Abstract
Objective
The objective of this study was to compare the performance of an RA algorithm developed and trained in 2010 utilizing natural language processing and machine learning, using updated data containing ICD10, new RA treatments, and a new electronic medical records (EMR) system.
Methods
We extracted data from subjects with ≥1 RA International Classification of Diseases (ICD) codes from the EMR of two large academic centres to create a data mart. Gold standard RA cases were identified from reviewing a random 200 subjects from the data mart, and a random 100 subjects who only have RA ICD10 codes. We compared the performance of the following algorithms using the original 2010 data with updated data: (i) a published 2010 RA algorithm; (ii) updated algorithm, incorporating ICD10 RA codes and new DMARDs; and (iii) published algorithm using ICD codes only, ICD RA code ≥3.
Results
The gold standard RA cases had mean age 65.5 years, 78.7% female, 74.1% RF or antibodies to cyclic citrullinated peptide (anti-CCP) positive. The positive predictive value (PPV) for ≥3 RA ICD was 54%, compared with 56% in 2010. At a specificity of 95%, the PPV of the 2010 algorithm and the updated version were both 91%, compared with 94% (95% CI: 91, 96%) in 2010. In subjects with ICD10 data only, the PPV for the updated 2010 RA algorithm was 93%.
Conclusion
The 2010 RA algorithm validated with the updated data with similar performance characteristics as the 2010 data. While the 2010 algorithm continued to perform better than the rule-based approach, the PPV of the latter also remained stable over time.
Keywords: rheumatoid arthritis, electronic medical record, machine learning
Rheumatology key messages
Many electronic medical record-based rheumatoid arthritis algorithms have not been tested with contemporary data.
Validation of an RA algorithm from 2010 on 2017 data resulted in similar high performance.
The RA algorithm developed in 2010 was temporally robust despite diagnosis code and medication changes.
Introduction
Increasingly, the clinical data housed in electronic medical records (EMRs) are also being utilized for clinical research studies. While the EMR contains a wealth of clinical data, a major challenge has been integrating the large amount of diverse data to study specific conditions and outcomes. To address this bottleneck, investigators have applied both rule-based algorithms and machine learning methods to classify patients with specific conditions for study [1–3]. For RA, various algorithms exist. The simplest are the rule-based approaches using only structured data, e.g. ≥3 RA International Classification of Diseases (ICD) codes [4]. Our group published an algorithm developed using machine learning in 2010 that included RA ICD9 codes and data extracted from the narrative notes using natural language processing (NLP) [5–9]. This algorithm was successfully ported and validated at two independent academic institutions [7]. The RA cohorts developed using this algorithm served as a foundation for a wide range of studies, including GWAS and PheWAS investigating RA risk alleles and associated conditions, as well as cohort studies investigating RA comorbidities [10–19]. While EMR-based algorithms are now increasingly being used, the impact of secular changes on the performance of these algorithms has not been closely examined.
The past decade has seen significant changes in both the management of RA and with the EMR data itself. Changes include the adoption of ICD 10th edition, and adoption of new EMR systems. Since 2010, there have been six new biologic DMARDs approved by the US Food and Drug Administration, and therefore not considered as a potential variable in the original RA algorithms [20–25]. Our institution has also adopted a new EMR system introducing different data types.
The objective of this study was: (i) to evaluate the temporal validity of the 2010 RA algorithm developed using ICD9 codes by testing the previously published algorithm on current EMR data; (ii) to investigate whether the performance of the original 2010 RA algorithm can be improved updating variables; and (iii) to study the impact and performance of ICD10 on the classification of RA using these algorithms.
