Significance
Presbycusis, or age-related hearing loss, is a major public health issue and the principal potentially modifiable risk factor for dementia. It is caused by environmental factors and largely uncharacterized genetic factors. We compared DNA sequences across genomic coding regions between familial or sporadic cases of severe presbycusis and controls with normal hearing. The frequency of ultrarare predicted pathogenic variants in genes known to cause dominant early-onset forms of deafness was significantly higher in both familial and sporadic cases than in controls. Pathogenicity of many of these variants was established with complementary analyses. Ultrarare variants have a large effect size and are known to cause monogenic disorders. These findings open up possibilities for curing these forms of presbycusis by gene therapy.
Keywords: presbycusis, age-related hearing loss, monogenic disorder, ultrarare variants, Tmc1
Abstract
Presbycusis, or age-related hearing loss (ARHL), is a major public health issue. About half the phenotypic variance has been attributed to genetic factors. Here, we assessed the contribution to presbycusis of ultrarare pathogenic variants, considered indicative of Mendelian forms. We focused on severe presbycusis without environmental or comorbidity risk factors and studied multiplex family age-related hearing loss (mARHL) and simplex/sporadic age-related hearing loss (sARHL) cases and controls with normal hearing by whole-exome sequencing. Ultrarare variants (allele frequency [AF] < 0.0001) of 35 genes responsible for autosomal dominant early-onset forms of deafness, predicted to be pathogenic, were detected in 25.7% of mARHL and 22.7% of sARHL cases vs. 7.5% of controls (P = 0.001); half were previously unknown (AF < 0.000002). MYO6, MYO7A, PTPRQ, and TECTA variants were present in 8.9% of ARHL cases but less than 1% of controls. Evidence for a causal role of variants in presbycusis was provided by pathogenicity prediction programs, documented haploinsufficiency, three-dimensional structure/function analyses, cell biology experiments, and reported early effects. We also established Tmc1N321I/+ mice, carrying the TMC1:p.(Asn327Ile) variant detected in an mARHL case, as a mouse model for a monogenic form of presbycusis. Deafness gene variants can thus result in a continuum of auditory phenotypes. Our findings demonstrate that the genetics of presbycusis is shaped by not only well-studied polygenic risk factors of small effect size revealed by common variants but also, ultrarare variants likely resulting in monogenic forms, thereby paving the way for treatment with emerging inner ear gene therapy.
Age-related hearing loss (ARHL), also known as presbycusis, is the most prevalent sensory impairment in the adult population and is 1 of the top 10 health conditions most frequently associated with disability in the elderly. According to the World Health Organization, 320 million people worldwide aged 65 and over currently suffer from hearing disability (1). Presbycusis is a progressive late-onset hearing impairment that begins after the age of 40 and mostly affects the perception of high-frequency sounds (2). Individuals initially encounter difficulties following conversations in noisy environments and localizing sound sources spatially, until they can no longer understand speech even in quiet environments (3). This disability leads to social isolation and desocialization, profoundly altering quality of life (4). Individuals face the threat of depression and cognitive decline (5, 6), resulting in a progressive loss of autonomy. There is growing evidence to suggest that hearing loss in and after middle age is the main modifiable risk factor for dementia (7).
Patient management is currently based on prostheses—hearing aids and cochlear implants—which restore hearing to an acceptable level for understanding speech in relatively quiet environments, in most individuals (8). However, these devices are not very effective in noisy environments (9).
Presbycusis was long thought to be a natural and inevitable consequence of aging, but like other aging processes, it actually depends on genetic and environmental factors and their interactions (10). Various environmental factors, such as noise overexposure, the use of ototoxic drugs and solvents, and smoking, have been identified as risk factors, as have several comorbid conditions, such as diabetes and hypertension (11, 12). The most prevalent environmental cause is noise overexposure, the impact of which is steadily increasing with the proportion of the world’s population living in overcrowded cities and inadequate controls over sound intensity, extending to recreational settings (13). Environmental factors are thought to account for about half the phenotypic variance of presbycusis (10). Heritability indices of 0.35 to 0.70 have been reported in studies based on hearing thresholds in twins (14), families (15, 16), or individuals with self-reported hearing impairment (17).
Genetic traits and diseases represent a continuum, with the effect size of the variants involved being inversely related to the frequency of the disease-causing variant (18, 19). The spectrum ranges from monogenic diseases (attributable to one extremely rare variant with a very large effect size—100% penetrance), via near-Mendelian traits due to variants with incomplete penetrance, and conditions due to moderately rare variants of intermediate effect size to highly polygenic traits (hundreds of common variants involved, each having a very small effect size) (20). Genetic analyses have implicated several genes in presbycusis. Association studies have been performed with candidate genes selected on the basis of a putative or demonstrated role of the encoded proteins in the cochlea. For example, given the well-established role of oxidative stress induced by overexposure to noise, associations have been highlighted between presbycusis and null alleles or single-nucleotide polymorphisms (SNPs) of certain genes involved in redox homeostasis, such as GSTT1, GSTM1 (21, 22), NAT2 (21, 23, 24), CYP1A1 (21), and UCP2 (21). Similarly, the observation that women have better hearing than men, at least until menopause, led to the discovery of an association between ESRRG and ARHL in women (25). Association studies have also been conducted with genes responsible for early-adulthood deafness displaying autosomal dominant inheritance, autosomal dominant deafness (DFNA) forms of deafness, or congenital hearing impairment displaying autosomal recessive inheritance, autosomal recessive deafness (DFNB) forms of deafness. Indicative associations have been reported with KCNQ4 (DFNA2) (26), GRHL2 (DFNA28) (27, 28), ILDR1 (DFNB42), and EYA4 (DFNA10) (29). Pangenomic association studies were then performed in large cohorts of sporadic cases of presbycusis (30, 31). Genome-wide association studies (GWAS) were performed, assuming that presbycusis, as a common disorder, would probably involve common variants (minor allele frequency [AF] > 5%) with a small effect size. GWAS detected significant associations (P ≤ 5 × 10−8) between presbycusis and SNPs close to ISG20/ACAN (29), PCDH20 (32), ARHGEF28 (30), and SLC28A3 (32) and SNPs within TRIOBP (29), SIK3 (33), NID2 (30), CLRN2 (30), ARHGEF28 (30), EYA4 (30), and very recently, ILDR1 (30, 31). Furthermore, in a large GWAS study based on the UK Biobank (including data for more than 250,000 individuals between the ages of 40 and 69 y), 36 new loci were reported to be significantly associated with self-reported hearing difficulties or with the use of hearing aids (30). A recent analysis conducted and replicated in about 10,000 affected individuals identified associations with four additional genes (31). These candidate presbycusis-predisposing (CPP) genes were mostly identified on the basis of associations of ARHL with intragenic (mostly intronic) SNPs or SNPs located in close proximity to the gene. Finally, next-generation sequencing has identified monoallelic mutations predicted to be deleterious in NHERF1, SPATC1L (34, 35), and MYO6 (DFNB37) (36) in a few sporadic and familial cases of presbycusis. It has also been suggested that mutations affecting micro ribonucleic acids (miRNAs) (37, 38) and the mitochondrial genome (39, 40) are involved in presbycusis. Together, these studies suggest that presbycusis has a large polygenic component, involving many common genetic variants of small effect size, whereas a few reports have described monogenic or monogenic-like components involving very rare variants with a large effect size.
Studies of common small-effect size variants are generally poorly efficient for achieving a mechanistic understanding of pathogenesis (41) unlike approaches based on large-effect size variants underlying Mendelian transmission. Furthermore, rapid progress has been made toward the development of inner ear gene therapy approaches, which are particularly suitable for treating monogenic forms of hearing impairment. We therefore investigated ultrarare variants as potential large-effect determinants likely to underlie monogenic forms of presbycusis. We used an approach focusing on severe ARHL with no identifiable risk factors or comorbidities. We selected variants in silico, based on both existing functional annotations and standard case–control comparisons of whole-exome sequencing (WES) (42) data, combined with various in silico and experimental functional tests to gain additional evidence of pathogenicity.
Results
Characterization of a Cohort of Multiplex and Simplex Cases of Severe ARHL.
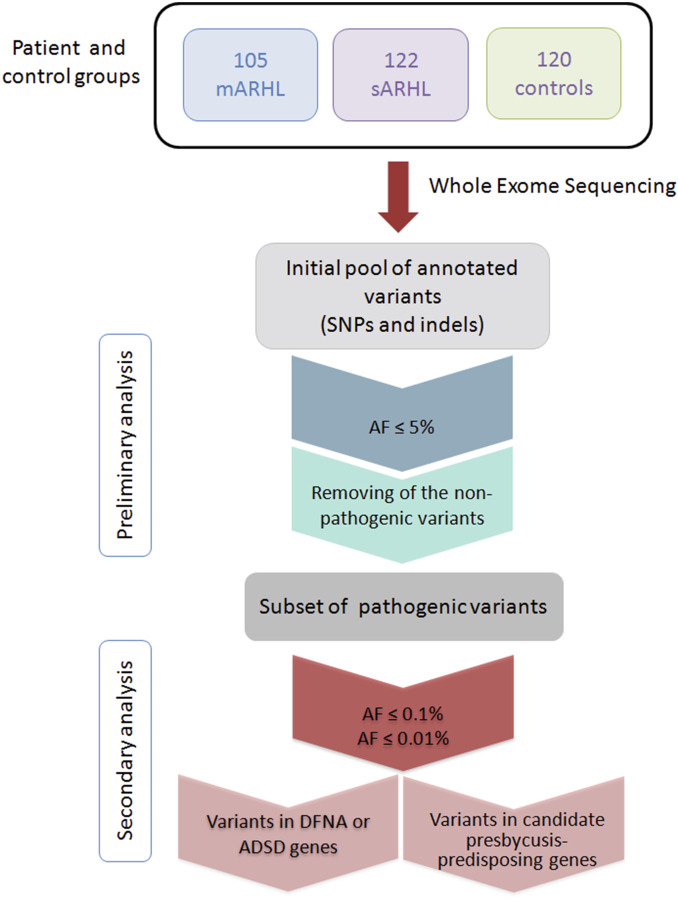
We initiated a collaborative multicenter project (SI Appendix, Patients and Methods) involving the recruitment of affected and control individuals from the French population for investigations of the genetics of severe ARHL (SI Appendix, Table S1). Patients were considered to have severe ARHL if their hearing thresholds exceeded the reference age- and sex-matched thresholds for individuals 20 y older in the general population, according to the pure-tone threshold average of the Bureau International d’Audio-phonologie (http://www.biap.org/fr/) (SI Appendix, Patients and Methods). We further restricted the clinical heterogeneity of patients by enrolling only patients with no known ARHL risk or comorbidity factors (SI Appendix, Patients and Methods and Table S2) self-reporting progressive hearing loss beginning after the age of 40 y. Epidemiological studies indicate that about 8% of individuals in each 10-y age band satisfy these criteria (43). The vast majority of patients and controls (91.3%) were known to have ancestors of French origin only (SI Appendix, Patients and Methods and Table S2). All patients and controls underwent audiological testing. The inclusion and exclusion criteria were checked, and 281 patients were enrolled: 159 multiplex family age-related hearing loss (mARHL) cases from 105 affected families (105 index mARHL cases) with at least two first-degree relatives reported to have severe presbycusis and 122 simplex/sporadic age-related hearing loss (sARHL) cases (SI Appendix, Patients and Methods). We also recruited 120 individuals with normal hearing as controls (Fig. 1 and SI Appendix, Patients and Methods). Most of the individuals in the three groups were aged between 60 and 79 y, and the proportions of men and women did not differ significantly between the various groups (Table 1). The self-reported age at which patients became aware of their hearing loss was 53.1 y in the mARHL group and 54.4 y in the sARHL group (Table 1).
