Abstract
Drosophila eggs are highly polarised cells that use RNA–protein complexes to regulate storage and translational control of maternal RNAs. Ribonucleoprotein granules are a class of biological condensates that form predominantly by intracellular phase separation. Despite extensive in vitro studies testing the physical principles regulating condensates, how phase separation translates to biological function remains largely unanswered. In this perspective, we discuss granules in Drosophila oogenesis as a model system for investigating the physiological role of phase separation. We review key maternal granules and their properties while highlighting ribonucleoprotein phase separation behaviours observed during development. Finally, we discuss how concepts and models from liquid–liquid phase separation could be used to test mechanisms underlying granule assembly, regulation and function in Drosophila oogenesis.
Keywords: developmental biology, phase separation, RNP granules
Introduction
Intracellular localisation of messenger RNAs (mRNAs) is a conserved mechanism for achieving compartmentalised protein expression in polarised cells such as neurons and fibroblasts [1–5]. To generate precise protein synthesis and prevent ectopic expression, mRNA localisation is often coupled to translational regulation. One way to achieve this regulation is through binding of trans-acting RNA binding proteins (RBP) to cis-acting sequences in the mRNA, which together form micron-sized compartments called ribonucleoprotein (RNP) granules [6–10]. These granules function in the packaging, transport and translational control of mRNAs, and can rapidly respond to cellular and external stimuli. With the ability to control spatial and temporal gene expression, understanding how RNP granules form and disassemble is a key question in cell biology.
RNP granules belong to a class of organelles which lack a physical membrane that separate their contents from the cytoplasm. Different from the commonly known membrane-bound organelles such as the endoplasmic reticulum and Golgi apparatus, membrane-less granules (also referred to as biomolecular condensates) constitute an additional level of macromolecular organization in the cell [11–13]. Most commonly forming via liquid–liquid phase separation (LLPS), these condensates function as microenvironments for cellular reactions [14–16]. This process of ‘de-mixing’ allows RNA and protein molecules to condense into a concentration dependent dense phase which coexists with the soluble cytoplasmic phase [17–20]. The idea that cellular contents exhibit liquid-like characteristics was proposed by multiple groups over the past century, but received renewed interest when P granules in the C. elegans embryo were shown to exhibit liquid-like behaviour [21,22]. This discovery has since led to a dramatic increase in the research of biomolecular condensates [18,23–35]. For a more extensive discussion on the physics of condensates, we refer readers to several excellent reviews [36–42].
Our current understanding of the physicochemical principles regulating condensates has been primarily elucidated through in vitro studies of purified RNP components under idealised conditions [43]. This approach has been instrumental in describing the role of non-equilibrium features of living cells including post-translational modifications and ATP driven processes, in addition to identifying sequence and structural determinants that control condensate phase behaviour [14,23,44–49]. However, why cells need these compartments, when are they utilised, and what their biochemical and biological functions are remain largely unanswered.
RNP granules, which typically form in response to accumulation of RNAs, are abundant in diverse oocytes including C. elegans, Drosophila and Xenopus [50–53]. Oocytes are highly specialised cells which often rely on RNP granules to localise maternal transcripts for pattern formation in the early embryo [5,54–56]. Oocytes therefore offer a unique opportunity to test the physicochemical principles of phase separation in a living system and further explore the biological role of RNP condensation.
Drosophila oocytes rely on maternal RNAs and proteins produced in the adjacent, supporting nurse cells which are subsequently deposited into the oocyte [1,2,7,57]. To support egg development in the absence of transcription in the oocyte, many RNP granules are highly optimised to ensure long term storage and translational repression of maternal mRNAs until fertilisation [7,58–60]. Importantly, homologues and orthologues of Drosophila RNPs have been shown to phase separate in other systems, including yeast, C.elegans and zebrafish [15,61–63]. With many experimental advantages, Drosophila eggs offer a powerful system to investigate the physical principles and biological role of RNP condensation in early development.
In this perspective, we provide a brief overview of RNP granules in Drosophila oogenesis and highlight examples of liquid-like behaviour observed during early development. We then discuss how key concepts and models from LLPS could be used to understand the physical, structural and molecular principles regulating granule assembly and function during oogenesis. We conclude by highlighting how a multi-disciplinary approach using in vitro and in vivo studies, along with modelling, could better illustrate the physiological role of biomolecular condensates.
