Significance
Many genes important for vertebrate development are surrounded by multiple series of remote enhancer sequences. Such regulatory landscapes and their target genes are usually located within the same chromatin domains, which appears to constrain the action of these regulatory sequences and hence to facilitate enhancer–promoter recognition and gene expression. We used the HoxD locus to assess the impact of modifying the regulatory topology upon gene activation in space and time. A series of chromosomal rearrangements involving deletions and inversions reveals that the enhancer topology plays a role in the timing of gene activation. However, gene expression was often recovered subsequently, illustrating the intrinsic capacity of some enhancers to find their target promoters despite an apparently adverse chromatin topology.
Keywords: enhancers, chromatin architecture, TAD boundary, gene regulation, Hox clusters
Abstract
The HoxD gene cluster is critical for proper limb formation in tetrapods. In the emerging limb buds, different subgroups of Hoxd genes respond first to a proximal regulatory signal, then to a distal signal that organizes digits. These two regulations are exclusive from one another and emanate from two distinct topologically associating domains (TADs) flanking HoxD, both containing a range of appropriate enhancer sequences. The telomeric TAD (T-DOM) contains several enhancers active in presumptive forearm cells and is divided into two sub-TADs separated by a CTCF-rich boundary, which defines two regulatory submodules. To understand the importance of this particular regulatory topology to control Hoxd gene transcription in time and space, we either deleted or inverted this sub-TAD boundary, eliminated the CTCF binding sites, or inverted the entire T-DOM to exchange the respective positions of the two sub-TADs. The effects of such perturbations on the transcriptional regulation of Hoxd genes illustrate the requirement of this regulatory topology for the precise timing of gene activation. However, the spatial distribution of transcripts was eventually resumed, showing that the presence of enhancer sequences, rather than either their exact topology or a particular chromatin architecture, is the key factor. We also show that the affinity of enhancers to find their natural target genes can overcome the presence of both a strong TAD border and an unfavorable orientation of CTCF sites.
During embryonic development, the precise control of gene activation in both time and space largely relies on the activity of cis-regulatory sequences. Such regulatory elements include insulators, enhancers, and repressive sequences that are either located in close proximity to the target gene or further away (1). Large regulatory distances can be overcome by the three-dimensional (3D) organization of chromatin that takes place at different levels (2). In this context, topologically associating domains (TADs) were defined as genomic intervals where chromatin interactions tend to take place more frequently than with adjacent regions (3, 4), and such domains are frequently understood as functional units that host enhancers and their target promoters (5). Indeed, some key developmental genes are found under the control of regulatory domains that are contained within TADs, which harbor tissue-specific regulatory sequences or multiple acting enhancers that confer robustness and resilience (6–10).
TADs and chromatin loops are thought to result from a loop extrusion mechanism that relies on the loading of the cohesin multiprotein ring. This protein complex allows the extrusion of the chromatin fiber until it is stopped or retained by CTCFs bound with convergent orientations, or by the stalling of two forming loops (11). The precise role(s) of these architectural proteins in gene expression have not yet been completely elucidated, and genome-wide depletion, either of CTCF or of the cohesin complex, did not have a pervasive effect on gene expression levels; and changes were clearly observed only at some genomic loci (12–15). Altogether, the relationship between chromatin topology and gene expression seems to be context-dependent and locus-specific. In some cases, indeed, deleting CTCF binding sites led to an alteration of the genes nearby (16–19) whereas other studies failed to reveal any obvious effects (20). Most of these studies nevertheless did not monitor gene expression using time course protocols in a physiological situation in embryo.
Hox clusters have been used as a paradigm of long-range regulation. The HoxD cluster is localized between two large TADs and acts itself as a boundary region, due to the high concentration of CTCF binding sites and their opposed orientations (21, 22). The centromeric domain (C-DOM) controls Hoxd gene transcription during the late, second phase of limb development, which accompanies the emergence of digits (7, 23). The telomeric domain (T-DOM) controls the early phase of transcription in limb buds (21, 24, 25), as well as in the cecum (26) and the mammary buds (27). T-DOM is divided into two sub-TADs by a chromatin boundary (CS38-40) containing three bound CTCFs, all oriented toward the HoxD cluster (22) (Fig. 1A).
Fig. 1.
The HoxD locus and its regulatory landscapes. (A) Scheme of the HoxD locus showing the gene cluster surrounded by limb enhancers (blue and yellow circles) and neighboring genes (gray boxes). A ChIP of CTCF in E12.5 proximal forelimbs is shown, as well as the orientation of their binding sites (blue and red arrowheads). The regulatory domains C-DOM (blue) and T-DOM (green, split in two sub-TADs) are represented as triangles. On the left, an E12.5 limb diagram shows the tissue where C-DOM and T-DOM are active, respectively. (B) Magnification of HoxD and the T-DOM region. Top shows a Hi-C map of E12.5 proximal limb (data from ref. 22). The arrow shows the sub-TAD division. The green tracks are ChIP datasets of H3K27ac in E12.5 proximal limb (PFL) and E9 forelimb buds at the 24- to 29-somite and 18- to 22-somite stage. The black tracks at the bottom are ChIP tracks of RAD21 and CTCF of E9 forelimbs at the 20- to 28-somite stage. The y axis shows ChIP signal coverage. (C) LacZ staining of mouse embryos showing limb (arrowheads) and mesodermal enhancer (arrows) activity of several transgenic constructs at E10.5 (ELCR2) and E.9.5 (ELCR3, CS39, and CS65). Picture was taken with a 2× magnification.
In the early limb bud, T-DOM is activated at E9.0 (embryonic day 9), leading to the first wave of colinear transcription, coinciding with the establishment of chromatin interactions between the newly activated genes (Hoxd9 to Hoxd11) and part of T-DOM (21). At E12.5, cells transcribing these genes are found in the proximal part of the limb buds, which will generate the arm and the forearm. At this stage, the distribution of interactions with T-DOM shows a clear topological segregation, with 3′-located genes (Hoxd1 to Hoxd8) interacting mostly with the first sub-TAD, whereas 5′-located genes (Hoxd9 to Hoxd11) associate in priority with the more distant sub-TAD (21, 22), suggesting a functional compartmentalization of T-DOM. All limb-specific enhancers were thus far associated to the distant sub-TAD, starting at the sub-TAD boundary and extending up to Hnrnpa3, including the CS65 and CS93 enhancers (21, 28).
In this work, we set up to assess whether a correlation exists between the precise temporal and spatial transcriptional activation of Hoxd genes in proximal limbs on the one hand, and a fine topological organization of its regulatory landscape on the other hand, or whether the mere presence of series of enhancers within T-DOM is necessary for HoxD regulation, regardless of their intrinsic organization. We show that, while the overall chromatin architecture determines the correct timing of gene activation, enhancer–promoter communication can be successfully established along with limb bud development, even after the engineering of major topological modifications, including the positioning of a strong TAD border in between them.
Results
Multiple Early Limb Enhancers in T-DOM.
Hi-C profiles from several cell types have previously revealed that the HoxD cluster is positioned at the boundary between two TADs. T-DOM (i.e., the TAD located telomeric to the gene cluster) is necessary for the transcription of Hoxd genes, both during limb budding and, subsequently, in the formation of the proximal segment of the prospective arm. Instead, the C-DOM controls Hoxd gene expression in developing digits, at later time points (Fig. 1A). From E9.5 to E12.5, T-DOM shows specific activation and decommissioning dynamics (21), which correlates with its 3D conformation, as only the more distant T-DOM sub-TAD (Fig. 1A, sub-TAD2) remains active at late (E12.5) embryonic stages. Most limb enhancers described thus far are located within this chromatin domain, in particular the CS39, CS65, and CS93 sequences (21, 28, 29).
In order to characterize the onset of activation of T-DOM in the incipient limb bud, we microdissected E9 forelimb buds and pooled them into two groups corresponding to embryos either between 18 and 22 somites (or early E9), or between 24 and 29 somites (or late E9). Chromatin immunoprecipitation (ChIP) of H3K27ac, a histone mark associated with enhancer activity and gene expression, revealed that most of the acetylated regions were located in sub-TAD2, which seemed particularly active in 24- to 29-somite-old limb buds (Fig. 1B). Two H3K27ac-positive regions were nevertheless identified in sub-TAD1 in E9 limb buds, which were not present in E12.5 proximal limb cells. In contrast to CS39, CS65, and CS93, however, these two early limb control regions (ELCR2 and ELCR3) were not found fully conserved in chicken, albeit they are present in all mammals (SI Appendix, Fig. S1) (21, 28). Transgenic analysis of both ELCR2 and ELCR3 showed strong LacZ expression in E9 limb buds, which coincides with the expression of CS39 and CS65 transgenes (Fig. 1C, arrowheads), as well as in other mesoderm derivatives (Fig. 1C, arrows).