Fig. 1.
Schematic workflow for the identification of genetic variants involved in presbycusis. WES data from mARHL and sARHL cases and from controls were filtered on the basis of AF in the European population: below 5, 0.1, and 0.01%. Only the variants predicted to be pathogenic by CADD, MutationTaster, PolyPhen-2, and SIFT, with a maximal score, were selected for further analysis. Indel, insertion/deletion.
Table 1.
Cases studied
| Cases | No. of independent individuals | N | Sex ratio (M/F) | Mean age (median ± SD) | Mean age at onset (median ± SD) |
| Multiplex (mARHL) | 105 | 159 affected | 77/82 | 65.1 (65 ± 11.2) | 53.1 (52 ± 8.9) |
| Simplex (sARHL) | 122 | 122 affected | 51/70 | 65.49 (66 ± 9.1) | 54.4 (54 ± 10.1) |
| Controls | 120 | 120 with normal hearing | 47/73 | 66.3 (67 ± 11.8) | — |
M/F, sex ratio (male/female); N, total number of individuals studied.
WES Analysis of Severe ARHL and Control Cases.
Genome analysis was performed by WES for all enrolled ARHL cases and controls. We carried out bioinformatic analyses to identify SNPs and insertions/deletions present in the heterozygous or homozygous state. We retained only those with an AF ≤ 0.05 in both the European non-Finnish gnomAD populations and a total of 250,000 exomes (from gnomAD release 2.0, DiscovEHR and Bravo, referred to as gnomAD+ DiscovEHR+ Bravo) (SI Appendix, Patients and Methods). The variants matching these criteria were further sorted to exclude those located in duplicated regions of the genome and highly polymorphic genes, which are known to generate false-positive hits in WES (Fig. 1 and SI Appendix, Patients and Methods). We retained only loss-of-function (LOF) variants (i.e., gain or loss of a stop codon, frameshifts, or splice-site mutations) and missense mutations with the highest pathogenicity scores in all of the following in silico pathogenicity predictor programs: MutationTaster, SIFT (Sorting Intolerant from Tolerant), PolyPhen-2, and CADD (Combined Annotation-Dependent Depletion). All variants predicted to be benign or likely benign by ClinVar and the Deafness Variation Database (DVD; deafnessvariationdatabase.org/) were excluded. We identified a total of 9,970 heterozygous and 108 homozygous variants in index mARHL and sARHL cases (227 patients) and 4785 heterozygous and 65 homozygous variants in normal-hearing individuals (120 controls). None of these variants were recurrent in the patients except the c.101T > C; p.(Met34Thr) variant in GJB2; three sARHL cases carried biallelic GJB2 mutations: one was homozygous for the c.101T > C; p.(Met34Thr) variant, and the other two were compound heterozygous for the c.35delG; p.(Gly12Valfs*2) and c.101T > C; p.(Met34Thr) variants, the pathogenicity of these two variants being well established (44, 45). The prevalence of GJB2 mutations responsible for DFNB1 and DFNA3 is notoriously high in many populations, including the French population (44).
Multiplex Cases of Severe ARHL Carry Ultrarare Mutations of Genes Responsible for Autosomal Dominant Early-Onset Forms of Deafness.
In 100 of the 105 families of the mARHL group (i.e., 95%), ARHL segregation was consistent with an autosomal dominant mode of inheritance. We determined which of the variants with the highest pathogenicity scores (see above) displayed the expected pattern of familial segregation (heterozygous in the index cases and their affected relatives [n = 54]). We detected three very rare variants (0.0001 ≤ AF < 0.001) in three DFNA genes in three unrelated mARHL cases (SI Appendix, Table S3). We then focused on ultrarare predicted pathogenic variants (AF < 0.0001). We identified 28 such variants in 19 DFNA genes and 1 in a gene responsible for an autosomal dominant syndromic form of deafness (ADSD) in 27 unrelated mARHL cases (Table 2). Nine of the 19 DFNA genes had been reported to cause only DFNA forms, whereas the other 10 had been implicated in both DFNA and DFNB forms. Variants of some of these genes (MYO6, MYO7A, PTPRQ, and TECTA) were detected in several unrelated mARHL cases, accounting for about half the index cases (11 of 27) carrying ultrarare variants of DFNA/ADSD genes.
Table 2.
Ultrarare variants of DFNA genes in mARHL cases
| Patient (x) | Gene/protein | Deafness form | Protein activity or function/location | Variants |
| 33029 (3) | CDHR23/cadherin-related 23 | DFNB12/DFNA†/ARSD:USH1D | Ca2+-dependent cell adhesion, component of the MET machinery/IHCs, OHCs | c.5131G > A; p.(Val1711Ile) |
| c.9932C > T; p.(Ser3311Leu) | ||||
| PAR132 | COL11A1/collagen type XI α1-chain | DFNA37/ARSD: STL2 | Extracellular matrix component/tectorial membrane, inner sulcus, Claudius' cells, Boettcher’s cells | c.5401G > T; p.(Gly1801Cys) |
| PAR136 | DIAPH1/protein diaphanous homolog 1 | DFNA1 | Actin nucleation and elongation factor/IHCs and OHCs, inner and outer pillar hair cells, spiral ganglion neurons | c.3128A > G; p.(His1043Arg) |
| PAR089 | ESPN/espin | DFNA/DFNB36 | Multifunctional actin-bundling protein/IHCs, OHCs | c.2515C > A; p.(Pro839Thr) |
| PAR057 | EYA4/eyes absent homolog 4 | DFNA10 | Transcriptional coactivator/cochlear sensory epithelia | c.(84-20_146)_(*2410_?)del |
| PAR032 (3) | GRHL2/grainyhead-like 2 | DFNA28 | Transcription factor/IHCs, OHCs, supporting cells, spiral ganglion neurons | c.931_932del; p.(Asp311*) |
| PAR016 (4) | KCNQ4/voltage-gated K+ channel subfamily KQT member 4 | DFNA2 | K+ channel plasma membrane/OHCs | c.1012C > G; p.(Arg338Gly) |
| PAR047 | c.1316G > A; p.(Arg439His) | |||
| PAR007 (2) | MITF/microphthalmia-associated transcription factor | ADSD:WS2 | Transcription factor/IHCs, OHCs | c.666 + 1G > T |
| 33016 (2) | MYH14/myosin-14 | DFNA4 | Actin-based motor protein/IHCs, OHCs, and the SV, cells lining the cochlear duct (Hensen and Claudius’ cells, external sulcus cells, and spiral prominence epithelial cells) | c.3745G > A; p.(Glu1249Lys) |
| PAR062 | MYO6/unconventional myosin-VI | DFNA22/DFNB37 | Actin-based motor protein/IHCs, OHCs | c.866_869del; p.(Lys289Argfs*17) |
| PAR013 (2) | c.1465C > G; p.(Leu489Val) | |||
| 33006 (2) | c.1926C > A; p.(Ser642Arg) | |||
| PAR010 | c.3791del; p.(Asn1264Metfs*21) | |||
| PAR023 | MYO7A/unconventional myosin-VIIa | DFNA11/DNFB2/ARSD:USH1B | Actin-based motor protein/IHCs, OHCs | c.3750 + 5G > T |
| 33008 | c.3956T > C; p.(Val1319Ala) | |||
| PAR051 | OSBPL2/oxysterol-binding protein-related protein 2 | DFNA67 | Intracellular lipid-binding receptor/IHCs, OHCs, SV, spiral ganglion neurons, and spiral ligament | c.337C > G; p.(His113Asp) |
| PAR031 (2) | PTPRQ/protein tyrosine phosphatase receptor type Q | DFNA73/DFNB84 | Plasma membrane tyrosine phosphatase receptor/IHC and OHC hair bundles | c.1148G > A; p.(Gly383Glu) |
| PAR125 | c.2521C > T; p.(Arg841Trp) | |||
| PAR126 (2) | SCD5/stearoyl-CoA desaturase 5 | DFNA | Integral membrane protein of the ER, catalyzes the formation of monounsaturated fatty acids from saturated fatty acids/IHCs, OHCs, SV, spiral ganglion neurons | c.680G > A; p.(Arg227His) |
| 33002 | TECTA/α-tectorin | DFNA8/12/DFNB21 | Extracellular matrix component/tectorial membrane | c.1376G > T; p.(Gly459Val) |
| PAR070 (2) | c.1951G > A; p.(Gly651Ser) | |||
| PAR054 (2) | c.2670_2671del; p.(Asp891Profs*8) | |||
| PAR079 | TJP2/tight junction protein ZO-2 | DFNA51 | Subplasma membrane protein of tight and adherens junctions/hair cell-supporting cell junctions | c.2428G > A; p.(Val810Met) |
| PAR093 (2) | TMC1/TM channel-like protein 1 | DFNA36/DFNB7/11 | Plasma membrane TM protein, component of the MET channel/IHCs and OHCs | c.980A > T; p.(Asn327Ile) |
| PAR094 | TRRAP/trafficking protein particle complex | DFNA75 | Adapter protein in chromatin complexes/spiral ligament | c.2086C > T; p.(Arg696Cys) |
| PAR009 (2) | WFS1/wolframin | DFNA6/14/38/ARSD:WFS1 | TM protein of the ER; putative ion channel; Ca2+ signaling and ER stress pathways/IHCs, OHCs spiral ganglion neurons | c.2150dup; p.(Ser718Valfs*41) |
x is the number of ARHL-affected individuals studied in each family. CoA, coenzyme A; ER, endoplasmic reticulum; IHC, inner hair cell; STL, Stickler syndrome; SV, stria vascularis; USH, Usher syndrome; WFS1, Wolfram syndrome; WS, Waardenburg syndrome.
