Abstract
Wilson’s disease (WD) is an autosomal recessive inherited disorder of chronic copper toxicosis with high mortality and disability. Recent evidence suggests a correlation between dysbiosis in gut microbiome and multiple diseases such as genetic and metabolic disease. However, the impact of intestinal microbiota polymorphism in WD have not been fully elaborated and need to be explore for seeking some microbiota benefit for WD patients. In this study, the 16S rRNA sequencing was performed on fecal samples from 14 patients with WD and was compared to the results from 16 healthy individuals. The diversity and composition of the gut microbiome in the WD group were significantly lower than those in healthy individuals. The WD group presented unique richness of Gemellaceae, Pseudomonadaceae and Spirochaetaceae at family level, which were hardly detected in healthy controls. The WD group had a markedly lower abundance of Actinobacteria, Firmicutes and Verrucomicrobia, and a higher abundance of Bacteroidetes, Proteobacteria, Cyanobacteria and Fusobacteria than that in healthy individuals. The Firmicutes to Bacteroidetes ratio in the WD group was significantly lower than that of healthy control. In addition, the functional profile of the gut microbiome from WD patients showed a lower abundance of bacterial groups involved in the host immune and metabolism associated systems pathways such as transcription factors and ABC-type transporters, compared to healthy individuals. These results implied dysbiosis of gut microbiota may be influenced by the host metabolic disorders of WD, which may provide a new understanding of the pathogenesis and new possible therapeutic targets for WD.
Subject terms: Microbiology, Endocrinology
Introduction
Wilson’s (WD), also known as hepatolenticular degeneration, is a hereditary disorder characterized by accumulation of copper (Cu2+) in multiple vital organs such as brain, liver, etc. with an estimated prevalence of approximately 0.03–0.02‰ in the USA, Europe and Asia1. WD is also a genetic disease with complex phenotypes regulated by molecular genetics and epigenetic mechanisms2. The typically pathogenic mutations are in the copper-transporting gene, ATP7B, resulting in excess copper excretion through the biliary tract2. Recently published studies indicated that the gene expression in WD can be potentially modified by environmental and dietary factors. For example, regulation of the gene expression involved in methionine metabolism and epigenetic mechanisms could be affected by dietary factors, such as iron and methyl group donors3. Additionally, Penicillamine, a metabolic by-product of penicillin originated from the microbiota that avidly chelates copper, was the first choice for the treatment of WD4. We speculated that the intestinal microecology may contribute to the success action of Penicillamine, which prompted our interest in the interaction between WD and gut microbiome.
The gastrointestinal (GI) tract is the primary site of copper entry into the body, while genetic risk of autoimmunity has been showed to be related to distinct changes in the human gut microbiome. The human microbiome is determined by thousands of environmental, genetic and clinical factors and stochastic ecological processes, resulting in vast individual variation and biogeographic differences5. The composition of human microbial colonization is heritable beginning in the postpartum period and developing in an incremental manner. Moreover, the gut microbiota can influent the human health by providing crucial benefits to the development of the immune system, prevention of infections, acquisition of nutrients, and functionality of the nervous system6,7. Considering the intricacies of the interaction between the gut microbiota and the human host, it is essential to explore correlation between composition of human gut microbiota and WD. Meanwhile, the potential bacterial biomarkers need to be explored to develop the possible novel treatment strategies. Therefore, 16 s rRNA sequencing analysis of the gut microbiome was performed to profile fecal samples in relation to WD.
Results
Taxonomic analysis of 16S rRNA V4 amplicon sequence data
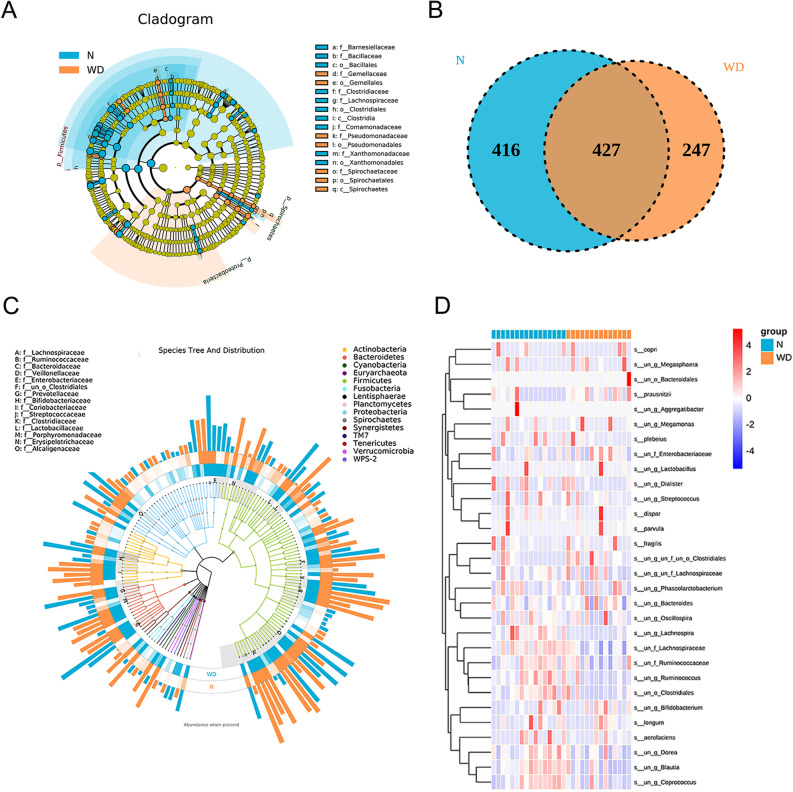
The clinical index such as age, sex and body mass and BMI were comparable between two groups and summarized in Table1. To explore the characteristics of gut microbial community in WD patients, the microbiota relative taxon abundance was compared with that in healthy individuals. A total of 1604 operational taxonomic units were annotated for subsequent taxon-dependent analysis including 15 phyla, 85 families and 153 genera of gut microbes inferred from V4 amplicon sequencing with 97% similarity among the samples (Fig. 1A). The predominant genera were defined as one bacteria genus comprising greater than 1% of the total gut microbiota. The Venn diagram reflecting the difference between two groups as shown in Fig. 1B, exhibited 674 and 843 in WD and control group, respectively. The bacterial taxonomy distribution and heatmap of the WD group showed decreased density and clustering compared to healthy controls (Fig. 1C,D).