Overview of maternal RNP granules in oogenesis
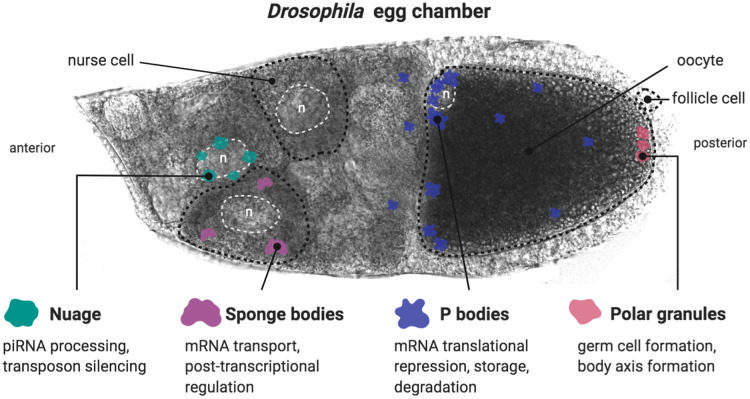
Body axis patterning of Drosophila depends on the localisation, storage, translational control and degradation of maternal RNAs throughout oogenesis and early embryogenesis [1,2,58,64]. Several aspects of RNA metabolism during development are known to be regulated by membrane-less organelles, primarily RNP granules. Based on the presence of specific RNP components, both cytoplasmic (e.g. Balbiani bodies and U bodies) and nuclear granules (e.g. Cajal bodies, histone-locus bodies and induced nuclear bodies) have been described in Drosophila egg chambers [65–69]. As a comprehensive discussion of all RNPs identified in oogenesis is beyond the scope of this perspective, we summarise key similarities and differences among the well-studied cytoplasmic maternal RNP granules, namely; nuage, sponge bodies, processing bodies (P bodies) and polar granules (Figure 1). We acknowledge that the nomenclature in this field is not always consistent and that the contents of certain ‘bodies’ can be contentious. Here, we discuss the granules based on existing structural and compositional evidence.
Figure 1. Schematic and role of maternal granules in egg chambers.
Nuage is localised around the nurse cell nuclei, while sponge bodies are dispersed throughout the cytoplasm of the nurse cells. P bodies are enriched at the anterior margin of the oocyte (especially in the dorso-anterior corner). They are also observed throughout the oocyte and nurse cell cytoplasm. Polar granules are present at the posterior pole of the oocyte. Fifteen nurse cells, positioned to the anterior, produce the components (mRNAs, proteins, etc.) required for the development of a single oocyte. These germline-derived cells are interconnected through cytoskeletal bridges, allowing for cytoplasmic movement between them, and are surrounded by somatic-derived layer of follicle cells. (Representative cell types of the egg chamber are outlined with black dotted lines. Representative nuclei are outlined with white dotted lines and marked with an ‘n’). Created in BioRender.
RNP granule similarities and differences
The first similarity is the absence of an outer membrane, thereby allowing RNP granules to rapidly and reversibly alter their composition in response to changes in cellular conditions such as pH, temperature and osmolarity. At egg activation, for example, a change in osmolarity is thought to cause P bodies in the mature oocyte to rapidly dissolve and release stored mRNAs for translation [69–71]. A second similarity is internal structuring within granules which creates an additional level of macromolecular organisation. Certain maternal mRNAs and RBPs for instance, are shown to be differentially partitioned within P bodies [62,69,72]. A third, obvious, similarity is that they all classify as RNP granules due to the presence of both proteins and RNAs. Therefore, physical principles underlying their biogenesis and regulation could be similar.
Despite these similarities, maternal RNP granules have unique functions at different stages of development. Nuage, the earliest visible RNP granules localised around nurse cell nuclei, are proposed to be sites of piwi-interacting RNA (piRNA) processing and transposon silencing while sponge bodies package and transport translationally repressed maternal mRNAs [65,73–76]. P bodies and polar granules are unique as their roles extend beyond oogenesis into embryogenesis. While P bodies help facilitate RNA storage and translational regulation, polar granules at the posterior of the oocyte function to sequester factors required for the formation of the embryonic germ cells [77–79]. A second key difference is compositional diversity among the different granules (Table 1). Importantly, granules such as sponge bodies, P bodies and polar granules, are not static in their protein composition, but rather are able to dramatically alter their composition to facilitate specific functions. Sponge bodies, for example, change their composition and dynamics immediately upon entry into the oocyte from the adjacent nurse cells [65]. Other notable differences such as size and morphology between maternal granules may exist, however, these features are less well characterised. It is likely that the morphological and compositional differences between granules dictate their material states. Even with these differences, structural and molecular similarities suggest that common underlying physical principles regulate the properties of maternal RNP granules.
Table 1. Compositional diversity and location of selected maternal RNP granules.
| Granule type | Proteins enriched | Location |
|---|---|---|
| Nuage | Ago3, Armitage, Aubergine, Krimper, Maelstrom, Me31B, Papi, Qin, Spindle-E, Squash, Tejas, Trailer hitch, Tudor, Vasa, Zucchini | nurse cells |
| Polar granules | Aubergine, Dcp-1, eiF4A, Me31B, Oskar, Piwi, Pyruvate kinase, 6-phosphofructokinase, Staufen, Ter 94, Tudor, Vasa | oocyte, embryo |
| P bodies | Ago-3, Bruno, Cup, Dcp-1, Dcp-2, Edc3, eiF4E, Exuperentia, Growl, Hpat, Hrb27C, Me31B, Orb, Pacman, Staufen, Squid, Trailer hitch | nurse cells, oocyte, embryo, adult neurons |
| Sponge bodies | BicC, Bruno, Btz, Cup, Dcp-1, Dcp-2, eiF4E, Exuperentia, Gus, Hrb27C, Me31B, Orb, Oskar, Squid, Trailer hitch | nurse cells, oocyte |
Proteins shown to be enriched/localised in selected maternal granules. In bold are proteins associated with more than one granule. Whilst this is not an exhaustive list of proteins, those included are the most well-understood relative to each granule. With many shared proteins, it is important to consider testing a combination of different markers when studying RNP granules in development.