Eight of these 28 variants were LOF, and 20 were missense variants. The 28 variants included 16 (57%) that had never been reported (private variants) and had an AF < 0.000002 [based on their absence from gnomAD+ DiscovEH+ Bravo (46); hereafter referred to as UR*]. The 16 UR* comprised seven LOF variants and nine missense variants. One of the 12 nonprivate ultrarare variants (hereafter referred to as URnp) was an LOF variant, and 11 were missense variants. Three were SNPs (SI Appendix, Table S4), and another two had been demonstrated to cause deafness: the c.866_869del; p.(Lys289Argfs*17) frameshift deletion in MYO6 (47) and the c.5131G > A; p.(Val1711Ile) variant of CDHR23 (48). The other seven were missense variants listed as variants of uncertain significance (VUS) in the DVD, as no clear link to hearing impairment had been established (SI Appendix, Table S4).
Although the frequency of variants in the population is an important criterion for their interpretation, additional evidence supporting their pathogenicity can be gleaned from studies of their effect on messenger ribonucleic acid (mRNA) (splicing variants) and protein folding/stability/activity, the coherence between their functional effect and the documented mechanism of their dominance as established in early DFNA and ADSD forms (haploinsufficiency and/or negative dominance), and occasionally, their previously reported causal role in early-onset DFNA and ADSD forms.
We first confirmed the splicing defects of the two splicing variants (Table 2) using hybrid minigenes; the (c.3750 + 5G > T) variant of MYO7A affecting a noncanonical splicing site is shown in SI Appendix, Fig. S1 A, B, D, and F. We then further documented the pathogenicity of the missense UR* variants by exploring their effect on the known three-dimensional (3D) structure/function of some of the encoded proteins. Based on the 3D structure of myosin-VI (Protein Data Bank [PDB] ID code 2V26) (49), the p.(Leu489Val) variant (UR*) affects a key site in the relay domain of the motor head (SI Appendix, Fig. S2A). The transmission of conformation changes, due to adenosine triphosphate (ATP) hydrolysis by the motor domain, to the converter and lever arm would therefore be predicted to decrease myosin motility. Another MYO6 variant, p.(Ser642Arg) (UR*), located in the motor head between the relay domain and the SH1 helix, resulted in the replacement of a short serine side chain by a long arginine side chain, introducing a cationic charge. This would be expected to induce steric clashes between this residue and its neighbors, destabilizing the conformation of the myosin-VI motor head (SI Appendix, Fig. S2B). Based on the 3D structure of KCNQ4 (PDB ID code 6B8P), the p.(Arg338Gly) variant, leading to the replacement of a long charged arginine side chain with a small uncharged glycine side chain, resulted in the loss of hydrogen π/aromatic bonds between the arginine residue and both the phenyl group of the phenylalanine residue in position 549 and the hydroxyl group of the threonine residue in position 552. It would, therefore, be expected to disrupt the packing between helices A (330 to 357) and B (528 to 554), thus greatly modifying the conformation of this channel (SI Appendix, Fig. S3). The ESPN c.2515C > A; p.(Pro839Thr) variant affected a residue located in the actin-binding domain of espin, the 3D structure of which has not yet been established. However, transfection experiments showed that this variant resulted in defective targeting of the coexpressed myosin-IIIa to the tips of the filopodia of LLC-PK1CL4 cells (SI Appendix, Fig. S4).
The pathogenicity of two of the eight missense URnp variants [p.(Asn327Ile) in TMC1 (see further) and p.(His1043Arg) in Diaphanous 1] was further documented. Based on the 3D structure of diaphanous 1 (PDB ID code 1V9D), the variant resulted in the replacement of the imidazole side chain of histidine with a long charged arginine side chain predicted to interact with serine and alanine residues at positions 866 and 962, respectively, with likely effects on the overall folding and stability of the α6-helix in the formin homology-2 domain (SI Appendix, Fig. S5).
We then assessed the causal link between these various monoallelic variants and the auditory phenotype by investigating whether their predicted effects—haploinsufficiency or negative dominance—had already been established for other variants of the same genes responsible for DFNA or ADSD forms. Four variants of EYA4, GRHL2, MYO6, and TECTA led to a nonsense mutation in the first half of the coding mRNA predicted to result in haploinsufficiency (by nonsense-mediated mRNA decay), which has already been implicated in DFNA forms involving these genes. All but two (variants of ESPN and TRRAP) of the missense variants, together with the C-terminal nonsense mutations (MYO6 and WFS1) predicted to result in truncated proteins, were found in genes encoding proteins known to form oligomers with expected dominant negative effects. Furthermore, missense mutations of several of these genes have already been shown to cause DFNA forms through a dominant negative effect (SI Appendix, Table S5). For the splice variant of MITF, either haploinsufficiency or negative dominance could account for the dominance of presbycusis inheritance. For MYO7A, variants displaying negative dominance (but not haploinsufficiency) cause DFNA forms (SI Appendix, Table S5). Consistent with this, the (c.3750 + 5G > T) splice variant led to the skipping of exon 29, resulting in an in-frame deletion of 40 amino acids in the first MyTH4 domain of the myosin tail (SI Appendix, Fig. S1). Overall, the 28 ultrarare variants detected in mARHL cases had characteristics consistent with a causal role in dominant forms of presbycusis.
In summary, ultrarare variants of the causal genes for DFNA and ADSD forms of deafness, with high scores for pathogenicity prediction supported by additional analyses for some of them, were identified in 27% (27/100) of the mARHL cases, all of whom presented hearing loss with a dominant mode of inheritance.
Similar Proportions of mARHL and sARHL Cases Involve Genes Responsible for Autosomal Dominant Early-Onset Forms of Deafness.
We investigated whether sARHL cases carried very rare or ultrarare variants, predicted to be pathogenic, in causal genes for DFNA or ADSD forms. We excluded the three sARHL cases accounted for by biallelic variants of GJB2 and analyzed WES data for the remaining 119 sARHL cases. Using the same filters and pathogenicity criteria as above, we identified two very rare predicted pathogenic variants of two DFNA genes in two patients (SI Appendix, Table S3). We found 31 ultrarare predicted pathogenic variants of 23 different causal genes for dominant forms of deafness, 19 DFNA and 4 ADSD genes, in 27 sARHL cases (i.e., 22.7%; 27 of 119). Four cases carried two ultrarare variants (present in two different DFNA genes in three cases and in two different ADSD genes in one case) (Table 3). Some genes (COL4A3, MYO6, MYO7A, PTPRQ, and TBC1D24) carried ultrarare variants in several sARHL cases. Eight of the 31 ultrarare variants were LOF, and 23 were missense variants. Twelve of these ultrarare variants (38.7%) were UR*: four were LOF, and eight were missense variants. From the 19 variants URnp detected in databases, 4 were LOF, and 15 were missense. Eight of the 19 URnp were SNPs present in the gnomAD database (SI Appendix, Table S4). Two have been reported as causal for deafness: p.(Gly148Val) in COL4A3 (an ADSD gene) for the dominant Alport syndrome 2/3 (50) and p.(Arg991*) in MYO6 for a DFNA form (51). The other nine variants were all located in genes responsible for DFNA forms and listed as VUS in DVD (SI Appendix, Table S4).
Table 3.
