Significance
Multiple resistance and pH adaptation (Mrp) complexes are the most sophisticated known cation/proton exchangers and are essential for the survival of a vast variety of alkaliphilic and/or halophilic microorganisms. Moreover, this family of antiporters represents the ancestor of cation pumps in nearly all known redox-driven transporter complexes, including the complex I of the respiratory chain. For the Mrp complex, an experimental structure is lacking. We now report the structure of Mrp complex at 3.0-Å resolution solved using the single-particle cryo-EM method. The structure-inspired functional study of Mrp provides detailed information for further biophysical and biochemical investigation of the intriguingly pumping mechanism and physiological functions of this complex, as well as for exploring its potential as a therapeutic drug target.
Keywords: Na+/H+ antiporter, Mrp complex, cryo-EM, complex I, energy coupling
Abstract
Multiple resistance and pH adaptation (Mrp) complexes are sophisticated cation/proton exchangers found in a vast variety of alkaliphilic and/or halophilic microorganisms, and are critical for their survival in highly challenging environments. This family of antiporters is likely to represent the ancestor of cation pumps found in many redox-driven transporter complexes, including the complex I of the respiratory chain. Here, we present the three-dimensional structure of the Mrp complex from a Dietzia sp. strain solved at 3.0-Å resolution using the single-particle cryoelectron microscopy method. Our structure-based mutagenesis and functional analyses suggest that the substrate translocation pathways for the driving substance protons and the substrate sodium ions are separated in two modules and that symmetry-restrained conformational change underlies the functional cycle of the transporter. Our findings shed light on mechanisms of redox-driven primary active transporters, and explain how driving substances of different electric charges may drive similar transport processes.
In all life kingdoms, homeostasis of Na+ and H+ is essential for many aspects of cell physiology, including maintaining appropriate osmotic pressure, intracellular pH, and electrostatic membrane potential (ΔΨ) (1). A variety of Na+/H+ exchangers (also called antiporters) are directly involved in regulating the homeostasis of Na+ and H+, and form a major part of the superfamily of monovalent cation/proton antiporters (CPA) (2, 3). Members of the CPA superfamily have been identified and characterized in fungi, plants, and mammals (4). Usually, Na+/H+ antiporters utilize the electrochemical potential of protons, commonly called proton motive force (PMF), to catalyze efflux of Na+ as well as other monovalent cations. The stoichiometric ratio of protons to sodium ions may vary from transporter to transporter, primarily depending on the strength of PMF in which the transporter has been evolved to function optimally (see discussion in SI Appendix, Supplementary Material). One family of Na+/H+ antiporters, namely, the NhaA-like single-subunit antiporters, has been well studied (5, 6), and was shown to represent the canonical type of secondary active transporters, in which the proton influx and the Na+ efflux most likely share the same pathway. In contrast, multiple resistance and pH adaptation (Mrp) complexes are the most sophisticated known secondary active transporters. They alone form a unique family of atypical antiporters, termed CPA3 (7–10), in which protons and Na+ ions are likely to flow through distinct physical paths. Under physiological conditions, the driving force for Mrp transport can only come from protons but not Na+ ions (SI Appendix, Supplementary Material); therefore, protons are referred to as the driving substance. Moreover, Mrp complexes are essential for alkaliphilic and/or halophilic microorganisms to adapt to their extreme environments (11).
A member of the Mrp family usually contains one copy of each of seven subunits called MrpA-F, all of which contribute to a total of ∼50 predicted transmembrane helices (TMs) (SI Appendix, Fig. S1). In the so-called group II Mrp complexes, subunit MrpB is fused to the C terminus of MrpA while keeping other subunits essentially unchanged, and the remaining nonfused complexes are termed group I (12). Amino acid sequence analysis showed that MrpA and MrpD are homologous (13), and their corresponding three-dimensional (3D) folding pattern is referred to as Mrp antiporter/pump folding. In addition, every subunit of the Mrp complex was shown to be important for the Na+/H+ antiporter activity (14). This notion raises an interesting question as to why a seemingly simple task requires such a complexed protein machinery.
Furthermore, a number of important energy-converting protein complexes, including those from bacteria (15), plants (16), and mammalian animals (17), are phylogenetically related to the Mrp complex. For instance, components of the respiratory chain complex I (henceforth called complex I) are highly homologous to MrpA and MrpD subunits. In the membrane arm of complex I, three Mrp pump subunits are aligned in a head-to-tail fashion, and presumably carry out proton efflux, and their driving forces come from the redox reaction between NADH (nicotinamide adenine dinucleotide) and quinone (18). In some anaerobic archaea lacking complex I, the membrane-bound hydrogenase (MBH) complex replaces the functional role of complex I (19). In this type of ion-pumping complex, a subcomplex containing Mrp homologous subunits functions as a Na+ pump, and the driving force for this pump comes from the redox reaction between reduced ferredoxin and protons, subsequently generating hydrogen gas. In complex I, MBH complex, as well as other related redox-driven transporters, cellular redox energy (i.e., differential redox potential between electron donors and acceptors) is converted to transmembrane electrochemical potential of either protons or Na+; thus, these primary active transporters play essential roles in cellular energy homeostasis (20). It is generally accepted that, in these redox-driven ion pumps, electrostatic interactions between the transferred electrons and pumps drive the conformational changes that are required for the cation export (21, 22). While a number of 3D structures have been reported in recent years for such redox-driven transporters (15, 16, 23), the detailed mechanisms responsible for the energy conversion remain under debate. Taken together, the Mrp complex likely shares a common ancestor with the ion-pumping module(s) in a variety of redox-driven transporters, and thus understanding of the mechanism of Mrp antiporter complex should shed light on the energy-coupling mechanisms of all Mrp antiporter-containing transporters.
Here, we report the cryoelectron microscopy (cryo-EM) structure of the group II Mrp complex from Dietzia sp. DQ12-45-1b, a strain of Gram+ bacteria isolated from the production water of a deep subterranean oil reservoir and which has also been found in a number of high-salinity environments (24). Our results from structural and functional analyses show that this Mrp complex contains two modules responsible for proton transport and Na+ pumping, respectively. On the basis of these findings, we propose a hitherto unknown mechanism for energy coupling between the two modules.
Results
Overall Structure.
Recombinant Mrp complex of Dietzia sp. (molecular weight: ∼220 kDa) was expressed in the Escherichia coli C43 (DE3) strain, and purified using n-dodecyl-β-d-maltopyranoside (DDM) (SI Appendix, Fig. S2). Our single-particle cryo-EM analysis of the Mrp complex (SI Appendix, Fig. S3) resulted in a 3D density map of an overall resolution of 3.0 Å, based on the Fourier shell correlation (FSC) (Fig. 1 and SI Appendix, Figs. S3C and S4A). Of a total of 2,000 residues of the Mrp complex, 1,872 were built into the final model with good geometry; in addition, we included 28 (partial) lipid/detergent molecules (SI Appendix, Fig. S4B). The unobserved parts include residues from a small number of loops in MrpA and MrpD (marked in SI Appendix, Fig. S1), which are characterized by their poor map quality. The statistics of the 3D reconstruction and model refinement are summarized (SI Appendix, Table S1).