Examples of phase separation during early Drosophila development
Despite sharing several proteins, how maternal RNP granules regulate their composition is a long-standing question. Our understanding of the biophysical and biochemical principles that govern granule diversity, assembly and disassembly has recently benefited from new conceptual frameworks. LLPS has emerged as an attractive model to explain the observed properties of membrane-less organelles, including RNP granules [11,48,80–83]. The earliest example of maternal RNP structures shown to exhibit liquid-like behaviour were induced ‘bodies’ found in the Drosophila oocyte nucleus. These bodies are highly dynamic, with frequent fusion events and exchange of molecules between the bodies and the nucleoplasm. Interestingly, their formation was induced by changes in the salt concentration, indicating that weak electrostatic interactions may govern their assembly [84,85].
Another example from oogenesis occurs in the cytoplasm when axis patterning maternal mRNAs, bcd and oskar (osk), enter the oocyte at the anterior margin from the adjacent nurse cells and independently coalesce into larger particles. These separate RNP associations localise to opposite poles of the oocyte, where they are anchored [86,87]. While the biological importance of coalescence and the impact on granule properties is not clear, it is plausible that coalescence leads to increased interactions that stabilise over time, likely to assist in anchorage.
In the early embryo, a key nuclear protein associated with heterochromatin assembly and function, Heterochromatin Protein 1 alpha (HP1α), phase separates to form dynamic liquid-like individual heterochromatin modules that become less dynamic, more stable with time [88]. This phase transition is accompanied by changes in the morphology and material state of HP1α, likely enabling stronger DNA compaction. Similarly, polar granule components such as Osk protein, exists as phase separated compartments exhibiting liquid-like and hydrogel-like properties [89]. Together, these examples suggest that RNP liquid-like properties and LLPS are a common phenomenon in Drosophila development. This is an appealing prediction as RNP granules can be regulated by developmental cues and dynamic molecular interactions. Below we ascribe the current knowledge of condensate properties for investigating RNP granules to elucidate their physiological role in development.
Compositional control
Establishing a condensed network of interacting macromolecules is an essential step in granule assembly [90]. According to the ‘scaffold and client’ model, scaffolds are essential proteins that help promote granule assembly, while clients are proteins that transiently interact with scaffolds and regulate condensate properties [91–93]. While this model has primarily been explored in vitro, RNP granules in the developing egg are a powerful in vivo system to test the model and have the advantage of overlapping RBPs associated with different granules. This is exemplified by the piRNA binding protein Aubergine (Aub), which behaves as a scaffold or client depending on the granule it is associated with. While aub mutants result in a partial loss of nuage in the nurse cells, these mutants completely disrupt polar granule formation at the posterior of the oocyte [94,95]. Identifying and testing scaffold and client proteins in vivo with genetics would be challenging since many RNPs are essential for egg chamber development in Drosophila. Therefore, reconstituting maternal granules in vitro through a minimal system of scaffold and client proteins, under physiological conditions, is an important alternative strategy [96]. This approach will provide insights into how RNP interactions regulate granule composition and enable systematic experimentation to identify the underlying sequence and structural determinants of scaffolding proteins.
Material properties
RNP granules can exist in diverse material states, such as liquid, gel or solid, each of which has a distinct functional consequence [22,62,96–99]. Balbiani bodies, for example, exhibit solid-like material state likely facilitating stable storage of organelles and macromolecules during oocyte dormancy [61,96]. Material states of RNPs have been largely explored in vitro, but how these properties impact biological function is less well understood.
Mature Drosophila oocytes can be stored for multiple days without affecting RNA levels [100]. This efficient storage of RNAs is likely through RNP granules adopting a stable material state. P bodies are an example of storage sites for maternal mRNAs during oogenesis. However, P bodies are more complex as grk mRNA associated with P bodies is translated during mid-oogenesis while bcd mRNA is stored in P bodies until egg activation [69]. How P bodies change material states to perform different functions in development is key to understanding their role in translational regulation. One clue comes from experiments on Maternal expression at 31B (Me31B), a conserved RNA helicase found in many storage granules including C. elegans germ granules and mammalian somatic P bodies [62,66,101]. While knockdown of Me31B shows premature translation of stored mRNAs during early oogenesis [66], whether Me31B mutants affect P body material state remains unknown. However, it is exciting to consider that these mutant P bodies could have less stable material properties resulting in premature mRNA release and subsequent translation. Interestingly, P bodies from arrested C. elegans oocytes adopt a semi-liquid, viscoelastic material state which allows both stability and flexibility for RNA regulation [62]. Considering the similarities in P body components between these systems, we speculate that Drosophila P bodies would adopt a similar material state. Comprehensive characterisation of material properties using a combination of genetics and quantitative live imaging of RNP components will provide key insights into how granule physical states are regulated in response to cellular and developmental cues.
Multilayered organisation
Although RNP condensates contain thousands of diverse macromolecules, for a long time they were considered homogenous in organisation. High resolution microscopy revealed that condensates can possess structured internal organisation on multiple scales. The nucleolus, for example, shows multiple liquid phases coexisting in the same granule giving rise to its heterogenous internal organisation [102,103]. While multi-phase organisation has also been reported in stress granules, P bodies, and P granules, the biological significance remains less clear [62,69,72,104].
