Significance
This work demonstrates that oncogenic activation of the PI3K-AKT-mTOR signaling pathway suppresses ferroptosis via the sterol regulatory element-binding protein-mediated lipogenesis, and that inhibition of this pathway potentiates the cancer therapeutic effect of ferroptosis induction. This finding unveils mechanisms for the regulation of ferroptosis and provides a potential cancer therapeutic approach for treating cancer patients bearing tumorigenic mutations in the PI3K-AKT-mTOR pathway.
Keywords: ferroptosis, mTOR, SREBP1, lipogenesis, cancer
Abstract
Ferroptosis, a form of regulated necrosis driven by iron-dependent peroxidation of phospholipids, is regulated by cellular metabolism, redox homeostasis, and various signaling pathways related to cancer. In this study, we found that activating mutation of phosphatidylinositol 3-kinase (PI3K) or loss of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) function, highly frequent events in human cancer, confers ferroptosis resistance in cancer cells, and that inhibition of the PI3K-AKT-mTOR signaling axis sensitizes cancer cells to ferroptosis induction. Mechanistically, this resistance requires sustained activation of mTORC1 and the mechanistic target of rapamycin (mTOR)C1-dependent induction of sterol regulatory element-binding protein 1 (SREBP1), a central transcription factor regulating lipid metabolism. Furthermore, stearoyl-CoA desaturase-1 (SCD1), a transcriptional target of SREBP1, mediates the ferroptosis-suppressing activity of SREBP1 by producing monounsaturated fatty acids. Genetic or pharmacologic ablation of SREBP1 or SCD1 sensitized ferroptosis in cancer cells with PI3K-AKT-mTOR pathway mutation. Conversely, ectopic expression of SREPB1 or SCD1 restored ferroptosis resistance in these cells, even when mTORC1 was inhibited. In xenograft mouse models for PI3K-mutated breast cancer and PTEN-defective prostate cancer, the combination of mTORC1 inhibition with ferroptosis induction resulted in near-complete tumor regression. In conclusion, hyperactive mutation of PI3K-AKT-mTOR signaling protects cancer cells from oxidative stress and ferroptotic death through SREBP1/SCD1-mediated lipogenesis, and combination of mTORC1 inhibition with ferroptosis induction shows therapeutic promise in preclinical models.
Accumulation of phospholipid peroxides, byproducts of cellular metabolism, can lead to an iron-dependent form of cell death, ferroptosis (1, 2). In mammalian cells, phospholipid peroxides are effectively neutralized by glutathione peroxidase-4 (GPX4), and blockage of this enzyme can trigger ferroptosis (3). As GPX4 requires the reducing agent glutathione to function, deprivation of cysteine, the essential building block of glutathione, via approaches such as cystine starvation or pharmacological inhibition of system xc-cystine/glutamate antiporter, can also trigger ferroptosis (1). In addition to GPX4, it was shown recently that FSP1, a CoQ reductase, suppresses ferroptosis by generating reduced form of CoQ to trap phospholipid peroxides (4, 5).
Although the physiological function of ferroptosis is still obscure, its involvement in various pathological conditions, including ischemic organ injury, neurodegeneration, and cancer has been demonstrated (1, 6–8). Particularly, mounting evidence indicates that ferroptosis plays an important role in tumor suppression (9–12), and activation of ferroptosis contributes to various cancer treatments, such as immune checkpoint blockade (13) and radiotherapy (14–16). Importantly, cancers of mesenchymal property or harboring E cadherin-NF2-Hippo pathway mutations have been shown to be highly susceptible to cell death by ferroptosis due to the alterations of in redox/iron homeostasis and metabolic processes related to ferroptosis (17–19). Therefore, ferroptosis induction is a promising approach for the treatment of cancers with certain specific genetic backgrounds. However, the role that individual tumorigenic mutations play in the regulation of the sensitivity of a given cancer to ferroptosis is just beginning to be investigated.
Sterol regulatory element-binding protein 1 (SREBP1) is a master regulator of lipid homeostasis and glucose metabolism (20, 21). As a transendoplasmic reticular membrane protein, SREBP1 senses various metabolic signals and is activated by specific proteolytic processing. The processed SREBP1 translocates into the nucleus to stimulate the transcription of its target genes, which include genes encoding critical enzymes in the lipogenesis pathway (such as ACLY, ACACA, FASN, and SCD) as well as those mediating gluconeogenesis and the pentose phosphate pathway (such as PKLR, PCK1, G6PC, and G6PD) (22, 23). More recently, it has been reported that the PI3K-AKT-mTOR signaling pathway can also regulate SREBP1 (24, 25).
In this study, we demonstrate that sustained activation of the PI3K-AKT-mTORC1 signaling pathway enables cancer cells resistant to ferroptosis through up-regulating the downstream SREBP1-mediated lipogenesis. Importantly, combing mechanistic target of rapamycin (mTOR)C1 inhibition with ferroptosis induction elicited a significant tumor regression in xenograft mouse models for breast cancer with activating PI3K mutation and prostate cancer with phosphatase and tensin homolog deleted on chromosome 10 (PTEN)-deficiency.
Results
Oncogenic Activation of the PI3K-AKT-mTOR Signaling Pathway Confers Resistance to Ferroptosis.
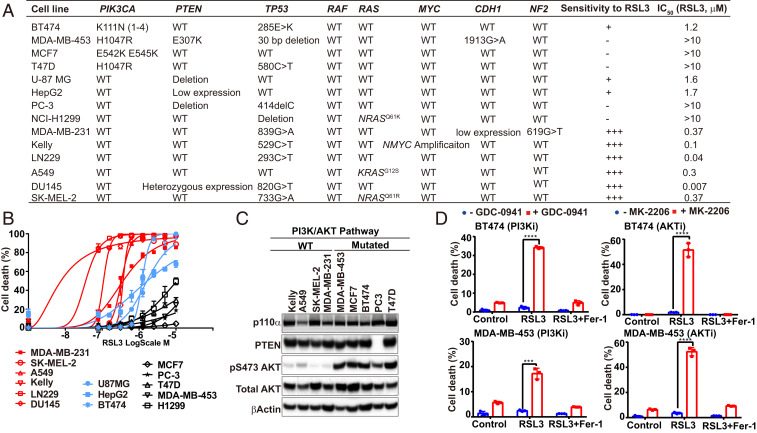
To investigate the functional interplay between ferroptosis and signaling pathways frequently mutated in cancer, we analyzed a panel of human cancer cell lines with defined genetic mutations. Ferroptosis was triggered by RSL3, a pharmacological inhibitor of GPX4. The sensitivity to ferroptosis induction varied among tested lines. Notably, we found that cancer cells carrying PIK3CA activating mutation or PTEN deletion appeared to be more resistant to RSL3 (Fig. 1 A–C). These mutations lead to the activation of the oncogenic PI3K-AKT signaling pathway, which is one of the most frequently altered signaling pathways in human cancers (26–28).