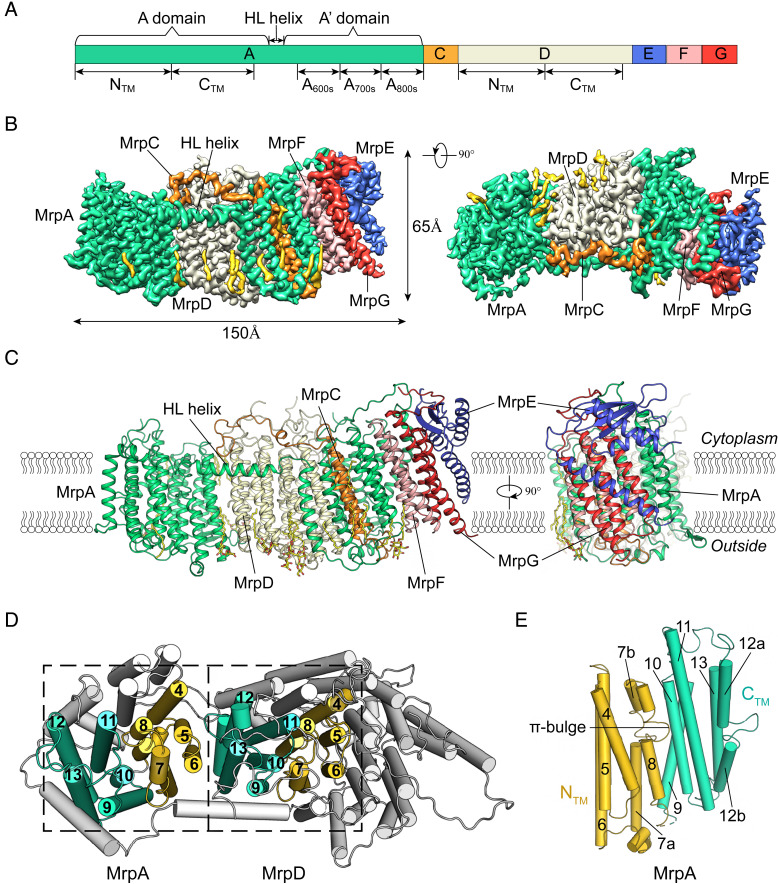
Fig. 1.
Overall structure of the Mrp complex. (A) Schematic diagram of the organization of the Mrp genes. (B) Cryo-EM map of the Mrp complex segmented by subunits (contoured at 6 σ) in front view (Left) and top view (Right). Subunits are colored the same as in A. Densities for lipid molecules are shown in yellow. (C) Two orthogonal views of the Mrp structure in ribbon presentation. Lipid/detergent molecules are included as yellow stick models. (D) The NTM (yellow) and CTM (cyan) domains of subunits A and D emphasized by dashed boxes. (E) Breaks in TM7 and TM12 as well as a π-bulge in TM8 in the Mrp pump subunit. Colored as in D.
The Mrp complex consists of six membrane-embedded subunits, including subunits A (containing 25 TMs), C (3 TMs), D (14 TMs), E (1 TM), F (3 TMs), and G (3 TMs). With a total of 49 TM helices, the Mrp complex model adopts a curved claviform shape, with a length of approximately 150 Å along the long axis and a width of approximately 65 Å in the membrane plane (Fig. 1B). Because of the absence of PMF during collection of the structural data, we considered this in vitro structure as representing the ground state of the Mrp complex. In the following discussion, we will use the “standard” orientation adopted in the complex I research field as a reference system, in which the cytosol is at the top, the extracellular side is at the bottom, and the characteristic amphipathic horizontal-and-long (HL) helix is in the front (Fig. 1C).
MrpA can be divided into a major N-terminal domain (TMs 1 to 15, A domain for short) and a minor C-terminal domain (TMs 16 to 25, A′ domain for short), with both domains connected by the HL helix. The A′ domain (residues 565 to 958) can be further divided into three segments, termed A600s, A700s, and A800s. The A800s segment, that is, TMs 23 to 25 from MrpA, corresponds to MrpB of the group I Mrp. The A domain (as well as MrpD) shows a canonical Mrp antiporter fold containing TMs 1 to 14, which was first observed in the complex I subunits that are homologous to MrpA/MrpD in amino acid sequences (18). Such a domain can be further divided into an NTM subdomain (TMs 1 to 8) and a CTM subdomain (TMs 9 to 14). In addition, TMs 4 to 8 of NTM and TMs 9 to 13 of CTM, each forming a five-TM helix bundle, are structurally related by a pseudo twofold inversion symmetry (also called inverted repeat), with the screw axis being parallel to the membrane plane (Fig. 1D). Both TM7 and TM12 are discontinuous in their middle region, which harbors a seven-residue insertion loop at the break, resulting in the formation of helical segments TM7a, TM7b, TM12a, and TM12b. These two helix breaks are signature features of the Mrp antiporter fold (Fig. 1 D and E). Moreover, TM8 of MrpA possesses a π-bulge at Ala240 (denoted as A240A8, in which the superscript A8 stands for TM8 in MrpA). As shown in Fig. 1E, this bulge is at the NTM–CTM interface. A similar bulge was also observed in TM8 of MrpD.
In the Mrp complex, from left to right end, the multiple subunits appear in the order of MrpA (A domain), MrpD, MrpC, MrpA′, MrpF, MrpG, and MrpE. Among them, MrpA contacts every other subunit to varied extents. In particular, the MrpA subunit clamps MrpD and MrpC from both sides with its MrpA and MrpA′ domains, and contacts MrpF and MrpG with two other helices (TMs 22 and 23). MrpA touches MrpE only briefly between the cytosolic end of TM22 of the MrpA′ domain and a β-sheet edge from MrpE. In addition, MrpC, MrpD, and the MrpA′ domain contact each other through multiple long loops. In contrast, MrpE is located at the right end of the elongated complex. Its N terminus contributes only one TM helix which is loosely packed with TM helices of MrpG. The remaining structure of MrpE forms a ferredoxin-like amphiphilic domain binding to the cytosolic side of MrpG (Fig. 1C), although the enzymatically essential Cys residues required for coordinating an iron−sulfur cluster are absent. In agreement with previous biochemical data (14), the MrpA′ domain as well as the subunits MrpD and MrpC appear to constitute a compact subcomplex, whereas subunits MrpF, MrpG, and MrpE form an additional subcomplex. On both sides of the membrane, only minor contacts through loops exist between the two subcomplexes.
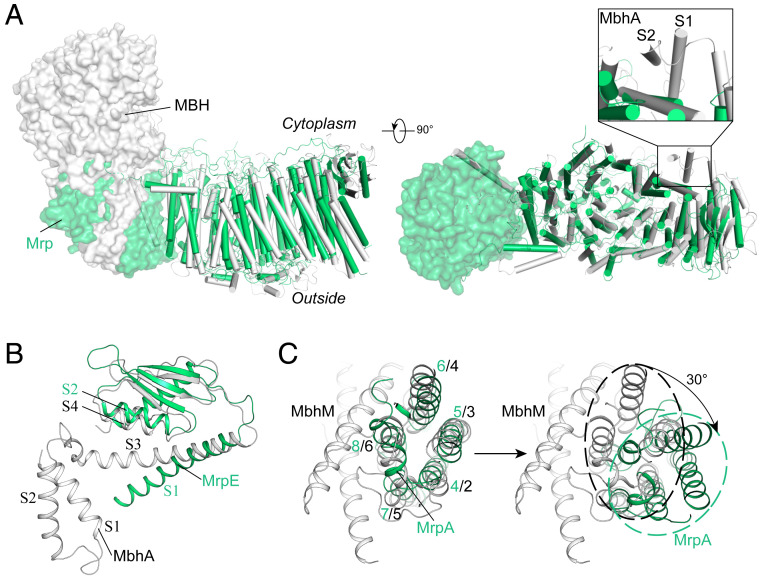
The overall structure of Mrp complex resembles the transporter part of the MBH complex (Protein Data Bank [PDB] ID code 6CFW). However, a major difference is that the A domain of the MrpA replaces the redox reaction-required MbhM subunit located at the left end of the MBH complex (Fig. 2A). Moreover, at the right end of these complexes, MrpE and its counterpart MbhA subunit share the same fold in their amphipathic domains and interact with the remaining complex in a similar manner. Nevertheless, in contrast to the MrpE subunit in our structure, MbhA contains two TM helices (Fig. 2). Further sequence analysis of Mrp complexes from different species indicated that the N terminus of MrpE from Dietzia sp. DQ12-45-1b misses ∼50 residues compared to MrpE from other species (SI Appendix, Fig. S1F), suggesting that the TM helices in MrpE/MbhA-like subunits play only auxiliary roles, probably anchoring the amphipathic domain. Overall, except for the A domain and the TM helix of MrpE, components of the transporter module from Mrp complex are superimposable with the counterparts in MBH (Fig. 2), with a root-mean-square deviation (rmsd) of 1.8 Å for 882 Cα pairs (using a 3-Å cutoff) or 2.5 Å for 1,136 Cα pairs (using a 6-Å cutoff). The MBH complex subunits MbhH, MbhG, MbhF, MbhE, MbhD/I, MbhB, MbhC, and MbhA, from left to right, correspond to the subunits MrpD, MrpC, MrpA800s, MrpA700s, MrpA600s, MrpF, MrpG, and MrpE in our Mrp structure, respectively. This similarity in 3D structures is consistent with the homology of the two types of complexes in their amino acid sequences (SI Appendix, Fig. S1) (23). Furthermore, the MbhM subunit from the MBH redox module contains eight TM helices (TMs 1 to 8), including a five-TM helix bundle characteristic of the Mrp antiporter folding (Fig. 2C). As isolated structural elements, the five-TM bundles from MbhM and MrpA NTM, including the break and inserted loop in the fourth helices (but not the π-bulge in the fifth TM helix), are superimposable, with an rmsd of 1.9 Å for 99 Cα pairs (using a 3-Å cutoff) (Fig. 2C). However, in the context of superposition of the remaining complexes, MrpA would require an additional ∼30° rotation to match its NTM five-TM bundle with that present in MbhM (Fig. 2C). See also SI Appendix for further discussion.
