Abstract
Microtubules are biopolymers that perform diverse cellular functions. Microtubule behavior regulation occurs in part through post-translational modification of both the α- and β-subunits of tubulin. One class of modifications is the heterogeneous addition of glycine and/or glutamate residues to the disordered C-terminal tails (CTTs) of tubulin. Because of their prevalence in stable, high-stress cellular structures such as cilia, we sought to determine if these modifications alter microtubules’ intrinsic stiffness. Here, we describe the purification and characterization of differentially modified pools of tubulin from Tetrahymena thermophila. We found that post-translational modifications do affect microtubule stiffness but do not affect the number of protofilaments incorporated into microtubules. We measured the spin dynamics of nuclei in the CTT backbone by NMR spectroscopy to explore the mechanism of this change. Our results show that the α-tubulin CTT does not protrude out from the microtubule surface, as is commonly depicted in models, but instead interacts with the dimer’s surface. This suggests that the interactions of the α-tubulin CTT with the tubulin body contributes to the stiffness of the assembled microtubule, thus providing insight into the mechanism by which polyglycylation and polyglutamylation can alter microtubule mechanical properties.
Significance
Microtubules are regulated in part by post-translational modifications, including the heterogeneous addition of chains of glycine and glutamate residues to the C-terminal tails. By producing and characterizing differentially modified tubulin, this work provides insight into the molecular mechanisms of how these modifications alter intrinsic microtubule properties, such as flexibility. These results have broader implications for revealing how ciliary structures are able to function under high stress.
Introduction
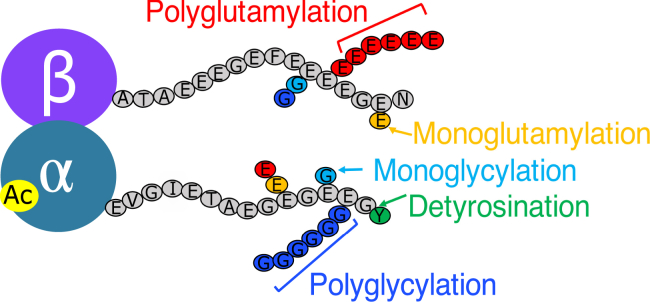
Microtubules are biopolymers involved in diverse cellular processes, including mitosis, transport and signaling, assembly of cilia and flagella for movement, and the cellular cytoskeleton structure (1, 2, 3, 4, 5). This diversity of function is enabled by post-translational modifications (6,7). Modifications can occur on both the ordered core of the tubulin molecules or the disordered C-terminal tails (CTTs) of the α- and β-tubulin monomers (summarized in Fig. 1). Functionally different subsets of microtubules vary in the degree and type of post-translational modification. For example, acetylation on the lumen of microtubules is common in stable microtubules (9) and is thought to protect these long-lived microtubules from buckling (10). Microtubule stability is also correlated with detyrosination, which removes of the C-terminal tyrosine from α-tubulin (11,12). Microtubules undergoing the dynamic process of mitosis contain mainly tyrosinated tubulin (13), whereas the stable and longer-lived microtubules in neurons and cardiomyocytes contain predominantly detyrosinated tubulin (14).
Figure 1.
Summary of the primary post-translational modifications on tubulin, which occur on both the ordered tubulin body (e.g., acetylation) and on the primarily disordered CTTs. Displayed are the portions of the T. thermophila CTT sequences that are sufficiently dynamic to be visible by NMR (8), starting with α-tubulin E434 and β-tubulin A428. To see this figure in color, go online.
Post-translational tubulin modifications can tune intrinsic microtubule properties, such as flexibility, stability, and polymerization and depolymerization rates. Acetylation increases the mechanical resilience and flexibility of microtubules protecting long-lived microtubules from damage (10). Axonemal microtubules have slower growth and lower catastrophe frequency compared with bovine brain microtubules (15). However, samples with distinct patterns of post-translational modifications prepared by fractionating axonemes show indistinguishable rates of microtubule polymerization (15).
A poorly understood class of modifications is the heterogeneous additions of glycine and glutamate residues to the CTTs. These modifications result from the stepwise addition of amino acids to the terminal subset of glutamate residues in the CTTs by tubulin tyrosine ligase-like (TTLL) enzymes (Fig. 1). Polyglycylation and polyglutamylation are predominantly found on stable microtubule structures, including cilia. Polyglutamylation is also present in the long, stable microtubules in the axons of neurons (16), and the misregulation of tubulin polyglutamylation is linked to neurodegeneration (17). Polyglutamylation ensures the proper regulation of microtubule turnover in part through the recruitment of the microtubule-severing enzymes, such as katanin and spastin (18, 19, 20). Although polyglycylation is primarily found in cilia, its functional role is not well understood, and little is known about the binding partners specific to this modification (21, 22, 23).
Ciliary microtubules are heavily modified by polyglycylation and polyglutamylation. Polyglycylation is present over the length of mature cilia and contributes to overall cilia stability (24). In addition, polyglycylation is present in microtubule structures that anchor the basal bodies to the cell cortex (25). In contrast, polyglutamylation in cilia is essential for early ciliary maturation (26) and proper ciliary beating (27). These two modifications compete for the same sites on the CTTs; both are attached to the terminal glutamate residues (28). The deletion or depletion of glycine ligases in cells results in an increase in polyglutamylation (28,29). Mutations of the modification sites on one of the tubulin subunits affect the levels of polyglycylation and polyglutamylation on the opposing nonmutated CTT (26,30). The perturbation of the balance of these modifications causes shorter cilia (29), destabilization of already assembled cilia (28), and the reduced frequency of primary cilia (31). Given the important role of polyglycylation and polyglutamylation in cellular structures with particularly high stresses on microtubules (such as cilia), we questioned whether these modifications might directly increase the stiffness of microtubules.
To determine whether and how polyglycylation and polyglutamylation affect the intrinsic flexibility of microtubules, we purified tubulin from three Tetrahymena thermophila strains that have altered microtubule post-translational modifications, T. thermophila ciliary function, and swimming behavior. Tubulin purified from T. thermophila allows simultaneous microscopy and NMR experiments on the same sample (8). The three resulting samples were primarily polyglycylated (wild-type), polyglutamylated (TTLL3(A-F)-KO), or polyglycylated only on the β-tubulin CTT (ATU1-6D). Microtubules polymerized under identical conditions differed in the distribution of the measured stiffness depending on whether the primary modification was polyglycylation or polyglutamylation. Moreover, the addition of the divalent cation magnesium significantly increased the stiffness of polyglycylated but not polyglutamylated microtubules. Using electron microscopy, we found that changes in rigidity are not correlated with changes in protofilament numbers. Our NMR experiments, which indicated interactions between the CTTs and the tubulin core, helped to explain these changes in microtubule stiffness.
Materials and Methods
Growth and purification of tubulin from mutant T. thermophila strains
The T. thermophila strains containing modifications to the CTT sequence (ATU1-6D (26)) and knockouts of the TTLL-3 family of enzymes (TTLL3(A-F)-KO (29)) were generously gifted by Jacek Gaertig (University of Georgia, Athens, GA). T. thermophila strains, TTLL3(A-F)-KO and ATU1-6D, were cultured in SPP-rich media. The cultures were grown in a shaking incubator at 30°C at 100 rpm in 2.8 L Fernbach flasks. The cells were harvested at maximal density (∼1 × 106 cells/mL) by two 5-min centrifugations at 2800 × g. Cell pellets were frozen at −70°C until further purification. Live cell videos were taken using Nikon Widefield with a 4× objective in a 100-μm well on a glass coverslip. The frame rate was 0.118 s/frame with an exposure time of 354 μs. Tubulin was purified from each strain as previously described (8). In short, the protein was purified using a TOG affinity column, as described by published methods (32).
