Abstract
Background
LncRNA has been widely investigated for decades and plays critical roles in the progression of cancer. However, lncRNA NLIPMT, as a novel non-coding RNA, only was studied in breast cancer. This study aimed to explore the role of NLIPMT in esophageal squamous-cell carcinomas (ESCC).
Materials and Methods
NLIPMT, miR320 and survivin mRNA in ESCC tissues (or non-tumor tissue) were detected by qRT-PCR. Dual-luciferase reporter assay was performed to assess the relationship between miR-320 and survivin. In ESCC cell lines KYSE510 and ECA109, miR-320 mimic and expression vectors carrying NLIPMT and survivin were used. Cell cycle, apoptosis, proliferation and migration were detected by flow cytometry, CCK-8, transwell assay, respectively. NIPMT, miR-320 and survivin expression were measured by qRT-PCR and Western blotting.
Results
NLIPMT was downregulated in ESCC and predicted poor survival of ESCC patients. NLIPMT was positively correlated with miR-320 and negatively correlated with survivin in ESCC tumor tissues. Dual-luciferase reporter assay showed that miR-320 directly regulated survivin. qRT-PCR and Western blotting showed that NLIPMT promoted miR-320 expression and inhibited survivin expression via up-regulating miR-320. Moreover, both NLIPMT and miR-320 overexpression inhibited cell proliferation and migration and promoted cell cycle arrest and apoptosis in ESCC cells, while their effects were abolished by survivin overexpression.
Conclusion
We demonstrate that NLIPMT inhibits cell proliferation and migration and promotes cell cycle arrest and apoptosis in ESCC cells by regulating the miR-320/survivin axis. NLIPMT may be a novel prognosis biomarker in ESCC patients.
Keywords: ESCC, NLIPMT, survivin, miR-320, tumorigenesis
Introduction
According to the latest GLOBOCAN statistics, esophageal cancer (EC) affected about 572,000 new cases in the year of 2018, which made it the seventh most common type of malignancy.1 Moreover, during this period, EC caused 509,000 deaths, which account for 5% of all cancer deaths.1 It is worth noting that the mortality rate of EC is close to the incidence rate.1–3 As the most common type of EC, esophageal squamous-cell carcinomas (ESCC) account for about 90% of all EC cases. Smoking, alcohol abuse and hot water drinking are the major risk factors of ESCC.4,5 However, the pathogenesis of ESCC is still hardly to completely known.6
Genetic and epigenetic factors are the main contributors to ESCC.7 Identification of novel genetic factors involved in ESCC may provide guidance to the development of novel targeted therapies.8 Survivin, also referred to BIRC5 (baculoviral inhibitor of apoptosis repeat-containing 5), inhibits programmed cell death to promote tumor growth and progression.9 In recent years, survivin-targeted therapies have attracted more and more attention.9,10 In effect, some tumor-suppressive miRNAs can target survivin to suppress cancer development.11 It is known that survivin can be downregulated by miR-320 to play the roles of insulin against myocardial ischemia.12 However, the interaction between them is unclear in cancer biology. NLIPMT (RP11-115N4.1) is a recently characterized tumor-suppressive lncRNA in breast cancer.13 It has been found that NLIPMT suppresses cancer progression by inactivating glycogen synthase kinase 3β (GSK3β), which has crosstalk with survivin in breast cancer.14 However, the involvement of NLIPMT in ESCC is unknown. Our preliminary RNA-seq analysis revealed that the expression of NLIPMT was downregulated in ESCC tumor tissues. Moreover, we found it embedded miR-320 through the bioinformatics search of whole NLIPMT genome sequence. Therefore, this study aimed to investigate the interactions among NLIPMT, miR-320 and survivin in ESCC.
Methods
ESCC Patients
From April 2011 to May 2014, Xingtai People’s Hospital admitted a total of 98 cases of ESCC. From these ESCC patients, this study enrolled 58 cases (32 males and 26 females; 39 to 67 years, 56.1 ± 6.3 years). This study obtained the ethics approval from the institutional review board of Ethics Committee of Xingtai People’s Hospital with approval number (2011-XPH-19). Inclusion criteria: 1) newly diagnosed ESCC; 2) confirmed by histopathological exams. Exclusion criteria: 1) recurrent cases; 2) patients complicated with other clinical disorders; 3) therapies were performed. All the patients were informed of the design of this project, and informed consent was signed by all patients.
Specimen Collection
Before the initiation of therapies, all patients were subjected to biopsy, which was performed using endoscopic submucosal dissection (ESD) under endoscopy following the guidance of MRI. During biopsy, ESCC (tumor) and non-tumor tissues were collected from each patient. All tissue specimens were confirmed by histopathological exams.
