Significance
The Arg/N-degron pathway targets proteins for degradation by recognizing their N-terminal or internal degradation signals. In the present study, we compared wild-type human cells to their double-knockout (2-KO) counterparts that lacked the UBR1/UBR2 ubiquitin ligases of the Arg/N-degron pathway. We found that a number of specific genes were either strongly induced or strongly repressed in 2-KO cells. In addition, specific transcription factors, including glucocorticoid receptor, were identified here as physiological substrates of the Arg/N-degron pathway. We discuss the emerged role of this proteolytic system as a regulator of mammalian gene expression.
Keywords: transcription, degradation, UBR1, GR, PREP1
Abstract
The Arg/N-degron pathway targets proteins for degradation by recognizing their N-terminal or internal degrons. Our previous work produced double-knockout (2-KO) HEK293T human cell lines that lacked the functionally overlapping UBR1 and UBR2 E3 ubiquitin ligases of the Arg/N-degron pathway. Here, we studied these cells in conjunction with RNA-sequencing, mass spectrometry (MS), and split-ubiquitin binding assays. 1) Some mRNAs, such as those encoding lactate transporter MCT2 and β-adrenergic receptor ADRB2, are strongly (∼20-fold) up-regulated in 2-KO cells, whereas other mRNAs, including those encoding MAGEA6 (a regulator of ubiquitin ligases) and LCP1 (an actin-binding protein), are completely repressed in 2-KO cells, in contrast to wild-type cells. 2) Glucocorticoid receptor (GR), an immunity-modulating transcription factor (TF), is up-regulated in 2-KO cells and also physically binds to UBR1, strongly suggesting that GR is a physiological substrate of the Arg/N-degron pathway. 3) PREP1, another TF, was also found to bind to UBR1. 4) MS-based analyses identified ∼160 proteins whose levels were increased or decreased by more than 2-fold in 2-KO cells. For example, the homeodomain TF DACH1 and the neurofilament subunits NF-L (NFEL) and NF-M (NFEM) were expressed in wild-type cells but were virtually absent in 2-KO cells. 5) The disappearance of some proteins in 2-KO cells took place despite up-regulation of their mRNAs, strongly suggesting that the Arg/N-degron pathway can also modulate translation of specific mRNAs. In sum, this multifunctional proteolytic system has emerged as a regulator of mammalian gene expression, in part through conditional targeting of TFs that include ATF3, GR, and PREP1.
Regulated protein degradation protects cells from abnormal (e.g., misfolded or aggregated) proteins and also modulates the levels of proteins that evolved to be short-lived in vivo. The bulk of intracellular protein degradation is mediated by the ubiquitin (Ub)-proteasome system (UPS) and by the autophagosome-endosome-lysosome system, with molecular chaperones playing essential roles in both processes (1–6). UPS comprises pathways that have in common two classes of enzymes, Ub ligases and deubiquitylases. An E3-E2 ligase recognizes a protein substrate through its feature called a degradation signal (degron) and conjugates a small protein Ub, usually in the form of a poly-Ub chain, to an amino acid residue of a substrate, usually its internal lysine. Deubiquitylases mediate, in particular, deubiquitylation of Ub-conjugated proteins (1–7). The 26S proteasome is a multisubunit ATP-dependent protease that binds to a poly-Ub of a ubiquitylated protein, unfolds the protein, and cleaves it to peptides that range from ∼3 to ∼25 residues (8–10).
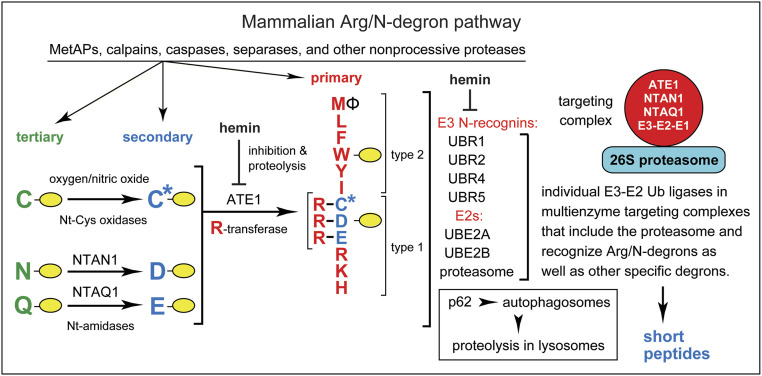
N-degron pathways (they were previously called “N-end rule pathways”) are proteolytic systems that can recognize proteins containing N-terminal (Nt) degrons called N-degrons. The targeted proteins are destroyed by the 26S proteasome and/or autophagy pathways in eukaryotes, and by the ClpS-ClpAP protease in bacteria (Fig. 1 and SI Appendix, Fig. S1) (2, 11–32). Specific determinants of an N-degron include a destabilizing Nt-residue of a protein, its internal lysine (or lysines) that functions as a site of polyubiquitylation, and a segment used by the proteasome to initiate degradation (2, 12).
Fig. 1.
The mammalian Arg/N-degron pathway (2, 15, 16). Single-letter abbreviations denote Nt-residues. The rest of a protein substrate is denoted by a yellow oval. The Arg/N-degron pathway targets proteins for degradation either by the 26S proteasome (via UBR1, UBR2, UBR4, and UBR5 E3s) or by the autophagy-lysosome pathway (via p62 N-recognin). The E3s cited above can recognize not only the indicated (primary destabilizing) Nt-residues of protein substrates but also specific non-N-terminal degrons in proteins that lack Arg/N-degrons. The terms “primary,” “secondary,” and “tertiary” denote specific classes of destabilizing Nt-residues. The Nt-amidases NTAN1 and NTAQ1 convert, respectively, Nt-Asn and Nt-Gln to the Nt-residues Asp and Glu. C* denotes an oxidized N-terminal Cys residue, either Cys-sulfinate or Cys-sulfonate. These derivatives of Nt-Cys can be produced in vivo through reactions that involve oxygen and NO. The Arg-tRNA-protein transferase (R-transferase) ATE1 conjugates Arg (with Arg-tRNA as a cosubstrate) to the Nt-Asp, Nt-Glu, or (oxidized) Nt-Cys residues. Hemin (Fe3+-heme) down-regulates both the activity of R-transferase and its in vivo stability. Hemin also interacts with UBR1/UBR2. The terms “type 1” and “type 2” denote, respectively, the basic (Arg, Lys, and His) and bulky hydrophobic residues (Leu, Phe, Trp, Tyr, Ile, and also Met, if the latter is followed by a bulky hydrophobic residue [Ф]). Type 1 and type 2 primary destabilizing Nt residues are recognized by distinct substrate-binding sites of the pathway’s E3 N-recognins. The UBR1 and UBR2 E3 Ub ligases are sequelogous to each other and to S. cerevisiae UBR1. Specific components of the human Arg/N-degron pathway have been shown to form a targeting complex (34). The S. cerevisiae Arg/N-degron pathway is also mediated by an analogous multienzyme complex (34). A “generic” targeting complex of the mammalian Arg/N-degron pathway (red circle on the Right) denotes a hypothetical set of analogous complexes. Only one of these complexes (containing UBR1 or UBR2 E3) was identified so far (34). Other, analogous targeting complexes may contain, in particular, either UBR4 or UBR5 E3s. A targeting complex is likely to include the 26S proteasome as well (34). The Ub-activating (E1) enzyme is expected to be a transient component of the complex, since E1 is a ligand of E2 enzyme. See Introduction for additional references.
The currently known eukaryotic N-degron pathways comprise the Arg/N-degron pathway (it targets, in particular, specific unacetylated Nt-residues); the Ac/N-degron pathway (it targets, in particular, the Nα-terminally acetylated [Nt-acetylated] Nt residues); the Pro/N-degron pathway (it targets, in particular, the Nt-Pro residue); the Gly/N-degron pathway (it targets the unmodified Nt-Gly residue); and the fMet/N-degron pathway (it targets Nt-formylated proteins) (Fig. 1 and SI Appendix, Fig. S1) (2, 11–28, 30–34).
Initially, most N-degrons are not active (pro-N-degrons). Active N-degrons are produced either constitutively (for example, cotranslationally) or via regulated steps. Many nonprocessive intracellular proteases, including aminopeptidases, caspases, calpains, separases, and cathepsins, function as “upstream” components of N-degron pathways that generate active N-degrons, since a cleavage of a protein can produce a C-terminal (Ct) fragment bearing a destabilizing Nt-residue (2, 23, 30). N-degrons can also be formed (activated) through enzymatic Nt-acetylation, Nt-deamidation, Nt-oxidation, Nt-arginylation, Nt-leucylation, and Nt-formylation of specific proteins or their Ct-fragments (Fig. 1 and SI Appendix, Fig. S1) (2, 16, 17, 19, 28). Recognition components of N-degron pathways are called N-recognins. They are E3 Ub ligases or other proteins (for example, mammalian p62 or bacterial ClpS) that can recognize N-degrons (2, 16, 25, 27, 29). In cognate sequence contexts, all 20 amino acids of the genetic code can act as destabilizing Nt-residues (SI Appendix, Fig. S1). Consequently, many proteins in a cell are conditionally short-lived substrates of N-degron pathways, either as full-length proteins or as Ct-fragments. An additional and functionally important feature of most E3 N-recognins is that they can target, through their multiple binding sites, not only N-degrons but other degradation signals as well. This ability of N-recognin E3s further expands the range of substrates targeted by N-degron pathways (Fig. 1) (2, 26, 35, 36).
Regulated degradation of proteins and their natural fragments by N-degron pathways has been shown to mediate a remarkably broad range of biological processes, including the sensing of oxygen, nitric oxide (NO), heme, and short peptides; the elimination of misfolded proteins and of proteins retrotranslocated to the cytosol from other compartments; the control of subunit stoichiometries in protein complexes; a suppression of neurodegeneration and regulation of apoptosis; the control of DNA repair, transcription, replication, and chromosome cohesion/segregation; the regulation of chaperones, cytoskeletal proteins, G proteins, autophagy, gluconeogenesis, peptide transport, meiosis, circadian rhythms, cell migration, fat metabolism, adaptive and innate immunity, the cardiovascular system, neurogenesis and spermatogenesis; and also plant defenses against pathogens, plant cell differentiation, the sensing of oxygen and NO, and many other processes in plants (2, 11–38 and refs. therein).
