Background
Gut microbes play a crucial role in the maintenance of human health. Components in the diet of the host affect their metabolism and diversity. Here, we investigated the influences of three commonly used non-caloric artificial sweeteners-aspartame, acesulfame K and sucralose-on the growth and metabolism of an omnipresent gut microbe Escherichia coli K-12. Methods: Growth of E. coli in the presence of aspartame, acesulfame K and sucralose in media was assessed and the influences of these artificial sweeteners on metabolism were investigated by relative expression analysis of genes encoding the rate limiting steps of important metabolic pathways as well as their global metabolomic profiles. Results: As a whole, E. coli growth was inhibited by aspartame and induced by acesulfame potassium, while the effect of sucralose on growth was less prominent. Although the expressions of multiple key enzymes that regulate important metabolic pathways were significantly altered by all three sweeteners, acesulfame K caused the most notable changes in this regard. In multivariate analysis with the metabolite profiles, the sucralose-treated cells clustered the closest to the untreated cells, while the acesulfame potassium treated cells were the most distant. These sweeteners affect multiple metabolic pathways in E. coli, which include propanoate, phosphonate, phosphinate and fatty acid metabolism, pentose phosphate pathway, and biosynthesis of several amino acids including lysine and the aromatic amino acids. Similar to the gene expression pattern, acesulfame potassium treated E. coli showed the largest deviation in their metabolite profiles compared to the untreated cells.
Keywords: Non-caloric artificial sweeteners, Aspartame, Acesulfame potassium, Sucralose, Escherichia coli, Metabolism
Highlights
-
•
Aspartame, acesulfame K and sucralose modulate growth and metabolism of the common gut microbe Escherichia coli K-12.
-
•
These sweeteners affect multiple metabolic pathways in E. coli.
-
•
Sucralose appears to have the least deviating impact on E. coli growth and metabolism under in vitro condition.
-
•
Acesulfame potassium appears to have the largest deviating impact on E. coli growth and metabolism under in vitro condition.
1. Introduction
Non-caloric artificial sweeteners, also popularly known as non-nutritive sweeteners, have gained much popularity over the past few decades for their ability to provide sweet taste without associated high caloric content [1]. This rise in popularity got more thrust from reports that linked high sugar content in diet with different physiological conditions like insulin resistance and type 2 diabetes mellitus (T2DM), cardiovascular diseases (CVDs), fatty liver, dyslipidemia and hyperuricemia [[1], [2], [3]] and showed a positive role of non-caloric artificial sweeteners (NAS) in weight reduction [4,5]. Artificial sweeteners are currently used in varieties of processed foods, soft drinks, powdered drinks, baking goods, etc.
However, the non-caloric artificial sweeteners may not be without caveats. Several studies in the recent years reported that artificial sweetener consumption may be associated with T2DM [6], obesity [7], CVDs [8], psychotic conditions [9], headaches [10], vestibular neuronitis and hearing loss [11], oxidative stress [12], and even cancer [13]. Although these artificial sweeteners were previously thought to be harmless due to their lack of interaction within the gastrointestinal tract [14], these newer findings have raised concerns about the mechanism behind such effects.
Human gastrointestinal tract is inhabited by a dense and diverse microbiota [15]. Their role in human physiology is so prominent that these are considered to be a virtual organ [16] and our second genome [17]. Their diversity is a result of host genotype and factors like lifestyle and diet [18,19]. Diet influences the metabolic pathways in these gut microbes [20,21]. The composition of gut microbiome can also be altered by artificial sweeteners [15,22], which in turn might result in the physiological abnormalities in their hosts [23,24]. Some studies in the recent years have reported that non-caloric artificial sweeteners can induce glucose intolerance [22] and weight gain [25] by altering the composition of gut microbiota. Such reports have formed a base to establish an indirect connection between the intake of these sweeteners and certain metabolic disorders via the alteration of gut microbiota.
Three most popular non-nutritive sweeteners are acesulfame potassium, aspartame and sucralose. Acesulfame potassium, which is approximately 200 times as sweet as sucrose, is the potassium salt of acesulfame (6-methyl-1,2,3-oxathiazine-4(3H)-one 2,2-dioxide), an acidic cyclic sulfonamide [26]. Aspartame (N-l-α-aspartyl-l-phenylalanine-1-methyl ester) is a methyl ester of phenylalanine and aspartic acid and it is also 200 times sweeter than sucrose [26]. Sucralose, the most widely used FDA-approved artificial sweetener, is a synthetic molecule generated by substitution of three hydroxyl groups in sucrose [14]. It is 600 times sweeter than sucrose.
Escherichia coli, a member of the Enterobacteriaceae family, is one of the first gut colonizers in human that persist throughout the lifetime [16,27]. Being a facultative anaerobe, E. coli helps creating an anaerobic environment by consuming the remaining oxygen in the gut [27]. It plays beneficial roles in human health by producing vitamin K [28] and conferring resistance to invading pathogens [29,30]. Despite its lower abundance in the human gut compared to several other major gut bacteria such as Bifidobacterium, Bacteroides, Clostridium, etc., it is one of the most common gut colonizers [16,27]. It can become pathogenic in immunocompromized individuals [31]. Alterations in the abundance of gut E. coli have been reported in diseases like type 2 diabetes [32], asthma [33] and inflammatory bowel diseases [34]. As a species, E. coli is one of the most important and best understood model organisms [35,36]. In addition, unlike most other gut bacteria, which are largely obligate anaerobes, E. coli can be grown in aerobic conditions in vitro.
In the present study, we investigated and compared the influence of acesulfame potassium, aspartame and sucralose on the growth and metabolism of E. coli. In a previous study, we found that a commercially available artificial sweetener preparation containing a combination of aspartame and acesulfame potassium can influence E. coli growth and modulate the expression of some of its key regulatory genes associated with glucose, nucleotide and fatty acid metabolism [37]. In the present study, we investigated and compared the influence of acesulfame potassium, aspartame and sucralose per se on the growth and metabolism of E. coli.
2. Materials and methods
2.1. Assessment of E. coli growth with different NAS in media
E. coli K-12 strain in log-phase were inoculated in Luria Bertani (LB) medium (610084, Liofilchem®) at pH 5.2. Different concentrations of acesulfame potassium, aspartame and sucralose were supplemented to the media to assess their influences on E. coli growth. NAS stock solutions in water were filter sterilized (0.22 μm) to ensure that the sweeteners were stable and functional in culture media and added after the media were autoclaved. Bacteria were incubated at 37 °C and 135 rpm in 96-well microtiter plates. Blank cultures corresponding to each sweetener concentration were also prepared. At least two biological replicates each with four technical replicates for each of the NAS were used in the study. Optical density (OD) at 630 nm was measured at 30 min time intervals using a microplate reader (Gentaur/GDMS, Belgium) for 5 h. The average optical density of the blank from each reading was subtracted from the corresponding OD of the bacterial culture. Bacterial growth curves were generated by plotting the average OD of the cultures at different time intervals.
2.2. Relative gene expression analysis
E. coli cells in log phase were inoculated in LB medium containing either 0 or 6 mg/mL of aspartame, acesulfame potassium or sucralose and incubated at 37 °C at 135 rpm for 5 h. The pH of the media was kept at 5.2. After 5 h of incubation, 2 mL aliquots of bacterial culture were taken into nuclease-free micro-centrifuge tubes and centrifuged at 5000 rpm for 3 min to collect the cell pellets. Cells were washed with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4) at 10,000×g for 2 min, re-suspended in 100 μL of lysozyme (62971, Sigma-Aldrich) solution in PBS and incubated at 37 °C for 15 min. Cells were disrupted using syringe and 21-gauge needle. Total RNA was extracted from the bacterial cell lysates using FavorPrep™ Tissue Total RNA Mini Kit (FATRK001, Favorgen®) following the manufacturer’s protocol. RNA was treated with RNase-free DNase I (18068015, Invitrogen™) solution in column to eliminate genomic DNA contamination. RNA was eluted in nuclease free water. Concentration and purity of RNA were measured using OneDrop Micro-Volume Spectrophotometer (Biometrics Technologies). The integrity of RNA was checked following electrophoresis in a 1% agarose gel in 0.5x TAE buffer. Fusion Pulse 6 gel documentation system (Vilber) was later used to visualize the RNA bands.