Staging, Treatment and Follow-Up
According to the AJCC staging system, the 58 patients were graded, and the results revealed 17, 20 and 21 cases at stage II, III and IV, respectively. According to patients’ conditions, surgical resection combined with chemotherapy (PF formula: Paclitaxel Plus Fluorouracil), radiotherapy (dose >50Gy) or chemotherapy and radiotherapy together was performed. From the day of admission, patients were followed up in a monthly manner through telephone for 5 years to record survival conditions. In survival analysis, patients who died of other causes or lost before the end of follow-up were excluded.
ESCC Cells and Transient Transfection
Human ESCC cell lines KYSE510 and ECA109 (Cancer Hospital of the Chinese Academy of Medical Sciences, Beijing, China) were used in this study. The cell culture medium was a mixture of 10% FBS (Gibco, USA) and 90% RPMI-1640 medium (Gibco, USA). Cell culture was in a chamber with 5% CO2, 95% humidity at 37 °C. NLIPMT and survivin expression vectors were constructed using pcDNA3 vector (Sangon, Shanghai, China). Primers used to clone NLIPMT were: 5ʹ-TGAACATTTGCTGCTGCTTCC-3ʹ (forward) and 5ʹ- TGCAAGGTCATAGCCAATGC-3ʹ (reverse). Negative control (NC) miRNA and miR-320 mimic were from Sigma-Aldrich (USA). Tumor cell lines were collected at confluence of 80%. Cells were counted, and 106 cells were transfected with 10 nM vector (empty vector as NC group) or 50 nM miRNA (NC miRNA as NC group) using lipofectamine 2000 (Sigma-Aldrich, USA). Untransfected cells were used as control (C) cells. Cells were harvested at 48 h post-transfection.
RNA Extraction and qRT-PCR
RNAzol (Sigma-Aldrich, USA) was used to extract total RNAs from 105 cells or 30 mg tissues. Cells were harvested at 48 h post-transfection, and tissues were kept in liquid nitrogen before use. To harvest miRNA, 85% ethanol was used to precipitate and wash RNAs. All RNA samples were digested at 37 °C with DNase I for 100 min to remove genomic DNA. To measure the mRNA expression of NLIPMT and survivin, all reverse transcriptions were performed using Tetro Reverse Transcriptase (Bioline), and all qRT-PCR assays were performed using QuantiTect SYBR Green PCR Kit (Qiagen). With GAPDH as endogenous control, gene expression was normalized. The expression of mature miR-320 was measured using All-in-One™ miRNA qRT-PCR Detection Kit (Genecopoeia). All steps were completed following the instructions from Genecopoeia.
The 2−ΔΔCT method was used to process Ct values. Each experiment performed three times.
Western Blotting
RIPA solution (Sigma-Aldrich) containing protease inhibitor cocktails was used to extract total protein from 105 cells (harvested at 48h post-transfection). BCA assay (Sigma-Aldrich) was performed to measure protein concentration. Following denaturing at 95 °C for 10 min, protein samples were subjected to electrophoresis (10% SDS-PAGE gel), followed by gel transfer to PVDF membrane. Blocking was performed in PBS containing 5% fat-free milk. Following incubation with primary rabbit anti-GAPDH (1:1800, ab22555, Abcam) and anti-survivin (1:1600, ab469, Abcam) antibodies at 4 °C for 12 h, HRP (IgG) (1:2000; ab6721; Abcam) goat secondary antibody was incubated for 2h at 24°C. Signals were produced using ECL Western Blotting Substrate (ab65623, Abcam), and data were processed using Image J v1.46 software.
Cell Proliferation Analysis
CCK-8 assay was performed at 48h post-transfection to analyze the effects of transfection on the proliferation of KYSE510 and ECA109 cells. Single-cell suspension was prepared by resuspending pellets containing 3×104 cells in 1 mL mixture containing 90% RPMI-1640 medium and 10% FBS. Cell culture plates (96-well, 0.1 mL per well) were used to cultivate cells under aforementioned methods, and 3 replicate wells were set for each transfection group. Each well was added with 10 µL CCK-8 solution (Sigma-Aldrich, USA) at 2 h before the termination of cell culture. After cell culture was ended, OD values were measured at 450nm absorbance.
Flow Cytometry
Flow cytometry was performed at 48h post-transfection to analyze the effects of transfection on cell cycle and apoptosis of tumor cells. Cells were stained with FITC labeled Annexin V and propidium iodide (PI) using an apoptosis detection kit (BioLegend, USA) or stained with PI after cell fixed by ice-cold ethanol (70%) overnight, to detect the cell apoptosis and cell cycle, respectively. The results were analyzed by flow cytometry (BD Biosciences), each experiment was repeated at least three times.
