Significance
Recent studies showed that cellular senescence, previously considered an irreversible biological phenomenon, may represent a reversible state, but the mechanism for the reversion is not well elucidated. To identify targets the inhibition of which would convert senescent cells into quiescent cells, we constructed a molecular regulatory network model of cellular senescence. From the model simulation, we identified PDK1 as a promising target that can revert the senescent state into a quiescent state. We validated this prediction in experiments with human dermal fibroblasts, which showed that PDK1 inhibition eradicates senescence hallmarks by suppressing both nuclear factor κB and mTOR signaling through the inactivation of a positive feedback loop composed of PDK1, AKT, IKBKB, and PTEN, resulting in restored skin regeneration capacity.
Keywords: cellular senescence, systems biology, network modeling, PDK1, skin aging
Abstract
Cellular senescence is defined as a stable, persistent arrest of cell proliferation. Here, we examine whether senescent cells can lose senescence hallmarks and reenter a reversible state of cell-cycle arrest (quiescence). We constructed a molecular regulatory network of cellular senescence based on previous experimental evidence. To infer the regulatory logic of the network, we performed phosphoprotein array experiments with normal human dermal fibroblasts and used the data to optimize the regulatory relationships between molecules with an evolutionary algorithm. From ensemble analysis of network models, we identified 3-phosphoinositide–dependent protein kinase 1 (PDK1) as a promising target for inhibitors to convert the senescent state to the quiescent state. We showed that inhibition of PDK1 in senescent human dermal fibroblasts eradicates senescence hallmarks and restores entry into the cell cycle by suppressing both nuclear factor κB and mTOR signaling, resulting in restored skin regeneration capacity. Our findings provide insight into a potential therapeutic strategy to treat age-related diseases associated with the accumulation of senescent cells.
Cellular senescence is defined as a stable, persistent exit from the cell cycle in response to stresses such as telomere shortening, oxidative stress, oncogene activation, and DNA damage (1, 2). A benefit of cellular senescence is prevention of tumorigenesis by blocking proliferation of damaged cells that may undergo malignant transformation (2, 3). However, senescent cells accumulate in tissues during aging and secrete proinflammatory cytokines, which can contribute to aging and age-related diseases, including cancer (2, 3). In studies with animal models, elimination of senescent cells prevents, alleviates, or reverses symptoms of aging (4, 5) and various age-related diseases (6, 7), such as osteoarthritis and atherosclerosis.
Cell-cycle arrest alone is not cellular senescence; cellular senescence requires additional signals that convert transient cell-cycle arrest into persistent exit from the cell cycle so that the cells fail to proliferate in response to growth signals, a process called geroconversion (8, 9). Terminally differentiated, nonmitotic cells can also undergo senescence; thus, cell-cycle exit is only one aspect of the senescent phenotype. Cellular senescence is a complex biological mechanism regulated by various signaling pathways (10, 11). Signaling pathways that mediate cellular senescence can be divided into three major categories. The first category includes the pathways that cause cell-cycle arrest in response to DNA damage, such as p53/p21CIP1 and p16INK4a/pRb pathways (11–14). The second category consists of the pathways mediating cell growth and energy metabolism, such as PI3K/AKT/mTOR and SIRT1/AMPK pathways (15–18). Activation of mTOR in cells arrested by persistent DNA damage represents a second stimulus that can convert transiently arrested cells into senescent cells that exhibit hypertrophy and an expanded lysosomal compartment (19). The last category consists of the pathways mediating the senescence-associated secretory phenotype (SASP) (3, 10, 20). The SASP is a characteristic feature of senescent cells and reflects their secretion of proinflammatory cytokines and chemokines. These cytokines and chemokines maintain cellular senescence through positive autoregulatory feedbacks, affect nonsenescent nearby cells, and promote aging and age-related diseases, including cancer (3, 21). Nuclear factor κB (NF-κB) activity is important for SASP, and suppression of NF-κB prevents age-related diseases and delays aging in mice (22, 23).
Spontaneous reversion from senescence to proliferation is extremely rare, but the reversion through manipulations is not. Some studies reported that senescent cells can reenter the cell cycle (24). The current understanding of senescence is as a dynamic multistep process that is reversible under some conditions (25). About 70 to 90% of cells with low p16INK4a levels in replicative senescence, which is senescence related to the finite number of divisions a cell can perform before telomeres become too short, resume proliferation following p53 inactivation (26). Inactivation of p53 also enables cells to escape from therapy-induced senescence, caused by the chemotherapeutic agent Adriamycin (27). Cells with oncogene-induced senescence can also escape from the senescent state. For instance, about 50% of mouse embryo fibroblasts with high Ras levels reenter the cell cycle upon inactivation of all three Rb family members (28), and about 70% of the fibroblasts reenter upon activation of H3K9 demethylases (29).
