SUMMARY
Stimulator of interferon genes (STING) is a key regulator of type I interferon and pro-inflammatory responses during infection, cellular stress, and cancer. Here, we reveal a mechanism for how STING balances activation of IRF3- and NF-κB-dependent transcription and discover that acquisition of discrete signaling modules in the vertebrate STING C-terminal tail (CTT) shapes downstream immunity. As a defining example, we identify a motif appended to the CTT of zebrafish STING that inverts the typical vertebrate signaling response and results in dramatic NF-κB activation and weak IRF3-interferon signaling. We determine a co-crystal structure that explains how this CTT sequence recruits TRAF6 as a new binding partner and demonstrate that the minimal motif is sufficient to reprogram human STING and immune activation in macrophage cells. Together, our results define the STING CTT as a linear signaling hub that can acquire modular motifs to readily adapt downstream immunity.
Graphical Abstract
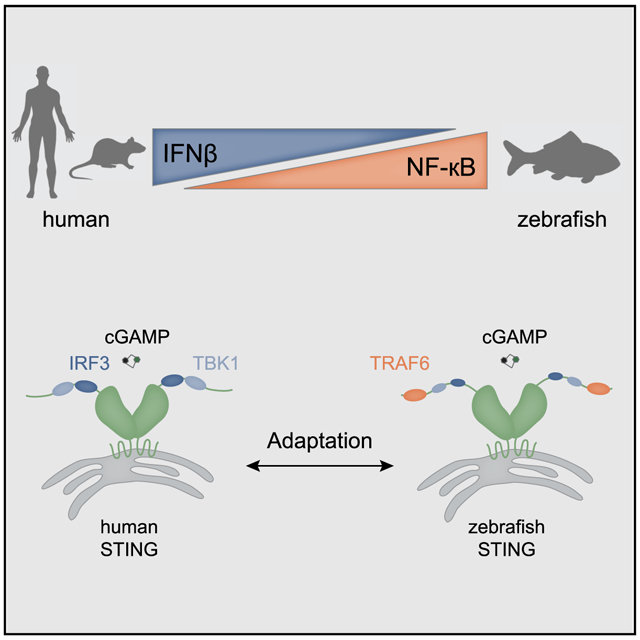
In Brief
de Oliveira Mann et al. define a mechanism that allows emergence of a signaling response in an innate immune pathway. Modular motifs in the STING CTT control the strength and specificity of downstream responses, and evolutionary acquisition of new signaling elements is facilitated by the linear arrangement of the CTT.
INTRODUCTION
A principal component of the human innate immune system is a series of receptors that detect mislocalized or modified nucleic acids and signal through discrete adaptor proteins. One key example is a cytosolic double-stranded DNA-sensing pathway controlled by the receptor cyclic GMP-AMP synthase (cGAS) (Sun et al., 2013). Following complex formation with DNA, cGAS is activated to enzymatically synthesize a 2′-5′, 3′-5′ cyclic GMP-AMP second messenger (2′3′ cGAMP). 2′3′ cGAMP binds to the adaptor stimulator of interferon genes (STING) and triggers a signaling cascade that culminates in activation of a downstream transcriptional program (Wu and Chen, 2014). STING signaling allows cells to mount a potent immune response to pathogen-, tumor-, and cell stress-derived cytosolic DNA, and STING is rapidly emerging as a key target for manipulation of antitumor immunity (Corrales et al., 2015; Fu et al., 2015). However, the transcriptional responses following STING activation are complex, and the mechanisms controlling STING signaling remain poorly understood.
STING forms a homodimeric complex that recognizes 2′3′ cGAMP and undergoes a large conformational change, enabling it to traffic from the endoplasmic reticulum through the Golgi apparatus. STING translocation permits recruitment of TANK-binding kinase (TBK1) and activation of the transcription factors IRF3 and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) (Haag et al., 2018; Tan et al., 2018). The mechanism for IRF3 recruitment, phosphorylation, and activation can be attributed to a specific sequence motif within the C-terminal tail of STING that is conserved among vertebrate STING species (Liu et al., 2015; Zhao et al., 2016). Consistent with robust TBK1 and IRF3 activation, STING signaling in human and mouse immune cells results in a predominant type I interferon response. However, evolution of the STING gene (TMEM173) predates the emergence of interferon signaling in vertebrates, and the STING motif responsible for IRF3 recruitment is not conserved in lower metazoans (Kranzusch et al., 2015). This downstream response is therefore a relatively recent evolutionary adaptation despite the importance of STING-dependent interferon activation in human cells. STING signaling in human cells also drives a weaker pro-inflammatory response through the transcription factor NF-κB, and it has been postulated that ancestrally related cGAS-STING pathways might have relied on distinct mechanisms of host defense involving NF-κB and autophagy (Goto et al., 2018; Kranzusch et al., 2015; Liu et al., 2018; Margolis et al., 2017; Martin et al., 2018). These models therefore suggest that plasticity is an important feature of the downstream responses of cGAS-STING immunity and that unknown mechanisms tune the strength and specificity of immunity across evolutionary time to match species-specific pathogen burdens.
Here we developed a phylogenetic screen for STING downstream signaling and discover a mechanism that allows emergence of a new signaling response in an innate immune pathway. We show that the STING C-terminal tail (CTT) is composed of discrete elements, with this modular architecture allowing acquisition of new motifs that rapidly adapt downstream signaling. We discover that a CTT extension conserved in the STING alleles of ray-finned fishes elicits a dramatic enhancement of NF-κB activation and that this motif is alone sufficient to remodel STING-dependent signaling in mammalian cells. A high-resolution co-crystal structure of the zebrafish STING CTT in complex with TRAF6 explains how this motif allows direct recruitment of a new signaling partner to remodel downstream immune activation. Our results define the STING CTT as a linear signaling hub and reveal how acquisition of modular motifs can adapt cGAS-STING immune responses throughout evolution.
RESULTS
STING C-Terminal Modules Control the Balance of Downstream IRF3 and NF-κB Signaling
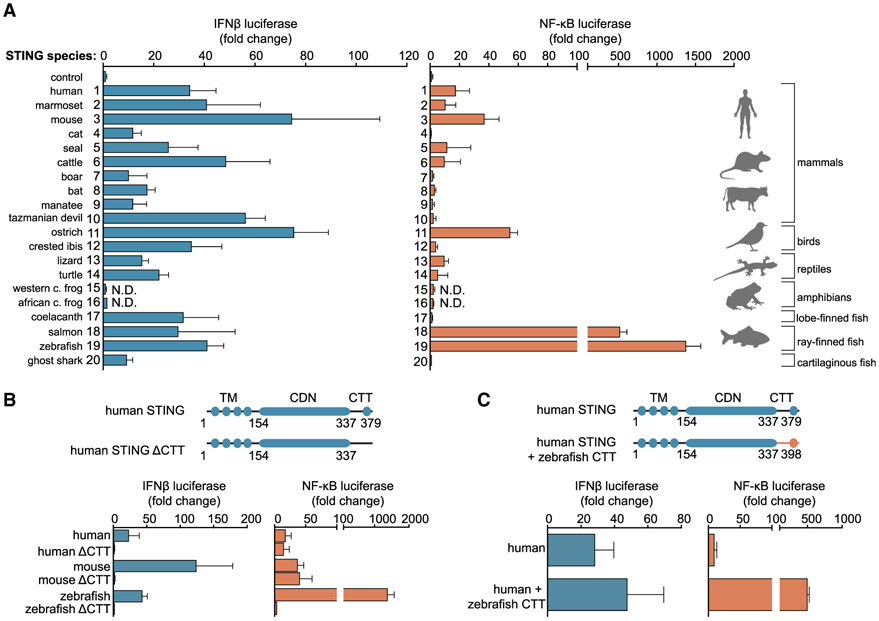
To define the STING protein motifs responsible for controlling downstream immune activation, we cloned a phylogenetically diverse panel of 20 vertebrate STING (TMEM173) genes and screened for altered signaling in human cells. Nearly all STING variants tested are expressed and able to induce both IRF3- and NF-κB-driven promoters in a cGAS-dependent manner (Figure 1A; Figures S1, S2A, and S2B). Levels of IRF3-interferon signaling induction were constant in 18 of the 20 STING genes tested, with the lone exception being amphibian STING proteins from Xenopus tropicalis and Xenopus laevis, for which no expression or downstream activation could be detected. The broad conservation of IRF3-interferon signaling demonstrates that the elements required for STING localization, response to the cGAS product 2′3′ cGAMP, and activation of downstream kinase and transcription factor components are conserved in vertebrate biology. Unexpectedly, the levels of NF-κB reporter induction were highly variable between different animal STING proteins (Figure 1A). Consistent with observations in primary immune cells (Fang et al., 2017; Ishikawa and Barber, 2008; Zhong et al., 2008), human and mouse STING alleles induced a strong IRF3 response (~40- to 60-fold reporter signaling) and a comparatively weak NF-κB response (~15-fold reporter signaling). In contrast, activation of STING alleles from the fish species Danio rerio (zebrafish) and Salmo salar (salmon) in human cells results in robust activation of a primarily NF-κB-driven response with more than 100-fold higher stimulation of the NF-κB reporter compared with the IRF3 reporter.