To evaluate potential changes in the global architecture of T-DOM along with developmental timing, we looked at the binding profiles of both CTCF and the cohesin subunit RAD21. The ChIP profiles of these architectural proteins using limb buds from 20- to 28-somite embryos did not substantially differ from the profiles obtained in E12.5 proximal limb (Fig. 1B; and figure 2 A and B of ref. 22). Most of the CTCF binding sites had a convergent orientation in relation to the HoxD cluster, including the three bound CTCFs found within the CS38-40 region, the boundary region that divides T-DOM into its two sub-TADs (Fig. 1B, arrow).
Deletion of the T-DOM Sub-TAD Boundary.
We asked whether such a partitioning of T-DOM into two subdomains was mandatory for this early limb bud regulation to be properly implemented. We tried to merge both domains by deleting the CS38-40 region (SI Appendix, Fig. S2A), which contains three CTCF binding sites as well as the CS39 limb enhancer and the transcription start site (TSS) of the Hog and Tog long noncoding RNA (lncRNAs) (22, 26), which coincides with a CpG island. We performed circular chromosome conformation capture sequencing (4C-seq) experiments in E9.5 mutant forelimb buds using several viewpoints distributed both along T-DOM and inside the HoxD cluster (Fig. 2 and SI Appendix, Fig. S2B). We then compiled all data from the different viewpoints into a virtual Hi-C matrix using the 4Cin software (30), which we adapted to plot the relative distances in a linear manner according to the real genomic coordinates. Because the 4Cin processing has an inherent variability that leads to the generation of different models, we assessed the correlation between 20 iterations and clustered them, thus displaying a merged average (Materials and Methods).
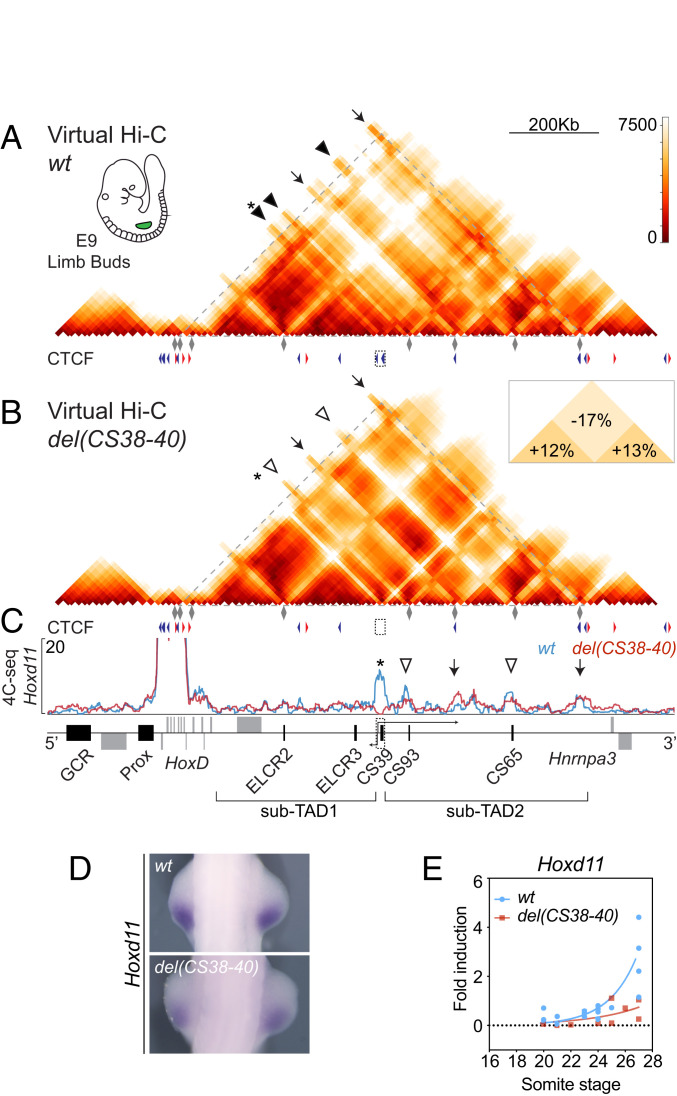
Fig. 2.
Deletion of region CS38-40. (A and B) Virtual Hi-C maps of wild-type (A) and del(CS38-40) (B) as reconstituted from 4C-seq datasets of E9 forelimb buds. The 4C viewpoints used to compose the matrices (gray diamonds) as well as the positions and orientations of the CTCF sites are depicted below. The scale on the right represents the computed distances between the different bins. An asterisk indicates the diagonal corresponding to the deleted region. Arrowheads and arrows mark the position of some enhancers and CTCF binding sites, respectively. The Inset in B shows the percentage change (mutant vs. wild type [wt]) in distances inside and between sub-TADs. (C) The 4C-seq profiles of Hoxd11 of E9 of wt and mutant forelimb buds (blue and red lines, respectively). The deleted region is shown as a dashed box around CS39, from which two noncoding transcripts are generated in the wild type. Arrows arising from the dashed box around CS39 represent the Hog and Tog lncRNAs in the wild type. Asterisk, arrowheads, and arrows indicate the same positions as in A and B. (D) WISH analysis of Hoxd11 in E9 forelimbs (∼26 to 27 somites) (6.3× magnification). (E) Comparison of individual qRT-PCR values (wt n = 17 and mutant n = 11) of Hoxd11 in E9 forelimb buds at different somite stages. An exponential fit is represented out of the real qRT-PCR values.
When applied to control limbs, the 4Cin approach generated a map of computed distances at T-DOM that resembled the expected contact distribution of a wild-type Hi-C matrix, including the subdivision of the domain in two sub-TADs, as well as specific contacts between CTCF-bound regions and enhancer–promoter interactions (Fig. 2A, arrows and arrowheads, respectively). Using the same viewpoints, we confirmed that the deletion of region CS38-40 affected the spatial organization of this regulatory domain. We observed a substantial increase in the interactions established between the two sub-TADs, with a 17% decrease in computed distances between the two subdomains (P value = 3e−49) leading to their fusion into a single domain (Fig. 2B). This was accompanied by an apparent loosening of the interactions inside each subdomain, seen as an increase in distances within sub-TAD1 (12%; P value = 0.008) and sub-TAD2 (13%; P value = 2e−12) (Fig. 2 B, Inset). The increase in contacts between the two sub-TADs could be observed when comparing any of the derived cluster representations (SI Appendix, Fig. S3 A and B). Also, the interactions established by the HoxD cluster throughout the regulatory domain seemed to decrease (Fig. 2 A and B, arrowheads).
More specifically, we analyzed the interaction profile of the Hoxd11 gene, whose expression in the posterior part of the E9.5 developing limb bud is maintained until E12.5, in the proximal limb. The 4C-seq data revealed that, in wild-type E9.5 limb buds, Hoxd11 strongly interacted with both region CS38-40 and the more distant sub-TAD2 (Fig. 2C). Upon deletion of the sub-TAD border, a modest increase in interactions was detected in the bound CTCF sites located 3′ to region CS38-40 and at the telomeric TAD border close to Hnrnpa3 (Fig. 2C, arrows). However, the contacts did not increase substantially along the region initially corresponding to sub-TAD2. Instead, interactions of Hoxd11 were reduced with the CS93 and CS65 limb enhancers, representing 65% and 81%, respectively, when compared to wild type (Fig. 2C, open arrowheads and SI Appendix, Table S1). These changes were specific to Hoxd11 as neither Hoxd9 nor Hoxd4 showed the same trend regarding enhancer interactions (SI Appendix, Fig. S2B and Table S1).
We assessed whether these alterations in contact distribution translated into changes in gene expression pattern. Whole-mount RNA in situ hybridization (WISH) showed a slight but visible decrease in Hoxd11 expression at E9.5 (Fig. 2D). To verify this observation, we performed qRT-PCR on forelimb buds dissected from embryos aged between 20 and 28 somites and plotted their individual values (Fig. 2E). The dynamics of Hoxd11 expression in control forelimb buds followed a strong increase right after the 24-somite stage. On the contrary, this dynamics in Hoxd11 messenger RNA (mRNA) was not observed in the mutant limb buds where the increase was not as fast (Fig. 2E). This was further confirmed by RNA-seq experiments showing that Hoxd10 and Hoxd12 had a delayed onset of transcription while more anterior genes (i.e., Hoxd4) did not seem to be affected at all (SI Appendix, Fig. S3C), in agreement with the lack of modification in enhancer contacts for these genes (SI Appendix, Fig. S2B). Altogether, the specific down-regulation of Hoxd11 could either be a consequence of the distinct spatial reorganization of T-DOM or be due to the removal of the CS39 enhancer.