Transwell Assay
Transwell assay was performed to detect the tumor cell migration. Cells were harvested at 24h post-transfection and then continue to be cultivated in 1 mL RPMI-1640 culture medium (serum-free) with 5×104 cells. Before migration assay, Matrigel (Millipore, USA) was used to block membrane for 6–8 h at 37 °C. Then, upper chamber filled with cell suspensions, and lower chamber filled with culture medium containing 20% FBS. After 12 h culture, 0.5% crystal violet (Sigma-Aldrich, USA) was used to stain membrane for 20 min. Finally, an optical microscope (Olympus) was used to measure the migration cells.
Dual-Luciferase Reporter Assay
KYSE510 cells were seeded into a 24-well plate 1 day before transfection, the cells were transfected with 1 µg pmiRGLO vector (Promega) containing wild-type (WT) or mutant (MUT) survivin 3ʹUTR and 50 nM miR-320 mimic with lipofectamine 2000. PRL-TK plasmid which encodes renilla luciferase was also transfected into cells together. After 48 h transfection, a dual-luciferase reporter system (Promega) was used to detect the firefly luciferase activity normalized to renilla. Three replicates were conducted in each experiment, and every experiment repeated three times.
Statistical Analysis
The experiments were performed in three replicates. All data analyses were performed using the mean values. Differences were explored between ESCC and non-tumor tissues by paired t-test. Survival analysis was performed by grouping the 58 ESCC patients into high and low NLIPMT level groups (n=29), followed by survival cure plotting and comparison by K-M plotter and Log rank test, respectively. Correlations were analyzed by Pearson’s correlation coefficient. Differences were explored among multiple groups by ANOVA (one-way) and Tukey’s test. Chi-square test was performed to analyze the associations between the expression of NLIPMT in ESCC and patient’s clinical data. P < 0.05 was statistically significant.
Results
NLIPMT Expression Was Downregulated in ESCC and Predict Poor Survival of ESCC Patients
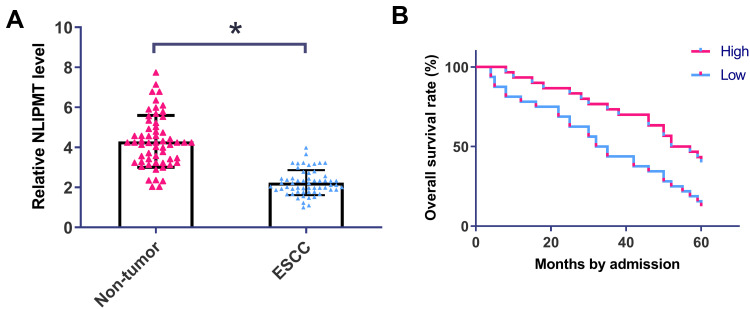
QRT-PCR was performed to detect the expression of NLIPMT in ESCC tumor tissues and non-tumor tissues. It was observed that expression of NLIPMT was significantly downregulated in ESCC tumor tissues compared with non-tumor tissues (Figure 1A, p<0.05). Chi-square test showed that expression of NLIPMT in ESCC was not closely correlated with patients’ age, gender, clinical stages, smoking and drinking habit (Table 1). Survival curves were plotted and compared. It was observed that the overall survival rate was significantly higher in high NLIPMT level group than in low NLIPMT level group (Figure 1B, p=0.007). Multivariate analysis confirmed that NLIPMT expression was an independent prognostic factor for overall survival in ESCC patients (p<0.01).
Figure 1.
NLIPMT expression was downregulated in ESCC and predict poor survival of ESCC patients. (A) QRT-PCR was performed to detect the expression of NLIPMT in ESCC and non-tumor tissues. Expression data were compared by performing paired t-test. (B) Survival analysis was performed by grouping the 58 ESCC patients into high and low NLIPMT level groups (n = 29), followed by survival cure plotting and comparison by K-M plotter and Log rank test, respectively. *p < 0.05.
Table 1.
Associations Between Expression Levels of NLIPMT in ESCC and Patient’s Clinical Data
| Items | Groups | Cases | NLIPMT High level |
NLIPMT Low level |
χ2 | p value |
|---|---|---|---|---|---|---|
| Age (years) | >55 | 30 | 14 | 16 | 0.28 | 0.60 |
| <55 | 28 | 15 | 13 | |||
| Gender | Male | 32 | 15 | 17 | 0.28 | 0.60 |
| Female | 26 | 14 | 12 | |||
| Stage | II | 17 | 7 | 10 | 0.78 | 0.68 |
| III | 20 | 11 | 9 | |||
| IV | 21 | 11 | 10 | |||
| Smoking | Yes | 27 | 15 | 12 | 0.62 | 0.43 |
| No | 31 | 14 | 17 | |||
| Drinking | Yes | 35 | 16 | 19 | 0.65 | 0.42 |
| No | 23 | 13 | 10 |
Abbreviations: ESCC, esophageal squamous-cell carcinomas; NLIPMT, lncRNA NLIPMT.