Fig. 1.
Oncogenic activation of the PI3K-AKT-mTOR signaling pathway confers resistance to ferroptosis. (A) Genetic background of the analyzed cancer cell lines and their sensitivity to RSL3. (B) Cells were seeded in 96-well plates, 2 × 104 cells per well, and incubated overnight. Cell death was induced by 24-h treatment of RSL3 with indicated concentrations. Cell death were measured by Sytox green staining, as detailed in Materials and Methods. (C) Indicated protein components in the PI3K-AKT pathway were detected by Western blot in indicated cell types. (D) Cells were treated with or without PI3K inhibitor GDC-0941 (2 μM), AKT inhibitor MK-2206 (2 μM), RSL3 (1 μM), or ferroptosis inhibitor Ferrostatin-1 (Fer-1, 1 μM) as indicated for 12 h (BT474) or 24 h (MDA-MB-453). Cell death was measured. Indicators for P values: ****P ≤ 0.0001, ***P ≤ 0.001.
To determine whether the resistance to ferroptosis is a result of PI3K-AKT signaling pathway activation, we tested whether pharmacological inhibition of this pathway could sensitize cancer cells to ferroptosis induction. Indeed, both the PI3K inhibitor (PI3Ki) GDC-0941 and AKT inhibitor (AKTi) MK-2206 sensitized BT474 and MDA-MB-453 cells (both harboring activating mutation of the pathway) to ferroptosis (Fig. 1D) and lipid peroxidation (SI Appendix, Fig. S1A). Moreover, inhibition of mTOR, a major downstream player of the PI3K-AKT pathway, by its catalytic inhibitor Torin also sensitized these cells to ferroptosis induced by RSL3 (SI Appendix, Fig. S1B). Therefore, sustained activation of the PI3K-AKT-mTOR pathway protects cancer cells from ferroptotic cell death. Notably, a role of mTORC1 in ferroptosis inhibition has been reported previously, such as in the context of cardiomyocytes (29), but the underlying mechanisms remain to be elusive.
mTORC1, Instead of mTORC2, Suppresses Ferroptosis.
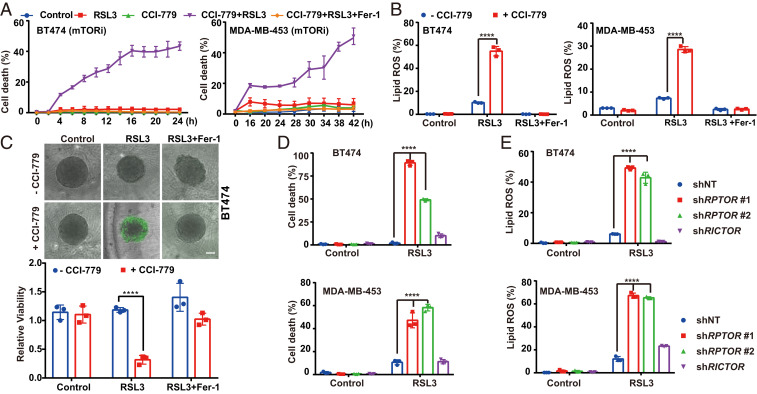
mTOR signaling is mediated by two branches, mTORC1 and mTORC2 (28). To determine which branch is responsible for the observed ferroptosis regulation, we first tested rapalog Temsirolimus (CCI-779), which inhibits the activity of mTORC1 but not that of mTORC2. Similar to Torin, CCI-779 could sensitize cancer cells to ferroptosis induction and lipid peroxidation (Fig. 2 A and B and SI Appendix, Fig. S1 C and D). Consistently, in a three-dimensional (3D) tumor spheroid system, mTOR inhibition also synergized with RSL3 in inducing ferroptosis in these mutant cancer cells (Fig. 2C and SI Appendix, Fig. S1 E and F). In contrast, inhibitors of ERK or BRAF failed to do so (SI Appendix, Fig. S1G).
Fig. 2.
mTORC1, instead of mTORC2, suppresses ferroptosis. (A) MDA-MB-453 and BT474 were treated with CCI-779 (0.5 μM), RSL3 (1 μM for MDA-MB-453 cells and 0.5 μM for BT474 cells), and Fer-1 (1 μM), as indicated. (B) Cells were seeded in six-well plates, 4 × 105 cells per well and incubated overnight. MDA-MB-453 and BT474 cells were treated as indicated. CCI-779, 0.5 μM; RSL3, 1 μM for MDA-MB-453 cells and 0.5 μM for BT474 cells; Fer-1, 1 μM. Cells were stained with 5 µM C11-BODIPY followed by flow cytometry after 8-h treatment. (C) Three-dimensional spheroids were treated as indicated. CCI-779, 0.5 μM; RSL3, 0.5 μM; Fer-1, 1 μM. (Upper) Dead cells were stained by SYTOX green. (Scale bar, 100 μm.) (Lower) Cell viability was assayed by measuring cellular ATP levels. (D) Cells expressing shRNAs targeting RPTOR or RICTOR were treated as indicated. CCI-779, 0.5 μM; RSL3, 0.5 μM; Fer-1, 1 μM. (E) Lipid peroxidation of samples as in D were measured; ****P ≤ 0.0001.
As both rapalog CCI-779 and mTOR catalytic inhibitor Torin can restore ferroptosis sensitivity, we hypothesized that mTORC1 instead of mTORC2 is responsible for the resistance of cancer cells with PI3K-AKT pathway mutation. Consistent with this notion, short-hairpin RNA (shRNA)-mediated silencing of RAPTOR (a component of mTORC1) but not that of RICTOR (a component of mTORC2) sensitized MDA-MB-453 and BT474 cells to RSL3 (Fig. 2 D and E and SI Appendix, Fig. S1H).