To explore these possibilities, we used a CRISPR/Cas9 approach to eliminate the binding of CTCF to the three motives positioned within region CS38-40. We initially deleted 26 base pairs (bp) of the CTCF binding site located in region CS38 [delCTCF(CS38)] (SI Appendix, Fig. S4A), preserving both the neighboring CpG island and the TSS of the Hog and Tog lncRNAs (SI Appendix, Fig. S2A). On top of this first editing, we generated a 1.5-kilobase (kb) large deletion that removed the two binding sites located around CS40, without removing the H3K27ac-enriched region localized around CS39 [delCTCFs(CS38;CS40)] (SI Appendix, Figs. S2A and S4A). We confirmed by ChIP that CTCF binding was no longer detected at any of these locations or elsewhere in this short DNA interval (SI Appendix, Fig. S2A). The deletion of the three CTCF sites led to a merge of the sub-TADs at E12.5 (SI Appendix, Fig. S4 B and C), thus confirming the importance of these bound proteins in the establishment of this specific topological structure. We analyzed gene expression and observed that Hoxd11 was briefly delayed in its activation, a lag that was rapidly resumed to generate a late pattern indistinguishable from wild type (SI Appendix, Fig. S4D). These results indicated that the merging of the two T-DOM sub-TADs moderately affected the onset of Hoxd gene expression in early limb buds, with a stronger effect observed in the absence of the CS39 enhancer.
Reinforcing a Sub-TAD Separation.
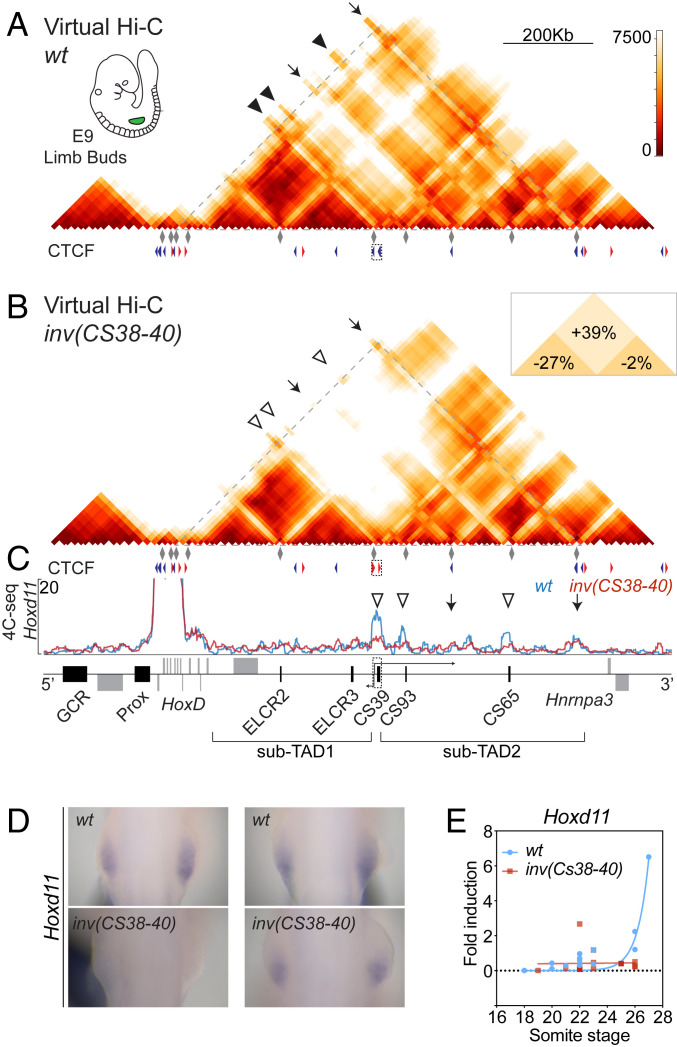
We next engineered the opposite situation to produce a more robust separation between the two sub-TADs such as to isolate them from one another as bona fide TADs. This was achieved by generating an inversion of the region comprising the three CTCF binding sites [the inv(CS38-40) allele]. In this configuration, the three CTCF sites now converged toward the strong telomeric TAD border at the 3′ end of the domain. In this allele, the three CTCF sites were still occupied, as expected (SI Appendix, Fig. S5A). A virtual Hi-C pattern of limb bud cells dissected from this mutant stock expectedly revealed that the inversion of this region had strengthened the segregation of the two sub-TADs. Indeed, a 39% increase in distances was observed (P value = 2.45e−163) while the compaction within sub-TAD1 decreased by 27% (P value = 1e−7) (Fig. 3 A and B, Inset). Concomitantly, it reduced the general contacts of the HoxD cluster with region CS38-40, as well as between some Hoxd genes and the CS93 and CS65 enhancers (Fig. 3 A and B and SI Appendix, Fig. S5B and Table S1). This was particularly well illustrated by using Hoxd11 as a 4C-seq viewpoint, showing a reduction of interactions over regions CS38-40, CS93, and CS65 (Fig. 3C), similar to what had been noted in the del(CS38-40) allele (Fig. 2C). These topological changes also correlated with a delay of Hoxd11 expression (Fig. 3 D and E), which was stronger than in the CTCF mutant alleles. This delay was nevertheless not pervasive for it did not affect all Hoxd mRNAs equally. For example, Hoxd9 did not show a clear transcriptional decrease, even at early stages (SI Appendix, Fig. S5C), whereas more “posterior” genes (like Hoxd11) seemed to be more affected. All these delays, however, were subsequently resumed, and, in E12.5 forelimb buds, changes in expression patterns could hardly be scored when comparing the inv(CS38-40) allele to wild-type littermates (SI Appendix, Fig. S5D).
Fig. 3.
Inversion of region CS38-40. (A and B) Virtual Hi-C maps of wild-type (A) and the inv(CS38-40) allele (B) from E9 forelimb buds, as reconstituted from several 4C-seq viewpoints (gray diamonds). The triangles showing CTCF orientation of region CS38-40 are inverted in the mutant. Arrowheads and arrows mark the position of some enhancers and CTCF binding sites, respectively. The Inset in B shows the percentage of the changes (mutant vs. wt) in distances inside and between sub-TADs. (C) The 4C-seq profiles of Hoxd11 for wt and mutant E9 forelimb buds (blue and red lines, respectively). The inverted region is shown as a dashed box around CS39. Open arrowheads show areas of decreased interaction in the inv(CS38-40) allele. Arrows indicate the position of some CTCF binding sites. (D) WISH analysis of Hoxd11 in E9 (18 to 22 somites) forelimb buds (earlier time point on the Left) (8× magnification). (E) Comparison of qRT-PCR values (wt n = 16 and mutant n = 17) of Hoxd11 in E9 forelimb buds at different somite stages. An exponential fit is represented out of the real qRT-PCR values.
Inversion of T-DOM.
Altogether, these genomic alterations did not produce long-lasting effects upon the transcription of Hoxd genes in limb buds. One possible explanation is that T-DOM contains several other CTCF binding sites, most of them displaying an orientation convergent to those numerous sites present in the telomeric part of the HoxD cluster itself. In this context, it is possible that such CTCF sites within T-DOM may assist remote enhancers reaching targets, regardless of small rearrangements occurring at their vicinity. We thus inverted the entire T-DOM and produced two novel configurations, one containing a strong TAD border between the inverted T-DOM and the HoxD cluster, and the other one lacking this TAD border (Figs. 4 and 5, Bd).