Materials and methods
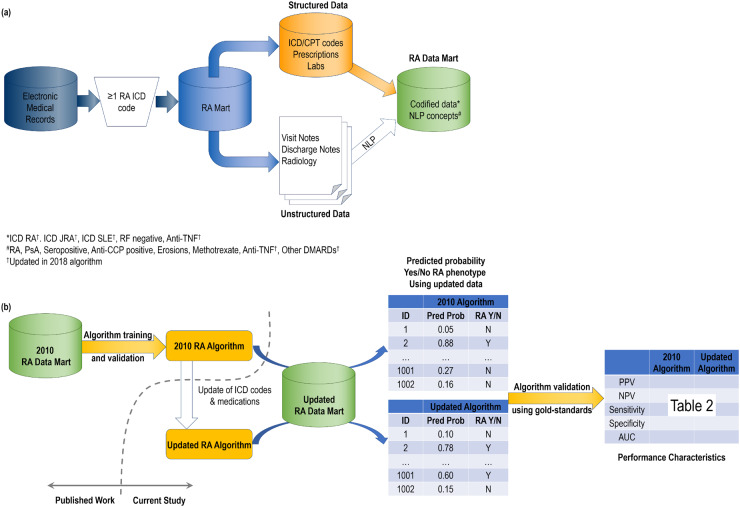
Our approach is outlined in Fig. 1, which also outlines the features included in the original 2010 RA algorithm vs the updated algorithm.
Fig. 1.
Overview of classification algorithms used for rheumatoid arthritis using electronic medical record data
(a) Development and feature curation into RA Data Mart. (b) Schematic for validation of the 2010 RA algorithm and updated algorithm using updated RA Data Mart. Anti-CCP: cyclic citrullinated peptide antibody; AUC: area under the receiver operator characteristic curve; CPT: current procedural terminology; DMARD: disease-modifying antirheumatic drug; ICD: International Classification of Diseases; NLP: natural language processing; NPV: negative predictive value; PPV: positive predictive value; PsA: psoriatic arthritis.
Data source
We used EMR data from two large academic hospitals in Boston, MA: the Brigham and Women’s Hospital (BWH) and Massachusetts General Hospital (MGH). BWH and MGH used the same locally-developed EMR system; the EMR was initiated at BWH on 1 October 1996 and at MGH on 3 October 1994. Epic EMR, a commercial EMR system was subsequently adopted in 2015 by BWH and 2016 by MGH. This commercial EMR system had a different provider-facing interface for note entry and ICD/procedure coding, which may result in differences in the data collected. As a first step, we applied a filter of ≥1 RA ICD9 or ICD10 code (714.x except 714.3x, M05.x, M06.x, M12.0x, M12.3x), and ≥2 notes with length >500 characters, to create a ‘RA data mart’. The RA data mart entry date was the inception of the locally-developed EMR system at the two hospitals. Using these criteria, 53 144 subjects were included in the RA data mart with data up to 3 November 2017, the date of creation for the RA data mart.
Codified data
We extracted codified data including ICD9 and ICD10 codes, electronic prescriptions, as well as RF and antibodies to cyclic citrullinated peptide (anti-CCP) laboratory results. The ICD9/10 codes included RA and related diseases (714.x except 714.3x, M05.x, M06.x except M06.1, M12.0x, M12.3x), SLE (710.0; M32.1x) and JIA/JRA (714.3x; M08.0x, M08.2x, M08.3, M08.4x). We mapped the ICD10 codes from the ICD9 codes included in the original 2010 algorithm using the Centers for Medicare and Medicaid General Equivalence Mappings (CMS GEMS) tool. The mappings were reviewed by a study rheumatologist to ensure accuracy and completeness.
Non-biologic DMARD included: methotrexate, azathioprine, leflunomide, sulfasalazine, hydroxychloroquine, penicillamine, ciclosporin and gold. Biologic DMARDs included: anti-TNF agents, infliximab, adalimumab, etanercept, certolizumab and golimumab; as well as abatacept, rituximab, anakinra, tocilizumab, sarilumab and tofacitinib. RF and anti-CCP positivity were determined by hospital laboratory cut-offs. Medication and serology data were coded as never vs ever present. Healthcare utilization was approximated using the total number of ICD code counts. The original 2010 RA algorithm used the total number of interactions with the healthcare system as a proxy for healthcare utilization. Carroll et al.’s study, which applied the RA algorithm at different institutions, used the total number of ICD code counts as a proxy for healthcare utilization [7]. Total ICD counts were found to vary less across institutions while still providing information on healthcare utilization. Thus, we employed the same approach to continue to facilitate portability.
