Abstract
The promotive effect of auxin on shoot cell expansion provided the bioassay used to isolate this central plant hormone nearly a century ago. While the mechanisms underlying auxin perception and signaling to regulate transcription have largely been elucidated, how auxin controls cell expansion is only now attaining molecular-level definition. The good news is that the decades-old acid growth theory invoking plasma membrane H+-ATPase activation is still useful. The better news is that a mechanistic framework has emerged, wherein Small Auxin Up RNA (SAUR) proteins regulate protein phosphatases to control H+-ATPase activity. In this review, we focus on rapid auxin effects, their relationship to H+-ATPase activation and other transporters, and dependence on TIR1/AFB signaling. We also discuss how some observations, such as near-instantaneous effects on ion transport and root growth, do not fit into a single, comprehensive explanation of how auxin controls cell expansion, and where more research is warranted.
Keywords: auxin, SAUR-PP2C.D, acid growth, cell expansion, PM H+-ATPase, elongation
1. INTRODUCTION
The plant hormone auxin controls diverse aspects of plant growth and development by regulating the fundamental cellular processes of expansion, division, and differentiation. One of auxin’s most striking effects is to rapidly mediate changes in cell expansion. Indeed, this property was the basis of the bioassay leading to the chemical discovery of auxin, and the name itself is derived from the Greek word, auxein, meaning “to grow.” In subsequent years, auxin-regulated cell expansion has been found to play crucial roles in numerous processes, including organ growth (105, 126), tropic bending (46), apical hook and root hair development (10, 66), and shoot elongation in response to temperature (43) and light cues (118, 126). Our goal here is to provide an overview of auxin-mediated cell expansion with particular emphasis on recent studies that have provided crucial mechanistic insights into this long-studied process.
The origin of scientific interest in auxin’s ability to promote elongation growth of plant shoots can be traced back to The Power of Movement in Plants by Charles and Francis Darwin (24), in which they postulated that a shoot apex-derived “influence” mediated phototropic bending in canary grass. Some 50 years later, Frits Went demonstrated that he could capture this still unknown chemical signal by incubating excised Avena coleoptile tips on gelatin blocks. When transferred to one side of decapitated coleoptile apices, the signal in these loaded gelatin blocks promoted cell expansion resulting in bending of the coleoptile (134). After a couple of notable false starts, subsequent biochemical studies exploiting Went’s bioassay identified auxin isolated from human urine, Rhizopus exudates, and eventually plant material, as indole-3-acetic acid (IAA) (135). The 80-plus years since this discovery have witnessed remarkable progress in our understanding of how auxin regulates cell expansion. That said, we still only have a rudimentary understanding of the mechanistic details of this process, and new discoveries continue to identify novel ways by which this rather remarkable hormone controls different aspects of plant cell expansion.
2. AUXIN SIGNALING
Over the last 25 years, impressive advances have been made regarding the molecular mechanisms of auxin signal transduction. The vast majority of auxin-mediated changes in plant growth and development can be accounted for by the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN (TIR1/AFB) auxin receptors and their cognate AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) coreceptors. Other auxin receptor systems have also been identified that mediate specific auxin effects.
2.1. Canonical SCFTIR1/AFB-Mediated Signaling
Auxin regulates numerous growth processes via the control of gene transcription by SCFTIR1/AFB-Aux/IAA-ARF nuclear signaling modules. This mechanism involves the integration of three core components, the F-box proteins TIR1/AFB1–AFB5, which are receptor subunits of SCF-type E3 ubiquitin ligases; the Aux/IAA transcriptional repressors, which are both auxin coreceptors with TIR1/AFB proteins and substrates of SCFTIR1/AFB ubiquitin ligases; and the activator class of AUXIN RESPONSE FACTOR (ARF) transcription factors, whose transcriptional activities are repressed when bound by Aux/IAA proteins. As SCFTIR1/AFB-mediated transcriptional regulation was the subject of a recent review in this journal (133), we provide only a short summary of this process and refer readers to the above review.
In Arabidopsis, the nuclear auxin signaling pathway is represented by 6 TIR1/AFBs, 29 Aux/IAAs, and 23 ARFs, which allow production of diverse transcriptional outputs depending on the cellular and environmental context (104). In general, when auxin levels are low, Aux/IAA proteins interact with ARFs and repress their transcriptional activity. This repression involves Aux/IAA-mediated recruitment of a corepressor called TOPLESS (TPL) to the chromatin of ARF target genes (122). Upon an auxin stimulus, auxin promotes the interaction between Aux/IAA and TIR1/AFB proteins, acting as a molecular glue between these two components of the auxin coreceptor (26, 57, 125). The auxin-dependent recruitment of Aux/IAA proteins to SCFTIR1/AFB complexes results in Aux/IAA ubiquitination and subsequent proteasome-mediated degradation (26, 42, 57, 125). Aux/IAA proteolysis is a critical event in auxin signaling, as it results in the derepression of ARF transcriptional activity, leading to changes in gene expression. Auxin-mediated Aux/IAA degradation occurs very rapidly. Although a few noncanonical Aux/IAA proteins are relatively long-lived, half-life measurements of several Aux/IAA proteins range from 5 to 15 min (1, 29, 42). Consequently, this system enables extremely rapid transcriptional changes with some auxin-induced transcripts increasing within 5 min of auxin treatment (2, 77).
2.2. Additional Auxin Receptor Systems
For years, AUXIN BINDING PROTEIN1 (ABP1) was hypothesized to function as an auxin receptor responsible for rapid signaling events at the cell surface, including the regulation of cell expansion, endocytosis, and cytoskeletal rearrangements. However, the recent identification of abp1 null alleles in Arabidopsis, and the surprising finding that these mutants exhibit no apparent phenotypic differences from wild-type plants (35, 40), calls into serious question the large body of prior work implicating ABP1 in cell expansion or any other aspect of plant growth and development. Indeed, two recent studies examining auxin-mediated hypocotyl elongation detected no apparent differences in either the growth kinetics or response magnitude between wild-type and abp1 null mutants, indicating that ABP1 is not required for this response (22, 35). A reduction in auxin-induced protoplast swelling was observed in abp1 mutant cells (22), but the relevance of this phenotype to in planta cell expansion remains unclear.
Additional auxin sensors include S-PHASE KINASE-ASSOCIATED PROTEIN2A (SKP2A) and the ARF transcription factor, ETTIN (ETT). Like TIR1, SKP2A is an F-box protein subunit of an SCF ubiquitin ligase. SCFSKP2A controls cell cycle progression by targeting the cell cycle repressors, E2FC and DPB, for ubiquitin-mediated degradation, and auxin binding to SKP2A promotes this process (55). While potential roles for auxin signaling through SKP2A in the regulation of cell expansion have not been rigorously examined, leaf cells in Arabidopsis plants carrying an RNAi knockdown construct targeting E2FC were smaller and exhibited lower ploidy levels than wild-type controls (25). However, these effects are likely a consequence of an accelerated rate of cell division rather than direct regulation of cell expansion. ETT/ARF3 is a noncanonical ARF transcription factor that lacks the Aux/IAA interaction domain. Thus, it is perhaps not surprising that ETT utilizes a unique auxin-sensing mechanism. ETT functions as a master regulator of gynoecium development (83). IAA modulates interactions between ETT and additional transcription factors, including INDEHISCENT (IND), to control the expression of ETT target genes (112, 113). While many of these genes encode regulators of organ development and hormone dynamics, within the context of cell expansion it is intriguing that multiple pectin methylesterases (PMEs) and pectin methylesterase inhibitors (PMEIs) were also identified as putative ETT targets. A recent follow-up study concluded that ETT promotes expansion of cells in the developing gynoecium by increasing PME expression and repressing PMEI expression, leading to reductions in pectin methylesterification and cell wall stiffness (4).
3. THE ACID GROWTH THEORY—A BRIEF HISTORY
It has long been recognized that turgor pressure drives cell expansion at a rate that is limited by mechanical properties of the cell wall (70, 103). Therefore, for auxin to increase the rate of cell expansion, the hormone must increase cell wall extensibility while maintaining turgor pressure. First proposed nearly 50 years ago, the acid growth theory provided an explanatory model for how auxin might accomplish these tasks (45, 96, 98).
At its core, the acid growth theory posits that auxin activates plasma membrane (PM) H+-ATPases, resulting in proton efflux. The ensuing drop in apoplastic pH alters the activity of cell wall modification proteins, including expansins (78), xyloglucan endotransglycosylase/hydrolases (XTHs) (84), and PMEs (51), leading to changes in wall extensibility. Furthermore, elevated PM H+-ATPase activity hyperpolarizes the PM, increasing the energy for solute uptake, which is necessary to maintain water uptake, and therefore the turgor pressure that forces expansion of the wall.
The acid growth model was proposed based on a long series of classical experiments employing a modified version of Went’s bending assay that involves a uniform, rather than asymmetric, application of auxin (Figure 1). Typically, apical segments of maize/oat coleoptiles or epicotyls/hypocotyls of pea, soybean, or more recently Arabidopsis are excised from young seedlings. Removing the shoot apex, a primary source of auxin, results in the depletion of IAA following a short preincubation period in buffer or solid media, and the segment ceases growth. Upon subsequent treatment with IAA, the segment elongates following a typical lag phase of 10–20 min, which correlates with acidification of the surrounding medium, thus implicating H+ pump activation (17, 18). Furthermore, cotreatment with neutral or basic buffers attenuates auxin’s ability to promote elongation, as do ATPase inhibitors. Early (16) as well as more recent studies (35) have also demonstrated that inhibitors of transcription or translation also prevent elongation growth, indicating that de novo protein synthesis is required for auxin-mediated growth. In contrast, segments treated with acidic buffers or fusicoccin (FC) elongate in the absence of exogenous auxin (19, 45, 95). FC is a fungal toxin that directly and irreversibly activates PM H+-ATPases. It promotes greater increases in H+ efflux than IAA and exhibits a lag phase of only 1–2 min (73). For an additional recent review of acid growth, we refer readers to Arsuffi & Braybrook (5).
Figure 1.