To keep notations uniform, human (Homo sapiens, hs) genetic terms (all-uppercase letters) are used below to denote both human, mouse (Mus musculus, mm) and yeast (Saccharomyces cerevisiae, sc) genes and proteins. scUBR1 encodes the 225-kDa RING-type E3, the sole N-recognin of the S. cerevisiae Arg/N-degron pathway. Unmodified N-terminal Arg, Lys, His, Leu, Phe, Tyr, Trp, Ile, and Met (if Nt-Met is followed by a bulky hydrophobic residue) are termed “primary” destabilizing Nt-residues in that they can be bound by the type-1 and type-2 sites of scUBR1 (2, 15, 16). In contrast, Nt-Asp and Nt-Glu are destabilizing, owing to their Nt-arginylation by scATE1 arginyltransferase (R-transferase). The Nt-conjugated Arg can be recognized by scUBR1. Nt-Asn and Nt-Gln are destabilizing because scNTA1 Nt-amidase can convert them to Nt-arginylatable, Nt-Asp and Nt-Glu (SI Appendix, Fig. S1G) (2, 32).
In contrast to S. cerevisiae, the mammalian Arg/N-degron pathway is mediated by at least four E3 N-recognins: the 200-kDa UBR1 and UBR2; the 570-kDa UBR4 (p600, BIG); and the 300-kDa UBR5 (EDD1, HYD) (Fig. 1) (2, 16, 25). Another, non-E3 N-recognin of this pathway is p62, an autophagy-regulating protein (27). hsUBR1 and hsUBR2 E3s are sequelogous† [similar in sequence (39)] to each other and to S. cerevisiae scUBR1. In contrast, sequelogies (39) (sequence similarities) between hsUBR1/hsUBR2 and hsUBR4 or hsUBR5 are confined largely to their ∼80-residue folded UBR domains, which bind to N-terminal Arg, Lys, or His (2, 16). In S. cerevisiae, scNTA1, an Asn/Gln/Nt-amidase, can deamidate either Nt-Asn or Nt-Gln, whereas animals and plants contain two Nt-amidases, the Nt-Asn-specific NTAN1 and Nt-Gln-specific NTAQ1 (Fig. 1 and SI Appendix, Fig. S1G) (2, 16).
In multicellular eukaryotes, Nt-arginylation encompasses not only Nt-Asp and Nt-Glu but also Nt-Cys, after its oxygen/NO-dependent oxidation to Nt-arginylatable Nt-Cys-sulfinate or Nt-Cys-sulfonate (Fig. 1). As a result, the Arg/N-degron pathway functions as a sensor of oxygen/NO, through the conditional and arginylation-dependent degradation of Nt-Cys-bearing transcription factors and regulators of G proteins (13, 18, 28). Five enzymes of the human Arg/N-degron pathway (UBR1 or UBR2 E3, UBE2A or UBE2B E2, ATE1 R-transferase, NTAN1 Asn/Nt-amidase, and NTAQ1 Gln/Nt-amidase) form a targeting complex (34) (Fig. 1). An analogous multienzyme complex mediates the S. cerevisiae Arg/N-degron pathway (34).
Homozygous inactivation of human hsUBR1 (with retention of other E3 Arg/N-recognins; Fig. 1) causes a birth defect called Johanson-Blizzard syndrome (JBS). Its symptoms include exocrine pancreatic insufficiency and inflammation, anatomical malformations, mental retardation, and deafness (2, 40). mmUBR1−/− mice have a milder version of JBS (40). Abnormal phenotypes of mmUBR2−/− mice include infertility of males, owing to apoptosis of mmUBR2−/− spermatocytes (2, 16). Mouse (or human) UBR1 and UBR2 E3s are 47% identical and overlap functionally (2, 15, 16). In contrast to viability of single-mutant mmUBR1−/− and mmUBR2−/− mouse strains, mice that lack both mmUBR1 and mmUBR2 die as midgestation embryos, with massive neural and cardiovascular defects (41).
Our previous work described construction of conditional double-knockout (2-KO) adult (mmUBR1−/− mmUBR2−/−) mice and also unconditional 2-KO (hsUBR1−/− hsUBR2−/−) human HEK293T cell lines that lacked both hsUBR1 and hsUBR2 (42). Employing these tools, we identified ATF3, a stress-inducible basic leucine zipper (bZIP) transcription factor (TF) that regulates hundreds of genes, as a short-lived substrate of the Arg/N-degron pathway (42).
In the present study, we used split-Ub protein-binding assays, RNA-sequencing (RNA-seq), and quantitative mass spectrometry (MS) (42–45) to search for other TFs that interact with hsUBR1/hsUBR2 and also to compare the levels of specific mRNAs and proteins between wild-type human HEK293T cells and their 2-KO (hsUBR1−/− hsUBR2−/−) mutants. The results described here include identification of the glucocorticoid receptor (hsGR) TF and hsPREP1 TF as putative substrates of the Arg/N-degron pathway, in part because both TFs interact with hsUBR1. The levels of some mRNAs and proteins were found to strongly differ between 2-KO and wild-type human cells. For example, we observed ∼20-fold increases of specific mRNAs (and encoded proteins) in 2-KO cells or an essentially complete repression of other mRNAs (and encoded proteins) in 2-KO cells. For instance, a TF called DACH1 and the main neurofilament subunits NF-L and NF-M were robustly expressed in wild-type cells but were virtually absent in 2-KO cells. Tellingly, a disappearance or near disappearance of some proteins in 2-KO cells took place despite increases in the levels of their mRNAs, strongly suggesting that the UBR1/UBR2-mediated Arg/N-degron pathway can also modulate translation of specific mRNAs. In sum, this multifunctional proteolytic system has emerged as a regulator of mammalian gene expression, in part through conditional targeting of TFs that include ATF3, GR, and PREP1.
Results and Discussion
Human 2-KO (hsUBR1−/− hsUBR2−/−) HEK293T cell lines that lacked both hsUBR1 and hsUBR2, two sequelogous (39) and functionally overlapping E3 N-recognins (Fig. 1), were constructed and characterized in our preceding study (42). We describe here the use of 2-KO cells and other tools to identify specific substrates and functions of this pathway.
RNA-Seq Analyses of Wild-Type Versus 2-KO (hsUBR1−/− hsUBR2−/−) Human Cells.
Genome-wide quantitative RNA-seq was carried out with RNA preparations from wild-type and 2-KO human HEK293T cells (Fig. 2 and SI Appendix, Figs. S2–S7). Our initial aim was to determine whether the ablation of hsUBR1/hsUBR2 would cause not only moderate alterations of gene expression but also significantly higher than twofold changes in the levels of specific human mRNAs. If large effects in either direction would be observed, we planned to verify them by independent methods. A focus on strong changes stemmed from the expectation that a major effect of ablating hsUBR1/hsUBR2 on the level of a specific mRNA would facilitate the understanding of a link between that effect and the Arg/N-degron pathway.
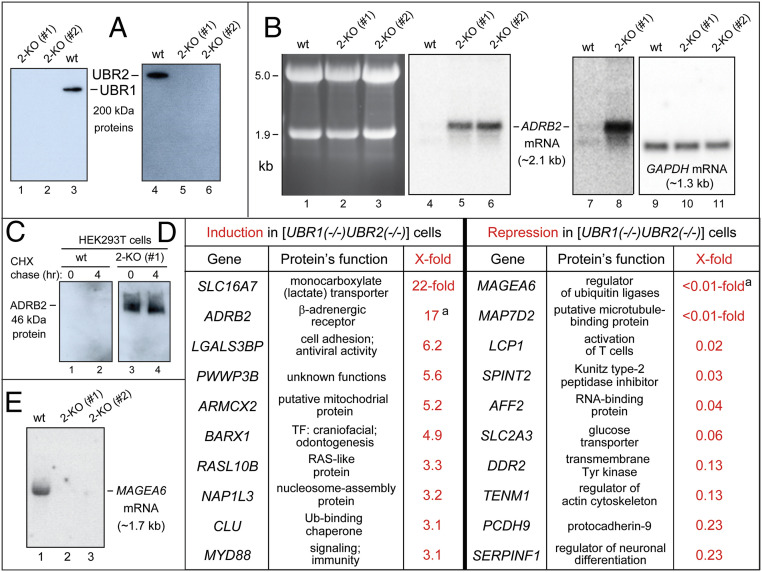
Fig. 2.
Summary of RNA-seq and related analyses of wild-type (wt) vs. 2-KO (hsUBR1−/− hsUBR2−/−) human HEK293T cells. (A) IB analyses of the wild-type (parental) HEK293T cell line and two independently produced 2-KO cell lines, #1 and #2, using anti-hsUBR1 and anti-hsUBR2 antibodies. (B) Lanes 1 through 3, fractionated total RNA, stained with ethidium bromide, from wild-type HEK293T cells and 2-KO cell lines #1 and #2. The bands of 18S and 28S rRNAs are indicated on the Left. Lanes 4 through 6, Northern hybridization, using a 32P-labeled RNA probe specific for the ∼2.1-kb hsADRB2 mRNA, of blotted RNAs in lanes 1 through 3. Lanes 7 and 8, same as lanes 4 and 5, but a 7-fold longer autoradiographic exposure to detect hsADRB2 mRNA in wild-type cells (lane 7). Lanes 9 through 11, same as lanes 4 through 6, but hybridization with a 32P-labeled RNA probe specific for the ∼1.3-kb hsGAPDH mRNA (loading control). (C) Lanes 1 and 2, IB-based (using anti-hsADRB2 antibody) CHX chase, for 0 and 4 h, of hsADRB2 in wild-type HEK293T cells. Lanes 3 and 4, same as lanes 1 and 2, but with 2-KO cell line #1, in which hsADRB2 mRNA was up-regulated by 17-fold (see B and the main text). (D) The list of 10 human mRNAs that were up-regulated by at least 3.1-fold (from 22-fold to 3.1-fold) in 2-KO cell line #1, and the analogous list of 10 mRNAs that were down-regulated by at least 4.3-fold (from more than 100-fold to 4.3-fold) in the above 2-KO cells. Superscript "a" in the right column denotes a minimal estimate of the extent of observed repression. (E) 32P-Northern analysis (see B) of the repression of hsMAGE6 mRNA (see D) in 2-KO cell lines #1 and #2 (lanes 2 and 3) vs. robust expression of hsMAGE6 in wild-type cells (lane 1).