Fig. 2.
Comparison of the Mrp and MBH complexes. (A) Overlay of Mrp (green) with the P. furiosus MBH (gray, PDB ID code 6CFW) in different views. Subunits MrpA and MbhM are shown in transparent surface presentation, and the remaining parts of the complexes are shown in cartoon presentation (helices and ropes). The overall structures were aligned without the A domain of MrpA and MbhM. Inset shows the structural difference between MrpE and MbhA. Insets show regions of large local conformational differences between MrpE and MbhA. (B) Structural alignment of MrpE and MbhA. (C) Structural comparison between MrpA and MbhM subunits. Left shows structural alignment of TMs 4 to 8 of MrpA and MbhM. Right shows the same structural elements in the context of alignment of the remaining transmembrane parts of Mrp and MBH complexes. It would require an additional ∼30° rotation for the TMs 4 to 8 of MrpA to match their counterparts in MbhM. Black dashed oval circle indicates five-TM bundle of MbhM and green dashed oval circle five-TM bundle of MrpA.
Structural Twist of the Complex.
Next, we analyzed the curvature of the complex structure to better understand its internal stress. The projection of the Mrp complex on the membrane plane is arc shaped (as shown in SI Appendix, Fig. S5A). In addition, if one end of the Mrp complex was “comfortably” positioned in the lipid bilayer (i.e., with its TM helices approximately perpendicular to the membrane plane), the other end became tilted about 25 Å toward the cytosol. In geometrical terms, the centerline of the complex along the long axis is not a straight line, but exhibits clearly visible curvature and torsion (SI Appendix, Fig. S5A). Furthermore, the 49 TM helices are arranged in 12 rows along the long axis. Among them, the A domain and MrpD each formed four rows of TM helices. MrpC, segment A600s (TMs 17 to 19), subunit F, and G each contributes one row. Each of the latter four rows is locked by an additional TM helix from the C-terminal region of the A′ domain. In this helix arrangement, the axes of TM helices present a right-handed helical twist along the central curve of the complex, with an average twist of about 5° per row (SI Appendix, Fig. S5B). Taken together, the TM portion of the detergent-solubilized Mrp complex was neither a linear nor a planar assembly; instead, it underwent simultaneous distortion in all three dimensions.
Mrp Antiporter Subunits and Their Interactions.
The A and A′ domains of MrpA are connected by the amphiphilic HL helix (residues 522 to 561), which is presumably located on the cytosolic surface of the membrane. The HL helix exhibits a nick in the region of residues 535 to 540, resulting in a twist conformation. The N terminus of the HL helix is connected to the CTM domain of A domain via TM15, whereas its C terminus is anchored to the NTM domain of MrpD via TM16 from the A′ domain. Additionally, a number of noncovalent interactions are visible between the HL helix and MrpD. Despite this structural constraint, the gap between subunits A and D is the largest among all intersubunit interfaces. A few pieces of clear density are found within the A−D gap (SI Appendix, Fig. S4B) and are laterally accessible to the lipid bilayer, indicating that lipid molecules may dynamically occupy the interface. Similar loose packing and lipid binding are also observed in the interface between the Mrp pump subunit (MbhH) and the redox module (MbhM) in the MBH complex (23). Furthermore, near the outer surface of the membrane, a series of β-hairpins and short helices form another elongated amphipathic structure termed βH belt (SI Appendix, Fig. S5C) (25). Both the HL helix and diagonally located βH belt stabilize the arc-shaped overall structure of Mrp. No other connecting loop between the A domain and MrpD is found on either side of the membrane. If the complex deviated from its observed curved conformation under external forces, the HL helix and βH belt would be either compressed or stretched, thereby storing a part of the conformational energy. These loose structural constraints allow conformational changes to occur between the NTM and CTM domains in both subunits A and D, which may be necessary for the transport process (21).
Central Polar Axis and the Transport Pathways.
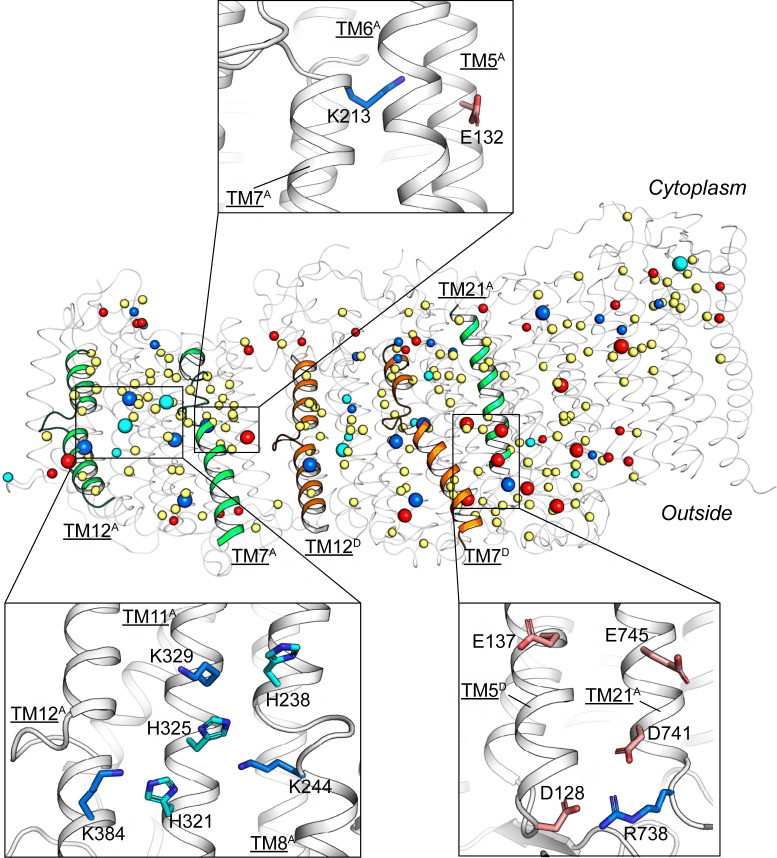
The middle layer of the TM region of Mrp complex possesses a central polar axis consisting of polar and charged residues. In particular, in the Mrp pump regions (i.e., domain A of MrpA and MrpD), a pair of acidic−basic residues (e.g., E132A5−K213A7a) in NTM and a hydrophilic core in CTM are arranged along the central polar axis (Fig. 3). These two structural features are highly conserved in all known Mrp antiporter-containing protein complexes (including MBH and complex I) (SI Appendix, Fig. S6) (23, 25). For instance, three conserved Lys residues (e.g., K244A8, K329A11, and K384A12b) surround a conserved His residue (H325A11) in the hydrophilic core of MrpA CTM domain (Fig. 3 and SI Appendix, Figs. S1 and S6A). The central polar axis probably assumes the role of the binding sites for substrate ions (such as Na+) and driving substance (such as H+). It is noted that many of the acidic residues along the central polar axis show poor density in the cryo-EM map in contrast to, for example, the basic residues (SI Appendix, Fig. S4A). This observation is likely to reflect the deprotonation states of these negatively charged residues (26).
Fig. 3.
Buried polar residues of the Mrp complex. Polar or charged residues which are buried and conserved are shown as spheres (in main figure) or stick models (Insets). Residues that are both buried and highly conserved (SI Appendix, Fig. S1) are emphasized as larger spheres. Basic residues are shown in blue; acidic residues are in red; His residues are in cyan; other polar residues (Asn, Gln, Ser, Thr, Cys, Tyr, and Trp) are in yellow.