Table1.
The baseline data of all participates.
| Groups (n) | Gender (M/F) | Age (y) | Weight (kg) | Urinary copper excretion(mg/L) | |||
|---|---|---|---|---|---|---|---|
| Mean ± SD | P value | Mean ± SD | P value | Mean ± SD | P value | ||
| WD (n = 14) | 9/5 | 32.2 ± 13.02 |
t = 1.042 P > 0.05 |
54.2 ± 10.33 |
t = 1.387 P > 0.05 |
726.44 ± 383.62 |
t = 6.966 P < 0.001 |
| Control (n = 16) | 9/7 | 37.8 ± 15.70 | 59.1 ± 9.05 | 58.65 ± 23.23 | |||
Figure. 1.
(A) LEfSe comparison of gut microbiota among WD and control groups, (B) Venn diagrams shared OTUs between different groups, (C) the species tree and distribution of gut microbial community, (D) the heatmap of gut microbiome among WD and control groups at species level.
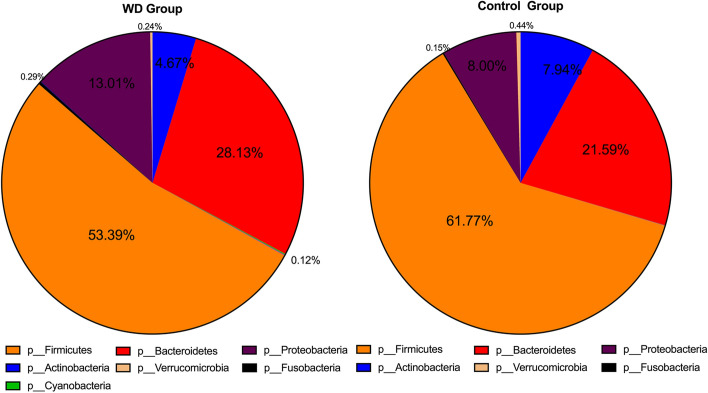
At the phylum level, Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Verrucomicrobia and Fusobacteria were identified as six common phyla in both the WD group and control group, accounting for 99.73% and 99.89% of each whole gut bacteri (Fig. 2). The WD group had a markedly lower abundance of Actinobacteria (4.67% vs. 7.94%, P < 0.05), Firmicutes (53.39% vs. 61.77%, P < 0.05) and Verrucomicrobia (0.24% vs. 0.44%, P < 0.05), and a higher abundance of Bacteroidetes (28.13% vs. 21.59%, P < 0.05), Proteobacteria (13.01% vs. 8.00%, P < 0.05) and Fusobacteria (0.29% vs. 0.15%, P < 0.05), compared to healthy controls. The WD group also showed a higher abundance of Cyanobacteria, compared to that of healthy controls (0.12% vs. 0, P < 0.05) (Fig. 2). The ratio of Firmicutes to Bacteroidetes was decreased in the WD group, compared to healthy controls (1.90 vs. 2.86, P < 0.05).
Figure. 2.
In the taxonomic profile, the OTUs were assigned to prevalent microbiome components of Firmicutes (53.39% vs. 61.77%, P < 0.05), Bacteroidetes (28.13% vs. 21.59%, P < 0.05), Acidobacteria (4.67% vs. 7.94%, P < 0.05), Proteobacteria (13.01% vs. 8.00%, P < 0.05), Verrucomicrobia (0.24% vs. 0.44%, P < 0.05) and Fusobacteria (0.29% vs. 0.15%, P < 0.05) at the phylum level.