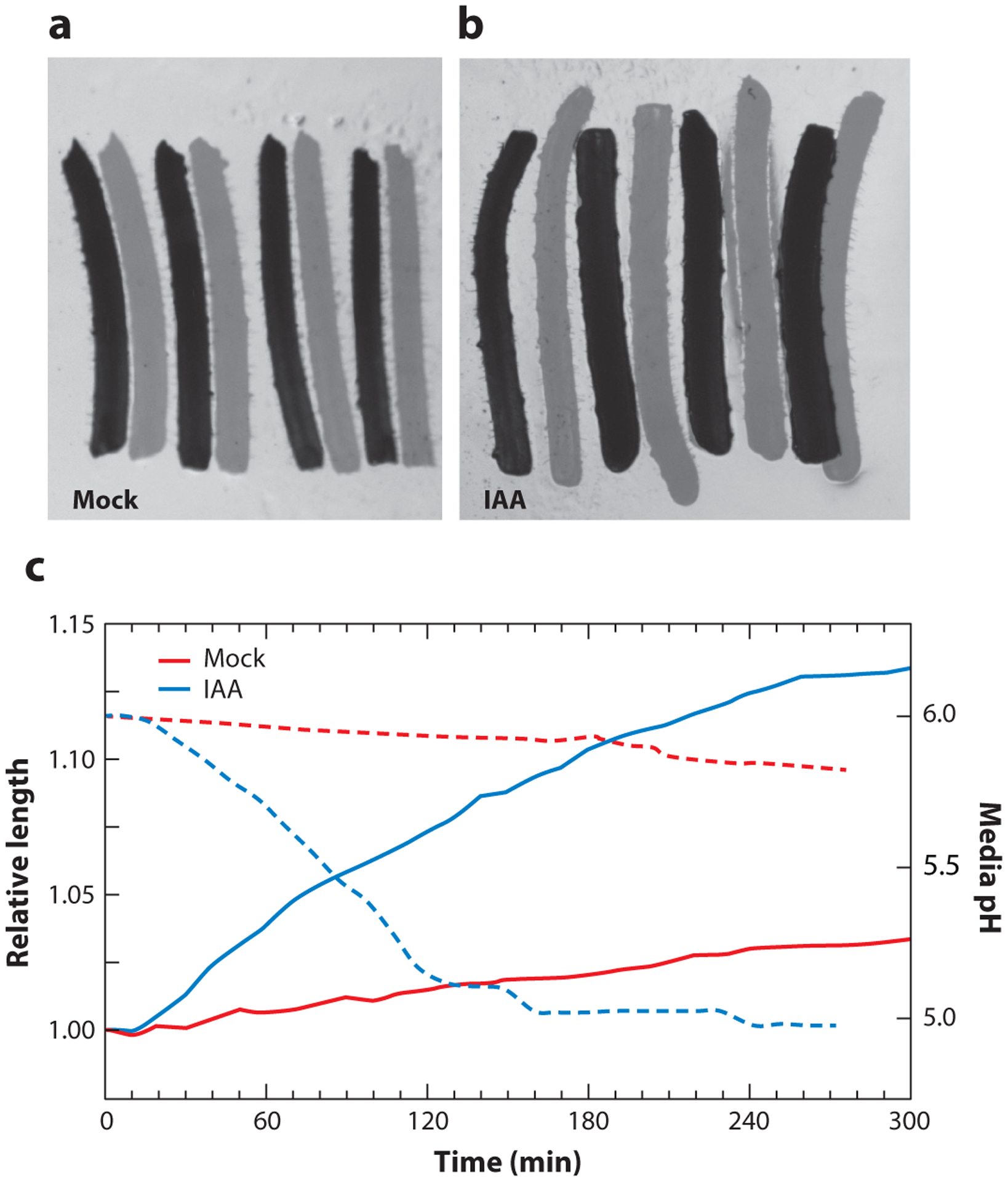
Auxin promotion of hypocotyl segment elongation. Apical hypocotyl segments were dissected from 5-day-old etiolated wild-type (M82) tomato seedlings. Segments were incubated for 1 h on KPS nutrient plates, transferred to KPS plates (a) lacking (mock) or (b) containing 10-μM indole-3-acetic acid (IAA), and imaged over time. Pictures depict overlays of the t = 0 min (dark) and t 300 min (light) time points. See Reference 115 for additional details. (c) Relative length (solid lines) of the=above hypocotyl segments over the 300-min time course. Dashed lines depict media acidification caused by auxin treatment of Avena coleoptiles over approximately the same time course. Data obtained from the Gray lab and Reference 95.
Plant cell growth requires expansion of the cell wall (20). Therefore, efforts to understand auxin-stimulated growth have included studying the effects of auxin on cell wall properties. Biochemical studies found that auxin caused changes in the size of wall matrix polysaccharides, but these did not correlate well with changes in growth (124). On the other hand, mechanical studies of killed stem sections, which are essentially cell wall samples, showed that low pH increased extensibility as did an auxin treatment prior to killing (65, 96, 97). A productive line of inquiry started with the finding that cucumber cell wall samples subjected to a constant, unidirectional force elongated (mechanical creep) when the pH of the surrounding solution was lowered. Removing proteins from the cell wall samples abolished this creep response to low pH. Adding back an essential protein fraction restored creep and led to the eventual identification of a cucumber protein named expansin (78). Consistent with the acid growth model, expansin-dependent creep is maximal at low pH (~4). Expansins do not appear to cleave covalent bonds within cell wall polymers, but rather appear to cause wall loosening by disrupting noncovalent interactions between cellulose microfibrils and matrix polysaccharides (21).
In addition to promoting pH-dependent cell wall remodeling, PM H+-ATPase activation also has energetic consequences. The difference in electric potential across the PM, also called the membrane potential or membrane voltage (Vm), is the result of active transporters moving ions against their electrochemical potential gradients, and passive transporters allowing the reverse to happen. In oat coleoptiles, auxin affects Vm within minutes, first in the positive direction (depolarization) and then in the negative direction (hyperpolarization) (8). The hyperpolarization is considered to be an electrophysiological consequence of the increased PM H+-ATPase activity (101) that causes apoplastic acidification over approximately the same time course (8) and was taken as independent biophysical evidence of auxin activating the proton pump, as opposed to other possible means of acid excretion. The preceding depolarization was interpreted as evidence of a symporter using energy released by more than one H+ moving inward to drive the uptake of an auxin anion against its electrochemical potential gradient (33). Recent work (28) described in Section 5.1 below supports this contention, albeit in Arabidopsis root hairs rather than coleoptiles. However, because Vm is the result of all ionic currents crossing the PM, any change in Vm could have multiple causes, and the above interpretations are not the only possible explanations. For example, auxin activation of a channel that exports anions or imports cations could cause the depolarization rather than, or in addition to, H+-IAA− symport. In fact, the patch-clamp technique, which can isolate specific ionic currents under rigorously controlled conditions, provided evidence that auxin activates anion channels in guard cells (74) and mesophyll cells (143). The affected channels were electrophysiologically characterized but were not molecularly identified in those early studies. More recently, auxin-induced Ca2+ influx has also been observed in Arabidopsis root cells (28, 82).
Subsequent PM hyperpolarization as a result of H+-ATPase activation, in addition to acidifying the apoplast, also increases the thermodynamic gradient that drives K+ uptake through channels. K+ uptake appears to contribute to the necessary process of maintaining a solute potential sufficiently negative to draw in water and thereby sustain the turgor pressure that forces cell expansion. The maize ZMK1 channel probably plays a role in auxin-mediated K+ uptake during the cell expansion response. First, it is more likely to enter its open, conducting state when the Vm is more negative and the apoplastic pH is low (two effects of proton pump activation), and second, its transcription is also induced by auxin (9, 90). Similar findings have been obtained with the Arabidopsis KAT1/KAT2 channels (91). K+ uptake cannot proceed alone because a rule of circuits is that all currents must sum to zero. Symporters use the proton gradient, enhanced by proton pump activation, to take up Cl− and NO3−, for example. These and other ions along with K+ probably contribute to the balance of solutes needed to maintain the water flux that drives auxin-induced cell expansion. Thus, the acid growth mechanism may be broadened to include proton pump activation, wall loosening by acid-activated apoplastic proteins, uptake of K+ through existing and newly made voltage-gated channels, and anions taken up by proton-driven symporters. The overall result is a faster, sustainable flux of water into a cell with a more extensible wall. These additional growth-sustaining ramifications of proton pump activation may explain why it has not always been possible to copy the action of auxin by simply acidifying the apoplast.
Despite the large body of work supporting an acid growth mechanism underlying auxin-mediated cell expansion, several justified criticisms of the model have been raised (61, 64). For example, many of the above experiments were conducted with abraded segments or even peeled segments that removed the epidermis. While these manipulations are not required for IAA- or FC-induced elongation, they are largely essential for acidic buffers to produce similar growth effects, presumably because the buffers cannot readily breach the waxy cuticle and enter the apoplast. Nonetheless, such manipulations raise concerns about whether or not this is a natural growth response, an artifact of the tissue damage incurred, or a greatly exaggerated response resulting from damage or removal of the epidermis, which has long been thought to constrain the growth of inner tissues (63, 75). Likewise, while surface pH measurements of intact segments detected clear acidification following IAA treatment, the degree of reduction in pH was suggested to be insufficient to explain the growth effects (64, 109). Again, however, the cuticle poses a barrier to excreted H+ ions, such that surface pH is likely not an accurate reflection of apoplastic pH. Perhaps most importantly, however, the acid growth theory lacked both significant genetic support and a clear molecular mechanism for how auxin might control H+ pump activity.
4. MECHANISTIC INSIGHTS INTO ACID GROWTH
Within the last few years, several studies have added substantial genetic and biochemical support to the acid growth theory, and a mechanistic basis for how auxin might regulate PM H+-ATPase activity has emerged. Thus, the decades-old acid growth model for auxin-mediated cell expansion is progressing from a physiological phenomenon to a molecularly defined process, spawning renewed interest in the topic and establishing a basic molecular framework of the signaling pathway from auxin perception to proton pump activation.
4.1. Auxin Promotes PM H+-ATPase Phosphorylation
How auxin might activate PM H+-ATPases is a question that dates back to the origins of the acid growth theory. Early work in maize coleoptiles detected increased PM H+-ATPase levels in PM fractions within 10 min of auxin treatment (44), reportedly as a result of increased exocytosis. Auxin also inhibits PM H+-ATPase endocytosis (88), which could also play a contributing role. Supporting this contention, a recent study reported that, in Arabidopsis hypocotyls, auxin represses expression of SYP132, which encodes an essential PM-localized SNARE protein that promotes PM H+-ATPase internalization (140). An additional study detected a threefold increase in MAIZE H+ ATPASE1 (MHA1) transcript levels in nonvascular coleoptile tissues following a 40-min auxin treatment (38). However, as PM H+-ATPase activity is intensively regulated by phosphorylation (47), it remained unclear as to whether or not these auxin-mediated increases in H+-ATPase abundance translated into increases in activity.
