Correction to: Scientific Reports 10.1038/s41598-017-05859-z, published online 19 July 2017
This Article contains an error in the order of panels of Figure 1. Panels a, b, c, d and e should appear in the order d, e, a, b, c. The correct Figure appears below as Figure 1.
Figure 1.
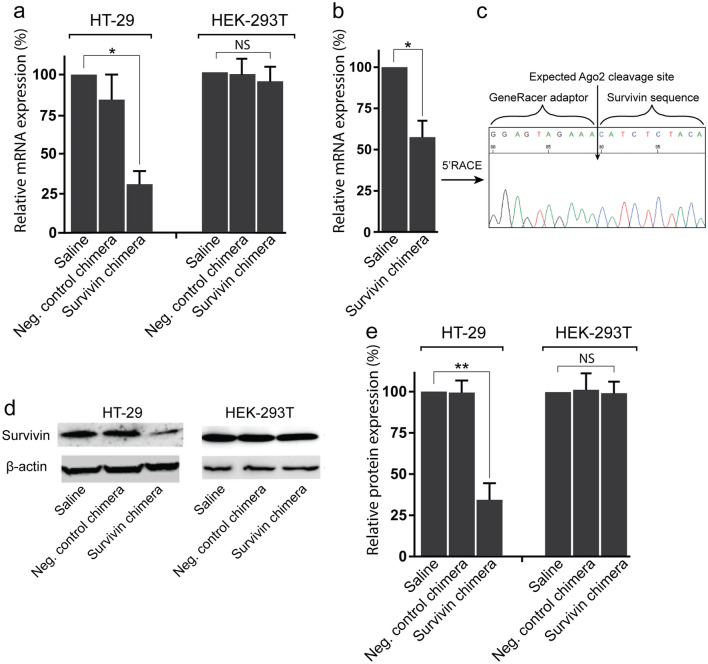
EpCAM aptamer-guided RNAi effectively silenced survivin. (a) Specificity and efficacy of EpCAMaptamer guided RNAi in knocking down survivin mRNA. Chimera or negative control chimera were incubated with HT-29 or HEK-2913T cells for 24 hours and the total RNA was extracted for qRT-PCR analysis of survivin mRNA levels. GAPDH was used as an internal control. (b, c) HT-29 Tumour-bearing mice were treated with 2 nmol/mouse of PEG-labelled chimera for 48 hours. The tumours were collected for RNA extraction followed by qRT-PCR analysis of survivin mRNA expression (b) and 5′RACE assay (c). (d) Effective downregulation of survivin protein via EpCAM aptamer-guided RNAi. Chimera or negative control chimera were incubated with HT-29 or HEK-2913T cells for 48 hours and the survivin protein levels were analyzed using Western blot analysis. β-actin was used as a loading control. (e) The bar graph shows the survivin protein levels in various treatment groups. Data shown are means ± SEM, n = 3. *p < 0.05, **p < 0.005. NS, no statistically significant difference..
Additionally, the panels in Figures 3 and 4 are missing labels. The correct Figures appear below as Figures 2 and 3 respectively.
Figure 2.
