Significance
While transmembrane proteins are essential for cellular function, it is extremely challenging to identify their interactions using traditional biochemical methods. Here we combine proximity labeling and single molecule binding assays to discover transmembrane protein interactions in cells. We initially screen for transmembrane proteins that are located proximal to a protein of interest (the “bait”), by tagging both the extracellular and cytoplasmic regions of the bait with the proximity labeling enzyme, BioID. We then directly test binding interactions between proximal proteins and the bait using single molecule atomic force microscopy. Using this approach, we identify previously unknown direct interactions between a number of transmembrane proteins and the extracellular region of E-cadherin, an essential cell–cell adhesion protein.
Keywords: cadherin, heterophilic binding, BioID, proteomics, atomic force microscopy
Abstract
We combine proximity labeling and single molecule binding assays to discover transmembrane protein interactions in cells. We first screen for candidate binding partners by tagging the extracellular and cytoplasmic regions of a “bait” protein with BioID biotin ligase and identify proximal proteins that are biotin tagged on both their extracellular and intracellular regions. We then test direct binding interactions between proximal proteins and the bait, using single molecule atomic force microscope binding assays. Using this approach, we identify binding partners for the extracellular region of E-cadherin, an essential cell–cell adhesion protein. We show that the desmosomal proteins desmoglein-2 and desmocollin-3, the focal adhesion protein integrin-α2β1, the receptor tyrosine kinase ligand ephrin-B1, and the classical cadherin P-cadherin, all directly interact with E-cadherin ectodomains. Our data shows that combining extracellular and cytoplasmic proximal tagging with a biophysical binding assay increases the precision with which transmembrane ectodomain interactors can be identified.
Transmembrane proteins play essential roles in coupling cells and in sensing mechanical and biochemical signals from the environment. However, it is extremely challenging to identify membrane protein interactions using traditional methods like affinity pulldown because of the hydrophobic nature of membrane spanning regions, harsh extraction conditions that disrupt protein interactions, and the inability of these methods to discriminate between cytoplasmic and extracellular protein interactions. Consequently, membrane protein interactomes have been mapped using several proximity-tagging systems such as BioID (1), APEX (2), and PUP-IT (3), where a protein of interest (the “bait”) is genetically fused to a proximity-based labeling enzyme which activates a substrate like biotin or Pup, and then releases the activated substrate to label proximal proteins. Recently, a micromapping platform using a photocatalytically induced carbene was also introduced for proximity labeling of membrane bound proteins (4). While enormously powerful, these assays merely report on proteins that are proximal to the bait; testing direct interactions between proximate proteins and the bait requires proximity-tagging techniques to be integrated with a complementary biophysical method.
Here we integrate the widely used proximity-tagging scheme, BioID, with single molecule atomic force microscope (AFM) binding assays, to identify transmembrane proteins that bind to the extracellular region of a transmembrane bait. In BioID, a promiscuous, mutant biotin ligase is fused to a bait protein and covalently tags lysine residues on nearby proteins with exogenously supplied biotin (1, 5); the biotin-tagged proteins are subsequently detected using mass spectrometry (MS). Unfortunately, a typical BioID screen results in numerous “false positive” hits and the number of proximate proteins reported are enormous (hundreds of candidate proteins), which limits the number of binding interactions that can be directly tested. We reasoned that transmembrane proteins that directly interact with a transmembrane bait would be positioned in close proximity to both the bait’s ectodomain and cytoplasmic regions. Consequently, if the proximity-based labeling enzyme was fused to both the bait’s extracellular and cytoplasmic regions, identifying transmembrane proteins that are biotinylated in both regions would enable us to narrow down the list of possible binding partners for subsequent AFM binding measurements. Furthermore, an integrated extracellular and cytoplasmic BioID measurement would significantly increase the precision of the screen by dramatically reducing the number of false positive hits. However, the BioID method has been mostly utilized to identify intracellular binding partners (6), including to the cytoplasmic region of cadherin cell adhesion proteins (7–9) and other junctional proteins (10). Recently, proteins secreted by rat neurons were also identified using BioID fused to ectodomains of N-cadherin (Ncad) (11). However, BioID has not been used to screen for extracellular binding partners of transmembrane proteins.
Here, as transmembrane bait, we use E-cadherin (Ecad), a ubiquitous cell–cell adhesion protein that plays an essential role in tissue morphogenesis, in maintaining tissue integrity, and in facilitating the collective migration of cells (12, 13). While over 170 proteins, including signaling molecules, scaffolding proteins, and cytoskeletal regulators, have been reported to associate directly or indirectly with the Ecad cytoplasmic tail (14), Ecad ectodomains are primarily believed to interact with identical Ecad ectodomains from opposing cells. Although previous studies suggest interactions between Ecad ectodomains and other adhesion proteins and cell-surface receptors (15–17), heterophilic Ecad binding partners have not been systematically catalogued.
We therefore fused BioID to the ectodomain and cytoplasmic regions of Ecad in epithelial cells in order to map Ecad transmembrane binding partners. By comparing the extracellular and cytoplasmic interactomes, we identified 11 transmembrane proteins that are proximal to Ecad. Single molecule AFM binding assays revealed that out of these target proteins, the extracellular regions of the desmosomal proteins desmoglein-2 (Dsg2) and desmocollin-3 (Dsc3), the focal adhesion protein integrin-α2β1 (Intα2β1), receptor tyrosine kinase ligand ephrin-B1 (EfnB1), and the classical cadherin P-cadherin (Pcad) all directly interact with Ecad ectodomains. Our results demonstrate that Ecad ectodomains do not merely engage in homophilic binding, but instead, like the Ecad cytoplasmic region, also bind to a range of junctional proteins.
Results and Discussion
Ecad Tagged with BioID on Their Extracellular and Cytoplasmic Regions Localize to Cell–Cell Junctions and Biotinylate Proteins at Intercellular Contacts.