The only two potential pathways from the central polar axis to cytosol are located at the NTM−CTM interfaces within subunits A and D, consisting of polar residues (Fig. 3). Such inward-facing pathways have been found to be conserved in all reported Mrp pump subunits of related complex structures (15, 17, 23, 27), although all of these pathways are likely to be present in their closed form.
The transport pathways from the central polar axis to the extracellular side appear to be more complicated, and vary with the composition of complexes. Our analysis of the Mrp complex structure indicates that D128D5, D741A21, E745A21, and E137D5 are forming what appears to be an ion pathway (Fig. 3). These residues are conserved in MBH (SI Appendix, Fig. S1) but not in complex I; the pathway formed by these acidic residues is proposed to function as a part of the channel for cationic substrate in MBH (23). Another potential pathway appears to be present at the left end of the Mrp complex, near the interface between the CTM lateral surface of MrpA and the lipid bilayer (Fig. 3, Bottom-Right Inset). This potential pathway connects the hydrophilic core of CTM to the extracellular space via polar residues in TM12b. An analogous pathway is located on the CTM lateral surface of the MbhH subunit of MBH complex. In complex I, a similar pathway appears on the CTM lateral surface of the distal (left end) NuoL subunit. This terminal pathway was recently proposed to serve as the exit of a long-range proton transport highway (21).
Structure-Based Mutagenesis Analysis.
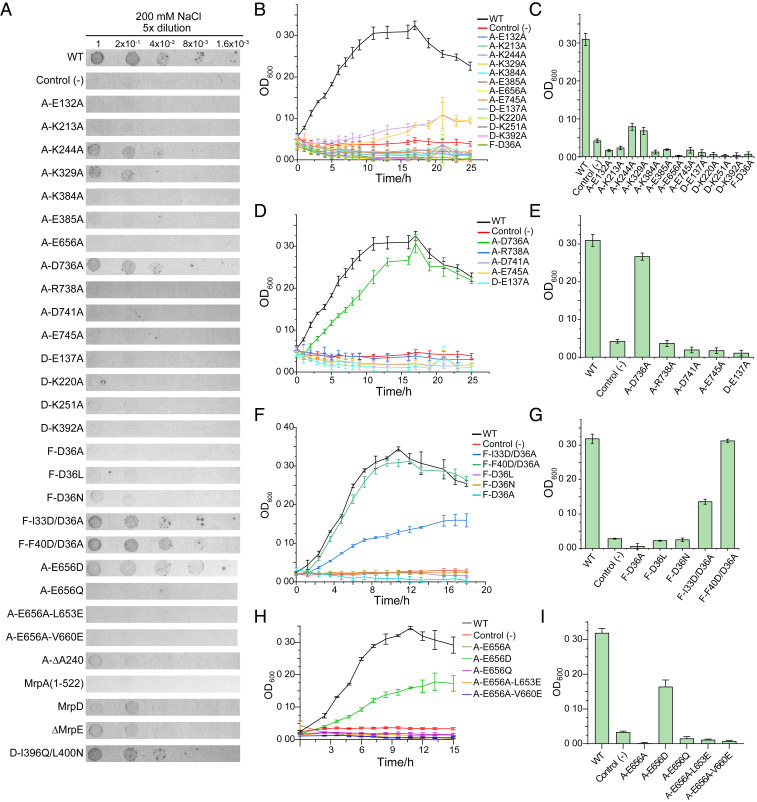
To identify and verify amino acid residues critically involved in the mechanism responsible for salt resistance, a total of 24 site-directed point mutations (marked in SI Appendix, Fig. S1) were introduced at a series of positions in potential proton/Na+ transport pathways. Functional effects of these Mrp mutants were analyzed in the E. coli KNabc strain [which is salt sensitive due to deletion of nhaA, nhaB, and chaA genes (28)] using growth assays in the presence of 200 mM NaCl, in both solid and liquid LBK (potassium-modified Luria-Bertani) media (29) (Fig. 4 and SI Appendix, Figs. S7 and S8). As shown in Fig. 4, the heterologous expression of the wild-type Mrp can functionally complement the salt-sensitive phenotype. We first introduced mutations of Ala substitution, followed by making additional mutations to verify or to rescue the primary mutations. All Mrp variants were expressed from pUC18-based plasmids. The results of functional study on these variants are summarized (SI Appendix, Fig. S10 and Table S2).
Fig. 4.
Salt-resistance assay in E. coli KNabc cells. (A) Colony formation on solid medium containing 200 mM NaCl. (B–I) Cell growth in cultures containing 200 mM NaCl. The histograms represent cell concentrations at 16 h (B–E) or at 12 h (F–I). Variants include mutations along the central polar axis (B and C), in a potential Na+ pathway (D and E), near Asp36 of MrpF (F and G), and near Glu656 of MrpA (H and I). The vector used for protein expression in all cases was pUC18. The experiments were repeated in triplicate, each including three biological repeats. The presented results represent the mean of at least six determinations, and SDs are shown as error bars. (See SI Appendix, Figs. S5 and S6 for results from the salt-free control assay and from more mutations.)
The mutated polar or charged residues along the central polar axis, from left to right end, include the following: K329A11, K244A8, K213A7, E132A5, K392D12, K220D7, K251D8, E137D5, E745A21, E656A19, and D36F2. Nearly all of these residues are highly conserved among Mrp subunits from different species and homologous subunits of MBH (SI Appendix, Fig. S1). All Ala substitutions at these positions lost the capacity to complement the salt-sensitive phenotype of KNabc, except that K244A8A and K329A11A maintained a fraction of the activity (Fig. 4 B and C). Interestingly, the K244A8A mutation in MrpA is equivalent to K251D8A in MrpD, yet their effects on salt-resistance activity appeared to be distinct from each other, especially when tested in the solid medium assay (Fig. 4A). In addition, K384A12 and E385A12 are located in the left-side path connecting the central polar axis in MrpA to the extracellular space, and their Ala substitutions appeared to be detrimental. Most mutations from this group (namely K213A7, E132A5, K220D7, E137D5, E745A21, E656A19, and D36F2) are equivalent to those of previous reports (13, 30–32), and our results are in agreement with the previous ones (SI Appendix, Table S2). Together, these results confirmed the essential roles of these polar and charged residues along the central polar axis in Na+/H+ antiport activity.
As mentioned above, charged residues at the MrpD−MrpA′ interface form another potential outward pathway (Fig. 3). These residues are E137D5, D741A21, E745A21, R738A21, and D736A21, arranged from the central axis to the extracellular side. Ala substitution mutations at these positions showed that the closer the residues are to the central polar axis, the more detrimental effects their mutation may have on the NaCl tolerance (Fig. 4). In previous reports, mutations equivalent to our R738A21A and E745A21A variants, together with other mutations nearby (e.g., those equivalent to Q69C3A and T74C3A), also support the importance of this putative pathway (13, 30).
Moreover, the π-bulge in the middle of TM8 of MrpA is a conserved structural feature of the Mrp antiporter fold, for example, in the corresponding subunits of complex I and MBH (16, 17, 23, 25, 27). It is likely that such a local bulge remains energetically unfavorable during protein folding (33), thus strongly suggesting that a helix bulge in the Mrp antiporter fold fulfills a conserved functional role. A deletion mutation in the π-bulge position A240A8 resulted in complete loss of the NaCl-resistance ability (SI Appendix, Fig. S8 F and H). Since such a single-residue deletion is unlikely to have extensive effects on protein folding (34), its loss-of-function phenotype indicates that the π-bulge in TM8 plays an important functional role.
