the forces that drive the motion of atoms in a molecular dynamics simulation with all atom resolution. The bond, angle and dihedral potentials are the intramolecular interactions that define molecule structure and the interactions between covalently bound atoms. Each atom has a partial charge that interacts with all other atoms through electrostatic forces to model electronegativity and H-bonding behavior; short range attractive and repulsive forces due to the Van-der-Waals and Pauli Exclusion Principle, respectively, are modeled through the Lennard-Jones interaction.
Several competing potential sets exist and for simulating any system with new molecules that have never been simulated before, often the case in pharmaceutical as opposed to biological research since we deal with unique man-made molecules, quantum chemistry calculations must be performed; choosing and building potential sets for the model to obtain the correct result requires significant expertise. While molecular dynamics simulation with an all atom model has seen significant success in the study of a wide range of biophysical systems, it is still limited to a length scales of ∼15 nm and time scales of ∼1–2 μs: too small to obtain insight into several phenomena we wish to study. Here we return to the aforementioned coarse grained models (Figure 2). While several phenomena cannot be observed as they are dependent on specific interatomic interactions, e.g., salt bridges and H-bonds, with good judgment such models allow for the metaphorical camera to zoom out and study behavior on larger length and timescales, however, with reduced resolution.
FIGURE 2.
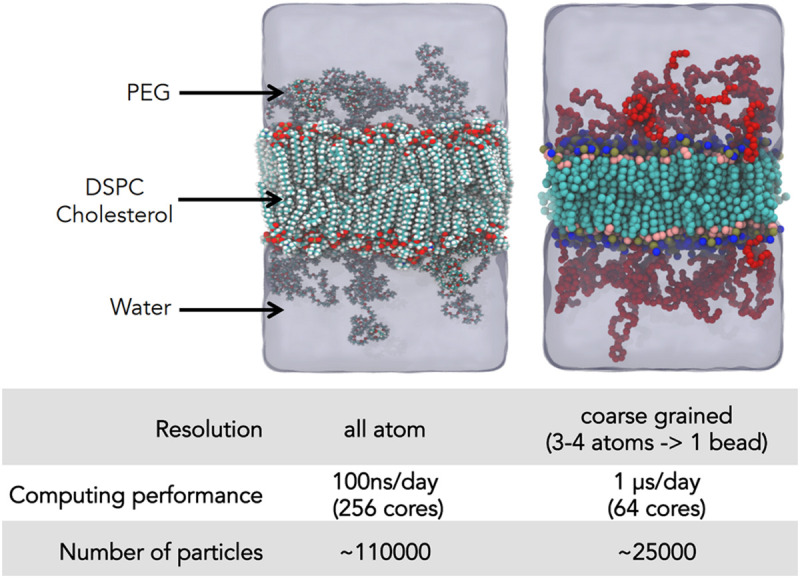
Illustration of coarse graining: the same system, a PEGylated membrane, modeled with an all atom and a MARTINI model is shown along with the decrease in number of particles and acceleration in the simulation time. Figure taken from Bunker: (Bunker et al., 2016) with permission.
While several schemes for the development of coarse grained models have been proposed (Miyazaki et al., 2020) the two that have been most frequently used are the MARTINI potential set (Marrink et al., 2007), where the coarse grained particles are groups of ∼3 atoms with the potential sets developed based on the solubility parameters of these groups and Dissipative Particle Dynamics (DPD) (Groot and Warren, 1997; Español and Warren, 2017) where the degree of coarse graining is greater still, where the particles are soft “momentum carriers” and temperature is controlled through a thermostat designed to conserve local momentum as the effects of hydrodynamics become important at this larger length and time scale. Another scheme is incorporating the effect of the solvent through adjustment to the interactions between particles in the molecules of interest, i.e., simulating with adjusted potentials in a vacuum; this is known as the “implicit solvent” model (Murtola et al., 2009). An ideal that is often sought and discussed is “multiscale simulation”—combining the insight from simulations carried out with different methodologies on different length and time scales (Haddish-Berhane et al., 2007; Murtola et al., 2009; Meier et al., 2013); in 2013 The Nobel Prize in Chemistry was awarded to Arieh Warshel, Martin Karplus and Michael Levitt for “development of multiscale models for complex chemical systems” (The Nobel Prize in Chemistry 2013, 2013). From the literature search for this review, it can, however, be surmised that this ideal, for the most part, remains an ideal: for the recent original research found, in our literature search for this review, that applied MD simulation in the field of drug delivery, the number of publications that use more than one methodology remain a small minority. Several reviews cover the use of coarse grained methods for the simulation of systems composed of lipids, polymers and proteins (Bennun et al., 2009; Loverde, 2014; Cascella and Vanni, 2016; Brancolini and Tozzini, 2019).
Now that we have this three-dimensional movie of our system, known as a trajectory, beyond just visualization there are several techniques to analyze this result and obtain useful insight into the system. Here we provide a few examples of frequently calculated properties from the trajectory. Considering pharmaceutical applications of MD simulations, a description of a binding mode (hydrogen bonds, salt bridges, stacking interaction, and hydrophobic interactions) of a drug in the protein binding cavity is the first key information to examine. Unlike binding modes obtained from experimental structural studies or docking predictions, MD simulations provide a dynamic description of the interaction between drug and protein (e.g., Kaszuba et al., 2012; Chen J. et al., 2019); this allows additional insight regarding the importance of individual interactions. Moreover, simulations provide explicit information concerning water participation in the binding mode (e.g., Kaszuba et al., 2010; Postila et al., 2013; Aguayo-Ortiz and Dominguez, 2019; Figure 3A), typically not resolved in structural studies and not considered in docking calculations. Analysis of intermolecular interactions is not limited to drug-protein interactions but can also be performed for any type of molecule/macromolecule studied, e.g., drug-lipid interactions are frequently studied (Cramariuc et al., 2012; Mayne et al., 2016; Pasenkiewicz-Gierula et al., 2016; Postila and Róg, 2020).
FIGURE 3.
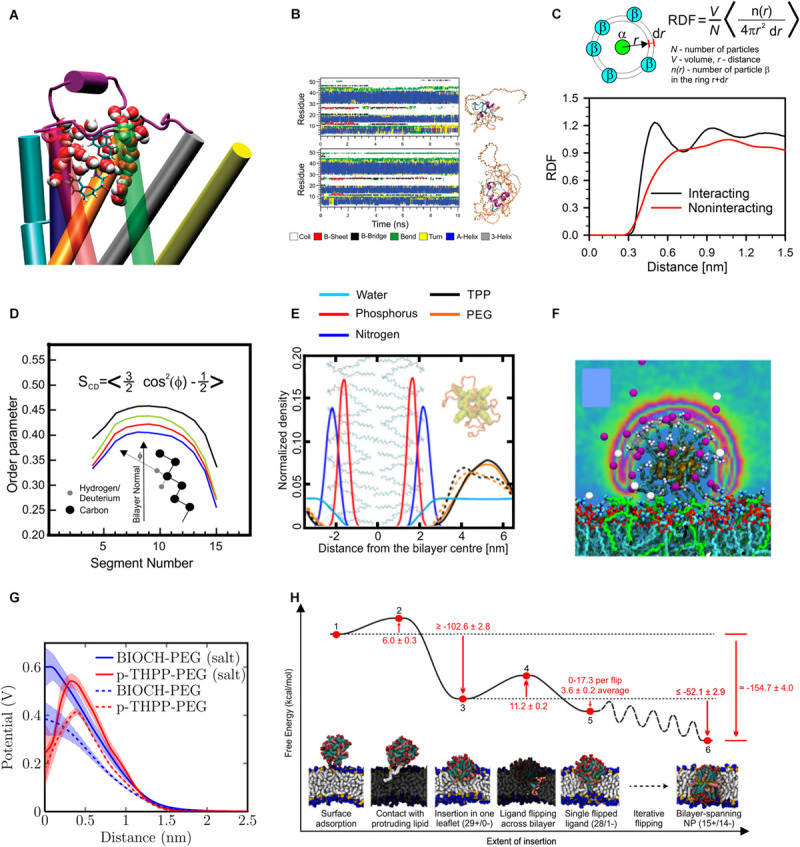
Results of MD simulations. (A) Snapshot showing involvement of water in binding mode of nebivolol to β2-adrenergic receptor, reproduced with permission from Kaszuba et al. (2010), Copyright (2010) American Chemical Society. (B) Time evolution of secondary structure of PEGylated insulin molecules, reproduced with permission from Yang et al. (2011), Copyright (2011) American Chemical Society. (C) An example of radial distribution functions (RDF) for interacting particles (black line) and non-interacting particles, data taken from Roìg and Pasenkiewicz-Gierula (2004). (D) Example of order parameter profile along the lipids acyl chain, reproduced from Mobarak et al. (2018) (CC BY 4.0). (E) An example of density profile showing position of atoms of lipid headgroups (phosphorus and nitrogen) and PEGylated tetra-phenyl-porphyrin (PEG and porphyrin densities are shown separately), at the presence (dashed line) and absence (solid line) of salt in solution, reproduced with permission from Rissanen et al. (2014), Copyright (2014) American Chemical Society. (F) Distribution of counter ions around gold nanoparticle functionalized with hydrocarbons capped with amine group, reproduced with permission from Heikkilä et al. (2014a), Copyright (2014) American Chemical Society, (G) electrostatic potential profile around PEGylated Biochanin (BIOH) and tetra-phenyl-porphyrin (p-THPP) in the presence and absence of salt in solution, reproduced with permission from Rissanen et al. (2014), Copyright (2014) American Chemical Society; (H) free energy landscape for the process of insertion of dendrimer into lipid bilayer, reproduced from Van Lehn and Alexander-Katz (2019), Copyright: 2019 Van Lehn, Alexander-Katz.
Additional observable properties used to evaluate intermolecular interactions are the radial distribution function (RDF) (Figure 3C) and the number of contacts. The RDF for the pairs of particles P1 and P2 gives us the normalized density of particle P2 at a given distance from particle P1. For the shortest distances the RDF value is 0 due to steric repulsion and converges to a constant value in the limit of infinite distance; for homogenous systems this value will always be 1. For an interacting pair of two particles, the RDF value initially rises with increasing distance to a maximum followed by a subsequent minimum (Figure 3C). E.g., for a pair of heavy atoms that form an H-bond, the maximum position is at ∼0.25 nm, and the minimum at ∼0.325 nm (Pasenkiewicz-Gierula et al., 1997). The number of contacts is the number of pairs of heavy atoms of two molecules located at a distance shorter than the selected cutoff. The most frequent choice for a cutoff length is the position of maximum or minimum at the RDF for carbon atoms in the liquid hydrocarbons. Calculations of numbers of contact are useful to evaluate equilibration in the simulations where self-assembly is studied. When a stable number of contacts is reached one can assume the end of the self-assembly process.
For interactions of larger molecules, MD simulations provide an area of contact (Acont). To obtain this, the solvent accessible surface area (SASA) (Connolly, 1983) for the considered molecule is first calculated separately (Amol1 and Amol2), and next, the same calculations are performed for the dimer (Adimer); this results in an area of contact:
| (1) |
Extensive MD simulation, either performed over a long time (Hurst et al., 2010; Dror et al., 2011) or as many multiple parallel simulations (Lolicato et al., 2020), are capable of elucidating the process of ligand entry into the binding pocket, however, the most frequent steered MD simulation methods (Izrailev et al., 1997) or randomly accelerated MD (RAMD) simulations (Lüdemann et al., 2000; Kokh et al., 2018) are used to reveal the entry/exit patch as they are more computationally efficient. For the case of functionalized proteins, their stability can be evaluated via calculations of secondary protein structure (Figure 3B). Other standard measurable properties provided by MD simulations include root mean square deviation (RMSD) and root mean square fluctuations (RMSF). The RMSD describes the similarity between the structures at the given time with the initial structure; thus, a large increase of this parameter can indicate a lack of protein stability. In studies of the interaction of drugs and nanoparticles with lipid bilayers, one can obtain insight into the xenobiotic degree of membrane perturbation.
The most frequently used tools to study lipid bilayer properties are surface area per lipid molecule, bilayer thickness and the order parameter. The most frequently calculated order parameters are the deuterium order parameter, SCD and molecular order parameter Smol (Vermeer et al., 2007; Figure 3D). The position in the membrane of any given xenobiotic molecule is quantitatively described by so-called density plots, which show the density of selected atoms, atom groups or whole molecules, along the bilayer normal. As a reference point, selected atoms of lipid molecules can be used, e.g., headgroups, glycerol moiety, or the last carbon of the acyl tails (Figure 3E). The next parameter describing drug behavior in the lipid bilayer is the drug molecule orientation with respect to the bilayer normal. Location and orientation of the drug in the lipid bilayer can be important for the entry of the drug into a protein binding cavity (e.g., Magarkar et al., 2018). Next, simulations describe the physicochemical properties of nanoparticles, including their size (quantitatively measured as the radius of gyration), nanoparticle hydration, interaction with ions (see Figure 3F) and electrostatic potential at the given distance from the nanoparticle center (Figure 3G). Finally, one should consider the statistical significance of results to avoid over-interpretation (Gapsys and Groot, 2020), carefully validate results against experimental data (Botan et al., 2015; Ollila and Pabst, 2016), and be critical as simulations are prone to methodological artifacts (Wong-ekkabut and Karttunen, 2016).
Sometimes the unbiased trajectory is not sufficient to obtain the insight we seek. The phenomenon we wish to study may occur in a region that is not sampled so frequently or we wish to calculate the free energy difference between two separate conformations of the system. For this we need the ability to apply a bias to the simulation to push it artificially toward a certain region of conformation space that we wish to examine. From calculating the bias needed along a path between two conformations one can obtain the free energy difference between then, an important measure of such quantities as the binding affinity of a drug for a specific active site of a protein (Michel and Essex, 2010). Two methods to calculate this free energy are umbrella sampling (Roux, 1995; Frenkel and Smit, 2001; Neale and Pomès, 2016; Figure 3H), where the path taken is through conformation space and what is known as a potential of mean force (PMF) (Roux, 1995) is calculated along this path and thermodynamic integration (Matos et al., 2017), an analogous calculation but where the path is through parameter space. The free energy calculations are computationally demanding and sensitive to force field details. Also one should consider possible artifacts due to a bias force, e.g., deformation of the lipid bilayers was observed in a few studies during umbrella sampling calculation of the profile of PMF of the studied compound along the bilayer normal (Neale et al., 2011; Filipe et al., 2014; Neale and Pomès, 2016). Metadynamics (Bussi and Laio, 2020) is an adaptive means to explore conformation space in an enhanced fashion by constantly biasing the system away from the regions of conformation space that have already been explored.
The remainder of this review paper will cover examples of how this tool, molecular dynamics simulation, has been and can continue to be, used in the context of drug delivery research (pharmaceutics). We will discuss applications across the breadth of the field, including obtaining insight relevant to dissolution and solubility, however, the majority of the discussion will cover the recent explosion in publications that use molecular dynamics simulation to study the more advanced drug delivery mechanisms, collectively known as nanomedicine.
Mechanistic Insight Into Drug Dissolution and Solubility From MD Simulation
The simplest application of MD simulation in drug delivery is gaining mechanistic insight into the universal processes of dissolution and solvation (Figure 4). Drugs often enter the body in crystalline form and dissolution of these crystals is the first step. Larsen et al. (2017a,b, 2019) have used MD simulation to study alteration to the crystal structure with varying levels of hydration. For systems with long range order such as this, a more accurate and computationally intensive COMPASS force field (Sun, 1998) is required, instead of the potential sets normally used for simulations of systems in the liquid state. The Ouyang group has studied the dissolution of drug molecules complexed with solid dispersions as a remedy for poor solubility using MD (Chen and Ouyang, 2017; Chan and Ouyang, 2018; Han et al., 2019a) in addition to machine learning techniques (Han et al., 2019b). Coarse grained simulations using the DPD protocol have been used by Otto et al. to study the release of the drug quercetin from poly(ethylene-glycol) (PEG) solid dispersions (Otto et al., 2013).
FIGURE 4.
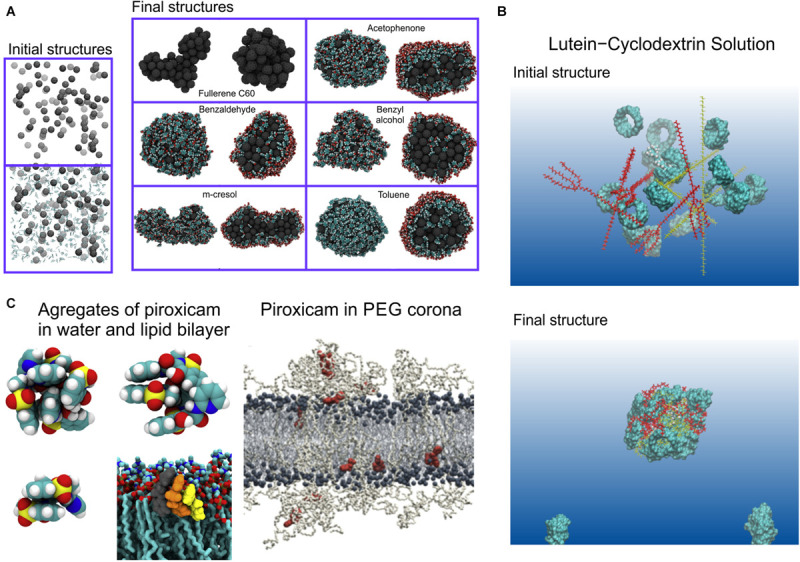
Dissolution and solubility. (A) Formation of clusters of fullerene with organic solvents, for the final structures clusters full view (left side) and its cross section (right side) are shown, organic solvent covers fullerene from outside and are present in small quantities inside the cluster, reproduced from Lehto et al. (2014), Copyright: 2014; (B) formation of lutein and cyclodextrin complexes (Zhao et al., 2018), Copyright (2018) American Chemical Society; (C) Aggregates of piroxicam formed in water and lipid bilayer, and piroxicam molecules dispersed in PEG corona of PEGylated lipid bilayer, reproduced with permission from Wilkosz et al. (2017).
As stated above, simplified QSAR/QSPR (Mathieu, 2020) or related machine learning models (Hutchinson and Kobayashi, 2019) are generally used to correlate drug structure to solubility using pattern recognition to relate structure to experimental solubility data; MD simulation can, however, be used to obtain both a more accurate result and, additionally, provide mechanistic understanding. The partition coefficient between water and octanol can be calculated for the specific molecule through MD simulation (Bannan et al., 2016) using the aforementioned techniques for free energy calculation, either by (1) using umbrella sampling to physically pull the candidate drug molecule structure through the boundary between a water and an octanol phase and calculate the free energy change along this path, the aforementioned PMF (example of a PMF calculation shown in Figure 3H) or (2) performing thermodynamic integration between the drug solvated in water and the drug solvated in octanol. Such a calculation is not the mechanistic insight advertised in the introduction; here we are using MD simulation as a tool to obtain a numerical estimate of a quantitative result. It is possible, however, to examine the MD simulation output further to obtain mechanistic insight regarding the relation between the structure of the molecule and the solvent; for example, Zhang et al. have investigated the H-bond network of the drug ibuprofen in water and ethanol (Zhang M. et al., 2020). Erlebach et al. (2020) have used a different technique combining simulations with atomistic resolution with solubility calculations based on Flory-Huggins theory. Other examples of MD used for solubility prediction also exist (Lüder et al., 2007, 2009; Westergren et al., 2007; Patel et al., 2010a; Gupta et al., 2011; Paluch et al., 2015; Matos et al., 2017; Matos and Mobley, 2019; Dasari and Mallik, 2020). To aid in the delivery of drugs that are otherwise too lipophilic, they are administered not alone but in a formulation with other molecules, known as excipients. Optimizing this drug formulation can be performed through combining screening experiments with pattern recognition and optimization algorithms, however, here too, MD simulation can play a powerful role in complementing other computational methods (Mehta et al., 2019), for example MD simulations of cyclodextrin-drug complexes (Zhao et al., 2018; Huang et al., 2019); cyclodextrin is a common agent for assisting the delivery of poorly soluble drugs. Persson et al. (2013) have used MD simulation to study drug solubility in excipient formulations and MD has been used to study polymeric excipients. Benson and Pleiss (2014) have used MD to study self-emulsifying drug delivery systems and Hathout et al. have modeled drug loading in the gelatin matrix (Ahmad et al., 2010; Warren et al., 2013; Jha and Larson, 2014; Hathout et al., 2020). Several comprehensive review papers have been written on the synergistic use of MD with other computational techniques to determine the solubility and dissolution characteristics of drugs and drug formulations (Johnson and Zheng, 2006; Bergström and Larsson, 2018; Li et al., 2018; Hossain et al., 2019; Das et al., 2020).
Describing the ease with which a drug travels through the body to reach its target through this one parameter, solubility, alone, is of course an extreme oversimplification: in addition to dissolving in the blood, drugs must traverse a variety of biological barriers, in particular cell membranes and perfect solubility will not insure this (Smith et al., 2018). Building systems to deliver drugs through these barriers requires an extra level of complexity; we now cross from simple formulation with the goal to optimize solubility into nanomedicine: nanoscale vectors designed to transport the drug through the bloodstream while protecting it from the body’s defense mechanisms and targeting the desired tissue.
Nanomedicine
Nanomedicine is officially defined as pharmaceutical applications of nanotechnology. Since “nanotechnology” is a meaningless buzzword quickly fading from fashion (Park, 2019) this is not a concise definition; in practical terms this encompasses all drug delivery systems that involve packaging the drug in structures with diameters =100 nm but larger than a single drug molecule: one or more drug molecules combined with one or more carrier molecules. For example, even merely grinding a crystal of the drug into pieces smaller than this size officially fits this definition, the result known as “nanocrystals” (Song et al., 2011) and recognized as the simplest form of nanomedicine. A very broad range of mechanisms have been developed that fit this definition and the nomenclature is cluttered, i.e., the language used to define different varieties, and how components are described is inconsistent; we will now describe the nomenclature and definitions we intend to use, but be warned: when you read the cited publications, the nomenclature may not be consistent.
When the drug and carrier are combined, the result is referred to as a nanoparticle. Nanoparticles are formed in one of two ways: (1) directly functionalizing a molecule to the drug, i.e., chemically bonding a molecule to the drug to alter its behavior in the bloodstream (Ekladious et al., 2019) or (2) combining one or more drug molecules with one or more carrier molecules that self-assemble to form the nanoparticle; I will refer to this as the functionalization and self-assembly routes of nanoparticle formation. The functionalization route to nanoparticle creation can also lead to the formation of a nanoparticle composed of more than one drug molecule, for example functionalizing a hydrophobic drug with a polymer could result in the formation of micelles with the drugs at the core. In most cases the direct functionalization is to a polymer, a long unstructured molecule, that, as a result, forms a protective sheath around the drug molecule in the bloodstream, however functionalization to a smaller molecule is also possible, for example, folic acid (Wolski et al., 2018; Alinejad et al., 2020) or glycine (Ghadri et al., 2020). A particularly ingenious idea is functionalization to amphiphilic “molecular umbrellas” that aid the transfection of hydrophilic drugs through the hydrophobic core of cell membranes (Janout et al., 2001, 2002, 2005, 2014; Jing et al., 2003; Janout and Regen, 2005, 2009; Ge et al., 2009). Drugs functionalized to polymers where the drug is activated by enzyme cleavage of the polymer are also referred to as “prodrugs” (Luo et al., 2019). Functionalization to peptides or small proteins can result in very specific fine tuning of the behavior of the drug as it interacts with its environment (Lu et al., 2015). Functionalization of lipids for a variety of applications is reviewed by Kepczynski and Róg (2016) and specifically for drug delivery by Kohli et al. (2014). Regarding nanoparticles formed via the self-assembly route, a rigorous literature search leads to a subdivision of the majority according to topology and choice of carrier molecule into roughly the following 9 categories: (1) solid inorganic, (2) micelles, (3) vesicles (Figure 5A), (4) lipoprotein based structures (Figure 5C), (5) other lipid-polymer structures, (6) carbon architectures (Figure 5B), (7) dendrimers (Figure 5D), (8) protein/peptide, and (9) the aforementioned nanocrystals. Bobo et al. (2016) have compiled the list of FDA-approved forms of nanomedicine, as of 2016.
FIGURE 5.
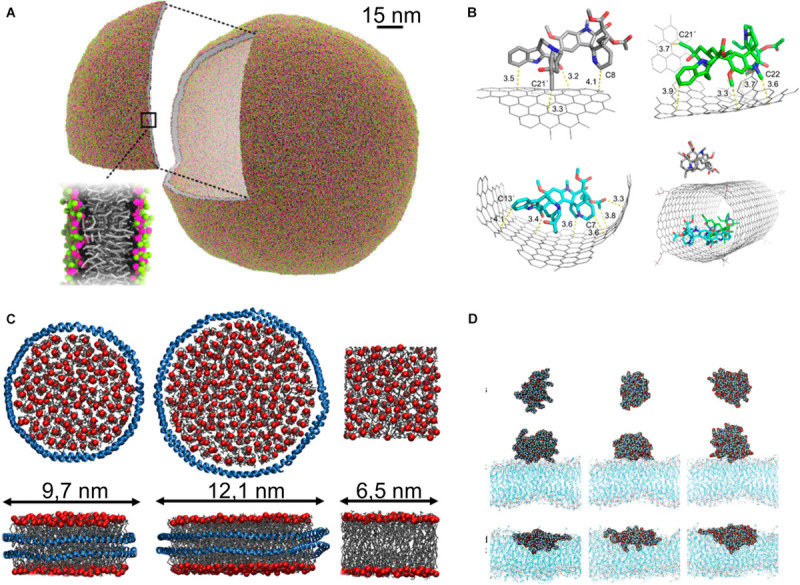
Nanoparticles. (A) Liposome simulated with dry MARTINI model, reproduced with permission from Arnarez et al. (2015), Copyright (2015) American Chemical Society; (B) carbon nanotube used for delivery of vinblastine (Li et al., 2016a), Copyright (2016) American Chemical Society; (C) nanodiscs formed of POPC and membrane scaffold protein MSP1D1 (Left), MSP1E3D1 (middle), and lipid bilayer (right), protein is shown as blue ribbon, phosphate groups of lipids shown as red sphere, and acyl tail as gray sticks, reproduced with permission from Stepien et al. (2020); (D) PAMAM dendrimer in water phase (top), at the lipid bilayer in gel phase (middle), and at the lipid bilayer in fluid phase (down), reproduced with permission from Kelly et al. (2008), Copyright (2008) American Chemical Society.
Solid inorganic nanoparticles are rigid structures formed from inorganic substances. These include gold (Ghosh et al., 2008; Charchar et al., 2016; Rossi and Monticelli, 2016), silver (Eckhardt et al., 2013; BurduŞel et al., 2018), titanium dioxide (Aranha et al., 2020), silica (Santos et al., 2014) nanoparticles, and boron nitride oxide nanoflakes (Duverger and Picaud, 2020). Gold and silver nanoparticles are solid structures that can be associated with drugs, or can be functionalized themselves to perform a specific function: the nanoparticle itself is a drug. For the case of silica nanoparticles they can be porous and contain drugs and can even have complex multi-compartment structure, carrying many different drug molecules (Torchilin, 2007; Pattni et al., 2015; Bulbake et al., 2017; El-Hammadi and Arias, 2019; Bhardwaj et al., 2020; Crommelin et al., 2020), for example for applications like theragnostics (Janib et al., 2010). In the same fashion as solid inorganic nanoparticles, carbon architectures are contiguous solid structures, however, due to its unique chemistry, composition from carbon allows for a wide variety of forms, including carbon dots (Peng et al., 2017; Ghosal and Ghosh, 2019), nanotubes (Sun et al., 2014), nanodiamonds (Barnard, 2016; Ge and Wang, 2017), nanographene (Zhang L. et al., 2013; Sun et al., 2014; Sgarlata et al., 2016; Ghadari and Kashefi, 2017; Hasanzade and Raissi, 2017; Moradi et al., 2018; Alinejad et al., 2020; Mahdavi et al., 2020), and graphene oxide (Duverger and Picaud, 2020; Shahabi and Raissi, 2020).
Micelles and vesicles are both formed from amphiphilic organic molecules but differ in topology: micelles have a hydrophobic core surrounded by a hydrophilic shell while in vesicles the amphiphilic molecules form a bilayer that itself forms into an enclosed pocket. In both cases they can be formed from a wide range of molecules, usually surfactants, lipids or diblock copolymers, however, other amphiphilic molecules are possible, for example, janus dendrimers (Nummelin et al., 2017; Yang Y.-L. et al., 2019). The most common micellar nanoparticle is the polymeric micelle (Cagel et al., 2017), composed of diblock co-polymers with hydrophobic drugs carried in the micelle core. The most common form of vesicular nanoparticle is the liposome (Bunker et al., 2016), a vesicle formed from naturally occurring phospholipids. Other amphiphilic molecules formed into vesicles are, however, also used in drug delivery, including ethosomes (Touitou et al., 2000), niosomes (Marianecci et al., 2014; Khan and Irchhaiya, 2016; Chen S. et al., 2019; Kapoor et al., 2019; Khalkhali et al., 2019; Inglut et al., 2020), polymersomes (Aibani et al., 2020; Khan et al., 2020), exosomes (Antimisiaris et al., 2018; Villa et al., 2019; Chung et al., 2020; Rahmati et al., 2020), ufasomes (Han, 2013), and drimersomes (Nummelin et al., 2017), comprehensive reviews have been written about vesicle formation (Šegota and Durdica, 2006) and application in drug delivery (Kapoor et al., 2019) in a general context. Polymers and lipids can be formed into other structures than micelles or vesicles, for example two different polymers can be used to form core-shell structures (Ramli et al., 2013; Abbott et al., 2017; Chen G. et al., 2018), for example a solid outer shell with a liquid polymer with drug encapsulated inside; solid lipid nanoparticles (Beloqui et al., 2016; Gordillo-Galeano and Mora-Huertas, 2018; Subramaniam et al., 2020), chitosan (Bernkop-Schnürch and Dünnhaupt, 2012), lipoplex (Scheideler et al., 2020) and other lipid-polymer nanoparticles (Date et al., 2018) have also been proposed. Another form of polymer based nanoparticle is dendrimers (Tomalia et al., 1990; Fatemi et al., 2020) and pseudodendrimers (Ghadari and Sabri, 2019), hyper-branched polymers with a fractal structure that results in a molecule that is, qualitatively, in the form of a fuzzy ball and can store molecules in their interior or bind nucleic acids to form a dendrimerplex. A particularly common form of dendrimer that has been proposed for drug delivery is poly(amidoamine) (PAMAM) dendrimers (Xiao et al., 2020).
