Significance
Infection with Zika virus can cause severe consequences in a subset of the patient population. When infection occurs during pregnancy, congenital abnormalities can occur. Additionally, the onset of Guillain–Barré syndrome, encephalitis, and myelitis are associated with Zika virus infection. We have identified several small molecules with favorable clinical profiles, including the five-lipoxygenase–activating protein inhibitor, MK-591, and the tetracycline antibiotic, methacycline, as inhibitors of Zika virus infection, the latter of which reduced neurological deficits in a Zika virus mouse model. These compounds have the potential to be used as prophylactics or for the treatment of the neurological complications of Zika virus infection.
Keywords: serine protease, Zika virus, flavivirus, high-throughput screening, encephalitis
Abstract
When Zika virus emerged as a public health emergency there were no drugs or vaccines approved for its prevention or treatment. We used a high-throughput screen for Zika virus protease inhibitors to identify several inhibitors of Zika virus infection. We expressed the NS2B-NS3 Zika virus protease and conducted a biochemical screen for small-molecule inhibitors. A quantitative structure–activity relationship model was employed to virtually screen ∼138,000 compounds, which increased the identification of active compounds, while decreasing screening time and resources. Candidate inhibitors were validated in several viral infection assays. Small molecules with favorable clinical profiles, especially the five-lipoxygenase–activating protein inhibitor, MK-591, inhibited the Zika virus protease and infection in neural stem cells. Members of the tetracycline family of antibiotics were more potent inhibitors of Zika virus infection than the protease, suggesting they may have multiple mechanisms of action. The most potent tetracycline, methacycline, reduced the amount of Zika virus present in the brain and the severity of Zika virus-induced motor deficits in an immunocompetent mouse model. As Food and Drug Administration-approved drugs, the tetracyclines could be quickly translated to the clinic. The compounds identified through our screening paradigm have the potential to be used as prophylactics for patients traveling to endemic regions or for the treatment of the neurological complications of Zika virus infection.
Zika virus is a positive-sense, single-stranded RNA virus in the Flavivirus genus. Other viruses in this genus include West Nile virus, dengue virus, yellow fever virus, and tick-borne encephalitis (1). Most people who contract Zika virus are asymptomatic; however, in a subset of patients, infection with Zika virus can have catastrophic consequences. When infection occurs during pregnancy, the virus may cross the placental barrier and infect the fetus, resulting in congenital abnormalities, most notably microcephaly (2). Additionally, an increased risk of Guillain–Barré syndrome is associated with Zika virus infection (3). Some patients develop an encephalitis or myelitis (3). Although significant advances in the field have occurred since the South American outbreak of 2015/2016 (4), no drugs or vaccines are approved for the treatment of, or protection from, Zika virus. Additionally, no small molecules have advanced to clinical trials. The global at risk population continues to expand; the first local transmission of Zika virus in Europe was reported in October of 2019 (5). Due to the potential for serious outcomes, optimized treatment strategies and prophylactic measures are needed.
Flaviviruses require proteolytic processing of polyprotein precursors to yield a functional viral particle. These cleavages are catalyzed by both host and virally encoded proteases (6). Inhibition of the virus-encoded protease is a strategy for drug development that has proven effective for other viruses. Several small-molecule inhibitors of the Zika virus protease were discovered over the past 3 y (7); however, a combination of factors make the pursuit of additional, specific inhibitors of the Zika virus protease a necessary undertaking. A split-luciferase complementation-based assay identified erythrosin B as an allosteric inhibitor of the Zika virus protease. Erythrosin B inhibited Zika virus replication in human placental and neural progenitor cells (8); however, it is a red food coloring dye (FD&C Red No. 3), and at concentrations higher than the acceptable daily intake (0.1 mg/kg per day) can prevent dopamine transport (9). Separately, an in silico screen predicted that novobiocin, an off-market antibacterial agent, would be a competitive inhibitor of the Zika virus protease. Novobiocin inhibited Zika virus infection in Vero cells, Huh-7 cells, and immunosuppressed mice (10); however, several properties of novobiocin make it unfavorable for the treatment of Zika virus. Novobiocin treatment can result in gastrointestinal discomfort, various forms of skin rashes, and leucopenia, a rare but potentially serious complication (11). Furthermore, serum concentrations are variable following oral administration (11), its penetration into the cerebrospinal fluid is poor (12), and treatment of infants with novobiocin caused inhibition of bilirubin metabolism, resulting in jaundice (13).
Flavivirus polyprotein processing occurs in the endoplasmic reticulum by the two-component protease, which consists of the small hydrophobic integral membrane protein, NS2B, and the cytosolic NS3 protein. The importance of the NS2B domain in the formation of an active protease complex was first shown for yellow fever virus and dengue virus (14), and subsequently for Zika virus (15). Using fluorogenic or chromogenic peptide substrates, in vitro assays were established by connecting the NS2B hydrophilic cofactor peptide and NS3 protease domain by a noncleavable, flexible polypeptide linker. Alternatively, the NS2B and NS3 proteins can be expressed on different promoters and allowed to complex in solution. Whether one construct should be used in preference to the other for drug discovery is an open question. Increased activity of the unlinked construct has been observed (15), however, one inhibitor exhibited equivalent IC50 values (16).
We report the use of both the linked and unlinked protease constructs in a high-throughput drug screening effort. An artificial intelligence (AI)-driven virtual screen was utilized to increase the inhibitor hit rate. Compounds were then validated in several orthogonal cell-based infection assays. We identified two compounds as potent in vitro Zika virus inhibitors, MK-591 and JNJ-40418677. A third compound, the tetracycline antibiotic methacycline, reduced the amount of Zika virus present in the brain and the severity of Zika virus-induced motor deficits in an immunocompetent Zika virus mouse model.
Results
Pilot Screen of the Spectrum Collection.
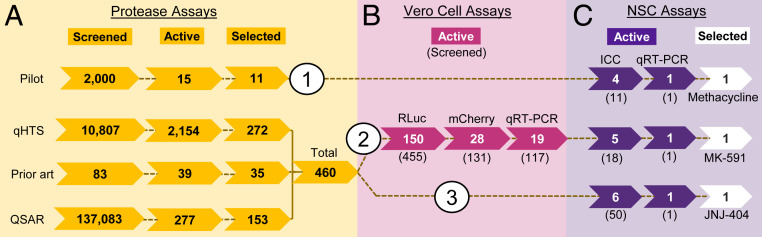
A small-molecule library, consisting of 2,000 bioactive compounds (final concentration of 10 µM), was screened for inhibitory activity against the linked, NS2B-NS3 Zika virus protease (384-well; Ac-VKTGKR-AMC [Ac: acetyl; AMC: 7-amino-4-methylcoumarin]), (see SI Appendix, Fig. S1 for assay details and substrate design) (17). Three classes of compounds were identified: The flavonoids, anthraquinones, and tetracyclines (SI Appendix, Fig. S2). Due to their extensive clinical use, oral availability (18), and ability to cross the placental barrier (19), we chose to focus our initial validation efforts on the family of tetracyclines and tested 11 commercially available tetracyclines in a human neural stem cell (NSC) assay (Fig. 1, Pilot, strategy 1). Simultaneously, we designed a 1,536-well quantitative high-throughput compound screen (qHTS) for protease inhibitors (Fig. 1A, qHTS, Prior art, and QSAR [quantitative structure-activity relationship]). Hits from the qHTS were first tested in a series of Vero cell infection assays (Fig. 1B); those that were active were then tested in the NSC assay (Fig. 1C, strategy 2). Subsequently, additional inhibitors were selected from the qHTS and tested in the NSC assay (Fig. 1, strategy 3).
Fig. 1.