NLIPMT Inhibited Survivin Expression by Up-Regulating miR-320 Expression
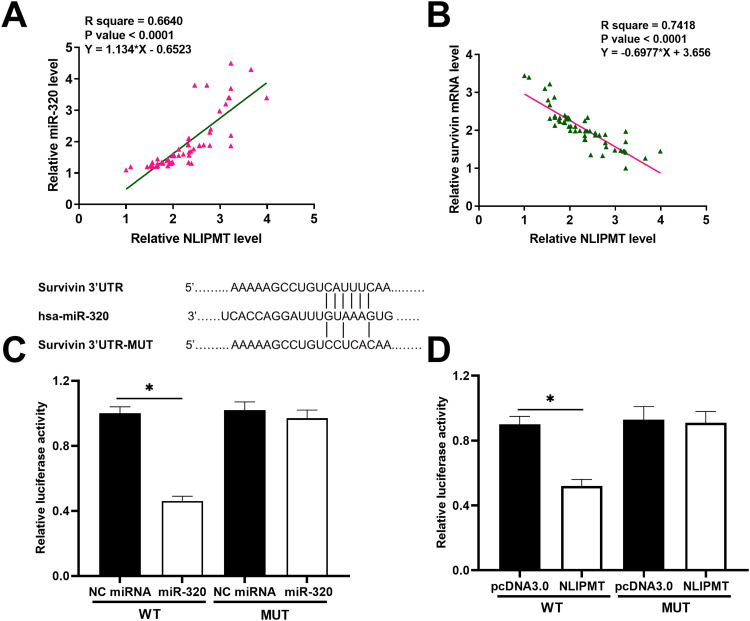
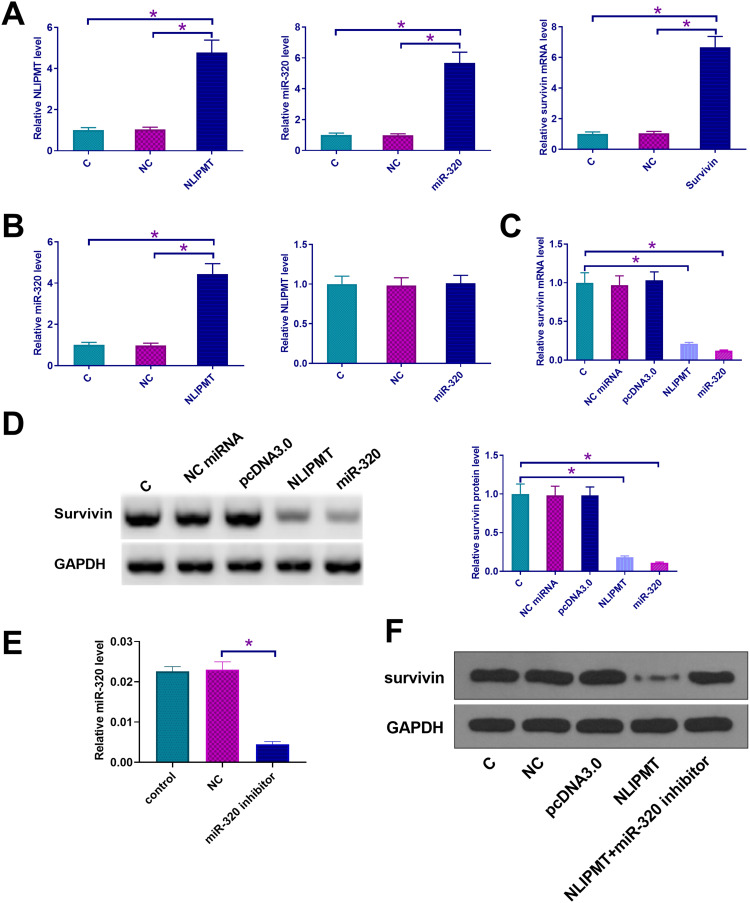
Since we predicted the whole genome sequence of NLIPMT embedded miR-320, and survivin 3ʹUTR had a pseudo binding site with miR-320 via Targetscan software analysis (http://www.targetscan.org/vert_72/), the mRNA expression of miR-320 and survivin in ESCC tissues was also measured by qRT-PCR. The correlation between mRNA expression of NLIPMT and miR-320/survivin was analyzed by Pearson’s correlation coefficient. It was observed that the expression levels of NLIPMT were significantly and positively correlated with the expression levels of miR-320 in ESCC tissues (Figure 2A). However, the expression levels of NLIPMT were significantly and inversely correlated with the expression levels of survivin mRNA in ESCC tissues (Figure 2B). Dual-luciferase reporter assay showed that the overexpression of miR-320 and NLIPMT reduced the luciferase activity in KYSE510 cells co-transfected with a reporter plasmid containing WT survivin 3ʹUTR, while it did not affect the luciferase activity in KYSE510 cells co-transfected with a reporter plasmid containing MUT survivin 3ʹUTR (Figure 2C and D). It indicates that survivin is a target of miR-320 and NLIPMT. NLIPMT and survivin expression vectors as well as miR-320 mimic were transfected into KYSE510 cells. The overexpression of NLIPMT, miR-320 and survivin was confirmed by qRT-PCR (Figure 3A, p<0.005). We found that NLIPMT overexpression led to the upregulation of miR-320 expression, while LIPMT expression was not significantly altered after miR-320 overexpression (Figure 3B, p<0.05). In addition, NLIPMT and miR-320 overexpression led to downregulation of survivin at both mRNA (Figure 3C, p<0.05) and protein levels (Figure 3D, p<0.05). MiR-320 inhibitor was also used. We found that miR-320 inhibitor significantly reduced miR-320 level in KYSE510 cells (Figure 3E). Then, NLIPMT expression vector and miR-320 inhibitor were co-transfected into KYSE510 cells. Results indicating that NLIPMT overexpression inhibited the expression of survivin, while its effect was abolished by miR-320 inhibitor (Figure 3F). Taken together, these data indicate that NLIPMT inhibits survivin expression by up-regulating miR-320.
