Abstract
Background:
Microorganisms are known to be involved in the formation of biofilm. These biofilms are often seen in chronic wound infections, surgical site infections, implants etc., These are capable of causing recalcitrant infections and most of them are also known to possess high antibiotic resistance.
Objectives:
This study was conducted to detect the biofilm formation in bacterial isolates from chronic wound infections.
Materials and Methods:
In the present study, ninety two isolates from chronic wound infections were identified by MALDI-TOF-MS (bioMerieux) and VITEK-2-MS (bioMerieux). These isolates were further screened for biofilm formation by three methods i. e., Tissue Culture Plate method (TCP), Tube Method (TM) and Congo Red Agar (CRA) method. Impact of biofilm production was correlated with the antibiotic resistant pattern.
Statistical Analysis:
Statistical analysis was done for all three methods considering TCP as Gold Standard and parameters like senitivity and specificity of TM i.e. 47.2 and 100% respectively.
Results:
Out of 92 isolates, biofilm formation was seen in 72 isolates (78.2%) by TCP method. 64 isolates were strong biofilm producers, 8 isolates were moderate biofilm producers and 20 isolates were nonbiofilm producing. High prevalence of biofilm formation was seen in nonhealing ulcers infected with Staphylococcus aureus followed by Klebsiella pneumoniae.
Conclusion:
Among three screening methods used for detection of biofilm production, TCP method is considered to be a standard and most reliable for screening of biofilm formation in comparison to TM and CRA.
Keywords: Biofilm screening, diabetic foot ulcer, nonhealing ulcer
INTRODUCTION
Biofilm is defined as the microbial population consisting of groups of bacterial cells which are adherent to a surface and are enclosed within a self-produced extracellular matrix.[1] Biofilm acts as an important virulence factor and provides protective environment for the organisms to survive.[2] The adaption of organisms to the surface attached growth within a biofilm is accompanied by significant changes in protein metabolism and gene expression which leads to resistance in antimicrobial therapy and host immune response.[3] The mechanism for antibiotic resistance also includes delayed penetration of antimicrobial agents through the biofilm matrix, altered growth rate of biofilm forming organisms and other physiological changes that occurs during the growth of biofilm.[4] Many pathogenic and nosocomial bacteria have been associated with the increase in antibiotic resistance and chronic recurrent infections especially in patients with diabetes.[5] Both pathogenic and nosocomial bacteria have been observed to exist as biofilms producers in natural environment as well as in infected tissues as polymicrobial communities.[1] The biofilm formation found to be the main cause of many chronic infections such as diabetic foot ulcers, cellulitis, necrotizing fasciitis which lead to the re-emergence of multidrug resistant strains and result in treatment failure.[5] Biofilms have a huge impact on the health care settings and associated with 65% of nosocomial infections.[3] Polysaccharide intracellular adhesin is the gene product of icaADBC which mediates cell to cell adhesion and regulates the biofilm formation by expressing itself.[6]
The chronic wound is a wound that is arrested in the inflammatory phase of wound healing and cannot progress further. The presence of necrotic tissue, foreign material and or bacteria regulates the ability of wound to heal by producing excessive neutrophils and proinflammatory cytokines. This environment allows the bacteria to proliferate and colonize the wound forming biofilms. Biofilm plays a significant role in nonhealing of chronic wounds. About 90% of chronic wounds suggest the formation of biofilm whereas only 6% of acute wounds are known to harbor the biofilm producing bacteria.[7] These chronic wounds or ulcers are commonly known to harbor infections with Gram-positive organisms like S. aureus, Enterococcus species and Gram-negative organisms like Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, and Acinetobacter baumannii complex.[5] The aim of the present study was to screen for biofilm production and its impact on antibiotic resistance profile of isolates from chronic wound infection.
MATERIALS AND METHODS
Place and duration of the study
The study was conducted in Department of Microbiology, Kasturba Medical College, Manipal, Karnataka from February 2017 to April 2017.