Narrative EMR data
We extracted narrative data using NLP from health care provider notes, radiology reports, pathology reports, discharge summaries and operative reports. NLP was performed using the Narrative Information Linear Extraction (NILE) package [26]. The NLP concepts extracted from the narrative data were: RA, psoriatic arthritis (PsA), SLE, seropositive, anti-CCP positive, erosions, methotrexate, anti-TNF, and all other DMARDs were included in a category called ‘other DMARDs’ (abatacept, anakinra, azathioprine, cyclophosphamide, ciclosporin, hydroxychloroquine, gold, leflunomide, penicillamine, sarilumab, sulfasalazine, tocilizumab, tofacitinib, rituximab). The NLP mentions for each disease (RA, PsA, SLE) were summed; whereas medication and serology data were coded as never vs ever present.
Gold standard set
The gold standard validation set was established by randomly selecting 200 subjects from the RA Mart, and reviewing their records for the presence of RA. Based on the phenotyping methods described by Zhang et al. [27], n = 200 was determined to be sufficient for the validation set, as algorithm retraining was not required. We additionally sampled a subset of n = 100 subjects who have RA ICD10 codes only. These are subjects who have RA follow-up in the EMR only after 2015, when the billing codes were switched from ICD9 to ICD10. In the chart review, subjects were identified as definite, probable and not RA based on RA diagnosis by a rheumatologist. The presence of the 2010 ACR/EULAR classification criteria for RA was also documented [28]. Subjects classified as definite RA were considered as ‘cases’ and subjects with possible or no RA were considered as ‘controls’.
Evaluation of the classification algorithms
We assessed the performance characteristics of three different algorithms using the most recent data in the: (i) original 2010 published logistic regression algorithm [5]; (ii) 2017 updated algorithm using the same model coefficients while incorporating ICD10 codes and updated medications to include newer anti-TNFs (golimumab and certolizumab), anti-IL-6 (tocilizumab, sarilumab) and JAK inhibitors (tofacitinib); and (iii) rule-based ICD RA code ≥ 3 (ICD9, ICD10, ICD9 + 10) [4]. Please see details on variables used and regression coefficients in Table 1.
Table 1.
Variables and component updates for the 2010 and 2017 updated RA logistic regression algorithm
| Variable | Regression coefficient | 2010 algorithm | 2017 updated algorithma |
|---|---|---|---|
| Positive predictors | |||
| NLP RA | 0.970 | ||
| NLP seropositive | 2.77 | ||
| ICD RA normalizedb | 66.0 | 714.xx excluding 714.3x | M05.x, M06.x excluding M06.1, M12.0x, M12.3x |
| ICD RAc | 0.639 | 714.xx excluding 714.3x | M05.x, M06.x excluding M06.1, M12.0x, M12.3x |
| NLP erosions | 1.26 | ||
| Codified RF negative | 0.851 | ||
| NLP methotrexate | 0.632 | ||
| Codified anti-TNF | 0.959 | infliximab, etanercept | adalimumab, certolizumab pegol, golimumab |
| NLP anti-CCP positive | 1.31 | ||
| NLP anti-TNF | 0.521 | infliximab, etanercept, adalimumab | certolizumab pegol, golimumab |
| NLP other DMARDs | 0.298 | cDMARDs: azathioprine, leflunomide, sulfasalazine, hydroxychloroquine, penicillamine, cyclosporine, gold. | bDMARDs: tocilizumab, sarilumab, tofacitinib. |
| bDMARDs: abatacept, rituximab, anakinra. | |||
| Negative predictors | |||
| ICD JRAc | −2.25 | 714.3x | M06.1, M08.0x, M08.2x, M08.3, M08.4x |
| ICD SLEc | −0.959 | 710.0 | M32.1x |
| NLP PsA | −0.856 | ||
Codified and NLP concepts included in addition to those listed in the 2010 algorithm. bICD RA normalized = number of ICD RA codes per subject normalized by number of ‘facts’.. cComputed as log(1+ICD counts). ICD counts computed 7 days apart. anti‐CCP: anti–cyclic citrullinated peptide; b: biologic; c: conventional; EMR: electronic medical record; ICD: International Classification of Diseases; NLP: natural language processing; PsA: psoriatic arthritis.
