Abstract
Plasmodium ovale is a benign tertian malaria parasite that morphologically resembles Plasmodium vivax. P. ovale also shares similar tertian periodicity and can cause relapse in patients without a radical cure, making it easily misidentified as P. vivax in routine diagnosis. Therefore, its prevalence might be underreported worldwide. The present study aimed to quantify the prevalence of P. ovale misidentified as P. vivax malaria using data from studies reporting confirmed P. ovale cases by molecular methods. Studies reporting the misidentification of P. ovale as P. vivax malaria were identified from three databases, MEDLINE, Web of Science, and Scopus, without language restrictions, but the publication date was restricted to 1993 and 2020. The quality of the included studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS). The random-effects model was used to estimate the pooled prevalence of the misidentification of P. ovale as P. vivax malaria by the microscopic method when compared to those with the reference polymerase chain reaction method. Subgroup analysis of participants was also performed to demonstrate the difference between imported and indigenous P. ovale cases. The heterogeneity of the included studies was assessed using Cochran's Q and I2 statistics. Publication bias across the included studies was assessed using the funnel plot and Egger’s test, and if required, contour-enhanced funnel plots were used to identify the source(s) of funnel plot asymmetry. Of 641 articles retrieved from databases, 22 articles met the eligibility criteria and were included in the present study. Of the 8,297 malaria-positive cases identified by the PCR method, 453 P. ovale cases were confirmed. The pooled prevalence of misidentification of P. ovale as P. vivax malaria by the microscopic method was 11% (95% CI: 7–14%, I2: 25.46%). Subgroup analysis of the participants demonstrated a higher prevalence of misidentification in indigenous cases (13%, 95% CI: 6–21%, I2: 27.8%) than in imported cases (10%, 95% CI: 6–14%, I2: 24.1%). The pooled prevalence of misidentification of P. vivax as P. ovale malaria by the microscopic method was 1%, without heterogeneity (95% CI: 0–3%, I2: 16.8%). PCR was more sensitive in identifying P. ovale cases than the microscopic method (p < 0.00001, OR: 2.76, 95% CI: 1.83–4.15, I2: 65%). Subgroup analysis of participants demonstrated the better performance of PCR in detecting P. ovale malaria in indigenous cases (p: 0.0009, OR: 6.92, 95% CI: 2.21–21.7%, I2: 68%) than in imported cases (p: 0.0004, OR: 2.15, 95% CI: 1.41–3.29%, I2: 63%). P. ovale infections misidentified as P. vivax malaria by the microscopic method were frequent and led to underreported P. ovale cases. The molecular identification of P. ovale malaria in endemic areas is needed because a higher rate of P. ovale misidentification was found in endemic or indigenous cases than in imported cases. In addition, updated courses, enhanced training, and refreshers for microscopic examinations, particularly for P. ovale identification, are necessary to improve the microscopic identification of Plasmodium species in rural health centres where PCR is unavailable.
Subject terms: Malaria, Epidemiology
Introduction
Plasmodium ovale is one of the five Plasmodium species that can infect humans, namely: P. falciparum, P. vivax, P. malariae, P. knowlesi, and P. ovale1. It is endemic to tropical western Africa and the Southwest Pacific but rarely occurs outside of these regions, with less than 1% isolates2. The rarity of P. ovale infections in published studies might be due to the low species-specific parasitaemia and the short duration of patient infections3. Compared to the other four Plasmodium species that can infect humans, P. ovale has morphological characteristics and the ability to cause relapse, similar to P. vivax. P. ovale malaria does not usually cause severe malaria, but a study of 1365 P. ovale malaria cases demonstrated that 3% of severe malaria cases were caused by P. ovale infection, including jaundice (1.1%), severe anaemia (0.88%), and pulmonary impairments (0.59%)4. The ability to cause relapse by way of hypnozoites in the liver is recognized in both P. ovale and P. vivax malaria and leads to the reactivation of dormant liver stages weeks or months later1.
The routine diagnostic method of patients suspected of having P. ovale infection is the visualization of thick and thin blood smears by microscopy and/or rapid diagnostic tests (RDT), if available5. It is well documented that microscopy is an inexpensive and rapid quantification of parasites and is a relatively sensitive method; nevertheless, it has several limitations; it is time consuming, it misdiagnoses Plasmodium species, it requires expert or well-trained microscopists, and it misses Plasmodium species in case of a low parasite density in mixed infection6,7. Since P. ovale infects only young erythrocytes, the parasite density is low, and infection with other Plasmodium species is mixed8, resulting in missed identification by microscopists. The advantages of the recent molecular technique in identifying P. ovale, polymerase chain reaction (PCR), which amplifies 18S small subunit ribosomal RNA (18S rRNA) target genes offers high sensitivity and high specificity9. Moreover, two distinct P. ovale subspecies, P. ovale curtisi (classic type) and P. ovale wallikeri (variant type), occur globally10. No differences in the clinical or laboratory characteristics or other demographic data were summarized. Some studies demonstrated that higher parasite density, platelet counts, and latency periods were reported in P. ovale curtisi infection than in P. ovale wallikeri infection11,12.
Routine identification of P. ovale relies on blood smear examination, which can lead to underestimation of the true prevalence of P. ovale globally, with little clinical consequence of the misidentification of P. ovale as P. vivax. Molecular techniques such as PCR were used to identify P. ovale to prevent confusion with P. vivax or to prevent missed identification of P. ovale mixed infection with other Plasmodium species, such as mixed infection with P. falciparum or with P. knowlesi. Mixed infections of Plasmodium spp. could lead to severe malaria if the treatment was inadequate or incorrect13; therefore, it is also necessary to detect sub-microscopic mixed infection of P. ovale by molecular methods. The present study aimed to quantify the microscopic misidentification of P. ovale as P. vivax to support and promote the use of molecular techniques for the accurate identification of P. ovale malaria and promote a training course on P. ovale microscopic identification by health governors to increase the accuracy of P. ovale identification by microscopic methods.
Methods
Report guideline of the systematic review
The protocol of this study was registered at the International Prospective Register of Systematic Reviews (PROSPERO)14 with registration number CRD42020204049. The report of this systematic review and meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Checklist S1)15.
The search strategy
Searches were performed in three databases that included MEDLINE, Scopus, and Web of Science, without language restrictions, but the publication date was restricted to 1993 and 2020. The search terms used are provided in Table S1. Searches of the reference list of the final included studies and review articles were also performed to reduce the chance of missing relevant studies.