SuperScript™ III First-Strand Synthesis kit (18080051, Invitrogen™) was used to synthesize the first-strand cDNA following the manufacturers’ protocol. 5 μg of total RNA from each sample was used with 1 μL of random hexamers to prime cDNA synthesis. The reaction mixtures were finally treated with 1 μL of RNase H (18080051, Invitrogen™) to remove RNA.
Nine key regulatory enzymes (aceE, adk, fabI, glgC, lpd, pfkA, pfkB, tdk1 and thyA) that control important metabolic pathways [37] in E. coli were selected from the Rate-Limiting Enzyme database (RLEdb) [38] for corresponding gene expression analysis. The gene encoding DNA gyrase (gyrA) was used as the reference to normalize gene expression data. Relevant information about the primers is given in Refs. [37]. Gene expression levels in the presence of different NAS were compared using relative quantitative PCR with PowerUp™ SYBR™ Green Master Mix (A25780, Thermo Fisher Scientific) in StepOnePlus™ Real-Time PCR machine (Applied Biosystems). Reaction mixtures were set in final volume of 15 μL with equal quantity of template cDNA in each well of MicroAmp™ Fast 8-Tube Strip (4358293, Applied Biosystems™). Overall the composition of each reaction mixture was as follows: PowerUp™ SYBR™ Green Master Mix 7.5 μL, 0.6 μL of forward and reverse primer mix (5 μM each), PCR grade-water 4.9 μL, sample cDNA 2.0 μL. The amplification programme was as follows: UDG activation at 50 °C for 2 min, DNA polymerase activation at 95 °C for 2 min, then 40 cycles each with denaturation at 95 °C for 15 s followed by annealing and extension at 60 °C for 1 min. Amplification data were collected at the end of each extension step. Amplification specificities were assessed by melt curve analysis after the end of each amplification programme. Specificities of the amplicons were also assessed in 1% agarose gel following electrophoresis. Primer efficiencies were calculated from the slopes of the standard curves and the relative gene expression levels were corrected for primer efficiencies following the method described by Pfaffl [39]. Cycle threshold (Ct) value of amplification for each gene sample was set manually.
2.3. Metabolic profiling and pathway identification
500 μL of the same bacterial culture, which was used for the gene expression analysis, was added to a micro-centrifuge tube and quenched by adding twice the amount (1 mL) of cold (−48 °C) 60% methanol (HPLC grade) to quench internal cellular processes without leeching metabolites in to the medium. Samples were centrifuged at 4800×g for 10 min at −8 °C to pellet cellular mass. Cell media was removed and re-centrifuged at 4800×g for 10 min at −8 °C followed by the removal of the residual quenching media. The cell biomass was suspended in 1 mL of cold (−48 °C) 80% methanol. The dissolved pellet was transferred to a pre-chilled Eppendorf tube, snap frozen in liquid nitrogen for 1 min and then allowed to melt on ice. Upon thawing, the solution was vortexed for 30 s and then the snap freezing step was repeated 2 more times. The solution was centrifuged at −8 °C at 14,500×g for 5 min and the supernatant was transferred into a fresh pre-chilled Eppendorf tube. 500 μL of extraction solvent (80% methanol at −80 °C) was added and the steps described above were repeated, followed by addition of the extracted solution to a 2 mL Eppendorf tube. The extracted metabolites were stored at −80 °C following snap freezing in liquid nitrogen.
Extracted metabolites were diluted using a 1:1 HPLC-grade methanol and water mixture. 0.1% formic acid was added to the mixture. The samples were 100 times diluted by mixing 20 μL extracted sample with 1980 μL of the dilution solution. A reference solution was made using Pierce™ Reserpine Standard for LC-MS (88326, ThermoFisher Scientific). A stock reference contained 1 pg/μL of reserpine in 50% methanol and 0.1% formic acid. This stock was diluted further to create 0.1 pg/μL and 0.01 pg/μL working solution. Diluted samples were kept submerged in ultrasonic water bath for 10 min to remove the soluble gases in the solution. The degassed samples were injected into the 3200MD QTRAP MS/MS System (SCIEX) using direct infusion method (at 10 μL/min flow rate) and ion-spray ionization (IonSpray Voltage 4500) for global Q1 mass analysis.
Metabolic data were processed and analyzed using the tools at the online platform MetaboAnalyst 4.0 [40]. All the m/z peak intensities were filtered by the corresponding values in the blank. Data from each sample were normalized by total ion current followed by log transformation and autoscaling. The global metabolomic profiles of different NAS treatments were compared using supervised and un-supervised multivariate analyses- Principal Component Analysis (PCA) and Partial Least Squares Discriminant Analysis (PLS-DA). Metabolites with significantly different intensities (False discovery rate <0.05) between paired groups (untreated vs aspartame treated, untreated vs acesulfame potassium treated, and untreated vs sucralose treated) were used to identify the associated pathways in Escherichia coli K-12 MG1655 based on the KEGG pathway database using Mummichog program via Metaboanalyst.
2.4. Statistical analysis
Raw data were processed using Microsoft Excel. Bacterial growth curves and gene expression bar graphs were generated and statistically analyzed using GraphPad Prism 6. Primer efficiencies were calculated from plots of serial dilution of templates. Metabolic data were analyzed using tools at MetaboAnalyst 4.0 [40].
3. Results and discussion
3.1. In vitro analysis of the effects of acesulfame potassium, aspartame and sucralose on E. coli growth
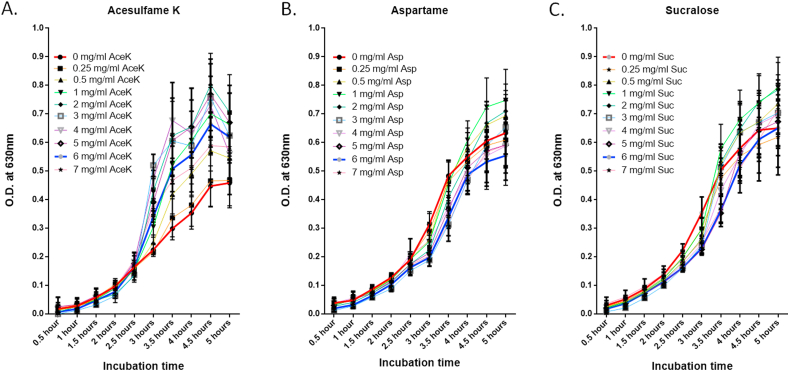
To assess the effects of non-cloric artificial sweeteners on E. coli growth, we incubated the bacteria in the presence of either aspartame, acesulfame potassium or sucralose for 5 h. E. coli showed different growth patterns in the presence of different sweeteners (Fig. 1). For the first 2.5 h, none of the sweeteners seemed to affect E. coli growth much. After 2.5 h, the presence of aspartame and sucralose inhibited E. coli growth compared to the control. But after 3.5 h, there appeared to be a decrease in the inhibitory effect of sucralose and after 5 h no difference could be observed between growth in the sucralose treated and the control groups. Aspartame continued to inhibit bacterial growth for rest of the incubation period. On the other hand, acesulfame potassium continued to increase E. coli growth after 2.5 h of incubation period. But growth seemed to slow down after 4.5 h of incubation. As a whole, E. coli growth was largely inhibited by aspartame and induced by acesulfame potassium, while the effect of sucralose on growth was less prominent.
