Abstract
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to an unprecedented effort toward the development of an effective and safe vaccine. Aided by extensive research efforts into characterizing and developing countermeasures towards prior coronavirus epidemics, as well as recent developments of diverse vaccine platform technologies, hundreds of vaccine candidates using dozens of delivery vehicles and routes have been proposed and evaluated preclinically. A high demand coupled with massive effort from researchers has led to the advancement of at least 31 candidate vaccines in clinical trials, many using platforms that have never before been approved for use in humans. This review will address the approach and requirements for a successful vaccine against SARS-CoV-2, the background of the myriad of vaccine platforms currently in clinical trials for COVID-19 prevention, and a summary of the present results of those trials. It concludes with a perspective on formulation problems which remain to be addressed in COVID-19 vaccine development and antigens or adjuvants which may be worth further investigation.
Keywords: Adjuvant, mRNA vaccine, Lipid Nanoparticles, Protein Nanoparticles, Virus-like Particles, DNA Vaccine, Protein and DNA Vaccine, Polyplexes
Graphical abstract
1. Introduction
Coronaviruses are a family of single-stranded RNA viruses that infect many animal species including bats and humans [1]. Before 2003, only twelve animal or human coronaviruses were identified [2]. In the last eighteen years, three deadly and novel strains have spilled over into humans [3]. In 2003, severe acute respiratory syndrome coronavirus (SARS-CoV) had an official 8096 cases and 774 deaths, with individuals with pre-existing conditions suffering from the highest mortality. In 2012, Middle East respiratory syndrome coronavirus (MERS-CoV) was first reported in Saudi Arabia and since then has infected 2,442 persons and killed 842 [5,6]. Finally, in December 2019, in Wuhan, China, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged into the human population, causing an outbreak of coronavirus infectious disease 2019 (COVID-19), which has since exploded into a global pandemic [7].
SARS-CoV was the first major domino in a trend that lead to the SARS-CoV-2 crisis, with all outbreaks likely linked to a similar animal host. It is not exactly known in which species the SARS-CoV originated, but evidence indicates that it was zoonotic transfer from bats. Similarly, MERS-CoV is known to be closely related to other bat viruses, and it is hypothesized that bats are a reservoir although this has not been confirmed. SARS-CoV-2 is 79% genetically similar to SARS-CoV but is 98% similar to the bat coronavirus RaTG13 as well as a virus found in pangolins [8]. Based on this homology, it is hypothesized, that bats or pangolins are the natural reservoir for SARS-CoV-2.
The interplay between bats and humans has been heightened, in part, by the drastic economic growth in China which has resulted in an increased consumption of animal protein. Protein consumption in southern China includes animals such as civets and bats. These animals are often part of ‘wet markets’ where the animals are sold alive with the thought that the meat is fresher when purchased in this manner. These wet markets often consist of a large variety and number of animals in overcrowded cages, and the animals are rudimentarily processed in these same markets, making this environment a perfect storm for potential animal spillover into the human population.
As of the writing of this review, the World Health Organization reports that there have been approximately 67 million cases of COVID-19 and over 1.54 million deaths worldwide due to the disease [9]. The symptoms of COVID-19 can include fever, dry cough, general weakness, dizziness, headache, vomiting, and diarrhea [10]. Cases can range from rather mild to significant hypoxia with acute respiratory distress syndrome, and severe cases can result in a diverse and incompletely characterized range of problems from immune disregulation to prolonged coagulopathy [[11], [12], [13]]. Reported mortality rates are higher in the elderly (14.8% for over 80 years old) and patients with pre-existing conditions including cardiovascular disease (10.5%), diabetes (7.3%), chronic respiratory disease (6.3%), hypertension (6.0%), and cancer (5.6%), while in the general population mortality is less than 1% [14]. This is similar to what was observed with the 2003 SARS-CoV infection, where individuals with pre-existing conditions had a higher mortality than those without [15].
1.1. A path to a vaccine for SARS-CoV-2
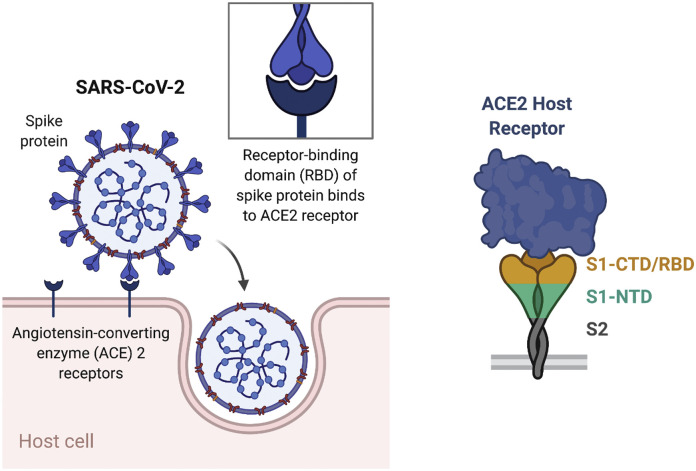
Infection of humans by SARS-CoV-2 usually occurs through inhalation where the virus initially infects the epithelial cells in the nasal cavity. The virus attaches to the cell through the membrane-bound receptor angiotensin-converting enzyme 2 (ACE2) (Fig. 1 ) [16]. This receptor converts angiotensin II to generate angiotensin1-7 which has many anti-inflammatory effects including decreasing hypertension, cardiac hypertrophy, heart failure and other cardiac diseases [17]. ACE2 is widely expressed throughout the body, including in the small intestine, heart, kidneys, and, surprisingly, to a lesser extent in the lung, where expression is highest on type II alveolar cells and macrophages.
Fig. 1.
Schematic of binding regions of SARS-CoV-2 to angiotensin-converting enzyme 2 (ACE2) of host cell. The S1 subunit of the S protein includes the C-terminal domain (CTD) and the N-terminal domain (NTD). The receptor-binding domain (RBD) is located in the S1 CTD.
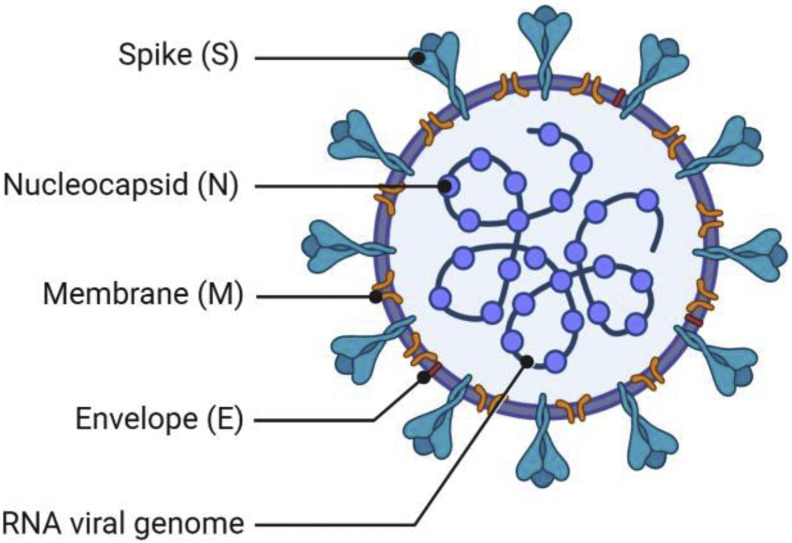
Attachment of the virus to the mammalian ACE2 receptor is mediated by the virus’ spike protein (S-protein), similar to how SARS-CoV infects cells [16,18]. The S-protein from SARS-CoV and SARS-CoV-2 share roughly 80% homology and have similar binding affinity to ACE2. For SARS-CoV entry, after binding to the ACE2 protein, the S-protein is primed by cellular surface proteases, such as TMPRSS2, inducing fusion of the viral and cellular membranes. The exact mechanism is not known as to how SARS-CoV-2 gains entry after ACE2 binding, and whether there are other co-receptors that the virus uses for cell entry.
The critical role of S-protein in cell entry makes it an ideal target for SARS-CoV vaccines. Indeed, prior research with SARS-CoV has shown the utility of the S-protein as a target for vaccine development. When inactivated SARS-CoV was used to vaccinate mice and rabbits it generated a high antibody titer against the S-protein [19]. Antibodies binding residues 318-510 in the S1 region, which includes the receptor-binding domain (RBD), accounted for a large fraction of the neutralizing antibodies. With this in mind, many vaccines for SARS-CoV were developed using the S-protein as a target antigen [18,[20], [21], [22]].
With demonstrated preclinical success and despite the fact that there are currently no approved SARS-CoV vaccines [23], the homologous S-protein is an obvious target in SARS-CoV-2. Similar to SARS-CoV, the S-protein in SARS-CoV-2 is composed of two subunits [24]. The S1 subunit is composed of the C terminal domain (CTD) and N terminal domain (NTD). Located in the CTD, the RBD consists of residues 319–541 and has a similar sequence to the RBD in SARS-CoV [25,26]. As in SARS-CoV, the SARS-CoV-2 RBD also binds ACE2. Prior research has shown that antibodies generated against the RBD correlate well with viral neutralization in humans [27]. While the RBD is thus the primary target for a neutralizing antibody response, neutralizing antibodies targeting the NTD have also been reported in humans following SARS-CoV-2 infection [28]. The S2 subunit is responsible for fusion of the viral envelope to the host cellular membrane. Therefore, based on its prominent display on the viral particle surface, essential role in viral host cell binding and entry, and preclinical results from SARS-CoV vaccine development, the S-protein is a promising antigen for SARS-CoV-2 vaccination.
In addition to antigen selection, vaccine safety and effectiveness is a concern. With SARS-CoV and SARS-CoV-2, immunopathology results from an overwhelming immune response which can lead to death. Immunopathology is a spectrum of conditions caused by the host’s immune system in response to infection. SARS-CoV pathogenesis can include hyper-immune responses involving a cytokine storm that leads to immune cell infiltration of the lungs and alveolar damage that may culminate in pulmonary failure due to acute respiratory distress syndrome [29]. In some cases, anti-SARS-CoV antibodies may potentiate these outcomes, as evidenced by pre-clinical macaque studies, where virus-specific IgG prevented the wound healing response and induced monocyte chemoattractant protein-1 (MCP-1) and interleukin 8 (IL-8) which resulted in monocyte and macrophage recruitment [30]. Through normal humoral responses, an antibody’s binding region (Fab) binds to the virus. The fragment crystallizable region (Fc region) of antibodies then binds to Fc receptors (such as FcᵞR) of immune cells like macrophages, dendritic cells (DCs), neutrophils, natural killer (NK) cells and B cells. Studies have indicated that when FcᵞR is blocked, cytokine production is reduced in humans, underscoring the role of this receptor [30]. This initiates cytokine signaling and should lead to phagocytosis and clearance of the pathogen; however, in the case of SARS-CoV, there is some evidence suggesting that this can lead to antibody-dependent enhancement (ADE) [31]. It should be noted that ADE is not universally accepted as a mechanism for SARS-CoV-2 infection, as it has not been conclusively demonstrated in humans [32]. In general, the exact method of ADE has not been fully elucidated, but it is thought to occur once the virus laden antibody binds the Fc receptor of immune cells, facilitating infection of the immune cell. Once infected, the virus continues to subvert the immune response by suppressing antiviral interferon signaling. Furthermore, this adverse response can lead to a highly inflammatory programmed cell death called pyroptosis, further exasperating the aberrant inflammatory response. Taken together the binding of non-neutralizing antibodies could lead to increased viral association with immune cell Fc receptors, an adverse inflammatory response, and pulmonary failure, not only highlighting the seriousness of this infection, but the need to have precise and effective vaccines to combat it.
An important consideration for the design of SARS-CoV-2 vaccines is the promotion of a type-1 helper T cell (Th1) response, to avoid immunopathology. Several groups demonstrated that inactivated SARS-CoV vaccines could elicit Th2 response-associated immune pathology in the lungs following either viral challenge or challenge in immune-senescent mice [33,34]. Similar results were observed in mice vaccinated with the SARS-CoV nucleocapsid (N) protein, which failed to protect from SARS-CoV replication and induced eosinophilic infiltrates into the lungs [35]. Th1/Th2 responses are a model that immunologists use to characterize helper T cell responses. A Th1 response is characterized by the production of cytokines such as interferon gamma (IFN-γ) and IL-12, whereas a Th2 response usually involves the production of IL-4 and IL-6. Many pathogens promote a Th2 response to help usurp clearance by the immune system, and indeed, serum samples from patients who are infected with SARS-CoV-2 have higher levels of Th2-associated cytokines compared to non-infected patients, suggesting that SARS-CoV-2 induces a Th2 response [36], although a Th2 response has also been shown to be largely abrogated in lymph nodes of patients who have died from COVID-19 [37]. Therefore, in developing a vaccine against the new SARS-CoV-2 virus, it may be important to skew the immune response towards a Th1 immune response, but evidence for this strategy to avoid ADE in humans has yet to emerge.
In this review, we highlight various vaccine technologies that have been announced to be in clinical trials, are in the process of recruiting volunteers for a clinical trial or have unique formulations in collaboration with an establish company and are working towards clinical trials. The companies and technologies are not exhaustive, but they represent the COVID-19 vaccine candidates that have had peer-reviewed publications from the company or have a method that has been referenced in literature. Our general approach will be to first describe the concept of each vaccine platform, and then seminal reports on that platform. In the case that the platform has been previously evaluated for immunization against another infectious disease, we will review those reports as they are pertinent. Finally, we will describe the platform as it is currently being applied in clinical trials for prevention of COVID-19, and report on any published results of these trials.
In reviewing these results it is important to understand the purpose and general structure of each phase of the clinical trial process [38,39]. Results reported in this review include preclinical, Phase 1, and Phase 2 results. Preclinical results are those collected from animal models. Phase 1 trials are relatively small trials performed with ~20-100 healthy volunteers, which are carried out by administering a range of doses to the patients and monitoring them to establish safety. While potential correlates of protection such as neutralizing antibodies and T cell responses are often collected and reported in the Phase 1 results reported below, these readouts are not the primary objective of a Phase 1 trial, which is simply to establish safety. Phase 2 studies are then performed to establish a dosage that is most likely to achieve the desired endpoint of the vaccine (e.g. protective efficacy). This is generally done by examining the effect of the dose on different biomarker correlates of protection, like the neutralizing antibody and T cell responses. Finally, Phase 3 trials involve evaluation of the vaccine to achieve a certain clinical endpoint. In the case of vaccines for COVID-19, this endpoint is protection against COVID-19 from SARS-CoV-2 exposure. As the state of these trials is rapidly evolving, the reader is encouraged to refer to a regularly updated list of vaccine candidates in preclinical and clinical stages of evaluation, maintained by the WHO [40].
1.2. Inactivated viral vaccines
Many FDA approved vaccines are inactivated viruses, including those to prevent influenza, polio, hepatitis A, and Japanese Encephalitis virus [41]. Using influenza as an example, the most frequently given influenza vaccines use virus grown in fertilized chicken eggs. In a process that takes approximately one week, eggs are first inoculated with the chosen strain of virus, which replicates in the egg for 72 hours, then the amniotic fluid is isolated and treated with formaldehyde, β-propiolactone, or similar acting chemical to inactivate the virus [42,43]. For most current influenza vaccines, the inactivated virus is subsequently split into its constituent antigens using a detergent such as Triton X-100 or deoxycholate.
During amplification, viral subpopulations with sequence differences better suited to replication in host cells may emerge. This phenomenon has been shown to occur in eggs and lead to diminished or altered antigenicity, which is reduced through the use of mammalian cell cultures such as Madin-Darby Canine Kidney (MDCK) or Vero (from African Green Monkey) cells as viral amplification host cells [44].The inactivated virus acts as an antigen to target the immune response, and residual pathogen associated molecular patterns (PAMPs) from the virus serve as adjuvants that promote potent immune responses. Indeed, prior research has shown that inactivated viruses can activate DCs and can enhance immune responses [45].
As a common FDA approved vaccine formulation method, it is not surprising that clinical trials have begun with an inactivated SARS-CoV-2 vaccine. The Beijing Institute of Biological Products Company reported preclinical results with an inactivated version of SARS-CoV-2 [46]. To generate the vaccine, Wang et al. isolated three SARS-CoV-2 strains from hospitalized patients. These three strains were similar in sequence to other virus strains isolated from humans. Of the three isolated strains, the HBO2 strain showed optimal replication and had the highest virus yield when cultured in vitro in Vero cells [47]. Vero cells have been certified by the WHO for use in vaccine production and have previously been used to generate polio and rabies virus for vaccines. In comparison to virus from other patients, the HBO2 strain had 100% homology in the S-protein. HBO2 was passaged ten times in Vero cells to induce adaptation to the host Vero cells. The tenth passage was deep sequenced and showed a 99.95% homology to the 7th passage, with a 100% homology to the amino acid sequence of the S-protein of the 7th passage, indicating that the virus had adapted and reached a stable genetic sequence, rendering it suitable for further scale up. This strain was mass produced in Vero cells using a novel basket reactor and inactivated by the addition of β-propionolactone. The resulting inactivated virus was then mixed with aluminum hydroxide (alum) adjuvant in bulk prior to administration.
To evaluate the alum-adjuvanted inactivated virus as a vaccine candidate, Wang et al. immunized mice, rabbits, rats, guinea pigs, cynomolgus monkeys, and rhesus macaques, resulting in 100% of the animals having detectable antibodies (seroconversion) 21 days after immunization. Additionally, rhesus macaques immunized with the alum-adjuvanted inactivated virus showed no viral load in the lungs. In other organs, viral load in the vaccinated group was much lower compared to the unvaccinated controls. Additionally, there was no detectable ADE after infection.
The safety and immunogenicity of this formulation in humans were subsequently evaluated in Phase 1 and 2 clinical trials [48]. Interestingly, neutralizing antibody titer as measured by the Plaque Reduction Neutralization Tests (PRNT) assay, as well as SARS-CoV-2-specific IgG titers, did not show a dose-dependence between the low, medium, and high dosages administered during Phase 1. Adverse events also did not show a dose dependence, and included fatigue, fever, nausea, and pain and swelling at the injection site. Higher neutralizing and antigen-specific IgG titers were elicited from groups receiving a boost vaccination 21 or 28 days after the prime injection as compared to those boosted 14 days after prime injection. While this study did not include comparisons to convalescent serum, the results indicated that a significant neutralizing and SARS-CoV-2 specific antibody response could be raised in humans with this formulation. Phase 3 clinical evaluation of this candidate is now underway (ChiCTR2000034780).
Another Chinese company, Sinovac, has published on their inactivated SARS-CoV-2 virus with similar results, eliciting 92.4% seroconversion using a day 0 and 14 prime-boost schedule and 97.4% seroconversion by a Day 0 and 28 prime-boost schedule [49,50]. Despite the evidence for greater seroconversion with a boost at day 28 rather than day 14, they have since begun evaluating a day 14 boost schedule in Phase 3 clinical trials in Indonesia (INA-WXFM0YX), Turkey (NCT045823440), and Brazil [51] (NCT04456595).
2. Subunit formulations
Subunit formulations are a common vaccine type where one or more elements of the pathogen are used as an antigen. These elements can be proteins, peptides, or sugars of the pathogen. Additionally, nucleic acids such as mRNA or DNA can be administered to use the body’s protein expression machinery to express subunit elements of the pathogen. Because subunit vaccines are only elements of the pathogen, they are considered the safest type of vaccine. One issue, however, is that they are often poorly immunogenic and often require an adjuvant to be co-administered. Adjuvants serve as immunostimulants that can add additional safety concerns. Individually FDA approved adjuvants include aluminum salts (alum), the squalene emulsion adjuvant MF59, and non-methylated DNA (CpG). Combined adjuvants include adjuvant system 4 (AS04) which is comprised of monophosphoryl lipid A (MPL) and alum [63]. AS01, a combination of MPL and the saponin QS-21, is also FDA approved. Table 1 presents a summary of vaccine components, most with potential adjuvant properties, which are included in formulations currently undergoing clinical evaluation as COVID-19 vaccine candidates.
Table 1.