Eligibility criteria
Observational studies that reported the prevalence of both P. vivax and P. ovale malaria by microscopy and PCR or molecular methods were screened for the misidentification of P. ovale as P. vivax malaria by a microscopic method. The inclusion criteria were (1) studies reporting the prevalence of the misidentification of P. ovale as P. vivax malaria by a microscopic method and (2) studies using PCR or molecular methods to confirm the Plasmodium species. The exclusion criteria were (1) case reports or case series that reported a small number of patients, which can lead to reporting bias for meta-analysis, (2) studies using both microscopic and molecular methods to identify Plasmodium species where the data could not be extracted, (3) studies carried out on the performance of tests as those tests attempted to develop new techniques or new tests for the detection of Plasmodium species, (4) experimental studies that aimed to explore the new finding related to Plasmodium species, (5) studies with no misidentification of P. ovale to P. vivax, or no P. vivax malaria was observed as those studies did not provide the evidence of the misidentification of P. ovale as P. vivax malaria, (6) review articles, (7) studies without the full text, (8) clinical trials, guidelines, studies using the same participants, and other studies without relevant data.
Study selection and data extraction
Two authors (MK and FRM) selected potentially relevant studies according to the eligibility criteria. Any discordance in the study selection was resolved by consensus. Data selection from relevant studies was managed using Endnote software X7 (Clarivate Analytics, Philadelphia, USA). Data extraction was also performed by two authors (MK and FRM) and crosschecked by the third author (KUK). The following data were extracted: name of first author, year of publication, study area, years of the study, study design, age range (years), gender (male, %), participants (imported or indigenous), PCR for identified Plasmodium spp., target gene for PCR, number of malaria cases identified by microscopy and PCR methods, number of P. vivax cases identified by microscopy and PCR methods, number of P. ovale cases identified by microscopy and PCR methods, number of misidentifications of P. ovale as P. vivax malaria by microscopy, and number of misidentifications of P. vivax as P. ovale malaria by microscopy. The data were extracted to pilot-standardized sheets created using Microsoft Excel 2010 (Microsoft Corporation, Washington, USA) before meta-analyses.
Quality of the included studies
The quality of the individual studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) (Table S2)16. The tool was comprised of 4 domains: patient selection, index test, reference standard, and flow and timing. Each domain was assessed in terms of risk of bias and concerns of applicability16. The index test was microscopy, while the reference standard was PCR method. The results of the QUADAS of all included studies were summarized in the methodological quality graph and summary.
Outcomes
The primary outcome of the present study was the prevalence of misidentification of P. ovale as P. vivax, and also P. vivax as P. ovale malaria by the microscopic method. The secondary outcome was the performance of the PCR test to identify P. ovale malaria compared to that of the microscopic method.
Data synthesis
For the primary outcome, the pooled prevalence of misidentification of P. ovale as P. vivax, and also P. vivax as P. ovale malaria by the microscopic method was estimated using a random-effects model provided by STATA Statistical Software version 15.0 (StataCorp. 2017. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC). The number of P. ovale misidentified as P. vivax by microscopy and the total number of true P. ovale malaria identified by PCR were computed using the “metaprop case total case” command provided in STATA Statistical Software version 15.0. For the secondary outcome, the performance of PCR to identify P. ovale malaria compared to that of the microscopic method, was estimated using the random-effects model provided by Review Manager 5.3 (The Cochrane Collaboration, London, UK) available at https://training.cochrane.org/. The results of primary and secondary outcomes were demonstrated as pooled prevalence or odds ratios (ORs) with their 95% confidence intervals (CIs) in the forest plot. Heterogeneity across the included studies was assessed using Cochran's Q and I2 statistics. Subgroup analyses of participants (imported or indigenous cases) were performed to identify the difference in the population that might affect the misidentification of P. ovale as P. vivax malaria.
Publication bias
Publication bias among the included studies was assessed by visualization of the funnel plot asymmetry. Funnel plot asymmetry was also assessed by Egger's test to quantify publication bias if it existed.
Consent for publication
All authors have read the manuscript and consent to its publication.
Results
Search results
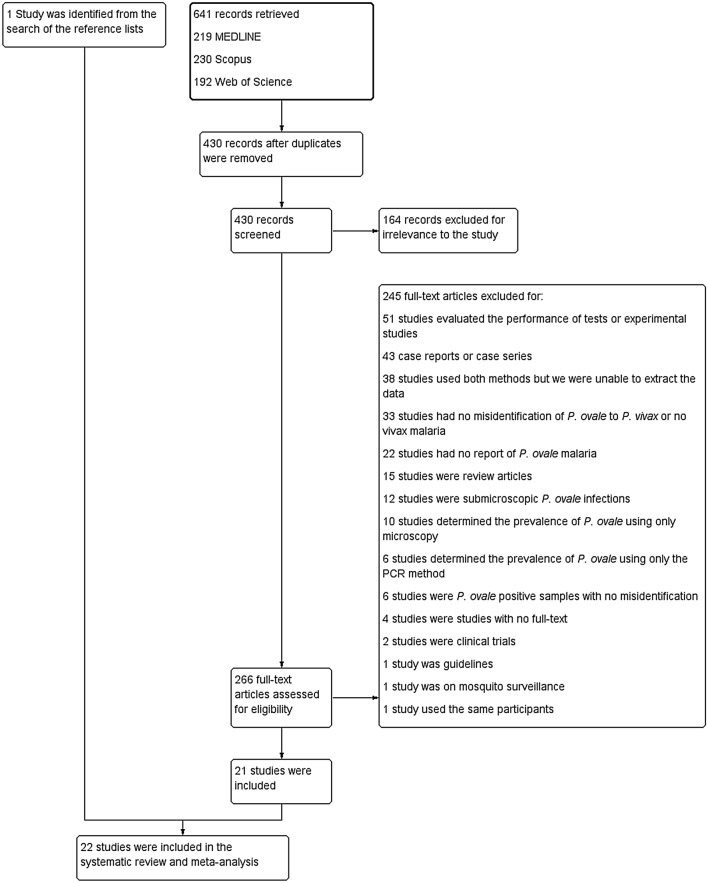
The literature search identified 641 records through three different databases: 219 for MEDLINE, 230 for Scopus, and 192 for Web of Science (Fig. 1). After the removal of duplicate articles, the remaining 430 studies were screened. In total, 164 records were excluded because they were irrelevant. The 266 potentially relevant studies were assessed in detail and 245 studies were excluded for the following reasons: 51 studies evaluated the performance of tests or experimental studies, 43 were case reports or case series, 38 studies used both methods but we were unable to extract the data, 33 had no misidentification of P. ovale to P. vivax or no vivax malaria, 22 had no report of P. ovale malaria, 15 were review articles, 12 were sub-microscopic P. ovale infections, 10 determined the prevalence of P. ovale using only microscopy, 6 determined the prevalence of P. ovale using only the PCR method, 6 were P. ovale positive samples with no misidentification, 4 were studies had no full-text, 2 were clinical trials, 1 was guidelines, 1 was on mosquito surveillance, and 1 study used the same participants. Twenty-one studies17–37 were included because they met the eligibility criteria, and one additional study38 was selected after reviewing the reference lists of the 20 included studies and review articles. Finally, a total of 22 studies17–38 were included for qualitative and quantitative syntheses.
Figure 1.
Flow chart for study selection.