Selection of isolates
A total of 92 isolates were tested for the detection of Biofilm. Organisms were selected based on the following criteria: bacterial isolates from chronic wound infections for over a period ranging from 1 to 3 months. All the patients included in the study were above 18 years of age.
Isolates obtained from the chronic wounds were identified by MALDI-TOF- Vitek MS followed by antimicrobial susceptibility profile by VITEK 2 system (BioMérieux, Inc, Durham, NC).
Reference strain of strong biofilm producer S. aureus MTCC 1430 and nonbiofilm producing strain E. coli ATCC 25922 were used as controls.
Ethical committee approval
Institutional ethical committee approval was obtained from Kasturba Hospital, Manipal (Reference number – IEC 138/2017). Patient's Informed consent was taken prior to collection of the data and clinical isolates. Biofilm detection was done by following methods:
Tissue Culture Plate method
Tissue Culture Plate (TCP) assay as described by Christensen's et al., 1995 is the most widely used method and is considered as the standard method for detection of biofilm formation.[8] The isolates from fresh agar plates were inoculated in 5 ml of Trypticase soy broth and were kept for incubation at 37°C for 24 h. The cultures were diluted in 1:100 with fresh medium.
Individual wells of 96 well–flat bottom polystyrene TCPs (Thermo Fisher Scientific, Shanghai, China) were filled with 0.2 ml aliquots of diluted culture. The uninoculated broth was added to the wells to check sterility and nonspecific binding of the media. The plates are then incubated at 37°C for 24 h and after the incubation, contents were removed from plates by tapping gently. Plates were washed twice with 0.2 ml of phosphate buffer saline (pH 7.2) and incubated at 37°C for an hour. The plates were stained with 0.2 ml of 0.1% crystal violet for 10 min.[9]
Excess stain was removed by washing twice with deionized water and the plates were kept for drying. 200 μl of 33% glacial acetic acid was added to the wells. Optical density (OD) of the isolates were determined using micro ELISA auto reader (BIORAD 680) at a wavelength of 570 nm (OD 570 nm). The experiment was performed in triplicates. Biofilm formation was classified into Strong, moderate and weak/nonbiofilm producers as shown in Table 1. The interpretation of biofilm production was done according to criteria of Stepanovic et al.[10]
Table 1.
Interpretation of biofilm production based on optical density values of Tissue Culture Plate method
| Average OD value* | Biofilm production |
|---|---|
| <0.17 | Negative |
| 0.17-0.34 | Weak positive |
| 0.35-0.68 | Moderate positive |
| >0.68 | Strong positive |
*ODc=Average OD of negative control + 3 × SD of negative control. OD: Optical density, ODc: OD cut off value, SD: Standard deviation
Tube Method
This is a qualitative method used for the detection of biofilm as described by Christensen et al., 1995.[8] 5 ml of Trypticase soy broth was inoculated with a loopful of organism and incubated for 24 h at 37° C. Tubes were then decanted and washed with phosphate buffer saline (pH 7.2) and were allowed to air dry. Tubes were then stained with (0.1%) crystal violet for 10 min and washed with deionized water. The tubes were then left to air dry in inverted position.
The scoring of biofilm formation was done based on the control strains used. The organisms were considered to be biofilm producing when there is formation of visible layer on walls and at the base of the tube while the formation of ring at the interference of the liquid medium indicated that the organism was nonbiofilm producing. The experiment was performed in triplicates. The amount of biofilm formed was scored as (1) negative; (2) weak positive; (3) -moderate positive; (4) strong positive.