Fig. 1.
Growth of E. coli in LB media supplemented with different NAS. (A) Acesulfame K, (B) Aspartame and (C) Sucralose.
An earlier study also reported less inhibitory effect of sucralose on E. coli compared to acesulfame potassium and aspartame [41]. Splenda, which is a combination of sucralose and maltodextrin, was reported to promote E. coli growth in SAMP mice [42]. However, a recent study showed strong inhibitory effects of both sucralose and acesulfame potassium at concentrations of 12.5 and 25 mg/mL [43]. We did not use such high concentrations in our study and differences in artificial sweetener dose can generate different results about their effects on gut microbiome [44]. Wang et al. [43] reported a drastic reduction in OD at 600 nm when the concentration of sucralose was doubled from 12.5 mg/mL to 25 mg/mL. It clearly indicated increased inhibition of growth with increasing sucralose concentration. The study also reported 98% decrease in E. coli K-12 growth at 25 mg/mL of acesulfame potassium after 5 h of incubation. In our study, we observed reduced growth of E. coli K-12 after 5 h of incubation in 7 mg/mL acesulfame potassium compared to 6 mg/mL of acesulfame potassium (Fig. 1A). So, it is possible that inhibitory effects of acesulfame potassium on E. coli K-12 becomes prominent at high concentrations. Hence, in case of both sucralose and acesulfame potassium, the strong inhibitory effects on E. coli growth could be due to their high concentrations.
Changes in the abundance of gut E. coli are associated with multiple diseases. Its increased abundance is associated with T2DM [24], Hashimoto’s thyroditis [45], cystic fibrosis [46], inflammatory bowel diseases [47], Crohn’s Disease [34], atheroscleorsis [48], asthma [33], etc. Decrease in its abundance is associated with Helicobacter pylori infection [49]. Altered abundance of E. coli also plays an important role in the development of chronic kidney disease, although the exact pattern of alteration is not yet clear [50,51]. So, enrichment of E. coli in the gut may be associated with more diseases than its reduced abundance.
In the present study, acesulfame potassium was the only sweetener that strongly stimulated E. coli growth during the incubation period. Long-term consumption of acesulfame potassium may accelerate atherosclerosis and senescence [52]. It may play a strong role in intestinal glucose uptake [52]. Acesulfame potassium consumption has been associated with glucose intolerance and adipose tissue dysfunction in pregnant mice [53]. The effects of acesulfame potassium on the abundance of E. coli in the gut may explain some of its health effects. However, under in vivo conditions, cecal and colonic bacteria are unlikely to be exposed to high concentrations of acesulfame potassium due to its fast absorption and urinary excretion [54]. This must be taken into account while assessing the roles of acesulfame potassium in diseases in this context.
In our study, sucralose did not strongly affect E. coli growth. This is also in agreement with the previous findings on the effects of sucralose on human health. Its consumption does not affect glucose homeostasis and short-term glucose control in diabetic patients [55]. It may not affect glucose homeostasis and short-term hunger signaling in non-diabetic individuals either [56].
In this study, aspartame inhibited E. coli growth. Aspartame does not affect satiety, energy intake, postprandial glucose and insulin mostly in healthy individuals [57,58]. It is well-tolerated by non-insulin-dependent diabetic patients [59]. But it may be associated with greater obesity-related glucose intolerance [60]. Obese individuals suffer from gut dysbiosis [61]. So, possible aspartame-mediated inhibition of E. coli growth may have complex effects on an already imbalanced gut microbiota.
3.2. Acesulfame potassium, aspartame and sucralose modulate expression of key metabolic genes in E. coli
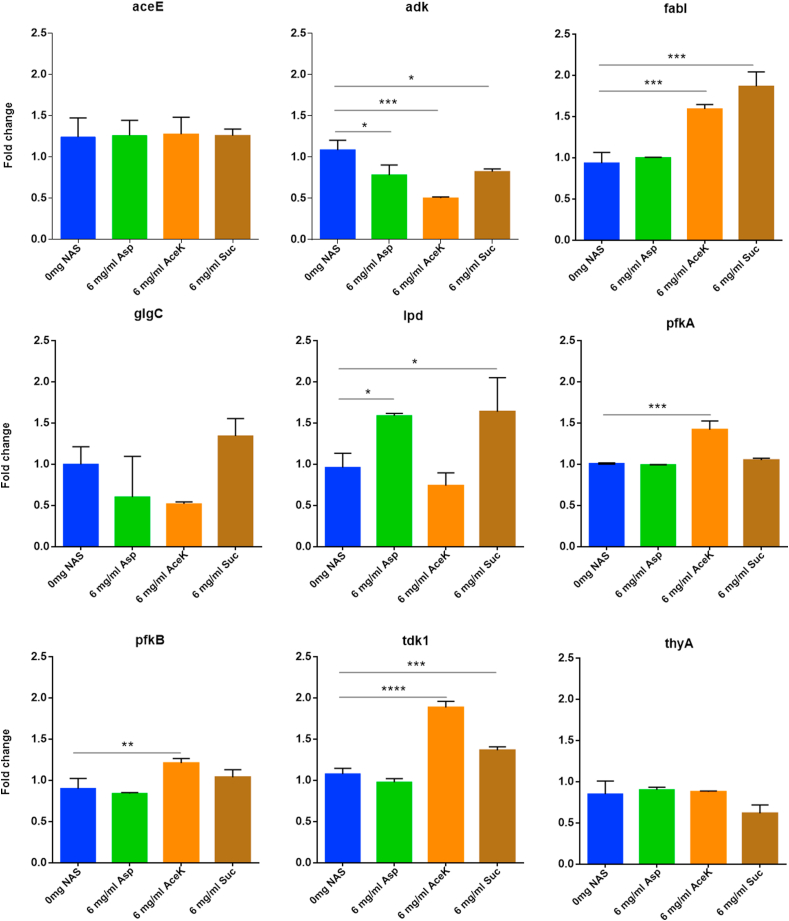
We assessed the relative expression of nine genes encoding the key enzymes that regulate important metabolic pathways [37]. Among these the relative expressions of adk, fabI, lpd, pfkA, pfkB and tdk were significantly altered (P < 0.05) by at least one of the artificial sweeteners used in this study (Fig. 2 and Table 1). Acesulfame potassium had the most prominent effects on E. coli gene expressions. Expressions of five (adk, fabI, pfkA, pfkB and tdk) out of nine regulatory enzymes were significantly altered (P < 0.05) by acesulfame potassium. Adenylate kinase (adk), thymidine kinase (tdk) and thymidylate synthase (thyA) are involved in nucleotide biosynthesis. Significant reduction (P < 0.001) in adk expression and significant increase (P < 0.0001) in tdk expression in the presence of acesulfame potassium may indicate reduced involvement of de novo and increased involvement of the salvage pathway of nucleotide biosynthesis. ThyA expression did not significantly differ with respect to the control. 6-phosphofructokinase -1 (pfkA) and −2 (pfkB), lipoamide dehydrogenase (lpd), E1component of pyruvate dehydrogenase (aceE) and glucose-1-phosphate adenylyltrasnferase (glgC) are involved in carbohydrate metabolism and FabI protein (encoded by fabI), an enoyl-ACP reductase, plays a determinant role in the completion of fatty acid biosynthesis [62]. Both Pfk-1 and -2 can catalyze the phosphorylation of fructose-6-phosphate, but these do not share sequence similarity [63]. Besides, Pfk-1 catalyzes >90% of this reaction, while Pfk-2 is responsible for <5% of total phosphofructokinase activity [64]. Although the expression of both enzymes were increased by acesulfame potassium, expression of pfkA was increased at a higher level (P < 0.001) than that of pfkB (P < 0.01) compared to the control. So, there was an increased production of fructose-1,6-bisphosphate, which is indicative of increased breakdown of glucose to pyruvate through the glycolytic pathway. Glycolysis and Entner-Doudoroff (ED) pathway are necessary for intestinal colonization by E. coli [65]. Expression of glgC was also slightly reduced by acesulfame potassium. This enzyme is responsible for glycogen accumulation, inversely related to growth rate and most active during stationary phase [66]. Glycogen synthesis and accumulation may be reduced by acesulfame potassium as it affects the glucose transport system of bacteria [67].