Characteristics of the included studies
All characteristics of the included studies can be found in Table 1. Twenty-two studies reported evidence of P. ovale misidentification as P. vivax by a microscopic method between 1993 and 2020. Of the 22 included studies, 8 studies20,23,26,27,29,33,35,37 reported evidence of P. vivax misidentification as P. ovale by the microscopic method. Most of the included studies were observational cross-sectional studies (17/22, 77.3%). Most of the studies (8/22, 36.4%) were conducted in Asia (2 in China34,36, 2 in Thailand25,30, Singapore38, Israel23, Sri Lanka24, and Malaysia35), Europe (7/22, 31.8%) (3 Italy18,28,29, 2 Belgium27,37, Germany22, and Switzerland32), America (4/22, 18.2%) (3 United States19,20,31, Canada26), Africa (2/22, 9.1%) (2 Ethiopia17,21), and Oceania (1/22, 4.55%) (1 Australia33). Most of the included studies (13/22, 59.1%) identified P. ovale using blood samples from patients suspected of having malaria. Almost half of the included studies (10/22, 45.5%) reported using nested PCR targeting 18S rRNA for identifying Plasmodium parasites, while the remaining studies used real-time PCR or did not specify the type of PCR. Twenty-two studies reported that a total of 8079 malaria cases were identified by microscopic methods, while a total of 8297 malaria cases were identified by PCR. A total of 453 P. ovale cases were confirmed by the PCR method, while 204 P. ovale cases were first identified by the microscopic method and subsequently confirmed as P. ovale by the PCR method. The misidentification of P. vivax and other Plasmodium species is shown in Table 2.
Table 1.
Characteristics of the included studies.
| No | Author, year | Study area (years of the survey) | Study design | Age range (years) | Gender (male, %) | Participants | Molecular techniques for Plasmodium sp. | Target gene | Microscopy (include mixed infection) | PCR/Molecular techniques (include mixed infection) | No. of P. ovale as P. vivax | No. of P. vivax as P. ovale | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. malaria | No. of P. vivax | No. of P. ovale (n/N)* | No. malaria | No. of P. vivax | No. of P. ovale | |||||||||||
| 1 | Alemu et al., 2014 | Ethiopia (2013) | Cross sectional study | NS | NS | 297 patients with suspected malaria | Nested PCR | 18S rRNA | 183 | 51 | 0 | 217 | 68 | 9 | 4 | 0 |
| 2 | Calderaro et al., 2013 | Italy (2000–2012) | Retrospective cross-sectional study | NS | PCR positive, 82 (64%) | 398 patients with suspected malaria | Real-time PCR | 18S rRNA | 126 | 9 | 8 (7/8) | 128 | 7 | 14 | 1 | 0 |
| 3 | Chavatte et al., 2015 | Singapore (2001–2014) | Retrospective cross-sectional study | NS | NS | 1053 malaria positive | Real-time PCR | 18S rRNA | 1053 | NS | 0 | 1053 | NS | 11 | 11 | 0 |
| 4 | Cullen et al., (2014) | USA (2012) | Retrospective cross-sectional study | NS | NS | 104 malaria positive for genetic markers | NS | 18S rRNA | 104 | 9 | 7 (5/7) | 104 | 14 | 12 | 1 | 0 |
| 5 | Cullen et al., (2016) | USA (2013) | Retrospective cross-sectional study | NS | NS | 137 malaria positive for genetic markers | NS | 18S rRNA | 137 | 8 | 5 (2/5) | 137 | 15 | 14 | 1 | 1 |
| 6 | Díaz et al., 2015 | Ethiopia (2010–2011) | Cross sectional study | Mean 13.4 (1–80), median 10 | 1507 | 3060 patients with suspected malaria, for microscopy and 1209 for PCR | Semi-nested multiplex PCR | Cytochrome b | 736 | 436 | 0 | 788 | 398 | 24 | 2 | 0 |
| 7 | Frickmann et al., 2019 | Germany (2010–2019) | Retrospective cross-sectional study | 31.6 ± 14.8 | 56, 72.7% | 77 P. ovale positive cases | Real-time PCR | 18S rRNA and Po-ldh | 77 | 16 | 25 (25/25) | 77 | 0 | 77 | 3 | 0 |
| 8 | Grossman et al., 2016 | Israel (2009–2015) | Cross-sectional study | NS | NS | 357 patients with suspected malaria | Real-time PCR | 18S rRNA | 307 | 73 | 7 (2/7) | 288 | 104 | 23 | 3 | 4 |
| 9 | Gunasekera et al., 2018 | Sri Lanka (2014–2017) | Cross-sectional study | PCR positive 37 (1–66) | PCR positive 159, 91.9% | 350 patients with suspected malaria | Nested PCR | 18S rRNA | 164 | 77 | 9 (9/9) | 173 | 77 | 10 | 1 | 0 |
| 10 | Han et al., 2007 | Thailand | Retrospective cross-sectional study | NS | NS | 121 malaria positive and negative cases | Nested PCR | 18S rRNA | 68 | 34 | 5 (5/5) | 71 | 10 | 8 | 2 | 0 |
| 11 | Humar et al., 1997 | Canada (1993–1995) | Cross-sectional study | NS | NS | 182 patients with suspected malaria | Nested PCR | 18S rRNA | 159 | 87 | 11 (10/11) | 159 | 88 | 15 | 3 | 1 |
| 12 | Loomans et al., 2019 | Belgium (2013–2017) | Cross-sectional study | Median (36, 1–84) | 610, 64.4% | 947 malaria positive and negative cases | Real-time PCR | 18S rRNA | 927 | 77 | 46 (27/46) | 893 | 81 | 63 | 8 | 3 |
| 13 | Maltha et al., 2010 | Belgium (1996–2009) | Retrospective cross-sectional study | 35 (1–84) | 2.16:1 | 590 malaria positive and negative cases | NS | 18S rRNA | 495 | 79 | 73 (69/73) | 495 | 76 | 76 | 7 | 4 |
| 14 | Paglia et al., 2012 | Italy (1998–2003) | Cross-sectional study | Malaria positive 38 ± 12 | 2:1 | 1226 patients with suspected malaria | Semi-nested PCR | 18S rRNA | 187 | 17 | 4 (3/4) | 196 | 20 | 7 | 2 | 0 |
| 15 | Perandin et al., 2004 | Italy | Retrospective study | NS | NS | 122 patients with suspected malaria | Nested PCR | 18S rRNA | 61 | 12 | 3 (2/3) | 60 | 8 | 10 | 5 | 1 |
| 16 | Putaporntip et al., 2009 | Thailand (2006–2007) | Cross-sectional study | Median (23, 1–81) | 2.25:1 | 1874 patients with suspected malaria | Nested PCR | 18S rRNA | 1695 | 1013 | 0 | 1751 | 1192 | 18 | 1 | 0 |
| 17 | Reller et al., 2013 | USA (2004–2012) | Cohort study | NS | NS | 148 malaria positive | Multiplex quantitative real-time PCR | 18S rRNA | 146 | 38 | 17 (17/17) | 157 | 37 | 20 | 2 | 0 |
| 18 | Rougemont et al., 2004 | Switzerland (2002–2003) | Prospective study | NS | NS | 60 patients with suspected malaria | Real-time PCR | 18S rRNA | 31 | 4 | 3 (2/3) | 34 | 5 | 4 | 1 | 0 |
| 19 | Whiley et al., 2004 | Australia | Prospective study | NS | NS | 279 patients with suspected malaria | Nested PCR | 18S rRNA | 219 | 131 | 6 (5/6) | 225 | 131 | 6 | 1 | 1 |
| 20 | Xu et al., 2016 | China (2012–2014) | Cross-sectional study | 20–54 (96.8%) | 92.5:1 | 374 patients with suspected malaria | Nested PCR | 18S rRNA | 374 | 40 | 14 (14/14) | 364 | 44 | 16 | 2 | 0 |
| 21 | Yusof et al., 2014 | Malaysia (2012–2013) | Retrospective study | NS | 77.9% | 457 malaria positive | Nested PCR | 18S rRNA | 457 | 137 | 1 (0/1) | 543 | 144 | 2 | 2 | 1 |
| 22 | Zhou et al., 2014 | China (2008–2012) | Cross-sectional study | NS | NS | 562 patients with suspected malaria | Nested PCR | 18S rRNA | 373 | 275 | 0 | 384 | 288 | 14 | 4 | 0 |
NS not specified, *n/N number of P. ovale cases confirmed by PCR/number of P. ovale cases detected by microscopy.