Ultrarare variants of DFNA genes in sARHL cases
| Patient | Gene/protein | Deafness form | Protein activity or function/location | Variants |
| 5738 | CCDC50/coiled-coil domain-containing protein 50 | DFNA44 | EGFR signaling/pillar cells, SV | c.167T > G; p.(Leu56Arg) |
| B00APG2 | COCH/cochlin | DFNA9 | Extracellular matrix component/spiral ligament and spiral limbus | c.730C > T; p.(Arg244Cys) |
| DMXL2/Dmx-like protein 2 | DFNA71 | Scaffolding protein of synaptic vesicle membrane/IHCs, OHCs, spiral ganglion neurons | c.1523T > C; p.(Met508Thr) | |
| 6586 | c.6566A > G; p.(Glu2189Gly) | |||
| 4106 | COL11A2/collagen type XI α2-chain | DFNA13/DFNB53 | Extracellular matrix component/tectorial membrane, inner sulcus, Claudius' cells, Boettcher’s cells | c.4540C > T; p.(Arg1514Cys) |
| 5426 | COL2A1/collagen type II α1-chain | ADSD: STL1 | Extracellular matrix component/tectorial membrane | c.4433G > A; p.(Gly1478Asp) |
| EDNRB/endothelin receptor type B | ADSD: WS4 | G-coupled receptor protein and associated signaling pathway/spiral ganglion neurons | c.1147T > A; p.(Tyr383Asn) | |
| 4011 | COL4A3/collagen type IV α3-chain | ARSD/ADSD: ATS 2/3 | Extracellular matrix component/basilar membrane | c.443G > T; p.(Gly148Val) |
| 6590 | c.3691G > A; p.(Gly1231Ser) | |||
| 6673 | c.4648_4649insAC; p.(Val1550Aspfs*10) | |||
| 3870 | DIAPH1/protein diaphanous homolog 1 | DFNA1 | Actin nucleation and elongation factor/IHCs and OHCs, inner and outer pillar hair cells, spiral ganglion neurons | c.1478C > T p.(Thr493Ile) |
| 6708 | DSPP/dentin sialophosphoprotein | DFNA39/ADSD: DGl1 | Extracellular matrix component/IHCs, OHCs | c.776C > T; p.(Ser259Phe) |
| 6403 | KITLG/Kit ligand | DFNA69 | Ligand for the receptor-type protein-tyrosine kinase KIT/unknown location | c.715-2A > G |
| MYH9/myosin-9 | DFNA17 | Actin-based motor protein/IHCs, OHCs | c.5339G > A; p.(Arg1780Gln) | |
| 5784 | MCM2/DNA replication licensing factor | DFNA70 | DNA replication/IHCs, OHCs | c.40C > T; p.(Pro14Ser) |
| 3920 | MYH14/myosin-14 | DFNA4 | Actin-based motor protein/IHCs, OHCs, SV, cells lining the cochlear duct (Hensen and Claudius’ cells, external sulcus cells, and the spiral prominence) | c.5599C > T, p.(Arg1867Cys) |
| 5718 | MYO3A/myosin-IIIA | DFNA/DFNB30 | Actin-based motor protein/IHCs, OHCs | c.3112-2A > G |
| P2RX2/P2X purinoceptor 2 | DFNA41 | ATP-gated cation channel/IHCs, OHCs | c.864dup; p.(Asp289*) | |
| 6505 | MYO6/unconventional myosin-VI | DFNA22/DFNB37 | Actin-based motor protein/IHCs, OHCs | c.1415G > C; p.(Cys472Ser) |
| 6489 | c.2971C > T; p.(Arg991*) | |||
| 6587 | MYO7A/unconventional myosin-VIIa | DFNA11/DNFB2/ARSD: USH1B | Actin-based motor protein/IHCs, OHCs | c.1232T > C p.(Val411Ala) |
| 6706 | c.2335T > C; p.(Arg779Trp) | |||
| 3948 | c.2368-1G > A | |||
| 5413 | PAX3/paired-box protein 3 | ADSD: WS3 | Transcription factor/SV | c.353T > C; p.(Ile118Thr) |
| 3961 | PTPRQ/protein tyrosine phosphatase receptor type Q | DFNA73/DFNB84 | Plasma membrane tyrosine phosphatase receptor/IHCs, OHCs, hair bundle | c.3066C > A; p.(Ser1022Arg) |
| 4392 | c.4286-1G > T | |||
| B00APFU | c.2636del; p.(Leu879Argfs*20) | |||
| 6596† | TBC1D24/TBC1 domain family member 24 | DFNA65/DFNB86 | Putative GTPase-activating protein for Rab proteins, ROS homeostasis/IHCs, OHCs, spiral ganglion neurons | c.734T > C; p.(Leu245Pro) |
| B00APFP | TECTA/α-tectorin | DFNA8/12/DFNB21 | Extracellular matrix component/tectorial membrane | c.907G > A; p.(Glu303Lys) |
| 3974 | TNC/tenascin | DFNA56 | Extracellular matrix component/basilar membrane, osseous spiral lamina of the cochlea | c.2314G > C; p.(Gly772Arg) |
| 5536 | TRRAP/trafficking protein particle complex | DFNA75 | Adapter protein in chromatin complexes with histone acetyltransferase activity/spiral ligament | c.5478C > A; p.(Asp1826Glu) |
ATS, Alport syndrome; DGI, dentogenesis imperfecta syndrome; EGFR, epidermal growth factor receptor; GTPase, guanosine triphosphatase; IHC, inner hair cell; ROS, reactive oxygen species; STL, Stickler syndrome; SV, stria vascularis; USH, Usher syndrome; WS, Waardenburg syndrome.
This patient also carried a heterozygous mutation in TBC1D24 (c.118C > T; p.Arg40Cys) that has been implicated in recessive DOORS (deafness, onychodystrophy, osteodystrophy, mental retardation, and seizures) syndrome.
We validated the effect of the four splicing variants by a minigene strategy. The c.2368-1G > A splicing variant of MYO7A leads to the skipping of exon 21, resulting in the deletion of isoleucine-glutamine (IQ) domains 3 to 5 (SI Appendix, Fig. S1 A, C, E, G, and H). This deletion abolishes the apocalmodulin-binding site, an effect similar to that proposed for the p.(Arg853Cys) mutation (affecting the fifth IQ domain) in a family with an early-onset form of dominant deafness. The p.(Cys472Ser) substitution in MYO6, located in the motor head according to the 3D structure (PDB ID code 2V26), may alter the strong thiol S-H/π phenylalanine interaction, with expected effects on the conformation/stability of this domain. Indeed, the cysteine 472 residue interacts with the phenylalanine 647 residue, these two residues being located in spatially close helices from the motor head (SI Appendix, Fig. S2B). We validated the pathogenicity of this mutation by transfection experiments in HEK cells, showing that the level of expression of this MYO6 variant and the subcellular distribution of the myosin-VI it encodes both differed from those of normal myosin-VI (SI Appendix, Fig. S6). We checked all of the proteins encoded by DFNA/ADSD carrying variants for posttranslational modifications in ARHL cases and found that the p.(Ser259Phe) variant of dentin sialophosphoprotein (DSPP) affected the serine 259 residue (equivalent to the serine 265 residue in the porcine ortholog) acting as a glycan attachment site (52).
We then investigated the causal link between the ultrarare monoallelic variants and the auditory phenotype, as above (SI Appendix, Table S5). All but two variants (in MYO3A and P2RX2 genes carried by the same patient) were present in genes for which haploinsufficiency or dominant negative mutations are known to lead to a dominant form of deafness (SI Appendix, Table S5) and could, therefore, be causal for a dominant form of presbycusis.
As sARHL cases may carry recessive mutations, we performed an in silico analysis searching for homozygous or compound heterozygous ultrarare predicted pathogenic variants of deafness genes responsible for autosomal recessive forms of hearing impairment (DFNB or autosomal recessive syndromic deafness [ARSD]). No homozygous or compound heterozygous ultrarare variants of any of these genes were found in the sARHL group, with the exception of the biallelic mutations of GJB2 present in three sARHL cases. Moreover, in sARHL patients carrying ultrarare or very rare monoallelic variants, we detected no exons of the genes concerned lacking heterozygous polymorphisms suggestive of exon deletion and thus, of autosomal recessive forms of presbycusis.
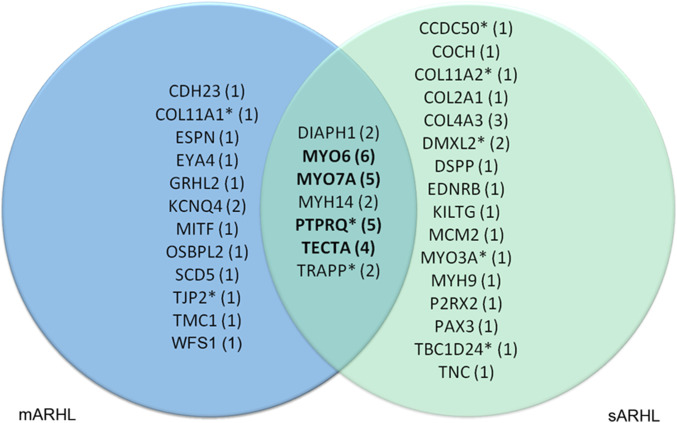
Altogether, we identified ultrarare predicted pathogenic variants of causal genes for DFNA or ADSD in 22.7% of the sARHL patients (27 of 119), a proportion similar to that in mARHL cases (25.7%). Seven DFNA genes (DIAPH1, MYH14, MYO6, MYO7A, PTPRQ, TECTA, and TRRAP) carried ultrarare predicted pathogenic variants in both mARHL and sARHL cases (Fig. 2). Remarkably, predicted pathogenic variants of four of these DFNA genes (MYO6, MYO7A, PTPRQ, and TECTA) accounted for more than one-third (38.5%) of ARHL cases (20/52) carrying such variants of DFNA or ADSD genes (Fig. 2 and SI Appendix, Table S4).
Fig. 2.
Distribution of ultrarare variants of DFNA and ADSD genes in mARHL and sARHL cases. Venn diagram of the variants (number in parentheses) present in DFNA and ADSD genes in sARHL (green) and mARHL (blue) cases. The variants in the intersection correspond to genes affected in both mARHL and sARHL cases. Variants of the genes in bold accounted for one-third of mARHL cases. *An ultrarare variant of the corresponding gene was also detected in the control group (SI Appendix, Table S5).
We then analyzed the WES data for the 120 controls with the same in silico prediction tools. We found a single very rare predicted pathogenic variant of DFNA gene (listed as VUS in DVD) in one individual (SI Appendix, Table S3). We detected 12 ultrarare predicted pathogenic variants of DFNA genes in nine individuals (SI Appendix, Tables S4 and S6). The 12 variants were missense variants. Three of these variants were UR*. The other nine variants were URnp and comprised eight variants listed as VUS in DVD and one SNP present in the gnomAD database. Three controls carried two variants, one in the same DFNA gene and two in two different DFNA genes. The proportion of individuals carrying ultrarare variants of genes responsible for dominant forms of deafness was significantly lower in the controls (7.5%; 9/120 individuals) than in mARHL (25.7%) and sARHL (22.7%) cases (Fisher’s exact test: P [control/mARHL] = 0.002 and P [control/sARHL] = 0.006, respectively). From the contingency table of mARHL and sARHL as against controls that recapitulates the aforementioned numbers of cases with and without ultrarare variants of interest, it is possible to compute an attributable risk of severe ARHL relating to the presence of these variants. Using standard approximations for a rare disorder (prevalence <10%) described by Taylor in 1977 (53) and Kuritz and Landis in 1988 (54), the estimation of this attributable risk falls roughly around 18%.
Remarkably, ultrarare variants of MYO6, MYO7A, PTPRQ, and TECTA were identified in 8.9% of mARHL and sARHL cases but in only one control (i.e., <1% of the controls; Fisher’s exact test: P [control/mARHL + sARHL] = 0.003) (Fig. 2). URnp or UR* LOF variants concluded to be causal for ARHL based on documented dominant effect of haploinsufficiency or dominant negative effect in the corresponding genes accounted for 15 of the 54 variants (28%); they were present in 15 of 224 ARHL cases but in none of the 120 controls (P = 0.003). Additional evidence supporting the pathogenicity of URnp and UR* variants (variants previously reported to be causal for early-onset dominant deafness and the pathogenicity of variants deduced from 3D structure/function relationships or posttranslation modification, cell biology experiments, and recombinant mice [see below]) was obtained in 10 other ARHL cases. Thus, about half the ARHL cases (25/54) were found to carry an ultrarare variant of DFNA/ADSD genes with pathogenicity documented by complementary approaches.
miRNAs and Mitochondrial Mutations in ARHL Cases.