Fig. 2.
Relative gene expression in E. coli following treatment with 6 mg/ml of aspartame, acesulfame K and sucralose. Expression levels were compared using Dunnett’s multiple comparison test (∗, ∗∗, ∗∗∗ and ∗∗∗∗ denote p < 0.05, p < 0.01, p < 0.001 and p < 0.0001, respectively).
Table 1.
Genes with significant difference in relative expression level in the non-caloric artificial sweetener treated cells compared to the untreated cells and associated pathways.
| Gene with significant difference in relative expression | |||||
|---|---|---|---|---|---|
| Affected pathways | |||||
| Aspartame |
Acesulfame potassium |
Sucralose |
|||
| Genea | Pathways | Genea | Pathways | Genea | Pathways |
| adk | Purine metabolism (eco00230) | adk | Purine metabolism (eco00230) | Adk | Purine metabolism (eco00230) |
| Thiamine metabolism (eco00730) | Thiamine metabolism (eco00730) | Thiamine metabolism (eco00730) | |||
| Nicotinate and nicotinamide metabolism (eco00760) | Nicotinate and nicotinamide metabolism (eco00760) | Nicotinate and nicotinamide metabolism (eco00760) | |||
| _ | _ | fabI | Fatty acid biosynthesis (eco00061) | fabI | Fatty acid biosynthesis (eco00061) |
| _ | Biotin metabolism (eco00780) | Biotin metabolism (eco00780) | |||
| Lpd | Valine, leucine and isoleucine degradation (eco00280) | _ | _ | Lpd | Valine, leucine and isoleucine degradation (eco00280) |
| Citrate cycle (TCA cycle) (eco00020) | _ | Citrate cycle (TCA cycle) (eco00020) | |||
| Glycolysis/Gluconeogenesis (eco00010) | _ | Glycolysis/Gluconeogenesis (eco00010) | |||
| Glycine, serine and threonine metabolism (eco00260) | _ | Glycine, serine and threonine metabolism (eco00260) | |||
| Glycerophospholipid metabolism (eco00564) | _ | Glycerophospholipid metabolism (eco00564) | |||
| Pyruvate metabolism (eco00620) | _ | Pyruvate metabolism (eco00620) | |||
| Glyoxylate and dicarboxylate metabolism (eco00630) | _ | Glyoxylate and dicarboxylate metabolism (eco00630) | |||
| Propanoate metabolism (eco00640) | _ | Propanoate metabolism (eco00640) | |||
| _ | _ | pfkA | Glycolysis/Gluconeogenesis (eco00010) | _ | _ |
| _ | Biosynthesis of amino acids (eco01230) | _ | |||
| _ | Pentose phosphate pathway (eco00030) | _ | |||
| _ | Fructose and mannose metabolism (eco00051) | _ | |||
| _ | Galactose metabolism (eco00052) | _ | |||
| _ | Methane metabolism (eco00680) | _ | |||
| _ | RNA degradation (eco03018) | _ | |||
| _ | _ | pfkB | Glycolysis/Gluconeogenesis (eco00010) | _ | _ |
| _ | Biosynthesis of amino acids (eco01230) | _ | |||
| _ | Pentose phosphate pathway (eco00030) | _ | |||
| _ | Fructose and mannose metabolism (eco00051) | _ | |||
| _ | Galactose metabolism (eco00052) | _ | |||
| _ | Methane metabolism (eco00680) | _ | |||
| _ | RNA degradation (eco03018) | _ | |||
| _ | tdk1 | Pyrimidine metabolism (eco00240) | tdk1 | Pyrimidine metabolism (eco00240) | |
Gene with significant difference in relative expression.
Aspartame and sucralose significantly reduced (P < 0.05) adk expression. But their effects on its expression were less prominent than acesulfame potassium. Tdk expression was significantly increased (P < 0.001) by sucralose, but not significantly altered by aspartame. ThyA expression was not significantly altered by either of the sweeteners. These expression patterns are consistent with the observation that E. coli growth was continuously increased and decreased by sucralose and aspartame, respectively, after 3.5 h compared to the control. Expression patterns also indicate that the salvage pathway of nucleotide biosynthesis may play a greater role than the de novo pathway in E. coli growth in the presence of sucralose. Although aceE expression was not significantly altered by either aspartame or sucralose, lpd expression was significantly increased (P < 0.05) by both of these. Besides the pdh promoter (which regulates expression of pyruvate dehydrogenase components), lpd expression is co-regulated with sdh promoter (which regulates the expression of 2-oxoglutarate dehydrogenase components) [68]. ArcA mediates this co-regulation [68], but does not strongly control the pdh promoter [68]. This may impact lysine metabolism [69]. Expression of the other genes associated with carbohydrate metabolism was not altered significantly by aspartame and sucralose. It suggests that these two artificial sweeteners may have less impact on E. coli carbohydrate metabolism than acesulfame potassium.
fabI expression was significantly increased (P < 0.001) by acesulfame potassium and sucralose, but was unaltered by aspartame. Its expression is associated with growth rate of bacteria [70]. Although not statistically significant, its apparently higher expression in presence of sucralose than acesulfame potassium may be explained by steeper growth at 5 h compared to the growth at 4.5 h. Over-expression of fabI in E. coli may cause production of butyrate [71]. Butyrate may play protective roles against conditions like obesity, cancer, diabetes, inflammatory bowel diseases, etc., cause epigenetic changes in the host and may also modulate behavior of the host [61,72,73].
3.3. Acesulfame potassium, aspartame and sucralose modulate metabolic pathways in E. coli
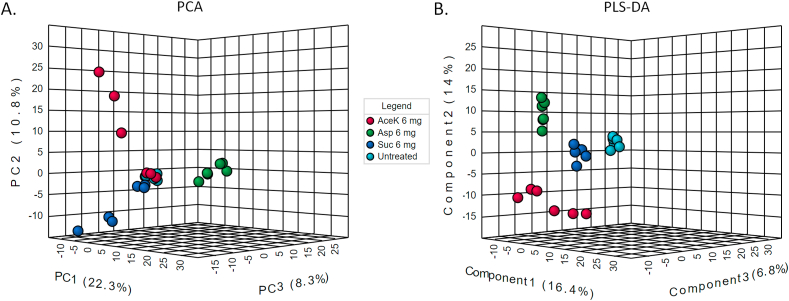
Global metabolic profiles (of Q1 mass) of 6 mg/ml aspartame, acesulfame potassium and sucralose treated E. coli cells were generated and compared using both supervised and unsupervised multivariate methods (Fig. 3). The cells in PLS-DA plot appear to form more distinct clusters as these exploit the information added as class identifier in the input file. In both PCA and PLS-DA, sucralose treated cells appeared to cluster closer to the untreated cells. Sucralose is highly stable and resistant to hydrolysis and unlikely to cause metabolic adaptations in gut microbiota [54]. Acesulfame potassium treated cells clustered farthest away from the control, which may be consistent with the finding that it significantly upregulated or downregulated (P < 0.05) five of the nine rate limiting regulatory enzymes of E. coli assessed in this study (Fig. 2). The aspartame treated cell cluster was closer to the sucralose treated ones. It is in agreement with the observation that alterations in gene expressions by aspartame was more similar to those casued by sucralose (Fig. 2).