A conserved acidic residue, D36F2 in MrpF, is located near the central polar axis. Intriguingly, when this Asp residue was substituted to Ala (D36A), Leu (D36L), or Asn (D36N), the mutant failed to recover NaCl resistance found in the Mrp complex in KNabc (Fig. 4). In a previous report, an Ala substitution mutation in Bacillus subtilis Mrp equivalent to our D36A variant exhibited a loss-of-function phenotype, whereas a D36N-like mutant remained functional (SI Appendix, Table S2) (32). In our case, an additional mutation F40D at the position one helix turn below D36F2 effectively eliminated the detrimental effect of the D36A mutation. Cells expressing the double-mutation variant D36A/F40D exhibited a growth rate identical to that harboring the wild-type complex (P < 0.05). Similarly, additional mutation I33D positioned one helix turn above D36F2 partially rescued NaCl resistance. Together, these results suggested that maintaining a negatively charged residue in this region is essential for the antiporter activity of the Mrp complex, although the precise location of such an acidic residue may not be critical for transport function. Thus, its effect is likely to be of long-range electrostatic nature. In addition, in TM19 of MrpA, E656 appears to be located in an environment similar to D36F2. The E656A19A mutant also lost the activity. However, in contrast to the double mutation of D36A/F40D in MrpF, additional mutations near E656A19 could not eliminate the detrimental effect of the E656A mutation. These results suggested that E656A19 has a functional role different from D36F2.
In addition, we expressed the MrpD subunit and the A domain (residues 1 to 522 of the MrpA subunit) individually in E. coli KNabc. Their expression was confirmed by immunoblotting (SI Appendix, Fig. S9). Neither of these constructs rescued NaCl resistance (Fig. 4 and SI Appendix, Figs. S7 and S8), indicating that both MrpA and MrpD are required in combination with other subunits for the antiporter activity. Next, we deleted the MrpE subunit, which loosely packs with the MrpG subunit at the right end of the complex (Fig. 1). The resulting ΔMrpE complex consisting of MrpACDFG also lacked Na+/H+ antiport activity (SI Appendix, Figs. S7B and S8 B and D). Its expression was confirmed by immunoblotting (SI Appendix, Fig. S9). This result is in agreement with previous findings showing that the presence of MrpE is critical for maintaining complex stability (14). We also attempted to create a new pathway from the central polar axis toward the extracellular side in the location between pump domains of MrpA and MrpD by introducing a double-mutation I369Q/L400N in the TM12b helix of MrpD. However, this construct failed to show detectable effects (SI Appendix, Figs. S7B and S8 B and D), suggesting that the existing outward pathways are more refined for the purpose of ion translocation.
Discussions
We showed that the Mrp complex shares a high homology with the redox-driven Na+-pumping MBH complex, both for the 3D structure and at the level of amino acid sequence. One major difference between Mrp and MBH complexes, however, is that, in the evolutionary process, the redox module of MBH (i.e., MbhM, etc., at the left end) has been exchanged for an MrpA subunit in the Mrp antiporter. On the basis of this finding, we postulate that the MrpA subunit constitutes the primary driving force for the Na+ pumping occurring in the remaining subunits. The Mrp antiporter domain of MrpA is likely to be the main transport path for the multiple protons which are the sole driving substance in Mrp, although the precise number of protons passing through each subunit remains unknown (see further discussion in SI Appendix, Supplementary Material). The NuoL subunit in complex I is homologous to MrpA and is known to function as a proton transporter. Previous reports showed that NuoL is able to compensate MrpA functionally (31, 35). In the redox module of MBH, the first loaded ligands are likely to be the electron acceptors, that is, positively charged protons, which are subsequently neutralized by electrons to generate hydrogen gas. Such a functional cycle found in MBH appears to be conceptually identical to that of proton loading and release in MrpA, suggesting that electrostatic interactions between the driving module and the cation pump are critical for the exporting of Na+ in both complexes. In agreement with this argument, in complex I, the first loaded ligands are likely to be the electrons and the electroneutral quinone (i.e., formation of semiquinone), and the driving module is located at the right end of the elongated membrane arm (15). Interestingly, the transmembrane subunit of the redox module, MbhM, shares similar folding with the NTM domain of MrpA (23), indicating that an ancient phylogenetic relationship exists between the two driving modules. Furthermore, the MbhH subunit in the MBH complex is likely to function as a Na+ pump, consistent with the physiological role of MBH in establishing transmembrane electrochemical potential of Na+. On the bases of structure homology, therefore, we hypothesize that its counterpart MrpD in the H+/Na+ antiporter Mrp functions as a Na+ pump as well.
Although MrpA and MrpD share similar protein folding architectures, they are likely to function differently. For instance, the K244A8A mutant form of MrpA showed effects distinct from that of K251D8A at the equivalent position in MrpD. The outward-facing pathway in MrpA is located at the CTM (left) side, whereas the outward-facing pathway in MrpD is located at the NTM (right) side (SI Appendix, Fig. S10). Due to the large gap between the Mrp pump domains in MrpA and MrpD in the ground state, we did not find direct polar interaction (<8.5 Å) between these two TM regions. Thus, MrpA is likely to possess its own inward- and outward-facing proton paths from the central polar axis. An interesting question is whether subunits of the same Mrp antiporter folding can function as pumps specific to distinct cationic ions, and whether they can even move in opposite directions. From a chemical kinetics point of view, all molecular machines may operate reversibly, depending on the direction from which the free energy is descending. Furthermore, considering that a protonated water molecule (H3O+) shares a similar size and electric property with a sodium ion, a Na+ transporter may adopt a role of proton translocation without drastic remodeling. Even without a water medium, protons and sodium ions may share the same translocation pathway, as illustrated in many simpler Na+/H+ antiporter structures (6).
Although PMF constitutes the sole energy source for the Mrp complex, we determined the structure of the detergent-solubilized Mrp complex in the absence of PMF. Therefore, we assume that the observed structure represents the low-energy ground state. However, such a state may also differ from the bona fide ground state in vivo. One supporting finding for this assumption is that the structure of the Mrp complex deviates from the ideal planar form of a membrane protein, by showing curvature (within the membrane plane), torsion (tilting away from the plane), and twist (rotating along the central axis). Under physiological conditions, such a “nonideal” structure would cause significant disorder in the surrounding lipid bilayer, a phenomenon known as frustration of the membrane (36). Thus, it is most likely that the interaction between the membrane protein and lipid bilayer will (partially) correct the geometric deviations, resulting in an approximately planar form of the complex parallel to the membrane. In such a corrected form, most TM helices will be aligned in a more parallel manner to each other as well to the membrane normal, instead of twisting by 60° from one end to the other as observed in the detergent-solubilized form of Mrp. Such a large-scale conformational change of the complex will require energy input from the membrane environment, which settles the complex in its in vivo ground conformation.
At the right end of the elongated Mrp complex, the F−G−E subcomplex appears to function as a “ballast stone” during minimization of the hydrophobic mismatch. More specifically, the amphipathic domain of MrpE at the right end is likely to anchor the subcomplex to the membrane plane and to impede displacement of the subcomplex in the direction of plane normal. A previous report showed that absence of MrpE destabilizes the complex (14), resulting in loss of function (SI Appendix, Figs. S7 and S8). These observations suggest that MrpF and MrpG fail to position themselves properly within the lipid bilayer in the absence of MrpE. Moreover, the conserved acidic residue D36F2 in the TM region of MrpF appears to be essential for the antiport activity of Mrp (Fig. 4). One possibility for the requirement of such an acidic residue is that the negative charge of D36F2 senses the ΔΨ and consequently adjusts the vertical position of MrpF inside the membrane (37). In agreement with this idea, we found that, while the D36A, D36N, and D36L mutant forms are inactive, the D36A/F40D double-mutant variant remains fully active. Another possible explanation for the requirement of an acidic residue is that such a negative charge in MrpF will impose an electrostatic field on the pump subunits on its left side, thus affecting their conformational change through long-range electrostatic interactions.
In the left-side region (i.e., MrpA end) of the complex, both the amphipathic HL helix and βH belt facilitate the structural correction, by interacting with the two surfaces of the lipid bilayer. While reducing the stress of the surrounding lipid bilayer, adjustment from a distorted form to a planar conformation is likely to generate stress within the protein complex itself. In other words, compared to the quasi ground state structure surrounded by a detergent micelle, the lipid bilayer environment places the Mrp complex in a “stressed” conformation, which may lower the transition barrier between the inward- and outward-facing conformations.