We next examined cancer cells that express the wild-type PI3K-AKT-mTOR pathway and are sensitive to ferroptosis induction. We tested in these wild-type cells how basal activity of the pathway allowed ferroptosis induction, and whether pharmacological inhibition of the pathway could further enhance ferroptosis. We used two cell lines with the wild-type PI3K-AKT-mTOR pathway, HT1080 and MDA-MB-231. Although low concentration of RSL3 alone was sufficient to induce ferroptosis in these cells, addition of mTOR inhibitors further enhanced ferroptosis (SI Appendix, Fig. S1I). Notably, mTORC1 activity was inhibited by RSL3 over time in these wild-type cells, as measured by S6K phosphorylation (SI Appendix, Fig. S1J). In contrast, cells harboring pathway mutation retained active mTORC1 upon RSL3 treatment for the same time period (SI Appendix, Fig. S1J). Interestingly, treatment with the lipid peroxide-trapping agent ferrostatin-1 (Fer-1) prevented RSL3-triggered inactivation of mTORC1 activity in wild-type cells (SI Appendix, Fig. S1J), suggesting lipid peroxidation is responsible for, and precedes, mTORC1 inactivation in response to RSL3. However, in cancer cells harboring PI3K-AKT-mTOR pathway mutation, RSL3-induced lipid peroxidation became clear only when the pathway was inhibited (Fig. 2 and SI Appendix, Fig. S1A), suggesting mTOR inhibition is responsible for substantial accumulation of lipid peroxides in these mutant cells. These results indicate that mTORC1 activity acts to prevent the toxic effect of lipid peroxides, whereas the accumulation of reactive oxygen species (ROS) and lipid peroxides in cells attenuates mTORC1 activity. In such a way, upon ferroptosis induction, lower basal mTORC1 activity in wild-type cells allows lipid peroxide accumulation, which in turn leads to the inhibition of mTORC1 activity and accelerated lipid peroxidation; but in mutant cancer cells, the more potent and sustained mTORC1 activity prevents lipid peroxide accumulation, thus causing resistance to ferroptosis.
NRF2 Is Not the Major Mediator of the Ferroptosis-Suppressing Activity of mTORC1.
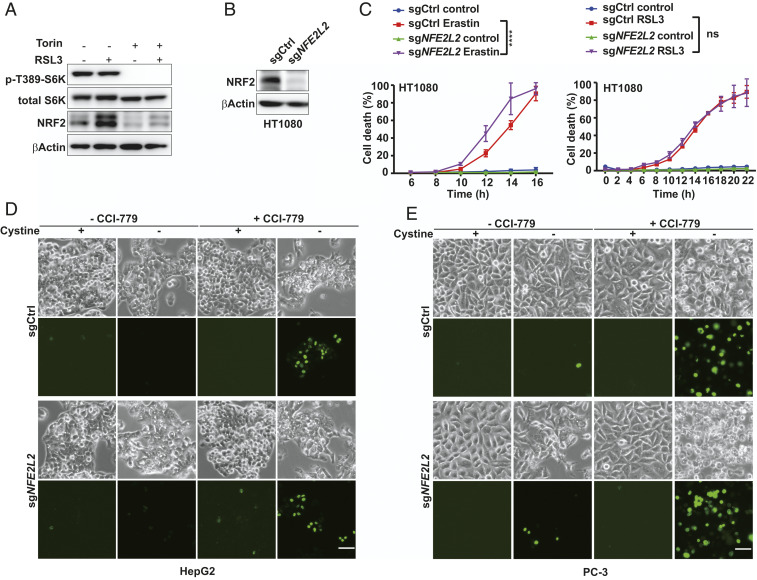
High levels of cellular ROS, a feature of ferroptosis (1), will trigger the antioxidant pathway by suppressing Keap1-mediated proteasomal degradation of the NRF2 transcription factor (30). It has been reported that mTORC1 promotes the association of p62 with Keap1 by phosphorylating p62, leading to the degradation of Keap1 and hence NRF2 accumulation (31). This p62-Keap1-NRF2 axis was reported to protect hepatocellular carcinoma cells from ferroptosis (32). Furthermore, NRF2 signaling has also been suggested to mediate the effect of mTOR on ferroptosis inhibition (33). However, although we found that RSL3-induced NRF2 accumulation was ablated by Torin treatment (Fig. 3A), NRF2-knockout in HT1080 cells only modestly enhanced ferroptosis induced by erastin (a chemical inhibitor of system xc-cystine/glutamate antiporter) and had no measurable effect on RSL3-induced ferroptosis (Fig. 3 B and C); in multiple cell lines with PI3K pathway mutation, ferroptosis induction could still be potently sensitized by mTOR inhibitors even after NRF2 was knocked out (Fig. 3 D and E and SI Appendix, Fig. S2 A–C). Furthermore, Keap1 knockout and consequent NRF2 accumulation in BT474 cells only resulted in a modest reduction of ferroptosis sensitization triggered by mTORC1 inhibition (SI Appendix, Fig. S2D). These results indicate that ferroptosis sensitization by mTORC1 inhibition is mainly through NRF2-independent mechanisms.
Fig. 3.
NRF2 is not the major mediator of the ferroptosis-suppressing activity of mTORC1. (A) BT474 cells were treated as indicated for 8 h. RSL3, 0.5 μM; Torin, 1 μM. Western blot was performed to measure p-T389 S6K, total S6K and NRF2. (B) NRF2 was depleted by CRISPR/Cas9 technology in HT1080 cells. NRF2 level was measured by Western blot. (C) Control or NRF2-depleted cells were treated as indicated. Erastin, 0.5 μM; RSL3, 25 nM. (D) HepG2 cells with or without NRF2 depletion were treated with cystine starvation with or without CCI-779 (0.5 μM) as indicated. Sytox green were added after 48 h for cell death staining. (Scale bar, 100 μm.) (E) PC-3 cells with or without NRF2 depletion were treated with cystine starvation with or without CCI-779 (0.5 μM) as indicated. Sytox green were added after 48 h for cell death staining. (Scale bar, 100 μm.)
mTORC1 Activation Suppresses Ferroptosis by Up-Regulating SREBP1.