Figure 2.
NLIPMT was positively correlated with miR-320 and negatively correlated with survivin. (A) The correlations between expression levels of NLIPMT and miR-320. (B) The correlations between expression levels of NLIPMT and survivin mRNA. (C) Dual-luciferase reporter assay implicated that survivin could be downregulated by miR-320. (D) Dual-luciferase reporter assay implicated that survivin could be downregulated by NLIPMT. *p < 0.05.
Figure 3.
NLIPMT upregulated survivin expression in tumor cells through miR-320. (A–D) NLIPMT and survivin expression vectors as well as miR-320 mimic were transfected into KYSE510 cells. (A) The overexpression of NLIPMT, miR-320 and survivin was confirmed by qRT-PCR. (B) The interaction between NLIPMT and miR-320 was analyzed by qRT-PCR. (C) The mRNA level of survivin was detected by qRT-PCR. (D) The protein level of survivin was detected by Western blotting. (E) miR-329 inhibitor was transfected into KYSE510 cells. miR-320 level was detected by qRT-PCR. (F) NLIPMT expression vector and miR-329 inhibitor were co-transfected into KYSE510 and ECA109 cells, and survivin, protein was measured by Western blotting. *p < 0.05.
NLIPMT Inhibited Cell Proliferation and Migration Through miR-320 and Survivin
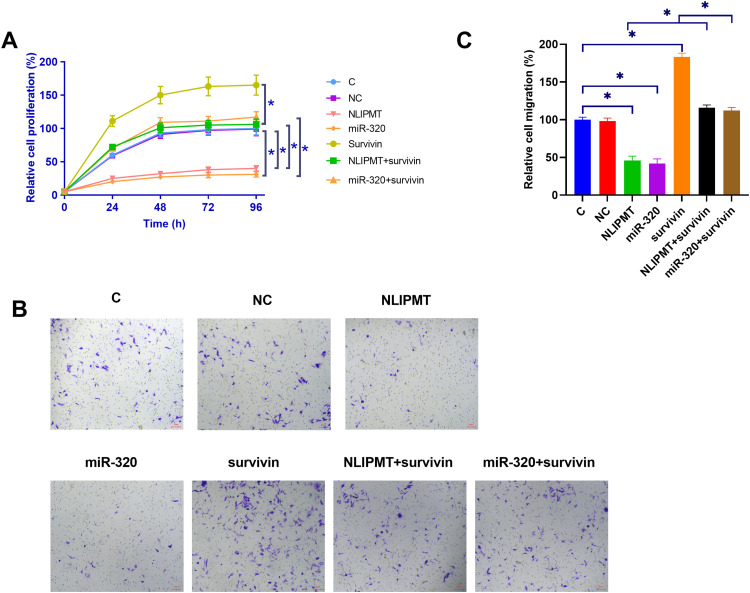
The effects of NLIPMT, miR-320 and survivin overexpression on the proliferation of KYSE510 and ECA109 cells were analyzed by CCK-8 assay. Compared with C and NC (empty pcDNA3.0 vector or NC miRNA) groups, NLIPMT and miR-320 overexpression led to decreased cell proliferation rate. Overexpression of survivin played an opposite role and reduced the effects of NLIPMT and miR-320 overexpression on cell proliferation (Figure 4A, S1A, p<0.05). Moreover, transwell assay illustrated that the expression of NLIPMT, miR-320 could inhibit the tumor cell migration, while this role would be reversed by the overexpression of survivin (Figure 4B and C, S1B, S1C, p<0.05).