Lipoproteins are used as the body’s mechanism for lipid transport. These are structures of several different lipids with proteins that control the form of the structure and the composition of the lipid types within the structure. As they transport lipids they undergo structural change upon deposition of their cargo from a spherical structure to a disk-like structure. Taking these structures as a starting point and modifying them to work as drug carriers, or building structures inspired by lipoproteins, is a novel avenue of nanomedicine that is currently being explored (Bricarello et al., 2011; Huang et al., 2015; Kuai et al., 2016a; Simonsen, 2016; Aranda-Lara et al., 2020; Chuang et al., 2020). The disk-like form of lipoprotein, known as nanodiscs have proven to be an extremely useful structure for a variety of applications, including nanomedicine (Denisov and Sligar, 2017). Nanodiscs were successfully used as a drug delivery vehicles to treat viral lung infections (Numata et al., 2013) and were used as a platform accommodating antigens and adjuvants in personalized cancer vaccines (Kuai et al., 2016b). Use of nanodiscs for simultaneous delivery of antigen and adjuvant has been found to increase the response of the immunological system by orders of magnitude in comparison to traditional vaccines. Due to the variety of possible applications of nanodiscs, their properties are the subject of intensive study (Debnath and Schäfer, 2015; Siuda and Tieleman, 2015; Stepien et al., 2015, 2020; Martinez et al., 2017; Pourmousa and Pastor, 2018; Bengtsen et al., 2020; Schachter et al., 2020); they are tuned via modification of their lipid composition (Augustyn et al., 2019) or alterations to the sequence, thus structure, of the scaffold proteins (Denisov et al., 2004; Nasr et al., 2016).
All of these structures can have their properties fine-tuned by being functionalized to polymers or smaller molecules themselves, in the same fashion as described above for the drug molecule itself. For example functionalizing poly(ethylene glycol) (PEG) (Israelachvili, 1997), a process known as “PEGylation” (Bunker, 2012, 2015; Pasut and Veronese, 2012; Bunker et al., 2016; Zhang Z. et al., 2020) has been proposed and studied for virtually all of these nanoparticle forms and, as we will discuss in further detail in the next section, alternate polymers to PEG are under investigation. The extent to which these systems can be fine-tuned is limitless, for example formulation alteration of liposomes offer an extremely broad pallet (Bunker et al., 2016; Li et al., 2019). We are thus left with several variables for their formulation in addition to the extremely complex environment of human physiology with which they interact, the topic that we will now discuss.
Nanoparticle Design and Function
Nanoparticles have been developed to assist in drug delivery in a very broad range of pharmaceutical contexts, for example treating atherosclerosis (Lobatto et al., 2011; Chen J. et al., 2020; Ramalho et al., 2020) and other neurodegenerative diseases (Goldsmith et al., 2014), cardiovascular disease (Godin et al., 2010), diabetes (Veiseh et al., 2015), infections disease (Zhu et al., 2014; Zazo et al., 2016), protein drugs (Qin et al., 2019), and vaccine delivery (Pison et al., 2006; Zhao et al., 2014) in fact vaccine adjuvant development involves many of the same mechanisms as nanomedicine (Copland et al., 2005; Wang et al., 2019); it can be argued that it is only for historical reasons that it is not referred to as nanomedicine. The main application of nanomedicine is, however, cancer therapy (Tong and Kohane, 2016; Youn and Bae, 2018), particularly chemotherapy agent delivery, as this involves drugs with extremely high toxicity; targeted delivery, where the drug is kept from the rest of the body and the greatest possible fraction is delivered to the target tissue, in this case the tumor, is extremely desirable. The nanoparticle is designed to have features that protect the drug, in the context of nanomedicine commonly referred to as the “payload” of the nanoparticle. Targeting is achieved through either active or passive means. Active targeting (Nag and Delehanty, 2019) involves a specific ligand functionalized to the nanoparticle exterior that binds to receptors that are overexpressed in the outer cell membrane of cells of the target tissue and passive targeting involves global properties (Ogawara et al., 2013) of the nanoparticle that lead to a greater percentage becoming lodged in the target tissue in comparison to other tissues. An example of passive targeting is what is referred to as the enhanced permeability and retention (EPR) effect (Maeda et al., 2013); liposomes can be designed to take advantage of the leaky vasculature of tumor tissue to become preferentially lodged there; PEGylation is a common means to achieve this. It must, however, be stated that whether or not the EPR effect is an effective passive targeting strategy in practical nanomedicine applications, has recently been brought into question (Danhier, 2016).
The nanoparticle thus carries and protects its drug payload through the bloodstream and preferentially delivers it to its target tissue. In the bloodstream, foreign particles in the size range of nanoparticles are removed (uptaken) by the mononuclear phagocyte system (MPS) (Chow et al., 2011); this involves an extremely complex and specific cascade of proteins: complement activation (Ricklin et al., 2010; Sarma and Ward, 2011). The efficiency with which a nanoparticle is removed through complement activation is determined by its surface properties. The nanoparticle can be designed to have a surface that inhibits uptake, thus prolonging circulation in the bloodstream and, as a result, the amount of the drug that reaches the target tissue per administered dose; such a nanoparticle surface is referred to as a “stealth sheath” and the aforementioned PEGylation is the gold standard to achieve a this (Pasut and Veronese, 2012; Bunker, 2015; Parray et al., 2020). While PEGylation is an extremely successful strategy, it is not perfect and the investigation of alternate polymers to PEG is an active field of research (Knop et al., 2010). Alternatives that have been proposed and studied include polyoxazolines (Sedlacek et al., 2012; Lorson et al., 2018), PASylation® (Viegas et al., 2011; Schlapschy et al., 2013; García et al., 2014; Binder and Skerra, 2017; Gebauer and Skerra, 2018), zwitterionic polymers (García et al., 2014), hydroxyethyl starch (Liebner et al., 2014), and polypeptides (Hou and Lu, 2019).
PEGylation, or the creation of an alternate polymer stealth sheath, is achieved though functionalizing the polymer to a component of the nanoparticle. For the case of the functionalization route to nanoparticle creation, functionalization to the protective polymer itself can be the nanoparticle. It is also possible to functionalize the drug to a copolymer where one of the copolymer constituents is the hydrophilic stealth sheath and the other performs another function, e.g., a hydrophobic polymer that encapsulates the drug. Examples of this include PEGylated boron nitride (Farzad and Hashemzadeh, 2020), folic acid (Wolski et al., 2017b; Alinejad et al., 2020), interferon (Xu et al., 2018), insulin (Yang et al., 2011; Figure 6E), other PEGylated peptides (Xue et al., 2011; Hamed et al., 2015; Ma et al., 2016; Figure 6F) and protein drugs (Katre, 1993; Jevševar et al., 2010; Yang et al., 2011; Zhang et al., 2012; Mu et al., 2013; Wu et al., 2014; Lawrence et al., 2014; Nischan and Hackenberger, 2014; Lawrence and Price, 2016; Xu et al., 2018; Wilding et al., 2018; Gupta et al., 2019; Zaghmi et al., 2019; Munasinghe et al., 2019; Kaupbayeva and Russell, 2020; Figure 6B); the broader context of polymer-protein drug molecules is covered in several reviews (Pelegri-Oday et al., 2014; Wang et al., 2019). As far back as 1977, long before “nano” was a word, functionalizing PEG to proteins was proposed to alter their immunological properties (Abuchowski et al., 1977). For the self-assembly route to nanoparticle creation the polymers are functionalized to constituent molecules of the nanoparticle. PEGylation has been proposed for virtually every one of the nanoparticle types described in the previous section. This includes PEGylated carbon nanotubes (Pennetta et al., 2020), gold nanoparticles (Oroskar et al., 2016; Lin et al., 2017; Sun et al., 2019), silver nanoparticle (Pinzaru et al., 2018), silver-graphene nanoparticles (Habiba et al., 2015), nano-graphene (Zhang et al., 2014; Zhang Z. et al., 2020; Mahdavi et al., 2020), lipid micelles (Arleth et al., 2005; Viitala et al., 2019; Figure 6D), nanodiscs (Zhang et al., 2014), dendrimers (Kojima et al., 2000; Lee and Larson, 2009, 2011; Zhang et al., 2014), and a topic covered comprehensively in our previous review, liposomes (Bunker et al., 2016; Figures 6A,C).
FIGURE 6.
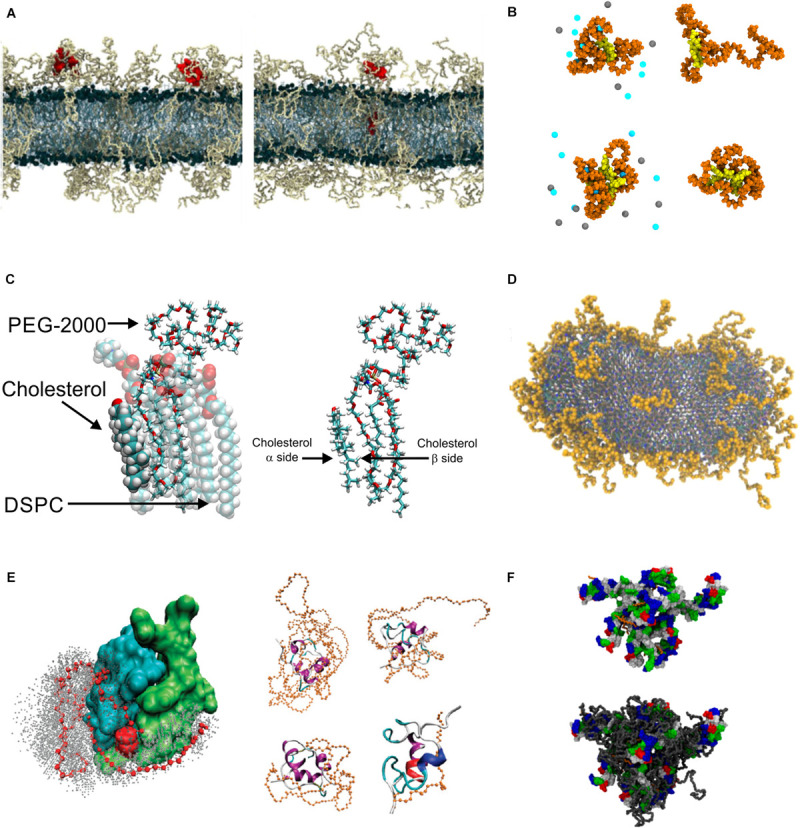
PEGylation. (A) Snapshot showing PEGylated lipid bilayer, reproduced with permission from Dzieciuch et al. (2015), Copyright (2015) American Chemical Society; (B) PEGylated biochanin (upper) and tetra-phenyl-porphyrin (lower), with salt (left) and without salt (right), reproduced with permission from Rissanen et al. (2014), Copyright (2014) American Chemical Society; (C) Snapshots showing DSPC, cholesterol and DSPE-PEG molecules, reproduced with permission from Magarkar et al. (2014), Copyright (2014) American Chemical Society; (D) PEGylated bicelle containing 10.5 mol % DSPE-PEG, reproduced from Viitala et al. (2019), Copyright: 2019; (E) PEGylated insulin, left panel shows position of PEG atoms during simulations, right panel shows snapshots of insulin PEGylated with PEG of various length, reproduced with permission from Yang et al. (2011), Copyright (2011) American Chemical Society.
For the case of inorganic nanoparticles, in particular gold nanoparticles, various alternatives to PEG coatings have been considered. Gold nanoparticles can be functionalized via a thiol group with hydrocarbons capped with a methyl group (Bolintineanu et al., 2014; Potdar and Sammalkorpi, 2015; Giri and Spohr, 2018), hydroxyl group (Potdar and Sammalkorpi, 2015; Villarreal et al., 2016; Yamanaka et al., 2019), carboxylic group (Heikkilä et al., 2014b; Giri and Spohr, 2018; Figure 7A), amine group (Heikkilä et al., 2014a,b; Giri and Spohr, 2018; Das et al., 2019; Lolicato et al., 2019), choline sulfate (Yamanaka et al., 2019), or a para-mercaptobenzoic acid (Figure 7B; Salorinne et al., 2016). Also, bulky branched coatings have been used to functionalize gold nanoparticles (Giri and Spohr, 2018; Yamanaka et al., 2019). The alternative coating can also be used to direct the nanoparticle to a selected environment, e.g., Potdar and Sammalkorpi proposed using a hydrophobic coating to cause the particle to locate to the hydrophobic core of the bilayer and a coating ended with a hydroxyl group to anchor the particle to the lipid headgroups (Potdar and Sammalkorpi, 2015). A coating composed of two types of moieties one a hydrophobic 1-octanethiol and the other a negatively charged 11-mercapto-1-undecanesulfonate causes the nanoparticle to locate to the center of the bilayer with its polar sulfonate groups exposed to the water at both membrane interfaces; this induces a local thinning of the bilayer (Van Lehn et al., 2013; Van Lehn and Alexander-Katz, 2014a, 2019; Simonelli et al., 2015) (Fi), or possibly even large scale deformation (Salassi et al., 2017). With the same coating moieties with polar coating placed on one-half of the particle and non-polar on the other, one can form an amphiphilic gold nanoparticle that will locate to the boundary between the water phase and the hydrophobic membrane core, i.e., the position of the lipid headgroups (Ou et al., 2020; Figure 7D). Such coating of other solid inorganic nanoparticles has also been considered, e.g., silver nanoparticles were coated with hydrophilic polymer poly(N-vinyl-2-pyrrolidone) (Kyrychenko et al., 2015), graphene nanoflakes with ssDNA (Moore et al., 2019), and silica nanoparticle with hydrocarbons (Peters et al., 2012).
FIGURE 7.
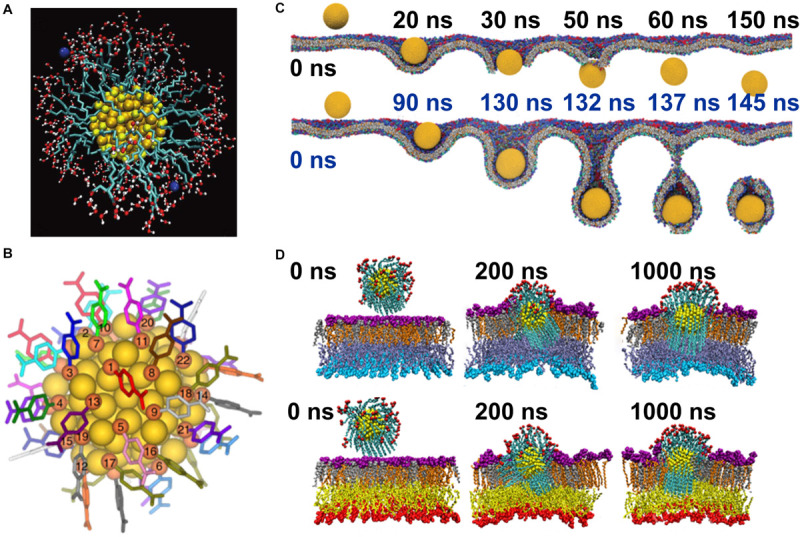
Gold nanoparticles. (A) Gold nanoparticle coated with dodecanoic acid, hydrating water and ions are shown, reproduced with permission from Heikkilä et al. (2012), Copyright (2012) American Chemical Society; (B) gold nanoparticle coated with para-mercaptobenzoic acid, gold is shown in yellow and sulfur in orange, reproduced from Salorinne et al. (2016), Copyright: 2016; (C) snapshots of (upper panel) internalizations of a neutral gold nanoparticle and (lower panel) uptake of a positively charged gold nanoparticle, reproduced with permission from Lunnoo et al. (2019), Copyright (2019) American Chemical Society; (D) snapshots of the MD trajectory of the insertion of amphipathic janus nanoparticle into lipid bilayers, reproduced with permission from Ou et al. (2020), Copyright (2020) American Chemical Society.
Proteins will agglomerate to any foreign particle in the bloodstream in the approximate size range of a nanoparticle resulting in a shell of proteins surrounding them, known as the “protein corona” (Walkey and Chan, 2012; Xiao et al., 2013; del Pino et al., 2014; Kharazian et al., 2016; Mahmoudi, 2016; Hadjidemetriou and Kostarelos, 2017; Pederzoli et al., 2017; Brancolini and Tozzini, 2019; Casalini et al., 2019; Nienhaus and Nienhaus, 2019; Zhadanov, 2019; Berrecoso et al., 2020; Gupta and Roy, 2020). The stealth sheath modulates the formation of this corona in a fashion that is not completely understood and has been a point of contention in the field for several decades. Regarding PEGylation, it was originally thought that it inhibits protein adhesion (Du et al., 1997; Bradley et al., 1998) then others found evidence that it actually accelerates protein corona formation (Szebeni et al., 2002) and yet others argued that they found evidence it had no effect (Price et al., 2001). It has been argued that the PEG sheath preferentially binds the common bloodstream protein albumin (Vert and Domurado, 2000) creating an albumin protein corona that, itself, acts as the stealth sheath that inhibits complement activation (Caracciolo, 2015). Alternate protective mechanisms unrelated to the protein corona have also been proposed, including direct inhibition of absorption by macrophages (Price et al., 2001). Most recently, evidence has been found that the formation of the protein corona is essential for the stealth properties of PEG (Schöttler et al., 2016). The most recent reviews of this much discussed topic are found here (Nienhaus and Nienhaus, 2019; Zhadanov, 2019; Li Z. et al., 2020).
Complement activation and the formation of a protein corona is only one aspect of the environment that the nanoparticle must traverse; in addition to the body’s defenses the nanoparticle must navigate the hydrodynamic environment of the bloodstream and, in most cases, deliver the payload drug through the cell membrane. While the surface properties of the nanoparticle play a role, both of these are heavily influenced by its size, shape (Truong et al., 2015) and rigidity/elasticity (Geng et al., 2007; Lee S.-Y. et al., 2009; Toy et al., 2011). Once the nanoparticle reaches the bloodstream its environment can be approximated as laminar flow in a cylinder. In this environment, in addition being pushed in the direction of the flow, a particle is subject to a force perpendicular to the flow that causes the particle to move toward the cylinder wall, a phenomenon known as margination (Gentile et al., 2008). Evolution has taken advantage of this: red blood cells are relatively rigid and have a disk-like form in order to minimize margination, as disk-like and more rigid particles experience a lesser extent of this force in comparison to spherical and more elastic particles; leukocytes have evolved to have the opposite structure, spherical and elastic, as margination to the blood vessel wall plays an essential role in their function (Lee S.-Y. et al., 2009). This is one of the reasons why the previously mentioned nanodiscs are a very promising form of nanoparticle, however, it is not the only reason: size, shape and elasticity of the particle also affect the interaction between nanoparticle and cell membranes (Lin X. et al., 2010; Zhang et al., 2012; Banerjee et al., 2016). Several design features of the nanoparticle are involved in tuning its properties to deliver the payload through biological barriers (Blanco et al., 2015) like the cell membrane, into the target cell and in some cases to a specific organelle within the cell; both surface properties and the size (Lin X. et al., 2010; Lunnoo et al., 2019), shape (Lin X. et al., 2010; Zhang et al., 2012; Lunnoo et al., 2019), and elasticity of the nanoparticles play a role in this. Also presence of negatively charged lipids affects intake of functionalized, cationic gold nanoparticles (Lolicato et al., 2019). There are several mechanisms through which this is possible; nanoparticles can directly permeate the membrane (Song et al., 2011), in many cases disrupting its structure (Figure 7C). For the case of liposomes and micelles (De Nicola et al., 2014), the payload can be delivered through membrane fusion and the nanoparticle can also be designed to induce endocytosis (Vácha et al., 2011). As mentioned previously, nanoparticles can be functionalized with targeting ligands that trigger preferential uptake by target cells (Bazak et al., 2015). It, however, must be said that active targeting, while a popular topic for research, has so far seen limited success; as far as the authors are aware there is only one approved therapy that features active targeting: Denileukin Diftox (Turturro, 2014). Finally, nanoparticles can be designed to release their drug payload when there is a certain external trigger, a scheme known as controlled release. This trigger can be pH change that occurs during endocytosis or an externally applied trigger used to cause the drug to release in the tissue to which this trigger is applied, for example a locally applied optical magnetic or thermal trigger (Table 1).
TABLE 1.
Triggers used to release drug payload.
| Trigger type and references |
| pH change (Guo et al., 2010; Zheng et al., 2011; Nie et al., 2013, 2014; Wang et al., 2015a, 2016, Luo Z. et al., 2016; Rungrotmongkol and Poo-arporn, 2016; Min et al., 2017; Wang Y. et al., 2017, Wang Z. et al., 2017; Wolski et al., 2017b, 2018; Quan et al., 2017; Gao et al., 2019; Wu W. et al., 2019; Wu Z. et al., 2019; Maleki et al., 2020) |
| Optical (Lajunen et al., 2016, 2018; Massiot et al., 2017) |
| Magnetic (Panczyk et al., 2013; Yang C. et al., 2020; Zhang X. et al., 2020) |
| Thermal (Dhawan et al., 2004; Pérez-Sánchez et al., 2020) |
Altogether, we see that the landscape of nanomedicine is extremely complex, both with a wide range of directions that nanoparticle design can take and the extremely complex environment of human physiology and the body’s natural defenses. While in vitro experimental insight and clinical studies can make some progress, one quickly reaches a dead end in a sea of complexity without the rational design approach made possible by a mechanistic understanding. The next section shows how molecular dynamics simulation, alongside complementary experimental analysis techniques, to some extent provide this.
Molecular Dynamics Simulation Applied to Nanomedicine
Now that we have outlined the different forms of nanomedicine and the issues encountered by nanoparticles in their context as drug delivery agents, we can proceed to showcase many examples where molecular dynamics simulation, using different degrees of coarse graining, have provided mechanistic insight that complements the research program to develop new nanoparticle based drug delivery mechanisms. The amount of work carried out in this area has exploded in the past decade, with molecular dynamics studies being applied to virtually every variety of nanoparticle discussed above in their context as drug delivery vehicles, including dendrimers, gel nanoparticles, polymeric micelles, other polymeric forms of nanoparticles, solid lipid nanoparticles, other micelles, nanocrystals, carbon dots, carbon nanotubes, nanographene, DNA nanotubes, nanodiamonds, peptide nanoparticles, gold nanoparticles, silver nanoparticles, silica nanoparticles, latex nanoparticle and vesicles, of which the application of molecular modeling to liposome based drug delivery systems is covered comprehensively in our previous review (Bunker et al., 2016); there has, however, been a significant amount of work performed since its publication, and molecular modeling has now been applied to the study of other vesicle based drug delivery systems including niosomes, ufasomes, polymeric vesicles (polymersomes), and glyceryl monostearate vesicles. A list of publications that feature the use of molecular dynamics modeling to study each of these systems if found in Table 2. One intriguing omission by the scientific community is lipoprotein based nanoparticles, including nanodiscs. Nanodiscs have been studied in the context of their possible use as a drug delivery mechanism and have been studied, in a general context, using molecular dynamics simulation, however, molecular dynamics simulation has never been applied in the context of their possible use in drug delivery.
TABLE 2.
Drug delivery vehicles studied using molecular modeling methods.
Regarding the functionalization route to nanoparticle development, there has also been a considerable amount of computational study carried out using molecular dynamics modeling. Protein structures can be downloaded and their potentials have already been parameterized; attach a polymer to the protein, solvate in water and you can study its behavior. Both PASylated (Hedayati et al., 2017) and PEGylated (Cohan et al., 2011) human recombinant erythropoietin have been simulated; Munasinghe et al. (2019) used molecular dynamics simulation to study conjugation of PEG to a hydrophobic pocket of bovine serum albumin using a model with atomistic resolution and Wilding et al. (2018) used a coarse grained model to study site specific PEGylation of the protein lysozyme. Atomistic MD has been used to study the effect of PEGylation on the stability and potency of interferon (Xu et al., 2018) and insulin (Yang et al., 2011) and the steric shielding effect that results from the PEGylation of Staphlokinase (Mu et al., 2013). A recent comprehensive overview of the application of molecular simulation to the study of protein-polymer conjugation has been written by Lin and Colina (2019).
In terms of the delivery of specific drugs using nanomedicine, a very large number have been simulated incorporated into a wide variety of nanoparticle types. These drugs include Alzheimer’s medication, anti-worm drugs, antibiotics, anti-cancer drugs, including chemotherapy agents, anti-viral agents, antifungal drugs, anti-inflammatory drugs, antimicrobial peptides, drug used for diabetes treatment, immunomodulators and immunosuppressants, local anesthetics, and others; a list is found, with citations, in Table 3. Altogether, it becomes clear that there is simply too much work that has been carried out to concisely summarize in its entirety in this review. We will instead focus on a few key areas where MD modeling has provided important insight and discuss review papers that focus on certain aspects of the use of molecular dynamics in the context of nanomedicine and some key examples of original research that demonstrate the power of the technique. The discussion will include key examples where we show concrete insight gained my molecular dynamics simulation. We will focus on three areas: (1) behavior of the nanoparticle in the bloodstream and the protective polymer corona, (2) drug loading and release and (3) nanoparticle interaction with lipid membranes and entry into the cell. We would like to here alert the reader to the fact that there are other reviews of aspects of the use of computational modeling for nanoparticle design (Angioletti-Uberti, 2017b; Bouzo et al., 2020).
TABLE 3.
List of drugs studied with MD simulations in context of drug delivery.
MD Insight Examples
Behavior in the Bloodstream and Protective Polymer Corona
As we discussed previously, when the nanoparticle enters the bloodstream it encounters hydrodynamic forces and a corona of bloodstream proteins forms on its surface; a subset of these proteins form the highly specific complement activation reaction that leads to removal by macrophages. Regarding behavior in the bloodstream and the effect of size and shape (Shah et al., 2011), the most suitable method is not MD, but rather a combination of theoretical calculation (Decuzzi et al., 2005) and a discretized continuum model known as computational fluid dynamics (CFD), described and used to model this by Li et al. (2014b), Gupta (2016), and Gao et al. (2020) to model nanoparticle transport in the faulty tumor vasculature (Gao et al.). As we have mentioned, the formation of the protein corona is an extremely complex process that still remains poorly understood. What is clear, however, is that the surface properties of the nanoparticle affect this and the mechanism through which the protective polymer corona increases the bloodstream lifetime is modulation of the interaction with bloodstream proteins. Schafer et al. (2017) and Settanni et al. (2017a,b, 2018) combined experimental analysis with MD to study the interaction between two protective polymers, PEG and poly-sarcosine, with a set of proteins found in the bloodstream. They found evidence that the interactions are not amino acid specific but rather a general tendency dependent on the charge and polarity of the amino acid and the nature of the interaction between the polymer and water, in addition to the direct polymer-protein interaction. Their methodology, synergistically combined with experimental work, could provide a route to a rational design approach to the development of new polymer materials being developed that may have superior performance as a protective polymer corona. Lee et al. used the coarse grained MARTINI model to directly simulate the effect of PEGylation and PEGylation density on the interaction between the liposome and blood stream proteins; Lee also used MD simulation with atomistic resolution to study the effect of nanoparticle electrostatics in protein corona formation (Lee, 2020a). There are other examples of the use of MD modeling to study the protein corona of nanoparticles (Dell’Orco et al., 2010; Vilaseca et al., 2013; Lopez and Lobaskin, 2015; Shao and Hall, 2016).
In our previous review publication, focused on liposome based delivery systems (Bunker et al., 2016), we surveyed the work that had been carried out using molecular dynamics modeling, particularly with a model with all atom resolution, on the interaction between the protective PEG corona and the lipid bilayer (Stepniewski et al., 2011; Magarkar et al., 2012, 2014). Since this time the methodology has been used to study the effect of exchanging PEG with two different poly-oxazolines, poly-ethoxazoline (PEOZ) and poly-methoxazoline (PMOZ), with the result indicating that several properties of PEG are highly specific and related to its amphiphilic nature and the ease with which it acts as a polymer electrolyte (Magarkar et al., 2017). We also simulated the effect of change in PEG length, branched structures, and functionalizing PEG to the cholesterol or cholane in the membrane rather than phospholipids and our results complemented both in vivo and in vitro experiments carried out on these novel liposome based delivery systems (Mastrotto et al., 2020).
PEGylation, in the context of other nanoparticle forms than liposomes, has also been studied extensively using MD simulation. Ambrosio et al. (2018) complemented experimental study by demonstrating, using MD simulation with all atom resolution, that a 2:1 ratio or greater of PEG-cholane molecules to the VIP-palm peptide being delivered, is required to form supramolecular assemblies; these assemblies were shown to effectively cover the VIP- peptide with a protective corona of PEG. In previous work we have used MD simulation to study the PEGylation of small drug molecules (Li Y.-C. et al., 2012). Two recent reviews, written by Lee, very comprehensively cover MD simulation work, using coarse grained in addition to all atom models, to study the structure and behavior of PEGylated nanoparticles, one covering PEGylated biomolecules, liposomes and solid nanoparticles (Lee, 2020b) and the other covering PEGylated peptides dendrimers and carbon nanotubes (Lee, 2014). Li et al. carried out a coarse grained MARTINI model simulation to investigate the effect of PEG chain length on the shielding effect of PEGylated nanoparticles (Li and Hu, 2014). A comprehensive review of computational modeling of PEGylation has been written by Souza et al. (2018).