Congo Red Agar method
Freeman et al., 1989 described a simple qualitative method to detect the biofilm formation by Congo Red Agar (CRA) method.[11] This method involves use of special media that is Brain Heart Infusion (BHI) agar with Sucrose and Congo red in the following composition: BHI agar-52 g/L; sucrose-36 g/L; agar-10 g/L; congo red-0.8 g/L. Congo red was prepared as concentrated solution and autoclaved. It is added to the medium when agar is cooled to 55°C and poured into petriplates. Plates were inoculated and incubated for 24–48 h at 37°C. Black colonies with dry crystalline morphology was considered positive for biofilm producing organisms while weak or nonbiofilm producing organisms appeared to be pink in colour. The experiment was performed in triplicates.
RESULTS
Among 92 bacterial isolates from chronic wounds, the standard method TCP detected 64 (69.5%) isolates as strong and 8 (8.6%) isolates as moderate biofilm producers and remaining 20 (21.7%) isolates were nonbiofilm producing bacteria [Figure 1]. By Tube Method (TM), the number of organisms that showed strong biofilm formation was 13 (14%) and 21 (23%) organisms showed moderate and about 58 (63%) isolates showed weak or no biofilm formation [Figure 2].
Figure 1.
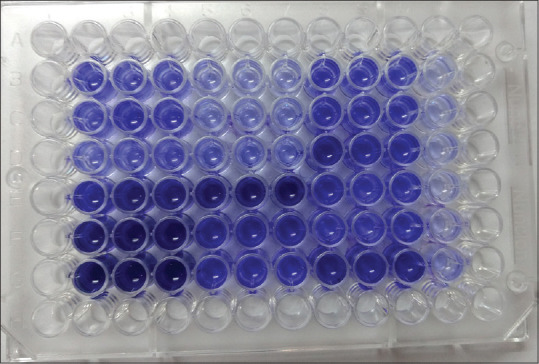
Screening of biofilm producing organisms by Tissue Culture Plate method
Figure 2.
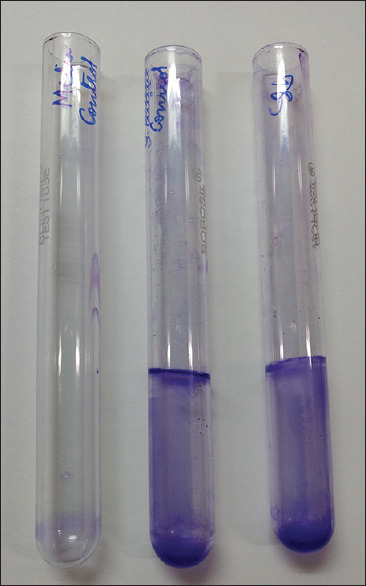
Screening of biofilm formation by Tube Method
Only 51 (55%) isolates were biofilm producers by CRA method [Figure 3 and Table 2].
Figure 3.

Screening of biofilm by Congo Red Agar method
Table 2.
Screening of 92 bacterial isolates for biofilm formation by Tissue Culture Plate, Tube Method and Congo Red Agar methods
| Biofilm production | TCP, n (%) | TM, n (%) | CRA, n (%) |
|---|---|---|---|
| Strong positive | 64 (69.5) | 13 (14) | 51 (55) |
| Moderate positive | 8 (8.6) | 21 (23) | - |
| Weak positive | 11 (12) | 33 (36) | - |
| Negative | 9 (10) | 25 (27) | 37 (40) |
TCP: Tissue Culture Plate, TM: Tube Method, CRA: Congo red
Statistical analysis was done for all the three methods to assess the sensitivity and specificity of these tests for detection of biofilm among bacterial isolates. TCP method was considered as Gold Standard for this study.[5,6] Comparative statistical analysis was done between TM and CRA with TCP. The parameters like sensitivity, specificity, positive predictive value, negative predictive value and accuracy were calculated for both TM and CRA method. True positives were biofilm producers by TCP, TM and CRA methods [Table 3]. False positive were biofilm producers by TM and CRA methods and not by TCP method. False negative were the isolates which were nonbiofilm producers by TM and CRA but were biofilm producing by TCP method. True negatives are those which were nonbiofilm producers by all the three methods.