Fig. 3.
Multivariate analysis with metabolomic data. A. Principal component analysis. B. Partial least squares discriminant analysis.
Several pathways of E. coli (fatty acid metabolism, pentose phosphate pathway, propanoate metabolism, phosphonate and phosphinate metabolism and biosynthesis of several amino acids including lysine and aromatic amino acids) were affected by all three sweeteners (Table 2). Acesulfame potassium and sucralose significantly increased (P < 0.001) fabI expression (Fig. 2). So, fatty acid metabolism are supposed to be affected by these [74]. Although aspartame did not significantly alter fabI expression, it significantly increased (P < 0.05) lpd expression. So, alterations in TCA cycle, which is linked to fatty acid metabolism, may be observed. Since fatty acids are essential in producing phospholipids for cell membrane, alterations in its metabolism can have effects on bacterial growth rate. Pentose phosphate pathway is, however, not essential for intestinal colonization [65]. Aspartame, acesulfame potassium and sucralose affected metabolism of propanoate, which is one of the most abundant short-chain fatty acids (SCFA) produced by gut micrbiota [75]. SCFAs (<8 carbon atoms) produced by gut microbiome through dietary carbohydrate metabolism immensely influence host’s intestinal integrity, energy homeostasis, glucose and lipid metabolism, insulin sensitivity, appetite and immune responses [[76], [77], [78]]. It was previously shown that artificial sweeteners may alter fibre fermentation capacity of gut microbiome [79]. Level of serum lipids may depend on acetate to propanoate ratio [78]. Propanoate protects host from pathogens probably by altering the intracellular pH of pathogens [80]. It also plays an important role in blood pressure regulation [81]. So, alterations in its metabolism may have substantial impact on an individual’s health. Biosyntheses of lysine and aromatic amino acids appeared to be affected by all artificial sweeteners. Lysine biosynthesis is closely related to energy metabolism [82]. Changes in aromatic amino acid metabolism of gut microbiome are associated with diseases of the liver, renal system, cardiovascular system, central nervous system, and inflammatory bowel diseases [83].
Table 2.
The pathways affected by aspartame, acesulfame K and sucralose.
| Affected pathways | ||
|---|---|---|
| Aspartame | Acesulfame potassium | Sucralose |
| Biotin metabolism | Chlorocyclohexane and chlorobenzene degradation | Arginine and proline metabolism |
| Fatty acid biosynthesis | Cysteine and methionine metabolism | Biotin metabolism |
| Fatty acid degradation | Fatty acid biosynthesis | Chlorocyclohexane and chlorobenzene degradation |
| Glyoxylate and dicarboxylate metabolism | Fatty acid degradation | Fatty acid degradation |
| Lysine biosynthesis | Lysine biosynthesis | Lysine biosynthesis |
| Pentose phosphate pathway | Pentose phosphate pathway | Pentose phosphate pathway |
| Phenylalanine, tyrosine and tryptophan biosynthesis | Phenylalanine, tyrosine and tryptophan biosynthesis | Phenylalanine, tyrosine and tryptophan biosynthesis |
| Phosphonate and phosphinate metabolism | Phosphonate and phosphinate metabolism | Phosphonate and phosphinate metabolism |
| Porphyrin and chlorophyll metabolism | Propanoate metabolism | Propanoate metabolism |
| Propanoate metabolism | Riboflavin metabolism | Riboflavin metabolism |
| Starch and sucrose metabolism | Starch and sucrose metabolism | |
| Toluene degradation | ||
Biotin metabolism was affected by aspartame and sucralose (Table 2). Its synthesis depends on fatty acid biosynthesis pathways [84]. Since fatty acid biosynthesis did not appear to be affected by sucralose, increased expression of fabI in its presence could actually increase biotin synthesis by E. coli [85]. Gut microbiome is an important source of biotin [86]. Alterations in its metabolism are associated with multiple human conditions including obesity and diabetes [87]. In fact, the lack of biotin-associated microbiome may be one of the most robust prostate cancer-associated indicators [88]. Acesulfame potassium and sucralose affected riboflavin metabolism. It is related to purine metabolism and pentose phosphate pathway, since it requires GTP and ribulose-5-phosphate [89]. Changes in riboflavin metabolism of gut microbiota may modulate proinflammatory responses in host [89].
Some pathways were affected by only one of the sweeteners. Aspartame affected porphyrin, glyoxylate and dicarboxylate metabolism. Glyoxylate bypass is essential for bacterial growth on acetate or fatty acids as carbon sources [90]. Changes in porphyrin metabolism of gut microbiota may be associated with allergic rhinitis [91]. Acesulfame potassium affected cysteine and methionine metabolism. Sulfur-containing metabolites produced through gut microbial metabolism of cysteine and methionine are associated with both male and female reproductive fitness [91]. Possible role of acesulfame potassium in the development of maternal metabolic dysfunction has been shown [53]. More studies are needed to fully elucidate the effects of this NAS on the reproductive health of parents and their offspring. In the present study, sucralose affected arginine and proline metabolism. Downregulation of the metabolism of these amino acids by gut microbiota was shown to be associated with T2DM [92].
Xenobiotic metabolism by E. coli may also be altered by non-caloric artificial sweeteners (for example, chlorocyclohexane and chlorobenezene degradation by acesulfame potassium and sucralose, toluene degradation by acesulfame potassium). Microbiome plays a crucial role in xenobiotics metabolism of the host [93]. There is currently an incomplete understanding of this process [94]. More studies are needed to explore the effects of non-claoric artificial sweeteners on the ability of gut microbiome to degrade xenobiotic compounds. Finally, non-claoric artificial sweeteners also seemed to affect the ability of E. coli to synthesize natural products (phosphonate and phosphinate metabolism). Besides, fatty acid metabolism and polyketide biosynthesis pathways are interrelated [95]. So, effects of non-claoric artificial sweeteners on the synthesis of natural products by microbiome should be further studied.
4. Conclusion
The gut commensals are closely associated with human health and well-being. Composition as well as metabolism of these diverse microorganisms are influenced by the dietary intakes. The non-caloric artificial sweeteners like aspartame, acesulfame potassium and sucralose have gained much global popularity, especially among the diabetic and obese individuals, due to their often over-pronounced health benefits. Studies in the recent years have, however, reported alteration of gut microbiota by these artificial sweeteners. Here, we investigated the effect of aspartame, acesulfame potassium and sucralose on the growth and metabolism of omnipresent gut habitant Escherichia coli by analyzing the relative expression levels of the key genes that encode the rate limiting enzymes of important metabolic pathways as well as the global metabolite profiles. Among these three popular non-caloric artificial sweeteners, sucralose seems to have the least deviating impact on E.coli growth and metabolism under in vitro condition. On the contrary, acesulfame potassium treated E. coli showed the largest deviation in the gene expression and metabolite profiles compared to the untreated cells. The data presented in this study may help understanding the influence of aspartame, acesulfame potassium and sucralose on the metabolism of gut microbes. Further studies are needed to assess the effects of these non-caloric artificial sweeteners on the abundance and growth of E. coli along with other gut bacteria under in vivo condition. In the context of the data presented in this study and previous findings regarding the effects of artificial sweeteners [41,53,55,96], sucralose appears to have less pronounced effects on gut microbiome and human health and thus may be safer for consumption.
Statement of ethics
This study did not involve any human or animal, and hence no ethical approval was required.
Funding sources
Special allocation in science and technology from the Ministry of Science and Technology, Bangladesh.