Several miRNAs have been implicated in ARHL, including microRNA (miR)-34a and miR-29b, involved in the miR-34a/SIRT1/p53 and miR-29b/SIRT1/PGC1α pathways (37, 38). We identified no variants of genes encoding ARHL-related miRNAs. Mitochondrial genome mutations have also been implicated in hearing impairment, including presbycusis (39, 40, 55). Whole-mitochondrial genome sequencing identified a single mutation, m.1555A > G, in the MT-RNR1 (mitochondrially encoded 12S ribosomal RNA) gene, conferring susceptibility to aminoglycoside ototoxicity and isolated deafness (56). This mutation was detected in 1 of the 27 mARHL cases displaying ARHL transmission consistent with maternal inheritance. One control also carried the m.1555A > G mutation. In the mARHL case, but not the control, the m.1555A > G mutation was associated with an ultrarare predicted pathogenic variant of a DFNA gene: p.(Leu489Val) in MYO6 (SI Appendix, Fig. S2A). One sARHL case carried the m.7444G > A mutation of the MT-TS1 gene (transfer RNA serine [tRNASer]), which has been implicated in myopathy-associated deafness (57), in addition to an ultrarare predicted pathogenic variant [p.(Ser546Phe)] of PHLDB1 (or LL5A), a CPP gene encoding pleckstrin homology-like domain family B member 1 (30) (SI Appendix, Table S7). In these two ARHL cases, the mitochondrial mutations may exacerbate the effect of ultrarare nuclear gene variants, as both myosin-VI and PHLDB1 have adenosine triphosphatase-dependent activity.
TMC1 Is a Presbycusis Gene.
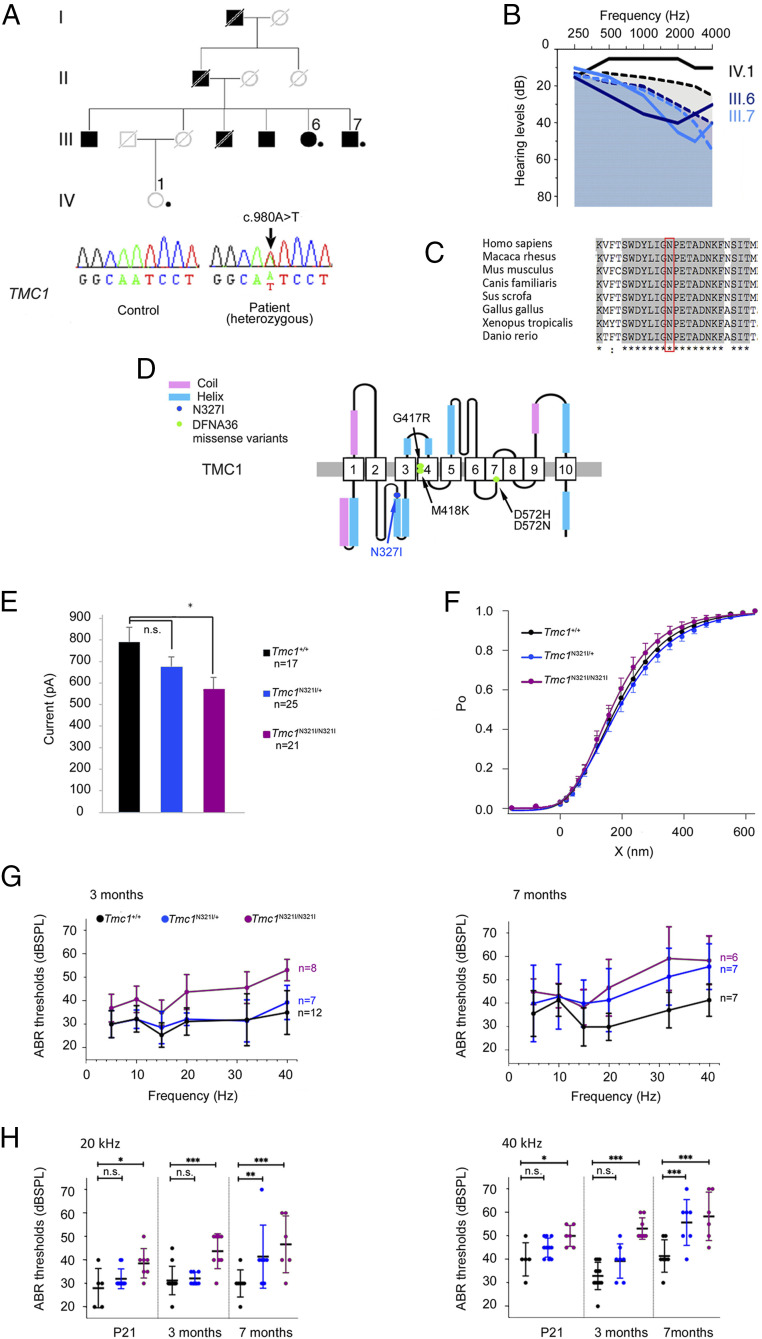
The best way to prove that ultrarare variants of DFNA and ADSD genes underlie Mendelian forms of presbycusis, due to the lack of significant linkage for such variants, is to determine whether the variant concerned underlies Mendelian forms of presbycusis in a mammalian animal model. We focused on the TMC1 variant [c.980A > T; p.(Asn327Ile)] detected in the heterozygous state in one mARHL case (Fig. 3 A and B) and previously classified as a VUS in DVD (Table 2) for the following reasons. 1) Transmembrane channel-like protein 1, TMC1, is an essential component of the basal part of the mechanoelectrical transduction (MET) machinery of the sensory hair cells, and there is growing evidence to suggest that it is a component of the MET channel (58, 59), making it possible to test the physiological effect of the variant by ex vivo MET current recording. 2) Only four variants have been described as responsible for DFNA36 (60): two are neighboring mutations, c.1249G > A; p.(Gly417Arg) and c.1253T > A; p.(Met418Lys), the latter of which is equivalent to the p.(Met412Lys) mutation underlying the deafness phenotype in Beethoven mice, whereas the other two, c.1714G > A; p.(Asp572Asn) and c.1714G > C; p.(Asp572His), affect the same amino acid residue. Based on the predicted structure of TMC1, these four mutations alter an ion permeation pathway of the MET channel (61), and probably the pore channel (59), whereas the c.980A > T; p.(Asn327Ile) variant affects a highly conserved asparagine residue in the first cytoplasmic loop between transmembrane (TM) domains TM2 and TM3 (59) (Fig. 3 C and D), a region of the protein of unknown function, casting doubts on the importance of its role.
Fig. 3.
Functional validation of the Tmc1N321I mutation as responsible for ARHL. (A) Pedigree of the PAR093 family (dots indicate individuals included in the genetic analysis) and DNA sequencing chromatograms of unaffected (Lower Left) and affected (Lower Right) individuals showing the variant in the heterozygous state in the ARHL patient (arrow). Nucleotide positions are based on the NM_138691.2 transcript used as a reference. (B) Audiograms of PAR093 family members: individuals with ARHL (III.6 and III.7: 63 and 59 y old, respectively) and an individual with normal hearing (IV.1: 51 y old). Dashed lines show the hearing threshold defining severe ARHL in age- and sex-matched members of the general population (in black for 51-y-old women, in dark blue for 63-y-old women, and in light blue for 59-y-old men) (SI Appendix, Patients and Methods). (C) Interspecies conservation of the asparagine residue in position 327 (boxed in red) through evolution. (D) Schematic diagram of the predicted human TMC1 protein with 10 TM domains (1 to 10), according to Pan et al. (59). (E–H) Results from Tmc1 mutant mice (in blue, Tmc1N321I/+; in purple, Tmc1N321I/N321I; in black, Tmc1+/+). (E) MET current amplitude of OHCs from Tmc1+/+ (n = 17), Tmc1N321I/+ (n = 25), and Tmc1N321I/N321I (n = 21) mice recorded on P8. The amplitude (mean ± SD) of the MET current in response to stimulation is significantly lower in cells from Tmc1N321I/N321I mice than in cells from Tmc1+/+mice. (F) MET current as a function of hair bundle displacement for Tmc1+/+, Tmc1N321I/+, and Tmc1N321I/N321I mice. For sensitivity measurements, the mean value (± SD) of the three-state derivative of the Boltzmann equation was calculated for displacements corresponding to Po values between 0.2 and 0.8. The Po(X) curves were superimposed, indicating that MET sensitivity to hair bundle displacement is unaffected in Tmc1N321I/+ and Tmc1N321I/N321I mutants. (G) Pure tone-evoked ABR thresholds (mean ± SD) in Tmc1+/+, Tmc1N321I/+, and Tmc1N321I/N321I mice at 3 (Left) and 7 mo (Right). (H) ABR thresholds (mean ± SD) at 20 kHz (Left) and 40 kHz (Right) in mice aged P21 (Tmc1+/+, n = 5; Tmc1N321I/+, n = 10; Tmc1N321I/N321I, n = 7), 3 mo (Tmc1+/+, n = 12; Tmc1N321I/+, n = 7; Tmc1N321I/N321I, n = 8), and 7 mo (Tmc1+/+, n = 7; Tmc1N321I/+, n = 7; Tmc1N321I/N321I, n = 6). n.s., nonsignificant; dBSPL, décibel sound pressure level. *P < 0.05; **P < 0.005; ***P < 0.0005.
The affected members of the family concerned, III.6 and III.7, carried the variant, which was absent from IV.1, who had a normal hearing threshold (Fig. 3 A and B). We generated Tmc1N321I/N321I mutant mice carrying Tmc1:c.962A > T; p.(Asn321Ile), the same mutation as in humans p.(Asn327Ile), in the FVB/NRJ background, which lacks the Cdh23ahl allele responsible for ARHL (SI Appendix, Patients and Methods and Fig. S7).