Here, we applied a systems biology approach to identify mechanisms underlying cell-cycle arrest, cell growth, and the SASP with the goal of finding inhibitable targets to convert the senescent state to the quiescent state. We studied normal human dermal fibroblasts (NHDFs), which can be experimentally induced into the senescent state (8). We constructed a molecular signaling network of cellular senescence using information in the literature and network databases to identify the relevant molecules, experimental data from time series of phosphorylated proteins in NHDFs to define the input–output relationships that reflect cellular states upon each input condition, and an evolutionary algorithm to determine the regulatory logic of the network (SI Appendix, Fig. S1 A–C). By analyzing the regulatory signaling network, we predicted that PDK1 was an inhibitor target that can convert senescent fibroblasts to quiescent fibroblasts (SI Appendix, Fig. S1D). To validate this prediction, we conducted experiments with NHDFs exposed to PDK1 inhibitors (SI Appendix, Fig. S1E), which eliminated hallmarks of cellular senescence, restored the proliferation of the cells in response to growth factors, and restored skin regeneration capacity in two-dimensional (2D) culture and a three-dimensional (3D) skin equivalent model. Our findings provide insight into a potential therapeutic strategy to treat aging and age-related diseases.
Results
Construction of a Prior Knowledge Network of Cellular Senescence.
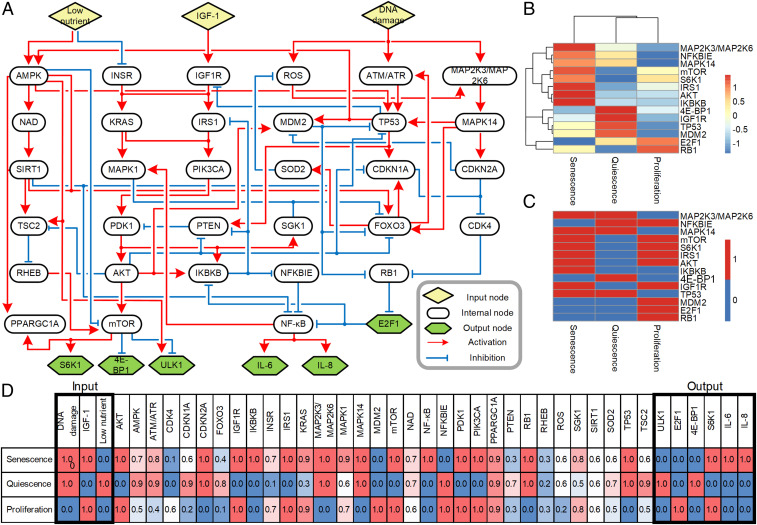
To understand the complex interactions underlying the signaling pathways of cellular senescence, we constructed a prior knowledge network by integrating information from the literature and network databases, including Kyoto Encyclopedia of Genes and Genomes (KEGG), STRING, and Omnipath (30–32). The network is composed of 41 nodes and 77 links, including 3 input nodes and 6 output nodes (Fig. 1A). The input nodes of the network represent three external stimuli: DNA damage, growth factor stimulation (insulin-like growth factor 1, IGF1), and low availability of nutrients (Low nutrient). Together, these stimuli influence whether a cell enters the senescent state. The network consists of interconnected signaling pathways regulated by the external stimuli: the p53/p21CIP1 (TP53/CDKN1A) pathway and p16INK4a/pRb (CDKN2A/RB1) pathway (DNA damage-response pathways); PI3K (PIK3CA)/AKT/mTOR pathway (nutrient-sensing and growth factor-response pathway); AMPK/SIRT1 pathway (nutrient-sensing pathway); and NF-κB pathway (stress response pathway). The output nodes represent indicators of SASP (interleukin [IL]-6 and IL-8), cell-cycle arrest (E2F1), and cell growth (S6K1, 4E-BP1, and ULK1). This network represents the core structure and includes key proteins that regulate the DNA damage response, cell-cycle arrest, cell growth, and the SASP.
Fig. 1.
Inference of multiple Boolean network models. (A) A schematic diagram of the cellular senescence network. The network consists of 41 nodes and 77 links: 48 links are activation links and 29 links are inhibition links. Not all links are direct; some intervening nodes are not included in the network. Shape and color define the 3 input nodes, 6 output nodes, and 32 internal nodes. Detailed information for each node and link is shown in SI Appendix, Table S1 and S2. (B) Hierarchical clustering heat map of phosphoprotein array data. AUC values of the indicated phosphorylated proteins were visualized using a hierarchical clustering heat map upon three conditions. (C) Heat map of binarized phosphoprotein arrays. Binarized AUC values of the indicated phosphorylated proteins were visualized using a heat map. (D) Average node activities of Boolean network models after the models reached their respective attractors. Input nodes and output nodes are highlighted by bold-lined boxes.
Time-Series Phosphoprotein Array of Human Dermal Fibroblasts.
To estimate the molecular regulatory logic of the senescence network, we performed a time-series phosphoprotein array of NHDFs under proliferation, quiescence, and senescence conditions (SI Appendix, Table S3). A nutrient-rich environment supplemented with IGF1, a primary mitogenic factor for NHDFs, and a high proportion of fetal bovine serum (FBS) (10%) was the “proliferation” condition. Because geroconversion is induced by the occurrence of DNA damage in the presence of growth stimuli (8, 9), the “senescence” condition was the proliferation condition plus the DNA-damaging chemical doxorubicin (100 ng/mL). Because the DNA damage without growth stimuli is not sufficient to induce geroconversion (8, 9), we defined the “quiescence” condition as a nutrient-poor environment lacking added IGF1 and serum plus the DNA-damaging chemical doxorubicin (100 ng/mL).
