Abstract
Seasonal human influenza viruses continually change antigenically to escape from neutralizing antibodies. It remains unclear how genetic variation in the intrahost virus population and selection at the level of individual hosts translates to the fast-paced evolution observed at the global level because emerging intrahost antigenic variants are rarely detected. We tracked intrahost variants in the hemagglutinin and neuraminidase surface proteins using longitudinally collected samples from 52 patients infected by A/H3N2 influenza virus, mostly young children, who received oseltamivir treatment. We identified emerging putative antigenic variants and oseltamivir-resistant variants, most of which remained detectable in samples collected at subsequent days, and identified variants that emerged intrahost immediately prior to increases in global rates. In contrast to most putative antigenic variants, oseltamivir-resistant variants rapidly increased to high frequencies in the virus population. Importantly, the majority of putative antigenic variants and oseltamivir-resistant variants were first detectable four or more days after onset of symptoms or start of treatment, respectively. Our observations demonstrate that de novo variants emerge, and may be positively selected, during the course of infection. Additionally, based on the 4–7 days post-treatment delay in emergence of oseltamivir-resistant variants in six out of the eight individuals with such variants, we find that limiting sample collection for routine surveillance and diagnostic testing to early timepoints after onset of symptoms can potentially preclude detection of emerging, positively selected variants.
Keywords: influenza virus, within-host evolution, next-generation sequencing
1. Introduction
The error-prone replication of influenza viruses gives rise to the genetic variation upon which selection pressures act. Natural selection of viruses that best escape neutralizing antibodies results in rapid turnover of the global virus population, necessitating regular vaccine reformulation. Selection likely acts differently on the global level and within individual infected hosts, but global evolution must start with selection of de novo variants that emerge during infection. How novel variants develop within an infected host is incompletely understood. Recent studies have found little intrahost variation associated with antibody escape (Dinis et al. 2016; Debbink et al. 2017; McCrone et al. 2018). Despite absence of compelling data on intrahost emergence of antigenic variants, emergence of drug-resistant variants during antiviral treatment shows that viruses with advantageous substitutions can be positively selected during a single infection (Kiso et al. 2004; Stephenson et al. 2009; Inoue et al. 2010; Lina et al. 2018).
Whether or not a specific de novo variant emerges within a patient and can be identified in a clinical specimen may depend on multiple factors. The presence or absence of humoral immunity likely governs selective outgrowth of antibody escape variants, distal parts of the human airways may hold genetically different virus populations, and variants may emerge more readily in specific patient populations due to higher and/or more prolonged viral replication (e.g. immunocompromised patients). Because only a handful of studies have examined intrahost variation of influenza viruses, the patient population that is most relevant for intrahost emergence of novel antigenic variants may not have been identified. Identification of emerging antigenic variants may also have been hindered by timing of specimen collection during infection, as previous studies analyzed diagnostic clinical specimens collected at single or at most two timepoints during the acute stage of infection and emergence of variants later during the course of infection cannot be ruled out.
Although the transient nature of typical influenza virus infections limits the time frame for de novo variants to emerge, we investigated whether intrahost evolution of antigenic variants can be detected during prolonged courses of acute influenza infection by analyzing longitudinal samples. Specifically, we analyzed the within-host virus populations of a cohort of individuals infected by A/H3N2 influenza virus who participated in a large randomized controlled trial of oseltamivir dosing in Southeast Asia (South East Asia Infectious Disease Clinical Research Network 2013). At the time of sampling, these individuals were mostly young children, in whom influenza virus infection may be prolonged compared to adults (Ng et al. 2016; Maier et al. 2018) Next-generation sequencing (NGS) was used to sequence the hemagglutinin (HA) and neuraminidase (NA) gene segments of the influenza virus populations found in the sequentially collected clinical specimens during the course of infection. The emergence and dynamics of intrahost variants of HA and NA were studied as these surface glycoproteins are the dominant targets of antibody-mediated immunity in humans and target for anti-viral therapy (Krammer 2019). Plasma samples collected at enrollment and during convalescence were used to identify immune and non-immune individuals. Because all patients in this study received oseltamivir for at least 5 days, we had the unique opportunity to compare evolutionary dynamics of variants associated with antigenic change to the emergence of oseltamivir-resistant variants.
2. Methods
2.1 Patients and samples
Clinical specimens were collected as part of a multi-center randomized controlled trial of standard dose versus double dose oseltamivir in hospitalized patients with laboratory confirmed influenza virus infection performed between April 2007 and February 2010. Details on patients, methods, and results of this trial have been described elsewhere (South East Asia Infectious Disease Clinical Research Network 2013). Briefly, patients ≥1 year of age with laboratory confirmed influenza virus infection and duration of symptoms ≤10 days were included. All patients received oseltamivir for at least 5 days. Nose and throat swabs and blood samples were obtained daily on days 0–10 and on day 14. For the current study, all patients that were influenza A/H3N2 virus positive for ≥3 days and samples with cycle threshold (Ct) values ≤35 were included. Ct values for all patients and timepoints are indicated in Supplementary Fig. S1. Information on single or double dose oseltamivir is available from Supplementary Table S1. The current study included 52 patients from hospitals in Thailand (n = 7) and Vietnam (n = 45).
2.2 Library preparation and deep sequencing
Total RNA was extracted from clinical specimens using the High Pure RNA isolation kit (Roche, 11828665001) according to the manufacturer’s instructions. For each timepoint, we used the sample (nose or throat swab) with the lowest Ct value. Influenza virus RNAs were reverse transcribed and amplified using the superscript III One-Step RT PCR Platinum with Taq High Fidelity DNA Polymerase (ThermoFisher, 12574030) using A/H3N2 virus HA and NA segment-specific primers (Supplementary Table S2). We performed six independent PCR reactions in total, resulting in three partly overlapping amplicons for the HA and NA segments each. Making use of shorter, overlapping amplicons increase efficiency in amplification and also ensure that the whole viral genome is sufficiently covered if there were any RNA degradation in the clinical specimen. PCR products were pooled in equimolar concentrations for each sample, and were subsequently purified using Agencourt Ampure XP beads (Beckman Coulter, A63882) and quantified using the Qubit dsDNA HS assay kit (ThermoFisher, Q32854). Pooled and cleaned amplicons were used for subsequent library preparation.