Western blot of tubulin from T. thermophila strains
10% SDS-PAGE gels containing 0.5 μg of tubulin purified from each of the strains were transferred to a polyvinylidene fluoride membrane. Antibodies and respective concentrations were as previously published (29). The primary antibodies and dilutions used were as follows: 1) mouse anti-α-tubulin (Developmental Studies Hybridoma Bank clone 12G10; Iowa City, IA) at 1:250, 2) rabbit anti-polyglutamylation (1:5000) (a generous gift from Jacek Gaertig (26)), 3) mouse anti-monoglycylated tubulin (clone TAP 952; EMD Millipore, Burlington, MA) at 1:5000, and 4) mouse anti-pan polyglycylated tubulin (clone AXO 49; EMD Millipore) at 1:5000. Secondary antibodies 1) anti-mouse IgG (H + L) AP conjugate (Promega, Madison, WI) and 2) anti-rabbit IgG (Fc) AP conjugate (Promega) were used at a dilution of 1:7500. Western blots were developed using the Pierce 1-Step NBT-BCIP reagent (Pierce Biotechnology, Waltham, MA) as recommended by the manufacturer and imaged with a Canon LiDE 210 Scanner.
Mass spectrometry of tubulin from T. thermophila
Purified tubulin samples were reduced, alkylated, and digested with trypsin (α) or chymotrypsin (β). Samples were resolved using a Waters nanoAcquity UPLC. Chromatographic separation was performed with a BEH C18 reversed-phase column (250 mm × 75 μm, 1.7 μm, 130 Å; Waters, Milford, MA), using a linear gradient from 95% Buffer A (0.1% formic acid) imo 35% Buffer B (0.1% formic acid, 99.9% acetonitrile) over 60 min at a flow rate of 300 nL/min. Tandem mass spectrometry (MS/MS) was performed using an linear trap quadrupole (LTQ) Orbitrap mass spectrometer, scanning between 150 and 2000 m/z (60,000 resolution), and the top six most intense precursor ions were selected for MS/MS sequencing using monoisotopic precursor selection, rejecting singly charged ions. Dynamic exclusion was used with a repeat count of one, a repeat duration of 30 s, an exclusion duration of 180 s, and an exclusion mass width of 20 ppm. The maximal injection time for Orbitrap parent scans was 500 ms with a target automatic gain control of 1 × 106. The maximal injection time for LTQ MS/MS scans was 250 ms, with a target automatic gain control of 1 × 104. The normalized collision energy was 35% with an activation Q of 0.25 for 30 ms.
Microtubule assembly
Microtubules were assembled in PIPES-based BRB80 (80 mM PIPES (pH 6.7), 1 mM MgCl2, and 1 mM EGTA) supplemented with 2 mM GTP (Sigma-Aldrich, St. Louis, MO) at 37°C for 1 h and then incubated at room temperature for at least 30 min. Starting tubulin concentration ranged from 1 to 10 μM. Samples for fluorescent bending measurements were supplemented with 10% rhodamine-labeled porcine brain tubulin (Cytoskeleton, Denver, CO). The resulting microtubules were spun down at 16,300 × g in a table-top microcentrifuge. Pelleted microtubules were resuspended in BRB80 supplemented with 10 μM taxol. High magnesium conditions were supplemented to 5, 10, or 50 mM MgCl2.
Electron microscopy of microtubules
After assembly, the microtubules were spun at 16,300 × g in a table-top microcentrifuge. The microtubules were resuspended in BRB80 supplemented with 10 μM taxol. Approximately 4 μL of undiluted or diluted (1:10) microtubules were adsorbed to glow discharged holey-carbon C-flat grids (Protochips, Morrisville, NC) for 10–30 s, blotted with Whatman filter paper, and immediately plunge frozen into liquid ethane using a homemade plunge-freezing device. Frozen samples were transferred under liquid nitrogen to a Gatan-626 cryo-holder (Gatan, Pleasanton, CA). Cryo-electron microscopy data were collected on an FEI Tecnai F20 FEG transmission electron microscope (FEI, Hillsboro, CO) operating at 200 kV. Images were collected at a magnification of 29,000× and a defocus of −4.0 μm using a total dose of 33 electrons/Å2. Images from wild-type microtubules were recorded binned by two on a 4K × 4K Gatan Ultrascan 895 CCD camera (Gatan). With this camera at a microscope magnification of 29,000×, the resulting pixel size corresponds to 7.6 Å on the specimen. Images from the TTLL3(A-F)-KO microtubules were taken on the same microscope, which was fitted with a 24 megapixel, 5.7K × 4.1K Gatan K3 camera (Gatan). Images were collected at a microscope magnification of 11,500× and a defocus of −3.5 μm using a total dose of 38 electrons/Å2. With this camera, the images were recorded binned by two with a resulting pixel size on the specimen of 6.2 Å. Each image was captured as 50 frame videos (0.76 electrons/Å2/frame) and aligned using SerialEM software (33). This software was also used to automate the data acquisition and minimize exposure of the specimen to the electron beam.
Fluorescence microscopy
All microtubules were assembled in BRB80, as described above. Microtubules were diluted at least 20-fold for imaging in the relevant buffer, with the addition of an oxygen scavenging system (0.5% β-mercaptoethanol, 4.5 mg/mL glucose, 0.2 mg/mL glucose oxidase, and 0.035 mg/mL catalase), 10 μM taxol to stabilize the microtubules (Sigma Paclitaxel), and 1% pluronic F-127 to inhibit sticking to the slide or coverslip (Sigma). 0.25 μL of the dilution was sealed with epoxy between a glass coverslip and glass slide and kept in the dark unless being imaged. Slides and coverslips were cleaned in a bath sonicator in 100% ethanol for 45 min and soaked in 100% ethanol until use. The slides and coverslips were flamed to dry immediately before use. Fluorescence bending images were acquired using a Nikon Confocal Spinning Disk with a 1.45 NA 100× oil objective (Nikon) and Andor 888 Ultra EMCCD camera. Videos were acquired for at least 100 frames. There was a 1 s delay between frames with a 100 ms exposure time.
Analysis of flexural rigidity
All videos were cropped to contain a single microtubule using ImageJ (34). Less than 10% of the total number of frames were removed if the microtubule drifted out of the z-plane or if any other particles obstructed the microtubule during imaging. Single microtubules were analyzed by previous methods (35, 36, 37). In short, the variances in the magnitude of the first 25 normal mode were measured using a customized MATLAB (The MathWorks, Natick, MA) script. To determine the uncertainty in the measured variance, bootstrapping statistics was employed using R (38) as previously described (35). Average persistence lengths were determined by fits to the cumulative distribution function (Fig. S5; Table S1).
15N-relaxation NMR (R1/R2)
Tubulin was purified from wild-type T. thermophila grown in minimal bacterized media, as previously described for isotopic labeling (8). GST-CTT peptides were expressed and purified as previously described (8). T1 and T2 relaxation experiments were performed using the standard 15N-HSQC experiment from the Varian BioPac (gNhsqc) on an 800 MHz magnet. For measurement of T1, relaxation delays used were as follows: 0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, and 0.9 ms. For the measurement of T2, relaxation delays used were as follows: 0.01, 0.03, 0.05, 0.07, 0.09, 0.11, 0.13, 0.15, 0.17, 0.19, 0.21, 0.23, and 0.25 ms. The data were processed using standard scripts in NMRPipe and analyzed using CCPNmr Analysis software (39).