Nuage during early Drosophila oogenesis exhibits levels of internal structuring with at least two sub-domains, one with Aub and another with Aub and Argonaute-3 [105]. Each internal level regulates a different step in the piRNA processing pathway in the nuage, supporting a model where proteins in different layers of an RNP granule can execute different functions. Drosophila P bodies are another example of granules shown to possess structured internal organisation, in this case a shell and core architecture is proposed. Specific mRNAs and RBPs are shown to be enriched in different layers of the P body, thereby facilitating differential translational regulation [69].
To resolve how different components contribute to the overall material state and function of RNP granules, a combination of super resolution imaging and quantitative single molecule assays should be used [106]. This would reveal finer details and localisation of specific molecules along with the material state of the RNP in question.
The role of RNA
RNA storage and translational control is likely a major function of RNP granules in oogenesis as they form in response to high levels of untranslated mRNAs and disperse at a time when many mRNAs are translated. While proteins are typically considered to be the key scaffolds for granule assembly, more recently RNA has also been shown to both phase separate and drive the assembly of RNP condensates [80,83,107,108].
In the oocyte, certain localised mRNAs appear to coalesce into larger, less dynamic particles at their destination. This apparent change in the physical state is also accompanied by their association with RNP granules, such as P bodies or polar granules. Separate studies have also demonstrated that RNAse treatment results in the breakdown of RNP granules, highlighting the importance of RNA in maintaining the integrity of RNP granules [66,78].
These observations suggest a model whereby ‘sticky’ mRNAs promote granule nucleation by concentrating key scaffold proteins through sequence specific binding and subsequently regulate stability and material property. Testing this model in vivo requires developing techniques to selectively disrupt mRNAs while observing the effect on the RNP granules [80]. Complementarily, in vitro transcribed RNAs could be used in combination with specific scaffold proteins to reconstitute RNP granules. If successful, in vitro studies would be amenable to testing RNA sequences, for example in the untranslated regions, in the formation and regulation of RNP granules.
Concluding remarks
The field of phase separation has made clear progress towards deciphering the physicochemical rules governing biomolecular condensates. Since the majority of the data are derived from in vitro reconstitution and cell culture studies, there is still much to learn about the functional role of phase separation at a biological level. Here, we have highlighted how granule properties impact function and discussed the potential for Drosophila oogenesis to be used for investigating fundamental principles of phase separation in RNP granule assembly, organisation and material properties (Figure 2).
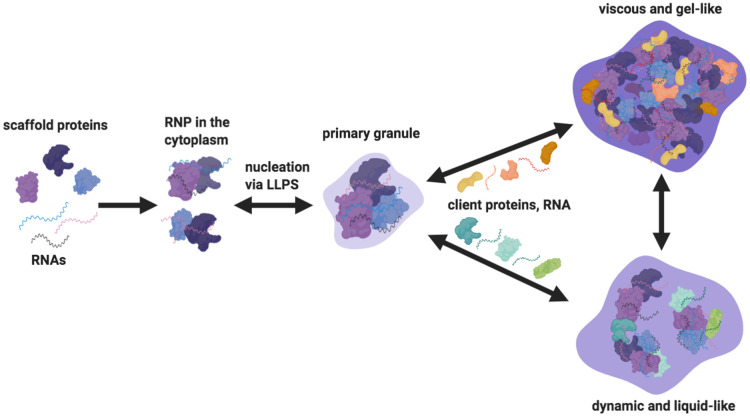
Figure 2. Model for RNP granule assembly and maturation in Drosophila egg chambers.
In the cytoplasm, key scaffolding proteins and mRNAs, through multivalent interactions, come together to form RNP complexes. Multiple RNP complexes nucleate to assemble a primary granule via LLPS. Depending on the partitioning of specific client proteins and RNAs, granule diversity may be achieved. Although client-scaffold interactions may already be present during primary granule assembly, our model proposes that higher partitioning of clients regulate granule material states by modulating the strength of the resulting molecular interactions. While liquid-like and gel-like physical states are more commonly observed in vivo, other material states can exist based on specific developmental and environmental cues. Created in BioRender.
Fundamentally, it is extremely challenging to control all of the variables and factors that regulate condensates in vivo. Therefore, in vitro studies, including RNP granule purification, are important for identifying key features, such as sequence determinants, the role of non-equilibrium factors and multivalent RNP interactions [109,110]. Together, a combination of in vitro studies, modelling and in vivo assays will be required to fully comprehend the physiological functions of biomolecular condensates in cell and developmental biology.
Summary
Liquid–liquid phase separation is an emerging paradigm to understand biomolecular condensates and their roles in regulating cellular processes.
RNP granules are highly conserved biomolecular condensates involved in regulating RNA metabolism.
Diverse maternal mRNAs are regulated by RNP granules during Drosophila oogenesis.
Studying the physicochemical principles of RNP granules in the Drosophila egg chamber could provide insights into the biological role of phase separation.