Identification of Zika virus inhibitor workflow. Three screening strategies were used to identify Zika virus inhibitors. Strategy 1: Hits from the Pilot protease screen were directly tested in NSCs. Strategy 2: Hits from the qHTS, prior art library, and virtual screen using QSAR screens were tested in Vero cells, then those compounds that were active were tested in NSCs. Strategy 3: Hits from the qHTS, prior art library screen, and QSAR were directly tested in NSCs. (A) Inhibition of the Zika virus NS2B-NS3 protease. The number of compounds tested in the primary screen is indicated, followed by the number of active compounds in each assay, and finally the number of compounds that were selected from these assays (based on potency and confirmation). (B) Inhibition of Zika virus in Vero cells using Zika virus-RLuc, Zika virus-mCherry, and qRT-PCR. The number of active compounds (and the number of compounds tested) are indicated. (C) Inhibition of Zika virus in NSCs. The number of compounds that confirmed in each NSC infection assay and names of our top candidate inhibitors identified through each strategy are indicated. Of the 11 tetracyclines tested, 4 inhibited Zika virus without toxicity (ICC: immunocytochemistry); 1 was chosen to confirm by qRT-PCR. Of the 18 Vero cell assay inhibitors, 5 inhibited Zika virus without toxicity; 1 was chosen to confirm by qRT-PCR. Of the 50 additional biochemical inhibitors selected, 6 inhibited Zika virus without toxicity; 1 was chosen to confirm by qRT-PCR.
qHTS for Small-Molecule Protease Inhibitors.
qHTS generates concentration-response curves and IC50 values at the primary screening stage, enabling the prioritization of active compounds (20). To optimize the biochemical assay for qHTS, we miniaturized the 384-well assay to a 1,536-well format (20). We red-shifted the fluorophore on the substrate from AMC (Bz-Nle-KRR-AMC [Bz: benzoyl], excitation/emission [Ex/Em]: 340/450 nm) to rhodamine110 [(Bz-Nle-KRR)2-Rd, Ex/Em: 480/540 nm] (21) (SI Appendix, Fig. S1). This reduced quenching of the fluorophore by compounds in the screening libraries, many of which absorb in the UV range (22). Next, the linked and unlinked protease constructs were tested against a chemically diverse library of 4,922 compounds. Possible differences in potency and efficacy were initially observed in 79 compounds (SI Appendix, Fig. S3A); however, during confirmation testing, we observed no significant difference in the activity of these inhibitors (SI Appendix). We chose the linked preparation for large-scale screening due to its higher catalytic activity, enabling the use of a lower enzyme concentration.
A collection of 10,807 bioactive, annotated, investigational, or approved compounds (Fig. 1A, qHTS) were screened against the linked protease in qHTS format [Bz-Nle-KRR)2-Rd]. A large number of compounds exhibited inhibition (2,154) (SI Appendix, Fig. S4 and Dataset S1); therefore, we applied filters to eliminate electrophiles and other problematic compounds. We retested 378 candidate inhibitors and confirmed that 272 (72%) were active compounds (Dataset S1; see SI Appendix for triage details). Additionally, we tested a custom collection of 83 protease inhibitors previously reported in the literature, 35 of which were active in our assay (Fig. 1A, Prior art, and Dataset S1).
AI-Driven QSAR Model.
In parallel to the qHTS screening effort, we employed an AI-based QSAR model to reduce the number of library compounds subjected to protease qHTS (Fig. 1A, QSAR). We used qHTS results from a subset of the above screening collection as a training set for our QSAR model. Quantitative neighborhoods of atoms (QNA) descriptors, together with deep-learning techniques (see SI Appendix for model performance), were used for the model building (23). The AI-based QSAR model was used to virtually screen the remaining 137,083 compounds in the National Center for Advancing Translational Sciences (NCATS) small-molecule library; 277 predicted inhibitors (Dataset S2) were selected for experimental verification. These predicted inhibitors were structurally distinct from the training set compounds (Materials and Methods). An additional 43 compounds from the original training set were included as positive and negative controls, totaling 320 compounds. Samples were tested for inhibition of the linked protease [(Bz-Nle-KRR)2-Rd] and 186 compounds exhibited activity, with IC50 values ranging from 0.5 to 89 µM (Datasets S2 and S3). Of the 186 active compounds, 153 were from the AI-based QSAR model (hit rate of 55%) and 33 from the training set controls. Among the 153 active QSAR compounds, 45 were active at <10 µM (hit rate of 29%). By comparison, the traditional approach of testing every library compound (10,807 compounds) yielded 442 active hits at <10 µM (hit rate of 4%), a greater than sevenfold improvement.
Vero Cell Assays.
To assess the 460 confirmed biochemical inhibitors in a cellular context, we chose the African green monkey kidney cell line, Vero, as it supports optimal virus proliferation (Fig. 1B). In an effort to identify inhibitors that are effective against several strains of Zika virus, we used two different Zika virus lineages in the three Vero cell assays: Two recombinant viruses generated using an isolate from the 2010 Cambodian outbreak and the original African strain (MR766, ATCC VR-84). We triaged compounds for inhibition of viral replication using a Renilla luciferase (Rluc, Cambodian strain) reporter system (72-h incubation, 1,536-well) (24) (Dataset S4), an mCherry reporter system (Cambodian strain, 66- or 72-h incubation, 384-well) (Dataset S5), and a qRT-PCR assay (MR766/African strain, 24-h incubation, 384-well) (Dataset S6; see SI Appendix for Vero cell assay performance). After applying hit criteria, 19 compounds (including three from prior art: Niclosamide, temoporfin, and selamectin) were identified with favorable screening profiles (Dataset S6).
NSC Assays.
Zika virus infects and disrupts development in NSCs (25–27) and fetal infection can lead to congenital Zika syndrome and microcephaly (2). The increased risk of microcephaly was first quantified in a retrospective study of the 2013 French Polynesian outbreak (28). Therefore, compounds were tested for their ability to inhibit Zika virus in human NSCs using an isolate from French Polynesia (48- or 72-h incubation, 24-well). We tested the 11 tetracyclines from the Pilot screen (Fig. 1C, strategy 1) and 18 of the 19 compounds that exhibited activity in all three Vero cells assays (including the three prior art compounds) (Fig. 1C, strategy 2). Based on the results of the biochemical and cellular HTS assays, and an earlier observation that some tetracyclines had significantly reduced activity in Vero cells versus NSCs, an additional 50 compounds were chosen for testing (Fig. 1C, strategy 3). Immunocytochemistry was used to determine the percentage of cells infected with Zika virus. An anti-Flavivirus envelope antibody (4G2) was used to quantify the rate of Zika virus infection, which was normalized to the total cell count using Hoechst nuclear stain. Percent inhibition was calculated relative to DMSO-treated, Zika virus-infected cells (Dataset S7).
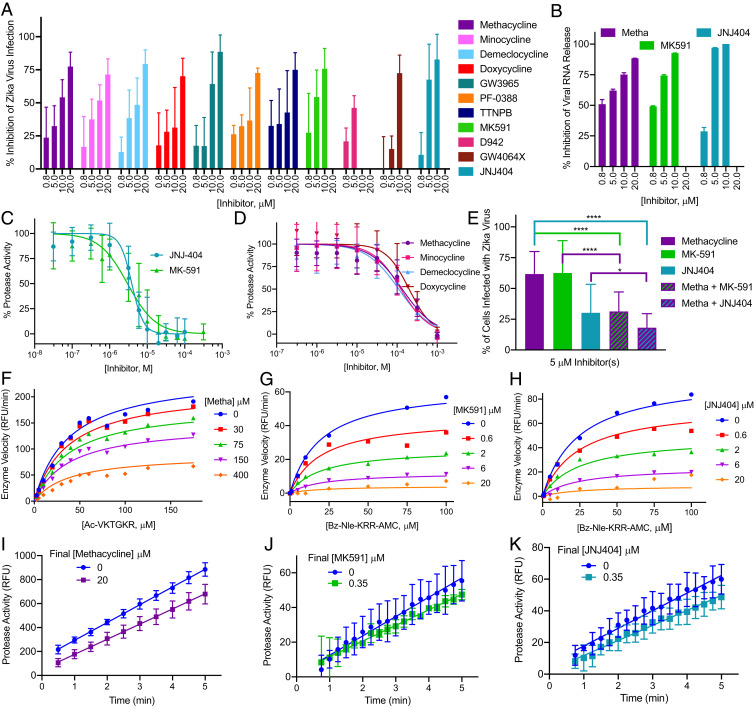
Of the 79 total compounds tested, 40 displayed some activity (defined as any reduction in Zika virus immunostaining, without toxicity, at a given dose), 32 of which inhibited Zika virus at two or more concentrations. All 11 tetracyclines inhibited Zika virus infection; however, sancycline, meclocycline, tigecycline, and demeclocycline displayed some cytotoxicity (SI Appendix, Fig. S5 and Dataset S7). Methacycline and minocycline were not toxic at the concentrations tested, had complete dose–response curves, and displayed IC50’s of ∼8 µM (Figs. 2A and 3). Seven nontetracycline compounds (D-942, PF-03882845, GSK-3965, MK-591, TTNPB, JNJ-40418677, GW-4064X) exhibited dose-dependent inhibition (Figs. 2A and 3). GSK-3965, PF-03882845, and TTNPB were not toxic at the concentration range tested and displayed IC50 values of 8.4 µM, 10.8 µM, and 9.1 µM, respectively. While MK-591 and JNJ-40418677 exhibited toxicity at 20 µM, they were potent against the Zika virus with IC50 values of ∼3 µM (Figs. 2A and 3). Based on these results, we used qRT-PCR to test the ability of methacycline, MK-591, and JNJ-404 to reduce the amount of viral RNA released from infected NSCs (Fig. 1C). All three compounds displayed dose-dependent inhibition of Zika virus (Fig. 2B).