A major breakthrough was obtained with the demonstration that auxin promotes rapid phosphorylation of the penultimate threonine residue within the C-terminal autoinhibitory domain of PM H+-ATPases (123). Phosphorylation of this critical regulatory residue, corresponding to Thr947 of Arabidopsis AUTOINHIBITED H+ ATPASE2 (AHA2), coincides with 14–3–3 protein binding and activation of the pump (39, 53, 59). Furthermore, FC treatment also results in increased Thr947 phosphorylation, with FC forming a ternary complex with the phosphosite and 14–3–3 protein, locking the pump in the activated state (85, 121, 139). Employing auxin-depleted Arabidopsis hypocotyl segments, Takahashi et al. (123) observed auxin-induced Thr947 phosphorylation within 10 min, with peak phosphorylation occurring by 20 min. Consistent with prior findings, this increase in Thr947 phosphorylation correlated with elevated PM H+-ATPase activity. Furthermore, the kinetics of auxin-induced Thr947 phosphorylation, the associated increase in ATPase activity, and auxin-induced segment elongation all closely mirrored one another, suggesting causality. Consistent with this possibility, preincubation of hypocotyl segments with phosphatase inhibitors abolished both auxin-induced Thr947 phosphorylation and segment elongation (123), presumably by interfering with regulation of the kinase activity responsible for Thr947 phosphorylation.
4.2. Acid Growth Requires SCFTIR1/AFB-Mediated Signaling
The TIR1/AFB-Aux/IAA auxin coreceptors mediate the vast majority of auxin responses. Nonetheless, genetic redundancy and the severe morphological defects of higher-order receptor mutants (27) have complicated investigations into the role of this pathway in acid growth. For example, hypocotyl segment elongation assays of tir1 afb2 (123) and tir1 afb1 afb2 afb3 (107) mutants failed to detect clear differences from wild-type segments in response to IAA, leading both studies to conclude that rapid auxin-mediated growth occurred independent of TIR1/AFB-Aux/IAA receptor systems. More recent studies, however, have dispelled this notion. First, by exploiting the finding that unlike TIR1, AFB5 exhibits high binding affinity for the synthetic auxin picloram (11, 131), Fendrych et al. (35) demonstrated that afb5 mutants display significantly reduced picloram-mediated hypocotyl elongation compared to wild-type controls. Secondly, definitive evidence implicating SCFTIR1/AFB signaling in acid growth was provided by an elegant study employing an engineered auxin-TIR1 receptor pair (128). Based on the TIR1 crystal structure (125), a bump- and-hole strategy was used to design a novel synthetic auxin containing an aryl group (bump) on the indole ring of IAA, and a mutation in the auxin binding pocket of TIR1 to carve space (hole) to accommodate this larger IAA derivative. The synthetic auxin cvxIAA neither induces auxin-responsive gene expression nor elicits auxinic growth effects when applied to wild-type plants. However, when applied to plants expressing the engineered ccvTIR1 receptor, cvxIAA behaves as an active auxin. The authors found that Arabidopsis hypocotyl segments from ccvTIR1 seedlings exhibited rapid PM H+-ATPase phosphorylation and elongation in response to treatment with cvxIAA. These responses were indistinguishable from those observed in wild-type segments treated with IAA, thus demonstrating conclusively that TIR1/AFB receptors mediate acid growth.
4.3. SAUR-PP2C.D Regulation of PM H+-ATPase Activity
The finding that auxin activates PM H+-ATPases at least in part by elevating Thr947 phosphorylation suggested that auxin must either activate protein kinase or repress protein phosphatase activity. Recent work on the SMALL AUXIN UP RNA (SAUR) gene family has provided compelling evidence for the latter.
Early molecular approaches to isolate auxin-induced genes identified SAUR genes, many of which are induced within 5 min of auxin application (76, 77). SAUR genes are present as large gene families in diverse plant species. The Arabidopsis genome encodes 79 SAURs, whose predicted molecular weights range from 9.3 to 21.4 kDa. Other angiosperm genomes contain similarly large numbers of SAUR genes (15, 52, 138), and even basal plants such as Physcomitrella (89) and Marchantia contain 18 and 13 SAUR genes, respectively. Several SAUR proteins localize to the PM (13, 114, 120), but other SAURs appear to exhibit nuclear or cytosolic localization (99).
The role of SAURs in auxin action remained enigmatic for years, as the proteins contain no obvious motifs or domains suggestive of a molecular function, and the likely genetic redundancy within the large gene family complicated standard genetic analyses. To the best of our knowledge, no SAUR loss-of-function mutant has ever been identified via traditional genetic screening approaches. Several SAUR proteins have extremely short half-lives and appear to be subject to ubiquitin-mediated proteolysis (13, 60, 114). In vitro calmodulin binding has also been demonstrated for some SAUR proteins (60, 141), but the biological significance of this activity remains uncertain. Curiously, at least some SAURs are stabilized when expressed as fusion proteins with GFP or even small epitope tags (13, 114), thus facilitating gain-of-function genetic approaches for elucidating SAUR functions. Overexpression of stabilized SAUR19 fusion proteins confers numerous phenotypes indicative of increased cell expansion, including increases in hypocotyl length and leaf size, and altered tropic growth (114, 129, 130). Similar findings were obtained with plants expressing SAUR63 fusion proteins (13), suggesting that SAURs may promote elongation growth. Consistent with this notion, many SAUR genes are highly expressed in expanding organs (13, 37, 76, 114, 117, 120). Intriguingly, the expression of many SAUR19-related family members is highly enriched in the epidermal layer of hypocotyls. Expression of these SAURs is further upregulated upon shade-induced hypocotyl elongation (93), consistent with the epidermal growth control hypothesis (63). While there are certainly exceptions within the large gene family, in general, SAUR gene expression is upregulated by treatments/conditions that promote growth (e.g., IAA, brassinosteroids, shade) and downregulated by factors that repress growth (e.g., abscisic acid, jasmonic acid, abiotic stress). For more detailed coverage of SAUR genes, we refer readers to recent reviews (99, 119).
Since SAUR19 family proteins are positive effectors of cell expansion and localize to the PM (114, 115), Spartz and colleagues (116) proposed that SAURs may link auxin signaling to PM H+-ATPase activation and acid growth. Supporting this contention, in addition to the aforementioned cell expansion phenotypes, plants overexpressing SAUR19 fusion proteins exhibit increased PM H+-ATPase activity and reduced apoplastic pH (35, 116). Additionally, these plants display reduced drought tolerance due to improper guard cell regulation and hypersensitivity to charged, toxic compounds (116), both of which can be attributed to increased H+ pump activity. Indeed, GFP-SAUR19 overexpression plants display the same suite of phenotypes as elicited by FC-treatment and exhibited by the OPEN STOMATA2 (ost2D) mutant, which contains a dominant, gain-of-function mutant allele of the AHA1 PM H+-ATPase that exhibits constitutive activity (79, 116). Furthermore, GFP-SAUR19 overexpression results in elevated AHA Thr947 phosphorylation, the same critical regulatory residue previously found to be phosphorylated in response to IAA and FC treatments (116).
The effects of SAUR19 overexpression on PM H+-ATPase phosphorylation and activity, and by extension auxin-mediated cell expansion, are mediated by a subset of type 2C protein phosphatases known as D-clade PP2Cs (PP2C.D) that dephosphorylate Thr947 of AHA proteins. SAUR19, as well as several additional SAURs, specifically interact with PP2C.D phosphatases and inhibit their enzymatic activity (115, 116, 120). Several PP2C.D family members have been shown to physically interact with PM H+-ATPases in planta and dephosphorylate Thr947 (100, 116, 136). Furthermore, when heterologously coexpressed with AHA2 in yeast, these PP2C.D proteins also dephosphorylate Thr947, resulting in diminished H+ pump activity. These recent findings are consistent with prior work implicating Mg2+/Mn2+-dependent phosphatases in controlling PM H+-ATPase phosphorylation and activity (50).
The finding that SAURs inhibit PP2C.D phosphatases, together with the differential effects of SAUR19 and PP2C.D phosphatases on PM H+-ATPase phosphorylation status and activity, suggested that these two families of proteins act antagonistically to control cell expansion. This possibility has been confirmed genetically. The PP2C.D gene family of Arabidopsis consists of nine family members (PP2C.D1–PP2C.D9) that exhibit distinct subcellular localizations, including PM, nuclear, cytosolic, mitochondrial, and other compartments (100, 127). Initial genetic support suggesting that PP2C.D proteins act to repress cell expansion was provided by PP2C.D1 overexpression studies (116). Arabidopsis seedlings harboring a 35S:PP2C.D1 transgene exhibit severe cell expansion defects resulting in a dramatic dwarf phenotype. These growth defects appear to be largely the result of diminished PM H+-ATPase activity, as 35S:PP2C.D1 seedlings exhibit reduced levels of both AHA Thr947 phosphorylation and H+ pump activity. Furthermore, the severe growth defects and reduction in Thr947 phosphorylation conferred by PP2C.D1 overexpression are suppressed by both coexpression of GFP-SAUR19 and FC treatment, the former demonstrating SAUR-PP2C.D antagonism and the latter demonstrating PM H+-ATPase dependence.
Like all gain-of-function genetic approaches, these findings should be interpreted with caution. Indeed, more recent loss-of-function genetic studies of pp2c.d1 mutants suggest that this phosphatase does not play a major role in regulating cell expansion associated with organ growth (100). Rather, three additional PP2C.D family members appear to be the primary effectors of PM-localized SAUR proteins that modulate PM H+-ATPase activity to govern growth. Unlike other family members, PP2C.D2, PP2C.D5, and PP2C.D6 localize exclusively to the PM. While single mutants of these genes exhibit no obvious phenotype, the pp2c.d2 pp2c.d5 pp2c.d6 triple mutant presents a near perfect phenocopy of 35S:GFP-SAUR19 plants, including increases in hypocotyl length, leaf size, and PM H+-ATPase Thr947 phosphorylation (100). These phosphatases were also found to physically interact with AHA2.
Perhaps unsurprising given the large gene family, genetic support on the SAUR side of the equation has been more difficult to obtain. Sun et al. (120) identified several SAURs, including SAUR16 and SAUR50, whose expression is induced by light in cotyledons and repressed by light in hypocotyls—a pattern that mimics how light affects the growth of these two organs. Consistent with this notion, etiolated saur16 saur50 double mutants display slightly shorter hypocotyls than wild type, and light-grown mutants exhibit modestly smaller cotyledons. A similar modest reduction in temperature-induced hypocotyl elongation is seen in saur19–saur24 sextuple mutants (A. Hovland & W.M. Gray, unpublished data), and the SAUR26–SAUR28 gene cluster defines a QTL controlling temperature-mediated changes in rosette architecture (132). Presumably these weak growth defects reflect considerable redundancy within the large SAUR gene family. In an effort to bypass the complications of SAUR redundancy, Wong et al. (136) recently isolated pp2c.d2 and pp2c.d5 mutants that are defective in SAUR binding and therefore immune to SAUR inhibition of phosphatase activity. When expressed in Arabidopsis from their native promoters, these SAUR-immune derivatives behave as constitutively active phosphatases, conferring severe cell expansion defects, including auxin-insensitive hypocotyls and dramatic reductions in organ size and fertility due to impaired stamen filament elongation, and constitutively low levels of Thr947 phosphorylated PM H+-ATPase.