Results and Discussion
Creation and characterization of differentially modified pools of tubulin
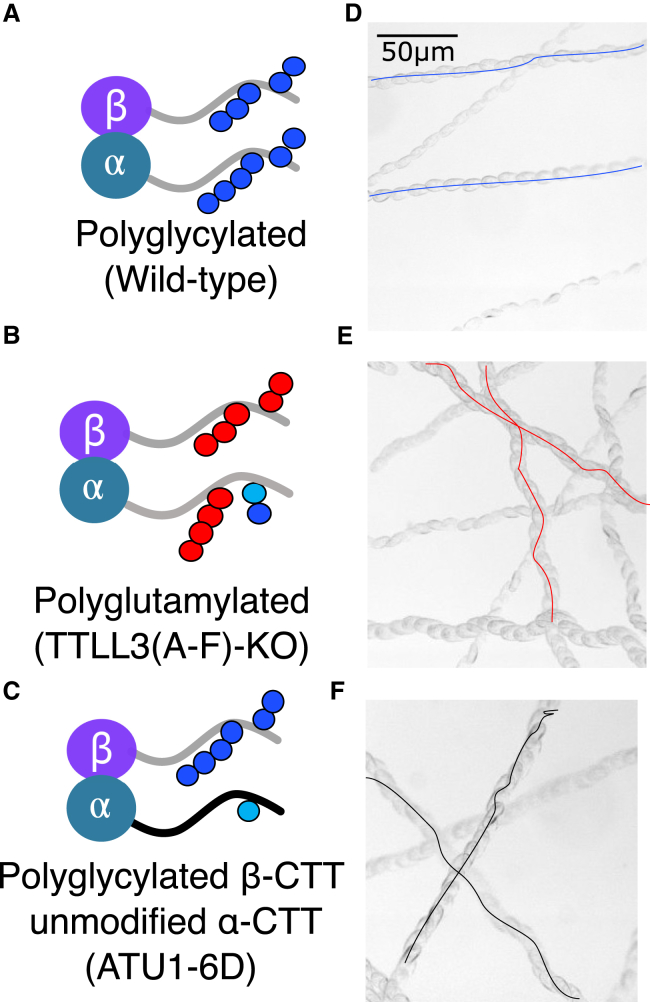
To obtain samples with different modifications, we took advantage of existing T. thermophila strains created to study the biological role of CTT modifications (26,29,40). We purified tubulin from three strains, giving primarily polyglycylated tubulin (wild-type), primarily polyglutamylated tubulin (TTLL3(A-F)-KO), and tubulin with polyglycylation only on the β-tubulin CTT (ATU1-6D) (Fig. 2, A–C). As we published previously, tubulin purified from our wild-type strain is primarily polyglycylated (8).
Figure 2.
Overview of the strains used and resulting predominant post-translational modifications observed in tubulin purified from each strain. (A–C) The samples were primarily polyglycylated (wild-type), polyglutamylated (TTLL3(A-F)-KO), or polyglycylated only on the β-tubulin CTT (ATU1-6D). (D–F) Time-lapse images showing the trajectory of typical swimming T. thermophila cells are shown. The two mutant strains have ciliary defects and abnormal swimming patterns. To see this figure in color, go online.
To obtain polyglutamylated tubulin, we purified tubulin from a strain with all members of the TTLL3 family of enzymes deleted (29). We refer to this strain as TTLL3(A-F)-KO or polyglutamylated. Strains lacking the glycine ligases showed increased levels of polyglutamylation (29). This strain has shorter cilia, slower growth rates, and increased resistance to stabilization by taxol (29). We observed a disrupted swimming pattern (Fig. 2 E), which was most likely because of changes in the ciliary length and organization.
To obtain tubulin that was polyglycylated on only β-tubulin, we purified tubulin from a strain in which the last six glutamate residues of α-tubulin were mutated to aspartate (ATU1-6D) (26). We previously mapped the sites of polyglycylation to these residues and did not observe polyglycylation on the other glutamate residues in the α-tubulin CTT (8). The aspartate substitutions retain the overall charge of the tail but are not an efficient substrate for the TTLL enzymes (26). This strain has slower swimming speeds (26) and disrupted swimming patterns (Fig. 2 F), although they are able to grow well enough for the purposes of purification of tubulin.
The levels of polyglutamylation in each preparation were measured by Western blot using an antibody specific to polyglutamylation. We detected polyglutamylation in tubulin purified from all three strains tested by Western blot (Fig. S2), including the wild-type strain in which polyglutamylation was not previously observed by either mass spectrometry or NMR (8). The tubulin purified from TTLL3(A-F)-KO has significantly higher levels of polyglutamylation on both tails than the other two strains, which is as expected because of previous observations of hyperglutamylation in whole cell lysates of TTLL3(A-F)-KO cells (23,29). We detected little polyglutamylation on α-tubulin purified from the ATU1-6D strain, as expected because all modification sites were mutated. However, the levels of polyglutamylation detected on β-tubulin from ATU1-6D cells is similar to the wild-type. Polyglutamylated peptides were difficult to detect by mass spectrometry but were observed in samples purified from TTLL3(A-F)-KO cells.
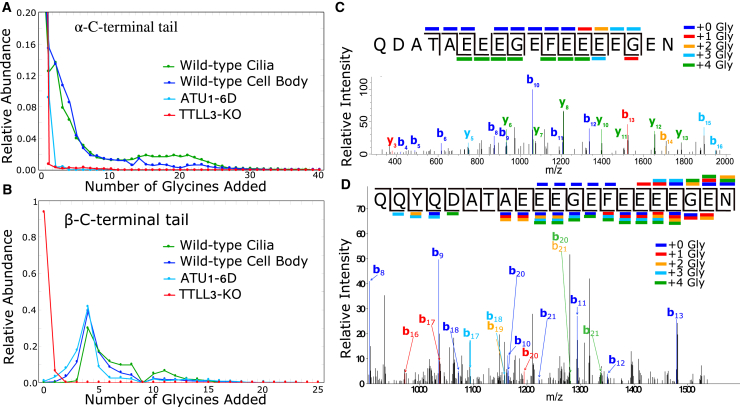
We then measured the degree of polyglycylation using both Western blot and mass spectrometry. We used antibodies to monoglycylated tubulin (TAP952) and polyglycylated tubulin (AXO49), which are specific to mono- or polyglycylation, respectively (Fig. S2). As previously determined by mass spectrometry, the tubulin purified from wild-type T. thermophila contained both mono- and polyglycylation (8). Both tails on α- and β-tubulin have significant polyglycylation. Because modification levels are thought to be different on the cilia relative to the rest of the cell, we deciliated cells and purified tubulin separately from the ciliary and cell body fractions. Unexpectedly, as measured by mass spectrometry, the levels of polyglycylation were similar between tubulin purified from cell bodies and from shed cilia (Fig. 3, A and B). However, there were subtle differences. We observed a higher prevalence of peptides containing many (∼20) glycine additions in tubulin samples purified from cilia than from the cell body.
Figure 3.
Mass spectrometry characterization of the polyglyclation patterns on differentially modified tubulin samples. For the wild-type samples, we purified tubulin from both ciliary and cell body fractions. (A and B) We measured the total ion current corresponding to trypsin (α) or chymotrypsin (β) C-terminal peptides, which include the disordered CTTs. We found a peak in the β-tubulin tail sample of four additional glycines. We mapped the glycyine additions of this parent peptide using MS/MS (41,42). (C) An example MS/MS scan shows fragmentation consistent with a single glycine on each of the four terminal glutamates. (D) Shown is a different MS/MS scan containing fragments corresponding to a wide rage of glycine localizations. Fragmentation of the modification itself is thought to contribute to the diversity of y ions. We did not observe b ions consistent with modifications upstream of the four terminal glutamates. To see this figure in color, go online.