We measured senescence-associated beta-galactosidase (SA-β-gal), p16, p21, 5-ethynyl-2′-deoxyuridine (EdU), and 53BP1 under both senescence and quiescence conditions up to day 14 (SI Appendix, Fig. S2). We also measured EdU of senescent cells and quiescent cells after 4 d in Dulbecco’s modified Eagle’s medium (DMEM) with 0% FBS and 10% FBS (SI Appendix, Fig. S3). Under the quiescence condition, p16 and SA-β-gal were not detected. Also, after 4 d, when doxorubicin was removed and 10% FBS was added, the cells resumed normal growth. On the other hand, under the senescence condition, p16 expression was increased and SA-β-gal was detected. After 4 d, even when doxorubicin and IGF1 were replaced with 10% FBS, the cells did not resume growth. These experimental observations show that irreversible cell-cycle arrest or senescence needs simultaneously activating DNA damage signals (doxorubicin) and growth signals (IGF1 + serum), which is well in accord with geroconversion (8, 9).
Based on experimental observations (SI Appendix, Figs. S2 and S3), we assumed that NHDFs reach steady states within 96 h under senescence and quiescence conditions and measured phosphoprotein abundance from 1 h to 96 h. For the proliferation condition, we used short time intervals (1, 6, and 24 h) based on the NHDF doubling time of ∼24 h (33). We normalized sample intensities with quantile normalization and obtained mean fold change values from phosphoprotein array (n = 4) (Dataset S1 and see Materials and Methods for details). For each condition and each time point, we selected genes with log2-transformed quantile-normalized raw intensities greater than 13 for Gene Ontology (GO) enrichment analysis (Dataset S2). From GO enrichment analysis we confirmed that gene ontologies related to cell proliferation and transcription are enriched under the proliferation condition, whereas in the quiescence condition gene ontologies related to both negative and positive regulation of transcription are enriched at the same time. For the senescence condition, gene ontologies related to cellular responses to cytokine and cellular responses to reactive oxygen species are significant. For all conditions, negative regulations of programmed cell death are enriched. We focused on the dynamics of 14 key proteins based on the previous studies (a list of references is included in SI Appendix, Table S4) showing their different expression levels under proliferation, quiescence, and senescence conditions and found that these proteins exhibited distinct expression patterns in our phosphoprotein array across the given conditions and time periods (SI Appendix, Table S4 and Fig. S4).
Boolean Network Modeling of the Cellular Senescence Network.
To analyze the dynamic characteristics of the cellular senescence network we used a discrete value Boolean network modeling approach rather than continuous value dynamic models (for example, an ordinary differential equation model). The Boolean approach is better for this network because of the limited experimental data on molecular regulation kinetics (34). Although the Boolean model is an abstract and simplified model that considers only the activation/inactivation (on/off) state of each node, this type of model is useful for analyzing key dynamic features of cellular behavior and can simulate well-known biological phenomena and predict potential drug effects with a high degree of confidence (35, 36).
To develop Boolean network models, we quantified the phosphoprotein array data based on normalized area under the curve (AUC), which represents the activity of key proteins across time points, and binarized the AUC values into 0 or 1 (Fig. 1 B and C and SI Appendix, Table S4 and see Materials and Methods for details). The similarities between the quiescence and senescence conditions were MDM2 (off), E2F1 (off), RB1(on), and TP53 (on). These molecules control cell-cycle arrest and the DNA damage response. The proliferation and senescence conditions showed similar patterns for mTOR (on), IGF1R (on), 4E-BP1 (off), IRS1 (on), S6K1 (on), and AKT (on) in the pathways related to cell growth, and MAPK14 (on) and MAP2K3/MAP2K6 (on) in the DNA damage response. Activation of the SASP-related molecules IKBKB (on) and NFKBIE (off) was unique to the senescence condition. To further validate these binarized AUC values, we measured quantified protein expression levels of some signaling proteins that represent the major signaling axes of the network model such as p53, IKK-beta, E2F1, and IGF1R through immunoblot experiments (SI Appendix, Fig. S5). The immunoblot results show a higher expression of p53 in the quiescence and senescence conditions, a higher expression of IGF1R in senescence and proliferation, a higher expression of IKK-beta in senescence, and a higher expression of E2F1 in proliferation. These results are in accord with the binarized AUC values.