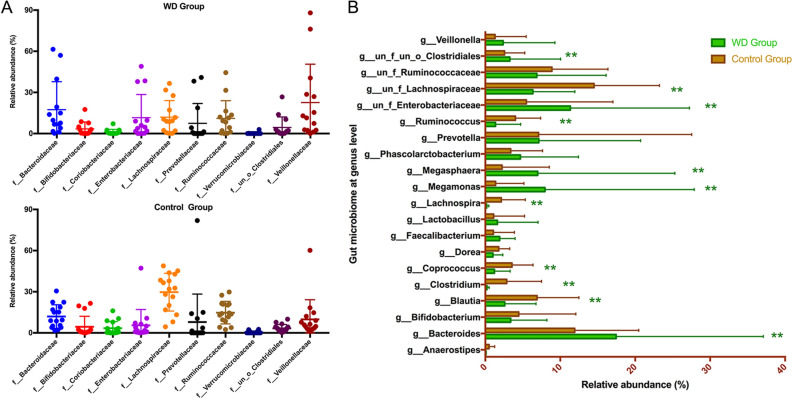
The internal individual variation in taxonomic structure was higher and more different dominant taxa were identified between two groups at the family level (Fig. 3A). Among the total families identified in the gut bacteria, 63 and 73 of the dominant families were respectively detected in the WD group and control group, respectively. Lachnospiraceae (phylum Firmicutes), Ruminococcaceae (phylum Firmicutes), Bacteroidaceae (phylum Bacteroidetes), Veillonellaceae (phylum Firmicutes), Prevotellaceae (phylum Bacteroidetes), Enterobacteriaceae (phylum Proteobacteria), Bifidobacteriaceae (phylum Actinobacteria) and Coriobacteriaceae (phylum Actinobacteria) represented eight most abundant components of the microbiome in WD group (Fig. 3A). While Veillonellaceae, Bacteroidaceae, Lachnospiraceae, Enterobacteriaceae, Ruminococcaceae, Prevotellaceae, Clostridiales (phylum Firmicutes) and Bifidobacteriaceae represented the eight most relative abundant components of the microbiome in control group.
Figure. 3.
(A) Comparison of abundant components of the microbiome at the family level between the two groups. (B) Taxonomic summary of the gut microbiota of each group at the genus level. Bacteroides, Enterobacteriaceae, Megamonas and Megasphaera were significantly more abundant in the WD group than the control group (P < 0.05). Lachnospiraceae, Ruminococcaceae, Blautia, Ruminococcus, Coprococcus Clostridium and Lachnospira of WD group were significantly less abundant in the WD group than the control group (P < 0.05). Starred samples (*) showed significant differences between the two groups.
The genus-level characterizations were more complex, with a number of bacterial genera showing significant differences between the two groups (Fig. 3B). In the WD group, Bacteroides (phylum Bacteroidetes), Enterobacteriaceae (phylum Proteobacteria), Megamonas (phylum Firmicutes), Prevotella (phylum Bacteroidetes), Megasphaera (phylum Firmicutes), Ruminococcaceae (phylum Firmicutes), Lachnospiraceae (phylum Firmicutes), Phascolarctobacterium (phylum Firmicutes), Bifidobacterium (phylum Actinobacteria), and Clostridiales (phylum Firmicutes) were the 10 most dominant components of 121 microbiota genera. While, the 10 most dominant components of the microbiota in 131 genera in the healthy control group were Lachnospiraceae, Bacteroides, Ruminococcaceae, Prevotella, Blautia (phylum Firmicutes), Enterobacteriaceae, Bifidobacterium, Ruminococcus (phylum Firmicutes), Coprococcus (phylum Firmicutes) and Phascolarctobacterium.
Alpha diversity analysis of the gut microbiota
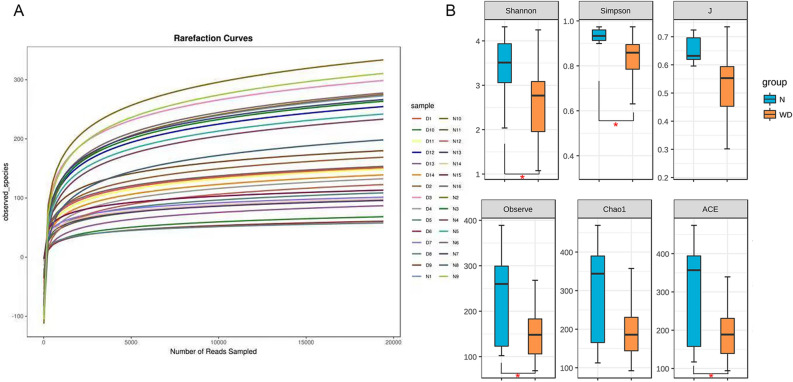
To evaluate differences in the microbiota community structure between the two groups, the alpha diversities were analyzed as shown in Fig. 4. The rarefaction curve indicated the quality of samples is high and size is reasonable. The α diversity of the gut microbiota in the WD group was lower than that in the healthy control group in terms of the index of Shannon and Simpson (Shannon index − 28.509 P = 0.0369; Simpson index − 28.607, P = 0.0386). The results of Observe and ACE index were also statistically different between the WD group and control group (− 24.513, P = 0.0493 and − 24.902, P = 0.0357, respectively). There were no significant differences between the WD group and healthy control group in terms of richness index includes J and Chao1 (− 28.696, P = 0.061; − 21.848, P = 0.0784).
Figure. 4.
(A) The rarefaction curve was in a flat shape presenting reasonable quantity of the samples. (B) Comparison of the alpha diversity of the gut microbiota between the two groups, including species richness (represented by Chao1, Observed species (Observe), ACE and J) and evenness (represented by Shannon and Simpson index). The microbial community of the WD group had less diversity, richness and evenness than the control group. Starred samples (*) showed significant differences between the two groups.