Figure 1. STING C-Terminal Modules Control the Balance of Downstream IRF3 and NF-κB Signaling.
(A) Reconstitution of the cGAS-STING pathway in human cells using a phylogenetically diverse panel of vertebrate STING alleles. Luciferase reporters were used to monitor cGAS-STING dependent interferon β (IFNβ blue) and NF-κB (orange) responses. Species shown are as follows: 1, human (Homo sapiens); 2, marmoset (Callithrix jacchus); 3, mouse (Mus musculus); 4, cat (Felis catus); 5, seal (Leptonychotes weddellii); 6, cattle (Bos taurus); 7, boar (Sus scrofa); 8, bat (Rousettus aegyptiacus); 9, manatee (Trichechus manatus latirostris); 10, Tasmanian devil (Sarcophilus harrisii); 11, ostrich (Struthio camelus australis); 12, crested ibis (Nipponia nippon); 13, lizard (Anolis carolinensis); 14, turtle (Chelonia mydas); 15, western clawed frog (Xenopus tropicalis); 16, African clawed frog (Xenopus laevis); 17, coelacanth (Latimeria chalumnae); 18, salmon (Salmo salar); 19, zebrafish (Danio rerio); and 20, ghost shark (Callorhinchus milii).
(B) Schematics of STING domain organization (TM, transmembrane domain; CDN, cyclic dinucleotide binding domain; CTT, C-terminal tail). Cellular reporter assay as in (A), mapping the motif responsible for enhanced NF-κB signaling to the CTT of zebrafish STING.
(C) Cellular reporter assay and schematics as in (B), comparing downstream signaling outputs of human STING with human STING containing the zebrafish STING CTT sequence. The zebrafish STING CTT is sufficient to enhance NF-κB signaling.
Cellular reporter assay data are representative of at least three independent experiments. Data are plotted with error bars representing the SD of the mean.
To map the zebrafish STING determinant required for altering the balance of NF-κB and IRF3 responses, we next created a series of STING truncations and measured signaling of each pathway (Figure 1B). The STING protein includes four predicted transmembrane segments, followed by a cytosolic cyclic-dinucleotide (CDN) binding domain that forms a homodimeric V-shaped receptor. At the extreme C terminus of the CDN-binding domain of human and mouse STING is an ~40 amino acid CTT that includes the IRF3 binding site (Liu et al., 2015; Zhao et al., 2016). In agreement with previous human STING studies, deletion of the CTT prevented IRF3-interferon activation for all alleles in our panel, including human and zebrafish STING (Figure 1B; Figure S2C) (Fang et al., 2017; Liu et al., 2015). Additionally, CTT deletion prevented zebrafish STING-dependent NF-κB activation (Figure 1B), suggesting that hyperactivation of NF-κB signaling may be due to a function present in zebrafish STING that is not present in human STING. In support of this model, human-zebrafish STING chimeras confirmed that the zebrafish STING CTT is sufficient to hyperactivate NF-κB reporters when fused to human STING (Figure 1C). Additionally, the STING allele in our screen from the most primitively diverged vertebrate lineage (Callorhinchus milii, ghost shark) contains a human-like CTT and does not exhibit heightened NF-κB activation (Figure 1A). Together, these results suggest that STING-dependent IRF3-interferon and NF-κB signaling is controlled through independent modules in the CTT that can be gained or lost to balance downstream immune activation.
Zebrafish STING Contains a Unique CTT Module that Boosts NF-κB Signaling
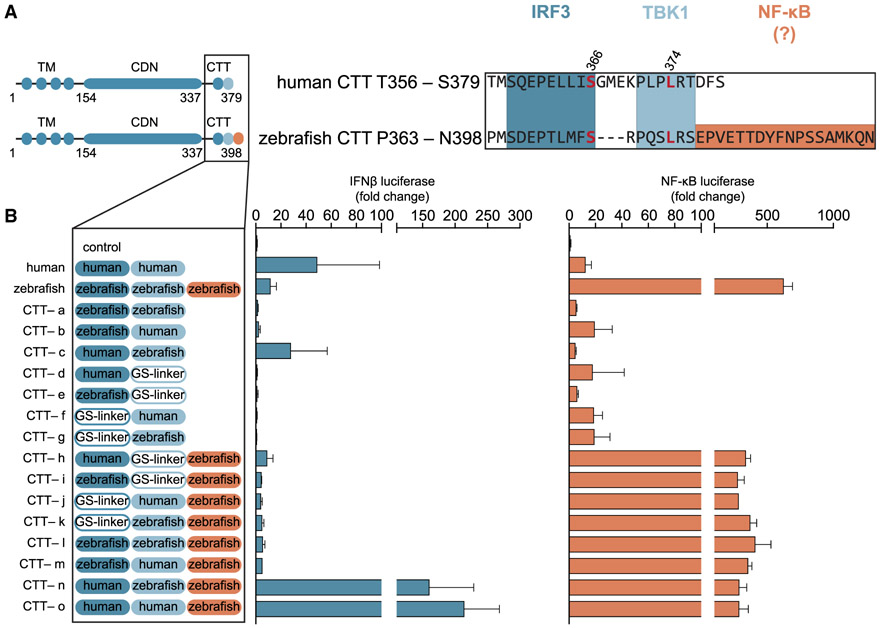
The STING CTT is an unstructured stretch of ~40 amino acids that contains sequence motifs required for STING phosphorylation and recruitment of IRF3. Previous experiments defined the human STING residue S366 as a primary TBK1 phosphorylation site that is part of an LxIS motif shared between innate immune adaptor proteins that activate interferon signaling (Liu et al., 2015; Tanaka and Chen, 2012; Zhao et al., 2016). Additionally, the human STING CTT contains a second PxPLR motif that includes the residue L374, which is required for TBK1 binding (Tanaka and Chen, 2012; Zhang et al., 2019). The LxIS and PxPLR sequences are highly conserved in all vertebrate STING alleles, and we refer to these as the IRF3 and TBK1 binding motifs, respectively (Figure 2A). The zebrafish STING CTT contains a further extension not present in human and mammalian STING alleles (Figure 2A). To determine whether this fish-specific STING sequence is responsible for the elevated NF-κB reporter activation, we constructed a series of CTT chimeras fused to the core human STING protein, designed to test the effect of inactivation of individual motifs (mutation to a glycine-serine-rich linker sequence, called the GS-Linker), and the importance of species specificity of motif conservation (human STING or zebrafish STING sequence) (Figure 2B; Figure S3B). We confirmed that the human STING chimeras retained robust expression in human cells (Figure S3A). All chimeras containing the new fish-specific STING CTT module DPVETTDY induced more than 100-fold activation of the NF-κB reporter, demonstrating that this region of the CTT sequence is both necessary and sufficient to direct enhanced NF-κB signal activation (Figure 2B). Additionally, enhanced NF-κB signaling is independent of IRF3-interferon activation. Mutation of the IRF3 and TBK1 binding modules blocks all interferon activation, but inactivation of these modules has no effect on zebrafish STING CTT-induced NF-κB signaling (Figure 2B). Interestingly, the zebrafish STING CTT module can also mediate hyperactivation of IRF3 reporter signaling, but only in the presence of the human STING IRF3 binding module (Figure 2B), suggesting that cross-talk between individual CTT modules affects the balance of overall STING signaling.
Figure 2. The Zebrafish STING Contains a Unique CTT Module that Boosts NF-κB Signaling.
(A) Schematics of STING chimera CTT constructs and sequences of human STING T356-S379 and zebrafish STING P363-N398 aligned using Jalview and with three highlighted signaling motifs mapped for type I interferon (dark and light blue) and NF-κB (orange). The human STING residues S366 and L374 required for interferon signaling are marked in red.
(B) Reconstitution of the cGAS-STING pathway in human cells using different human-zebrafish STING chimera constructs. Interferon (blue) and NF-κB (orange) reporter activity was measured as in Figure 1A. All STING CTT chimeras consist of a human core domain with three variable CTT modules (dark blue, light blue, and orange) from human and zebrafish STING, respectively (constructs CTT-a to CTT-o).
Cellular reporter assay data are representative of at least three independent experiments. Data are plotted with error bars representing the SD of the mean.