Fig. 2.
Inhibition of Zika virus infection and Zika virus protease. (A and B) Drugs were preincubated for 1 h with NSCs prior to Zika virus infection (French Polynesian strain). At 48- or 72-hpi, supernatant was collected, and cells were fixed with 4% PFA. Percent inhibition is relative to DMSO-treated, Zika virus-infected cells. (A) Immunostaining with anti-Flavivirus envelope 4G2 antibody was used to quantify the rate of infection, which was normalized to the total number of cells (Hoechst nuclear stain). Drug-induced toxicity was observed at 20 µM of MK591 and JNJ404 and 10 and 20 µM of D942. Data represent mean ± SD from three independent experiments. (B) qRT-PCR for Zika virus RNA was used to quantify the rate of infection in cell-free supernatant. (C and D) Inhibition of the linked Zika virus protease. Each experiment was performed in quadruplicates. Data represent mean ± SEM of three independent experiments. (C) MK-591 and JNJ-404 (30 nM to 100 µM) inhibit Zika virus protease; Bz-Nle-KRR-AMC (Bz: benzoyl; AMC: 7-amino-4-methylcoumarin) was used as a substrate. (D) Tetracyclines (300 nM to 1 mM) weakly inhibit Zika virus protease; Ac-VKTGKR-AMC was used as a substrate. (E) NSCs were treated with a combination of 5 µM of methacycline and either 5 µM of MK-591 or 5 µM of JNJ-404 followed by infection with Zika virus. Infection was monitored by immunostaining. Data represent mean ± SD from three independent experiments. Significance was calculated using an unpaired, two-tailed t test: ****P < 0.0001; *P < 0.05. (F–K) Kinetic experiments with the linked Zika virus protease (Ac-VKTGKR-AMC [Ac: acetyl] or Bz-Nle-KRR-AMC); values were normalized to no-enzyme and no-inhibitor controls. (F) Methacycline-inhibited, (G) MK-591-inhibited, and (H) JNJ-404-inhibited enzyme velocity could not be restored by additional substrate. The data were best fit to the noncompetitive inhibition model. Jump dilution analysis indicated (I) methacycline (velocity ∼85%), (J) MK-591 (velocity ∼81%), and (K) JNJ-404 (velocity ∼85%) are reversible inhibitors with a fast dissociation rate (velocity is closer to 90% of the uninhibited enzyme than 10%). Data represents mean ± SD of triplicate samples.
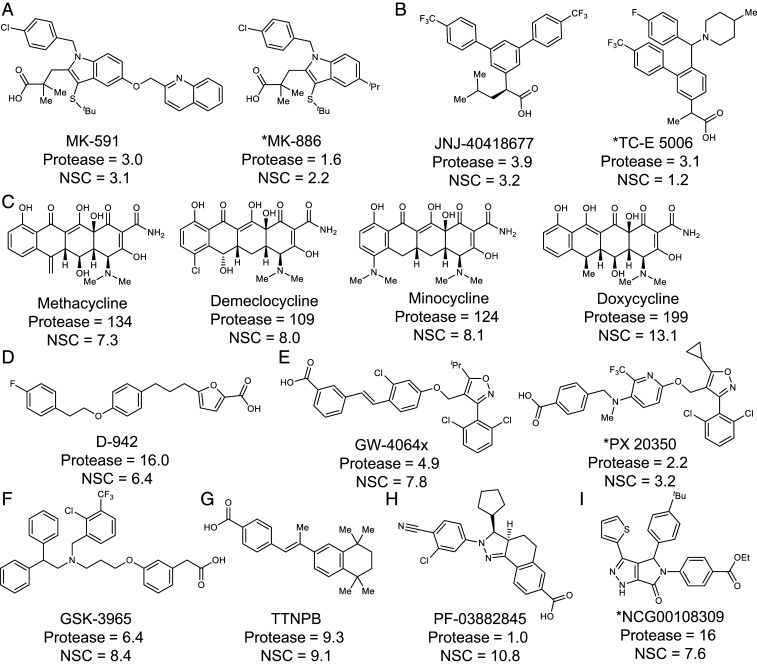
Fig. 3.
Chemical structures and IC50 (μM) values for selected hits in the linked Zika virus protease (Protease) and Zika virus infection assay in NSCs. (A) FLAP inhibitors, (B) γ-secretase modulators, (C) tetracyclines, (D) AMPK activator, (E) farnesoid X receptor agonists, (F) liver X receptor agonist, (G) retinoic acid analog, (H) mineralocorticoid receptor antagonist, (I) compound from QSAR screen. An asterisk (*) indicates IC50 values are a single data point from qHTS. All other values are an average of at least three independent experiments from the confirmation testing of commercial compounds. Protease and NSC IC50 values were calculated in GraphPad Prism using the equation: Log(inhibitor) vs. normalized response–variable slope.
Protease Inhibition by Methacycline, MK-591, and JNJ-404.
Commercial sources of methacycline, MK-591, and JNJ-404 were tested in the linked protease assay. MK-591 and JNJ-404 displayed similar inhibitory results to the 1,536-well screen (Figs. 2C and 3). In contrast to the HTS result, the tetracyclines exhibited reduced activity against the Zika virus protease (Figs. 2D and 3 and SI Appendix, Fig. S5). An impurity was identified in the methacycline obtained from Microsource (SI Appendix, Fig. S5G), which is the source of tetracyclines for multiple screening libraries, including the Spectrum library used in our Pilot screen. Our results suggest the presence of this fluorescent artifact resulted in an apparent increase in potency. Methacycline’s IC50 for viral inhibition in the NSCs (7.3 µM) is lower than its IC50 for protease inhibition (134 µM), suggesting the tetracyclines may exert their effects against Zika virus through multiple mechanisms. Therefore, we demonstrated that in our NSC assay, the combination of methacycline with either of the two potent protease inhibitors, MK-591 or JNJ-404, resulted in increased efficacy (Fig. 2E).
We next used kinetic biochemical experiments to determine the mechanism of enzyme inhibition by methacycline, MK-591, and JNJ-404. These initial experiments indicated that they are all allosteric, reversible inhibitors of the Zika virus protease (Fig. 2 F–K). Regardless of potency, increasing concentrations of substrate (Ac-VKTGKR-AMC or Bz-Nle-KRR-AMC) did not restore enzyme activity in the presence of methacycline, MK-591, or JNJ-404 (Fig. 2 F–H), suggestive of a noncompetitive inhibition model (29). Jump dilution analysis was performed to examine the reversibility of protease inhibition (30). One-hundred times the required concentration of Zika virus protease was incubated with each inhibitor at concentrations 10 times its IC50 (∼90% inhibition). After incubation, the reaction was diluted 100-fold in assay buffer (inhibitor concentration that would result in ∼10% inhibition). In the presence of an irreversible inhibitor, the initial velocity is expected to remain inhibited by ∼90% (no dissociation upon dilution). A reversible inhibitor is expected to dissociate, allowing enzymatic rates to return to close to vehicle-only controls (∼10% inhibition). All three compounds displayed an initial velocity close to that of the uninhibited enzyme (∼10% inhibition), indicating reversible inhibition (Fig. 2 I–K). The initial enzyme velocity in the presence of fixed concentrations of MK-591 and JNJ-404 at the original (∼90% inhibition) and diluted (∼10% inhibition) concentrations are shown for comparison in the SI Appendix, Fig. S5 J and K.
Zika Virus Infection in an Immunocompetent Mouse Model.
Studies of Zika virus infection were performed in a C57BL/6 mouse model. At postnatal day 1, a Zika virus isolate from the 2015 Brazilian outbreak was subcutaneously injected at 5 × 105 PFU/mL. At 9 d postinfection (dpi), Zika virus-infected mice developed motor deficits including tremors, stiffness of limbs, difficulty in staying upright, loss of balance, and flaccid paralysis of the limbs. Zika virus-infected mice were lower in body weight, developed seizures, and met the requirements for euthanasia between 13 and 16 dpi (SI Appendix, Fig. S6 and Movies S1–S3). Histological examination of brain samples showed gliosis and loss of cortical and Purkinje neurons. Expansion of blood vessels, vacuolation, inflammatory cell infiltration, and an accumulation of cell debris all contributed to damage in the brain parenchyma (SI Appendix, Fig. S7).
Methacycline was prioritized for in vivo studies because it is a Food and Drug Administration-approved drug that can be rapidly moved into the clinic, it is not toxic and inhibits Zika virus in NSCs, and tetracyclines are known to cross the placental barrier (19). Furthermore, many are off-patent, making them affordable to low-income, at-risk populations. Exposure data are available for mice (31), which allowed us to choose our dose of methacycline without first screening for toxicity.