An analysis of MET currents based on patch-clamp electrophysiological recordings on postnatal day 8 (P8) revealed current amplitudes in outer hair cells (OHCs) 27% lower in Tmc1N321I/N321I mice than in Tmc1+/+ mice (Tmc1N321I/N321I [n = 21]: 571 ± 249 pA; Tmc1+/+ [n = 17]: 791 ± 274 pA, unpaired t test, P = 0.014) (Fig. 3E), with no difference in the mean sensitivity of this current to hair bundle displacement (Fig. 3F). Auditory brainstem responses (ABRs) to bursts of pure tones (5 to 40 kHz) showed that ABR thresholds across all frequencies were higher in Tmc1N321I/N321I mice than in Tmc1+/+ mice from P21 onward (the time of the first evaluation) (Fig. 3 G and H). On P21, threshold values reached 40 ± 0.94 dB in Tmc1N321I/N321I mice (n = 8) vs. 32 ± 2 dB in Tmc1+/+ mice (n = 5) at 5 kHz (mean threshold ± SEM; t test, P = 0.0018) and 49 ± 1.8 dB in Tmc1N321I/N321I mice and 40 ± 3.16 dB in Tmc1+/+ mice at 40 kHz (t test, P = 0.025). By the age of 3 mo, threshold responses to high frequencies (40 kHz) were more strongly affected: 53 ± 1.6 dB in Tmc1N321I/N321I mice (n = 8) vs. 35 ± 2.6 dB in Tmc1+/+ mice (n = 12); t test, P < 0.0001 (Fig. 3G). At the age of 7 mo, the ABR threshold reached 58 ± 4.2 dB at 40 kHz in Tmc1N321I/N321I mice (n = 6) vs. 41 ± 2.6 dB in Tmc1+/+ mice (n = 7; P < 0.0001) (Fig. 3G). By contrast, the hearing threshold in Tmc1+/+ mice remained constant.
In Tmc1N321I/+ mice, neither the maximal amplitude of the MET current (677 ± 218 pA in Tmc1N321I/+ mice [n = 25] and 791 ± 274 pA in Tmc1+/+ mice [n = 17], unpaired t test, P = 0.144) nor its sensitivity were significantly affected on P8 (Fig. 3 E and F). Until the age of 3 mo, the ABR thresholds of Tmc1N321I/+ mice and Tmc1+/+ mice were similar (30 ± 2.2 dB in Tmc1N321I/+ mice [n = 7] and 30 ± 1.6 dB for Tmc1+/+ mice [n = 12] at 5 kHz; 39 ± 2.8 dB in Tmc1N321I/+ mice and 35 ± 2.6 dB in Tmc1+/+ mice at 40 kHz; unpaired t test, P > 0.05) (Fig. 3 G and H). However, at the age of 7 mo, substantial differences in ABR thresholds were observed between Tmc1N321I/+ and Tmc1+/+ mice at high frequencies (ABR thresholds: 41.4 ± 5.1 dB in Tmc1N321I/+ mice [n = 7] vs. 30 ± 2.2 dB in Tmc1+/+ mice [n = 7] at 20 kHz, t test, P < 0.005; 56 ± 3.7 dB in Tmc1N321I/+ mice [n = 7] vs. 41 ± 2.6 dB in Tmc1+/+ mice [n = 7] at 40 kHz, t test, P = 0.0005) (Fig. 3H). At this age, hearing thresholds at 40 kHz were similar in Tmc1N321I/+ and Tmc1N321I/N321I mice. Microscopy studies showed neither abnormalities of the hair bundles nor losses of spiral ganglion neurons in Tmc1N321I/+ and Tmc1N321I/N321I mice (SI Appendix, Fig. S8). These results demonstrate that the Tmc1:c.962A > T; p.(Asn321Ile) variant leads to a late-onset form of progressive dominant deafness more pronounced at high frequencies.
Variants of CPP Genes in Cases of Severe ARHL.
The implication of the same genes in both early-onset dominant deafness and severe presbycusis suggests a breakdown of the genetic boundaries between early-onset forms of deafness and presbycusis. We thus considered a potential further extension of this situation: the possibility that some CPP genes cause monogenic forms of severe ARHL. Consistent with this hypothesis, two CPP genes, ILDR1 and TRIOBP, are responsible for congenital monogenic recessive forms of deafness (DFNB42 and DFNB28, respectively) but have not been implicated in DFNA forms. We searched for ultrarare and very rare predicted pathogenic variants in the 76 CPP genes reported as presenting an indicative or a significant association with presbycusis (SI Appendix, Table S7). Using the same in silico prediction tools as above, we identified a total of 58 variants in the mARHL (SI Appendix, Table S8A), sARHL (SI Appendix, Table S8B), and control (SI Appendix, Table S8C) groups. Ultrarare CPP variants predicted to be pathogenic were detected in 9.5% of mARHLs, 14.7% of sARHLs, and 10.9% of controls, and very rare variants were detected in 2.8% of mARHLs, 7.3% of sARHLs, and 4.1% of controls (SI Appendix, Table S8). These results do not support a causal role of these variants in monogenic forms. We further analyzed these variants, focusing on those neither associated with ultrarare variants of DFNA or ADSD genes in ARHL cases nor located in genes not carrying an ultrarare predicted pathogenic variant in controls. One such variant, an LOF variant of SLC28A3 in an sARHL case, raised the possibility of severe presbycusis being due to haploinsufficiency of this gene. We also found ultrarare predicted pathogenic missense variants of CPP genes (AGBL2, CLRN2, NAT2, SLC28A3, and ZNF318 in seven mARHL cases and ACADVL, DCLK1, GRM7, GRM8, ILDR1, SLC28A3, and ZNF318 in seven sARHL cases), which are also attractive candidate causal genes for monogenic forms of presbycusis (four of these variants were UR*) (SI Appendix, Fig. S9 and Table S9). Interestingly, for two of these CPP genes, ZNF318 and SLC28A3, ultrarare variants were found in both the mARHL and sARHL groups. The inclusion of CPP genes carrying very rare predicted pathogenic variants in both sARHL and mARHL cases extended the list of candidate causal genes for Mendelian forms of presbycusis to ILDR1 and AGBL2 (SI Appendix, Fig. S9 and Tables S8 A and B).
Discussion
Attempts to decipher the genetic landscape of presbycusis are increasingly focusing on its polygenic component through GWAS analyses, the statistical power of which is steadily increasing with the growing size of the cohorts of patients studied. While GWASs will continue contributing to the characterization of presbycusis genetic risk, they have some limitations. In particular, they identify common variants spread across the genome, each associated with a modest effect on presbycusis risk. As a result, the identification of these variants generally provides limited information about the underlying pathogenic process. Moreover, GWAS variants are usually not causal ones but are tagging uncharacterized causal variants. Here, we investigated the putative monogenic component of presbycusis. The methodological requirements of this approach differ substantially from those for GWAS. Far fewer patients and controls are required, and unlike GWAS, this approach focuses on ultrarare variants because the probability of pathogenicity is highest for such variants. Some of the concerns of GWAS, such as matching for ethnicity, also vanish for those of the ultrarare variants that are private variants (46). Moreover, ultrarare pathogenic variants are causal and provide direct insight into the underlying pathogenic mechanisms. However, demonstrating the pathogenicity of such variants, especially missense variants, remains highly challenging, particularly for late-onset disorders, given the limited family information available.
By analyzing the coding genome, together with exon-flanking sequences and noncoding exons, we were able to show that ultrarare (AF ≤ 0.0001) pathogenic variants of known causal genes for early-onset dominant deafness, DFNA and ADSD, underlie severe forms of presbycusis. About half these variants were absent from all databases, including the DVD (AF < 0.000002). We combined several lines of evidence to obtain an accurate interpretation of these variants. First, only the 59 ultrarare variants with the highest possible score for each of the four pathogenicity prediction programs used were taken into account. Second, for some of these variants, causality in hearing loss could be deduced from previous studies. Some of these variants had already been reported to be responsible for early-onset forms of dominant deafness. For all variants expected to result in haploinsufficiency, it was possible to infer their functional impact from similar types of variants of the same genes reported to be responsible for early-onset dominant deafness. The pathogenicity of predicted truncating variants was also inferred from similar variants located in the immediate vicinity that had been reported to cause early-onset dominant deafness. Third, the effect of these variants, particularly the missense variants, on the encoded proteins could be documented from the 3D structure/function relationship or posttranslational modifications. Furthermore, in vitro transfection experiments revealing abnormalities in the level of the protein or its subcellular distribution provided support for variant pathogenicity. Finally, the pathogenicity and monogenic inheritance of the ARHL are best demonstrated in vivo in animal models; the TMC1 [c.980A > T; p.(Asn327Ile)] variant, for example, was shown to be responsible for a monogenic form of ARHL, when present in the heterozygous state, in mice. Further support for the causality of the detected ultrarare variants of genes responsible for early-onset dominant deafness in presbycusis is provided by their frequency in ARHL cases: 25.7 and 22.7% of mARHL and sARHL cases, respectively, significantly higher than their frequency in normal-hearing individuals (7.5% of control cases). Their gene distributions also differed significantly between ARHL patients and controls, ultrarare variants of MYO6, MYO7A, PTPRQ, and TECTA, the genes most frequently implicated in ARHL, being present in 8.9% of ARHL cases but in less than 1% of controls. Overall, our results provide strong support for the existence of monogenic forms of severe presbycusis. They also indicate that half the nonprivate ultrarare variants previously classified as VUS in DVD (16/31 URnp, carried by 14 DFNA/ADSD genes in ARHL cases) (62–64) should be reclassified as likely pathogenic.
Another WES study reported new variants of early-onset deafness genes detected in 10 sporadic ARHL cases (aged from 60 to 79 y) (65). By contrast, we studied more than 220 ARHL cases (about half of which were multiplex cases) and specifically, severe forms. Furthermore, our study is a case–control study, including 110 phenotyped and age-matched controls with normal hearing, a format required for the interpretation of the variants observed in ARHL cases.