We detected no mono- or polyglycylation in tubulin purified by the same protocol from the TTLL3(A-F)-KO strain by Western blot. This is as expected because the enzymes responsible for the ligation of glycine residues were deleted from the micronucleus (29). However, by mass spectrometry, we detected peptides corresponding to both mono- and polyglycylation on the α-tubulin CTT. The low levels of polyglycylation are presumed to be from residual DNA coding for the TTLL3 enzymes in the macronucleus or could potentially be due to another polyglycylating enzyme. As measured by mass spectrometry, TTLL3(A-F)-KO tubulin has no detectable polyglycylation on the β-tubulin CTT.
Tubulin purified from the ATU1-6D strain and levels of mono- and polyglycylation on the β-tubulin subunit were very similar to those seen in our wild-type cells. We observed low levels of monoglycylation on the α-tubulin subunit by Western blot. This was unexpected because all glutamate residues that we had previously determined to be sites of modification on the α-tubulin CTT were mutated in this strain to aspartates (Fig. S2; (8)). By mass spectrometry, we detected monoglycylation on the introduced aspartate residues and not on any residues further upstream on the α-CTT (data not shown). Therefore, the TTLL3 glycine ligases appear to be able to modify the aspartate residues, albeit at a very low level. Interestingly, we detected tyrosinated ATU1-6D α-tubulin in addition to detyrosinated ATU1-6D α-tubulin; we have not detected the peptide containing the terminal tyrosine residue in any of our other samples.
We measured the total ion current corresponding to tryptic or chymotryptic peptides with varying numbers of glycine additions for all three strains and found a strong preference for the addition of four glycine residues to the β-tubulin CTT in wild-type and ATU1-6D cells (Fig. 3, A and B; (41,42)). We then analyzed the distribution of glycine residues on this species. The fragmentation pattern in MS/MS experiments of the peptides with four added glycine residues was very heterogeneous (Fig. 3, C and D; Fig. S3). We obtained strong evidence for several different arrangements of the four glycine residues. For example, we were able to identify fragments that strongly support the presence of a species with one glycine on each of the last four glutamates from the peptide from ATU1-6D (Fig. 3 C). We also identified MS/MS fragments corresponding to several other heterogeneous arrangements in this ATU1-6D tubulin peptide (Fig. 3 C), including all four glycine residues being on the last glutamate residue. However, the fragmentation of not just the tail peptide but also the modification itself confounded our analysis. Although we found y ions giving evidence of fewer than four additional glycines in the MS/MS fragments, we did not find b ions containing positive evidence of glycine modifications upstream of the final five glutamates. We conclude that the primary sites of modification are the five C-terminal glutamates, which is similar to the previous mapping of wild-type tubulin (8).
From both the Western blot and mass spectrometry analysis, we conclude that we have three distinct subsets of differentially modified tubulin: 1) polyglycylated tubulin from wild-type T. thermophila, 2) polyglutamylated tubulin from TTLL3(A-F)-KO T. thermophila, and 3) tubulin polyglycylated on only the β-tubulin CTT, with an unmodified α-tubulin CTT from ATU1-6D T. thermophila. These modifications are summarized in Fig. 2. With these tools in place, we sought to determine the role of polyglutamylation and polyglycylation on microtubule mechanical properties.
Microtubule flexibility depends on the post-translational modifications present on the CTTs
To test the hypothesis that microtubule stiffness varies between differentially modified microtubules, we measured the persistence length (Lp) of freely fluctuating microtubules from each of our three tubulin pools using fluorescence microscopy, as was done previously (35, 36, 37,43). We analyzed the distribution of contributions of each normal mode to the microtubule shape (43). We acquired frames every second for 8 min. The frames were approximately uncorrelated. We used bootstrapping to ensure good error estimation and for the robustness of the analysis (35). From the measurements, we determined the average microtubule persistence length, which is a measure of the stiffness of a filament. A stiffer filament has a higher persistence length, whereas a lower persistence length means a more flexible filament. Consistent with previous work, we found no dependence of persistence length on contour length (35,37,43,44), which ranged from 10 to 45 μM (Fig. 4; Fig. S4).
Figure 4.
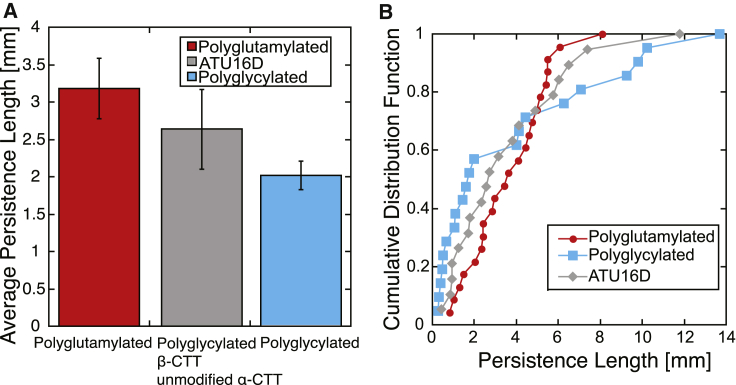
Measured microtubule persistence lengths of our three differentially modified tubulins. (A) The mean of the persistence lengths measured as and the data are well described by a log-normal distribution. (B) Cumulative distribution functions for our different microtubule populations are shown. To see this figure in color, go online.
We tested whether differences in CTT post-translational modifications, in particular polyglycylation and polyglutamylation, affected the stiffness of microtubules. As done previously, we fit the range of persistence length values to the cumulative distribution functions (36). We used the nonparametric Kolmogorov-Smirnov statistical test (45) and found that all of our data were better described by a log-normal distribution than a normal distribution. The fit parameters and pairwise p-values for all of the data are listed in Tables S1 and S2. In addition, we determined the average persistence length as (Fig. 4), with the most significant difference being between polyglutamylated and polyglycylated microtubules (p = 0.045).
Microtubules from T. thermophila tubulin (in 1 mM MgCl2) ranged in average persistence lengths from Lp = 2.0 ± 0.19 mm for polyglycylated microtubules (n = 21) to Lp = 3.18 ± 0.41 mm for polyglutamylated microtubules (n = 23). As expected, microtubules with polyglycylated β-tubulin CTTs have average persistence length values situated between the values of polyglytamylated and polyglycylated at 2.63 ± 0.53 mm (n = 19).
In addition, the spread of the data (σ) is larger for polyglycylated as compared with polyglutamylated tubulin (Fig. 4 B). For polyglycylated microtubules, σ = 1.47 ± 0.07 more than twice the value for polyglutamylated microtubules σ = 0.60 ± 0.04. Continuing the trend, the microtubules with polyglycylated β-tubulin CTTs are intermediate between the two other pools of tubulin with σ = 0.94 ± 0.04.
Polyglycylated microtubules, the most flexible that we measured, are almost twofold stiffer than taxol-stabilized rhodamine-labeled porcine tubulin (1.19 ± 0.04 mm) (35, 36, 37). The differences between porcine and T. thermophila tubulin could be attributed to sequence and structural differences between the two species of tubulin. In addition, porcine brain tubulin contains a mixture of post-translational modifications that are different from our samples, including the potential acetylation, which is known to reduce microtubule stiffness (9,10). However, our samples do not have significantly less acetylation than reported values for bovine brain tubulin (∼30% acetylated) (46).
Using mass spectrometry data, we semiquantitatively compared the ratio of ion currents to determine the level of acetylation in our samples. The level of acetylation in our wild-type sample was 50%, our ATU1-6D sample was ∼16%, and our TTLL3(A-F)-KO was at 37%. There does not appear to be a strong correlation between stiffness and acetylation levels in our samples, in contrast to previous work (10).