Acknowledgements
We thank Dr Ethan Greenblatt, Dr Baskar Bakthavachalu, Mrs Swetha Anandhan, and Mr Ben Wood for advice and manuscript feedback.
Abbreviations
- Aub
Aubergine
- HP1α
heterochromatin protein 1 alpha
- LLPS
liquid–liquid phase separation
- Me31B
maternal expression at 31B
- mRNAs
messenger RNAs
- Osk
Oskar
- P bodies
processing bodies
- piRNA
piwi-interacting RNA
- RBP
RNA binding proteins
- RNP
ribonucleoprotein
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
Our research is supported by the Wellcome Trust 200734/Z/16/Z (to T.T.W.), University of Cambridge ISSF grant number 097814 (to T.T.W.), and the INLAKS-Cambridge Trust Studentship (to M.S.).
Open Access
Open access for this article was enabled by the participation of University of Cambridge in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Author Contributions
M.S. produced the original draft of the text, edited the text and figures, and referenced manuscript. T.T.W. edited the text and the produced original drafts of the figures.
References
- 1.St. Johnston D. (2005) Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6, 363–375 10.1038/nrm1643 [DOI] [PubMed] [Google Scholar]
- 2.Becalska A.N. and Gavis E.R. (2009) Lighting up mRNA localization in Drosophila oogenesis. Development 136, 2493–2503 10.1242/dev.032391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin K.C. and Ephrussi A. (2010) mRNA localization : gene expression in the spatial dimension. Cell 136, 719–730 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holt C.E. and Bullock S.L. (2009) Subcellular mRNA localization in animal cells and why it matters. Science 326, 1212–1216 10.1126/science.1176488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medioni C., Mowry K. and Besse F. (2012) Principles and roles of mRNA localization in animal development. Development 139, 3263–3276 10.1242/dev.078626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kindler S., Wang H., Richter D. and Tiedge H. (2005) RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21, 223–245 10.1146/annurev.cellbio.21.122303.120653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato Y. and Nakamura A. (2012) Roles of cytoplasmic RNP granules in intracellular RNA localization and translational control in the Drosophila oocyte. Dev. Growth Differ. 54, 19–31 10.1111/j.1440-169X.2011.01314.x [DOI] [PubMed] [Google Scholar]
- 8.Harvey R.F., Smith T.S., Mulroney T., Queiroz R.M.L., Pizzinga M., Dezi V. et al. (2018) Trans-acting translational regulatory RNA binding proteins. Wiley Interdiscip. Rev. RNA 9, 1–19 10.1002/wrna.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebauer F., Preiss T. and Hentze M.W. (2012) From cis-regulatory elements to complex. Cold Spring Harb. Perspect. Biol. 4, a012245 10.1101/cshperspect.a012245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnstone O. and Lasko P. (2001) Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35, 365–406 10.1146/annurev.genet.35.102401.090756 [DOI] [PubMed] [Google Scholar]
- 11.Mitrea D.M. and Kriwacki R.W. (2016) Phase separation in biology; functional organization of a higher order short linear motifs—The unexplored frontier of the eukaryotic proteome. Cell Commun. Signal. 14, 1–20 10.1186/s12964-015-0125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banani S.F., Lee H.O., Hyman A.A. and Rosen M.K. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilera-Gomez A. and Rabouille C. (2017) Membrane-bound organelles versus membrane-less compartments and their control of anabolic pathways in drosophila. Dev. Biol. 428, 310–317 10.1016/j.ydbio.2017.03.029 [DOI] [PubMed] [Google Scholar]
- 14.Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L. et al. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler J.R., Matheny T., Jain S., Abrisch R. and Parker R. (2016) Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delarue M., Brittingham G.P., Pfeffer S., Surovtsev I V., Pinglay S., Kennedy K.J. et al. (2018) mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349.e20 10.1016/j.cell.2018.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin Y. and Brangwynne C.P. (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- 18.Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A. et al. (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 10.1016/j.molcel.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberti S. (2017) Phase separation in biology. Curr. Biol. 27, R1097–R1102 10.1016/j.cub.2017.08.069 [DOI] [PubMed] [Google Scholar]
- 20.Hyman A.A., Weber C.A. and Jülicher F. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 21.Walter H. and Brooks D.E. (1995) Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 361, 135–139 10.1016/0014-5793(95)00159-7 [DOI] [PubMed] [Google Scholar]
- 22.Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J. et al. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- 23.Patel A., Malinovska L., Saha S., Wang J., Alberti S., Krishnan Y. et al. (2017) Biochemistry: ATP as a biological hydrotrope. Science 356, 753–756 10.1126/science.aaf6846 [DOI] [PubMed] [Google Scholar]
- 24.Al-Husini N., Tomares D.T., Bitar O., Childers W.S. and Schrader J.M. (2018) α-Proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell 71, 1027–1039.e14 10.1016/j.molcel.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguzzi A. and Altmeyer M. (2016) Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 26, 547–558 10.1016/j.tcb.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 26.Abbondanzieri E.A. and Meyer A.S. (2019) More than just a phase: the search for membraneless organelles in the bacterial cytoplasm. Curr. Genet. 65, 691–694 10.1007/s00294-018-00927-x [DOI] [PubMed] [Google Scholar]