Activation of the interferon β (IFNβ) promoter involves recruitment of transcription factors to distinct positive regulatory domains (PRDs), including PRD III-I (IRF3 binding sites), PRD II (NF-κB binding site), and PRD IV (ATF-2-c-Jun binding site) (Fitzgerald et al., 2003). To further define the mechanism of enhanced signaling driven by the zebrafish STING CTT DPVETTDY motif, we next tested the ability of STING chimeras to activate transcription from individual regulatory elements in the IFNβ promoter (Figure S3C; Table S1). In agreement with our IFNβ and NF-κB promoter results, human STING and zebrafish STING primarily drive IRF3 and NF-κB responses that are only dependent on the PRD III-I (IRF3) and PRD II (NF-kB) binding sites. In contrast, the chimera STING CTT-o, containing both human IRF3 and TBK1 binding modules and the zebrafish NF-κB module, was now capable of activating promoters containing either the IRF3- or NF-κB-responsive elements.
The results from these chimeras confirm a conserved function for the IRF3 and TBK1 binding motifs in IRF3 activation and define two new rules that govern the role of the STING CTT in signal induction. First, the STING CTT functions as an assembly of modular elements, and new motifs can be acquired to reshape downstream signaling. The fish-specific STING CTT readily enhances signaling in all tested STING backgrounds and can function independently of already existing STING CTT motifs. Second, STING CTT motifs can be tuned with species-specific substitutions to refine the potency of downstream signal activation. Hyperactivation by the zebrafish STING CTT extends to IRF3 recruitment only in the presence of the human STING IRF3 binding motif, suggesting that the atypical I365 (human STING) > F372 (zebrafish STING) substitution in the zebrafish IRF3 binding motif may have evolved as a compensatory mutation to fine-tune IRF3-mediated signaling following acquisition of a NF-κB hyperactivating module. These results reveal a modular organization to the STING CTT and a low evolutionary bar to adaptations that transform STING downstream innate immune responses.
The Zebrafish STING CTT Module Directly Recruits TRAF6 to Activate NF-κB Signaling
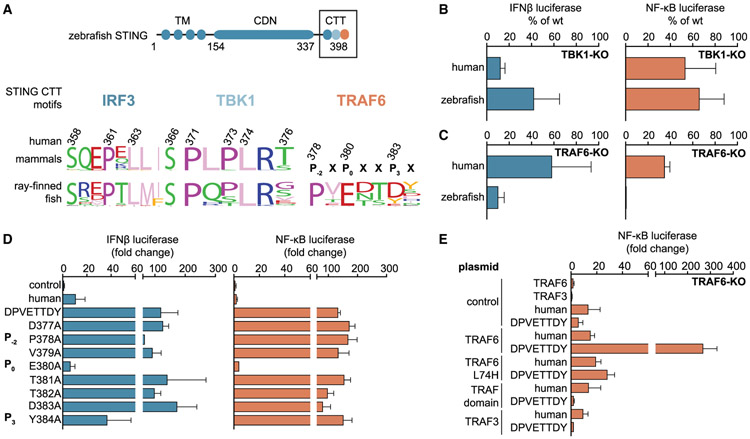
Following recognition of the cGAS product 2′3′ cGAMP, human STING activation requires recruitment of the kinase TBK1. TBK1 is required for STING-dependent IRF3-interferon signaling and has also been implicated in STING NF-κB signaling (Abe and Barber, 2014; Fang et al., 2017). Therefore, we next asked whether the elevated NF-κB response directed by the zebrafish STING CTT was dependent on TBK1. CRISPR/Cas9-directed knockout of TBK1 inhibited nearly all interferon signaling for human STING but had only a partial effect on zebrafish STING, reducing interferon signaling to ~40% of wild-type cell levels (Figure 3B; Figure S3D) However, NF-κB signaling persisted with both human and zebrafish STING alleles, revealing that another unknown downstream factor is required for zebrafish STING-dependent NF-κB signaling (Figure 3B).
Figure 3. The Zebrafish STING CTT Module Directly Recruits TRAF6 to Activate NF-κB Signaling.
(A) Schematics of zebrafish STING and sequence logo of the highlighted STING CTT signaling factor recruitment motifs of mammals compared with ray-finned fish species (9 amino acids [aa]; 6 aa for interferon signaling and an additional 7 aa in ray-finned fish for NF-κB signaling). Amino acids are numbered according to the human STING sequence.
(B) Reporter assay for IFNβ (blue) or NF-κB (orange) using human and zebrafish STING in TBK1 knockout (KO) HEK293T cells. IFNβ signaling is lost in TBK1 KO cells. However, the NF-κB signaling responses for human and zebrafish STING persist in TBK1 KO cells.
(C) Reporter assay as in (B), performed in TRAF6 KO HEK293T cells. All zebrafish STING-dependent NF-κB signaling is abolished in TRAF6 KO cells.
(D) Reporter assay as in (B), performing alanine scan mutagenesis of the TRAF6 binding motif in the zebrafish STING CTT chimera containing a human STING core domain. Consistent with other TRAF6 recruitment motifs, only the P0 position is essential for STING-TRAF6 complex formation.
(E) Cellular assay as in (B), monitoring STING-dependent signaling in the presence of alternative TRAF6 alleles. Endogenous TRAF6 was removed (TRAF6 KO), and signaling was reconstituted with plasmids encoding alternative TRAF6 and TRAF3 alleles as indicated. Disruption of the TRAF6 E3 ligase domain (L74H) or expression of only the TRAF domain prevents zebrafish STING-dependent NF-κB signaling.
Cellular reporter assay data are representative of at least three independent experiments. Data are plotted with error bars representing the SD of the mean.
To determine the missing factor responsible for zebrafish STING NF-κB activation, we used phylogenetic analysis to search for a new CTT motif present in the STING alleles of ray-finned fish species. Sequence logos generated for each region of the STING CTT revealed that, in addition to conservation of the IRF3 and TBK1 binding motifs, all ray-finned fish species STING proteins contain a highly conserved PxExxD motif at the extreme C terminus that is not found in mammalian STING (Figure 3A). Homology searches revealed that the consensus fish PxExxD motif shares similarity with tumor necrosis factor receptor-associated factor (TRAF) binding sites (McWhirter et al., 1999; Park et al., 1999; Shi et al., 2015b; Ye et al., 2002). In agreement with a potential role in STING NF-κB signaling, TRAFs are adaptor proteins involved in a variety of NF-κB signaling transduction events in adaptive immunity, innate immunity, and tissue homeostasis (Wu, 2004). Comparison of the zebrafish STING sequence with known TRAF-interacting proteins revealed the closest homology with TRAF6 recruitment motifs (Ye et al., 2002). Using CRISPR/Cas9-mediated knockout, we confirmed that TRAF6 is essential for zebrafish STING-induced NF-κB reporter activation (Figure 3C; Figures S3D-S3F). Both NF-κB and interferon signaling downstream of zebrafish STING are dependent on TRAF6, consistent with previous reports that TRAF6 has unique sequence preferences not shared by the other TRAF proteins (Darnay et al., 1999; Pullen et al., 1998). In contrast, human STING NF-κB and IRF3 reporter activation is reduced by only ~60% and ~40% compared with induction in wild-type cells, indicating that the role of TRAF6 in cGAS-dependent human STING signaling is non-essential (Figure 3C). Interestingly, TRAF6 was recently described in a cGAS-independent pathway of STING-mediated NF-κB signaling (Dunphy et al., 2018), suggesting that utilization of TRAF proteins in STING signaling is both species- and context-specific. Together, our results define an NF-κB motif in the STING alleles of fish species acquired to specifically recruit TRAF6 and alter downstream signaling responses and reveal that TRAF6 plays distinct species-specific roles in STING signaling and NF-κB activation.
The zebrafish STING NF-κB motif exhibits all key features identified previously through analysis of TRAF6-interacting peptides from the human and mouse immune proteins cluster of differentiation 40 (CD40), receptor activator of NF-κB (RANK), and mitochondrial antiviral signaling adaptor protein (MAVS) (Shi et al., 2015b; Ye et al., 2002). Accordingly, we denote the conserved residues in the zebrafish STING TRAF6 motif, P378 (P−2), E380 (P0), and D383 (P3) (Figure 3A), and used alanine mutagenesis to confirm that the anchor position, E380 (P0), is essential for downstream NF-κB activation (Figure 3D). TRAF6 signaling relies on an N-terminal RING domain that acts as an E3 ubiquitin ligase and catalyzes formation of the K63-linked polyubiquitin chains required to recruit downstream NF-κB signaling components (Yin et al., 2009). To test the role of this E3 ligase activity in zebrafish STING signaling, we reconstituted TRAF6 knockout cells with full-length TRAF6 and TRAF6 mutants. Expression of the TRAF domain alone does not permit zebrafish STING-dependent downstream signaling, indicating that the TRAF6 N-terminal RING domain is required for enhanced NF-κB activation (Figure 3E). Likewise, a single point mutation in the N terminus of TRAF6 (L74H) that prevents interaction with E2-conjugating enzymes is sufficient to ablate zebrafish STING NF-κB signaling (Yin et al., 2009; Figure 3E). Expression of wild-type TRAF3 is unable to restore NF-κB signaling, further confirming that the zebrafish STING CTT is specific for recruitment of TRAF6 (Figure 3E). Together, these data demonstrate that zebrafish STING gained the ability to initiate elevated NF-κB responses through recruitment of the canonical TRAF6 signaling complex.