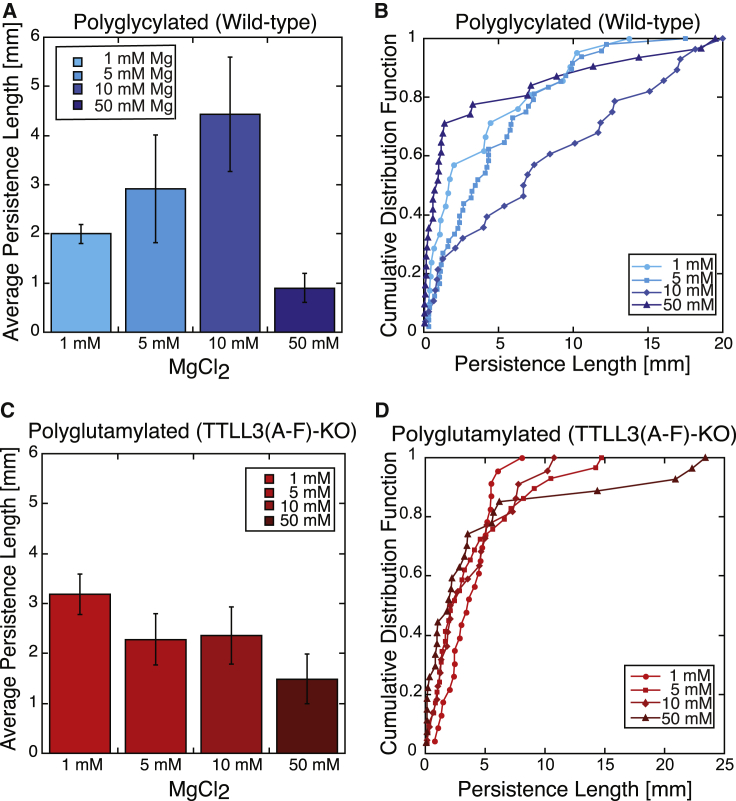
We sought to determine whether magnesium would affect bending rigidity differently for our polyglycylated and polyglutamylated microtubules. Divalent salts have been shown to affect microtubule bending rigidity in a CTT-dependent manner, with 5 mM MgCl2 increasing the stiffness of mammalian tubulin by ∼50% (47). The high negative charge of the tails is thought to be important for their contribution to the salt dependence. Microtubule polyglutamylation adds significant charge to the CTTs, whereas polyglycylation does not and may instead act as a steric barrier to prevent charges from approaching each other. Furthermore, divalent salt concentration affects the flexibility of microtubules, which may be a result of changes in the conformation of the CTTs (47). We performed similar measurements as above of persistence length of polyglycylated versus polyglutamylated microtubules in varying concentrations of magnesium (1, 5, 10, and 50 mM MgCl2). All microtubules were assembled in 1 mM MgCl2, and then, additional magnesium was added before measuring the bending stiffness.
We saw an increase in persistence length upon the addition of moderate concentrations of magnesium for polyglycylated microtubules (Fig. 5), consistent with previous observations (47). Particularly notable is the strong increase in stiffness of polygylcylated microtubules at 10 mM MgCl2, which increases to a persistence length of 4.43 ± 1.16 mm (n = 31). However, the stiffness of polyglutamylated microtubules was not significantly changed in 5 or 10 mM MgCl2 as compared with 1 mM MgCl2. As a result, in 10 mM MgCl2, the polyglycylated microtubules were significantly stiffer than the polyglutamylated ones.
Figure 5.
Measured persistence lengths of our differentially modified microtubules at different concentrations of magnesium. Shown are the mean persistence length (A and C) and cumulative distribution plots (B and D) of tubulin purified from the wild-type (polyglycylated) tubulin (A and B) and TTLL3 knockout (polyglutamylated) tubulin (C and D). To see this figure in color, go online.
When the magnesium concentration was further increased to 50 mM, the stiffness decreased, and the persistence lengths became statistically similar between polyglutamylated and polyglycylated microtubules. Interestingly, a small subset of both polyglycylated and polyglutamylated microtubules had persistence lengths of greater than 10 mm in 50 mM MgCl2 (Fig. 5). We could not identify a biological reason for these stiffer filaments; they did not appear to be bundled or to incorporate a different concentration of fluorescently labeled tubulin. Contrary to our expectation, the polyglycylated microtubules showed more significant changes in bending rigidity upon increasing magnesium than did the polyglutamylated ones. The differences in the effect of polyglycylation and polyglutamylation on microtubule stiffness may be due to the conformation of the CTTs on the surface. Polyglycylated tails are hypothesized to adopt a collapsed conformation (48). Very high magnesium concentrations are proposed to disfavor this conformation and promote a more extended conformation, reducing interactions with the surface (47).
Differentially modified microtubules incorporate similar numbers of protofilaments
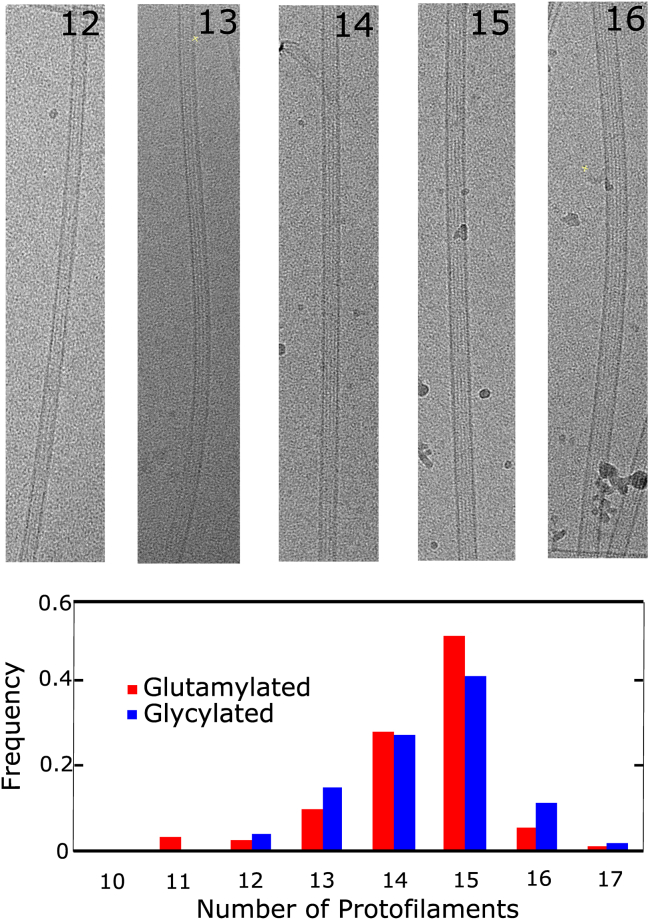
Next, we tested whether differences in bending rigidity of the different microtubule pools could be explained by structural differences in assembled microtubules. In mammalian tubulin, microtubules containing different numbers of protofilaments showed different microtubule flexural rigidity (36). We used electron microscopy to determine if tubulin modifications caused changes in the number of protofilaments incorporated into microtubules. We considered only the wild-type and TTLL3(A-F)-KO strains, which are primarily polyglycylated and polyglutamylated, respectively. The number of protofilaments per microtubule is typically 13 in cells but can vary from 9 to 17 under in vitro polymerization conditions (49). In electron cryo-microscopy images, the number of protofilaments in each microtubule can be determined from the unique helical pitch and moire patterns of each microtubule type (49). We counted the number of protofilaments for wild-type (glycylated) and TTLL3(A-F)-KO (glutamylated) (n = 145 and n = 96 microtubules, respectively) (Fig. 6).