We prepared sequencing libraries using the Nextera XT DNA Library Preparation kit (Illumina, FC-131-1096) according to instructions of the manufacturer. Briefly, for each sample 5 µl of diluted amplicons were enzymatically fragmented and Illumina adapters were ligated to the fragments. Subsequently each sample was purified twice using Agencourt Ampure XP beads. Library size distribution was evaluated using the High sensitivity dsDNA kit on a 2100 Bioanalyzer (Agilent, 5067-4626) and qPCR-based library quantification was performed using the KAPA Library Quantification kit for Illumina platforms (KAPA Biosystems, KK4824) on a LightCycler480 (Roche). The normalized library pools were sequenced using the Illumina MiSeq 600-cycle MiSeq Reagent Kit v3 (Illumina, MS-102-3003).
2.3 Quality control, variant detection, and data analysis
The Maximum Information quality filtering approach of the Trimmomatic tool (version 0.36, parameters; leading: 3, trailing: 3, maxinfo: 80:0.4, crop: 280) was used for quality trimming of Illumina MiSeq reads (Bolger, Lohse, and Usadel 2014). Merging, mapping, and coverage analysis was done using the BBmerge, BBwrap, and pileup scripts from the BBMap bioinformatics toolkit version 36.27 (https://sourceforge.net/projects/bbmap/; last accessed 1 November 2020). Read pairs with inappropriate orientation and reads with a Q-score below 25 were discarded. We used FastQC version v0.11.5 to monitor quality control (http://www.bioinformatics.babraham.ac.uk/projects/fastqc; last accessed 1 November 2020). FASTQ files of the trimmed merged reads that were used for mapping are available from https://github.com/mirdesign/NGS-FluAnalyzer/tree/master/Raw_Data_For_Intrahost.AH3N2.Virus.HANA.Variants (last accessed 1 November 2020). All reads were mapped to the HA and NA gene segments of A/Stockholm/40/2013 (EPI_ISL_155657).
Subsequent steps were performed using a set of custom scripts (available from https://github.com/mirdesign/NGS-FluAnalyzer; last accessed 1 November 2020). Mapped reads were translated and prepared for variant calling by identification of appropriate reading frames and conversion of read numbering to protein numbering for HA and NA. All HA amino acid positions reported in this study are according to H3 numbering without signal peptide (Burke and Smith 2014). We next applied a number of filtering steps to identify amino acid variants. Only variants outside of the primer regions were identified as part of stringent quality controls. However, we made an exception for oseltamivir-resistant variant NA E119V, which is in the reverse primer region for NA amplicon 2, that emerged in patient 1682 (Fig. 4 and Supplementary Fig. S6). Unlike most other uncharacterized HA and NA variants identified, NA E119V is a widely known resistant mutation. It was found in one sample collected from a single individual only and reached moderate proportions (42%) after the patient was treated with oseltamivir. As such, it is more likely that the mutation was selected for as opposed to it being spuriously introduced by the primer sequence. Each nucleotide position in the codon encoding the amino acid variant must have a minimum coverage of 100×. The amino acid variant must be found in at least 1 per cent of the virus population. If coverage at the codon level is less than 500×, a minimum of five observations is required for the variant to be called. Finally, for a variant to be included in our longitudinal analyses, the variant proportion must be 5 per cent or higher for at least one timepoint during the course of infection. We identified global prevalence for the variants in our study that were present at >1 per cent of total sequences in the GISAID database since the start of specimen collection for this study in 2007. To make the global variation plots in Supplementary Fig. S2, HA and NA sequences between the years 2000 and 2019 were obtained from the GISAID database using global frequency statistics via FluSurver, with duplicate strain names and H3N2v removed before analysis.
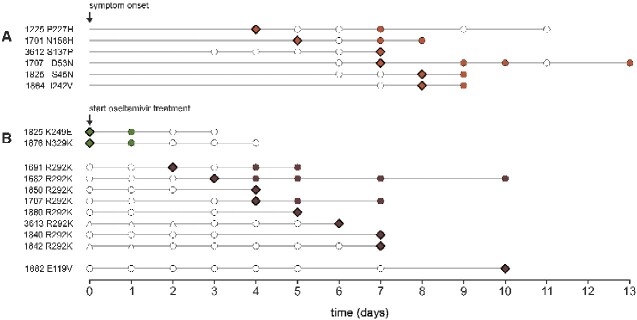
Figure 4.
Initial day of variant detection. Diamond shapes specify the first observation in the virus population. Circles indicate the days of sample collection. Open circles indicate days where the variant proportion did not reach the minimum sample inclusion threshold of 1 per cent. Columns on the left show patient ID and amino acid variant. (A) Antigenic site variants (orange). (B) Reduced inhibition variants (green) and highly reduced inhibition variants (brown). Open triangles indicate days of oseltamivir treatment prior to enrollment.
2.4 Serology
Hemagglutination inhibition (HI) assays were performed as described previously (Hirst 1943). Briefly, plasma samples were treated overnight with receptor destroying enzyme (Denka Seiken, 370013) according to manufacturer’s instructions, and incubated at 56 °C for 1 h. Two-fold serial dilutions of plasma starting at a 1:40 dilution were mixed with 25 µl virus (A/Brisbane/10/07) containing four hemagglutinating units and were incubated at 37 °C for 30 min. Next, 25 µl 1 per cent turkey erythrocytes was added and the mixture was incubated at 4 °C for 1 h before HI patterns were read. HI titers were expressed as the reciprocal value of the highest plasma dilution at which agglutination of turkey erythrocytes was completely inhibited.
2.5 Ethics statement
The protocol of the original trial (NCT00298233) was approved by the ethics committees of all recruiting hospitals, the National Institute of Allergy and Infectious Diseases, and the Oxford University tropical research ethics committee (OxTREC), and informed consent was given by all patients (or proxies). Further molecular analysis of viral nucleic acids and immune response was part of this protocol. Additional approvals to use these samples outside of the period of approval of the original trial were given by the IRBs of the six hospitals in Vietnam and Thailand included in this analysis (NHTD: 05/HDDD-NDTU, VNCH: NHP-RICH-15-010, HTD: CS/ND/14/25, CH1: CS/N1/15/12, CH2: expedited review, no code, Siriraj: 052/2559(EC2)).