Table 2.
Misidentification of any Plasmodium species such as P. ovale by microscopic method.
| No | Author, year | Microscopy | PCR/molecular techniques | ||
|---|---|---|---|---|---|
| No. of P. ovale | True P. ovale cases | Number of misidentifications | Misidentified Plasmodium species | ||
| 1 | Alemu et al., 2014 | 0 | – | – | – |
| 2 | Calderaro et al., 2013 | 8 (7/8) | 7 | 1 | P. falciparum |
| 3 | Chavatte et al., 2015 | 0 | – | – | – |
| 4 | Cullen et al., 2014 | 7 (5/7) | 5 | 2 | 1 P. falciparum, 1 P. malariae |
| 5 | Cullen et al., 2016 | 5 (2/5) | 2 | 3 | 2 P. falciparum, 1 P. vivax |
| 6 | Díaz et al., 2015 | 0 | – | – | – |
| 7 | Frickmann et al., 2019 | 25 (25/25) | 25 | 0 | – |
| 8 | Grossman et al., 2016 | 7 (2/7) | 2 | 5 | 4 P. vivax, 1 P. falciparum + P. vivax |
| 9 | Gunasekera et al., 2018 | 9 (9/9) | 9 | 0 | – |
| 10 | Han et al., 2007 | 5 (5/5) | 5 | 0 | – |
| 11 | Humar et al., 1997 | 11 (10/11) | 10 | 1 | 1 P. vivax |
| 12 | Loomans et al., 2019 | 46 (27/46) | 27 | 19 | 10 P. falciparum, 3 P. vivax, 6 P. malariae |
| 13 | Maltha et al., 2010 | 73 (69/73) | 69 | 4 | 4 P. vivax |
| 14 | Paglia et al., 2012 | 4 (3/4) | 3 | 1 | 1 P. falciparum |
| 15 | Perandin et al., 2004 | 3 (2/3) | 2 | 1 | 1 P. vivax |
| 16 | Putaporntip et al., 2009 | 0 | – | – | – |
| 17 | Reller et al., 2013 | 17 (17/17) | 17 | 0 | – |
| 18 | Rougemont et al., 2004 | 3 (2/3) | 2 | 1 | 1 P. falciparum |
| 19 | Whiley et al., 2004 | 6 (5/6) | 5 | 1 | 1 P. vivax |
| 20 | Xu et al., 2016 | 14 (14/14) | 14 | 0 | – |
| 21 | Yusof et al., 2014 | 1 (0/1) | 0 | 1 | 1 P. vivax |
| 22 | Zhou et al., 2014 | 0 | – | – | – |
Quality of the included studies
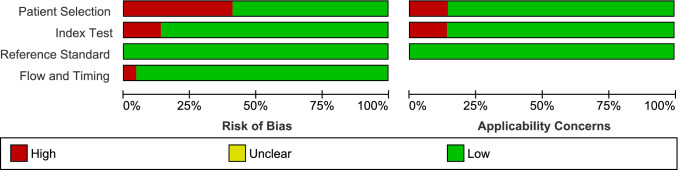
The quality of the individual studies assessed using QUADAS can be referenced in Fig. 2 and Supplementary Fig. 1.
Figure 2.
Methodological quality graph.
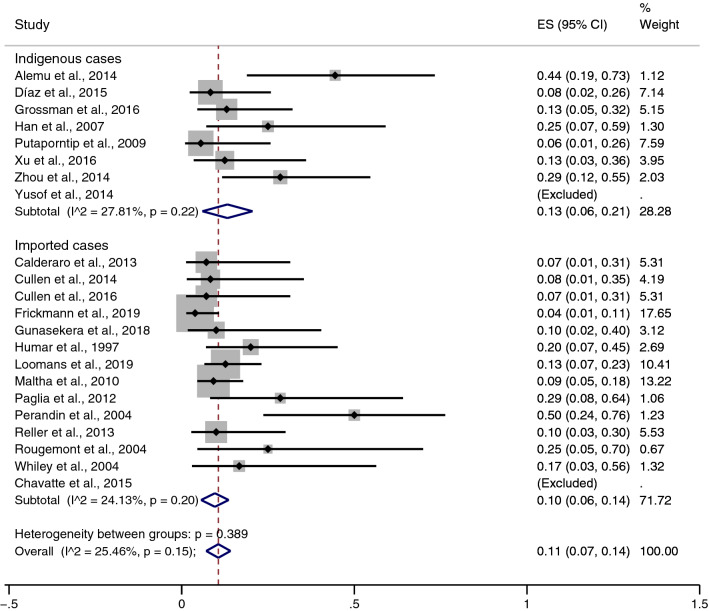
The pooled prevalence of the misidentification of P. ovale as P. vivax malaria
The pooled prevalence of the misidentification of P. ovale as P. vivax malaria was estimated using all 22 included studies (Fig. 3). Overall, the pooled prevalence of misidentification of P. ovale as P. vivax malaria by the microscopic method was 11% without heterogeneity (95% CI: 7–14%, I2: 25.5%). The prevalence of misidentification of P. ovale as P. vivax malaria in two studies35,38 was not estimated because both studies reported 100% misidentification. Subgroup analysis of participants demonstrated a higher prevalence of misidentification in indigenous cases (13%, 95% CI: 6–21%, I2: 27.8%) than in imported cases (10%, 95% CI: 6–14%, I2: 24.1%). The highest rate of misidentification in indigenous cases (44%, 95% CI: 79–43%) was demonstrated in the study by Alemu et al.17, while the highest rate of misidentification in imported cases (50%, 95% CI: 24–76%) was demonstrated in the study by Perandin et al.29.
Figure 3.