- 27.Freeman Rosenzweig E.S., Xu B., Kuhn Cuellar L., Martinez-Sanchez A., Schaffer M., Strauss M. et al. (2017) The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162.e19 10.1016/j.cell.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A. et al. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huizar R.L., Lee C., Boulgakov A.A., Horani A., Tu F., Marcotte E.M. et al. (2018) A liquid-like organelle at the root of motile ciliopathy. eLife 7, 1–24 10.7554/eLife.38497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson B.A., Doolittle L.K., Schneider M.W.G., Jensen L.E., Gamarra N., Henry L. et al. (2019) Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 10.1016/j.cell.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franzmann T.M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A.S., Nüske E. et al. (2018) Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654 10.1126/science.aao5654 [DOI] [PubMed] [Google Scholar]
- 32.Dine E., Gil A.A., Uribe G., Brangwynne C.P. and Jared E. (2019) Protein phase separation provides long-term memory of transient spatial stimuli. Cell Syst. 6, 655–663 10.1016/j.cels.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brangwynne C.P., Mitchison T.J. and Hyman A.A. (2011) Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. U.S.A. 108, 4334–4339 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P. and Weitz D.A. (2015) RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl Acad. Sci. U.S.A. 112, E5237–E5245 10.1073/pnas.1509317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee P.R., Milin A.N., Moosa M.M., Onuchic P.L. and Deniz A.A. (2017) Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew. Chem. Int. Ed. Engl. 56, 11354–9 10.1002/anie.201703191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry J., Brangwynne C.P. and Haataja M. (2018) Physical principles of intracellular organization via active and passive phase transitions. Reports Prog. Phys. 81, 046601 10.1088/1361-6633/aaa61e [DOI] [PubMed] [Google Scholar]
- 37.Choi J.-M., Holehouse A.S. and Pappu R V. (2020) Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133 10.1146/annurev-biophys-121219-081629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosowski K.A., Sai T., Vidal-Henriquez E., Zwicker D., Style R.W. and Dufresne E.R. (2020) Elastic ripening and inhibition of liquid–liquid phase separation. Nat. Phys. 16, 422–425 10.1038/s41567-019-0767-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C.F. and Wurtz J.D. (2018) Novel physics arising from phase transitions in biology. J. Phys. D Appl. Phys. 52, 023001 10.1088/1361-6463/aae510 [DOI] [Google Scholar]
- 40.Söding J., Zwicker D., Sohrabi-Jahromi S., Boehning M. and Kirschbaum J. (2020) Mechanisms for active regulation of biomolecular condensates. Trends Cell Biol. 30, 4–14 10.1016/j.tcb.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 41.Perry S.L. (2019) Phase separation: bridging polymer physics and biology. Curr. Opin. Colloid Interface Sci. 39, 86–97 10.1016/j.cocis.2019.01.007 [DOI] [Google Scholar]
- 42.Brangwynne C.P., Tompa P. and Pappu R V. (2015) Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904 10.1038/nphys3532 [DOI] [Google Scholar]
- 43.Alberti S., Saha S., Woodruff J.B., Franzmann T.M., Wang J. and Hyman A.A. (2018) A user's guide for phase separation assays with purified proteins. J. Mol. Biol. 430, 4806–4820 10.1016/j.jmb.2018.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C. et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y., Protter D.S.W., Rosen M.K. and Parker R. (2015) Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C.C.H., Eckmann C.R., Myong S. et al. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. U.S.A. 112, 7189–7194 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hofweber M. and Dormann D. (2019) Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150 10.1074/jbc.TM118.001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M. et al. (2018) Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 10.1016/j.cell.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 49.Kim T.H., Tsang B., Vernon R.M., Sonenberg N., Kay L.E. and Forman-Kay J.D. (2019) Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829 10.1126/science.aax4240 [DOI] [PubMed] [Google Scholar]
- 50.Voronina E., Seydoux G., Sassone-Corsi P. and Nagamori I. (2011) RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 3, a002774 10.1101/cshperspect.a002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patterson J.R., Wood M.P. and Schisa J.A. (2011) Assembly of RNP granules in stressed and aging oocytes requires nucleoporins and is coordinated with nuclear membrane blebbing. Dev. Biol. 353, 173–185 10.1016/j.ydbio.2011.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eulalio A., Behm-Ansmant I., Schweizer D. and Izaurralde E. (2007) P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27, 3970–3981 10.1128/MCB.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schisa J.A. (2012) New insights into the regulation of RNP granule assembly in oocytes. Int. Rev. Cell Mol. Biol. 295, 233–289 10.1016/B978-0-12-394306-4.00013-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kloc M. and Etkin L.D. (2005) RNA localization mechanisms in oocytes. J. Cell Sci. 118, 269–282 10.1242/jcs.01637 [DOI] [PubMed] [Google Scholar]
- 55.Jansova D., Tetkova A., Koncicka M., Kubelka M. and Susor A. (2018) Localization of RNA and translation in the mammalian oocyte and embryo. PLoS ONE 13, 1–25 10.1371/journal.pone.0192544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du T.G., Schmid M. and Jansen R.P. (2007) Why cells move messages: the biological functions of mRNA localization. Semin. Cell Dev. Biol. 18, 171–177 10.1016/j.semcdb.2007.01.010 [DOI] [PubMed] [Google Scholar]