Summary of non-antigen components for SARS-CoV-2 vaccines
| Component | Clinical Vaccine Candidates Containing Adjuvant/Component (Antigen Type) | Description | Effect/Skew | Mechanism |
|---|---|---|---|---|
| Advax-SM | Vaxine Pty/Medytox (Recombinant Protein) | Delta-inulin (water-insoluble polysaccharide) microparticles mixed with CpG 1018 | Adjuvant, Th1 skew (No skew without CpG) [67] | Unknown, antigen-presenting cell-dependent [68] |
| Alum | Sinovac (Inactivated Virus), Sinopharm (Inactivated Virus), Bharat Biotech (Inactivated Virus), Clover (With CpG 1018, Recombinant Protein), FBRI SRC VB VECTOR (Peptide Subunit), West China Hospital/Sichuan University (Recombinant Protein) | Aluminum salts (aluminum hydroxide or aluminum phosphate) | Adjuvant, Th2 Skew | Multifaceted [69] [70] |
| AS03 | Clover (Recombinant Protein), Medicago (VLP), Sanofi/GSK (Recombinant Protein) | Squalene and DL-α-tocopherol oil-in-water emulsion stabilized by polysorbate 80 | Adjuvant, Th2 skew | Unknown, potentially innate immune recruitment and activation [71] |
| CpG 1018 | Vaxine Pty/Medytox (Included in Advax-SM, Recombinant Protein), Medicago (VLP), Clover (With Alum, Recombinant Protein), Medigen/NIAID/Dynavax (Recombinant Protein) | Unmethylated oligodeoxynucleotide (ODN) | Adjuvant, Th1 skew | TLR9 stimulation [72] |
| Ionizable Lipid (various proprietary versions) | Moderna/NIAID (mRNA), Pfizer (mRNA and replicon RNA), Arcturus (replicon RNA), PLA Academy of Military Sciences/Walvax Biotech | Lipid molecules containing amino groups which become cationic at acidic pH. | Complexes anionic macromolecules (e.g. RNA) and promotes cytosolic delivery. Adjuvant, Th2 skew in absence of other adjuvants [73] | Unknown, potentially TLR2/TLR4 stimulation [74] |
| Matrix M | Novavax (Recombinant Protein) | Lipid nanoparticles containing cholesterol and immunostimulatory Quillaja triterpenoid saponins Matrix-A and Matrix-C in an 85:15 ratio [75] | Adjuvant, Balanced Th1/Th2 skew | Unknown, potentially innate immune recruitment and activation [76] [77] |
| MF59 | Anhui Longcom (Recombinant Protein), Queensland/Seqirus/CSL (Recombinant Protein) | Squalene oil-in-water emulsion stabilized by polysorbate 80 and sorbitan trioleate | Adjuvant, Th2 skew | Unknown, potentially innate immune recruitment and activation [78] [79] |
| Polysorbate 80 | Novavax (Recombinant protein) | Nonionic surfactant, a.k.a. Tween 80 | Inhibits aggregation of emulsions and hydrophobic proteins | Stabilizes interfaces in emulsions, prevents protein adsorption to potentially denaturing interfaces, multimerizes transmembrane proteins [80,81]. |
| RNA | Moderna/NIAID (mRNA), Pfizer/BioNTech (mRNA and replicon RNA), Curevac (mRNA), Arcturus (replicon RNA) | genetic material which encodes antigenic constructs and stimulates immune responses | Th1 skew | TLRs 3, 7, 8, 9, and 13 stimulation [82] |
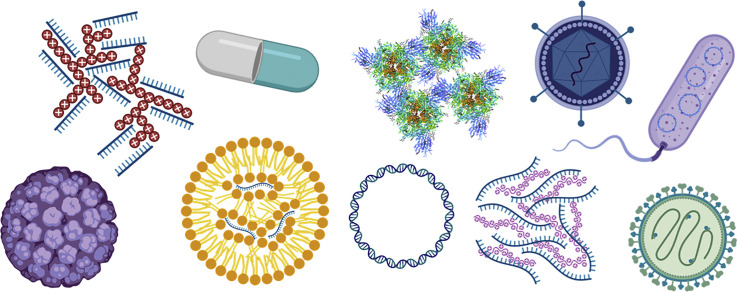
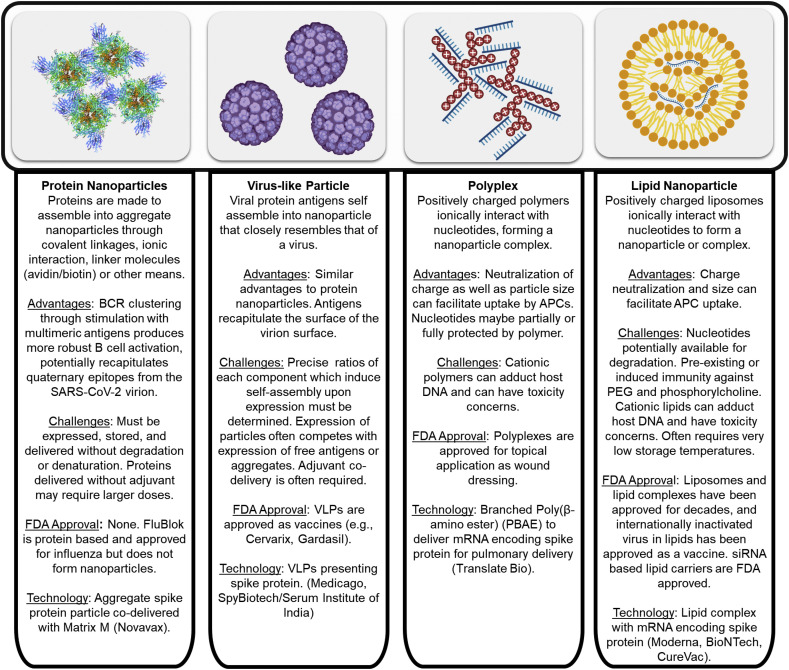
With respect to SARS-CoV-2 nanoparticulate subunit vaccine formulations, there are protein, VLP (virus-like particles) and nucleic acid-based vaccines (Fig. 2 ). (See Fig. 3.)
Fig. 2.
Overview of nanoparticulate subunit technologies applied to SARS-CoV-2 vaccines [[52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62]].
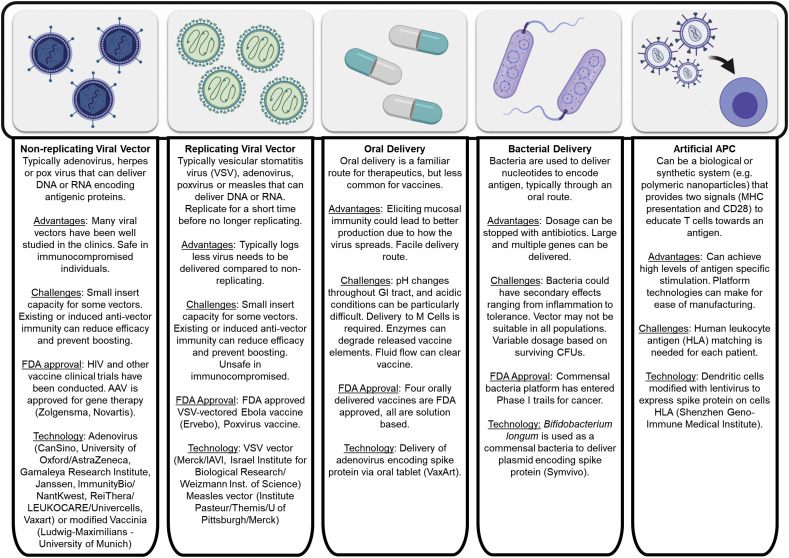
Fig. 3.
Overview of vector-based SARS-CoV-2 vaccines. [62,[158], [159], [160], [161], [162], [163]], NCT04299724, NCT04334980
2.1. Recombinant protein antigens
Clover Biopharmaceuticals Inc. has entered Phase 1 trials in Australia with a stabilized spike protein trimer (NCT04405908). Clover uses Trimer-Tag technology to express stabilized S-protein at high levels from CHO cells. The tag portion consists of human C-propeptide of α1(I) collagen that has been shown to stabilize other proteins such as TNF-related apoptosis-inducing ligand (TRAIL) through forming disulfide bond-linked homotrimers of the protein [64]. This technology is a subunit based vaccine adjuvanted with Dynavax Technologies’ CpG 1018 adjuvant with alum or with AS03 [59,61].
A similar but distinct approach to multimerization and stabilization of the spike protein trimer is being employed for the vaccine candidate from the University of Queensland in partnership with CSL and Seqirus. The antigen in this vaccine is the SARS-CoV-2 spike stabilized in its prefusion form by a so-called ‘molecular clamp’. This clamp is composed of a pair of complementary heptad repeat regions derived from the HIV-1 GP160 protein [65,66]. Registration for a Phase 1 clinical trial of this antigen in combination with the MF59 adjuvant has begun in Australia (ACTRN12620000674932).
Another form of multimerized recombinant protein antigen is the RBD-dimer [83]. This antigen was initially developed upon observation that a recombinant RBD of MERS existed in an equilibrium state between monomers and dimers in solution, and that the dimer exhibited significantly greater immunogenicity compared to the monomer upon immunization in BALB/c mice. To create a stable form of this dimer, cysteines which usually formed a disulfide bridge at the C’ terminus of each of the RBD constructs were truncated and then connected in tandem as a single-chain construct. An analogous strategy was subsequently used to generate RBD-dimers of SARS-CoV and SARS-CoV-2. The resulting single chain constructs exhibited higher RBD-specific IgG and pseudovirus neutralization titers relative to their monomers after immunization of BALB/c mice in combination with AddaVax (MF59-like) adjuvant. However, in the case of the SARS-CoV-2 RBD-dimer, splenocytes isolated 45 days after the last of three vaccinations did not demonstrate significant secretion of IFN-γ, IL-2, TNF-α, or IL-4 after stimulation with an RBD-derived peptide pool, indicating a poor induction of cellular immunity. The authors also demonstrated the potential for high-yield expression of their RBD-dimers in clinical-grade CHO cell lines, paving the way for producing material for clinical application. This dimer concept is now being evaluated by Anhui Zhifei Longcom Biopharmaceutical and the Institute of Microbiology at the Chinese Academy of Sciences in a Phase 1 clinical trial (NCT04445194), with recruitment already begun for Phase 3 evaluation (NCT04466085).
Vaxine Pty Ltd. have partnered with Medytox in the development of an adjuvanted subunit protein vaccine. To our knowledge, the only publicly available information regarding the antigen being employed is that it is a recombinant spike protein [40], however, the adjuvant being employed is Advax-SM. Advax is composed of the immunostimulatory polysaccharide delta-inulin, which has been demonstrated to amplify the immune response without affecting Th1/Th2 skew when co-administered with diverse antigens such as those from influenza and hepatitis B [84]. In addition, intramuscular prime-boost immunization with SARS-CoV spike protein and Advax or Advax and CpG induced spike-protein specific neutralizing antibody titers in BALB/c mice [85]. Using SARS-CoV spike protein to stimulate splenocytes harvested 1 year after vaccination, mice immunized with Advax alone demonstrated significantly greater secretion of IFN-γ, IL-2, and IL-4 compared to those immunized with spike protein alone, or those immunized with spike protein, Advax, and CpG. In the same experiment, the Advax and CpG adjuvanted group demonstrated significantly greater secretion of IL-17 compared to all other groups. In addition, both Advax-containing groups were protected from lethal challenge with mouse-adapted SARS-CoV and exhibited reduced eosinophilic immunopathology in the lung compared to mice immunized with spike protein and alum. This raises the possibility that a Th1 and/or Th17-skewed T cell responses may help to reduce immunopathology upon SARS-CoV infection, which merits further investigation with SARS-CoV-2. Despite the prime-boost schedule employed in this preclinical study, the Phase 1 clinical trial will use a prime-only intramuscular vaccination (NCT04453852).
Medigen Vaccine Biologics Corporation, the National Institute of Allergy and Infectious Disease (NIAID), and Dynavax are collaborating to advance a vaccine candidate employing a spike protein with a double proline (2P) substitution known to stabilize coronavirus spike proteins in their prefusion conformations [86,87]. This antigen was evaluated preclinically in mice in combination with alum and CpG 1018, and shown to generate anti-spike and pseudovirus-neutralizing titers from a prime-boost regimen [88]. The T cell response to this antigen with alum, CpG, or the combination also indicated that the addition of CpG to alum reduced the Th2 skew compared to alum alone.
2.2. Nanoparticulate protein formulations
While soluble antigens represent a relatively simple form of antigen which is employed in many currently-approved vaccines, arranging protein antigens into a particulate form offers the potential advantages of greater B cell activation by increased B cell receptor crosslinking, increased cross-presentation of particulate antigens, and a greater likelihood of receiving T cell help with the codelivery of multiple proteins with potential T cell epitopes.
2.2.1. Protein nanoparticles
Novavax is developing a vaccine for COVID-19 using protein nanoparticles that is based on technology from a 1978 publication focused on protein micelles [89]. Simons et al. developed protein micelles from amphiphilic membrane proteins. Antigenic proteins were expressed by infection of corresponding host cells with the target antigen-expressing viruses and isolated with detergent, resulting in soluble aggregates which assembled into the protein nanoparticles upon detergent removal. Recently, Novavax has developed both MERS-CoV and SARS-CoV S-protein nanoparticles. Similar to the Simons et al. work, insect cells are infected with a baculovirus to express antigenic proteins in the membrane of the cell. The proteins are isolated by standard purification techniques. During the purification process, the detergents are removed, resulting in the S trimers forming micellular protein nanoparticles [90]. In this work, they do not do a direct comparison to soluble protein antigens to underscore the advantage of the protein nanoparticles. They do show a statistical increase in neutralizing antibodies when adding the adjuvant alum, and even a greater increase with the adjuvant Matrix M1. Matrix M1 is a proprietary adjuvant owned by Novavax [91] wherein Quillaja saponins are formulated into nanoparticles along with cholesterol and phospholipids. It has previously been used in clinical trials, and has been deemed to have an acceptable safety profile [92].
Novavax recently reported the development and evaluation of this protein nanoparticle technology for a COVID-19 vaccine [93]. They again used baculovirus for expression of their target proteins in Spodoptera frugiperda (Sf9) insect cells. The spike protein expressed in this case contained the same 2P mutation mentioned for the Medigen recombinant protein vaccine above, while also containing a mutation of the furin cleavage site 682-RRAR-685 to 682-QQAQ-685. This furin cleavage site mutation was introduced to increase stability against proteases [94]. The resulting protein was purified from the Sf9 cells and resuspended in a phosphate buffer in the presence of the nonionic surfactant polysorbate 80, with the reported mass ratio of polysorbate 80 to protein ranging from 2-1.33. The extent to which this surfactant is incorporated into the protein nanoparticles, or the role that it plays in stabilization of the nanoparticles, is not made clear in this manuscript. However, transmission electron microscopy (TEM) images indicate that the S trimer is anchored to the surface of distinct polysorbate 80 micelles, as confirmed by further TEM in a subsequent investigation of the structural characteristics of this formulation [81].
The stability of the engineered antigen in stressed storage conditions was also assessed. This is a highly important facet to investigate, as stability of a vaccine formulation can significantly diminish logistical hurdles, especially if the formulation can demonstrate stability outside of cold chain conditions [95]. Stability was assessed by exposing the antigens to either prolonged agitation, elevated temperature (25 or 37 °C), pH (4 or 9), or oxidating conditions by hydrogen peroxide, each for 48 hours. Of these conditions, only oxidizing conditions affected the binding affinity of the stabilized spike trimer to hAce2 in an ELISA experiment, while the trimer lacking the stabilizing 2P modification had reduced binding affinity from multiple stress conditions. This indicated the stabilizing effect of the 2P modification and gives an initial indication of stability of this formulation, although much more rigorous stability studies must be undertaken to understand the stability of any vaccine formulation approved for human use [96].
Vaccination of mice with this stabilized spike trimer nanoparticle antigen along with Matrix-M adjuvant generated a high spike-specific titer, significant CD4+ and CD8+ antigen-specific response and a Th1 dominant phenotype and protected against mouse-adapted viral challenge. Similarly, vaccination of olive baboons showed generation of high titers of anti-spike IgG, in groups which received both the protein nanoparticle antigen and the adjuvant. A subsequent study demonstrated that prime-boost vaccination with this formulation inhibited SARS-CoV-2 replication and pathology in the upper and lower airways after administration of the virus by intranasal and intratracheal instillation [97].
The Novavax vaccine formulation was then evaluated in a Phase 1/2 clinical trial (NCT04368988). Using a prime-boost schedule with the boost occurring at day 21 after the prime, they detected significant formation of spike-specific and viral neutralizing antibodies [98]. This result was not significantly dose-dependent, but inclusion of the Matrix-M adjuvant significantly enhanced overall anti-spike IgG titer and viral neutralizing titer. In the subset of patients who were evaluated for a T cell response, significant Th1-skewed spike-specific responses in CD4+ T cells were noted 7 days after the boost in groups receiving the adjuvanted vaccine. Also, of note was the fact that this formulation was stored at 2-8 °C, an easier condition to maintain than subfreezing conditions stipulated by other candidate vaccines.
Novavax is conducting a second Phase 2b clinical trial in South Africa in collaboration with the Bill and Melinda Gates foundation (NCT045333990). Notably, this trial will recruit approximately 240 HIV-positive patients to evaluate the safety and immunogenicity of the vaccine in this highly vulnerable immunocompromised population, in addition to gathering further safety, immunogenicity, and preliminary efficacy data in healthy HIV-negative participants. Novavax has begun a Phase 3 clinical trial in the United Kingdom (2020-004123-16) and is slated to begin further Phase 3 trials in the United States and Mexico.
2.2.2. Virus like particles
A VLP is a viral particle that displays protein antigens, that lacks any DNA or RNA, and maintains the structure of the original virus particle [99]. Based on their surface, size, and shape, VLPs can display epitopes in a highly dense fashion which allows for potent stimulation of the immune system [100]. Most VLPs are made recombinantly in mammalian cells. However, the company Medicago uses plants as a source to produce recombinant proteins that self-assemble into VLPs. Growing proteins in plants for vaccine applications is inexpensive, and inherently has a low risk of contamination with mammalian pathogens, or endotoxin [99]. Researchers use the bacterial vector, Agrobacterium tumefaciens, to transiently infect plants by forcing a bacterial suspension into the extracellular space of the leaf tissue [101]. Nicotiana benthamiana, a close relative of the tobacco plant indigenous to Australia [102], is used extensively to produce recombinant proteins since it allows a wide range of pathogens to infect it. Medicago has previously published a Phase 2 clinical trial using their system for a quadrivalent plant-derived VLP influenza vaccine [103]. They showed that the incidence of pain at the injection site is higher than compared to placebo. However, most of the local symptoms were mild and resolved within a day. The addition of alum with VLPs did not increase the antibody titers. In measuring the antibody response, they did not use a positive control based on an inactivated influenza vaccine, so it is hard to determine how effective the VLP technology is compared to traditional methods. Medicago was able to generate cross-reactive antibodies against heterologous strains of influenza and illustrate T-cell responses, which potentially would allow for broad protection. Based on this technology, Medicago has announced that they are in the process of going into Phase 1 Clinical Trials for a COVID-19 vaccine (NCT04450004) [62].
2.3. Nucleic acid based
Nucleic acid based vaccines were first identified in the early 1990’s when plasmid was injected intramuscularly and a humoral response to the encoded antigen was noted [104]. Quickly thereafter, clinical trials began for cancer and infectious disease vaccines, wherein plasmid was introduced via electroporation or injector gun. With DNA based vaccines, the plasmid must be delivered to the nucleus of the host cells to induce antigen expression and the resulting immune response. Shortly after DNA vaccines were identified, mRNA-based vaccines were developed [105]. In contrast to DNA based vaccines that require nuclear delivery, mRNA vaccines only need to be delivered to the cytoplasm. Moreover, although it has never been reported, DNA vaccines do theoretically have the potential to incorporate into the host genome, a problem that is avoided with mRNA vaccines. On a molar basis, DNA and mRNA vaccines are theoretically similarly efficacious; however, mRNA can lead to more rapid expression than DNA [105]. Overall, the first generation of DNA and RNA based vaccines did not illustrate strong protection in humans. Advancements in the use nucleic acid vaccines have focused on better identification of antigens, modifications to improve nucleic acid stability and translation, improved delivery, and inclusion of adjuvants to generate more protective responses.
With respect to delivery of RNA and DNA based vaccines, neutralization of the negative charge of the nucleotides can facilitate delivery through the cell membrane. To this end, cationic polymers, proteins, lipids, or other elements have been used to form complexes with anionic nucleotides to generate non-viral vectors for RNA or DNA delivery. For the portion of negative to positive charges, often an N/P ratio is reported to relate the number of amine groups on the cationic material that can be positively charged to the number of nucleotide phosphate groups on the that can be negatively charged. Changing the N/P ratio of the carrier and the nucleic acid can influence many other properties such as the stability, size, and net surface charge. Usually increasing the N/P ratio increases activity. However, there is a trade-off with an increase in activity, which is an increase in toxicity. An increase in the N/P ratio helps particles enter the cell, open the phagosome/lysosome, and allows the nucleic acids escape to enter the cytosol for RNA, or the nucleus for DNA [106]. One mechanism by which this occurs is the ‘proton sponge’ effect, where a weakly basic molecule causes the phagosome to leak and perhaps rupture [107].
Of all the technologies applied for SARS-CoV-2, nucleic acid vaccines have seen the most rapid advancement to clinical evaluation. This is indicative of one significant advantage to this technology, the ease and speed of development. If a company already has a gene delivery platform, then as soon as an antigen’s genetic sequence is known, a vaccine can be developed. One drawback with this technology is that although there has been research on DNA or RNA vaccines for upwards of 30 years, there has yet to be an FDA approved formulation that is available clinically.