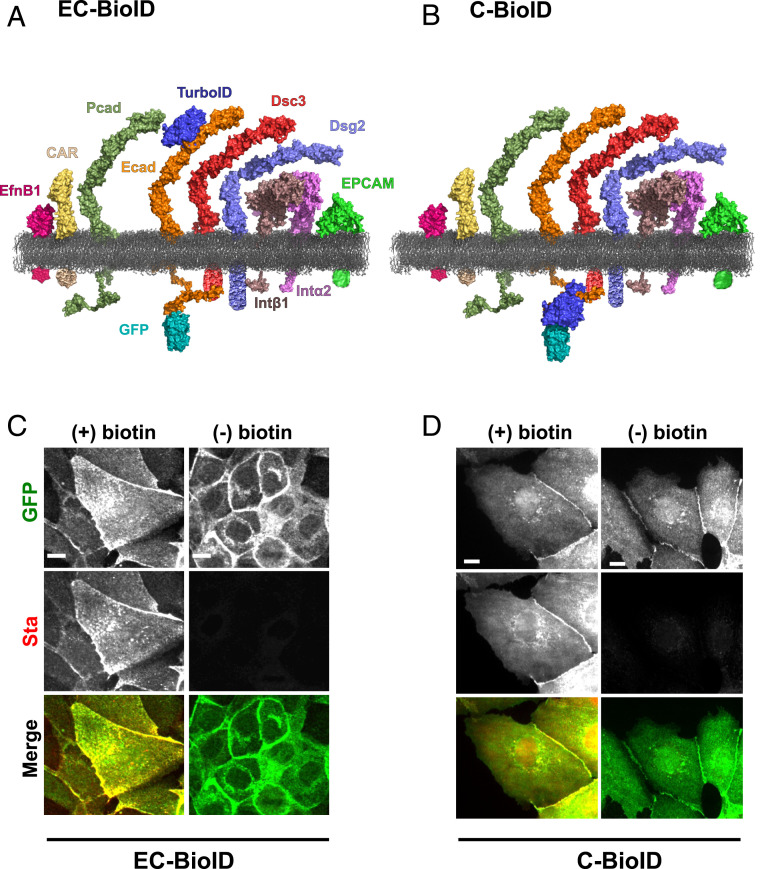
Ecad ectodomains comprise five tandemly arranged extracellular domains (EC1 to EC5). In our experiments, we fused the biotin ligase TurboID which labels proteins with a labeling radius of ∼35 nm in 10 min (compared to ∼18 h for previously used ligases) (18, 19) to either the EC2 domain (EC-BioID) or to the cytoplasmic region (C-BioID) of Ecad and expressed these fusion constructs in epithelial Madin-Darby Canine Kidney (MDCK) cells. Due to posttranslational cleavage of the N-terminal signal and propeptide, inserting BioID at the Ecad N terminus was not feasible. Furthermore, since the Ecad homophilic binding site is located on the protein’s N-terminal EC1 domain, modifications in this region abolish Ecad adhesion. We therefore generated the EC-BioID, by inserting TurboID on the EC2 domain, between amino acids 152 and 153 from the N terminus of mature Ecad (Fig. 1A); this location was previously used to insert a green fluorescent protein (GFP) without affecting cadherin function (20). C-BioID was generated by fusing TurboID to the cytoplasmic C terminus of Ecad (Fig. 1B). A GFP was inserted at the C terminus of both the EC-BioID and C-BioID constructs (Fig. 1 A and B and SI Appendix, Fig. S1 A and C). Because of high levels of endogenous Ecad in parental MDCK cells, we rescued Ecad knockout (KO) MDCK cells with EC-BioID and C-BioID and generated stable cell lines. Both EC-BioID and C-BioID localized to cell–cell junctions, verifying that the insertion of TurboID did not disrupt the incorporation of Ecad into intercellular junctions (Fig. 1 C and D, Top row). Next, to demonstrate that the TurboID was functional, we incubated the cells with free biotin and tagged the resulting biotinylated proteins with fluorescently labeled streptavidin (Fig. 1 C and D, Middle row). In the absence of exogenous biotin, we observed very little fluorescent streptavidin signal. Western blots (SI Appendix, Fig. S1 B and D) also confirmed that the TurboID required an external source of biotin to efficiently label proteins. Merging GFP and streptavidin images (Fig. 1 C and D, Bottom row) showed that most of the biotinylated proteins were localized to intercellular junctions.
Fig. 1.
Characterizing Ecad BioID constructs. TurboID was fused to (A) EC2 domain of Ecad (EC-BioID) and (B) C-terminal of Ecad (C-BioID). Proteomic analysis of both constructs identified ephrin-B1 (EfnB1), coxsackie and adenovirus receptor (CAR), P-cadherin (Pcad), desmocollin-3 (Dsc3), desmoglein-2 (Dsg2), integrin β1 (Intβ1), integrin α2 (Intα2), epithelial cell adhesion molecule (EPCAM), chloride intracellular channel protein (CLIC1), and voltage-dependent anion channel 1 (VDAC1) as proteins that were proximal to Ecad. CLIC1 and VDAC1 are primarily found in nuclei and mitochondria, respectively, and are therefore not shown. Proteins are depicted using deposited Protein Data Bank (PDB) structures or predicted from homology modeling using Modeller. The following PDB IDs were used in generating the images: EfnB1, 6P7Y; CAR, 3J6N; TurboID, 4WF2; GFP, 1GFL; Ecad and Pcad, 3Q2V; Dsc3, 5IRY; Dsg2, 5ERD; Intα2, 3K6S; Intβ1, 3IJE; EPCAM, 4MZV; and membrane, POPE. Images were reconstructed using PyMOL. All cytoplasmic domains are schematic representations. GFP localized to intercellular junction in (C) EC-BioID and (D) C-BioID confirming that Ecad was functional. Fluorescent streptavidin (Sta) staining showed that while BioID biotinylated proteins in the presence (+) of exogenous biotin, low levels of biotinylation was observed in the absence (−) of exogenous biotin. Merged images of GFP and Sta show most of the biotinylation occurs near the junction. (Scale bar, 10 µm.)
Mass Spectrometry Analysis Identifies Transmembrane Proteins Proximal to Ecad.
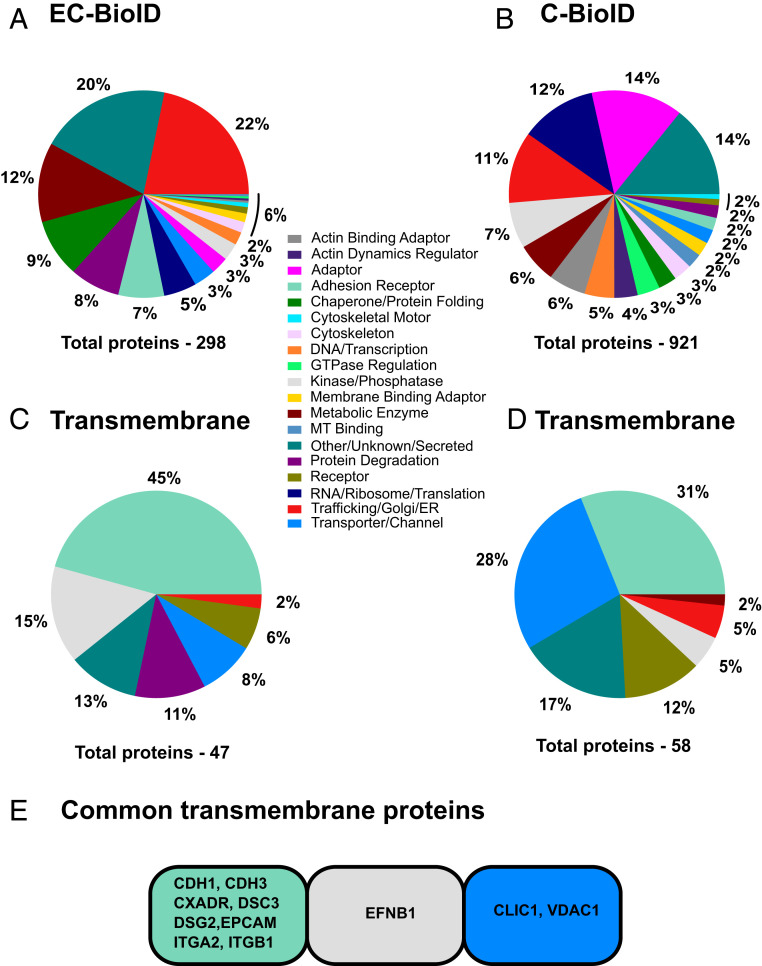
To identify Ecad-proximate proteins, we incubated the EC-BioID and C-BioID cells with exogenous biotin and captured the biotinylated proteins from cell lysates using streptavidin-coated beads. Trypsin digestion of the beads released protein fragments that were analyzed using MS. We selected proteins found in all replicates with more than 4.8% peptide coverage (ratio between detected peptides and predicted peptides from trypsin digestion) which resulted in 298 proteins in EC-BioID and 921 proteins in C-BioID [MS data attached in SI (Dataset S1)]. We manually annotated these proteins into 19 functional categories (Fig. 2 A and B) similar to previous cadherin interactomes (7, 8) using the available information from databases Uniprot, GeneCards, and Entrez. Since our primary focus was on determining Ecad extracellular binding partners, we identified proteins that localize to the plasma membrane and are denoted to have a transmembrane domain in Uniprot and classified them as transmembrane proteins. These transmembrane proteins were grouped into similar functional categories (Fig. 2 C and D). Adhesion receptors represented the largest fraction of transmembrane proteins with 45% coverage in EC-BioID (21 of 47 transmembrane proteins) and 31% coverage in C-BioID (18 of 58 transmembrane proteins). Comparing both EC-BioID and C-BioID resulted in 10 common transmembrane proteins in addition to Ecad, out of 62 common proteins [Fig. 2E, MS data attached in SI (Dataset S1)]. These proteins (along with their gene IDs) are: desmoglein-2 (DSG2), desmocollin-3 (DSC3), integrin alpha 2 (ITGA2), integrin beta 1 (ITGB1), ephrin-B1 (EFNB1), coxsackie and adenovirus receptor (CXADR; CAR), P-cadherin (CDH3), epithelial cell adhesion molecule (EPCAM), chloride intracellular channel protein (CLIC1), and voltage-dependent anion channel 1 (VDAC1). These proteins were not detected at a significant level in the absence of biotin.