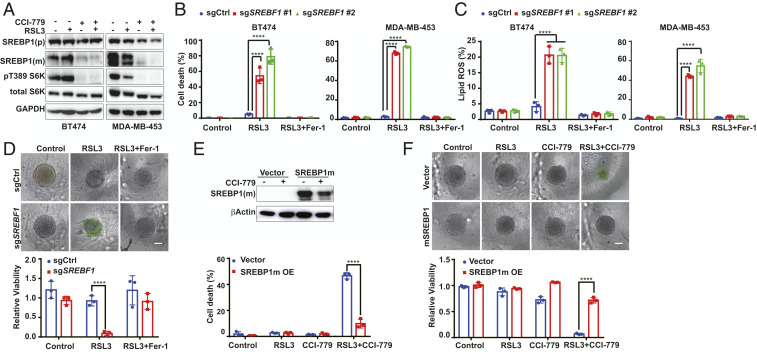
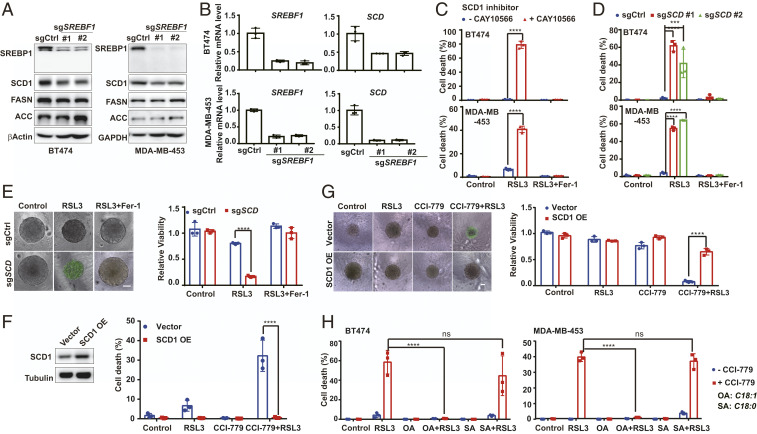
Ferroptotic cell death requires phospholipid peroxidation. To investigate if mTORC1 regulates ferroptosis through modulating cellular lipid metabolism, we examined a central regulator of lipid synthesis, SREBP1 (20, 21), which was recently demonstrated as a downstream target of mTORC1 activity (22, 25, 34, 35). Indeed, in multiple types of RSL3-resistant cancer cells, mTORC1 inhibitor CCI-779 decreased the level of the mature form of SREBP1 (SREBP1m) that can translocate into the nucleus to regulate its downstream transcriptional targets (Fig. 4A and SI Appendix, Fig. S3A). Functionally, pharmacological inhibition of SREBP activity by Fatostatin A (36) or genetic deletion of the SREBF1 gene by CRISPR/cas9 sensitized ferroptosis and lipid peroxidation in these cells (Fig. 4 B–D and SI Appendix, Fig. S3 B–D). Moreover, in these SREBF1-knockout cells, mTOR inhibitors (Torin and CCI-779), PI3K inhibitor (GDC-0941), and AKT inhibitor (MK-2206) all failed to further enhance RSL3-induced ferroptosis (SI Appendix, Fig. S3E). Conversely, ectopic expression of the constitutively active nuclear form of SREBP1 (SREPB1m), as tested in both two-dimensional (2D) cell culture and 3D tumor spheroid experiments, rendered 1) the otherwise RSL3-sensitive A549 cells resistant, and 2) multiple RSL3-resistant cell lines not responsive any longer to the combination of CCI-779 with RSL3 (Fig. 4 E and F and SI Appendix, Fig. S3F). In conclusion, mTORC1 promotes cancer cell resistance to ferroptosis induction through the up-regulation of SREBP1 function.
Fig. 4.
mTORC1 activation suppresses ferroptosis by up-regulating SREBP1. (A) BT474 and MDA-MB-453 cells were treated as indicated. RSL3, 0.5 μM; CCI-779, 0.5 μM. Cell lysates were collected after 8 h and 24 h of treatment for BT474 cells and MDA-MB-453 cells, respectively, for Western blot detecting p-T389 S6K, total S6K, unprocessed SREBP1 [SREBP1(p)] and processed, [SREBP1(m)]. (B) Cells were treated as indicated. RSL3, 0.5 μM for BT474 cells and 1 μM for MDA-MB-453 cells; Fer-1, 1 μM. (C) Cells were treated as in B. Lipid peroxidation was measured. (D) Three-dimensional spheroids derived from BT474 cells harboring control or SREBF1 sgRNA were treated as indicated. CCI-779, 0.5 μM; RSL3, 0.5 μM; Fer-1, 1 μM. (Upper) Cell death stained by Sytox green. (Scale bar, 100 μm.) (Lower) Cell viability. (E) SREBP1m was overexpressed in BT474 cells and determined by Western blot. Cells were treated as indicated. RSL3, 0.5 μM; CCI-779, 0.5 μM. Cell death was measured (Lower). (F) Three-dimensional spheroids derived from BT474 cells were treated as indicated. RSL3, 0.5 μM; CCI-779, 0.5 μM; Fer-1, 1 μM. (Upper) Cell death stained by Sytox green. (Scale bar, 100 μm.) (Lower) Cell viability. ****P ≤ 0.0001.
SREBP1 Protects Cells from Ferroptosis through SCD1 Activity.
SREBP1 is a transcription factor that regulates, among other metabolic genes, multiple lipid synthesis-related genes, including ACLY, ACACA, FASN, and SCD (SI Appendix, Fig. S6A) (20). In the tested cell lines bearing PI3K-AKT-mTOR pathway mutation, SREBF1 knockout decreased the expression of SCD1 (both mRNA level and protein level) more significantly than that of other targets (Fig. 5 A and B and SI Appendix, Fig. S4). This result and the recently reported antiferroptotic function of SCD1 (37) prompted us to examine whether SCD1 is the major downstream target of SREBP1 that mediates the resistance to ferroptosis induction. Pharmacologically, SCD1 inhibitor CAY10566 sensitized the effect of RSL3 on the induction of ferroptosis (Fig. 5C) and lipid peroxidation (SI Appendix, Fig. S5A). Genetically, CRISPR/Cas9-mediated SCD knockout also sensitized cells to ferroptosis induction and lipid peroxidation (Fig. 5 D and E and SI Appendix, Fig. S5 B–D). Furthermore, upon SCD knockout, inhibition of mTORC1, PI3K, or AKT could not further sensitize cancer cells to ferroptosis (SI Appendix, Fig. S5E). Conversely, SCD1 overexpression protected cancer cells from ferroptosis induced by the combination of RSL3 with mTOR inhibition or with SREBF1 knockout (Fig. 5 F and G and SI Appendix, Fig. S5 F and H).
Fig. 5.
SREBP1 protects cells from ferroptosis through SCD1 activity. (A) The expression of SREBP1, and its targets SCD1, FASN, and ACACA, in control and SREBF1-sgRNA cells were detected by Western blot. (B) The mRNA level of SREPF1 and its targets gene SCD were measured by RT-PCR. (C) Cells were pretreated with or without 5 μM CAY10566 overnight, and then subjected to the indicated treatments. RSL3, 0.5 μM for BT474 cells and 1 μM for MDA-MB-453 cells; CAY10566, 5 μM; Fer-1, 1 μM. (D) Cells expressing control or SCD-sgRNA were treated as indicated. RSL3, 1 μM for MDA-MB-453 cells and 0.5 μM for BT474 cells; Fer-1, 1 μM. (E) Three-dimensional spheroids derived from BT474 cells harboring control or SCD-sgRNA were treated as indicated. RSL3, 0.5 μM; CCI-779, 0.5 μM; Fer-1, 1 μM. (Left) Cell death staining. (Scale bar, 100 μm.) (Right) Cell viability. (F) SCD1 was overexpressed in BT474 cells and determined by Western blot. Cells were treated as indicated. RSL3, 0.5 μM; CCI-779, 0.5 μM. (G) Three-dimensional spheroids derived from BT474 cells with control or SCD1 overexpression were treated as indicated. RSL3, 0.5 μM; CCI-779, 0.5 μM; Fer-1, 1 μM. (Left) Cell death staining. (Scale bar, 100 μm.) (Right) Cell viability. (H) Cells were treated as indicated. RSL3, 0.5 μM for BT474 and 1 μM for MDA-MB-453; CCI-779, 0.5 μM; OA, 0.5 mM; SA, 0.5 mM. ****P ≤ 0.0001; ***P ≤ 0.001.
