Abstract
The novel human betacoronavirus SARS-CoV-2 has caused an unprecedented pandemic in the 21st century. Several studies have revealed interactions between SARS-CoV-2 viral proteins and host nucleoporins, yet their functions are largely unknown. Here, we demonstrate that the open-reading frame 6 (ORF6) of SARS-CoV-2 can directly manipulate localization and functions of nucleoporins. We found that ORF6 protein disrupted nuclear rim staining of nucleoporins RAE1 and NUP98. Consequently, this disruption caused aberrant nucleocytoplasmic trafficking and led to nuclear accumulation of mRNA transporters such as hnRNPA1. Ultimately, host cell nucleus size was reduced and cell growth was halted.
Keywords: SARS-CoV-2 ORF6, RAE1, NUP98, hnRNPA1
Highlights
-
•
ORF6 overexpression displaced nuclear pore localization of RAE1 and NUP98.
-
•
ORF6 overexpression induced hnRNPA1 nuclear accumulation.
-
•
ORF6 can directly manipulate localization and functions of nucleoporins.
Abbreviations
- COVID-19
Corona Virus Disease 2019
- hnRNPA1
Heterogeneous nuclear ribonucleoprotein A1
- NUP98
Nucleoporin 98
- ORF6
Open-reading frame 6
- RAE1
RNA export factor 1
- SARS-CoV-2
Severe Acute Respiratory Syndrome Corona Virus 2
1. Introduction
Since late 2019, the overwhelming effects of a novel coronavirus disease (named COVID-19 for “coronavirus disease 2019″) have spread rapidly and developed into a global pandemic. An overriding exponential rate of new cases and deaths has urged scientists across the world to develop vaccines and antiviral drugs against the causative pathogen of COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, many unknowns about the molecular invasion and replication pathways of SARS-CoV-2 components present major obstacles for COVID-19 studies. The SARS-CoV-2–30-kb positive-strand RNA genome contains 14 potential open reading frames (ORFs) and encodes replicase (ORF1a and ORF1b), structural proteins (spike, nucleocapsid, membrane, and envelope), and accessory proteins (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, and ORF9b) of poorly understood function. These accessory proteins may be important for virus replication [1,2].
Recently, two efforts utilizing immunoprecipitation coupled with mass spectrometry (IP-MS) of epitope-tagged viral proteins to map SARS-CoV-2–host protein-protein interactions (PPIs) identified >1000 putative virus-host PPIs in HEK293T cells [3,4]. Remarkably, the SARS-CoV-2 ORF6 interactome is uniquely enriched in nuclear pore complex (NPC) components, especially nucleoporins RNA export factor 1 (RAE1) and 98 (NUP98) [3,5]. RAE1 (also known as Gle2 or Mnrp41), initially identified as an mRNA export protein [[6], [7], [8]], is a homologue of spindle checkpoint protein of BUB3 [9,10]. We and others showed that RAE1 binds to the GLEBS motif of NUP98, allowing them to interact and function together in mRNA export or spindle assembly to prevent chromosome mis-segregation [[10], [11], [12], [13], [14], [15]]. RAE1 and NUP98, which are kinetically partitioned among other components in both cytoplasmic and nuclear compartments, also act as regulators for BMAL1 shuttling between the cytoplasm and nucleus, as well as maintenance of the correct pace of the circadian clock [16]. In addition, RAE1 was found to mediate ZEB1 expression by promoting epithelial-mesenchymal transition in breast cancer [17].
Notably, RAE1 and NUP98 are the key host cell targets for the matrix (M) protein of vesicular stomatitis virus (VSV) [18,19]. As a canonical seven-bladed β-propeller, RAE1 is a likely binding platform for other proteins [20]. Unsurprisingly, VSV promotes heterogeneous ribonucleoprotein A1 (hnRNPA1) relocalization in a RAE1-dependent manner; hnRNPs are thought to serve as mRNA export factors [[21], [22]]. Similar to VSV, ORF6 also contains a methionine in its C-terminal region, which may facilitate interactions with RAE1 and NUP98 3. However, unlike HIV capsid entering into the nuclear pore [23,24] it remains largely unknown why SARS-CoV2 ORF6 needs to interact with nucleoporins after infection because SARS-CoV2 hijacks the protein-making machinery of the host cell to translate its RNA directly into new copies of the virus.
Here, we found that overexpression of GFP-ORF6 induced mislocalization of RAE1 and NUP98. We also found that the nuclear size of ORF6-transfected cells was severely reduced, potentially due to hnRNPA1 nuclear accumulation and subsequent deferment of mRNA export. Finally, we showed that overexpression of RAE1 partially rescued this phenomenon.
2. Methods
2.1. Plasmids and siRNA
Constructs for overexpressing SARS-CoV-2 ORF6 were generated by ligation of synthetic DNA into the pEGFP-N1 vector (Clontech #6085–1) using EcoRI (New England Biolabs, R0101) and BamHI (TOYOBO, BAH-111) restriction sites. The FLAG-RAE1 plasmid was previously described [15]. For RAE1 siRNA, stealth RNAi targeting RAE1 was customarily synthesized (Invitrogen) to target the sequence corresponding to nucleotides 343–367 of human RAE1 mRNA (GenBank Accession No. AK292247). Universal negative control siRNA (SIC001) was purchased from Sigma-Aldrich.