Relative mRNA levels were the averages of three independent RNA-seq measurements for wild-type and 2-KO HEK293T cell lines. The scatter of RNA-seq data among three datasets (for each of these cell lines), particularly for mRNAs whose levels changed by more than twofold in either direction, was quite low (<5%) (SI Appendix, Fig. S2). Some mRNAs analyzed by RNA-seq were also quantified by 32P-Northern hybridization and/or RT-qPCR. The results were in at least qualitative agreement with RNA-seq data (Fig. 2 and SI Appendix, Figs. S2–S5). In these measurements, a second, independently constructed 2-KO HEK293T cell line, denoted as 2-KO #2, was also used as additional control (Fig. 2 A, B, and E and SI Appendix, Fig. S3).
RNA-seq analyses (Fig. 2 and SI Appendix, Figs. S2, S4–S7) quantified mRNA levels for ∼18,000 human genes. Among them, 161 (<1%) genes exhibited at least twofold increases of mRNAs in 2-KO cells. Conversely, 211 (<2%) genes were decreased by twofold or more (Fig. 2 and SI Appendix, Figs. S2 and S5 A–C). We confined further analyses to genes for which the maximal level of a specific mRNA, in either 2-KO or wild-type cells, comprised at least 50 sequence “reads.” Each read is an alignment of a sequenced, RNA-seq-detected segment of mRNA to the reference human genome (SI Appendix, Figs. S6 and S7). Consequently, the above 161 genes up-regulated by twofold or higher became 39 genes, chosen for high-confidence (at least 50 sequence reads) increases of mRNA in 2-KO cells. Analogously, the above 211 down-regulated genes became 93 genes, selected for high-confidence twofold or stronger decreases of mRNA levels in 2-KO cells (Fig. 2 and SI Appendix, Figs. S2–S7).
The 132 (39 + 93) altered-level mRNAs, that were chosen using criteria described above for explicit citation, comprise a high-confidence subset of all mRNAs whose levels were altered, in either direction, between 2-KO and wild-type cells (Fig. 2 and SI Appendix, Figs. S2–S7). SI Appendix, Fig. S4 cites 39 human mRNAs whose levels were increased in 2-KO cells by at least 2-fold (from 22-fold to 2-fold). The fold values, measured by RNA-seq and also for some mRNAs by 32P-Northerns and RT-qPCRs, are shown in black, red, and green, respectively (SI Appendix, Fig. S4). Descriptions of encoded proteins that are TFs or components of the Ub system are in red and blue, respectively (SI Appendix, Fig. S4). SI Appendix, Fig. S5 uses the same notations, citing 93 mRNAs that were decreased by at least 2-fold (from ∼100-fold to 2.0-fold).
Up-Regulation of Specific Human mRNAs in 2-KO Cells.
In the above sets of mRNAs, our analyses concentrated on 10 up-regulated and 10 down-regulated mRNAs for which the effects of ablating hsUBR1/hsUBR2 were particularly strong (Fig. 2D). It should be emphasized that alterations in the levels of other mRNAs, the ones not cited in Fig. 2D (SI Appendix, Figs. S4 and S5), are also worth exploring further, since many of these changes are likely to be as significant functionally as the strongest effects.
In the 10-gene set of up-regulated mRNAs, two of them, hsADRB2, encoding β-adrenergic receptor-2, and hsMCT2 (hsSLC16A7), encoding a monocarboxylate transporter, were increased by 17-fold and 22-fold, respectively (Fig. 2 B and D and SI Appendix, Fig. S4). In the former case, which was quantified independently by 32P-Northerns, the detection of hsADRB2 mRNA in wild-type cells required a strong overexposure of 32P-autoradiograms (Fig. 2B, lanes 7 and 8; compare to lanes 4 and 5). Four mRNAs, encoding hsLGALS3BP (it functions in cell adhesion), hsPWWP3B (a protein of unknown function), hsARMCX2 (a putative mitochondrial protein), and hsBARX1 (a TF that mediates craniofacial development and odontogenesis), were increased, in 2-KO cells, by 6.2-fold, 5.6-fold, 5.2-fold, and 4.9-fold, respectively (Fig. 2D and SI Appendix, Figs. S3A and S4).
In the 10-gene set of down-regulated mRNAs, 3 of them encoding hsMAGEA6 (a regulator of Ub ligases), hsMAP7D2 (a putative microtubule-binding protein), and hsLCP1 (an actin-binding protein), were decreased by at least 50-fold, from robust expression in wild-type cells to undetectable or nearly undetectable levels in 2-KO cells (Fig. 2 D and E and SI Appendix, Fig. S5A). mRNAs encoding hsSPINT2 (a peptidase inhibitor), hsAFF2 (an RNA-binding protein), and hsSLC2A3 (a glucose transporter) were decreased, in 2-KO cells, by 33-fold, 25-fold, and 16-fold, respectively (Fig. 2D and SI Appendix, Figs. S5A and S7).
hsADRB2 mRNA, Encoding β-Adrenergic Receptor-2, Is Increased by 17-Fold in 2-KO Cells.
The mammalian ADRB2 β-adrenergic receptor-2, a multispanning transmembrane protein, is expressed in most tissues. ADRB2 recognizes catecholamines (including epinephrine), is coupled to a subset of G proteins and regulates in particular the levels of cyclo-AMP (cAMP) (46–48). The 17-fold up-regulation of hsADRB2 mRNA in 2-KO cells was accompanied by a robust expression of hsADRB2 protein in these cells, as indicated by immunoblotting (IB) with antibody to hsADRB2 (Fig. 2 B–D and SI Appendix, Fig. S4). In contrast, hsADRB2 was undetectable by IB in wild-type cells, in agreement with near-zero levels of hsADRB2 mRNA in those cells (Fig. 2B).
Comparisons of Transcriptional Promoters to Address Overexpression of hsADRB2 in 2-KO Cells.
The hsADRB1 gene, encoding β-adrenergic receptor-1, was not up-regulated in 2-KO (hsUBR1−/− hsUBR2−/−) HEK293T cells, in contrast to hsADRB2, which encodes a protein highly sequelogous to hsADRB1 (Fig. 2 B–D and SI Appendix, Figs. S3 and S4). (Levels of hsADRB1 mRNA were too low for detection by RNA-seq in both 2-KO and wild-type cells.) While the 17-fold increase of hsADRB2 mRNA in 2-KO cells (Fig. 2D and SI Appendix, Fig. S4) might result in part from a metabolic stabilization of this mRNA, a parsimonious interpretation is that at least the bulk of this increase is caused by transcriptional induction of hsADRB2 in the absence of hsUBR1/hsUBR2.
For reasons that include large DNA spans and complexity of mammalian transcriptional control, our comparisons of hsADRB2 and hsADRB1 promoters were confined to ∼2-kb DNA segments upstream of transcription start sites. Our aim was to identify experimentally tractable differences in the patterns of specific TF-binding sites between these promoters, with the possibility of expanding, later, a search for differences beyond initially examined DNA segments (SI Appendix, Fig. S8). hsADRB1 and hsADRB2 have been previously mapped using expression assays, mutagenesis, gel-shift DNA binding, and other methods (47, 48). hsADRB1 and hsADRB2 promoters contain binding sites for a number of TFs, including hsNF-κB, hsAP2, and hsSP1. Most of these and other TF-binding sites are shared between hsADRB1 and hsADRB2. Nevertheless, we detected two possible (nonalternative) causes of “up-regulation” difference between these otherwise similar genes.
First, hsADRB2 promoter contains two sites that can bind to hsSP1, an activator TF that up-regulates many genes (SI Appendix, Fig. S8) (49). In contrast, the analogous region of hsADRB1 contains one hsSP1-binding site (SI Appendix, Fig. S8). Second, the single hsSP1-binding site in hsADRB1 overlaps with a site recognized by hsEGR1, a transcriptional repressor (47, 48). Thus, interactions of hsSP1 and hsEGR1 TFs with hsADRB1 promoter are likely to be mutually exclusive (SI Appendix, Fig. S8). Significantly, the hsADRB2 promoter lacks a binding site for the hsEGR1 repressor, in contrast to hsADRB1. Finally, the hsEGR1 repressor may be, at least in part, a substrate of the Arg/N-degron pathway; this remains to be verified. If so, hsEGR1 would be up-regulated in 2-KO cells and would act to inhibit transcription of hsADRB1, but not of hsADRB2. This mechanistically specific and testable model (SI Appendix, Fig. S8) is ready to be verified in future experiments.
hsMCT2 mRNA, Encoding a Monocarboxylate Transporter, Is Increased by 22-Fold in 2-KO Cells.
hsMCT2 (hsSLC16A7) is a proton-linked transporter of monocarboxylates such as lactate, pyruvate, and ketone bodies (50, 51). hsMCT2 mRNA was increased by 22-fold in 2-KO cells, as discovered by RNA-seq and confirmed by RT-qPCR (the latter method suggested an even higher, 35-fold increase of hsMCT2 mRNA in 2-KO cells) (Fig. 2D and SI Appendix, Fig. S4). The family of mammalian SLC16 (MCT) transporters comprises 14 proteins. Substrates and partial functions are known for only seven of these transporters (50, 51). Mammalian MCT2 is expressed in most tissues and can mediate either influx or efflux of its substrates, depending on substrate levels and a pH gradient across a membrane (either the plasma membrane or specific intracellular membranes). Functions of MCT2 include lactate transport, particularly in the brain and skeletal muscle (50).
In contrast to hsMCT2 mRNA, which was increased by 22-fold in 2-KO cells (Fig. 2D and SI Appendix, Fig. S4), RNA-seq did not detect mRNAs of 11 hsSLC16-family genes in either wild-type or 2-KO cells. Two hsSLC16 family members other than hsMCT2, specifically hsMCT1 (hsSLC16A7) and hsMCT8 (hsSLC16A2) mRNAs, were increased by 1.1-fold and 1.4-fold, respectively, in 2-KO cells. Furthermore, MS-based protein analyses (see below) of 2-KO cell lines #1 and #2 vs. wild-type cells indicated, respectively, 2.4-fold and 4-fold decreases of the hsMCT8 protein in 2-KO cells (SI Appendix, Fig. S12B), a direction of changes that is opposite to 22-fold induction of hsMCT2 mRNA (Fig. 2D) and up-regulation of hsMCT2 protein (see below).