Mice were treated with methacycline (10 mg/kg, subcutaneously) prior to and daily after Zika virus exposure, and the phenotypic and antiviral responses were quantified (Fig. 4). Methacycline treatment reduced the extent of Zika virus infection in the brain and the severity of Zika virus-induced motor deficits; however, it did not prevent weight loss in the infected animals (Fig. 4B). Trunk control ability, tested by surface righting, the time to turn back over when flipped on its back, was significantly improved in methacycline-treated mice (Fig. 4C). The hindlimb suspension test revealed stronger leg muscles in treated mice (Fig. 4 D and E). Treated animals also showed improved balance of movement in the hindlimb foot angle measurement (Fig. 4F). The endpoint for euthanasia and collection of brain samples (11 dpi) was chosen based on the time the mice survived without extreme signs in the pilot study (13 to 16 dpi) (SI Appendix, Fig. S6A). Mice showed large amounts of Zika virus RNA and protein in the brain tissue. Reduction of Zika virus RNA in the brains of mice treated with methacycline was visualized using RNAscope (Fig. 4G) and quantified by qRT-PCR (SI Appendix, Fig. S8A). A reduction in Zika virus protein expression was apparent in several brain regions [anti-Flavivirus envelope 4G2 antibody: Chromogenic immunohistochemistry (Fig. 4H); fluorescent immunohistochemistry (Fig. 4I)]. Quantification of Fig. 4L revealed a significant reduction in Zika virus protein levels upon treatment with methacycline (>40% reduction). Calbindin staining demonstrated a loss of cerebellar Purkinje cells in response to Zika virus infection. Methacycline treatment had no detrimental effect in mock-infected mice and protected against Purkinje cell loss in Zika virus-infected mice (Fig. 4J). Furthermore, methacycline treatment reduced Zika virus-induced neuronal cell death and glial cell activation (Fig. 4K and SI Appendix, Fig. S8 B–D). These results indicate that prophylactic methacycline treatment prior to Zika virus exposure lessens the extent of infection and resulting toxicity in newborn mice.
Fig. 4.
Methacycline reduces Zika virus (ZIKV) infection in a mouse model. (A) Overview of in vivo protocol. Mice were treated with methacycline daily from birth, infected with Zika virus at postnatal day 1 (PND1), and killed at PND12. (B) Zika virus-infected mice gain weight at a slower rate. Methacycline treatment improved (C) trunk control, (D and E) muscle strength, and (F) balance. Methacycline reduced (G) Zika virus RNA levels, visualized using a Zika virus-specific probe that targets the 219–5443 fragment of the consensus sequence and quantified by qRT-PCR (SI Appendix, Fig. S8A), and Zika virus protein levels, visualized using (H) chromogenic immunohistochemistry and (I) fluorescent immunohistochemistry images and quantified with MetaMorph software. (J) Methacycline prevented Purkinjie cell loss. (K) Methacycline decreased astrocyte activation (GFAP) and increased neuronal cell staining (NeuN). Quantified with MetaMorph software (SI Appendix, Fig. S8 B–D). All data were analyzed with GraphPad Prism. Differences between means were assessed by one-way ANOVA followed by post hoc tests. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. The mean is displayed, and all error bars represent SE. We analyzed 10 mock, 7 mock and methacycline, 11 Zika, and 11 Zika and methacycline animals.
Discussion
Zika virus poses a continued threat in endemic regions and there is the potential for reemergence and spread to new locations. The lessons and responses presented in this report can be used to provide a blueprint on how to collaborate and respond with parallel efforts for other viral outbreaks. Since the public health emergency notice, there has been an outpouring of publications on small-molecule Zika virus inhibitors (4). As shown by our prior art screening library (Dataset S4), we tested an extensive number of these compounds, some of which confirmed in several, but not in all of our assays (for example, niclosamide was efficacious in Vero cells but toxic in NSCs), and others that did not confirm in any of our assays (for example, bromocriptine). Other viral protease inhibitors, such as the hepatitis C protease inhibitors danoprevir and vaniprevir, exhibited double-digit micromolar (weak) inhibition in our biochemical assays. Danoprevir was additionally tested in the Vero cell luciferase screen but was inactive based on our hit criteria.
We employed two HTS strategies: First, a traditional brute-force screen of a collection of chemically diverse, approved, annotated, and prior art compounds; second, selection of compounds using a deep-learning model. For the traditional approach, we observed a high hit rate of 20%, not surprising given the composition of the compounds tested and wavelength detection. After applying hit criteria, we decreased the hit rate to a more manageable 4% for confirmatory testing, where we observed 72% confirmation. While more than acceptable, this approach would require significant time and resources to test our entire screening collection. The use of qHTS on a diverse collection of compounds had the benefit of serving as a training set. This enabled us to use deep learning for the construction of a QSAR model and employ it to reduce the number of compounds required for physical screening by more than 130,000. Using the bioactivity of ∼4,900 compounds, we employed an AI-based QSAR model to examine the remaining 137,083 compounds in our collection. This approach identified 277 compounds for biochemical testing and improved the screening hit rate to 55%. Additionally, the AI-based QSAR model improved the hit rate at both the 30-μM and 10-μM potency cutoffs: 49% vs. 14% and 29% vs. 4%, respectively. The resources needed to identify these candidate inhibitors was far less using the deep-learning model, saving reagents, consumables, and time. The inclusion of an AI-based QSAR model as a hybrid approach to HTS efforts could also allow access to chemical space not available in standard screening libraries and extend the structural diversity of compounds ready for medicinal chemistry optimization.
Our ability to employ multiple substrates in combination with the linked and unlinked proteases allowed us to select the best screening conditions to identify small-molecule inhibitors. We used the linked protease to compare the rhodamine110 [(Bz-Nle-KRR)2-Rd] and AMC (Bz-Nle-KRR-AMC) substrates and showed that the rhodamine110 substrate not only reduced false-positives, but false-negatives as well, indicating a more sensitive and robust assay. We observed excellent reproducibility between the 1,536-well and 384-well rhodamine assays (95%). We chose the native Ac-VKTGKR-AMC substrate for orthogonal validation, which resulted in a confirmation rate of 82% (Dataset S5). When comparing the linked and unlinked protease constructs, we observed very few differences in inhibitor identification and chose to use the linked preparation (SI Appendix, Fig. S3).
The hits from the qHTS protease assays were characterized in a rigorous panel of follow-up assays, an approach that provided well-validated candidate compounds, with a faster screening turnaround. HTS campaigns typically identify a large number of compounds with activity; however, many early hits are false-positives or assay artifacts (32). Using multiple orthogonal screening approaches and biological characterization methods allowed us to rule out false-positives and identify robust in vitro inhibitors that were active in several assays and cell types.
We first assessed the ability of our candidate compounds to inhibit the virus in Vero cell infection assays. We chose to use an HTS-amenable, recombinant Zika virus with a luciferase reporter to indirectly measure viral replication. This cell-based assay was successful in removing two-thirds of our candidate inhibitors from the biochemical screen. Biochemically active compounds can lose potency in cellular assays due to solubility, stability, permeability, target promiscuity, and cytotoxicity. We further triaged our hits with an mCherry fluorescence-based imaging assay, which can detect viral replication in real time using recombinant virus, making it a more sensitive reporter system. While most compounds appeared to confirm, ∼75% were removed by the counterscreen due to potential cytotoxicity. The decreased concordance with the Rluc assay likely reflects the stability of the enzymatic reporter, which may persist even in the presence of cytotoxic compounds. For the final Vero cell assay, we employed qRT-PCR to quantify the reduction of viral RNA present in the infected cells. The African Zika virus strain (MR766; ATCC VR-84) has likely undergone many passages since its deposition in 1953 and may have acquired mutations that increase its toxicity (33). Despite this, we felt confident using this Zika virus isolate for our final Vero cell assay because we had already tested the compounds against the more recent Cambodian isolates, the goal of the qRT-PCR experiment was not to measure virus-induced toxicity, and the protease region is highly conserved between the African and Asian lineages (98% sequence match, GenBank: KX830960.1 and KJ776791.2). The qRT-PCR assay eliminated ∼50% of the remaining compounds, leaving 16 newly identified candidate inhibitors. Excluding prior art compounds, our Vero cell panel of assays removed ∼95% of our biochemical hits, allowing us to focus on a smaller number of active compounds in lower-throughput downstream assays. Fifteen were identified from the traditional brute-force qHTS and one (NCGC00108309) from the deep-learning model.