The above findings provide both strong genetic support for the acid growth theory and a mechanistic framework (Figure 2). In response to auxin, SAUR gene expression is rapidly upregulated through the canonical SCFTIR1/AFB signaling pathway. The resulting increase in SAUR protein abundance leads to the repression of PP2C.D2/PP2C.D5/PP2C.D6 phosphatase activity. This inhibition prevents PM H+-ATPase Thr947 dephosphorylation, trapping H+ pumps in the activated state, resulting in the apoplastic acidification and PM hyperpolarization that promote cell expansion.
Figure 2.
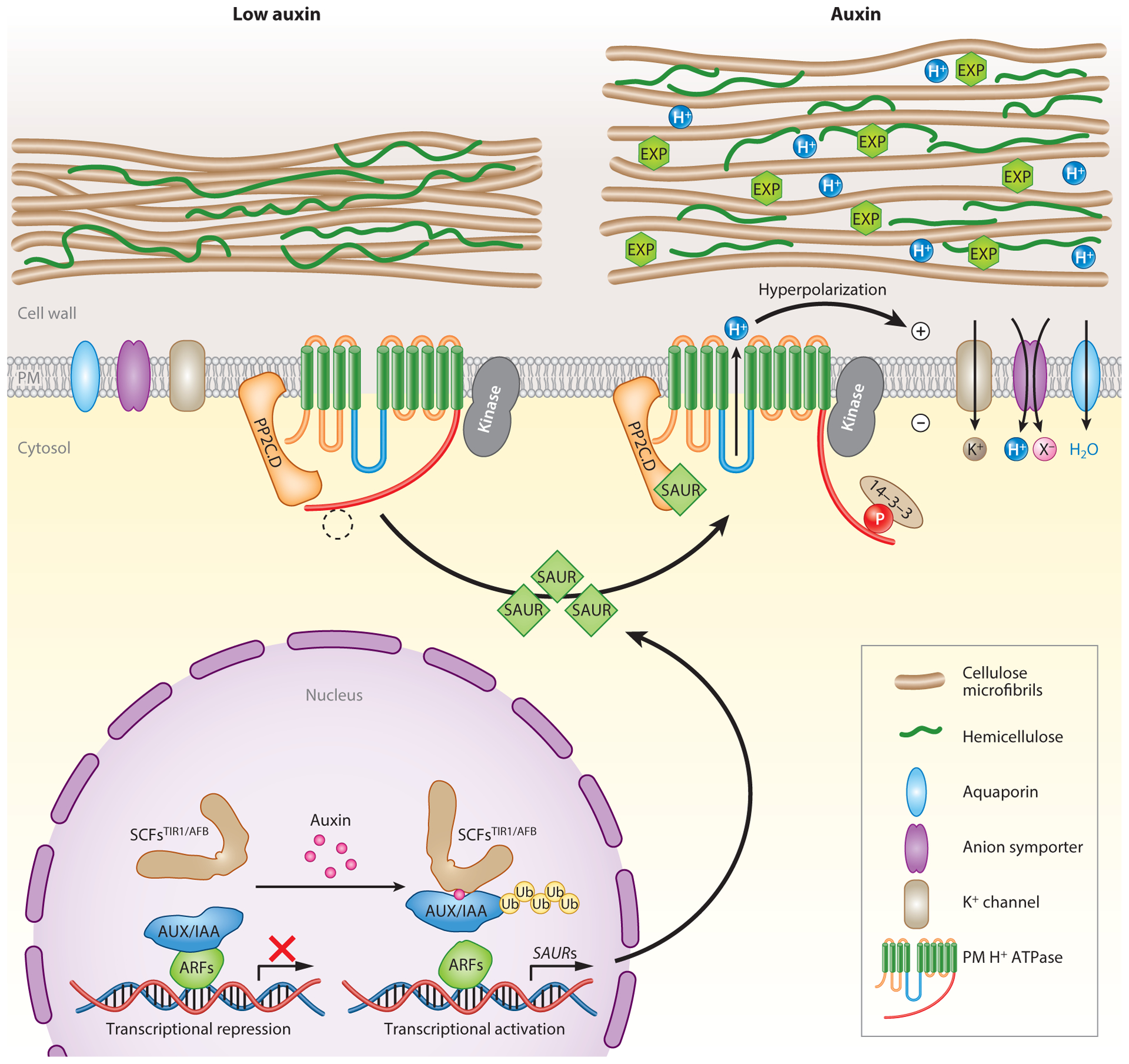
A model for auxin-mediated acid growth. Under low auxin conditions (left), basal plasma membrane (PM) H+-ATPase activity is maintained through the balance of phosphorylation via an unknown protein kinase and dephosphorylation by PP2C.D protein phosphatases. In response to auxin (right), canonical SCFTIR1/AFB-mediated signaling promotes SAUR gene expression. Following protein synthesis, SAUR proteins directly interact with and inhibit PM-localized PP2C.D2/PP2C.D5/PP2C.D6, preventing these phosphatases from dephosphorylating Thr947 and perhaps additional residues of PM H+-ATPases, thus keeping these proton pumps in the phosphorylated, activated state. This increase in PM H+-ATPase activity acidifies the apoplast, activating expansins (EXPs) and cell wall-remodeling enzymes to loosen the cell wall. Proton pumping also hyperpolarizes the PM, which activates inward-rectifying K+ channels and energizes H+-coupled anion symporters (X−). These transport activities maintain solute uptake needed for sustained water uptake and turgor pressure maintenance.
Given this model, a key question is whether or not auxin-induced SAUR expression is sufficient to trigger expansion growth. Initial findings suggest that this may be the case. Auxin-depleted hypocotyl segments require auxin to elongate in segment assays employing machine vision methods. However, hypocotyl segments prepared from Arabidopsis or tomato seedlings in which GFP-SAUR19 was constitutively expressed from the 35S promoter instead of its normal auxin-regulated promoter exhibited auxin-independent growth, elongating equally well whether auxin was present or absent (35, 115) (Supplemental Videos 1, 2). Furthermore, compared to wild-type controls, tomato GFP-SAUR19 segments displayed increased cell wall extensibility (115), which was not enhanced further by auxin treatment. Thus, constitutive SAUR expression results in constitutive increases in both growth and wall extensibility, suggesting that the only function of IAA that is required to promote cell expansion is the induction of SAUR expression.
A following related question is whether or not PM H+-ATPase activation is sufficient to trigger cell expansion. Here, the answer is less certain. Unlike 35S:GFP-SAUR19, Arabidopsis hypocotyl segments harboring the constitutively active ost2D allele of AHA1 do not exhibit auxin-independent growth. Interpretation is complicated, however, by the 11-member Arabidopsis AHA family and the likelihood that hexameric PM H+-ATPases are hetero-oligomers (56). Consistent with this possibility, when the ost2D mutation was introduced into an aha2 null mutant background, auxin-independent elongation of excised hypocotyl segments was observed (35). However, the amount of elongation was only about 50% of that obtained with 35S:GFP-SAUR19 segments, and a lag phase of >1 h was observed. While it remains possible that the diminished magnitude and delayed kinetics of the ost2D aha2 hypocotyls are due to contributions of the remaining wild-type AHA proteins, it is perhaps more plausible that PM H+-ATPases are not the only targets of SAUR-PP2C.D regulatory modules. For example, if additional phosphoproteins involved in ion and water transport, cell wall modification, and cell wall biosynthesis and vesicle trafficking to the cell surface were regulated by auxin via SAUR-PP2C.D modules, this might enable a coordinated growth response that is both faster and more robust than that obtained by PM H+-ATPase activation alone. This possibility is also attractive in terms of explaining the basis for large SAUR and PP2C.D gene families and the distinct subcellular localization patterns reported for individual SAUR and PP2C.D proteins, which could lead to complex, combinatorial regulation of protein phosphorylation patterns in a compartment-specific manner. Regardless of whether or not H+ pump activation is sufficient for growth, SAUR-PP2C.D modules, or additional auxin signaling outputs, seem almost certain to regulate supporting functions such as increased production of cell wall components to accommodate the increased growth. Supporting this contention, rapid changes in cell wall biosynthesis (62) as well as elevated transcription of genes encoding wall biosynthetic and modification enzymes have been reported in the literature (72). To date, however, PM H+-ATPases are the only proteins known to be regulated by SAUR-PP2C.D modules.
5. RAPID AUXIN GROWTH EFFECTS ON ROOTS
The effects of auxin on cell expansion are both concentration and tissue dependent. In general, IAA concentrations within the physiological range promote cell expansion of shoot tissues and inhibit cell expansion in roots. Notable exceptions to this generalization exist, however. For example, during apical hook development in hypocotyls, IAA accumulates on the concave side of the hook and inhibits cell expansion (137, 142). Reciprocally, auxin promotes root hair elongation (54, 92). Root growth, however, is inhibited by IAA concentrations in the nanomolar range and above. Interestingly, application of picomolar IAA concentrations have been reported to promote root elongation, suggesting that endogenous IAA levels may be suboptimal for growth (32). In studies examining auxin regulation of root cell expansion, it is important to remember that root growth is not a direct proxy for cell expansion, as auxin also plays important roles in the regulation of cell division and differentiation events at the root tip.
Given that auxin has a general inhibitory effect on root growth, is the acid growth theory relevant to this organ? Apoplastic acidification has been reported to both promote (80) and inhibit (87) root cell expansion, and exogenous auxin has been found to trigger apoplastic alkalization (6, 41, 82) as well as acidification (6). Such contradictory findings suggest complicated and dynamic roles for auxin in controlling root apoplastic pH homeostasis and cell expansion and point to the need for kinematic studies that specifically assess elongation apart from meristematic effects of IAA on root growth.