Figure 6.
Electron microscopy representative images of varying protofilament number (top). Shown is the normalized distribution of protofilament numbers for polyglycylated (blue) versus polyglutamylated (red) microtubules (bottom). The two distributions are similar, giving a p-value of 0.97 by the Kolmogorov-Smirnov test. To see this figure in color, go online.
The two differentially modified microtubule subsets have indistinguishable distributions of protofilament number (Kolmogorov-Smirnov test p-value = 0.97). Most contain 14–15 protofilaments as is typically seen in in vitro polymerization (49). We conclude that the observed differences in flexibility are due to the molecular-level interactions of the CTTs rather than differences in the overall size or structure of assembled microtubules.
Spin dynamics suggest interactions of the α-tubulin CTT with the tubulin body
To probe the molecular-level interactions of the CTTs, we used NMR spectroscopy, which is sensitive to transient interactions characteristic of intrinsically disordered domains. In particular, differences in NMR relaxation measurements in different conditions can be indicative of changes in chain dynamics and interactions (50). As measured by NMR, the highly dynamic regions of the tails are 15 amino acids long for the α-tubulin CTTs (EVGIETAEGEGEEEG) and 15 for the β-tubulin CTTs (ATAEEEGEFEEEEGEN) (8). This is a larger region than expected for both the α-tubulin and β-tubulin CTTs based on sequence conservation (51) and previous x-ray crystal structures (52,53), in which electron density has been observed in some, but not all, structures of tubulin for the residues corresponding to the first three amino acids visible in our NMR spectroscopy experiments.
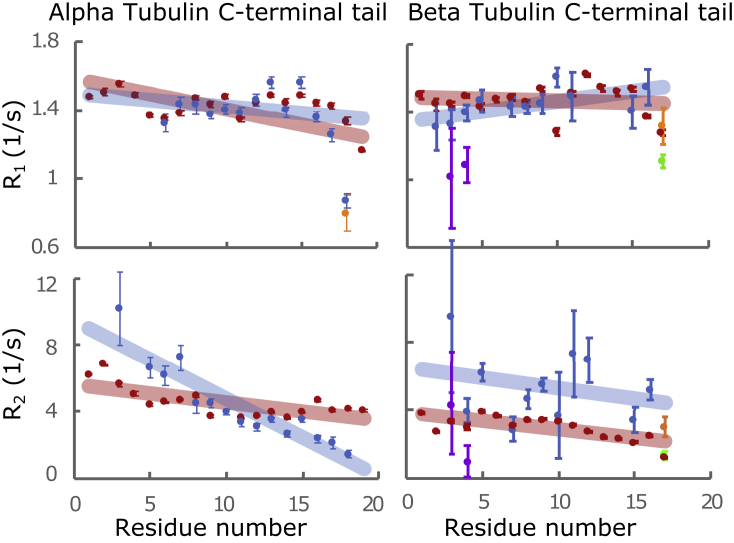
We performed R1 and R2 NMR 15N-relaxation experiments to probe the local dynamics of the CTTs in the presence of the tubulin dimer (54,55). NMR spin relaxation experiments give information about the dynamics of each residue of the CTTs. We compared the NMR spin dynamics of the atoms in the CTTs of the tubulin heterodimer with the same residues on peptides expressed attached to GST homodimers (the α and β GST-fusion constructs were measured in different experiments) (Fig. 7). Our low sample concentration values, coupled with typically low values for disordered domains, reduced the utility of heteronuclear 1H-15N experiments, which were all below noise (NOE values <0.6). As a result, we did not attempt to apply quantitative interpretations based on the combination of the spin relaxation measurements. Fig. 7 shows the values of the R1 and R2 relaxation rates of the two CTTs plotted as a function of the amino acid position. The shaded lines are linear fits to guide the eye. We compared the measured values for the CTTs as part of the tubulin dimer in our wild-type or primarily polyglycylated sample and of the CTTs as unmodified peptides, each individually attached to GST dimers, to mimic their N-terminal attachment. The latter gives an indication of the intrinsic differences due to the amino acid sequence and not to any interactions as part of the full dimer or as a result of post-translational modifications.
Figure 7.
NMR spin relaxation measurements of CTTs. Shown are the R1 (top row) and R2 (bottom row) relaxation rates for α- (left) and β- (right) tubulin CTTs as peptides attached to GST (red) or as part of the full tubulin dimer (blue, orange, yellow, and green). Several residues in our NMR spectra are present in two different chemical environments (8). For residues near the C-termini, these correspond to the modification state (orange: polyglycylation of the i-1 residue, green: monoglycylation of the i-1 residue). For residues near the N-termini, one set of peaks is brighter (blue) and the other dimmer (purple). To see this figure in color, go online.
We see a clear trend as a function of distance from the tubulin body in the α-tubulin CTT values of R2, which is not apparent in either R1 of α- or R1 or R2 of β-tubulin. Increased values of R2 are typically taken as an indication of increased interactions of the appropriate timescale, and so we conclude that the more N-terminal portions of the α-tubulin CTT interact more significantly with the tubulin body than do residues on the tail further from the surface. The values of R2 for the β-tubulin CTT were in general higher than those for the peptide, indicating the presence of some interaction. However, the overall trend was similar to that of the peptide, which indicates that these values are dominated by local interactions, rather than interactions with the tubulin dimer surface. In contrast, the R1 relaxation values, typically attributed to the overall motion of individual bonds, are almost identical between the peptide and CTT of the α-tubulin.
Our NMR spectrum contains more than one resonance for several of the residues in the CTT. At the C-terminal extreme, we can distinguish species according to the modification state of the adjacent residue. For example, the terminal asparagine on β-tubulin is present in our sample in three species of differing modification states (Fig. 7; Fig. S6). The measured relaxation rates differ based on the modification state, with both R1 and R2 being larger for polyglycylated residues relative to either the unmodified or monogylcylated residues.
Residues of the tail near the tubulin body are also present in two environments, which we attributed to differences in the environment of the underlying tubulin body, although we have not been able to determine the source of these differences (8). For the β-tubulin CTT, the segment from residues β-tubulin 428–432 (ATAE) is present in two groups corresponding to two different environments for this segment. Interestingly, the relaxation values were easy to measure for one of the groups and are shown in blue with the rest of the chain in Fig. 7. The other group is the dimmer and more overlapped with other peaks in the spectrum. The apparent difference in relaxation rates between the two groups is further evidence of these residues having different interactions with the tubulin body.
Conclusions
Post-translational modifications of tubulin dimers have been linked to both intrinsic and extrinsic properties of microtubules (6,7). In this study, we investigated the effect of polyglycylation and polyglutamylation on the intrinsic flexural rigidity of microtubules. We purified tubulin out of three different T. thermophila strains, producing three pools of differentially modified tubulin: 1) from wild-type cells, we purified primarily polyglycylated tubulin; 2) from TTLL3 knockout cells lacking all tubulin mono- and polyglycylases, we purified primarily polyglutamylated; and 3) from ATU16D cells, we purified protein that is polyglycylated on the β-tubulin CTT and primarily unmodified on the α-tubulin CTT. We confirmed the extent of these modifications by mass spectrometry and Western blot. An advantage of our method of purifying differential-modified samples of tubulin is that our approach allows for sufficient quantities for NMR experiments (8,56,57). We determined that polyglycylation and polyglutamylation do not change the number of protofilaments incorporated into the microtubule lattice and therefore do not significantly affect the size of microtubules.