SCD1 is an enzyme that converts saturated fatty acids to monounsaturated fatty acids (MUFAs) (SI Appendix, Fig. S6A). It has been reported that MUFAs can inhibit ferroptosis (38), providing a mechanistic explanation to our observation. Indeed, supplementation of MUFA palmitoleic acid (16:1, PO) or oleate acid (18:1, OA), but not saturated fatty acid palmitic acid (16:0, PA) or stearic acid (18:0, SA), resulted in ferroptosis resistance upon treatment of CCI-779 plus RSL3 (Fig. 5H and SI Appendix, Fig. S6 B and C). Collectively, these results indicate that SREBP1 protects cancer cells from ferroptosis mainly by up-regulating SCD1. Remarkably, SCD1 is an iron-dependent enzyme that catalyzes fatty acid desaturation, which is by nature an oxidative reaction; and we found here that this iron-dependent, oxidative enzymatic reaction can mitigate ferroptosis, an iron-dependent, oxidative form of cell death.
Combination of mTORC1 Inhibition with Ferroptosis Induction Leads to Tumor Regression In Vivo.
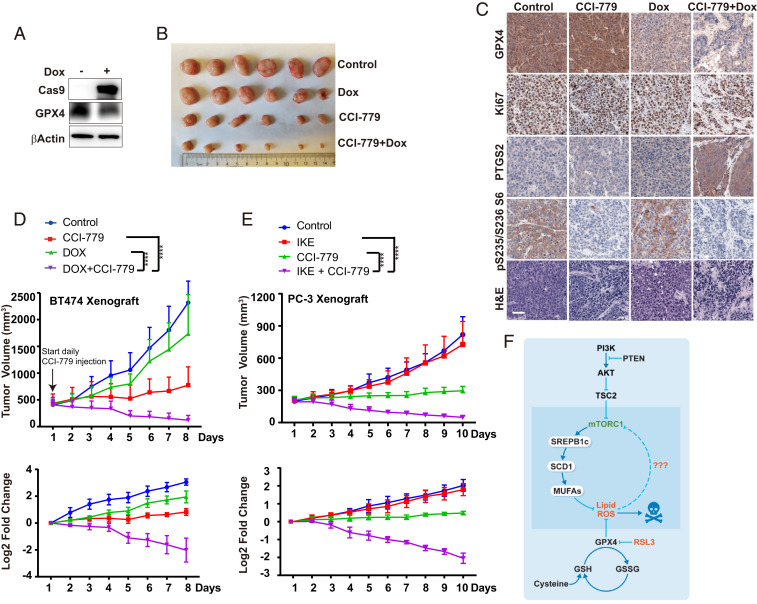
To explore the cancer therapeutic potential of combining mTORC1 inhibition with ferroptosis induction, we analyzed two mouse xenograft models for human cancer. In the first model, we generated CRISPR/Cas9-mediated GPX4 knockout in a doxycycline (Dox)-inducible manner in PI3K-mutated BT474 breast cancer cells (Fig. 6A). In these cells, only the combination of GPX4 knockout with mTORC1 inhibition, but not either alone, induced potent ferroptosis (SI Appendix, Fig. S7A). In mice xenografted with these cells, we allowed the average volume of tumors to reach ∼400 mm3, and then started mTORC1 inhibition by CCI-779 administration (Dox administration was started 2 d earlier). While CCI-779 administration decelerated tumor growth, strikingly, the combination of Dox treatment with CCI-779 caused a near-complete regression of tumors (Fig. 6 B and D and SI Appendix, Fig. S7B). Immunohistochemical analysis of PTGS2, a marker of oxidative stress and ferroptosis (3), supported such synergistic effect of the combining inhibition of GPX4 and mTORC1 in inducing tumor ferroptosis in vivo (Fig. 6C). In the other mouse model, we used imidazole ketone erastin (IKE), a potent and metabolically stable analog of erastin that has been validated for in vivo use (39), instead of genetic deletion of GPX4, to induce tumor cell ferroptosis. PTEN-defective PC-3 prostate cancer cells were used to generate xenograft tumors in mice [PTEN deficiency predicts poor prognosis in prostate cancer (40)]. Similar to what we observed in the BT474 xenograft experiment, here IKE alone has no effects on tumor growth, but its combination with CCI-779 resulted in dramatic tumor regression (Fig. 6E and SI Appendix, Fig. S7 C–E). Collectively, these two in vivo experiments demonstrate that the combination of mTORC1 inhibition with ferroptosis induction is a promising therapeutic approach for the treatment of cancer-harboring activating mutation of the PI3K-AKT-mTORC1 pathway.
Fig. 6.
Combination of mTORC1 inhibition with ferroptosis induction leads to tumor regression in vivo. (A) CRISPR/Cas9-mediated, Dox-induced GPX4 knockout (GPX4-iKO) in BT474 cells, monitored by Western blot. (B) Images of resected tumors from mice xenografted with GPX4-iKO BT474 cells. Groups of mice were treated with CCI-779 and/or Dox as indicated (n = 6 per group). See Materials and Methods for detail. (C) Representative H&E and immunostaining images of GPX4, Ki67, PTGS2, and pS235/236 S6, all counterstained with hematoxylin (blue), are shown from sections of xenografted tumors. (Scale bar, 50 μm.) (D) Growth curves of BT474 tumors of each group. Data are plotted as mean ± SD, n = 6, on the linear scale for actual tumor size (Upper) or the log2 scale for the fold-change of tumors (Lower). (E) Growth curves of PC-3 tumors of each group (n = 6), on the linear scale for actual tumor size (Upper) or the log2 scale for the fold change of tumors (Lower). (F) Model depicting that oncogenic activation of PI3K-AKT-mTORC1 signaling suppresses ferroptosis via SREBP1/SCD1-mediated lipogenesis.
Discussion
In conclusion, this work reveals a ferroptosis-regulatory mechanism in that oncogenic activation of the PI3K-AKT-mTORC1 pathway suppresses ferroptosis in cancer cells via downstream SREBP1/SCD1-mediated lipogenesis (Fig. 6F). Previously, it has been reported that various metabolic processes, such as glutaminolysis and the mitochondrial TCA cycle (11, 41), contribute to ferroptosis. This study unravels the function of SREBP-mediated lipid metabolism in ferroptosis regulation. Therefore, cellular metabolism, including that of amino acids, carbohydrates, and lipids, can all regulate ferroptosis. Cancer cells usually endure elevated oxidative stress due to increased metabolic and proliferative burden (42, 43). Consequently, pathways countering oxidative stress, particularly NRF2 signaling (30), are often activated in cancer cells. This work shows that mTORC1 is also an important modulator of cellular redox homeostasis, and it does so through both NRF2-mediated signaling and SREBP1/SCD1-mediated MUFA synthesis. The latter mechanism is crucial for the suppression of a probably more devastating type of oxidative stress, ferroptosis-inducing lipid peroxidation.