Drug Loading and Controlled Release
The ability of nanoparticles to hold drugs and release them with an external trigger has been studied for several nanoparticle forms by several groups. In most cases the drugs being considered are hydrophobic and sit within a non-polar region. Nanoparticles that have been simulated carrying their drug payload include carbon nanotubes, nanographene, peptide carriers, PAMAM dendrimers, polymeric nanoparticles, polymeric micelles, hydrophobic drugs within the membrane of liposomes, other issues related to drug loading of liposomes (Cern et al., 2014) and polymersomes (Grillo et al., 2018) (further citations found in Table 4). Drug cargoes studied include cucurbitacin, carmustine, 5-flouracil (Barraza et al., 2015), chacone, picoplatin, porphyrins, ibuprofen, paclitaxel, and albendazole, however, the most popular drug for these model systems is doxorubicin (see Table 4 for citations). In many cases these nanoparticles are designed to release their drug payload in response to a pH change trigger (see Table 1); MD simulation is able to model the effect of pH change. In an MD simulation pH is modeled through the partial charges on the atoms, so the system can be equilibrated with the partial charges corresponding to neutral pH and then the partial charges can be changed to model pH change and the behavior of the system in response to this, i.e., the drug release, can be modeled. One interesting aspect of the work carried out using MD simulation in this area is that use of all three levels of coarse graining is represented: atomistic MD, MARTINI model and DPD (Table 4). Reading this literature with this in mind provides a very good case study of the strengths and weaknesses of each model and the aspects of the system each are most ideally suited to investigate.
TABLE 4.
Nanoparticles, cargo molecules, and methods used to study drug loading and release.
Studies of itraconazole in a liposome, combining MD simulation with experiment, provides an example of where MD simulation was able to provide concrete insight not obtainable experimentally. Itraconazole is an antifungal drug characterized by low solubility, which limits its bioavailability. One possible solution to overcome low solubility is incorporating drugs into liposomes, which was achieved in a few independent studies. To optimize the liposome properties, cholesterol is frequently used as a molecule known to increase the stability of lipid bilayers (Róg and Vattulainen, 2014). In fact, cholesterol is used in 9 out of 15 liposome-based formulations approved for clinical use (Bulbake et al., 2017). Thus, the incorporation of cholesterol into the liposome-itraconazole formulation was the next step. MD simulations showed that this is not the right choice because cholesterol and itraconazole do not mix well and separate into small domains (Poojari et al., 2020). This observation was next validated in experimental studies, which showed decreased affinity of itraconazole toward liposomes containing cholesterol (Poojari et al., 2020). The observed behavior of the itraconazole may be explained by its structure: it is a long rigid molecule with a few weakly polar groups distributed along the molecule backbone. Due to this structure, itraconazole molecules locate to the water membrane interface oriented parallel to the membrane surface and, in turn, orient other molecules in the same fashion (Poojari et al., 2019). On the other hand, cholesterol has strong preferences to adopt an orientation perpendicular to the membrane surface and affect the orientation of neighboring molecules; these opposite preferences lead to the observed demixing of drugs and cholesterol.
Nanoparticle Interaction With the Lipid Membrane
Once the nanoparticle has reached the cell, surviving the journey through the bloodstream with its payload still contained and intact, there is one final barrier that must be crossed for the drug delivery system to be efficacious: the cell membrane (Smith et al., 2018). It is possible for nanoparticles, particularly if they are hydrophobic, to directly transfect, also referred to as translocation, through the cell membrane and many nanoparticles enter the cell through this route. There is, however, an alternative: the nanoparticle can be designed to cross the membrane via receptor mediated endocytosis (Gao et al., 2005). When a nanoparticle is taken up via endocytosis it is possible to take advantage of pH triggered release due to the lowered pH environment on the interior of the endosome (Hu et al., 2015). Valuable insight in both the context of direct membrane transfection and endocytosis have been obtained through MD simulation. As is the case with drug loading and release, MD simulation of nanoparticle-lipid membrane interactions have been carried out for different nanoparticle forms, including carbon dots, graphene, dendrimers, gold nanoparticles (shown in Table 5), and nanocrystals (Song et al., 2011); examples can be found of all levels of model resolution being used including atomistic MD, MARTINI model, DPD, and implicit solvent (Table 5). An overview of MD simulation of nanoparticle – lipid membrane interactions has been written by Tian et al. (2014a).
TABLE 5.
Nanoparticles and methods used to study theirs interactions with membranes.
| Nanoparticles: |
| Carbon Dots (Erimban and Daschakraborty, 2020) |
| Graphene (Raczyński et al., 2020), dendrimers (Lee and Larson, 2008; Kanchi et al., 2018; He et al., 2020) |
| Gold Nanoparticles (Lin et al., 2011; Gkeka et al., 2014; Mhashal and Roy, 2014; Mhasal and Roy, 2016; Oroskar et al., 2016; Quan et al., 2017; Das et al., 2019) |
| Nanocrystals (Song et al., 2011) |
| Methods: |
| Atomistic MD (Mhashal and Roy, 2014; Van Lehn and Alexander-Katz, 2014b; Mhasal and Roy, 2016; Erimban and Daschakraborty, 2020) |
| MARTINI model (Lin X. et al., 2010; Lin X. et al., 2020; Song et al., 2011, 2012; Lin and Gu, 2014; Oroskar et al., 2015; Shimizu et al., 2016; Quan et al., 2017; Su et al., 2017; Zhang Z. et al., 2017; Bai et al., 2018; Das et al., 2019; Salassi et al., 2019; He et al., 2020) |
| DPD (Lee and Larson, 2008; Yang and Ma, 2010; Ding and Ma, 2012, 2014a; Ding et al., 2012; Tian et al., 2014b; Liu et al., 2016; Bai et al., 2018), |
| Implicit Solvent (Vácha et al., 2011; Schubertová et al., 2015) |
For nanoparticles that enter the cell through direct transfection, the issue is the direct physical reaction between the nanoparticle and the membrane; this phenomenon can be studied directly through an MD simulation of the nanoparticle interacting with the membrane (Yang and Ma, 2010; Ding et al., 2012; Liang, 2013; Van Lehn and Alexander-Katz, 2014b; Shimizu et al., 2016; Zhang Z. et al., 2017; Erimban and Daschakraborty, 2020; Gupta et al., 2020). When nanoparticles translocate through the membrane, the membrane structure can be disrupted and leakage and even pore formation can occur; this has been studied directly using MD (Song et al., 2012; Mhashal and Roy, 2014; Van Lehn and Alexander-Katz, 2014b; Oroskar et al., 2015; Ding and Ma, 2018). The effect of size (Lin X. et al., 2010), shape (Li Y. et al., 2012; Liu et al., 2016; Yang Y. et al., 2019), and surface properties (Ding and Ma, 2016) of the nanoparticle on membrane transfection has also been studied, including effect of PEGylation (Oroskar et al., 2016; Bai et al., 2018), and other polymer coatings (Liang, 2013; Xia et al., 2020) as well as protein (Ding and Ma, 2014a) and, for the study of inhaled nanoparticles, pulmonary surfactant corona (Bai et al., 2018) and other issues related to translocation across the pulmonary surfactant monolayer (Chen P. et al., 2018). Additionally, Gupta et al. used MD simulations to study transdermal delivery of interferon-alpha using gold nanoparticles (Gupta et al., 2018).
Regarding receptor mediated endocytosis, the interaction is more complex; while direct simulation of nanoparticle endocytosis has been performed and gained important insight (Vácha et al., 2011; Ding and Ma, 2012; Li et al., 2014a) this only tells part of the story as many aspects of the specific interaction between the ligand and the receptors are not elucidated by such a simulation. Nanoparticles can be designed to actively target specific cell types through functionalizing targeting ligands onto the nanoparticle surface. These targeting ligands bind to the specific receptors that induce endocytosis. There are two issues that govern the efficacy of this binding: (1) the distribution of the targeting ligands on the surface, i.e., the pattern of where they are located and (2) the orientation and, as a result of orientation, extent of exposure at the nanoparticle surface and thus availability to the target receptors. The effect of ligand distribution has been studied by Liu et al. (2016) through direct MD simulation of nanoparticle-membrane interactions and ligand density has been studied through a different computational modeling technique: Monte Carlo simulation (Martinez-Veracoechea and Frenkel, 2011; Wang and Dormidontova, 2012; Angioletti-Uberti, 2017a).
Regarding the orientation, and thus exposure, of the targeting ligand to the receptor that induces receptor mediated endocytosis, one needs chemically accurate atomistic simulations of the nanoparticle surface to investigate the degree to which the targeting ligand is exposed and available to the receptor. We have performed such simulations for the case of liposome based delivery systems, with targeting ligands, in several previous publications, for example our determination of the cause of failure of the new AETP moiety (Lehtinen et al., 2012). These involved simulating a section of the liposome membrane with the targeting ligands and, in some cases, the protective polymer corona as well. Our study regarding the AETP moiety was another example of a specific topic where we were able to obtain concrete insight, not obtainable experimentally. The AETP moiety was found to be successful, when its binding affinity for the target receptor was tested experimentally, however, when functionalized to a PEGylated liposome the targeting moiety failed to show any effect. In comparison to the more hydrophilic RGD peptide, that has been shown to be an effective targeting moiety for a PEGylated liposome, the AETP moiety is more hydrophobic; it could be hypothesized, from the experiment alone, that the moiety is obscured within the membrane core; our MD simulation, however, showed this not to be the case: it was rather the PEG corona itself that was obscuring the AETP moiety; as PEG is soluble in both polar and non-polar solvents it was thus a more comfortable, i.e., less hydrophilic, environment for the AETP moiety than the polar solvent (Lehtinen et al., 2012). Since it was the PEG corona itself that was the culprit we could propose a solution: replace PEG with a more hydrophilic polymer that has been approved for internal use. Just such a polymer exists: Poly-methoxazoline (PMOZ); in a subsequent study we performed a simulation with the PEG corona replaced by a PMOZ corona and we saw increased exposure of the AETP moiety (Magarkar et al., 2017). We have also studied liposomes functionalized with stearylamine arginine ligands (Pathak et al., 2016). A comprehensive review of the theoretical and computational investigation of nanoparticle interactions with biomembranes has been written by Ding and Ma (2014b).
Conclusion
In this review, we have attempted to summarize the role that molecular dynamics modeling can play as a tool in drug delivery research in a fashion that is hopefully comprehensible to both those with an expertise in molecular modeling who wish to pursue pharmaceutical applications of their research and pharmaceutical researchers interested in what insight this new tool can provide. All aspects of the journey that the drug molecule takes, from dissolution to solvation or transit through the bloodstream inside a nanoparticle, to finally crossing the plasma membrane of the target cell, is a story of molecular interactions. The interactions involved, however, are not all interactions. Any chemist reading this review will have noticed an omission: chemical reactions; these play a very small role in the story, one dominated by intermolecular interactions. For this reason, MD simulation is the perfect tool to obtain molecular level insight as precisely the variety of interaction it is best able to study are those which play the dominant role: formation of structure based on lipophilicity and H-bonding. Whether it is the hydration that occurs during dissolution, interaction between drug molecule and excipients, behavior of molecules at the surface of the nanoparticle in the bloodstream, or the interaction between the nanoparticle and the plasma membrane of the target cell, these are the interactions that determine the most important aspects of behavior.
Molecular dynamics modeling is still a new tool in the design of drug delivery mechanisms; only in the past decade have we seen the explosion in the number of publications that make use of this tool. Widespread adoption is hindered by the fact that, unlike computational drug design tools like ligand docking and QSAR/QSPR, the calculations involved are, as of yet, for the most part too expensive to be carried out anywhere other than national level supercomputing resources. As the widely available computational power continues to grow exponentially, this barrier may dissipate. Looking toward the future and the role that molecular dynamics modeling will be able to play in the development of drug delivery systems, the analogy that we feel is most apt is that of computationally assisted design (CAD) (Narayan et al., 2008), in mechanical and civil engineering. Before the advent of computational technology, engineers were forced to build scale models of systems and experiment with them, testing every aspect with real experimental models and sometimes varying parameters empirically. Now, with widespread computational resources available to all engineers, CAD allows every aspect of a new machine, or structure, to be examined and tested in silico with all aspects of mechanical stress, heat dissipation etc. of the system visible, and the change resulting from any design alteration straightforward to analyze entirely virtually. While we clearly do not mean to imply that human physiology is no more complex than designing a car or a bridge, we foresee that, in the future, drug delivery devices will be designed in an analogous fashion, with molecular dynamics modeling playing the role in pharmaceutics that CAD plays in mechanical and civil engineering. Our studies of the AETP targeting moiety and itraconazole in liposome based delivery systems show clear examples of how the design approach can be applied, using in silico modeling to test aspects of the delivery system design in an analogous approach to CAD. Alongside cutting edge experimental techniques that complement it, molecular dynamics simulation has the potential to lead the way to a new era of rational design in the development of drug delivery systems. Finally, other complementary reviews that cover similar material can also be found (Thota and Jiang, 2015; Ramezanpour et al., 2016; Thewalt and Tieleman, 2016; Katiyar and Jha, 2018; Sen et al., 2018; Shamsi et al., 2019).
Author Contributions
AB and TR wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abbott A. P., Ahmed E. I., Prasad K., Qader I. B., Ryder K. S. (2017). Liquid pharmaceuticals formulation by eutectic formation. Fluid Phase Equilib. 448 2–8. 10.1016/j.fluid.2017.05.009 [DOI] [Google Scholar]
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Abuchowski A., Van Es T., Palczuk N. C., Davis F. F. (1977). Alteration of immunological properties of bovine serum albumin by covalent attachment of polyethylene glycol. J. Biol. Chem. 252 3578–3581. [PubMed] [Google Scholar]
- Acharya C., Coop A., Polli J. E., Mackerell A. D., Jr. (2011). Recent advances in ligand-based drug design: relevance and utility of the conformationally samples pharmacophore approach. Curr. Comput. Drug Des. 7 10–22. 10.2174/157340911793743547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnan A., Lam R., Chen H., Lee J., Schaffer D. J., Barnard A. S., et al. (2011). Atomistic simulation and measurement of pH dependent cancer therapeutic interactions with nanodiamond carrier. Mol. Pharm. 8 368–374. 10.1021/mp1002398 [DOI] [PubMed] [Google Scholar]
- Aguayo-Ortiz R., Dominguez L. (2019). APH-1A component of γ-secretase forms an internal water and ion-containing cavity. ACS Chem. Neurosci. 10 2931–2938. 10.1021/acschemneuro.9b00150 [DOI] [PubMed] [Google Scholar]
- Ahmad S., Johnston B. F., Mackay S. P., Schatzlein A. G., Gellert P., Sengupta D., et al. (2010). In silico modelling for drug-polymer interactions for pharmaceutical formulations. J. R. Soc. Interf. 7 S423–S433. 10.1098/rsif.2010.0190.focus [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibani N., Khan T. N., Callan B. (2020). Liposome mimicking polymersomes; a comparative study of the merits of polymersomes in terms of formulation and stability. Int. J. Pharm. X 2:100040. 10.1016/j.ijpx.2019.100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano J. M. R., Grillo D., Facelli J. C., Ferraro M. B., Pickholz M. (2019). Study of the lammellar and micellar phases of pluronic F127: a molecular dynamics approach. Processes 7:606 10.3390/pr7090606 [DOI] [Google Scholar]
- Alinejad A., Raissi H., Hashemzadeh H. (2020). Understanding co-loading of doxorubicin and camptothecin on graphene and folic acid-conjugated graphene for targeting drug delivery: classical MD simulation and DFT calculation. J. Biomol. Struct. Dyn. 38 2737–2745. 10.1080/07391102.2019.1645044 [DOI] [PubMed] [Google Scholar]
- Allen M. P., Tildesley D. J. (1989). Computer Simulation of Liquids. New York, NY: Clarendon Press. [Google Scholar]
- Alonso H., Bliznyuk A. A., Gready J. E. (2006). Combining docking and molecular dynamics simulations in drug design. Med. Res. Rev. 26 531–568. 10.1002/med.20067 [DOI] [PubMed] [Google Scholar]
- Alves V. M., Hwang D., Muratov E., Sokolsky-Papkov M., Varlamova E., Vinod N., et al. (2019). Cheminformatics-driven discovery of polymeric micelle formulations for poorly soluble drugs. Sci. Adv. 5:eaav9784. 10.1126/sciadv.aav9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio E., Podmore A., Gomes Dos Santos A. L., Magarkar A., Bunker A., Caliceti P., et al. (2018). Control of peptide aggregation and fibrillation by physical PEGylation. Biomacromolecules 19 3958–3969. 10.1021/acs.biomac.8b00887 [DOI] [PubMed] [Google Scholar]
- Angioletti-Uberti S. (2017a). Exploiting receptor competition to enhance nanoparticle binding selectivity. Phys. Rev. Lett. 118:68001. 10.1103/PhysRevLett.118.068001 [DOI] [PubMed] [Google Scholar]
- Angioletti-Uberti S. (2017b). Theory simulations and design of functionalized nanoparticles for biomedical applications: a soft matter perspective. NPJ Comput. Mater. 3:48 10.1038/s41524-017-0050-y [DOI] [Google Scholar]
- Ansari M., Moradi S., Shahlaei M. (2018). A molecular dynamics simulation study on the mechanism of loading of gemcitabine and camptothecin in poly lactic-co-glycolic acid as a nano drug delivery system. J. Mol. Liq. 269 110–118. 10.1016/j.molliq.2018.08.032 [DOI] [Google Scholar]
- Antimisiaris S. G., Mourtas S., Marazioti A. (2018). Exosomes and exosome-driven vesicles for targeted drug delivery. Pharmaceutics 10:218. 10.3390/pharmaceutics10040218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Lara L., Morales-Avila E., Luna-Gutiérrez M. A., Olivé-Alvarez E., Isaac-Olivé K. (2020). Radiolabeled liposomes and lipoproteins as lipidic nanoparticles for imaging and therapy. Chem. Phys. Lipids 230:104934. 10.1016/j.chemphyslip.2020.104934 [DOI] [PubMed] [Google Scholar]
- Aranha M. P., Mukherjee D., Petridis L., Khomami B. (2020). An atomistic molecular dynamics study of titanium dioxide adhesion to lipid bilayers. Langmuir 36 1043–1052. 10.1021/acs.langmuir.9b03075 [DOI] [PubMed] [Google Scholar]
- Arleth L., Ashok B., Onyuksel H., Thiyagarajan P., Jacob J., Hjelm R. P. (2005). Detailed structure of hairy mixed micelles formed by phosphatidylcholine and PEGylated phospholipids in aqueous media. Langmuir 21 3279–3290. 10.1021/la047588y [DOI] [PubMed] [Google Scholar]
- Arnarez C., Uusitalo J. J., Masman M. F., Ingólfsson H. I., De Jong D. H., Melo M. N., et al. (2015). Dry martini, a coarse-grained force field for lipid membrane simulations with implicit solvent. J. Chem. Theory Comput. 11 260–275. 10.1021/ct500477k [DOI] [PubMed] [Google Scholar]
- Asadzadeh H., Moosavi A., Arghavani J. H. (2020). The effect of chitosan and PEG polymers on stabilization of GF-17 structure: a molecular dynamics study. Carbohydr. Polym. 237:116124. 10.1016/j.carbpol.2020.116124 [DOI] [PubMed] [Google Scholar]
- Augustyn B., Stepien P., Poojari C., Mobarak E., Polit A., Wisniewska-Becker A., et al. (2019). Cholesteryl hemisuccinate is not a good replacement for cholesterol in lipid nanodiscs. J. Phys. Chem. B 123 9839–9845. 10.1021/acs.jpcb.9b07853 [DOI] [PubMed] [Google Scholar]
- Aziz Z. A. B. A., Ahmad A., Mohd-Setapar S. H., Hassan H., Lokhat D., Kamal M. A., et al. (2017). Recent advances in drug delivery of polymeric nano-micelles. Curr. Drug Metab. 18 16–29. 10.2174/1389200217666160921143616 [DOI] [PubMed] [Google Scholar]
- Badalkhani-Khamseh F., Ebrahim-Habibi A., Hadipour N. L. (2017). Atomistic computer simulations on multi-loaded PAMAM denrimers: a comparison of amine- and hydroxyl-terminated dendrimers. J. Comput. Aid. Mol. Des. 31 1097–1111. 10.1007/s10822-017-0091-9 [DOI] [PubMed] [Google Scholar]
- Badalkhani-Khamseh F., Ebrahim-Habibi A., Hadipour N. L. (2019). Influence of dendrimer surface chemistry and pH on the binding and release pattern of chalcone studied by molecular dynamics simulations. J. Mol. Recognit. 32:e2757. 10.1002/jmr.2757 [DOI] [PubMed] [Google Scholar]
- Bai X., Xu M., Liu S., Hu G. (2018). Computational investigations of the interaction between the cell membrane and nanoparticles coated with pulmonary surfactant. ACS Appl. Mater. Interf. 10 20368–20376. 10.1021/acsami.8b06764 [DOI] [PubMed] [Google Scholar]
- Banerjee A., Qi J., Gogoi R., Wong J., Mitragorti S. (2016). Role of nanoparticle size, shape and surface chemistry in oral drug delivery. J. Control. Release 238 176–185. 10.1016/j.jconrel.2016.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannan C. C., Calabró G., Kyu D. Y., Mobley D. L. (2016). Calculating partition coefficients of small molecules in octanol/water and cyclohexane/water. J. Chem. Theory Comput. 12 4015–4024. 10.1021/acs.jctc.6b00449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard A. S. (2016). Challenges in modelling nanoparticles for drug delivery. J. Phys. Condens. Matter 28:23002 10.1088/0953-8984/28/2/023002 [DOI] [PubMed] [Google Scholar]
- Barraza L. F., Jiménez V. A., Alderete J. B. (2015). Effect of PEGylation on the structure and drug loading capacity of PAMAM-G4 dendrimers: a molecular modelling approach on the complexation of 5-flourouracil with native and PEGylated PAMAM-G4. Macromol. Chem. Phys. 216 1689–1701. 10.1002/macp.201500179 [DOI] [Google Scholar]
- Bazak R., Houri M., Achy S. E., Kamel S., Refaat T. (2015). Cancer active targeting by nanoparticles: a comprehensive review of literature. J. Cancer res. Clin. Oncol. 141 769–784. 10.1007/s00432-014-1767-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfo J. P., Lemos J. M. (eds). (2021). “Pharmacokinetic and pharmacodynamic models,” in Optimal Impulsive Control for Cancer Therapy, (Cham: Springer; ), 11–18. 10.1007/978-3-030-50488-5_2 [DOI] [Google Scholar]
- Beloqui A., Solinís M. Á, Rodríguez-Gascón A., Almeida A. J., Préat V. (2016). Nanostructured lipid carriers: promising drug delivery systems for future clinics. Nanomed Nanotechnol. 12 143–161. 10.1016/j.nano.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Belubbi T., Shevade S., Dhawan V., Sridhar V., Majumdar A., Nunes R., et al. (2018). Lipid architectonics for superior oral bioavailability of nelfinavir mesylate: comparative it in vitro and in vivo assessment. AAPS Pharm. Sci. Tech. 19 3584–3598. 10.1208/s12249-018-1156-3 [DOI] [PubMed] [Google Scholar]
- Bengtsen T., Holm V. L., Kjølbye L. R., Midtgaard S. R., Johansen N. T., Tesei G., et al. (2020). Structure and dynamics of a nanodisc by integrating NMR, SAXS and SANS experiments with molecular dynamics simulations. eLife 9:e56518. 10.7554/elife.56518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennun S. V., Hoopes M. I., Xing C., Faller R. (2009). Coarse-grained modeling of lipids. Chem. Phys. Lipids 159 59–66. 10.1016/j.chemphyslip.2009.03.003 [DOI] [PubMed] [Google Scholar]
- Benson S. P., Pleiss J. (2014). Molecular dynamics simulations of self-emulsifying drug-delivery systems (SEDDS): influence of excipients on droplet nanostructure and drug localization. Langmuir 30 8471–8480. 10.1021/la501143z [DOI] [PubMed] [Google Scholar]
- Bergström C. A. S., Larsson P. (2018). Computational prediction of drug solubility in water based systems: qualitative and quantitative approaches used in the current drug discovery and development settings. Int. J. Pharm. 540 185–193. 10.1016/j.ijpharm.2018.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernkop-Schnürch A., Dünnhaupt S. (2012). Chitosan-based drug delivery systems. Eur. J. Pharm. Biopharm. 81 463–469. 10.1016/j.ejpb.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Berrecoso G., Crecente-Campo J., Alonso M. J. (2020). Unveiling the pitfalls of the protein corona of polymeric drug nanocarriers. Drug Deliv. Transl. Res. 10 730–750. 10.1007/s13346-020-00745-0 [DOI] [PubMed] [Google Scholar]
- Bhardwaj P., Tripathi P., Gupta R., Panday S. (2020). Niosomes: a review on niosomal research in the last decade. J. Drug Deliv. Sci. Tech. 56:101581 10.1016/j.jddst.2020.101581 [DOI] [Google Scholar]
- Binder U., Skerra A. (2017). PASylation: a versatile technology to extend drug delivery. Curr. Opin. Colloid Interf. Sci. 31 10–17. 10.1016/j.cocis.2017.06.004 [DOI] [Google Scholar]
- Blanco E., Shen H., Ferrari M. (2015). Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 33 941–951. 10.1038/nbt.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazhynska M. M., Kyrychenko A., Kalugin O. N. (2018). Molecular dynamics simulation of the size-dependent morphological stability of cubic shape silver nanoparticles. Mol. Simulat. 44 981–991. 10.1080/08927022.2018.1469751 [DOI] [Google Scholar]
- Bobo D., Robinson K. J., Islam J., Thurecht K. J., Corrie S. R. (2016). Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm. Res. 33 2373–2387. 10.1007/s11095-016-1958-5 [DOI] [PubMed] [Google Scholar]
- Bohacek R. S., McMartin C., Guida W. C. (1996). The art and practice of structure-based drug design: a molecular modeling perspective. Med. Res. Rev. 16 3–50. [DOI] [PubMed] [Google Scholar]
- Bolintineanu D. S., Lane J. M. D., Grest G. S. (2014). Effects of functional groups and ionization on the structure of alkanethiol-coated gold nanoparticles. Langmuir 30 11075–11085. 10.1021/la502795z [DOI] [PubMed] [Google Scholar]