Fig. 2.
Ecad proximal transmembrane proteins identified from EC-BioID and C-BioID interactomes. Proteins identified from MS analysis of (A) EC-BioID and (B) C-BioID were manually grouped into 19 functional categories based on reported functions from protein databases. From these interactomes, transmembrane proteins in (C) EC-BioID and (D) C-BioID were separated into their corresponding functional groups. (E) Transmembrane proteins common to both EC-BioID and C-BioID.
It is important to note that since the proximity labeling experiments were performed with stably transfected cells grown to confluency, they do not identify whether junctional proteins bind to Ecad in trans (from opposing cells) or cis (from the same cell) orientations. It is also important to note that only ∼20% of the transmembrane proteins in the EC-BioID and C-BioID were common to both interactomes. These differences in EC-BioID and C-BioID transmembrane protein hits arise due to fundamental BioID limitations including: 1) differential biotin labeling of cytoplasmic and extracellular regions of a protein due to differences in availability of Lys residues; 2) increased biotin labeling efficiency of C-BioID compared to EC-BioID, because the activated biotins in C-BioID are confined within the cell and can tag proteins that are present in low copy numbers; and 3) different MS detection efficiencies of target ectodomains and cytoplasmic regions due to their different tryptic digestion profiles. Furthermore, not all Ecad transmembrane binding partners were captured in our assay since proteins that are present in low copy numbers are not readily tagged and detected. Consequently, previously identified Ecad binding partners (17, 21) such as Ncad (gene ID: CDH2) and Epidermal Growth Factor Receptor (gene ID: EGFR) were not detected in EC-BioID because of their low expression levels in MDCK cells (22, 23). Interestingly, both CDH2 and EGFR were detected in the C-BioID screens because these low copy number proteins could presumably be labeled by “confined” activated biotin.
Single Molecule AFM Binding Assays.
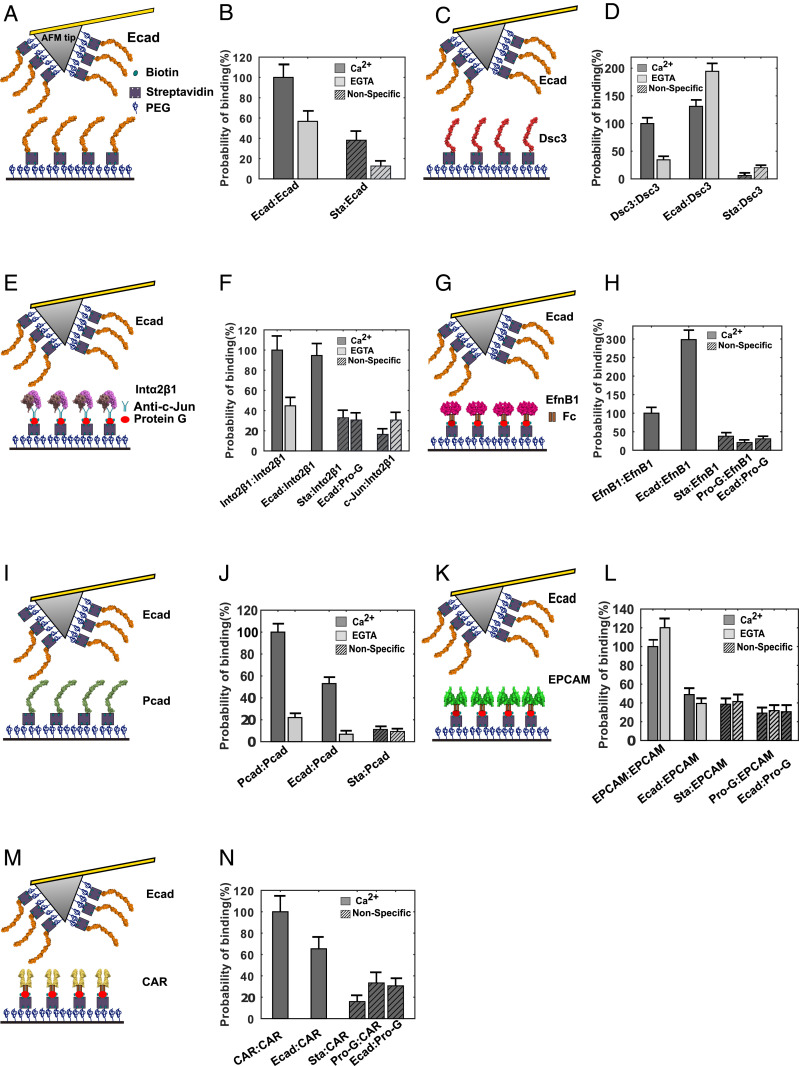
We next tested the direct binding of ectodomains of the adhesion and phosphatase/kinase protein hits (Dsc3, Intα2-Intβ1 heterodimers, EfnB1, Pcad, EPCAM, and CAR) with Ecad ectodomains, using single molecule AFM binding assays. We have previously shown, using single molecule and cellular structure-function experiments, that the ectodomain of the adhesive protein Dsg2 directly interacts with Ecad ectodomains (15). We did not measure Ecad interactions with the transporter proteins CLIC1 and VDAC1 since they primarily localize to the nucleus and mitochondria, respectively, although both proteins have been reported to be present in small amounts on the plasma membrane (24, 25). In our AFM experiments, we used the complete extracellular region of all of the target proteins expressed and purified from mammalian cells. The C terminus of the proteins were either tagged with human Fc-dimers (EfnB1, EPCAM, and CAR), biotin (Dsc3, Pcad, and Ecad), or human c-Jun and c-Fos (Intα2β1 heterodimer). The biotin, c-Jun and Fc tags were used to immobilize the proteins on AFM tips and glass coverslips (Fig. 3 A, C, E, G, I, K, and M and Methods). Briefly, AFM cantilevers and coverslips were functionalized with a monolayer of polyethylene glycol (PEG) decorated with streptavidin. Biotinylated Ecad, Dsc3, and Pcad were directly attached to the streptavidin (Fig. 3 A, C, and I), while Fc-tagged EfnB1, EPCAM, and CAR were linked to streptavidin using biotinylated protein G (Fig. 3 G, K, and M) (Methods). Heterodimeric Intα2β1 was linked to streptavidin decorated with biotinylated protein G using an antibody against c-Jun (Fig. 3E).