The application of the original 2010 algorithm to the 2017 updated data mart was achieved by using the same model coefficients as well as variable definitions as previously published, i.e. using ICD9 and previously published list of medications. The 2017 updated algorithm incorporated ICD10 codes (without roll-up) to existing variable fields as well as updated medication list, while keeping the same published model coefficients without retraining.
The positive predictive value (PPV), negative predictive value (NPV), sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) were compared across algorithms. Algorithms 1 and 2 had a specificity of 95%. Based on applications of these algorithms in prior studies [2], PPVs were used to compare the accuracy between the algorithms.
We calculated the descriptive statistics of the subjects classified as RA using the EMR-based algorithm. We further compared the descriptive statistics to subjects in the RA cohort with data after 1 January 2016 to allow a more direct comparison with recently published data from the Corrona RA registry [29].
All analyses were conducted with the R Version 3.6.1 (The R project for Statistical Computing, online at: www.r‐project.org/). All aspects of this study were approved by the Partners Healthcare Institutional Review Board.
Results
We identified 53 144 subjects with ≥1 RA ICD code and ≥2 visit notes. From chart review of 200 randomly selected subjects, the gold standard validation set identified n = 75 (37.5%) of the subjects had RA. Within the RA cases, 54 (72.0%) fulfilled the ACR/EULAR 2010 classification criteria [28]. The RA cases had a mean age of 65.5 years, 78.7% female, with 74.1% seropositive, reflecting a typical RA population (Table 2).
Table 2.
Characteristics of the validation set with RA cases defined by chart review (n = 200)
| RA cases, n = 75 | Controls (possible + no RA), n = 125 | P-value | |
|---|---|---|---|
| Age (mean (s.d.)) | 65.45 (16.10) | 66.44 (17.40) | 0.690 |
| Female (n, %) | 59 (78.7) | 89 (71.2) | 0.318 |
| Seropositivea (n, %) | 20 (74.1) | 5 (41.7) | 0.113 |
| Methotrexate (n, %) | 37 (49.3) | 13 (10.4) | <0.001 |
| Anti-TNF (n, %) | 21 (28.0) | 8 (6.4) | <0.001 |
% computed using available data. Anti-CCP: anti-cyclic citrullinated peptide; Anti-TNF: anti-tumour necrosis factor agents.
Performance of the classification algorithms
Using EMR data up to 2017, the AUC of the updated algorithms were nearly identical to the 2010 algorithm (Fig. 2) [5]. The PPV of the 2010 algorithm was 91%, with specificity 95%. The updated algorithm had a slightly higher sensitivity calculated within the gold standard set (77% vs 76%) (Table 3). When applied to the entire RA Mart, the updated algorithm classified an additional 1046 subjects that were not classified as RA using the 2010 algorithm (Table 3).
Fig. 2.