The selective and massive up-regulation of hsMCT2 mRNA upon the ablation of hsUBR1/hsUBR2 (Fig. 2D and SI Appendix, Fig. S4) opens up the same strategy of comparing transcriptional promoters that yielded a verifiable model of selective up-regulation of the hsADRB2 gene in 2-KO cells (SI Appendix, Fig. S8). Analogous promoter comparisons are also possible with other genes that are up-regulated or down-regulated in 2-KO cells (Fig. 2D). In the present paper, such comparisons are confined to hsADRB2 vs. hsADRB1 (SI Appendix, Fig. S8), inasmuch as verifications of resulting models are still in the future.
Double-KO HEK293T Cells, Which Overexpress hsMCT2 mRNA, Are Hypersensitive to Dimethyloxalylglycine.
MS-based protein analyses (see below) confirmed an up-regulation of hsMCT2 protein in 2-KO cells, with, respectively, 6.0-fold and 2.7-fold increases of hsMCT2 in 2-KO cell lines #1 and #2 (SI Appendix, Fig. S12A).
To address this question in a different way, we asked whether the 22-fold increase of hsMCT2 mRNA in 2-KO cells (Fig. 2D and SI Appendix, Fig. S4) and the resulting increase of encoded protein cause a phenotype that can be traced to the level of hsMCT2. The metabolite α-ketoglutarate (αKG) is a cosubstrate (together with oxygen) of αKG-dependent dioxygenases (αKGDDs), a family of enzymes that include prolyl hydroxylases (PHDs). The oxygen-dependent hydroxylation by PHDs of specific Pro residues in, for example, HIFα TF (which mediates responses to oxygen), activates its degron and thereby regulates, through oxygen-modulated degradation of HIFα, the expression of genes controlled by HIFα (52, 53).
N-oxalylglycine (NOG), a synthetic analog of αKG, is a cytotoxic inhibitor of αKGDD enzymes, but NOG cannot enter cells (51). Dimethyloxalylglycine (DMOG), a derivative of NOG, is rapidly hydrolyzed in aqueous solutions to methyloxalylglycine (MOG). Upon its entry into cells, MOG is converted to NOG. The import of MOG is mediated largely by the MCT2 transporter (51). Consequently, an up-regulation of hsMCT2 in 2-KO cells would predict their hypersensitivity to extracellular DMOG, in comparison to wild-type cells.
To compare sensitivities of cells to DMOG, we used a cell mass accumulation assay (51). In the absence of DMOG, 2-KO cell cultures grew at rates similar to those of parental HEK293T cells (SI Appendix, Fig. S9A). In contrast, proliferation of 2-KO cells was found to be at least 5.4-fold more sensitive to DMOG than proliferation of wild-type cells (SI Appendix, Fig. S9B). “At least” refers to the fact that parameters of the cell accumulation assay were not varied to maximize the measured difference in sensitivity to DMOG between two cell lines.
These results (SI Appendix, Fig. S9), together with previous demonstration that hsMCT2 is the main MOG importer (51), confirm the above prediction that 2-KO cells, which overexpress both hsMCT2 mRNA and the encoded protein (Fig. 2D and SI Appendix, Figs. S4 and S11A), would be hypersensitive to growth suppression by DMOG (specifically by NOG, which DMOG is converted to, via MOG).
Other Genes That Are Up-Regulated in 2-KO Cells.
In addition to hsADRB2 and hsMCT2 mRNAs (increased by 17-fold and 22-fold, respectively), several other mRNAs (hsLGALS3BP, hsPWWP3B, hsARMCX2, and hsBARX1) were also up-regulated by more than 4-fold in 2-KO cells (Fig. 2D and SI Appendix, Fig. S4). hsLGALS3BP is a protein that promotes cell adhesion (https://www.uniprot.org/uniprot/Q08380). hsPWWP3B is a broadly expressed protein of unknown function (https://www.uniprot.org/uniprot/Q5H9M0) (Fig. 2D and SI Appendix, Fig. S4). hsARMCX2 is a largely uncharacterized protein (https://www.uniprot.org/uniprot/Q7L311) (Fig. 2D and SI Appendix, Fig. S4). hsBARX1 is a homeodomain TF that is important for development of craniofacial bones, teeth, cartilage, muscle, spleen, stomach, and esophagus (54). The increase of hsBARX1 mRNA in 2-KO HEK293T cells (4.9-fold by RNA-seq; 7.3-fold by RT-qPCR) (Fig. 2D and SI Appendix, Figs. S3A and S4) may be caused by a decreased degradation, in the absence of hsUBR1/hsUBR2, of an unknown activator TF whose stabilization accelerates transcription of hsBARX1. A nonalternative possibility is that hsBARX1 TF is a positive regulator of its own gene and a short-lived substrate of the Arg/N-degron pathway.
Repression of Specific Genes in 2-KO Cells.
hsMAGEA6 is a member of the family of human MAGE genes. Some MAGE proteins, including hsMAGEA6, are components and modulators of specific Ub ligases. MAGE genes, including hsMAGEA6, are often ectopically expressed in cancer cells, in which specific MAGEs can act as oncogenic drivers (55). We found that hsMAGEA6 mRNA was expressed in wild-type cells, but was decreased by at least 100-fold, i.e., was in effect shut off in 2-KO cells (Fig. 2 D and E and SI Appendix, Fig. S5A). hsLCP1 mRNA, which encodes an actin-binding protein (https://www.uniprot.org/uniprot/P13796), was decreased, in 2-KO cells by at least 50-fold to <2% of its level in wild-type cells (Fig. 2D and SI Appendix, Fig. S5A). hsSLC2A3 mRNA, which encodes a glucose transporter (https://www.uniprot.org/uniprot/P11169), was decreased in 2-KO cells to <6% of its level in wild-type cells (Fig. 2D and SI Appendix, Fig. S5A).
Mass Spectrometric Analyses of Proteins in Wild-Type and 2-KO HEK293T Cells.
MS-based protein surveys employed the quantitative TMT-SPS-MS3 (targeted mass tags-based sample multiplexing-mass spectrometric-3) technique (45) and were carried out with protein preparations from wild-type vs. 2-KO HEK293T cells (SI Appendix, Fig. S10). The TMT-SPS-MS3 method is summarized in SI Appendix, Fig. S11. TMT-SPS-MS3 analyses of wild-type and two 2-KO cell lines (#1 and #2) encompassed 83,513 peptides and corresponded to 7,714 different human proteins. Cited below are the main MS-based results:
-
1)
A total of 45 proteins were found to be up-regulated, relative to wild-type HEK293T cells, by at least 2-fold in the 2-KO #1 cell line. In most (though not all) cases, these proteins were also classed as up-regulated in the 2-KO cell line #2 (SI Appendix, Fig. S12 A and B).
-
2)
A total of 111 proteins were found to be down-regulated relative to wild-type cells by at least 2-fold in 2-KO cells (Fig. 3 and SI Appendix, Fig. S13 A–D).
-
3)
The above proteins (156 = 45 + 111), whose levels were significantly increased or decreased in 2-KO HEK293T cells, relative to wild-type cells, encompassed a vast range of functions or putative functions (SI Appendix, Figs. S12 A and B and S13 A–D).
-
4)
Changes in levels of the above 156 proteins (156 = 45 + 111) in 2-KO cells, as determined by MS analyses, will be gradually verified in future studies by independent methods, including quantitative IB. Verifications were initiated here with three proteins, hsDACH1, hsNF-L (hsNFEL), and hsNF-M (hsNFEM). According to MS, these proteins were down-regulated, respectively, by ∼5-fold, ∼16-fold, and ∼16-fold, in 2-KO cells, relative to wild-type cells (SI Appendix, Fig. S13A).
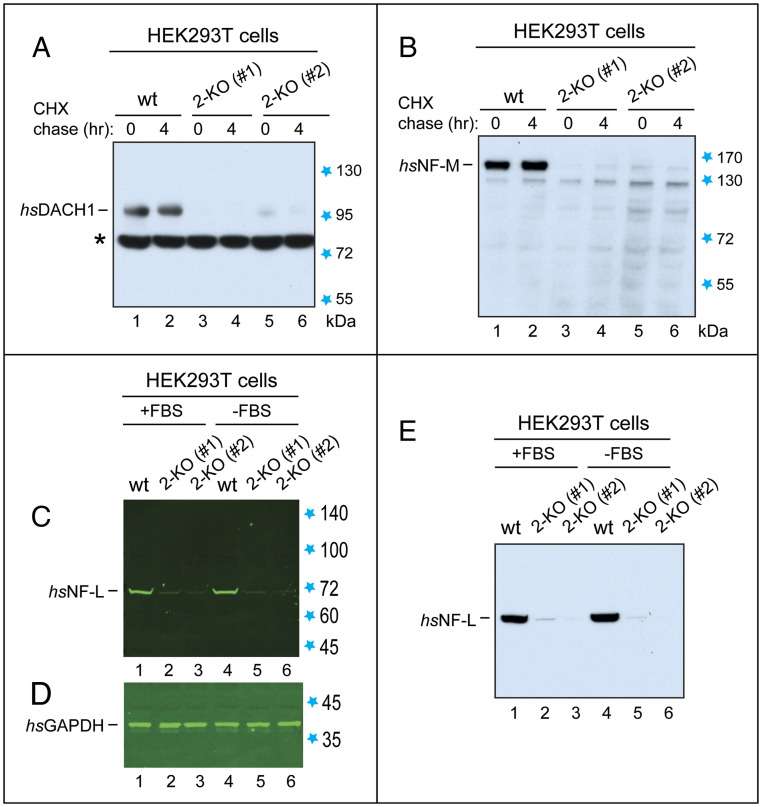
Fig. 3.