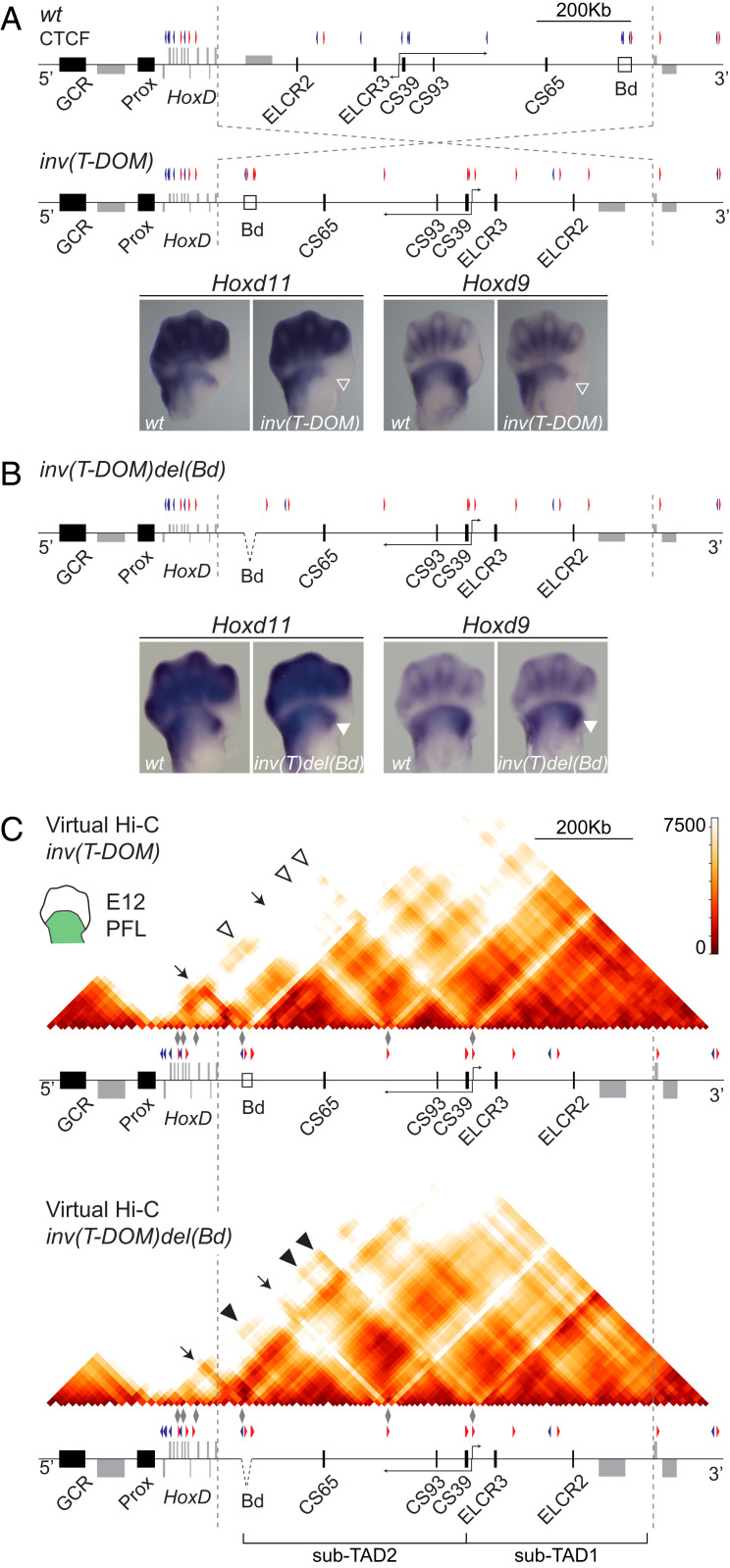
Fig. 4.
Inversion of T-DOM in the presence of a TAD border. (A and B) Virtual Hi-C maps of wild-type (A) and inv(T-DOM) mutant mice (B) from E9 forelimb buds, as reconstituted from several 4C-seq viewpoints (gray diamonds). Dashed lines indicate the inverted region. The distant boundary is marked as an empty box (Bd) at the end of T-DOM in the wild type and close to the HoxD cluster in the mutant allele. The CTCF orientations in the inv(T-DOM) allele are inverted accordingly. In the mutant allele, open arrowheads represent lost interactions that can be scored in the wt (filled arrowheads). Arrows indicate the position of some CTCF binding sites, which are located close to the HoxD cluster after the inversion of T-DOM. (C) qRT-PCR values (wt n = 17 and mutant allele n = 21) of Hoxd11 in E9 forelimb buds at different somite stages. An exponential fit is represented out of the real qRT-PCR values. (D) WISH images of forelimb buds at E9.5 (Top, ∼20 somites, 6.3× magnification) and E10.5 (Bottom, 4× magnification) of wt and the inv(T-DOM) mutant allele. The loss of expression in the mutant is indicated as open arrowheads.
Fig. 5.
Inversion of T-DOM in the absence of a TAD border. (A) Scheme of the rearranged HoxD locus after inversion of T-DOM. The orientation of CTCF sites is represented. WISH images of Hoxd11 and Hoxd9 expression pattern in E12.5 forelimbs are shown for both wt and inv(T-DOM) mutant embryos. An open arrowhead demarcates the loss of expression in the anterior part of the proximal limb domain for both Hoxd9 and Hoxd11 (3.2× magnification). (B) Scheme showing the deletion of the TAD boundary (Bd) on top of the inv(T-DOM) allele. WISH of Hoxd11 and Hoxd9 using E12.5 wt and inv(T-DOM)del(Bd) -inv(T)del(Bd)- mutant forelimbs shows the same patterns with the reappearance of the anterior domain missing in the inv(T-DOM) allele (arrowhead) (3.2× magnification). (C) Virtual Hi-C maps of E12 proximal limb buds using either inv(T-DOM), or inv(T-DOM)del(Bd) mutant limb bud cells. Both matrices were mapped on the artificial inv(T-DOM) genome. Dashed vertical lines demarcate the extension of the inverted region in both mutant lines. Filled arrowheads indicate the increase in contacts with enhancers CS65, CS93, and CS39 [indicated with open arrowheads in inv(T-DOM)] after the deletion of the TAD boundary (Bd).
The inversion of T-DOM was induced by targeting CRISPR guides at both sides of this regulatory domain. To be as inclusive as possible, the break points of this inversion were selected close to the 3′ end of the Hoxd1 gene and 5 kb upstream of the TSS of the Hnrnpa3 gene, respectively (Fig. 4 A and B, dashed lines). Due to the position of the latter break point, the Hnrnpa3 TAD border (Fig. 4A, Bd) was inverted along with T-DOM and placed just between the HoxD cluster and the inverted T-DOM. Upon inversion, a substantial loss of contacts was scored by using 4Cin on E9.5 limb bud cells, all along the T-DOM and particularly in the region containing the ELCR2 and ELCR3 enhancers, which were now located further away (Fig. 4 A and B).
In contrast, the CS65, CS93, and CS39 enhancers, which were initially located further telomeric, were now repositioned much closer to the HoxD cluster in the inverted allele. These enhancers were nevertheless separated from their natural Hoxd target genes by a very efficient TAD border (Fig. 4B, Bd). Of note, however, some interactions could still be observed between Hoxd genes and these regulatory regions although clearly diminished when compared to the control (Fig. 4 A and B and SI Appendix, Fig. S6C and Table S1). These interactions took place despite this TAD border (Bd), which otherwise tightly isolated the inverted T-DOM from the HoxD cluster, creating a new and well identified TAD. Again, these changes were observed regardless of the model that was generated in our virtual Hi-C approach (SI Appendix, Fig. S6 A and B). While the interactions between the HoxD cluster and the T-DOM enhancers in sub-TAD2 were remarkably weaker, they were observed, in particular when looking at the 4C-seq profiles (SI Appendix, Fig. S6C). In contrast, the gene cluster was no longer interacting with the enhancers located in sub-TAD1, which had been located further away upon inversion (SI Appendix, Fig. S6C, ELCR2 and ELCR3).
The inversion of the whole regulatory domain was accompanied by a severe delay in the onset of Hoxd11 expression, which was not detectable before E10.5 at the most posterior aspect of the growing bud (Fig. 4 C and D), likely due to the weak interactions with T-DOM enhancers. This strong variation in the timing of expression was subsequently translated into an absence of both Hoxd9 and Hoxd11 transcripts in the most anterior part of the proximal expression domain at E12.5: i.e., when the transcript domains have reached their final spatial deployments (Fig. 5A, open arrowheads). Altogether, these results demonstrated that, in this case, a delay in Hox gene activation impacted upon the spatial distribution of their transcripts. They also illustrate that enhancers seem to be able to still contact their natural target genes, despite the presence of a strong ectopic TAD border in between.
Enhancer Tropism over Chromatin Topology.
To see whether the altered expression timing and patterns of Hoxd9 and Hoxd11 were due either to the mere inversion of T-DOM or to the introduction of a strong new TAD border between the gene cluster and T-DOM, we further deleted the boundary region on top of the inverted allele to produce the inv(T-DOM)del(Bd) mutant line (Fig. 5). The deleted 20-kb-large boundary region, which normally tightly isolates T-DOM from its more telomeric TAD, contained three CTCF binding sites. Fetuses carrying this additional deletion fully recovered wild-type expression patterns for both Hoxd9 and Hoxd11, with expression domains in the limb buds undistinguishable from their wild-type counterparts (Fig. 5B). In particular, the proximal-anterior transcript domain lacking in inv(T-DOM) embryos (Fig. 5A, open arrowheads) was fully rescued after deletion of the ectopic TAD border (Fig. 5B, filled arrowheads).