3. Results
3.1 Putative intrahost antigenic variants occur infrequently in young children
The patient population consisted of 49 children between 1 and 7 years of age (median 2 years), and 3 adults (43, 58, and 73 years of age) infected by A/H3N2 influenza virus, from whom at least three respiratory specimens with a Ct value of 35 or less were available (Table 1, Supplementary Figs S1 and S3). Patients were enrolled 0–7 days (median 5 days) after the onset of illness. All patients received 5 days of oseltamivir after enrollment. Six patients had received 1–3 days of oseltamivir prior to inclusion. We performed NGS on 226 longitudinally collected samples available from the 52 patients and deep sequenced the HA and NA gene segments of the viral populations infecting these individuals, achieving an average read depth of for both genes across all samples (Supplementary Fig. S4).
Table 1.
Patient characteristics.
| Vietnam |
Thailand | Total | |||
|---|---|---|---|---|---|
| 2007 | 2008 | 2009 | 2008 | ||
| n = 1 | n = 12 | n = 32 | n = 7 | n = 52 | |
| Sex | |||||
| Male | 1 | 8 | 20 | 6 | 35 |
| Female | – | 4 | 12 | 1 | 17 |
| Age group | |||||
| 1–2 | – | 7 | 17 | 2 | 26 |
| 2–3 | 1 | 5 | 8 | 1 | 15 |
| 3–4 | – | – | 2 | 3 | 5 |
| 4–5 | – | – | 1 | – | 1 |
| 5–6 | – | – | – | 1 | 1 |
| 6–7 | – | – | 1 | – | 1 |
| 40–80 | – | – | 3 | – | 3 |
| Median (range) | 2 (72) | ||||
| Clinical outcome | |||||
| Average length of stay (range) | 7 (–) | 7 (3) | 9.6 (12) | 10.3 (21) | 9.1 (23) |
| ICU (cases) | – | 4 | 0 | 1 | 5 |
| Died | – | 1 | – | – | 1 |
| Oseltamivir dosing | |||||
| Single | 1 | 8 | 16 | 4 | 29 |
| Double | – | 4 | 16 | 3 | 23 |
| Days between onset and enrollment | |||||
| <2 | – | 1 | 3 | – | 4 |
| 2–4 | – | 6 | 13 | 3 | 22 |
| 5–7 | 1 | 5 | 16 | 4 | 26 |
The viral load of all analyzed specimens (Ct values of all samples are between 22 and 35 cycles; 104 out of 226 specimens (46%) have Ct values < 30) should provide ample viral genetic material for sequencing such that variant proportions do not become significantly skewed during amplification (Xue et al. 2018). To estimate the precision of our reported variant proportions, we plotted the proportions of nucleotide polymorphic sites that were covered by the independent reads sequenced from two different, but overlapping amplicons against each other (Supplementary Fig. S5). Such amplicons are independently amplified using different RT-PCR primers. The median number of such sites is 29.5 per sample (interquartile range (IQR) = 12–70.5). We found that most variant proportions computed from the sequenced reads of one amplicon are highly similar to those sequenced from another overlapping template (median 6.5% (IQR = 4.4%–13.2%) of such sites differs in proportions by >5% between the two amplicons), even for samples with low viral load (i.e. high Ct values, Supplementary Fig. S5). Differences between different templates were even alleviated when we compared the variant proportions computed by combining over all amplicons (i.e. the proportions used for further analyses in this work) against those from individual amplicon (median 4.7% (IQR = 4.2%–5.4%) of sites where the proportions from combined amplicons differ by >5% relative to individual amplicon). Therefore, we were able to conclude that detected variants were representative of the variants present in the patient specimens.
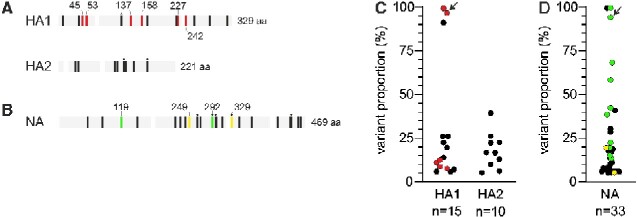
Genetic diversity of intrahost influenza virus population is generally low as we found only 1–4 amino acid variants in both genes that reached proportions of at least 5 per cent in the intrahost virus population for each individual (Supplementary Fig. S6). HA variants were found in 18 of 52 patients (Supplementary Fig. S6). Fifteen variants, found in 14 patients, were in the immunodominant HA1 subunit while another 10 variants were observed in the HA2 subunit, in a total of 10 patients. Amino acid variation per position was similar in both HA1 and HA2 subunits (0.046 and 0.045 variants per amino acid position, respectively). Only three HA variants reached majority proportions (>50%) in at least one timepoint during the course of infection: D53N, S169P, and I242V (Fig. 1C) while the rest were only detected in minority proportions. Six variants were in antigenic sites that may affect antibody recognition. Putative antigenic variants were identified in antigenic sites A (S137P), B (N158H), C (S45N and D53N), and D (P227H and I242V), and occurred in six patients of 2–4 years of age. Variant S137P was observed in a last-day specimen only (Fig. 2), the remaining five antigenic site variants were detected on multiple days. Apart from D53N and I242V which became a majority allele at some point, the highest proportions observed for the remaining four minority antigenic site variants (8–12%) were similar or lower than peak proportions of non-antigenic site variants (6–26%) and variants in HA2 (5–39%). The amino acid changes in two variants, S8N and S45N (an antigenic site variant), lead to addition of N-glycosylation patterns in the HA of these variants.
Figure 1.
Intrahost diversity in the HA and NA proteins. Black vertical lines in panels (A) and (B) represent variable amino acid positions. White vertical bars indicate primer regions. (A) Overview of variable amino acid positions in HA1 and HA2. Red vertical lines indicate amino acid variations in antigenic sites (Wiley et al. 1981; Wilson and Cox 1990). All HA positions are based on H3 numbering without signal peptide. (B) Overview of variable amino acid positions in NA. Yellow vertical lines indicate substitutions associated with reduced inhibition by oseltamivir, green vertical lines indicate variable positions associated with highly reduced inhibition by oseltamivir (Lina et al. 2018). Asterisks indicate positions varying in multiple patients; HA: 119 (two patients), 453 (two), and 498 (two); NA: 264 (two), 292 (two), 307 (two), 329 (two), and 442 (two). (C) Maximum proportions reached by HA1 and HA2 variants. (D) Maximum proportions reached by NA variants. Color coding as in panels (A) and (B). Arrows indicate HA D53N and NA R292K from patient 1707 that co-emerge (see main text).