Structural Basis of Zebrafish STING-TRAF6 Complex Formation
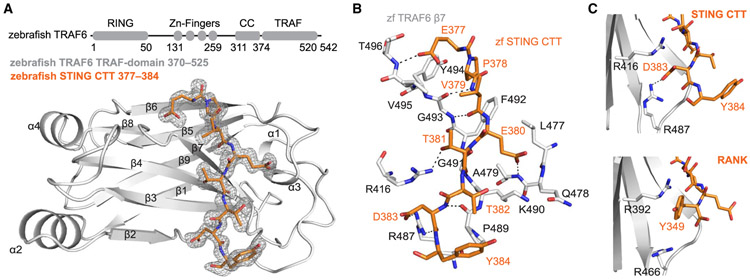
Structure-based alignments of TRAF6-interacting protein complexes have demonstrated that TRAF6 recognizes peptide sequences that contain three conserved positions: proline at P−2, glutamate at P0, and an aromatic or acidic residue at P3 (Wu, 2004). However, all structures of TRAF6-peptide complexes to date contain peptides with an aromatic residue at position P3. The zebrafish STING CTT NF-κB motif contains an acidic residue, D383, at position P3, and it remains unknown how TRAF6 is able to recognize this class of interacting motif. To define the structural basis of the zebrafish STING-TRAF6 interaction and the mechanism of P3 acidic residue recognition, we determined a 1.4-Å co-crystal structure of the TRAF-C domain of zebrafish TRAF6 bound to the zebrafish STING NF-κB peptide 377-EPVETTDY-384 (Table S2). The TRAF-C domain of zebrafish TRAF6 shares 66% amino acid sequence identity with human TRAF6 and adopts the canonical eight-stranded anti-parallel β sandwich observed for all other TRAF structures (Figure 4A; Figure S4A). The zebrafish STING NF-κB peptide makes 13 intermolecular hydrogen bonds between side chains and along the TRAF6 b7 strand to extend the core TRAF6 second β sheet and is located in the same shallow groove as observed previously for TRAF6-interacting peptides containing an aromatic position at P3. The zebrafish STING P378 (P−2) residue binds in a hydrophobic pocket specific to TRAF6 proteins formed by Y494, F492, and M471 (human Y473, F471, and M450). Mutagenesis of the human TRAF6 residues F471 and Y473 leads to decreased NF-κB signaling, supporting the importance of these hydrophobic interactions with the conserved proline at position P−2 (Figure S4B). Zebrafish STING E380 at position P0 is recognized by hydrogen bonds with the main chain of TRAF6 residues A479 and Q478 (human A458 and L456), holding the loop between β6 and β7 in place (Figure 4B). The acidic D383 P3 position of zebrafish STING is recognized by TRAF6 R487 (human R466), which re-orients to form a hydrogen bond interaction with the acidic side chain. This interaction is distinct from the previously observed pi-stacking interaction between TRAF6 R416 (human R392) and the aromatic residues at the P3 position of the RANK and MAVS complexes (Figure 4C). Importantly, all zebrafish TRAF6 residues that interact with the zebrafish STING NF-κB peptide are conserved among vertebrate TRAF6 proteins. These results define the molecular basis of how TRAF6 adapters recognize sequences with an acidic residue at the P3 position and confirm the specificity of TRAF6-recruitment in zebrafish STING signaling adaptation.
Figure 4. Structural Basis of Zebrafish STING-TRAF6 Complex Formation.
(A) Schematics of zebrafish TRAF6 domain organization (RING domain, zinc fingers, coiled coil [CC], and TRAF domain) and the co-crystal structure of the zebrafish TRAF6 C-domain (light gray) in complex with the zebrafish STING CTT NF-κB motif (orange) at 1.4Å. The STING CTT peptide 2Fo–Fc electron density is shown in gray and contoured at 1.0 σ.
(B) Interaction map showing the interface between TRAF6 and the zebrafish STING CTT peptide. Main-chain hydrogen bonds between the STING CTT (orange) and the TRAF6 β7 residues (light gray) are shown as dotted lines.
(C) Comparison of TRAF6 (light gray) interactions with aromatic or acidic residues at peptide position P3 (orange). STING has an acidic D383 at position P3 that is recognized by an arginine residue at the bottom of the pocket (human TRAF6 R466, zebrafish TRAF6 R487), whereas RANK (PDB: 1LB5) has an aromatic residue, Y349, at the same position that interacts with a distinct arginine at the top of the pocket (human TRAF6 R392, zebrafish TRAF6 R416).
Acquisition of the STING TRAF6 Module Remodels Downstream Immune Responses
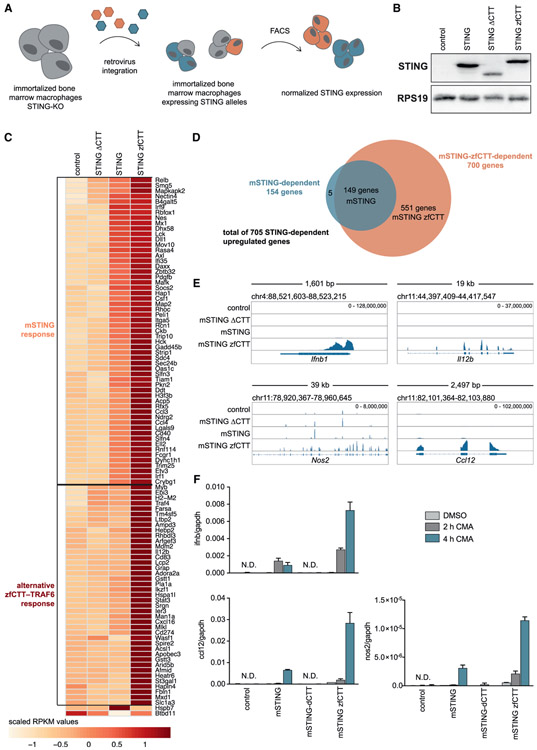
To define the downstream signaling effect of acquisition of a new motif in the STING CTT, we next analyzed the effect of chimeric STING alleles on transcription induction in macrophage immune cells. We first verified that the zebrafish TRAF6 recruitment motif stimulates NF-κB signaling when fused to mouse STING (Figure S5A) and then transduced STING−/− murine macrophages with mouse STING alleles encoding no CTT, the wild-type mouse CTT, or a chimeric CTT including the zebrafish TRAF6-recruitment motif (Figures 5A and 5B). RNA sequencing results of the different macrophage cell lines stimulated with the STING-specific agonist CMA (10-carboxymethyl-9-acridanone) (Cavlar et al., 2013) revealed 705 upregulated genes divided into two major sets (Figures 5C and 5D; Table S3). The first major set includes 154 genes that are induced in the presence of the wild-type STING CTT and further upregulated in macrophages encoding the chimeric STING allele with the zebrafish TRAF6-interacting motif (Figures 5C and 5D). These genes include the IFNb transcript and are highly enriched in known STING-responsive interferon-stimulated genes (Abe et al., 2013; Surpris et al., 2016), demonstrating that one effect of acquisition of the zebrafish CTT motif is enhancement of the existing basal STING signaling response (Figures 5C-5E). In addition to enhancing the basal STING response, addition of the zebrafish TRAF6 recruitment motif results in upregulation of a second set of 551 genes that are not induced upon wild-type STING activation (Figures 5C and 5D; Figure S5C). This second set includes many known immune-related cytokines and chemokines, including Il12b, Il1b, Ccl7, and Nos2. Consistent with the zebrafish TRAF6 recruitment motif enhancing engagement of NF-κB signaling, analysis of the promoters associated with this second gene set demonstrates significant enrichment for NF-κB (RelA, Rel, NFKB1, and NF-κB) family member binding sites (Figure S5B). Furthermore, known targets like Il12b have been shown previously to be specifically dependent on TRAF6 activation (Han et al., 2017; Mason et al., 2004). We confirmed these results for three upregulated genes (Ifnb, Nos2, and Ccl12) using qRT-PCR (Figures 5E and 5F). Therefore, acquisition of the zebrafish TRAF6 recruitment motif results in two distinct effects on downstream immune signaling. First, appending the new motif increases the magnitude of the basal STING interferon response. Second, direct recruitment of TRAF6 to the CTT results in induction of an additional set of NF-κB-dependent genes that are not normally induced upon wild-type STING activation. These results define a mechanism where the STING signaling response is controlled through CTT motifs that discretely activate specific gene sets and synergistically combine to control the strength of the downstream signaling response.
Figure 5. Acquisition of the STING TRAF6 Module Remodels Downstream Immune Responses.