Our results establish that variants of numerous deafness genes lead to a phenotypic continuum of Mendelian forms of hearing impairment encompassing congenital deafness (mostly DFNB forms), early-onset forms of dominant deafness, and severe forms of presbycusis. This continuum can be accounted for by the heterogeneity of the variants, as exemplified by the variants of TMC1. The four TMC1 mutations previously reported to be responsible for dominant hearing loss displayed a postlingual early onset, with high frequencies initially affected (60, 66). However, the initial degree of hearing loss at high frequencies and the rate of its progression differ between variants, ranging from slow progression for carriers of the c.1249G > A; p.(Gly417Arg) variant to much faster progression for carriers of c.1714G > A; p.(Asp572Asn) and c.1714G > C; p.(Asp572His) (60). Nevertheless, all these variants lead to severe to profound hearing loss, affecting all frequencies before the age of 15 y, with a limited sparing of the lowest frequencies. According to recent models, Tmc1 is predicted to form a dimer (58), with each protomer containing a large cavity formed from the helices of the TM domains TM4 to TM7: TM4 containing the first two mutations [p.(Gly417Arg) and p.(Met418Lys)] and TM7 the last two [p.(Asp572His) and p.(Asp572Asn)] (59, 61). Based on the same model, the c.980A > T; p.(Asn327Ile) variant affects the cytoplasmic loop extending between TM2 and TM3, immediately upstream from the α-helix of this loop. By contrast to the other four DFNA mutations, no function, molecular interaction, or deleterious missense mutation has yet been associated with this loop, which is not conserved in the TMEM16 proteins used to generate the models of TMC1 structure (59, 61). The hearing loss resulting from the p.(Asn327Ile) variant in the heterozygous state also preferentially affects high frequencies, but it differs markedly from that resulting from the other four dominant variants: it has a later onset, between the ages of 40 and 50, and is less severe. By contrast, our finding, in ARHL cases, of ultrarare variants previously reported in early-onset forms of dominant deafness (three such variants in total) indicates that these variants alone cannot account for differences in auditory phenotype. For example, the MYO6 [p.(Arg991*)] variant, detected here in the heterozygous state in an sARHL case, was previously reported to cause a dominant form of mild to moderate hearing loss with onset during the teenage years (51). Similarly, the GJB2:c.101T > C; p.(Met34Thr) variant detected here in the homozygous state in an sARHL case and the c.101T > C; p.(Met34Thr)/c.35delG; p.(Gly12Valfs*2) variants found here in two sARHL cases were previously reported to underlie moderate hearing loss beginning before the age of 10 y (67–69). This suggests that effective early protective or compensatory mechanisms were involved in these ARHL cases or that aggravating factors of genetic or environmental origin are associated with early-onset forms.
Two-thirds of the genes (23 of 35) identified here as carrying ultrarare predicted pathogenic variants in ARHL cases encode proteins playing crucial roles in the inner hair cells and OHCs. These genes display particular enrichment in genes encoding myosins (MYO6, MYO7A, MYH14, MYO3A, and MYH9), ion channels (KCNQ4, TMC1, and WFS1), and transcription factors or coactivators (GRHL2, MITF, PAX3, and EYA4). Variants of several genes encoding components of extracellular matrices, the tectorial (TECTA, COL11A1, COL11A2, and COL2A1) and basilar (COL4A3) membranes, were also identified. Four genes (DIAPH1, EDNRB, GRHL2, and TBC1D24) are also expressed in the auditory neurons. Variants of DFNA/ADSD genes involved in ionic (EYA4, KCNQ4, WFS1), metabolic (KITLG, WFS1), and/or redox (TBC1D24, WFS1) homeostasis in the cochlea were fewer in number (Tables 2 and 3). The contribution of genes involved in redox homeostasis is probably underestimated, as these genes are underrepresented among the causal genes for early-onset DFNA forms.
It is not surprising that variants of some DFNA/ADSD genes have deleterious late effects on hearing. This is particularly true for genes for which defects enhance cochlear fragility to mechanical stimulation or metabolic stress, both of which are likely to be aggravated by long-term sound stimulation. Aging processes may also potentiate the effect of some ARHL-causing variants. Recent efforts to decipher common aging processes have demonstrated the importance of genome instability, telomere attrition, epigenetic alterations, losses of proteostasis, and mitochondrial dysfunction (70). By decreasing the ATP supply, mitochondrial dysfunction would be expected to aggravate the functional defects caused by mutated proteins with ATP-dependent activity. This effect is expected to apply, in particular, to myosins, the most prominent class of proteins encoded by variant-positive DFNA/ADSD genes in ARHL cases. Moreover, by increasing reactive oxygen species production, mitochondrial dysfunction is also expected to reduce tolerance to oxidative stress (71, 72), whatever its cause. Finally, mitochondrial ATP production is required for the generation of endocochlear potential (EP), the driving force impelling endolymphatic K+ ions through the MET channels, and there is compelling evidence to suggest that EP decreases with aging (73). Such effects on EP may be additive with those of variants in genes encoding MET components, potentially exacerbating the effect of TMC1 c.980A > T; p.(Asn327Ile) on hearing (74).
The finding of a similar proportion of ultrarare pathogenic variants of DFNA/ADSD genes present in the heterozygous state in the sARHL and mARHL groups (22.7 and 25.7%, respectively) is intriguing. Although de novo mutations in sARHL cannot be excluded in the absence of analyses of the parents’ DNA, this suggests masked heritability in the sARHL group. An absence of family history, possibly due to the early death of the proband’s parents, small family size, or a lack of awareness of the hearing status of relatives, may partly account for this result. The lower penetrance or expressivity of the ultrarare variants of sARHL than of mARHL cases may also account for masked heritability. However, the distribution of variant types (predicted LOF vs. missense variants) provided no support for a lower expressivity of the sARHL variants (70.3 and 74% of the ultrarare variants were missense variants in the mARHL and sARHL groups, respectively). The involvement of additional genetic or epigenetic factors would also provide a plausible explanation for lower expressivity. In this respect, it is worth noting that four sARHL cases carried two ultrarare variants in two different deafness genes (Table 3), a situation not observed in mARHL cases and not explained by the method used to capture the deafness gene variants in the index case (no changes in the identified DFNA/ADSD gene variants were introduced as a result of the comparison of WES from index mARHL cases and related affected family members). In these sARHL cases, the absence of heritability may be accounted for by the contribution of two ultrarare variants to the phenotype. Based on current knowledge of the functions of the proteins encoded by these genes, the variants associated in individual ARHL cases are involved in different cellular pathways, suggesting a cumulative effect of the resulting dysfunctions on the auditory phenotype, rather than a synergistic effect in a given cellular pathway or function.
The finding that a substantial fraction of severe presbycusis cases is due to monogenic defects caused by ultrarare variants reveals the genetic landscape of presbycusis to consist of both common variants with individually small impacts on phenotype and ultrarare variants that are particularly deleterious. These results, which require replication in other populations, should stimulate efforts worldwide to improve the functional validation of ultrarare predicted pathogenic variants. Accurate interpretation of the ultrarare variants is, indeed, a critical challenge in the translation of personal genomic data into precision medicine and is particularly difficult for missense variants. Protein analyses are currently underexploited. However, characterization of the 3D structure/function of the proteins encoded by these genes, posttranslational modifications, and protein–protein interactions, together with the associated signaling pathways, are steadily improving; they have the potential to make a major contribution to the assessment of variant pathogenicity (75) and thus, to foster the development of a reference framework of standardized methods.
The results presented here should also prompt searches for new genes underlying Mendelian forms of severe presbycusis; CPP genes should be considered, even though the proportion of LOF and UR* variants of these genes in ARHL cases is lower than that for DFNA genes. These results should also encourage evaluations of the respective contributions of ultrarare variants and common variants to the various forms of presbycusis. In this perspective, much more detailed audiological analyses than have been carried out to date should be performed (very subtle associated features in ARHL cases caused by ultrarare variants in ADSD genes should be investigated). Moreover, polygenic risk scores (derived from genome-wide analyses on large datasets) for ARHL should be developed, as already done for a number of common diseases.
Finally and above all, our findings are important with a view to treatment; the contribution of monogenic forms to severe presbycusis opens up prospects for the alleviation or cure of severe presbycusis by gene therapy.
Patients and Methods
A detailed description of the materials and methods (patients, inclusion and exclusion criteria, auditory tests, WES, animal experiments, and splicing mutation validation) is presented in SI Appendix, Patients and Methods. This study was performed in accordance with the Declaration of Helsinki and institutional guidelines, with the approval of the local ethics committee (Comité de Protection des Personnes dans la Recherche Biomédicale). Informed consent was obtained from all participants before the clinical examination and genetic testing. Animal experiments were performed in accordance with French and European guidelines for the protection of animals used for scientific purposes, with the approval of the animal ethics committee of the Pasteur Institute (project authorization Committee for Ethics in Animal Experimentation: 2014-0005, reference no. 02440.02).
Supplementary Material
Acknowledgments
We thank the patients for participating in this study and Céline Trébeau for technical assistance. S.B. received funding from the University of Angers (Medical School), the University Hospital of Angers, and the Collège Français d’oto-rhino-laryngologistes. This work was supported by a grant from Fondation pour l’Audition (to C.P.), LabEx Lifesenses Grant ANR-10-LABX-65, and Light4deaf Grant ANR-15-RHUS-0001.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010782117/-/DCSupplemental.
Data Availability.
All of the relevant data are available from the article and SI Appendix.
References
- 1.World Health Organization , Addressing the rising prevalence of hearing loss (2018). https://apps.who.int/iris/handle/10665/260336. Accessed 13 November 2020.