Because an evolutionary algorithm can find solutions to problems of which the objective functions are discontinuous or nonlinear (37, 38), we used an evolutionary algorithm to optimize the Boolean network logics by fitting the simulation results to the binarized phosphoprotein array data. We set a cost function as the sum of mean-squared error between simulated attractor states of key proteins and their binarized AUC values for all three input conditions (SI Appendix, Table S4 and see Materials and Methods for details). Attractor states represent stable states toward which a dynamic system tends to evolve. In biology, an attractor state corresponds to a particular cellular phenotype (39, 40). Because we could infer multiple sets of Boolean logic that produce zero cost function, we constructed multiple Boolean network models with different Boolean logic that all satisfy the phosphoprotein array data. We found that 5,000 optimized Boolean network models reach attractors for senescence, quiescence, or proliferation within 30 time steps (SI Appendix, Fig. S6A). The cost function decreased as time steps increased and converged to zero when all Boolean network models reached respective attractors (SI Appendix, Fig. S6B). Average attractor states for the optimized Boolean network models showed that most average attractor states of internal nodes are skewed or even fixed to either on or off states (Fig. 1D and SI Appendix, Table S5) and have positive correlations with normalized AUC values of phosphoprotein array data (SI Appendix, Fig. S7 and Table S6).
Ensemble Model Simulation to Identify Targets that Can Reverse Senescence to Quiescence.
Our goal is to identify targets in the network that can cause a transition from the senescence state to the quiescence state while avoiding the proliferation state. To identify such targets while minimizing bias that a single Boolean network model may have, we created an ensemble of Boolean network models to achieve robust and accurate results (41). We calculated a score for each node , where represents a ratio of Boolean network models for which inhibition of the [node] converts the senescence attractor to the quiescence attractor, and represents a ratio of Boolean network models for which inhibition of the [node] converts the senescence attractor to the proliferation attractor (see Materials and Methods for details). A positive score indicated that inhibition of the node was more likely to cause a transition from senescence to quiescence; a negative score indicated that inhibition was more likely to cause a transition from senescence to proliferation.
To determine how many Boolean network models are required for robust ensemble model simulation, we evaluated the rank correlation between ensemble models of the same number of different Boolean network models. As the number of Boolean network models increases, the simulation results became similar and the rank correlation increased. The rank correlation coefficients became saturated at ∼0.9 when the number of models exceeded 700 (SI Appendix, Fig. S8). Finally, we conducted extensive simulation analysis on our network models modified with randomly added nodes. We measured weighted rank correlations of the results obtained from the modified networks against the results from the original network in order to examine the robustness of our results. Consequently, we confirmed that our results remain robust against a certain range of perturbations on the network model (SI Appendix, Fig. S9). Thus, we concluded that the ensemble model with >700 Boolean network models would give robust results regardless of the different set of the optimized Boolean network models as well as the added nodes over a significant range as far as the backbone structure of the network is maintained.
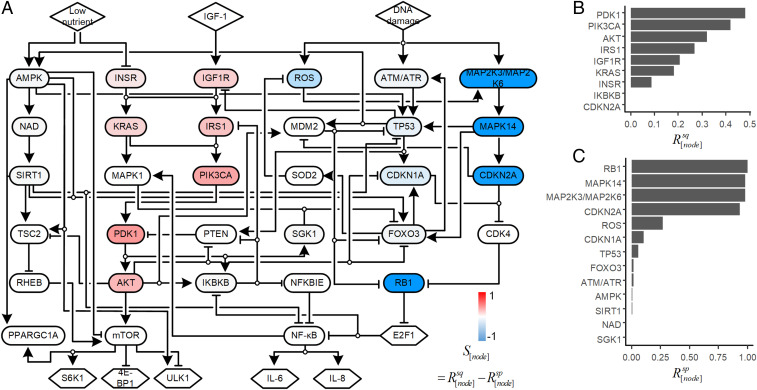
To identify senescence-reversing targets, we conducted simulations with one inhibited node (single-node inhibition) for 5,000 Boolean network models and integrated the simulation results into the score, which we mapped onto the network (Fig. 2A). We found that positively scoring targets are located in pathways related to cell growth (Fig. 2A) and negatively scoring targets are located in pathways related to cell-cycle arrest and DNA damage response (Fig. 2A). Consistent with these scores, the values for simulations in which PDK1, PIK3CA, or AKT was inhibited showed that more than 30% of the models produced the transition from senescence to quiescence (Fig. 2B). In contrast, the values showed that inhibition of RB1, MAPK14, or MAP2K3/MAP2K6 resulted in nearly all simulations converting the senescence attractor to the proliferation attractor (Fig. 2C). Although CDKN2A had a positive value for the , its value for was much greater, resulting in inhibition of CDKN2A having a score associated with the conversion of the model from the senescence attractor to the proliferation attractor.
Fig. 2.
Ensemble model simulation to identify senescence-reversing targets. (A) Nodes were colored according to their scores indicating if inhibition resulted in a transition of the network output from senescence attractor to a proliferation attractor (blue, negative values) or from a senescence attractor to a quiescence attractor (red, positive values). The equation for determining the score is shown. (B) A bar graph of the ratio of Boolean network models for which inhibition of the listed node converts the senescence state into the quiescence state (). The values for IKBKB and CDKN2A are close to zero, but not zero. (C) A bar graph of the ratio of Boolean network models for which inhibition of the listed node converts the senescence state into the proliferation state ().