- Bolla P. K., Meraz C. A., Rodriguez V. A., Deaguero I., Singh M., Yellepeddi V. K., et al. (2019). Clotrimazole loaded ufosomes for topical delivery: formulation development and in-vitro studies. Molecules 24:3139. 10.3390/molecules24173139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botan A., Favela-Rosales F., Fuchs P. F. J., Javanainen M., Kanduè M., Kulig W., et al. (2015). Toward atomistic resolution structure of phosphatidylcholine headgroup and glycerol backbone at different ambient conditions. J. Phys. Chem. B 119 15075–15088. 10.1021/acs.jpcb.5b04878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzo B. L., Calvelo M., Martín-Pastor M., García-Fandiño R., De La Fuente M. (2020). In vitro- in silico modeling approach to rationally designed simple and versatile drug delivery systems. J. Phys. Chem. B 124 5788–5800. 10.1021/acs.jpcb.0c02731 [DOI] [PubMed] [Google Scholar]
- Bradley A. J., Devine D. V., Ansell S. M., Janzen J., Brooks D. E. (1998). Inhibition of liposome-induced complement activation by incorporated poly(ethylene glycol)-lipids. Arch. Biochem. Biophys. 357 185–194. 10.1006/abbi.1998.0798 [DOI] [PubMed] [Google Scholar]
- Brancolini G., Tozzini V. (2019). Multiscale modeling of proteins interaction with functionalized nanoparticles. Curr. Opin. Colloid Interf. Sci. 41 66–73. 10.1016/j.cocis.2018.12.001 [DOI] [Google Scholar]
- Bricarello D. A., Smilowitz J. T., Zivkovic A. M., German J. B., Parikh A. N. (2011). Reconstituted lipoprotein: a versatile nanostructures. ACS Nano 5 42–57. 10.1021/nn103098m [DOI] [PubMed] [Google Scholar]
- Brooks B. R., Brooks C. L., III, Mackerell A. D., Nilsson L., Petrella R. J., Roux B., et al. (2009). CHARMM: the biomolecular simulation program B. J. Comput. Chem. 30 1545–1614. 10.1002/jcc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulbake U., Doppalapudi S., Kommineni N., Khan W. (2017). Liposomal formulations in clinical use: an updated review. Pharmaceutics 9:12. 10.3390/pharmaceutics9020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker A. (2012). Poly(ethylene glycol) in drug delivery, why does it work, and can we do better? All atom molecular dynamics simulation provides some answers. Phys. Proc. 34 24–33. 10.1016/j.phpro.2012.05.004 [DOI] [Google Scholar]
- Bunker A. (2015). “Molecular modeling as a tool to understand the role of Poly(Ethylene) glycol in drug delivery,” in Computational pharmaceutics: Applications of Molecular modelling in Drug Delivery, eds Ouyang D., Smith S. C. (Hoboken, NJ: John Wiley & Sons; ), 217–234. 10.1002/9781118573983.ch11 [DOI] [Google Scholar]
- Bunker A., Magarkar A., Viitala T. (2016). Rational design of liposomal drug delivery systems, a review: combined experimental and computational studies of lipid membranes, liposomes and their PEGylation. Biochim. Biophys. Acta Biomembr. 1858 2334–2352. 10.1016/j.bbamem.2016.02.025 [DOI] [PubMed] [Google Scholar]
- BurduŞel A.-C., Gherasim O., Grumezescu A. M., Mogoantã L., Ficai A., Andronescu E. (2018). Biomedical applications of silver nanoparticles: an up-to-date overview. Nanomaterials 8:681. 10.3390/nano8090681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G., Laio A. (2020). Using metadynamics to explore complex free-energy landscapes. Nat. Rev. Phys. 2 200–212. 10.1038/s42254-020-0153-0 [DOI] [Google Scholar]
- Buxton G. A. (2014). Simulating the co-encapsulation of drugs in a “Smart” core-shell-shell polymer nanoparticle. Eur. Phys. J. E 37:14. 10.1140/epje/i2014-14014-5 [DOI] [PubMed] [Google Scholar]
- Cagel M., Tesan F. C., Bernabeu E., Salgueiro M. J., Zubillaga M. B., Moretton M. A., et al. (2017). Polymeric mixed micelles as nanomedicines: achievements and perspectives. Eur. J. Pharm. Biopharm. 113 211–228. 10.1016/j.ejpb.2016.12.019 [DOI] [PubMed] [Google Scholar]
- Caracciolo G. (2015). Liposome-protein corona in a physiological environment: challenges and opportunities for targeted delivery of nanomedicines. Nanomed. Nanotechnol. Biol. Med. 11 543–557. 10.1016/j.nano.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Casalini T., Limongelli V., Schmutz M., Som C., Jordan O., Wick P., et al. (2019). Molecular modeling for nanomaterial-biology interactions: opportunities, challenges, and perspectives. Front. Bioeng. Biotech. 7:268. 10.3389/fbioe.2019.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Vanni S. (2016). Toward accurate coarse-graining approaches for protein and membrane simulations. Chem. Model. 12 1–52. 10.1039/9781782622703-00001 [DOI] [Google Scholar]
- Case D. A., Cheatham T. E., III, Darden T., Gohlke H., Luo R., Merz K. M., et al. (2005). The Amber biomolecular simulation programs. J. Comput. Chem. 26 1668–1688. 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cern A., Barenholz Y., Tropsha A., Goldblum A. (2014). Computer-aided design of liposomal drugs: in silico prediction and experimental validation of drug candidates for liposomal remote loading. J. Control. Release 173 125–131. 10.1016/j.jconrel.2013.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T., Ouyang D. (2018). Investigating the molecular dissolution process of binary solid dispersions by molecular dynamics simulations. Asian J. Pharm. Sci. 13 248–254. 10.1016/j.ajps.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-Y., Ju S.-P., Wang L.-F., Chen C.-C., Chuang Y.-C., Wu H.-L., et al. (2017). Investigation of the self-assembly of CS and PCL copolymers with different molecular weights in water solution by coarse-grained molecular dynamics simulation. J. Mol. Model. 23:151. 10.1007/s00894-017-3294-z [DOI] [PubMed] [Google Scholar]
- Charchar P., Christofferson A. J., Todorova N., Yarovsky I. (2016). Understanding and designing the gold-bio interface: insights from simulations. Small 12 2395–2418. 10.1002/smll.201503585 [DOI] [PubMed] [Google Scholar]
- Chen G., Wang Y., Xie R., Gong S. (2018). A review on core-shell structured unimolecular nanoparticles for biomedical applications. Adv. Drug Deliv. Rev. 130 58–72. 10.1016/j.addr.2018.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Zhang Z., Gu N., Ji M. (2018). Effect of the surface charge density of nanoparticles on their translocation across pulmonary surfactant monolayer: a molecular dynamics simulation. Mol. Simulat. 44 85–93. 10.1080/08927022.2017.1342118 [DOI] [Google Scholar]
- Chen J., Wang J., Yin B., Pang L., Wang W., Zhu W. (2019). Molecular mechanism of binding selectivity of inhibitors toward BACE1 and BACE2 revealed by multiple short molecular dynamics simulations and free-energy predictions. ACS Chem. Neurosci. 10 4303–4318. 10.1021/acschemneuro.9b00348 [DOI] [PubMed] [Google Scholar]
- Chen S., Hanning S., Falconer J., Locke M., Wen J. (2019). Recent advances in non-ionic surfactant vesicles (niosomes): fabrication, characterization, pharmaceutical and cosmetic applications. Eur. J. Pharm. Biopharm. 144 18–39. 10.1016/j.ejpb.2019.08.015 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang X., Millican R., Creutzmann J. E., Martin S., Jun H.-W. (2020). High density lipoprotein mimicking nanoparticles for atherosclerosis. Nano Converg. 7:6. 10.1186/s40580-019-0214-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Guan G., Deng F., Yang D., Wu P., Kang S., et al. (2020). Anisotropic active ligandations in siRN-loaded hybrid nanodisks lead to distinct carcinostatic outcomes by regulating nano-bio interactions. Biomaterials 251:120008. 10.1016/j.biomaterials.2020.120008 [DOI] [PubMed] [Google Scholar]
- Chen M., Pierstorff E. D., Lam R., Li S.-Y., Huang H., Osawa E., et al. (2009). Nanodiamond-mediated delivery of water-insoluble therapeutics. ACS Nano 3 2016–2022. 10.1021/nn900480m [DOI] [PubMed] [Google Scholar]
- Chen W., Ouyang D. (2017). Investigation of molecular dissolution mechanism of ketoprofen binary and ternary solid dispersions by molecular dynamics simulations. Mol. Simulat. 43 13–16. 10.1080/08927022.2017.1321755 [DOI] [Google Scholar]
- Chow A., Brown B. D., Merad M. (2011). Studying the mononuclear phagocyte system in the molecular age. Nat. Rev. Immunol. 11 788–798. 10.1038/nri3087 [DOI] [PubMed] [Google Scholar]
- Chuang S. T., Cruz S., Narayanaswami V. (2020). Reconfiguring nature’s cholesterol accepting lipoproteins as nanoparticle platforms for transport and delivery of therapeutic and imaging agents. Nanomaterials 10:906. 10.3390/nano10050906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun B. J., Choi J. I., Jang S. S. (2015). Molecular dynamics simulation of sodium dodecyl sulfate micelle: water penetration and sodium dodecyl sulfate dissociation. Colloid Surf. A 474 36–43. 10.1021/jp013616z [DOI] [Google Scholar]
- Chung I.-M., Rajakumar G., Venkidasamy B., Subramanian U., Thiruvengadam M. (2020). Exosomes: current use and future applications. Clin. Chim. Acta 500 226–232. 10.1016/j.cca.2019.10.022 [DOI] [PubMed] [Google Scholar]
- Cohan R. A., Madadkar-Sobhani A., Khanahmad H., Roohvand F., Aghasadeghi M. R., Hedayati M. H., et al. (2011). Design, modeling, expression, and chemoselective PEGylation of a new nanosize cysteine analog of erythropoietin. Int. J. Nanomed. 6 1217–1227. 10.2147/IJN.S19081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly M. L. (1983). Solvent-accessible surfaces of proteins and nucleic acids. Science 221 709–714. 10.1126/science.6879170 [DOI] [PubMed] [Google Scholar]
- Contreras M. L., Torres C., Villarroel I., Rozas R. (2019). Molecular dynamics assessment of doxorubicin-carbon nanotubes molecular interactions for the design of drug delivery systems. Struct. Chem. 30 369–384. 10.1007/s11224-018-1210-5 [DOI] [Google Scholar]
- Copland M. J., Rades T., Davies N. M., Baird M. A. (2005). Lipid based particulate formulations for delivery of antigen. Immunol. Cell Biol. 83 97–105. 10.1111/j.1440-1711.2005.01315.x [DOI] [PubMed] [Google Scholar]
- Craig W. A. (1998). Pharmacokinetics/pharmacodynamics parameters: rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 26 1–12. 10.1086/516284 [DOI] [PubMed] [Google Scholar]
- Cramariuc O., Rogl T., Vattulainen I. (2012). Drug-lipid membrane interaction mechanisms revealed through molecular simulations. Curr. Phys. Chem. 2 379–400. 10.2174/1877946811202040379 [DOI] [Google Scholar]
- Cramer C. J. (2002). Essentials of Computational Chemistry. Hoboken, NJ: John Wiley & Sons Ltd. [Google Scholar]
- Crommelin D. J. A., van Hoogevest P., Storm G. (2020). The role of liposomes in clinical nanomedicine development. What now? Now what? J. Control. Release 318 256–263. 10.1016/j.jconrel.2019.12.023 [DOI] [PubMed] [Google Scholar]
- Csongradi C., du Plessis J., Aucamp M. E., Gerber M. (2017). Topical delivery of roxithromycin solid-state forms entrapped in vesicles. Eur. J. Pharm. Biopharm. 114 96–107. 10.1016/j.ejpb.2017.01.006 [DOI] [PubMed] [Google Scholar]
- da Silva Costa A. C., Carvalho S. C., de Farias Silva N., Do Nascimento-Júnior A. E. S., Cruz J. N., Maia de Jesus Chaves Neto A. (2020). Effect of chitosan/albendazole nanocarriers’ solvation by molecular dynamics. Theor. Chem. Acc. 139:105 10.1007/s00214-020-02620-0 [DOI] [Google Scholar]
- Damiati S., Scheberl A., Zayni S., Damiati S. A., Schuster B., Kompella U. B. (2019). Albumin-bound nanodisks as delivery vehicle candidates: development and characterization. Biophys. Chem. 251:106178. 10.1016/j.bpc.2019.106178 [DOI] [PubMed] [Google Scholar]
- Danhier F. (2016). To exploit the tumor microenvironment: since the EPR effect fails in the clinic, what is the future of nanomedicine? J. Control. Release 244 108–121. 10.1016/j.jconrel.2016.11.015 [DOI] [PubMed] [Google Scholar]
- Das M., Dahal U., Mesele O., Liang D., Cui Q. (2019). Molecular dynamics simulation of interaction between functionalized nanoparticles with lipid membranes: analysis of coarse-grained models. J. Phys. Chem. B 123 10547–10561. 10.1021/acs.jpcb.9b08259 [DOI] [PubMed] [Google Scholar]
- Das T., Mehta C. H., Nayak U. Y. (2020). Multiple approaches for achieving drug solubility: an it in silico perspective. Drug Discov. Today 25 1206–1212. 10.1016/j.drudis.2020.04.016 [DOI] [PubMed] [Google Scholar]
- Dasari S., Mallik B. S. (2020). Solubility and solvation free energy of a cardiovascular drug, LASSBio-294, in ionic liquids: a computational study. J. Mol. Liq. 301:112449 10.1016/j.molliq.2020.112449 [DOI] [Google Scholar]
- Date T., Nimbalkar V., Kamat J., Mittal A., Mahato R. I., Chitkara D. (2018). Lipid-polymer hybrid nanocarriers for delivering cancer therapeutics. J. Control. Release 271 60–73. 10.1016/j.jconrel.2017.12.016 [DOI] [PubMed] [Google Scholar]
- De Nicola A., Hezaveh S., Zhao Y., Kawakatsu T., Roccatano D., Milano G. (2014). Micellar drug nanocarriers and biomembranes: how do they interact? Phys. Chem. Chem. Phys. 16 5093–5105. 10.1039/c3cp54242d [DOI] [PubMed] [Google Scholar]
- Debnath A., Schäfer L. V. (2015). Structure and dynamics of phospholipid nanodiscs from all-atom and coarse-grained simulations. J. Phys. Chem. B 119 6991–7002. 10.1021/acs.jpcb.5b02101 [DOI] [PubMed] [Google Scholar]
- Decuzzi P., Lee S., Bhushan B., Ferrari M. (2005). A theoretical model for the margination of particles within blood vessels. Ann. Biomed. Eng. 33 179–190. 10.1007/s10439-005-8976-5 [DOI] [PubMed] [Google Scholar]
- Dehneshin N., Raissi H., Hasanzade Z., Farzad F. (2019). Using molecular dynamics simulation to explore the binding of the three potent anticancer drugs sorafenib streptozotocin and sunitinib to functionalized carbon nanotubes. J. Mol. Model. 25:159. 10.1007/s00894-019-4024-5 [DOI] [PubMed] [Google Scholar]
- del Pino P., Pelaz B., Zhang Q., Maffre P., Nienhaus G. U., Parak W. J. (2014). Protein corona formulation around nanoparticles - from the past to the future. Mater. Horiz. 1 301–313. 10.1039/C3MH00106G [DOI] [Google Scholar]
- Dell’Orco D., Lundqvist M., Oslakovic C., Cedervall T., Linse S. (2010). Modeling the time evolution of the nanoparticle-protein corona in a body fluid. PLoS One 5:e10949. 10.1371/journal.pone.0010949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov I. G., Grinkova Y. V., Lazarides A. A., Sligar S. G. (2004). Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126 3477–3487. 10.1021/ja0393574 [DOI] [PubMed] [Google Scholar]
- Denisov I. G., Sligar S. G. (2017). Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 117 4669–4713. 10.1021/acs.chemrev.6b00690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan M., Price D., Trump D., Kanter P., Shore N., Needham D., et al. (2004). New drug delivery approach for the treatment of prostate cancer (preclinical results and phase I study results). J. Clin. Oncol. 22:4661 10.1200/jco.2004.22.90140.4661 [DOI] [Google Scholar]
- Dhawan V., Magarkar A., Joshi G., Makhija D., Jain A., Shah J., et al. (2016). Stearylated cycloargenine nanosystems for intracellular delivery - simulations, formulation and proof of concept. RSC Adv. 6 113538–113550. 10.1039/C6RA16432C [DOI] [Google Scholar]
- Ding H. M., Ma Y. Q. (2014a). Computer simulation of the role of protein corona in cellular delivery of nanoparticles. Biomaterials 35 8703–8710. 10.1016/j.biomaterials.2014.06.033 [DOI] [PubMed] [Google Scholar]
- Ding H. M., Ma Y. Q. (2014b). Theoretical and computational investigations of nanoparticle-biomembrane interactions in cellular delivery. Small 11 1055–1071. 10.1002/smll.201401943 [DOI] [PubMed] [Google Scholar]
- Ding H., Ma Y. (2012). Role of physicochemical properties of coating ligands in receptor-mediated endocytosis of nanoparticles. Biomaterials 33 5798–5802. 10.1016/j.biomaterials.2012.04.055 [DOI] [PubMed] [Google Scholar]
- Ding H., Ma Y. (2016). Design strategy of surface decoration for efficient delivery of nanoparticles by computer simulation. Sci. Rep. 6:26783. 10.1038/srep26783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H., Ma Y. (2018). Computational approaches to cell-nanomaterial interactions: keeping balance between therapeutic efficiency and cytotoxicity. Nanoscale Horiz. 3 6–27. 10.1039/C7NH00138J [DOI] [PubMed] [Google Scholar]
- Ding H., Tian W., Ma Y. (2012). Designing nanoparticle translocation through membranes by computer simulations. ACS Nano 6 1230–1238. 10.1021/nn2038862 [DOI] [PubMed] [Google Scholar]
- Dowlatabadi M., Jahangiri M., Farhadian N. (2019). Prediction of chlortetracycline adsorption on the FE3O4 nanoparticle using molecular dynamics simulation. J. Biomol. Struct. Dyn. 37 3616–3626. 10.1080/07391102.2018.1521746 [DOI] [PubMed] [Google Scholar]
- Dror R. O., Pan A. C., Arlow D. H., Borhani D. W., Maragakis P., Shan Y., et al. (2011). Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 108 13118–13123. 10.1073/pnas.1104614108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Chandaroy P., Hui S. W. (1997). Grafted poly-(ethylene glycol) on lipid surfaces inhibits protein adsorption and cell adhesion. Biochim. Biophys. Acta 1326 236–248. 10.1016/S0005-2736(97)00027-8 [DOI] [PubMed] [Google Scholar]
- Durbin E. W., Buxton G. (2010). A Coarse-grained model of targeted drug delivery from responsive polymer nanoparticles. Soft Matter 6 762–767. 10.1039/B918476G [DOI] [Google Scholar]
- Duverger E., Picaud F. (2020). Theoretical study of ciprofloxacin antibiotic trapping on graphene or boron nitride oxide nanoflakes. J. Mol. Model. 26:135. 10.1007/s00894-020-04410-8 [DOI] [PubMed] [Google Scholar]
- Dzieciuch M., Rissanen S., Szydłowska N., Bunker A., Kumorek M., Jamróz D., et al. (2015). Pegylated liposomes as carriers of hydrophobic porphyrins. J. Phys. Chem. B 119 6646–6657. 10.1021/acs.jpcb.5b01351 [DOI] [PubMed] [Google Scholar]
- Dzieciuch-Rojek M., Poojari C., Bednar J., Bunker A., Kozik B., Nowakowska M., et al. (2017). Effects of membrane PEGylation on entry and location of antifungal drug itraconazole and their pharmacological implications. Mol. Pharmacol. 14 1057–1070. 10.1021/acs.molpharmaceut.6b00969 [DOI] [PubMed] [Google Scholar]
- Eckhardt S., Brunetto P. S., Gagnon J., Priebe M., Giese B., Fromm K. M. (2013). Nanobio silver: its interactions with peptides and bacteria, and its uses in medicine. Chem. Rev. 113 4708–4754. 10.1021/cr300288v [DOI] [PubMed] [Google Scholar]
- Ekladious I., Colson Y. L., Grinstaff M. W. (2019). Polymer-drug conjugate therapeutics: advances, insights and prospects. Nat. Rev. Drug Discov. 18 273–294. 10.1038/s41573-018-0005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hammadi M. M., Arias J. L. (2019). An update on liposomes in drug delivery: a patent review (2014 - 2018). Expert. Opin. Ther. Pat. 29 891–907. 10.1080/13543776.2019.1679767 [DOI] [PubMed] [Google Scholar]
- Erimban S., Daschakraborty S. (2020). Translocation of a hydroxyl functionalized carbon dot across a lipid bilayer: an all-atom molecular dynamics simulation study. Phys. Chem. Chem. Phys. 22 6335–6350. 10.1039/C9CP05999G [DOI] [PubMed] [Google Scholar]
- Erlebach A., Muljajew I., Chi M., Bückmann C., Weber C., Schubert U. S., et al. (2020). Predicting solubility of small molecules in macromolecular compounds for nanomedicine applications from atomistic simulations. Adv. Theory Simul. 3:2000001 10.1002/adts.202000001 [DOI] [Google Scholar]
- Esalmi M., Nikkhah S. J., Hashemianzadeh S. M., Sajadi S. A. S. (2016). The compatibility of tacrine molecule with poly(n-butylcyanoacrylate) and chitosan as efficient carriers for drug delivery: a molecular dynamics study. Eur. J. Pharm. Sci. 82 79–85. 10.1016/j.ejps.2015.11.014 [DOI] [PubMed] [Google Scholar]
- Español P., Warren P. B. (2017). Perspective: dissipative particle dynamics. J. Chem. Phys 146:150901 10.1063/1.4979514 [DOI] [PubMed] [Google Scholar]
- Exner K. S., Ivanova A. (2020). Identifying a gold nanoparticle as a proactive carrier for transport of a doxorubicin-peptide complex. Colloid Surf. B 194:111155. 10.1016/j.colsurfb.2020.111155 [DOI] [PubMed] [Google Scholar]
- Farmanzadeh D., Ghaderi M. (2018). A computational study of PAMAM dendrimer interaction with trans isomer of picoplatin anticancer drug. J. Mol. Graph. Model. 80 1–6. 10.1016/j.jmgm.2017.12.010 [DOI] [PubMed] [Google Scholar]
- Farzad F., Hashemzadeh H. (2020). Probing the effect of polyethylene glycol on the adsorption mechanisms of Gem on the hexagonal boron nitride as a highly efficient polymerbased drug delivery system: DFT, classical MD and well-tempered metadynamics simulations. J. Mol. Graph. Model. 98:107613. 10.1016/j.jmgm.2020.107613 [DOI] [PubMed] [Google Scholar]
- Fatemi S. M., Fatemi S. J., Abbasi Z. (2020). PAMAM dendrimer-based macromolecules and their potential applications: recent advances in theoretical studies. Polym. Bull. 77 6671–6691. 10.1007/s00289-019-03076-4 [DOI] [Google Scholar]
- Ferreira L. G., dos Santos R. N., Oliva G., Andricopulo A. D. (2015). Molecular docking and structure-based drug design strategies. Molecules 20 13384–13421. 10.3390/molecules200713384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe H. A. L., Moreno M. J., Ròg T., Vattulainen I., Loura L. M. S. (2014). How to tackle the issues in free energy simulations of long amphiphiles interacting with lipid membranes: convergence and local membrane deformations. J. Phys. Chem. B 118 3572–3581. 10.1021/jp501622d [DOI] [PubMed] [Google Scholar]
- Fox L. J., Richardson R. M., Briscoe W. H. (2018). PAMAM dendrimer - cell membrane interactions. Adv. Colloid Interf. Sci. 257 1–18. 10.1016/j.cis.2018.06.005 [DOI] [PubMed] [Google Scholar]
- Frenkel D., Smit B. (2001). Understanding Molecular Simulation: From Algorithms to Applications. Cambridge, MA: Academic Press. [Google Scholar]
- Gao H., Shi W., Freund L. B. (2005). Mechanics of receptor-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 102 9469–9474. 10.1073/pnas.0503879102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang P., Wang Z., Li C., Sun S., Hu S. (2019). Self-assembly of DCPD-loaded cross-linked micelle from triblock copolymers and its pH-responsive behavior: a dissipative particle dynamics study. Chem. Eng. Sci. 195 325–334. 10.1016/j.ces.2018.09.028 [DOI] [Google Scholar]
- Gao Y., Shi Y., Fu M., Feng Y., Lin G., Kong D., et al. (2020). Simulation study of the effects of interstitial fluid pressure and blood flow velocity on transvascular transport of nanoparticles in tumor microenvironment. Comput. Meth. Prog. Biol. 193:105493. 10.1016/j.cmpb.2020.105493 [DOI] [PubMed] [Google Scholar]
- Gapsys V., Groot B. L. D. (2020). On the importance of statistics in molecular simulations for thermodynamics, kinetics and simulation box size. eLife 9:e57589. 10.7554/eLife.57589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García K. P., Zarschler K., Barbaro L., Barreto J. A., O’Malley W., Spiccia L., et al. (2014). Zwitterionic-coated “stealth” nanoparticles for biomedical applications: recent advances in countering biomolecular corona formation and uptake by the mononuclear phagocyte system. Small 13 2516–2529. 10.1002/smll.201303540 [DOI] [PubMed] [Google Scholar]
- Ge D., Wu D., Wang Z., Shi W., Wu T., Zhang A., et al. (2009). Cellular uptake mechanism of molecular umbrella. Bioconjug. Chem. 20 2311–2316. 10.1021/bc9003074 [DOI] [PubMed] [Google Scholar]
- Ge Z., Wang Y. (2017). Estimation of nanodiamond surface charge density from zeta potential and molecular dynamics simulations. J. Phys. Chem. B 121 3394–3402. 10.1021/acs.jpcb.6b08589 [DOI] [PubMed] [Google Scholar]
- Gebauer M., Skerra A. (2018). Prospects of PASylation for the design of protein and peptide therapeutics with extended half-life and enhanced action. Bioorg. Med. Chem. 26 2882–2887. 10.1016/j.bmc.2017.09.016 [DOI] [PubMed] [Google Scholar]
- Geng Y., Dalhaimer P., Cai S., Tsai R., Tewari M., Minko T., et al. (2007). Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2 249–255. 10.1038/nnano.2007.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile F., Curcio A., Indolfi C., Ferrari M., Decuzzi P. (2008). The margination propensity of spherical particles for vascular targeting in microcirculation. J. Nanobiotechnol. 6:9. 10.1186/1477-3155-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadari R., Kashefi A. (2017). A computational study of the usability of amino acid-functionalized nitrogen doped graphene oxides as temperature-responsive drug delivery systems. Int. J. Hyperth. 33 785–795. 10.1080/02656736.2017.1308020 [DOI] [PubMed] [Google Scholar]
- Ghadari R., Mohammedzadeh Y. (2018). MD simulation studies on the effect of the temperature and protonation state on the imide-linked amino acid-based dendrimers. Comput. Mater. Sci. 151 124–131. 10.1016/j.commatsci.2018.05.011 [DOI] [Google Scholar]
- Ghadari R., Sabri A. (2019). In silico study on core-shell pseudodendrimeric glycodise structures in drug delivery related usages. Polyhedron 160 10–19. 10.1016/j.poly.2018.12.013 [DOI] [Google Scholar]
- Ghadri Z., Raissi H., Shahabi M., Farzad F. (2020). Molecular dynamics simulation study of glycine tip-functionalisation of single-walled carbon nanotubes as emerging nanovectors for the delivery of anticancer drugs. Mol. Simulat. 46 111–120. 10.1080/08927022.2019.1679363 [DOI] [Google Scholar]
- Ghasemi F., Mehridehnavi A., Pérez-Garrado A., Pérez-Sánchez H. (2018). Neural network and deep-learning algorithms used in QSAR studies: merits and drawbacks. Drug Discov. Today 23 1784–1790. 10.1016/j.drudis.2018.06.016 [DOI] [PubMed] [Google Scholar]
- Ghitman J., Stan R., Vlasceanu G., Vasile E., Iovu H. (2019). Predicting the drug loading efficiency into hybrid nanocarriers based on PLGA-vegetable oil using molecular dynamics simulation approach and Flory-Huggins theory. J. Drug Deliv. Sci. Tech. 53:101203 10.1016/j.jddst.2019.101203 [DOI] [Google Scholar]
- Ghosal K., Ghosh A. (2019). Carbon dots: the next generation platform for biomedical applications. Mater. Sci. Eng. C 96 887–903. 10.1016/j.msec.2018.11.060 [DOI] [PubMed] [Google Scholar]
- Ghosh M., Ren G., Simonsen J. B., Ryan R. O. (2014). Cationic lipid nanodisks as an siRNA delivery vehicle. Biochem. Cell Biol. 92 200–205. 10.1139/bcb-2014-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Singh A. T. K., Xu W., Sulchek T., Gordon L. I., Ryan R. O. (2011). Curcumin nanodisks: formulation and characterization. Nanomed. Nanotechnol. 7 162–167. 10.1016/j.nano.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Han G., De M., Kim C. K., Rotello V. M. (2008). Gold nanparticles in delivery applications. Adv. Drug Deliv. Rev. 60 1307–1315. 10.1016/j.addr.2008.03.016 [DOI] [PubMed] [Google Scholar]
- Giri A. K., Spohr E. (2018). Influence of chain length and branching on the structure of functionalized gold nanoparticles. J. Phys. Chem. C 122 26739–26747. 10.1021/acs.jpcc.8b08590 [DOI] [Google Scholar]
- Gkeka P., Angelikopoulos P., Sarkisov L., Cournia Z. (2014). Membrane partitioning of anionic, ligand-coated nanoparticles is accompanied by ligand snorkeling, local disordering, and cholesterol depletion. PLoS Comput. Biol. 10:e1003917. 10.1371/journal.pcbi.1003917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin B., Sakamoto J. H., Serda R. E., Grattoni A., Bouamrani A., Ferrari M. (2010). Emerging applications of nanomedicine for the diagnosis and treatment of cardiovascular diseases. Trends Pharmacol. Sci. 31 199–205. 10.1016/j.tips.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golda-Cepa M., Riedlová K., Kulig W., Cwiklik L., Kotarba A. (2020). Functionalization of the parylene c surface enhances the nucleation of calcium phosphate: combined experimental and molecular dynamics simulations approach. ACS Appl. Mater. Interf. 12 12426–12435. 10.1021/acsami.9b20877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith M., Abramovitz L., Peer D. (2014). Precision nanomedicine in neurodegenerative diseases. ACS Nano 8 1958–1965. 10.1021/nn501292z [DOI] [PubMed] [Google Scholar]