We then tested our candidate inhibitors in a more physiologically relevant system: Infection of human NSCs with a Zika virus isolate known to cause microcephaly (French Polynesian 2013). Five of the 18 compounds that were active in Vero cells displayed dose–response inhibition in NSCs: MK-591, a five-lipoxygenase–activating protein (FLAP) inhibitor (34); GSK3965, a liver X receptor agonist; TTNPB, a retinoic acid analog; PX-20350, a farnesoid X receptor agonist; and compound NCGC00108309, from the deep-learning model. Ten compounds that would have otherwise been excluded due to inactivity in the Vero cell assays displayed dose–response inhibition in NSCs: JNJ-40418677 and TC-E 5006 (BIIB042), both γ-secretase modulators (35); D-942, an AMPK activator; GW-4064X, a farnesoid X receptor agonist; PF-03882845, a mineralocorticoid receptor antagonist; MK-886, a FLAP inhibitor (34); and methacycline, demeclocycline, minocycline, and doxycycline, all tetracycline antibiotics. While many factors could contribute to the differences in activity between cell lines, the increased expression of permeability glycoprotein (P-gp) in Vero cells is likely contributing (36). Many compounds, including the tetracyclines, are known substrates of P-gp, which cause their expulsion from the cell, resulting in decreased access to intracellular targets and reduced activity (37).
Based on the results of the initial NSC screen, we chose to validate the tetracyclines and seven additional compounds: GSK-3965, MK-591, TTNPB, D-942, PF-03882845, JNJ-40418677, and GW-4064X. MK-591 and JNJ-404 were the most potent, with IC50s of ∼3 µM. For these two compounds, the IC50s for protease inhibition corresponds to the IC50s in the NSC assay, suggesting that MK-591 and JNJ-404 are exerting antiviral effects primarily through inhibition of the viral protease (Figs. 2 and 3). Additional compounds from the FLAP inhibitor (MK-886 and AM-679) and γ-secretase modulator (TC-E 5006) classes were also Zika virus protease inhibitors, validating the potential of these two compounds classes. Compounds that modulate γ-secretase were developed as Alzheimer’s treatments; however, all have failed in clinical trials because of either inefficacy or toxicity. Development of JNJ-404 was halted after observing liver toxicity in a canine model (35). While these results prevent the immediate clinical use of JNJ-404, other compounds in the γ-secretase modulating family could be investigated as Zika virus protease inhibitors. FLAP activity is required for the production of the inflammatory mediators, leukotrienes. Leukotriene production inhibitors are being pursued as drugs for a range of inflammatory diseases (34). MK-591 was tested in several clinical trials [for example, asthma (34), ulcerative colitis (38), and glomerulonephritis (39)] and displayed excellent safety profiles. A phase II clinical trial for asthma was largely successful and development was only stopped in favor of the advancement of montelukast, a leukotriene receptor antagonist (34). Furthermore, FLAP inhibitors show good penetration into the central nervous system in models of brain injury (40). Thus MK-591 is a promising candidate treatment of Zika virus infection.
Several factors suggest that methacycline could be used safely and effectively to prevent or treat Zika virus infection. Methacycline displayed activity against two strains of the virus known to cause neurological complications, one isolate from the French Polynesian 2013 outbreak and another from the Brazilian 2015 outbreak. It inhibited Zika virus infection in NSCs without toxicity, reduced the extent of Zika virus infection in mouse brains, and improved the motor deficits caused by Zika virus infection. Although unable to restore weight loss in the time frame measured, the beneficial effects are significant. The extensive clinical data available for the use of tetracyclines would eliminate the need for a phase I safety trial in healthy adults, thus accelerating the path to efficacy trials. Despite being P-gp substrates, many tetracyclines are able to cross the blood–brain barrier and successfully treat infections of the nervous system (18, 41). Tetracyclines cross the placental barrier into the fetal compartment (19), which would be necessary to treat a fetal infection. Early generation tetracyclines, however, are not recommended for use past the third month of gestation due to fetal tooth discoloration (42). While infection with Zika virus at any time during pregnancy has the potential to cause birth defects, the risk for severe congenital disorders is highest when infection occurs during the first 3 mo (43). Additionally, the use of later generation tetracyclines, especially doxycycline (42, 44), are not definitively linked to teratogenic effects. Pending further investigation and a thorough discussion of the potential side-effects with a medical professional, it is possible that the benefits of treatment with certain tetracyclines during pregnancy could outweigh the potential risks. In the absence of other treatment options, testing methacycline, or other tetracyclines such as doxycycline or minocycline, in an interventional (neurological complications, such as Guillain–Barré syndrome, encephalitis, and myelitis) or prophylactic (to prevent infection and slow transmission) clinical trial could be beneficial.
Materials and Methods
Study Design.
The hypothesis guiding this study was that previously developed small molecules contained in the NIH screening libraries could inhibit the NS2B-NS3 Zika virus protease and Zika virus infection. We planned to develop a fluorescence-based protease assay and use HTS to identify and select inhibitors. First, we screened a small library at a single dose and selected hit compounds to test in NSCs and one to validate in a mouse model. Second, we screened a larger compound library, used computational modeling to select additional compounds for testing, added a panel of Vero cell infection assays to further narrow down compounds, then proceeded to testing in NSCs. For the mouse experiments, the sample size for each group was chosen at the beginning of the study by estimating the minimum number of animals required to significantly differentiate two groups at a P value of 0.05 and confidence interval of 95% (45). “LaMorte’s Power Calculator” (http://www.uml.edu/Images/LaMorte%20calculator_tcm18-37802.xls) indicated eight animals per group were needed for the specified statistical requirements. We analyzed 10 mock, 7 mock and methacycline, 11 Zika, and 11 Zika and methacycline animals. We used mice from similar litter sizes and randomized them into groups. The mice underwent behavioral analysis on the indicated days. The endpoint for euthanasia and collection of brain samples (11 dpi) was chosen based on the time the mice survived without severe signs in the pilot study (13 to 16 dpi). No data or outliers were excluded. This study was not blinded. The number of replicates for each experiment is given in the corresponding figure legends. Most experiments were performed in biological triplicates, with some exceptions, including the screening data, which was only a single replicate. The mouse behavioral studies were performed in order of least stressful to most stressful and were repeated one to three times.
Compound Sources.
The pilot screen was performed using the Microsource Discovery Spectrum collection, http://www.msdiscovery.com/spectrum.html. All other screening compounds were from the NCATS/NIH (46, 47), https://ncats.nih.gov/preclinical/core/compound. Additional compounds and substrates were obtained from Sigma, Abcam, Cayman Chemicals, Genscript, Bachem, or Cambridge Research Biochemicals.
1536-Well Protease (Rhodamine110 Substrate) Assay.
Enzyme (3 µL, 1 nM and 25 nM for linked and unlinked, respectively) in assay buffer (10 mM Tris⋅HCl pH 8.0 with 0.01% Tween 20) were dispensed into a 1,536-well solid-bottom black plate (Greiner Bio One). Compounds (23 nL, 7.28 nM to 114 µM) or control Aprotinin (3.47 nM to 114 µM) were transferred via Wako Pin-tool (Wako Automation). Samples were incubated (room temperature, protected from light) for 15 min followed by addition of 1 µL rhodamine110 substrate (1 or 2 µM). Plates were immediately read on a ViewLux high-throughput CCD imager (PerkinElmer) equipped with FITC fluorescence optics (480/20-nm excitation, 540/25-nm emission), then incubated (room temperature, protected from light) for 15 min (∼15% conversion). The change in fluorescence intensity over the 15-min reaction period was normalized against no-inhibitor and no-enzyme controls. The percent inhibition data were fitted to a four-parameter Hill equation. Further details are given in SI Appendix.
AI-Based QSAR Approach.
For the development of the AI-based QSAR model we used QNA descriptors together with 10 whole-molecular descriptors and a deep-learning technique for the model building. The QNA descriptors (23) were calculated using command line program QNA2015toCSV. The qHTS data of 4,915 compounds (rhodamine110 substrate with the unlinked protease; PubChem AID 1347160) were used for the construction of QSAR. The QNA-based deep-learning model was used for virtual screening of the remainder of the NCATS screening library. The predicted compounds were distinct in structure from the training set compounds in the Morgan fingerprints space (the closest predicted compound to the training set has only 0.5 Tanimoto similarity; open-source cheminformatics, http://www.rdkit.org/). Further details are given in SI Appendix.
Dose–Response of Select Compounds in Linked Protease Assay.
Fresh aliquots of the commercially available hit compounds were tested in the linked protease assay. Compounds (3 µL, 300 nM to 1 mM) were transferred to the assay plate (384-well) and incubated with protease (25 nM) for ∼15 min. The substrate, Ac-VKTGKR-AMC or Bz-Nle-KRR-AMC (Bachem; DMSO, 6 mM) was further diluted into assay buffer to a concentration of 180 μM. A multichannel repeat pipettor was used to add 5 μL of substrate (final concentration of 30 μM) to the assay plate. The fluorescence intensity (excitation: 355 nm, emission: 460 nm) was measured every 30 s for 30 min and a time point within initial velocity conditions was normalized to no-enzyme and no-inhibitor controls.