Fig. 3.
Single molecule AFM force measurements confirm binding partners for Ecad. (A) Schematic of single molecule AFM force measurement experiment. The AFM tip and substrate were functionalized with PEG linkers, some of which were decorated with streptavidin (Sta). Biotinylated Ecad was directly attached to Sta. (B) Ecad showed Ca2+-dependent homophilic interactions. (C) Biotinylated Dsc3 directly immobilized on Sta (D) showed Ca2+-dependent homophilic interactions and Ca2+-independent heterophilic interactions with Ecad. (E) Intα2β1 heterodimer tagged with c-Jun was immobilized using an anti-c-Jun antibody and biotinylated protein G (Pro-G). (F) Intα2β1 formed Ca2+-dependent homophilic complexes and also formed heterophilic complexes with Ecad. (G) Fc-tagged EfnB1 was attached to Sta using biotinylated Pro-G (H) interacted homophilically and also bound heterophilic to Ecad. (I) Biotinylated Pcad attached to Sta (J) showed Ca2+-dependent homophilic and heterotypic interactions. (K) EPCAM-Fc attached to Sta using biotinylated Pro-G (L) formed Ca2+-independent homophilic dimers. However, EPCAM-Fc did not interact heterophilically with Ecad. (M) CAR-Fc immobilized using biotinylated Pro-G (N) showed low probability of heterophilic interactions with Ecad. Error bars are SE calculated using bootstrap with replacement.
At the start of each experiment, the AFM cantilever and substrate were brought into contact to allow opposing proteins to interact and the tip was then withdrawn from the substrate. As previously described (15), interaction of proteins resulted in unbinding events characterized by nonlinear stretching of the PEG tethers which served as a molecular fingerprint for single molecule unbinding. We measured the homophilic interaction of every candidate protein, the heterophilic interaction of every candidate protein with Ecad, and nonspecific interactions under each measurement condition. Nonspecific interactions were determined between an Ecad functionalized AFM tip and a coverslip lacking the candidate protein and also using an AFM tip lacking Ecad and a coverslip functionalized with the candidate protein. For ease of comparison, all binding probabilities were normalized by the corresponding homophilic binding rates in Ca2+. We confirmed that the Ecad was functional by measuring homophilic binding (Fig. 3B). Since Ecad is a Ca2+-dependent adhesion protein, we confirmed that homophilic binding probability was reduced in the presence of ethylene glycol tetraacetic acid (EGTA), a Ca2+ chelating agent (Fig. 3B and SI Appendix, Table S1).
Dsc3 dimerizes with Ecad independent of Ca2+.
One of the most abundant proteins found in both EC-BioID and C-BioID were different isoforms of the desmosome-associated proteins, Dsc and Dsg, which mediate robust cell–cell adhesion in tissues like the epidermis and heart that are exposed to significant levels of mechanical stress. While Dsg2 and Dsc3 were identified in both EC-BioID and C-BioID, Dsc2 and Dsg1 were only identified in C-BioID. As predicted by previous biophysical studies (26), our AFM measurements showed that Dsc3 interacts homophilically in a Ca2+-dependent fashion (Fig. 3D and SI Appendix, Table S1). Surprisingly, Dsc3 ectodomains also interacted heterophilically with Ecad ectodomains in a Ca2+-independent fashion (Fig. 3D and SI Appendix, Table S1). It is important to point out that we have already demonstrated that Ecad and Dsg2 directly bind via a conserved Leu175 on the Ecad, which promotes desmosome assembly.
The interaction of Dsc3 with Ecad provides a molecular explanation for previous studies showing that blocking Ecad adhesion with antibodies or knocking down Ecad, delays desmosome formation in cells (27, 28), and that classical cadherin-deficient mice show defective desmosome assembly (29). While we have shown that Dsc2 does not directly bind to Ecad (15), it still appears as a “hit” in our C-BioID assay because Dsg2 interacts with Dsc2 (15, 30).
Intα2β1 binds to Ecad ectodomains.
Interestingly, both EC-BioID and C-BioID showed that the focal adhesion proteins, Intα2 and Intβ1, were proximate to Ecad ectodomains. Since integrins are composed of noncovalently associated αβ heterodimers and Intα2 forms a heterodimer exclusively with Intβ1, we used Intα2β1 in our AFM binding measurements. Similar to Intα3β1 that interacts homophilically in a cation-dependent manner (31), our AFM measurements demonstrated homophilic Ca2+-dependent Intα2β1 adhesion (Fig. 3F and SI Appendix, Table S1). Furthermore, as anticipated from the BioID results, we measured heterophilic adhesion between Ecad ectodomains and Intα2β1 ectodomains in Ca2+ (Fig. 3F and SI Appendix, Table S1).
The discovery of interactions between Ecad and Intα2β1 ectodomains is particularly exciting since although there is a large body of evidence supporting the existence of cross-talk between integrins and cadherins (32–34), this cross-talk is primarily believed to be mediated by cytoplasmic proteins. Heterotypic adhesion has previously been described between Ecad and integrin-αEβ7 that mediates binding of intraepithelial lymphocytes to epithelial cells (35, 36). Furthermore, Ecad and integrin-α6 interaction and colocalization have been reported in liver metastasis of colorectal cancer cells (37). Even though Intα2β1 is a receptor for the ECM proteins collagen and laminin (38), immunofluorescence studies have shown that Intα2β1 localizes to cell–cell contacts in keratinocytes (39) and with Ecad and Ncad in melanoma cells (40). Furthermore, cell-spreading assays suggest that interaction between cells and an Ecad-coated surface is abolished by monoclonal antibodies that target Intα2β1 (41). Our demonstration of homophilic binding and heterotypic adhesion with Ecad suggests that Intα2β1 could also play a functional role in cell–cell adhesion and in collective cell migration.
EfnB1 forms heterotypic complexes with Ecad.
Next, we measured the homophilic and heterophilic binding of EfnB1, a ligand for erythropoietin-producing hepatocellular carcinoma (Eph), the largest subfamily of receptor tyrosine kinases (42). In agreement with previous structural reports (43), our measurements showed that EfnB1 adheres homophilically (Fig. 3H and SI Appendix, Table S1). Interestingly, the heterophilic binding between Ecad monomers and EfnB1-Fc dimers was almost threefold higher than EfnB1 homophilic binding (Fig. 3H and SI Appendix, Table S1).
While EfnB1 is known to interact with the tight junction protein claudin (44), direct binding of EfnB1 and Ecad has not been previously reported. It has been shown that EphB2 receptors interact with Ecad and the metalloproteinase ADAM10 at sites of adhesion, and their activation induces shedding of Ecad by ADAM10 at interfaces with EfnB1-expressing cells, which suppress tumor progression in colorectal cancer (45, 46). It is possible that Ecad ectodomains interact with EfnB1 and structurally hinder ADAM10-mediated Ecad cleavage. Consequently, when EfnB1 dissociates from Ecad and binds to EphB2, the ADAM10 cleavage site on Ecad becomes accessible. Interestingly, both EphB2 and ADAM10 appear as hits in the EC-BioID. Finally, we note that while our AFM force measurement shows that EfnB1 forms homodimers, previous analytical ultracentrifugation measurements suggest that EfnB1 is monomeric (47). This discrepancy may arise from the higher single molecule sensitivity of our AFM binding measurements.