CRediT authorship contribution statement
Shayan Shahriar: Formal analysis, Data curation, Acquisition, analysis, and interpretation of data. Tamim Ahsan: Formal analysis, Data curation, Writing - original draft, Writing - review & editing, Manuscript preparation and reviewing, Acquisition, analysis, and interpretation of data. Abira Khan: Formal analysis, Data curation, Acquisition, analysis, and interpretation of data. Sharif Akhteruzzaman: Writing - original draft, Writing - review & editing, Manuscript preparation and reviewing. Saadlee Shehreen: Writing - original draft, Writing - review & editing, Design of the work, Manuscript preparation and reviewing. Abu Ashfaqur Sajib: Writing - original draft, Writing - review & editing, Design of the work, Manuscript preparation and reviewing.
Declaration of competing interest
The authors have no conflict of interest to declare.
Acknowledgement
This study was supported by a grant from the Ministry of Science and Technology, Bangladesh. The authors are thankful for the support.
Contributor Information
Saadlee Shehreen, Email: saadleeshehreen@gmail.com.
Abu Ashfaqur Sajib, Email: abu.sajib@du.ac.bd.
References
- 1.Lohner S., Toews I., Meerpohl J.J. Health outcomes of non-nutritive sweeteners: analysis of the research landscape. Nutr J. 2017;16:55. doi: 10.1186/s12937-017-0278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siervo M., Montagnese C., Mathers J.C., Soroka K.R., Stephan B.C., Wells J.C. Sugar consumption and global prevalence of obesity and hypertension: an ecological analysis. Publ Health Nutr. 2014;17:587–596. doi: 10.1017/S1368980013000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanhope K.L. Sugar consumption, metabolic disease and obesity: the state of the controversy. Crit Rev Clin Lab Sci. 2016;53:52–67. doi: 10.3109/10408363.2015.1084990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Ruyter J.C., Olthof M.R., Seidell J.C., Katan M.B. A trial of sugar-free or sugar-sweetened beverages and body weight in children. N Engl J Med. 2012;367:1397–1406. doi: 10.1056/NEJMoa1203034. [DOI] [PubMed] [Google Scholar]
- 5.Tordoff M.G., Alleva A.M. Effect of drinking soda sweetened with aspartame or high-fructose corn syrup on food intake and body weight. Am J Clin Nutr. 1990;51:963–969. doi: 10.1093/ajcn/51.6.963. [DOI] [PubMed] [Google Scholar]
- 6.Fagherazzi G., Vilier A., Saes Sartorelli D., Lajous M., Balkau B., Clavel-Chapelon F. Consumption of artificially and sugar-sweetened beverages and incident type 2 diabetes in the Etude Epidemiologique aupres des femmes de la Mutuelle Generale de l’Education Nationale-European Prospective Investigation into Cancer and Nutrition cohort. Am J Clin Nutr. 2013;97:517–523. doi: 10.3945/ajcn.112.050997. [DOI] [PubMed] [Google Scholar]
- 7.Fowler S.P., Williams K., Resendez R.G., Hunt K.J., Hazuda H.P., Stern M.P. Fueling the obesity epidemic? Artificially sweetened beverage use and long-term weight gain. Obesity. 2008;16:1894–1900. doi: 10.1038/oby.2008.284. [DOI] [PubMed] [Google Scholar]
- 8.Andersson C., Sullivan L., Benjamin E.J., Aragam J., Jacques P., Cheng S., Vasan R.S. Association of soda consumption with subclinical cardiac remodeling in the Framingham heart study. Metabolism. 2015;64:208–212. doi: 10.1016/j.metabol.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindseth G.N., Coolahan S.E., Petros T.V., Lindseth P.D. Neurobehavioral effects of aspartame consumption. Res Nurs Health. 2014;37:185–193. doi: 10.1002/nur.21595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipton R.B., Newman L.C., Cohen J.S., Solomon S. Aspartame as a dietary trigger of headache. Headache. 1989;29:90–92. doi: 10.1111/j.1526-4610.1989.hed2902090.x. [DOI] [PubMed] [Google Scholar]
- 11.Pisarik P., Kai D. Vestibulocochlear toxicity in a pair of siblings 15 years apart secondary to aspartame: two case reports. Cases journal. 2009;2:1–4. doi: 10.1186/1757-1626-0002-0000009237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashok I., Poornima P.S., Wankhar D., Ravindran R., Sheeladevi R. Oxidative stress evoked damages on rat sperm and attenuated antioxidant status on consumption of aspartame. Int J Impot Res. 2017;29:164. doi: 10.1038/ijir.2017.17. [DOI] [PubMed] [Google Scholar]
- 13.Soffritti M., Belpoggi F., Esposti D.D., Lambertini L., Tibaldi E., Rigano A. First experimental demonstration of the multipotential carcinogenic effects of aspartame administered in the feed to Sprague-Dawley rats. Environ Health Perspect. 2005;114:379–385. doi: 10.1289/ehp.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts A., Renwick A.G., Sims J., Snodin D.J. Sucralose metabolism and pharmacokinetics in man. Food Chem Toxicol. 2000;38:31–41. doi: 10.1016/s0278-6915(00)00026-0. [DOI] [PubMed] [Google Scholar]
- 15.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grice E.A., Segre J.A. The human microbiome: our second genome. Annu Rev Genom Hum Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mountzouris K.C., McCartney A.L., Gibson G.R. Intestinal microflora of human infants and current trends for its nutritional modulation. Br J Nutr. 2002;87:405–420. doi: 10.1079/BJNBJN2002563. [DOI] [PubMed] [Google Scholar]
- 19.Zoetendal E.G., Akkermans A.D.L., Vliet W.M.A., de Visser J.A.G.M., de Vos W.M. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis. 2001;13:129–134. [Google Scholar]
- 20.David L.A., Maurice M.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muegge B.D., Kuczynski J., Knights D., Clemente J.C., González A., Fontana L. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suez J., Korem T., Zeevi D., Zilberman-Schapira G., Thaiss C.A., Maza O. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 23.Bäckhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 25.Bian X., Chi L., Gao B., Tu P., Ru H., Lu K. The artificial sweetener acesulfame potassium affects the gut microbiome and body weight gain in CD-1 mice. PloS One. 2017;12 doi: 10.1371/journal.pone.0178426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suvarna K., Stevenson D., Meganathan R., Hudspeth M.E. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J Bacteriol. 1998;180:2782–2787. doi: 10.1128/jb.180.10.2782-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter T.K.S., Michalski J.M., Zanetti L., Tennant S.M., Chen W.H., Rasko D.A. Responses of the human gut Escherichia coli population to pathogen and antibiotic disturbances. mSystems. 2018;3 doi: 10.1128/mSystems.00047-18. 00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christofi T., Panayidou S., Dieronitou I., Michael C., Apidianakis Y. Metabolic output defines Escherichia coli as a health-promoting microbe against intestinal Pseudomonas aeruginosa. Sci Rep. 2019;9 doi: 10.1038/s41598-019-51058-3. Article number: 14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katouli M. Population structure of gut Escherichia coli and its role in development of extra-intestinal infections. Iran J Microbiol. 2010;2:59–72. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong H., Ren H., Lu Y., Fang C., Hou G., Yang Z. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naïve type 2 diabetics. EBioMedicine. 2019;47:373–383. doi: 10.1016/j.ebiom.2019.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okba A.M., Saber S.M., Abdel-Rehim A.S., Amin M.M., Mohamed D.A. Fecal microbiota profile in atopic asthmatic adult patients. Eur Ann Allergy Clin Immunol. 2018;50:117–124. doi: 10.23822/EurAnnACI.1764-1489.48. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Medina M., Aldeguer X., Gonzalez-Huix F., Acero D., Garcia-Gil L.J. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 35.Idalia V.M.N., Bernardo F. Escherichia coli as a model organism and its application in biotechnology, recent advances on physiology, pathogenesis and biotechnological applications. Tech Open. 2017:253–274. [Google Scholar]