(A) Schematic representing generation of mouse macrophage cell lines with different STING gene alleles.
(B) Western blot analysis of STING expression in different mouse macrophage cell lines compared with the RPS19 loading control. Data are representative of three biological experiments.
(C) Heatmap of reads per kilobase of transcript per million mapped reads (RPKM) of 100 differentially upregulated genes in mouse macrophage cell lines: control, mouse STING (mSTING) ΔCTT, mSTING, and mSTING zebrafish C-terminal tail (zfCTT). Acquisition of the zebrafish STING CTT enhances the STING-dependent interferon response and additionally activates alternative NF-κB responses.
(D) Venn diagram depicting upregulated genes, the normal mouse STING response, and the alternative mSTING zfCTT response.
(E) Visualization of RNA sequencing (RNA-seq) gene coverage and transcript levels for the selected differentially expressed genes Ifnb, Il12b, nos2, and ccl12 in macrophage cell lines: control, mSTING ΔCTT, mSTING, and mSTING zfCTT using an integrative genomics viewer (IGV) (Robinson et al., 2011).
(F) Transcript-level analysis of selected genes by qRT-PCR. qRT-PCR data are representative of three independent experiments.
Data are plotted with error bars representing the SD of the mean.
DISCUSSION
Our results define a mechanism of STING signal activation where modular motifs in the receptor CTT control the strength and specificity of downstream immune responses. The linear arrangement of motifs in the STING CTT tunes the balance between IRF3 and NF-κB signaling and permits evolutionary acquisition of new signaling elements. We demonstrate that an extension in the zebrafish STING CTT encodes a noncanonical TRAF6 recruitment motif that dramatically upregulates STING-dependent NF-κB signaling. Transfer of this zebrafish STING motif to human or mouse STING is sufficient to recruit the canonical TRAF6 signaling complex and expand the range of genes induced upon STING activation (Figure 5).
Cyclic dinucleotide binding triggers a conformational change in STING, but it remains unclear how activation of the dimeric receptor core allows accessibility of the unstructured CTT for selective recruitment of downstream signaling factors (Gao et al., 2013; Huang et al., 2012; Kranzusch et al., 2015; Ouyang et al., 2012; Shang et al., 2012; Shi et al., 2015a; Shu et al., 2012; Yin et al., 2012; Zhang et al., 2013). A current model in the field is that STING conformational change initiates receptor trafficking and re-localization of STING to a perinuclear compartment that then allows clustering and signal activation (Haag et al., 2018; Tan et al., 2018). A recent cryoelectron microscopy (cryo-EM) structure of a hybrid chicken STING-human TBK1 complex provides direct data demonstrating that oligomerization of both proteins is required for STING CTT phosphorylation (Zhang et al., 2019). Our data provide further support of this model and suggest that the mechanism of STING clustering is evolutionarily conserved. First, nearly all tested vertebrate STING alleles readily activate immune signaling in human cells, demonstrating that the molecular mechanism of STING signaling is shared in vertebrates. Second, TRAF6 recruitment is known to be specifically dependent on motif clustering (Baud et al., 1999; David et al., 2018; Hu et al., 2017; Lin et al., 2010; Liu et al., 2013; Takamatsu et al., 2013), and the ability of zebrafish STING to directly recruit TRAF6 to the CTT is consistent with acquisition of a linear motif into an immune signaling pathway that already undergoes ligand-mediated receptor clustering. Notably, the IRF3 and TRAF6 recruitment motifs in the zebrafish STING CTT are located in close proximity, and it is not physically possible for IRF3 and TRAF6 to bind to the same STING CTT. STING clustering provides a mechanism to re-localize multiple CTTs in close proximity and allows simultaneous activation of downstream signaling factors.
Acquisition of a TRAF6 recruitment element in STING and retention in all currently sequenced extant ray-finned fish species suggests a genetic event where an alternative STING response provided a major selective advantage. One possible explanation is that elevated NF-κB activation shifts the STING response to protect from a pathogen not normally susceptible to cGAS-STING immunity. Consistent with this model, STING signaling is typically associated with a canonical type I interferon response, whereas Toll-like receptor (TLR)-dependent activation of TRAF6 induces a pro-inflammatory response (Scumpia et al., 2017). Interestingly, several groups have previously noted that primitive STING receptors in insects and early metazoans completely lack the CTT signaling elements (Goto et al., 2018; Kranzusch et al., 2015; Liu et al., 2018; Margolis et al., 2017; Martin et al., 2018). Emergence of the unstructured STING CTT in vertebrates coincides with development of interferon signaling. Interestingly, in addition to our discovery of gain of a signaling module in the CTT in fish species, deletion or mutation of CTT modules appears to have driven evolutionary loss of STING-dependent interferon signaling in amphibians and bats (Figure 1; Xie et al., 2018). Therefore, a key feature of the STING CTT is a modular signaling platform that readily accommodates both gain or loss of discrete signaling motifs.
Innate adaptor immune signaling is defined by two major components: activation of distinct transcription factors followed by refinement of the respective immune signaling strength. STING is one of three major immune adaptor proteins, including MAVS and TIR domain-containing adaptor-inducing IFNβ (TRIF). Following upstream receptor activation, these adaptors function as a point of convergence that provides a foundation for kinase and transcription factor recruitment and downstream immune activation of type I interferon and proinflammatory cytokine signaling (Wu and Chen, 2014). Our discovery of the modular architecture of the STING CTT explains how adaptor proteins in general can gain discrete motifs over evolutionary time to activate pre-existing pathways to rapidly re-wire immune responses tailored to the invading pathogen.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Philip J. Kranzusch (philip_kranzusch@dfci.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Escherichia coli Strains
Recombinant zebrafish TRAF6 protein was expressed in E. coli BL21-RIL DE3 (Agilent) bacteria harboring a pRARE2 tRNA plasmid. Transformations and starter cultures were grown in MDG media (0.5% glucose, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5mMNa2SO4, 2 mM MgSO4, 0.25% aspartic acid, 100 mg mL−1 ampicillin, 34 mg mL−1 chloramphenicol, and trace metals) overnight at 37 °C and used to seed 1 L cultures grown in M9ZB media (0.5% glycerol, 1% Cas-Amino Acids, 47.8 mM Na2HPO4, 22 mM KH2PO4, 18.7 mM NH4Cl, 85.6 mM NaCl, 2 mM MgSO4, 100 mg mL−1 ampicillin, 34 mg mL−1 chloramphenicol, and trace metals) (Studier, 2005). M9ZB cultures were cultivated at 37 °C until OD600 of 1.5–2.5, cooled on ice for 20 min, and then recombinant protein synthesis was induced by supplementation with 0.5 mM IPTG. Cultures were incubated at 16 °C with shaking for approximately 16 hours before harvest.
Cell Culture and Recombinant Cell Lines
HEK293Ts and Immortalized bone marrow derived macrophages (iBMDMs) were cultured in DMEM supplemented with 10% FBS at 37°C in 5% CO2. HEK293T cells were passaged at a 1:10 dilution by washing with HBSS and lifted with 0.05% trypsin. iBMDMs were lifted using HBSS supplemented with 2.5 mM EDTA and passaged at a 1:10 dilution. Sting−/− iBMDMs were derived as part of a previous study (Tan and Kagan, 2019), the sex of the cell line is not known.
METHOD DETAILS
Cloning
STING genes from different species were chemically synthesized (IDT) with≥18 base pairs of homology flanking the insert sequence and ligated into a BamHI / NotI linearized custom pcDNA4 vector (Kranzusch et al., 2013) by Gibson assembly. STING CTT chimeras were cloned by primer-annealing into the pcDNA4 vector and single point mutations were generated by standard mutagenesis techniques using Q5 DNA Polymerase (NEB).
CRISPR/Cas9-mediated knockout cell line generation
HEK293T cells were transfected with two different gRNAs in the U6-SpyM1-sgRNA vector expressing SpyCas9 (kind gift from L. Harrington and J. Doudna, UC-Berkeley). Guides were designed specific for early coding exons of the human TBK1 and TRAF6 genes. After 48 h cells were selected with puromycin (1 μg mL−1) and serial dilution method was performed to seed single cells into 96-well plates. After 10 days, visible growing clones were selected by bright-field microscopy and transferred into 24-wells. TBK1 and TRAF6 knockout efficiency were tested by western blot analysis. Each experiment was performed in biological triplicates with two selected clones.
Generation of STING mouse macrophage cell lines
mSTING, mSTING DCTT and mSTING zfCTT genes were cloned into the MSCV2.2 retroviral expression construct upstream of an internal ribosome entry site (IRES)-GFP. Retrovirus was generated by transfection of HEK293T cells with pCL-Eco, VSV-G and the respective MSCV2.2-STING-IRES-GFP vector. As a control retrovirus with empty pMSCV2.2 IRES-GFP vector was generated. After 48 h virus-containing cell supernatants were collected and filtered over 0.2 μm before transducing Sting−/− immortalized mouse bone marrow macrophages. Stable STING expressing macrophages were sorted for GFP expression. To normalize STING expression levels across different STING-KI cell lines, macrophages were sorted a second time for equal levels of GFP expression.