Relevant to cancer, this study indicates that oncogenic alterations in PI3K-AKT-mTOR signaling, one of the most mutated pathways in human cancer (26–28), render cancer cells more resistant to ferroptosis induction. Intriguingly, it has been reported recently that oncogenic mutations in the E cadherin-NF2-Hippo-YAP pathway, also a frequently mutated signaling pathway in human cancer, instead sensitize cancer cells to ferroptosis (18). Therefore, whether and how a specific cancer-driving mutation enhances or suppresses cancer cell ferroptosis depends on how the gene product regulates cellular processes related to ferroptosis, such as metabolism and redox and iron homeostasis. In either case, these mechanistic findings provide highly valuable insights into future ferroptosis-inducing cancer therapies: Malignant mutations in the E cadherin-NF2-Hippo-YAP pathway can be used as biomarkers that predict cancer cell responsiveness to ferroptosis induction as a monotherapy; and patients bearing tumorigenic mutations in the PI3K-AKT-mTOR pathway might be treated effectively by therapies combining ferroptosis induction with inhibitors of mTORC1 or other components of the pathway, many of which are available clinically. As both pathways are highly mutated in various types of malignancy, ferroptosis induction holds enormous potential for cancer treatment.
Materials and Methods
Reagents.
Reagents used were RSL3 (1219810-16-8, Cayman), Torin (10997, Cayman), Temsirolimus (CCI-779, NSC 683864, Selleck), Ferostatin-1 (17729, Caymen), MK-2206 (S1078, Selleck Chemicals), GDC-0941 (S1065, Selleck Chemicals), CAY10566(10012562, Cayman Chemicals), Fatostatin A (4444, Tocris), SYTOX green (S7020, Thermo Fisher), propidium iodide (556463, BD Biosciences), BODIPY 581/591 C11 (Thermo Fisher, Cat #D3861), OA (O1383, Sigma-Aldrich), SA (S4751, Sigma), PA (P0500, Sigma-Aldrich), palmitoleic acid (P9417, Sigma), and IKE (HY-114481, MCE).
Cell Culture.
The Kelly cell line was obtained from Sigma-Aldrich. Mouse embryonic fibroblasts, HT1080, MDA-MB-231, MDA-MB-453, BT474, MCF7, T47D, U87MG, HepG2, PC-3, DU145, A549, NCI-H1299, LN229, and SK-MEL-2 were obtained from the American Tissue Culture Collection (ATCC) and cultured in media conditions recommended by the ATCC in a humidified atmosphere containing 5% CO2 at 37 °C. Medium was prepared by the Memorial Sloan Kettering Cancer Center (MSKCC) Media Preparation Core Facility. All cell lines were subjected to short tandem repeat authentication through ATCC or MSKCC IGO Core Facility.
Generation of 3D Spheroids.
Spheroids were generated by plating tumor cells at 103/well into U-bottom Ultra Low Adherence 96-well plates (Corning). Optimal 3D structures were achieved by centrifugation at 600 × g for 5 min followed by addition of 2.5% Matrigel (Corning). Plates were incubated for 72 h at 37 °C, 5% CO2, 95% humidity for formation of a single spheroid of cells. Spheroids were then treated with RSL3 in fresh medium containing Matrigel for the indicated time.
Cell Death Quantification and Cell Viability Measurement.
Cells were seeded in plates at appropriate cell density and incubated overnight at 37 °C containing 5% CO2, and then subjected to treatments as described in individual experiments. Cells were stained with hoechst 33342 (0.1 μg/mL) to monitor total cell number, and with Sytox green (5 nM) to monitor cell death. Culture plates were read by Cytation 5 at indcated time points. Percentage of cell death was calculated as Sytox green+ cell number over total cell number. For 3D spheroids, cell viability was determined by using the CellTiter-Glo 3D Cell Viability Assay (Promega) following the manufacturer’s instructions. Viability was calculated by normalizing ATP levels of samples to that of negative controls (spheroids in normal full media without treatment).
Measurement of Lipid Peroxidation.
Lipid peroxidation was analyzed by flow cytometry. Cells were seeded at appropriate density in a six-well plate and grown overnight in DMEM. Cells were stained with 5 μM BODIPY C11 (Thermo Fisher, Cat #D3861) for 30 min after indicated treatment. Labeled cells were trypsinized, resuspended in PBS plus 2% FBS, and then subjected to flow cytometry analysis.
Western Blot.
Cell lysates were resolved on SDS/PAGE gels and transferred to a nitrocellulose membrane. The membranes were incubated in 5% skim milk for 1 h at room temperature and then incubated with primary antibodies diluted in blocking buffer at 4 °C overnight. The following primary antibodies were used: PTEN (9559L, CST), Phospho-Akt Ser473 (4060, CST), Akt (2920, CST), β-actin (Sigma-Aldrich, A1978), GAPDH (SC-47724, Santa Cruz), Raptor (2280, CST), Rictor (9476, CST), Total S6K (2708, CST), Phospho-p70 S6 Kinase Thr389 (9205, CST), ATG5 (A0731, Sigma), LC3 I/II (L7543, Sigma), SREBP1(SC-13551, Santa Cruz), SCD1 (ab39969, Abcam), FASN (3180, CST), ACACA (3662, CST), NRF2 (16396-1-AP, Proteintech Group), Keap1 (8047S, CST), GPX4 (ab125066, Abcam), and Cas9 (14697S, CST). After three washes, membranes were incubated with goat anti-mouse HRP-conjugated antibody or donkey anti-rabbit HRP-conjugated antibody (Invitrogen) at room temperature for 1 h and subjected to chemiluminescence using Clarity Western ECL Substrate (Bio-Rad). An Amersham Imager 600 (GE Healthcare Life Sciences) were used for the final detection.
RT-PCR.
Total RNA was prepared with the TRIzol reagent (Invitrogen). Twenty percent chloroform was added to each sample. The samples were shaken vigorously for 15 s and incubated at room temperature for 15 min. Samples were then centrifuged at 12,000 × g for 15 min at 4 °C. The aqueous phase was transferred to a new tube and an equal volume of isopropanol was added. Samples were incubated at room temperature for 10 min, followed by centrifugation at 12,000 × g for 10 min at 4 °C. mRNA pellets were washed in 75% ethanol, dried, and resuspended in nuclease-free water. mRNA was reverse transcribed into cDNA with an iScript Reverse Transcription Supermix (Bio-Rad). cDNA was amplified with the iQ SYBR Green Supermix (Bio-Rad) in a CFX Connect Real-Time PCR Detection System (Bio-Rad). The PCR program was as follows: 95 °C, 30 s; 40 cycles (for each cycle 95 °C, 15 s; 55 °C, 40 s). All primers synthesized by Invitrogen are shown in Table 1.