In the ensemble model simulation result, PDK1 scored the highest of all single targets that when inhibited in the simulations produced the senescence-to-quiescence transition (Fig. 2B and SI Appendix, Table S7). PDK1 is an oncogene that supports tumor cell survival and proliferation during hypoxia (42). However, the role of PDK1 in cellular senescence is unknown. Other targets that produced a score and values greater than 0.25 were PIK3CA, AKT, and IRS1. Inhibition or loss of function of these three proteins (or their encoding genes), along with IGFR1 and INSR, are all reported to prolong the lifespan in mice (SI Appendix, Table S7). Identification of these high-scoring targets with connections to longevity provided validation that the simulation results of the ensemble model are biologically relevant, enabling us to predict that PDK1 and KRAS may be targets for inhibition to reverse cellular senescence. In particular, we hypothesized that inhibition of PDK1, as the highest-scoring target, reverses cellular senescence.
We also performed simulations with inhibition of two nodes (double-node inhibition). Combined inhibition of mTOR and NF-κB was the highest scoring dual target (SI Appendix, Fig. S10). Both mTOR and NF-κB are key regulators of cellular senescence (19, 43), and both are downstream of PDK1 in the cellular senescence network. Thus, we predicted that the senescence reversion through PDK1 inhibition results from the subsequent inhibition of both mTOR and NF-κB.
To evaluate the effects of optimized logic on , we simulated Boolean network models with simplified network logical parameter values (1, −1, or 0). For , mTOR was the top ranking, and half of network nodes, including PDK1, had similar values (SI Appendix, Fig. S11). Unlike with the continuous model simulations, in this simplified analysis there was no target that converted the senescence attractor to the proliferation attractor.
Analysis of the Mechanism by Which PDK1 Inhibition Reverses Senescence.
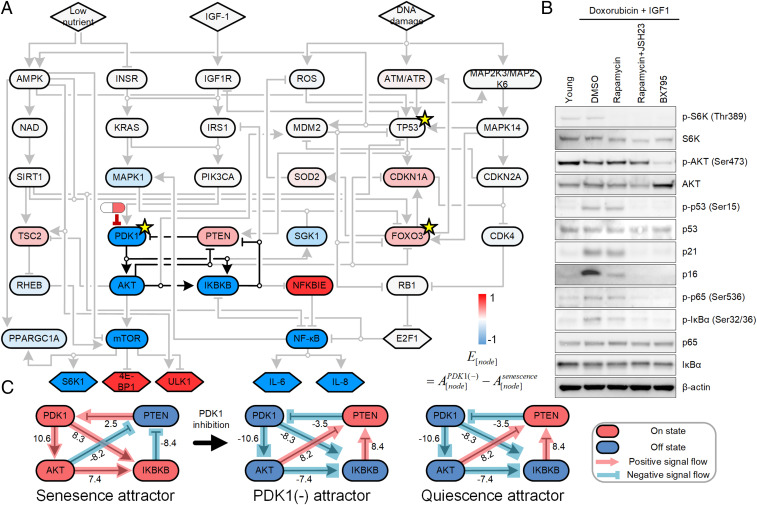
Both network structure and dynamics play important roles in controlling complex networks (44). To explore the potential mechanism by which inhibition of PDK1 promoted the transition from senescence to quiescence, we applied a topological analysis of our network by identifying the feedback vertex set (FVS), a set of vertices (nodes) the removal of which leaves the network without cycles (45). FVS is useful when establishing a robust control strategy in a large and complex network using only the structure (topology) of the network (46, 47). FVS is called minimal FVS (mFVS) if none of its subsets is an FVS. In the cellular senescence network, PDK1, FOXO3, and TP53 correspond to an mFVS (Fig. 3A). PDK1 controls various positive feedback loops, including PDK1-AKT-IKBKB-PTEN, PDK1-AKT-MDM2-TP53-PTEN, and PDK1-AKT-FOXO3-ATM/ATR-TP53-PTEN, any of which can be a key module to control the senescence phenotype. This topological analysis result provides a hint about why PDK1 was the highest-scoring target for controlling the transition from senescence to quiescence: Either one or a combination of these feedback loops are critically important for maintaining the senescent state.
Fig. 3.
Mechanistic analysis of PDK1 inhibition as a method of converting the senescence state to the quiescent state. (A) Nodes were colored according to whether PDK1 inhibition increased node activity (red) or decreased activity (blue) using the value of , which was calculated using the equation shown (see Materials and Methods for more details). Three nodes of mFVS are marked by yellow stars. The pill shape represents inhibition of PDK1. (B) Immunoblot showing the abundance of the indicated proteins or phosphorylated proteins in NHDFs. Young NHDFs were not subjected to senescence conditions or drug treatment. Samples from young NHDFs were collected when cultures reached ∼80% confluence at PDL 12 (Materials and Methods). The remaining samples were from NHDFs induced to senescence by treatment with doxorubicin (100 ng/mL) plus IGF1 (100 ng/mL) for 7 d. The senescent cells were then exposed to DMSO, rapamycin (100 nM), rapamycin plus JSH23 (2.5µM), or BX795 (100 nM) for 7 more days. Cell lysates were subjected to immunoblot analysis using antibodies against the indicated proteins. Data are representative of at least two independent experiments. (C) A diagram of the positive feedback loop that is deactivated by PDK1 inhibition. Each regulatory connection is labeled with its signal strength in the indicated condition (see Materials and Methods for details).