Barbez et al. (6) recently examined the relationship between extracellular pH and Arabidopsis root epidermal cell expansion using the fluorescent, pH-sensitive dye HPTS. Supporting a role for auxin-mediated acidification in the promotion of root cell expansion, epidermal cells in the elongation zone exhibited a lower apoplastic pH than flanking cells in the meristematic and differentiation zones, and this pattern correlated with the expression of auxin response reporters. Furthermore, epidermal cells in the distal meristem were longer or shorter, respectively, when seedlings were grown on low- or high-pH medium. Genetic perturbations that reduced IAA levels or impaired SCFTIR1/AFB-mediated signaling also resulted in elevated apoplastic pH and diminished epidermal cell length. In contrast, ost2D mutants and GFP-SAUR19 seedlings exhibited increased cell length. Together, these findings provide compelling support for an acid growth mechanism governing root cell expansion in response to endogenous auxin. However, as seen in prior studies, exogenous auxin promoted rapid and transient apoplastic alkalization that correlated with reduced cell expansion, pointing to a complex concentration-dependent and temporally regulated interplay between auxin, pH, and root cell size (6).
5.1. Auxin Elicits Near-Instantaneous Membrane Effects
Studies examining root extracellular pH with real-time fluorescent pH reporters have revealed that auxin elicits alkalization within 15 s (41, 82). Coinciding with this extremely rapid alkalization is an increase in cytosolic Ca2+ ([Ca2+]cyt) levels. The increase in extracellular pH is both inhibited by pretreatment with the Ca2+ channel blocker La3+ (81, 82) and mimicked by treatment with Ca2+ ionophores (81), suggesting that elevated [Ca2+]cyt functions as a second messenger to mediate auxin-induced alkalization. Furthermore, mutations in Arabidopsis CYCLIC NUCLEOTIDE-GATED CHANNEL14 (CNGC14) completely abolish both the auxin-induced increases in [Ca2+]cyt and extracellular pH (110). Physiological evidence implicating Ca2+-coupled alkalization in auxin inhibition of root cell expansion was provided by the demonstration that both [Ca2+]cyt and extracellular pH increase along the lower (nonelongating) flank of gravistimulated roots, and cngc14 mutants exhibit reduced gravitropic bending, as well as delayed root growth inhibition in response to exogenous IAA (81, 82).
Complementing the above findings is a recent study of auxin-triggered PM depolarization in Arabidopsis root hairs (28). The depolarization began within seconds, much faster than the response originally recorded in coleoptiles (8). The authors describe a dose-dependent rise in Vm, of up to 70 mV following a 1-s pulse of 10-μM IAA. Because the depolarization was not observed in aux1 influx carrier mutants, it was attributed to electrogenic symport of H+ and IAA anions. It is surprising that a symporter provided with only micromolar concentrations of substrate has such a large effect on Vm. Another surprising feature of these results is that membrane depolarization persisted for several minutes after the brief auxin pulse. Sustained depolarization would not be expected after the symporter substrate was removed unless the changes in Vm had a more complicated basis, such as IAA-dependent activation of ion channels to allow a large, depolarizing current, such as anion efflux or cation influx. Indeed, simultaneous Ca2+ influx was also recorded, resulting in a transient increase in [Ca2+]cyt that lasted for ~5 min and propagated as a wave into neighboring cells (28). Furthermore, direct injections of IAA into root hair cells triggered the rise in [Ca2+]cyt and a delayed depolarization response. Together, these findings suggest that H+-coupled auxin uptake rapidly depolarizes the PM, and that cytosolic IAA then promotes an inward depolarizing Ca2+ current. The resulting rise in [Ca2+]cyt could secondarily change the activity of other transporters, including PM H+-ATPases (58). Once again, CNGC14 was found to be essential for the auxin-induced increase in [Ca2+]cyt. Surprisingly, the initial PM depolarization response was also completely abolished in cngc14 mutants, suggesting a possible role for CNGC14 in regulating AUX1 activity.
These extremely rapid changes in extracellular pH, Vm, and [Ca2+]cyt are undoubtedly too fast to be accounted for by SCFTIR1/AFB-mediated transcriptional changes. Nonetheless, both membrane depolarization and the [Ca2+]cyt increase were substantially attenuated in tir1 afb2 afb3 mutants and in wild-type seedlings pretreated with the auxin antagonist auxinole, which binds TIR1/AFB proteins but precludes docking of Aux/IAA coreceptors (28, 49), suggesting SCFTIR1/AFB signaling may have nontranscriptional outputs in addition to the well-characterized regulation of ARF transcriptional activity. While highly suggestive, the possibility that steady-state levels of key auxin signaling components are different between wild-type and tir1 afb2 afb3 mutants due to impaired response to endogenous IAA cannot be eliminated. It will be interesting to see if these near-instantaneous auxin effects require the Aux/IAA coreceptors and whether or not they are evoked by the synthetic cvxIAA-ccvTIR1 hormone receptor system. In contrast, Monshausen et al. (82) reported that tir1 afb2 afb3 triple mutants exhibited wild-type-like changes in extracellular pH in response to IAA. At present, it remains to be determined whether this is simply the result of residual receptor activity provided by the remaining AFB proteins, or if rapid apoplastic alkalization represents a distinct, TIR1/AFB-independent auxin response. A summary of these extremely rapid auxin effects in roots is provided in Table 1.
Table 1.
Rapid auxin-triggered phenomena in Arabidopsis roots
| Response | Elective [IAA] | Lag time | Root location | TIR1/AFB-dependent | Reference |
|---|---|---|---|---|---|
| Inhibition of elongation | 1 μM | 45 s | Throughout the entire elongation zone | Not tested | 110 |
| CNGC14-dependent [Ca2+]cyt increase and surface alkalization | 1 μM | 10 s | Elongation zone | Not tested | 110 |
| Inhibition of elongation | Low nM | 30 s | Location of effect was not determined | Yes | 34 |
| Surface alkalization (global auxin application) | 1 nM-1 μM | 15 s | Elongation zone | Noa | 82 |
| [Ca2+]cyt increase (global auxin application) | 0.1 μM | 7–14 s | Growing root tip | Not tested | 82 |
| Surface alkalization (auxin application to root tip) | 1 μM | 49 ± 7 s | 200 μm from tip, propagates shootward | Not tested | 82 |
| [Ca2+]cyt increase (auxin application to root tip) | 1 μM | 45 ± 8 s | 200 μm from tip, propagates shootward | Not tested | 82 |
| H+ and Ca2+ influx, PM depolarization | 10 nM-10 μM | <5 s | Young root hairs | Yesb | 28 |
Abbreviations: IAA, indole-3-acetic acid; PM, plasma membrane.
Response observed in tir1 afb2 afb3 AFB1 AFB4 AFB5 seedlings.
Attenuated response in tir1 afb2 afb3 mutants and in wild-type root hairs treated with auxinole.
5.2. Nontranscriptional Inhibition of Root Elongation
Inhibition of root growth by the sustained presence of exogenous auxin was the basis for genetic screens leading to the isolation of the auxin-resistant mutants that defined the SCFTIR1/AFB signaling mechanism (67, 68, 94, 102). Thus, since their discovery, it has been apparent that SCFTIR/AFB components and regulators are essential for auxin-mediated root growth inhibition. Lacking from these studies, however, were highly precise kinetic data for this response. Inhibition was known to occur within 10–15 min of IAA treatment (31, 106), but this timing falls well within the existing paradigm of SCFTIR1/AFB-mediated transcriptional regulation. Recent findings, however, demonstrate that auxin inhibits root growth much more rapidly than previously appreciated and point toward novel aspects of TIR1/AFB-mediated signaling.
Employing an impressive microfluidics growth system mounted on a confocal microscope, Fendrych et al. (34) demonstrated that nanomolar concentrations of IAA begin to inhibit Arabidopsis root growth within 30 s (Table 1). Furthermore, this growth inhibition was rapidly reversible, as roots resumed growth within 2 min of auxin removal. Conceivably, such rapid and reversible control of root elongation could be advantageous in gravitropic growth during seedling establishment or following soil disturbance. Similar to the electrophysiological responses described above, the rapidity with which root growth responds to IAA effectively eliminates transcriptional reprogramming as a mechanism, which the authors confirmed by demonstrating that growth rate declined significantly before any detectable changes in auxin-inducible gene expression.
Growth inhibition in response to physiological IAA concentrations required AUX1, pointing toward the involvement of an intracellular auxin receptor. While tir1 afb2 afb3 mutants still exhibited significant growth inhibition, the rapid reduction in growth rate was less dramatic than observed in wild type, and the dose-response curve was shifted toward higher IAA concentrations in the mutant. Definitive evidence implicating TIR1/AFB coreceptors in this rapid, nontranscriptional inhibition of root growth was obtained using the synthetic cvxIAA-ccvTIR1 receptor system. Whereas cvxIAA application to wild-type control seedlings elicited no detectable response, cvxIAA inhibited the root growth of ccvTIR1 seedlings nearly identically to inhibition seen with IAA-treated controls. These findings clearly point toward novel, nontranscriptional outputs of the TIR1/AFB coreceptors. The molecular basis of this regulation as well as possible connections to the coincident changes in Vm, [Ca2+]cyt, and extracellular pH await elucidation.
5.3. Coordination of Growth and Intracellular Dynamics
While coupled changes in ion transport across the PM and cell wall extensibility are generally thought to be the primary drivers of auxin-mediated cell expansion, several intracellular events also occur that may facilitate the hormone’s control of cell growth. For example, cortical microtubule rearrangements occur within minutes of IAA treatment (14). However, these rearrangements are generally thought to regulate the directionality of expansion rather than the actual growth rate (3, 7). In contrast, recent findings provide an intriguing case linking auxin-induced changes in vacuolar morphogenesis to the inhibition of root cell expansion.
Unlike the volume of the cytosol, vacuolar volumes positively correlate with cell size (30, 86). To explore potential regulatory consequences of this correlation, Löfke et al. (71) examined the effects of auxin on cell expansion and vacuole morphogenesis in late meristematic epidermal cells of Arabidopsis roots. Inhibitory concentrations of auxin diminished both vacuole size and cell length, and intriguingly, the effect on vacuoles slightly preceded that on cell size, with a clear reduction in vacuole size being evident within 15 min. Likewise, an increase in endogenous IAA levels conferred by overexpression of the YUC6 biosynthetic enzyme conferred similar effects, whereas pharmacological inhibition of IAA synthesis resulted in increases in both vacuole size and cell length. The effect of auxin on vacuole morphogenesis was TIR1/AFB dependent and appears to involve posttranscriptional increases in the abundance of vacuole-localized SNARE proteins (71) and auxin-induced reorganization of actin filaments (108). While the abundance of multiple vacuolar SNAREs increased in response to auxin, mutation of a single SNARE, VTI11, partially attenuated the effects of auxin on both vacuole and cell size (71). Similar findings were obtained by treating wild-type roots with Wortmannin—a phosphatidylinositol kinase inhibitor believed to act upstream of SNAREs in vacuole morphogenesis. Notably, however, while both Wortmannin-treated cells and vti11 mutants contained larger vacuoles than wild-type controls, cell size was not increased. Thus, while reducing vacuole size may be sufficient to restrict root cell expansion, an increase in vacuole size alone does not promote cell elongation.