The well-accepted alternating access model (38) states that each transporter possesses two terminal states/conformations, denoted as Cin and Cout, permitting the substrate binding site(s) alternating access to the two sides of the membrane. A considerable number of studies found that conformational changes in a transporter comply with the same symmetry of its structure, which ultimately originates from the symmetry of the lipid bilayer (39). In the two conformations, the transporter is likely to show distinct affinity to the substrate(s), and this difference is associated with an energy term ΔGD (º RT × ln(Kd,1/Kd,0)), which is essential for energy coupling (40). In cases where transport of the driving substance and the substrate are carried out by two separate modules (as in the Mrp complex), communication and energy coupling between the modules become crucial for an efficient and cooperative transport process. Based on the ground-state structure of the Mrp complex, we hypothesize that the modules for transporting driving substance and substrate are synchronized in Cin−Cout transition, instead of opposing each other. For this type of antiporters, the driving substance (i.e., H+ for Mrp) first drives the transporter from its Cout ground state to the Cin excited state, waiting for the binding of substrate (Na+). Part of the input energy from the driving substance is stored in the excited state as conformational energy, ΔGC. Only upon substrate binding will the driving substance dissociate from the transporter, and ΔGC is released to drive the substrate-carrying, Cin-to-Cout conformational change. Only upon dissociation of the substrate in the Cout state can binding of the driving substance be renewed to initiate the next cycle of transport. This order of binding and release events of the reactants is essential for an effective antiporter; otherwise, the cellular energy source in the form of transmembrane electrochemical potential will be wasted (40).
In the ground state of the Mrp complex, four five-TM bundles from the NTM and CTM domains of both MrpA and MrpD subunits are arranged in a linear head-to-tail string (Fig. 1C). Structures of these helix bundles are superimposable (25), suggesting that they form rigid-body elements that are involved in the conformational changes required for the transport process. In this string of helix bundles, adjacent domains are restrained by a pseudo twofold screw symmetry, suggesting a possibility that the same symmetry also underlies the conformational changes required for transport. Structure and functional analyses suggest that the interface between NTM and CTM within each Mrp pump subunit constitutes the inward-facing pathway, and the two outward-facing pathways are located at the CTM (left) side of MrpA and the NTM (right) side of MrpD (SI Appendix, Fig. S10). Our more detailed structural analysis revealed two types of interactions between these helix bundles. For type I, the interdomain packings within both MrpA and MrpD are relatively tight. In the ground state, such packing represents the closed form of the inward-facing pathway. For type II, the packing between MrpA and MrpD is relatively loose. Repacking between these helix bundles with a subsequent switch between types I and II upon binding of protons will result in a number of changes in the Mrp pump subunits. First, due to the insertion loops in both TM7 and TM12, the gap between the five-TM bundles will change significantly upon repacking. For example, switching the packing at the NTM−CTM interface within MrpA or MrpD from type I to type II will adequately open the pathway from the central polar axis toward cytosol. Such repacking between NTM and CTM can be achieved simply by a relative rotation around an axis parallel to the membrane normal. Second, the π-bulge in TM 8 may contact either side of TM 11, thus serving as a toggle switch by participating in either type I or type II packing. As mentioned above, no major structural constraint exists within the string of helix bundles, except the HL helix and βH belt. Under external forces from PMF, both the HL helix and βH belt are likely to serve as structural elements for ΔGC storage and for mechanical coupling between the proton- and Na+-transport modules. Hypothetically, in the absence of the HL helix and/or the βH belt, conformational changes in either MrpA or MrpD would function independently from each other, and the elongated complex would quickly fall apart, a process primarily driven by the input energy of PMF. In addition, the NTM and CTM domains of MrpD are covered by three flexible loops, including a double pass of the loop connecting TMs 13 and 14 of MrpD and the C-terminal tail of MrpC contacting the two domains of MrpD. However, none of these flexible loops seems rigid enough to inhibit the domain repacking in MrpD.
Regarding repacking between the NTM domain of MrpA and the CTM domain of MrpD, from type II to type I, the distance between a conserved ion pair, E132A5−K392D12b, is likely to be shortened, making the central polar axis continuous. Such an interaction may cause E132A5 to dissociate from K213A7a, and the freed Lys residue will further perturb the pKa value (the negative log of the acid dissociation constant) of the proton binding site(s) in MrpA. Such dynamic connection along the central polar axis would constitute the structural basis for information exchange between neighboring subunits. A similar interdomain communication mechanism has been proposed for the complex I proton-pumping system and termed a wave spring mechanism (22, 41).
On the basis of our structural analysis presented here, we propose a model that describes the functional cycle of Na+/H+ antiport in the Mrp complex (SI Appendix, Fig. S11). In the ground state, multiple protons are bound to the central polar axis in the pump domain of MrpA from the extracellular side via the left-end, outward-facing proton path. Because of the ΔΨ-related electrostatic force on the protons, the pump domain undergoes a Cout-to-Cin transition, that is, closing the outward-facing path and opening the inward-facing path. In the Cin state, the protons remain associated due to high pKa relative to cytosolic pH, and thus the pump domain maintains its Cin state. Because of structural coupling between MrpA and MrpD, MrpD will then undergo a similar Cout-to-Cin transition and store ΔGC. The substrate Na+ is subsequently loaded to the central polar axis via the inward-facing path that is formed in the Cin state of MrpD. This cationic substrate electrostatically repels K392D12b in the hydrophilic core of CTM, thus prompting formation of an ionic pair with E132A5 in MrpA. The E132A5 residue will then abandon its ion pair partner K213A7a of the ground state. The released positive charge of K213A7a will reduce pKa of the proton binding site(s) in MrpA and trigger proton release, thereby achieving a long-distance competition between bindings of the substrate (Na+) and driving substance (H+). It was suggested, in a previous study, that such a competitive relationship is essential for the functional cycle of any canonical antiporter (40). Dissociation of the protons eliminates the electrostatic force that maintains the Cin state, leading to the release of ΔGC and consequently to the Cin-to-Cout transition in both MrpA and MrpD. The substrate-carrying conformational change in MrpD is powered by ΔGC, resulting in an increase of the electrochemical potential of the substrate Na+ (i.e., moving against both the concentration gradient and ΔΨ). Consistent with this mechanistic model, the redox-driven MBH complex contains only one Mrp antiporter domain, MbhH, which is the counterpart of MrpD, and the role of MrpA is replaced by the redox module.
Whether all proton influx in the Mrp complex is carried out exclusively through MrpA remains an open question. Because of the energetically uphill movement of Na+, a simple antiporter with a low stoichiometry ratio of H+:Na+ appears to be insufficient to drive proton uptake, especially in a highly alkaline environment (see discussion in SI Appendix, Supplementary Material). Thus, more-powerful proton pump subunits might be beneficial to a transport cycle in the desired direction. If MrpD shares part of the proton transport, a similar mechanism of the Mrp pump described for MrpA may also be applicable to MrpD. Such an antiporter mechanism is proposed for the homologous Na+ transporter module in the MBH complex (23). Another possibility would be that MrpA and MrpD exhibit inverse cooperativity. Here, MrpA and MrpD would alternate between Cin and Cout but in opposite phases. In the present ground state structure, however, both Mrp pumps of MrpA and MrpD appear to adopt the same Cout conformation. Therefore, data from the current structural study do not support an inverse cooperativity model. Whether the existence of both lipid bilayer and electrochemical potential would promote distinct ground-state structure of the Mrp complex remains to be further investigated.
Method
Cloning and Mutation Constructs.
The gene encoding Mrp was amplified from the genomic DNA of the Dietzia sp. DQ12-45-1b strain, which was obtained from the enriched cultures by phenol chloroform extracting method (42). The PCR products were cloned into the plasmid pUC18 with XbaI and HindIII restriction sites (29), and transformed into E. coli C43 (DE3) and E. coli KNabc for expression and functional assay, respectively. The wild-type Mrp complex was expressed and purified with a double-strep (strep II) tag at the N terminus of MrpA and a green fluorescence protein (GFP) at the C terminus of MrpG being preceded with a Human Rhinovirus (HRV) 3C protease cleavage site.
Expression and Purification of Mrp.