Figure 2.
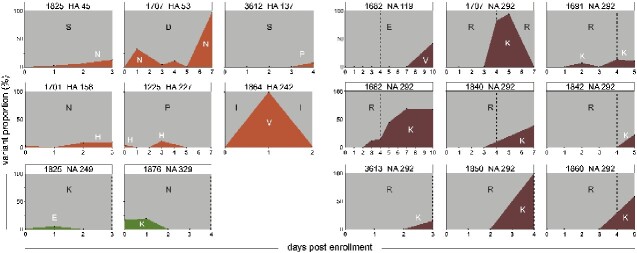
Variant proportion changes over time. Orange, green, and brown plots show evolutionary dynamics of antigenic site variants, reduced inhibition variants, and highly reduced inhibition variants, respectively. Patient ID, protein, and amino acid position are specified above each plot. Amino acid changes are indicated in black and white capital letters, where white letters indicate the minority variant enrollment. A vertical dashed line in the NA plots specifies the last day of oseltamivir treatment. Supplementary Fig. S6 shows evolutionary dynamics of all HA and NA variants.
We also found premature stop codons in two amino acid sites in two different samples (patient 3612; HA position 367, and patient 3615; NA position 6). In both cases, these stop codons only emerged on the last day of sampling of the two patients in low proportions (HA-367 stop codon: 14.5% on day 4 post enrolment of patient 3612; NA-6 stop codon: 11.1% on day 7 post-enrolment of patient 3615). At the same time, no minority amino acid variant was positively selected to become a majority allele in either patient, even during later timepoints (Supplementary Fig. S6). Instead, all of them persisted at low proportions, never amounting to more than 20 per cent, suggesting that purifying selection may be purging deleterious mutants within host. However, the presence of lethal premature stop codons also suggest that purifying selection may not be acting strongly enough in these individuals to reduce the proportions of deleterious mutations completely (Xue and Bloom 2020). Furthermore, incomplete influenza virus genomes have been frequently observed at the cellular level and these defective genomes can be reactivated by complementation through reassortment (Jacobs et al. 2019). As such, it may not be surprising to find the presence of premature stop codons in genes of intrahost viral populations. It is, however, difficult to generalize these observations further based on this study as premature stop codons were only found in two individuals.
3.2 Evolutionary dynamics of ha variants provide few signs for selective outgrowth or fixation
We next investigated the dynamics of intrahost HA variants. Proportions of HA variants often fluctuated over time. Differences in variant proportions of 5–20 per cent between specimens collected at consecutive days frequently occurred (Fig. 2 and Supplementary Fig. S6). We observed fluctuating dynamics for intrahost minority variants present at low proportions in the virus population (e.g. P227H, T436I), as well as in variants that reach high or even majority proportions (e.g. D53N, S169P, I242V, R361I). Antigenic site variants first appeared in the virus populations 4–8 days after onset of symptoms (Fig. 4), and in five of six patients antigenic site variants were present in last-day clinical specimens. The dynamics of antigenic and non-antigenic site variants seemed mostly similar. The rapid increase in variant proportion of HA D53N in the virus population after day 5 (from <1% to 97%) coincided with NA K292R (from 6% to 94%) that emerged after discontinuation of oseltamivir treatment, and that is responsible for reversion to an oseltamivir susceptible phenotype (Fig. 3 and Supplementary Fig. S7). Apart from 169P, a non-antigenic site variant that seemed to fixate in the virus population in a period of 4–s5 days, and antigenic site variant S45N, which steadily increased in the intrahost virus population during four consecutive days, there were few other signs of fixation or selective outgrowth in HA variants (Supplementary Fig. S6).
Figure 3.
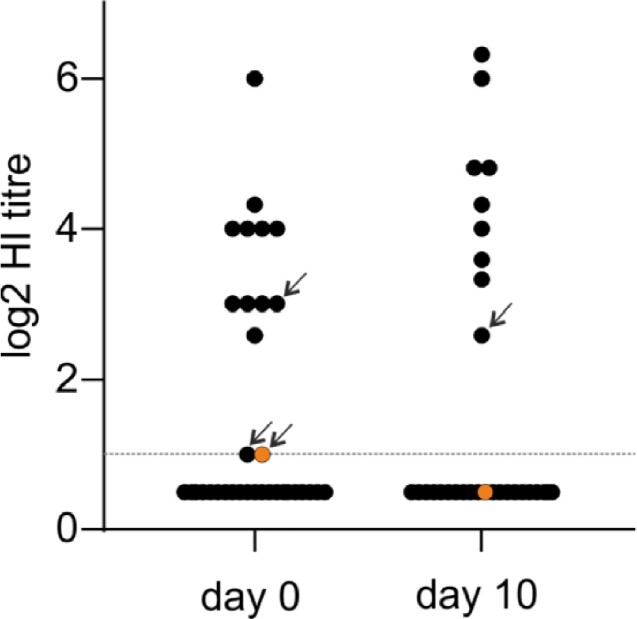
Serologic analysis of day 0 and day 10 plasma samples. The dashed horizontal line indicates the detection limit of the HI assay at the starting dilution of 1/40 used here. Arrows in the day 0 column indicate plasma samples for which HI titers dropped below detection at day 10. The arrow in the day 10 column indicates a plasma sample for which there was no titer above the detection limit at day 0. Orange filled symbols are the log2 HI titers for the patient with antigenic site variant HA S45N.
Five intrahost HA variants reached proportions of 44–100 per cent in the globally circulating influenza virus population (Supplementary Fig. S6). Antigenic site variants S45N and D53N emerged in patients included in this study immediately prior to increases in global rates to 96 per cent and 44 per cent, respectively. N225D disappeared from the globally circulating virus population in 2008, when prevalence declined to 2 per cent from 98 per cent in 2004. Variant R361I was also identified immediately after global prevalence declined to 1 per cent globally, but persisted as minority in the intrahost virus population. S169P was predominantly S at enrollment, but changed to 100 per cent P over 6 days, thereby conforming to the 20-year global consensus. The HA variants that were observed in multiple patients (E119K (n = 2), K453R/N (n = 2), and N498K (n = 2)) did not reach high proportions in the global virus population.