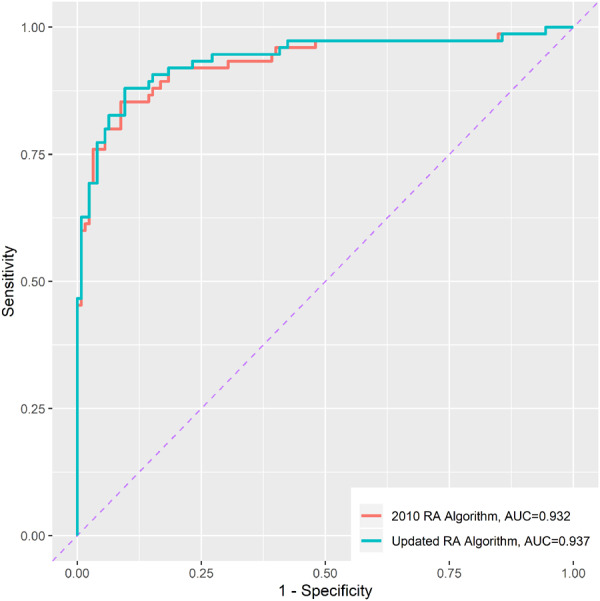
Receiver operating characteristic (ROC) curve of 2010 RA algorithm and updated RA algorithm
ROC curve and area under the ROC curve (AUC) calculated using the gold standard set (n = 200). AUC: area under the receiver operating characteristic curve.
Table 3.
Comparison of performance characteristics for RA classification algorithm
| General RA mart (n = 200) | ||||||
|---|---|---|---|---|---|---|
| AUC | PPV | NPV | Specificity | Sensitivity | # classified w/ RA by algorithm (n)c | |
| NLP-based algorithms | ||||||
| 2010 RA algorithma | 0.932 | 0.905 | 0.869 | 0.952 | 0.760 | 15 312 |
| Updated RA algorithma | 0.937 | 0.906 | 0.875 | 0.952 | 0.773 | 16 358 |
| Rule-based algorithms | ||||||
| ≥3 ICD9 RA codesb | __ | 0.536 | 0.822 | 0.592 | 0.787 | 25 707 |
| ≥3 ICD9 or ICD10 RA codesb | __ | 0.558 | 0.900 | 0.576 | 0.893 | 28 445 |
| RA subjects with ICD10 codes and no RA ICD9 codes (n = 100) | ||||||
|---|---|---|---|---|---|---|
| AUC | PPV | NPV | Specificity | Sensitivity | ||
| NLP-based algorithm | ||||||
| Updated RA algorithma | 0.784 | 0.926 | 0.600 | 0.954 | 0.472 | |
| Rule-based algorithms | ||||||
| ≥3 ICD10 RA codesb | __ | 0.585 | 0.500 | 0.500 | 0.585 | |
Specificity set at 0.95. bBinary classification, no AUC shown. cNumber computed by applying the algorithm on RA Mart. AUC: area under the receiver operating characteristic curve; ICD9/10: International Classification of Diseases; NLP: natural language processing; NPV: negative predictive value; PPV: positive predictive value.
The RA algorithms utilizing both codified and NLP data performed significantly better than the rule-based algorithm using ICD9 or ICD10 data alone (Table 3, Fig. 3). The algorithm using ≥3 RA ICD codes achieved a PPV of 56%.
Fig. 3.
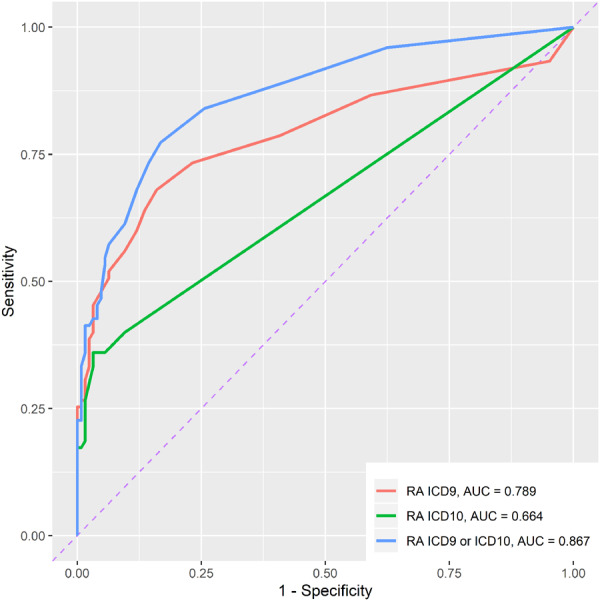
Receiver operating characteristic (ROC) curve of RA ICD codes
ROC curve and area under the ROC curve (AUC) calculated using the gold standard set (n = 200). AUC: area under the receiver operating characteristic curve; ICD: International Classification of Diseases.