2.3.1. Intradermal DNA delivery
Electroporation is a method that Inovio Pharmaceuticals is pursuing for COVID-19 vaccines [53]. In general, electroporation is the application of brief electric pulses to cells and tissue which transiently and reversibly permeabilize the cell membrane. This disruption allows for the entry of large molecules, including plasmid DNA to enter the cell. Using electroporation can drastically enhance the protein expression generated by the injection of a plasmid [108]. Enhancement of the immune response using electroporation could be in part due to the induction of a local inflammatory process. Electroporation induces the production of inflammatory cytokines and chemokines along with the recruitment of immune cells [109].
Previously Inovio Pharmaceuticals used electroporation to deliver a DNA vaccine against MERS in a Phase 1 clinical trial [110]. Vaccination involved using the plasmid, GLS-5300, a DNA vaccine expressing a full-length MERS coronavirus S-protein. Vaccination involved injecting the plasmid in one mL intramuscularly in the deltoid followed by intramuscular electroporation at the site of injection to enhance plasmid entry, using a Cellectra-5P Adaptive Constant Current Electroporation Device, which is made by Inovio Pharmaceuticals. This device emits square-wave electric pulses with an adjustable electric field [111]. In their Phase 1 clinical trial against MERS, 50% of the vaccinated participants generated detectable neutralizing antibodies at one or more timepoints in the study. Additionally, 88% of the patients had T cells that produced IFN-γ in the presence of the S-protein.
With regards to COVID-19, Inovio Pharmaceuticals generated preclinical data showing the efficacy of their vaccine [112]. A plasmid was generated encoding the SARS-CoV-2 S-protein along with a N-terminal IgE leader sequence. The plasma generated contained a human cytomegalovirus (CMV) immediate-early promoter and a bovine growth hormone polyadenylation signal. Using their system to vaccinate BALB/c mice and guinea pigs resulted in the generation of neutralizing antibodies against SARS-CoV-2, showing the immunogenicity of their system. They also reported generation of neutralizing antibodies and IFN-γ-secreting spike-specific T cells in rhesus macaques after a prime-boost immunization, leading to a significant reduction in viral load, relative to naïve controls, in bronchoalveolar lavage fluid and nasal swab samples collected after intratracheal and intranasal challenge with SARS-CoV-2 17 weeks after vaccination [113]. On April 6, 2020, Inovio Pharmaceuticals announced they will start Phase 1 clinical trials [52,53].
Another company, Genexine, has also reported the start of a Phase 1/2 clinical trial against COVID-19 [114]. Genexine has previously published results from Phase 1 and Phase 2 clinical trials using electroporation for HPV vaccination [115,116]. The plasmid encoded the virus antigens HPV E6 and E7. Additionally, unique to the other nucleic acid approaches listed in this review, the plasmid encoded an adjuvant: Fms-like tyrosine kinase-3 ligand (FLT3L). A previous study showed that a nucleic acid cancer vaccine can be boosted with the addition of FLT3L [117]. Systemic delivery of FLT3L ligand prior to injecting RNA increased T-cell homing to the tumor and the vaccine’s therapeutic efficacy. Overall, the cure rate was enhanced by the addition of FLT3L. In the Phase 2 clinical trial that Genexine ran using their electroporation system, 63% of the patients showed histopathological regression. Unfortunately, no placebo was used, and no historical controls were discussed in the publication. In a recently released preprint, Genexine detailed that they compared intramuscular vaccination of BALB/c mice with DNA coding for the full-length S protein (pGX27-S) or the S protein without the S2 portion (pGX27-S ΔTM), and found that pGX27-S ΔTM induced higher anti-S protein titers, leading them to choose this antigen for further evaluation in vaccination of rhesus macaques. Using a prime-boost-boost model in rhesus macaques, they saw induction of anti-S and neutralizing IgG in all animals after one vaccination, with titers increasing after the boosts. They also saw induction of S-specific CD4 and CD8 t cells. They saw an insignificant reduction in viral loads relative to controls after viral challenge ten weeks after the last vaccination, but saw a significant reduction in airway tissue pathology 4 days after infection relative to unvaccinated controls [118].
An alternative strategy for DNA transfection of the skin is being employed in a collaboration composed of Osaka University, AnGes, and Takara Bio. While publicly available information on the antigen and adjuvant is unavailable, it is evident that they are employing plasmid DNA delivered by a pyro-drive jet injector (PJI), which employs the detonation of small amounts of explosive powder to propel plasmids in a jet into the skin at variable, controllable depths [119]. In a preclinical evaluation of this technology to vaccinate against the model antigen ovalbumin in mice and rats, they demonstrated that plasmid DNA delivered by PJI induced significantly greater antigen expression and resultant antibody titers compared to needle-injected plasmid DNA [120]. This technology presents the advantage of needle-free, single step delivery with the potential to optimize the depth of delivery to induce the optimal immune response. Clinical Phase 1/2 evaluation of the PJI system delivering a COVID-19 vaccine is currently underway (NCT04463472).
2.3.2. Polyplex
TranslateBio and Sanofi have entered an agreement to develop a vaccine that will enter Phase 1 clinical trials in the fourth quarter of 2020 [121]. Currently, TranslateBio has no published preclinical or clinical results with respect to COVID-19. However, they have published work on delivery of mRNA to the lungs with polyplexes [122]. Patel et al. were the first to report the effective delivery of mRNA to the lungs using various cationic poly(beta amino esters) (PBAEs) they had synthesized for mRNA delivery. PBAEs have an advantage over the ubiquitously used polyethyleneimine (PEI) in delivering nucleic material since they are fully biodegradable, and a large library of PBAEs can be made with simple starting materials [123]. By mixing libraries of commercially available amine containing compounds with acrylates, a library of polymers with degradable ester bonds can be synthesized in a facile manner. For pulmonary delivery, Patel et al. chose a hyperbranched PBAE since at high concentrations, the hyperbranched PBAE is resistant to aggregation compared to a linear polymer, allowing for the facile delivery using a vibrating mesh nebulizer. They showed that covalent attachment of polyethylene glycol (PEG) to the polyplex did not increase stability of the particle, and therefore they did not move forward with in vivo delivery using a PEGylated formulation. Using their system, Patel et al. were able to obtain expression in all lobes of the lung in mice, and a majority of cells transfected were lung epithelial cells. Protein expression plateaued at 24 hours but quickly dissipated at 48 hours. Using this system, TranslateBio and Sanofi will hopefully develop a vaccine that will enter Phase 1 clinical trials soon for SARS-CoV-2.
2.3.3. mRNA-lipid nanoparticles (LNPs)
Due to its hydrolytic instability, poor membrane permeability, and the abundance of RNAses in the body, mRNA must generally be complexed for otherwise protected to ensure delivery to the cytosol where it can result in translate of its encoded antigen. One method to accomplish this is through LNPs. Nucleic acid containing LNPs can be formed by mixing mRNA, various ionizable and neutral lipids. The lipids and nucleic acids are often mixed with a microfluidic mixer or other similar mixing apparatus [54]. Ionizable lipids have a pKa such that they are positively charged at a low pH, allowing for complexation of anionic RNA, but neutral at neutral pH, reducing their toxicity relative to cationic lipids. Moderna is a company which has been working to use this delivery method for the treatment of a broad spectrum of diseases. Injection of LNPs encoding full-length hemagglutinin H10, either intradermal or intramuscular, into rhesus macaques, resulted in the development of serum hemagglutination inhibition (HAI) activity against influenza. At the site of mRNA LNP injection, there was a general increase in CD45+ cells (a marker of differentiated hematopoietic cells, not including erythrocytes and plasma cells), including an increase in neutrophils and monocytes. This increase was irrespective of the presence of RNA, implying that the lipids induce immune cell infiltration. Their studies also indicated that the mRNA LNPs can induce type I interferon expression in DCs, demonstrating they can induce a potent antiviral response. Even though the LNPs alone induced infiltration of immune cells, the mRNA in the LNPs was required to induce activation of DCs. There were, however, no controls comparing the mRNA vaccine to traditional inactivated or subunit vaccine which complicates the analysis of the immunogenicity of this approach versus more traditional techniques.
Expanding on this research, Moderna has tested mRNA LNPs in mice to explore their immunogenicity against SARS-CoV-2 [57]. Corbett et al. administered LNPs that encapsulate mRNA encoding a modified S-protein. As noted in the protein subunit section above, typically the native S-protein is highly unstable during expression, resulting in a significant challenge in producing the protein effectively enough to generate a protective antibody response. Therefore, the S-protein encoded by the Moderna vaccine candidate includes the 2P stabilizing mutation, which results in a higher immunogenicity [86,87,124]. Using LNPs encapsulating mRNA encoding this modified S-protein, Corbett et al. were able to generate neutralizing antibody in three different mouse strains. Depending on the dose given, the Th1/Th2 ratio changed but did not appear to be significant with dose. Additionally, they show at suboptimal doses of vaccination there is no immune pathology induced by the vaccine. Alum controls had a different Th1/Th2 ratio, with more antibodies that were of the Th2 subtype, which is characteristic of this adjuvant. As noted above, a Th2 skewed vaccine could result in undesirable immunopathology for SARS-CoV-2.
The first report of Phase 1 clinical data from Moderna’s vaccine candidate came in the form of a dose-escalation study of 45 adults aged 18 to 55 who received a prime and boost (day 0 and 28), at a dose of 25, 100 or 250 μg of mRNA [125]. Titers increased with the boost and the upper 50% of the neutralizing titer were comparable to a panel of convalescent serum samples. Adverse events increased with the boost, with 20% of the participants in the 250 μg dose reporting one or more adverse events. Subsequently, this trial was expanded to include 40 participants over the age of 55 [126]. These patients were only administered the lower (25 μg and 100 μg) dosages due to their lower incidence of adverse events in the younger cohort. No serious adverse events occurred in this trial [127], and the most common adverse events were headache, fatigue, myalgia, chills, and injection-site pain. Most of these adverse events were elicited after the boost of the vaccine. Encouragingly, anti-S binding and neutralizing antibodies, as well as anti-S CD4 T cell responses, were elicited in all participants and reached comparable levels to those in the younger cohort. Moderna, working with the Biomedical Advanced Research and Development Authority (BARDA) and NIAID, has begun both Phase 2 (NCT04405076) and Phase 3 (NCT04470427) clinical trials to evaluate this vaccine candidate. In November of 2020, it was reported that Moderna’s vaccine was 94.5% effective in preventing symptomatic COVID-19 in patients [128]. Moderna’s trial Phase 3 Trial has been noted for its diversity with 37% of the study representing communities of color (e.g. African American, Hispanic). In addition, to greater than 42% of individuals in the study are from at least one high-risk group (e.g. obese, diabetic, 65+) [129].
As stated previously, an advantage of the LNP system is the speed at which it can be developed. The day after SARS-CoV-2 sequence was released publicly, the modified prefusion sequence was determined, and synthesis was started. Twenty-five days after the sequence was published, clinical grade LNPs were sent for mouse experiments and Moderna started Phase 1 clinical trials 66 days after the release of the sequence of SARS-CoV-2, with Phase 2 Trials starting 160 days and Phase 3 Trials 193 days after release of the sequence. This accelerated production of vaccines to enter clinical trial is extremely fast compared to traditional viral culture or recombinant protein techniques. According to the WHO, it takes five to six months to generate a vaccine against a pandemic influenza [130]. This is required based on the complicated sequential steps that are needed to produce a new vaccine. However, using LNP to deliver mRNA, Moderna could make a vaccine to be injected into humans in approximately two months. In the long term this type of technology would be highly beneficial in combating emergent pathogens. Additionally, the White House has selected Moderna in their Operation Warp Speed program to decrease the amount of time required for FDA approval [131] and indeed Moderna has already filed for emergency FDA approval of their vaccine [132].
Another LNP vaccine is being developed via a partnership between Pfizer and BioNTech [133]. BioNTech has previously published a preclinical study in animal models of Zika virus [134]. Similar to Moderna, they use a proprietary nanoparticle, with BioNTech’s platform consisting of an ionizable lipid/phosphatidylcholine/cholesterol/PEG-lipid nanoparticle in a 50:10:38.5:1.5 molar ratio. The LNP is made when mRNA in a pH 4.0 buffer is mixed rapidly with the lipids dissolved in ethanol. For their previous study they used the Zika envelope (E) protein as an antigen. In mice, antibody response peaked at eight weeks and maintained the same level through week 20. In rhesus macaques, immunized monkeys were challenged with virus five weeks after vaccination. All control macaques had a high level of viremia while four out of five vaccinated macaques demonstrated no detectable viremia.
Based on this technology, Pfizer and BioNTech published a Phase 1/2 clinical trial for a COVID-19 vaccine. This vaccine candidate, BNT162b1, consisted of mRNA encoding for the spike RBD fused to a T4 fibritin-derived trimerization domain, and incorporating 1-methyl-pseudouridine in place of uridine to reduce innate immune sensing while increasing overall translation [[135], [136], [137]]. After vaccinations on day 0 and 21, volunteers had neutralizing antibody levels higher than patients who were recovering from COVID-19. 100% percent of the volunteers had local pain at the injection site, while patients who received the placebo had roughly 35% injection site pain. Overall, reactogenicity was higher after the second dose was administered, but symptoms resolved after a few days. A second non-randomized open-label clinical trial of a prime-boost immunization with this formulation yielded similar humoral results, as well as demonstrating that a large fraction of the CD4+ and CD8+ T cells elicited by immunization secreted IFN-γ, indicating a Th1 skew. The strong CD8+ T cell response in 29 of 36 vaccinated patients contrasted with the lower CD8+ T cell response observed in Moderna’s trial results.
A head-to-head Phase 1 clinical trial comparing BNT162b1 to another Pfizer and BioNTech candidate vaccine, BNT162b2, indicated the BNT162b2, which encodes for 2P-stabilized spike protein rather than trimerized RBD but otherwise uses mRNA in a similar stabilizing formulation, resulted in fewer systemic reactions compared to BNT162b1 [138]. Both candidate vaccines induced similar humoral responses. At the time of this writing the evaluation of the cellular response to BNT162b2 has not been published. Based on this data this group is has advanced BNT162b2 to Phase 3 Trials (NCT04368728). In November of 2020, Pfizer and BioNTech initially reported a greater than 90% interim vaccine efficacy with their formulation [139], which was later revised to be 95% effective in preventing symptomatic COVID-19 [140]. Recently, Pfizer/BioNTech has been approved for application in the United Kingdom [141].
Another mRNA lipid nanoparticle under development as a COVID-10 vaccine is formulated by CureVac. Classically, CureVac has reported the complexation of mRNA with protamine. Using this protamine approach Curevac was the first to show the use of an mRNA vaccine nanoparticle in humans [58]. For this, they chose rabies as a proof of concept in humans since most humans are naïve to the virus and there are validated methods to access the efficacy of a vaccine. In their Phase 1 clinical trial, 77% of the patients receiving the rabies vaccine through intramuscular administration induced measurable neutralizing titers, while 50% produced neutralizing titers considered by the World Health Organization (WHO) to protect against exposure to rabies. Intradermal administration resulted in 71% of the participants achieving productive neutralizing titers with 57% having protective titers after one year. In the clinical trial, no controls were used to compare the efficacy of this vaccination technique to standard inactivated virus or subunit vaccines, however, there are historical trials that may be used in comparison. In a clinical trial conducted in 1987 [142], patients received the standard human diploid cell rabies vaccine on day 0, 7, and 28 intradermally (similar to the schedule for the Curevac mRNA rabies vaccine trial). Unlike the RNA vaccination, 100% of the volunteers had effective neutralizing antibody titers after 1 year, while 89% had protective antibody titers for over two years. Curevac has announced that they will begin Phase 1/2a clinical trials in June of 2020 for a SARS-CoV-2 vaccine, with this vaccine utilizing the same lipid formulation as Pfizer/BioNtech with mRNA encoding S protein of the virus [58,143,144]. This change from a protamine based carrier to a lipid based particle may be due to the fact that LNPs are shown to work better when given via standard intramuscular injection [145], whereas the protamine based formulation only induced neutralizing titers when administered with less common needle-free injection mechanisms similar to those used in previous studies of DNA vaccination [58,120]. Another consideration might be the enhanced storage they announced with their new LNP formulation, noting that they have a formulation suitable for storage at three months at standard refrigeration temperature (5 °C) [146].
Another RBD-encoding mRNA LNP formulation was developed by Zhang et al. [147]. In this formulation, a simple mRNA construct encoding the RBD amino acid sequence was encapsulated in a lipid nanoparticle containing an ionizable lipid, phospholipid, cholesterol, and a PEGylated lipid. The authors did not specify the identity of the ionizable lipid or PEGylated lipid used. The resulting LNPs effectively transfected RBD, and prime-boost intramuscular immunization of mice and cynomolgus monkeys induced high levels of RBD-specific and virus-neutralizing titers, as well as robust, Th1-skewed T cell responses. Vaccinated mice challenged with a SARS-COV-2 mouse-adapted strain [148] demonstrated full protection from infection and sterilizing immunity. In addition, the group did a preliminary thermostability study indicating that their formulation retained 100% transfection ability after seven days of storage at 25 °C. However, in the same time period at 37°C they saw significant decrease in transfection. This formulation is in Phase 1 clinical evaluation by the Chinese People's Liberation Army (PLA) Academy of Military Sciences in collaboration with Walvax Biotech (ChiCTR2000034112).
Additionally, there are other LNP systems for delivering mRNA. An Imperial College London laboratory has avoided collaborating with any major pharmaceutical company in order to deliver a vaccine to the United Kingdom and developing countries for a reasonable price [149]. This group is using a self-amplifying RNA approach. Self-amplifying (or replicating) mRNA (also called a replicon) encodes the antigen of interest as well as proteins which lead to replication of the subgenomic RNA encoding for the antigen. Rather than just the sequence encoding the antigen, the mRNA sequence is a single strand that is able to self-replicate in the cytoplasm of the cell [150]. Accordingly, less self-amplifying mRNA is theoretically needed compared to traditional mRNA. Imperial College London’s RNA vaccine is formulated into lipid nanoparticles with cationizable lipids similarly to Moderna and Pfizer, using a cationizable lipid patented by Acuitas Therapeutics [151]. They have recently published their preclinical results, wherein a prime-boost immunization induced high levels of Th1-biased virus-specific and neutralizing titers and SARS-CoV-2 peptide-specific T cells in mice [152]. The Imperial College London group is currently conducting a Phase 1 clinical trial with this formulation using a prime-boost schedule (ISRCTN17072692).
Duke-National University of Singapore and Arcturus Therapeutics are also developing a replicon RNA-based vaccine [153]. They are delivering a replicon encoding the spike protein using their proprietary Lipid-enabled and Unlocked Nucleomonomer Agent modified RNA (LUNAR) formulation. This formulation contains common components of LNPs for mRNA delivery such as cholesterol, phospholipids, PEGylated lipids and a proprietary lipid with an ionizable amino head group [[154], [155], [156]]. In addition, this lipid has an ester group incorporated into it to facilitate rapid degradation after RNA delivery. The U-N-A portion of the acronym refers to unlocked nucleomonomer agents, which are nucleotides lacking a carbon-carbon bond between their 2’ and 3’ carbons, and potentially including rearrangement of the location of the phosphoester bond relative to those carbons, or substitution with new functional groups at those carbons. These modifications are undertaken to alter the physicochemical, and potentially translational, properties of RNA molecules containing UNAs [157]. However, there is no published data available on the extent to which these UNAs are incorporated into the Arcturus vaccine candidate RNA sequence. While preclinical data has not yet been reported for this formulation, Phase 1/2 clinical evaluation has begun (NCT04480957).
3. Viral and vector based formulations
Attenuated virus vaccines were the first vaccine type reported by Jenner for Smallpox [164]. In essence, they are a virus capable of replicating in the vaccinated host and eliciting an immunogenic response but are altered relative to the target virus such that they are minimally pathogenic, if at all. However, there are safety concerns when attenuated viral vaccines are given to those that are immunocompromised, including individuals who are HIV+, TB+, pregnant, very young or very old. In these populations, the virus can become pathogenic, resulting in serious side effects and perhaps even death due to the viral infection. Despite this limitation, there are several FDA approved live attenuated vaccines including those for Polio (Sabin), Rotavirus, Varicella zoster, Yellow Fever and the FluMist formulation for Influenza. A live attenuated virus of SARS-CoV was developed by introducing mutations in the 2′O methyltransferase NSP16 as well as the exonuclease NSP14 [165]. Mutation of NSP16 alone resulted in a virus with some virulence in aged mice as well as potential to revert to the unmutated virus, and the NSP14 mutation alone diminished viral replication. The combination of the two strategies resulted in a virus which induced robust immunity in mice, including aged mice, to heterologous CoV challenge while also resulting in no pathology. Some groups have also preliminarily reported the isolation of attenuated SARS-CoV-2 mutants [166], and intranasal inoculation of Syrian golden hamsters with this virus resulted in significant viral replication in the nasal turbinates but not in the lungs, milder lung tissue pathology than a SARS-CoV-2 control, development of SARS-CoV-2 neutralizing serum antibodies, and sterilizing immunity against SARs-CoV-2 infection [167]. At least three other groups are working on the generation of attenuated SARS-CoV-2 vaccines through the use of codon deoptimization to create viruses attenuated through reduced mRNA stability and translation efficiency [40,168].