Cell-based Interferon β and NF-κB Luciferase Assay
Cellular reporter assays were performed as previously described (Kranzusch et al., 2015). Briefly, HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) in a 96-well format with Renilla luciferase (2 ng), a reporter plasmid expressing either an interferon β or NF-κB inducible firefly luciferase (20 ng), cGAS (20 ng) and different STING variants (15 ng). Luciferase activity was assessed 16 h post transfection and firefly luciferase activity was normalized to Renilla. As a control empty vector (15 ng) instead of STING was used. For Figure S3C HEK293T cells were transfected as before using the individual enhancer element reporter plasmids: pLuc-IFNβ, pLuc-PRD(III-I)3, pLuc-PRD(II)2 and pLuc-PRD(IV)6 (kind gift from K. Fitzgerald, University of Massachusetts Medical School) (Fitzgerald et al., 2003). For Figure S3E HEK293T cells were transfected with Renilla luciferase (2 ng), a reporter plasmid expressing NF-κB inducible firefly luciferase (20 ng) and empty vector (15 ng). 14 h post transfection cells were treated with 10 ng μL−1 or 20 ng μL−1 TNF-α (Biolegend) for 6 h before measuring luciferase activity. Data are mean ± standard deviation from 3 replicates and are representative of 3 independent experiments.
Antibodies and Western Blot analysis
Cells were lysed in Laemmli buffer and denatured at 95 °C for 10 min. Cell lysates were separated by 10% or 12% SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Western blot analysis was performed using the following antibodies: high affinity anti-hemagglutinin (Sigma, anti-HA-Peroxidase, rat mAb 3F10), rabbit anti-TBK1/NAK (Cell Signaling-3013), rabbit anti-TRAF6 (abcam-EP591Y ab33915), rabbit anti-STING (Cell Signaling-D2P2F), anti-RPS19 (Bethyl-A304-002A) and as secondary antibody anti-rabbit-HRP (GE Healthcare-NA934).
Protein Expression and Purification
Zebrafish TRAF6 TRAF-C domain (370–525) gene sequence was synthesized (IDT) and cloned into a modified pET16 vector with an N-terminal 6 × His-MBP-SUMO2-tag. Expression was performed in E. coli BL21-RIL DE3 (Agilent) bacteria harboring a pRARE2 tRNA plasmid using MDG media in pre-cultures and M9ZB media in large-scale cultures (2 × 1 L). Cells were induced with 0.5 mM IPTG after reaching an OD600 1.0–1.5 and protein was expressed at 18 °C for 18 h. Harvested cells were re-suspended in lysis buffer (20 mM HEPES-KOH pH 7.5, 400 mM NaCl, 30 mM imidazole, 10% glycerol, 1 mM DTT) and disrupted by sonication. Lysate was clarified by centrifugation and subsequent filtration through glass wool. Recombinant TRAF6 TRAF-C domain protein was purified by nickel-affinity chromatography (QIAGEN) and the His-MBP-SUMO2-tag was subsequently removed by addition of human SENP2-protease (250 mg, fragment D364-L589 with M497A mutation) and dialyzed over night at 4 °C in 20 mM HEPES-NaOH pH 7.5, 200mMNaCl, 1mMDTT. The protein was further purified by size exclusion chromatography using a Superdex S75 column (GE Healthcare) equilibrated with 20mMHEPES-NaOH pH 7.5, 150mMNaCl, 1mMTCEP. Final TRAF6 TRAF-C domain protein was concentrated to 5–6 mg mL−1 before being flash-frozen in liquid nitrogen for storage at −80 °C.
Crystallization and Structure Determination
For crystallization, purified zebrafish TRAF6 TRAF-C domain 370–525 was mixed with the zebrafish STING peptide 377-EPVETTDY-384 in a molar ratio 1:10 and incubated for 30 min on ice. Crystals were obtained by hanging drop vapor diffusion in 0.1MHEPES pH 7.5, 1.5 M LiSO4 in drops mixed 2:1 after a week at 20 °C. Crystals were cryo-protected using reservoir solution containing 20% ethylene glycol and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at Advanced Photon Source (Beamline 24-ID-E). Diffraction data were processed using XDS and AIMLESS (Kabsch, 2010) using the SSRL autoxds script (A. Gonzalez, Stanford SSRL). Crystals were indexed in spacegroup P 21 containing two copies of the TRAF6–STING complex in the asymmetric unit. The structure was determined by molecular replacement using PHASER in PHENIX (Adams et al., 2010) and human TRAF6 TRAF-C domain structure (PDB: 1LB5) (Ye et al., 2002) as a search model. Structure determination was completed with iterative model building and refinement using Coot (Emsley and Cowtan, 2004) and PHENIX, respectively. Data collections and refinement statistics are listed in Table S1. Figures were created with PyMOL (DeLano, 2002).
Macrophage Stimulation and RNA-sequencing
Mouse macrophages stably expressing different STING proteins were cultured in DMEM supplemented with 10% (v/v) FBS and seeded 4×106 cells per 10 cm dish one day prior to stimulation with 500 μg mL−1 CMA (10-carboxymethyl-9-acridanone from Sigma) for 4 h. Cells were harvested by incubation with 2.5 mM EDTA in PBS for 5 min at 37 °C, washed with PBS and pelleted by centrifugation at 1,000 × g for 5 min at 4°C. Cell pellet was re-suspended in three volumes of NP40 lysis buffer (50 mM HEPES-KOH pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5% Nonidet P-40 alternative, 0.5 mM DTT), incubated for 10 min on ice and cleared by centrifugation at 13,000 × g for 10 min at 4 °C. RNAs were purified by phenol-chloroform extraction and ethanol precipitation. Single-end Illumina sequencing libraries from polyadenylated mRNA were performed using a standard protocol (Lee et al., 2015). cDNA libraries were sequenced using an Illumina NextSeq 500. Two biological replicates of each stimulated macrophage sample for RNA-sequencing were performed.
RNA-seq computational analysis
Quality filtering was performed using the FASTX Toolkit. Cutadapt was used to trim adapters from original reads and to discard reads shorter than 15 nucleotides (Martin, 2011). Reads were mapped to the mouse GRCm38 (mm10) genome assembly using HiSAT2 (Kim et al., 2015) and gene-level quantification was performed using R/Bioconductor. Differential gene expression analysis was performed with DESeq2 (Love et al., 2014) and only genes with a sum across samples higher than 10 counts were considered for statistical analysis. P value correction was applied using the fdrtool package (Strimmer, 2008). Average RPKM values for differentially expressed genes with a fold change higher than 2 were calculated (Table S2) and used to generate a heatmap using pheatmap.
qRT-PCR
Mouse macrophages stably expressing different STING variants were stimulated with DMSO as a control or 500 μg mL−1 CMA (10-carboxymethyl-9-acridanone, Sigma) for 2 or 4 h. Cells were scraped into HBSS, pelleted by centrifugation at 1,000 g for 5 min at 4 °C. The pellet was re-suspended in three volumes of NP40 lysis buffer (50 mM HEPES-KOH pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5% Nonidet P-40 alternative, 0.5 mM DTT), incubated for 10 min on ice and cleared by centrifugation at 13,000 × g for 10 min at 4 °C. RNAs were isolated by phenol-chloroform extraction and ethanol precipitation. cDNA was reverse transcribed using random hexamers and MMLV M5 reverse transcriptase (Arezi and Hogrefe, 2009). qPCR was performed using the NEB Luna qPCR master mix and the following oligonucleotides: GAPDH-Forward, 5′-GGAGATTGTTGCCATCAACGACC-3′; GAPDH-Reverse, 5′-GTGGGGTCTCGCTCCTGG-3′; IFNB-Forward, 5′-CTCCAGCTCCAAGAAAGGAC-3′; IFNB-Reverse, 5′-TGGCAAAGGCAGTGTAACTC-3′ (Gulen et al., 2017); NOS2-Forward, 5′-CACCTTGGAGTTCACCCAGT-3′; NOS2-Reverse, 5′-TGGTCACCTCCAACACAAGA-3′; CCL12-Forward, 5′-TCCTCAGGTATTGGCTGGAC-3′; CCL12-Reverse, 5′-TGGCTGCTTGTGATTCTCCT-3′. GAPDH was used as an endogenous normalization control and quantitative RT-qPCR was performed in duplicate biological samples.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis was performed using GraphPad Prism7 software. Technical replicates were plotted as representative of at least three independent experiments. Statistical details for each experiment can be found in the figure legends, and outlined in the corresponding methods details section.