Table 1.
Synthesized primers
| Gene | Forward primer 5′-3′ | Reverse primer 5′-3′ |
| βActin | CTCTTCCAGCCTTCCTTCCT | AGCACTGTGTTGGCGTACAG |
| SREBF1 | GCTGCTGACCGACATCGAA | GGGTGGGTCAAATAGGCCAG |
| FASN | AACTCCAAGGACACAGTCACCAT | CAGCTGCTCCACGAACTCAA |
| ACACA | GGATGGGCGGAATGGTCTCTTT | GCCAGCCTGTCGTCCTCAATGTC |
| ACLY | AACGCCAGCGGGAGCACATC | TTGCAGGCGCCACCTCATCG |
| SCD | CCGGACACGGTCACCCGTTG | CGCCTTGCACGCTAGCTGGT |
Lentiviral-Mediated shRNA Interference.
MISSION lentiviral shRNA clones targeting RPTOR and RICTOR were purchased from Sigma-Aldrich. The clone IDs for the shRNA are: RPTOR #1 (TRCN0000039770), RPTOR #2 (TRCN0000039770), and RICTOR (TRCN0000074290). Lentiviruses were produced by the cotransfection of the lentiviral vector with the Delta-VPR envelope and CMV vesicular-stomatitis virus (VSV)-G packaging plasmids into 293T cells using PEI. Medium was changed 8 h after transfection. The supernatant was collected 48 h after transfection and passed through a 0.45-µm filter. Cells were incubated with infectious particles in the presence of 4 μg/mL polybrene (Sigma-Aldrich) overnight and cells were given fresh complete medium. After 48 h, cells were placed under the appropriate antibiotic selection.
Retroviral-Mediated Gene Overexpression.
For inducible expression of SREBP1 and SCD1, cDNAs were obtained from DNASU plasmid repository and were subcloned into a modified version of the retroviral vector pTRE-Tight (Clontech) (44). Retrovirus was produced by cotransfection of the retroviral vector with gag/pol (Addgene) and VSV-G (Addgene) into 293T cells using PEI. Virus was collected and passed through a 0.45-µm filter. Infected cells were selected in medium containing hygromycin. Gene expression was induced by addition of 100 ng/mL Dox to culture medium.
Inducible CRISPR/Cas9-Mediated GPX4 Knockout.
The lentiviral Dox-inducible pCW-Cas9 vector and pLX single-guide RNA (sgRNA) were from Eric Lander (Massachusetts Institute of Technology (MIT), Cambridge, MA; Broad Institute of MIT and Harvard, Cambridge, MA; Harvard Medical School, Boston, MA) and David Sabatini (Massachusetts Institute of Technology, Cambridge, MA; Broad Institute of MIT and Harvard, Cambridge, MA; Whitehead Institute for Biomedical Research, Cambridge, MA; David H. Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA; Howard Hughes Medical Institute, Massachusetts Institute of Technology, Cambridge, MA) (#50661 and #50662 respectively, Addgene) for inducible gene knockout (iKO). The sgRNA sequence targeting human GPX4 is CACGCCCGATACGCTGAGTG. Lentivirus was packaged in 293T cells using Lipofectamine 2000 (Life Technologies). Medium was changed 8 h after transfection, and the virus-containing supernatant was collected and filtered 48 h after transfection. BT474 cells in six-well tissue culture plates were infected with pCW-Cas9 viral supernatant containing 4 µg/mL polybrene. Cells were selected with 2 μg/mL puromycin 48 h after infection. Single clones were screened for Dox-inducible Cas9 expression. Single clones with Cas9 expression were infected with the GPX4 sgRNA virus-containing supernatant with 4 μg/mL polybrene. Cells were selected with 10 μg/mL blasticidin 48 h after infection. Single clones with Dox-inducible Cas9 expression and GPX4 knockout were amplified and used.
Generation of Constitutive CRISPR/Cas9-Mediated Knockout.
Keap1, NRF2, and SREBP1 depleted cells were generated with CRISPR/Cas9-mediated knockout system, using the LentiCRISPRV2 vector (Addgene). sgRNA sequences were designed with the Benchling CRSPR tool, and cloned into LentiCRISPRV2. SCD1 depleted cells were generated with CRISPR/Cas9 mediated knockout system, using stable Cas9 expression cells and sanger CRISPR clone from Sigma (HS5000004019 and HS5000004020). Lentivirus was produced by cotransfection of the lentiviral vector with psPAX2 (Addgene) and VSV-G (Addgene) into 293T cells using PEI. Infected cells were selected in puromycin-containing medium before proceeding to experiments. sgRNA sequences used in this study are listed in Table 2.
Table 2.
sgRNA sequences used in this study
| Gene | sgRNA sequence |
| KEAP1 | AGCCGCCCGCGGTGTAGATC |
| NFE2L2 | GCGACGGAAAGAGTATGAGC |
| SREBF1 | #1: GAGACCTGCCGCCTTCACAG |
| #2: GGATCTGGTGGTGGGCACTG | |
| SCD | #1: GAGACGATGCCCCTCTACTTGG |
| #2: TACTATTTTGTCAGTGCCCTGG |
In Vivo Xenograft Mouse Model.
For the in vivo xenograft mouse model, 17-β-estradiol 60-d release pellets (Innovative Research of America) were implanted subcutaneously into the left flank 7 d before tumor inoculation. GPX4 iKO BT474 cells were inoculated by injecting 5 × 106 cells in 50% Matrigel subcutaneously in the right flank of 6- to 8-wk-old female athymic nu/nu mice (Envigo). Tumor growth was monitored regularly via external caliper measurements. When tumors reached intended size, mice were divided randomly into four groups: 1) Vehicle group (daily intraperitoneal vehicle and normal diet), 2) CCI-779 group (daily intraperitoneal 2 mg/kg of CCI-779 and normal diet), 3) Dox group (daily intraperitoneal vehicle and Dox diet), and 4), Dox+CCI-779 group (daily intraperitoneal 2 mg/kg of CCI-779 and DOX diet). Mice were given intraperitoneal injections of 0.9% sterile saline or Dox (daily 100 mg/kg body weight, intraperitoneally) for 2 d, right before CCI-779 treatment. Subsequently, mice were provided with daily Dox diet for Dox group and Dox+CCI-779 group, with or without CCI-779 treatment, as indicated. CCI-779 were dissolved in ethanol and diluted with a solution of 5% Tween 80 and 5% PEG400 in sterile water and administered by intraperitoneal injection. The maximum width (X) and length (Y) of the tumor were measured every day and the volume (V) was calculated using the formula: V = (X2Y)/2. Tumor growth was monitored over time. For all experiments, mice were killed at a predetermined endpoint. If any tumor exceeded a volume of 2,000 mm3, 1.5 cm in diameter, or 10% of body weight, the mice would immediately be killed. At the end of the study, mice were killed with CO2 and tumors were taken for measurement of weight, followed by immunohistochemical staining. Results are presented as mean tumor volume ± SD. All protocols for animal experiments were approved by the MSKCC Institutional Animal Care and Use Committee.