To investigate the detailed PDK1 inhibition mechanism, we collected the Boolean network models (n = 2,401) for which inhibition of PDK1 changed the attractor state from senescence to quiescence (SI Appendix, Fig. S12) and investigated their transition trajectories under PDK1 inhibition (see Materials and Methods for details). For each node, we determined the effect of PDK1 inhibition , where a positive value is increased activity compared to the activity in the absence of PDK1 inhibition and a negative value is decreased activity. The value of is calculated for each model producing the senescence attractor state from the difference the average node state in the presence of the PDK1 inhibitor and the average node state without the PDK1 inhibitor . The transition trajectory analysis showed that PDK1 inhibition deactivates a positive feedback loop composed of PDK1, AKT, IKBKB, and PTEN, which then reduces mTOR and NF-κB activity (Fig. 3A and SI Appendix, Table S8). Moreover, the role of the positive feedback loop in maintaining the senescent state was confirmed; the resilience of output node states to input node (e.g., IGF1) was lost when the feedback loop was disabled (SI Appendix, Table S9). Thus, this positive feedback loop appeared to be the critical one for maintaining the senescent state instead of reversion to the quiescent state.
To confirm this pathway in senescent NHDFs, we exposed the cells to doxorubicin and IGF1 to induce senescence, added various inhibitors, and monitored the phosphorylation and abundance of relevant proteins. We compared the responses of the senescent cells to those exposed to an inhibitor of PDK1 (BX795), an inhibitor of mTOR (rapamycin), and rapamycin with an inhibitor of NF-κB (JSH23) (Fig. 3B and SI Appendix, Fig. S13A). With the exception of phosphorylated AKT (p-AKT), inhibition of PDK1 or combined inhibition of mTOR and NF-κB produced similar changes, consistent with the predictions of ensemble single-target inhibition or dual-target inhibition simulations and the ability of PDK1 inhibition to disrupt the PDK1-AKT-IKBKB-PTEN loop and reduce activation of NF-κB. To confirm whether BX795 is properly inhibiting the PDK1 expression in senescent NHDFs, we tested the expression level of PDK1 and Akt phosphorylation under the senescence condition. As a result, we found that down-regulation of PDK1 enzyme activity can inhibit the expression of phospho-Akt (SI Appendix, Fig. S14). We also confirmed the expression of SA-β-gal (SI Appendix, Fig. S15), PDK1, and Akt phosphorylation (SI Appendix, Fig. S14 A and B) of two other primary cell lines under the senescence condition. This implicates that the effect of PDK1 inhibition would be the same in these two senescent NHDFs.
Using the simulation data, we explored the signal flow among these four nodes in networks producing the senescence attractor, networks in the presence of the PDK1 inhibitor, and networks producing the quiescence attractor (Materials and Methods). We found that the deactivated feedback loop has the same signal flow dynamics as the feedback loop in the quiescence condition (Fig. 3C and SI Appendix, Table S10).
Collectively, these simulation results suggest that PDK1 inhibition down-regulates the activity of both mTOR and NF-κB, which implies a reduction in cell growth and SASP, key processes in maintaining the senescence phenotype, and does so by deactivating the key positive feedback loop composed of PDK1, AKT, IKBKB, and PTEN. The resulting change in signaling restores the quiescence phenotype.
Cell-Cycle Reentry and Eradication of Senescence-Associated Phenotypes by PDK1 Inhibition.