Recombinant Mrp proteins with MrpA strep II/MrpG GFP tags were overexpressed in the E. coli C43 (DE3) strain. Cells were grown in Terrific Broth supplemented with 100 µg/mL ampicillin at 37 °C for 16 h. Cells were harvested and resuspended in buffer A (50 mM Hepes [pH 7.5], 250 mM NaCl, and 5 mM β-mercaptoethanol) and subjected to three runs of homogenization at 10,000 to 15,000 p.s.i. (pounds per square inch) using a JN-R2C homogenizer (JNBio, Guangzhou, China). Cell debris was removed by centrifugation at 16,000 × g for 10 min, and the supernatant was ultracentrifuged at 100,000 × g for 45 min. The pellet membrane fraction was solubilized in buffer A supplemented with 1% (wt/vol) DDM (Anatrace) for 2 h at 4 °C. After second ultracentrifugation at 100,000 × g for 30 min, the supernatant was loaded onto Streptactin Beads 4FF (Smart Lifesciences) by gravity flow. Resin was further washed with buffer A containing 0.025% DDM, and the protein sample was eluted with buffer A containing 0.025% DDM and 2.5 mM d-desthiobiotin. Eluted protein sample was concentrated using 100-kDa cutoff concentrator (Millipore) and loaded onto a Superose-6 10/300 GL column (GE Healthcare) running in buffer B (50 mM Hepes [pH 7.5], 250 mM NaCl, 2 mM Tris [2-carboxyethyl] phosphine, and 0.025% DDM). Peak fractions were collected, and the pooled protein sample was concentrated to 7 mg/mL before cryo-EM sample preparation.
Cryo-EM Sample Preparation and Data Acquisition.
A droplet of 4 μL of 8 µg/µL purified Mrp complex was placed on Quantifoil R1.2/1.3 holey carbon grids glow-discharged for 60 s using 50% H2 and 50% O2. The grids were then blotted for 6 s at 4 °C under a blot force of level 0 at 100% humidity and flash frozen in liquid ethane using a Mark IV Vitrobot (FEI).
Cryo-EM data were collected on a 300-kV Titan Krios microscope (FEI) using a K2 camera positioned after a GIF quantum energy filter. The energy filter slit width was set to 20 eV. Micrographs were recorded in superresolution counting mode, at 130,000× magnification, corresponding to a calibrated physical pixel size of 1.04 Å, with the defocus ranging from –1.8 μm to –3.0 μm. A 6.4-s exposure was fractionated into 32 frames, each exposed for 0.2 s at a dose rate of 10 e− per pixel per s, resulting in a total dose of 60 e−/Å2.
Image Processing.
A total of 1,835 micrographs were corrected for beam-induced drift and binned by 2× using MotionCor2 with 5 × 5 patches (43). The contrast transfer function parameters of dose-weighted micrographs were determined by the software Gctf (44) and reference free particle picking was carried out using the software Gautomatch. Several rounds of 2D classification in Relion were used to remove ice contamination, micelles, and other false positives. From an initial set of 293,049 putative particles, 137,082 particles were selected for initial model generation and 3D classification. The initial model was prepared in Cryosparc (45), consisting of 49 TMs, which is consistent with our understanding of the Mrp complex. Consequently, this initial model served as a reference model for 3D classification in Relion. The subset of particles was categorized into five classes. The most populated class contained 93,505 particles, and this class was used for subsequent 3D refinement, resulting in a 3.5-Å-resolution map as estimated by FSC between two independently refined half-maps. The particle stack from Relion was further locally refined using cisTEM (Computational Imaging System for Transmission Electron Microscopy), which improved the density map features and the FSC resolution to 3.0 Å. To assess the quality of reconstruction, the FSCs between two half-maps and the angular distribution of particles used for refinement were plotted, and local resolution throughout the various maps was calculated using ResMap.
Model Building.
Homology models were extracted from the structure of T. thermophilus complex-I (PDB ID code 4HEA) (15) and of P. furiosus MBH complex (PDB ID code 6CFW) (23) as templates. The homology models were fitted as rigid bodies with the UCSF (University of California, San Francisco) Chimera software (46) and subsequently manually adjusted and rebuilt using the Coot software (47). The cryo-EM density map showed clear densities for most side chains, which provided a reliable guide to inspect and adjust the model. Refinement of the Mrp complex model against the cryo-EM map in real space was performed using the phenix.real_space_refine in the PHENIX (Python-based Hierarchical ENvironment for Integrated Xtallography) software package (48). The refined model was converted into a density map to calculate the FSC coefficients with the experimental density map using the phenix.mtriage software utility. The model stereochemistry was evaluated using MolProbity (49). All figures were prepared with Chimera or PyMOL (Schrödinger, LLC).
Salt-Resistance Assays.
To analyze the mutational effects of Mrp variants on the antiport activity, colony growth and cell culture assay were performed. Plasmids containing the variant gene were transformed into E. coli KNabc strain. In both types of assays, pUC18-mrp was used as the positive control, and the empty pUC18 vector was used as the negative control.
In the NaCl-resistance colony assay, cell cultures were first grown to 1.0 OD600 (the optical density of a sample measured at a wavelength of 600 nm) in LBK medium, and a series of dilutions (1- to 54-fold, 1 µL each) were spotted onto the solid medium of LBK supplemented with 200 mM NaCl, 100 µg/mL ampicillin, and 50 µg/mL kanamycin. After 16 h incubation at 37 °C, colony formation was recorded. For the salt-free control experiments, cell culture was first grown to 1.0 OD600, and a series of dilutions (10- to 105-fold, 1 µL each) were spotted onto the solid medium of LBK supplemented with 100 µg/mL Ampicillin and 50 µg/mL Kanamycin, but without NaCl.
In the cell culture assay, 120 µL of the abovementioned LBK media were used in each well of a flat-bottom 96-well microplate. All initial concentrations were adjusted to 0.05 OD600. The plate was incubated at 37 °C with a shaking rate of 600 rounds per min. Time courses for cell growth were recorded using a microplate reader.
High-Performance Liquid Chromatography Assays.
We utilized high-performance liquid chromatography to analyze the mutational effects of Mrp variants on the complex assemblage. Five milliliters of cells were harvested and resuspended in buffer A and subjected to homogenization using a JY92-IIN Ultrasonic homogenizer (Scientz). The membrane fractions were collected by ultracentrifugation at 100,000 × g for 30 min. The pellet membrane fractions were solubilized in buffer A supplemented with 1% (wt/vol) DDM for 1 h at 4 °C. After second ultracentrifugation at 100,000 × g for 30 min, the supernatants were loaded onto a Superose-6 10/300 GL column running in buffer B.
Western Blot Assays.
Western blotting was used to determine the expression of Mrp complex’s subunits in E. coli KNabc. Briefly, cells grown in LBK medium were harvested and resuspended in buffer A and subjected to homogenization using a JY92-IIN Ultrasonic homogenizer. The membrane fraction was collected by ultracentrifugation at 100,000 × g for 30 min. The pellet membrane fraction was solubilized in buffer A supplemented with 1% (wt/vol) DDM for 1 h at 4 °C. After second ultracentrifugation at 100,000 × g for 30 min, the supernatants were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis on a 15% polyacrylamide gel and then transferred to a polyvinylidene fluoride membrane at 20 V for 16 min using Semi Dry Transfer. Immunoblotting was performed using anti-strep II antibody (Bioworld Technology) with 1:5,000-fold dilution, horseradish peroxidase-conjugated secondary antibody with 1:10,000-fold dilution (Bioworld Technology).
Supplementary Material
Acknowledgments
We thank Prof. Lei Wang of China Agricultural University for kindly providing E. coli KNabc, and the staff in the Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences for help in cryo-EM data collection. We thank Dr. Xianjin Ou of Institute of Biophysics, Chinese Academy of Sciences for technical assistance in protein expression and purification. We thank Dr. T. Juelich (the University of Chinese Academy of Sciences, Beijing) for linguistic assistance during the preparation of this manuscript. The work was supported by National Key R&D Program of China (Grants 2018YFA0902100 and 2018YFA0902103 to X.-L.W.), Chinese Academy of Sciences Strategic Priority Research Program (Grants XDB08020301, XDB37030304, and XDB37030301 to X.C.Z.), and National Natural Science Foundation of China (Grant 31770120 to X.-L.W. and Grant 31971134 to X.C.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006276117/-/DCSupplemental.
Data Availability.