Table 3.
Diagnostic parameters of Tube Method and Congo Red Agar method for biofilm detection
| Screening methods | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | Accuracy (%) |
|---|---|---|---|---|---|
| Tube Method | 47.2 | 100 | 100 | 34.4 | 58.6 |
| CRA | 56.9 | 50 | 80.3 | 24.3 | 55.4 |
CRA: Congo Red Agar
Sensitivity and specificity of TM were 47.2% and 100% respectively, while the sensitivity and specificity of CRA were 56.9% and 50% respectively.
Majority of biofilm producing organisms were S. aureus (37.5%), followed by K. pneumoniae (33.3%), P. aeruginosa (25%) and A. baumanii (8%). These organisms were isolated from diabetic foot ulcers (18.4%), nonhealing ulcers (38%), necrotizing fasciitis (16.3%), Fournier's gangrene (8.7%), miscellaneous (18.4%) [Tables 4 and 5]. Strong biofilm producing S. aureus were sensitive to cotrimoxazole, tetracycline, vancomycin and linezolid [Table 6]. Among biofilm producing Gram-negative bacteria, K. pneumoniae was sensitive to colistin and tigecycline which were isolated from the patients having nonhealing ulcers [Table 7]. We have observed increased antimicrobial resistance in biofilm producing bacteria than nonbiofilm producing bacteria.
Table 4.
Correlation of biofilm production of Gram-positive isolates and the antibiotics resistant pattern with the patient’s clinical conditions
| Organism | Clinical condition | Number of isolates (n) % | Biofilm formation |
Antibiotic Resistance Pattern | |
|---|---|---|---|---|---|
| Strong positive | Moderate positive | ||||
| Staphylococcus aureus (n=27) | Nonhealing ulcer | 12 | 12 | - | Ciprofloxacin, Erythromycin |
| Diabetic foot ulcer | 8 | 7 | 1 | Ciprofloxacin, Erythromycin, Oxacillin | |
| Necrotizing fasciitis | 2 | 2 | - | Ciprofloxacin | |
| Fournier’s gangrene | 1 | 1 | - | Ciprofloxacin, Erythromycin, Gentamicin | |
| Cellulitis | 1 | 1 | - | Ciprofloxacin, Oxacillin, Erythromycin | |
| Folliculitis | 1 | 1 | - | Ciprofloxacin | |
| Venous ulcer | 1 | 1 | - | Ciprofloxacin, Oxacillin, Erythromycin | |
| Furunculosis | 1 | 1 | - | Ciprofloxacin | |
Table 5.
Correlation of biofilm production of Gram-negative isolates and the antibiotic resistant pattern with the patient’s clinical conditions
| Organism | Clinical condition | Number of isolates | Biofilm formation |
Antibiotic Resistance Pattern | |
|---|---|---|---|---|---|
| Strong positive | Moderate positive | ||||
| Pseudomonas aeruginosa | Nonhealing ulcer | 8 | 7 | 1 | Ciprofloxacin, Meropenem, Cefoperazone - Sulbactum |
| Diabetic foot ulcer | 2 | 1 | 1 | ||
| Gangrene | 2 | 2 | NA | ||
| Necrotizing fasciitis | 3 | 3 | NA | ||
| Cellulitis | 4 | 4 | NA | ||
| Klebsiella pneumoniae | Nonhealing ulcer | 8 | 5 | 3 | Amoxicillin - Clavulanic acid, Ceftriaxone, Amikacin |
| Diabetic foot infection | 5 | 4 | 1 | ||
| Necrotizing fasciitis | 2 | 2 | NA | ||
| Gangrene | 4 | 4 | NA | ||
| Cellulitis | 2 | 2 | NA | ||
| Acinetobacter baumanii | Necrotizing fasciitis | 2 | 2 | NA | Amoxicillin - Clavulanic acid, ciprofloxacin |
| Nonhealing ulcer | 2 | 1 | 1 | ||
| Diabetic foot ulcer | 1 | 1 | NA | ||
| Cellulitis | 1 | 1 | NA | ||
Table 6.