Pcad interacts with Ecad in the presence of Ca2+.
The next candidate protein identified in our BioID assay was Pcad, a type I classical cadherin that shares high sequence homology with Ecad (48), and that is proposed to play important roles as both a tumor promotor and a tumor suppressor (49). In agreement with previous structural studies (48, 50), our single molecule AFM binding assays revealed homophilic Pcad adhesion (Fig. 3J and SI Appendix, Table S1) which was Ca2+ dependent as expected for classical cadherins. Interestingly, single molecule binding assays also demonstrated that Pcad heterophilically interacts with Ecad in a Ca2+-dependent manner (Fig. 3J and SI Appendix, Table S1). These data are in agreement with previous biophysical measurements and coimmunoprecipitation studies showing heterophilic interactions of Ecad and Pcad (48, 51).
Ecad and Pcad are known to have unique cellular roles depending on physiological context. It has been proposed that Pcad controls intercellular tension while Ecad controls the rate at which this tension changes over time. However, in the absence of Ecad, Pcad is believed to fill the role of Ecad (52). This is not surprising since both Ecad and Pcad share the same conserved residues that mediate homophilic binding and they are structurally homologous (48). Therefore, it is possible that the same homophilic binding interface may participate in Pcad-Ecad heterodimer formation as well. In fact cells expressing Ecad and Pcad completely intermix in coaggregation assays, suggesting heterotypic binding (53).
EPCAM does not form a heterodimer complex with Ecad.
EPCAM, a transmembrane glycoprotein which serves as a tumor marker, was identified as a candidate protein in both EC-BioID and C-BioID interactomes. In agreement with previous cell aggregation assays which show that EPCAM forms Ca2+-independent intercellular adhesion (54), our AFM binding assay also shows that EPCAM homophilically interacts in a Ca2+-independent manner (Fig. 3L and SI Appendix, Table S1). However, our AFM data show that the heterophillic interaction of EPCAM with Ecad is comparable to nonspecific binding, suggesting that EPCAM and Ecad do not heterodimerize (Fig. 3L and SI Appendix, Table S1). Consistent with these data, previous coaggregation assays also showed that cells expressing Ecad and EPCAM do not participate in heterotypic interactions (55). Previous studies show that overexpression of EPCAM disrupts Ecad anchorage to the cytoskeleton and that the cytoplasmic domain of EPCAM is required to break Ecad-cytoskeletal linkages (55). It is therefore likely that Ecad and EPCAM interact via their cytoplasmic domains (either directly or using an adaptor protein). Consequently, while EPCAM is a hit in both EC-BioID and C-BioID assays, we do not detect interactions between EPCAM and Ecad extracellular regions in the AFM binding measurements.
CAR does not interact directly with Ecad.
The last candidate protein we tested using AFM force measurements was Fc-dimers of CAR, a transmembrane virus receptor that localizes to tight junctions. In agreement with structural and sedimentation analysis (56), our AFM data also showed homophilic CAR interactions (Fig. 3N and SI Appendix, Table S1). However, the heterophilic binding probability of CAR and Ecad was only ∼30% greater than nonspecific binding (Fig. 3N and SI Appendix, Table S1), suggesting that CAR ectodomains either do not interact with Ecad or bind to Ecad with a low probability. Previous studies show that while Ecad and CAR are not colocalized in stable cell–cell junctions, CAR localization is observed with internalized Ecad in vesicles (57). This suggests that CAR can be biotinylated in internalized vesicles with both EC-BioID and C-BioID, even though it might not directly interact with Ecad on the cell surface. Another possible explanation for the low Ecad-CAR heterophilic binding measured in our AFM experiments is that CAR and Ecad interactions are primarily mediated by their respective cytoplasmic domains.
Our strategy of only evaluating hits common to both EC-BioID and C-BioID interactomes, decreases the number of false positive hits and increases the precision (fraction of true positive identifications) of our assay. Precision in identifying transmembrane Ecad ectodomain interactors for the combined EC-BioID/C-BioID assay is 64% (7 true positives confirmed by AFM out of a total of 11 transmembrane interactors that were captured by the assay). In contrast, stand-alone EC-BioID has a precision of only 30% (14 true positives out of 47 transmembrane protein hits). Stand-alone C-BioID has an even lower precision of 17% (10 true positives out of 58 transmembrane protein hits). It is important to note that these estimated precisions are only approximate since all hits were not directly tested using a binding assay. We estimated true positives in stand-alone EC-BioID and stand-alone C-BioID by supplementing the results of our AFM binding assays with a list of potential Ecad binding partners curated from the literature. In addition to the seven ectodomain binding partners that we measured with AFM, the gene IDs of the published Ecad binding partners that appear among the 47 transmembrane proteins in EC-BioID are CDCP1 (58), EPHB2 (45), ERBB3 (59), TGFBR1 (60), ADAM10 (61), ADAM9 (62), and MMP15 (63). Similarly, the additional published gene IDs of Ecad binding partners that appear among the 58 transmembrane proteins in C-BioID are CDH2 (21), EGFR (17), and ERBB2 (64). A noteworthy caveat is that, since many of these Ecad binding partners were identified in the literature using immunoprecipitation and cell binding assays which do not measure direct protein–protein interactions and also do not discriminate between ectodomain and cytoplasmic domain interactions, the precision of stand-alone EC-BioID and C-BioID assays may be even lower than we report.
In conclusion, our discovery of several previously unknown Ecad ectodomain interactors, such as Dsc3 and EfnB1, demonstrate that Ecad ectodomains do not merely engage in homophilic binding, but instead, like the Ecad cytoplasmic region, also bind to a range of proteins. We anticipate that integrated BioID-AFM experiments can be similarly used to initially screen and subsequently characterize heterophilic transmembrane binding partners for other junctional proteins with high precision.
Methods
Molecular Cloning of EC-BioID and C-BioID.
We inserted TurboID in Ecad at a site where GFP had previously been inserted in mouse Ncad using in vitro transposition (20). We identified from sequence alignment, a similar sequence at position 152 from N-terminal sequence in canine Ecad and inserted TurboID with a peptide linker on both ends; the linker was composed of the same amino acids (AYSILT-LSLIHIWRA-V5-TurboID-GRARADVYKRQ- QDPLLP; canine Ecad sequence is in bold italicized font while linker sequence is in normal font) used previously (20).
We PCR amplified Ecad using pEGFP-N1-Ecad plasmid (65) and V5-TurboID using mutant BirA R118S (TurboID) (Addgene) (18) using PCR primers (SI Appendix, Table S2). The PCR products were inserted between EcoRV and BglII using Gibson assembly (New England Biolabs) in pEGFP-N1-Ecad vector. We PCR amplified the EC-BioID sequence and inserted it into the PiggyBac-GFP vector using the NheI site (PB533A, System Biosciences). For C-BioID, we PCR amplified using PCR primers (SI Appendix, Table S2) the complete Ecad using pEGFP-N1-Ecad plasmid. The PCR products were inserted at the BamHI site in a PiggyBac vector.
The PiggyBac EC-BioID-GFP and C-BioID-EGFP plasmids were transfected into Ecad-KO cells (66) using Lipofectamine 2000 (Invitrogen) along with PiggyBac transposase expression plasmid (System Biosciences) and selected with 500 μg/mL G418 (Invitrogen). G418-resistant cells were subcloned and selected by confocal microscopy and Western blotting to obtain homogeneous cell populations.