2.2. Antibodies
Primary antibodies used for immunocytofluorescence, immunoprecipitation, and immunoblotting were as follows: anti-RAE1 (Santa Cruz Biotechnology, sc-393252), anti-NUP98 (Santa Cruz Biotechnology, sc-74553), anti-hnRNPA1 (Cell Signaling Technology, 8443), anti-Lamin B1 (MBL, PM064), anti-FLAG (MERCK, F3165; MBL, PM020; and Wako, 018–22381), anti-GFP (Invitrogen, A6455; and Wako, 012–20461), anti-NUP153 (Santa Cruz Biotechnology, sc-101544), anti-TPR (Santa Cruz Biotechnology, sc-101294), and anti-beta actin (Cell Signaling Technology, 4967). Secondary antibodies were as follows: Alexa Fluor 488 goat anti-mouse (Invitrogen, A11029), Alexa Fluor 488 goat anti-rabbit (Invitrogen, A11034), rhodamine red goat anti-mouse (Invitrogen, R6393), rhodamine red goat anti-rabbit (Invitrogen, R6394), Alexa Fluor 633 goat anti-mouse (Invitrogen, A21050), horseradish peroxidase (HRP)-linked anti-rabbit IgG (Cell Signaling Technology, 7074), HRP-linked anti-rabbit IgG (Cell Signaling Technology, 7076), and HRP-linked anti-rat IgG (Cell Signaling Technology, 7077).
2.3. Cell culture and transfection
HEK293T cells were propagated in Dulbecco’s Modified Eagle’s Medium (Wako, 043–30085) supplemented with 10% (v:v) fetal bovine serum (Life Technologies, 10082147) and 50 U/mL penicillin-streptomycin (Nacalai Tesque, 26253–84), as previously described [25]. Cells were cultured at 37 °C and 5% CO2.
DNA transfection was performed in six-well plates using Lipofectamine 2000 (Invitrogen, 11668019) with 1000 ng of plasmid DNA or 50 nM siRNA per well according to the manufacturer’s protocol, as previously described [26].
2.4. Immunocytofluorescence
HEK293T cells were cultured on glass coverslips in six-well plates. Cells on coverslips were washed in phosphate-buffered saline [PBS; 137 mM NaCl (Nacalai Tesque, 31320–34), 2.7 mM KCl (Sigma, P9541), 10 mM Na2HPO4 (Wako, 198–05955F), 1.8 mM KH2PO4 (Nacalai Tesque, 28736–75)], fixed for 30 min with 4% paraformaldehyde (Nacalai Tesque, 26123–55) in PBS, washed again with PBS, and permeabilized with 0.3% Triton X-100 (Nacalai Tesque, 35501–15) in PBS for 3 min. Cells were then washed and blocked in PBS containing 4% bovine serum albumin (BSA; Wako, 015–23295) for 30 min at room temperature. Coverslips were then incubated overnight in 4% BSA/PBS containing primary antibodies (1:500 dilution). Cells were then rinsed and incubated with secondary antibodies in 4% BSA/PBS (1:500 dilution) for 2 h at room temperature. After washing with PBS, coverslips were mounted on slides using ProLong Diamond Antifade reagent with DAPI (Invitrogen, P36966), and observed on a confocal laser-scanning microscope with 60X PlanApo/1.45NA DIC objective (Olympus, FV10i-LIV), as previously described [27].
2.5. Immunoblotting
For protein extraction, 6 × 105 cells were solubilized in 100 μL of lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% Nonidet P-40, 2 mM EDTA, and 10% glycerol) supplemented with EDTA-free protease inhibitor cocktail (Nacalai Tesque, 03969). After centrifugation at 14,000×g for 10 min at 4 °C, supernatants were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes (Millipore, IPVH00010). Membranes were incubated with primary antibodies and exposed to secondary HRP-conjugated antibodies. Proteins were visualized by chemiluminescence using Immobilon Western Chemilum HRP Substrate (Millipore, WBKLS0500). Images were acquired with a LAS-4000 image analyzer (Fujifilm).
Nuclear/cytoplasmic fractionation analysis was performed using nuclear and cytoplasmic extraction reagents according to the manufacturer’s protocol (Thermo Scientific, 78840), as previously described [25,27]. Cytoplasmic and nuclear extracts were then subjected to immunoblotting analysis.
2.6. Immunoprecipitation
Cells were transiently transfected with pEGFP-ORF6 and/or FLAG-RAE1 plasmids, and harvested 48 h after transfection. Cells were centrifuged at 500×g for 5 min, lysed in lysis buffer supplemented with EDTA-free protease inhibitor cocktail. Next, cell lysates were centrifuged at 14,000×g for 10 min at 4 °C. The resulting supernatants were mixed with 5 μg of the indicated antibodies and Dynabeads protein A/G (Veritus, DB10001/10003), and rotated at 4 °C overnight. The following day, beads were washed three times with lysis buffer. Whole-cell lysates and immunoprecipitates were analyzed by immunoblotting, as previously described [27].