Beta diversity analysis of the gut microbiota
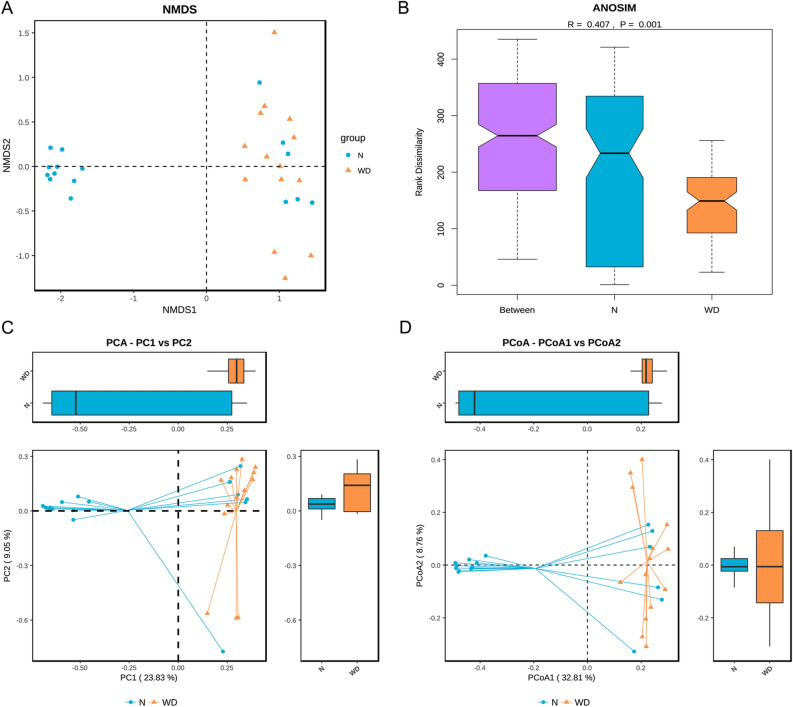
Non-metric multi-dimensional scaling (NMDS) analysis revealed that the microbial genera were significant difference between WD patients and healthy controls based on the Bray–Curtis distances (Fig. 5A). The structure of the gut microbiota evaluated by ANOSIM was significantly different between the two groups (R = 0.407, P = 0.001) (Fig. 5B). Principal component analysis (PCA: PC1 23.83% vs. PC2 9.05%) and principal coordinates analysis (PCoA: PCoA1 32.81% vs. PCoA2 8.76%) based on unweighted UniFrac distance (Fig. 5C) and weighted UniFrac distance (Fig. 5D) indicated that the gut microbiome of WD patients clustered significantly separated from healthy controls.
Figure. 5.
Comparison of the beta diversity of the gut microbiota between two groups. (A) Non-metric multi-dimensional scaling, (B) analysis of similarities (ANOSIM, R = 0.407, P = 0.001), (C) principal component analysis (PC1 23.83% vs. PC2 9.05%), and (D) principal coordinates analysis (PCoA1 32.81% vs. PCoA2 8.76%) illustrating the grouping patterns of the WD group and control group based on the Bray–Curtis and unweighted UniFrac distances. Each dot represents a sample, and the corresponding group can be found in the legend. Distances between any pair of samples represent the dissimilarities between each sample. There was a significant difference in β-diversity between the two groups.
Functional profile of the gut microbiome
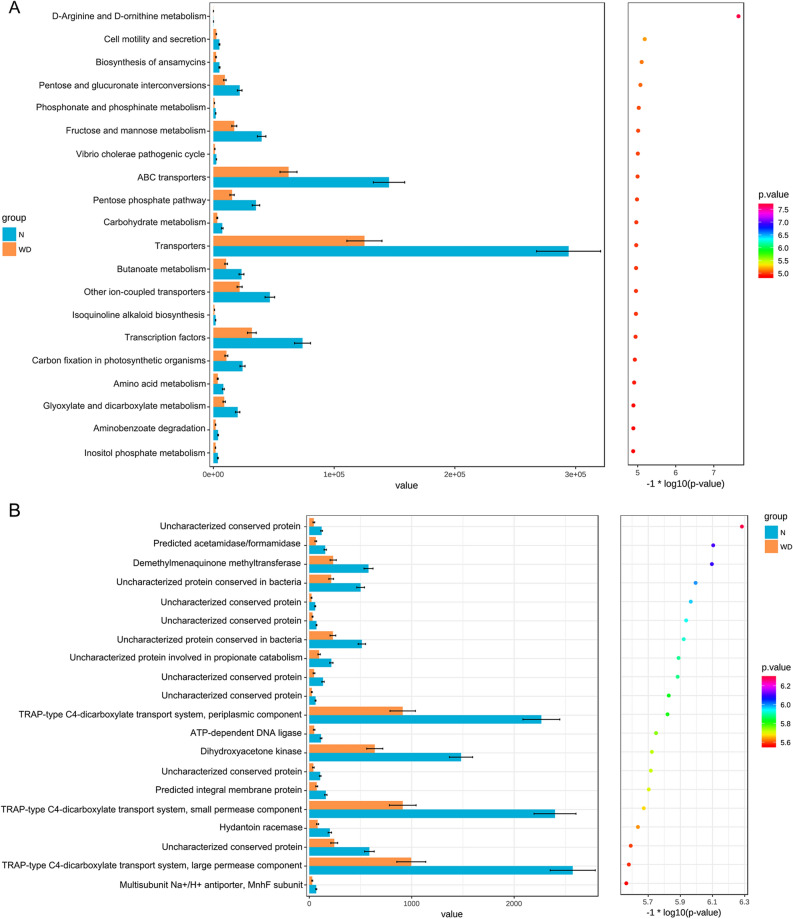
The functional composition of the gut microbiome was compared between WD patients and controls by KEGG and COG pathway analyses. Although the functional compositions of the two groups were highly similar, the gut microbiome in WD patients showed a less richness in pathways of transcription factors and ABC-type transporters than healthy controls (F = 31.745, P < 0.001). The analysis indicated the clustering of metabolic modules were decreased in WD group including metabolism of fructose (F = 31.2291, P < 0.001), mannose (F = 28.745, P < 0.001), butanoate (F = 31.385, P < 0.001), glyoxylate and dicarboxylate (F = 30.7384, P < 0.001), interconversions of pentose and glucuronate (F = 32.6781, P < 0.001), other ion-coupled transporters (F = 31.4571, P < 0.001), and carbon fixation in photosynthetic organisms (F = 30.6242, P < 0.001) (Fig. 6A). In addition, the TRAP-type C4-dicarboxylate transport system, periplasmic component (F = 38.397, P < 0.001), dihydroxyacetone kinas (F = 37.2417, P < 0.001), TRAP-type C4-dicarboxylate transport system, small permease component (F = 37.5488, P < 0.001), and TRAP-type C4-dicarboxylate transport system, large permease component (F = 36.4327, P < 0.001) were all significantly less abundant in the WD group than in the healthy controls. Similarly, demethylmenaquinone methyltransferase (F = 41.6979, P < 0.001) and uncharacterized protein conserved in bacteria (F = 41.9507, P < 0.001) and uncharacterized protein conserved in bacteria (F = 38.5353, P < 0.001) were also less abundant in WD patients than in healthy controls (P < 0.05) (Fig. 6B).
Figure. 6.
(A) Abundance in the KEGG pathway was lower in the WD group than in healthy controls (P < 0.001). (B) Abundance in the COG pathway was less in the WD group than in healthy controls (P < 0.001).