Immunofluorescence.
EC-BioID- and C-BioID-expressing MDCK cells were grown to confluency and switched to serum-free Dulbecco’s modified Eagle’s medium (DMEM) cell culture media and supplemented with 50 µM biotin overnight. To selectively detect extracellular biotinylation, Alexa Fluor 568-conjugated streptavidin (Invitrogen) was added for 30 min before cell fixation and permeabilization. Cells were fixed using 3% paraformaldehyde and 0.3% Triton X-100 in phosphate-buffered saline (PBS) for 10 min and blocked with 1% bovine serum albumin (BSA) and 0.3% Triton X-100 in PBS for 30 min. Anti-GFP (rabbit polyclonal, Invitrogen) antibody and Alexa Fluor 488-conjugated anti-rabbit antibody were used to detect GFP-tagged proteins. For C-BioID-expressing cells, biotinylated proteins were detected by incubating the fixed cells for 30 min with Alexa Fluor 568-conjugated streptavidin. Cells were imaged using a Zeiss AxioObserver equipped with a Yokogawa CSU-10 spinning disk confocal system, 40× objective, 488- and 561-nm solid-state lasers, a Photometrics CoolSNAP HQ2 camera, and Slidebook software (Intelligent Imaging Innovations). Images were reconstructed using ImageJ.
Purification of Biotinylated Proteins for MS Analysis.
Four replicates of EC-BioID and two replicates of C-BioID were used for MS analysis. Cells expressing EC-BioID and C-BioID were seeded on p150 dishes and cultured to 70 to 90% confluency in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Gibco), penicillin (Invitrogen), streptomycin (Invitrogen), and 200 μg/mL G418 (Invitrogen). Cells were then incubated overnight with serum-free media supplemented with 50 μM biotin. After three PBS washes, cells were scraped and centrifuged. Pelleted cells were resuspended in lysis buffer (67, 68) (50 mM Tris pH 7.5, 150 mM NaCl, 0.4% sodium dodecyl sulfate (SDS), 1% octylphenoxy poly(ethyleneoxy)ethanol (IGEPAL CA-630), 1.5 mM MgCl2, 1 mM EGTA) with 2 μL/mL of protease inhibitor mixture (Sigma-Aldrich), and 1 μL/mL benzonase (250 U/μL) (EMD-Millipore). Cell lysate was incubated for 30 min at 4 °C and then sonicated at 10 to 30% duty ratio for 1 min and centrifuged at 13,000 rpm for 30 min at 4 °C. The supernatant was collected, and their concentrations were measured with RC/DC protein assay kit (Bio-Rad). Supernatant (1 mL at ∼4 to 5 mg/mL) was incubated with 50 μL of superparamagnetic Dynabeads Streptavidin C1 (Invitrogen) and rotated overnight at 4 °C. First, beads were washed with the lysis buffer and transferred to new tubes. The beads were washed again with the 2% SDS in 50 mM Tris pH 7.4, then washed twice with lysis buffer. Next, the beads were washed three times with 100 mM ammonium bicarbonate (NH4HCO3) and resuspended in 30 μL of digestion buffer containing 1 µg/µL of trypsin gold in 50 mM NH4HCO3 for overnight digestion at 37 °C. To stop the digestion, the reaction mixture was acidified with 1% trifluoroacetic acid (TFA). The resulting peptides were recovered from the beads using a magnet and then acidified with 0.5% TFA. The eluted tryptic peptides were dried in a vacuum centrifuge and reconstituted in 0.1% formic acid. Tryptic peptides were analyzed using nano-scale liquid chromatographic tandem mass spectrometry (nLC-MS/MS).
Nano-Scale Liquid Chromatography.
nLC was performed on an ultra-high pressure nano-flow Easy nLC system (Bruker Baltonics). Liquid chromatography was performed at 40 °C with a constant flow rate of 400 nL/min on an in-house packed reversed-phase column (25 cm × 75 µm inner diameter) with a pulled emitter tip, packed with 1.9 µm C18-coated porous silica beads (Dr. Maisch). Mobile phases A and B were water with 0.1% formic acid (v/v) and 80/20/0.1% acetonitrile/water/formic acid (v/v/v), respectively. Peptides were separated using a 33-min gradient from 5 to 38% B within 25 min, followed by an increase to 95% B within 1 min, a 1 min washing step at 95% B, ending with 8 min of reequilibration at 5% B.
Mass Spectrometry.
MS was performed on a hybrid trapped ion mobility spectrometry-quadrupole time of flight mass spectrometer (timsTOF Pro, Bruker Daltonics) with a modified nano-electrospray ion source (CaptiveSpray, Bruker Daltonics). In the experiments described here, the mass spectrometer was operated in Parallel Accumulation Serial Fragmentation (PASEF) mode. All experiments were acquired with a 100 ms ramp and 10 PASEF MS/MS scans per topN acquisition cycle. Low-abundance precursors with an intensity below a “target value” were repeatedly scheduled for PASEF-MS/MS scans until the summed ion counts reached a target value of 20,000 a.u. MS and MS/MS spectra were recorded from m/z 100 to 1,700. A polygon filter was applied to the m/z and ion mobility plane to select features most likely representing peptide precursors rather than singly charged background ions. The quadrupole isolation width was set to 2 Thomson (Th) for m/z under 700 and 3 Th for m/z larger than 700, and the collision energy was ramped stepwise as a function of increasing ion mobility: 52 eV for 0 to 19% of the ramp time, 47 eV from 19 to 38%, 42 eV from 38 to 57%, 37 eV from 57 to 76%, and 32 eV for the remainder.
MS Data Analysis.
Mass spectrometry raw files were processed with MsFragger (69). For all searches, a protein sequence database of reviewed canine proteins (accessed 11/27/2019 from UniProt; 1,886 entries including decoys and 115 common contaminant sequences) was used. Decoy sequences were generated and appended to the original database for MSFragger. A maximum of two missing cleavages were allowed, the required minimum peptide sequence length was 7 amino acids, and the peptide mass was limited to a maximum of 5,000 Da. Carbamidomethylation of cysteine residues was set as a fixed modification, and methionine oxidation and acetylation of protein N termini as variable modifications. The initial maximum mass tolerances were 50 ppm for precursor and fragment ions. A reversed sequence library was generated/used to control the false discovery rate (FDR) at less than 1% for peptide spectrum matches and protein group identifications. Decoy database hits, proteins identified as potential contaminants, and proteins identified exclusively by one-site modification were excluded from further analysis. Label-free protein quantification was performed with the IonQuant algorithm (69). All other MsFragger parameters were kept at their default values.
Cloning, Purification, and Biotinylation of Ecad, Dsc3, and Pcad Monomers for Single Molecule AFM Experiments.
The extracellular region of the Dsc3 was PCR amplified from full-length human Dsc3 purchased from DNASU plasmid repository (clone ID: HsCD00821324) using primers listed in SI Appendix, Table S2. Avi-Tev-6x His (ATH) sequence was amplified using pcDNA3.1(+) Ecad-pATH (70). The Dsc3 extracellular region and ATH fragments were cloned into pcDNA3.1(+) Ecad-pATH between EcoRV and XhoI sites. Similarly, the extracellular region of Pcad was PCR amplified using primers listed in SI Appendix, Table S2 from full-length Pcad purchased from DNASU (clone ID: HsCD00296273) and cloned into pcDNA3.1(+) Dsc3 at EcoRV and NheI sites. Cloning of Ecad-ATH plasmids has been described elsewhere (71).