Kinetics of Methacycline, MK-591, and JNJ-404 Inhibition.
Assay buffer (75 μL) was added to all wells of a 96-well black plate. The protease was diluted to 166 nM in assay buffer and 15 μL was dispensed into the assay plate (25 nM). Inhibitor was diluted in the buffer and transferred to the assay plate (5 μL, concentrations indicated on graph, Fig. 2 F–H). After a 15-min incubation at room temperature, substrate (Ac-VKTGKR-AMC or Bz-Nle-KRR-AMC) was added. The fluorescence intensity (excitation: 355 nm, emission: 460 nm) was measured every 30 s for 30 min and a timepoint within initial velocity conditions was normalized to no-enzyme and no-inhibitor controls.
For jump dilution analysis, the protease was diluted to 3 µM in assay buffer and dispensed into a 96-well PCR plate (10 µL, 1.5 μM). Methacycline (10 µL, 2 mM), MK-591 (10 µL, 35 µM), or JNJ-404 (10 µL, 35 µM) was added to the protease solution and incubated for 25 min at room temperature. Assay buffer and substrate (Ac-VKTGKR-AMC or Bz-Nle-KRR-AMC) were added to a 96-well black assay plate. The inhibitor/protease mixture (2.5 µL, final concentrations of 15 nM of protease and 20 µM of methacycline, 0.35 µM of MK-591, and 0.35 µM JNJ-404), was added to the assay buffer/substrate mixture (final total volume per well was 250 µL, final substrate concentration was 30 µM). The fluorescence intensity (excitation: 355 nm, emission: 460 nm) was measured every 15 to 30 s for 10 min and normalized to no-enzyme controls.
Cell Lines and Viral Stocks.
Vero cell line was purchased from ATCC (catalog #CCL-81) and maintained in DMEM plus 4% FBS. Zika virus (ZIKV; wild-type MR766) was purchased from ATCC (VR-84; GenBank: KX830960.1). The two 2010 Cambodian origin (FSS13025, passage #2, amplification in Vero cells) recombinant Zika viruses with Renilla luciferase (ZIKV-Rluc) (24) and mCherry reporters (ZIKV-mCherry) (48) were obtained from P. Y. Shi, University of Texas Medical Branch, Galveston, TX. An aliquot of Zika virus from the French Polynesian outbreak of 2013 was obtained at passage #2 (a single amplification in Vero cells) from Steve Whitehead, National Institute of Allergy and Infectious Diseases, North Bethesda, MD. This aliquot was subsequently amplified an additional four times in Vero cells and aliquots at passage #5 were used in the NSC assay. NSCs were obtained from Tongguang Wang, National Institute of Neurological Disorders and Stroke, Bethesda, MD: Human induced pluripotent stem cells (iPSC) were transformed from adult hematopoietic progenitor cells (CD34 cells from a healthy donor, in accordance with the NIH Institutional Review Board) following our previously published protocol (49). iPSCs were differentiated to NSCs using the protocol outlined for Gibco PSC neural induction media (ThermoFisher, A164780). After conversion to NSCs was complete and nestin immunostaining was confirmed (49), the media was switched to Gibco StemPro NSC Serum Free Media (NSC SFM, ThermoFisher, A10509-01). An aliquot of Zika virus from the Brazilian outbreak of 2015, at passage #4 (amplification in Vero cells), from Steve Whitehead at the National Institute of Allergy and Infectious Diseases, was used in the in vivo experiments. Cell lines were tested for mycoplasma periodically using MycoAlert (Lonza).
Vero Cell Infections Assays.
We adapted (24) a high-throughput assay in 1,536-well plate format to use the recombinant virus ZIKV-Rluc (Cambodian). Vero cells (3 µL) were dispensed using a Multidrop Dispenser into a solid bottom white 1,536-well plate at a concentration of 1,000 cells per well. The plate was incubated overnight in a CO2 incubator at 37 °C. Compounds were delivered as a 23 nL DMSO solution by Kalypsys pintool transfer. ZIKV-Rluc (2 µL) was added at a multiplicity of infection (MOI) of 1 to 2 and incubated in a CO2 incubator for 48 to 72 h at 37 °C. After incubation of infected cells in test plates, live-cell Renilla luciferase substrate (Promega) was added. The luminescence signal was measured after 1- to 1.5-h exposure to substrate in ViewLux reader (Perkin-Elmer).
We used recombinant virus ZIKV-mCherry to test 131 compounds in a 384-well plate format (Dataset S5); 108 were from the qHTS or virtual screens, while 9 were previously reported to have antiviral activity. Vero cells (30 µL) were dispensed by Multidrop Dispenser into a clear solid bottom black 384-well plate (ultracarrier, Perkin-Elmer LifeSciences) at a concentration of 5,000 cells per well. The plate was incubated overnight in a CO2 incubator at 37 °C. Compounds were delivered as a 126 nL DMSO solution by Kalypsys pintool transfer. ZIKV-mCherry (10 µL) was added at an MOI of 1 and incubated in a CO2 incubator for 48 to 72 h at 37 °C. After sufficient incubation of infected cells in test plates, 8 µL of 32% paraformaldehyde (PFA) was dispensed into assay plates to fix cells and incubated for 20 min at room temperature. Plates were washed three times with PBS using BioTek EL406 microplate washer/dispenser. After washing, 30 µL of PBS with 0.01% TX-100 containing Hoechst dye was dispensed into the each well of the plates for nuclear staining. Next, 384-well plates were imaged on an automated, widefield high-content imager (INCell 2200, GE Healthcare) using a 10×/0.45 NA lens and standard DAPI (nuclear stain) and mCherry (cy5) excitation and emission filters. The TIFFs were quantitated using the canned Multi Target Analysis protocol (GE Investigator Workstation software v3.7.2). Nuclei from the DAPI channel were identified using top hat segmentation and a sensitivity setting of 96 and minimum size area of 35 µm2. A recombinant Zika viral infection was monitored in the Cy5 channel (cells) using a 2-µm collar dilation from the nuclear bitmap. mCherry objects with an average nuclear RFU intensity above 450 (3 SDs above mean negative control wells) were considered “mCherry positive” and data were analyzed by percent infection rate.
For the qRT-PCR assay, Vero cells in 384-well plates were treated with the indicated compounds at different concentrations, infected with ZIKV (MR766, MOI = 1), and incubated at 37 °C for 3 h to permit virus invasion. After 3 h, supernatant was removed, and cells were washed once to remove extracellular virus. After a further 21-h incubation, infected cells were washed three times with cold PBS (4 °C). Plates were processed using the TaqmanGene Expression Cells-to-Ct Kit (ThermoFisher Scientific) following the manufacture’s protocol. Primers directed to the NS5 sequence (Forward-GCTGTACCTCAAGGATGGGAGAT; Reverse-GCTCGGCCAATCAGTTCA) along with Taqman probe (FAM-ATTGTGGTCCCTTGCCGCCACC-BHQ; Biosearch Technologies) were used to quantify drug inhibition of viral load. Reactions were run at initial 95 °C for 10 min, then 40 cycles of 95 °C for 20 s and 60 °C for 40 s using a LightCycler 480 (Roche). Relative viral copy numbers were generated by normalizing to cells treated with vehicle.
qHTS Data Analysis, Statistics, and Availability.
Data from each assay were normalized plate-wise to corresponding intraplate controls (neutral control DMSO and positive control as noted). The same controls were used for the calculation of the Z′ factor, a measure of assay quality control. Concentration-response curves were fitted and classified as described previously (20, 50). IC50s were calculated using Prism software (v7.04, GraphPad Software), sigmoidal dose–response (variable slope). We used the Palantir Technologies (Washington, DC) data integration platform, which is configured to ingest all HTS results generated at NCATS, and harmonized this data with other sources such as ChEMBL and OrthoMCL. All qHTS screening results are publicly available at PubChem (AIDs 1347149, 1347150, 1347151, 1347152, 1347153, 1347154, 1347155, 1347156, 1347157, 1347158, 1347159, 1347160,1347161). The chemical structures were standardized using the LyChI (Layered Chemical Identifier) program (51) (v20141028, https://github.com/ncats/lychi).
NSC Infection Assays.