Figure 4.
NLIPMT promoted cell proliferation and migration through miR-320 and survivin in KYSE510 cells. (A) The effects of NLIPMT, miR-320 and survivin overexpression on the proliferation of KYSE510 cells were analyzed by CCK-8 assay. (B) After transfection, transwell assay was used to assess the cell migration. The captured photos were compared between different groups. (C) Statistical analysis of transwell assay results. *p < 0.05.
Abbreviations: C, control cells; NC, control cells transfected empty vector.
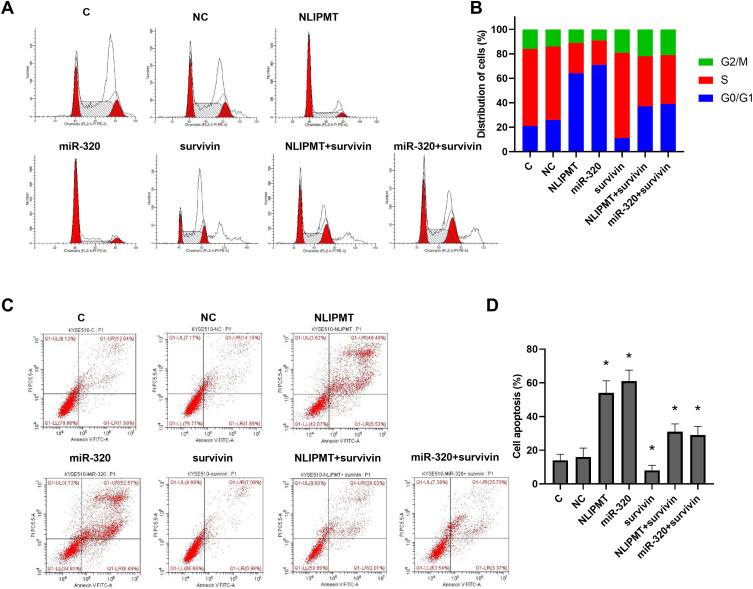
NLIPMT Promoted Tumor Cell Cycle Arrest and Cell Apoptosis by Regulating miR-320 and Survivin
To further detect the role of NLIPMT and miR-320 in tumor cells, flow cytometry was performed to analyze tumor cell cycle and apoptosis. The results showed that overexpression of NLIPMT and miR-320 in KYSE510 and ECA109 cells increased the G0/G1 phase ratio compared to S phase, and this effect was reversed by the expression of survivin (Figure 5A and B, S2A, S2B). However, apoptosis analysis in KYSE510 and ECA109 cells also found that the expression of NLIPMT and miR-320 could promote tumor cell apoptosis (Figure 5C and D, S2C, S2D, p<0.05). Therefore, these results suggested that NLIPMT could be considered as a tumor suppressor in ESCC.
Figure 5.
NLIPMT increased cell cycle arrest and apoptosis through miR-320 and survivin in KYSE510 cells. Flow cytometry analyzed the effects of NLIPMT, miR-320 and survivin overexpression on the cell cycle and apoptosis of tumor cells. (A) Representative pictures of cell cycle distributions in KYSE510 cells. (B) Statistical analysis of cell cycle distributions. (C) Representative pictures of cell apoptosis. (D) Statistical analysis of cell apoptosis. *p < 0.05.