As viruses and viral vectors have advanced, particularly for application in gene therapy, platforms have been developed that can be used to plug-and-play genetic information for treatment of a specific disease or development of a vaccine. In much the same way that non-viral vector vaccines can be accelerated into development once the genetic code for the pathogen is known, viral vector-based formulations have the same flexibility. There are several types of common viral vectors that classically cause mild to no symptoms upon infection in humans. These vectors include adeno-associated virus (AAV), adenoviruses and pox viruses, such as canary pox. The viral vectors can be replicating viral vectors or non-replicating viral vectors. The safety of non-replicating viral vectors is generally greater than that of their replicating counterparts. Both replicating and non-replicating viral vectors have been applied in the development of a SARS-CoV-2 vaccine.
In addition to viral vectors, other biological vectors such as bacteria and mammalian cells can be used as carriers for vaccine elements. Historically attenuated bacteria have been used for vaccines, as exemplified by the attenuated bacteria tuberculosis vaccine Bacille Calmette-Guérin (BCG). This technology has advanced, similarly to viral vectors, wherein bacteria is used as platform that can plug-and-play antigens to form new vaccines. This includes using attenuated pathogenic bacteria, such as Salmonella enterica, and expressing antigens on the bacteria surface, in this way the bacteria can serve as both an antigen and adjuvant source due to the PAMPs present on the bacteria’s surface [169]. Similarly, commensal bacteria have been used as platforms for delivery of antigens but since they lack immune stimulating PAMPs, they can be considered more safe than pathogenic vectors. The most recently developed vector for vaccines has been mammalian cell-based vaccines. Although growth of virus in cells has been reported for some time, particularly for influenza, [170] these constructs are delivering the cells as a platform for vaccine elements. Whereas attenuated pathogens initiate an immune response due to their PAMPs, delivery of mammalian cells can activate the immune system through non-self sugars, MHCs and other elements that would result in rejection of the cell. Rejection of these cells often limits their use to autologous cells. Both bacterial and mammalian cells have been used to develop platforms that have been applied to SARS-CoV-2 vaccines.
3.1. Viral vector
Viral vectors, like Adeno-associated virus (AAV), have had successful products for gene therapy reach FDA approval and clinical application. For vaccine applications they have chiefly been used for cancer and HIV vaccines. Canarypox and adenovirus vectors have also been used in these areas of study and are often given as part of a schedule where the prime or boost is a protein or DNA subunit vaccine. These viral vectors are typically not given as both the prime and boost in a vaccination schedule because neutralizing antibodies can significantly attenuate the second administration of the vector. Indeed, since most individuals are not naïve to these vectors, extensive screening is often done prior to the vectors use to ensure that significant neutralizing titers are not present. For gene therapy applications, methods to attenuate a neutralizing response, using variants of virus that are less immunogenic, or delivering the vector to areas that are more tolerant (e.g. the liver) can help to mitigate some of these issues. Several of these concepts can be applied to viral vector-based vaccines. Non-replicating and replicating adeno virus have been developed for SARS-CoV-2 vaccines.
3.1.1. Adenovirus non-replicating viral vectors
CanSino has previously shown the efficacy of using a replication defective adenovirus type 5 (Ad5) vector as an Ebola vaccine in a Phase 2 clinical trial [171]. The results of that trial concluded that 53% of participants in the high dose group, 48% in the low dose group, and 43% in the placebo group had adverse effects. Approximately 100% of the patients produced antibodies against Ebola on day 14 and 28. By day 168, approximately 70% of the patients were still making antibodies against Ebola, however, on average, these antibodies were not deemed protective against Ebola. This data contradicted CanSino’s Phase 1 clinical trial that was performed in China (the Phase 2 clinical trial was done with patients in Sierra Leone). Others have shown that memory responses against Ebola last longer in Europeans compared to volunteers who live in endemic regions of Ebola infection [172]. Another concern about using adenovirus is preexisting immunity against the virus in the general population. Of the patients enrolled in this clinical trial, 85% had preexisting antibodies against Ad5. Even though these antibodies existed, vaccination with the platform generated antibodies against Ebola on day 14, 28, and 168 when the study was conducted in China.
With these results, CanSino is developing a vaccine against SARS-CoV-2, and they were the first company to publish the results of their Phase 1 clinical trial [173]. For this trial, they developed an Ad5 virus expressing the S-protein from SARS-CoV-2. Patients were administered a single (prime-only) intramuscular injection of the viral vector. Similar to the Ebola vaccine, patients in the trial had a preexisting immune response against Ad5. However, for patients who received the high dose of virus particles, 94% still produced an immune response on day 14, while 100% produced an immune response on day 28. Furthermore, 75% of the high dose group had a four-fold increase in neutralizing antibody by day 28 compared to patients who did not receive the vaccine. It was noted that high preexisting antibodies did lower the immune response against the S-protein. These patients will be continued to be monitored to measure long term memory responses generated by this platform.
The results of a Phase 2 trial have also been reported for the CanSino Ad5 vaccine candidate [174]. Patients were again administered a prime-only immunization consisting of one of two different doses (5 x 1010 and 1 x 1011 viral particles) injected intramuscularly. RBD-specific and virus-specific neutralizing antibodies were elicited at similar levels in both groups by day 28 after injection. However, individuals with pre-existing anti-Ad5 antibodies (52% of all participants), and those older than age 55 both demonstrated significantly lower titers. Spike-specific T cell responses were determined using an ELISPOT for IFN-γ secreting T cells. Significant increases in these cells were detected by day 28 in ~90% of participants, with no significant differences between those with and without preexisting anti-Ad5 antibodies, or between age groups. In addition, no serious adverse events were observed, with common adverse events such as pain, fatigue, and fever being observed in both dose groups. These results have led CanSino to pursue evaluation of this candidate in a Phase 3 trials (NCT04526990, NCT04540419), again using a prime-only immunization schedule. It remains to be seen whether the reduced humoral response in patients with preexisting anti-Ad5 immunity will hamper the protective efficacy of this candidate.
Other companies are also using adenovirus for vaccination against COVID-19. Oxford is collaborating with AstraZeneca in developing an COVID-19 using an adenoviral vector derived from a chimpanzee adenovirus and encoding the S protein (ChAdOx1 nCoV-19) [175]. Because the vector is derived from chimpanzees rather than a human adenovirus, pre-existing ant-vector immunity is very low in the general population. Preliminary results of the Phase 1/2 clinical trials that started April 29, 2020 have been reported, wherein ChAdOx1 nCoV-19 was compared with a meningococcal conjugate vaccine as control [176]. The vaccine was administered on a prime and boost (day 0, 28) schedule with prophylactic acetaminophen given, which reduced local and systemic reaction to the vaccine. After a single vaccination, 91% (32/35) patients had neutralizing antibody titers, and with boost, 100% of participants had neutralizing responses. Neutralizing titers correlated well with total anti-spike IgG antibody levels. They report T-cell response to the spike protein, noting it peaked at day 14, prior to the boost [176]. In November of 2020, AstraZeneca reported a 70% efficacy with their vaccine. Interestingly, two different doses were given at their two trial sites with increased efficacy noted at the site which administered a lower initial dose. The half-dose prime followed by a full-dose boost resulted in a 90% efficacy, whereas patients which received a prime-boost with a full dose reported a 62% efficacy. This disparity in efficacy could be due to an adverse immune response to the viral vector, rendering the boost less effective due to neutralization generated from the prime vaccination [177].
Janssen’s AdVac® technology uses a nonreplicating version of adenovirus type 26 (Ad26) and has been used as a platform against the Ebola virus [178]. Neutralizing antibody titers against Ad26 are much lower than those against Ad5 in populations in North America, South America, sub-Saharan Africa, and Southeast Asia [179], reducing the potential impact of pre-existing immunity against the vector on the success of this candidate. As of September 2020, the AstraZeneca vaccine is in Phase 3 clinical trials [180] (ISRCTN89951424, NCT04516746), and a Phase 3 trial for the Janssen candidate has also been registered (NCT04505722). Both Janssen’s and AstraZeneca’s vaccine candidates are part of the United States Warp Speed vaccine development program against COVID-19 [177].
Another method to circumvent pre-existing immunity to the Ad5 vector is being employed by ImmunityBio in collaboration with NatKwest. They have developed a modified Ad5 vector, Ad5 [E1-, E2b-, E3-], which contains a number of gene deletions which result in reduced late gene expression by the vector, inducing less host response to the vector than unmodified Ad5 [[181], [182], [183]]. They evaluated this vector used to deliver the SARS-CoV-2 S protein, as well as the N protein [184]. The justification for inclusion of the nucleoprotein antigen in the encoded sequence was to induce T cell-mediated immunity to N protein epitopes in addition to S protein epitopes. The N protein in this case was coupled with what they call and Enhanced T Cell Stimulation Domain (ETSD). The precise structure of this domain was not disclosed by the group, but they disclose that it is designed to direct the protein to induce endosomal processing and MHC Class II presentation. Other groups have accomplished this task with SARS-CoV N by fusing the N antigen to lysozome-associated membrane protein (LAMP) [185]. In the preclinical evaluation of this platform, they reported the generation of anti-S and N antibodies, neutralizing antibodies, and CD4 and CD8 T cell responses against both S and N peptide pools after subcutaneous prime-boost vaccination of CD1 mice. Efficacy in protection against viral challenge was not reported. A Phase 1 clinical trial of this vaccine candidate is currently underway, using a prime-boost schedule and subcutaneous injection (NCT045917170).
Another adenoviral vector using a simian adenovirus encoding the spike protein is reportedly in development by a collaboration of Reithera, LEUKOCARE, and Univercells. The trial provides for a prime-only vaccination, which is logistically preferable and circumvents the issue of developing vector-specific immunity to inhibit the effectiveness of a boost. However, it is unclear what may be unique to this formulation to allow it to generate significant immunity from a prime-only schedule. LEUKOCARE’s contribution to the collaboration will be developing a formulation to stabilize the viral vectors. They have recently published and extensively patented methods using algorithmic methods to identify mixtures of excipients which can stabilize diverse types of biopharmaceuticals. With this method they used Design of Experiments principles combined with accelerated stability testing to determine mixtures of excipients which most effectively preserved the stability of adenoviral vectors [186,187]. It remains to be seen what degree of stability this method may be able to impart to a vector-based vaccine against SARS-CoV-2. The formulation is currently in a Phase 1 clinical trial (NCT04528641).
3.1.2. Vesicular stomatitis virus (vsv) vector vaccines
Vaccines using vesicular stomatitis virus (VSV) as a vector have been developed for a variety of viral pathogens. This technology was initially developed for application as an influenza vaccine, where genes encoding for influenza HA were inserted into the VSV genome, resulting in a replication-competent vector which induced protective immunity against influenza A challenge in mice after intranasal administration [188]. This vector was subsequently improved by altering or deleting the VSV glycoprotein gene from the viral sequence to reduce vector-induced pathogenesis and abrogate the generation of anti-vector immunity [189]. Use of this platform with the VSV glycoprotein deleted, and encoding for the Ebola glycoprotein (rVSV-ZEBOV) was shown to induce protection against an Ebola virus challenge in animal models [190]. Clinical evaluation demonstrated the safety and immunogenicity of this platform, and it was subsequently the first licensed Ebola vaccine and given the brand name Ervebo (Merck) [191,192].
Similar efforts were also undertaken to use VSV as a vector for vaccines against various coronaviruses. For example, a vaccine for MERS-CoV was developed through replacement of the VSV glycoprotein with the MERS spike protein. Single-dose Intramuscular or intranasal administration of this vaccine to rhesus macaques induced a robust neutralizing antibody and T cell response. For vaccination against SARS-CoV, a VSV vector was designed by inserting a gene encoding for the SARS S protein into the VSV sequence. Intranasal immunization with this vaccine prevented SARS-CoV replication in mice challenged intranasally, and protection was determined to be antibody-mediated [193]. An alternative strategy of fully replacing the VSV glycoprotein with the SARS-CoV S protein, resulting in a replication-incompetent vector, was compared to the replication-competent vector that retained the glycoprotein for use as an intramuscularly-administered vaccine [194]. A greater humoral response against the S protein was observed in mice given the replication-incompetent vector compared to the replication-competent vector.
Two studies described the generation of VSV vectors encoding the S protein of SARS-CoV-2 for use as vaccines. In one study, VSV encoding S protein as well as green fluorescent protein (GFP), and trace amounts of VSV glycoprotein, was produced. This vector was initially developed to provide a tool to identify inhibitors of SARS-CoV-2 S-protein-mediated entry [195]. In a subsequent study, VSV glycoprotein was added to the vector in order to permit glycoprotein-mediated cell entry and a single round of replication upon administration to mice, as murine Ace2 differs from human [196]. Intranasal vaccination of BALB/c mice using a prime-only or prime-boost regimen induced anti-S titers and neutralizing titers, with a significantly greater overall humoral response from the prime-boost regimen. Mice were transfected with hACE2 and administered an anti-IFNAR1 monoclonal antibody in a disease model which was shown to recapitulate lung pathology associated with human infection [197]. Mice vaccinated with the VSV vector encoding GFP and S protein demonstrated reduced viral lung titers and lung pathology after a SARS-CoV-2 challenge, with the prime-boost receiving mice having significantly lower viral loads and lung pathology.
In another study investigating VSV as a vector to vaccinate against the S protein, a VSV vector initially bearing both S protein and the VSV glycoprotein was used to infect Vero cells [198]. The resulting vector was then serially passaged through Vero cells to increase viral titer and infectivity. As this also resulted in mutations to the vector S protein, the authors verified antigenic similarity of the vector S protein to that of SARS-CoV-2 by comparing the capacity of COVID-19 patient convalescent serum to neutralize their vector and SARS-CoV-2. They then subcutaneously immunized Syrian golden hamsters with this vector on a prime-only schedule and challenged them with SARS-CoV-2 vector 25 days later. Immunized mice demonstrated significantly lower viral titers, weight loss, and lung tissue pathology relative to unimmunized controls, as well as robust induction of SARS-CoV-2 neutralizing titers. This vaccine, developed by the Israel Institute for Biological Research, is slated to begin clinical trials in Israel in November 2020. Another Phase 1 clinical trial instituted by a collaboration between Merck and the International AIDS Vaccine Initiative (IAVI) using VSV with its glycoprotein replaced by the S protein will be conducted in the United States (NCT04569786).
3.1.3. Attenuated measles vector vaccine
Themis has reported a measles virus based vaccine that expresses the SARS-CoV-2 S protein. The measles vector is based on the Schwarz vaccine strain which has 10 amino acids different from the Edmonston B strain, a strain abandoned as a measles vaccine a quarter century ago [200]. Previously, Themis’ platform has shown efficacy in Phase 1 and 2 clinical trials for Chikungunya [201,202]. In a preclinical evaluation of a similar SARS-CoV-2 formulation, IFNAR-/- 577 -CD46Ge mice were used because these mice can become infected with measles [199]. Soluble SARS-CoV-2 S-protein with alum was used as a control. Using a prime and boost schedule (Day 0 and 28), total antibody concentration was lower on day 28 and 49 than observed for measles and the alum concentration had the highest amount of antibodies of the SARS-CoV-2 groups. However, virus neutralizing titers were only present in the measles virus vector expressing the S-protein, wherein three of the six mice responded at day 49. The vector and S-protein vaccine responded similarly for measles neutralization where 6/6 mice had responses above background. In evaluation of IFN-γ production from cells (via ELISpot) upon restimulation, it was noted that with this indicator of a cellular response the splenocytes isolated from mice vaccinated with the S protein expressing virus had significantly greater response than controls. Similarly, this group showed proliferation of T-cells, and multifunctional T cell responses higher than the response to the measles vector. Additionally, splenocytes from mice vaccinated with the S protein expressing virus had significantly greater cell killing upon restimulation than any other of the vaccine groups. These assays strongly conclude a potent cellular response, which indicates Th1 response and may indicate reduced immunopathology with vaccination [199]. The preclinical responses of this vaccine appear promising because the generation of neutralizing antibodies over non-neutralizing antibodies is higher than the alum group and a strong Th1 biased cellular response is noted. Institut Pasteur, CEPI, Themis, The University of Pittsburgh Center for Vaccine Research, and Merck Sharp & Dohme are now collaborating on a Phase 1 Clinical Trial using this platform (NCT04497298) [162].
3.1.4. Orally delivered adenoviral vector vaccine
Vaxart is one of the few companies pursuing a COVID-19 vaccine that can be administered orally. Oral delivery can be advantageous for application in resource limited areas, and if effective can provoke a potent mucosal response to prevent infection. Vaxart’s technology is based on a non-replicating adenovirus where expression is driven by a CMV promoter. The adenovirus also expresses a TLR3 adjuvant driven by a different promoter. The vector is mixed with various excipients and lyophilized. After lyophilization, it is then tableted with microcrystalline cellulose/starch mixture. The tablet is then coated with Eudragit L100. The first Eudragit coating was developed by Röhm & Haas GmbH in Darmstadt in 1953, where the coating was resistant to stomach pH, and became soluble when exposed to the higher pH environment of the intestines, releasing the encapsulated compounds [203]. Eudragit is synthesized by the polymerization of acrylic and methacrylic acids or their equivalent esters. There now exists a whole library of Eudragit polymers commonly used for oral drug delivery. Twenty-four years after the original polymer, Eudragit L100 was developed. The polymer dissolves at a pH above 6.0, resulting in it being soluble in the intestine and thereby releasing the encapsulates in this region. The Eudragit L100 coating on Vaxart’s tablets allow for the adenovirus to therefore be released in the intestine.
Vaxart previously has performed a clinical trial using their technology for the development of a norovirus vaccine [204]. In Vaxart’s clinical trial for norovirus, 61% of the patients in the low dose group made a 2-fold increase over the placebo group, while the high dose group had 78% increase in antibodies over the placebo group. Additionally, by delivering an adenovirus orally, they were able to generate IgA antibodies with a 16-fold increase compared to the controls. Based on this data, Vaxart has announced that they are developing a vaccine against SARS-CoV-2 [159]. They plan on starting Phase 1 Clinical Trials in the second half of 2020. Vaxart is one of the few companies that have been selected for Operation Warp Speed, selected by the US Government, to decrease overall the time for approval of the vaccine [205].
3.2. Cell based delivery systems
Cell based delivery systems are an emerging area of vaccine research that has accomplished great advances and similar platforms already approved may help to advance this technology further. Although commensal bacteria delivery is common as supplements and attenuated or inactivated bacterial vaccines do exist clinically and preclinically, gene therapy or delivery of nucleic acids via bacteria has not reached full FDA approval. Similarly, with the FDA approval of CAR T-cell therapy, there is a clear path forward for approval of mammalian cell-based platforms, but there are still significant hurdles to overcome, particularly when the cell is not from the host. Both bacterial based delivery of plasmid and use of a mammalian cell as an artificial antigen presenting cell have been applied as COVID-19 vaccines.
3.2.1. Plasmid delivery with bacteria
Symvivo’s method to vaccinate is using commensal bacteria to deliver plasmid DNA to the host.[206] The plasmid for this method has a bacterial backbone with a mammalian expression cassette. The bacterial component has a bacterial origin of replication with antibiotic resistance while the mammalian component has a eukaryotic promoter for gene expression. Usually with this system, a recombinant bacterium is used where any pathogenetic features are removed to protect the host from pathogenic assault. Using commensal bacteria, this vaccine can be delivered to mucosal routes which allows for the induction of both mucosal and systemic immune responses. In an in vitro study, they used commensal bacteria Lactococcus lactis, and showed delivery of plasmids effectively to DCs to induce gene expression as well as DC activation [207]. Symvivo is filing for a Phase 1 clinical trial (NCT04334980) to evaluate this technology further. Based on their website, they are developing a vaccine against the S-protein, nucleocapsid protein and the matrix glycoprotein [208]. The type of bacteria that they are using is not known.