Pooled prevalence of misidentification of P. ovale as P. vivax malaria. ES: Estimated prevalence. The pooled prevalence was estimated using STATA Statistical Software version 15.0 (StataCorp. 2017. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC).
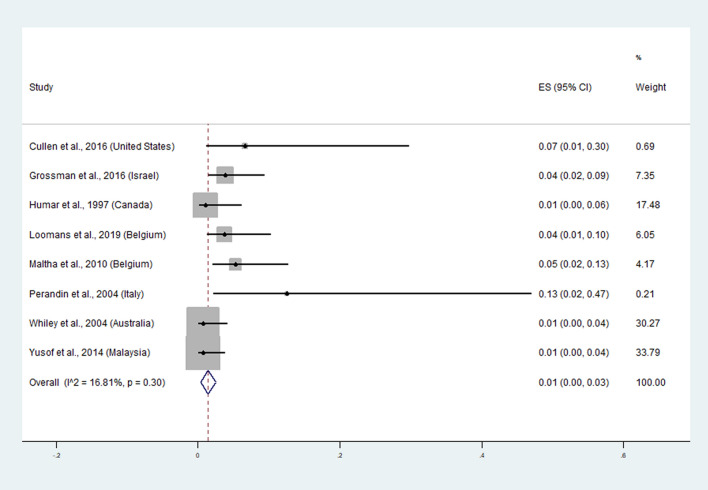
The pooled prevalence of the misidentification of P. vivax as P. ovale malaria
The pooled prevalence of the misidentification of P. vivax as P. ovale malaria was estimated using the data from 8 studies20,23,26,27,29,33,35,37. Overall, the pooled prevalence of misidentification of P. vivax as P. ovale malaria by the microscopic method was 1% without heterogeneity (95% CI: 0–3%, I2: 16.8%) (Fig. 4). A high rate of misidentification was reported in imported cases in Italy (13%, 95% CI: 2–47%)29, the United States (7%, 1–30%)20, Belgium (5%, 2–13%)27, Belgium (4%, 1–10%)37, and Israel (4%, 2–9%)23.
Figure 4.
Pooled prevalence of misidentification of P. vivax as P. ovale malaria. ES: Estimated prevalence. The pooled prevalence was estimated using STATA Statistical Software version 15.0 (StataCorp. 2017. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC).
The performance of PCR to identify P. ovale malaria compared to that of the microscopic method
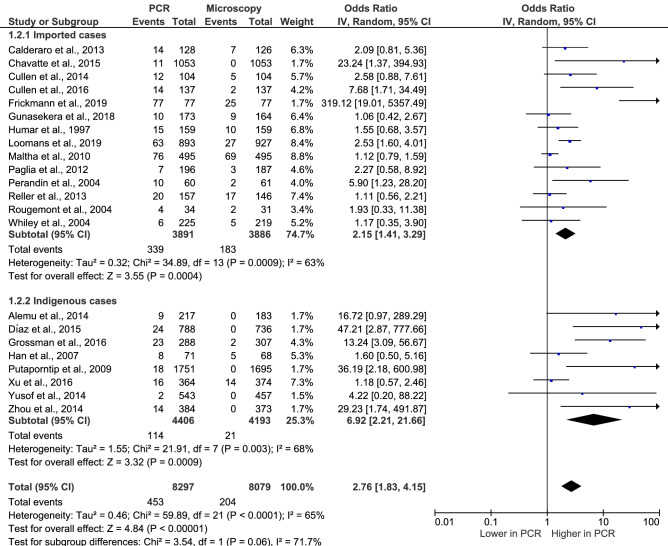
The performance of PCR to identify P. ovale malaria versus that of the microscopic method was estimated using the random-effects model (Fig. 5). The number of P. ovale cases identified using microscopic method that were subsequently confirmed by PCR method (204 cases) and the number of P. ovale identified by PCR were used in the present analysis. The results demonstrated a higher performance of PCR in identifying P. ovale cases than that of the microscopic method, with substantial heterogeneity (p < 0.00001, OR: 2.76, 95% CI: 1.83–4.15, I2: 65%). Subgroup analysis of participants demonstrated a higher performance of PCR in detecting P. ovale malaria in indigenous cases (p: 0.0009, OR: 6.92, 95% CI: 2.21–21.6, I2: 68%) than in imported cases with substantial heterogeneity (p: 0.0004, OR: 2.15, 95% CI: 1.41–3.29, I2: 63%). No difference between the two subgroups was found (p: 0.06, I2: 71.7%). Among indigenous cases, six included studies17,21,30,35,36,38 demonstrated no cases of P. ovale by the microscopic method and were the same cases that were confirmed by the PCR method.
Figure 5.
The performance of PCR to identify P. ovale IV: Inverse Variance, CI: Confidence Interval, Event: number of patients with P. ovale, random: random effects model, Total: number of all P. ovale cases, Lower in PCR: the proportion of P. ovale cases detected by PCR was lower than those detected by the microscopic method. Higher in PCR: the proportion of P. ovale cases detected by PCR was higher than those detected by the microscopic method. The performance of PCR to identify P. ovale malaria was analysed using Review Manager 5.3 (The Cochrane Collaboration, London, UK)
available at https://training.cochrane.org/.
Sensitivity test
The robustness of the pooled prevalence of the misidentification of P. ovale as P. vivax malaria was determined using the trim and fill method by excluding low-quality studies from the pooled analysis. The result of the trim and fill method by removing the three studies19,20,37 with low qualities demonstrated that the pooled prevalence of the misidentification of P. ovale as P. vivax was similar to that of the pooled prevalence of 22 studies (11%, 95% CI: 7–16%, I2: 34.6%).
Publication bias
Visual inspection of the funnel plots demonstrated some small study effects that caused an asymmetric distribution of studies in the plots between the OR and SE (logOR) (Fig. 6). The asymmetric distribution of the funnel plots was quantified with Egger's test. Egger's test showed a significant asymmetric distribution due to the small-study effects across the 22 included studies (p: 0.001, coefficient: 12.5, standard error: 3.17, t: 3.95). The contour-enhanced funnel plot was further evaluated if the asymmetric distribution was due to publication bias or other factors. The results showed that most of the included studies were located in the significant area of the plot (p < 0.01), indicating that publication bias was the cause of the asymmetric distribution of the funnel plot (Fig. 7).
Figure 6.
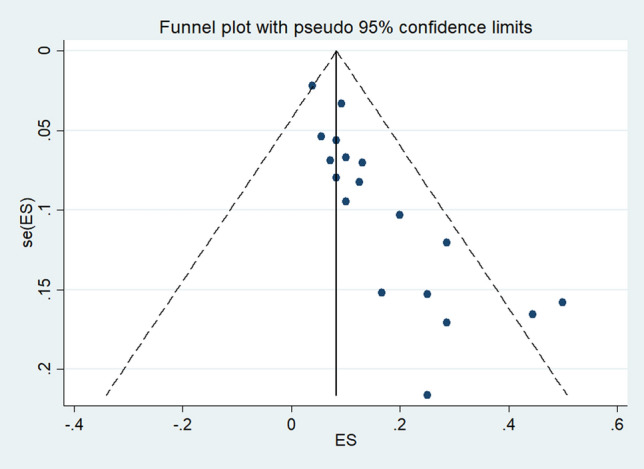
Publication bias among the included studies. Publication bias was determined using Review Manager 5.3 (The Cochrane Collaboration, London, UK)
available at https://training.cochrane.org/.