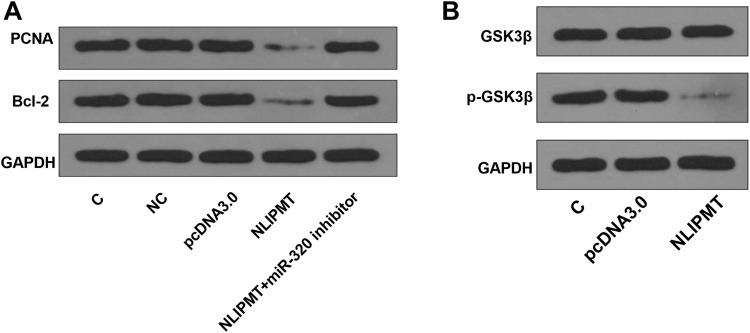
NLIPMT Reduced PCNA and Bcl-2 Protein Expression and Inhibited the Phosphorylation of GSK3β
We also explored the effect of NLIPMT on the expression of proliferation marker PCNA and apoptosis marker Bcl-2. MiR-320 inhibitor was used to knockdown miR-320 expression (Figure S3A). We found that NLIPMT overexpression inhibited the expression of PCNA and Bcl-2, while miR-320 inhibitor abolished NLIPMT overexpression-induced inhibition on these two proteins (Figure 6A, S3B). Moreover, we also assessed the effect of NLIPMT on GSK3β, an effector protein of NLIPMT. Results indicated that NLIPMT overexpression had no effect on total GSK3β, while reduced the level of phosphorylated GSK3β (Figure 6B, S3C).
Figure 6.
NLIPMT reduced PCNA and Bcl-2 proteins and inhibited the phosphorylation of GSK3β in KYSE510 cells. (A) NLIPMT expression vector and miR-329 inhibitor were co-transfected into KYSE510 cells. The protein levels of PCNA and Bcl-2 were detected by Western blotting. (B) NLIPMT expression vector was transfected into KYSE510 cells. The phosphorylated and total GSK3β expression levels were detected by Western blotting.
Discussion
IncRNA has been identified to be critical regulators in many cancers. For example, LncRNA PVT1 induces angiogenesis in gastric cancer, and LncRNA GAS5 inhibits the development of ovarian cancer.15,16 Recently, a study has revealed that LncRNA NLIPMT represses the metastasis in breast cancer.13 However, whether it has a role in ESCC is unknown. In this study, we found that NLIPMT was significantly down-regulated in ESCC tissues and predicted poor survival of ESCC patients. In two ESCC cell lines, we revealed that NLIPMT overexpression inhibited cell proliferation and migration and promoted cell cycle arrest and apoptosis in ESCC cells. Therefore, NLIPMT may be a tumor suppressor in ESCC.
Through the bioinformatics search of whole NLIPMT genome sequence, we found that NLIPMT embedded miR-320. MiR-320 has been characterized as a tumor-suppressive miRNA in many types of cancers, and it suppresses cancer progression mainly by targeting oncogenes, such as neuropilin 1 and Mcl-1.17,18 However, the function of miR-320 in ESCC is unclear. Here, we found that miR-320 overexpression inhibited cell proliferation and migration and promoted cell cycle arrest and apoptosis in ESCC cells, suggesting that miR-320 could inhibit tumorigenesis in ESCC. Interestingly, we observed that miR-320 level was positively correlated with NLIPMT level in ESCC tissues. In ESCC cells, we uncovered that NLIPMT overexpression remarkably increased miR-320 level, while miR-320 overexpression did not affect the expression of NLIPMT. Therefore, we speculated that NLIPMT repressed the development of ESCC by up-regulating miR-320.
It has been reported that miR-320 can target survivin to play the roles of insulin against myocardial ischemia.12 In this study, we also verified that survivin was a direct target of miR-320. In addition, both NLIPMT and miR-320 overexpression reduced the mRNA and protein levels of survivin, while miR-320 knock-down abolished NLIPMT-inhibited expression of survivin. It suggests that NLIPMT inhibits survivin expression by up-regulating miR-320. Furthermore, we revealed that survivin overexpression attenuated NLIPMT and miR-320 overexpression-mediated inhibition on tumorigenesis in ESCC cells. Therefore, survivin is also implicated in the regulatory effects of NLIPMT and miR-320 in ESCC.
We also explored other possible mechanisms by which NLIPMT regulated the tumorigenesis in ESCC. We found that NLIPMT overexpression reduced PCNA and Bcl-2 proteins by up-regulating miR-320 expression. Therefore, NLIPMT/miR-320 may also affect proliferation and apoptosis in ESCC cells via regulating these two proteins. Moreover, we found that NLIPMT overexpression enhanced the activity of GSK3β by reducing its phosphorylation. Previous study has revealed that inhibition of GSK3β activity can abolish NLIPMT overexpression-inhibited proliferation, migration and invasion in breast cancer cells.13 Hence, we speculated that NLIPMY might also affect proliferation and migration in ESCC cells via regulating the activity of GSK3β. Further studies are needed to verify our speculation.