DATA AND SOFTWARE AVAILABILITY
The accession number for coordinates of the STING-TRAF6 complex reported in this paper is PDB: 6MYD. The accession number for the RNA-seq data reported in this paper is GEO: GSE128363. Gene expression data are in included in Table S3.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-HA-Peroxidase (3F10) | Roche | Cat# 12013819001; RRID:AB_390917 |
| Rabbit polyclonal anti-TBK1/NAK | Cell Signaling Technology | Cat# 3013S; RRID:AB_2199749 |
| Rabbit monoclonal anti-TRAF6 [EP591Y] | Abcam | Cat# ab33915, RRID:AB_778572 |
| Rabbit monoclonal anti-STING (D2P2F) | Cell Signaling Technology | Cat# 13647; RRID:AB_2732796 |
| Rabbit polyclonal anti-RPS19 | Bethyl | Cat# A304-002A, RRID:AB_2620351 |
| Donkey anti-rabbit IgG, HRP Conjugated | GE Healthcare | Cat# NA934, RRID:AB_772206 |
| Bacterial and Virus Strains | ||
| E. coli BL21-RIL DE3 | Agilent | Cat# 230245 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668030 |
| Recombinant Human TNF-α | Biolegend | Cat# 570102 |
| 10-carboxymethyl-9-acridanone (CMA) | Sigma-Aldrich | Cat# 17927 |
| Ni-NTA Agarose | QIAGEN | Cat# 30250 |
| HEPES | VWR | Cat# 97061-824 |
| Lithium sulfate | Sigma-Aldrich | Cat# 62609 |
| Ethylene glycol | VWR | Cat# 97061 |
| Critical Commercial Assays | ||
| Dual-Luciferase Reporter Assay System | Promega | Cat# E1960 |
| Luna® Universal qPCR Master Mix | New England Biolabs | Cat# M3003E |
| Deposited Data | ||
| TRAF6–STING complex | This paper | PDB: 6MYD |
| Raw and analyzed data | This paper | GEO: GSE128363 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL-3216 |
| Tbk1−/− HEK293T | This paper | N/A |
| Traf6−/− HEK293T | This paper | N/A |
| Sting−/− Immortalized bone marrow-derived macrophages (iBMDMs) | Jonathan Kagan Laboratory | Tan and Kagan, 2019 |
| Sting−/− Immortalized bone marrow-derived macrophages (iBMDMs) expressing GFP | This paper | N/A |
| Sting−/− Immortalized bone marrow-derived macrophages (iBMDMs) expressing mSTING | This paper | N/A |
| Sting−/− Immortalized bone marrow-derived macrophages (iBMDMs) expressing mSTING-DCTT | This paper | N/A |
| Sting−/− Immortalized bone marrow-derived macrophages (iBMDMs) expressing mSTING-zfCTT | This paper | N/A |
| Oligonucleotides | ||
| qRT-PCR primers see Table S5 | This paper | N/A |
| TBK1 gRNA-1: 5′-TCCACGTTATGATTTAGACG-3′ (sense) | This paper | N/A |
| TBK1 gRNA-2: 5′-TGTGGGAGTTTATACACTGT-3′ (antisense) | This paper | N/A |
| TRAF6 gRNA-1: 5′-GAAGCAGTGCAAACGCCATG-3′ (sense) | This paper | N/A |
| TRAF6 gRNA-2: 5′-CCAGTCACACATGAGAATGT-3′ (sense) | This paper | N/A |
| Recombinant DNA | ||
| STING plasmid constructs see Table S4 | This paper | N/A |
| pcDNA4-human-cGAS | Philip Kranzusch Laboratory | N/A |
| pGL3-2 × IFNβ-FLuc | Jae Jung Laboratory | N/A |
| pRL-TK | Promega | Cat# E2231 |
| pGL2-ELAM-NF-κB-FLuc | Russell Vance Laboratory | N/A |
| pLuc-IFNβ-110 | Kate Fitzgerald Laboratory | N/A |
| pLuc-PRD(III-I)3 | Kate Fitzgerald Laboratory | N/A |
| pLuc-PRD(II)2 | Kate Fitzgerald Laboratory | N/A |
| pLuc-PRD(IV)6 | Kate Fitzgerald Laboratory | N/A |
| pCMV-VSV-G | Jonathan Kagan Laboratory | N/A |
| pCL-Eco | Jonathan Kagan Laboratory | N/A |
| pMSCV 2.2 IRES-GFP | Jonathan Kagan Laboratory | N/A |
| pMSCV 2.2 IRES-GFP mSTING | This paper | N/A |
| pMSCV 2.2 IRES-GFP mSTING dCTT | This paper | N/A |
| pMSCV 2.2 IRES-GFP mSTING-zfTRAF6 | This paper | N/A |
| pET16MBP-SUMO2-zebrafish-TRAF6 370-525 | This paper | N/A |
| pcDNA4-human-TRAF6 | This paper | N/A |
| pcDNA4-human-TRAF6 333-508 | This paper | N/A |
| pcDNA4-human-TRAF6 L74H | This paper | N/A |
| pcDNA4-human-TRAF6 Y473A | This paper | N/A |
| pcDNA4-human-TRAF6 F471A | This paper | N/A |
| pcDNA4-human-TRAF6 M450A | This paper | N/A |
| pcDNA4-human-TRAF6 R466A | This paper | N/A |
| pcDNA4-human-TRAF3 | This paper | N/A |
| U6-SpyM1-sgRNA1(anti-TBK1)-Cas9-Puro | This paper | N/A |
| U6-SpyM1-sgRNA2(anti-TBK1)-Cas9-Puro | This paper | N/A |
| U6-SpyM1-sgRNA1(anti-TRAF6)-Cas9-Puro | This paper | N/A |
| U6-SpyM1-sgRNA2(anti-TRAF6)-Cas9-Puro | This paper | N/A |
| Software and Algorithms | ||
| Prism v7.0d | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Clustal omega | EMBL-EBI | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Jalview v2.10.1 | Waterhouse et al., 2009 | http://www.jalview.org |
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de/ |
| Phenix v1.13-2998 | Adams et al., 2010 | https://www.phenix-online.org |
| Coot v0.8.9 | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Pymol v1.7.4.4 | Schrödinger, LLC | https://pymol.org/2/ |
| FASTX Toolkit v0.0.13 | N/A | http://hannonlab.cshl.edu/fastx_toolkit/ |
| cutadapt v1.16 | Martin, 2011 | https://pypi.org/project/cutadapt/ |
| HiSAT2 v2.1.0 | Kim et al., 2015 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| DESeq2 package v1.20.0 | Love et al., 2014 | https://bioconductor.org |
| fdrtool package v1.2.15 | Strimmer, 2008 | https://bioconductor.org |
| pheatmap package v1.0.10 | N/A | https://bioconductor.org |
| IGV v2.4.14 | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv/ |
Highlights.