- Gordillo-Galeano A., Mora-Huertas C. E. (2018). Solid lipid nanoparticles and nanostructured lipid carriers: a review emphasizing on particle structure and drug release. Eur. J. Pharm. Biopharm. 133 285–308. 10.1016/j.ejpb.2018.10.017 [DOI] [PubMed] [Google Scholar]
- Grillo D. A., Albano J. M. R., Mocskos E. E., Facelli J. C., Pickholz M., Ferraro M. B. (2018). Mechanical properties of drug loaded diblock copolymer bilayers: a molecular dynamics study. J. Chem. Phys. 148:214901 10.1063/1.5028377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot R. D., Warren P. B. (1997). Dissipative particle dynamics: bridging the gap between atomistic and mesoscopic simulation. J. Chem. Phys. 107 4423–4435. 10.1063/1.474784 [DOI] [Google Scholar]
- Guo X. D., Qian Y., Zhang C. Y., Nie S. Y., Zhang L. J. (2012a). Can drug molecules diffuse into the core of micelles? Soft Matter 8 9989–9995. 10.1039/C2SM26200B [DOI] [Google Scholar]
- Guo X. D., Zhang L. J., Qian Y. (2012b). Systematic multiscale method for studying the structure-performance relationship of drug-delivery systems. Ind. Eng. Chem. Res. 51 4719–4730. 10.1021/ie2014668 [DOI] [Google Scholar]
- Guo X. D., Tan J. P. K., Kim S. H., Zhang L. J., Zhang Y., Hedrick J. L., et al. (2009a). Computational studies on self-assembled paclitaxel structures: templates for heirarchical block copolymer assemblies and sustained drug release. Biomaterials 30 6556–6563. 10.1016/j.biomaterials.2009.08.022 [DOI] [PubMed] [Google Scholar]
- Guo X. D., Tan J. P. K., Zhang L. J., Khan M., Liu S. Q., Yang Y. Y., et al. (2009b). Phase Behavior study of paclitaxel loaded amphiphilic copolymer in two solvents by dissipative particle dynamics simulations. Chem. Phys. Lett. 473 336–342. 10.1016/j.cplett.2009.04.009 [DOI] [Google Scholar]
- Guo X. D., Zhang L. J., Wu Z. M., Qian Y. (2010). Dissipative particle dynamics studies on microstructure of pH-sensitive micelles for sustained drug delivery. Macromolecules 43 7839–7844. 10.1021/ma101132n [DOI] [Google Scholar]
- Gupta A. S. (2016). Role of particle size, shape, and stiffness in design of intravascular drug delivery systems: insights from computations, experiments, and nature. WIREs Nanomed. Nanobiotechnol. 8 255–270. 10.1002/wnan.1362 [DOI] [PubMed] [Google Scholar]
- Gupta J., Nunes C., Vyas S., Jonnalagadda S. (2011). Prediction of solubility parameters and miscibility of pharmaceutical compounds by molecular dynamics simulation. J. Phys. Chem. B 115 2014–2023. 10.1021/jp108540n [DOI] [PubMed] [Google Scholar]
- Gupta M. N., Roy I. (2020). How corona formation impacts nanomaterials as drug carriers. Mol. Pharm. 17 725–737. 10.1021/acs.molpharmaceut.9b01111 [DOI] [PubMed] [Google Scholar]
- Gupta R., Badhe Y., Mitragorti S., Rai B. (2020). Permeation of nanoparticles across the intestinal lipid membrane: dependence on shape and surface chemistry studied through molecular simulations. Nanoscale 12 6318–6333. 10.1039/C9NR09947F [DOI] [PubMed] [Google Scholar]
- Gupta R., Kashyap N., Rai B. (2017). Transdermal cellular membrane penetration of proteins with gold nanoparticles: a molecular dynamics study. Phys. Chem. Chem. Phys. 19 7537–7545. 10.1039/C6CP08775B [DOI] [PubMed] [Google Scholar]
- Gupta R., Kashyap N., Rai B. (2018). Molecular mechanism of transdermal co-delivery of interferon-alpha protein with gold nanoparticle - a molecular dynamics study. Mol. Simulat. 44 274–284. 10.1080/08927022.2017.1367094 [DOI] [Google Scholar]
- Gupta R., Rai B. (2016). Penetration of gold nanoparticles through human skin: unraveling its mechanisms at the molecular scale. J. Phys Chem. B 120 7133–7142. 10.1021/acs.jpcb.6b03212 [DOI] [PubMed] [Google Scholar]
- Gupta R., Rai B. (2017). Effect of size and surface charge of gold nanoparticles on their skin permeability: a molecular dynamics study. Sci. Rep. 7:45292. 10.1038/srep45292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Biswas P. (2018). Effect of pH on size and internal structure of poly(propylene imine) dendrimers: a molecular dynamics simulation study. J. Phys. Chem. B 122 9250–9263. 10.1021/acs.jpcb.8b04653 [DOI] [PubMed] [Google Scholar]
- Gupta V., Bhavanasi S., Quadir M., Singh K., Ghosh G., Vasamreddy K., et al. (2019). Protein PEGylation for cancer therapy: bench to bedside. J. Cell Commun. Signal. 13 319–330. 10.1007/s12079-018-0492-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habiba K., Bracho-Rincon D. P., Gonzalez-Feliciano J. A., Villalobos-Santos J. C., Makarov V. I., Ortiz D., et al. (2015). Synergistic antibacterial activity of PEGylated silver-graphene quantum dots nanocomposites. Appl. Mater. Today 1 80–87. 10.1016/j.apmt.2015.10.001 [DOI] [Google Scholar]
- Haddish-Berhane N., Rickus J. L., Haghighi K. (2007). The role of multiscale computational approaches for rational design of conventional and nanoparticle oral drug delivery systems. Int. J. Nanomed. 2 315–331. [PMC free article] [PubMed] [Google Scholar]
- Hadjidemetriou M., Kostarelos K. (2017). Evolution of the nanoparticle corona. Nat. Nanotechnol. 12 288–290. 10.1038/nnano.2017.61 [DOI] [PubMed] [Google Scholar]
- Hamed E., Ma D., Keten S. (2015). Multiple PEG chains attached onto the surface of a helix bundle: conformations and implications. ACS Biomater. Sci. Eng. 1 79–84. 10.1021/ab500088b [DOI] [Google Scholar]
- Han R., Huang T., Liu X., Yin X., Li H., Lu J., et al. (2019a). Insight into the dissolution molecular mechanism of ternary solid dispersions by combined experiments and molecular simulations. AAPS Pharm. Sci. Tech. 20:274. 10.1208/s12249-019-1486-9 [DOI] [PubMed] [Google Scholar]
- Han R., Xiong H., Ye Z., Yang Y., Huang T., Jing Q., et al. (2019b). Predicting physical stability of solid dispersions by machine learning techniques. J. Control. Release 311–312 16–25. 10.1016/j.jconrel.2019.08.030 [DOI] [PubMed] [Google Scholar]
- Han S. (2013). Molecular dynamics simulation of oleic acid/oleate bilayers: an atomistic model for ufasome membrane. Chem. Phys. Lipids 175–176 79–83. 10.1016/j.chemphyslip.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Hao L., Lin L., Zhou J. (2019). pH-responsive zwitterionic copolymer DHA-PBLG-PCB for targeted drug delivery: a computer simulation study. Langmuir 35 1944–1953. 10.1021/acs.langmuir.8b00626 [DOI] [PubMed] [Google Scholar]
- Hasanzade Z., Raissi H. (2017). Investigation of graphene-based nanomaterial as nanocarrier for adsorption of paclitaxel anticancer drug: a molecular dynamics simulation study. J. Mol. Model. 23:36. 10.1007/s00894-017-3207-1 [DOI] [PubMed] [Google Scholar]
- Hashemzadeh H., Raissi H. (2017). The functionalization of carbon nanotubes to enhance the efficacy of the anticancer drug paclitaxel: a molecular dynamics simulation study. J. Mol. Model. 23:222 10.1007/s00894-017-3391 [DOI] [PubMed] [Google Scholar]
- Hathout R. M., Metwally A. A. (2016). Towards better modelling of drug loading in solid lipid nanoparticles: molecular dynamics, docking experiments and gaussian processes machine learning. Eur. J. Pharm. Biopharm. 108 262–268. 10.1016/j.ejpb.2016.07.019 [DOI] [PubMed] [Google Scholar]
- Hathout R. M., Metwally A. A., Woodman T. J., Hardy J. G. (2020). Prediction of drug loading in the gelatin matrix using computational methods. ACS Omega 5 1549–1556. 10.1021/acsomega.9b03487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Gu Z., Wang L., Qu Z., Xu F. (2020). Coarse-grained molecular dynamics simulation of dendrimer transmembrane transport with temperature-dependent membrane phase states. Int. J. Heat Mass Transf. 155:119797 10.1016/j.ijheatmasstransfer.2020.119797 [DOI] [Google Scholar]
- Hedayati M. H., Norouzian D., Aminian M., Teimourian S., Ahangari Cohan R., Sardari S., et al. (2017). Molecular design, expression and evaluation of PASylated human recombinant erythropoietin with enhanced functional properties. Protein J. 36 36–48. 10.1007/s10930-017-9699-9 [DOI] [PubMed] [Google Scholar]
- Heikkilä E., Gurtovenko A. A., Martinez-Seara H., Häkkinen H., Vattulainen I., Akola J. (2012). Atomistic simulations of functional Au 144(SR) 60 gold nanoparticles in aqueous environment. J. Phys. Chem. C 116 9805–9811. 10.1021/jp301094m [DOI] [Google Scholar]
- Heikkilä E., Martinez-Seara H., Gurtovenko A. A., Javanainen M., Häkkinen H., Vattulainen I., et al. (2014a). Cationic Au nanoparticle binding with plasma membrane-like lipid bilayers: potential mechanism for spontaneous permeation to cells revealed by atomistic simulations. J. Phys. Chem. C 118 11131–11141. 10.1021/jp5024026 [DOI] [Google Scholar]
- Heikkilä E., Martinez-Seara H., Gurtovenko A. A., Vattulainen I., Akola J. (2014b). Atomistic simulations of anionic Au144(SR)60 nanoparticles interacting with asymmetric model lipid membranes. Biochim. Biophys. Acta Biomembr. 1838 2852–2860. 10.1016/j.bbamem.2014.07.027 [DOI] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008). GROMACS 4: algorithms for highly efficient, load-balanced and scalable molecular simulation. J. Chem. Theory Comput. 4 435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Hlaváč D., Klushina D., Tokarský J. (2018). Interaction of antitumoral drug erlotinib with biodegradable triblock copolymers: a molecular modeling study. Chem. Pap. 72 2023–2034. 10.1007/s11696-018-0413-y [DOI] [Google Scholar]
- Hoffmann T., Gastreich M. (2019). The next level in chemical space navigation: going far beyond enumerable compound libraries. Drug Discov. Today 24 1148–1156. 10.1016/j.drudis.2019.02.013 [DOI] [PubMed] [Google Scholar]
- Hossain S., Kabedev A., Parrow A., Bergström C. A. S., Larsson P. (2019). Molecular simulation as a computational pharmaceutics tool to predict drug solubility, solubilization processes and partitioning. Eur. J. Pharm. Biopharm. 137 46–55. 10.1016/j.ejpb.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Lu H. (2019). Protein PEPylation: a new paradigm of protein-polymer conjugation. Bioconjug. Chem. 30 1604–1616. 10.1021/acs.bioconjchem.9b00236 [DOI] [PubMed] [Google Scholar]
- Hu W., Mao A., Wong P., Larsen A., Yazaki P. J., Wong J. Y. C., et al. (2017). Characterization of 1,2-Distearol-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycerol)-2000] and its complex with doxorubicin using nuclear magnetic resonance spectroscopy and molecular dynamics. Bioconjug. Chem. 28 1777–1790. 10.1021/acs.bioconjchem.7b00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. B., Dammer E. B., Ren R. J., Wang G. (2015). The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 4 1–10. 10.1186/s40035-015-0041-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Cruz W., Chen J., Zheng G. (2015). Learning from biology: synthetic lipoproteins for drug delivery. Interdiscip. Rev. Nanomed. Nanobiotechnol. 7 298–314. 10.1002/wnan.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Zhao Q., Su Y., Ouyang D. (2019). Investigation of molecular aggregation mechanism of glipizide/cyclodextrin complexation by combined experimental and molecular modeling approaches. Asian J. Pharm. Sci. 14 609–620. 10.1016/j.ajps.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D. P., Grossfield A., Lynch D. L., Feller S., Romo T. D., Gawrisch K., et al. (2010). A lipid pathway for ligand binding is necessary for a cannabinoid G protein-coupled receptor. J. Biol. Chem. 285 17954–17964. 10.1074/jbc.M109.041590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson S. T., Kobayashi R. (2019). Solvent-specific featurization for predicting free energies of solvation through machine learning. J. Chem. Inf. Model. 59 1338–1346. 10.1021/acs.jcim.8b00901 [DOI] [PubMed] [Google Scholar]
- Huynh L., Neale C., Pomès R., Allen C. (2012). Computational approaches to the rational design of nanoemulsions, polymeric micelles, and dendrimers for drug delivery. Nanomed. Nanotechnol. 8 20–36. 10.1016/j.nano.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Inglut C. T., Sorrin A. J., Kuruppu T., Vig S., Cicalo J., Ahmad H., et al. (2020). Immunological and toxicological considerations for the design of liposomes. Nanomaterials 10:190. 10.3390/nano10020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson H. I., Lopez C. A., Uusitalo J. J., de Jong D. H., Gopal S. M., Periole X., et al. (2014). The power of coarse graining in biomolecular simulations. Wiley Interdiscip. Rev. Comput. Mol. Sci. 4 225–248. 10.1002/wcms.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelachvili J. (1997). The different faces of poly(ethylene glycol). Proc. Natl. Acad. Sci. U.S.A. 94 8378–8379. 10.1073/pnas.94.16.8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelachvili J. N. (1985). Intermolecular and Surface Forces. Cambridge, MA: Academic Press. [Google Scholar]
- Izadyar A., Farhadian N., Chenarani N. (2016). Molecular dynamics simulation of doxorubicin adsorption on a bundle of functionalized CNT. J. Biomol. Struct. Dyn. 34 1797–1805. 10.1080/07391102.2015.1092475 [DOI] [PubMed] [Google Scholar]
- Izrailev S., Stepaniants S., Balsera M., Oono Y., Schulten K. (1997). Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 72 1568–1581. 10.1016/S0006-3495(97)78804-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V., Maingi V., Maiti P. K., Bharatam P. V. (2013). Molecular dynamics simulations of PPI dendrimer-drug complexes. Soft Matter 9 6482–6496. 10.1039/C3SM50434D [DOI] [Google Scholar]
- Jain V., Maiti P. K., Bharatam P. V. (2016). Atomic level insights into realistic molecular models of dendrimer-drug complexes through MD simulations. J. Chem. Phys. 145:124902 10.1063/1.4962582 [DOI] [PubMed] [Google Scholar]
- Janib S. M., Moses A. S., MacKay J. A. (2010). Imaging and drug delivery using theranostic nanoparticles. Adv. Drug Deliv. Rev. 62 1052–1063. 10.1016/j.addr.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janout V., Cline L. L., Feuston B. P., Klein L., O’Brien A., Tucker T., et al. (2014). Molecular umbrella conjugate for the ocular delivery of siRNA. Bioconjug. Chem. 25 197–201. 10.1021/bc400506m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janout V., Jing B., Regen S. L. (2002). Molecular umbrella-assisted transport of thiolated AMP and ATP across phospholipid bilayers. Bioconjug. Chem. 13 351–356. 10.1021/bc015564m [DOI] [PubMed] [Google Scholar]
- Janout V., Jing B., Regen S. L. (2005). Molecular umbrella-assisted transport of an oligonucleotide across cholesterol-rich phospholipid bilayers. J. Am. Chem. Soc. 127 15862–15870. 10.1021/ja053930x [DOI] [PubMed] [Google Scholar]
- Janout V., Regen S. L. (2005). A needle-and-thread approach to bilayer transport: permeation of a molecular umbrella-oligonucliotide conjugation across a phospholipid membrane. J. Am. Chem. Soc. 127 22–23. 10.1021/ja044257z [DOI] [PubMed] [Google Scholar]
- Janout V., Regen S. L. (2009). Bioconjugate-based molecular umbrellas. Bioconjug. Chem. 20 183–192. 10.1021/bc800296g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janout V., Zhang L., Staina I. V., Di Georgio C., Regen S. L. (2001). Molecular umbrella-assisted transport of glutathione across a phospholipid membrane. J. Am. Chem. Soc. 123 5401–5406. 10.1021/ja010124r [DOI] [PubMed] [Google Scholar]
- Jevševar S., Kunstelj M., Porekar V. G. (2010). PEGylation of therapeutic proteins. Biotechnol. J. 5 113–128. 10.1002/biot.200900218 [DOI] [PubMed] [Google Scholar]
- Jha P. K., Larson R. G. (2014). Assessing the efficiency of polymeric excipients by atomistic molecular dynamics simulations. Mol. Pharm. 11 1676–1686. 10.1021/mp500068w [DOI] [PubMed] [Google Scholar]
- Jiang W., Luo J., Nangia S. (2015). Multiscale approach to investigate self-assembly of telodendrimer based nanocarriers for anticancer drug delivery. Langmuir 31 4270–4280. 10.1021/la503949b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing B., Janout V., Regen S. L. (2003). Fully detachable molecular umbrellas as peptide delivery agents. Bioconjug. Chem. 14 1191–1196. 10.1021/bc034074m [DOI] [PubMed] [Google Scholar]
- Johnson S. R., Zheng W. (2006). Recent progress in the computational prediction of aqueous solubility and absorption. AAPS J. 8 E27–E40. 10.1208/aapsj080104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A., Swope W. C., Jordan K. E., Warren P. B., Noro M. G., Bray D. J., et al. (2016). Toward a standard protocol for micelle simulation. J. Phys Chem. B 120 6337–6351. 10.1021/acs.jpcb.6b03075 [DOI] [PubMed] [Google Scholar]
- Kacar G. (2019). Molecular understanding of interactions, structure, and drug encapsulation efficiency of pluronic micelles from dissipative particle dynamics simulations. Colloid Polym. Sci. 297 1037–1051. 10.1007/s00396-019-04535-0 [DOI] [Google Scholar]
- Kacar G. (2020). Thermodynamic stability of ibuprofen loaded poloxamer micelles. Chem. Phys. 533:110713 10.1016/j.chemphys.2020.110713 [DOI] [Google Scholar]
- Kamel M., Raissi H., Morsali A. (2017). Theoretical study of solvent and co-solvent effects on the interaction of flutamide anticancer drug with carbon nanotube as a drug delivery system. J. Mol. Liq. 248 490–500. 10.1016/j.molliq.2017.10.078 [DOI] [Google Scholar]
- Kanchi S., Gosika M., Ayappa K. G., Maiti P. K. (2018). Dendrimer interactions with lipid bilayer: comparison of force field and effect of implicit vs explicit solvation. J. Chem. Theory Comput. 14 3825–3839. 10.1021/acs.jctc.8b00119 [DOI] [PubMed] [Google Scholar]
- Kapoor B., Gupta R., Gulati M., Singh S. K., Khursheed R., Gupta M. (2019). The why, where, who, how and what of the vesicular delivery systems. Adv. Colloid Interf. 271:101985. 10.1016/j.cis.2019.07.006 [DOI] [PubMed] [Google Scholar]
- Karjiban R. A., Basri M., Basyaruddin M., Rahman A., Salleh A. B. (2012). Molecular dynamics simulation of palmitate ester self-assembly with diclofenac. Int. J. Mol. Sci. 13 9572–9583. 10.3390/ijms13089572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnati K. R., Wang Y. (2018). Understanding the co-loading and releasing of doxorubicin and paclitaxel using chitosan functionalized single walled carbon nanotubes by molecular dynamics simulations. Phys. Chem. Chem. Phys. 20 9389–9400. 10.1039/C8CP00124C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. (2002). Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 9 646–652. 10.1038/nsb0902-646 [DOI] [PubMed] [Google Scholar]
- Kasomova A. O., Pavan G. M., Danani A., Mondon K., Cristiani A., Scapozza L., et al. (2012). Validation of a novel molecular dynamics simulation approach for lipophilic drug incorporation into polymer micelles. J. Phys. Chem. B 116 4338–4345. 10.1021/jp2104819 [DOI] [PubMed] [Google Scholar]
- Kaszuba K., Roìg T., Bryl K., Vattulainen I., Karttunen M. (2010). Molecular dynamics simulations reveal fundamental role of water as factor determining affinity of binding of β-blocker nebivolol to β-adrenergic receptor. J. Phys. Chem. B 114 8374–8386. 10.1021/jp909971f [DOI] [PubMed] [Google Scholar]
- Kaszuba K., Roìg T., Danne R., Canning P., Fülöp V., Juhász T., et al. (2012). Molecular dynamics, crystallography and mutagenesis studies on the substrate gating mechanism of prolyl oligopeptidase. Biochimie 94 1398–1411. 10.1016/j.biochi.2012.03.012 [DOI] [PubMed] [Google Scholar]
- Katiyar R. S., Jha P. K. (2018). Molecular simulations in drug delivery: opportunities and challenges. WIREs Comput. Mol. Sci. 8:e1358 10.1002/wcms.1358 [DOI] [Google Scholar]
- Katre N. V. (1993). The conjugation of proteins with polyethylene glycol and other polymers altering properties of proteins to enhance their therapeutic potential. Adv. Drug Deliv. Rev. 10 91–114. 10.1016/0169-409X(93)90005-O [DOI] [Google Scholar]
- Kaupbayeva B., Russell A. J. (2020). Polymer-enhanced biomacromolecules. Prog. Polym. Sci. 101:101194 10.1016/j.progpolymsci.2019.101194 [DOI] [Google Scholar]
- Kavyani S., Amjad-Iranagh S., Dadvar M., Modarress H. (2016). Hybrid dendrimers of PPI(core)-PAMAM(shell): a molecular dynamics study. J. Phys. Chem. B 120 9564–9575. 10.1021/acs.jpcb.6b05142 [DOI] [PubMed] [Google Scholar]
- Kavyani S., Dadvar M., Modarress H., Amjad-Iranagh S. (2018a). A coarse grained molecular dynamics simulation study on the structural properties of carbon nanotube - dendrimer composites. Soft Matter 14 3151–3163. 10.1039/C8SM00253C [DOI] [PubMed] [Google Scholar]
- Kavyani S., Dadvar M., Modarress H., Amjad-Iranagh S. (2018b). Molecular perspective mechanism for drug loading on carbon nanotube-dendrimer: a coarse-grained molecular dynamics study. J. Phys. Chem. B 122 7956–7969. 10.1021/acs.jpcb.8b04434 [DOI] [PubMed] [Google Scholar]
- Kelly C. V., Leroueil P. R., Orr B. G., Holl M. M. B., Andricioaei L. (2008). Poly(amidoamine) dendrimers on lipid bilayers II: effects of bilayer phase and dendrimer termination. J. Phys. Chem. B 112 9346–9353. 10.1021/jp8013783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C. V., Liroff M. G., Triplett L. D., Leroueil P. R., Mullen D. G., Wallace J. M., et al. (2009). Stoichiometry and structure of poly(amidoamine) dendrimer- Lipid complexes. ACS Nano 3 1886–1896. 10.1021/nn900173e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepczynski M., Róg T. (2016). Functionalized lipids and surfactants for specific applications. Biochim. Biophys. Acta Biomembr. 1858 2362–2379. 10.1016/j.bbamem.2016.02.038 [DOI] [PubMed] [Google Scholar]
- Khalkhali M., Mohammadinejad S., Khoeini F., Rostamizadeh K. (2019). Vesicle-like structure of lipid based nanoparticles as drug delivery system revealed by molecular dynamics simulation. Int. J. Pharm. 559 173–181. 10.1016/j.ijpharm.2019.01.036 [DOI] [PubMed] [Google Scholar]
- Khan R., Irchhaiya R. (2016). Niosomes: a potential tool for novel drug delivery. J. Pharm. Investig. 46 165–204. 10.1007/s40005-016-0249-9 [DOI] [Google Scholar]
- Khan S., McCabe J., Hill K., Beales P. A. (2020). Biodegradable hybrid block copolymer - lipid vesicles as potential drug delivery systems. J. Colloid Interf. Sci. 562 418–428. 10.1016/j.jcis.2019.11.101 [DOI] [PubMed] [Google Scholar]
- Kharazian B., Hadipour N. L., Ejtehadi M. R. (2016). Understanding the nanoparticle - protein corona complexes using computational and experimental methods. Int. J. Biochem. Cell Biol. 75 162–174. 10.1016/j.biocel.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Kłos J. S., Paturej J. (2020). Charged dendrimers with finite-size counterions. J. Phys. Chem. B. 124 7957–7968. 10.1021/acs.jpcb.0c05092 [DOI] [PubMed] [Google Scholar]
- Kłos J. S., Sommer J.-U. (2013). Coarse grained simulations of neutral and charged dendrimers. Polym. Sci. Ser. C 55 125–153. 10.1134/S1811238213070023 [DOI] [Google Scholar]
- Knop K., Hoogenboom R., Fischer D., Schubert U. (2010). Poly(ethylene glycol) in drug delivery: pros and cons as well as potential alternatives. Angew. Chem. Int. Edn. 49 6288–6308. 10.1002/anie.200902672 [DOI] [PubMed] [Google Scholar]
- Kohli A. G., Kierstead P. H., Venditto V. J., Walsh C. L., Szoka F. C. (2014). Designer lipids for drug delivery: from heads to tails. J. Control. Release 190 274–287. 10.1016/j.jconrel.2014.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivuniemi A., Vattulainen I. (2012). Revealing structural and dynamical properties of high density lipoproteins through molecular simulations. Soft Matter 8 1262–1267. 10.1039/C1SM06742G [DOI] [Google Scholar]
- Kojima C., Kono K., Maruyama K., Takagishi T. (2000). Synthesis of polyamidoamine dendrimers having poly(ethylene glycol) grafts and their ability to encapsulate anticancer drugs. Bioconjug. Chem. 11 910–917. 10.1021/bc0000583 [DOI] [PubMed] [Google Scholar]
- Kokh D. B., Amaral M., Bomke J., Grädler U., Musil D., Buchstaller H. P., et al. (2018). Estimation of drug-target residence times by τ-random acceleration molecular dynamics simulations. J. Chem. Theory Comput. 14 3859–3869. 10.1021/acs.jctc.8b00230 [DOI] [PubMed] [Google Scholar]
- Koochaki A., Moghbeli M. R., Nikkhah S. J., Ianiro A., Tuinier R. (2020). Dual responsive PMEEECL-PAE block copolymers: a computational self assembly and doxorubicin uptake study. RSC. Adv. 10 3233–3245. 10.1039/C9RA09066E [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordzadeh A., Amjad-Iranagh S., Zarif M., Modarress H. (2019). Adsorption and encapsulation of the drug doxorubicin on covalent functionalized carbon nanotubes: a scrutinized study by using molecular dynamics simulation and quantum mechanics calculation. J. Mol. Graph. Model. 88 11–22. 10.1016/j.jmgm.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Kuai R., Li D., Chen Y. E., Moon J. J., Schwendeman A. (2016a). High-density lipoproteins: nature’s multifunctional nanoparticles. ACS Nano 10 3015–3041. 10.1021/acsnano.5b07522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai R., Ochyl L. J., Bahjat K. S., Schwendeman A., Moon J. J. (2016b). Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat. Mater. 16 489–496. 10.1038/nmat4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G., Kant A., Kumar M., Masram D. T. (2019). Synthesis, characterizations and kinetic study of metal organic framework nanocomposite excipient used as extended release delivery vehicle for an antibiotic drug. Inorgan. Chim. Acta 496:119036 10.1016/j.ica.2019.119036 [DOI] [Google Scholar]
- Kuramochi H., Andoh Y., Yoshii N., Okazaki S. (2009). All-atom molecular dynamics study of a spherical micelle composed of N-acetylated poly(ethylene glycol)-poly(γ-benzyl L-glutamate) block copolymers: a potential carrier of drug delivery systems for cancer. J. Phys. Chem. B 113 15181–15188. 10.1021/jp906155z [DOI] [PubMed] [Google Scholar]
- Kyrychenko A., Karpushina G. V., Bogatyrenko S. L., Kryshtal A. P., Doroshenko A. O. (2011). Preparation, structure, and a coarse-grained molecular dynamics model for dodecanethiol-stabilized gold nanoparticles. Comput. Theor. Chem. 977 34–39. 10.1016/j.comptc.2011.09.003 [DOI] [Google Scholar]
- Kyrychenko A., Korsun O. M., Gubin I. I., Kovalenko S. M., Kagulin O. N. (2015). Atomistic simulations of coating of silver nanoparticles with poly(vinylpyrrolidone) oligomers: effect of oligomer chain length. J. Phys. Chem. C 119 7888–7899. 10.1021/jp510369a [DOI] [Google Scholar]
- Lajunen T., Kontturi L. S., Viitala L., Manna M., Cramariuc O., Roìg T. (2016). Indocyanine green-loaded liposomes for light-triggered drug release. Mol. Pharm. 13 2095–2107. 10.1021/acs.molpharmaceut.6b00207 [DOI] [PubMed] [Google Scholar]
- Lajunen T., Nurmi R., Wilbie D., Ruoslahti T., Johansson N. G., Korhonen O., et al. (2018). The effect of light sensitizer localization on the stability of indocyanine green liposomes. J. Control Release 284 213–223. 10.1016/j.jconrel.2018.06.029 [DOI] [PubMed] [Google Scholar]
- Lammers T., Ferrari M. (2020). The success of nanomedicine. Nano Today 31:100853 10.1016/j.nantod.2020.100853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A. S., Olsen M. A., Moustafa H., Larsen F. H., Sauer S. P. A., Rantanen J., et al. (2019). Determining short-lived solid forms during phase transformations using molecular dynamics. Cryst. Eng. Commun. 21 4020–4024. 10.1039/C9CE00460B [DOI] [Google Scholar]
- Larsen A. S., Rantanen J., Johansson K. E. (2017a). Computational dehydration of crystalline hydrates using molecular dynamics simulations. J. Pharm. Sci. 106 348–355. 10.1016/j.xphs.2016.10.005 [DOI] [PubMed] [Google Scholar]
- Larsen A. S., Ruggiero M. T., Johansson K. E., Zeitler J. A., Rantanen J. (2017b). Tracking dehydration mechanisms in crystalline hydrates with molecular dynamics simulations. Cryst. Growth Des. 17 5017–5022. 10.1021/acs.cgd.7b00889 [DOI] [Google Scholar]