A recent follow-up study suggests that growth-associated vacuole morphogenesis is functionally coupled to cell wall pH (30). Direct acidification of the rhizosphere by growth on low-pH media as well as genetic (inducible GFP-SAUR19) and pharmacological (FC treatment) approaches all promoted increases in vacuole size in epidermal cells from the late meristematic region. The authors proposed that this coupling represents a cell wall-sensing mechanism involving FERONIA (FER), as fer mutants exhibited enlarged vacuoles and epidermal cells and were previously found to be defective in auxin-induced rapid apoplastic alkalization (6). FER encodes a Catharanthus roseus receptor-like kinase (CrRLK) family member previously implicated in cell wall mechanosensing (36, 111). It functions as a receptor for certain RAPID ALKALINIZATION FACTOR (RALF) peptides, triggering autophosphorylation, a transient [Ca2+]cyt increase, and phosphorylation of Ser899 of the AHA2 PM H+-ATPase, which is believed to inhibit H+ pumping activity (48). RALF-independent functions for FER have also been suggested (69), including a pectin-binding activity hypothesized to function in cell wall integrity sensing (36). Mechanistic insight was provided by demonstrating that the FER extracellular domain interacts with a subset of LEUCINE RICH EXTENSINs (LRXs)—protein components of cell walls (30). lrx3 lrx4 lrx5 triple mutants phenocopy the vacuole and growth defects of fer plants, and genetic analysis suggests that these LRXs and FER act in a common pathway to restrict root cell growth by coordinating changes in wall pH with vacuolar morphogenesis. Thus, while TIR1/AFB-mediated auxin signaling clearly impinges upon membrane trafficking processes, vacuolar morphology, and other intracellular processes, a mechanistic understanding of how these changes relate to auxin’s control of cell size awaits elucidation.
6. FUTURE PERSPECTIVES
Recent advances have extended our understanding of how auxin promotes cell expansion, resulting in the outlines of a basic molecular framework involving SAUR-PP2C.D regulatory modules that control PM H+-ATPase activity. That said, major questions remain. First, the kinase(s) that phosphorylate the penultimate Thr residue of PM H+-ATPases remain to be identified. On the one hand, auxin regulation of kinase activity could work hand-in-glove with SAUR-PP2C.D2/PP2C.D5/PP2C.D6 modules to activate H+ pumps. Alternatively, the kinase activity could simply be constitutive, with auxin exerting all of its control on the back end via inhibition of PP2C.D activity through the control of SAUR expression. Secondly, regulation of H+ pump activity is far more complex than simply phosphorylation status of the penultimate Thr residue. Several additional regulatory phosphosites, conferring either positive or negative effects on activity, have been reported (47), as well as auxin’s effects on PM H+-ATPase endocytosis (140). It will be interesting to see if, and how, auxin might control the phosphostatus of these sites and whether phosphorylation plays a role in trafficking. Additionally, the increase in outward H+ currents resulting from PM H+-ATPase activation must be balanced by an increase in a current of the opposite sign (cation influx or anion efflux). The changes in Vm and Ca2+ flux following auxin application are likely indications of necessary compensatory ion fluxes. Future studies should investigate their role in auxin-induced apoplast acidification and subsequent changes in cell expansion.
Additionally, plant genomes contain numerous SAUR genes. Which SAURs act when, where, and how remains largely unknown. Interestingly, not all SAURs and PP2C.D proteins localize to the PM (99, 100). It is intriguing to speculate that non-PM-localized SAUR-PP2C.D modules may control the phosphorylation status of cytosolic, nuclear, and organellar proteins to couple intracellular changes with cell expansion to generate a coordinated growth response. Likewise, the functional importance and regulatory mechanisms underlying the posttranscriptional regulation of SAUR expression and the calmodulin-binding activity of these proteins remain to be determined.
The elucidation of the molecular underpinnings of acid growth may enable new strategies for tailoring plant organ growth. Studies to date suggest that SAUR-PP2C.D2/PP2C.D5/PP2C.D6 regulatory modules comprise an output of the SCFTIR1/AFB signaling pathway that specifically controls cell expansion. No major developmental abnormalities are apparent in either the SAUR overexpression plants or gain- or loss-of-function PP2C.D plants that have been characterized. Rather the cells and the corresponding organs of these plants are simply larger or smaller, suggesting that SAUR-PP2C.D modules do not play major roles in auxin-mediated patterning and differentiation. Consequently, transgenic approaches involving the expression of SAUR or PP2C.D2/PP2C.D5/PP2C.D6 proteins from cell- and/or organ-type specific promoters may provide a tunable strategy for manipulating the sizes of agriculturally important plant organs without the detrimental side effect of developmental perturbation.
Recent findings also point toward novel auxin signaling mechanisms (12, 113) (see the sidebar titled TMK1-Mediated Cell Expansion) and rapid, nontranscriptional outputs of the TIR1/AFB pathway (28, 34) that regulate cell expansion. The recent demonstration that auxin reversibly inhibits root elongation within seconds in a TIR/AFB-dependent manner has revealed a whole new aspect of auxin signaling through these receptors. Perhaps some Aux/IAA proteins promote root growth independent of their role in regulating ARF activity, and SCFTIR1/AFB-mediated degradation of these Aux/IAAs results in rapid growth repression. Alternatively, novel auxin coreceptors may partner with TIR1/AFB proteins to mediate this rapid, nontranscriptional regulation of growth. Regardless of these possibilities, the downstream effectors await discovery. Likewise, confirming if, and how, TIR1/AFB-mediated signaling regulates the extremely rapid auxin-mediated ionic effects described in Section 5.1 above, and ultimately how this may result in root growth inhibition, will undoubtedly be a major emphasis of future studies. While recent years have witnessed major advances in our understanding of auxin-regulated cell expansion, clearly the fascinating effects of this hormone that garnered the attention of Went and his contemporaries nearly 100 years ago will continue to capture the interests of future generations of plant biologists.
TMK1-MEDIATED CELL EXPANSION.
Auxin plays a central role in governing apical hook development (10). During hook formation, an auxin maximum on the concave side of the hook inhibits cell expansion. Later, gradual decrease of the auxin maximum promotes expansion, leading to a reversal of the growth rate differential across the hypocotyl, and thus hook opening. Recently, the receptor-like kinase TMK1 was found to mediate auxin inhibition of concave apical hook cells (12). High auxin levels trigger cleavage of the TMK1 kinase domain, which subsequently migrates to the nucleus and phosphorylates IAA32 and IAA34 to primarily repress transcription. Unlike canonical Aux/IAA proteins, IAA32/IAA34 lack the degron motif that mediates TIR1/AFB interactions and are therefore not degraded in response to auxin. Rather, auxin increased the stability of IAA32/IAA34 in a TMK1-dependent manner. Furthermore, both auxin-mediated cleavage of TMK1 and the increase in IAA32/IAA34 stability appear to be TIR1/AFB independent, suggesting a novel auxin signaling pathway may be involved. TMK1 belongs to a four-member family, and higher-order mutants exhibit multiple growth defects (23). It will be interesting to see if a similar signaling mechanism is involved in these other TMK-mediated processes and how the auxin signal is perceived and transmitted to the TMKs.
Supplementary Material
SUMMARY POINTS.
Auxin regulates cell expansion in a tissue- and concentration-dependent manner. In general, physiological IAA concentrations promote expansion of shoot cells and inhibit expansion of root cells.
Rayle & Cleland’s 1992 statement (98) that “the Acid Growth Theory of auxin-induced cell elongation is alive and well” is perhaps even more true today, as recent findings have begun to elucidate the molecular underpinnings of this decades-old and often controversial theory.
Plasma membrane (PM)-localized SAUR-PP2C.D modules regulate PM H+-ATPase phosphorylation status and activity. Auxin-induced SAUR expression via the SCFTIR1/AFB pathway leads to the inhibition of PP2C.D activity, thus trapping PM H+-ATPases in the phosphorylated, activated state.
Increases in SAUR expression are sufficient to promote cell expansion. Constitutive overexpression of GFP-SAUR19 confers auxin-independent increases in cell expansion, apoplastic acidification, and cell wall extensibility, implicating SAURs as the primary effectors of auxin-mediated cell expansion.
Auxin inhibition of root growth occurs within seconds. This requires the TIR1/AFB receptors, implicating novel, nontranscriptional, and as yet undiscovered outputs of this signaling pathway.
Influx of IAA into root cells promotes a transient rise in cytosolic Ca2+ levels within seconds. This contributes to sustained PM depolarization, requires the CNGC14 channel protein, and also appears to be TIR1/AFB dependent.
ACKNOWLEDGMENTS
Research in the authors’ lab is supported by grants from the National Institutes of Health (GM067203 to W.M.G.) and the National Science Foundation (MCB-1613809 to W.M.G. and IOS-1360751 to E.P.S.). The authors wish to thank Dr. Lihao Lin for assistance with figure preparation and members of the Gray lab for helpful comments.
Glossary
- IAA
indole-3-acetic acid
- SCF
Skp1/Cullin1/F-box protein ubiquitin ligase
- PM
plasma membrane
- Fusicoccin (FC)
a wilting toxin produced by Fusicoccum amygdali
- Vm
membrane voltage
- SNARE
SNAp REceptor proteins that mediate vesicle fusion
- SAUR
SMALL AUXIN UP RNA
- GFP
green fluorescent protein
- 35S
Cauliflower mosaic virus 35S promoter
- QTL
quantitative trait locus
- HPTS
8-hydroxypyrene-1,3,6-trisulfonic acid trisodium salt
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abel S, Oeller PW, Theologis A. 1994. Early auxin-induced genes encode short-lived nuclear proteins. PNAS 91:326–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abel S, Theologis A. 1996. Early genes and auxin action. Plant Physiol. 111:9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamowski M, Li L, Friml J. 2019. Reorientation of cortical microtubule arrays in the hypocotyl of Arabidopsis thaliana is induced by the cell growth process and independent of auxin signaling. Int. J. Mol. Sci 20:e3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andres-Robin A, Reymond MC, Dupire A, Battu V, Dubrulle N, et al. 2018. Evidence for the regulation of gynoecium morphogenesis by ETTIN via cell wall dynamics. Plant Physiol. 178:1222–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arsuffi G, Braybrook SA. 2018. Acid growth: an ongoing trip. J. Exp. Bot 69:137–46 [DOI] [PubMed] [Google Scholar]
- 6.Barbez E, Dünser K, Gaidora A, Lendl T, Busch W. 2017. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. PNAS 114:E4884–93 [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that auxin-mediated apoplastic acidification promotes root cell elongation and high auxin levels trigger FERONIA-dependent root apoplastic alkalization.