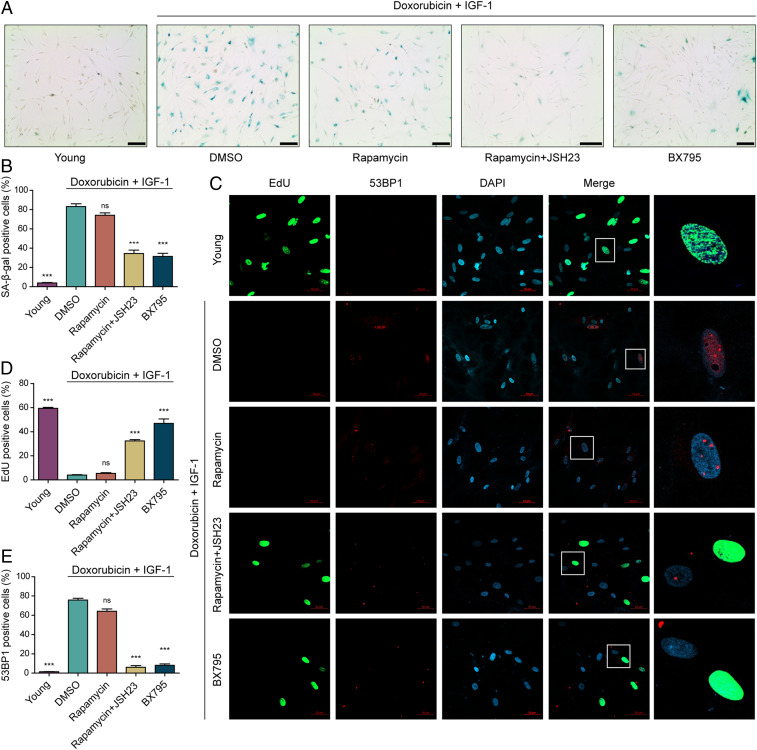
To further validate the predictions from our model simulations, we assessed cellular states of NHDFs associated with senescence, in particular the number of cells positive for SA-β-gal activity, the number of proliferating cells, and activity of the DNA damage response as assessed by the number of cells positive for 53BP1. Because our simulation results suggested that PDK1 inhibition or dual inhibition of mTOR and NF-κB can convert the cellular state from senescence to quiescence, we examined these two conditions and compared them with single inhibition of mTOR. We induced senescence with combined exposure to doxorubicin and IGF1 for 7 d, then exposed the cells to rapamycin (mTOR inhibitor), rapamycin plus JSH23 (mTOR and NF-κB inhibitors), or BX795 (PDK1 inhibitor) for 7 d, then subcultured the cells in normal growth media without the inhibitors. Either PDK1 inhibition or dual inhibition of mTOR and NF-κB reduced the SA-β-gal–positive cell population from >80% to <40% (Fig. 4 A and B and SI Appendix, Fig. S16 A and B), whereas mTOR inhibition alone did not significantly alter the percent of SA-β-gal–positive cells. We found that the percent of proliferating cells increased after either PDK1 inhibition or dual inhibition of mTOR and NF-κB, but not after only mTOR inhibition (Fig. 4 C and D and SI Appendix, Figs. S13B and S16 C and D). Proliferation of these reverted cells stopped in the low-nutrient environment with serum starvation (SI Appendix, Fig. S17), consistent with these cells responding properly to nutrient and growth signals. PDK1 inhibition or dual inhibition of mTOR and NF-κB reduced the number of cells positive for 53BP1, indicating that these conditions suppressed the DNA damage response (Fig. 4E). Notably, mTOR single inhibition did not alter these features of senescence, suggesting that the ability of PDK1 inhibition to convert senescence cells to quiescent cells involves reduced signaling and activation of both mTOR and NF-κB (Fig. 4).
Fig. 4.
Senescence-associated phenotypes following PDK1 inhibition in NHDFs. Young NHDFs were not subjected to senescence conditions or drug treatment. Young NHDFs were analyzed when cultures reached ∼80% confluence at PDL 12. The remaining NHDFs were induced to senescence by 7-d exposure to doxorubicin (100 ng/mL) plus IGF1 (100 ng/mL). The senescent cells were then treated with DMSO, rapamycin (100 nM), rapamycin plus JSH (2.5µM), or BX795 (100 nM) for 7 more days. (A) Representative images of SA-β-gal staining. (Scale bars, 200 μm.) (B) Bar diagram showing percentage SA-β-gal–positive cells. (C) Representative immunofluorescent images of NHDFs stained for EdU (proliferating cells) and 53BP1 (cells with active DNA damage response). (Scale bars, 50 μm.) Last column shows a representative cell’s nucleus for each condition. (D) Quantification of EdU as percent of cells with positive staining. (E) Quantification of 53BP1-positive cells as percent of cells with positive nuclear staining. For B, D, and E, data are shown as mean ± SD (n = 5), ***P < 0.001 compared with the DMSO control senescent cells by one-way analysis of variance. ns, not statistically significant.
Restoration of Regenerative Capacity upon PDK1 Inhibition.
To determine if true regenerative capacity and not just a reduction in hallmarks of senescence was achieved by PDK1 inhibition or dual mTOR and NF-κB inhibition, we examined capacity of the reverted cells for 2D wound healing and 3D skin regeneration. PDK1 inhibition or dual inhibition of mTOR and NF-κB, but not mTOR single inhibition, facilitated the closure of scratch wounds of NHDF monolayers (SI Appendix, Fig. S18 A and B).
This system also provided an opportunity to evaluate the effect of PDK1 inhibition on the SASP, which our network simulations predicted was reduced through PDK1 inhibition-mediated NF-κB suppression. We examined the messenger RNA (mRNA) for several key SASP components, including IL-6, IL-8, CXCL-1, and MCP-1. Senescent cells showed a significantly increased expression of these SASP-associated genes (SI Appendix, Fig. S18C). Either exposure to the NF-κB inhibitor JSH23 or the PDK1 inhibitor BX795 reduced the expression of these genes. These proinflammatory cytokines are frequently observed in senescence-associated microenvironments, where they promote and reinforce cellular senescence in paracrine and autocrine ways (20). To examine the role of SASP in wound healing, we applied the PDK1 inhibitor in conditioned medium produced by the senescent cells, which blocked the enhanced wound closure mediated by PDK1 inhibition (SI Appendix, Fig. S18 A and B) and increased mRNA expression of the SASP-associated genes (SI Appendix, Fig. S18C).