This recovery in expression was concomitant to a clear increase in interactions between the HoxD cluster and various T-DOM limb enhancers when looking both at the virtual Hi-C matrices (Fig. 5C and SI Appendix, Fig. S7A) and to the Hoxd gene 4C-seq profiles (SI Appendix, Fig. S7B and Table S1). The reestablishment in the spatial deployment of transcripts at day E12.5 was, however, not observed at the earliest stages analyzed, which still showed an important time lag in target gene activation, even though the ectopic TAD border had been removed (SI Appendix, Fig. S7C). These results showed that the mere inversion of the regulatory domain had an impact upon the onset of HoxD expression. The observed delay could nevertheless be caught up in a few days, a recovery that was not completely possible when the telomeric TAD border was present between the enhancers and the target Hoxd genes.
Discussion
The fine-tuned regulation of genes involved in developmental processes is often achieved by complex regulatory landscapes, which can extend up to megabases around the target gene(s). Such regulatory landscapes generally match the extents of TADs and contain all of the enhancers necessary for the various expression specificities. Even though a clear causal relationship is difficult to establish, the prevalent model is that TADs somehow restrict the sphere of operation for such regulations by providing a spatial unit where genes can be properly controlled, in isolation from their neighbors. The action of enhancers is thought to depend on their 3D spatial proximity to the target promoters they regulate, a hypothesis supported by several lines of evidence (reviewed in refs. 1 and 2). Recent reports, however, have challenged this view, showing that transcriptional activity does not always correlate with a direct promoter–enhancer physical interaction (31–33).
In this work, we used the HoxD locus and one of its two flanking regulatory landscapes as a paradigm to look at the effect of modifying the regulatory topology upon the precisely orchestrated transcription of this series of genes. We engineered several rearrangements within the regulatory domain to determine the impact of both the distribution of enhancer sequences, the presence and orientation of CTCF binding sites, and the ectopic introduction of a TAD border between the promoters and the corresponding enhancers. We conclude that, while the global TAD architecture may serve to properly implement the regulatory modalities in time, major rearrangements do not critically modify the regulatory outcome at a later stage, making enhancer–promoter contacts very resilient and somewhat poorly dependent from the architectural context.
A Split Regulatory Landscape.
T-DOM is normally divided into two sub-TADs at the level of region CS38-40 (21), a region that contains three CTCF sites with an orientation convergent to that of numerous sites within the HoxD cluster (22). The deletion of this border region expectedly led to the fusion of the two sub-TADs. However, rather than reenforcing contacts between enhancers and promoters in the de novo created single TAD, enhancer–promoter contacts tended to decrease. Therefore, the presence of these two subdomains within T-DOM favors maximal efficiency in the regulatory outcome (see summary scheme in Fig. 6). One potential explanation is that it is not the global structure itself that is important but, instead, the presence of three CTCF binding sites that may trigger part of the necessary interactions, in particular due to their shared orientation toward the HoxD cluster. In this deleted allele, Hoxd genes were expressed rather normally, but with a clear delay in their activation.
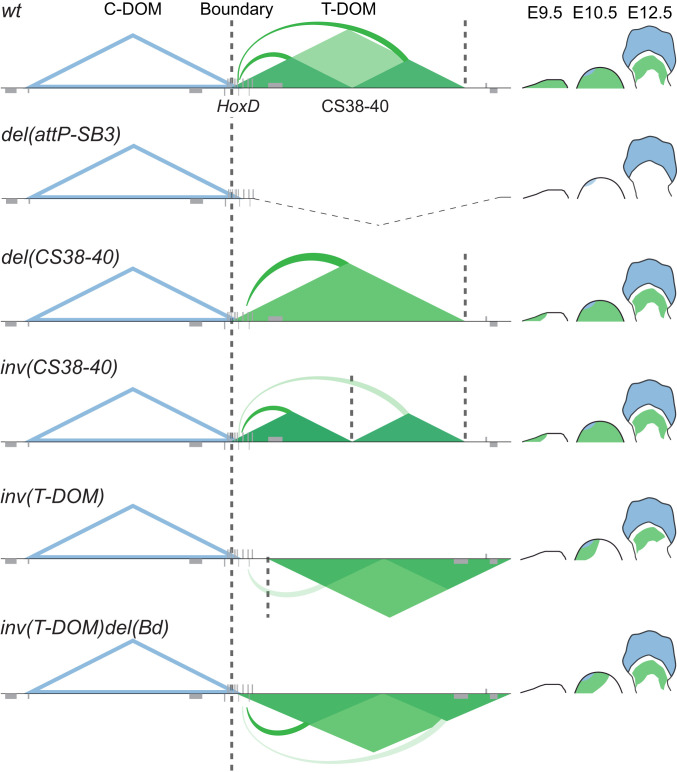
Fig. 6.
Summary of the regulatory effects observed in the various alleles. The C-DOM TAD (blue) is only active in distal cells during late limb development, and hence it does not interfere with the current alleles. T-DOM (green) controls the transcriptional onset of Hoxd genes during early development (E9.5) and maintenance in the proximal limb bud until day 12.5. On Top, the deletion of T-DOM abolished HoxD gene expression in the proximal limb (21). Deletion of region CS38-40 leads to the merging of the two sub-TADs and a short delay in Hoxd gene activation. Inversion of the same region increases the segregation of the distant sub-TAD, while also delaying expression, which is subsequently rapidly recovered. The inversion of T-DOM and introduction of an ectopic TAD boundary hampers access of Hoxd genes to the domain. The delay in the activation is more severe. There is also an impact on the spatial distribution of transcripts in the anterior aspect of the proximal domain. The further deletion of this TAD border restores promoter–enhancer interactions at a later stage whereas a strong impact is still visible during the earliest stages of limb development. Thick dashed lines mark the position of TAD boundaries.
A more precise deletion strategy removing these three CTCF sites led to a similar fusion between the subdomains. However, the effect upon Hoxd gene transcription was even milder than in the deletion of the boundary, likely because the full boundary deletion also included the CS39 enhancer, which was left in place in the CTCF deletion allele. In the latter case, mRNA accumulation was also delayed, but even less than in the first allele. Therefore, it seems that the presence of these CTCF sites, rather than the global structure that they help to organize, as well as the full collection of limb enhancers are the key elements to properly activate the target genes in time.
The importance of CTCF sites and/or of their orientation for chromatin interactions was previously predicted in silico and illustrated experimentally at a variety of specific loci (20, 34, 35). In developing tissues, the presence of bound CTCF in a specific locus favored the ectopic action of some clustered enhancers when placed in a different TAD (36). Here, we show that, when the entire boundary region containing the three sites was inverted, the isolation of the two sub-TADs became much stronger due to the convergence of these inverted sites with the natural telomeric 3′ TAD border, giving rise to two qualified TADs. Despite this accentuated split, which further isolated many limb enhancers from the target Hoxd genes, the transcriptional impact was once again restricted to the onset of their expression, similar to the effect of deleting the sub-TAD boundary. These results confirmed that the partition of T-DOM into sub-TADs may not respond to any particular regulatory necessity, at least in limb cells. Instead, it may be a consequence of CTCF being engaged into facilitating enhancer–promoter contacts.
Enhancer Topology and Regulatory Heterochrony.
It is often argued that chromatin architecture is instrumental to ensure the proper temporal dynamics of gene activation (see, e.g., ref. 5). However, in the case where regulatory landscapes contain multiple enhancers with either identical (37) or related (7, 8) specificities, it is less clear whether the respective positions of these enhancers and their distances to one another are critical factors for target gene activation. T-DOM contains multiple limb bud enhancers over 800 kb, which tend to be distributed far from the gene cluster, interspersed with CTCF sites. The large engineered inversion of T-DOM lacking the TAD border gave us a rather clear answer to this question, at least regarding this particular locus. While Hoxd genes were importantly delayed in their activation at E9.5, their expression patterns at E12.5 were indistinguishable from control limb buds. This once more points to the separation between two distinct regulatory aspects. On the one hand, the full series of enhancers will end up delivering their integrated information, regardless of their global organization within the landscape. On the other hand, an appropriate order and/or chromatin organization will help to properly orchestrate this process.