- 7.Baskin TI. 2015. Auxin inhibits expansion rate independently of cortical microtubules. Trends Plant Sci. 20:471–72 [DOI] [PubMed] [Google Scholar]
- 8.Bates GW, Goldsmith MH. 1983. Rapid response of the plasma-membrane potential in oat coleoptiles to auxin and other weak acids. Planta 159:231–37 [DOI] [PubMed] [Google Scholar]
- 9.Becker D, Hedrich R. 2002. Channelling auxin action: modulation of ion transport by indole-3-acetic acid In Auxin Molecular Biology, ed. Perrot-Rechenmann C, Hagen G, pp. 349–56. Dordrecht, Neth.: Springer Neth. [PubMed] [Google Scholar]
- 10.Béziat C, Kleine-Vehn J. 2018. The road to auxin-dependent growth repression and promotion in apical hooks. Curr. Biol 28:R519–25 [DOI] [PubMed] [Google Scholar]
- 11.Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, et al. 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol 8:477–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao M, Chen R, Li P, Yu Y, Zheng R, et al. 2019. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568:240–43 [DOI] [PubMed] [Google Scholar]; Demonstrates auxin-dependent cleavage of the TMK1 kinase domain and its regulation of noncanonical Aux/IAAs during apical hook development.
- 13.Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, et al. 2012. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 71:684–97 [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Grandont L, Li H, Hauschild R, Paque S, et al. 2014. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 516:90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Hao X, Cao J. 2014. Small auxin upregulated RNA (SAUR) gene family in maize: identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J. Integr. Plant Biol 56:133–50 [DOI] [PubMed] [Google Scholar]
- 16.Cleland R. 1971. Instability of the growth-limiting proteins of the Avena coleoptile and their pool size in relation to auxin. Planta 99:1–11 [DOI] [PubMed] [Google Scholar]
- 17.Cleland R. 1973. Auxin-induced hydrogen ion excretion from Avena coleoptiles. PNAS 70:3092–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleland RE. 1975. Auxin-induced hydrogen ion excretion: correlation with growth, and control by external pH and water stress. Planta 127:233–42 [DOI] [PubMed] [Google Scholar]
- 19.Cleland RE. 1976. Fusicoccin-induced growth and hydrogen ion excretion in Avena coleoptiles: relation to auxin responses. Planta 128:201–6 [DOI] [PubMed] [Google Scholar]
- 20.Cosgrove DJ. 2005. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol 6:850–61 [DOI] [PubMed] [Google Scholar]
- 21.Cosgrove DJ. 2015. Plant expansins: diversity and interactions with plant cell walls. Curr. Opin. Plant Biol 25:162–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahlke RI, Fraas S, Ullrich KK, Heinemann K, Romeiks M, et al. 2017. Protoplast swelling and hypocotyl growth depend on different auxin signaling pathways. Plant Physiol. 175:982–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai N, Wang W, Patterson SE, Bleecker AB. 2013. The TMK subfamily of receptor-like kinases in Arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLOS ONE 8:e60990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darwin C, Darwin FE. 1880. The Power of Movement in Plants. New York: D. Appleton [Google Scholar]
- 25.del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. 2006. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18:2224–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435:441–45 [DOI] [PubMed] [Google Scholar]
- 27.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, et al. 2005. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9:109–19 [DOI] [PubMed] [Google Scholar]
- 28.Dindas J, Scherzer S, Roelfsema MRG, von Meyer K, Müller HM, et al. 2018. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat. Commun 9:1174. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that IAA influx triggers Ca2+-coupled depolarization in root hairs and this response is diminished in tir1/afb and cngc14 mutants.
- 29.Dreher KA, Brown J, Saw RE, Callis J. 2006. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18:699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dünser K, Gupta S, Herger A, Feraru MI, Ringli C, Kleine-Vehn J. 2019. Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J. 38:e100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans ML. 1976. A new sensitive root auxanometer: preliminary studies of the interaction of auxin and acid pH in the regulation of intact root elongation. Plant Physiol. 58:599–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans ML, Ishikawa H, Estelle MA. 1994. Responses of Arabidopsis roots to auxin studies with high temporal resolution: Comparison of wild type and auxin-response mutants. Planta 194:215–22 [Google Scholar]
- 33.Felle H, Peters W, Palme K. 1991. The electrical response of maize to auxins. Biochim. Biophys. Acta Biomembr 1064:199–204 [DOI] [PubMed] [Google Scholar]
- 34.Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, et al. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat. Plants 4:453–59 [DOI] [PMC free article] [PubMed] [Google Scholar]; Reveals rapid nontranscriptional roles of the TIR1/AFB receptor system in auxin-mediated root growth inhibition.
- 35.Fendrych M, Leung J, Friml J. 2016. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5:e19048. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Reference 115, reveals that constitutive GFP-SAUR19 expression confers auxin-independent hypocotyl elongation and apoplastic acidification.
- 36.Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, et al. 2018. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol 28:666–75.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, et al. 2011. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. PNAS 108:20231–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frias I, Caldeira MT, Perez-Castineira JR, Navarro-Avino JP, Culianez-Macia FA, et al. 1996. A major isoform of the maize plasma membrane H+-ATPase: characterization and induction by auxin in coleoptiles. Plant Cell 8:1533–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, et al. 1999. Binding of 14–3–3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem 274:36774–80 [DOI] [PubMed] [Google Scholar]
- 40.Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. 2015. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. PNAS 112:2275–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gjetting KS, Ytting CK, Schulz A, Fuglsang AT. 2012. Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot 63:3207–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414:271–76 [DOI] [PubMed] [Google Scholar]
- 43.Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. 1998. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. PNAS 95:7197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hager A, Debus G, Edel HG, Stransky H, Serrano R. 1991. Auxin induces exocytosis and the rapid synthesis of a high-turnover pool of plasma-membrane H+-ATPase. Planta 185:527–37 [DOI] [PubMed] [Google Scholar]
- 45.Hager A, Menzel H, Krauss A. 1971. Experiments and hypothesis concerning the primary action of auxin in elongation growth. Planta 100:47–75 [DOI] [PubMed] [Google Scholar]
- 46.Harmer SL, Brooks CJ. 2018. Growth-mediated plant movements: hidden in plain sight. Curr. Opin. Plant Biol 41:89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haruta M, Gray WM, Sussman MR. 2015. Regulation of the plasma membrane proton pump (H+-ATPase) by phosphorylation. Curr. Opin. Plant Biol 28:68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343:408–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, et al. 2012. Rational design of an auxin antagonist of the SCFTIR1 auxin receptor complex. ACS Chem. Biol 7:590–98 [DOI] [PubMed] [Google Scholar]
- 50.Hayashi Y, Nakamura S, Takemiya A, Takahashi Y, Shimazaki K, Kinoshita T. 2010. Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol. 51:1186–96 [DOI] [PubMed] [Google Scholar]
- 51.Hocq L, Pelloux J, Lefebvre V. 2017. Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 22:20–29 [DOI] [PubMed] [Google Scholar]
- 52.Jain M, Tyagi AK, Khurana JP. 2006. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88:360–71 [DOI] [PubMed] [Google Scholar]
- 53.Jelich-Ottmann C, Weiler EW, Oecking C. 2001. Binding of regulatory 14–3–3 proteins to the C terminus of the plant plasma membrane H+-ATPase involves part of its autoinhibitory region. J. Biol. Chem 276:39852–57 [DOI] [PubMed] [Google Scholar]
- 54.Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, et al. 2009. Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol 11:78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jurado S, Abraham Z, Manzano C, López-Torrejón G, Pacios LF, Del Pozo JC. 2010. The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22:3891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanczewska J, Marco S, Vandermeeren C, Maudoux O, Rigaud JL, Boutry M. 2005. Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14–3–3 proteins converts a dimer into a hexamer. PNAS 102:11675–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435:446–51 [DOI] [PubMed] [Google Scholar]
- 58.Kinoshita T, Nishimura M, Shimazaki K. 1995. Cytosolic concentration of Ca2+ regulates the plasma membrane H+-ATPase in guard cells of Fava bean. Plant Cell 7:1333–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kinoshita T, Shimazaki K. 1999. Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J. 18:5548–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knauss S, Rohrmeier T, Lehle L. 2003. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J. Biol. Chem 278:23936–43 [DOI] [PubMed] [Google Scholar]
- 61.Kutschera U. 1994. The current status of the acid-growth hypothesis. New Phytol. 126:549–69 [Google Scholar]
- 62.Kutschera U, Briggs WR. 1987. Rapid auxin-induced stimulation of cell wall synthesis in pea internodes. PNAS 84:2747–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kutschera U, Niklas KJ. 2007. The epidermal-growth-control theory of stem elongation: an old and a new perspective. J. Plant Physiol 164:1395–409 [DOI] [PubMed] [Google Scholar]
- 64.Kutschera U, Schopfer P. 1985. Evidence against the acid-growth theory of auxin action. Planta 163:483–93 [DOI] [PubMed] [Google Scholar]
- 65.Kutschera U, Schopfer P. 1986. Effect of auxin and abscisic acid on cell wall extensibility in maize coleoptiles. Planta 167:527–35 [DOI] [PubMed] [Google Scholar]
- 66.Lee RD, Cho HT. 2013. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front. Plant Sci 4:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. 1993. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364:161–64 [DOI] [PubMed] [Google Scholar]
- 68.Leyser HM, Pickett FB, Dharmasiri S, Estelle M. 1996. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10:403–13 [DOI] [PubMed] [Google Scholar]
- 69.Liao H, Tang R, Zhang X, Luan S, Yu F. 2017. FERONIA receptor kinase at the crossroads of hormone signaling and stress responses. Plant Cell Physiol. 58:1143–50 [DOI] [PubMed] [Google Scholar]
- 70.Lockhart JA. 1965. An analysis of irreversible plant cell elongation. J. Theor. Biol 8:264–75 [DOI] [PubMed] [Google Scholar]
- 71.Löfke C, Dünser K, Scheuring D, Kleine-Vehn J. 2015. Auxin regulates SNARE-dependent vacuolar morphology restricting cell size. eLife 4:e05868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Majda M, Robert S. 2018. The role of auxin in cell wall expansion. Int. J. Mol. Sci 19:E951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marre E. 1979. Fusicoccin: A tool in plant physiology. Annu. Rev. Plant Physiol 30:273–88 [Google Scholar]
- 74.Marten I, Lohse G, Hedrich R. 1991. Plant growth hormones control voltage-dependent activity of anion channels in plasma membrane of guard cells. Nature 353:758–62 [Google Scholar]
- 75.Masuda Y, Yamamoto R. 1972. Control of auxin-induced stem elongation by the epidermis. Physiol. Plant 27:109–15 [Google Scholar]
- 76.McClure BA, Guilfoyle T. 1987. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol 9:611–23 [DOI] [PubMed] [Google Scholar]