2.7. Cell proliferation assay
Cells were seeded into 96-well plates at a density of 3000 cells per well and cultured for the indicated times. Cell viability was assessed using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Tokyo Chemical Industry, D0801) method. Briefly, 10 μL of 12 mM MTT solution was added to each well, incubated for 3 h, and stopped by adding 100 μL of STOP solution [2% acetic acid, 16% SDS (Wako, 194–13985), and 42% N,N-dimethyl formamide (Nakalai tesque, 13016–65)], as previously described [28]. Samples were mixed thoroughly and measured at 570 nm for absorbance.
2.8. Nuclear size measurement
Nuclear area and intensity measurements were normalized to controls. Averaging and statistical analyses were performed for independently repeated experiments [29]. Two-tailed Student’s t-test assuming equal variances was performed using GraphPad PRISM 7 software to evaluate statistical significance. P-values, number of independent experiments, number of nuclei quantified, and error bars are denoted in the figure legends.
2.9. Statistical analysis
Statistical analysis was performed using PRISM 7 software. Data are presented as mean ± standard deviation. Statistically significant differences in mean or median values between respective groups were tested by Student’s t-test or one-way ANOVA analysis. P values < 0.05 were considered to indicate a statistically significant difference.
3. Results
3.1. Overexpression of SARS-CoV-2 ORF6 displaced RAE1 from the nuclear membrane
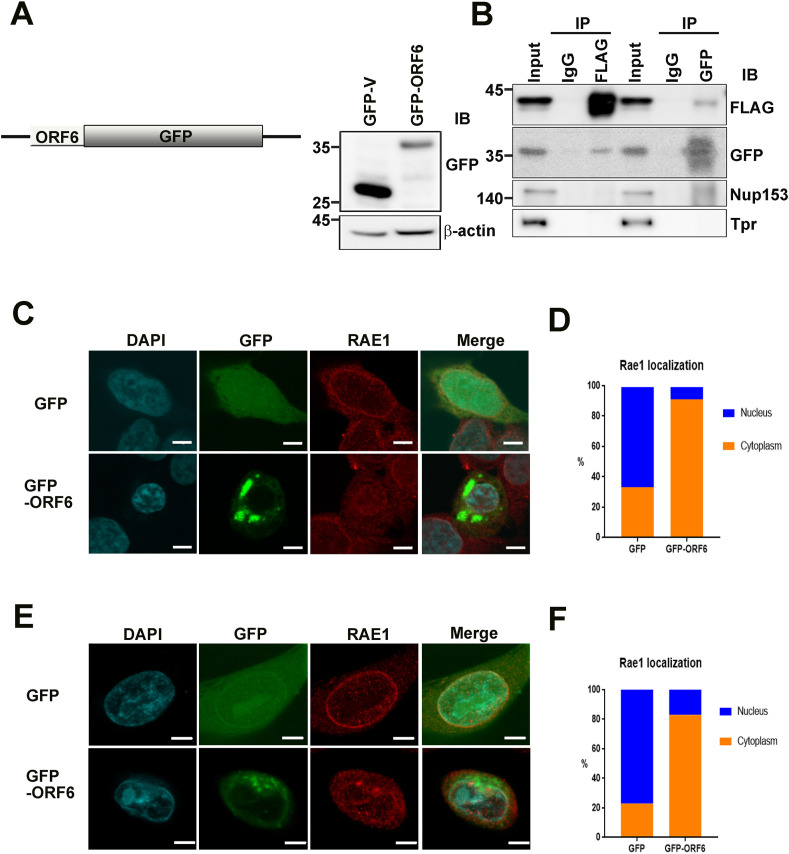
Given that RAE1 localizes around the nuclear rim and nucleoplasm, as well as in the cytoplasm, we hypothesized that there might be manifold forms of RAE1 and that ORF6 protein spatiotemporally interacts with RAE1 to form several ORF6-RAE1 protein complexes involved in reticence of host gene expression. To confirm their interaction, we first generated a fluorescent GFP-ORF6 construct and transfected it into HEK293T cells, using GFP vector as control (Fig. 1 A).
Fig. 1.