While similar chromosomal rearrangements have been engineered at other developmental loci, it is difficult to propose a synthetic view of the results for several parameters are usually involved and mixed with one another, such as the presence or absence of TAD borders and/or CTCF sites, as well as the presence of enhancers, their relative distribution, or their displacement related to their target genes. Deletions and duplications at the Ihh locus disrupted the communication with multiple enhancers, leading to limb malformations (10). Also, rearrangements of the TAD containing Shh and its enhancers led to deleterious effects on gene activation and concurrent phenotypes (38). Yet, at this specific locus, moderate topological modifications did not elicit any severe limb defects, suggesting that enhancer–promoter communication may not rely only on a sustained 3D structure and that sporadic interactions may be sufficient (31–33). Expectedly, stronger phenotypes were obtained by deleting the ZRS enhancer region as this sequence is the only known limb enhancer in this landscape (39–41). The transitory effects observed when modifying T-DOM could either reflect a normal enhancer deployment delayed by changes in the 3D context or, alternatively, a novel organization in enhancer–promoter interaction due to the known cooperative capacity that neighboring enhancers can display during embryonic development (6, 8, 42). Similarly, the deletion of the TAD border and inversion of the regulatory domain at the Sox9/Kcnj2 locus only had mild effects on gene expression (43).
The Resilience of Enhancer–Promoter Interactions.
In its initial form, the inversion of T-DOM introduced a strong ectopic TAD border between the enhancers and their target genes, in addition to the reorientation of all CTCF binding sites. In this allele (Fig. 6), accordingly, the access of Hoxd genes to their cognate limb enhancers was dramatically reduced. While a severe delay was scored in transcriptional activation, some interactions surprisingly remained between the gene cluster and the regulatory domain, despite the latter being clearly in a distinct TAD. These contacts could even resume the expression of Hoxd genes in proximal limb cells, although with a truncated spatial distribution. This observation is slightly at odds with the view of TAD borders restricting the access to neighboring enhancers and delimiting regulatory interactions (4, 22, 44–46). Here, despite the presence of a strong TAD border and the inversion of CTCF sites, which clearly led to the formation of a new TAD excluding the HoxD cluster, some enhancer–promoter interactions could still occur at a sufficient level to eventually produce detectable mRNAs in the expected proximal domain, thereby indicating that such contacts have intrinsic driving forces and do not entirely depend upon an instructive 3D context.
Colinear Regulation and Phenotypic Effects.
During early limb bud development, Hoxd genes are activated in a time sequence that follows their respective positions along the gene cluster (24, 47). The mechanism underlying this temporal colinearity process has been studied by intensive chromosome engineering whereby the order and/or presence of genes was modified, as well as their physical relationships with the adjacent regulatory landscapes (see references in ref. 48). However, a potential involvement of the regulatory topology, rather than the target end, remained to be assessed. In this study, by inverting the entire T-DOM, we could rule out the possibility that the physical order of various distant enhancers could play a major role in this mechanism, other than introducing a transcriptional delay, particularly visible in late-expressed Hoxd genes.
Finally, it is legitimate to wonder whether such moderate differences in the timing of gene activation could be detrimental to the development of the limb, considering that a close-to-normal expression pattern was resumed in E12.5 limb buds, except for the inversion of T-DOM containing the ectopic boundary where the anterior part of the domain remained absent even at later stages. This particular question was not addressed in this paper since the detection of any loss of function phenotype would likely be hampered by the cooperative function of both the HoxA and HoxD clusters in developing limbs. Indeed, while their combined deletion led to very severe limb truncations, their deletion in isolation triggered much milder phenotypes (49). While this functional complementation between these two gene clusters makes phenotypic analyses very complex in the mouse (it obliges one to systematically remove the other gene cluster), it has allowed one to study the underlying regulatory mechanisms in some detail due to the persistence of a rather normal structure even after drastic chromosomal interventions.
In the above-mentioned alleles, it is thus difficult to anticipate whether or not any phenotype would be observable in the absence of the HoxA cluster. In the case of T-DOM inversion including the TAD border, it is, however, clear that the lack of transcripts at the anterior margin of the proximal expression domain would lead to an abnormal formation of the intermediate part of the limbs, as was shown in mice carrying a double inactivation of Hoxa11 and Hoxd11, which displayed severely ill-formed forearms (50).
Materials and Methods
Mouse Strains.
The HoxDdel(CS38-40) or del(CS38-40) allele was described in ref. 27. The HoxDdelCTCFs(CS38;CS40) or del(CTCFs), HoxDinv(CS38-40) or inv(CS38-40), HoxDinv(T−DOM) or inv(T-DOM), and the HoxDinv(T−DOM)del(Bd) or inv(T-DOM)del(Bd) alleles were generated through CRISPR/Cas9 editing technology using electroporation of mouse zygotes. The del(CTCFs) allele was derived from the HoxDdelCTCF(CS38) or delCTCF(CS38) allele and was also generated for this study using a guide RNA (gRNA) directed against the consensus CTCF binding site located in region CS38, which generated a 26-bp large deletion. Subsequently, two gRNAs flanking region CS40 were designed to produce a 1,533-bp-large deletion encompassing both CTCF binding sites at region CS40. For the inv(CS38-40) allele, two gRNAs were designed flanking the region CS38-40. Mice were genotyped either for a deletion or for an inversion of the region. Out of 48 specimens, only one mouse had an inversion, which we used as founder of the mutant line. For the inv(T-DOM) allele, two different gRNAs were simultaneously directed at each end of the T-DOM regulatory domain. Out of 43 mice, only one had a full inversion, which was subsequently used to establish the mutant line. The break points for the inversion were located 3,433 bp downstream of the Hoxd1 gene and 2,557 bp upstream of the Hnrnpa3 gene, inducing an inversion of 888,111 bp. This line was subsequently used to generate the inv(T-DOM)del(Bd) mice. For this latter allele, two gRNAs were designed flanking the boundary region now relocated in inv(T-DOM) close to Hoxd1, deleting a 17,303-bp-large region. The delCTCF(CS38) and inv(T-DOM) alleles were generated after cloning the gRNAs into the pX330:hSpCas9 (Addgene ID 42230) vector and DNA injection into pronuclei. All other alleles were generated by electroporation of one-cell embryos with transcribed RNAs. The gRNAs and genotyping primers are listed in SI Appendix, Table S2. All break points were validated through Sanger sequencing, and this information was used to generate the artificial mutant genomes, which can be found in https://zenodo.org/record/3826913#.X6QqXS9h2L4 and a diagram in SI Appendix, Fig. S8.
4C-Seq.
The 4C-seq experiments were carried out as described in ref. 22. Samples were microdissected from E12.5 or E9.5 forelimbs and placed in 10% fetal bovine serum (FBS)/phosphate-buffered saline (PBS) and incubated with collagenase at 37 °C for 40 or 15 min, respectively. Cell suspensions were then strained and fixed in 2% formaldehyde (FBS/PBS) for 10 min. For E12.5 experiments, between 10 and 12 pairs of distal or proximal forelimbs were used while between 90 and 150 pairs of forelimbs were dissected for the E9.5 experiments. All E9.5 and the E12.5 del(CTCFs) 4C-seqs were conducted in embryos obtained from homozygous crosses while all others were obtained from heterozygous crosses. All 4C-seq primers used in this study are listed in SI Appendix, Table S3. The fastq from 4C-seq were demultiplexed, mapped, and analyzed using a local version of the pipeline that was present in HTSstation (51) on the wild-type mm10 mouse genome (Figs. 2, 3, and 4A and SI Appendix, Figs. S2–S4) or on the inv(T-DOM) mm10 mouse genome (Figs. 4B and 5C and SI Appendix, Figs. S5 and S6). During the demultiplexing step, reads were assigned to a viewpoint and a barcode, and then reads were truncated to 30 bp starting at CATG. Reads corresponding exactly to self-ligated or undigested sequences were identified and removed. During the analysis step, mapped reads were attributed to NlaIII-NlaIII fragments when overlapping a valid NlaIII-DpnII interval and transformed to scores taking into account the multimapping and the repeat masker. Scores of individual replicates were normalized to the number of million mapped reads before average. The distribution of all 4C-seq viewpoints used in this study and their associated statistics are listed in SI Appendix, Table S4. The detected NlaIII-NlaIII valid fragments were defined as the number of fragments with a nonzero score. To evaluate the proportion of trans contacts, the scores were summed on all chromosomes except chromosome 2 and divided by the sum of all genome-wide scores. The percentage of signal in the plotted region (chr2:74401941–75800320) was defined as the sum of scores in this region relative to the sum of scores genome-wide. The number of plotted fragments before smoothing represents the number of fragments with nonzero score in the plotted region. The profiles were smoothened using a window size of 11 fragments. The track profiles were obtained using pygenometracks (52). To quantify the contacts with enhancers, a region of interactions around each of them was defined based on the peak calling (wSize = 11, qWd = 1.5, qWr = 0.6) obtained by peakC (53) on the wild-type 4C-seq tracks of Hoxd11 (CS39, chr2:75128401–75162753; CS93, chr2:75194928–75211234, and CS65, chr2:75420048–75444960), Hoxd9 (ELCR2, chr2:74925621–74939634), and Hoxd4 (ELCR3, chr2:75081283–75094718). The above-mentioned coordinates were also adapted to the inv(T-DOM) genome for analysis of the inverted mutant lines. The sum of scores in the smoothed profile was computed from the above-mentioned enhancer regions. The sum was also performed on the HoxD cluster (chr2:74639799–74765142) and on T-DOM (for wild-type mm10 genome, chr2:74768600–75604841; for inv(T-DOM) in silico genome, chr2:74820438–75656679), which correspond to the full extension of sub-TAD1 and sub-TAD2 shown in Figs. 4 and 5 and SI Appendix, Figs. S6 and S7. All quantifications were performed using the facilities of the Scientific IT and Application Support Center of Ecole Polytechnique Fédérale de Lausanne (EPFL) and are shown in SI Appendix, Table S1.