- 57.Bastock R. and St Johnston D. (2008) Drosophila oogenesis. Curr. Biol. 18, 1082–1087 10.1016/j.cub.2008.09.011 [DOI] [PubMed] [Google Scholar]
- 58.Lasko P. (2012) mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 4, a012294 10.1101/cshperspect.a012294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Derrick C.J. and Weil T.T. (2017) Translational control of gurken mRNA in Drosophila development. Cell Cycle 16, 23–32 10.1080/15384101.2016.1250048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piccioni F., Zappavigna V. and Verrotti A.C. (2005) Translational regulation during oogenesis and early development: the cap-poly(A) tail relationship. Comptes Rendus Biol. 328, 863–881 10.1016/j.crvi.2005.05.006 [DOI] [PubMed] [Google Scholar]
- 61.Roovers E.F., Kaaij L.J.T., Redl S., Bronkhorst A.W., Wiebrands K., de Jesus Domingues A.M. et al. (2018) Tdrd6a regulates the aggregation of buc into functional subcellular compartments that drive germ cell specification. Dev. Cell 46, 285–301.e9 10.1016/j.devcel.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hubstenberger A., Noble S.L., Cameron C. and Evans T.C. (2013) Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev Cell. 27, 161–173 10.1016/j.devcel.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hondele M., Sachdev R., Heinrich S., Wang J., Vallotton P., Fontoura B.M.A. et al. (2019) DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 10.1038/s41586-019-1502-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weil T.T. (2014) mRNA localization in the Drosophila germline. RNA Biol. 11, 1010–1018 10.4161/rna.36097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snee M.J. and Macdonald P.M. (2009) Dynamic organization and plasticity of sponge bodies. Dev. Dyn. 238, 918–930 10.1002/dvdy.21914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakamura A., Amikura R., Hanyu K. and Kobayashi S. (2001) Me31b silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128, 3233–3242 PMID: [DOI] [PubMed] [Google Scholar]
- 67.Liu J.L., Murphy C., Buszczak M., Clatterbuck S., Goodman R. and Gall J.G. (2006) The Drosophila melanogaster Cajal body. J. Cell Biol. 172, 875–884 10.1083/jcb.200511038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cox R.T. and Spradling A.C. (2003) A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130, 1579–1590 10.1242/dev.00365 [DOI] [PubMed] [Google Scholar]
- 69.Weil T.T., Parton R.M., Herpers B., Soetaert J., Veenendaal T., Xanthakis D. et al. (2012) Drosophila patterning is established by differential association of mRNAs with P bodies. Nat. Cell Biol. 14, 1305–1313 10.1038/ncb2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hara M., Lourido S., Petrova B., Lou H.J., Von Stetina J.R., Kashevsky H. et al. (2018) Identification of PNG kinase substrates uncovers interactions with the translational repressor TRAL in the oocyte-to-embryo transition. eLife 7, e33150 10.7554/eLife.33150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.York-Andersen A.H., Parton R.M., Bi C.J., Bromley C.L., Davis I. and Weil T.T. (2015) A single and rapid calcium wave at egg activation in Drosophila. Biol. Open. 4, 553–560 10.1242/bio.201411296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A. and Parker R. (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang F., Wang J., Xu J., Zhang Z., Koppetsch B.S., Schultz N. et al. (2012) UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151, 871–884 10.1016/j.cell.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilsch-Bräuninger M., Schwarz H. and Nüsslein-Volhard C. (1997) A sponge-like structure involved in the association and transport of maternal products during drosophila oogenesis. J. Cell Biol. 139, 817–829 10.1083/jcb.139.3.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R. et al. (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ai K.L. and Kai T. (2007) Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster (Proceedings of the National Academy of Sciences of the United States of America (2007) 104, 16, (6714-6719) DOI: 10.1073/pnas.0701920104). Proc. Natl Acad. Sci. U.S.A. 104, 20143 10.1073/pnas.0710102104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang M., Ly M., Lugowski A., Laver J.D., Lipshitz H.D., Smibert C.A. et al. (2017) ME31B globally represses maternal mRNAs by two distinct mechanisms during the Drosophila maternal-to-zygotic transition. eLife 6, e27891 10.7554/eLife.27891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin M.D., Jiao X., Grima D., Newbury S.F., Kiledjian M. and Bin C.T. (2008) Drosophila processing bodies in oogenesis. Dev. Biol. 322, 276–288 10.1016/j.ydbio.2008.07.033 [DOI] [PubMed] [Google Scholar]
- 79.Trcek T. and Lehmann R. (2019) Germ granules in Drosophila. Traffic 20, 650–660 10.1111/tra.12674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garcia-Jove Navarro M., Kashida S., Chouaib R., Souquere S., Pierron G., Weil D. et al. (2019) RNA is a critical element for the sizing and the composition of phase-separated RNA–protein condensates. Nat. Commun. 10, 3230 10.1038/s41467-019-11241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.George-Hyslop P S., Lin J.Q., Miyashita A., Phillips E.C., Qamar S., Randle S.J. et al. (2018) The physiological and pathological biophysics of phase separation and gelation of RNA binding proteins in amyotrophic lateral sclerosis and fronto-temporal lobar degeneration. Brain Res. 1693, 11–23 10.1016/j.brainres.2018.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nielsen F.C., Hansen H.T. and Christiansen J. (2016) RNA assemblages orchestrate complex cellular processes. BioEssays 38, 674–681 10.1002/bies.201500175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Langdon E.M. and Gladfelter A.S. (2018) A new lens for RNA localization: liquid-liquid phase separation. Annu. Rev. Microbiol. 72, 255–271 10.1146/annurev-micro-090817-062814 [DOI] [PubMed] [Google Scholar]