STING-dependent activation of IRF3 and NF-κB signaling varies between species
A motif in zebrafish STING CTT causes dramatic enhancement of NF-κB signaling
STING CTT-TRAF6 structure explains the mechanism of NF-κB enhancement
Zebrafish CTT module is sufficient to reprogram STING signaling in mammalian cells
ACKNOWLEDGMENTS
The authors acknowledge K. Chat, R. Vance, S. Margolis, and members of the Kranzusch lab and Lee lab for helpful comments and discussion. We thank H. Rivas for cloning assistance and R. Tay and L. Ferrari de Andrade for assistance with cell sorting. Sequencing was performed at the Harvard Biopolymers Facility. The work was funded by the Claudia Adams Barr Program for Innovative Cancer Research (to P.J.K.), the Charles H. Hood Foundation (to P.J.K.), a Cancer Research Institute CLIP grant (to P.J.K.), a V Foundation V Scholar Award (to P.J.K.), the DFCI-Novartis Drug Discovery Program (to P.J.K.), the Searle Scholars Program (to A.S.Y.L.), the Pew Biomedical Scholars Program (to A.S.Y.L.), and a Sloan Research Fellowship (to A.S.Y.L.). C.C.d.O.M. is supported as a Cancer Research Institute/Eugene V. Weissman Fellow. J.C.K. is supported by NIH grants AI133524, AI093589, AI116550, and P30DK34854 and holds an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund. M.H.O. was supported by NIH grants T32AI007512 and K99AI30258. X-ray data were collected at the Northeastern Collaborative Access Team beamlines 24-ID-C and 24-ID-E, which are funded by the NIGMS (P41 GM103403) and an NIH-ORIP HEI grant (S10 RR029205) and used resources of the DOE Argonne National Laboratory Advanced Photon Source (under contract DE-AC02-06CH11357).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.03.098.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abe T, and Barber GN (2014). Cytosolic DNA-Mediated, STING-Dependent Pro-Inflammatory Gene Induction Necessitates canonical NF-κB activation Through TBK1. J. Virol 88, 5328–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, and Barber GN (2013). STING recognition of cytoplasmic DNA instigates cellular defense. Mol. Cell 50, 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arezi B, and Hogrefe H (2009). Novel mutations in Moloney Murine Leukemia Virus reverse transcriptase increase thermostability through tighter binding to template-primer. Nucleic Acids Res. 37, 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud V, Liu Z-G, Bennett B, Suzuki N, Xia Y, and Karin M (1999). Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13, 1297–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavlar T, Deimling T, Ablasser A, Hopfner KP, and Hornung V (2013). Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 32, 1440–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo S-R, Lemmens E, Banda T, Leong JJ, et al. (2015). Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnay BG, Ni J, Moore PA, and Aggarwal BB (1999). Activation of NF-kappaB by RANK requires tumor necrosis factor receptor-associated factor (TRAF) 6 and NF-kappaB-inducing kinase. Identification of a novel TRAF6 interaction motif. J. Biol. Chem 274, 7724–7731. [DOI] [PubMed] [Google Scholar]
- David L, Li Y, Ma J, Garner E, Zhang X, and Wu H (2018). Assembly mechanism of the CARMA1-BCL10-MALT1-TRAF6 signalosome. Proc. Natl. Acad. Sci. USA 115, 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002). The PyMOL molecular graphics system. https://pymol.org/2/. [Google Scholar]
- Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jønsson KL, Jakobsen MR, Nevels MM, Bowie AG, and Unterholzner L (2018). Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-κB Signaling after Nuclear DNA Damage. Mol. Cell 71, 745–760.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fang R, Wang C, Jiang Q, Lv M, Gao P, Yu X, Mu P, Zhang R, Bi S, Feng J-M, and Jiang Z (2017). NEMO-IKKβ Are Essential for IRF3 and NF-κB Activation in the cGAS-STING Pathway. J. Immunol 199, 3222–3233. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao S-M, and Maniatis T (2003). IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol 4, 491–496. [DOI] [PubMed] [Google Scholar]
- Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, Mechette K, Leong JJ, Lauer P, and Liu W (2015). STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med 7, 283ra252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, et al. (2013). Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Okado K, Martins N, Cai H, Barbier V, Lamiable O, Troxler L, Santiago E, Kuhn L, and Paik D (2018). The Kinase IKKβ Regulates a STING-and NF-κB-Dependent Antiviral Response Pathway in Drosophila. Immunity 49, 225–234.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulen MF, Koch U, Haag SM, Schuler F, Apetoh L, Villunger A, Radtke F, and Ablasser A (2017). Signalling strength determines proapoptotic functions of STING. Nat. Commun 8, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, Heymann M, van der Goot FG, Turcatti G, Behrendt R, and Ablasser A (2018). Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273. [DOI] [PubMed] [Google Scholar]
- Han D, Walsh MC, Kim KS, Hong S-W, Lee J, Yi J, Rivas G, Choi Y, and Surh CD (2017). Dendritic cell expression of the signaling molecule TRAF6 is required for immune tolerance in the lung. Int. Immunol 29, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Xu J, Xie X, Zhou Y, Tao P, Li H, Han X, Wang C, Liu J, Xu P, et al. (2017). Oligomerization-primed coiled-coil domain interaction with Ubc13 confers processivity to TRAF6 ubiquitin ligase activity. Nat. Commun 8, 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-H, Liu X-Y, Du X-X, Jiang Z-F, and Su X-D (2012). The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol 19, 728–730. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, and Barber GN (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS-Y, Berger JM, and Doudna JA (2013). Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 3, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Wilson SC, Lee AS, Berger JM, Doudna JA, and Vance RE (2015). Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, and Cate JH (2015). eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-C, Lo Y-C, and Wu H (2010). Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen J, Cai X, Wu J, Chen X, Wu Y-T, Sun L, and Chen ZJ (2013). MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife 2, e00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu Y-T, Grishin NV, and Chen ZJ (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu Y, Gordesky-Gold B, Leney-Greene M, Weinbren NL, Tudor M, and Cherry S (2018). Inflammation-Induced, STING-Dependent Autophagy Restricts Zika Virus Infection in the Drosophila Brain. Cell Host Microbe 24, 57–68.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SR, Wilson SC, and Vance RE (2017). Evolutionary origins of cGAS-STING signaling. Trends Immunol. 38, 733–743. [DOI] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Martin M, Hiroyasu A, Guzman RM, Roberts SA, and Goodman AG (2018). Analysis of Drosophila STING Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep. 23, 3537–3550.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason NJ, Fiore J, Kobayashi T, Masek KS, Choi Y, and Hunter CA (2004). TRAF6-dependent mitogen-activated protein kinase activation differentially regulates the production of interleukin-12 by macrophages in response to Toxoplasma gondii. Infect. Immun 72, 5662–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Pullen SS, Holton JM, Crute JJ, Kehry MR, and Alber T (1999). Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc. Natl. Acad. Sci. USA 96, 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. (2012). Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YC, Burkitt V, Villa AR, Tong L, and Wu H (1999). Structural basis for self-association and receptor recognition of human TRAF2. Nature 398, 533–538. [DOI] [PubMed] [Google Scholar]
- Pullen SS, Miller HG, Everdeen DS, Dang TT, Crute JJ, and Kehry MR (1998). CD40-tumor necrosis factor receptor-associated factor (TRAF) interactions: regulation of CD40 signaling through multiple TRAF binding sites and TRAF hetero-oligomerization. Biochemistry 37, 11836–11845. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Bio-technol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scumpia PO, Botten GA, Norman JS, Kelly-Scumpia KM, Spreafico R, Ruccia AR, Purbey PK, Thomas BJ, Modlin RL, and Smale ST (2017). Opposing roles of Toll-like receptor and cytosolic DNA-STING signaling pathways for Staphylococcus aureus cutaneous host defense. PLoS Pathog. 13, e1006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, and Gu L (2012). Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol 19, 725–727. [DOI] [PubMed] [Google Scholar]
- Shi H, Wu J, Chen ZJ, and Chen C (2015a). Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING. Proc. Natl. Acad. Sci. USA 112, 8947–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Zhang Z, Zhang Z, Wang Y, Li C,Wang X, He F, Sun L, Jiao S, and Shi W (2015b). Structural Insights into Mitochondrial Antiviral-Signaling Protein (MAVS)–Tumor Necrosis Factor Receptor-Associated Factor 6 (TRAF6) Signaling. J. Biol. Chem 290, 26811–26820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, and Li P (2012). Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol 19, 722–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K (2008). fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461–1462. [DOI] [PubMed] [Google Scholar]
- Studier FW (2005). Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif 41, 207–234. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, and Chen ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpris G, Chan J, Thompson M, Ilyukha V, Liu BC, Atianand M, Sharma S, Volkova T, Smirnova I, Fitzgerald KA, and Poltorak A (2016). Cutting edge: novel Tmem173 allele reveals importance of STING N terminus in trafficking and type I IFN production. J. Immunol 196, 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu S, Onoguchi K, Onomoto K, Narita R, Takahasi K, Ishidate F, Fujiwara TK, Yoneyama M, Kato H, and Fujita T (2013). Functional characterization of domains of IPS-1 using an inducible oligomerization system. PLoS ONE 8, e53578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, and Kagan JC (2019). Innate Immune Signaling Organelles Display Natural and Programmable Signaling Flexibility. Cell, Published online February 28, 2019. 10.1016/j.cell.2019.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Sun L, Chen J, and Chen ZJ (2018). Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu. Rev. Microbiol 72, 447–478. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, and Chen ZJ (2012). STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal 5, ra20–ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H (2004). Assembly of post-receptor signaling complexes for the tumor necrosis factor receptor superfamily In Advances in Protein Chemistry, Garcia KC, ed. (Elsevier; ), pp. 225–279. [DOI] [PubMed] [Google Scholar]
- Wu J, and Chen ZJ (2014). Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol 32, 461–488. [DOI] [PubMed] [Google Scholar]
- Xie J, Li Y, Shen X, Goh G, Zhu Y, Cui J, Wang L-F, Shi Z-L, and Zhou P (2018). Dampened STING-dependent interferon activation in bats. Cell Host Microbe 23, 297–301.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, et al. (2002). Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418, 443–447. [DOI] [PubMed] [Google Scholar]
- Yin Q, Lin S-C, Lamothe B, Lu M, Lo Y-C, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, et al. (2009). E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol 16, 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, and Wu H (2012). Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, and Chen ZJ (2013). Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Shang G, Gui X, Zhang X, Bai XC, and Chen ZJ (2019). Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Shu C, Gao X, Sankaran B, Du F, Shelton CL, Herr AB, Ji J-Y, and Li P (2016). Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc. Natl. Acad. Sci. USA 113, E3403–E3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang Y-Y, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, and Shu HB (2008). The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for coordinates of the STING-TRAF6 complex reported in this paper is PDB: 6MYD. The accession number for the RNA-seq data reported in this paper is GEO: GSE128363. Gene expression data are in included in Table S3.