- 77.McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ. 1989. Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1:229–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McQueen-Mason S, Durachko DM, Cosgrove DJ. 1992. Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, et al. 2007. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26:3216–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moloney MM, Elliott MC, Cleland RE. 1981. Acid growth effects in maize roots: evidence for a link between auxin-economy and proton extrusion in the control of root growth. Planta 152:285–91 [DOI] [PubMed] [Google Scholar]
- 81.Monshausen GB, Bibikova TN, Weisenseel MH, Gilroy S. 2009. Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21:2341–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Monshausen GB, Miller ND, Murphy AS, Gilroy S. 2011. Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J. 65:309–18 [DOI] [PubMed] [Google Scholar]
- 83.Nemhauser JL, Feldman LJ, Zambryski PC. 2000. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127:3877–88 [DOI] [PubMed] [Google Scholar]
- 84.Nishitani K, Vissenberg K. 2007. Roles of the XTH protein family in the expanding cell In The Expanding Cell, ed. Verbelen JP, Vissenberg K, pp. 89–116. Heidelberg, Ger.: Springer-Verlag [Google Scholar]
- 85.Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C. 1998. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14–3–3 binding. Plant Physiol. 118:551–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Owens T, Poole RJ. 1979. Regulation of cytoplasmic and vacuolar volumes by plant cells in suspension culture. Plant Physiol. 64:900–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pacheco-Villalobos D, Díaz-Moreno SM, van der Schuren A, Tamaki T, Kang YH, et al. 2016. The effects of high steady state auxin levels on root cell elongation in Brachypodium. Plant Cell 28:1009–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, et al. 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435:1251–56 [DOI] [PubMed] [Google Scholar]
- 89.Paponov IA, Teale W, Lang D, Paponov M, Reski R, et al. 2009. The evolution of nuclear auxin signalling. BMC Evol. Biol 9:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Philippar K, Fuchs I, Luthen H, Hoth S, Bauer CS, et al. 1999. Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. PNAS 96:12186–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Philippar K, Ivashikina N, Ache P, Christian M, Luthen H, et al. 2004. Auxin activates KAT1 and KAT2, two K+-channel genes expressed in seedlings of Arabidopsis thaliana. Plant J. 37:815–27 [DOI] [PubMed] [Google Scholar]
- 92.Pitts RJ, Cernac A, Estelle M. 1998. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16:553–60 [DOI] [PubMed] [Google Scholar]
- 93.Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J. 2016. The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 30:1529–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Quint M, Ito H, Zhang W, Gray WM. 2005. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 43:371–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rayle DL. 1973. Auxin-induced hydrogen-ion secretion in Avena coleoptiles and its implications. Planta 114:63–73 [DOI] [PubMed] [Google Scholar]
- 96.Rayle DL, Cleland R. 1970. Enhancement of wall loosening and elongation by acid solutions. Plant Physiol. 46:250–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rayle DL, Cleland RE. 1980. Evidence that auxin-induced growth of soybean hypocotyls involves proton excretion. Plant Physiol. 66:433–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rayle DL, Cleland RE. 1992. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 99:1271–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ren H, Gray WM. 2015. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 8:1153–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ren H, Park MY, Spartz AK, Wong JH, Gray WM. 2018. A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLOS Genet. 14:e1007455. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies PP2C.D2/D5/D6 as the primary effectors of PM-localized SAURs that regulate PM H+-ATPase phosphorylation/activity.
- 101.Ruck A, Palme K, Venis MA, Napier RM, Felle HH. 1993. Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J. 4:41–46 [Google Scholar]
- 102.Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12:198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sachs J, Vines SH. 1882. Text-Book of Botany, Morphological and Physiological. Oxford, UK: Clarendon [Google Scholar]
- 104.Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sandalio LM, Rodríguez-Serrano M, Romero-Puertas MC. 2016. Leaf epinasty and auxin: a biochemical and molecular overview. Plant Sci. 253:187–93 [DOI] [PubMed] [Google Scholar]
- 106.Scheitz K, Lüthen H, Schenck D. 2013. Rapid auxin-induced root growth inhibition requires the TIR and AFB auxin receptors. Planta 238:1171–76 [DOI] [PubMed] [Google Scholar]
- 107.Schenck D, Christian M, Jones A, Lüthen H. 2010. Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol. 152:1183–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scheuring D, Löfke C, Krüger F, Kittelmann M, Eisa A, et al. 2016. Actin-dependent vacuolar occupancy of the cell determines auxin-induced growth repression. PNAS 113:452–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schopfer P. 1989. pH-dependence of extension growth in Avena coleoptiles and its implications for the mechanism of auxin action. Plant Physiol. 90:202–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shih HW, DePew CL, Miller ND, Monshausen GB. 2015. The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in Arabidopsis thaliana. Curr. Biol 25:3119–25 [DOI] [PubMed] [Google Scholar]; The cyclic nucleotide-gated channel, CNGC14, is required for rapid Ca2+ signaling and extracellular alkalization following gravitropic stimulation of roots.
- 111.Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. 2014. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr. Biol 24:1887–92 [DOI] [PubMed] [Google Scholar]
- 112.Simonini S, Bencivenga S, Trick M, Østergaard L. 2017. Auxin-induced modulation of ETTIN activity orchestrates gene expression in Arabidopsis. Plant Cell 29:1864–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simonini S, Deb J, Moubayidin L, Stephenson P, Valluru M, et al. 2016. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 30:2286–96. Corrigendum. 2017. Genes Dev. 31:1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, et al. 2012. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70:978–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Spartz AK, Lor VS, Ren H, Olszewski NE, Miller ND, et al. 2017. Constitutive expression of Arabidopsis SMALL AUXIN UP RNA19 (SAUR19) in tomato confers auxin-independent hypocotyl elongation. Plant Physiol. 173:1453–62 [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that constitutive GFP-SAUR19 expression confers increased cell wall extensibility and, together with Reference 35, auxin-independent hypocotyl elongation.
- 116.Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, et al. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26:2129–42 [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies SAUR-PP2C.D regulatory modules that modulate PM H+-ATPase phosphorylation/activity to regulate auxin-responsive cell expansion.
- 117.Stamm P, Kumar PP. 2013. Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 32:759–69 [DOI] [PubMed] [Google Scholar]
- 118.Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, et al. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–91 [DOI] [PubMed] [Google Scholar]
- 119.Stortenbeker N, Bemer M. 2019. The SAUR gene family: the plant’s toolbox for adaptation of growth and development. J. Exp. Bot 70:17–27 [DOI] [PubMed] [Google Scholar]
- 120.Sun N, Wang J, Gao Z, Dong J, He H, et al. 2016. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. PNAS 113:6071–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, et al. 1999. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14–3–3 protein. Plant Cell 11:2379–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384–86 [DOI] [PubMed] [Google Scholar]
- 123.Takahashi K, Hayashi K, Kinoshita T. 2012. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 159:632–41 [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates auxin-induced increases in PM H+-ATPase C-terminal phosphorylation.
- 124.Talbott LD, Ray PM. 1992. Changes in molecular size of previously deposited and newly synthesized pea cell wall matrix polysaccharides: effects of auxin and turgor. Plant Physiol. 98:369–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, et al. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–45 [DOI] [PubMed] [Google Scholar]
- 126.Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tovar-Mendez A, Miernyk JA, Hoyos E, Randall DD. 2014. A functional genomic analysis of Arabidopsis thaliana PP2C clade D. Protoplasma 251:265–71 [DOI] [PubMed] [Google Scholar]
- 128.Uchida N, Takahashi K, Iwasaki R, Yamada R, Yoshimura M, et al. 2018. Chemical hijacking of auxin signaling with an engineered auxin-TIR1 pair. Nat. Chem. Biol 14:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]; Develops a synthetic auxin receptor system to demonstrate conclusively that TIR1/AFB receptors mediate acid growth.
- 129.Vanhaeren H, Gonzalez N, Coppens F, De Milde L, Van Daele T, et al. 2014. Combining growth-promoting genes leads to positive epistasis in Arabidopsis thaliana. eLife 3:e02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vi SL, Trost G, Lange P, Czesnick H, Rao N, et al. 2013. Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. PNAS 110:13994–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, et al. 2006. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 142:542–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Z, Yang L, Liu Z, Lu M, Wang M, et al. 2019. Natural variations of growth thermo-responsiveness determined by SAUR26/27/28 proteins in Arabidopsis thaliana. New Phytol. 224:291–305 [DOI] [PubMed] [Google Scholar]
- 133.Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol 67:539–74 [DOI] [PubMed] [Google Scholar]
- 134.Went FW. 1926. On growth accelerating substances in the coleoptile of Avena sativa. Proc. Kon. Akad. Wetensch 30:10–19 [Google Scholar]
- 135.Wildman SG. 1997. The auxin-A, B enigma: scientific fraud or scientific ineptitude? Plant Growth Regul. 22:37–68 [Google Scholar]
- 136.Wong JH, Spartz AK, Park MY, Du M, Gray WM. 2019. Mutation of a conserved motif of PP2C.D phosphatases confers SAUR immunity and constitutive activity. Plant Physiol. 181:353–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu G, Cameron JN, Ljung K, Spalding EP. 2010. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 62:179–91 [DOI] [PubMed] [Google Scholar]
- 138.Wu J, Liu S, He Y, Guan X, Zhu X, et al. 2012. Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 509:38–50 [DOI] [PubMed] [Google Scholar]
- 139.Würtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C. 2003. Structural view of a fungal toxin acting on a 14–3–3 regulatory complex. EMBO J. 22:987–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xia L, Marquès-Bueno MM, Bruce CG, Karnik RA. 2019. Unusual roles of secretory SNARE SYP132 in plasma membrane H+-ATPase traffic and vegetative plant growth. Plant Physiol. 180:837–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang T, Poovaiah BW. 2000. Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem 275:3137–43 [DOI] [PubMed] [Google Scholar]
- 142.Žádníková P, Wabnik K, Abuzeineh A, Gallemi M, Van Der Straeten D, et al. 2016. A model of differential growth-guided apical hook formation in plants. Plant Cell 28:2464–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zimmermann S, Thomine S, Guern J, Barbier-Brygoo H. 1994. An anion current at the plasma membrane of tobacco protoplasts shows ATP-dependent voltage regulation and is modulated by auxin. Plant J. 6:707–16 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