Discussion
WD is an autosomal-recessive disorder characterized by abnormal copper metabolism leading to copper excretion disorder and deposition in target organs. Penicillamine is one of the standard therapies which was introduced as a treatment for WD by Walshe in 19568. Penicillamine can combine with copper deposited in the tissue to form a soluble complex, and then excreted from the urine. Recently published study found that oral penicillin can induce gut microbiota dysbiosis in mice, which may associate with host lipid metabolism dysfunction and low-grade inflammation9. As all known, the human gut microbiome has been confirmed to play a causal role in the development of pathologies in animal models of metabolic disease such as obesity, Alzheimer’s disease and Type 1 diabetes (T1D)10–12. The human gut microbiome participates in nutrient metabolism, inhibition of pathogenic growth, angiogenesis stimulation, and the maturation and maintenance of the immunological system to ensure a balance or homeostasis in the host6,13,14. In light of the multiple roles of the gut microbiome played in a number of diseases and the satisfactory efficacy of Penicillamine on WD, it is necessary to explore the possible correlation between the gut microbiome and WD. Actually, the features of the gut microbiome in WD patients has been explored by Geng15. The results of two studies both indicated the disruption of the equilibrium in the gut microbiota of WD patients, exhibiting elimination, decreased density and loss of bacterial diversity in the microbial ecosystem. However, Geng only analyzed the features of gut microbiome in WD patients at phylum level. Though α and β diversity between WD and healthy controls were compared, the variation in details of different levels were lacked. The present study revealed some special and detailed characterizations of the gut microbiome in WD.
At the phylum level, the gut microbiome of the WD group showed a significantly lower abundance of Firmicutes compared to the healthy controls, covering the majority of butyrate-producing bacteria in human bacterial communities. However, the abundance of Firmicutes in WD patients reported by Geng was significantly higher than that of healthy controls (26.18% vs. 19.83%, respectively, P < 0.05)15. Firmicutes is known for its function in transforming undigested proteins and carbohydrates into acetic acid, and producing energy for the organism of host. The crucial short-chain fatty acids (SCFAs) butyrate participates in activating multiple physiological signal pathways, such as anti-inflammatory activities, and the differentiation and proliferation of regulatory T cells16,17. We speculated that the alteration of bacterial composition may cause a low concentration of intestinal SCFAs, leading to skewed physiological functions in WD patients. Subsequent KEGG and COG pathway analysis further confirmed reduced transport and metabolism functional groups in the gut microbiome of the WD group, which intensify the correlation between WD and the gut microbiome. We thus speculate that the WD triggers dysfunction of gut microbiota and increases intestinal permeability, interfering with the intestinal microbiota by transplanting fecal bacteria rich in SCFAs, and supplementing with butyric acid may be effective therapies for WD.