Figure 7.
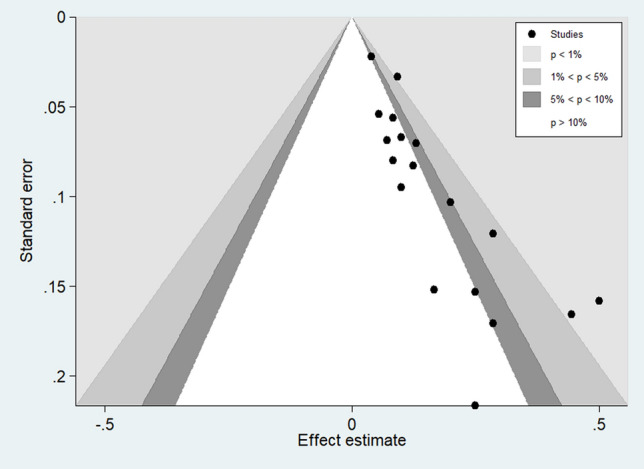
The contour enhanced funnel plot. The contour enhanced funnel plot was estimated using STATA Statistical Software version 15.0 (StataCorp. 2017. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC).
Discussion
The microscopic method for identifying Plasmodium species is still considered the gold standard method for malaria diagnosis in clinical laboratories. However, its limitation is its low sensitivity to detect malaria parasites that are present with a low parasite density39–41. The sensitivity of microscopy under optimal conditions is limited to approximately 10–50 parasites/μl of blood42. In contrast to microscopy, PCR has the advantage of higher sensitivity and is capable of detecting less than 10 parasites/µl of blood43–45. In addition to low sensitivity, microscopy also has a low specificity or inability to distinguish the morphologically similar P. vivax and P. ovale malaria even by a well-trained or expert microscopist examining blood films.
This is the first systematic review and meta-analysis to quantify the misidentification of P. ovale as P. vivax by microscopic methods. The prevalence of misidentification of P. ovale as P. vivax malaria was high (11%), particularly in P. ovale endemic countries (13%). A previous study suggested that misidentification of P. vivax with P. ovale was likely due to the infection of P. ovale resulting in a low parasite density compared to that of P. falciparum18, and the misidentification of P. ovale as P. vivax is more frequent than the misidentification of P. ovale as P. falciparum29. A previous study included laboratories in hospitals and demonstrated that participants had much more difficulty identifying P. ovale with 100% failure rates, while difficulty identifying P. malariae (22.5% failure) and P. vivax (21.7% failure) was lower46. The difficulty in identifying P. ovale malaria was also observed by a study on laboratories in the United Kingdom47. In addition, a survey study of 19 provincial laboratories in China with a total of 168 staff members also demonstrated that P. ovale was likely to be misdiagnosed as P. vivax by microscopy48. The external quality assessment (EQA) conducted in Senegal, which was part of the national malaria control program (NMCP), demonstrated the misidentification of a P. ovale slide as P. vivax by experts49. Moreover, microscopists participating in post training on the proficiency of laboratory technicians in Plasmodium species identification could misidentify P. vivax as P. ovale malaria50. Although the prevalence of misidentification of P. ovale as P. vivax was high, the treatment of these two species with chloroquine and radical cure with primaquine to eliminate the liver stages were similar.
Imported malaria in non-endemic countries continues to be reported worldwide as international travel or immigration from endemic zones has increased51–54. Therefore, malaria diagnostic tests with high sensitivity and specificity to identify Plasmodium species among travellers are necessary. Due to the low sensitivity of RDTs to identify P. ovale27,55,56, molecular techniques such as PCR are recommended to identify P. ovale cases, although they have been shown to miss some P. ovale cases57. Moreover, molecular methods such as semi-nested PCR more accurately detected P. ovale mixed infections than microscopy28. With more advances in molecular methods for the detection and identification of malaria parasites, real-time PCR methods using fluorescent labels for detecting and quantifying DNA targets have been developed with high sensitivity and specificity23,58,59. Interestingly, multiplex real-time PCR failed to detect P. falciparum and P. ovale mixed infection in a previous study because of the high difference in the ratio between P. falciparum and P. ovale (> 1000:1)32. These results suggested that the misidentification of P. ovale as P. vivax in several studies might be caused by bias across the considerable prevalence of the main endemic Plasmodium species, by changes in the morphology of the parasite during specimen storage or treatment, or by very low parasitaemia levels. In addition, there is a strong perception that P. vivax is rare in subtropical Africa60. Therefore, the identification bias in retrospective studies of imported malaria, where the patients are considered to have acquired their infection in a subtropical African country, is because there is a clear bias in these cases to identify any non-falciparum or non-malariae case as P. ovale can occur. Thus, the number of P. vivax cases misidentified as P. ovale was quantified. The results showed that the misidentification of P. vivax as P. ovale in imported countries was very low (1%). This result indicated a lower possibility of P. vivax being misidentified as P. ovale (1%) than those of P. ovale being misidentified as P. vivax (11%) in imported cases.
The major concern of the misidentification of P. ovale as P. vivax in imported cases should be addressed. Therefore, the molecular method can make a decisive contribution to the identification of a less common Plasmodium species or two Plasmodium species with similar morphologies. Taking into account the high sensitivity and specificity of molecular methods for identifying malaria parasites, molecular methods are labour intensive and have a greater potential for contamination and long turnaround times for routine diagnosis, and are not convenient for use in remote settings. Updated courses and intensified training of microscopic examinations are necessary to improve the microscopic identification of Plasmodium species in rural health centres, where molecular techniques are unavailable.
The present study have limitations. First, limited numbers of P. ovale were identified and reported, as it is a neglected Plasmodium species. Therefore, the pooled prevalence of the misidentification of P. ovale as P. vivax malaria might not represent all misidentification that occurred. Second, several studies performed microscopy and PCR to confirm malaria infection, but could not be included in the present study as the necessary data cannot be extracted and the comparison between microscopy and PCR was either not clearly presented or not provided.
Conclusion
Misidentification of P. ovale infections as P. vivax malaria by microscopic methods are frequent and lead to the underreported status of P. ovale cases worldwide. The molecular identification of P. ovale malaria in endemic areas is necessary to provide data for malaria elimination because a higher rate of P. ovale misidentification was found in endemic cases than in imported cases. In addition, updated courses and intensified training of microscopic examinations, particularly for P. ovale identification, are required to improve the microscopic identification of Plasmodium species in rural health centres and other resource-limited territories where PCR is unavailable.