SARS-CoV-2 ORF6 bound to RAE1 and changed RAE1 localization in HEK293T and PC9 cells. (A) Schematic diagram and expression of ORF6-green fluorescent protein fusion protein (pEGFP-ORF6). Right panel: pEGFP-N1 and pEGFP-ORF6 were transiently expressed in HEK293T cells. Lysates were analyzed by immunoblotting using a GFP antibody. (B) Immunoprecipitation of pEGFP-ORF6 and FLAG-RAE1 co-transfected HEK293T cell extracts with anti-FLAG, anti-GFP, or nonspecific rabbit antibodies (IgG) as analyzed by SDS-PAGE, and immunoblotting with antibodies against FLAG, GFP, NUP153, and Tpr. (C, E) Confocal images of GFP and RAE1 localization in HEK293T (C) and PC9 (E) cells transfected with pEGFP-N1 or pEGFP-ORF6 (Scale bars, 5 μm). Green, anti-GFP; red, anti-RAE1; blue, chromatin (DAPI). (D, F) Quantification (relative percentage) of cytoplasmic and nuclear RAE1 localization in HEK293T cells (D) transfected with pEGFP-N1 (n = 57) or pEGFP-ORF6 (n = 57), and PC9 cells (F) transfected with pEGFP-N1 (n = 43) or pEGFP-ORF6 (n = 58). GFP, green fluorescent protein; NUP153, nucleoporin 153; ORF6, open-reading frame 6; RAE1, RNA export factor 1; SARS-CoV-2, severe acute respiratory syndrome corona virus 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Immunoprecipitation analysis of HEK293T cells co-transfected with GFP-ORF6 and FLAG-RAE1 suggested that ORF6 unambiguously interacts with RAE1, but not other nucleoporins such as NUP153 or TPR (Fig. 1B). These results are consistent with recent IP-MS and BioID-based interactome data [3,5]. To examine RAE1 localization in ORF6-overexpressing cells, we performed confocal microscopy. As shown in Fig. 1C and D, ORF6 overexpression induced marked displacement of RAE1 from the nuclear rim to the cytoplasm in HEK293T cells and the human lung carcinoma cell line PC9 (Fig. 1E and F). These results suggest that disturbance of mRNA nucleocytoplasmic transport proteins related to RAE1 function and the inhibition of host gene expression by SARS-CoV-2 ORF6 are correlated with alterations in RAE1 distribution. Interestingly, we also observed a nuclear size reduction in ORF6-transfected cells compared with controls [Supplemental Fig. 1A (n = 50)]. As shown in Supplemental Fig. 1B, montages of representative DAPI-stained nuclei exhibited ORF6 overexpression that led to reduced nuclear size. Taken together, these results indicate that modulation of ORF6 levels affects localization of the NPC protein RAE1, suggesting that changes in nuclear RNA export capacity underlie observed changes in nuclear size.
3.2. Overexpression of SARS-CoV-2 ORF6 contributed to displacement of the RAE1 binding partner NUP98 from the nuclear membrane
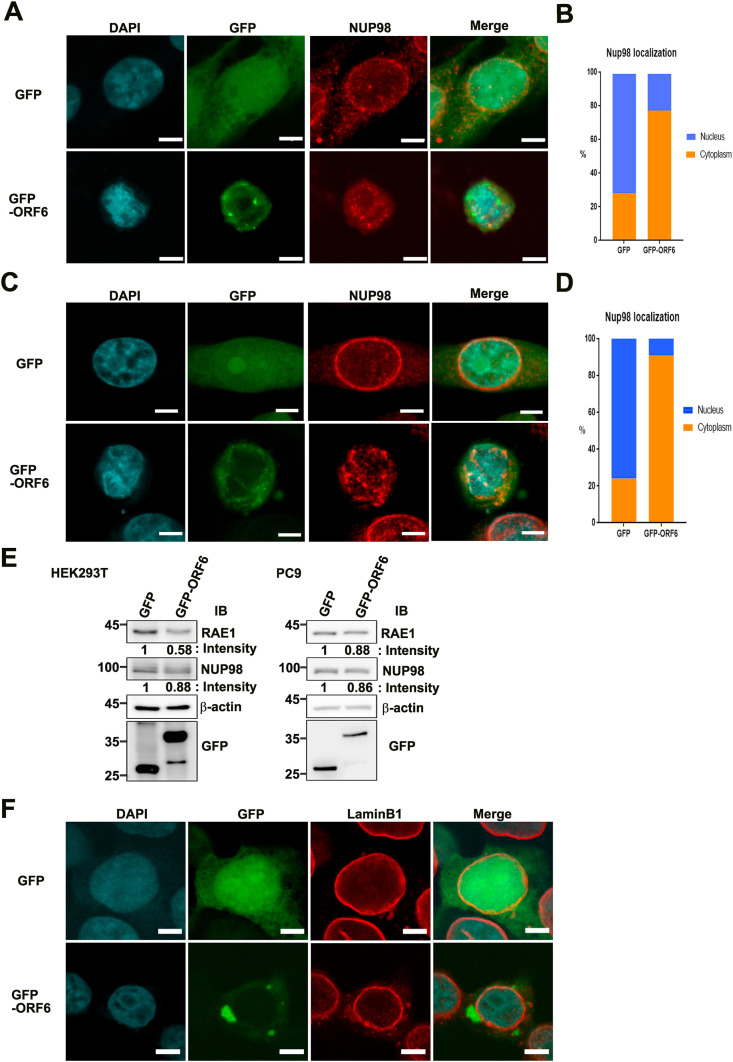
We next examined the RAE1 binding partner NUP98 using co-immunofluorescence staining. The results demonstrated that GFP-ORF6 overexpression induced marked displacement of NUP98 from the nuclear rim to the cytoplasm in HEK293T cells Fig. 2 A and B (n = 70) and PC9 cells Fig. 2C and D (n = 54 and 21, respectively) compared with GFP vector control cells. Moreover, we found that ORF6 overexpression affected RAE1 and NUP98 protein levels, as estimated by immunoblotting experiments (Fig. 2E). Further double staining with Lamin B1, a nuclear membrane marker, indicated that ORF6 overexpression had no effect on nuclear rim staining, as it retained its usual punctuate distribution (Fig. 2F). These results provide cellular evidence that ORF6 expression alone is sufficient to disturb the nucleocytoplasmic mRNA transport factor RAE1 and NUP98, contributing to mislocalization of host endogenous mRNA export machinery and ultimately driving nuclear size reduction.
Fig. 2.