Despite having the same distribution in the number of protofilaments, we found significant differences in the measured persistence lengths of polyglycylated and polyglutamylated microtubules in the presence of both 1 and 10 mM magnesium (p = 0.045 and p = 0.021, respectively). At intermediate concentrations, the addition of magnesium caused the polyglycylated microtubules to become stiffer, but then at very high concentrations, the microtubule’s rigidity decreased. This effect may be due to the proposed collapsed conformation of the CTTs upon polyglycylation (48). In this case, the collapsed tail would interact more strongly with the tubulin body, an interaction that is reduced in the presence of a high concentration of magnesium.
A clear dependence of the R2 15N-relaxation rates of the α-tubulin CTTs as a function of distance along the tail from the tubulin body surface suggests that the α-tubulin CTT residues interact with the tubulin body, consistent with previous simulation (58). This interaction could explain the importance of polyglycylation of the α-tubulin CTT on microtubule stiffness. The α-tubulin CTT projects toward the adjacent dimer, supporting the view that interdimer rather than intradimer interactions dominate the bending rigidity (59).
Polyglycylation on the CTTs contributes to the overall stiffness and structural integrity of ciliary microtubules. The depletion of TTLL3 enzymes leads to destabilization of already assembled cilia in T. thermophila (29) and reduced frequency of primary cilia in mammalian cells (24,31). In Tetrahymena, the TTLL3(A-F)-KO strain (lacking polyglycylation) has poor growth, shorter cilia, and failure to elongate cilia in the presence of taxol (29). Polyglycylation is thought to be localized primarily on the B-tubules within cilia, whereas the A-tubules are predominantly unmodified (60). The tips of cilia contain microtubules with no polyglycylation (60). Although polyglutamylation is localized to the outer doublets in cilia and missing from the central pair (61), excessive polyglutamylation results in unstable and short cilia (62). The depletion of glycine ligases results in hyperglutamylation and vice versa, making interpretation of any phenotype difficult. For example, hyperglutamylation stimulates katanin and spastin activity (19) and causes misregulation of tubulin turnover (40). Extrinsic factors such as MAPs contribute to ciliary stiffness (44,63). The intrinsic differences in flexibility solely because of the modifications suggest a possible role for polyglycylation, especially in cellular structures undergoing high stresses such as the cilia.
Post-translational modifications play a significant role in the regulation of microtubules. We demonstrate that post-translational modifications can alter the intrinsic flexibility of microtubules. Although the α- and β-tubulin CTTs often contain similar modifications, they show different spin dynamics and may contribute differently to the stiffness of the microtubule. Our results provide insight into the complex mechanism of how polyglycylation and polyglutamylation alter microtubule mechanical properties.
Author Contributions
K.P.W., T.L.H., and L.H. designed research, performed research, analyzed data, and wrote the manuscript. H.H. analyzed flexural rigidity data. T.L. collected and analyzed mass spectrometry data. C.P. collected and analyzed electron microscopy data.
Acknowledgments
We thank Jacek Gaertig (University of Georgia) for his generous gift of T. thermophila strains and polyglutamylation antibody. We thank the University of Colorado Boulder Central Analytical Mass Spectrometry Core Facility for performing LC MS/MS experiments. We thank Joe Dragavon (BioFrontiers Institute, University of Colorado Boulder) for his training and expertise. Electron microscopy was done at the University of Colorado Boulder EM Services Core Facility, with the technical assistance of Gerry Morgan. We would also like to acknowledge J. Richard McIntosh (University of Colorado Boulder), Chad Pearson (University of Colorado, Anschutz Medical Campus), and Dan Sackett (National Institute of Health) for advice.
This work was performed with support from National Institutes of Health R35 GM119755 and T32 GM065103. This work utilized Thermo Fisher Scientific LTQ Orbitrap velos that was purchased with funding from W.M. Keck Foundation. The imaging work was performed at the BioFrontiers Institute Advanced Light Microscopy Core. Spinning disk confocal microscopy was performed on Nikon Ti-E microscope supported by the BioFrontiers Institute and the Howard Hughes Medical Institute. The 800 MHz spectrometer was purchased and supported by the National Institutes of Health (RR011969 and RR16649) and the National Science Foundation (DBI-0230966 and 960241).
Editor: David Sept.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.09.040.
Supporting Material
References
- 1.Desai A., Mitchison T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Inoué S. Mitotic organization and force generation by assembly/disassembly of microtubules. Cell Struct. Funct. 1996;21:375–379. doi: 10.1247/csf.21.375. [DOI] [PubMed] [Google Scholar]
- 3.Kapitein L.C., Hoogenraad C.C. Building the neuronal microtubule cytoskeleton. Neuron. 2015;87:492–506. doi: 10.1016/j.neuron.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 4.Barlan K., Gelfand V.I. Microtubule-based transport and the distribution, tethering, and organization of organelles. Cold Spring Harb. Perspect. Biol. 2017;9:a025817. doi: 10.1101/cshperspect.a025817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haimo L.T., Rosenbaum J.L. Cilia, flagella, and microtubules. J. Cell Biol. 1981;91:125s–130s. doi: 10.1083/jcb.91.3.125s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadadhar S., Bodakuntla S., Janke C. The tubulin code at a glance. J. Cell Sci. 2017;130:1347–1353. doi: 10.1242/jcs.199471. [DOI] [PubMed] [Google Scholar]
- 7.Roll-Mecak A. Intrinsically disordered tubulin tails: complex tuners of microtubule functions? Semin. Cell Dev. Biol. 2015;37:11–19. doi: 10.1016/j.semcdb.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wall K.P., Pagratis M., Hough L.E. Molecular determinants of tubulin’s C-terminal tail conformational ensemble. ACS Chem. Biol. 2016;11:2981–2990. doi: 10.1021/acschembio.6b00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szyk A., Deaconescu A.M., Roll-Mecak A. Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell. 2014;157:1405–1415. doi: 10.1016/j.cell.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Z., Schaedel L., Nachury M.V. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science. 2017;356:328–332. doi: 10.1126/science.aai8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gundersen G.G., Khawaja S., Bulinski J.C. Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. J. Cell Biol. 1987;105:251–264. doi: 10.1083/jcb.105.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webster D.R., Wehland J., Borisy G.G. Detyrosination of alpha tubulin does not stabilize microtubules in vivo. J. Cell Biol. 1990;111:113–122. doi: 10.1083/jcb.111.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geuens G., Gundersen G.G., DeBrabander M. Ultrastructural colocalization of tyrosinated and detyrosinated alpha-tubulin in interphase and mitotic cells. J. Cell Biol. 1986;103:1883–1893. doi: 10.1083/jcb.103.5.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerr J.P., Robison P., Ward C.W. Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nat. Commun. 2015;6:8526. doi: 10.1038/ncomms9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orbach R., Howard J. The dynamic and structural properties of axonemal tubulins support the high length stability of cilia. Nat. Commun. 2019;10:1838. doi: 10.1038/s41467-019-09779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audebert S., Desbruyères E., Eddé B. Reversible polyglutamylation of alpha- and beta-tubulin and microtubule dynamics in mouse brain neurons. Mol. Biol. Cell. 1993;4:615–626. doi: 10.1091/mbc.4.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magiera M.M., Singh P., Janke C. Tubulin posttranslational modifications and emerging links to human disease. Cell. 2018;173:1323–1327. doi: 10.1016/j.cell.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Lacroix B., van Dijk J., Janke C. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J. Cell Biol. 2010;189:945–954. doi: 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valenstein M.L., Roll-Mecak A. Graded control of microtubule severing by tubulin glutamylation. Cell. 2016;164:911–921. doi: 10.1016/j.cell.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin S.C., Im S.-K., Kim E.E. Structural and molecular basis for katanin-mediated severing of glutamylated microtubules. Cell Rep. 2019;26:1357–1367.e5. doi: 10.1016/j.celrep.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Bré M.H., Redeker V., Levilliers N. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J. Cell Sci. 1996;109:727–738. doi: 10.1242/jcs.109.4.727. [DOI] [PubMed] [Google Scholar]