Antibiotic resistance pattern of biofilm producing Staphylococcus aureus
| Antimicrobial agent | Biofilm producing Staphylococcus aureus (%) |
|---|---|
| Ciprofloxacin | 89 |
| Erythromycin | 41 |
| Co-trimoxazole | 4 |
| Clindamycin | 14 |
| Gentamicin | 14 |
| Cefoxitin | 55 |
Table 7.
Antibiotic resistance pattern of biofilm producing Gram-negative bacteria
| Antimicrobial agent | Biofilm producing Gram-negative organisms (%) (n=45) | Nonbiofilm producing Gram-negative organisms (%) (n=20) |
|---|---|---|
| Ciprofloxacin | 53 | 95 |
| Co-trimoxazole | 33 | 60 |
| Piperacillin - Tazobactum | 80 | 25 |
| Amikacin | 22 | 15 |
| Ceftriaxone | 46 | 75 |
| Cefoperazone - Sulbactum | 75 | 30 |
| Meropenem | 71 | 15 |
DISCUSSION
Bacteria are known to cause a variety of infections in humans. The pathogenicity of most of these bacteria is due to certain virulence factors they are known to possess. These virulence factors include pili, fimbriae, toxin production, biofilm formation etc., Out of all these virulence factors, the biofilm formation is responsible for most of the recalcitrant infections and it is found to be difficult to eradicate as the organisms involved in biofilm formation are highly resistant to antimicrobial agents.[5]
There are different methods that can be used for screening the biofilm formation. In the present study, 92 bacterial isolates from chronic wound infections were screened for biofilm formation using TCP, TM and CRA method. We have found that majority of biofilm producing bacteria were from nonhealing ulcers (38%). In TCP method, number of isolates showing biofilm formation was 72 (78.2%) and non or weak biofilm producers were 20 (21.7%). Addition of 1% Glucose to Trypticase Soy Broth enhances the biofilm production.[5,12,13] This was also reported by Mathur et al., and Bose et al.[6,14]
TM detected 37% of isolates as biofilm producers and 63% as nonbiofilm producers. By this method, none of the isolates gave false positive results but 38 (41.3%) isolates have given false negative results. The sensitivity of TM was found to be 47.2%, specificity was 100% and accuracy was 58.7% for biofilm detection. This method correlated well with TCP in identifying strong biofilm producers, but it was difficult to differentiate between moderate, weak and nonbiofilm producers. We have observed that most of the P. aeruginosa and K. pneumoniae isolates have shown strong biofilm production in TCP method where as they were observed to be weak or nonbiofilm producers by TM and CRA methods.
With CRA method, 51 isolates were found to be biofilm producing whereas 41 isolates were nonbiofilm producers. CRA method has shown a significant correlation with other two methods of Gram-positive isolates when compared to the Gram-negative isolates. Other parameters such as sensitivity (57%), specificity (50%) and accuracy (55%) were low compared to TM and TCP. By this method, only 10 isolates were found to be false positive while 31 isolates were found to be false negative. Knobloch et al., didn't recommend the CRA method for biofilm detection in their study.[15] Out of 128 isolates of S. aureus, CRA detected only 3.8% as biofilm producing as compared to TCP method which detected 57.1% as biofilm producing bacteria.[5,15]
Studies suggest that 4%–10% of foot ulcers are seen in patients with diabetes mellitus. These ulcers if left untreated can lead to severe complications and amputation. In our study, 29% of the isolates were Gram-positive isolates while 70.6% were Gram-negative isolates. This data correlates with Bansal et al., in which 76% isolates were Gram-negative while only 24% were Gram-positive.[2,16] In the present study, S. aureus (29%) and K. pneumoniae (26%) were most commonly isolated organisms followed by P. aeruginosa (20%).