3.2.2. Artificial antigen presenting cells
Shenzhen Geno-Immune Medical Institute has registered for a Phase 1 clinical trial (NCT04299724) using an Artificial Antigen Presenting Cell (aAPC) (Fig. 4 ). aAPCs are a technology that was first reported in 1997 where Vaccinia virus was constructed to express MHC II and a co-stimulatory signal, resulting in antigen specific T cell responses [209]. Since their development MHC and co-stimulatory signals have been added to a variety of additional substrates including those which are protein, polymer, inorganic material and cell based [210]. These platforms have been applied for cancer and infectious disease vaccines as well as to create tolerance [210,211]. However, in one of the first clinical trials with the technology, the trial was delayed due to the inability to generate GMP quality K562 cells and although cancer co-therapy trials have been performed [212], there are some concerns about the platform because the K562 is a malignant cancer clone, which is irradiated to halt replication, but nonetheless there are reservations about giving it to cancer patients [213]. Synthetic aAPCs have been used ex vivo to stimulate CAR T cells in clinical trials but have yet to have clinical trials involving their use [213,214].
Fig. 4.
Artificial antigen presenting cell under development for a SAR-CoV-2 vaccine by Shenzhen Geno-Immune Medical Institute.
Some barriers to the development of aAPC in the clinic are likely due to the number of HLA and peptides that would be required in an outbred population, particularly for cancer where immunogenic cancer antigen can be difficult to identify [214]. Indeed, the K562 trial had multiple HLAs prepared for their constructs [213]. For infectious disease vaccines, antigen selection may be easier due to the simplicity of viruses compared to cancer, but HLA matching would still be critical to avoid rejection of the cells and allow them to effectively present antigen. This application then may give aAPCs an application to shine and allow further development of this vaccine technology.
Shenzhen Geno-Immune Medical Institute used lentivirus, NHP/TYF, infect cells and encode various proteins from SARS-CoV-2. It is not clear the exact technology being used with this method; however the leadership of this company, published a methods paper in 2010 on using the NHP/TYF lentivirus to infect DCs for vaccine applications [215]. In their methods paper they describe how unlike other viruses, lentiviral transduction in DCs can occur with a relatively high efficiency. Prior research has shown that a lentivirus can be used to deliver the gene for IL-12 or siRNA suppressing IL-10 to induce a DC that can promote Th1 responses [216]. The NHP/TYF expresses both a reporter gene, and siRNA, and can transduce human immature DCs by over 90%. Patient’s PBMCs are drawn, and then DCs are expanded by culturing the PBMCs with IL-4 and GM-CSF. After a five-day culture, the DCs are transfected with the lentivirus and then can be injected back into the patient. For trials involving this technology, patients will be administered five million aAPCs. There is no published data on how effective this method is on generating an immune response, but it can be hypothesized that this method is highly expensive and would be difficult to apply to vaccinate a large population.
4. Future directions
From a formulation perspective, a major aspect of COVID-19 vaccine development which still needs to be addressed is use of storage outside of the cold chain, antigens other than S-protein, and alternative routes of vaccination.
4.1. Storage considerations
Modification of the spike protein to increase its expression has also been shown to increase its stability [88,217], and some preliminary results also have shown a degree of thermostability for RBD-encoding mRNA LNPs [147]. However, the RNA LNP vaccine candidates from Pfizer/BioNTech and Moderna both stipulate storage at extremely low temperature. Pfizer’s candidates require storage at -70 °C and Moderna’s at -20 °C, with Moderna also specifying storage at -70 °C in their Phase 2 trial protocol, and expiration of thawed vaccine after 8 hours at room temperature [125]. This is an area where LNP RNA vaccines have a significant logistical disadvantage, although CureVac reports an LNP which has three-month thermostability at 5 °C [58,146,218]. As noted above, Novavax performed some preliminary stability analysis of its protein nanoparticle formulation on the timescale of 48 hr. Each of these studies have been for only short periods of time and some do not necessarily recapitulate the potential for high humidity and heat conditions that may be seen when shipping vaccines outside of the cold chain. Breakages in the cold chain are common and a major impediment to distribution of currently approved vaccines, while it has been shown that vaccines which can be administered in the ‘Controlled Temperature Chain’ rather than the cold chain can demonstrate increased rates of vaccine coverage with a lower economic burden [219,220]. The use of biomaterial delivery vehicles and excipients are currently being explored as means to stabilize biologic drugs and vaccines [221], and exploring these strategies offers a critical and seemingly underexplored opportunity to increase the effective and equitable distribution of any vaccine which may be approved.
4.2. Alternative antigens
For most vaccine candidates currently in clinical trials, S-protein or regions of S-protein are the main target antigen. This can be attributed to the fact that all potently-neutralizing antibodies recovered from patient convalescent sera have targeted parts of the spike, whether the RBD or elsewhere [222,223]. However, for future vaccine development, there are other potential targets for protection (Fig. 5 ). For example, a microarray screen of patient convalescent plasma for antibodies against various SARS-CoV-2 proteins exhibited antibodies specific for the N, ORF9b, and NSP5 proteins in addition to S [224]. Although antibodies against these proteins may be non-neutralizing, it is known that non-neutralizing antibodies can play a role in control of infection against diverse viral pathogens including influenza [[225], [226], [227]], HIV [228,229], Ebola [230], and Marburg [231] through mechanisms including cooperativity and antibody-dependent cellular cytotoxicity, although they also bear the risk of inducing ADE as detailed above.
Fig. 5.
Structure and proteins of SARS-CoV-2.
An extensive investigation of adaptive immune response to COVID-19 infection in humans by Moderbacher et al. demonstrated that prevention of disease generally correlates well with neutralizing antibody levels, while resolution of disease generally correlates well with CD4 T cell response rather than neutralizing antibody titers [232]. Indeed, following infection, there is an association of higher titers with worse disease [233,234]. Thus, it could be important for a vaccine designed to prevent disease to induce a strong CD4 T cell response, especially in the case that vaccine approval is only contingent on 50% reduction in disease incidence [235,236]. Kiyotani et al. [237] screened in silico for epitopes that could bind to common HLA present in a Japanese population. They determined that peptides derived from the M-protein had high potential for presentation on HLA class I and class II. Grifoni et al. [238] showed that patients with COVID-19 had a high level of CD4 T cells that were specific for the S-protein, which correlated well with IgG and IgA antibodies against SARS-CoV-2. Additionally, infected patients mounted CD4 T cell responses to the M, N, NSP3, NSP4, ORF3a, and ORF8, while for CD8 T-cells, S-protein and M-protein were targets for humans. Surprisingly, Grifoni et al. showed that 40-60% of unexposed individuals have CD4 T-cells that are reactive to SARS-CoV-2 epitopes. A subsequent study revealed that a substantial proportion of these pre-existing reactive T cells were memory T cells with cross-reactivity to common cold coronaviruses (HCoV)-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1. Others have shown that the E, Replicate polyprotein 1ab, Protein 3a, Non-structural Protein 3b, Protein 7a, and Protein 9b could be potential epitopes for T cell stimulation [239,240]. The important role of CD4 T cell-mediated immunity in reducing disease severity, and the large number of T cell epitopes present in proteins besides S in SARS-CoV-2, suggest inclusion of these proteins in future vaccine formulations may confer a protective benefit.
An additional consideration in the design of emerging vaccine platforms is the use of adjuvants. A Th1 response with minimal non-neutralizing antibodies are known to mitigate immunotoxicity. PAMP-based adjuvants can promote a strong Th1 response, especially in contrast to Th2 adjuvants like alum, MF59 and AS03 that would fall short. Many of the emerging technologies are non-adjuvanted, which will likely require multiple boosts to achieve protective immunity. In considering the viral vectors that cannot be easily used multiple times, an adjuvanted subunit vaccine may be an alternative to include as the prime or boost. A similar approach has been seen previously with a non-adjuvanted DNA prime and adenovirus boost as a HIV vaccine (HVTN 505).
4.3. Alternative routes of vaccination
Another area where formulations can address major logistical problems is in the need for sterility and trained medical professionals in the administration of vaccines. Many of the formulations mentioned in this review require either intramuscular or intradermal injection using sterile equipment by a trained professional. Especially given the need for both a prime and a boost injection for most formulations, this constitutes a massive logistical burden to achieve the amount of coverage to achieve herd immunity. Formulations using alternative delivery routes may provide some relief from this burden. This includes orally administered vaccines such as the aforementioned vaccine in development by Vaxart or the intranasal Merck/IAVI VSV vaccine. Another potential solution to this problem could come in the form of microneedle patches. These patches have been evaluated preclinically and clinically, where they have been shown to stabilize antigen and adjuvant components while safely generating robust immune responses, while having the major advantages of being small, relatively thermostable, and potentially allowing for self-administration [[241], [242], [243]]. A microneedle formulation for SARS-CoV-2 vaccination has even been evaluated preclinically by Kim et al. [244], and is proceeding towards clinical evaluation.
In the case of pathogens which infect through a mucosal surface, sterilizing immunity is often associated with mucosal immune responses. It is important to consider the distinction between induction of sterilizing and nonsterilizing immunity. A sterilizing immune response will inhibit viral infection and replication within the host, while a non-sterilizing immune response will permit infection in the host, while still potentially preventing disease. A host with nonsterilizing immunity may become infected by a pathogen and spread it to others despite being asymptomatic. Given the substantial role of asymptomatic carriers in the spread of SARS-CoV-2, induction of sterilizing immunity by a vaccine would be a highly desirable characteristic because it could limit viral transmission in addition to directly preventing disease.
For a respiratory pathogen like SARS-CoV-2, induction of high levels of secretory IgA is considered to have the strongest potential for inducing sterilizing immunity, as this class of antibody is considered to play the greatest role in protection of the upper respiratory tract where viral laden droplets are most likely to make first contact. In contrast, the lower respiratory tract has a greater proportion of IgG. While natural infection with SARS-CoV-2 has been shown to induce both secretory IgA and IgG, vaccination by injection, especially intramuscular, is often ineffective in inducing secretory IgA. Given that most of the clinical trials mentioned above use intramuscular or intradermal injection, it is possible that sterilizing immunity will not be achieved by many candidates.
For vaccines against respiratory pathogens, the most common method for inducing a mucosal response is through intranasal vaccine administration. For example, Flumist® is an attenuated influenza virus vaccine licensed by the FDA for intranasal administration. Intranasally-administered subunit vaccine formulations can also be used to induce a robust mucosal response, and a number of adjuvants have been developed to increase their immunogenicity [245,246]. There is thus a compelling case for development of intranasal vaccine formulations against SARS-CoV-2 [247].
Indeed, a recent study by Hassan et al. demonstrated that a prime-only intranasal immunization with a chimpanzee adenovirus encoding the 2P-stabilized S protein could protect against disease and induce significant S- and RBD-specific levels of serum IgA in an hACE-2 receptor transgenic mouse model. They observed neither infectious SARS-CoV-2 virus nor viral RNA in the lungs of the mice four days after viral challenge, but still observed some viral RNA in the nasal wash and turbinates. This RNA could have been from the original challenge dose rather than viral replication. They investigated anti-N IgM and IgG serum titers eight days after viral challenge, finding no significant difference in titers against this antigen, which is not encoded for by the vaccine vector. This experiment was meant to demonstrate that there was no viral replication and thus no generation of NP against which a significant humoral response could be raised, although it still does not provide definitive proof of sterilizing immunity.
While injected vaccines are conventionally considered incapable of generating a mucosal response [248], there is some evidence for induction of mucosal immunity to vaccines injected via diverse routes at nonmucosal surfaces. This has occurred using specific adjuvant compounds or unconventional routes of administration [[249], [250], [251], [252]]. As intranasal immunization can result in reduced levels of systemic antibody induction relative to an intramuscular injection [253], use of a one of these methods to induce a combination of robust mucosal and systemic immunity through intramuscular injection merits further investigation.
4.4. What success may mean for A SARS-Cov-2 vaccine
Current clinical trial designs and recommendations from the WHO assess vaccine efficacy as the prevention of disease caused by infection of SARS-CoV-2, rather than infection itself (sterilizing immunity). Thus, these trials will not thoroughly assess the capacity of each vaccine candidate to induce sterilizing immunity or prevent disease transmission. It is thus possible that, even with an effective vaccine, we will not achieve herd immunity, and instead COVID-19 will become a recurrent or seasonal disease [254,255]. While there are practical limitations on the direct assessment of sterilizing immunity during trials involving community transmission, quantification of S-protein specific and/or virus-neutralizing IgA in serum and/or nasal midturbinates should be feasible and could provide more insight into the potential of each vaccine to limit viral spread [256]. Other strategies involving consistent monitoring of viral load and in-depth contact and cluster tracing may also give some indication of successful prevention of infection and transmission [257]. It was recently announced that the United Kingdom company Open Orphan/hVivo will be initiating human challenge trials with SARS-CoV-2, while the governments of Belgium and the United States have also allocated funding for similar trials. While the ethics and value of such trials are controversial, human challenge trials could provide the potential to evaluate the induction, duration, and correlates of sterilizing immunity against SARS-CoV-2 in humans by administering a known dose of infectious virus at a defined time [258]. Similar insights could also be achieved at a lesser risk to trial subjects through development of a dosing regimen or attenuated virus which does not cause disease [259].
While comprehensively addressing the details of clinical trial design is beyond the scope of this review, it is critical that Phase 3 trials for each of these vaccines are undertaken in a manner to provide the greatest protection to humanity. Current FDA and WHO guidelines set the necessary protective efficacy as 50%, with a recommendation that this be defined as protection against symptomatic or severe COVID-19 [235,236]. While this endpoint has been reported by several vaccine candidates [128,139,140,177], it will still be important to evaluate candidates for more robust endpoints, including duration of immunity, potential for sterilizing immunity, and efficacy in pediatric, older, or immunocompromised individuals. The capacity to achieve these more demanding endpoints may be best assessed through trials which directly compare different candidates, which are being organized by organizations like the WHO [260]. It is also critical to ensure the recruitment of racial and ethnic minority populations in trial recruitment, as these populations have been disproportionately affected by COVID-19 [[261], [262], [263]]. The Moderna Phase 3 Trial highlights diversity can be achieved in the study’s participants and greater patient diversity across all trials should be sought [129]. Demonstration of efficacy in Phase 3, resulting in approval for distribution, could have deleterious effects on further evaluation of each candidate by slowing recruitment to other efficacy trials. It could also result in unblinding of the trial as the placebo participants could be justified to receive the treatment after it is proven efficacious. This would eliminate the a control group required for effective long-term efficacy and safety monitoring, and so must be carefully considered [264].
5. Conclusions
Overall, a wide variety of approaches are being taken to rapidly develop a vaccine against SARS-CoV-2. Due to the need to simply know the sequence of the desired antigen to begin manufacturing, viral and non-viral nucleic acid vaccines seem to be the quickest out of the gate. These plug-and-play platforms are advancing rapidly through clinical trials. Close behind are more traditional inactivated and subunit vaccines, followed by emerging technologies that apply cells or bacteria to generate potential protective responses. Spike- and RBD-binding, virus-neutralizing antibodies have been successfully raised using a myriad of approaches, and these antibodies have correlated strongly with protection from infection in multiple animal models. In most cases where it has been evaluated, vaccine candidates have been successful in inducing a Th1-skewed T cell response. While the determinants of immune pathology in COVID-19 have not been definitively determined, avoiding a Th2 skew has some theoretical basis for reducing immune pathology.
The overall effect of COVID-19 vaccine development has been a massive invigoration of the field of pandemic vaccine development. It has made real the theoretical promise of platforms which only require an antigen sequence, such as mRNA and vector-based platforms, and massively accelerated their development towards rapid Phase 3 evaluation, on a timeline never seen before for vaccines. However, it is important to note that, despite their rapid manufacturing timeline, these platforms encode for an antigen which was developed over a timeline of many years through basic research on coronavirus biology and protein engineering. Large scale investment and unprecedented mobilization of the research community have generated insight into design, manufacturing, formulation, and deployment of vaccine candidates that may pay dividends in the future when society will need to confront the next inevitable infectious disease outbreak.
Acknowledgements
This work has been funded in part by North Carolina Policy Collaboratory, and NIH NIAID R01AI147497. Images were in part generated by BioRender.