3.3 Five of six antigenic site variants emerged in patients without detectable HI titers
We performed HI assays using the A/Brisbane/10/07 strain that circulated immediately prior to the period of sample collection to test for anti-A/H3 antibodies. Plasma samples collected at days 0 and 10 were available for all and 48/52 study patients, respectively. Plasma antibodies against the test strain were detected in 25 per cent (n = 13) and 19 per cent (n = 9) of patients at day 0 and day 10 (Fig. 3). HI titers in seropositive patients ranged from 20 to 640 at day 0, and from 20 to 800 at day 10 (median 80 and 200). Antigenic site-variant HA S45N emerged in a patient that had a day 0 plasma HI titer of 20 (the detection limit of the assay we used), but had no detectable HI titer at day 10 (Fig. 3). Other antigenic site variants emerged in patients in whom no anti-H3 antibodies were detected.
3.4 Oseltamivir-resistant variants emerge in ten of fifty-two patients
We identified thirty-three NA variants in twenty-four patients that reached proportions of at least 5 per cent in the intrahost virus population (Fig. 1B). Because all patients had received oseltamivir, we next identified variants conferring reduced inhibition (RI) and highly reduced inhibition (HRI) by oseltamivir. RI and HRI are defined as 10–100 and >1000-fold increase in IC50 compared to homologous viruses without substitutions conferring oseltamivir resistance (i.e. normal inhibition, NI) (World Health Organisation 2012). We identified 10 patients (19%) in whom oseltamivir resistance emerged. RI variants K249E and N329K reached variant proportions of 5–20 per cent in the intrahost virus population (Fig. 1B and D) and were each identified in a single patient. HRI-variant R292K was found in eight patients (15% of all patients). R292K reached majority proportions of 58, 68, 94, and >99 per cent in four patients. In the remaining four patients, R292K variants were present at minority proportions (13–38%). HRI-variant E119V was identified in a single patient in whom R292K variants also emerged and reached 42 per cent in the virus population (Supplementary Fig. S6). NI variants reached proportions of 6–41 per cent, but I307M reached a peak proportion of >99 per cent alongside the peak of HA I242V in the same patient. It is tempting to speculate if HA-I242V, which is part of the H3 antigenic site D (Wiley, Wilson, and Skehel 1981; Wilson and Cox 1990), could have been epistatically linked to NA-I307M, resulting in their coordinated increase in proportions. However, HA-242V was eventually lost while NA-307M remained as a majority allele in the intrahost viral population. Amino acid variation per position in the NA was higher than that found for HA (0.070 variants per amino acid position). However, when excluding positions associated with RI or HRI, amino acid variation per position in NA was similar to observed variation in the HA1 subunit including antigenic site positions and the HA2 subunit (0.046, 0.046, and 0.045 variants per amino acid position, respectively).
3.5 Oseltamivir-resistant variants rapidly increase in the intrahost virus population
HRI variants E119V and R292K mostly showed a rapid increase in variant proportion (Fig. 2). However, first detection for R292K and E119V varied greatly among patients, ranging from 2 to 7 and 10 days after initiation of oseltamivir treatment, respectively (Fig. 4). HRI variants remained present in the virus population until the last day viral RNA was detectable in clinical specimens. For a single patient, R292K was present until the last day of oseltamivir treatment, after which the wild type re-emerged and became the majority variant (Fig. 2). No reversion to the wild type (292R) was observed in the two other patients that remained A/H3N2 positive after treatment ended. RI variants K249E and N329K were identified at minority levels at the day of enrollment, but disappeared from the virus population at later timepoints (Fig. 2). The fluctuating dynamics and lower variant proportions of RI variants, that confer a less pronounced reduction of inhibition by oseltamivir, were more similar to that of the NI variants identified in this study (Fig. 2. and Supplementary Fig. S6). There were no clear differences in emergence, or first detection, of RI or HRI variants between patients treated with standard and double dose of oseltamivir, but the number of patients in whom such variants emerged were small (Fig. 4 and Supplementary Table S1).
The prevalence of five NA variants in the global population ranged from 7 per cent to fixation of the amino acid in the population, but global frequencies for these NA variants were mostly lower than observed for HA variants (Supplementary Fig. S6). Two intrahost NA variants were present at majority proportions on the global level. In agreement with globally circulating strains, NA R150H was identified at a low variant proportion in a patient immediately after global prevalence declined from 100 per cent in 2004 to 1 per cent in 2008. Variant S367N was found at a low variant proportion intrahost and globally in 2008, preceding a steep increase in global frequency to 87 per cent in 2010 and 100 per cent in 2013. The remaining NA variants were found at maximum global proportions of 42 per cent, 17 per cent and 99 per cent, and 7 per cent (N329T, I307M and M307I, and K249E, respectively). Variants observed in multiple patients (H264N, R292K, I307M and M307I, N329K and N329T, S442N) were either RI or HRI variants or did not reach high proportions in the global virus population.
4. Discussion
Anti-HA antibody responses and oseltamivir treatment are likely to select for viruses that escape antibody neutralization or inhibition by oseltamivir. Amino acid substitutions responsible for escape from neutralizing antibodies on the global level are sufficient to provide a strong selective advantage in model systems (Li et al. 2016; Doud, Hensley, and Bloom, 2017; Doud, Lee, and Bloom 2018), but recent studies have found little evidence for selective outgrowth of putative antigenic variants during the course of infection in individual patients (Dinis et al. 2016; Debbink et al. 2017; McCrone et al. 2018). Most studies on intrahost variation lacked longitudinal data, thus providing snapshots of intrahost virus evolution, and mostly included clinical specimens collected from adult patients. All clinical specimens in these studies were obtained in the United States. However, potential antigenic variants may emerge later during the course of infection or in different study populations. In contrast to antigenic variants, viruses with highly reduced inhibition to oseltamivir frequently emerge during treatment, especially in young children (Kiso et al. 2004; Stephenson et al. 2009; Tamura et al. 2011; Whitley et al. 2013; Eshaghi et al. 2014; Lina et al. 2018). Here we used NGS on longitudinally collected clinical specimens from a cohort of mostly young Southeast Asian children who were treated with oseltamivir, to track within host emergence of HA and NA variants over the course of infection. We found that the evolutionary dynamics of oseltamivir-resistant NA variants strongly suggest positive selection, but few indications of similar strong selection for variants at antigenic sites in HA.