- 2.Gates G. A., Mills J. H., Presbycusis. Lancet 366, 1111–1120 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Anderson S., Parbery-Clark A., Yi H.-G., Kraus N., A neural basis of speech-in-noise perception in older adults. Ear Hear. 32, 750–757 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin F. R., Albert M., Hearing loss and dementia—who is listening? Aging Ment. Health 18, 671–673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deal J. A., et al. , Hearing treatment for reducing cognitive decline: Design and methods of the aging and cognitive health evaluation in elders randomized controlled trial. Alzheimers Dement. (N. Y.) 4, 499–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fortunato S., et al. , A review of new insights on the association between hearing loss and cognitive decline in ageing. Acta Otorhinolaryngol. Ital. 36, 155–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livingston G., et al. , Dementia prevention, intervention, and care. Lancet 390, 2673–2734 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Carniel C. Z., et al. , Implications of using the hearing aids on quality of life of elderly. CoDAS 29, e20160241 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Picou E. M., MarkeTrak 10 (MT10) survey results demonstrate high satisfaction with and benefits from hearing aids. Semin. Hear. 41, 21–36 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Eyken E., Van Camp G., Van Laer L., The complexity of age-related hearing impairment: Contributing environmental and genetic factors. Audiol. Neurotol. 12, 345–358 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Akinpelu O. V., Mujica-Mota M., Daniel S. J., Is type 2 diabetes mellitus associated with alterations in hearing? A systematic review and meta-analysis. Laryngoscope 124, 767–776 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Przewoźny T., Gójska-Grymajło A., Kwarciany M., Gąsecki D., Narkiewicz K., Hypertension and cochlear hearing loss. Blood Press. 24, 199–205 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Dubno J. R., Eckert M. A., Lee F.-S., Matthews L. J., Schmiedt R. A., Classifying human audiometric phenotypes of age-related hearing loss from animal models. J. Assoc. Res. Otolaryngol. 14, 687–701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan H., et al. , Heritability of age-related hearing loss in middle-aged and elderly Chinese: A population-based twin study. Ear Hear. 40, 253–259 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Hendrickx J.-J., et al. , Familial aggregation of pure tone hearing thresholds in an aging European population. Otol. Neurotol. 34, 838–844 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Kvestad E., Czajkowski N., Krog N. H., Engdahl B., Tambs K., Heritability of hearing loss. Epidemiology 23, 328–331 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Christensen K., Frederiksen H., Hoffman H. J., Genetic and environmental influences on self-reported reduced hearing in the old and oldest old. J. Am. Geriatr. Soc. 49, 1512–1517 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Manolio T. A., et al. , Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W. et al.; NHLBI Exome Sequencing Project , Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–220 (2013).Corrected in: Nature495, 270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClellan J., King M.-C., Genetic heterogeneity in human disease. Cell 141, 210–217 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Manche S. K., Jangala M., Putta P., Koralla R. M., Akka J., Association of oxidative stress gene polymorphisms with presbycusis. Gene 593, 277–283 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Ateş N. A., et al. , Glutathione S-transferase gene polymorphisms in presbycusis. Otol. Neurotol. 26, 392–397 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Van Eyken E., et al. , Contribution of the N-acetyltransferase 2 polymorphism NAT2*6A to age-related hearing impairment. J. Med. Genet. 44, 570–578 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unal M., et al. , N-acetyltransferase 2 gene polymorphism and presbycusis. Laryngoscope 115, 2238–2241 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Nolan L. S., et al. , Estrogen-related receptor gamma and hearing function: Evidence of a role in humans and mice. Neurobiol. Aging 34, 2077.e1–2077.e9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Eyken E., et al. , KCNQ4: A gene for age-related hearing impairment? Hum. Mutat. 27, 1007–1016 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Van Laer L., et al. , The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum. Mol. Genet. 17, 159–169 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Han B., et al. , Association of polymorphisms in grainyhead-like-2 gene with the susceptibility to age-related hearing loss: A systematic review and meta-analysis. Medicine (Baltimore) 98, e16128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann T. J., et al. , A large genome-wide association study of age-related hearing impairment using electronic health records. PLoS Genet. 12, e1006371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells H. R. R., et al. , GWAS identifies 44 independent associated genomic loci for self-reported adult hearing difficulty in UK Biobank. Am. J. Hum. Genet. 105, 788–802 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagtegaal A. P., et al. , Genome-wide association meta-analysis identifies five novel loci for age-related hearing impairment. Sci. Rep. 9, 15192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vuckovic D., et al. , Genome-wide association analysis on normal hearing function identifies PCDH20 and SLC28A3 as candidates for hearing function and loss. Hum. Mol. Genet. 24, 5655–5664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolber L. E., et al. , Salt-inducible kinase 3, SIK3, is a new gene associated with hearing. Hum. Mol. Genet. 23, 6407–6418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girotto G., et al. , Next generation sequencing and animal models reveal SLC9A3R1 as a new gene involved in human age-related hearing loss. Front. Genet. 10, 142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan A., et al. , Next-generation sequencing identified SPATC1L as a possible candidate gene for both early-onset and age-related hearing loss. Eur. J. Hum. Genet. 27, 70–79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oonk A. M. M., et al. , Progressive hereditary hearing impairment caused by a MYO6 mutation resembles presbyacusis. Hear. Res. 299, 88–98 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Xiong H., et al. , Activation of miR-34a/SIRT1/p53 signaling contributes to cochlear hair cell apoptosis: Implications for age-related hearing loss. Neurobiol. Aging 36, 1692–1701 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Xue T., et al. , miR-29b overexpression induces cochlear hair cell apoptosis through the regulation of SIRT1/PGC-1α signaling: Implications for age-related hearing loss. Int. J. Mol. Med. 38, 1387–1394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai U., Seidman M. D., Hinojosa R., Quirk W. S., Mitochondrial DNA deletions associated with aging and possibly presbycusis: A human archival temporal bone study. Am. J. Otol. 18, 449–453 (1997). [PubMed] [Google Scholar]
- 40.Manwaring N., et al. , Mitochondrial DNA haplogroups and age-related hearing loss. Arch. Otolaryngol. Head Neck Surg. 133, 929–933 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Gallagher M. D., Chen-Plotkin A. S., The post-GWAS era: From association to function. Am. J. Hum. Genet. 102, 717–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bamshad M. J., et al. , Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745–755 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Davis A.; MRC Institute of Hearing Research , Hearing in Adults: The Prevalence and Distribution of Hearing Impairment and Reported Hearing Disability in the MRC Institute of Hearing Research’s National Study of Hearing (Whurr Publishers, 1995). [Google Scholar]
- 44.Denoyelle F., et al. , Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: Implications for genetic counselling. Lancet 353, 1298–1303 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Shen J. et al.; ClinGen Hearing Loss Working Group , Consensus interpretation of the p.Met34Thr and p.Val37Ile variants in GJB2 by the ClinGen hearing loss expert panel. Genet. Med. 21, 2442–2452 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Povysil G., et al. , Rare-variant collapsing analyses for complex traits: Guidelines and applications. Nat. Rev. Genet. 20, 747–759 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Shearer A. E., et al. , Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 107, 21104–21109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagatsuma M., et al. , Distribution and frequencies of CDH23 mutations in Japanese patients with non-syndromic hearing loss. Clin. Genet. 72, 339–344 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Ménétrey J., et al. , The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature 435, 779–785 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malone A. F., et al. , Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int. 86, 1253–1259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon T.-J., et al. , The effect of novel mutations on the structure and enzymatic activity of unconventional myosins associated with autosomal dominant non-syndromic hearing loss. Open Biol. 4, 140107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamakoshi Y., Dentin sialophophoprotein (DSPP) and dentin. J Oral Biosci 50, 33–44 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor J. W., Simple estimation of population attributable risk from case-control studies. Am. J. Epidemiol. 106, 260 (1977). [DOI] [PubMed] [Google Scholar]
- 54.Kuritz S. J., Landis J. R., Summary attributable risk estimation from unmatched case-control data. Stat. Med. 7, 507–517 (1988). [DOI] [PubMed] [Google Scholar]
- 55.Falah M., et al. , Association of genetic variations in the mitochondrial DNA control region with presbycusis. Clin. Interv. Aging 12, 459–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bykhovskaya Y., et al. , Phenotype of non-syndromic deafness associated with the mitochondrial A1555G mutation is modulated by mitochondrial RNA modifying enzymes MTO1 and GTPBP3. Mol. Genet. Metab. 83, 199–206 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Usami S.-i., Nishio S.-y., “Nonsyndromic hearing loss and deafness, mitochondrial” in GeneReviews, Adam M. P., et al., Eds. (University of Washington, Seattle, WA, 1993). [Google Scholar]
- 58.Corey D. P., Akyuz N., Holt J. R., Function and dysfunction of TMC channels in inner ear hair cells. Cold Spring Harb. Perspect. Med. 9, a033506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan B., et al. , TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron 99, 736–753.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawashima Y., Kurima K., Pan B., Griffith A. J., Holt J. R., Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflugers Arch. 467, 85–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ballesteros A., Fenollar-Ferrer C., Swartz K. J., Structural relationship between the putative hair cell mechanotransduction channel TMC1 and TMEM16 proteins. Elife 7, e38433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Azaiez H., et al. , Genomic landscape and mutational signatures of deafness-associated genes. Am. J. Hum. Genet. 103, 484–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DiStefano M. T., et al. , ClinGen expert clinical validity curation of 164 hearing loss gene-disease pairs. Genet. Med. 21, 2239–2247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landrum M. J., et al. , ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis M. A., et al. , Whole exome sequencing in adult-onset hearing loss reveals a high load of predicted pathogenic variants in known deafness-associated genes and identifies new candidate genes. BMC Med. Genomics 11, 77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y., et al. , A novel DFNA36 mutation in TMC1 orthologous to the Beethoven (Bth) mouse associated with autosomal dominant hearing loss in a Chinese family. PLoS One 9, e97064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Löppönen T., et al. , Homozygous M34T mutation of the GJB2 gene associates with an autosomal recessive nonsyndromic sensorineural hearing impairment in Finnish families. Acta Otolaryngol. 132, 862–873 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Teek R., et al. , Prevalence of c.35delG and p.M34T mutations in the GJB2 gene in Estonia. Int. J. Pediatr. Otorhinolaryngol. 74, 1007–1012 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Dória M., Fernandes S., Moura C. P., Study of Met34Thr variant in nonsyndromic hearing loss in four Portuguese families. Porto Biomed. J. 1, 32–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seidman M. D., et al. , Age-related hearing loss and its association with reactive oxygen species and mitochondrial DNA damage. Acta Otolaryngol. Suppl. 552, 16–24 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Fetoni A. R., et al. , Cx26 partial loss causes accelerated presbycusis by redox imbalance and dysregulation of Nfr2 pathway. Redox Biol. 19, 301–317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmiedt R. A., Effects of aging on potassium homeostasis and the endocochlear potential in the gerbil cochlea. Hear. Res. 102, 125–132 (1996). [DOI] [PubMed] [Google Scholar]
- 74.Schmiedt R. A., Lang H., Okamura H. O., Schulte B. A., Effects of furosemide applied chronically to the round window: A model of metabolic presbyacusis. J. Neurosci. 22, 9643–9650 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glusman G., et al. , Mapping genetic variations to three-dimensional protein structures to enhance variant interpretation: A proposed framework. Genome Med. 9, 113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyagawa M., Nishio S. Y., Usami S., Prevalence and clinical features of hearing loss patients with CDH23 mutations: A large cohort study. PLoS One 7, e40366 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson K. R., et al. , Effects of Cdh23 single nucleotide substitutions on age-related hearing loss in C57BL/6 and 129S1/Sv mice and comparisons with congenic strains. Sci. Rep. 7, 44450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng Q. Y., et al. , Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum. Mol. Genet. 14, 103–111 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the relevant data are available from the article and SI Appendix.