4Cin.
To compose the virtual Hi-C matrices, we used the 4Cin package (30) and added an additional step to display the matrices with the linear distances. First, the profiles were preprocessed in four different ways to reduce the inherent variability of this computational approach. To this aim, the data values located at 0, <1, < 2, or <3 kb from the viewpoint were removed, and then the data were smoothed using a window size of 11 fragments. Only data corresponding to chr2:74,400,000–75,800,000 (mm10) on the wild-type genome were used as input for the 4Cin.py script package (30) with default parameters. In order to obtain the best fitted representations, 20 different models generated by the 4Cin package were taken into account (five times for each of the four different exclusion preprocessings). They were clustered according to the Spearman correlation established between them. A Ward clustering was then applied using Euclidean distances (SI Appendix, Fig. S9). The number of clusters was set so that the mean correlation of grouped models was <0.95 between clusters. For each cluster, an average was performed out of all of the models and shown as supplementary figures when the cluster included more than one single model. In the main figures, an average of all 20 models was shown to better integrate all possible models, only if the clusters included more than one model. For example, for the inv(CS38-40) allele, only cluster 1 was considered as a bona fide representation, encompassing 17 models, while the other three independent clusters were considered as outliers. In order to display the linear distances of the matrices, the virtual Hi-C output text file was converted to a cool file using a custom python script (https://github.com/lldelisle/scriptsForRodriguezCarballoEtAl2020). The track profiles were obtained using pygenometracks (52). The replicates and viewpoints used for each set of 4Cin are listed in SI Appendix, Table S4. The quantification of the distances for each sub-TAD (sub-TAD1, chr2:74768600–75133816; sub-TAD2, chr2:75153815–75604841) and between them shown in Figs. 2 and 3 and SI Appendix, Fig. S4 was performed using a custom python script. The range of the quantified sub-TADs is shown below the virtual Hi-C panels in Figs. 2 and 3 and SI Appendix, Fig. S4. Roughly, we took the value of each bin that overlapped the sub-TAD, and we computed the fold-change of mean distances and assessed the significance by a Mann–Whitney U test. The calculations were performed using the facilities of the Scientific IT and Application Support Center of EPFL.
RNA Extraction, RNA-Seq, and qRT-PCR.
Limb tissues were dissected and placed in individualized tubes containing RNAlater (Qiagen) and were frozen until genotyping and further processing. For E9.5 samples, once the limb buds were dissected, the rest of the embryo was fixed in 4% paraformaldehyde/PBS and stained with DAPI (Qiagen) for easy visualization of the somites under a microscope and characterization of the embryonic stage. All samples were processed following the RNeasy Microkit (Qiagen). RNA-seq libraries (one replicate per time point) were generated from 100 ng of total RNA following the TruSeq Stranded mRNA protocol and sequenced on a HiSeq 2500 machine (100-bp single read). The gtf file used for STAR was based on Ensembl version 92 annotations (https://zenodo.org/record/3827120 - .X6Q2qC9h2L5). Adapters were removed using cutadapt (v1.16; options -a GATCGGAAGAGCACACGTCTGAACTCCAGTCAC -m 15 -q 30) and aligned on mm10 using STAR version 2.6.0b-1 (54) with ENCODE parameters. Only uniquely mapped reads were kept, and coverage on each strand was obtained with bedtools genomecov v2.27.0. The track profiles were obtained using pygenometracks (52), and they show uniquely mapped reads normalized to the total number of uniquely mapped reads. For qRT-PCR (quantitative reverse transcription-PCR), RNA was retrotranscribed using the Promega GoScript Reverse Transcriptase (Promega). Custom SYBR probes were used for qRT-PCR in a QuantStudio5 384-well block machine. All primers were described in refs. 26 and 55. RNA-seq processing was done in a local instance of galaxy (galaxyproject.org) (56). For E9 samples, all values are relative to the average of the respective wild-type littermates. The number of replicates is mentioned in the figure legends. qRT-PCR results were plotted using Graphpad Prism8.
ChIP.
Limb tissues were dissected and fixed in 1% formaldehyde/PBS for 10 min at room temperature, and then incubated 3 min with Stop Solution from the ChIP-IT High Sensitivity Kit (Active Motif) and washed three times with PBS before being frozen at −80 °C until further use. E9 wild-type limb tissues were pooled prior to fixation according to their somite stage, which was determined under a bright-field microscope. Mutant samples were pooled according to their genotype prior to the experiment. Between 15 and 17 pairs of forelimb buds were used for each of the E9 ChIP experiments. For the E12.5 H3K27ac ChIP sample, two pairs of proximal forelimbs were dissected from wild-type mice. For E12 CTCF ChIPs, the four entire limbs coming from one embryo were used in each experiment. All ChIP experiments were conducted following the ChIPmentation method (57) as adapted in ref. 58. Briefly, samples were Polytron minced and homogenized by douncing in Prep Buffer (ChIP-IT High Sensitivity Kit; Active Motif). They were then sonicated in 100 µL of sonication buffer (0.1% sodium dodecyl sulfate [SDS], 50 mM Tris⋅HCl, pH 8, 10 mM ethylenediaminetetraacetic acid [EDTA], pH 8, and proteinase inhibitors) in a Bioruptor Pico sonicating device (Diagenode). All ChIPs were incubated overnight with the respective antibodies (CTCF 61311, Active Motif; H3K27me3 17-662, Millipore; H3K27ac C15410196, Diagenode; RAD21 ab992, abcam) and precipitated after a 2-h-long incubation with magnetic beads (Dynabeads Protein A, 10001D; Invitrogen). Washes were carried out in RIPA-LS (10 mM Tris·HCl pH 8, 140 mM NaCl, 1 mM EDTA pH 8, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and proteinase inhibitors), RIPA-HS (10 mM Tris·HCl pH 8, 500 mM NaCl, 1 mM EDTA pH 8, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and proteinase inhibitors), and RIPA-LiCl (10 mM Tris·HCl pH 8, 250 mM LiCl, 1 mM EDTA pH 8, 0.5% NP-40, 0.5% sodium deoxycholate, and proteinase inhibitors). Beads were then resuspended in tagmentation buffer and incubated at 37 °C for either 2 min (CTCF, RAD21, H3K27me3) or 10 min (H3K27ac) with 1 µL of Tn5 transposase (15027865, from Nextera DNA Library Prep Kit 15028212; Illumina). After washing with RIPA-LS and TE buffer (10 mM Tris·HCl pH 8, 1 mM EDTA pH 8), beads were incubated in elution buffer (10 mM Tris⋅HCl, pH 8, 5 mM EDTA, pH 8, 300 mM NaCl, 0.4% SDS) and proteinase K. DNA was then purified using the Qiagen MiniElute kit, and a qRT-PCR was performed to determine the number of cycles to be applied during library amplification. DNA libraries were purified, and size was selected with CleanNGS magnetic beads (CleanNA) and sequenced in a HiSeq 4000 machine as 50-bp or 100-bp reads. The sequencing output was mapped and processed as in ref. 58 without a normalization step. ChIP processing was done in a local instance of galaxy (usegalaxy.org) (56). The University of California, Santa Cruz (UCSC) genome browser was used to compose the tracks in SI Appendix, Figs. S2A and S5A while pygenometracks (52) was used for all of the ChIP tracks in other figures. In all figures, the orientations of the CTCF sites were obtained using the CTCFBSDB 2.0 database (insulatordb.uthsc.edu) based on the E9 CTCF ChIP data.
Whole-Mount In Situ Hybridization and LacZ Transgenes.