The ICD10 codes had modestly better accuracy than ICD9 codes. In subjects with ICD10 codes only, the PPV of ≥3 ICD10 codes was 59% (Table 3). In comparison, in subjects with both ICD9 and ICD10 data, the PPV of ≥3 ICD9 codes was 54% (Table 3).
To anticipate the potential impact of ICD10 on the updated algorithm, it was applied to subjects with ICD10 codes only. The updated RA algorithm in this group had a modestly lower sensitivity of 49%, however, maintaining a high PPV of 93% (Table 3).
Clinical characteristics of the EMR RA cohort as classified by the updated RA algorithm
The updated RA algorithm classified a total of n = 16 358 subjects. The average age was 66.2 (s.d. 15.5). The majority of subjects were female (75.5%) and seropositive (66.5%). Using codified electronic prescription data, approximately half of the cohort had ≥1 prescription for MTX, and one-third of the cohort had a prescription for anti-TNF therapy during the follow-up period (Table 4). When restricting the summary statistics to subjects with EMR follow-up after 1 January 2016, the proportion of subjects on methotrexate was 55% and anti-TNF was 39%. Over 80% of the subjects has had at least one DMARD prescription. The proportion of subjects taking non-biologic DMARD in our RA cohort was 75.3%, compared with 82% in Corrona, and 42% for biologic DMARDs compared with 40% in Corrona [29].
Table 4.
Characteristics of subjects classified as having RA by updated RA algorithm
| EMR cohort (n=16 358) | |
|---|---|
| Age (mean (s.d.)) | 66.18 (15.45) |
| Female (n, %) | 12 344 (75.5) |
| Seropositivea (n, %) | 4470 (66.5) |
| Methotrexate (n, %) | 8057 (49.3) |
| Anti-TNF (n, %) | 5294 (32.4) |
% computed using available data. Anti-TNF: anti-tumour necrosis factor agents; EMR: electronic medical record.
Discussion
As machine-learning trained algorithms are increasingly applied to EMR data for clinical studies, it is important to routinely reassess the performance of the algorithms. These algorithms work by identifying specific patterns in the data associated with RA. For the purposes of research, the goal of these algorithms was to achieve a PPV of 90% or higher. We observed that the algorithm developed in 2010 was robust to secular changes in the EMR data over the past 7 years, including a change in coding from ICD9 to ICD10, new RA treatments, as well as a new EMR system. The overall performance characteristics between the original 2010 RA algorithm and the updated version had a similar PPV of 91% in this contemporary dataset. However, the updated algorithm had a slightly higher sensitivity, and classified 1000 additional subjects into the EMR RA cohort [5]. These codified + NLP-based algorithms both outperformed the rule-based algorithms using codified data alone. The rule-based algorithm of using ≥3 RA ICD codes performed similarly with the updated data compared with the original publication, demonstrating that the accuracy of RA ICD codes has remained relatively stable over time [5].
Additionally, we studied patients who only had ICD10 RA codes. Interestingly, in this subset, the sensitivity of the updated RA algorithm was modestly lower at 49%, while maintaining the PPV at 93%. We believe this was due to the shorter follow up time and thus less availability of EMR data for classifying these patients as having RA. Indeed, the algorithms were designed to identify prevalent disease rather than early RA. These data suggest the need for reassessment of existing rule-based approaches relying on structured data for patients with only ICD10 data. A major difference between the machine learning vs rule-based algorithm was the incorporation of NLP concepts from unstructured data. This likely provided the key RA information despite the short follow-up time, allowing for improved classification of newly-diagnosed RA subjects.