A large portion of the HA globular head surface is covered by amino acid positions that are associated with antibody escape (Wiley, Wilson, and Skehel 1981; Wilson and Cox 1990). However, only few key positions close to the receptor binding site have contributed to large antigenic changes during A/H3N2 virus evolution on the global level (Koel et al. 2013). Of the six antigenic site variants we observed, only the variant at position 158 concerns a key position, while the variant at position 37 is immediately adjacent to the receptor binding site. The remaining four variants are located in antigenic sites away from the receptor binding site. Incidence of influenza virus infection in young children is relatively high (Tokars et al. 2018). Depending on age, a substantial proportion of young children may not yet have experienced influenza virus infection (Bodewes et al. 2011). In Vietnam, the influenza vaccine is not part of the National vaccination program and coverage is typically very low. We therefore expected that only a moderate percentage of patients enrolled in this study to have prior immunity to the A/H3N2 virus subtype. Indeed, we found that only 25 per cent of the patients analyzed had detectable circulating HI antibodies to a contemporary A/H3N2 virus at the day of enrollment. Notably, titers were at or below the detection limit of our assay in patients in whom antigenic site variants were identified. In other words, the limited number of antigenic HA variants detected in this within-host study could be attributed to a lack of antibody-mediated immunity against A/H3N2 viruses in this cohort of mostly naïve children. Children may need multiple exposures to influenza viruses before they are able to generate a robust antibody response (Neuzil et al. 2006), thus possibly explaining the lack of HI titer increase after they were infected and consequently the limited strength of antibody-mediated immunity to select for de novo antigenic variants. In contrast, even though immunocompromised adults have severely impaired immunity, some adaptive (memory) immune response must have remained to select for antigenic variants during prolonged infections (Xue et al. 2017).
In spite of the lack of antibody-mediated immunity in these children, we found several variants emerging in antigenic sites that remained present until the last available clinical specimen collected, including some that reached high global levels in the years after identification in patients included in this study. These outcomes show intrahost emergence of globally relevant virus variants, which may suggest that antigenic site variants identified in this study have some selective advantage. However, these variants were mostly present at low proportions, fluctuated in the virus population during the course of infection, or could have been associated with variation in NA. It therefore seems unlikely that the antigenic site variants identified here are sufficiently fitter than the other viruses in the intrahost virus population to ensure a large selective advantage.
Similar to previous studies in children, oseltamivir-resistant variants emerged in 19 per cent (n = 10) of our patients during treatment (Kiso et al. 2004). RI variants K249E and N329K at the start of oseltamivir treatment possibly represent the infecting wild-type virus population, as both variants were present at low frequencies at the global level at the time of specimen collection (http://flusurver.bii.a-star.edu.sg; last accessed 1 November 2020). In agreement with the small reductions in oseltamivir susceptibility associated with RI variants, K249E and N329K were not positively selected and disappeared from the population. In contrast, HRI variants E119V and R292K reached high proportions and remained present in the intrahost virus populations. The first HRI variants emerged 2–3 days after initiation of treatment. Although such early detection of oseltamivir-resistant variants has been reported by others, resistant variants usually emerge later, especially in adults (Kiso et al. 2004; Tamura et al. 2011; Whitley et al. 2013; Eshaghi et al. 2014; Lina et al. 2018). In six of eight patients in our young study population oseltamivir-resistant variants did not emerge until days 4–7 after the start of oseltamivir treatment. These outcomes suggest that the time required for generation and selection of novel variants may be several days, including those that emerge under strong selection pressures such as antiviral treatment.
Our findings indicate that a de novo amino acid variant can emerge and reach high proportions in the intrahost virus population during acute infection if its selective advantage is sufficiently large. Whereas HRI variants emerged to high proportions under oseltamivir treatment, antigenic variants in our patient population did not, possibly suggesting either that antibody selection may be a comparatively weak selection pressure on the individual level or that antigenic mutations may incur a significant fitness cost to the virus. Our results in a group of 13 immune individuals are in accordance with the apparent absence of outgrowth of antigenic variants in individual patients reported by other studies (Dinis et al. 2016; Debbink et al. 2017; McCrone et al. 2018). However, our results also indicate that timing of specimen collection is critical for the detection of emerging variants, even those under strong selection pressures. Clinical specimens are usually collected when symptom severity peaks, 2–3 days after infection, around the peak of viral replication (Carrat et al. 2008). Previous studies looking into intrahost evolution of influenza viruses primarily used samples collected at days 0–3 after symptom onset (Xue and Bloom 2020). If our samples were collected at only a single timepoint up to day 3 post start of oseltamivir treatment, we would have identified R292K variants in none or at most a single patient out of our fifty-two patients, depending on the day of sample collection. A detection rate of 12.5 per cent in the best-case scenario. Antigenic variants are not as likely as HRI variants to outcompete other viruses in the intrahost population if antibody pressure is indeed a weaker selective force than oseltamivir, and may not emerge as rapidly as the HRI variants identified in this study. As few studies have had the opportunity to analyze large numbers of late timepoint samples or sequentially collected samples, it may not be surprising that outgrowth of antigenic variants has not been observed to date. Hence, further studies sampling later timepoint samples are needed to not only detect for such delayed variants but also to understand the strength of positive antigenic selection and how it might differ between different individuals.
We identified HA and NA variants in the majority of the patients in our study, but the proportion of most variants remains low, fluctuates between successive samples, frequently drops below detection levels, and outgrowth of variants towards majority levels is rare. These findings are in line with various other within-host studies analyzing evolutionary dynamics of acute influenza infections where intrahost virus diversity is found to be low (Dinis et al. 2016; Debbink et al. 2017; McCrone et al. 2018). Nonetheless, several intrahost variants were identified in this study coincided with those found at the global population level. Two initial HA minority variants (53N and 169P) that seemed to fixate in the intrahost virus population at later timepoints were also observed at high global proportions. A third intrahost HA variant (S45N) that steadily increased over time during the course of infection also reached high global frequencies. Whether intrahost variation during acute infection has predictive value for global trends or is merely a reflection of contemporary variation should be addressed in further studies. We find occasional co-evolution of variants with substitutions that confer oseltamivir resistance, but nearly all variation in HA and NA observed in these patients seemed independent of oseltamivir treatment. Although we cannot rule out that some of the observed variation is linked to antiviral treatment or other selection pressures, our observations agree with the suggestion that much of the intrahost influenza virus evolution is determined by stochastic processes (McCrone et al. 2018).