WISH was performed as in ref. 59 with the following modifications. E10.5 and E12.5 embryos were bleached in a 3% H2O2/PBS solution. After rehydration, embryos were digested in a proteinase K solution (20 µg/mL, 10 to 12 min, for E12.5 embryos; 10 µg/mL, 5 min, for E10.5 embryos; and 5 µg/mL, 4 to 5 min, for E9.5 embryos). Digestion of E9.5 embryos was arrested by three quick washes in a 2-mg/mL glycine solution while E10.5 and E12.5 proteinase K digestions were stopped with a 10-min incubation in an acetic anhydride/triethanolamine solution. After mRNA probe hybridization and anti-DIG incubation, E9.5 embryos were washed several times overnight in maleic acid buffer while E10.5 and E12.5 embryos’ washes were extended for a day.
The ELCR2 and ELCR3 regions were amplified and cloned into the betaGlobin reporter plasmid as in ref. 60 to generate the corresponding LacZ transgenes. These enhancer regions were amplified using the primer sequences GATGCTTGGCCTTAGCTCCT (Fw) and CTGTGGAAACGGAGCCAGAA (Rv) for ELCR2:LacZ; and TCTCTGCCCATTCACTCTCATCA (Fw) and TTTTCTGTGCAGTGGCTGTGAC (Rv) for ELCR3:LacZ. The CS39:lacZ and CS65:LacZ transgenic lines were previously described (29). All images were taken with Leica MZFLIII and Leica M205 FA microscopes. A list with the number of replicates of each WISH is shown in SI Appendix, Table S5.
Animal Experimentation and Ethics Approval.
All experiments were performed in agreement with the Swiss law on animal protection (Loi Fédérale sur la Protection des Animaux [LPA]), under license GE81/14 (to D.D.).
Supplementary Material
Acknowledgments
We thank Thi Hanh Nguyen Huynh, Marie-Laure Gadolini, and Julien Codourey for their help with mice breeding, genotyping, and transgenesis; as well as Mylène Docquier, Didier Chollet, Brice Petit, and Christelle Barraclough (Genomics Platform, University of Geneva). We also thank Ibai Irastorza-Azcarate and Asier Ullate-Agote for their help in implementing the 4Cin pipeline. We thank Aurélie Hintermann and other members of the D.D. laboratories for helpful discussions and suggestions.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015083117/-/DCSupplemental.
Data Availability.
All new sequencing data are deposited in the Gene Expression Omnibus database and can be found under the accession no. GSE154189. All scripts used to generate the final outputs of Hi-C and 4C-seq (including figures) are available in GitHub, https://github.com/lldelisle/scriptsForRodriguezCarballoEtAl2020.
References
- 1.Long H. K., Prescott S. L., Wysocka J., Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoenfelder S., Fraser P., Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Dixon J. R., et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nora E. P., et al. , Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furlong E. E. M., Levine M., Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amândio A. R., Lopez-Delisle L., Bolt C. C., Mascrez B., Duboule D., A complex regulatory landscape involved in the development of mammalian external genitals. eLife 9, e52962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montavon T., et al. , A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Osterwalder M., et al. , Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sagai T., et al. , A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development 136, 1665–1674 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Will A. J., et al. , Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog). Nat. Genet. 49, 1539–1545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fudenberg G., et al. , formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nora E. P., et al. , Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao S. S. P., et al. , Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarzer W., et al. , Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soshnikova N., Montavon T., Leleu M., Galjart N., Duboule D., Functional analysis of CTCF during mammalian limb development. Dev. Cell 19, 819–830 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Cuartero S., et al. , Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat. Immunol. 19, 932–941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowen J. M., et al. , Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanssen L. L. P., et al. , Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 19, 952–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hnisz D., et al. , Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Wit E., et al. , CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Andrey G., et al. , A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340, 1234167 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Carballo E., et al. , The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 31, 2264–2281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lonfat N., Montavon T., Darbellay F., Gitto S., Duboule D., Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science 346, 1004–1006 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Tarchini B., Duboule D., Control of Hoxd genes’ collinearity during early limb development. Dev. Cell 10, 93–103 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Zákány J., Kmita M., Duboule D., A dual role for Hox genes in limb anterior-posterior asymmetry. Science 304, 1669–1672 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Delpretti S., et al. , Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Rep. 5, 137–150 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Schep R., et al. , Control of Hoxd gene transcription in the mammary bud by hijacking a preexisting regulatory landscape. Proc. Natl. Acad. Sci. U.S.A. 113, E7720–E7729 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yakushiji-Kaminatsui N., et al. , Similarities and differences in the regulation of HoxD genes during chick and mouse limb development. PLoS Biol. 16, e3000004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beccari L., et al. , A role for HOX13 proteins in the regulatory switch between TADs at the HoxD locus. Genes Dev. 30, 1172–1186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irastorza-Azcarate I., et al. , 4Cin: A computational pipeline for 3D genome modeling and virtual Hi-C analyses from 4C data. PLOS Comput. Biol. 14, e1006030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander J. M., et al. , Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 8, e41769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benabdallah N. S., et al. , Decreased enhancer-promoter proximity accompanying enhancer activation. Mol. Cell 76, 473–484.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukaya T., Lim B., Levine M., Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y., et al. , CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanborn A. L., et al. , Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 112, E6456–E6465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraft K., et al. , Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat. Cell Biol. 21, 305–310 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Hörnblad A., Langenfeld K., Bastide S., Langa Vives F., Spitz F., Dissection of the Fgf8 regulatory landscape by in vivo CRISPR-editing reveals extensive inter- and intra-enhancer redundancy. bioRxiv:2020.03.03.966796 (3 March 2020). [DOI] [PMC free article] [PubMed]
- 38.Symmons O., et al. , The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev. Cell 39, 529–543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lettice L. A., Devenney P., De Angelis C., Hill R. E., The conserved sonic hedgehog limb enhancer consists of discrete functional elements that regulate precise spatial expression. Cell Rep. 20, 1396–1408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paliou C., et al. , Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc. Natl. Acad. Sci. U.S.A. 116, 12390–12399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williamson I., et al. , Developmentally regulated Shh expression is robust to TAD perturbations. Development 146, dev.179523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolt C. C., Duboule D., The regulatory landscapes of developmental genes. Development 147, dev171736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Despang A., et al. , Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 51, 1263–1271 (2019). [DOI] [PubMed] [Google Scholar]
- 44.van Bemmel J. G., et al. , The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsix and Xist. Nat. Genet. 51, 1024–1034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franke M., et al. , Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Lupiáñez D. G., et al. , Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dollé P., Izpisúa-Belmonte J.-C., Falkenstein H., Renucci A., Duboule D., Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature 342, 767–772 (1989). [DOI] [PubMed] [Google Scholar]
- 48.Tschopp P., Duboule D., A genetic approach to the transcriptional regulation of Hox gene clusters. Annu. Rev. Genet. 45, 145–166 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Kmita M., et al. , Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435, 1113–1116 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Davis A. P., Witte D. P., Hsieh-Li H. M., Potter S. S., Capecchi M. R., Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375, 791–795 (1995). [DOI] [PubMed] [Google Scholar]
- 51.David F. P. A., et al. , HTSstation: A web application and open-access libraries for high-throughput sequencing data analysis. PLoS One 9, e85879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Delisle L., et al. , pyGenomeTracks: Reproducible plots for multivariate genomic data sets. Bioinformatics, 10.1093/bioinformatics/btaa692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geeven G., Teunissen H., de Laat W., de Wit E., peakC: A flexible, non-parametric peak calling package for 4C and Capture-C data. Nucleic Acids Res. 46, e91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montavon T., Le Garrec J. F., Kerszberg M., Duboule D., Modeling Hox gene regulation in digits: Reverse collinearity and the molecular origin of thumbness. Genes Dev. 22, 346–359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afgan Enis, et al. , The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidl C., Rendeiro A. F., Sheffield N. C., Bock C., ChIPmentation: Fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodríguez-Carballo E., Lopez-Delisle L., Yakushiji-Kaminatsui N., Ullate-Agote A., Duboule D., Impact of genome architecture on the functional activation and repression of Hox regulatory landscapes. BMC Biol. 17, 55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woltering J. M., et al. , Axial patterning in snakes and caecilians: Evidence for an alternative interpretation of the Hox code. Dev. Biol. 332, 82–89 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Guerreiro I., et al. , Reorganisation of Hoxd regulatory landscapes during the evolution of a snake-like body plan. eLife 5, 1–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All new sequencing data are deposited in the Gene Expression Omnibus database and can be found under the accession no. GSE154189. All scripts used to generate the final outputs of Hi-C and 4C-seq (including figures) are available in GitHub, https://github.com/lldelisle/scriptsForRodriguezCarballoEtAl2020.