- 36.Blount Z.D. The unexhausted potential of E. coli. eLife. 2015;4 doi: 10.7554/eLife.05826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahmud R., Shehreen S., Shahriar S., Rahman M.S., Akhteruzzaman S., Sajib A.A. Non-caloric artificial sweeteners modulate the expression of key metabolic genes in the omnipresent gut microbe Escherichia coli. Microbial Physiology. 2019;29:43–56. doi: 10.1159/000504511. [DOI] [PubMed] [Google Scholar]
- 38.Zhao M., Chen X., Gao G., Tao L., Wei L. RLEdb: a database of rate-limiting enzymes and their regulation in human, rat, mouse, yeast and E. coli. Cell Res. 2009;19:793. doi: 10.1038/cr.2009.61. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. e45-e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harpaz D., Yeo L.P., Cecchini F., Koon T.H.P., Kushmaro A., Tok A.I.Y. Measuring artificial sweeteners toxicity using a bioluminescent bacterial panel. Molecules. 2018;23 doi: 10.3390/molecules23102454. Article ID-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Palacios A., Harding A., Menghini P., Himmelman C., Retuerto M., Nickerson K.P. The artificial sweetener Splenda promotes gut proteobacteria, dysbiosis, and myeloperoxidase reactivity in Crohn’s disease–like ileitis. Inflamm Bowel Dis. 2018;24:1005–1020. doi: 10.1093/ibd/izy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q.P., Browman D., Herzog H., Neely G.G. Non-nutritive sweeteners possess a bacteriostatic effect and alter gut microbiota in mice. PloS One. 2018;13 doi: 10.1371/journal.pone.0199080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz-Ojeda F.J., Plaza-Diaz J., Saez-Lara M.J., Gil A. Effects of sweeteners on the gut microbiota: a review of experimental studies and clinical trials. Adv Nutr. 2019;10:S31–S48. doi: 10.1093/advances/nmy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishaq H.M., Mhammad I.S., Guo H., Shahzad M., Hou Y.J., Ma C. Molecular estimation of alteration in intestinal microbial composition in Hashimoto’s thyroiditis patients. Biomed Pharmacother. 2017;95:865–874. doi: 10.1016/j.biopha.2017.08.101. [DOI] [PubMed] [Google Scholar]
- 46.Miragoli F., Federici S., Ferrari S., Minuti A., Rebecchi A., Bruzzese E. Impact of cystic fibrosis disease on archaea and bacteria composition of gut microbiota. FEMS Microbiol Ecol. 2017;93 doi: 10.1093/femsec/fiw230. Article ID-fiw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knoll R.L., Forslund K., Kultima J.R., Meyer C.U., Kullmer U., Sunagawa S. Gut microbiota differs between children with Inflammatory Bowel Disease and healthy siblings in taxonomic and functional composition: a metagenomic analysis. Am J Physiol Gastrointest Liver Physiol. 2017;312:G327–G339. doi: 10.1152/ajpgi.00293.2016. [DOI] [PubMed] [Google Scholar]
- 48.Jie Z., Xia H., Zhong S.-L., Feng Q., Li S., Liang S. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00900-1. Article ID- 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang W., Jeoung N.H., Cho K.-H. Modified apolipoprotein (apo) A-I by artificial sweetener causes severe premature cellular senescence and atherosclerosis with impairment of functional and structural properties of apoA-I in lipid-free and lipid-bound state. Mol Cell. 2011;31:461–470. doi: 10.1007/s10059-011-1009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gryp T., Huys G.R.B., Joossens M., Biesen W.V., Glorieux G., Vaneechoutte M. Isolation and quantification of uremic toxin precursor-generating gut bacteria in chronic kidney disease patients. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21061986. Article ID-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang S., Xie S., Lv D., Wang P., He H., Zhang T., Zhou Y., Lin Q., Zhou Y., Jiang J., Nie J., Hou F., Chen Y. Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci Rep. 2017;7 doi: 10.1038/s41598-017-02989-2. Article ID-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Y., Sarr M.G. Effect of the artificial sweetener, acesulfame potassium, a sweet taste receptor agonist, on glucose uptake in small intestinal cell lines. J Gastrointest Surg. 2013;17:153–158. doi: 10.1007/s11605-012-1998-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plows J.F., Morton-Jones J., Bridge-Comer P.E., Ponnampalam A., Stanley J.L., Vickers M.H., Reynolds C.M. Consumption of the artificial sweetener acesulfame potassium throughout pregnancy induces glucose intolerance and adipose tissue dysfunction in mice. J Nutr. 2020;150:1773–1781. doi: 10.1093/jn/nxaa106. [DOI] [PubMed] [Google Scholar]
- 54.Magnuson B.A., Carakostas M.C., Moore N.H., Poulos S.P., Renwick A.G. Biological fate of low-calorie sweeteners. Nutr Rev. 2016;74:670–689. doi: 10.1093/nutrit/nuw032. [DOI] [PubMed] [Google Scholar]
- 55.Grotz V.L., Henry R.R., McGill J.B., Prince M.J., Shamoon H., Trout J.R., Pi-Sunyer F.X. Lack of effect of sucralose on glucose homeostasis in subjects with type 2 diabetes. J Am Diet Assoc. 2003;103:1607–1612. doi: 10.1016/j.jada.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 56.Brown A.W., Brown M.M.B., Onken K.L., Beitz D.C. Short-term consumption of sucralose, a nonnutritive sweetener, is similar to water with regard to select markers of hunger signaling and short-term glucose homeostasis in women. Nutr Res. 2011;31:882–888. doi: 10.1016/j.nutres.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Tey S.L., Salleh N.B., Henry J., Forde C.G. Effects of aspartame-, monk fruit-, stevia- and sucrose-sweetened beverages on postprandial glucose, insulin and energy intake. Int J Obes. 2017;41:450–457. doi: 10.1038/ijo.2016.225. [DOI] [PubMed] [Google Scholar]
- 58.Anton S.D., Martin C.K., Han H., Coulon S., Cefalu W.T., Geiselman P., Williamson D.A. Effects of stevia, aspartame, and sucrose on food intake, satiety, and postprandial glucose and insulin levels. Appetite. 2010;55:37–43. doi: 10.1016/j.appet.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stern S.B., Bleicher Sj, Flores A., Gombos G., Recitas D., Shu J. Administration of aspartame in non-insulin-dependent diabetics. J Toxicol Environ Health. 1976;2:429–439. doi: 10.1080/15287397609529444. [DOI] [PubMed] [Google Scholar]
- 60.Kuk J.L., Brown R.E. Aspartame intake is associated with greater glucose intolerance in individuals with obesity. Appl Physiol Nutr Metabol. 2016;41:795–798. doi: 10.1139/apnm-2015-0675. [DOI] [PubMed] [Google Scholar]
- 61.Singer-Englar T., Barlow G., Mathur R. Obesity, diabetes, and the gut microbiome: an updated review. Expet Rev Gastroenterol Hepatol. 2019;13:3–15. doi: 10.1080/17474124.2019.1543023. [DOI] [PubMed] [Google Scholar]
- 62.Bergler H., Fuchsbichler S., Högenauer G., Turnowsky F. The enoyl-[acyl-carrier-protein] reductase (FabI) of Escherichia coli, which catalyzes a key regulatory step in fatty acid biosynthesis, accepts NADH and NADPH as cofactors and is inhibited by palmitoyl-CoA. Eur J Biochem. 1996;242:689–694. doi: 10.1111/j.1432-1033.1996.0689r.x. [DOI] [PubMed] [Google Scholar]