- 84.Zhu L. and Brangwynne C.P. (2015) Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 34, 23–30 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singer A.B. and Gall J.G. (2011) An inducible nuclear body in the Drosophila germinal vesicle. Nucleus 2, 403–409 10.4161/nucl.2.5.17250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weil T.T., Parton R., Davis I. and Gavis E.R. (2008) Changes in bicoid mRNA anchoring highlight conserved mechanisms during the oocyte-to-embryo transition. Curr. Biol. 18, 1055–1061 10.1016/j.cub.2008.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Little S.C., Sinsimer K.S., Lee J.J., Wieschaus E.F. and Gavis E.R. (2015) Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 17, 558–568 10.1038/ncb3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Strom A.R., Emelyanov A V., Mir M., Fyodorov D V., Darzacq X. and Karpen G.H. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kistler K.E., Trcek T., Hurd T.R., Chen R., Liang F.X., Sall J. et al. (2018) Phase transitioned nuclear oskar promotes cell division of Drosophila primordial germ cells. eLife 7, e37949 10.7554/eLife.37949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanders D.W., Kedersha N., Lee D.S.W., Strom A.R., Drake V., Riback J.A. et al. (2020) Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28 10.1016/j.cell.2020.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jo Y. and Jung Y. (2020) Interplay between intrinsically disordered proteins inside membraneless protein liquid droplets. Chem. Sci. 11, 1269–1275 10.1039/C9SC03191J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ditlev J.A., Case L.B. and Rosen M.K. (2018) Who's in and who's out—compositional control of biomolecular condensates. J. Mol. Biol. 430, 4666–4684 10.1016/j.jmb.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R. et al. (2016) Compositional control of phase-separated cellular bodies. Cell 166, 651–663 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harris A.N. and Macdonald P.M. (2001) Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to elF2C. Development 128, 2823–2832 PMID: [DOI] [PubMed] [Google Scholar]
- 95.Snee M.J. and Macdonald P.M. (2004) Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J. Cell Sci. 117, 2109–2120 10.1242/jcs.01059 [DOI] [PubMed] [Google Scholar]
- 96.Boke E., Ruer M., Wühr M., Coughlin M., Lemaitre R., Gygi S.P. et al. (2016) Amyloid-like self-assembly of a cellular compartment. Cell 166, 637–650 10.1016/j.cell.2016.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y. et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 98.Putnam A., Cassani M., Smith J. and Seydoux G. (2019) A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 26, 220–226 10.1038/s41594-019-0193-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hubstenberger A., Cameron C., Noble S.L., Keenan S. and Evans T.C. (2015) Modifiers of solid RNP granules control normal RNP dynamics and mRNA activity in early development. J. Cell Biol. 211, 703–716 10.1083/jcb.201504044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Greenblatt E.J., Obniski R., Mical C. and Spradling A.C. (2019) Prolonged ovarian storage of mature drosophila oocytes dramatically increases meiotic spindle instability. eLife 8, e49455 10.7554/eLife.49455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ayache J., Bénard M., Ernoult-Lange M., Minshall N., Standart N., Kress M. et al. (2015) P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol. Biol. Cell 26, 2579–2595 10.1091/mbc.E15-03-0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boisvert F.M., Van Koningsbruggen S., Navascués J. and Lamond A.I. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- 103.Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M. et al. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sheth U., Pitt J., Dennis S. and Priess J.R. (2010) Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137, 1305–1314 10.1242/dev.044255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Webster A., Li S., Hur J.K., Wachsmuth M., Bois J.S., Perkins E.M. et al. (2015) Aub and Ago3 are recruited to nuage through two mechanisms to form a ping-pong complex assembled by krimper. Mol. Cell 59, 564–575 10.1016/j.molcel.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Trcek T., Lionnet T., Shroff H. and Lehmann R. (2017) mRNA quantification using single-molecule FISH in Drosophila embryos. Nat. Protoc. 12, 1326–1347 10.1038/nprot.2017.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jain A. and Vale R.D. (2017) RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Van Treeck B., Protter D.S.W., Matheny T., Khong A., Link C.D. and Parker R. (2018) RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl Acad. Sci. U.S.A. 115, 2734–2739 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Khong A., Matheny T., Jain S., Mitchell S.F., Wheeler J.R. and Parker R. (2017) The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820.e5 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Courel M., Clément Y., Bossevain C., Foretek D., Cruchez O.V., Yi Z. et al. (2019) Gc content shapes mRNA storage and decay in human cells. eLife 8, e49708 10.7554/eLife.49708 [DOI] [PMC free article] [PubMed] [Google Scholar]