NSCs at passage numbers 6 to 8 were plated into Geltrex-coated 24-well plates 2 to 4 d prior to infection. When the cells reached 30 to 65% confluency (∼65,000 to 100,000 cells per well), drugs were added to the desired concentrations and incubated for at least 1 h. All media was removed from each well and replaced with 250 µL of FBS-free DMEM Glutamax media containing Zika virus (French Polynesian, MOI 3). Mock-infected controls (250 µL of media without virus) were included on each plate. Drugs were added to desired concentrations, plates were rocked to mix, and incubated for 3.5 to 4.5 h. All virus-containing media was removed and replaced with 500 µL fresh NSC SFM containing drugs at desired concentrations. At 24 h postinfection (hpi), 250 µL of fresh NSC SFM containing drugs was added to each well. At 48 hpi, in plates with cells at less than 90% confluency, 350 µL supernatant was removed and replaced with 350 µL fresh NSC SFM containing drugs. At 48 or 72 hpi, plates with confluent cells were centrifuged at 1,000 rpm for 5 min. Supernatant was transferred to a 96-well plate and stored at −80 °C for later analysis by qRT-PCR. Cells were fixed for 15 min with ++PBS containing 4% PFA. Cells were washed 3× with ++PBS, then put into 0.5 mL of blocking buffer for 1 h (4% BSA, 0.1% Triton X-100). Primary antibody (mouse antiflavivirus envelope 4G2, generated from hybridoma cell line D1-4G2-4-15 [ATCC], 1:5 or 1:10) was added and incubated overnight at 4 °C. Cells were washed 3× with ++PBS, followed by a 1-h incubation at room temperature with secondary antibody (1:500, goat anti-mouse IgG [H + L] Alexa fluor 488, Abcam or ThermoFisher). Cells were washed 3× with ++PBS; in the second wash, Hoechst nuclear stain (ThermoFisher) was included at 1:2,000. Cells were stored in the dark, at 4 °C, in ++PBS until time of imaging. Stained cells were counted on a Cytation 5 (Biotek) imaging reader. Cell counts were generated with Gen 5 3.04 software (Biotek) using the software’s default settings for cell count analysis. Plates of cultured cells were loaded onto the imaging reader and total cell counts were determined with a filter with emission/excitation of 377 nm/447 nm, at 10× magnification. Zika virus infected cells were similarly quantified with a filter having an emission/excitation of 469 nm/525 nm. All cell count data were exported to Microsoft Excel for further analysis. The number of Zika-infected (4G2-positive) cells was divided by the total number of cells (Hoechst-positive) to give the percent infected cells. Reported inhibition values are relative to the intraplate DMSO-treated control, which was normalized to 100% infection. For the combination studies, significance was calculated in GraphPad Prism using an unpaired, two-tailed t test: ****P < 0.0001; *P < 0.05.
Select compounds that demonstrated inhibition of Zika virus staining were chosen for further analysis of the viral RNA present in the cell-free supernatant. The supernatant was thawed, and viral RNA was extracted using the Qiagen EZ1 Advanced XL according to manufacturer’s protocol. Viral RNA was converted to cDNA (SuperScript II first-strand synthesis SuperMix, Invitrogen 11752), which was quantified by qRT-PCR using Taqman probes directed against the Zika virus envelope. The change in Ct value between no-drug control and treated samples was used to determine the level of inhibition.
Zika Virus Mouse Model.
All mouse model procedures adhered to the NIH Guide for the Care and Use of Laboratory Animals (52). The National Institute of Neurological Disorders and Stroke/National Institute on Deafness and Other Communication Disorders/National Center for Complementary and Integrative Health Animal Care and Use Committee approved the animal protocol. All data were analyzed with GraphPad Prism. Differences between means were assessed by one-way ANOVA followed by post hoc tests. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. Further details are available in SI Appendix.
The newborn pups of three wild-type C57BL/6 mice were randomly divided into four groups: Mock-infected control (n = 10), mock-infected and methacycline-injected control (n = 7), Zika virus-infected (n = 11), and Zika virus and methacycline-injected (n = 11) groups. To identify groups, we tattooed mice on their paws with green paste. All mice weighed 1 g, 1 d after birth. Since litter size can significantly affect development, we used mice from similar litter sizes. At postnatal day 1, methacycline was injected subcutaneously, 1 h prior to subcutaneous injection with Zika virus (5 × 105 PFU/mL; Fortaleza, Brazil/2015, passage #4). For the following days, 10 mg/kg of methacycline was administered daily.
Animals were weighed, appearance was monitored, and nonintrusive behavioral tests for the evaluation of neonatal motor function were performed. To determine if Zika virus infection could delay general development in mice, eye opening, fur development, and body weight were measured on a daily basis. To determine if Zika virus infection could cause a delay in sensory-motor development, reflex tests were used to score developmental locomotor behavior. Behavioral motor tests modified from a cerebral palsy animal model (53) were conducted to compare motor abilities between groups. Animals were killed when they started to display severe signs, such as becoming unresponsive to touch, breathing slowly, bad fur condition, sudden weight loss (30% in a few days), and a hunched back. For postmortem analysis, cryoprotected brains were cut in the sagittal plane, including the cerebral cortex, hippocampus, basal ganglia, and cerebellum, into 40-μm-thick sections.
For immunofluorescence staining, the brain sections were washed in Tris-buffered saline (TBS) prior to incubation in blocking solution (TBS with 0.5% Triton-X and 2.5% donkey serum). Primary antibody 4G2 (Millipore Sigma) supernatant (1:10) was incubated overnight at room temperature. Primary antibodies were diluted in blocking solution as follows: 4G2 (flavivirus envelope protein, 1:10, Millipore Sigma), anti-NeuN (1:250, Chemicon), anti-GFAP (1:2,000; Dako), and anti-IBA1 (1:1,000; Wako) and incubated at 4 °C. After washing with TBS, sections were incubated with fluorescent conjugated secondary antibodies (1:250, Jackson ImmunoResearch), followed by washing and counterstaining with DAPI to label all nuclei. Stained sections were mounted on slides and viewed with a Zeiss confocal laser microscope. MetaMorph software was used to quantitate the total cell counts. The Zika virus infected (4G2, green channel-positive) cells was normalized to DAPI to quantitate the percentage of Zika virus infected cells.
For chromogen-based immunohistochemistry, sections were incubated in 0.3% hydrogen peroxide (H2O2) to remove endogenous peroxidase activity, then with blocking solution to prevent nonspecific protein binding. Primary antibody incubation was performed with mouse 4G2 supernatant (1:10) or Calbindin (1:500; Swant), overnight at room temperature. The secondary antibody, PowerVision polymeric horseradish peroxidase (HRP) anti-mouse (Leica Biosystems), was applied for 2 h at room temperature. Antibody binding was developed with 3,3′-diaminobenzidine (DAB; Vector Laboratories). Sections were counterstained with hematoxylin (Dako). Images were processed using an Aperio whole slide scanner (Leica Biosystems).
RNAscope in situ hybridization was performed according to the manufacturer’s instructions (Advanced Cell Diagnostics). Sections were dried for 1 h at 60 °C, deparaffinized and rehydrated. Sections were then hybridized with a probe against ZIKV for 2 h at 40 °C, and amplified sequentially with the 2.5 HD Detection Kit (following manufacture’s procedure) at 40 °C. To reveal the signal, slides were incubated in a red working solution for 10 min at room temperature. Slides were counterstained with 50% hematoxylin and mounted with EcoMount mounting medium (Biocare Medical). Images were captured using a whole-slide scanner (Aperio, Leica Biosystems).
To quantify the viral RNA present in the brain samples, sagittal brain sections were scraped off of the imaging slides using a clean razor blade. The slides were not paraffin-embedded, so the RNA extraction was performed according to the manufacturer’s protocol (Qiagen, RNeasy FFPE Handbook), except the step for deparaffination was skipped. Conversion to cDNA and quantification by qRT-PCR was performed as described above (NSC assay).
Statistical Analysis.
Data were analyzed with GraphPad Prism. For the drug combination studies in NSCs, significance was calculated using an unpaired, two-tailed t test: ****P < 0.0001; *P < 0.05. For the mouse studies, differences between means were assessed by one-way ANOVA followed by post hoc tests. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
Supplementary Material
Acknowledgments
We thank the National Center for Advancing Translational Studies (NCATS) Compound Management, Automation, and Analytical groups for their support; Early Translation Branch/NCATS Chemical Genomics Center trainees for their support; and Dr. Tongguang Wang, Marie Medynets, and Valerie McDonald for providing the neural stem cells. This work was funded in part by the Intramural Research Program of NCATS, National Institutes of Health Grant 1ZIATR000291-03 (to A.S.); The Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health (A.N.); and National Institutes of Health Grant R21AI109185 (to R.P.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005463117/-/DCSupplemental.
Data Availability.
All qHTS screening results are publicly available at PubChem (AIDs 1347149–1347161) (54). Processed data are available in Datasets S1–S15.