SARS-CoV-2 ORF6 altered localization of NUP98 but not lamin B1. (A, C) Confocal images of GFP and NUP98 localization in HEK293T (A) and PC9 (C) cells transfected with pEGFP-N1 or pEGFP-ORF6 (Scale bars, 5 μm). Green, anti-GFP; red, anti-NUP98; blue, chromatin (DAPI). (B, D) Quantification (relative %) of nuclear and cytoplasmic NUP98 localization in HEK293T cells (B) transfected with pEGFP-N1 (n = 70) or pEGFP-ORF6 (n = 70), and PC9 cells (D) transfected with pEGFP-N1 (n = 54) or pEGFP-ORF6 (n = 21). (E) Immunoblot analysis of RAE1 and NUP98 in pEGFP-ORF6-transfected HEK293T (left) and PC9 (right) cells. (F) Confocal images of GFP and lamin B1 localization in HEK293T cells transfected with pEGFP-N1 or pEGFP-ORF6. GFP, green fluorescent protein; hnRNPA1, heterogeneous nuclear ribonucleoprotein A1; NPC, nuclear pore complex; NUP98, nucleoporin 98; ORF6, open-reading frame 6; RAE1, RNA export factor 1; SARS-CoV-2, severe acute respiratory syndrome corona virus 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Overexpression of SARS-CoV-2 ORF6 induced strong nuclear accumulation of hnRNPA1
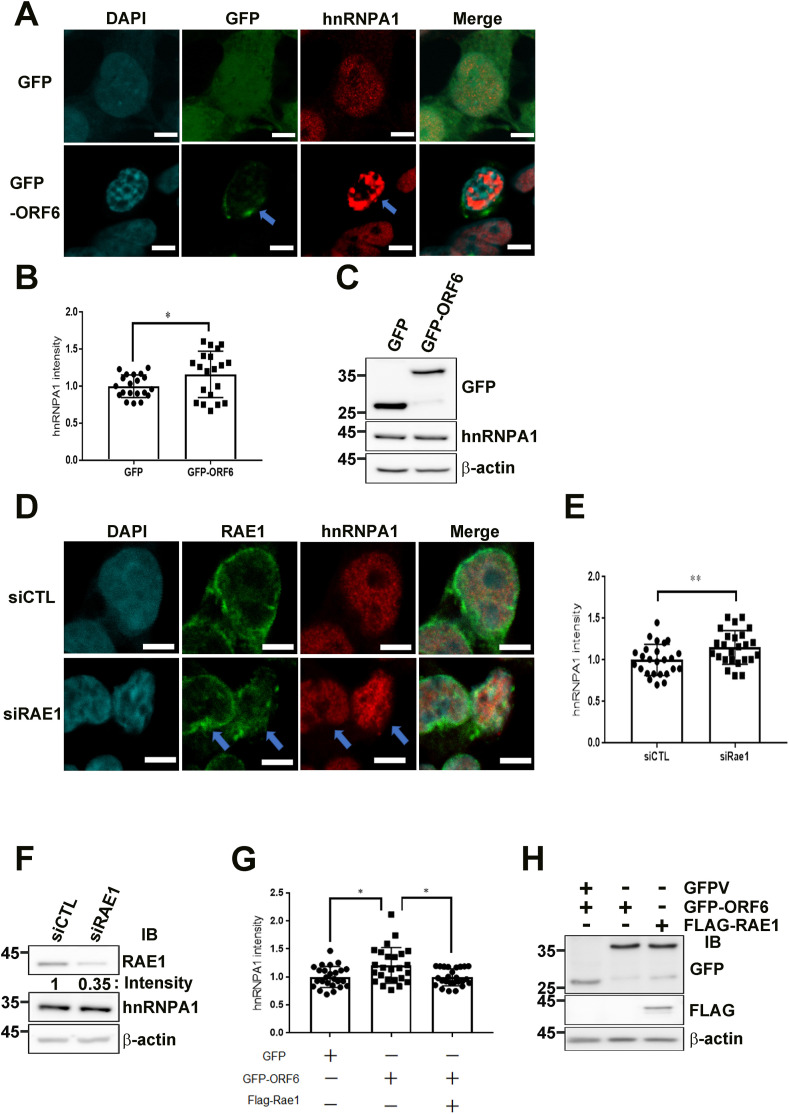
Export of mRNAs is a multistep process involving the packaging of mRNAs into messenger ribonucleoprotein particles (mRNPs), their transport through NPCs, and mRNP remodeling events prior to translation [22]. Thus, we next investigated hnRNPA1 expression and localization. Normally, hnRNPA1 is bound to RNAs during their processing in the nucleus, as well as some cytoplasmic mRNAs. hnRNPA1-containing mRNPs are thought to chaperone mRNAs through the NPC, which undergoes dynamic changes in structure and composition during transport. Importantly, relocalization of hnRNPA1 following VSV infection was previously correlated with inhibition of gene expression by VSV, and was found to be dependent on the host factor RAE1 [22].