Natural selection of influenza virus variants that most efficiently escape neutralizing antibodies is considered a major selective force driving global evolution of seasonal human influenza viruses. As previous studies have found few indications that de novo antigenic variants emerge to high proportions during the course of typical influenza virus infections it remains unclear how antigenic variants emerge on the global level. Previous studies have posited a number of hypotheses as to why emerging intrahost antigenic variants are seldom identified. Antigenic selection may be difficult to detect at the individual level because existing cross-reactive antibodies reduce the viral population size on which selection can act (Petrova and Russell 2017), intrahost influenza evolution is dominated by stochastic effects (McCrone et al. 2018), antigenic variants need compensatory co-mutations to increase replicative fitness (Koel et al. 2013; Dinis et al. 2016), or because selection for antigenically novel variants occurs between hosts rather than within infected individuals (Xue and Bloom 2020). It seems clear that intrahost emergence of novel antigenic variants is rare, and while each of these hypotheses may be correct, there could also be a simpler explanation. Our results suggest that insights into the evolutionary dynamics during infection may be hampered by incomplete sampling during the course of infection. We find that longitudinal sampling in different patient populations is required to maximize detection of novel variants, irrespective of the strength of the selective pressure.
Data availability
FASTQ files of the trimmed merged reads that were used for mapping are available from https://github.com/mirdesign/NGS-FluAnalyzer/tree/master/Raw_Data_For_Intrahost.AH3N2.Virus.HANA.Variants (last accessed 1 November 2020).
Supplementary data
Supplementary data are available at Virus Evolution online.
Supplementary Material
Acknowledgements
We thank the patients for enrolling and their guardians for granting permission to enroll their children into the study, the participating physicians and nurses of the Children's Hospital 1 (Ho Chi Minh City, Vietnam), Children’s Hospital 2 (Ho Chi Minh City, Vietnam), Hospital for Tropical Diseases (Ho Chi Minh City, Vietnam), National Hospital for Tropical Diseases (Hanoi, Vietnam), Vietnam National Children's Hospital (Hanoi, Vietnam), and Siriraj Hospital (Bangkok, Thailand) for their help in sample acquisition. The authors are grateful to Silvana Roos (Amsterdam UMC, The Netherlands) for assistance and technical support, and to Ron Fouchier and Sander Herfst (Erasmus MC, The Netherlands) for providing the turkey erythrocytes used in this study. We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAID’s EpiFlu™ Database used in this study (the list is available from Supplementary Table S3). B.F.K. and M.P. thank SURFsara (www.surfsara.nl) for the support in using the Lisa Compute Cluster.
Funding
This research was supported by ZonMW TOP grant 91213058, Wellcome Trust Major Overseas Programme Vietnam 089276/Z/09/Z, ESCMID research grant 113228, Amsterdam Infection and Immunity Institute Postdoc Stipend 1807018, and by the Amsterdam UMC (Amsterdam, The Netherlands).
Conflict of interest: None declared.
Contributor Information
B F Koel, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
R M Vigeveno, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
M Pater, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
S M Koekkoek, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
A X Han, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
H M Tuan, Children's Hospital 2.
T T N Anh, Children's Hospital 2.
N T Hung, Children's Hospital 1, Ho Chi Minh City, Vietnam.
L Q Thinh, Children's Hospital 1, Ho Chi Minh City, Vietnam.
L T Hai, Vietnam National Children's Hospital, Hanoi, Vietnam.
H T B Ngoc, Vietnam National Children's Hospital, Hanoi, Vietnam.
N V V Chau, Hospital for Tropical Diseases, Ho Chi Minh City, Vietnam.
N M Ngoc, Hospital for Tropical Diseases, Ho Chi Minh City, Vietnam.
K Chokephaibulkit, Siriraj Hospital, Bangkok, Thailand.
P Puthavathana, Siriraj Hospital, Bangkok, Thailand.
N V Kinh, National Hospital of Tropical Diseases, Hanoi, Vietnam.
T Trinh, National Hospital of Tropical Diseases, Hanoi, Vietnam.
R T C Lee, Bioinformatics Institute, Agency for Science Technology and Research, Singapore 138671 Singapore.
S Maurer-Stroh, Bioinformatics Institute, Agency for Science Technology and Research, Singapore 138671 Singapore; Department of Biological Sciences, National University of Singapore, Singapore 117558, Singapore; National Public Health Laboratory, National Centre for Infectious Diseases, Ministry of Health, Singapore 308442, Singapore.
D Eggink, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
T T Thanh, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
L V Tan, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam.
H R van Doorn, Oxford University Clinical Research Unit, Hanoi, Vietnam; Nuffield Department of Medicine, Centre for Tropical Medicine and Global Health, University of Oxford, Oxford, UK.
M D de Jong, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
References
- BBMap short read aligner, and other bioinformatic tools <https://sourceforge.net/projects/bbmap/> accessed 1 Nov 2020.