References
- 1.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramadan N., Shaib H. Middle East respiratory syndrome coronavirus (MERS-CoV): a review. Germs. 2019;9:35–42. doi: 10.18683/germs.2019.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnelly M.M., Elkholy A., Cauchemez S., Van Kerkhove M.D. Worldwide Reduction in MERS Cases and Deaths since 2016. Emerg. Infect. Dis. 2019;25:1758. doi: 10.3201/eid2509.190143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC . DHS. 2020. Cases in the U.S. [Google Scholar]
- 10.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin D.O., Jensen A., Khan M., Chin J., Chin K., Saad J., Parnell R., Awwad C., Patel D. Pulmonary embolism and increased levels of d-dimer in patients with coronavirus disease. Emerg. Infect. Dis. 2020;26:1941. doi: 10.3201/eid2608.201477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin L., Lu L., Cao W., Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection–a review of immune changes in patients with viral pneumonia. Emer. Microbes & Infect. 2020;9:727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martines R.B., Ritter J.M., Matkovic E., Gary J., Bollweg B.C., Bullock H., Goldsmith C.S., Silva-Flannery L., Seixas J.N., Reagan-Steiner S. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:2005. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bale B.F., Doneen A.L., Vigerust D.J. Microvascular disease confers additional risk to COVID-19 infection. Med. Hypotheses. 2020;144:109999. doi: 10.1016/j.mehy.2020.109999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau E.H., Hsiung C.A., Cowling B.J., Chen C.H., Ho L.M., Tsang T., Chang C.W., Donnelly C.A., Leung G.M. A comparative epidemiologic analysis of SARS in Hong Kong, Beijing and Taiwan. BMC Infect. Dis. 2010;10:50. doi: 10.1186/1471-2334-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kai H., Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19. Hypertens. Res. 2020;43(7):648–654. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y., Zhou Y., Siddiqui P., Jiang S. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 2004;325:445–452. doi: 10.1016/j.bbrc.2004.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaume M., Yip M.S., Kam Y.W., Cheung C.Y., Kien F., Roberts A., Li P.H., Dutry I., Escriou N., Daeron M., Bruzzone R., Subbarao K., Peiris J.S., Nal B., Altmeyer R. SARS CoV subunit vaccine: antibody-mediated neutralisation and enhancement. Hong Kong Med J. 2012;18(Suppl. 2):31–36. [PubMed] [Google Scholar]
- 21.Zeng F., Chow K.Y., Hon C.C., Law K.M., Yip C.W., Chan K.H., Peiris J.S., Leung F.C. Characterization of humoral responses in mice immunized with plasmid DNAs encoding SARS-CoV spike gene fragments. Biochem. Biophys. Res. Commun. 2004;315:1134–1139. doi: 10.1016/j.bbrc.2004.01.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R.Y., Wu L.Z., Huang B.J., Huang J.L., Zhang Y.L., Ke M.L., Wang J.M., Tan W.P., Zhang R.H., Chen H.K., Zeng Y.X., Huang W. Adenoviral expression of a truncated S1 subunit of SARS-CoV spike protein results in specific humoral immune responses against SARS-CoV in rats. Virus Res. 2005;112:24–31. doi: 10.1016/j.virusres.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roper R.L., Rehm K.E. SARS vaccines: where are we? Expert Rev Vacc. 2009;8:887–898. doi: 10.1586/erv.09.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y.J.C. 2020. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C.E., Weimer E., Scherer E.M., Rouphael N., Edupuganti S., Weiskopf D., Tse L.V., Hou Y.J., Margolis D., Sette A., Collins M.H., Schmitz J., Baric R.S., de Silva A.M. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., Wu T., Cheung K.W., Chan K.H., Alvarez X., Qin C., Lackner A., Perlman S., Yuen K.Y., Chen Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., Suthar M., Harkema J., Whitmore A., Pickles R., West A., Donaldson E., Curtis K., Johnston R., Baric R. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China, Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.08.025. 3652322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman L.M., Furberg C.D., DeMets D.L., Reboussin D.M., Granger C.B. Springer; 2015. Fundamentals of clinical trials. [Google Scholar]
- 39.Mahan V.L. Clinical trial phases. Int. J. Clin. Med. 2014;5:1374. [Google Scholar]
- 40.WHO . 2020. Draft Landscape of COVID-19 Candidate Vaccines. [Google Scholar]
- 41.FDA . 2020. Vaccines Licensed for Use in the United States. [Google Scholar]
- 42.Kon T.C., Onu A., Berbecila L., Lupulescu E., Ghiorgisor A., Kersten G.F., Cui Y.-Q., Amorij J.-P., Van der Pol L. Influenza vaccine manufacturing: effect of inactivation, splitting and site of manufacturing. comparison of influenza vaccine production processes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0150700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthews J.T. Egg-based production of influenza vaccine: 30 years of commercial experience. Bridge-Washington-National Academy Of Engineering. 2006;36:17. [Google Scholar]
- 44.Neuzil K.M., Bright R.A. Influenza vaccine manufacture: keeping up with change. J. Infect. Dis. 2009;200:835–837. doi: 10.1086/605507. [DOI] [PubMed] [Google Scholar]
- 45.Stoel M., Pool J., de Vries-Idema J., Zaaraoui-Boutahar F., Bijl M., Andeweg A.C., Wilschut J., Huckriede A. Innate responses induced by whole inactivated virus or subunit influenza vaccines in cultured dendritic cells correlate with immune responses in vivo. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., Liang H., Bao L., Xu Y., Ding L., Zhou W., Gao H., Liu J., Niu P., Zhao L., Zhen W., Fu H., Yu S., Zhang Z., Xu G., Li C., Lou Z., Xu M., Qin C., Wu G., Gao G.F., Tan W., Yang X. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182(3):713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Govorkova E.A., Murti G., Meignier B., De Taisne C., Webster R.G. African green monkey kidney (Vero) cells provide an alternative host cell system for influenza A and B viruses. J. Virol. 1996;70:5519–5524. doi: 10.1128/jvi.70.8.5519-5524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia S., Duan K., Zhang Y., Zhao D., Zhang H., Xie Z., Li X., Peng C., Zhang Y., Zhang W., Yang Y., Chen W., Gao X., You W., Wang X., Wang Z., Shi Z., Wang Y., Yang X., Zhang L., Huang L., Wang Q., Lu J., Yang Y., Guo J., Zhou W., Wan X., Wu C., Wang W., Huang S., Du J., Meng Z., Pan A., Yuan Z., Shen S., Guo W., Yang X. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. JAMA. 2020;324(10):951–960. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C. Rapid development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y.-J., Zeng G., Pan H.-X., Li C.-G., Kan B., Hu Y.-L., Mao H.-Y., Xin Q.-Q., Chu K., Han W.-X. Immunogenicity and safety of a SARS-CoV-2 inactivated vaccine in healthy adults aged 18-59 years: report of the randomized, double-blind, and placebo-controlled phase 2 clinical trial. MedRxiv. 2020 doi: 10.1101/2020.07.31.20161216. [DOI] [Google Scholar]
- 51.Palacios R., Patiño E.G., de Oliveira Piorelli R., Conde M.T.R.P., Batista A.P., Zeng G., Xin Q., Kallas E.G., Flores J., Ockenhouse C.F. Double-blind, randomized, placebo-controlled phase III clinical trial to evaluate the efficacy and safety of treating healthcare professionals with the adsorbed COVID-19 (inactivated) vaccine manufactured by Sinovac–PROFISCOV: a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21:1–3. doi: 10.1186/s13063-020-04775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.INOVIO . 2020. INOVIO and IVI Partner with Seoul National University Hospital to Start Phase 1/2 Clinical Trial of INOVIO's COVID-19 DNA Vaccine (INO-4800) in South Korea. [Google Scholar]
- 53.Technology P. 2020. Inovio Reports Promising Data from its Covid-19 Vaccine Studies. [Google Scholar]
- 54.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Rohss J., John S., Hassett K., Yuzhakov O., Bahl K., Brito L.A., Salter H., Ciaramella G., Lore K. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in Rhesus Macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park A. Inside the company that's hot-wiring vaccine research in the race to combat the coronavirus. Time. 2020 [Google Scholar]
- 56.NIAID . 2020. NIH Clinical Trial of Investigational Vaccine for COVID-19 Begins. [Google Scholar]
- 57.Corbett K.S., Edwards D., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., Dinnon K.H., Elbashir S.M., Shaw C.A., Woods A., Fritch E.J., Martinez D.R., Bock K.W., Minai M., Nagata B.M., Hutchinson G.B., Bahl K., Garcia-Dominguez D., Ma L., Renzi I., Kong W.-P., Schmidt S.D., Wang L., Zhang Y., Stevens L.J., Phung E., Chang L.A., Loomis R.J., Altaras N.E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M.K., Shi W., Leung K., Yang E.S., West A., Gully K.L., Wang N., Wrapp D., Doria-Rose N.A., Stewart-Jones G., Bennett H., Nason M.C., Ruckwardt T.J., McLellan J.S., Denison M.R., Chappell J.D., Moore I.N., Morabito K.M., Mascola J.R., Baric R.S., Carfi A., Graham B.S. SARS-CoV-2 mRNA vaccine development enabled by prototype pathogen preparedness. bioRxiv. 2020 doi: 10.1038/s41586-020-2622-0. 2020.2006.2011.145920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., Fotin-Mleczek M., Hoerr I., Clemens R., von Sonnenburg F. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 59.Novavax I. 2020. Novavax Identifies Coronavirus Vaccine Candidate; Accelerates Initiation of First-in-Human Trial to Mid-May. [Google Scholar]
- 60.Healthcare G. Novavax’s vaccines limited; 2020. Clover’s Adjuvant Choice for Covid-19 an Edge, but Comparison with IMV’s. [Google Scholar]
- 61.Arena C.T. 2020. Clover Biopharmaceuticals Starts Phase I Covid-19 Vaccine Trial. [Google Scholar]
- 62.Medicago . 2020. Medicago: Pipeline. [Google Scholar]
- 63.CDC . 2018. Adjuvants Help Vaccines Work Better. [Google Scholar]
- 64.Liu H., Su D., Zhang J., Ge S., Li Y., Wang F., Gravel M., Roulston A., Song Q., Xu W., Liang J.G., Shore G., Wang X., Liang P. Scientific Reports (Nature Publisher Group) Vol. 7. 2017. Improvement of pharmacokinetic profile of TRAIL via trimer-tag enhances its antitumor activity; pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.K.J. Chappell, D. Watterson, P.R. Young, Chimeric molecules and uses thereof, Google Patents, 2020.
- 66.U.o. Queensland’s . 2020. UQ Vaccine Scientists Report Positive Results From Pre-Clinical Testing. [Google Scholar]
- 67.Hayashi M., Aoshi T., Haseda Y., Kobiyama K., Wijaya E., Nakatsu N., Igarashi Y., Standley D.M., Yamada H., Honda-Okubo Y. Advax, a delta inulin microparticle, potentiates in-built adjuvant property of co-administered vaccines. EBioMedicine. 2017;15:127–136. doi: 10.1016/j.ebiom.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cooper P.D., Petrovsky N. Delta inulin: a novel, immunologically active, stable packing structure comprising β-d-[2→ 1] poly (fructo-furanosyl) α-d-glucose polymers. Glycobiology. 2011;21:595–606. doi: 10.1093/glycob/cwq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.HogenEsch H., O’Hagan D.T., Fox C.B. Optimizing the utilization of aluminum adjuvants in vaccines: you might just get what you want. npj Vaccines. 2018;3:51. doi: 10.1038/s41541-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hotez P.J., Corry D.B., Strych U., Bottazzi M.E. COVID-19 vaccines: neutralizing antibodies and the alum advantage. Nat. Rev. Immunol. 2020;20:399–400. doi: 10.1038/s41577-020-0358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garçon N., Vaughn D.W., Didierlaurent A.M. Development and evaluation of AS03, an Adjuvant System containing α-tocopherol and squalene in an oil-in-water emulsion. Expert Rev. Vacc. 2012;11:349–366. doi: 10.1586/erv.11.192. [DOI] [PubMed] [Google Scholar]
- 72.Campbell J.D. Springer; 2017. Development of the CpG Adjuvant 1018: A Case Study, Vaccine Adjuvants; pp. 15–27. [DOI] [PubMed] [Google Scholar]
- 73.Swaminathan G., Thoryk E.A., Cox K.S., Meschino S., Dubey S.A., Vora K.A., Celano R., Gindy M., Casimiro D.R., Bett A.J. A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccine. 2016;34:110–119. doi: 10.1016/j.vaccine.2015.10.132. [DOI] [PubMed] [Google Scholar]
- 74.Awasthi S., Hook L.M., Swaminathan G., Cairns T.M., Brooks B., Smith J.S., Ditto N.T., Gindy M.E., Bett A.J., Espeseth A.S. Antibody responses to crucial functional epitopes as a novel approach to assess immunogenicity of vaccine adjuvants. Vaccine. 2019;37:3770–3778. doi: 10.1016/j.vaccine.2019.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lovgren K., Morein B. The requirement of lipids for the formation of immunostimulating complexes (iscoms) Biotechnol. Appl. Biochem. 1988;10:161–172. [PubMed] [Google Scholar]
- 76.Reimer J.M., Karlsson K.H., Lövgren-Bengtsson K., Magnusson S.E., Fuentes A., Stertman L. Matrix-M™ adjuvant induces local recruitment, activation and maturation of central immune cells in absence of antigen. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bengtsson K.L., Karlsson K.H., Magnusson S.E., Reimer J.M., Stertman L. Matrix-M™ adjuvant: enhancing immune responses by ‘setting the stage’for the antigen. Expert Rev. Vacc. 2013;12:821–823. doi: 10.1586/14760584.2013.814822. [DOI] [PubMed] [Google Scholar]
- 78.O’Hagan D., Ott G.S., De Gregorio E., Seubert A. The mechanism of action of MF59–an innately attractive adjuvant formulation. Vaccine. 2012;30:4341–4348. doi: 10.1016/j.vaccine.2011.09.061. [DOI] [PubMed] [Google Scholar]
- 79.Tritto E., Mosca F., De Gregorio E. Mechanism of action of licensed vaccine adjuvants. Vaccine. 2009;27:3331–3334. doi: 10.1016/j.vaccine.2009.01.084. [DOI] [PubMed] [Google Scholar]
- 80.Kerwin B.A. Polysorbates 20 and 80 used in the formulation of protein biotherapeutics: structure and degradation pathways. J. Pharm. Sci. 2008;97:2924–2935. doi: 10.1002/jps.21190. [DOI] [PubMed] [Google Scholar]
- 81.Bangaru S., Ozorowski G., Turner H.L., Antanasijevic A., Huang D., Wang X., Torres J.L., Diedrich J.K., Tian J.-H., Portnoff A.D.J.S. Vol. 370. 2020. Structural Analysis of Full-Length SARS-CoV-2 Spike Protein from an Advanced Vaccine Candidate; pp. 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freund I., Eigenbrod T., Helm M., Dalpke A.H. RNA modifications modulate activation of innate toll-like receptors. Genes. 2019;10:92. doi: 10.3390/genes10020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., Yang M., Li Y., Cheng H., Yuan Y., Zhang W., Ke C., Wong G., Qi J., Qin C., Yan J., Gao G.F. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733. doi: 10.1016/j.cell.2020.06.035. e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petroski N. Advax adjuvant: a potent and safe immunopotentiator composed of delta inulin. Immunopoten. Modern Vacc. 2017:199–210. [Google Scholar]
- 85.Honda-Okubo Y., Barnard D., Ong C.H., Peng B.-H., Tseng C.-T.K., Petrovsky N. Severe acute respiratory syndrome-associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology. J. Virol. 2015;89:2995–3007. doi: 10.1128/JVI.02980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.P., Andres E.L., Kettenbach A.N., Denison M.R., Chappell J.D., Graham B.S., Ward A.B., McLellan J.S. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuo T.-Y., Lin M.-Y., Coffman R.L., Campbell J.D., Traquina P., Lin Y.-J., Liu L.T.-C., Cheng J., Wu Y.-C., Wu C.-C., Tang W.-H., Huang C.-G., Tsao K.-C., Shih S.-R., Chen C. Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. bioRxiv. 2020 doi: 10.1038/s41598-020-77077-z. 2020.2008.2011.245704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simons K., Helenius A., Leonard K., Sarvas M., Gething M.J. Formation of protein micelles from amphiphilic membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 1978;75:5306–5310. doi: 10.1073/pnas.75.11.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coleman C.M., Liu Y.V., Mu H., Taylor J.K., Massare M., Flyer D.C., Smith G.E., Frieman M.B. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32:3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Magnusson S.E., Altenburg A.F., Bengtsson K.L., Bosman F., de Vries R.D., Rimmelzwaan G.F., Stertman L. Matrix-M adjuvant enhances immunogenicity of both protein- and modified vaccinia virus Ankara-based influenza vaccines in mice. Immunol. Res. 2018;66:224–233. doi: 10.1007/s12026-018-8991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cox R.J., Pedersen G., Madhun A.S., Svindland S., Saevik M., Breakwell L., Hoschler K., Willemsen M., Campitelli L., Nostbakken J.K., Weverling G.J., Klap J., McCullough K.C., Zambon M., Kompier R., Sjursen H. Evaluation of a virosomal H5N1 vaccine formulated with Matrix M adjuvant in a phase I clinical trial. Vaccine. 2011;29:8049–8059. doi: 10.1016/j.vaccine.2011.08.042. [DOI] [PubMed] [Google Scholar]
- 93.Tian J.-H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., Lague J., Portnoff A.D., Norton J., Guebre-Xabier M., Zhou B., Jacobson K., Maciejewski S., Khatoon R., Wisniewska M., Moffitt W., Kluepfel-Stahl S., Ekechukwu B., Papin J., Boddapati S., Wong C.J., Piedra P.A., Frieman M.B., Massare M.J., Fries L., Bengtsson K. Lövgren, Stertman L., Ellingsworth L., Glenn G., Smith G. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 elicits immunogenicity in baboons and protection in mice. bioRxiv. 2020 doi: 10.1038/s41467-020-20653-8. 2020.2006.2029.178509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kumru O.S., Joshi S.B., Smith D.E., Middaugh C.R., Prusik T., Volkin D.B. Vaccine instability in the cold chain: mechanisms, analysis and formulation strategies. Biologicals. 2014;42:237–259. doi: 10.1016/j.biologicals.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 96.Schofield T.L. Vaccine stability study design and analysis to support product licensure. Biologicals. 2009;37:387–396. doi: 10.1016/j.biologicals.2009.08.009. discussion 421-383. [DOI] [PubMed] [Google Scholar]
- 97.Guebre-Xabier M., Patel N., Tian J.-H., Zhou B., Maciejewski S., Lam K., Portnoff A.D., Massare M.J., Frieman M.B., Piedra P.A., Ellingsworth L., Glenn G., Smith G. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. bioRxiv. 2020 doi: 10.1016/j.vaccine.2020.10.064. 2020.2008.2018.256578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J., McGrath M., Weston S., Piedra P.A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J.D., Griffin P., Wilkinson B., Glenn G.M. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marsian J., Lomonossoff G.P. Molecular pharming - VLPs made in plants. Curr. Opin. Biotechnol. 2016;37:201–206. doi: 10.1016/j.copbio.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 100.Bachmann M.F., Jennings G.T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 101.D'Aoust M.A., Lavoie P.O., Couture M.M., Trepanier S., Guay J.M., Dargis M., Mongrand S., Landry N., Ward B.J., Vezina L.P. Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 2008;6:930–940. doi: 10.1111/j.1467-7652.2008.00384.x. [DOI] [PubMed] [Google Scholar]
- 102.Bally J., Jung H., Mortimer C., Naim F., Philips J.G., Hellens R., Bombarely A., Goodin M.M., Waterhouse P.M. The rise and rise of nicotiana benthamiana: a plant for all reasons. Annu. Rev. Phytopathol. 2018;56:405–426. doi: 10.1146/annurev-phyto-080417-050141. [DOI] [PubMed] [Google Scholar]
- 103.Pillet S., Couillard J., Trepanier S., Poulin J.F., Yassine-Diab B., Guy B., Ward B.J., Landry N. Immunogenicity and safety of a quadrivalent plant-derived virus like particle influenza vaccine candidate-Two randomized Phase II clinical trials in 18 to 49 and >/=50 years old adults. PLoS One. 2019;14 doi: 10.1371/journal.pone.0216533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ingolotti M., Kawalekar O., Shedlock D.J., Muthumani K., Weiner D.B. DNA vaccines for targeting bacterial infections. Expert Rev. Vacc. 2010;9:747–763. doi: 10.1586/erv.10.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ulmer J.B., Mason P.W., Geall A., Mandl C.W. RNA-based vaccines. Vaccine. 2012;30:4414–4418. doi: 10.1016/j.vaccine.2012.04.060. [DOI] [PubMed] [Google Scholar]
- 106.Gary D.J., Min J., Kim Y., Park K., Won Y.-Y. The effect of N/P ratio on the in vitro and in vivo interaction properties of PEGylated poly[2-(dimethylamino)ethyl methacrylate]-based siRNA complexes. Macromol. Biosci. 2013;13:1059–1071. doi: 10.1002/mabi.201300046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sakae M., Ito T., Yoshihara C., Iida-Tanaka N., Yanagie H., Eriguchi M., Koyama Y. Highly efficient in vivo gene transfection by plasmid/PEI complexes coated by anionic PEG derivatives bearing carboxyl groups and RGD peptide. Biomed. Pharmacother. 2008;62:448–453. doi: 10.1016/j.biopha.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 108.Dupuis M., Denis-Mize K., Woo C., Goldbeck C., Selby M.J., Chen M., Otten G.R., Ulmer J.B., Donnelly J.J., Ott G., McDonald D.M. Distribution of DNA vaccines determines their immunogenicity after intramuscular injection in mice. J. Immunol. 2000;165:2850–2858. doi: 10.4049/jimmunol.165.5.2850. [DOI] [PubMed] [Google Scholar]
- 109.Babiuk S., Baca-Estrada M.E., Foldvari M., Middleton D.M., Rabussay D., Widera G., Babiuk L.A. Increased gene expression and inflammatory cell infiltration caused by electroporation are both important for improving the efficacy of DNA vaccines. J. Biotechnol. 2004;110:1–10. doi: 10.1016/j.jbiotec.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 110.Modjarrad K., Roberts C.C., Mills K.T., Castellano A.R., Paolino K., Muthumani K., Reuschel E.L., Robb M.L., Racine T., Oh M.D., Lamarre C., Zaidi F.I., Boyer J., Kudchodkar S.B., Jeong M., Darden J.M., Park Y.K., Scott P.T., Remigio C., Parikh A.P., Wise M.C., Patel A., Duperret E.K., Kim K.Y., Choi H., White S., Bagarazzi M., May J.M., Kane D., Lee H., Kobinger G., Michael N.L., Weiner D.B., Thomas S.J., Maslow J.N. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Diehl M.C., Lee J.C., Daniels S.E., Tebas P., Khan A.S., Giffear M., Sardesai N.Y., Bagarazzi M.L. Tolerability of intramuscular and intradermal delivery by CELLECTRA((R)) adaptive constant current electroporation device in healthy volunteers. Hum Vaccin Immunother. 2013;9:2246–2252. doi: 10.4161/hv.24702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., Xu Z., Walters J., Bhojnagarwala P., Yang M., Chokkalingam N., Pezzoli P., Parzych E., Reuschel E.L., Doan A., Tursi N., Vasquez M., Choi J., Tello-Ruiz E., Maricic I., Bah M.A., Wu Y., Amante D., Park D.H., Dia Y., Ali A.R., Zaidi F.I., Generotti A., Kim K.Y., Herring T.A., Reeder S., Andrade V.M., Buttigieg K., Zhao G., Wu J.M., Li D., Bao L., Liu J., Deng W., Qin C., Brown A.S., Khoshnejad M., Wang N., Chu J., Wrapp D., McLellan J.S., Muthumani K., Wang B., Carroll M.W., Kim J.J., Boyer J., Kulp D.W., Humeau L., Weiner D.B., Broderick K.E. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Patel A., Walters J., Reuschel E.L., Schultheis K., Parzych E., Gary E.N., Maricic I., Purwar M., Eblimit Z., Walker S.N., Guimet D., Bhojnagarwala P., Doan A., Xu Z., Elwood D., Reeder S.M., Pessaint L., Kim K.Y., Cook A., Chokkalingam N., Finneyfrock B., Tello-Ruiz E., Dodson A., Choi J., Generotti A., Harrison J., Tursi N.J., Andrade V.M., Dia Y., Zaidi F.I., Andersen H., Lewis M.G., Muthumani K., Kim J.J., Kulp D.W., Humeau L.M., Ramos S., Smith T.R.F., Weiner D.B., Broderick K.E. Intradermal-delivered DNA vaccine provides anamnestic protection in a rhesus macaque SARS-CoV-2 challenge model. bioRxiv. 2020 doi: 10.1016/j.xcrm.2021.100420. 2020.2007.2028.225649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.L.G. Ho D . 2020. South Korea’s Genexine Begins Phase I/IIa Trials for COVID-19 Vaccine. [Google Scholar]
- 115.Kim T.J., Jin H.T., Hur S.Y., Yang H.G., Seo Y.B., Hong S.R., Lee C.W., Kim S., Woo J.W., Park K.S., Hwang Y.Y., Park J., Lee I.H., Lim K.T., Lee K.H., Jeong M.S., Surh C.D., Suh Y.S., Park J.S., Sung Y.C. Clearance of persistent HPV infection and cervical lesion by therapeutic DNA vaccine in CIN3 patients. Nat. Commun. 2014;5:5317. doi: 10.1038/ncomms6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Choi Y.J., Hur S.Y., Kim T.J., Hong S.R., Lee J.K., Cho C.H., Park K.S., Woo J.W., Sung Y.C., Suh Y.S., Park J.S. A phase II, prospective, randomized, multicenter, open-label study of GX-188E, an HPV DNA vaccine, in patients with cervical intraepithelial neoplasia 3. Clin. Cancer Res. 2020;26:1616–1623. doi: 10.1158/1078-0432.CCR-19-1513. [DOI] [PubMed] [Google Scholar]
- 117.Kreiter S., Diken M., Selmi A., Diekmann J., Attig S., Husemann Y., Koslowski M., Huber C., Tureci O., Sahin U. FLT3 ligand enhances the cancer therapeutic potency of naked RNA vaccines. Cancer Res. 2011;71:6132–6142. doi: 10.1158/0008-5472.CAN-11-0291. [DOI] [PubMed] [Google Scholar]
- 118.Seo Y.B., Suh Y.S., Ryu J.I., Jang H., Oh H., Koo B.-S., Seo S.-H., Hong J.J., Song M., Kim S.-J. Soluble Spike DNA vaccine provides long-term protective immunity against SAR-CoV-2 in mice and nonhuman primates. bioRxiv. 2020 doi: 10.3390/vaccines9040307. bioRxiv 2020.10.09.334136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miyazaki H., Atobe S., Suzuki T., Iga H., Terai K. Development of pyro-drive jet injector with controllable jet pressure. J. Pharm. Sci. 2019;108:2415–2420. doi: 10.1016/j.xphs.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 120.Chang C., Sun J., Hayashi H., Suzuki A., Sakaguchi Y., Miyazaki H., Nishikawa T., Nakagami H., Yamashita K., Kaneda Y. Stable immune response induced by intradermal DNA vaccination by a novel needleless pyro-drive jet injector. AAPS PharmSciTech. 2019;21:19. doi: 10.1208/s12249-019-1564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sanofi . 2020. Sanofi and Translate Bio Expand Collaboration to Develop Mrna Vaccines Across all Infectious Disease Areas. [Google Scholar]
- 122.Patel A.K., Kaczmarek J.C., Bose S., Kauffman K.J., Mir F., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater. 2019;31 doi: 10.1002/adma.201805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Y., Li Y., Keskin D., Shi L. Poly(beta-amino esters): synthesis, formulations, and their biomedical applications. Adv Healthc Mater. 2019;8 doi: 10.1002/adhm.201801359. [DOI] [PubMed] [Google Scholar]
- 124.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cohen E. 2020. Moderna's Coronavirus Vaccine Is 94.5% Effective, According To Company Data. [Google Scholar]
- 129.Byrne J. 2020. Moderna signals Diversity of its US COVID-19 Vaccine trial Cohort. [Google Scholar]
- 130.WHO . 2009. Pandemic Influenza Vaccine Manufacturing Process and Timeline. [Google Scholar]
- 131.Weiland S.D. New York Times; 2020. Trump Administration Selects Five Coronavirus Vaccine Candidates as Finalists. [Google Scholar]
- 132.D. Grady, Moderna Applies for Emergency F.D.A. Approval for Its Coronavirus Vaccine, New York Times, 2020.