We next sought to determine whether ORF6 spatiotemporally regulated hnRNPA1 subcellular distribution by manipulating RAE1 localization. HEK293T cells transfected with GFP-ORF6 were labeled with an antibody to hnRNPA1 protein and stained with DAPI, which labels DNA to define the nuclear compartment. In Fig. 3 A, mock-transfected GFP vector control samples are shown in the top row for each protein, while GFP-ORF6-transfected cells are shown in the bottom row. In control cells, the signal for hnRNPA1 was coincident with DNA staining, indicating that these proteins were in the nucleus. We found that overexpression of SARS-CoV-2 ORF6 alone did not redistribute hnRNPA1 to the cytoplasm. Interestingly, the density of hnRNPA1 was significantly increased and consistently remained inside the nuclear region of GFP-ORF6-transfected cells. This result also suggests that ORF6 overexpression induced nuclear accumulation of hnRNPA1 (Fig. 3A). Moreover, if the imaging results described above are correct, overexpression of RAE1 siRNA would partially mimick the GFP-ORF6 phenotype, including reappearance of abnormal hnRNPA1 nuclear accumulation. Indeed, confocal microscopy (Fig. 3B) and biochemical fractionation assay (Fig. 3C and D) of siRNA-mediated RAE1-knockdown HEK293T cells revealed that hnRNPA1 protein levels were largely unaltered and hnRNPA1 was consistently accumulated inside nuclei, respectively. Moreover, if our hypothesis is correct, overexpression of FLAG-RAE1 and GFP-ORF6 would partially relieve hnRNPA1 stacking through sequestration of endogenous nuclear hnRNPA1. Consistent with our prediction, we found that hnRNPA1 nuclear accumulation was partially relieved by hnRNPA1 nuclear stacking (Fig. 3C and D), and inhibition of cell proliferation was also partially rescued (Supplemental Fig. 2). Thus, a highly plausible interpretation of these results is that FLAG-RAE1 binds to and sequesters endogenous hnRNPA1.
Fig. 3.
ORF6 overexpression resulted in nuclear accumulation of mRNA binding protein hnRNPA1 by hijacking RAE1 function. (A) Confocal images of GFP and hnRNPA1 localization in HEK293T cells transfected with pEGFP-N1 or pEGFP-ORF6 (Scale bars, 5 μm). Green, anti-GFP; red, anti-hnRNPA1; blue, chromatin (DAPI). (B) Quantification of relative nuclear hnRNPA1 intensity in pEGFP-N1 or pEGFP-ORF6 transfected HEK293T cells (n = 20, respectively). Unpaired t-test was performed using GraphPad QuickCalcs. ∗p < 0.05. (C) Immunoblot analysis of hnRNPA1 in GFP-ORF6-transfected HEK293T cells. (D) Confocal images of RAE1 and hnRNPA1 localization in HEK293T cells transfected with control or RAE1 siRNA (Scale bars, 5 μm). Green, anti-RAE1; red, anti-hnRNPA1; blue, chromatin (DAPI). (E) Quantification of relative nuclear hnRNPA1 intensity in HEK293T cells transfected with control or RAE1 siRNA (n = 25 per group). Unpaired t-test was performed using GraphPad QuickCalcs. ∗∗p < 0.01. (F) Immunoblot analysis of RAE1 and hnRNPA1 in HEK293T cells transfected with control or RAE1 siRNA. (G) Quantification of relative nuclear hnRNPA1 intensity in HEK293T cells transfected with GFP or GFP-ORF6, or co-transfected with GFP-ORF6 and FLAG-RAE1 (n = 25 per group). Dunnett’s multiple comparisons test was performed using GraphPad QuickCalcs. ∗∗p < 0.01. (H) Immunoblot analysis of GFP and FLAG in HEK293T cells transfected with pEGFP-N1 or pEGFP-ORF6, or co-transfected with pEGFP-ORF6 and FLAG-RAE1. GFP, green fluorescent protein; hnRNPA1, heterogeneous nuclear ribonucleoprotein A1; NPC, nuclear pore complex; ORF6, open-reading frame 6; RAE1, RNA export factor 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Here, we provide critical insights supporting the notion that SARS-CoV-2 ORF6 could be important for virus replication via dislocation of nuclear pore proteins RAE1 and NUP98. Our results delineate a cascade of molecular and cellular events initiated by the inflow of ORF6 into cells. We speculate that SARS-CoV-2 ORF6 hijacks and subverts the host mRNA export system. SARS-CoV-2 ORF6 appears to hijack host NPC proteins RAE1 and NUP98 to suppress host translational activity by regulating hnRNP trafficking dynamics. This hijacking of NPC proteins may be one of the essential mechanisms leading to COVID-19. Using PYMOL (http://pymol.org), we performed electrostatic mapping via the Adaptive Poisson-Boltzmann Solver. We also simulated two distinct predicted models of ORF6 three-dimensional structures (Supplemental Fig. 3; Supplemental Movie 1). Moreover, we recently developed a nanoscopic technique to visualize docking of individual influenza viral proteins to exosomes via high-speed atomic force microscopy [[30], [31], [32], [33]]. It will be useful to visualize ORF6 and NPC protein dynamics in real time in the near future.