Immunoblot analyses of proteins whose down-regulation in 2-KO (hsUBR1−/− hsUBR2−/−) human HEK293T cells was initially detected by MS. Unless stated otherwise, cells were grown in the presence of fetal bovine serum (FBS). Detection of bound antibodies was performed using chemiluminescence (in A, B, and E) or near-infrared fluorescence and an Odyssey-type scanner (in C and D). (A) Lanes 1 and 2, IB-based CHX chase (using anti-hsDACH1 antibody) for 0 and 4 h of hsDACH1 TF in extracts from wild-type HEK293T cells. Lanes 3 and 4, same as lanes 1 and 2, but with 2-KO cell line #1. Lanes 5 and 6, same as in lanes 3 and 4, but with 2-KO cell line #2. Blue stars, in this and other panels, indicate positions of molecular mass markers. Black asterisk indicates a cross-reacting protein. (B) Same as in A, but with the hsNF-M subunit of neurofilaments, using anti-hsNF-M antibody. The apparent (anomalous) Mr (molecular mass), upon sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE), of the 916-residue hsNF-M is ∼150 kDa. (C) Lanes 1 through 3, IB analyses of hsNF-L, using anti-hsNF-L antibody, in wild-type HEK293T cells, 2-KO cell line #1, and 2-KO cell line #2. The apparent Mr, upon SDS/PAGE, of the 543-residue hsNF-L is ∼70 kDa. Lanes 4 through 6, same as in lanes 1 through 3, but using extracts from cells preincubated in the absence of FBS. (D) Same as in C but IB with anti-hsGAPDH antibody (loading control). (E) Same as in C but detection of hsNF-L using anti-hsNF-L antibody and chemiluminescence.
Using IB, we confirmed these findings (Fig. 3). Moreover, IB analyses showed that these endogenous, untagged proteins, while robustly expressed in wild-type HEK293T cells, either disappeared or nearly disappeared in 2-KO cell lines #1 and #2 (Fig. 3). Thus, the extents of repression of these three proteins in 2-KO cells that were measured by quantitative IB were even greater than MS-suggested repression levels (Fig. 3 and SI Appendix, Fig. S13A; see also below). These findings suggest that MS analyses, while invaluable for discovering specific proteins that are down-regulated in 2-KO cells (SI Appendix, Fig. S13 A–D), may underestimate the extents of their repression. This circumstance encourages future analyses of MS-identified proteins that are down-regulated in 2-KO cells (SI Appendix, Fig. S12 A–D) using independent methods, including quantitative IB. Described below are our first IB results with hsDACH1, hsNF-L, and hsNF-M.
Near Disappearance of the hsDACH1 Transcription Factor in 2-KO Cells.
The 758-residue hsDACH1 is a TF whose functions include tumor suppression and regulation of organogenesis (56). According to TMT-SPS-MS3 data, the levels of hsDACH1 protein in 2-KO cells, in comparison to wild-type cells, were decreased by ∼5-fold and ∼3-fold, respectively, in 2-KO cell lines #1 and #2 (SI Appendix, Fig. S13A).
IB analyses of endogenous, untagged hsDACH1 using anti-hsDACH1 antibody and chemiluminescence-based IB showed a robust expression of hsDACH1 in wild-type HEK293T cells and an even stronger (more than 10-fold) repression of the hsDACH1 protein in 2-KO #1 and #2 cell lines than the extent of down-regulation estimated by MS (Fig. 3A and SI Appendix, Fig. S13A). Detection of proteins by IB using chemiluminescence is more sensitive than quantitative detection using near-infrared fluorescence dyes and an Odyssey-type scanner, but the former method is at best semiquantitative, hence the above minimal estimate rather than a measurement. With a near-infrared quantitative system, detection of hsDACH1 in 2-KO cells would have been impossible without a strong sample overload. Possible causes of the near disappearance of hsDACH1 in 2-KO cells (Fig. 3A) are mentioned below.
Near Disappearance of the Neurofilament Subunits NF-M and NF-L in 2-KO Cells.
Neurofilaments are a specific class of intracellular intermediate filaments. Four main subunits of mammalian neurofilaments are NF-L (encoded by NFEL), NF-M (encoded by NFEM), NF-H (encoded by NFEH), and either α-internexin (encoded by INA) or peripherin (encoded by PRPH) (57, 58). Neurofilaments are present largely in neurons. Most HEK293T (and related) cell lines (which were produced by transformation of primary cultures of human embryonic kidney [HEK] cells with fragments of adenoviral DNA) contain neurofilaments and express their subunits, suggesting that HEK cell lines are actually of neuronal origin (59). Our wild-type HEK293T cells contained at least the NF-L, NF-M, and α-internexin subunits (Fig. 3B, lanes 1 and 2; Fig. 3 C and E, lanes 1 and 4, and SI Appendix, Figs. S12A and S13A).
In neurons, neurofilaments are present in both perikarya and dendrites and are particularly abundant in axons, in which they play major roles. Neurofilaments are regulated by phosphorylation and other modifications, and interact with a number of intracellular proteins. Abnormal accumulations of neurofilaments are characteristic of many diseases, including amyotrophic lateral sclerosis, Charcot-Marie-Tooth disease, neurofilament inclusion disease, giant axonal neuropathy, spinal muscular atrophy, and both Alzheimer’s and Parkinson’s diseases. Aberrant overproduction of neurofilaments in these diseases apparently contributes in major ways to the death of affected neurons (57, 58).
According to MS, the levels of hsNF-M protein were decreased by ∼16-fold in both 2-KO cell lines, #1 and #2, relative to wild-type cells (SI Appendix, Fig. S13A). IB analyses (using anti-hsNF-M antibody) confirmed these results. Moreover, the endogenous, untagged hsNF-M protein, while robustly expressed in wild-type HEK293T cells, was nearly undetectable by IB in both 2-KO cell lines, suggesting an even greater than ∼16-fold repression of hsNF-M in 2-KO cells (Fig. 3B and SI Appendix, Fig. S13A). Cycloheximide (CHX) chases of hsNF-M indicated its stability over 4 h in wild-type cells (Fig. 3B), in agreement with earlier findings (57, 58).
Similar results were obtained with hsNF-L, another neurofilament subunit, which binds to hsNF-M. According to MS, the level of hsNF-L was decreased, respectively, by ∼16-fold and ∼20-fold in 2-KO cell lines #1 and #2, relative to wild-type cells (SI Appendix, Fig. S13A). IB analyses of hsNF-L (using anti-hsNF-L antibody) employed both the quantitative Odyssey IB system and chemiluminescence-based IB. These and other IB assays also used antibody to glyceraldehyde 3-phosphate dehydrogenase (hsGAPDH) to detect hsGAPDH as a loading control (Fig. 3 C–E).
IBs detected expression of the endogenous, untagged hsNF-L protein in wild-type HEK293T cells and also indicated a complete or nearly complete disappearance of hsNF-L in both 2-KO cell lines (Fig. 3 C–E). Quantification using the Odyssey system of the repression of hsNF-L in 2-KO cells yielded an ∼40-fold difference between the levels of hsNF-L in wild-type vs. 2-KO cells (Fig. 3 C and D; see also Fig. 3E). Thus, IB assays indicated an even stronger down-regulation of hsNF-L than the one suggested by MS data (SI Appendix, Fig. S13A).
In sum, the expression of hsNF-L and hsNF-M proteins was nearly completely abolished in 2-KO human cells, in contrast to robust expression of these proteins in wild-type cells (Fig. 3 B–E and SI Appendix, Fig. S13A). α-Internexin (encoded by hsINA) is present in some but not all neurofilaments. In contrast to hsNF-L and hsNF-M, α-internexin was increased by approximately twofold in 2-KO cells (SI Appendix, Fig. S12A). Thus, the nearly complete dependence of expression of hsNF-L and hsNF-M on the presence of hsUBR1/hsUBR2 (Fig. 3 B–E and SI Appendix, Fig. S13A) does not encompass all subunits of neurofilaments.
Repression in 2-KO Cells of NF-L and NF-M Takes Place Despite Up-Regulation of Their mRNAs.
Remarkably, the disappearance or near disappearance of hsNF-L and hsNF-M in 2-KO cells (in contrast to their robust expression in wild-type cells) took place despite up-regulation by ∼1.7-fold and ∼1.2-fold, respectively, of hsNF-L (hsNFEL) and hsNF-M (hsNFEM) mRNAs in 2-KO cells (Fig. 3 B–E and SI Appendix, Fig. S13A). Thus, the near absence of hsNF-L and hsNF-M proteins in 2-KO cells could not have been caused by transcriptional repression of the corresponding genes or by destabilization of hsNF-L and hsNF-M mRNAs.
A parsimonious interpretation of these results is that the absence of the hsUBR1/hsUBR2 Ub ligases causes repression of translation of hsNF-L and hsNF-M mRNAs in 2-KO cells. In one verifiable model, a selective translational repression would be caused by metabolic stabilization (and therefore up-regulation) of a normally short-lived (and remaining to be identified) translational repressor(s) of hsNF-L and hsNF-M mRNAs in 2-KO cells. The postulated repressor(s), presumably an RNA-binding protein(s) that recognizes hsNF-L and hsNF-M mRNAs, is normally down-regulated through degradation by the hsUBR1/hsUBR2-mediated Arg/N-degron pathway. Consequently, this repressor(s) becomes long-lived in 2-KO cells, and its level increases strongly enough to shut off translation of (at least) hsNF-L and hsNF-M mRNAs. Work to verify this model is under way.
In contrast to hsNF-L and hsNF-M proteins, whose near disappearance in 2-KO cells takes place despite up-regulation of their respective mRNAs in these cells (Fig. 3 B–E and SI Appendix, Fig. S13A), the near absence of hsDACH1 TF in 2-KO cells is accompanied by a 3.8-fold decrease of its mRNA (Fig. 3A and SI Appendix, Fig. S13A). Nevertheless, the observed down-regulation of hsDACH1 protein in 2-KO cells is much stronger than the down-regulation of hsDACH1 mRNA, suggesting that an hsUBR1/hsUBR2-dependent translational repression, described in the context of hsNF-L and hsNF-M proteins, may also apply, at least in part, to regulation of hsDACH1.
Split-Ubiquitin Binding Assays.