- Laudadido E., Mobbili G., Minelli C., Massaccesi L., Galeazzi R. (2017). Salts influence cathechins and flavonoids encapsulation in liposomes: a molecular dynamics investigation. Mol. Inform. 36:1700059. 10.1002/minf.201700059 [DOI] [PubMed] [Google Scholar]
- Lawrence P. B., Gavrilov Y., Matthews S. S., Langlois M. I., Shental-Bechor D., Greenblatt H. M., et al. (2014). Criteria for selecting PEGylation sites of proteins for higher thermodynamic and proteolytic stability. J. Am. Chem. Soc. 136 17547–17560. 10.1021/ja5095183 [DOI] [PubMed] [Google Scholar]
- Lawrence P. B., Price J. L. (2016). How PEGylation influences protein conformational stability. Curr. Opin. Chem. Biol. 34 88–94. 10.1016/j.cbpa.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. H., Hsin J., Sotomayor M., Comellas G., Schulten K. (2009). Discovery through the computational microscope. Structure 17 1295–1306. 10.1016/j.str.2009.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-Y., Ferrari M., Decuzzi P. (2009). Shaping nano-/micro-particles for enhanced vascular interaction in laminar flows. Nanotechnology 20:495101 10.1088/0957-4484/20/49/495101 [DOI] [PubMed] [Google Scholar]
- Lee H. (2014). Molecular modeling of PEGylated peptides, dendrimers, and single-walled carbon nanotubes for biomedical applications. Polymers 6 776–798. 10.3390/polym6030776 [DOI] [Google Scholar]
- Lee H. (2020a). Effects of nanoparticle electrostatics and protein - protein interactions on corona formation: conformation and hydrodynamics. Small 16:1906598. 10.1002/smll.201906598 [DOI] [PubMed] [Google Scholar]
- Lee H. (2020b). Molecular simulations of PEGylated biomolecules, liposomes, and nanoparticles for drug delivery applications. Pharmaceutics 12:533. 10.3390/pharmaceutics12060533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Choi J. S., Larson R. G. (2011). Molecular dynamics studies of the size and internal structure of the PAMAM dendrimer grafted with arginine and histidine. Macromolecules 44 8681–8686. 10.1021/ma2019396 [DOI] [Google Scholar]
- Lee H., Larson R. G. (2008). Coarse-grained molecular dynamics simulation of the concentration and size dependence of fifth- and seventh-generation PAMAM dendrimers on pore formation in DMPC bilayer. J. Phys. Chem. B 112 7778–7784. 10.1021/jp802606y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Larson R. G. (2009). Molecular dynamics study of the structure and interparticle interactions of polyethylene glycol-conjugated PAMAM dendrimers. J. Phys. Chem. B 113 13202–13207. 10.1021/jp906497e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Larson R. G. (2011). Effects of PEGylation on the size and internal structure of dendrimers: self-penetration of long PEG chains into the dendrimer core. Macromolecules 44 2291–2298. 10.1021/ma102482u [DOI] [Google Scholar]
- Lee I., Athey B. D., Wetzel A. W., Meixner W., Baker J. R. (2002). Structural molecular dynamics studies on polyamidoamine dendrimers for a therapeutic application: effects of pH and generation. Macromolecules 35 4510–4520. 10.1021/ma010354q [DOI] [Google Scholar]
- Lehtinen J., Magarkar A., Stepniewski M., Hakola S., Bergman M., Roìg T., et al. (2012). Analysis of cause of failure of new targeting peptide in PEGylated liposome: molecular modeling as rational design tool for nanomedicine. Eur. J. Pharm. Sci. 46 121–130. 10.1016/j.ejps.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Lehto M., Karilainen T., Roìg T., Cramariuc O., Vanhala E., Tornaeus J., et al. (2014). Co-exposure with fullerene may strengthen health effects of organic industrial chemicals. PLoS One 9:e114490. 10.1371/journal.pone.0114490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ying S., Ren H., Dai J., Zhang L., Liang L., et al. (2020). Molecular dynamics study of the encapsulation and release of anticancer drug doxorubicin by chitosan. Int. J. Pharm. 580:119241. 10.1016/j.ijpharm.2020.119241 [DOI] [PubMed] [Google Scholar]
- Li Z., Wang Y., Zhu J., Zhang Y., Zhang W., Zhou M., et al. (2020). Emerging well-tailored nanoparticulate delivery system based on in situ regulation of the protein corona. J. Control Release 320 1–18. 10.1016/j.jconrel.2020.01.007 [DOI] [PubMed] [Google Scholar]
- Li L., Totton T., Frenkel D. (2018). Computational methodology for solubility prediction: application to sparingly soluble organic/inorganic materials. J. Chem. Phys. 149:54102 10.1063/1.5040366 [DOI] [PubMed] [Google Scholar]
- Li M., Du C., Guo N., Teng Y., Meng X., Sun H., et al. (2019). Composition design and medical application of liposomes. Eur. J. Med. Chem. 164 640–653. 10.1016/j.ejmech.2019.01.007 [DOI] [PubMed] [Google Scholar]
- Li Y., Hu Y. (2014). Computational investigation of the influence of chain length on the shielding effect of PEGylated nanoparticles. RSC Adv. 4 51022–51031. 10.1039/c4ra11142g [DOI] [Google Scholar]
- Li Y., Kröger M., Liu W. K. (2014a). Endocytosis of PEGylated nanoparticles accompanied by structural and free energy changes of the grafted polyethylene glycol. Biomaterials 35 8467–8478. 10.1016/j.biomaterials.2014.06.032 [DOI] [PubMed] [Google Scholar]
- Li Y., Stroberg W., Lee T.-R., Kim H. S., Man H., Ho D., et al. (2014b). Multiscale modeling and uncertainty quantification in nanoparticle-mediated drug/gene delivery. Comput. Mech. 53 511–537. 10.1007/s00466-013-0953-5 [DOI] [Google Scholar]
- Li Y.-C., Rissanen S., Stepniewski M., Cramariuc O., Róg T., Mirza S., et al. (2012). Study of interaction between PEG carrier and three relevant drug molecules: piroxicam, paclitaxel, and hematoporphyrin. J. Phys. Chem. B 116 7334–7341. 10.1021/jp300301z [DOI] [PubMed] [Google Scholar]
- Li Y., Yue T., Yang K., Zhang X. (2012). Molecular modeling of the relationship between nanoparticle shape anisotropy and endocytosis kinetics. Biomaterials 33 4965–4973. 10.1016/j.biomaterials.2012.03.044 [DOI] [PubMed] [Google Scholar]
- Li Z., Tozer T., Alisaraie L. (2016a). Molecular dynamics studies for optimization of noncovalent loading of vinblastine on single-walled carbon nanotube. J. Phys. Chem. C 120 4061–4070. 10.1021/acs.jpcc.5b10646 [DOI] [Google Scholar]
- Li Z., Van Dyk A. K., Fitzwater S. J., Fichthorn K. A., Milner S. T. (2016b). Atomistic molecular dynamics simulations of charged latex particle surfaces in aqueous solution. Langmuir 32 428–441. 10.1021/acs.langmuir.5b03942 [DOI] [PubMed] [Google Scholar]
- Liang L., Shen J.-W., Wang Q. (2017). Molecular dynamic study on DNA nanotubes as drug delivery vehicle for anticancer drugs. Colloid Surf. B 153 168–173. 10.1016/j.colsurfb.2017.02.021 [DOI] [PubMed] [Google Scholar]
- Liang Q. (2013). Penetration of polymer-grafted nanoparticles through a lipid bilayer. Soft Matter 9 5594–5601. 10.1039/C3SM27254K [DOI] [Google Scholar]
- Liebner R., Mathaes R., Meyer M., Hey T., Winter G., Besheer A. (2014). Protein HESylation for half-life extension: synthesis, characterization and pharmacokinetics of HESylated anakinra. Eur. J. Pharm. Biopharm. 87 378–385. 10.1016/j.ejpb.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Lin J., Zhang H., Chen Z., Zheng Y. (2010). Penetration of lipid membranes by gold nanoparticles: insights into cellular uptake, cytotoxicity, and their relationship. ACS Nano 4 5421–5429. 10.1021/nn1010792 [DOI] [PubMed] [Google Scholar]
- Lin X., Li Y., Gu N. (2010). Nanoparticle’s size effect on its translocation across a lipid bilayer: a molecular dynamics simulation. J. Comput. Theor. Nanosci. 7 269–276. 10.1166/jctn.2010.1358 [DOI] [Google Scholar]
- Lin J., Zhang H., Morovati V., Dargazany R. (2017). PEGylation on mixed monolayer gold nanoparticles: effect of grafting density, chain length, and surface curvature. J. Colloid Interf. Sci. 504 325–333. 10.1016/j.jcis.2017.05.046 [DOI] [PubMed] [Google Scholar]
- Lin J.-Q., Zheng Y.-G., Zhang H.-W., Chen Z. (2011). A simulation study on nanoscale holes generated by gold nanoparticles on negative lipid bilayers. Langmuir 27 8323–8332. 10.1021/la201086u [DOI] [PubMed] [Google Scholar]
- Lin P., Colina C. M. (2019). Molecular simulation of protein-polymer conjugates. Curr. Opin. Chem. Eng. 23 44–50. 10.1016/j.coche.2019.02.006 [DOI] [Google Scholar]
- Lin W., Xue Z., Wen L., Li Y., Liang Z., Xu J., et al. (2019). Mesoscopic simulations of drug loaded diselenide crosslinked micelles: stability, drug loading and release properties. Colloid Surf. B 182:110313. 10.1016/j.colsurfb.2019.06.043 [DOI] [PubMed] [Google Scholar]
- Lin W. J., Nie S. Y., Chen Q., Qian Y., Wen X. F., Zhang L. J. (2014). Structure-property relationship of pH-sensitive (PCL) (PDEA-b-PPEGMA) Micelles: experiment and DPD simulation. AICHe J. 60 3634–3646. 10.1002/aic.14562 [DOI] [Google Scholar]
- Lin X., Gu N. (2014). Surface properties of encapsulating hydrophobic nanoparticles regulate the main phase transition temperature of lipid bilayers: a simulation study. Nano Res. 7 1195–1204. 10.1007/s12274-014-0482-3 [DOI] [Google Scholar]
- Lin X., Lin X., Gu N. (2020). Optimization of hydrophobic nanoparticles to better target lipid rafts with molecular dynamics simulations. Nanoscale 12 4101–4109. 10.1039/C9NR09226A [DOI] [PubMed] [Google Scholar]
- Lipinski C. A. (2004). Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 1 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46 3–26. 10.1016/S0169-409X(96)00423-1 [DOI] [PubMed] [Google Scholar]
- Liu P., Long W. (2009). Current mathematical methods used in QSAR/QSPR studies. Int. J. Mol. Sci. 10 1978–1998. 10.3390/ijms10051978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Peng B., Sohrabi S., Liu Y. Y. (2016). The configuration of copolymer ligands on nanoparticles affects adhesion and uptake. Langmuir 32 10136–10143. 10.1021/acs.langmuir.6b02371 [DOI] [PubMed] [Google Scholar]
- Lobatto M. E., Fuster V., Fayad Z. A., Mulder W. J. M. (2011). Perspectives and opportunities for nanomedicine in the management of atherosclerosis. Nat. Rev. Drug Discov. 10 835–852. 10.1038/nrd3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolicato F., Joly L., Martinez-Seara H., Fragneto G., Scoppola E., Baldelli Bombelli F., et al. (2019). The role of temperature and lipid charge on intake/uptake of cationic gold nanoparticles into lipid bilayers. Small 15:1805046. 10.1002/smll.201805046 [DOI] [PubMed] [Google Scholar]
- Lolicato F., Juhola H., Zak A., Postila P. A., Saukko A., Rissanen S., et al. (2020). Membrane-dependent binding and entry mechanism of dopamine into its receptor. ACS Chem. Neurosci. 11 1914–1924. 10.1021/acschemneuro.9b00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez H., Lobaskin V. (2015). Coarse-grained model of adsorption of blood plasma proteins onto nanoparticles. J. Chem. Phys. 143:243138 10.1063/1.4936908 [DOI] [PubMed] [Google Scholar]
- Lorson T., Lübtow M. M., Wegener E., Haider M. S., Borova S., Nahm D., et al. (2018). Poly(2-oxazoline)s based biomaterials: a comprehensive and critical update. Biomaterials 178 204–280. 10.1016/j.biomaterials.2018.05.022 [DOI] [PubMed] [Google Scholar]
- Loverde S. M. (2014). Computer simulation of polymer and biopolymer self-assembly for drug delivery. Mol. Simulat. 40 794–801. 10.1080/08927022.2014.898118 [DOI] [Google Scholar]
- Loverde S. M., Klein M. L., Discher D. E. (2011). Nanoparticle shape improves delivery: rational coarse grain molecular dynamics (rCG-MD) of Taxol in worm-like PEG-PCL micelles. Adv. Mater. 24 3823–3830. 10.1002/adma.201103192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Bennett W. F. D., Ding Y., Zhang L., Fan H. Y., Zhao D., et al. (2015). Design and characterization of a multi-functional pH-triggered peptide C8 for selective anticancer activity. Adv. Healthc. Mater. 4 2709–2718. 10.1002/adhm.201500636 [DOI] [PubMed] [Google Scholar]
- Lüdemann S. K., Lounnas V., Wade R. C. (2000). How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms. J. Mol. Biol. 303 797–811. 10.1006/jmbi.2000.4154 [DOI] [PubMed] [Google Scholar]
- Lüder K., Lindfors L., Westergren J., Nordholm S., Kjellander R. (2007). In silico prediction of drug solubility: 2. free energy of solvation in pure melts. J. Phys. Chem. B 111 7303–7311. 10.1021/jp0642239 [DOI] [PubMed] [Google Scholar]
- Lüder K., Lindfors L., Westergren J., Nordholm S., Persson R., Pedersen M. (2009). In silico prediction of drug solubility: 4. will simple potentials suffice? J. Comput. Chem. 30 1859–1871. 10.1002/jcc.21173 [DOI] [PubMed] [Google Scholar]
- Lundsten S., Hernández V. A., Gedda L., Sarén T., Brown C. J., Lane D. P., et al. (2020). Tumor-targeted delivery of the p53-activating VIP116 with PEG-stabilized lipodisks. Nanomaterials 10:783 10.1002/mats.201900026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnoo T., Assawakhajornasak J., Puangmali T. (2019). In silico study of gold nanoparticle uptake into a mammalian cell: interplay of size, shape, surface charge, and aggregation. J. Phys. Chem. C 123 3801–3810. 10.1021/acs.jpcc.8b07616 [DOI] [Google Scholar]
- Lunnoo T., Assawakhajornasak J., Ruangchai S., Puangmali T. (2020). Role of surface functionalization on cellular uptake of AuNPs characterized by computational microscopy. J. Phys. Chem. B 124 1898–1908. 10.1021/acs.jpcb.9b11600 [DOI] [PubMed] [Google Scholar]
- Luo S., Zhang Y., Cao J., He B., Li S. (2016). Arginine modified polymeric micelles as a novel drug delivery system with enhanced endocytosis efficiency. Colloid Surf. B 148 181–192. 10.1016/j.colsurfb.2016.07.023 [DOI] [PubMed] [Google Scholar]
- Luo Z., Li Y., Wang B., Jiang J. (2016). pH-sensitive vesicles formed by amphiphilic grafted copolymers with tunable membrane permeability for drug loading/release: a multiscale simulation study. Macromolecules 49 6084–6094. 10.1021/acs.macromol.6b01211 [DOI] [Google Scholar]
- Luo X., Wang S., Xu S., Lang M. (2019). Relevance of the polymeric prodrug and its drug loading efficiency: comparison between computer simulation and experiment. Macromol. Theory Simul. 28:1900026. [Google Scholar]
- Luo Z., Jiang J. (2012). pH-sensitive drug loading/releasing in amphiphilic copolymer PAE-PEG: integrating molecular dynamics and dissipative particle dynamics simulations. J. Control Release 162 185–193. 10.1016/j.jconrel.2012.06.027 [DOI] [PubMed] [Google Scholar]
- Ma D., DeBenedictis E. P., Lund R., Keten S. (2016). Design of polymer conjugated 3-helix micelles as nanocarriers with tunable shapes. Nanoscale 8 19334–19342. 10.1039/C6NR07125B [DOI] [PubMed] [Google Scholar]
- Macháèková M., Tokarskı J., Èapková P. (2013). A simple molecular modeling method for the characterization of polymeric drug carriers. Eur. J. Pharm. Sci. 48 316–322. 10.1016/j.ejps.2012.11.010 [DOI] [PubMed] [Google Scholar]
- Maeda H., Nakamura H., Fang J. (2013). The EPR effect for macromolecular drug delivery to solid tumors: improvement of tumor uptake, lowering of systemic toxicity and distinct tumor imaging in vivo. Adv. Drug Del. Rev. 65:71. 10.1016/j.addr.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Magarkar A., Karakas E., Stepniewski M., Roìg T., Bunker A. (2012). Molecular dynamics simulation of PEGylated bilayer interacting with salt ions: a model of the liposome surface in the bloodstream. J. Phys. Chem. B 116 4212–4219. 10.1021/jp300184z [DOI] [PubMed] [Google Scholar]
- Magarkar A., Parkkila P., Viitala T., Lajunen T., Mobarak E., Licari G., et al. (2018). Membrane bound COMT isoform is an interfacial enzyme: general mechanism and new drug design paradigm. Chem. Commun. 54 3440–3443. 10.1039/c8cc00221e [DOI] [PubMed] [Google Scholar]
- Magarkar A., Roìg T., Bunker A. (2014). Molecular dynamics simulation of PEGylated membranes with cholesterol: building toward the DOXIL formulation. J. Phys. Chem. C 118 15541–15549. 10.1021/jp504962m [DOI] [Google Scholar]
- Magarkar A., Roìg T., Bunker A. (2017). A computational study suggests that replacing PEG with PMOZ may increase exposure of hydrophobic targeting moiety. Eur. J. Pharm. Sci. 103 128–135. 10.1016/j.ejps.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Mahdavi M., Fattahi A., Tajkorshid E., Nouranian S. (2020). Molecular insights into the loading and dynamics of doxorubicin on PEGylated graphene oxide nanocarriers. ACS Appl. Biol. Mater. 3 1354–1363. 10.1021/acsabm.9b00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi M. (2016). Protein corona: the golden gate to clinical applications of nanoparticles. Int. J. Biochem. Cell Biol. 75 141–142. 10.1016/j.biocel.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Maiti P. K., Bagchi B. (2006). Structure and dynamics of DNA-dendrimer complexation: role of counterions, water, and base pair. Nano Lett. 6 2478–2485. 10.1021/nl061609m [DOI] [PubMed] [Google Scholar]
- Maleki R., Afrouzi H. H., Hosseini M., Toghraie D., Piranfar A., Rostami S. (2020). pH-sensitive loading/releasing of doxorubicin using single-walled carbon nanotube and multi-walled carbon nanotube: a molecular dynamics study. Comput. Meth. Prog. Biol. 186:105210. 10.1016/j.cmpb.2019.105210 [DOI] [PubMed] [Google Scholar]
- Marianecci C., Di Marzio L., Rinaldi F., Celia C., Paolino D., Alhaique F., et al. (2014). Niosomes from 80s to present: the state of the art. Adv. Colloid Interf. 205 187–206. 10.1016/j.cis.2013.11.018 [DOI] [PubMed] [Google Scholar]
- Marrink S. J., Risselada H. J., Yefimov S., Tieleman D. P., de Vries A. H. (2007). The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B 111 7812–7824. 10.1021/jp071097f [DOI] [PubMed] [Google Scholar]
- Martinez D., Decossas M., Kowal J., Frey L., Stahlberg H., Dufourc E. J. E. J., et al. (2017). Lipid internal dynamics probed in nanodiscs. Chem. Phys. Chem. 18 2651–2657. 10.1002/cphc.201700450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Veracoechea F. J., Frenkel D. (2011). Designing super selectivity in multivalent nano-particle binding. Proc. Natl. Acad. Sci. U.S.A. 108 10963–10968. 10.1073/pnas.1105351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho N., Florindo H., Silva L., Brocchini S., Zloh M., Barata T. (2014). Molecular modeling to study dendrimers for biomedical applications. Molecules 19 20424–20467. 10.3390/molecules191220424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwah M., Magarkar A., Ray D., Aswal V. K., Bunker A., Nagarsenker M. (2018). Glyceryl monostearate: probing the self assembly of a lipid amenable to surface modification for hepatic targeting. J. Phys. Chem. C 122 22160–22169. 10.1021/acs.jpcc.8b05931 [DOI] [Google Scholar]
- Massiot J., Makky A., Di Meo F., Chapron D., Trouillas P., Rosilio V. (2017). Impact of lipid composition and photosensitizer hydrophobicity on the efficiency of light-triggered liposomal release. Phys. Chem. Chem. Phys. 19 11460–11473. 10.1039/C7CP00983F [DOI] [PubMed] [Google Scholar]
- Mastrotto F., Brazzale C., Bellato F., De Martin S., Grange G., Mahmoudzadeh M., et al. (2020). In vitro and in vivo behavior of liposomes decorated with PEGs with different chemical features. Mol. Pharm. 17 472–487. 10.1021/acs.molpharmaceut.9b00887 [DOI] [PubMed] [Google Scholar]
- Mathieu D. (2020). QSPR versus fragment-based methods to predict octanol-air partition coefficients: revisiting a recent comparison of both approaches. Chemosphere 245:125584. 10.1016/j.chemosphere.2019.125584 [DOI] [PubMed] [Google Scholar]
- Matos G. D. R., Kyu D. Y., Loeffler H. H., Chodera J. D., Shirts M. R., Mobley D. L. (2017). Approaches for calculating solvation free energies and enthalpies demonstrated with an update of the FreeSolv database. J. Chem. Eng. Data 62 1559–1569. 10.1021/acs.jced.7b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos G. D. R., Mobley D. L. (2019). Challenges in the use of atomistic simulations to predict solubilities of drug-like molecules. F1000Research 7:686. 10.12688/f1000research.14960.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayne C. G., Arcario M. J., Mahinthichaichan P., Baylon J. L., Vermaas J. V., Navidpour L., et al. (2016). The cellular membrane as a mediator for small molecule interaction with membrane proteins. Biochim. Biophys. Acta Biomembr. 1858 2290–2304. 10.1016/j.bbamem.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazloom-Jalali A., Shariatinia Z. (2019). Polycaprilactone nanocomposite systems used to deliver ifosfamide anticancer drug: molecular dynamics simulations. Struct. Chem. 30 863–876. 10.1007/s11224-018-1233-y [DOI] [Google Scholar]
- Mehta C. H., Narayan R., Nayak U. Y. (2019). Computational modelling for formulation design. Drug Discov. Today 24 781–788. 10.1016/j.drudis.2018.11.018 [DOI] [PubMed] [Google Scholar]
- Meier K., Choutko A., Dolenc J., Eichenberger A. P., Riniker S., van Gunsteren W. F. (2013). Multi-resolution simulation of biomolecular systems: a review of methodological issues. Angew. Chem. Int. Edn. 52 2820–2834. 10.1002/anie.201205408 [DOI] [PubMed] [Google Scholar]
- Mhasal A. R., Roy S. (2016). Free energy of bare and capped gold nanoparticles permeating through a lipid bilayer. Chem. Phys. Chem. 17 3504–3514. 10.1002/cphc.201600690 [DOI] [PubMed] [Google Scholar]
- Mhashal A. R., Roy S. (2014). Effect of gold nanoparticle on structure and fluidity of lipid membrane. PLoS One 9:e114152. 10.1371/journal.pone.0114152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel J., Essex J. W. (2010). Prediction of protein-ligand binding affinity by free energy simulations: assumptions, pitfalls and expectations. J. Comput. Aid. Mol. Des. 24 639–658. 10.1007/s10822-010-9363-3 [DOI] [PubMed] [Google Scholar]
- Miller S. E., Yamada Y., Patel N., Suárez E., Andrews C., Tau S., et al. (2019). Electrostatically driven Guanidinium interaction domains that control hydrogel-mediated protein delivery in vivo. ACS Cent. Sci. 5 1750–1759. 10.1021/acscentsci.9b00501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W., Zhao D., Quan X., Sun D., Li L., Zhou J. (2017). Computer simulations on the pH-sensitive tri-block copolymer containing zwitterionic sulfobetaine as a novel anti-cancer drug carrier. Colloid Surf. B 152 260–268. 10.1016/j.colsurfb.2017.01.033 [DOI] [PubMed] [Google Scholar]
- Miyazaki Y., Okazaki S., Shinoda W. (2020). pSpica: a coarse-grained force field for lipid membranes based on a polar water model. J. Chem. Theory Comput. 16 782–793. 10.1021/acs.jctc.9b00946 [DOI] [PubMed] [Google Scholar]
- Mobarak E., Javanainen M., Kulig W., Honigmann A., Sezgin E., Aho N., et al. (2018). How to minimize dye-induced perturbations while studying biomembrane structure and dynamics: PEG linkers as a rational alternative. Biochim. Biophys. Acta Biomembr. 1860 2436–2445. 10.1016/j.bbamem.2018.07.003 [DOI] [PubMed] [Google Scholar]
- Mobasheri M., Attar H., Sorkhabadi S. M. R., Khamesipour A., Jaafari M. R. (2016). Solubilization behavior of polyene antibiotics in nanomicellar system: insights from molecular dynamics simulation of the amphotericin B and nystatin interactions with polysorbate 80. Molecules 21:6. 10.3390/molecules21010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghimi S. M., Simberg D., Anchordoquy T. J. (2020). Tuning the engines of nanomedicine. Mol. Ther. 28 693–694. 10.1016/j.ymthe.2020.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monpara J., Kanthou C., Tozer G. M., Vavia P. R. (2018). Rational design of cholesterol derivative for improved stability of paclitaxel cationic liposomes. Pharm. Res. 35:90. 10.1007/s11095-018-2367-8 [DOI] [PubMed] [Google Scholar]
- Moore T. C., Yang A. H., Ogungbesan O., Hartkamp R., Iacovella C. R., Zhang Q., et al. (2019). Influence of single-stranded DNA coatings on the Interaction between Graphene Nanoflakes and Lipid Bilayers. J. Phys. Chem. B 123 7711–7721. 10.1021/acs.jpcb.9b04042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi S., Taran M., Mohajeri P., Sadrjavadi K., Sarrami F., Karton A., et al. (2018). Study of dual encapsulation possibility of hydrophobic and hydrophilic drugs into a nanocarrier based on bio-polymer coated graphene oxide using density functional theory, molecular dynamics and experimental methods. J. Mol. Liq. 262 204–217. 10.1016/j.molliq.2018.04.089 [DOI] [Google Scholar]
- Mortazavifar A., Raissi H., Akbari A. (2019). DFT and MD investigations on the functionalized boron nitride nanotube as an effective drug delivery carrier for carmustine anticancer drug. J. Mol. Liq. 276 577–587. 10.1016/j.molliq.2018.12.028 [DOI] [Google Scholar]
- Mousavi M., Hakimian S., Mustafa T. A., Aziz F. M., Salihi A., Ale-Ebrahim M., et al. (2019). The interaction of silica nanoparticles with catalase and human mesenchymal stem cells: biophysical, theoretical and cellular studies. Int. J. Nanomed. 14 5355–5368. 10.2147/IJN.S210136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi S.-D., Maghsoodi F., Panahandeh F., Tazdian-Robati R., Reisi-Vanani A., Tafaghodi M. (2018). Doxorubicin delivery via magnetic nanomicelles comprising from reduction-responsive poly(ethylene glycol)-b-poly(s-caprilactone) (PEG-SS-PCL) and loaded with superparamagnetic iron oxide (SPIO) nanoparticles: preparation, characterization and loaded with. Mater. Sci. Eng. C 92 631–643. 10.1016/j.msec.2018.06.066 [DOI] [PubMed] [Google Scholar]
- Mu Q., Hu T., Yu J. (2013). Molecular insight into the steric shielding effect of PEG on the conjugated staphylokinase: biochemical characterization and molecular dynamics simulation. PLoS One 8:e68559. 10.1371/journal.pone.0068559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munasinghe A., Mathavan A., Mathavan A., Lin P., Colina C. M. (2019). PEGylation within a confined hydrophobic cavity of a protein. Phys. Chem. Chem. Phys. 21 25584–25596. 10.1039/C9CP04387J [DOI] [PubMed] [Google Scholar]
- Murtola T., Bunker A., Vattulainen I., Deserno M., Karttunen M. (2009). Multiscale modeling of emergent materials: biological and soft matter. Phys. Chem. Chem. Phys. 11 1869–1892. 10.1039/B818051B [DOI] [PubMed] [Google Scholar]
- Myint K. H., Brown J. R., Shim A. R., Wylouzil B. E., Hall L. M. (2016). Encapsulation of nanoparticles during polymer micelle formation: a dissipative particle dynamics study. J. Phys. Chem. B 120 11582–11594. 10.1021/acs.jpcb.6b07324 [DOI] [PubMed] [Google Scholar]
- Myung Y., Yeom S., Han S. (2016). A niosomal bilayer of sorbitan monostearate in complex with flavones: a molecular dynamics study. J. Liposome Res. 26 336–344. 10.3109/08982104.2016.1144204 [DOI] [PubMed] [Google Scholar]
- Nag O. K., Delehanty J. B. (2019). Active cellular and subcellular targeting of nanoparticles for drug delivery. Pharmaceutics 11:543. 10.3390/pharmaceutics11100543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandy B., Maiti P. K. (2011). DNA compaction by a dendrimer. J. Phys. Chem. B 115 217–230. 10.1021/jp106776v [DOI] [PubMed] [Google Scholar]
- Nandy B., Maiti P. K., Bunker A. (2013). Force biased molecular dynamics simulation study of effect of dendrimer generation on interaction with DNA. J. Chem. Theory Comput. 9 722–729. 10.1021/ct300678r [DOI] [PubMed] [Google Scholar]
- Nandy B., Santosh M., Maiti P. K. (2012). Interaction of nucleic acids with carbon nanotubes and dendrimers. J. Biosci. 37 457–474. 10.1007/s12038-012-9220-8 [DOI] [PubMed] [Google Scholar]
- Nantasenamat C., Isarankura-Na-Ayudhya C., Naenna T., Prachayasittikul V. (2009). A practical overview of qualitative structure-activity relationship. EXCLI J. 8 74–88. 10.17877/DE290R-690 [DOI] [Google Scholar]
- Narayan K. L., Rao K. M., Sarcar M. M. M. (2008). Computer Aided Design and Manufacturing. New Delhi: Prentice Hall of India. [Google Scholar]