The Ecad, Dsc3, and Pcad plasmids were transiently transfected into HEK 293T cells using polyethylenimine (PEI) (Polysciences, Inc.). Four days posttransfection, the conditioned media were collected for protein purification. As described previously (15), protein was purified using the NGC Chromatography Systems (Bio-Rad). At 4 °C, media containing cadherin were flowed through Ni-Nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen) packed on 1 mL Empty Bio-Scale Mini Cartridges (Bio-Rad). The column was then washed with buffer at pH 7.5 (25 mM HEPES, 5 mM NaCl, 1 mM CaCl2). Proteins bound to Ni-NTA were biotinylated by incubating the beads with BirA enzyme (BirA500 kit; Avidity) for 1 h in 30 °C followed by an overnight incubation at 4 °C. Next, the column was washed with 25 mM HEPES, 500 mM NaCl, 1 mM CaCl2 with 20 mM imidazole at pH 7.5 and then eluted with the same buffer containing 250 mM imidazole. Following purification, the protein was exchanged to a Tris 10 mM, NaCl 100 mM, KCl 10 mM, CaCl2 2.5 mM, buffer at pH 7.5.
Single Molecule AFM Force Measurements.
Biotinylated Ecad, biotinylated Dsc3, biotinylated Pcad, Intα2β1-c-Jun/c-Fos, EfnB1-Fc, EPCAM-Fc, and CAR-Fc were immobilized on coverslips (CSs) and AFM cantilevers (Olympus, model TR400PSA) using a previously described method (70). Briefly, the CSs and cantilevers were cleaned with 25% H2O2:75% H2SO4 and washed with deionized (DI) water. Then the CS was cleaned with 1 M KOH and washed with DI water. Both the CSs and cantilevers were washed with acetone and functionalized using 2% (v/v) 3-aminopropyltriethoxysilane (Sigma) dissolved in acetone. Next, N-hydroxysuccinimide ester functionalized PEG spacers (MW 5000, Lysan Bio) were covalently attached to the silanized AFM tip and coverslip; 7% of the PEG spacers were decorated with biotin groups. Prior to a measurement, the functionalized AFM cantilever and coverslip were incubated overnight with BSA (1 mg/mL) to further reduce nonspecific binding. The tip and surface were then incubated with 0.1 mg/mL streptavidin for 30 min and biotinylated cadherins were attached to the streptavidin. Recombinant EfnB1-Fc (R&D catalog no. 473-EB), recombinant CAR-Fc (R&D catalog no. 3336-CX), and recombinant EPCAM-Fc (R&D catalog no. 960-EP) were linked to streptavidin using biotinylated protein G as previously shown (71). Integrin α2β1 heterodimer with c-Jun and c-Fos (R&D catalog no. 5698-A2) was linked to biotinylated protein G by c-Jun antibody (R&D catalog no. MAB2670). Finally, the surfaces were incubated with 0.02 mg/mL biotin for 10 min to block the free biotin binding sites on streptavidin.
Force measurements were performed using an Agilent 5500 AFM with a closed loop scanner. The spring constants of the cantilevers were measured using the thermal fluctuation method (72). All of the experiments were performed in a pH 7.5 buffer containing 10 mM Tris, 100 mM NaCl and 10 mM KCl with either 2.5 mM Ca2+ or 2 mM EGTA. For each experiment 850 to 1,448 force curves were collected (SI Appendix, Table S1). Force curves with nonlinear polymer stretching greater than the contour length of a single PEG molecule, were counted as an interaction.
Supplementary Material
Acknowledgments
Research in the S.S. laboratory was supported in part by the National Institute of General Medical Sciences of the NIH (R01GM121885). S.Y.’s research was supported in part by NIH R03 EB021636 and NSF 1562095. We thank Dr. Gabriela Grigorean for performing LC-MS/MS and data analysis in the Proteomics Core Facility of the Genome Center, University of California, Davis.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010209117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee H.-W., et al. , Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Q., et al. , A proximity-tagging system to identify membrane protein-protein interactions. Nat. Methods 15, 715–722 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Geri J. B., et al. , Microenvironment mapping via Dexter energy transfer on immune cells. Science 367, 1091–1097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi-Rhee E., Schulman H., Cronan J. E., Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 13, 3043–3050 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D. I., Roux K. J., Filling the void: Proximity-based labeling of proteins in living cells. Trends Cell Biol. 26, 804–817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Z., et al. , E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci. Signal. 7, rs7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., et al. , The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics. J. Cell Sci. 132, jcs221606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Itallie C. M., et al. , Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J. Cell Sci. 127, 885–895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredriksson K., et al. , Proteomic analysis of proteins surrounding occludin and claudin-4 reveals their proximity to signaling and trafficking networks. PLoS One 10, e0117074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Na Y., et al. , Fbxo45 binds SPRY motifs in the extracellular domain of N-cadherin and regulates neuron migration during brain development. Mol. Cell. Biol. 40, e00539-e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia M. A., Nelson W. J., Chavez N., Cell-cell junctions organize structural and signaling networks. Cold Spring Harb. Perspect. Biol. 10, a029181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niessen C. M., Leckband D., Yap A. S., Tissue organization by cadherin adhesion molecules: Dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 91, 691–731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaidel-Bar R., Cadherin adhesome at a glance. J. Cell Sci. 126, 373–378 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Shafraz O., et al. , E-cadherin binds to desmoglein to facilitate desmosome assembly. eLife 7, e37629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reshetnikova G., Troyanovsky S., Rimm D. L., Definition of a direct extracellular interaction between Met and E-cadherin. Cell Biol. Int. 31, 366–373 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Fedor-Chaiken M., Hein P. W., Stewart J. C., Brackenbury R., Kinch M. S., E-cadherin binding modulates EGF receptor activation. Cell Commun. Adhes. 10, 105–118 (2003). [PubMed] [Google Scholar]
- 18.Branon T. C., et al. , Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.May D. G., Scott K. L., Campos A. R., Roux K. J., Comparative application of BioID and TurboID for protein-proximity biotinylation. Cells 9, 1070 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S. A., Tai C. Y., Mok L. P., Mosser E. A., Schuman E. M., Calcium-dependent dynamics of cadherin interactions at cell-cell junctions. Proc. Natl. Acad. Sci. U.S.A. 108, 9857–9862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsamba P., et al. , Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc. Natl. Acad. Sci. U.S.A. 106, 11594–11599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shih W., Yamada S., N-cadherin-mediated cell-cell adhesion promotes cell migration in a three-dimensional matrix. J. Cell Sci. 125, 3661–3670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagiyama M., et al. , Modest static pressure suppresses columnar epithelial cell growth in association with cell shape and cytoskeletal modifications. Front. Physiol. 8, 997 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., et al. , Plasminogen kringle 5 induces endothelial cell apoptosis by triggering a voltage-dependent anion channel 1 (VDAC1) positive feedback loop. J. Biol. Chem. 289, 32628–32638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valenzuela S. M., et al. , Molecular cloning and expression of a chloride ion channel of cell nuclei. J. Biol. Chem. 272, 12575–12582 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Spindler V., et al. , Desmocollin 3-mediated binding is crucial for keratinocyte cohesion and is impaired in pemphigus. J. Biol. Chem. 284, 30556–30564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gumbiner B., Stevenson B., Grimaldi A., The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J. Cell Biol. 107, 1575–1587 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michels C., Buchta T., Bloch W., Krieg T., Niessen C. M., Classical cadherins regulate desmosome formation. J. Invest. Dermatol. 129, 2072–2075 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Tinkle C. L., Pasolli H. A., Stokes N., Fuchs E., New insights into cadherin function in epidermal sheet formation and maintenance of tissue integrity. Proc. Natl. Acad. Sci. U.S.A. 105, 15405–15410 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison O. J., et al. , Structural basis of adhesive binding by desmocollins and desmogleins. Proc. Natl. Acad. Sci. U.S.A. 113, 7160–7165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sriramarao P., Steffner P., Gehlsen K. R., Biochemical evidence for a homophilic interaction of the alpha 3 beta 1 integrin. J. Biol. Chem. 268, 22036–22041 (1993). [PubMed] [Google Scholar]