The coordinates of the Mrp complex have been deposited into PDB database with the accession code 7D3U. The cryo-EM map is accessible with EMDB (Electron Microscopy Data Bank) number EMD-30567.
All study data are included in the article and SI Appendix.
Note Added in Proof
We first submitted this manuscript on April 3, 2020. During review and production, an article “Structure and mechanism of the Mrp complex, an ancient cation/proton antiporter” was submitted by Steiner and Sazanov on June 2020 and published in July 2020 (50). The article reported the structure of Group-I Mrp complex from Anoxybacillus flavithermus (AfMrp). However, in contrast to the six subunits in DqMrp analyzed in our group, the AfMrp complex contains seven subunits. Second, the AfMrp structure was determined in an elongated dimer form, with the MrpE subunit serving as the dimerization interface. Third, in the AfMrp complex, a highly negatively charged cavity is reported to be located between the MrpA and MrpF subunits; however, there is no significant cavity observed in corresponding position in our DqMrp complex. Our DqMrp complex seems to use a Na+-transport path distinct from the one proposed for AfMrp. To identify and verify amino acid residues critically involved in the mechanism responsible for salt resistance, we introduced a series of positions in potential proton/Na+ transport pathways. Further studies on both systems might help characterize the transition motion for all proteins in the family.
Change History
September 10, 2021: The text of this article, including the affiliations list and the references list, has been updated; please see accompanying Correction for details.
References
- 1.Krulwich T. A., Sachs G., Padan E., Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krulwich T. A., Hicks D. B., Ito M., Cation/proton antiporter complements of bacteria: Why so large and diverse? Mol. Microbiol. 74, 257–260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C., et al., A two-domain elevator mechanism for sodium/proton antiport. Nature 501, 573–577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battaglino R. A., et al., NHA-oc/NHA2: A mitochondrial cation-proton antiporter selectively expressed in osteoclasts. Bone 42, 180–192 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunte C., et al., Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Williams K. A., Three-dimensional structure of the ion-coupled transport protein NhaA. Nature 403, 112–115 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Ito M., Guffanti A. A., Oudega B., Krulwich T. A., mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181, 2394–2402 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kajiyama Y., Otagiri M., Sekiguchi J., Kosono S., Kudo T., Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis. J. Bacteriol. 189, 7511–7514 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosono S., et al., Characterization of a multigene-encoded sodium/hydrogen antiporter (sha) from Pseudomonas aeruginosa: Its involvement in pathogenesis. J. Bacteriol. 187, 5242–5248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putnoky P., et al., The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol. Microbiol. 28, 1091–1101 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Hamamoto T., et al., Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 14, 939–946 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Swartz T. H., Ikewada S., Ishikawa O., Ito M., Krulwich T. A., The Mrp system: A giant among monovalent cation/proton antiporters? Extremophiles 9, 345–354 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Morino M., Ogoda S., Krulwich T. A., Ito M., Differences in the phenotypic effects of mutations in homologous MrpA and MrpD subunits of the multi-subunit Mrp-type Na+/H+ antiporter. Extremophiles 21, 51–64 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Morino M., Natsui S., Swartz T. H., Krulwich T. A., Ito M., Single gene deletions of mrpA to mrpG and mrpE point mutations affect activity of the Mrp Na+/H+ antiporter of alkaliphilic Bacillus and formation of hetero-oligomeric Mrp complexes. J. Bacteriol. 190, 4162–4172 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baradaran R., Berrisford J. M., Minhas G. S., Sazanov L. A., Crystal structure of the entire respiratory complex I. Nature 494, 443–448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuller J. M., et al., Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science 363, 257–260 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Zhu J., Vinothkumar K. R., Hirst J., Structure of mammalian respiratory complex I. Nature 536, 354–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Efremov R. G., Baradaran R., Sazanov L. A., The architecture of respiratory complex I. Nature 465, 441–445 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Mayer F., Müller V., Adaptations of anaerobic archaea to life under extreme energy limitation. FEMS Microbiol. Rev. 38, 449–472 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Verkhovskaya M., Bloch D. A., Energy-converting respiratory complex I: On the way to the molecular mechanism of the proton pump. Int. J. Biochem. Cell Biol. 45, 491–511 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Zhang X. C., Li B., Towards understanding the mechanisms of proton pumps in Complex-I of the respiratory chain. Biophys. Rep. 5, 219–234 (2019). [Google Scholar]
- 22.Kaila V. R. I., Long-range proton-coupled electron transfer in biological energy conversion: Towards mechanistic understanding of respiratory complex I. J. R. Soc. Interface 15, 20170916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H., et al., Structure of an ancient respiratory system. Cell 173, 1636–1649.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X. B., et al., Degradation of petroleum hydrocarbons (C6-C40) and crude oil by a novel Dietzia strain. Bioresour. Technol. 102, 7755–7761 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Efremov R. G., Sazanov L. A., Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Wang J., On the appearance of carboxylates in electrostatic potential maps. Protein Sci. 26, 396–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zickermann V., et al., Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347, 44–49 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Hiramatsu T., Kodama K., Kuroda T., Mizushima T., Tsuchiya T., A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 180, 6642–6648 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang H., Qin X. Y., Zhang K. D., Nie Y., Wu X. L., Role of the Group 2 Mrp sodium/proton antiporter in rapid response to high alkaline shock in the alkaline- and salt-tolerant Dietzia sp. DQ12-45-1b. Appl. Microbiol. Biotechnol. 102, 3765–3777 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Morino M., et al., Single site mutations in the hetero-oligomeric Mrp antiporter from alkaliphilic Bacillus pseudofirmus OF4 that affect Na+/H+ antiport activity, sodium exclusion, individual Mrp protein levels, or Mrp complex formation. J. Biol. Chem. 285, 30942–30950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sperling E., Górecki K., Drakenberg T., Hägerhäll C., Functional differentiation of antiporter-like polypeptides in complex I; a site-directed mutagenesis study of residues conserved in MrpA and NuoL but not in MrpD, NuoM, and NuoN. PLoS One 11, e0158972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajiyama Y., Otagiri M., Sekiguchi J., Kudo T., Kosono S., The MrpA, MrpB and MrpD subunits of the Mrp antiporter complex in Bacillus subtilis contain membrane-embedded and essential acidic residues. Microbiology (Reading) 155, 2137–2147 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Cooley R. B., Arp D. J., Karplus P. A., Evolutionary origin of a secondary structure: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J. Mol. Biol. 404, 232–246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz D. W., et al., Accommodation of amino acid insertions in an alpha-helix of T4 lysozyme. Structural and thermodynamic analysis. J. Mol. Biol. 236, 869–886 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Moparthi V. K., Kumar B., Mathiesen C., Hägerhäll C., Homologous protein subunits from Escherichia coli NADH:quinone oxidoreductase can functionally replace MrpA and MrpD in Bacillus subtilis. Biochim. Biophys. Acta 1807, 427–436 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Marsh D., Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys. J. 93, 3884–3899 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X. C., Li H., Interplay between the electrostatic membrane potential and conformational changes in membrane proteins. Protein Sci. 28, 502–512 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jardetzky O., Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966). [DOI] [PubMed] [Google Scholar]
- 39.Shi Y., Common folds and transport mechanisms of secondary active transporters. Annu. Rev. Biophys. 42, 51–72 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Zhang X. C., Zhao Y., Heng J., Jiang D., Energy coupling mechanisms of MFS transporters. Protein Sci. 24, 1560–1579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Bacete J., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T., Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1: Conserved charged residues essential for energy-coupled activities. J. Biol. Chem. 282, 36914–36922 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Nie Y., Liang J., Fang H., Tang Y. Q., Wu X. L., Two novel alkane hydroxylase-rubredoxin fusion genes isolated from a Dietzia bacterium and the functions of fused rubredoxin domains in long-chain n-alkane degradation. Appl. Environ. Microbiol. 77, 7279–7288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Pettersen E. F., et al., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afonine P. V., et al., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen V. B., et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steiner J., Sazanov L., Structure and mechanism of the Mrp complex, an ancient cation/proton antiporter. eLife 9, e59407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the Mrp complex have been deposited into PDB database with the accession code 7D3U. The cryo-EM map is accessible with EMDB (Electron Microscopy Data Bank) number EMD-30567.
All study data are included in the article and SI Appendix.