Previous research has shown that RA classification algorithms based on ICD data alone can have limited performance [30–32]. Other machine-learning based algorithms using EMR data have also demonstrated improved performance by incorporating NLP data from the unstructured data [6]. However, none of these algorithms have been temporally validated. Our study demonstrated that the NLP-based RA classification algorithm developed in 2010 is temporally robust both with and without updating the structured and unstructured data fields. The absence of significant change in NLP-based algorithm performance with incorporation of ICD10 and new medications may be due to the short duration of ICD10 usage and utilization of novel RA therapies.
The strength of the study was the ability to perform the temporal validation using a large real-world EMR dataset where the data have undergone significant changes over time. This study highlights the need to reassess algorithms and provide a roadmap for reassessing algorithms at a future point in time. By porting a previously published RA phenotyping algorithm to current data, we demonstrated the temporal robustness of the EMR-based machine-learning algorithm that incorporated both structured and NLP data. We were also able to assess the potential future impact of ICD10 by examining a subset of patients with ICD10 data only.
The study was subject to a number of limitations. In our study, we demonstrated the temporal portability of an EMR algorithm in one chronic disease, RA. The medication information collected using codified data likely underestimates treatment use in earlier years as it reflects data from electronic prescriptions that were not initially mandatory for prescribing. Thus, other modes for prescribing, e.g. paper prescriptions, telephone orders, could be used and the information would not be available in the codified data. However, when comparing more recent data (after 2016) from this RA cohort to data from an independent RA registry Corrona, with data from a similar time period, the medication use was similar. Additionally, the clinical data were performed using data from tertiary care centres where the initial studies were performed to allow for a comparison. Future studies evaluating the temporal robustness of other EMR phenotyping algorithms are needed.
In conclusion, an existing RA algorithm trained using machine-learning approaches on EMR data was robust temporally, despite the introduction of new medical information and EMR systems. The published and updated RA algorithm continue to perform better than rule-based approaches using ICD data. At this time, including ICD10 had a minimal impact on classification, and the accuracy of the ICD10 codes for RA appear similar to ICD9.
Acknowledgements
K.P.L. is supported by the Harold and DuVal Bowen Fund. All authors were involved in drafting the article or revising it critically for important intellectual contact, and all authors approved the final version to be published. S.H. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study conception and design: S.H., J.H., T.-X.C., K.P.L. Acquisition of data: S.H., J.H., T.-R.C., K.D., A.C., Z.H., J.S., I.G., C.H., T.-X.C., K.P.L. Analysis and interpretation of data: S.H., J.H., T.-R.C., K.D., A.C., Z.H., J.S., I.G., C.H., T.-X.C., K.P.L.
Funding: This work was supported by the National Institutes of Health (grant numbers P30-AR072577 (VERITY), T32-AR007530).
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1. Shivade C, Raghavan P, Fosler-Lussier E. et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc 2014;21:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liao KP, Cai T, Savova GK. et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 2015;350:h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH.. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Ann Rev Biomed Data Sci 2018;1:53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schneeweiss S, Setoguchi S, Weinblatt ME. et al. Anti-tumor necrosis factor alpha therapy and the risk of serious bacterial infections in elderly patients with rheumatoid arthritis. Arthritis Rheum 2007;56:1754–64. [DOI] [PubMed] [Google Scholar]
- 5. Liao KP, Cai T, Gainer V. et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res 2010;62:1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carroll RJ, Eyler AE, Denny JC.. Naive electronic health record phenotype identification for rheumatoid arthritis. AMIA Annu Symp Proc 2011;2011:189–96. [PMC free article] [PubMed] [Google Scholar]