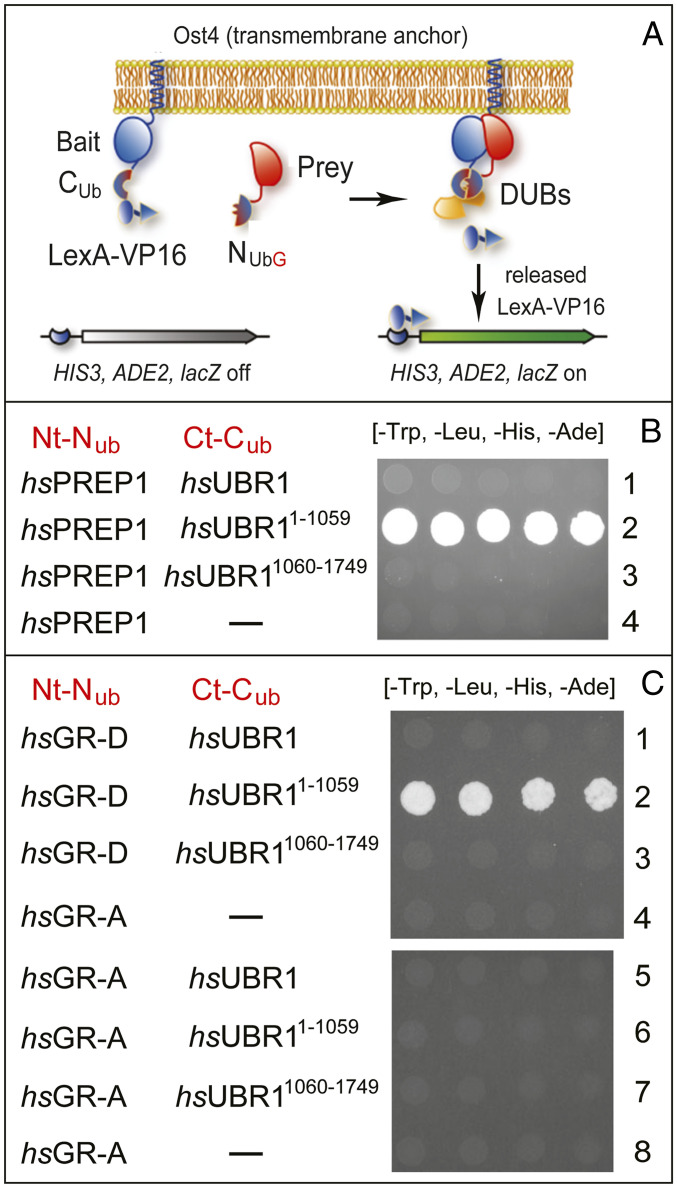
In this technique, two proteins are expressed in yeast as fusions to a Ct-half of Ub (CUb) and to its mutant Nt-half (NUb), respectively (Fig. 4A) (43, 60). An interaction between two examined proteins (which contain linked Ub halves) would reconstitute a Ub moiety from CUb and mutant NUb. Consequently, a CUb-containing test fusion would be cleaved by deubiquitylases at the last (Gly) residue of the reconstituted Ub moiety. This cleavage acts, through additional steps, as a set of assay’s readouts (Fig. 4A) (43). They comprise, in particular, an induction of scHIS3 and scADE2 genes (Fig. 4A), thereby making possible growth assays on media that lack either histidine (His) or both His and adenine (Ade) (Fig. 4). Control experiments included IBs to examine expression of split-Ub fusions and also verification of the absence of autoactivation, i.e., that binding-positive fusions did not remain positive in split-Ub assays with just one of two fusions. The results described below passed all of these controls.
Fig. 4.
Split-ubiquitin binding assays with transcription factors hsPREP1 and glucocorticoid receptor (hsGR) vs. hsUBR1 E3 ubiquitin ligase. (A) Design of split-Ub assays (also see the main text). (B) Row 1, hsPREP1 vs. full-length hsUBR1. Row 2, hsPREP1 vs. Nt-fragment of hsUBR1 (hsUBR11−1059). Row 3, hsPREP1 vs. Ct-fragment of hsUBR1 (hsUBR11060−1749). Row 4, hsPREP1 vs. vector alone. Note the binding of hsPREP1 solely to the Nt-fragment of hsUBR1. (C) See the main text and Fig. 5 for definitions and descriptions of the hsGR-D isoform. Row 1, hsGR-D vs. full-length hsUBR1. Row 2, hsGR-D vs. Nt-fragment of hsUBR1 (hsUBR11−1059). Row 3, hsGR-D vs. Ct-fragment of hsUBR1 (hsUBR11060−1749). Row 4, hsGR-D vs. vector alone. Row 5, hsGR-A vs. full-length hsUBR1. Row 6, hsGR-A vs. Nt-fragment of hsUBR1 (hsUBR11−1059). Row 7, hsGR-A vs. Ct-fragment of hsUBR1 (hsUBR11060−1749). Row 8, hsGR-A vs. vector alone.
Having previously detected, in part through split-Ub assays, the binding of the stress-inducible bZIP-class hsATF3 TF to an Nt-fragment of hsUBR1 (42), we examined here hsUBR1 interactions with two other TFs, the hsPREP1 TF and the glucocorticoid receptor (hsGR) TF, encoded by the hsNR3C1 gene (Fig. 4). We tested hsPREP1 (hsPKNOX1), a homeodomain TF (61) (Fig. 4B), owing to a significant sequelogy between hsPREP1 and S. cerevisiae scCUP9, a yeast transcriptional repressor. scCUP9 is a key part of a circuit in which the conditional (regulated by short peptides) degradation of scCUP9 by the Arg/N-degron pathway controls the rate of peptide import in yeast (see below) (2, 35, 36).
hsGR, a zinc-finger TF, is modulated by glucocorticoids and regulates processes that include immune responses as well as cell growth and differentiation (62). We tested hsGR for its binding to hsUBR1 (Fig. 4C) owing, in part, to MS-based data that the hsGR protein was up-regulated by 1.5-fold and 2.0-fold, respectively, in 2-KO cell lines #1 and #2, relative to wild-type cells. In addition, hsGR mRNA was up-regulated by 1.4-fold in 2-KO cell line #1, according to RNA-seq. These results are not mentioned in the corresponding protein and mRNA lists (SI Appendix, Figs. S4 and S12), inasmuch as the cited protein increases had been “ordered” according to data with 2-KO cell line #1 and had to be 2-fold or higher to make it into the list. We found later that hsGR protein is up-regulated by significantly more than 2-fold in 2-KO cells, in agreement with physical binding of an isoform of hsGR to an Nt-half of hsUBR1, as described below (Figs. 4C and 5).
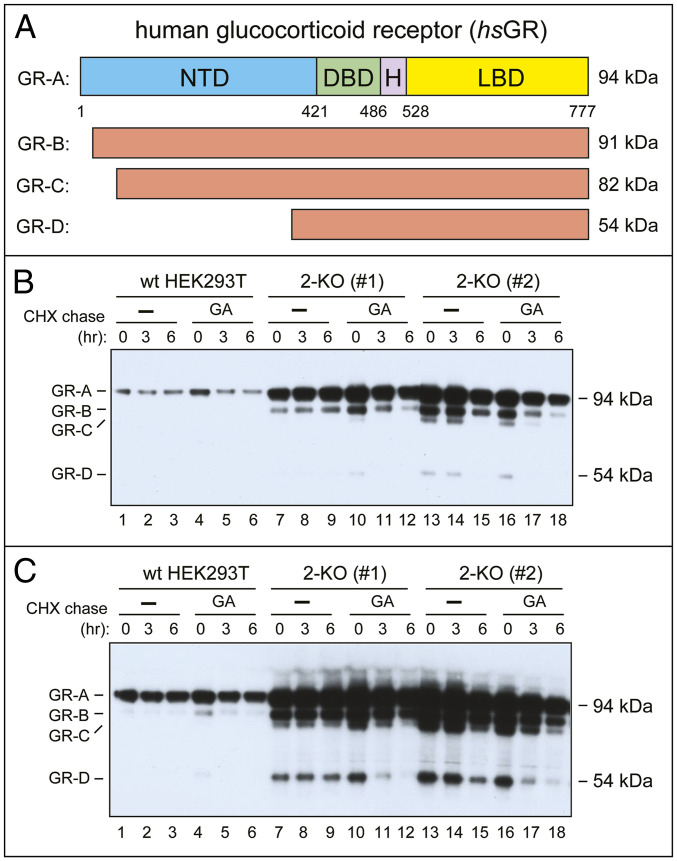
Fig. 5.
Up-regulation of the hsGR transcription factor in 2-KO HEK293T cells. (A) Main hsGR isoforms (detailed terminology of hsGR isoforms is more complex than the one shown here) (ref. 62 and refs. therein). (B) Lanes 1 through 3, IB-based CHX chase, for 0, 3, and 6 h, with wild-type HEK293T cells, using anti-hsGR antibody that recognizes hsGR isoforms shown in A. Lanes 4 through 6, same as lanes 1 through 3, but wild-type cells were incubated with GA at the beginning and during chase (SI Appendix, Materials and Methods). Lanes 7 through 9, same as lanes 1 through 3, but with 2-KO cell line #1. Lanes 10 through 12, same as lanes 4 through 6, but with 2-KO cell line #1. Lanes 13 through 18, same as lanes 7 through 12, but with 2-KO cell line #2. (C) Same as in B but a longer chemiluminescence exposure.
hsPREP1 Transcription Factor Binds to the N-Terminal Fragment of hsUBR1 but Not to Full-Length hsUBR1.
Split-Ub assays revealed the binding of hsUBR11−1059, the 123-kDa Nt-fragment of the 200 kDa hsUBR1 E3, to the full-length 48-kDa hsPREP1 (Fig. 4B, row 2). In contrast, hsPREP1 did not bind to either full-length hsUBR1 or its 77-kDa Ct fragment (hsUBR11060−1749) (Fig. 4B, rows 1 through 4). The binding of a physiological substrate of UBR1 to its Nt-fragment but not to full-length UBR1 has been encountered earlier in two different settings.
First, hsATF3, a bZIP TF, was found to interact, both in split-Ub assay and in another (conceptually different) binding assay, with the Nt-fragment of hsUBR1 but not with its full-length counterpart (42). Second, S. cerevisiae scCUP9 TF, a transcriptional repressor of a regulon that includes the scPTR2 peptide importer, was found to bind, unconditionally, to the analogous Nt-fragment of scUBR1, but would bind to full-length scUBR1 only in the presence of dipeptides bearing destabilizing (type-1/2) Nt-residues (2, 35, 36). As shown previously, Nt-residues of such dipeptides can interact with two cognate-binding sites of full-length scUBR1 (they are present in hsUBR1 as well; Fig. 1), thereby altering conformation of scUBR1 and enabling its binding to the scCUP9 repressor, followed by polyubiquitylation and degradation of scCUP9 (36). The resulting proteolysis-based scUBR1-scCUP9-scPTR2 circuit, regulated by short peptides, enables yeast cells to “sense” extracellular peptides and to accelerate their uptake (2, 35, 36). Despite physiologically plausible analogies between the conditional binding of scCUP9 to full-length scUBR1 and the current disposition with human hsATF3 and hsPREP1 (and with hsGR as well; see below), our binding assays did not suggest, so far, any “induction,” by short peptides, of interactions between full-length hsUBR1 and either hsATF3 or hsPREP1, which bind to the Nt-fragment of hsUBR1 but not to its full-length counterpart (Fig. 4B).