For PC-3 tumor models, male athymic nu/nu mice aged 5 to 6 wk were injected in the right flank with 5 × 106 PC-3 cells. Tumors were measured with calipers daily. When tumors reached a mean volume of 200 mm3, mice were randomized into four groups: 1) Vehicle group (daily intraperitoneal 65% D5W [5% dextrose in water], 5% Tween-80, 30% PEG-400); 2) IKE group (daily intraperitoneal 50 mg/kg IKE dissolved in 65% D5W [5% dextrose in water], 5% Tween-80, 30% PEG-400); 3) CCI-779 group (daily intraperitoneal 2 mg/kg of CCI-779 dissolved in ethanol and diluted with a solution of 5% Tween 80 and 5% PEG400 in sterile water); and 4) IKE + CCI-779 group (daily intraperitoneal 50 mg/kg IKE and 2 mg/kg of CCI-779). At the end of the study, mice were killed with CO2 and tumors were taken for measurement of weight. All protocols for animal experiments were approved by the MSKCC Institutional Animal Care and Use Committee.
Immunohistochemistry.
Formalin-fixed, paraffin-embedded specimens were collected, and a routine H&E slide was first evaluated. Antigen retrieval was performed with the Retrievagen A antigen retrieval system (550524, BD Biosciences) according to the manufacturer’s instructions. Immunohistochemical staining was performed on 5-μm-thick paraffin-embedded sections using rabbit anti-GPX4 (ab125066, Abcam), mouse anti-Ki67 (9449, Cell Signaling), rabbit anti-PTGS2 (12282, Cell Signaling), and rabbit-anti pS235/236 S6 (2211S, Cell signaling) antibodies with a standard avidin-biotin HRP detection system according to the instructions of the manufacturer (anti-mouse/rabbit HRP-DAB Cell & Tissue Staining Kit, R&D Systems). Tissues were counterstained with hematoxylin, dehydrated, and mounted.
Statistical Analyses.
All data, if applicable, were expressed as mean ± SD from at least three independent experiments. Group differences were performed using two-tailed t test or two-way ANOVA. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Elisa de Stanchina and Amber Bahr of the Antitumor Assessment Core of Memorial Sloan Kettering Cancer Center for help with mouse modeling experiments; members of the X.J. laboratory for critical reading and suggestions; and Drs. Yu Song and Jianchun Wu, who provided technical support. This work is supported by NIH R01CA204232 (to X.J.), NIH R01CA201318 (to C.B.T.), and a Leukemia and Lymphoma Society postdoctoral fellowship (to J.Z.). This work is also supported by National Cancer Institute Cancer Center Core Grant P30 CA008748 to the Memorial Sloan Kettering Cancer Center.
Footnotes
Competing interest statement: C.B.T. is a founder of Agios Pharmaceuticals and a member of its scientific advisory board. He is also a former member of the Board of Directors and stockholder of Merck and Charles River Laboratories, and holds patents related to cellular metabolism. X.J. holds patents related to autophagy and cell death.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017152117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Stockwell B. R., et al. , Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixon S. J., et al. , Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W. S., et al. , Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bersuker K., et al. , The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doll S., et al. , FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Gao M., Jiang X., To eat or not to eat-the metabolic flavor of ferroptosis. Curr. Opin. Cell Biol. 51, 58–64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedmann Angeli J. P., Krysko D. V., Conrad M., Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer 19, 405–414 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Hassannia B., Vandenabeele P., Vanden Berghe T., Targeting ferroptosis to iron out cancer. Cancer Cell 35, 830–849 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., et al. , BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 20, 1181–1192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L., et al. , Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao M., et al. , Role of mitochondria in ferroptosis. Mol. Cell 73, 354–363.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jennis M., et al. , An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 30, 918–930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., et al. , CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang X., et al. , Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 9, 1673–1685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye L. F., et al. , Radiation-induced lipid peroxidation triggers ferroptosis and synergizes with ferroptosis inducers. ACS Chem. Biol. 15, 469–484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei G., et al. , The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 30, 146–162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viswanathan V. S., et al. , Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J., et al. , Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature 572, 402–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hangauer M. J., et al. , Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimano H., Sato R., SREBP-regulated lipid metabolism: Convergent physiology–divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Horton J. D., Goldstein J. L., Brown M. S., SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Düvel K., et al. , Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz R., et al. , Sterol regulatory element-binding protein-1 (SREBP-1) is required to regulate glycogen synthesis and gluconeogenic gene expression in mouse liver. J. Biol. Chem. 289, 5510–5517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ricoult S. J., Yecies J. L., Ben-Sahra I., Manning B. D., Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene 35, 1250–1260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yecies J. L., et al. , Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 14, 21–32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., et al. , A pan-cancer proteogenomic Atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell 31, 820–832.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fruman D. A., et al. , The PI3K pathway in human disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Baba Y., et al. , Protective effects of the mechanistic target of rapamycin against excess iron and ferroptosis in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 314, H659–H668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rojo de la Vega M., Chapman E., Zhang D. D., NRF2 and the hallmarks of cancer. Cancer Cell 34, 21–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichimura Y., et al. , Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 51, 618–631 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Sun X., et al. , Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63, 173–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo Y., Lee H. J., Jung Y. M., Jung Y. J., mTOR-mediated antioxidant activation in solid tumor radioresistance. J. Oncol. 2019, 5956867 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owen J. L., et al. , Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc. Natl. Acad. Sci. U.S.A. 109, 16184–16189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porstmann T., et al. , SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8, 224–236 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamisuki S., et al. , A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16, 882–892 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Tesfay L., et al. , Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 79, 5355–5366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magtanong L., et al. , Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem. Biol. 26, 420–432.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., et al. , Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 26, 623–633.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jamaspishvili T., et al. , Clinical implications of PTEN loss in prostate cancer. Nat. Rev. Urol. 15, 222–234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X., Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell 59, 298–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoxhaj G., Manning B. D., The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuber J., et al. , Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 29, 79–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.