Meanwhile, the WD group showed significantly decreased phylum levels of Actinobacteria and Verrucomicrobia compared to the control group. The Actinobacteria is known to have the capability of surviving in extreme, even toxic environments18. Verrucomicrobia is mucin-degrading bacteria residing in the intestinal mucosa that contribute to intestinal health and glucose homeostasis, and plays as an interface between the human gut microbiome and host tissues19. Therefore, the decrease of these two probiotics may cause the disorder of physiological functions in WD patients. The gut microbiome in the WD group also showed a significantly higher abundance of Proteobacteria and Fusobacteria than in healthy individuals, consistent with the results reported by Geng15. The opportunistic pathogen of Proteobacteria created a major structural imbalance of gut microbiota in WD patients, while Fusobacteria have been widely recognized for the potential inducer of T regulatory cells or carcinogens promoting autophagic activation20. The WD group also showed higher abundance of Cyanobacteria, which has ability to perform nitrogen and carbon fixation and are involved in complex metabolic pathways with different mechanisms. Moreover, Cyanobacteria have an exceptionally high iron demand because of their involvement in the functions of a variety of crucial enzymes21. We speculated that microbiota-derived products may be act as triggers for promoting a proinflammatory and metabolically dysfunctional environment in WD patients. Microbiota-produced butyrate has been reported to provide energy for colonocytes and then prevent autophagy in the intestine of host17. Additionally, physiological homeostasis may be disrupted by gut microbiota, resulting in host metabolism disruption, and immune, neurological or cognitive system dysregulation and others22,23. Therefore, the decreased abundance of ABC transporters system signaling pathways further confirmed potential alteration of energy metabolism related to gut microbiota in WD. To our best knowledge, it is the first report to analyze the correlation between transporters system signaling pathways and gut microbiome of WD.
We also noticed that WD patients exhibited a lower ratio of Firmicutes to Bacteroidetes than healthy individuals. The dysbiosis of gastrointestinal tract metabolism was reported to be associated with low Firmicutes/Bacteroidetes ratio, causing a low concentration of circulating SCFAs, influencing elements of immune and inflammation system. These observations suggest that the gut microbiome has a potential impact on conserved functions, which may further inducing the pathogenesis of WD. Human lymphocyte antigen (HLA) gene alleles have been confirmed to have a significant effect on the composition of the gut microbiota in late infancy18. We thus speculated that some metabolic product of gut bacteria may be recognized by high risk genotypes in WD leading to alteration of the taxa in the human gut.
The features of gut microbiome in WD group at the family and genus levels were more complex and varied significantly from the control group, presenting a different pathogen microbiome and well characterized structures. The Bacteroides showed a greater abundance in the WD group than in healthy individuals. Bacteroides are correlated with diets high in animal protein and saturated fats. Moreover, the Bacteroides genus was reported to enhance the efficacy of the anti-CTLA4 immune checkpoint in mice and is speculated to contact and stimulate T cells and DCs directly by means of pathogen-associated molecular patterns in the host24. The Enterobacteriaceae were markedly more abundant in WD than control group. The significant opportunistic pathogen Enterobacteriaceae is present in the human gut without causing symptoms or diseases under normal circumstance. However, host immunity and environmental factors, such as redox state and availability of oxygen, may result in significant variation of Enterobacteriaceae. The richness of Enterobacteriaceae in WD further suggested the correlation between pathogen microbiome and WD. The development of targeting identified bacterial biomarkers of gut microbiome may be an alternative therapeutic strategy to WD in future. The gut microbiome of controls and WD patients has a comparable abundance of Prevotella genus, which is more universal in populations with high carbohydrate and sugar consumption, as observed in agrarian and vegetarian societies25. Furthermore, the WD group exhibited significantly lower abundance of Blautia, Ruminococcus and Coprococcus genus in comparison with control group, which are considered as important components of commensal microbiota and play important roles in several homeostatic functions such as immunity, neurohormones and metabolism. Blautia plays a role in digestion of complex carbohydrates and also shows decreased abundance in diabetes, irritable bowel syndrome, Crohn’s disease and and nonalcoholic fatty liver diseases26. Coprococcus is a genus of anaerobic cocci which is part of the human faecal microbiota that functions in producing butyrate27. The decreased of these probiotics indicated the defections of multiple physiological function presented in WD may be caused by the dysbiosis of gut microbiome. Additionally, Megamonas, Megasphaera and Clostridiales are genera of Firmicutes bacteria that showed increased abundance in the WD group. Clostridiales includes bacterial species that produce short-chain fatty acid, which are important for the equilibrium between regulatory T cells and helper T type17 (Th17) cells23. These results further indicated the structure of bacterial communities is more various than previous study and some potential gut microbiome may be identified as bacterial biomarkers. In addition, the analysis of KEGG and COG pathway indicated that these bacteria affect the host by shedding different microbial bioactive molecules, including transcription factors, metabolism of fructose, mannose, butanoate, glyoxylate and dicarboxylate, different transporters system such as ion-coupled transporters, ABC tranporters and TRAP-type C4-dicarboxylate transport system. Increasing evidences demonstrated that gut microbiota related transporters system appears to be key microbial bioactive signaling pathways. These transport systems regulate the energy system by promoting utilization of glucose, ribose/galactoside. We speculated that the dysfunction of gut microbiota in WD may influent the metabolic of copper via transporters system signaling pathways. In the next step, specific metabolites of the microbiome may be searched for as therapeutic chemicals like penicillamine in WD.
However, the main limitation of the study is the small samples size causing from rarity which may affect the accuracy of the results. Meanwhile, more advanced method including metagenomics and metabonomics need to be applied to explore the possible specific metabolic or microbiota for WD. To compensate for these weaknesses, cooperation among multi-medical center need to fully elucidate the intestinal flora and treatment of this disease.