Supplementary Information
Acknowledgements
The authors would like to acknowledge Mr David C Chang, who performed English language editing of this study. The authors would like to thank Walailak University for providing the funding, the New Strategic Research (P2P) project, for this study.
Author contributions
M.K., F.R.M., K.U.K., and G.D.M. participated in the study design, data analysis, and writing of the paper. All of the authors read and approved the final paper.
Data availability
The datasets used during the current study are demonstrated in the present manuscript along with additional files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-78691-7.
References
- 1.Collins WE, Jeffery GM. Plasmodium ovale: Parasite and disease. Clin. Microbiol. Rev. 2005;18:570–581. doi: 10.1128/CMR.18.3.570-581.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okafor, C. N. & Finnigan, N. A. in StatPearls (2020).
- 3.Mueller I, Zimmerman PA, Reeder JC. Plasmodium malariae and Plasmodium ovale—The "bashful" malaria parasites. Trends Parasitol. 2007;23:278–283. doi: 10.1016/j.pt.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotepui M, Kotepui KU, Milanez GD, Masangkay FR. Severity and mortality of severe Plasmodium ovale infection: A systematic review and meta-analysis. PLoS ONE. 2020;15:e0235014. doi: 10.1371/journal.pone.0235014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abanyie FA, Arguin PM, Gutman J. State of malaria diagnostic testing at clinical laboratories in the United States, 2010: A nationwide survey. Malar. J. 2011;10:340. doi: 10.1186/1475-2875-10-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mekonnen SK, Aseffa A, Medhin G, Berhe N, Velavan TP. Re-evaluation of microscopy confirmed Plasmodium falciparum and Plasmodium vivax malaria by nested PCR detection in southern Ethiopia. Malar. J. 2014;13:48. doi: 10.1186/1475-2875-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nega D, et al. Comprehensive competency assessment of malaria microscopists and laboratory diagnostic service capacity in districts stratified for malaria elimination in Ethiopia. PLoS ONE. 2020;15:e0235151. doi: 10.1371/journal.pone.0235151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doctor SM, et al. Low prevalence of Plasmodium malariae and Plasmodium ovale mono-infections among children in the Democratic Republic of the Congo: A population-based, cross-sectional study. Malar. J. 2016;15:350. doi: 10.1186/s12936-016-1409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snounou G, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol. Biochem. Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland CJ, et al. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J. Infect. Dis. 2010;201:1544–1550. doi: 10.1086/652240. [DOI] [PubMed] [Google Scholar]
- 11.Rojo-Marcos G, et al. Prospective comparative multi-centre study on imported Plasmodium ovale wallikeri and Plasmodium ovale curtisi infections. Malar. J. 2018;17:399. doi: 10.1186/s12936-018-2544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rojo-Marcos, G. et al. Comparison of imported Plasmodium ovale curtisi and P. ovale wallikeri infections among patients in Spain, 2005–2011. Emerg. Infect. Dis.20, 409–416, 10.3201/eid2003.130745 (2014). [DOI] [PMC free article] [PubMed]
- 13.Kotepui, M., Kotepui, K. U., De Jesus Milanez, G. & Masangkay, F. R. Plasmodium spp. mixed infection leading to severe malaria: a systematic review and meta-analysis. Sci Rep10, 11068, 10.1038/s41598-020-68082-3 (2020). [DOI] [PMC free article] [PubMed]
- 14.PROSPERO. PROSPERO International Prospective Register of Systematic Reviews, https://www.crd.york.ac.uk/prospero/ (2020).
- 15.Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G. & Group, P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med6, e1000097, 10.1371/journal.pmed.1000097 (2009). [DOI] [PMC free article] [PubMed]
- 16.Whiting PF, et al. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 17.Alemu A, et al. Comparison of Giemsa microscopy with nested PCR for the diagnosis of malaria in North Gondar, north-west Ethiopia. Malar. J. 2014;13:174. doi: 10.1186/1475-2875-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calderaro, A. et al. Accurate identification of the six human Plasmodium spp. causing imported malaria, including Plasmodium ovale wallikeri and Plasmodium knowlesi. Malar. J.12, 10.1186/1475-2875-12-321 (2013). [DOI] [PMC free article] [PubMed]
- 19.Cullen KA, Arguin PM. Malaria surveillance–United States, 2012. MMWR Surveill. Summ. 2014;63:1–22. [PubMed] [Google Scholar]
- 20.Cullen, K. A., Mace, K. E., Arguin, P. M., Centers for Disease, C. & Prevention. Malaria surveillance—United States, 2013. MMWR Surveill. Summ.65, 1–22, 10.15585/mmwr.ss6502a1 (2016). [DOI] [PubMed]
- 21.Diaz PB, et al. Quality of malaria diagnosis and molecular confirmation of Plasmodium ovale curtisi in a rural area of the southeastern region of Ethiopia. Malar. J. 2015;14:8. doi: 10.1186/s12936-015-0893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frickmann H, Wegner C, Ruben S, Loderstädt U, Tannich E. A comparison of two PCR protocols for the differentiation of Plasmodium ovale species and implications for clinical management in travellers returning to Germany: A 10-year cross-sectional study. Malar. J. 2019;18:272. doi: 10.1186/s12936-019-2901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossman T, et al. Contribution of real-time PCR to Plasmodium species identification and to clinical decisions: A nationwide study in a non-endemic setting. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:671–675. doi: 10.1007/s10096-016-2844-0. [DOI] [PubMed] [Google Scholar]
- 24.Gunasekera W, et al. Utility of pf/pan RDT for diagnosis in the prevention of re-establishment of malaria in Sri Lanka. Pathogens Global Health. 2018;112:360–367. doi: 10.1080/20477724.2018.1536855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han ET, et al. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J. Clin. Microbiol. 2007;45:2521–2528. doi: 10.1128/jcm.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humar A, Harrington MA, Kain KC. Evaluation of a non-isotopic polymerase chain reaction-based assay to detect and predict treatment failure of Plasmodium vivax malaria in travellers. Trans. R. Soc. Trop. Med. Hyg. 1997;91:406–409. doi: 10.1016/s0035-9203(97)90258-3. [DOI] [PubMed] [Google Scholar]