References
- 1.Wang A., Thurmond S., Islas L., Hui K., Hai R., Zika virus genome biology and molecular pathogenesis. Emerg. Microbes Infect. 6, e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mlakar J., et al. , Zika virus associated with microcephaly. N. Engl. J. Med. 374, 951–958 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Carod-Artal F. J., Neurological complications of Zika virus infection. Expert Rev. Anti Infect. Ther. 16, 399–410 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Abrams R. P. M., Solis J., Nath A., Therapeutic approaches for Zika virus infection of the nervous system. Neurotherapeutics 14, 1027–1048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady O. J., Hay S. I., The first local cases of Zika virus in Europe. Lancet 394, 1991–1992 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Drag M., Salvesen G. S., Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 9, 690–701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang C., Keller T. H., Luo D., Zika virus protease: An antiviral drug target. Trends Microbiol. 25, 797–808 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Li Z., et al. , Erythrosin B is a potent and broad-spectrum orthosteric inhibitor of the flavivirus NS2B-NS3 protease. Antiviral Res. 150, 217–225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafferman J. A., Silbergeld E. K., Erythrosin B inhibits dopamine transport in rat caudate synaptosomes. Science 205, 410–412 (1979). [DOI] [PubMed] [Google Scholar]
- 10.Yuan S., et al. , Structure-based discovery of clinically approved drugs as Zika virus NS2B-NS3 protease inhibitors that potently inhibit Zika virus infection in vitro and in vivo. Antiviral Res. 145, 33–43 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Dowling H. F., et al. , A clinical and bacteriologic evaluation of novobiocin in seventy-five patients. AMA Arch. Intern. Med. 98, 273–283 (1956). [DOI] [PubMed] [Google Scholar]
- 12.Brown R., Thomassen P. R., Singler J. M., The use of novobiocin for treament of infections of odontogenic origin; a preliminary clinical study. Oral Surg. Oral Med. Oral Pathol. 11, 598–602 (1958). [DOI] [PubMed] [Google Scholar]
- 13.Hargreaves T., Holton J. B., Jaundice of the newborn due to novobiocin. Lancet 1, 839 (1962). [DOI] [PubMed] [Google Scholar]
- 14.Falgout B., Pethel M., Zhang Y. M., Lai C. J., Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65, 2467–2475 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phoo W. W., et al. , Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 7, 13410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuiper B. D., et al. , Increased activity of unlinked Zika virus NS2B/NS3 protease compared to linked Zika virus protease. Biochem. Biophys. Res. Commun. 492, 668–673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei J., et al. , Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science 353, 503–505 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Agwuh K. N., MacGowan A., Pharmacokinetics and pharmacodynamics of the tetracyclines including glycylcyclines. J. Antimicrob. Chemother. 58, 256–265 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Nahum G. G., Uhl K., Kennedy D. L., Antibiotic use in pregnancy and lactation: What is and is not known about teratogenic and toxic risks. Obstet. Gynecol. 107, 1120–1138 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Inglese J., et al. , Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. U.S.A. 103, 11473–11478 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant S. K., Sklar J. G., Cummings R. T., Development of novel assays for proteolytic enzymes using rhodamine-based fluorogenic substrates. J. Biomol. Screen. 7, 531–540 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Simeonov A., et al. , Fluorescence spectroscopic profiling of compound libraries. J. Med. Chem. 51, 2363–2371 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Filimonov D. A., Zakharov A. V., Lagunin A. A., Poroikov V. V., QNA-based ‘Star Track’ QSAR approach. SAR QSAR Environ. Res. 20, 679–709 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Shan C., et al. , An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe 19, 891–900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chavali P. L., et al. , Neurodevelopmental protein Musashi-1 interacts with the Zika genome and promotes viral replication. Science 357, 83–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., et al. , Zika virus protease cleavage of host protein septin-2 mediates mitotic defects in neural progenitors. Neuron 101, 1089–1098.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Retallack H., et al. , Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. U.S.A. 113, 14408–14413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cauchemez S., et al. , Association between Zika virus and microcephaly in French Polynesia, 2013-15: A retrospective study. Lancet 387, 2125–2132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaelis L., Menten M. L., Johnson K. A., Goody R. S., The original Michaelis constant: Translation of the 1913 Michaelis-Menten paper. Biochemistry 50, 8264–8269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Copeland R. A., Basavapathruni A., Moyer M., Scott M. P., Impact of enzyme concentration and residence time on apparent activity recovery in jump dilution analysis. Anal. Biochem. 416, 206–210 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Rauthan M., Pilon M., A chemical screen to identify inducers of the mitochondrial unfolded protein response in C. elegans. Worm 4, e1096490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlin J. L., Walters M. A., The essential roles of chemistry in high-throughput screening triage. Future Med. Chem. 6, 1265–1290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duggal N. K., et al. , Mutations present in a low-passage Zika virus isolate result in attenuated pathogenesis in mice. Virology 530, 19–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gür Z. T., Çalışkan B., Banoglu E., Drug discovery approaches targeting 5-lipoxygenase-activating protein (FLAP) for inhibition of cellular leukotriene biosynthesis. Eur. J. Med. Chem. 153, 34–48 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Gijsen H. J. M., Mercken M., γ-Secretase modulators: Can we combine potency with safety? Int. J. Alzheimers Dis. 2012, 295207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiebaut F., et al. , Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc. Natl. Acad. Sci. U.S.A. 84, 7735–7738 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavallaris M., Madafiglio J., Norris M. D., Haber M., Resistance to tetracycline, a hydrophilic antibiotic, is mediated by P-glycoprotein in human multidrug-resistant cells. Biochem. Biophys. Res. Commun. 190, 79–85 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Roberts W. G., et al. , Leukotrienes in ulcerative colitis: Results of a multicenter trial of a leukotriene biosynthesis inhibitor, MK-591. Gastroenterology 112, 725–732 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Guasch A., Zayas C. F., Badr K. F., MK-591 acutely restores glomerular size selectivity and reduces proteinuria in human glomerulonephritis. Kidney Int. 56, 261–267 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Heidenreich K. A., Corser-Jensen C. E., “5-Lipoxygenase-activating protein inhibitors: Promising drugs for treating acute and chronic neuroinflammation following brain injury” in New Therapeutics for Traumatic Brain Injury, Heidenreich K. A., Ed. (Academic Press, San Diego, 2017), chap. 12, pp. 199–210. [Google Scholar]
- 41.Yim C. W., Flynn N. M., Fitzgerald F. T., Penetration of oral doxycycline into the cerebrospinal fluid of patients with latent or neurosyphilis. Antimicrob. Agents Chemother. 28, 347–348 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wormser G. P., Wormser R. P., Strle F., Myers R., Cunha B. A., How safe is doxycycline for young children or for pregnant or breastfeeding women? Diagn. Microbiol. Infect. Dis. 93, 238–242 (2019). [DOI] [PubMed] [Google Scholar]
- 43.França G. V. A., et al. , Congenital Zika virus syndrome in Brazil: A case series of the first 1501 livebirths with complete investigation. Lancet 388, 891–897 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Damkier P., Brønniche L. M. S., Korch-Frandsen J. F. B., Broe A., In utero exposure to antibiotics and risk of congenital malformations: A population-based study. Am. J. Obstet. Gynecol. 221, 648.e1–648.e15 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Dell R. B., Holleran S., Ramakrishnan R., Sample size determination. ILAR J. 43, 207–213 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang R., et al. , The NCGC pharmaceutical collection: A comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 3, 80ps16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kearney S. E., et al. , Canvass: A crowd-sourced, natural-product screening library for exploring biological space. ACS Cent. Sci. 4, 1727–1741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan C., et al. , A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat. Med. 23, 763–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T., et al. , Derivation of neural stem cells from human adult peripheral CD34+ cells for an autologous model of neuroinflammation. PLoS One 8, e81720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Jadhav A., Southal N., Huang R., Nguyen D. T., A grid algorithm for high throughput fitting of dose-response curve data. Curr. Chem. Genomics 4, 57–66 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stefaniak F., Prediction of compounds activity in nuclear receptor signaling and stress pathway assays using machine learning algorithms and low-dimensional molecular descriptors. Front. Environ. Sci. 3, 77 (2015). [Google Scholar]
- 52.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 53.Feather-Schussler D. N., Ferguson T. S., A battery of motor tests in a neonatal mouse model of cerebral palsy. J. Vis. Exp. 117, 53569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abrams R. P. M., et al. , Raw Screening Data PubChem. https://pubchem.ncbi.nlm.nih.gov/source/National%20Center%20for%20Advancing%20Translational%20Sciences%20(NCATS). Deposited 10 November 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All qHTS screening results are publicly available at PubChem (AIDs 1347149–1347161) (54). Processed data are available in Datasets S1–S15.