In our study, among Gram-positive isolates, 55% methicillin-resistant S. aureus (MRSA) and 45% methicillin-sensitive S. aureus (MSSA) were obtained where both MRSA and MSSA were equally involved in biofilm formation. MSSA exhibited high level resistance to ciprofloxacin (88.8%). There was a lower level of resistance to other antibiotics (erythromycin, co-trimoxazole, gentamicin etc.) that were tested. These S. aureus strains were susceptible to linezolid and vancomycin. Prevalence of MRSA strains in our study was higher which is similar to others studies in which it ranged from 40% to 69.8%.[2,16,17,18,19] The MRSA displayed high level of resistance to erythromycin (66%).
In our study, 78.2% isolates showed biofilm production which is similar when compared to prior studies in which it ranged from 73% to 77.1%.[20,21] With reference to Gram-negative bacteria, 43% of the organisms were extended-spectrum β-lactamases (ESBL) producers, with highest production by K. pneumonia 19 isolates (29%) out of which 17 isolates (89.4%) showed biofilm formation. This is in parallel with preexisting studies in which 44.7% to 57.4% are ESBL positive.[16,18] The biofilm forming Gram-negative organisms showed a high level of resistance to piperacillin – tazobactum (80%), cefoperazone – sulbactum (75%), meropenem (71%) while nonbiofilm producing organisms showed high level of resistance to ciprofloxacin (95%), cotrimoxazole (60%) and ceftriaxone (75%). The high degree of drug resistance to piperacillin – tazobactum, cefoperazone – sulbactum and meropenem could be due to prolonged hospitalization with mechanical ventilation and these strains could be hospital acquired.
We have observed that among 9 multidrug resistant isolates, 8 isolates were showing biofilm formation. Swarna et al., reported that 80.39% of multidrug resistance (MDR) isolates were biofilm formers and it is significantly larger number as compared to present study.[20] The mechanism of multidrug resistance in biofilm forming organisms is believed to be a direct result of close cell-cell contact in the biofilm, which allows for easy transfer of plasmids containing MDR genes.[2,22]
CONCLUSION
In this study we found that the biofilm formation might be one of the factors responsible for nonhealing of ulcers and their chronicity. The formation of biofilm might also be the reason for the emerging resistance of antimicrobial agents in patients with nonhealing ulcers and Diabetic foot ulcers.
Financial support and sponsorship
All the materials and equipments required for performing the experiment were provided by Department of Microbiology and Department of Virology, Kasturba Medical College, Manipal University, Manipal.
Conflicts of interest
There are no conflicts of interest.
Acknowledgement
We are grateful to Manipal Academy of Higher Education, Manipal, Karnataka for providing this opportunity and the constant support. We would like to thank Dr. Arun Kumar G., Professor and Head of Department of Virology (MCVR), Manipal Academy of Higher Education, Manipal, Karnataka for helping us with the resources required for completing the work. We would also like to thank the faculty of Department of Biostatistics, Manipal Academy of Higher Education for their valuable guidance in statistical analysis.