- 133.R C. 2020. BNT162 SARS-CoV-2 Vaccine. [Google Scholar]
- 134.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., Wagner W., Granados A., Greenhouse J., Walker M., Willis E., Yu J.S., McGee C.E., Sempowski G.D., Mui B.L., Tam Y.K., Huang Y.J., Vanlandingham D., Holmes V.M., Balachandran H., Sahu S., Lifton M., Higgs S., Hensley S.E., Madden T.D., Hope M.J., Kariko K., Santra S., Graham B.S., Lewis M.G., Pierson T.C., Haynes B.F., Weissman D. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S.P., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fonter-Garfias C., Shi P.-Y., Tuereci O., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Sahin U., Jansen K.U. Phase 1/2 study to describe the safety and immunogenicity of a covid-19 rna vaccine candidate (BNT162b1) in adults 18 to 55 years of age: interim report. medRxiv. 2020 2020.2006.2030.20142570. [Google Scholar]
- 136.Tai W., Zhao G., Sun S., Guo Y., Wang Y., Tao X., Tseng C.K., Li F., Jiang S., Du L., Zhou Y. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology. 2016;499:375–382. doi: 10.1016/j.virol.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wire B. 2020. Pfizer and BioNTech Announce Vaccine Candidate Against COVID-19 Achieved Success in First Interim Analysis from Phase 3 Study. [Google Scholar]
- 140.Thomas K. New York Times; 2020. New Pfizer Results: Coronavirus Vaccine Is Safe And 95% Effective. [Google Scholar]
- 141.Roberts M. BBC; 2020. Covid-19: Pfizer/BioNTech Vaccine Judged Safe for Use in UK. [Google Scholar]
- 142.Morrison A.J., Jr., Hunt E.H., Atuk N.O., Schwartzman J.D., Wenzel R.P. Rabies pre-exposure prophylaxis using intradermal human diploid cell vaccine: immunologic efficacy and cost-effectiveness in a university medical center and a review of selected literature. Am J Med Sci. 1987;293:293–297. doi: 10.1097/00000441-198705000-00003. [DOI] [PubMed] [Google Scholar]
- 143.CureVac . 2020. CureVac´s Optimized mRNA Platform Provides Positive Pre-Clinical Results at Low Dose for Coronavirus Vaccine Candidate. [Google Scholar]
- 144.Kremsner P., Mann P., Bosch J., Fendel R., Gabor J.J., Kreidenweiss A., Kroidl A., Leroux-Roels I., Leroux-Roels G., Schindler C., Schunk M., Velavan T.P., Fotin-Mleczek M., Müller S., Quintini G., Schönborn-Kellenberger O., Vahrenhorst D., Verstraeten T., Walz L., Wolz O.-O., Oostvogels L. Phase 1 assessment of the safety and immunogenicity of an mRNA- lipid nanoparticle vaccine candidate against SARS-CoV-2 in human volunteers. medRxiv. 2020 doi: 10.1007/s00508-021-01922-y. 2020.2011.2009.20228551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., Fotin-Mleczek M. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. npj Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.CureVac . 2020. CureVac’s COVID-19 Vaccine Candidate, CVnCoV, Suitable for Standard Fridge Temperature Logistics. [Google Scholar]
- 147.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., Guo Y., Sun S.H., Fan H., Zu S.L., Chen Q., He Q., Cao T.S., Huang X.Y., Qiu H.Y., Nie J.H., Jiang Y., Yan H.Y., Ye Q., Zhong X., Xue X.L., Zha Z.Y., Zhou D., Yang X., Wang Y.C., Ying B., Qin C.F. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182(5):1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X.-F., Li J., Zhang N., Yang X., Chen S., Zhao G., Wang X., Luo D., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.-F., Zhou Y. Rapid adaptation of SARS-CoV-2 in BALB/c mice: novel mouse model for vaccine efficacy. bioRxiv. 2020 2020.2005.2002.073411. [Google Scholar]
- 149.DD K. New York Times; 2020. U.K. Lab to Sidestep Drug Industry to Sell Potential Virus Vaccine. [Google Scholar]
- 150.He W., Evans A.C., Rasley A., Bourguet F., Peters S., Kamrud K.I., Wang N., Hubby B., Felderman M., Gouvis H., Coleman M.A., Fischer N.O. Cationic HDL mimetics enhance in vivo delivery of self-replicating mRNA. Nanomedicine: Nanotechnology Biol. Med. 2020;24:102154. doi: 10.1016/j.nano.2020.102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.X. Du, S.M. Ansell, Lipids and lipid nanoparticle formulations for delivery of nucleic acids, Google Patents, 2019.
- 152.McKay P.F., Hu K., Blakney A.K., Samnuan K., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine induces equivalent preclinical antibody titers and viral neutralization to recovered COVID-19 patients. bioRxiv. 2020 2020.2004.2022.055608. [Google Scholar]
- 153.Arcturus . 2020. Arcturus Reports Additional Supportive Preclinical Data for its COVID-19 Vaccine Candidate (LUNAR-COV19) [Google Scholar]
- 154.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.J.W. Hooper, E.M. Mucker, P. Chivukula, Nucleic acid vaccine composition comprising a lipid formulation, and method of increasing the potency of nucleic acid vaccines, Google Patents, 2020.
- 156.J.E. Payne, P. Chivukula, Ionizable cationic lipid for RNA delivery, Google Patents, 2018.
- 157.J. Wengel, UNA oligomers for therapeutics, Google Patents, 2015.
- 158.Kim L., Martinez C.J., Hodgson K.A., Trager G.R., Brandl J.R., Sandefer E.P., Doll W.J., Liebowitz D., Tucker S.N. Systemic and mucosal immune responses following oral adenoviral delivery of influenza vaccine to the human intestine by radio controlled capsule. Sci. Rep. 2016;6:37295. doi: 10.1038/srep37295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Newswire G. 2020. Vaxart Announces Selection of its Oral COVID-19 Vaccine Lead Candidate. [Google Scholar]
- 160.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., Jia S.-Y., Jiang H.-D., Wang L., Jiang T., Hu Y., Gou J.-B., Xu S.-B., Xu J.-J., Wang X.-W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cross R. Chemical & Engineering News; 2020. CanSino Publishes first COVID-19 Vaccine Data to Muted Response. [Google Scholar]
- 162.Cohen J. Science Magazine; 2020. Merck, One of Big Pharma’s Biggest Players, Reveals its COVID-19 Vaccine and Therapy Plans. [Google Scholar]
- 163.Philippidis A. 2020. Sanofi Expands COVID-19 Pipeline with Translate Bio Partnership. [Google Scholar]
- 164.Jenner E. Or Cow-Pox; Sampson Low, London: 1798. An Inquiry Into the Causes and Effects of the Variolæ Vaccinæ. [Google Scholar]
- 165.Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.H., Leist S.R., Yount B.L., McAnarney E.T., Graham R.L., Waters K.M., Baric R.S. Combination attenuation offers strategy for live attenuated coronavirus vaccines. J. Virol. 2018;92 doi: 10.1128/JVI.00710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lau S.-Y., Wang P., Mok B.W.-Y., Zhang A.J., Chu H., Lee A.C.-Y., Deng S., Chen P., Chan K.-H., Song W. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emer. Microbes & Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang P., Lau S.-Y., Deng S., Chen P., Mok B.W.-Y., Zhang A.J., Lee A.C.-Y., Chan K.-H., Song W., K.K.-W. To Pathogenicity, immunogenicity, and protective ability of an attenuated SARS-CoV-2 variant with a deletion at the S1/S2 junction of the spike protein. bioRxiv. 2020 bioRxiv 2020.08.24.264192. [Google Scholar]
- 168.Groenke N., Trimpert J., Merz S., Conradie A.M., Wyler E., Zhang H., Hazapis O.-G., Rausch S., Landthaler M., Osterrieder N. Mechanism of virus attenuation by codon pair deoptimization. Cell Rep. 2020;31:107586. doi: 10.1016/j.celrep.2020.107586. [DOI] [PubMed] [Google Scholar]
- 169.Clark-Curtiss J.E., Curtiss R., 3rd Salmonella vaccines: conduits for protective antigens. J. Immunol. 2018;200:39–48. doi: 10.4049/jimmunol.1600608. [DOI] [PubMed] [Google Scholar]
- 170.CDC . 2019. Cell-Based Flu Vaccines. [Google Scholar]
- 171.Zhu F.C., Wurie A.H., Hou L.H., Liang Q., Li Y.H., Russell J.B., Wu S.P., Li J.X., Hu Y.M., Guo Q., Xu W.B., Wurie A.R., Wang W.J., Zhang Z., Yin W.J., Ghazzawi M., Zhang X., Duan L., Wang J.Z., Chen W. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: a single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2017;389:621–628. doi: 10.1016/S0140-6736(16)32617-4. [DOI] [PubMed] [Google Scholar]
- 172.Agnandji S.T., Huttner A., Zinser M.E., Njuguna P., Dahlke C., Fernandes J.F., Yerly S., Dayer J.A., Kraehling V., Kasonta R., Adegnika A.A., Altfeld M., Auderset F., Bache E.B., Biedenkopf N., Borregaard S., Brosnahan J.S., Burrow R., Combescure C., Desmeules J., Eickmann M., Fehling S.K., Finckh A., Goncalves A.R., Grobusch M.P., Hooper J., Jambrecina A., Kabwende A.L., Kaya G., Kimani D., Lell B., Lemaitre B., Lohse A.W., Massinga-Loembe M., Matthey A., Mordmuller B., Nolting A., Ogwang C., Ramharter M., Schmidt-Chanasit J., Schmiedel S., Silvera P., Stahl F.R., Staines H.M., Strecker T., Stubbe H.C., Tsofa B., Zaki S., Fast P., Moorthy V., Kaiser L., Krishna S., Becker S., Kieny M.P., Bejon P., Kremsner P.G., Addo M.M., Siegrist C.A. Phase 1 Trials of rVSV Ebola Vaccine in Africa and Europe. N. Engl. J. Med. 2016;374:1647–1660. doi: 10.1056/NEJMoa1502924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhu F.C., Li Y.H., Guan X.H., Hou L.H., Wang W.J., Li J.X., Wu S.P., Wang B.S., Wang Z., Wang L., Jia S.Y., Jiang H.D., Wang L., Jiang T., Hu Y., Gou J.B., Xu S.B., Xu J.J., Wang X.W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J., Wu S.P., Wang Z., Wu X.H., Xu J.J., Zhang Z., Jia S.Y., Wang B.S., Hu Y., Liu J.J., Zhang J., Qian X.A., Li Q., Pan H.X., Jiang H.D., Deng P., Gou J.B., Wang X.W., Wang X.H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.AstraZeneca . 2020. AstraZeneca Advances Response To Global COVID-19 Challenge as it Receives First Commitments For Oxford’s Potential New Vaccine. [Google Scholar]
- 176.P.M. Folegatti, K.J. Ewer, P.K. Aley, B. Angus, S. Becker, S. Belij-Rammerstorfer, D. Bellamy, S. Bibi, M. Bittaye, E.A. Clutterbuck, C. Dold, S.N. Faust, A. Finn, A.L. Flaxman, B. Hallis, P. Heath, D. Jenkin, R. Lazarus, R. Makinson, A.M. Minassian, K.M. Pollock, M. Ramasamy, H. Robinson, M. Snape, R. Tarrant, M. Voysey, C. Green, A.D. Douglas, A.V.S. Hill, T. Lambe, S.C. Gilbert, A.J. Pollard, J. Aboagye, K. Adams, A. Ali, E. Allen, J.L. Allison, R. Anslow, E.H. Arbe-Barnes, G. Babbage, K. Baillie, M. Baker, P. Baker, I. Baleanu, J. Ballaminut, E. Barnes, J. Barrett, L. Bates, A. Batten, K. Beadon, R. Beckley, E. Berrie, L. Berry, A. Beveridge, K.R. Bewley, E.M. Bijker, T. Bingham, L. Blackwell, C.L. Blundell, E. Bolam, E. Boland, N. Borthwick, T. Bower, A. Boyd, T. Brenner, P.D. Bright, C. Brown-O'Sullivan, E. Brunt, J. Burbage, S. Burge, K.R. Buttigieg, N. Byard, I. Cabera Puig, A. Calvert, S. Camara, M. Cao, F. Cappuccini, M. Carr, M.W. Carroll, V. Carter, K. Cathie, R.J. Challis, I. Chelysheva, J.-S. Cho, P. Cicconi, L. Cifuentes, H. Clark, E. Clark, T. Cole, R. Colin-Jones, C.P. Conlon, A. Cook, N.S. Coombes, R. Cooper, C.A. Cosgrove, K. Coy, W.E.M. Crocker, C.J. Cunningham, B.E. Damratoski, L. Dando, M.S. Datoo, H. Davies, H. De Graaf, T. Demissie, C. Di Maso, I. Dietrich, T. Dong, F.R. Donnellan, N. Douglas, C. Downing, J. Drake, R. Drake-Brockman, R.E. Drury, S.J. Dunachie, N.J. Edwards, F.D.L. Edwards, C.J. Edwards, S.C. Elias, M.J. Elmore, K.R.W. Emary, M.R. English, S. Fagerbrink, S. Felle, S. Feng, S. Field, C. Fixmer, C. Fletcher, K.J. Ford, J. Fowler, P. Fox, E. Francis, J. Frater, J. Furze, M. Fuskova, E. Galiza, D. Gbesemete, C. Gilbride, G. Gorini, L. Goulston, C. Grabau, L. Gracie, Z. Gray, L.B. Guthrie, M. Hackett, S. Halwe, E. Hamilton, J. Hamlyn, B. Hanumunthadu, I. Harding, S.A. Harris, A. Harris, D. Harrison, C. Harrison, T.C. Hart, L. Haskell, S. Hawkins, I. Head, J.A. Henry, J. Hill, S.H.C. Hodgson, M.M. Hou, E. Howe, N. Howell, C. Hutlin, S. Ikram, C. Isitt, P. Iveson, S. Jackson, F. Jackson, S.W. James, M. Jenkins, E. Jones, K. Jones, C.E. Jones, B. Jones, R. Kailath, K. Karampatsas, J. Keen, S. Kelly, D. Kelly, D. Kerr, S. Kerridge, L. Khan, U. Khan, A. Killen, J. Kinch, T.B. King, King, J. King, L. Kingham-Page, P. Klenerman, F. Knapper, J.C. Knight, S. Koleva, A. Kupke, C.W. Larkworthy, J.P.J. Larwood, A. Laskey, A.M. Lawrie, A. Lee, K.Y. Ngan Lee, E.A. Lee, H. Legge, A. Lelliott, N.-M. Lemm, A.M. Lias, A. Linder, S. Lipworth, X. Liu, S. Liu, R. Lopez Ramon, M. Lwin, F. Mabesa, M. Madhavan, G. Mallett, K. Mansatta, I. Marcal, S. Marinou, E. Marlow, J.L. Marshall, J. Martin, J. McEwan, G. Meddaugh, A.J. Mentzer, N. Mirtorabi, M. Moore, E. Moran, E. Morey, V. Morgan, S.J. Morris, H. Morrison, G. Morshead, R. Morter, Y.F. Mujadidi, J. Muller, T. Munera-Huertas, C. Munro, A. Munro, S. Murphy, V.J. Muster, P. Mweu, A. Noé, F.L. Nugent, E. Nugent, K. O'Brien, D. O'Connor, B. Oguti, J.L. Oliver, C. Oliveira, P.J. O'Reilly, M. Osborn, P. Osborne, C. Owen, D. Owens, N. Owino, M. Pacurar, K. Parker, H. Parracho, M. Patrick-Smith, V. Payne, J. Pearce, Y. Peng, M.P. Peralta Alvarez, J. Perring, K. Pfafferott, D. Pipini, E. Plested, H. Pluess-Hall, K. Pollock, I. Poulton, L. Presland, S. Provstgaard-Morys, D. Pulido, K. Radia, F. Ramos Lopez, J. Rand, H. Ratcliffe, T. Rawlinson, S. Rhead, A. Riddell, A.J. Ritchie, H. Roberts, J. Robson, S. Roche, C. Rohde, C.S. Rollier, R. Romani, I. Rudiansyah, S. Saich, S. Sajjad, S. Salvador, L. Sanchez Riera, H. Sanders, K. Sanders, S. Sapaun, C. Sayce, E. Schofield, G. Screaton, B. Selby, C. Semple, H.R. Sharpe, A. Shea, H. Shelton, S. Silk, L. Silva-Reyes, D.T. Skelly, H. Smee, C.C. Smith, D.J. Smith, R. Song, A.J. Spencer, E. Stafford, A. Steele, E. Stefanova, L. Stockdale, A. Szigeti, A. Tahiri-Alaoui, M. Tait, H. Talbot, R. Tanner, I.J. Taylor, V. Taylor, R. Te Water Naude, N. Thakur, Y. Themistocleous, A. Themistocleous, M. Thomas, T.M. Thomas, A. Thompson, S. Thomson-Hill, J. Tomlins, S. Tonks, J. Towner, N. Tran, J.A. Tree, A. Truby, K. Turkentine, C. Turner, N. Turner, S. Turner, T. Tuthill, M. Ulaszewska, R. Varughese, N. Van Doremalen, K. Veighey, M.K. Verheul, I. Vichos, E. Vitale, L. Walker, M.E.E. Watson, B. Welham, J. Wheat, C. White, R. White, A.T. Worth, D. Wright, S. Wright, X.L. Yao, Y. Yau, Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial Lancet.