Isoform D of Glucocorticoid Receptor TF Binds to the N-Terminal Fragment of hsUBR1 but Not to Full-Length hsUBR1.
Human hsGR, encoded by hsNR3C1 and described above, exists in cells as a set of protein isoforms that include the major and largest (94 kDa) isoform hsGR-A, as well as smaller isoforms, including the 54 kDa hsGR-D (Fig. 5A). hsGR-A comprises the Nt domain, DNA-binding domain, hinge region, and the ligand-binding (glucocorticoid binding) domain (LBD) (Fig. 5A) (62).
Split-Ub assays revealed the binding of the Nt-fragment of hsUBR1 (hsUBR11−1059) to the smallest, 54-kDa isoform hsGR-D (Fig. 4C, row 2, and Fig. 5A). However, similarly to hsATF3 and hsPREP1 TFs, the hsGR-D did not bind to full-length hsUBR1 (Fig. 4C, rows 1 through 4). In addition and remarkably, the largest isoform, hsGR-A, despite encompassing the entire hsGR-D isoform (Fig. 5A), did not bind to either full-length hsUBR1 or its fragments (Fig. 4C, rows 5 through 8; compare with rows 1 through 4). Thus, the interaction, under the conditions of split-Ub assays, between hsGR and hsUBR1 was “restricted” for both ligands, in that the binding occurred solely between the Nt-fragment of hsUBR1 and the hsGR-D isoform, but was not observed (for any form of hsUBR1) with the largest isoform, hsGR-A (Figs. 4C and 5A).
Up-Regulation of Glucocorticoid Receptor in 2-KO Cells.
Cycloheximide (CHX)-based chase-degradation assays with untagged, endogenous hsGR used IB assays, anti-hsGR antibody, and wild-type HEK293T cells vs. 2-KO cells (Fig. 5). hsGR-A, the largest hsGR isoform, was largely stable during 6-h CHX chase, but became unstable in the presence of geldanamycin (GA), an inhibitor of the HSP90 chaperone (Fig. 5B, lanes 1 through 6). The latter finding was in agreement with hsGR being a client of HSP90, which assists the folding of hsGR and partially protects it from degradation (62). Remarkably, the steady-state levels of endogenous hsGR-A were at least 5-fold higher in both 2-KO cell lines, #1 and #2, than in wild-type cells (Fig. 5B, lanes 7 through 18; compare with lanes 1 through 6).
In addition, a higher-sensitivity detection (a longer chemiluminescence exposure) revealed, in 2-KO cell lines, a greatly increased level of hsGR-D, the smallest hsGR isoform (Fig. 5C), the one that has been found, above, to interact with the Nt-half of hsUBR1 (Fig. 4C). While undetectable in wild-type HEK293T cells even at the highest sensitivity of IB assays (Fig. 5C, lanes 1 through 6), the hsGR-D isoform was readily detectable and stable during a 6-h chase in the 2-KO cell line #1 (Fig. 5C, lanes 7 through 9), while being also up-regulated but less stable in the 2-KO cell line #2 (Fig. 5C, lanes 13 through 15).
Interestingly, the hsGR-D isoform became short-lived, in both 2-KO cell lines, in the presence of GA, despite the absence in these cells of both the hsUBR1 and hsUBR2 Ub ligases (Fig. 5C, lanes 10 through 12 and 16 through 18). Thus, the bulk of the Arg/N-degron pathway (its hsUBR1/hsUBR2 part) is not required for the degradation of either hsGR-A or hsGR-D isoforms (the latter isoform binds to the Nt-fragment of hsUBR1) that lost their protection by the HSP90 chaperone, in agreement with the known targeting of many specific TFs by more than one proteolytic pathway (63).
A parsimonious but not the only possible interpretation of these results is that the strong increase of both hsGR-A and hsGR-D isoforms (and other, “intermediate” hsGR isoforms as well) in 2-KO HEK293T cells (Fig. 5C) stems, at least in part, from a metabolic stabilization of these isoforms in the absence of hsUBR1/hsUBR2. The apparent stabilization of the hsGR-D isoform in this genetic background (Fig. 5C, lanes 7 through 9) suggests that the relevant (hsUBR1/hsUBR2 targeted) degron of hsGR resides in its LBD, which occupies the bulk of hsGR-D isoform (Fig. 5A).
In this interpretation, the largest (hsGR-A) isoform, the one that did not bind to any form of hsUBR1, in contrast to the hsGR-D isoform (Fig. 4C), is nevertheless recognized and destroyed by the Arg/N-degron pathway in vivo, in wild-type cells, similarly to hsGR-D. If so, a specific reason for the reproducible absence of binding of hsGR-A to any form of hsUBR1 in split-Ub assays, in contrast to the binding of hsGR-D to the Nt-half of hsUBR1 (Fig. 4C), remains to be understood. One possibility, to be addressed in future studies of hsGR vs. the Arg/N-degron pathway, is that this pathway mediates an early degradation of newly formed hsGR molecules, before their conformational maturation and/or interaction with other proteins, including hsGR itself. Yet another unknown in this setting, to be addressed by future experiments, is the possibility that a strong up-regulation of all isoforms of hsGR in 2-KO cells (Fig. 5C) may be caused, in part, by effects of the hsUBR1/hsUBR2 ablation on, for example, the efficacy of translation of hsGR mRNAs. In sum, while it is highly likely that at least hsGR-D (and possibly all isoforms of hsGR) are targeted by the Arg/N-degron pathway, the details of this circuit and whether the role of hsUBR1/hsUBR2 is solely degradative or has nonproteolytic aspects as well, remain to be understood.
Concluding Remarks.
It is still an unproven assumption that all functions of the Arg/N-degron pathway (Fig. 1) involve, in the end, the destruction of a targeted protein substrate, as distinguished, for example, from a nondegradative modification of substrate through its ubiquitylation by this pathway (2). On that assumption, the remarkably large alterations in the levels of some mRNAs and proteins that result from genetic ablation of the hsUBR1/hsUBR2 E3 N-recognins in human HEK293T cells (Figs. 2 and 3 and SI Appendix, Figs. S2–S13) are likely to be caused, at least in part, by metabolic stabilization (and therefore up-regulation) of specific TFs (repressors and/or activators) in 2-KO mutant cells that lack hsUBR1/hsUBR2. One such “promoter-based” model, for the strongly up-regulated hsADRB2 mRNA and its encoded protein, is described in this paper (Fig. 2 B–D and SI Appendix, Fig. S8). Analogous and verifiable models can also be developed, using the same logic, for other hsUBR1/hsUBR2-impacted genes described in the present study.
Our earlier work identified hsATF3, a stress-inducible TF that regulates hundreds of genes, as a short-lived substrate of the Arg/N-degron pathway in ref. 42. In the present study, we identified another major TF, the glucocorticoid receptor (hsGR), as a second TF that interacts with hsUBR1 in split-Ub assays and is strongly influenced, in ways described above, by the ablation of hsUBR1/hsUBR2 (Figs. 4C and 5). A TF called hsPREP1was also found to bind to hsUBR1 of the Arg/N-degron pathway (Fig. 4B), suggesting that hsPREP1 is yet another human TF that is impacted, through degradation and/or otherwise, by this proteolytic system.
All three TFs, while binding to the Nt-half of the 200-kDa hsUBR1 E3, did not interact in split-Ub assays with full-length hsUBR1 (Fig. 4). This binding pattern was first encountered in studies of S. cerevisiae scCUP9, a transcriptional repressor, which can bind unconditionally to the Nt-half of scUBR1 but would bind to full-length scUBR1 only in the presence of short peptides that bear destabilizing Nt residues and thereby can convert scUBR1, upon its binding to these peptides, into a conformer that can interact with scCUP9. As described in more detail above, these properties of scCUP9-scUBR1 interactions underlie the ability of the Arg/N-degron pathway to control peptide transport in yeast, through the regulated (by short peptides) degradation of the scCUP9 repressor (2, 35, 36). It remains to be determined whether a human TF, such as, for example, hsPREP1 (it is sequelogous to scCUP9) may regulate peptide transport in mammals by being a conditionally short-lived substrate of the Arg/N-degron pathway.
Given that three TFs of different structures were found to bind to the Nt-half of hsUBR1 in split-Ub assays (hsATF3 is a bZIP TF, hsGR is a zinc-finger TF, and hsPREP1 is a homeodomain TF) (Fig. 4) (42), it would be illuminating to identify all or most human TFs (∼1,600 distinct TFs total) (63) that are bona fide in vivo substrates of the Arg/N-degron pathway. Specific degrons (binding sites) that hsUBR1 recognizes in the three TFs remain to be determined as well. Yet another vista opened up by the present study (and discussed above) is the function of the Arg/N-degron pathway as a regulator of translation of specific mRNAs, such as those that encode the NF-L and NF-M subunits of neurofilaments (Fig. 3 and SI Appendix, Fig. S13A).
A large number of mRNAs and proteins that were found to be either significantly or very strongly up-regulated or down-regulated in 2-KO human cells that lacked hsUBR1/hsUBR2 has greatly exceeded our ability to follow up and explore these findings in the present study. Consequently, these detailed results (Figs. 2–5 and SI Appendix, Figs. S2–S13) are likely to serve for a long time as sources of mRNA and protein leads for analyzing circuits that involve the Arg/N-degron pathway. This multifunctional proteolytic system has emerged as a regulator of specific mammalian genes, in part through conditional targeting of TFs that include ATF3, GR, and PREP1.
Materials and Methods
For further information, see SI Appendix, Materials and Methods.
RNA-Seq Analyses.
RNA-seq was carried out with RNA preparations from wild-type vs. 2-KO (UBR1−/− UBR2−/−) HEK293T cell lines using methods described in SI Appendix, Materials and Methods.
Quantitative Mass Spectrometric (TMT-SPS-MS3) Protein Analyses.
MS-based analyses of extracts from wild-type and 2-KO HEK293T cells employed the TMT-SPS-MS3 technique, described in SI Appendix, Materials and Methods.
Cell Viability and Proliferation Assay.
Cell viability and proliferation were assayed using the Trypan Blue exclusion test, as described in SI Appendix, Materials and Methods.