- 22.Levilliers N., Fleury A., Hill A.M. Monoclonal and polyclonal antibodies detect a new type of post-translational modification of axonemal tubulin. J. Cell Sci. 1995;108:3013–3028. doi: 10.1242/jcs.108.9.3013. [DOI] [PubMed] [Google Scholar]
- 23.Rogowski K., Juge F., Janke C. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 2009;137:1076–1087. doi: 10.1016/j.cell.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 24.Gadadhar S., Dadi H., Janke C. Tubulin glycylation controls primary cilia length. J. Cell Biol. 2017;216:2701–2713. doi: 10.1083/jcb.201612050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Junker A.D., Soh A.W.J., Pearson C.G. Microtubule glycylation promotes attachment of basal bodies to the cell cortex. J. Cell Sci. 2019;132:jcs233726. doi: 10.1242/jcs.233726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wloga D., Rogowski K., Gaertig J. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot. Cell. 2008;7:1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikegami K., Sato S., Setou M. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc. Natl. Acad. Sci. USA. 2010;107:10490–10495. doi: 10.1073/pnas.1002128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosch Grau M., Masson C., Janke C. Alterations in the balance of tubulin glycylation and glutamylation in photoreceptors leads to retinal degeneration. J. Cell Sci. 2017;130:938–949. doi: 10.1242/jcs.199091. [DOI] [PubMed] [Google Scholar]
- 29.Wloga D., Webster D.M., Gaertig J. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell. 2009;16:867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Redeker V., Levilliers N., Bré M.-H. Mutations of tubulin glycylation sites reveal cross-talk between the C termini of alpha- and beta-tubulin and affect the ciliary matrix in Tetrahymena. J. Biol. Chem. 2005;280:596–606. doi: 10.1074/jbc.M408324200. [DOI] [PubMed] [Google Scholar]
- 31.Rocha C., Papon L., Janke C. Tubulin glycylases are required for primary cilia, control of cell proliferation and tumor development in colon. EMBO J. 2014;33:2247–2260. doi: 10.15252/embj.201488466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Widlund P.O., Podolski M., Drechsel D.N. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol. Biol. Cell. 2012;23:4393–4401. doi: 10.1091/mbc.E12-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawkins T.L., Mirigian M., Ross J.L. Perturbations in microtubule mechanics from tubulin preparation. Cell. Mol. Bioeng. 2012;5:227–238. [Google Scholar]
- 36.Harris B.J., Ross J.L., Hawkins T.L. Microtubule seams are not mechanically weak defects. Phys. Rev. E. 2018;97:062408. doi: 10.1103/PhysRevE.97.062408. [DOI] [PubMed] [Google Scholar]
- 37.Hawkins T.L., Sept D., Ross J.L. Mechanical properties of doubly stabilized microtubule filaments. Biophys. J. 2013;104:1517–1528. doi: 10.1016/j.bpj.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2011. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 39.Vranken W.F., Boucher W., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 40.Xia L., Hai B., Gaertig J. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 2000;149:1097–1106. doi: 10.1083/jcb.149.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biemann K., Martin S.A. Mass spectrometric determination of the amino acid sequence of peptides and proteins. Mass Spectrom. Rev. 1987;6:1–75. [Google Scholar]
- 42.Schweppe R.E., Haydon C.E., Ahn N.G. The characterization of protein post-translational modifications by mass spectrometry. Acc. Chem. Res. 2003;36:453–461. doi: 10.1021/ar020143l. [DOI] [PubMed] [Google Scholar]
- 43.Gittes F., Mickey B., Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mickey B., Howard J. Rigidity of microtubules is increased by stabilizing agents. J. Cell Biol. 1995;130:909–917. doi: 10.1083/jcb.130.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massey F.J., Jr. The Kolmogorov-smirnov test for goodness of fit. J. Am. Stat. Assoc. 1951;46:68–78. [Google Scholar]
- 46.Eshun-Wilson L., Zhang R., Nogales E. Effects of α-tubulin acetylation on microtubule structure and stability. Proc. Natl. Acad. Sci. USA. 2019;116:10366–10371. doi: 10.1073/pnas.1900441116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouxsein N.F., Bachand G.D. Single filament behavior of microtubules in the presence of added divalent counterions. Biomacromolecules. 2014;15:3696–3705. doi: 10.1021/bm500988r. [DOI] [PubMed] [Google Scholar]
- 48.Tran H.T., Mao A., Pappu R.V. Role of backbone-solvent interactions in determining conformational equilibria of intrinsically disordered proteins. J. Am. Chem. Soc. 2008;130:7380–7392. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 49.Ray S., Meyhöfer E., Howard J. Kinesin follows the microtubule’s protofilament axis. J. Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen M.R., Ruigrok R.W., Blackledge M. Describing intrinsically disordered proteins at atomic resolution by NMR. Curr. Opin. Struct. Biol. 2013;23:426–435. doi: 10.1016/j.sbi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Rostovtseva T.K., Sheldon K.L., Sackett D.L. Tubulin binding blocks mitochondrial voltage-dependent anion channel and regulates respiration. Proc. Natl. Acad. Sci. USA. 2008;105:18746–18751. doi: 10.1073/pnas.0806303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kikkawa M., Sablin E.P., Hirokawa N. Switch-based mechanism of kinesin motors. Nature. 2001;411:439–445. doi: 10.1038/35078000. [DOI] [PubMed] [Google Scholar]
- 53.Prota A.E., Danel F., Steinmetz M.O. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J. Mol. Biol. 2014;426:1848–1860. doi: 10.1016/j.jmb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Kay L.E., Nicholson L.K., Torchia D. Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson (1969) 1992;97:359–375. [Google Scholar]
- 55.Farrow N.A., Muhandiram R., Kay L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 56.Vemu A., Garnham C.P., Roll-Mecak A. Generation of differentially modified microtubules using in vitro enzymatic approaches. Methods Enzymol. 2014;540:149–166. doi: 10.1016/B978-0-12-397924-7.00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Souphron J., Bodakuntla S., Magiera M.M. Purification of tubulin with controlled post-translational modifications by polymerization-depolymerization cycles. Nat. Protoc. 2019;14:1634–1660. doi: 10.1038/s41596-019-0153-7. [DOI] [PubMed] [Google Scholar]
- 58.Freedman H., Luchko T., Tuszynski J.A. Molecular dynamics modeling of tubulin C-terminal tail interactions with the microtubule surface. Proteins. 2011;79:2968–2982. doi: 10.1002/prot.23155. [DOI] [PubMed] [Google Scholar]
- 59.Fedorov V.A., Orekhov P.S., Gudimchuk N.B. Mechanical properties of tubulin intra- and inter-dimer interfaces and their implications for microtubule dynamic instability. PLoS Comput. Biol. 2019;15:e1007327. doi: 10.1371/journal.pcbi.1007327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wloga D., Joachimiak E., Gaertig J. Posttranslational modifications of tubulin and cilia. Cold Spring Harb. Perspect. Biol. 2017;9:a028159. doi: 10.1101/cshperspect.a028159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suryavanshi S., Eddé B., Gaertig J. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr. Biol. 2010;20:435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wloga D., Dave D., Gaertig J. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot. Cell. 2010;9:184–193. doi: 10.1128/EC.00176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Felgner H., Frank R., Schliwa M. Domains of neuronal microtubule-associated proteins and flexural rigidity of microtubules. J. Cell Biol. 1997;138:1067–1075. doi: 10.1083/jcb.138.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.