- Bodewes R. et al. (2011) ‘ Prevalence of Antibodies against Seasonal Influenza A and B Viruses in Children in Netherlands’, Clinical and Vaccine Immunology, 18: 469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014) ‘ Trimmomatic: A Flexible Trimmer for Illumina Sequence Data’, Bioinformatics, 30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. F., Smith D. J. (2014) ‘ A Recommended Numbering Scheme for Influenza a HA Subtypes’, PLoS One, 9: e112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrat F et al. (2008) ‘ Time Lines of Infection and Disease in Human Influenza: A Review of Volunteer Challenge Studies’, American Journal of Epidemiology, 167: 775–85. [DOI] [PubMed] [Google Scholar]
- Debbink K. et al. (2017) ‘ Vaccination Has Minimal Impact on the Intrahost Diversity of H3N2 Influenza Viruses’, PLOS Pathogens, 13: e1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinis J. M. et al. (2016) ‘ Deep Sequencing Reveals Potential Antigenic Variants at Low Frequencies in Influenza a Virus-Infected Humans’, Journal of Virology, 90: 3355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud M. B., Hensley S. E., Bloom J. D. (2017) ‘ Complete Mapping of Viral Escape from Neutralizing Antibodies’, PLOS Pathogens, 13: e1006271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud M. B., Lee J. M., Bloom J. D. (2018) ‘ How Single Mutations Affect Viral Escape from Broad and Narrow Antibodies to H1 Influenza Hemagglutinin’, Nature Communications, 9: doi:10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A. et al. (2014) ‘ Multiple Influenza A (H3N2) Mutations Conferring Resistance to Neuraminidase Inhibitors in a Bone Marrow Transplant Recipient’, Antimicrobial Agents and Chemotherapy, 58: 7188–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FastQC <http://www.bioinformatics.babraham.ac.uk/projects/fastqc> accessed 1 Nov 2020.
- Hirst G. K. (1943) ‘ Studies of Antigenic differences among Strains of Influenza a by Means of Red Cell Agglutination’, Journal of Experimental Medicine, 78: 407–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- <http://flusurver.bii.a-star.edu.sg> accessed 1 Nov 2020.
- Inoue M. et al. (2010) ‘ Emergence of Oseltamivir-Resistant Pandemic (H1N1) 2009 Virus within 48 Hours’, Emerging Infectious Diseases, 16: 1633–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs N. T. et al. (2019) ‘ Incomplete Influenza a Virus Genomes Occur Frequently but Are Readily Complemented during Localized Viral Spread’, Nature Communications, 10: 3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiso M. et al. (2004) ‘ Resistant Influenza a Viruses in Children Treated with Oseltamivir: Descriptive Study’, The Lancet, 364: 759–65. [DOI] [PubMed] [Google Scholar]
- Koel B. F. et al. (2013) ‘ Substitutions near the Receptor Binding Site Determine Major Antigenic Change during Influenza Virus Evolution’, Science, 342: 976–9. [DOI] [PubMed] [Google Scholar]
- Krammer F. (2019) ‘ The Human Antibody Response to Influenza a Virus Infection and Vaccination’, Nature Reviews Immunology, 19: 383–97. [DOI] [PubMed] [Google Scholar]
- Li C. et al. (2016) ‘ Selection of Antigenically Advanced Variants of Seasonal Influenza Viruses’, Nature Microbiology, 1: 16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina B. et al. (2018) ‘ Five Years of Monitoring for the Emergence of Oseltamivir Resistance in Patients with Influenza a Infections in the Influenza Resistance Information Study’, Influenza and Other Respiratory Viruses, 12: 267–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H. E. et al. (2018) ‘ Obesity Increases the Duration of Influenza a Virus Shedding in Adults’, The Journal of Infectious Diseases, 218: 1378–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrone J. T. et al. (2018) ‘ Stochastic Processes Constrain the within and between Host Evolution of Influenza Virus’, eLife, 7: e35962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuzil K. M. et al. (2006) ‘ Immunogenicity and Reactogenicity of 1 versus 2 Doses of Trivalent Inactivated Influenza Vaccine in Vaccine-Naive 5–8-Year-Old Children’, The Journal of Infectious Diseases, 194: 1032–9. [DOI] [PubMed] [Google Scholar]
- Ng S. et al. (2016) ‘ The Timeline of Influenza Virus Shedding in Children and Adults in a Household Transmission Study of Influenza in Managua, Nicaragua’, The Pediatric Infectious Disease Journal, 35: 583–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova V. N. Russell C. A. (2018) ‘ The Evolution of Seasonal Influenza Viruses’, Nature Reviews Microbiology, 16: 47–60. 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- South East Asia Infectious Disease Clinical Research Network. (2013) ‘ Effect of Double Dose Oseltamivir on Clinical and Virological Outcomes in Children and Adults Admitted to Hospital with Severe Influenza: Double Blind Randomised Controlled Trial’, The BMJ, 346: f3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson I. et al. (2009) ‘ Neuraminidase Inhibitor Resistance after Oseltamivir Treatment of Acute Influenza A and B in Children’, Clinical Infectious Diseases, 48: 389–96. [DOI] [PubMed] [Google Scholar]
- Tamura D. et al. (2011) ‘ Frequency of Drug-Resistant Viruses and Virus Shedding in Pediatric Influenza Patients Treated with Neuraminidase Inhibitors’, Clinical Infectious Diseases, 52: 432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokars J. I., OlsenS. J., and Reed C. (2018) ‘ Seasonal Incidence of Symptomatic Influenza in the United States’, Clinical Infectious Diseases, 66: 1511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R. J. et al. (2013) ‘ Global Assessment of Resistance to Neuraminidase Inhibitors, 2008–2011: The Influenza Resistance Information Study (IRIS)’, Clinical Infectious Diseases, 56: 1197–205. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. (1981) ‘ Structural Identification of the Antibody-Binding Sites of Hong Kong Influenza Haemagglutinin and Their Involvement in Antigenic Variation’, Nature, 289: 373–8. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Cox N. J. (1990) ‘ Structural Basis of Immune Recognition of Influenza Virus Hemagglutinin’, Annual Review of Immunology, 8: 737–87. [DOI] [PubMed] [Google Scholar]
- World Health Organisation. (2012) ‘ Meetings of the WHO Working Group on Surveillance of Influenza Antiviral Susceptibility – Geneva, November 2011 and June 2012’, Relevé Épidémiologique Hebdomadaire, 87: 369–74. [PubMed] [Google Scholar]
- Xue K. S., Bloom J. D. (2020) ‘ Linking Influenza Virus Evolution within and between Human Hosts’, Virus Evolution, 6: 812016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K. S. et al. (2017) ‘ Parallel Evolution of Influenza across Multiple Spatiotemporal Scales’, eLife, 6: e26875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K. S. et al. (2018) ‘ Within-Host Evolution of Human Influenza Virus’, Trends in Microbiology, 26: 781–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
FASTQ files of the trimmed merged reads that were used for mapping are available from https://github.com/mirdesign/NGS-FluAnalyzer/tree/master/Raw_Data_For_Intrahost.AH3N2.Virus.HANA.Variants (last accessed 1 November 2020).