- 63.Hellinga H.W., Evans P.R. Nucleotide sequence and high-level expression of the major Escherichia coli phosphofructokinase. Eur J Biochem. 1985;149:363–373. doi: 10.1111/j.1432-1033.1985.tb08934.x. [DOI] [PubMed] [Google Scholar]
- 64.Kotlarz D., Garreau H., Buc H. Regulation of the amount and of the activity of phosphofructokinases and pyruvate kinases in Escherichia coli. Biochim Biophys Acta. 1975;381:257–268. doi: 10.1016/0304-4165(75)90232-9. [DOI] [PubMed] [Google Scholar]
- 65.Chang D.E., Smalley D.J., Tucker D.L., Leatham M.P., Norris W.E., Stevenson S.J. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Romeo T., Preiss J. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vitro effects of cyclic AMP and guanosine 5’-diphosphate 3’-diphosphate and analysis of in vivo transcripts. J Bacteriol. 1989;171:2773–2782. doi: 10.1128/jb.171.5.2773-2782.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfeffer M., Ziesenitz S.C., Siebert G., Acesulfame K. Cyclamate and saccharin inhibit the anaerobic fermentation of glucose by intestinal bacteria. Zeitschrift fur Ernahrungswissenschaft. 1985;24:231–235. doi: 10.1007/BF02023668. [DOI] [PubMed] [Google Scholar]
- 68.Cunningham L., Georgellis D., Green J., Guest J.R. Co-regulation of lipoamide dehydrogenase and 2-oxoglutarate dehydrogenase synthesis in Escherichia coli: characterisation of an ArcA binding site in the lpd promoter. FEMS Microbiol Lett. 1998;169:403–408. doi: 10.1111/j.1574-6968.1998.tb13347.x. [DOI] [PubMed] [Google Scholar]
- 69.Keseler I.M., Mackie A., Santos-Zavaleta A., Billington R., Bonavides-Martinez C., Caspi R. the EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017;45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goh S., Boberek J.M., Nakashima N., Stach J., Good K. Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PloS One. 2009;4 doi: 10.1371/journal.pone.0006061. Article ID-e6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vick J.E., Clomburg J.M., Blankschien M.D., Chou A., Kim S., Gonzalez R. Escherichia coli enoyl-acyl carrier protein reductase (FabI) supports efficient operation of a functional reversal of β-oxidation cycle. Appl Environ Microbiol. 2015;81:1406–1416. doi: 10.1128/AEM.03521-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stilling R.M., van de Wouw M., Clarke G., Stanton C., Dinan T.G., Cryan J.F. The neuropharmacology of butyrate: the bread and butter of the microbiota-gut-brain axis? Neurochem Int. 2016;99:110–132. doi: 10.1016/j.neuint.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 73.Felizardo R.J.F., de Almeida D.C., Pereira R.L., Watanabe I.K.M., Doimo N.T.S., Ribeiro W.R. Gut microbial metabolite butyrate protects against proteinuric kidney disease through epigenetic- and GPR109a-mediated mechanisms. Faseb J. 2019;33:11894–11908. doi: 10.1096/fj.201901080R. [DOI] [PubMed] [Google Scholar]
- 74.Heath R.J., Rock C.O. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J Biol Chem. 1995;270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- 75.Morrison D.J., Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microb. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun Y., O’Riordan M.X.D. Regulation of bacterial pathogenesis by intestinal short-chain fatty acids. Adv Appl Microbiol. 2013;85:93–118. doi: 10.1016/B978-0-12-407672-3.00003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Canfora E.E., Jocken J.W., Blaak E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11:577–591. doi: 10.1038/nrendo.2015.128. [DOI] [PubMed] [Google Scholar]
- 78.Wong J.M.W., de Souza R., Kendall C.W.C., Emam A., Jenkins D.J.A. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 79.Gerasimidis K., Bryden K., Chen X., Papachristou E., Verney A., Roig M. The impact of food additives, artificial sweeteners and domestic hygiene products on the human gut microbiome and its fibre fermentation capacity. Eur J Nutr. 2020;59:3213–3230. doi: 10.1007/s00394-019-02161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacobson A., Lam L., Rajendram M., Tamburini F., Honeycutt J., Pham T. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe. 2018;24:296–307. doi: 10.1016/j.chom.2018.07.002. e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microb. 2014;5:202–207. doi: 10.4161/gmic.27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y., Hou Y., Wang G., Zheng X., Hao H. Gut microbial metabolites of aromatic amino acids as signals in host-microbe interplay. Trends Endocrinol Metabol. 2020;S1043–2760:30054. doi: 10.1016/j.tem.2020.02.012. 30050. [DOI] [PubMed] [Google Scholar]
- 84.Cronan J.E., Lin S. Synthesis of the α,ω-dicarboxylic acid precursor of biotin by the canonical fatty acid biosynthetic pathway. Curr Opin Chem Biol. 2011;15:407–413. doi: 10.1016/j.cbpa.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin S., Hanson R.E., Cronan J.E. Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat Chem Biol. 2010;6:682–688. doi: 10.1038/nchembio.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spector R., Mock D.M. Biotin transport and metabolism in the central nervous system. Neurochem Res. 1988;13:213–219. doi: 10.1007/BF00971535. [DOI] [PubMed] [Google Scholar]
- 87.Sharma M., Li Y., Stoll M.L., Tollefsbol T.O. The epigenetic connection between the gut microbiome in obesity and diabetes. Front Genet. 2020;10 doi: 10.3389/fgene.2019.01329. Article ID-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liss M.A., White J.R., Goros M., Gelfond J., Leach R., Johnson-Pais T. Metabolic biosynthesis pathways identified from fecal microbiome associated with prostate cancer. Eur Urol. 2018;74:575–582. doi: 10.1016/j.eururo.2018.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Merletti R., Lo Conte L.R., Cisari C., Actis M.V. Age related changes in surface myoelectric signals. Scand J Rehabil Med. 1992;24:25–36. [PubMed] [Google Scholar]
- 90.Cronan J.E., Jr., Laporte D. Tricarboxylic acid cycle and glyoxylate bypass. EcoSal Plus. 2005;1 doi: 10.1128/ecosalplus.3.5.2. [DOI] [PubMed] [Google Scholar]
- 91.Dai Z., Wu Z., Hang S., Zhu W., Wu G. Amino acid metabolism in intestinal bacteria and its potential implications for mammalian reproduction. Mol Hum Reprod. 2015;21:389–409. doi: 10.1093/molehr/gav003. [DOI] [PubMed] [Google Scholar]
- 92.Okazaki F., Zang L., Nakayama H., Chen Z., Gao Z.-J., Chiba H. Microbiome alteration in type 2 diabetes mellitus model of zebrafish. Sci Rep. 2019;9 doi: 10.1038/s41598-018-37242-x. Article ID-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jandhyala S.M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Reddy D.N. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clarke G., Sandhu K.V., Griffin B.T., Dinan T.G., Cryan J.F., Hyland N.P. Gut reactions: breaking down xenobiotic-microbiome interactions. Pharmacol Rev. 2019;71:198–224. doi: 10.1124/pr.118.015768. [DOI] [PubMed] [Google Scholar]
- 95.Cronan J.E., Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mezitis N.H., Maggio C.A., Koch P., Quddoos A., Allison D.B., Pi-Sunyer F.X. Glycemic effect of a single high oral dose of the novel sweetener sucralose in patients with diabetes. Diabetes Care. 1996;19:1004–1005. doi: 10.2337/diacare.19.9.1004. [DOI] [PubMed] [Google Scholar]