- 177.Callaway E. Why Oxford’s positive COVID vaccine results are puzzling scientists. Nature. 2020;588:16–18. doi: 10.1038/d41586-020-03326-w. [DOI] [PubMed] [Google Scholar]
- 178.Janssen . 2020. Janssen Vaccine Technology. [Google Scholar]
- 179.Barouch D.H., Kik S.V., Weverling G.J., Dilan R., King S.L., Maxfield L.F., Clark S., Ng'ang'a D., Brandariz K.L., Abbink P., Sinangil F., de Bruyn G., Gray G.E., Roux S., Bekker L.G., Dilraj A., Kibuuka H., Robb M.L., Michael N.L., Anzala O., Amornkul P.N., Gilmour J., Hural J., Buchbinder S.P., Seaman M.S., Dolin R., Baden L.R., Carville A., Mansfield K.G., Pau M.G., Goudsmit J. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine. 2011;29:5203–5209. doi: 10.1016/j.vaccine.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Express I. 2020. Coronavirus (Covid-19) Vaccine Status Check: Oxford Vaccine Most Advanced, Says Who; Sanofi Accelerates Trials. [Google Scholar]
- 181.Gabitzsch E.S., Xu Y., Yoshida L.H., Balint J., Gayle R.B., Amalfitano A., Jones F.R. A preliminary and comparative evaluation of a novel Ad5 [E1-, E2b-] recombinant-based vaccine used to induce cell mediated immune responses. Immunol. Lett. 2009;122:44–51. doi: 10.1016/j.imlet.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gabitzsch E.S., Xu Y., Balint J.P., Jr., Hartman Z.C., Lyerly H.K., Jones F.R. Anti-tumor immunotherapy despite immunity to adenovirus using a novel adenoviral vector Ad5 [E1-, E2b-]-CEA. Cancer Immunol. Immunother. 2010;59:1131–1135. doi: 10.1007/s00262-010-0847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Osada T., Yang X.Y., Hartman Z.C., Glass O., Hodges B.L., Niedzwiecki D., Morse M.A., Lyerly H.K., Amalfitano A., Clay T.M. Optimization of vaccine responses with an E1, E2b and E3-deleted Ad5 vector circumvents pre-existing anti-vector immunity. Cancer Gene Ther. 2009;16:673–682. doi: 10.1038/cgt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rice A., Verma M., Shin A., Zakin L., Sieling P., Tanaka S., Adisetiyo H., Taft J., Patel R.S., Buta S. 2020. A Next Generation Bivalent Human Ad5 COVID-19 Vaccine Delivering Both Spike and Nucleocapsid Antigens Elicits Th1 Dominant CD4+, CD8+ T-cell and Neutralizing Antibody Responses, bioRxiv. [Google Scholar]
- 185.Yang K., Sun K., Srinivasan K., Salmon J., Marques E., Xu J., August J. Immune responses to T-cell epitopes of SARS CoV-N protein are enhanced by N immunization with a chimera of lysosome-associated membrane protein. Gene Ther. 2009;16:1353–1362. doi: 10.1038/gt.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Petropoulos K. American Society of Hematology Washington; DC: 2019. Effective Stabilization of Viral Vectors in Liquid Using an Algorithm-Based Development Approach. [Google Scholar]
- 187.Reinauer E.B., Grosso S.S., Henz S.R., Rabas J.A., Rodenstein C., Altrichter J., Scholz M., Kemter K.F. Algorithm-based liquid formulation development including a doe concept predicts long-term viral vector stability. J. Pharm. Sci. 2020;109:818–829. doi: 10.1016/j.xphs.2019.10.063. [DOI] [PubMed] [Google Scholar]
- 188.Roberts A., Kretzschmar E., Perkins A.S., Forman J., Price R., Buonocore L., Kawaoka Y., Rose J.K. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J. Virol. 1998;72:4704–4711. doi: 10.1128/jvi.72.6.4704-4711.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Roberts A., Buonocore L., Price R., Forman J., Rose J.K. Attenuated vesicular stomatitis viruses as vaccine vectors. J. Virol. 1999;73:3723–3732. doi: 10.1128/jvi.73.5.3723-3732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jones S.M., Feldmann H., Ströher U., Geisbert J.B., Fernando L., Grolla A., Klenk H.-D., Sullivan N.J., Volchkov V.E., Fritz E.A. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 2005;11:786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- 191.Regules J.A., Beigel J.H., Paolino K.M., Voell J., Castellano A.R., Hu Z., Muñoz P., Moon J.E., Ruck R.C., Bennett J.W. A recombinant vesicular stomatitis virus Ebola vaccine. N. Engl. J. Med. 2017;376:330–341. doi: 10.1056/NEJMoa1414216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Suder E., Furuyama W., Feldmann H., Marzi A., de Wit E. The vesicular stomatitis virus-based Ebola virus vaccine: from concept to clinical trials. Human Vacc. Immunother. 2018;14:2107–2113. doi: 10.1080/21645515.2018.1473698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kapadia S.U., Rose J.K., Lamirande E., Vogel L., Subbarao K., Roberts A. Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology. 2005;340:174–182. doi: 10.1016/j.virol.2005.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kapadia S.U., Simon I.D., Rose J.K. SARS vaccine based on a replication-defective recombinant vesicular stomatitis virus is more potent than one based on a replication-competent vector. Virology. 2008;376:165–172. doi: 10.1016/j.virol.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.-M., Zeng Q., Tahan S., Droit L. 2020. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Case J.B., Rothlauf P.W., Chen R.E., Kafai N.M., Fox J.M., Smith B.K., Shrihari S., McCune B.T., Harvey I.B., Keeler S.P. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe. 2020;28 doi: 10.1016/j.chom.2020.07.018. 465-474. e464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.011. 744-753. e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yahalom-Ronen Y., Tamir H., Melamed S., Politi B., Shifman O., Achdout H., Vitner E.B., Israeli O., Milrot E., Stein D. A single dose of recombinant vsv-∆ g-spike vaccine provides protection against sars-cov-2 challenge. bioRxiv. 2020 doi: 10.1038/s41467-020-20228-7. bioRxiv 2020.06.18.160655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hörner C., Schürmann C., Auste A., Ebenig A., Muraleedharan S., Herrmann M., Schnierle B., Mühlebach M.D. A Highly Immunogenic Measles Virus-based Th1-biased COVID-19 Vaccine. bioRxiv. 2020 doi: 10.1073/pnas.2014468117. 2020.2007.2011.198291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Combredet C., Labrousse V., Mollet L., Lorin C., Delebecque F., Hurtrel B., McClure H., Feinberg M.B., Brahic M., Tangy F. A molecularly cloned Schwarz strain of measles virus vaccine induces strong immune responses in macaques and transgenic mice. J. Virol. 2003;77:11546–11554. doi: 10.1128/JVI.77.21.11546-11554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ramsauer K., Schwameis M., Firbas C., Müllner M., Putnak R.J., Thomas S.J., Desprès P., Tauber E., Jilma B., Tangy F. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect. Dis. 2015;15:519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 202.Reisinger E.C., Tschismarov R., Beubler E., Wiedermann U., Firbas C., Loebermann M., Pfeiffer A., Muellner M., Tauber E., Ramsauer K. Immunogenicity, safety, and tolerability of the measles-vectored chikungunya virus vaccine MV-CHIK: a double-blind, randomised, placebo-controlled and active-controlled phase 2 trial. Lancet. 2019;392:2718–2727. doi: 10.1016/S0140-6736(18)32488-7. [DOI] [PubMed] [Google Scholar]
- 203.Nikam V., Kotade K.B., Gaware V.M. Eudragit a versatile polymer: a review. Pharmacologyonline. 2011;1:152–164. [Google Scholar]
- 204.Kim L., Liebowitz D., Lin K., Kasparek K., Pasetti M.F., Garg S.J., Gottlieb K., Trager G., Tucker S.N. Safety and immunogenicity of an oral tablet norovirus vaccine, a phase I randomized, placebo-controlled trial. JCI Insight. 2018;3 doi: 10.1172/jci.insight.121077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vaxart . 2020. Vaxart’s COVID-19 Vaccine Selected for the U.S. Government’s Operation Warp Speed. [Google Scholar]
- 206.Yurina V. Live bacterial vectors-a promising DNA vaccine delivery system. Med Sci (Basel) 2018;6 doi: 10.3390/medsci6020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.de Azevedo M., Meijerink M., Taverne N., Pereira V.B., LeBlanc J.G., Azevedo V., Miyoshi A., Langella P., Wells J.M., Chatel J.M. Recombinant invasive Lactococcus lactis can transfer DNA vaccines either directly to dendritic cells or across an epithelial cell monolayer. Vaccine. 2015;33:4807–4812. doi: 10.1016/j.vaccine.2015.07.077. [DOI] [PubMed] [Google Scholar]
- 208.Symvivo . 2020. COVID-19: Program Vision. [Google Scholar]
- 209.Oertli D., Marti W.R., Norton J.A., Tsung K. Artificial antigen-presenting cells engineered by recombinant vaccinia viruses expressing antigen, MHC class II, and costimulatory molecules elicit proliferation of CD4+ lymphocytes in vitro. Clin. Exp. Immunol. 1997;110:144–149. doi: 10.1046/j.1365-2249.1997.5061405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang C., Sun W., Ye Y., Bomba H.N., Gu Z. Bioengineering of artificial antigen presenting cells and lymphoid organs. Theranostics. 2017;7:3504–3516. doi: 10.7150/thno.19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rhodes K.R., Meyer R.A., Wang J., Tzeng S.Y., Green J.J. Biomimetic tolerogenic artificial antigen presenting cells for regulatory T cell induction. Acta Biomater. 2020;12:136–148. doi: 10.1016/j.actbio.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Butler M.O., Lee J.S., Ansen S., Neuberg D., Hodi F.S., Murray A.P., Drury L., Berezovskaya A., Mulligan R.C., Nadler L.M., Hirano N. Long-lived antitumor CD8+ lymphocytes for adoptive therapy generated using an artificial antigen-presenting cell. Clin. Cancer Res. 2007;13:1857–1867. doi: 10.1158/1078-0432.CCR-06-1905. [DOI] [PubMed] [Google Scholar]
- 213.Neal L.R., Bailey S.R., Wyatt M.M., Bowers J.S., Majchrzak K., Nelson M.H., Haupt C., Paulos C.M., Varela J.C. The basics of artificial antigen presenting cells in t cell-based cancer immunotherapies. J. Immunol. Res. Ther. 2017;2:68–79. [PMC free article] [PubMed] [Google Scholar]
- 214.Su Q., Igyártó B.Z. One-step artificial antigen presenting cell-based vaccines induce potent effector CD8 T cell responses. Sci. Rep. 2019;9:18949. doi: 10.1038/s41598-019-55286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chang L.J. Lentiviral vector transduction of dendritic cells for novel vaccine strategies. Methods Mol. Biol. 2010;614:161–171. doi: 10.1007/978-1-60761-533-0_11. [DOI] [PubMed] [Google Scholar]
- 216.Chen X., He J., Chang L.J. Alteration of T cell immunity by lentiviral transduction of human monocyte-derived dendritic cells. Retrovirology. 2004;1:37. doi: 10.1186/1742-4690-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., Chou C.-W., Byrne P.O., Hjorth C.K., Johnson N.V., Ludes-Meyers J., Nguyen A.W., Park J., Wang N., Amengor D., Lavinder J.J., Ippolito G.C., Maynard J.A., Finkelstein I.J., McLellan J.S. Vol. 369. 2020. Structure-Based Design of Prefusion-Stabilized SARS-CoV-2 Spikes; pp. 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Stitz L., Vogel A., Schnee M., Voss D., Rauch S., Mutzke T., Ketterer T., Kramps T., Petsch B. A thermostable messenger RNA based vaccine against rabies. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang L., Li J., Chen H., Li F., Armstrong G.L., Nelson C., Ze W., Shapiro C.N. Hepatitis B vaccination of newborn infants in rural China: evaluation of a village-based, out-of-cold-chain delivery strategy. Bull. World Health Organ. 2007;85:688–694. doi: 10.2471/BLT.06.037002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lydon P., Zipursky S., Tevi-Benissan C., Djingarey M.H., Gbedonou P., Youssouf B.O., Zaffran M. Economic benefits of keeping vaccines at ambient temperature during mass vaccination: the case of meningitis A vaccine in Chad. Bull. World Health Organ. 2013;92:86–92. doi: 10.2471/BLT.13.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kumru O.S., Joshi S.B., Smith D.E., Middaugh C.R., Prusik T., Volkin D.B. Vaccine instability in the cold chain: mechanisms, analysis and formulation strategies. Biologicals. 2014;42:237–259. doi: 10.1016/j.biologicals.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 222.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 223.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020 doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jiang H.-w., Li Y., Zhang H.-n., Wang W., Yang X., Qi H., Li H., Men D., Zhou J., Tao S.-c. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Dunand C.J.H., Leon P.E., Huang M., Choi A., Chromikova V., Ho I.Y., Tan G.S., Cruz J., Hirsh A., Zheng N.-Y. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tan G.S., Leon P.E., Albrecht R.A., Margine I., Hirsh A., Bahl J., Krammer F. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.DiLillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.J.A. Horwitz, Y. Bar-On, C.-L. Lu, D. Fera, A.A. Lockhart, J.C. Lorenzi, L. Nogueira, J. Golijanin, J.F. Scheid, M.S. Seaman, Non-neutralizing antibodies alter the course of HIV-1 infection in vivo, Cell, 170 (2017) 637-648. e610. [DOI] [PMC free article] [PubMed]
- 229.Forthal D., Hope T., Alter G. New paradigms for functional HIV-specific non-neutralizing antibodies. Curr. Opin. HIV AIDS. 2013;8:393. doi: 10.1097/COH.0b013e328363d486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Howell K.A., Brannan J.M., Bryan C., McNeal A., Davidson E., Turner H.L., Vu H., Shulenin S., He S., Kuehne A.J.C.R. Vol. 19. 2017. Cooperativity Enables Non-Neutralizing Antibodies to Neutralize Ebolavirus; pp. 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.P.A. Ilinykh, K. Huang, R.I. Santos, P. Gilchuk, B.M. Gunn, M.M. Karim, J. Liang, M.E. Fouch, E. Davidson, D.V.J.C.H. Parekh, Microbe, Non-neutralizing Antibodies from a Marburg Infection Survivor Mediate Protection by Fc-Effector Functions and by Enhancing Efficacy of Other Antibodies, (2020). [DOI] [PMC free article] [PubMed]
- 232.Moderbacher C.R., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Guthmiller J.J., Stovicek O., Wang J., Changrob S., Li L., Halfmann P., Zheng N.-Y., Utset H., Stamper C.T., Dugan H.L. 2020. SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike, bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang P., Liu L., Nair M.S., Yin M.T., Luo Y., Wang Q., Yuan T., Mori K., Solis A.G., Yamashita M. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emer. Microbes & Infect. 2020:1–15. doi: 10.1080/22221751.2020.1823890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.W.H. Organization, WHO target product profiles for COVID-19 vaccines, Version.
- 236.Food D. 2020. Administration, Development and licensure of vaccines to prevent Covid-19: guidance for industry. June. [Google Scholar]
- 237.Kiyotani K., Toyoshima Y., Nemoto K., Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.E. Fast, B. Chen, Potential T-cell and B-cell Epitopes of 2019-nCoV, bioRxiv, (2020) 2020.2002.2019.955484.
- 240.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kim Y.-C., Quan F.-S., Compans R.W., Kang S.-M., Prausnitz M.R. Formulation and coating of microneedles with inactivated influenza virus to improve vaccine stability and immunogenicity. J. Control. Release. 2010;142:187–195. doi: 10.1016/j.jconrel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Prausnitz M.R., Mikszta J.A., Cormier M., Andrianov A.K. Springer; 2009. Microneedle-Based Vaccines, Vaccines for Pandemic Influenza; pp. 369–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Van Damme P., Oosterhuis-Kafeja F., Van der Wielen M., Almagor Y., Sharon O., Levin Y. Safety and efficacy of a novel microneedle device for dose sparing intradermal influenza vaccination in healthy adults. Vaccine. 2009;27:454–459. doi: 10.1016/j.vaccine.2008.10.077. [DOI] [PubMed] [Google Scholar]
- 244.Kim E., Erdos G., Huang S., Kenniston T.W., Balmert S.C., Carey C.D., Raj V.S., Epperly M.W., Klimstra W.B., Haagmans B.L. Microneedle array delivered recombinant coronavirus vaccines: immunogenicity and rapid translational development. EBioMedicine. 2020;102743 doi: 10.1016/j.ebiom.2020.102743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Johnson-Weaver B.T., Abraham S.N., Staats H.F. Elsevier; 2020. Innate Immunity-Based Mucosal Modulators and Adjuvants, Mucosal Vaccines; pp. 167–183. [Google Scholar]
- 246.Kim E., Attia Z., Cormet-Boyaka E., Boyaka P.N. Elsevier; 2020. Toxin-Based Modulators for Regulation of Mucosal Immune Responses, Mucosal Vaccines; pp. 185–201. [Google Scholar]
- 247.Travis C.R. As plain as the nose on your face: the case for a nasal (mucosal) route of vaccine administration for covid-19 disease prevention. Front. Immunol. 2020;11:2611. doi: 10.3389/fimmu.2020.591897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020:1–16. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 249.Bouvet J.-P., Decroix N., Pamonsinlapatham P. Stimulation of local antibody production: parenteral or mucosal vaccination? Trends Immunol. 2002;23:209–213. doi: 10.1016/s1471-4906(02)02186-5. [DOI] [PubMed] [Google Scholar]
- 250.Thompson J.M., Nicholson M.G., Whitmore A.C., Zamora M., West A., Iwasaki A., Staats H.F., Johnston R.E. Nonmucosal alphavirus vaccination stimulates a mucosal inductive environment in the peripheral draining lymph node. J. Immunol. 2008;181:574–585. doi: 10.4049/jimmunol.181.1.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Clements J.D., Freytag L.C. Parenteral vaccination can be an effective means of inducing protective mucosal responses. Clin. Vaccine Immunol. 2016;23:438–441. doi: 10.1128/CVI.00214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Su F., Patel G.B., Hu S., Chen W. Induction of mucosal immunity through systemic immunization: phantom or reality? Human Vacc. Immunother. 2016;12:1070–1079. doi: 10.1080/21645515.2015.1114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Beyer W., Palache A., De Jong J., Osterhaus A. Cold-adapted live influenza vaccine versus inactivated vaccine: systemic vaccine reactions, local and systemic antibody response, and vaccine efficacy: a meta-analysis. Vaccine. 2002;20:1340–1353. doi: 10.1016/s0264-410x(01)00471-6. [DOI] [PubMed] [Google Scholar]
- 254.Peiris M., Leung G.M. What can we expect from first-generation COVID-19 vaccines? Lancet (London, England) 2020;396:1467–1469. doi: 10.1016/S0140-6736(20)31976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Stephens D.S., McElrath M.J. COVID-19 and the Path to Immunity. Jama. 2020;324:1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Walsh E.E., Falsey A.R. A simple and reproducible method for collecting nasal secretions in frail elderly adults, for measurement of virus-specific IgA. J. Infect. Dis. 1999;179:1268–1273. doi: 10.1086/314726. [DOI] [PubMed] [Google Scholar]
- 257.Lipsitch M., Dean N.E. Understanding COVID-19 vaccine efficacy. Science. 2020;370(6518):763–765. doi: 10.1126/science.abe5938. [DOI] [PubMed] [Google Scholar]
- 258.Shah S.K., Miller F.G., Darton T.C., Duenas D., Emerson C., Lynch H.F., Jamrozik E., Jecker N.S., Kamuya D., Kapulu M. Ethics of controlled human infection to address COVID-19. Science. 2020;368:832–834. doi: 10.1126/science.abc1076. [DOI] [PubMed] [Google Scholar]
- 259.Deming M.E., Michael N.L., Robb M., Cohen M.S., Neuzil K.M. Accelerating development of SARS-CoV-2 vaccines—the role for controlled human infection models. N. Engl. J. Med. 2020;383 doi: 10.1056/NEJMp2020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Krause P., Fleming T.R., Longini I., Henao-Restrepo A.M., Peto R., Dean N., Halloran M., Huang Y., Fleming T., Gilbert P. COVID-19 vaccine trials should seek worthwhile efficacy. Lancet. 2020;396:741–743. doi: 10.1016/S0140-6736(20)31821-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bui D.P., McCaffrey K., Friedrichs M., LaCross N., Lewis N.M., Sage K., Barbeau B., Vilven D., Rose C., Braby S. Racial and ethnic disparities among COVID-19 cases in workplace outbreaks by industry sector—Utah, March 6–June 5, 2020. Morb. Mortal. Wkly Rep. 2020;69:1133. doi: 10.15585/mmwr.mm6933e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Goyal M.K., Simpson J.N., Boyle M.D., Badolato G.M., Delaney M., McCarter R., Cora-Bramble D. 2020. Racial and/or Ethnic and Socioeconomic Disparities of SARS-CoV-2 Infection Among Children, Pediatrics, 146. [DOI] [PubMed] [Google Scholar]
- 263.C.f.D. Control . 2020. Prevention, COVID-19 Hospitalization and Death by Race/Ethnicity. [Google Scholar]
- 264.Krause P.R., Gruber M.F. Emergency use authorization of covid vaccines—safety and efficacy follow-up considerations. N. Engl. J. Med. 2020;383 doi: 10.1056/NEJMp2031373. e106. [DOI] [PubMed] [Google Scholar]