In summary, this study analyzed the features and possible functions of gut microbiota in WD patients, providing a new understanding of the pathogenesis of WD and identified potential bacterial biomarkers. The structure of the microbiota in WD patients showed an elimination, low-density, microbial ecosystem with loss of bacterial diversity. Both COG and KEGG pathway analysis indicated decreased abundance of some dominant metabolism and transport related pathways in WD patients. The dysbiosis of gut microbiota may contribute to host metabolic disorders of WD. The results may provide new insights and will facilitated future studies of the pathogenesis of WD and the development of novel treatment strategies. Under these conditions, fecal transplant and microbiota-related metabolites may hold promise for the prevention of WD.
Materials and methods
Samples
Fecal samples were obtained from 14 newly diagnosed WD patients and 16 healthy participants for subsequently sequencing of 16S rRNA. The diagnosis of WD was confirmed by ATP7B gene and satisfy clinical diagnostic criteria including family history, clinical manifestations, neurological examination, low level of serum ceruloplasmin, high urinary copper excretion in 24 h, examination of liver function, ultrasonography of the liver, and magnetic resonance imaging (MRI) of the brain. Patients with any of the following conditions were excluded: respiratory or renal failure, congestive cardiac disease, severe liver dysfunction, or being prescribed with probiotics or antibiotics within 1 month before admission. The control group was consisted of 16 healthy participants who did not take any probiotics or antibiotics within 1 month before inclusion. The clinical characteristics of all subjects were summarized in Table 1 and the parameters such as age, gender and weight were comparable between two groups (P > 0.05). The fresh fecal samples from all participates were collected into sterile eppendorf tubes and were frozen at − 80 °C refrigerator immediately, until the extraction of DNA. Each patient signed written informed consent before inclusion in this study. All experiments were approved and carried out in accordance with the guidelines of the Ethics Committee of the First Affiliated Hospital of Guangdong Pharmaceutical University.
DNA extraction, amplification, and sequencing of the V4 region of the bacterial 16S rRNA gene
Microbial DNA was extracted from 30 fecal samples using a PowerSoil DNA Stool Mini Kit (MoBio) according to the protocol recommended by manufacturer. Briefly, the V4 variable region of the bacterial 16S rRNA gene was used to amplify by polymerase chain reaction (PCR) with universal primers 806R (GGACTACHVGGGTWTCTAAT) and 341F (CCTACGGGNGGCWGCAG). The PCR conditions consisted of predenaturation step at 95 °C for 2 min followed by 30 cycles denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 45 s, with a final extension step at 72 °C for 5 min. The PCR products were visualized by the method of agarose gels electrophoresis. Finally, equimolar amounts of purified amplicons were pooled and subjected to paired-end sequencing on an Illumina HiSeq/MiniSeq benchtop sequencer (Illumina) for genome analysis28,29.
The analysis of Microbiome data
The raw FASTQ files were initially de-multiplexed, and then quality-filtered by Chimera check and merged using FLASH with the processed sequences according to the BIPES protocol.10 and Quantitative Insights Into Microbial Ecology (QIIME) 1.928,29. Briefly, forward and reverse bacterial 16S rRNA reads were merged with a minimum merge length of 200 bp, singletons and chimeras were simultaneously removed by filtering in UPARSE. Operational taxonomic units (OTUs) were defined as a sequence similarity level of 97% and assigned to taxon-dependent analysis through the Ribosomal Database Project (RDP) classifier by the Green Genes Database to explore WD-associated differences in the human gut microbiota. Based on OTUs analysis, the index of Ace, observed species, Shannon, Chao1, Simpson and J were used to calculate alpha diversity metrics. Principal component analysis (PCA) and principal coordinate analysis (PCoA) were then analyzed to compare the microbial composition between the samples. The unweighted-pair group method with arithmetic mean (UPGMA) trees was created based on unweighted UniFrac distance matrices. The statistical significance of difference between groups was evaluated by analysis of similarities (ANOSIM). The databases of Kyoto Encyclopedia of Genes and Genomes (KEGG) and Cluster of Orthologous Groups (COG) were used to analyze the pathway richness by PICRUSt30,31. Finally, the influence of each differentially abundant gut microbiota was evaluated by the method of linear discriminant analysis (LDA) effect size (LEfSe).
Statistical analysis
Statistical tests were performed using R 3.0.3 software (R Foundation for Statistical Computing) and Prism software (Graph Prism 7.0 Software Inc. CA, USA). The diversities between two groups were compared by Wilcoxon’s rank-sum test. The categorical variables were analyzed by Fisher’s exact test. The chi-square test was used for categorical variables. In all analyses, a value of P < 0.05 considered statistically significant difference in the compared groups.
Conference presentation
This article is present on a university repository website and can be accessed on https://www.researchsquare.com/article/rs-15131/v1.
Acknowledgements
We gratefully thank Guangzhou Health Care Co-innovation Major Plan Fund (No. 201803040014).
Author contributions
X.Y., F.L. and H.S. conceived and designed the study. X.C., L.D. and X.M. conducted the experiments. Y.G., Y.G., Y.H., M.L. and Z.F. participated in the completion of the experiments. J.D. and H.L. analyzed the data. X.C. wrote the manuscript. X.Y., F.L. and H.S. revised the manuscript. All the authors have read and approved the final version of this manuscript for publication.