REFERENCES
- 1.Sanchez CJ, Jr, Mende K, Beckins ML, Akers KS, Romano DR, et al. Biofilm formation by clinical isolates and implications in chronic infections. BMC Infect Dis. 2013;13:47. doi: 10.1186/1471-2334-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shashikala V, Ali F, Lokare N, Matew J. Diabetic foot ulcers and biofilm formation-The culprits. Int J Biomed Adv Res. 2016;7:428–33. [Google Scholar]
- 3.CharanKaur D, Khare AS. Biofilm formation and antibiotic susceptibility pattern in MRSA strains in a tertiary care hospital. Indian J Basic Appl Med Res. 2013;1:37–44. [Google Scholar]
- 4.Andhale JD, Misra RN, Gandham NR, Angadi KM, Jadhav SV, Vyawahare CR, et al. Incidence of Pseudomonas aeruginosa with special reference to drug resistance and biofilm formation from clinical samples in tertiary care hospital. J Pharm Biomed Sci. 2016;6:387–91. [Google Scholar]
- 5.Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in clinical isolates. Braz J Infect Dis. 2011;15:305–11. [PubMed] [Google Scholar]
- 6.Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococcus: An evaluation of three different screening methods. Indian J Med Microbiol. 2006;24:25–9. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- 7.Attinger C, Wolcott R. Clinically addressing biofilm in chronic wounds. Adv Wound Care. 2012;1:127–32. doi: 10.1089/wound.2011.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen GD, Simpson WA, Younger JA, Baddour LM, Barrett FF, Melton DM, et al. Adherence of coagulase negative staphylococci to plastic tissue cultures: A quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1995;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saify H, Patidar RK, Khare M, Sahare KN, Singh V. Difference in biofilm development capability of vancomycin and ciprofloxacin resistant Staphylococcus aureus clinical isolates. Res J Infect Dis. 2013. Available from: http://www.hoajonline.com/jou rnals/pdf/2052-5958-1-8 .
- 10.Stepanovic S, Vukovi D, Hola V, Bonaventura GD, Djukić S, Cirković I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practi- cal recommendations for assessment of biofilm production by Staphylococci. APMIS. 2007;115:891–9. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 11.Freeman J, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42:872–4. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim L. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efthikar F, Speert DP. Biofilm formation by persistent and non persistent isolates of Staphylococcus epidermidis from neonatal intensive care unit. J Hosp Infect. 2009;71:112–6. doi: 10.1016/j.jhin.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Bose S, Khodke M, Basak S, Mallick SK. Detection of biofilm producing staphylococci: Need of the hour. J Clin Diagn Res. 2009;3:1915–20. [Google Scholar]
- 15.Knobloch JK, Horsetkotte MA, Rohde H, Mack D. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med Microbiol Immunol. 2002;191:101–06. doi: 10.1007/s00430-002-0124-3. [DOI] [PubMed] [Google Scholar]
- 16.Bansal E, Garg A, Bhatia S, Attri AK, Chander J. Spectrum of microbial flora in diabetic foot ulcers. Indian J Pathol Microbiol. 2008;51:204–8. doi: 10.4103/0377-4929.41685. [DOI] [PubMed] [Google Scholar]
- 17.Gadepalli R, Dhawan B, Sreenivas V, Kapil A, Ammini AC, Chaudhary RA. Clinico microbiological study of diabetic foot ulcers in an Indian tertiary care hospital. Diabetes Care. 2006;29:1727–32. doi: 10.2337/dc06-0116. [DOI] [PubMed] [Google Scholar]
- 18.Rani V, Lakshmi JN. A comparative study of diabetic and Non diabetic wound infections with special reference to MRSA and ESBL. Int J Curr Microbiol Appl Sci. 2014;3:546–54. [Google Scholar]
- 19.Tentolouris N, Jude EB, Smirnof I, Knowles EA, Boulton AJ. Methicillin resistant Staphylococcus aureus: An increasing problem in diabetic foot. Diabet Med. 1999;1:767–71. doi: 10.1046/j.1464-5491.1999.00132.x. [DOI] [PubMed] [Google Scholar]
- 20.Swarna SR, Madhavan R, Gomathi S, Thamaraiselvi S. A study of biofilm on diabetic foot ulcer. Int J Res Pharm Biomed Sci. 2012;3:1809–14. [Google Scholar]
- 21.Zubair M, Malik A, Ahmad J, Rizvi M, Farooqui KT, Rizwi MW. A study of biofilm production by Gram negative organisms isolated from diabetic foot ulcer patients. Biol Med. 2011;3:147–57. [Google Scholar]
- 22.Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–9. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]