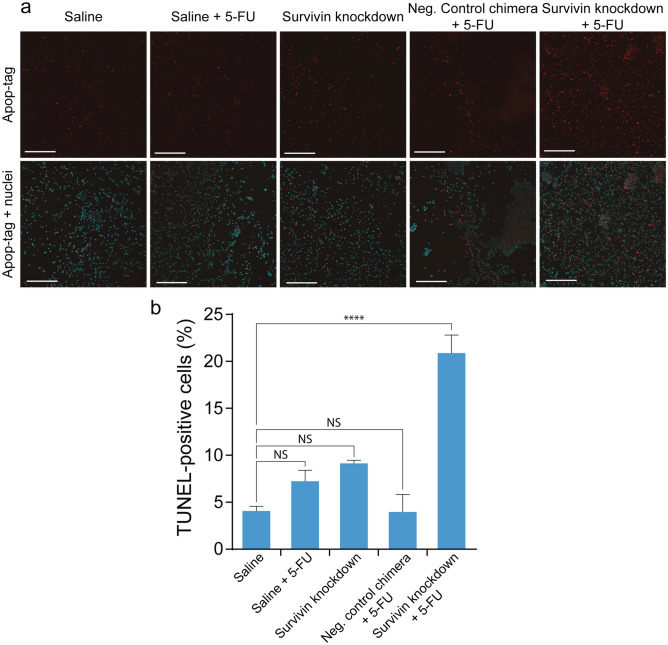
Survivin knockdown in vivo enhances 5-FU-induced apoptosis in HT-29 tumour cells. (a) Representative images of TUNEL apoptosis assay on dissociated HT-29 xenograft tumours after in vivo treatment with chimera and 5-FU. NOD/SCID mice bearing HT-29 tumours (60 mm3) were treated intravenously with 3 injections of 2 nmol/mouse of chimera with or without 3 additional treatment of 30 mg/kg of 5-FU. Two days after the final treatment, tumours were dissociated by collagenase digestion and subjected to TUNEL apoptosis assay. (b) Percentage of apoptotic cells in treated tumours. Data shown are means ± SEM, n = 3. ****p < 0.001. NS, no statistically significant difference.
Figure 3.
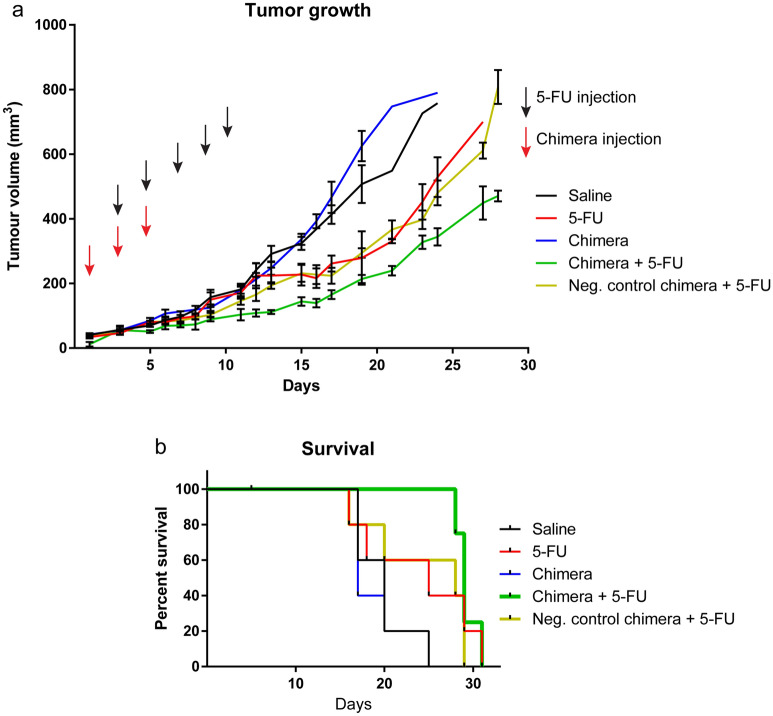
Combined chimera and 5-FU treatments improves therapeutic outcome in HT-29 tumour-bearing mice. (a) The graph represents the tumour volume in mice in response to various treatments as indicated. (b) Survival rate of mice observed over the course of various treatments. Data shown are mean ± SEM, n = 4–5.
Finally, the Supplementary Information file contains a typographical error.
“The chimera structure features an asymmetric structure to facilitate recognition by an endogenous Dicer enzyme to cleave the chimera which results in the release of the expected 21-mer siRNA sequence [251] (Figure S1b).”
should read:
“The chimera structure features an asymmetric structure to facilitate recognition by an endogenous Dicer enzyme to cleave the chimera which results in the release of the expected 21-mer siRNA sequence (Figure S1b).”