- 27.Maltha J, et al. Evaluation of a rapid diagnostic test (CareStart Malaria HRP-2/pLDH (Pf/pan) Combo Test) for the diagnosis of malaria in a reference setting. Malar. J. 2010;9:171. doi: 10.1186/1475-2875-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paglia MG, et al. Molecular diagnosis and species identification of imported malaria in returning travellers in Italy. Diagn. Microbiol. Infect. Dis. 2012;72:175–180. doi: 10.1016/j.diagmicrobio.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Perandin F, et al. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J. Clin. Microbiol. 2004;42:1214–1219. doi: 10.1128/jcm.42.3.1214-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putaporntip C, et al. Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J. Infect. Dis. 2009;199:1143–1150. doi: 10.1086/597414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reller ME, Chen WH, Dalton J, Lichay MA, Dumler JS. Multiplex 5' nuclease quantitative real-time PCR for clinical diagnosis of malaria and species-level identification and epidemiologic evaluation of malaria-causing parasites, including Plasmodium knowlesi. J. Clin. Microbiol. 2013;51:2931–2938. doi: 10.1128/jcm.00958-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rougemont M, et al. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J. Clin. Microbiol. 2004;42:5636–5643. doi: 10.1128/jcm.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whiley DM, et al. Detection and differentiation of Plasmodium species by polymerase chain reaction and colorimetric detection in blood samples of patients with suspected malaria. Diagn. Microbiol. Infect. Dis. 2004;49:25–29. doi: 10.1016/j.diagmicrobio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Xu C, et al. Characteristics of imported malaria and species of Plasmodium involved in Shandong Province, China (2012–2014) Korean J. Parasitol. 2016;54:407–414. doi: 10.3347/kjp.2016.54.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yusof, R. et al. High proportion of knowlesi malaria in recent malaria cases in Malaysia. Malar. J.13, 10.1186/1475-2875-13-168 (2014). [DOI] [PMC free article] [PubMed]
- 36.Zhou, X. et al. A molecular survey of febrile cases in malaria-endemic areas along China-Myanmar border in Yunnan province, People's Republic of China. Parasite (Paris, France)21, 27, 10.1051/parasite/2014030 (2014). [DOI] [PMC free article] [PubMed]
- 37.Loomans, L. et al. Accuracy of malaria diagnosis by clinical laboratories in Belgium. Malar. J.18, 10.1186/s12936-019-2731-0 (2019). [DOI] [PMC free article] [PubMed]
- 38.Chavatte JM, Tan SB, Snounou G, Lin RT. Molecular characterization of misidentified Plasmodium ovale imported cases in Singapore. Malar. J. 2015;14:454. doi: 10.1186/s12936-015-0985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fradejas I, et al. Prevalence of submicroscopic malaria infection in immigrants living in Spain. Malar. J. 2019;18:9. doi: 10.1186/s12936-019-2870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oyedeji SI, Awobode HO, Bassi PU. Molecular investigation of sub-microscopic and mixed Plasmodium species infection in North-Central Nigeria. Asian Pac. J. Trop. Dis. 2017;7:220–224. doi: 10.12980/apjtd.7.2017D6-415. [DOI] [Google Scholar]
- 41.Walker-Abbey A, et al. Malaria in pregnant Cameroonian women: the effect of age and gravidity on submicroscopic and mixed-species infections and multiple parasite genotypes. Am. J. Trop. Med. Hyg. 2005;72:229–235. doi: 10.4269/ajtmh.2005.72.229. [DOI] [PubMed] [Google Scholar]
- 42.Trampuz A, Jereb M, Muzlovic I, Prabhu RM. Clinical review: Severe malaria. Crit. Care. 2003;7:315–323. doi: 10.1186/cc2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanscheid T, Grobusch MP. How useful is PCR in the diagnosis of malaria? Trends Parasitol. 2002;18:395–398. doi: 10.1016/s1471-4922(02)02348-6. [DOI] [PubMed] [Google Scholar]
- 44.Patsoula, E., Spanakos, G., Sofianatou, D., Parara, M. & Vakalis, N. C. A single-step, PCR-based method for the detection and differentiation of Plasmodium vivax and P. falciparum. Ann. Trop. Med. Parasitol.97, 15–21, 10.1179/000349803125002535 (2003). [DOI] [PubMed]
- 45.Fuehrer HP, Noedl H. Recent advances in detection of Plasmodium ovale: implications of separation into the two species Plasmodium ovale wallikeri and Plasmodium ovale curtisi. J. Clin. Microbiol. 2014;52:387–391. doi: 10.1128/JCM.02760-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edson, D. C., Glick, T. & Massey, L. D. Detection and identification of malaria parasites: A review of proficiency test results and laboratory practices. Lab Med.41, 719–723 (2010).
- 47.Milne LM, Kyi MS, Chiodini PL, Warhurst DC. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J. Clin. Pathol. 1994;47:740–742. doi: 10.1136/jcp.47.8.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin JH, et al. Establishing a China malaria diagnosis reference laboratory network for malaria elimination. Malar. J. 2015;14:40. doi: 10.1186/s12936-015-0556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diallo MA, et al. Quality control of malaria microscopy reveals misdiagnosed non-falciparum species and other microscopically detectable pathogens in Senegal. Ann. Clin. Microbiol. Antimicrob. 2018;17:8. doi: 10.1186/s12941-018-0261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obare P, et al. Misclassification of Plasmodium infections by conventional microscopy and the impact of remedial training on the proficiency of laboratory technicians in species identification. Malar. J. 2013;12:113. doi: 10.1186/1475-2875-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Millet JP, et al. Imported malaria in a cosmopolitan European city: A mirror image of the world epidemiological situation. Malar. J. 2008;7:56. doi: 10.1186/1475-2875-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kendjo E, et al. Epidemiologic trends in malaria incidence among travellers returning to metropolitan France, 1996–2016. JAMA Netw. Open. 2019;2:e191691. doi: 10.1001/jamanetworkopen.2019.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norman FF, et al. Imported malaria in Spain (2009–2016): Results from the +REDIVI Collaborative Network. Malar. J. 2017;16:407. doi: 10.1186/s12936-017-2057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dakic Z, et al. Imported malaria in Belgrade, Serbia, between 2001 and 2009. Wien Klin. Wochenschr. 2011;123(Suppl 1):15–19. doi: 10.1007/s00508-011-0040-x. [DOI] [PubMed] [Google Scholar]
- 55.Van der Palen M, et al. Test characteristics of two rapid antigen detection tests (SD FK50 and SD FK60) for the diagnosis of malaria in returned travellers. Malar. J. 2009;8:90. doi: 10.1186/1475-2875-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marx A, et al. Meta-analysis: Accuracy of rapid tests for malaria in travelers returning from endemic areas. Ann. Intern. Med. 2005;142:836–846. doi: 10.7326/0003-4819-142-10-200505170-00009. [DOI] [PubMed] [Google Scholar]
- 57.Calderaro A, et al. Genetic polymorphisms influence Plasmodium ovale PCR detection accuracy. J. Clin. Microbiol. 2007;45:1624–1627. doi: 10.1128/JCM.02316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nijhuis RHT, van Lieshout L, Verweij JJ, Claas ECJ, Wessels E. Multiplex real-time PCR for diagnosing malaria in a non-endemic setting: A prospective comparison to conventional methods. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:2323–2329. doi: 10.1007/s10096-018-3378-4. [DOI] [PubMed] [Google Scholar]
- 59.Haanshuus CG, et al. Assessment of malaria real-time PCR methods and application with focus on low-level parasitaemia. PLoS ONE. 2019;14:e0218982. doi: 10.1371/journal.pone.0218982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Howes RE, et al. Global epidemiology of Plasmodium vivax. Am. J. Trop. Med. Hyg. 2016;95:15–34. doi: 10.4269/ajtmh.16-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the current study are demonstrated in the present manuscript along with additional files.