- 7. Carroll RJ, Thompson WK, Eyler AE. et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012;19:e162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirby JC, Speltz P, Rasmussen LV. et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016;23:1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wei WQ, Teixeira PL, Mo H. et al. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc 2016;23:e20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kurreeman F, Liao K, Chibnik L. et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet 2011;88:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liao KP, Kurreeman F, Li G. et al. Associations of autoantibodies, autoimmune risk alleles, and clinical diagnoses from the electronic medical records in rheumatoid arthritis cases and non-rheumatoid arthritis controls. Arthritis Rheum 2013;65:571–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diogo D, Kurreeman F, Stahl EA. et al. Rare, low-frequency, and common variants in the protein-coding sequence of biological candidate genes from GWASs contribute to risk of rheumatoid arthritis. Am J Hum Genet 2013;92:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liao KP, Diogo D, Cui J. et al. Association between low density lipoprotein and rheumatoid arthritis genetic factors with low density lipoprotein levels in rheumatoid arthritis and non-rheumatoid arthritis controls. Ann Rheum Dis 2014;73:1170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liao KP, Cai T, Gainer VS. et al. Lipid and lipoprotein levels and trend in rheumatoid arthritis compared to the general population. Arthritis Care Res 2013;65:2046–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin C, Karlson EW, Canhao H. et al. Automatic prediction of rheumatoid arthritis disease activity from the electronic medical records. PLoS One 2013;8:e69932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doss J, Mo H, Carroll RJ, Crofford LJ, Denny JC.. Phenome-wide association study of rheumatoid arthritis subgroups identifies association between seronegative disease and fibromyalgia. Arthritis Rheumatol 2017;69:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liao KP, Sparks JA, Hejblum BP. et al. Phenome-wide association study of autoantibodies to citrullinated and noncitrullinated epitopes in rheumatoid arthritis. Arthritis Rheumatol 2017;69:742–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu Z, Kim SC, Vanni K. et al. Association between inflammation and systolic blood pressure in RA compared to patients without RA. Arthritis Res Ther 2018;20:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kreps DJ, Halperin F, Desai SP. et al. Association of weight loss with improved disease activity in patients with rheumatoid arthritis: a retrospective analysis using electronic medical record data. Int J Clin Rheumtol 2018;13:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimzia (certolizumab pegol) [package insert]. UCB, Inc., Symrna, GA. 2018.
- 21.Simponi (golimumab) [package insert]. Janssen Biotech, Inc., Horsham, PA. 2011.
- 22.Actemra (tocilizumab) [package insert]. Genentech, Inc., South San Francisco, CA. 2013.
- 23.Kevzara (sarilumab) [package insert]. Sanofi-Aventis US, LLC, Bridgewater, NJ. 2017.
- 24.Xeljanz (tofacitinib) [package insert]. Pfizer Labs, NY, NY. 2018.
- 25.Olumiant (baricitinib) [package insert]. Lilly USA, LLC, Indianapolis, IN. 2018.
- 26. Yu S, Cai T, Cai T. NILE: fast natural language processing for electronic health records. 2019; arXiv:1311.6063 (https://celehs.hms.harvard.edu/packages/nile/).
- 27. Zhang Y, Cai T, Yu S. et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat Protoc 2019;14:3426–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aletaha D, Neogi T, Silman AJ. et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis 2010;69:1580–8. [DOI] [PubMed] [Google Scholar]
- 29. Lee YC, Kremer J, Guan H, Greenberg J, Solomon DH.. Chronic opioid use in rheumatoid arthritis: prevalence and predictors. Arthritis Rheumatol 2019;71:670–7. [DOI] [PubMed] [Google Scholar]
- 30. Singh JA, Holmgren AR, Noorbaloochi S.. Accuracy of Veterans Administration databases for a diagnosis of rheumatoid arthritis. Arthritis Rheum 2004;51:952–7. [DOI] [PubMed] [Google Scholar]
- 31. Kim SY, Servi A, Polinski JM. et al. Validation of rheumatoid arthritis diagnoses in health care utilization data. Arthritis Res Ther 2011;13:R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ng B, Aslam F, Petersen NJ, Yu HJ, Suarez-Almazor ME.. Identification of rheumatoid arthritis patients using an administrative database: a Veterans Affairs study. Arthritis Care Res 2012;64:1490–6. [DOI] [PMC free article] [PubMed] [Google Scholar]