SARS-CoV-2 encodes several interferon antagonists that delay host cell recognition of infection, innate immune sensing, and signaling pathways, as well as interferon-stimulated gene expression [34]. ORF6 also reportedly has critical roles in host innate immunity [[35], [36]]. Li et al. screened the viral proteins of SARS-CoV-2 and found that viral ORF6, ORF8, and nucleocapsid proteins were potential inhibitors of the type I interferon signaling pathway, a key component for antiviral responses of host innate immunity. ORF6 strongly inhibited interferon beta (IFN-β) and nuclear factor κB-responsive promoters, and was able to inhibit the interferon-stimulated response element (ISRE) after infection with Sendai virus; notably, only ORF6 and ORF8 proteins were able to inhibit the ISRE after treatment with IFN-β [35]. Lei et al. showed that ORF6 hinders both type I interferon production and downstream signaling, and that the C-terminus region of ORF6 is critical for its antagonistic effect [36].
To increase viral replication and transmission between hosts, many pathogenic RNA viruses encode proteins that specifically antagonize nuclear import to inhibit host innate immunity and other critical cellular macromolecular processes [37]. For instance, some cardiovirus L proteins directly interact with Ran-GTPases, which are required for the export of new nuclear mRNA [38]. The Nipah virus W protein prevents both virus- and Toll-like receptor 3-triggered signaling in infected cells by inhibiting phosphorylation and activation of STAT1, as well as subsequent downstream interferon-stimulated gene induction [39]. Similarly, Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear import [40]. However, whether inhibition of host innate immunity by ORF6 is related to RAE1 and NUP98 nuclear displacement and nuclear size reduction remains to be investigated.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Acknowledgements
We thank all members of the Wong laboratory. We also thank Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript. This work was supported by MEXT/JSPS KAKENHI grant numbers 19K07398 (A.K.), 17H05874 and 17K08655 (R.W) from MEXT Japan, and by grants from the NanoLSI Grant for Transdisciplinary Research Promotion (S.K), Kanazawa University against COVID-19 (R.W), the Kobayashi International Scholarship Foundation (R.W), and the Shimadzu Science Foundation (R.W).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2020.11.115.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon D.E., Jang G.M., Bouhaddou M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouhaddou M., Memon D., Meyer B. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.St-Germain J.R., Astori A., Samavarchi-Tehrani P. A SARS-CoV-2 BioID-based virus-host membrane protein interactome and virus peptide compendium: new proteomics resources for COVID-19 research. bioRxiv. 2020;28:269175. doi: 10.1101/2020.08.28.269175. [DOI] [Google Scholar]
- 6.Brown J.A., Bharathi A., Ghosh A. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J. Biol. Chem. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- 7.Kraemer D., Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakano H., Wang W., Hashizume C., Funasaka T., Sato H., Wong R.W. Unexpected role of nucleoporins in coordination of cell cycle progression. Cell Cycle. 2011;10:425–433. doi: 10.4161/cc.10.3.14721. [DOI] [PubMed] [Google Scholar]
- 9.Babu J.R., Jeganathan K.B., Baker D.J. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub 3 to prevent chromosome missegregation. J. Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong R.W., Blobel G., Coutavas E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19783–19787. doi: 10.1073/pnas.0609582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong R.W. Interaction between Rae1 and cohesin subunit SMC1 is required for proper spindle formation. Cell Cycle. 2010;9:198–200. doi: 10.4161/cc.9.1.10431. [DOI] [PubMed] [Google Scholar]
- 12.Wong R.W., Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15441–15445. doi: 10.1073/pnas.0807660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeganathan K.B., Baker D.J., van Deursen J.M. Securin associates with APCCdh1 in prometaphase but its destruction is delayed by Rae1 and Nup98 until the metaphase/anaphase transition. Cell Cycle. 2006;5:366–370. doi: 10.4161/cc.5.4.2483. [DOI] [PubMed] [Google Scholar]
- 14.Funasaka T., Balan V., Raz A., Wong R.W. Nucleoporin Nup98 mediates galectin-3 nuclear-cytoplasmic trafficking. Biochem. Biophys. Res. Commun. 2013;434:155–161. doi: 10.1016/j.bbrc.2013.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funasaka T., Nakano H., Wu Y., Hashizume C., Gu L., Nakamura T., Wang W., Zhou P., Moore M.A., Sato H., Wong R.W. RNA export factor RAE1 contributes to NUP98-HOXA9-mediated leukemogenesis. Cell Cycle. 2011;10:1456–1467. doi: 10.4161/cc.10.9.15494. [DOI] [PubMed] [Google Scholar]