In this study, we found that the expression of NLIPMT and survivin was downregulated, and the expression of miR-320 was upregulated in ESCC. In addition, follow-up studies showed that the low levels of NLIPMT in ESCC might be used as an indicator of the poor survival of ESCC patients. However, whether NLIPMT expression can be used as a prognostic marker for ESCC patients requires further clinical validation.
To our knowledge, this study is the first to report that NLIPMT, as a precursor of miR-320, plays an important role in the progression of ESCC. At the same time, our research suggested a novel NLIPMT/miR-320/survivin pathway involved in the pathological process of ESCC.
Conclusion
In conclusion, we demonstrate that NLIPMT is a tumor suppressor in ESCC by inhibiting cell proliferation and migration and promoting cell cycle arrest and apoptosis in ESCC cells. Moreover, we reveal that NLIPMT functions by regulation of survivin and miR-320 expression. NLIPMT may be a potential prognostic biomarker in ESCC.
Funding Statement
There is no funding to report.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of Xingtai Medical College. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients provided written informed consent prior to their inclusion within the study.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Zeng H, Zheng R, Zhang S, et al. Esophageal cancer statistics in China, 2011: estimates based on 177 cancer registries. Thorac Cancer. 2016;7(2):232–237. doi: 10.1111/1759-7714.12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Screening PDQ, Board PE. Esophageal Cancer Prevention (PDQ®). PDQ Cancer Information Summaries. National Cancer Institute (US); 2018. [Google Scholar]
- 4.Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360–373. doi: 10.1053/j.gastro.2017.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islami F, Poustchi H, Pourshams A, et al. A prospective study of tea drinking temperature and risk of esophageal squamous cell carcinoma. Int J Cancer. 2019. doi: 10.1002/ijc.32220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimonosono M, Idichi T, Seki N, et al. Molecular pathogenesis of esophageal squamous cell carcinoma: identification of the antitumor effects of miR‑145‑3p on gene regulation. Int J Oncol. 2019;54(2):673–688. [DOI] [PubMed] [Google Scholar]
- 7.Lin DC, Wang MR, Koeffler HP. Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients. Gastroenterology. 2018;154(2):374–389. doi: 10.1053/j.gastro.2017.06.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qie S, Yoshida A, Parnham S, et al. Targeting glutamine-addiction and overcoming CDK4/6 inhibitor resistance in human esophageal squamous cell carcinoma. Nat Commun. 2019;10(1):1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8(1):61–70. doi: 10.1038/nrc2293 [DOI] [PubMed] [Google Scholar]
- 10.Jaiswal PK, Goel A, Mittal RD. Survivin: a molecular biomarker in cancer. Indian J Med Res. 2015;141(4):389–397. doi: 10.4103/0971-5916.159250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Lyu H, Wang J, et al. MicroRNA regulation and therapeutic targeting of survivin in cancer. Am J Cancer Res. 2014;5(1):20–31. [PMC free article] [PubMed] [Google Scholar]
- 12.Yang N, Wu L, Zhao Y, et al. MicroRNA‐320 involves in the cardioprotective effect of insulin against myocardial ischemia by targeting survivin. Cell Biochem Funct. 2018;36(3):166–171. doi: 10.1002/cbf.3328 [DOI] [PubMed] [Google Scholar]
- 13.Jiang Y, Lin L, Zhong S, et al. Overexpression of novel lncRNA NLIPMT inhibits metastasis by reducing phosphorylated glycogen synthase kinase 3β in breast cancer. J Cell Physiol. 2019;234(7):10698–10708. doi: 10.1002/jcp.27738 [DOI] [PubMed] [Google Scholar]
- 14.Li J, Xing M, Zhu M, et al. Glycogen synthase kinase 3β induces apoptosis in cancer cells through increase of survivin nuclear localization. Cancer Lett. 2008;272(1):91–101. doi: 10.1016/j.canlet.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37(30):4094–4109. doi: 10.1038/s41388-018-0250-z [DOI] [PubMed] [Google Scholar]
- 16.Li J, Yang C, Li Y, et al. LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep. 2018;38(2):BSR20171150. doi: 10.1042/BSR20171150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu YY, Chen YL, Jao YC, et al. miR-320 regulates tumor angiogenesis driven by vascular endothelial cells in oral cancer by silencing neuropilin 1. Angiogenesis. 2014;17(1):247–260. doi: 10.1007/s10456-013-9394-1 [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Zou P, Wang T, et al. Down-regulation of miR-320 associated with cancer progression and cell apoptosis via targeting Mcl-1 in cervical cancer. Tumour Biol. 2016;37(7):8931–8940. doi: 10.1007/s13277-015-4771-6 [DOI] [PubMed] [Google Scholar]