- Nasr M. L., Baptista D., Strauss M., Sun Z. Y. J., Grigoriu S., Huser S., et al. (2016). Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 14 49–52. 10.1038/nmeth.4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale C., Bennett W. F. D., Tieleman D. P., Pomès R. (2011). Statistical convergence of equilibrium properties in simulations of molecular solutes embedded in lipid bilayers. J. Chem. Theory Comput. 7 4175–4188. 10.1021/ct200316w [DOI] [PubMed] [Google Scholar]
- Neale C., Pomès R. (2016). Sampling errors in free energy simulations of small molecules in lipid bilayers. Biochim. Biophys. Acta Biomembr. 1858 2539–2548. 10.1016/j.bbamem.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Nie S. Y., Lin W. J., Yao N., Guo X. D., Zhang L. J. (2014). Drug release from pH-sensitive polymeric micelles with different drug distributions: insight from coarse-grained simulations. ACS Appl. Mater. Interf. 6 17668–17678. 10.1021/am503920m [DOI] [PubMed] [Google Scholar]
- Nie S. Y., Sun Y., Lin W. J., Wu W. S., Guo X. D., Qian Y., et al. (2013). Dissipative particle dynamics studies of doxorubicin-loaded micelles assembled from four-arm star triblock polymers 4AS-PCL-b-PDEAEMA-b-PPEGMA and their pH-release mechanism. J. Phys. Chem. B 117 13688–13697. 10.1021/jp407529u [DOI] [PubMed] [Google Scholar]
- Nienhaus K., Nienhaus G. U. (2019). Towards a molecular-level understanding of the protein corona around nanoparticles - recent advances and persisting challenges. Curr. Opin. Biomed. Eng. 10 11–22. 10.1016/j.cobme.2019.01.002 [DOI] [Google Scholar]
- Nikfar Z., Shariatinia Z. (2019). The RGD tripeptide anticancer drug carrier: DFT computations and molecular dynamics simulations. J. Mol. Liq. 281 565–583. 10.1016/j.molliq.2019.02.114 [DOI] [Google Scholar]
- Nischan N., Hackenberger C. P. R. (2014). Site-specific PEGylation of proteins: recent developments. J. Organ. Chem. 79 10727–10733. 10.1021/jo502136n [DOI] [PubMed] [Google Scholar]
- Numata M., Grinkova Y. V., Mitchell J. R., Chu H. W., Sligar S. G., Voelker D. R. (2013). Nanodiscs as a therapeutic delivery agent: inhibition of respiratory syncytial virus infection in the lung. Int. J. Nanomed. 8 1417–1427. 10.2147/IJN.S39888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nummelin S., Selin M., Legrand S., Ropponen J., Seitsonen J., Nykänen A., et al. (2017). Modular synthesis of self-assembling Janus-dendrimers and facule preparation of drug-loaded dendrimersomes. Nanoscale 9 7189–7198. 10.1039/C6NR08102A [DOI] [PubMed] [Google Scholar]
- Ogawara K., Yoshizawa Y., Un K., Araki T., Kimura T., Higaki K. (2013). Nanoparticle-based passive drug targeting to tumors: considerations and implications for optimization. Biol. Pharm. Bull. 36 698–702. 10.1248/bpb.b13-00015 [DOI] [PubMed] [Google Scholar]
- Ollila O. H. S., Pabst G. (2016). Atomistic resolution structure and dynamics of lipid bilayers in simulations and experiments. Biochim. Bbiophys. Acta Biomembr. 1858 2512–2528. 10.1016/j.bbamem.2016.01.019 [DOI] [PubMed] [Google Scholar]
- Oroskar P. A., Jameson C. J., Murad S. (2015). Surface-functionalized nanoparticle permeation triggers lipid displacement and water and ion leakage. Langmuir 31 1074–1085. 10.1021/la503934c [DOI] [PubMed] [Google Scholar]
- Oroskar P. A., Jameson C. J., Murad S. (2016). Simulated permeation and characterization of PEGylated gold nanoparticles in a lipid bilayer system. Langmuir 32 7541–7555. 10.1021/acs.langmuir.6b01740 [DOI] [PubMed] [Google Scholar]
- Otto D. P., Otto A., De Villers M. M. (2013). Experimental and mesoscale computational dynamics studies of the relationship between solubility and release of quercetin from PEG solid dispersions. Int. J. Pharm. 456 282–292. 10.1016/j.ijpharm.2013.08.039 [DOI] [PubMed] [Google Scholar]
- Ou L., Corradi V., Tieleman D. P., Liang Q. (2020). Atomistic simulations on interactions between amphiphilic Janus nanoparticles and lipid bilayers: effects of lipid ordering and leaflet asymmetry. J. Phys. Chem. B 124 4466–4475. 10.1021/acs.jpcb.9b11989 [DOI] [PubMed] [Google Scholar]
- Ouyang D., Smith S. C. (2015). “Introduction to computational pharmaceutics,” in Computational Pharmaceutics: Application of Molecular Modeling in Drug Delivery, eds Ouyang D., Smith S. C. (Hoboken, NJ: Wiley; ), 27–31. [Google Scholar]
- Pakdel M., Raissi H., Shahabi M. (2020). Predicting doxorubicin drug delivery by single-walled carbon nanotube through cell membrane in the absence and presence of nicotine molecules: a molecular dynamics simulation study. J. Biomol. Struct. Dyn. 38 1488–1498. 10.1080/07391102.2019.1611474 [DOI] [PubMed] [Google Scholar]
- Paluch A. S., Parameswaran S., Liu S., Kolavennu A., Mobley D. L. (2015). Predicting the excess solubility of acetanilide, acetaminophen, phenacetin, benzocaine, and caffeine in binary water/ethanol mixtures via molecular simulation. J. Chem. Phys. 142:44508 10.1063/1.4906491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Segrest J. P. (2016). Computational studies of plasma lipoprotein lipids. Biochim. Biophys. Acta Biomembr. 1858 2401–2420. 10.1016/j.bbamem.2016.03.010 [DOI] [PubMed] [Google Scholar]
- Panczyk T., Jagusiak A., Pastorin G., Ang W. H., Narkiewicz-Michalek J. (2013). Molecular dynamics study of cisplatin release from carbon nanotubes capped by magnetic nanoparticles. J. Phys. Chem. C 117 17327–17336. 10.1021/jp405593u [DOI] [Google Scholar]
- Panczyk T., Wojton P., Wolski P. (2020). Molecular dynamics study of the interaction of carbon nanotubes with telomeric DNA fragment containing noncanonical G-quadruplex and i-motif forms. Int. J. Mol. Sci. 21:1925. 10.3390/ijms21061925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. (2016). Drug delivery of the future: chasing the invisible gorilla. J. Control Release 240 2–8. 10.1016/j.jconrel.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. (2019). The beginning of the end of nanomedicine. J. Control Release 305 221–222. 10.1016/j.jconrel.2019.05.044 [DOI] [PubMed] [Google Scholar]
- Parray Z. A., Hassan M. I., Ahmad F., Islam A. (2020). Amphiphilic nature of polyethylene glycols and their role in medical research. Polym. Test. 82:106316 10.1016/j.polymertesting.2019.106316 [DOI] [Google Scholar]
- Pasenkiewicz-Gierula M., Baczynski K., Markiewicz M., Murzyn K. (2016). Computer modelling studies of the bilayer/water interface. Biochim. Biophys. Acta Biomembr. 1858 2305–2321. 10.1016/j.bbamem.2016.01.024 [DOI] [PubMed] [Google Scholar]
- Pasenkiewicz-Gierula M., Takaoka Y., Miyagawa H., Kitamura K., Kusumi A. (1997). Hydrogen bonding of water to phosphatidylcholine in the membrane as studied by a molecular dynamics simulation: location, geometry, and lipid-lipid bridging via hydrogen-bonded water. J. Phys. Chem. A 101 3677–3691. 10.1021/jp962099v [DOI] [Google Scholar]
- Pasut G., Veronese F. M. (2012). State of the art in PEGylation: the great versatility achieved after 40 years of research. J. Control Release 161 461–472. 10.1016/j.jconrel.2011.10.037 [DOI] [PubMed] [Google Scholar]
- Patel S. K., Lavasanifar A. A., Choi P. (2010a). Molecular dynamics study of the encapsulation capability of a PCL-PEO based block copolymer for hydrophobic drugs with different spatial distributions of hydrogen bond donors and acceptors. Biomaterials 31 1780–1786. [DOI] [PubMed] [Google Scholar]
- Patel S. K., Lavasanifar A., Choi P. (2010b). Prediction of the solubility of cururbitacin drugs in self associationg poly(ethylene oxide)-b-poly(alpha-benzyl carboxylate epsilon-caprolactone) block copolymer with different tacticities using molecular dynamics simulation. Biomaterials 31 345–357. [DOI] [PubMed] [Google Scholar]
- Pathak P., Dhawan V., Magarkar A., Danne R., Govindarajan S., Ghosh S., et al. (2016). Design of cholesterol arabinogalactan anchored liposomes for asialoglycoprotein receptor mediated targeting to hepatocellular carcinoma: In silico modeling, in vitro and in vivo evaluation. Int. J. Pharm. 509 149–158. 10.1016/j.ijpharm.2016.05.041 [DOI] [PubMed] [Google Scholar]
- Pattni B. S., Chupin V. V., Torchilin V. P. (2015). New developments in liposomal drug delivery. Chem. Rev. 115 10938–10966. 10.1021/acs.chemrev.5b00046 [DOI] [PubMed] [Google Scholar]
- Pederzoli F., Tosi G., Vandelli M. A., Belletti D., Forni F., Ruozi B. (2017). Protein corona and nanoparticles: how can we investigate on? WIREs Nanomed. Nanobiotechnol. 9:e1467. 10.1002/wnan.1467 [DOI] [PubMed] [Google Scholar]
- Pelegri-Oday E. M., Lin E. W., Maynard H. D. (2014). Therapeutic protein-polymer conjugates: advancing beyond pegylation. J. Am. Chem. Soc. 136 14323–14332. 10.1021/ja504390x [DOI] [PubMed] [Google Scholar]
- Peng Z., Han X., Li S., Al-Youbi A. O., Bashammakh A. S., El-Shahawi M. S., et al. (2017). Carbon dots: biomacromolecule interaction, bioimaging and nanomedicine. Coord. Chem. Rev. 343 256–277. 10.1016/j.ccr.2017.06.001 [DOI] [Google Scholar]
- Pennetta C., Floresta G., Graziano A. C.-E., Cardile V., Rubino L., Galimberti M., et al. (2020). Functionalization of single and multi-walled carbon nanotubes with polypropylene glycol decorated pyrrole for the development of doxorubicin nano-conveyors for cancer drug delivery. Nanomaterials 10:1073. 10.3390/nano10061073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sánchez G., Vicente F. A., Schaeffer N., Cardoso I. S., Ventura S. P. M., Jorge M., et al. (2020). Unravelling the interactions between surface-active ionic liquids and triblock copolymers for the design of thermal responsive systems. J. Phys. Chem. B 124 7046–7058. 10.1021/acs.jpcb.0c02992 [DOI] [PubMed] [Google Scholar]
- Persson L. C., Porter C. J. H., Charman W. N., Bergström C. A. S. (2013). Computational prediction of drug solubility in lipid based formulation excipients. Pharm. Res. 30 3225–3237. 10.1007/s11095-013-1083-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. L., Lane J. M. D., Ismail A. E., Grest G. S. (2012). Fully atomistic simulations of the response of silica nanoparticle coatings to alkane solvents. Langmuir 28 17443–17449. 10.1021/la3023166 [DOI] [PubMed] [Google Scholar]
- Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., et al. (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem. 26 1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzaru I., Coricovac D., Dehelean C., Moacã E. A., Mioc M., Baderca F., et al. (2018). Stable PEG-coated silver nanoparticles-A comprehensive toxicological profile. Food Chem. Toxicol. 111 546–556. 10.1016/j.fct.2017.11.051 [DOI] [PubMed] [Google Scholar]
- Pison U., Welte T., Giersing M., Groneberg D. A. (2006). Nanomedicine for respiratory diseases. Eur. J. Pharmacol. 533 341–350. 10.1016/j.ejphar.2005.12.068 [DOI] [PubMed] [Google Scholar]
- Plimpton S. (1995). Fast parallel algorithms for short-range molecular dynamics. J. Comp. Phys. 117 1–19. 10.1006/jcph.1995.1039 [DOI] [Google Scholar]
- Poojari C., Wilkosz N., Lira R. B., Dimova R., Jurkiewicz P., Petka R., et al. (2019). Behavior of the DPH fluorescence probe in membranes perturbed by drugs. Chem. Phys. Lipids 223:104784. 10.1016/j.chemphyslip.2019.104784 [DOI] [PubMed] [Google Scholar]
- Poojari C., Zak A., Dzieciuch-Rojek M., Bunker A., Kepczynski M., Róg T. (2020). Cholesterol reduces partitioning of antifungal drug itraconazole into lipid bilayers. J. Phys. Chem. B 124 2139–2148. 10.1021/acs.jpcb.9b11005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postila P. A., Kaszuba K., Sarewicz M., Osyczka A., Vattulainen I., Róg T. (2013). Key role of water in proton transfer at the Qo-site of the cytochrome bc1 complex predicted by atomistic molecular dynamics simulations. Biochim. Biophys. Acta Bioenerg. 1827 761–768. 10.1016/j.bbabio.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Postila P. A., Róg T. (2020). A perspective: active role of lipids in neurotransmitter dynamics. Mol. Neurobiol. 57 910–925. 10.1007/s12035-019-01775-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potdar D., Sammalkorpi M. (2015). Asymmetric heat transfer from nanoparticles in lipid bilayers. Chem. Phys. 463 22–29. 10.1016/j.chemphys.2015.09.016 [DOI] [Google Scholar]
- Pourmousa M., Pastor R. W. (2018). Molecular dynamics simulations of lipid nanodiscs. Biochim. Bbiophys. Acta Biomembr. 1860 2094–2107. 10.1016/j.bbamem.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prhashanna A., Tan W. K., Khan S. A., Chen S. B. (2016). Co-micellization behavior of triblock copolymers in the presence of hydrophobic drug molecules: a simulation study. Colloid Surf. B 148 299–307. 10.1016/j.colsurfb.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Price M. E., Cornelius R. M., Brash J. L. (2001). Protein adsorption to polyethylene glycol modified liposomes from fibrinogen solution and from plasma. Biochim. Biophys. Acta 1512 191–205. 10.1016/S0005-2736(01)00330-3 [DOI] [PubMed] [Google Scholar]
- Qin X., Yu C., Wei J., Li L., Zhang C., Wu Q., et al. (2019). Rational design of nanocarriers for intracellular protein delivery. Adv. Mater. 31:1902791. 10.1002/adma.201902791 [DOI] [PubMed] [Google Scholar]
- Quan X., Zhao D., Li L., Zhou J. (2017). Understanding the cellular uptake of pH-responsive zwitterionic gold nanoparticles: a computer simulation study. Langmuir 33 14480–14489. 10.1021/acs.langmuir.7b03544 [DOI] [PubMed] [Google Scholar]
- Raczyński P., Górny K., Bełdowski P., Yuvan S., Dendzik Z. (2020). Application of graphene as a nanoindenter interacting with phospholipid membranes-computer simulation study. J. Phys. Chem. B 124 6592–6602. 10.1021/acs.jpcb.0c02319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmati S., Shojaei F., Shojaeian A., Rezakhani L., Dehkordi M. B. (2020). An overview of current knowledge in biological functions and potential theragnostic applications of exosomes. Chem. Phys. Lipids 226:104836. 10.1016/j.chemphyslip.2019.104836 [DOI] [PubMed] [Google Scholar]
- Ramalho M. J., Andrade S., Loureiro J. A., do Carmo Pereira M. (2020). Nanotechnology to improve the Alzheimer’s disease therapy with natural compounds. Drug Deliv. Transl. Res. 10 380–402. 10.1007/s13346-019-00694-3 [DOI] [PubMed] [Google Scholar]
- Raman A. S., Pajak J., Chiew Y. C. (2018). Interaction of PCL based self-assembled nano-polymeric micelles with model lipid bilayers using coarse-grained molecular dynamics simulations. Chem. Phys. Lett. 712 1–6. 10.1016/j.cplett.2018.09.049 [DOI] [Google Scholar]
- Ramezani M., Shamsara J. (2016). Application of DPD in the design of polymeric nano-micelles as drug carriers. J. Mol. Graph. Model. 66 1–8. 10.1016/j.jmgm.2016.01.010 [DOI] [PubMed] [Google Scholar]
- Ramezanpour M., Leung S. S. W., Delgado-Magnero K. H., Bashe B. Y. M., Thewalt J., Tieleman D. P. (2016). Computational and experimental approaches for investigating nanoparticle-based drug delivery systems. Biochim. Biophys. Acta 1858 1688–1709. 10.1016/j.bbamem.2016.02.028 [DOI] [PubMed] [Google Scholar]
- Ramli R. A., Laftah W. A., Hashim S. (2013). Core-shell polymers: a review. RSC Adv. 3 15543–15565. 10.1039/C3RA41296B [DOI] [Google Scholar]
- Ramos M. C., Horta V. A. C., Horta B. A. C. (2019). Molecular dynamics simulations of PAMAM and PPI dendrimers using the GROMOS-compatible 2016H66 force field. J. Chem. Inf. Model. 59 1444–1457. 10.1021/acs.jcim.8b00911 [DOI] [PubMed] [Google Scholar]
- Razmimanesh F., Amjad-Iranagh S., Modarress H. (2015). Molecular dynamics simulation study of chitosan and gemcitabine as a drug delivery system. J. Mol. Model. 21:165. 10.1007/s00894-015-2705-2 [DOI] [PubMed] [Google Scholar]
- Ricklin D., Hajishengallis G., Yang K., Lambris J. D. (2010). Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 11 785–797. 10.1038/ni.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehemann K., Schneider S. W., Luger T. A., Godin B., Ferrari M., Fuchs H. (2009). Nanomedicine-challenge and perspectives. Angew. Chem. Int. Edn. 48 872–897. 10.1002/anie.200802585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissanen S., Kumorek M., Martinez-Seara H., Li Y. C., Jamróz D., Bunker A., et al. (2014). Effect of PEGylation on drug entry into lipid bilayer. J. Phys. Chem. B 118 144–151. 10.1021/jp4105745 [DOI] [PubMed] [Google Scholar]
- Ritwiset A., Krongsuk S., Johns J. R. (2016). Molecular structure and dynamical properties of niosome bilayers with and without cholesterol incorporation: a molecular dynamics simulation study. Appl. Surf. Sci. 380 23–31. 10.1016/j.apsusc.2016.02.092 [DOI] [Google Scholar]
- Rodríguez-Hidalgo M.-R., Soto-Figueroa C., Vicente L. (2011). Mesoscopic simulation of the drug release mechanism on the polymeric vehicle P(ST-DVB) in an acid environment. Soft Matter 7 8224–8230. [Google Scholar]
- Rossi G., Monticelli L. (2016). Gold nanoparticles in model biological membranes: a computational perspective. Biochim. Biophys. Acta Biomembr. 1858 2380–2389. 10.1016/j.bbamem.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Roux B. (1995). The calculation of the potential of mean force using computer simulations. Comput. Phys. Commun. 91 275–282. 10.1016/0010-4655(95)00053-I [DOI] [Google Scholar]
- Rozman K. K., Doull J. (2001). Paracelsus, Haber and Arndt. Toxicology 160 191–196. 10.1016/s0300-483x(00)00447-9 [DOI] [PubMed] [Google Scholar]
- Roìg T., Pasenkiewicz-Gierula M. (2004). Non-polar interactions between cholesterol and phospholipids: a molecular dynamics simulation study. Biophys. Chem. 107 151–164. 10.1016/j.bpc.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Róg T., Vattulainen I. (2014). Cholesterol, sphingolipids, and glycolipids: what do we know about their role in raft-like membranes? Chem. Phys. Lipids 184 82–104. 10.1016/j.chemphyslip.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Ruiz-Garcia A., Bermejo M., Moss A., Casabo V. G. (2008). Pharmacokinetics in drug discovery. J. Pharm. Sci. 97 654–690. 10.1002/jps.21009 [DOI] [PubMed] [Google Scholar]
- Rungrotmongkol C. R. T., Poo-arporn R. P. (2016). pH-controlled doxorubicin anticancer loading and release from carbon nanotube noncovalently modified by chitosan: MD simulations. J. Mol. Graph. Model. 70 70–76. 10.1016/j.jmgm.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Salassi S., Canepa E., Ferrando R., Rossi G. (2019). Anionic nanoparticle-lipid membrane interactions: the protonation of anionic ligands at the membrane surface reduces membrane disruption. RSC. Adv. 9 13992–13997. 10.1039/C9RA02462J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salassi S., Simonelli F., Bochicchio D., Ferrando R., Rossi G. (2017). Au nanoparticles in lipid bilayers: a comparison between atomistic and coarse-grained models. J. Phys. Chem. C 121 10927–10935. 10.1021/acs.jpcc.6b12148 [DOI] [Google Scholar]
- Salorinne K., Malola S., Wong O. A., Rithner C. D., Chen X., Ackerson C. J., et al. (2016). Conformation and dynamics of the ligand shell of a water-soluble Au 102 nanoparticle. Nat. Commun. 7:10401. 10.1038/ncomms10401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos H. A., Mäkilä E., Airaksinen A. J., Bimbo L. M., Hirvonen J. (2014). Porous silicon nanoparticles for nanomedicine: preparation and biomedical applications. Nanomedicine 9 535–554. 10.2217/nnm.13.223 [DOI] [PubMed] [Google Scholar]
- Sarma J. V., Ward P. A. (2011). The complement system. Cell Tissue Res. 343 227–235. 10.1007/s00441-010-1034-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanell J. W., Blanckley A., Boldon H., Warrington B. (2012). Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11 191–200. 10.1038/nrd3681 [DOI] [PubMed] [Google Scholar]
- Schachter I., Allolio C., Khelashvili G., Harries D. (2020). Confinement in nanodiscs anisotropically modifies lipid bilayer elastic properties. J. Phys. Chem. B 124 7166–7175. 10.1021/acs.jpcb.0c03374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer T., Zhou J., Schmid F., Settanni G. (2017). “Blood proteins and their interactions with nanoparticles investigated using molecular dynamics simulation,” in High Performance Computing in Science and Engineering ‘17, eds Nagel W. E., Kröner D. H., Resch M. M. (Berlin: Springer; ), 5–19. 10.1007/978-3-319-68394-2 [DOI] [Google Scholar]
- Scheideler M., Vidakovic I., Prassl R. (2020). Lipid nanocarriers for microRNA delivery. Chem. Phys. Lipids 226:104837. 10.1016/j.chemphyslip.2019.104837 [DOI] [PubMed] [Google Scholar]
- Schlapschy M., Binder U., Börger C., Theobald I., Wachinger K., Kisling S., et al. (2013). PASylation: a biological alternative to PEGylation for extending the plasma half-life of pharmaceutically active proteins. Protein. Eng. Des. Sel. 26 489–501. 10.1093/protein/gzt023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler S., Becker G., Winzen S., Steinbach T., Mohr K., Landfester K., et al. (2016). Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)- coated nanocarriers. Nat. Nanotechnol. 11 372–377. 10.1038/nnano.2015.330 [DOI] [PubMed] [Google Scholar]
- Schubertová V., Martinez-Veracoechea F. J., Vácha R. (2015). Influence of ligand distribu- tion on uptake efficiency. Soft Matter 11 2726–2730. 10.1039/C4SM02815E [DOI] [PubMed] [Google Scholar]
- Sedlacek O., Monnery B. D., Filippov S. K., Hoogenboom R., Hruby M. (2012). Poly(2-oxazoline)s - are they more advantageous for biomedical applications than other polymers? Macromol. Rapid Commun. 33 1648–1662. 10.1002/marc.201200453 [DOI] [PubMed] [Google Scholar]
- Šegota S., Durdica T. (2006). Spontaneous formation of vesicles. Adv. Coll. Int. Sci. 121 51–75. 10.1063/1.465966 [DOI] [PubMed] [Google Scholar]
- Sen S., Han Y., Rehak P., Vuković L., Král P. (2018). Computational studies of micellar and nanoparticle nanomedicines. Chem. Soc. Rev. 47 3849–3860. 10.1039/C8CS00022K [DOI] [PubMed] [Google Scholar]
- Settanni G., Schäfer T., Muhl C., Barz M., Schmid F. (2018). Poly-sarcosine and poly(ethylene-glycol) interactions with proteins investigated using molecular dynamics simulations. Comput. Struct. Biotechnol. 16 543–550. 10.1016/j.csbj.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settanni G., Zhou J., Schmid F. (2017a). Interactions between proteins and poly(ethylene-glycol) investigated using molecular dynamics simulations. J. Phys. Conf. Ser. 921:12002 10.1088/1742-6596/921/1/012002 [DOI] [Google Scholar]
- Settanni G., Zhou J., Suo T., Schöttler S., Landfester K., Schmid F., et al. (2017b). Protein corona composition of poly(ethylene glycol)- and poly(phosphoester)-coated nanoparticles correlates strongly with the amino acid composition of the protein surface. Nanoscale 9 2138–2144. 10.1039/C6NR07022A [DOI] [PubMed] [Google Scholar]
- Sgarlata C., D’Urso L., Consiglio G., Grasso G., Satriano C., Forte G. (2016). pH sensitive functionalized graphene oxide as a carrier for delivering Gemcitabine: a computational approach. Comput. Theor. Chem. 1096 1–6. 10.1016/j.comptc.2016.09.026 [DOI] [Google Scholar]
- Shadrack D. M., Swai H. S. (2019). Solvent effects on molecular encapsulation of Toussantine-A by chiotosan nanoparticle: a metadynamics study. J. Mol. Liq. 292:111434 10.1016/j.molliq.2019.111434 [DOI] [Google Scholar]
- Shah S., Liu Y., Hu W., Gao J. (2011). Modeling particle shape-dependent dynamics in nanomedicine. J. Nanosci. Nanotechnol. 11 919–928. 10.1166/jnn.2011.3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahabi M., Raissi H. (2020). Payload delivery of anticancer drug Tegafur with the assistance of graphene oxide nanosheet during biomembrane penetration: molecular dynamics simulation survey. Appl. Surf. Sci. 517:146186 10.1016/j.apsusc.2020.146186 [DOI] [Google Scholar]
- Shamsi M., Mohammadi A., Manshadi M. K. D., Sanati-Nezhad A. (2019). Mathematical and computational modeling of nano-engineered drug delivery systems. J. Control Release 307 150–165. 10.1016/j.jconrel.2019.06.014 [DOI] [PubMed] [Google Scholar]
- Shan P., Shen J.-W., Xu D.-H., Shi L.-Y., Gao J., Lan Y., et al. (2014). Molecular dynamics study on the interaction between doxorubicin and hydrophobically modified chitosan oligosaccaride. RSC. Adv. 4 23730–23739. 10.1039/C4RA01199F [DOI] [Google Scholar]
- Shao Q., Hall C. K. (2016). Protein adsorption on nanoparticles: model development using computer simulation. J. Phys. Condens. Matter 28:414019 10.1088/0953-8984/28/41/414019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariatinia Z., Mazloom-Jalali A. (2019). Chitosan nanocomposite drug delivery systems designed for the ifosfamide anticancer drug using molecular dynamics simulations. J. Mol. Liq. 273 346–367. 10.1016/j.molliq.2018.10.047 [DOI] [Google Scholar]
- Shen J.-W., Li J., Zhao Z., Zhang L., Peng G., Liang L. (2017). Molecular dynamics study on the mechanism of polynucleotide encapsulation by chitosan. Sci. Rep. 7:5050. 10.1038/s41598-017-05197-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Sun Y., Wu H., Zhu C., Wei G., Li J., et al. (2016). Exploring the effect of hydrophilic and hydrophobic structure of grafted polymeric micelles on drug loading. Int. J. Pharm. 512 282–291. 10.1016/j.ijpharm.2016.08.054 [DOI] [PubMed] [Google Scholar]
- Shimizu K., Nakamura H., Watano S. (2016). MD simulation study of direct permeation of a nanoparticle across the cell membrane under an external electric field. Nanoscale 8:11897. 10.1039/C6NR02051H [DOI] [PubMed] [Google Scholar]
- Simonelli F., Bochicchio D., Ferrando R., Rossi G. (2015). Monolayer-protected anionic au nanoparticles walk into lipid membranes step by step. J. Phys. Chem. Lett. 6 3175–3179. 10.1021/acs.jpclett.5b01469 [DOI] [Google Scholar]
- Simonsen J. B. (2016). Evaluation of reconstituted high-density lipoprotein (rHDL) as a drug delivery platform - a detailed survey of rHDL particles ranging from biophysical properties to clinical implications. Nanomed. Nanotechnol. Biol. Med. 12 2161–2179. 10.1016/j.nano.2016.05.009 [DOI] [PubMed] [Google Scholar]
- Siuda I., Tieleman D. P. (2015). Molecular models of nanodiscs. J. Chem. Theory Comput. 11 4923–4932. 10.1021/acs.jctc.5b00668 [DOI] [PubMed] [Google Scholar]
- Sliwakosky G., Kothiwale S., Meiler J., Lowe E. W. (2014). Computational methods in drug discovery. Pharmacol. Rev. 66 334–395. 10.1124/pr.112.007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeijers A. F., Markvoort A. J., Pieterse K., Hilbers P. A. J. (2016a). Coarse-grained modelling of urea-adamantyl functionalized poly(propylene imine) dendrimers. Mol. Simulat. 42 882–895. 10.1080/08927022.2015.1096359 [DOI] [PubMed] [Google Scholar]
- Smeijers A. F., Markvoort A. J., Pieterse K., Hilbers P. A. J. (2016b). Coarse-grained simulations of poly(propylene imine) dendrimers in solution. J. Chem. Phys. 144:74903 10.1063/1.4941379 [DOI] [PubMed] [Google Scholar]
- Smith C. J., Wagle D. V., Bhawawet N., Gehrke S., Hollóczki O., Pingali S. V., et al. (2020). Combined small-angle neutron scattering, diffusion NMR, and Molecular dynamics study of a eutectogel: illuminating the dynamical behavior of glyceline confined in bacterial cellulose gels. J. Phys. Chem. B 124 7647–7658. 10.1021/acs.jpcb.0c04916 [DOI] [PubMed] [Google Scholar]
- Smith D. J., Leal L.-G., Mitragorti S., Shell M. S. (2018). Nanoparticle transport across model cellular membranes: when do solubility-diffusion models break down? J. Phys. D Appl. Phys. 51:294004 10.1088/1361-6463/aacac9 [DOI] [Google Scholar]