Data availability
All data used during the study are available from the corresponding author by request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiangsheng Cai, Lin Deng and Xiaogui Ma.
Contributor Information
Xiangsheng Cai, Email: xiangshengcai@gdpu.edu.cn.
Fang Liu, Email: liufangndyx@163.com.
Xiaorong Yang, Email: 122424818@qq.com.
References
- 1.Sandahl TD, et al. The prevalence of Wilson disease. An update. Hepatology (Baltim., MD.) 2019 doi: 10.1002/hep.30911. [DOI] [PubMed] [Google Scholar]
- 2.Medici V, LaSalle JM. Genetics and epigenetic factors of Wilson disease. Ann. Transl. Med. 2019;7:S58. doi: 10.21037/atm.2019.01.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medici V, Weiss KH. Genetic and environmental modifiers of Wilson disease. Handb. Clin. Neurol. 2017;142:35–41. doi: 10.1016/b978-0-444-63625-6.00004-5. [DOI] [PubMed] [Google Scholar]
- 4.Devassy DE, Harshad SR, Devarbhai H. Pompholyx-Like Eruptions induced by penicillamine in a patient with Wilson's disease. Indian J. Dermatol. 2019;64:321–323. doi: 10.4103/ijd.IJD_328_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell JT, et al. Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat. Commun. 2019;10:3621. doi: 10.1038/s41467-019-11460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenzie C, Tan J, Macia L, Mackay CR. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol. Rev. 2017;278:277–295. doi: 10.1111/imr.12556. [DOI] [PubMed] [Google Scholar]
- 7.Zhong H, et al. Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children. Microbiome. 2019;7:2. doi: 10.1186/s40168-018-0608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walshe JM. Penicillamine, a new oral therapy for Wilson's disease. Am. J. Med. 1956;21:487–495. doi: 10.1016/0002-9343(56)90066-3. [DOI] [PubMed] [Google Scholar]
- 9.Jin Y, et al. From the cover: exposure to oral antibiotics induces gut microbiota dysbiosis associated with lipid metabolism dysfunction and low-grade inflammation in mice. Toxicol. Sci. Off. J. Soc. Toxicol. 2016;154:140–152. doi: 10.1093/toxsci/kfw150. [DOI] [PubMed] [Google Scholar]
- 10.Mullaney JA, et al. Correction to: type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome. 2018;6:51. doi: 10.1186/s40168-018-0438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullaney JA, et al. Type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome. 2018;6:35. doi: 10.1186/s40168-018-0417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin CH, et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson's disease. J. Neuroinflam. 2019;16:129. doi: 10.1186/s12974-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirata SI, Kunisawa J. Gut microbiome, metabolome, and allergic diseases. Allergol. Int. Off. J. Jpn. Soc. Allergol. 2017;66:523–528. doi: 10.1016/j.alit.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Balakrishnan B, Taneja V. Microbial modulation of the gut microbiome for treating autoimmune diseases. Expert Rev. Gastroenterol. Hepatol. 2018;12:985–996. doi: 10.1080/17474124.2018.1517044. [DOI] [PubMed] [Google Scholar]
- 15.Geng H, et al. Association study of gut flora in Wilson's disease through high-throughput sequencing. Medicine. 2018;97:e11743. doi: 10.1097/md.0000000000011743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuik F, et al. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United Eur. Gastroenterol. J. 2019;7:897–907. doi: 10.1177/2050640619852255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagpal R, Neth BJ, Wang S, Craft S, Yadav H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer's disease markers in subjects with mild cognitive impairment. EBioMedicine. 2019 doi: 10.1016/j.ebiom.2019.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson JR, et al. A preliminary examination of gut microbiota, sleep, and cognitive flexibility in healthy older adults. Sleep Med. 2017;38:104–107. doi: 10.1016/j.sleep.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SN, et al. Fusobacterium pseudoperiodonticum sp. Nov., isolated from the human oral cavity. Curr. Microbiol. 2019;76:659–665. doi: 10.1007/s00284-019-01675-y. [DOI] [PubMed] [Google Scholar]
- 21.Jacan A, et al. Synergistic and antagonistic interactions between antibiotics and synbiotics in modifying the murine fecal microbiome. Eur. J. Nutr. 2019 doi: 10.1007/s00394-019-02035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, et al. Microbial intervention as a novel target in treatment of non-alcoholic fatty liver disease progression. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018;51:2123–2135. doi: 10.1159/000495830. [DOI] [PubMed] [Google Scholar]
- 23.Ticinesi A, Nouvenne A, Tana C, Prati B, Meschi T. Gut Microbiota and microbiota-related metabolites as possible biomarkers of cognitive aging. Adv. Exp. Med. Biol. 2019;1178:129–154. doi: 10.1007/978-3-030-25650-0_8. [DOI] [PubMed] [Google Scholar]
- 24.Yu SY, et al. Bacteroides faecalis sp. Nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2019 doi: 10.1099/ijsem.0.003690. [DOI] [PubMed] [Google Scholar]
- 25.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franzosa EA, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mobeen F, Sharma V, Tulika P. Enterotype variations of the healthy human gut microbiome in different geographical regions. Bioinformation. 2018;14:560–573. doi: 10.6026/97320630014560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics (Oxf., Engl.) 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used during the study are available from the corresponding author by request.