- 32.de Rooij J., Kerstens A., Danuser G., Schwartz M. A., Waterman-Storer C. M., Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J. Cell Biol. 171, 153–164 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yano H., et al. , Roles played by a subset of integrin signaling molecules in cadherin-based cell-cell adhesion. J. Cell Biol. 166, 283–295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borghi N., Lowndes M., Maruthamuthu V., Gardel M. L., Nelson W. J., Regulation of cell motile behavior by crosstalk between cadherin- and integrin-mediated adhesions. Proc. Natl. Acad. Sci. U.S.A. 107, 13324–13329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cepek K. L., et al. , Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the α E β 7 integrin. Nature 372, 190–193 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Shiraishi K., et al. , Critical role of the fifth domain of E-cadherin for heterophilic adhesion with α E β 7, but not for homophilic adhesion. J. Immunol. 175, 1014–1021 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Marchiò S., et al. , A complex of α6 integrin and E-cadherin drives liver metastasis of colorectal cancer cells through hepatic angiopoietin-like 6. EMBO Mol. Med. 4, 1156–1175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harburger D. S., Calderwood D. A., Integrin signalling at a glance. J Cell Sci. 122, 159–163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Symington B. E., Takada Y., Carter W. G., Interaction of integrins α 3 β 1 and α 2 β 1: Potential role in keratinocyte intercellular adhesion. J. Cell Biol. 120, 523–535 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siret C., et al. , Interplay between cadherins and α2β1 integrin differentially regulates melanoma cell invasion. Br. J. Cancer 113, 1445–1453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whittard J. D., et al. , E-cadherin is a ligand for integrin alpha2beta1. Matrix Biol. 21, 525–532 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Darling T. K., Lamb T. J., Emerging roles for Eph receptors and ephrin ligands in immunity. Front. Immunol. 10, 1473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toth J., et al. , Crystal structure of an ephrin ectodomain. Dev. Cell 1, 83–92 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Tanaka M., Kamata R., Sakai R., Phosphorylation of ephrin-B1 via the interaction with claudin following cell-cell contact formation. EMBO J. 24, 3700–3711 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solanas G., Cortina C., Sevillano M., Batlle E., Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling. Nat. Cell Biol. 13, 1100–1107 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Cortina C., et al. , EphB-ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat. Genet. 39, 1376–1383 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Nikolov D. B., Li C., Barton W. A., Himanen J. P., Crystal structure of the ephrin-B1 ectodomain: Implications for receptor recognition and signaling. Biochemistry 44, 10947–10953 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Vendome J., et al. , Structural and energetic determinants of adhesive binding specificity in type I cadherins. Proc. Natl. Acad. Sci. U.S.A. 111, E4175–E4184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vieira A. F., Paredes J., P-cadherin and the journey to cancer metastasis. Mol. Cancer 14, 178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kudo S., et al. , Identification and characterization of the X-dimer of human P-cadherin: Implications for homophilic cell adhesion. Biochemistry 53, 1742–1752 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Klingelhöfer J., Troyanovsky R. B., Laur O. Y., Troyanovsky S., Amino-terminal domain of classic cadherins determines the specificity of the adhesive interactions. J. Cell Sci. 113, 2829–2836 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Bazellières E., et al. , Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 17, 409–420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duguay D., Foty R. A., Steinberg M. S., Cadherin-mediated cell adhesion and tissue segregation: Qualitative and quantitative determinants. Dev. Biol. 253, 309–323 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Litvinov S. V., Velders M. P., Bakker H. A., Fleuren G. J., Warnaar S. O., Ep-CAM: A human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 125, 437–446 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Litvinov S. V., et al. , Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J. Cell Biol. 139, 1337–1348 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patzke C., et al. , The coxsackievirus-adenovirus receptor reveals complex homophilic and heterophilic interactions on neural cells. J. Neurosci. 30, 2897–2910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morton P. E., et al. , CAR regulates epithelial cell junction stability through control of E-cadherin trafficking. Sci. Rep. 3, 2889 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Law M. E., et al. , Glucocorticoids and histone deacetylase inhibitors cooperate to block the invasiveness of basal-like breast cancer cells through novel mechanisms. Oncogene 32, 1316–1329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Witta S. E., et al. , ErbB-3 expression is associated with E-cadherin and their coexpression restores response to gefitinib in non-small-cell lung cancer (NSCLC). Ann. Oncol. 20, 689–695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andl C. D., et al. , Coordinated functions of E-cadherin and transforming growth factor beta receptor II in vitro and in vivo. Cancer Res. 66, 9878–9885 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maretzky T., et al. , ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and β-catenin translocation. Proc. Natl. Acad. Sci. U.S.A. 102, 9182–9187 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirao T., et al. , Overexpression of ADAM9 enhances growth factor-mediated recycling of E-cadherin in human colon cancer cell line HT29 cells. Exp. Cell Res. 312, 331–339 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Gómez-Escudero J., et al. , E-cadherin cleavage by MT2-MMP regulates apical junctional signaling and epithelial homeostasis in the intestine. J. Cell Sci. 130, 4013–4027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Najy A. J., Day K. C., Day M. L., The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J. Biol. Chem. 283, 18393–18401 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamada S., Nelson W. J., Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J. Cell Biol. 178, 517–527 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koirala R., et al. , Inside-out regulation of E-cadherin conformation and adhesion. bioRxiv: 10.1101/2020.05.02.074187 (3 May 2020). [DOI] [PMC free article] [PubMed]
- 67.Hesketh G. G., Youn J. Y., Samavarchi-Tehrani P., Raught B., Gingras A. C., Parallel exploration of interaction space by bioid and affinity purification coupled to mass spectrometry. Methods Mol. Biol. 1550, 115–136 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Cheah J. S., Yamada S., A simple elution strategy for biotinylated proteins bound to streptavidin conjugated beads using excess biotin and heat. Biochem. Biophys. Res. Commun. 493, 1522–1527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu F., et al. Fast quantitative analysis of timsTOF PASEF data with MSFragger and IonQuant. Mol. Cell Proteomics. 19, 1575–1585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rakshit S., Zhang Y., Manibog K., Shafraz O., Sivasankar S., Ideal, catch, and slip bonds in cadherin adhesion. Proc. Natl. Acad. Sci. U.S.A. 109, 18815–18820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y., Sivasankar S., Nelson W. J., Chu S., Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc. Natl. Acad. Sci. U.S.A. 106, 109–114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hutter J. L., Bechhoefer J., Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64, 1868 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.