- Soltani S., Sardari S., Soror S. A. (2010). Computer simulation of a novel pharmaceutical silicon nanocarrier. Nanotechnol. Sci. Appl. 3 149–157. 10.2147/NSA.S8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somjid S., Krongsuk S., Johns J. R. (2018). Cholesterol concentration effect on the bilayer properties and phase formation of niosome bilayers: a molecular dynamics simulation study. J. Mol. Liq. 256 591–598. [Google Scholar]
- Song B., Yuan H., Jameson C. J., Murad S. (2011). Permeation of nanocrystals across lipid membranes. Mol. Phys. 109 1511–1526. 10.1080/00268976.2011.569511 [DOI] [Google Scholar]
- Song B., Yuan H., Pham S. V., Jameson C. J., Murad S. (2012). Nanoparticle permeation induces water penetration, ion transport and lipid flip-flop. Langmuir 28 16989–17000. 10.1021/la302879r [DOI] [PubMed] [Google Scholar]
- Sousa S. F., Fernandes P. A., Ramos M. J. (2006). Protein-ligand docking: current status and future challenges. Proteins 65 15–26. 10.1002/prot.21082 [DOI] [PubMed] [Google Scholar]
- Souza S. F., Peres J., Coelho M., Vieira T. F. (2018). Analyzing PEGylation through molecular dynamics. Chem. Select 3 8415–8427. 10.1002/slct.201800855 [DOI] [Google Scholar]
- Sridhar D. B., Gubta R., Rai B. (2018). Effect of surface coverage and chemistry on self-assembly of monolayer protected gold nanoparticles: a molecular dynamics simulations study. Phys. Chem. Chem. Phys. 20 25883–25891. 10.1039/c8cp04044c [DOI] [PubMed] [Google Scholar]
- Srinivas G., Mohan R. V., Kelkar A. D. (2013). Polymer micelle assisted transport and delivery of model hydrophilic components inside a biological lipid vesicle: a coarse grained simulation study. J. Phys. Chem. B 117 12095–12104. 10.1021/jp405381k [DOI] [PubMed] [Google Scholar]
- Stepien P., Augustyn B., Poojari C., Galan W., Polit A., Vattulainen I., et al. (2020). Complexity of seemingly simple lipid nanodiscs. Biochim. Bbiophys. Acta Biomembr. 1862:183420. 10.1016/j.bbamem.2020.183420 [DOI] [PubMed] [Google Scholar]
- Stepien P., Polit A., Wisniewska-Becker A. (2015). Comparative EPR studies on lipid bilayer properties in nanodiscs and liposomes. Biochim. Biophys. Acta Biomembr. 1848 60–66. 10.1016/j.bbamem.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Stepniewski M., Kepczynski M., Jamróz D., Nowakowska M., Rissanen S., Vattulainen I., et al. (2012). Interaction of hematoporphyrin with lipid membranes. J. Phys. Chem. B 116 4889–4897. 10.1021/jp300899b [DOI] [PubMed] [Google Scholar]
- Stepniewski M., Pasenkiewicz-Gierula M., Roìg T., Danne R., Orłowski A., Karttunen M., et al. (2011). Study of PEGylated lipid layers as a model for PEGylated liposome surfaces: molecular dynamics simulation and langmuir monolayer studies. Langmuir 27 7788–7798. 10.1021/la200003n [DOI] [PubMed] [Google Scholar]
- Styliari I. D., Taresco V., Theophilus A., Alexander C., Garnett M., Laughton C. (2020). Nanoformulation-by-design: an experimental and molecular dynamics study for polymer coated drug nanoparticles. RSC Adv. 10 19521–19533. 10.1039/D0RA00408A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C.-F., Merliz H., Rabbel H., Sommer J.-U. (2017). Nanoparticles of various degrees of hydrophobicity interacting with lipid membranes. J. Phys. Chem. Lett. 8 4069–4076. 10.1021/acs.jpclett.7b01888 [DOI] [PubMed] [Google Scholar]
- Su Y., Quan X., Li L., Zhao J. (2018). Computer simulation of DNA condensation by PAMAM dendrimer. Macromol. Theory Simul. 27:1700070 10.1002/mats.201700070 [DOI] [Google Scholar]
- Subramaniam B., Siddik Z. H., Nagoor N. H. (2020). Optimization of nanostructured lipid carriers: understanding the types, designs and parameters in the process of formulations. J. Nanopart. Res. 22:141 10.1007/s11051-020-04848-0 [DOI] [Google Scholar]
- Sun H. (1998). COMPASS: an ab initio force-field optimized for condensed-phase applications - overview with details on alkane and benzene compounds. J. Phys. Chem. B 102 7338–7346. 10.1021/jp980939v [DOI] [Google Scholar]
- Sun H., She P., Lu G., Xu K., Zhang W., Liu Z. (2014). Recent advances in the development of functionalized carbon nanotubes: a versatile vector for drug delivery. J. Mater. Sci. 49 6845–6854. 10.1007/s10853-014-8436-4 [DOI] [Google Scholar]
- Sun X., Riccardi L., De Biasi F., Rastrelli F., De Vivo M., Mancin F. (2019). Molecular-dynamics-simulation-directed rational design of nanoreceptors with targeted affinity. Angew. Chem. Int. Edn. 58 7702–7707. 10.1002/anie.201902316 [DOI] [PubMed] [Google Scholar]
- Sun Y., Xia Y. (2003). Gold and silver nanoparticles: a class of chromophores with colors tunable in the range from 400 to 750 nm. Analyst 128 686–691. 10.1039/B212437H [DOI] [PubMed] [Google Scholar]
- Szebeni J., Baranyi L., Savay S., Milosevits J., Bunger R., Laverman P., et al. (2002). Role of complement activation in hypersensitivity reactions to doxil and hynic PEG liposomes: experimental and clinical studies. J. Liposome Res. 12 165–172. 10.1081/lpr-120004790 [DOI] [PubMed] [Google Scholar]
- Tavanti F., Pedone A., Menziani M. C. (2019). Multiscale molecular dynamics simulation of multiple protein adsorptin on gold nanoparticles. Int. J. Mol. Sci. 20:3539. 10.3390/ijms20143539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Nobel Prize in Chemistry 2013. (2013). NobelPrize.org. Nobel Media AB. Available online at: https://www.nobelprize.org/prizes/chemistry/2013/summary/ [Google Scholar]
- Thewalt J., Tieleman D. P. (2016). Biophysical experiments and simulation in nanoparticle based drug delivery systems. J. Drug Target. 24 768–773. 10.1080/1061186X.2016.1221957 [DOI] [PubMed] [Google Scholar]
- Thiel W. (2014). Semiempirical quantim-chemical methods. WIREs Comput. Mol. Sci. 4 145–157. 10.1021/acs.jctc.8b01265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thota N., Hu Z., Jiang J. (2016). Ibuprofen loading and release in amphiphilic peptide FA32 and its derivatives: a coarse-grained molecular dynamics simulation study. Mol. Simulat. 42 679–687. 10.1080/08927022.2015.1079907 [DOI] [Google Scholar]
- Thota N., Jiang J. (2015). Computational amphiphilic materials for drug delivery. Front. Mater. 2:64 10.3389/fmats.2015.00064 [DOI] [Google Scholar]
- Tian F., Yue T., Li Y., Zhang X. (2014a). Computer simulation studies on the interactions between nanoparticles and cell membrane. Sci. China Chem. 57 1662–1671. 10.1007/s11426-014-5231-7 [DOI] [Google Scholar]
- Tian F., Zhang X., Dong W. (2014b). How hydrophobic nanoparticles aggregate in the interior of membranes: a computer simulation. Phys. Rev. E 90:52701. 10.1103/PhysRevE.90.052701 [DOI] [PubMed] [Google Scholar]
- Tian W., Ma Y. (2013). Theoretical and computational studies of dendrimers as delivery vectors. Chem. Soc. Rev. 42 705–727. 10.1039/c2cs35306g [DOI] [PubMed] [Google Scholar]
- Tokarský J., Andrýsek T., ČapkovŁ P. (2011). Molecular modeling of gel nanoparticles with cyclosporine A for oral drug delivery. Int. J. Pharm. 410 196–205. 10.1016/j.ijpharm.2011.03.026 [DOI] [PubMed] [Google Scholar]
- Tomalia D. A., Naylor A. M., Goddard W. A., III (1990). Starburst dendrimers: molecular-level control of size, shape, surface chemistry, topology, and flexibility from atoms to macroscopic matter. Angew. Chem. Int. Edn. Engl. 29 138–175. 10.1002/anie.199001381 [DOI] [Google Scholar]
- Tong R., Kohane D. S. (2016). New strategies in cancer nanomedicine. Annu. Rev. Pharmacol. Toxicol. 56 41–57. 10.1146/annurev-pharmtox-010715-103456 [DOI] [PubMed] [Google Scholar]
- Toporov A. A., Toporova A. P. (2020). QSPR/QSAR: state-of-the-art, wierdness, the future. Molecules 25:1292. 10.3390/molecules25061292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchilin V. P. (2007). Micellar nanocarriers: pharmaceutical perspectives. Pharm. Res. 24 1–16. 10.1007/s11095-006-9132-0 [DOI] [PubMed] [Google Scholar]
- Touitou E., Dayan N., Bergelson L., Godin B., Eliaz M. (2000). Ethosomes - novel vesicular carriers for enhanced delivery: characterization and skin penetration properties. J. Control Release 65 403–418. 10.1016/s0168-3659(99)00222-9 [DOI] [PubMed] [Google Scholar]
- Toy R., Hayden E., Shoup C., Baskaran H., Karathanasis E. (2011). Effect of particle size, density and shape on margination of nanoparticles in microcirculation. Nanotechnology 22:115101 10.1088/0957-4484/22/11/115101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong N. P., Whittaker M. R., Mak C. W., Davis T. P. (2015). The importance of nanoparticle shape in cancer drug therapy. Expert Opin. Drug Deliv. 12 129–142. 10.1517/17425247.2014.950564 [DOI] [PubMed] [Google Scholar]
- Tu C.-K., Chen K., Tian W., Ma Y. (2013). Computational investigations of a peptide-modified dendrimer interacting with lipid membranes. Macromol. Rapid Commun. 34 1237–1242. 10.1002/marc.201300360 [DOI] [PubMed] [Google Scholar]
- Turturro F. (2014). Denileukin diftox: a biotherapeutic paradigm shift in the treatment of lymphoid-derived disorders. Expert. Rev. Anticancer Ther. 7 11–17. 10.1586/14737140.7.1.11 [DOI] [PubMed] [Google Scholar]
- Vácha R., Martinez-Veracoechea F. J., Frenkel D. (2011). Receptor-mediated endocytosis of nanoparticles of various shapes. Nano Lett. 11 5391–5395. 10.1021/nl2030213 [DOI] [PubMed] [Google Scholar]
- van Gunsteren W. F., Dolenc J., Mark A. E. (2008). Molecular simulation as an aid to experimentalists. Curr. Opin. Struct. Biol. 18 149–153. 10.1016/j.sbi.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Van Lehn R. C., Alexander-Katz A. (2014a). Fusion of ligand-coated nanoparticles with lipid bilayers: effect of ligand flexibility. J. Phys. Chem. A 118 5848–5856. 10.1021/jp411662c [DOI] [PubMed] [Google Scholar]
- Van Lehn R. C., Alexander-Katz A. (2014b). Membrane-embedded nanoparticles induce lipid rearrangements similar to those exhibited by biological membrane proteins. J. Phys. Chem. B 118 12586–12598. 10.1021/jp506239p [DOI] [PubMed] [Google Scholar]
- Van Lehn R. C., Alexander-Katz A. (2019). Energy landscape for the insertion of amphiphilic nanoparticles into lipid membranes: a computational study. PLoS One 14:e0209492. 10.1371/journal.pone.0209492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lehn R. C., Atukorale P. U., Carney R. P., Yang Y. S., Stellacci F., Irvine D. J., et al. (2013). Effect of particle diameter and surface composition on the spontaneous fusion of monolayer-protected gold nanoparticles with lipid bilayers. Nano Lett. 13 4060–4067. 10.1021/nl401365n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasumathi V., Maiti P. K. (2010). Complexation of siRNA with dendrimer: a molecular modeling approach. Macromolecules 43 8264–8274. 10.1021/ma1012495 [DOI] [Google Scholar]
- Veiseh O., Tang B. C., Whitehead K. A., Anderson D. G., Langer R. (2015). Managing diabetes with nanomedicine: challenges and opportunities. Nat. Rev. Drug Discov. 14 45–57. 10.1038/nrd4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditto V. J., Szoka F. C. (2013). Cancer nanomedicines: so many papers and so few drugs! Adv. Drug Del. Rev. 65 80–88. 10.1016/j.addr.2012.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer L. S., de Groot B. L., Réat V., Milon A., Czaplicki J. (2007). Acyl chain order parameter profiles in phospholipid bilayers: computation from molecular dynamics simulations and comparison with 2H NMR experiments. Eur. Biophys. J. 36 919–931. 10.1007/s00249-007-0192-9 [DOI] [PubMed] [Google Scholar]
- Vert M., Domurado D. (2000). Poly(ethylene glycol): protein-repulsive or albumin-compatible? J. Biomater. Sci. Polym. Edn. 11 1307–1317. 10.1163/156856200744345 [DOI] [PubMed] [Google Scholar]
- Viegas T. X., Bentley M. D., Harris J. M., Fang Z., Yoon K., Dizman B., et al. (2011). Polyoxazoline: chemistry, properties, and applications in drug delivery. Bioconjug. Chem. 22 976–986. 10.1021/bc200049d [DOI] [PubMed] [Google Scholar]
- Viitala L., Pajari S., Gentile L., Määttä J., Gubitosi M., Deska J., et al. (2019). Shape and phase transitions in a PEGylated phospholipid system. Langmuir 35 3999–4010. 10.1021/acs.langmuir.8b03829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaseca P., Dawson K. A., Franzese G. (2013). Understanding and modulating the competitive surface-adsorption of proteins through coarse-grained molecular dynamics simulations. Soft Matter 9 6978–6985. 10.1039/C3SM50220A [DOI] [Google Scholar]
- Villa F., Quarto R., Tasso R. (2019). Extracellular vesicles as natural, safe and efficient drug delivery systems. Pharmaceutics 11:557. 10.3390/pharmaceutics11110557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal O. D., Rodriguez R. A., Yu L., Wambo T. O. (2016). Molecular dynamics simulations on the effect of size and shape on the interactions between negative Au18(SR)14, Au102(SR)44 and Au144(SR)60 nanoparticles in physiological saline. Colloids Surf. A Physicochem. Eng. Asp. 503 70–78. 10.1016/j.colsurfa.2016.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukoviæ L., Khatib F. A., Drake S. P., Madriaga A., Brandenburg K. S., Král P., et al. (2011). Structure and dynamics of highly PEG-ylated sterically stabilized micelles in aqueous media. J. Am. Chem. Soc. 133 13481–13488. 10.1021/ja204043b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkey C. D., Chan W. C. W. (2012). Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 41 2780–2799. 10.1039/C1CS15233E [DOI] [PubMed] [Google Scholar]
- Wang N., Chen M., Wang T. (2019). Liposomes used as a vaccine adjuvant-delivery system: from basicis to clinical immunization. J. Control Release 303 130–150. 10.1016/j.jconrel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Dormidontova E. E. (2012). Selectivity of ligand-receptor interactions between nanoparticles and cell surfaces. Phys. Rev. Lett. 109:238102. 10.1103/PhysRevLett.109.238102 [DOI] [PubMed] [Google Scholar]
- Wang X.-Y., Zhang L., Wei X.-H., Wang Q. (2013). Molecular dynamics of paclitaxel encapsulated by salicyclic acid-grafted chitosan oligosaccharide aggregates. Biomaterials 34 1843–1851. 10.1016/j.biomaterials.2012.11.024 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen B. Z., Liu Y. J., Wu Z. M., Guo X. D. (2017). Application of mesoscale simulation to explore the aggregate morphology of pH-senstitive nanoparticles used as the oral drug delivery carriers. Colloid Surf. B 151 280–286. 10.1016/j.colsurfb.2016.12.027 [DOI] [PubMed] [Google Scholar]
- Wang Z., Gao J., Ustach V., Li C., Sun S., Hu S., et al. (2017). Tunable permeability of cross-linked microcapsules from pH-responsive amphiphilic diblock copolymers: a dissipative particle dynamics study. Langmuir 33 7288–7297. 10.1021/acs.langmuir.7b01586 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li Q. Y., Liu X. B., Zhang C. Y., Wu Z. M., Guo X. D. (2015a). Mesoscopic simulations and experimental studies of pH-sensitive micelles for controlled drug delivery. ACS Appl. Mater. Interf. 7 25592–25600. 10.1021/acsami.5b08366 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhu D. D., Zhou J., Wang Q. L., Zhang C. Y., Liu Y. J., et al. (2015b). Mesoscopic simulation studies on the formation mechanism of drug loaded polymeric micelles. Colloid Surf. B 136 536–544. 10.1016/j.colsurfb.2015.09.049 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ren J. W., Zhang C. Y., He M. C., Wu Z. M., Guo X. D. (2016). Compatability studies between an amphiphilic pH-sensitive polymer and hydrophobic drug using multiscale simulations. RSC Adv. 6:101323 10.1039/C6RA15950H [DOI] [Google Scholar]
- Warren D. B., King D., Benameur H., Poulton C. W., Chalmers D. K. (2013). Glyceride lipid formulations: molecular dynamics modeling of phase behavior during dispersion and molecular interactions between drugs and excipients. Pharm. Res. 30 3238–3253. 10.1007/s11095-013-1206-1 [DOI] [PubMed] [Google Scholar]
- Wen X.-F., Lan J.-L., Cai Z.-Q., Pi P.-H., Xu S.-P., Zhang L., et al. (2014). Dissipative particle dynamics simulation on drug loading/release in polyester-PEG dendrimer. J. Nanopart. Res. 16:2403 10.1007/s11051-014-2403-5 [DOI] [Google Scholar]
- Westergren J., Lindfors L., Höglund T., Lüder K., Nordholm S., Kjellander R. (2007). In silico prediction of drug solubility: 1. free energy of hydration. J. Phys. Chem. B 111 1872–1882. 10.1021/jp064220w [DOI] [PubMed] [Google Scholar]
- Wilding K. M., Smith A. K., Wilkerson J. W., Bush D. B., Knotts T. A., IV, Bundy B. C. (2018). The locational impact of site-specific PEGylation: streamlines screening with cell-free protein expression and coarse-grain simulation. ACS Synth. Biol. 7 510–521. 10.1021/acssynbio.7b00316 [DOI] [PubMed] [Google Scholar]
- Wilkosz N., Rissanen S., Cyza M., Szybka R., Nowakowska M., Bunker A., et al. (2017). Effect of piroxicam on lipid membranes: drug encapsulation and gastric toxicity aspects. Eur. J. Pharm. Sci. 100 116–125. 10.1016/j.ejps.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Wolski P., Narkiewicz-Michalek J., Panczyk M., Pastorin G., Panczyk T. (2017a). Molecular dynamics modeling of the encapsulation and de-encapsulation of the carmustine anticancer drug in the inner volume of a carbon nanotube. J. Phys Chem. C 121 18922–18934. 10.1021/acs.jpcc.7b05229 [DOI] [Google Scholar]
- Wolski P., Nieszporek K., Panczyk T. (2017b). PEGylated and folic acid functionalized carbon nanotubes as pH controlled carriers of doxorubicin. Molecular dynamics analysis of the stability and drug release mechanism. Phys. Chem. Chem. Phys. 19 9300–9312. 10.1039/C7CP00702G [DOI] [PubMed] [Google Scholar]
- Wolski P., Nieszporek K., Panczyk T. (2018). Multimodal, pH sensitive, and magnetically assisted carrier of doxorubicin designed and analyzed by means of computer simulations. Langmuir 34 2543–2550. 10.1021/acs.langmuir.7b04211 [DOI] [PubMed] [Google Scholar]
- Wolski P., Nieszporek K., Panczyk T. (2020). Carbon nanotubes and short cytosine-rich telomeric DNA oligomers as platforms for controlled release of doxorubicin-a molecular dynamics study. Int. J. Mol. Sci. 21:3619. 10.3390/ijms21103619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolski P., Wojton P., Nieszporek K., Panczyk T. (2019). Interaction of human telomeric i-motif DNA with single-walled carbon nanotubes: insights from molecular dynamics simulations. J. Phys. Chem. B 123 10343–10353. 10.1021/acs.jpcb.9b07292 [DOI] [PubMed] [Google Scholar]
- Wong-ekkabut J., Karttunen M. (2016). The good, the bad and the user in soft matter simulations. Biochim. Biophys. Acta Biomembr. 1858 2529–2538. 10.1016/j.bbamem.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Wu J., Zhao C., Lin W., Hu R., Wang Q., Chen H., et al. (2014). Binding characteristics between polyethylene glycol (PEG) and proteins in aqueous solution. J. Mater. Chem. B 2 2983–2992. 10.1039/C4TB00253A [DOI] [PubMed] [Google Scholar]
- Wu W., Yi P., Zhang J., Cheng Y., Li Z., Hao X., et al. (2019). 4/6-hetero-arm and 4/6-mikto-arm star shaped block polymeric drug loaded micelles and their pH-responsive controlled release properties: a dissipative particle dynamics simulation. Phys. Chem. Chem. Phys. 21 15222–15232. 10.1039/C9CP02411E [DOI] [PubMed] [Google Scholar]
- Wu Z., Duan M., Xiong D., Zhang C. Y. (2019). Mesoscale simulations of pH-responsive amphiphilic polymeric micelles for oral drug delivery. Pharmaceutics 11:620. 10.3390/pharmaceutics11120620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q., Zhu T., Jiang Z., Ding H., Ma Y. (2020). Enhancing the targeting ability of nanoparticles via protected copolymers. Nanoscale 12 7804–7813. 10.1039/d0nr01176b [DOI] [PubMed] [Google Scholar]
- Xiao T., Li D., Shi X., Shen M. (2020). PAMAM dendrimer-based nanodevices for nuclear medicine applications. Macromol. Biosci. 20:1900282. 10.1002/mabi.201900282 [DOI] [PubMed] [Google Scholar]
- Xiao X.-F., Jiang X.-Q., Zhou L.-J. (2013). Surface modification of poly ethylene glycol to resist nonspecific adsorption of proteins. Chin. J. Anal. Chem. 41 445–453. 10.1016/S1872-2040(13)60638-6 [DOI] [Google Scholar]
- Xie X., Xu S., Pi P., Cheng J., Wen X., Liu X., et al. (2018). Dissipative particle dynamic simulation on the assembly and release of siRNA/polymer/gold nanoparticles based polyplex. AIChE J. 64 810–821. 10.1002/aic.15961 [DOI] [Google Scholar]
- Xu D., Smolin N., Shaw R. K., Battey S. R., Tao A., Huang Y., et al. (2018). Molecular insights into the improved clinical performance of PEGylated interferon therapeutics: a molecular dynamics perspective. RSC. Adv. 8 2315–2322. 10.1039/c7ra12480e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., O’Mara M. L., Surawski P. P. T., Trau M., Mark A. E. (2011). Effect of poly(ethylene glycol) (PEG) spacers on the conformational properties of small peptides: a molecular dynamics study. Langmuir 27 296–303. 10.1021/la103800h [DOI] [PubMed] [Google Scholar]
- Yahyaei M., Mehrnejad F., Naderi-manesh H., Rezayan A. H. (2017). Follicle-stimulating hormone encapsulation in the cholesterol-modified chitosan nanoparticles via molecular dynamics simulations and binding free energy calculations. Eur. J. Pharm. Sci. 107 126–137. 10.1016/j.ejps.2017.07.007 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., De Nicola A., Munaò G., Soares T. A. (2019). Effect of the ligand’s bulkiness on the shape of functionalized gold nanoparticles in aqueous solutions: a molecular dynamics study. Chem. Phys. Lett. 731:136576 10.1016/j.cplett.2019.07.004 [DOI] [Google Scholar]
- Yang C., Liu W., Xiao J., Yuan C., Chen Y., Hangbo Yue J. G., et al. (2020). pH-sensitive mixed micelles assembled from PDEAEMA-PPEGMA and PCL-PPEGMA for doxorubicin delivery: experimental and DPD simulation study. Pharmaceutics 12:170. 10.3390/pharmaceutics12020170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Lu D., Liu Z. (2011). How PEGylation enhances the stability and potency of insulin: a molecular dynamics simulation study. Biochemistry 50 2585–2593. 10.1021/bi101926u [DOI] [PubMed] [Google Scholar]
- Yang C., Sun Y., Zhang L., Zhu G., Zhang C., Qian Y. (2012). Dissipative particle dynamics study on aggregation of MPEG-PAE-PLA block polymer micelles loading doxorubicin. Chin. J. Chem. 30 1980–1986. 10.1002/cjoc.201200629 [DOI] [Google Scholar]
- Yang C., Yuan C., Liu W., Guo J., Feng D., Yin X., et al. (2019). DPD studies on mixed micelles self-assembled from MPEG-PDEAEMA and MPEG-PCL for controlled doxorubicin release. Colloid Surf. B 178 56–65. 10.1016/j.colsurfb.2019.02.043 [DOI] [PubMed] [Google Scholar]
- Yang Y.-L., Sheng Y.-J., Tsao H.-K. (2019). Bilayer membranes of Janus dendrimers with hybrid hydrogenated and fluorinated dendrons: microstructures and coassembly with lipids. Phys. Chem. Chem. Phys. 21 15400–15407. 10.1039/c9cp01635j [DOI] [PubMed] [Google Scholar]
- Yang Y., Nie D., Liu Y., Yu M., Gan Y. (2019). Advances in particle shape engineering for improved drug delivery. Drug Discov. Today 24 575–583. 10.1016/j.drudis.2018.10.006 [DOI] [PubMed] [Google Scholar]
- Yang K., Ma Y.-Q. (2010). Computer simulation of the translocation of nanoparticles with different shapes across a lipid bilayer. Nat. Nanotechnol. 5 579–583. 10.1038/NNANO.2010.14 [DOI] [PubMed] [Google Scholar]
- Yang P.-Y., Ju S.-P., Chuang Y.-C., Chen H.-Y. (2017). Molecular dynamics simulations of PAMAM dendrimer-encapsulated Au nanoparticles of different sizes under different pH conditions. Comput. Mater. Sci. 137 144–152. 10.1016/j.commatsci.2017.05.020 [DOI] [Google Scholar]
- Youn Y. S., Bae Y. H. (2018). Perspectives on the past, present, and future of cancer nanomedicine. Adv. Drug Deliv. Rev. 130 3–11. 10.1016/j.addr.2018.05.008 [DOI] [PubMed] [Google Scholar]
- Zaboli M., Raissi H. (2017). The influence of nicotine on pioglitazone encapsulation into carbon nanotube: the investigation of molecular dynamic and density functional theory. J. Biomol. Struct. Dyn. 35 520–534. 10.1080/07391102.2016.1152565 [DOI] [PubMed] [Google Scholar]
- Zaghmi A., Mendez-Villuendas E., Greschner A. A., Liu J. Y., de Haan H. W., Gauthier M. A. (2019). Mechanisms of activity loss for a multi-PEGylated protein by experiment and simulation. Mater. Today Chem. 12 121–131. 10.1016/j.mtchem.2018.12.007 [DOI] [Google Scholar]
- Zazo H., Colino C. I., Lanao J. M. (2016). Current applications of nanoparticles in infectious diseases. J. Control Release 224 86–102. 10.1016/j.jconrel.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Zhadanov V. P. (2019). Formation of a protein corona around nanoparticles. Curr. Opin. Colloid Int. 41 95–103. 10.1016/j.cocis.2018.12.002 [DOI] [Google Scholar]
- Zhang L., Peng G., Li J., Liang L., Kong Z., Wang H., et al. (2018). Molecular dynamics study on the configuration and arrangement of doxorubicin in carbon nanotubes. J. Mol. Liq. 262 295–301. 10.1016/j.molliq.2018.04.097 [DOI] [Google Scholar]
- Zhang L., Wang Z., Lu Z., Shen H., Huang J., Zhao Q., et al. (2013). PEGylated reduced graphene oxide as a superior ssRNA delivery system. J. Mater. Chem. B 1 749–755. 10.1039/C2TB00096B [DOI] [PubMed] [Google Scholar]
- Zhang M., Huang Y., Hao D., Ji Y., Ouyang D. (2020). Solvation structure and molecular interactions of ibuprofen with ethanol and water: a theoretical study. Fluid Phase Equilibr. 510:112454 10.1016/j.fluid.2019.112454 [DOI] [Google Scholar]
- Zhang X., Zheng L., Luo M., Shu C., Wang E. (2020). Evaluation of particle shape, size and magnetic field intensity for targeted delivery efficiency and plaque injury in treating atherosclerosis. Powder Technol. 366 63–72. 10.1016/j.powtec.2020.02.003 [DOI] [Google Scholar]
- Zhang Z., Zhang Y., Song S., Yin L., Sun D., Gu J. (2020). Recent advances in the bioanalytical methods of polyethylene glycols and PEGylated pharmaceuticals. J. Sep. Sci. 43 1978–1997. 10.1002/jssc.201901340 [DOI] [PubMed] [Google Scholar]
- Zhang W., Sun J., Liu Y., Tao M., Ai X., Su X., et al. (2014). PEG-stabilized bilayer nanodisks and carriers of doxorubicin delivery. Mol. Pharm. 11 3279–3290. 10.1021/mp400566a [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhao Q., Deng J., Hu Y., Wang Y., Ouyang D. (2017). Big data analysis of global advances in pharmaceutics and drug delivery 1980-2014. Drug Discov. Today 22 1201–1208. 10.1016/j.drudis.2017.05.012 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lin X., Gu N. (2017). Effects of temperature and PEG grafting density on the translocation of PEGylated nanoparticles across asymmetric lipid membrane. Colloid Surf. B 160 92–100. 10.1016/j.colsurfb.2017.09.013 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tekobo S., Tu Y., Zhou Q., Jin X., Dergunov S. A., et al. (2012). Permission to enter cell by shape: nanodisk vs. nanosphere. ACS Appl. Mater. Interf. 4 4099–4105. 10.1021/am300840p [DOI] [PubMed] [Google Scholar]
- Zhao L., Seth A., Wibowo N., Zhao C.-X., Mitter N., Yu C., et al. (2014). Nanoparticle vaccines. Vaccine 32 327–337. 10.1016/j.vaccine.2013.11.069 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Miriyala N., Su Y., Chen W., Gao X., Shao L., et al. (2018). Computer-aided formulation design for a highly soluble lutein-cyclodextrin multiple-component delivery system. Mol. Pharm. 15 1664–1674. 10.1021/acs.molpharmaceut.8b00056 [DOI] [PubMed] [Google Scholar]
- Zheng L. S., Yang Y. Q., Guo X. D., Sun Y., Qian Y., Zhang L. J. (2011). Mesoscopic simulations on the aggregation behavior of pH-responsive polymeric micelles for drug delivery. J. Colloid Interf. Sci. 363 114–121. 10.1016/j.jcis.2011.07.040 [DOI] [PubMed] [Google Scholar]
- Zhu X., Radovic-Moreno A. F., Wu J., Langer R., Shi J. (2014). Nanomedicine in the management of microbial infection - overview and perspectives. Nano Today 9 478–498. 10.1016/j.nantod.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]