- 16.Zheng X., Zhao X., Zhang Y. RAE1 promotes BMAL1 shuttling and regulates degradation and activity of CLOCK: BMAL1 heterodimer. Cell Death Dis. 2019;10:62. doi: 10.1038/s41419-019-1346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh J.H., Lee J.Y., Yu S. RAE1 mediated ZEB1 expression promotes epithelial-mesenchymal transition in breast cancer. Sci. Rep. 2019;9:2977. doi: 10.1038/s41598-019-39574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quan B., Seo H.S., Blobel G., Ren Y. Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 • Nup98) Proc. Natl. Acad. Sci. U.S.A. 2014;111:9127–9132. doi: 10.1073/pnas.1409076111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajani K.R., Kneller E.L.P., McKenzie M.O. Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakraborty P., Seemann J., Mishra R.K., Wei J.H., Weil L., Nussenzveig D.R., Heiber J., Barber G.N., Dasso M., Fontoura B.M. Vesicular stomatitis virus inhibits mitotic progression and triggers cell death. EMBO Rep. 2009;10:1154–1160. doi: 10.1038/embor.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren Y., Seo H.-S., Blobel G., Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10406–10411. doi: 10.1073/pnas.1005389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kneller E.L.P., Connor J.H., Lyles D.S. hnRNPs Relocalize to the cytoplasm following infection with vesicular stomatitis virus. J. Virol. 2009;83:770–780. doi: 10.1128/JVI.01279-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis A.C., Melikyan G.B. Single HIV-1 imaging reveals progression of infection through CA-dependent steps of docking at the nuclear pore, uncoating, and nuclear transport. Cell Host Microbe. 2018;23:536–548. doi: 10.1016/j.chom.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong R.W., Mamede J.I., Hope T.J. Impact of nucleoporin-mediated chromatin localization and nuclear architecture on HIV integration site selection. J. Virol. 2015;89:9702–9705. doi: 10.1128/JVI.01669-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazawa M., Sakai K., Kobayashi A., Yoshino H., Iga Y., Iwashima Y., Lim K.S., Voon D.C., Jiang Y., Horike S., Lin D.C., Wong R.W. Disease-specific alteration of karyopherin-a subtype establishes feed-forward oncogenic signaling in head and neck squamous cell carcinoma. Oncogene. 2020;39:2212–2223. doi: 10.1038/s41388-019-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi A., Hashizume C., Dowaki T., Wong R.W. Therapeutic potential of mitotic interaction the nucleoporin Tpr and aurora kinase A. Cell Cycle. 2015;14:1447–1458. doi: 10.1080/15384101.2015.1021518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dewi F.R.P., Jiapaer S., Kobayashi A., Hazawa M., Ikliptikawati D.K., Hartono, Sabit H., Nakada M., Wong R.W. Nucleoporin TPR (translocated promoter region, nuclear basket protein) upregulation alters MTOR-HSF1 trails and suppresses autophagy induction in ependymoma. Autophagy. 2020;24:1–12. doi: 10.1080/15548627.2020.1741318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazawa M., Lin D.C., Kobayashi A., Jiang Y.Y., Xu L., Dewi F.R.P., Mohamed M.S., Hartono, Nakada M., Meguro-Horike M., Horike S.I., Koeffler H.P., Wong R.W. ROCK-dependent phosphorylation of NUP62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 2018;19:73–88. doi: 10.15252/embr.201744523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jevtić P., Schibler A.C., Wesley C.C. The nucleoporin ELYS regulates nuclear size by controlling NPC number and nuclear import capacity. EMBO Rep. 2019;20 doi: 10.15252/embr.201847283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim K.S., Kodera N., Wang H., Mohamed M.S., Hazawa M., Kobayashi A., Yoshida T., Hanayama R., Yano S., Ando T., Wong R.W. High-speed afm reveals molecular dynamics of human influenza A hemagglutinin and its interaction with exosomes. Nano Lett. 2020 doi: 10.1021/acs.nanolett.0c01755. [DOI] [PubMed] [Google Scholar]
- 31.Lim K.S., Mohamed M.S., Wang H., Hartono, Hazawa M., Kobayashi A., Voon D.C., Kodera N., Ando T., Wong R.W. Direct visualization of avian influenza H5N1 hemagglutinin precursor and its conformational change by high-speed atomic force microscopy. Biochim. Biophys. Acta Gen. Subj. 2020;1864:129313. doi: 10.1016/j.bbagen.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed M.S., Hazawa M., Kobayashi A., Guillaud L., Watanabe-Nakayama T., Nakayama M., Wang H., Kodera N., Oshima M., Ando T., Wong R.W. Spatiotemporally tracking of nano-biofilaments inside the nuclear pore complex core. Biomaterials. 2020;256:120198. doi: 10.1016/j.biomaterials.2020.120198. [DOI] [PubMed] [Google Scholar]
- 33.Mohamed M.S., Kobayashi A., Taoka A., Watanabe-Nakayama T., Kikuchi Y., Hazawa M., Minamoto T., Fukumori Y., Kodera N., Uchihashi T., Ando T., Wong R.W. High-speed atomic force microscopy reveals loss of nuclear pore resilience as a dying code in colorectal cancer cells. ACS Nano. 2017;11:5567–5578. doi: 10.1021/acsnano.7b00906. [DOI] [PubMed] [Google Scholar]
- 34.Yuen C.K., Lam J.Y., Wong W.M. SARS-CoV-2 nsp 13, nsp 14, nsp 15 and orf6 function as potent interferon antagonists. Emerg. Microb. Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J.Y., Liao C.H., Wang Q. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei X., Dong X., Ma R. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sims A.C., Tilton S.C., Menachery V.D. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J. Virol. 2013;87:3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter F.W., Palmenberg A.C. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J. Virol. 2009;83:1941–1951. doi: 10.1128/JVI.01752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw M.L., Cardenas W.B., Zamarin D. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J. Virol. 2005;79:6078–6088. doi: 10.1128/JVI.79.10.6078-6088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid S.P., Leung L.W., Hartman A.L. Ebola virus VP24 binds karyopherin alpha 1 and blocks STAT1 nuclear accumulation. J. Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.