Cell Toxicity Assay with Dimethyloxalylglycine.
This assay, which uses DMOG to probe relative in vivo levels of the hsMCT2 transporter, is described in SI Appendix, Materials and Methods.
Immunoblotting and Chase-Degradation Assays.
IB analyses and chase-degradation assays were carried out largely as described previously (23, 26) and in SI Appendix, Materials and Methods.
Split-Ubiquitin Assay.
A version of split-Ub binding assay (Fig. 4A) (43) was carried out in S. cerevisiae as described previously (26, 42, 60) and in SI Appendix, Materials and Methods. Standard techniques were used for construction of S. cerevisiae strains and transformation by DNA.
Supplementary Material
Acknowledgments
We are grateful to I. Antoshechkin for RNA-seq analyses. T.T.M.V. and A.V. thank current and former members of the A.V. laboratory for their advice and assistance. D.C.M. and S.P.G. thank J. Paulo for assistance with mass spectrometry experiments. This work was supported by NIH grants 1R01DK039520 and 1R01GM031530 (A.V.) and R01GM067945 (S.P.G.).
Footnotes
The authors declare no competing interest.
†“Sequelog” denotes a sequence that is similar, to a specified extent, to another sequence (39). Derivatives of sequelog include sequelogy (sequence similarity) and sequelogous (similar in sequence). The usefulness of sequelog and derivative notations stems from the rigor and clarity of their evolutionary neutrality. By contrast, in settings that use “homolog,” “ortholog,” and “paralog” (they denote, respectively, common descent and functional similarity/dissimilarity), these terms are often interpretation laden and imprecise. Homolog, ortholog, and paralog are compatible with the sequelog terminology. The former terms can be used to convey understanding about common descent and biological functions if this additional information, distinct from sequelogy per se, is actually present (39).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020124117/-/DCSupplemental.
Data Availability.
All relevant data are included in the article and supporting information.
References
- 1.Hershko A., Ciechanover A., Varshavsky A., The ubiquitin system. Nat. Med. 6, 1073–1081 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A., N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U.S.A. 116, 358–366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finley D., Ulrich H. D., Sommer T., Kaiser P., The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vittal V., Stewart M. D., Brzovic P. S., Klevit R. E., Regulating the regulators: Recent revelations in the control of E3 ubiquitin ligases. J. Biol. Chem. 290, 21244–21251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pohl C., Dikic I., Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 366, 818–822 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Balchin D., Hayer-Hartl M., Hartl F. U., In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Zheng N., Shabek N., Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Schweitzer A., et al. , Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc. Natl. Acad. Sci. U.S.A. 113, 7816–7821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bard J. A. M., et al. , Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finley D., Prado M. A., The proteasome and its network: Engineering for adaptability. Cold Spring Harb. Perspect. Biol. 12, a033985 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachmair A., Finley D., Varshavsky A., In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986). [DOI] [PubMed] [Google Scholar]
- 12.Timms R. T., Koren I., Tying up loose ends: The N-degron and C-degron pathways of protein degradation. Biochem. Soc. Trans. 48, 1557–1567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu R.-G., et al. , The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Lee M. J., et al. , RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 15030–15035 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varshavsky A., The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasaki T., Sriram S. M., Park K. S., Kwon Y. T., The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ree R., Varland S., Arnesen T., Spotlight on protein N-terminal acetylation. Exp. Mol. Med. 50, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holdsworth M. J., Vicente J., Sharma G., Abbas M., Zubrycka A., The plant N-degron pathways of ubiquitin-mediated proteolysis. J. Integr. Plant Biol. 62, 70–89 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Hwang C. S., Shemorry A., Varshavsky A., N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shemorry A., Hwang C. S., Varshavsky A., Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S. J., Wu X., Wadas B., Oh J.-H., Varshavsky A., An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 355, 366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao S., et al. , Interconversion between anticipatory and active GID E3 ubiquitin ligase conformations via metabolically driven substrate receptor assembly. Mol. Cell 77, 150–163.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Piatkov K. I., Brower C. S., Varshavsky A., The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc. Natl. Acad. Sci. U.S.A. 109, E1839–E1847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brower C. S., Piatkov K. I., Varshavsky A., Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol. Cell 50, 161–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shearer R. F., Iconomou M., Watts C. K., Saunders D. N., Functional roles of the E3 ubiquitin ligase UBR5 in cancer. Mol. Cancer Res. 13, 1523–1532 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Oh J. H., Hyun J. Y., Varshavsky A., Control of Hsp90 chaperone and its clients by N-terminal acetylation and the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 114, E4370–E4379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo Y. D., et al. , N-terminal arginylation generates a bimodal degron that modulates autophagic proteolysis. Proc. Natl. Acad. Sci. U.S.A. 115, E2716–E2724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masson N., et al. , Conserved N-terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants. Science 365, 65–69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao X., Yeom J., Groisman E. A., The expanded specificity and physiological role of a widespread N-degron recognin. Proc. Natl. Acad. Sci. U.S.A. 116, 18629–18637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timms R. T., et al. , A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science 365, eaaw4912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J. S., et al. , Structural analyses on the deamidation of N-terminal Asn in the human N-degron pathway. Biomolecules 10, 163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim M. K., Oh S. J., Lee B. G., Song H. K., Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 113, 12438–12443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dougan D. A., Varshavsky A., Understanding the Pro/N-end rule pathway. Nat. Chem. Biol. 14, 415–416 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Oh J. H., Hyun J. Y., Chen S. J., Varshavsky A., Five enzymes of the Arg/N-degron pathway form a targeting complex: The concept of superchanneling. Proc. Natl. Acad. Sci. U.S.A. 117, 10778–10788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner G. C., Du F., Varshavsky A., Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405, 579–583 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Du F., Navarro-Garcia F., Xia Z., Tasaki T., Varshavsky A., Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. U.S.A. 99, 14110–14115 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbs D. J., Holdsworth M. J., Every breath you take: New insights into plant and animal oxygen sensing. Cell 180, 22–24 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Chui A. J., et al. , N-terminal degradation activates the NLRP1B inflammasome. Science 364, 82–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varshavsky A., ‘Spalog’ and ‘sequelog’: Neutral terms for spatial and sequence similarity. Curr. Biol. 14, R181–R183 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Zenker M., et al. , Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat. Genet. 37, 1345–1350 (2005). [DOI] [PubMed] [Google Scholar]
- 41.An J. Y., et al. , Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 6212–6217 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vu T. T. M., Varshavsky A., The ATF3 transcription factor is a short-lived substrate of the Arg/N-degron pathway. Biochemistry 59, 2796–2812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnsson N., Varshavsky A., Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U.S.A. 91, 10340–10344 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Berge K., et al. , RNA sequencing data: Hitchhiker’s guide to expression analysis. Annu. Rev. Biomed. Data Sci. 2, 139–173 (2019). [Google Scholar]
- 45.Navarrete-Perea J., Yu Q., Gygi S. P., Paulo J. A., Streamlined tandem mass tag (SL-TMT) protocol: An efficient strategy for quantitative (phospho)proteome profiling using tandem mass tag-synchronous precursor selection-MS3. J. Proteome Res. 17, 2226–2236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daaka Y., Luttrell L. M., Lefkowitz R. J., Switching of the coupling of the beta2-adrenergic receptor to different G proteins by protein kinase A. Nature 390, 88–91 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Jaeger A., Fritschka S., Ponsuksili S., Wimmers K., Muráni E., Identification and functional characterization of cis-regulatory elements controlling expression of the porcine ADRB2 gene. Int. J. Biol. Sci. 11, 1006–1015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahouth S. W., Beauchamp M. J., Vu K. N., Reciprocal regulation of beta(1)-adrenergic receptor gene transcription by Sp1 and early growth response gene 1: Induction of EGR-1 inhibits the expression of the beta(1)-adrenergic receptor gene. Mol. Pharmacol. 61, 379–390 (2002). [DOI] [PubMed] [Google Scholar]
- 49.O’Connor L., Gilmour J., Bonifer C., The Role of the ubiquitously expressed transcription factor Sp1 in tissue-specific transcriptional regulation and in disease. Yale J. Biol. Med. 89, 513–525 (2016). [PMC free article] [PubMed] [Google Scholar]
- 50.Halestrap A. P., The SLC16 gene family–Structure, role and regulation in health and disease. Mol. Aspects Med. 34, 337–349 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Fets L., et al. , MCT2 mediates concentration-dependent inhibition of glutamine metabolism by MOG. Nat. Chem. Biol. 14, 1032–1042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schödel J., Ratcliffe P. J., Mechanisms of hypoxia signalling: New implications for nephrology. Nat. Rev. Nephrol. 15, 641–659 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Kaelin W. G., Jr, The VHL tumor suppressor gene: Insights into oxygen sensing and cancer. Trans. Am. Clin. Climatol. Assoc. 128, 298–307 (2017). [PMC free article] [PubMed] [Google Scholar]
- 54.Jayewickreme C. D., Shivdasani R. A., Control of stomach smooth muscle development and intestinal rotation by transcription factor BARX1. Dev. Biol. 405, 21–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee A. K., Potts P. R., A comprehensive guide to the MAGE family of ubiquitin ligases. J. Mol. Biol. 429, 1114–1142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Popov V. M., et al. , The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol. Metab. 21, 41–49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szaro B. G., Strong M. J., Post-transcriptional control of neurofilaments: New roles in development, regeneration and neurodegenerative disease. Trends Neurosci. 33, 27–37 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Yuan A., Rao M. V., Nixon Veeranna, R. A., Neurofilaments and neurofilament proteins in health and disease. Cold Spring Harb. Perspect. Biol. 9, a018309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw G., Morse S., Ararat M., Graham F. L., Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16, 869–871 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Gisler S. M., et al. , Monitoring protein-protein interactions between the mammalian integral membrane transporters and PDZ-interacting partners using a modified split-ubiquitin membrane yeast two-hybrid system. Mol. Cell. Proteomics 7, 1362–1377 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blasi F., Bruckmann C., Penkov D., Dardaei L., A tale of TALE, PREP1, PBX1, and MEIS1: Interconnections and competition in cancer. BioEssays 39, 1600245 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Weikum E. R., Knuesel M. T., Ortlund E. A., Yamamoto K. R., Glucocorticoid receptor control of transcription: Precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 18, 159–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambert S. A., et al. , The human transcription factors. Cell 172, 650–665 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the article and supporting information.