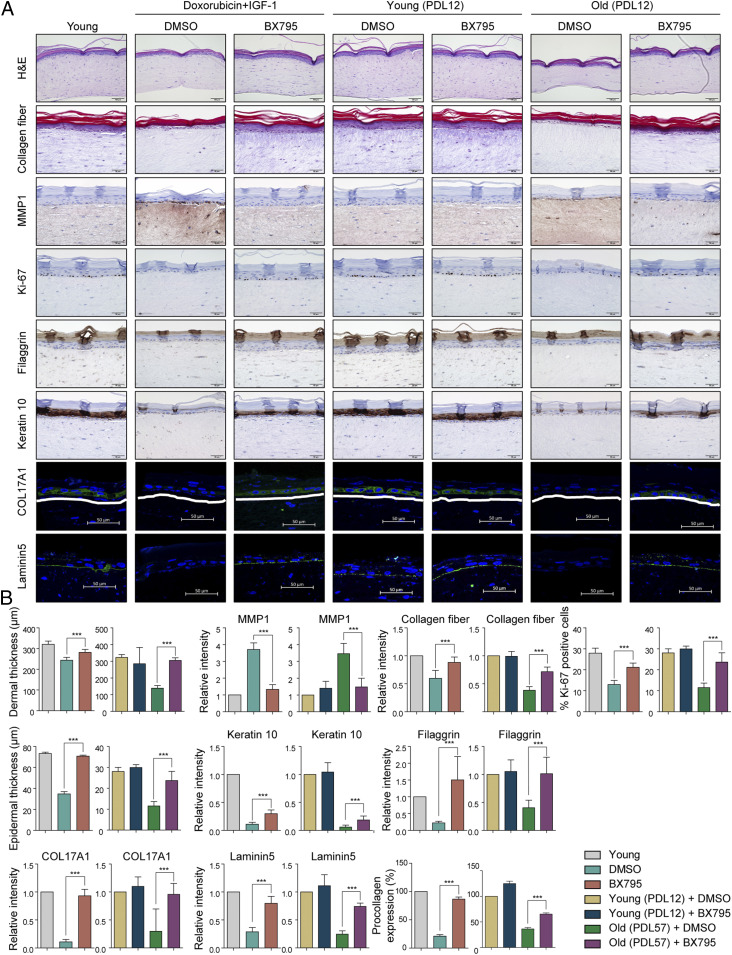
Senescent keratinocytes and fibroblasts accumulate with age in the skin and contribute to the loss of skin function and integrity during aging (48, 49). Therefore, shifting senescence into quiescence may suppress the age-associated deterioration of the skin, such as epidermal thinning, reduced dermal collagen content, increased looseness, and impaired wound healing (50). To identify the effects of PDK1 inhibition on skin aging, we employed the skin equivalent model prepared from NHDF chemically induced into senescence (doxorubicin + IGF1) or induced into replicative senescence through repeated division (Old). BX795 was added to the senescent dermal layers and maintained in the medium for 7 d prior to seeding keratinocytes. When compared to skin equivalents prepared from early-passage (young) NHDFs, skin equivalents prepared from either type of senescent NHDFs showed aged skin phenotypes: decreased epidermal and dermal thickness, impaired epidermal differentiation markers, and reduced proliferative capacity (Fig. 5 A and B). PDK1 inhibition reversed the aging of dermis associated with both replicative senescence and chemically induced senescence (Fig. 5 A and B), as evidenced by the attenuation of the increase in MMP1, of the decrease in collagen fibers, and of the reduction in secretion of procollagen. For epidermal homeostasis, the balance between keratinocyte proliferation and differentiation is regulated by dermal fibroblasts (51). Therefore, we investigated whether the reversion of senescent dermal fibroblast by PDK1 inhibition affects keratinocyte proliferation and differentiation. The Ki-67 proliferation marker was markedly elevated in the epidermis of BX795-treated skin equivalents (Fig. 5 A and B), indicating an increase in the proliferative capacity of keratinocytes, which preserved epidermal thickness in skin equivalents from senescence-induced NDHFs (Fig. 5B). In addition, we compared the intensity of staining for markers of keratinocyte differentiation (keratin 10 and filaggrin) and markers of the basement membrane in the dermal epidermal junction (COL17A1 and laminin5). PDK1 inhibition rescued the reduced abundance of these keratinocyte differentiation markers and components of the dermal epidermal junction found in skin equivalents from senescent NHDFs.
Fig. 5.
PDK1 inhibition on skin aging. Three-dimensional skin models (skin equivalents) were prepared from young NHDFs (PDL 12) or senescent NHDFs induced into senescence by doxorubicin plus IGF1 or repeated plating (replicative senescence, PDL 57) were exposed to DMSO or BX795. (A) Skin equivalent model sections were stained with hematoxylin and eosin (H&E) or Masson’s trichrome to detect collagen fibers. The indicated proteins were detected by immunohistochemistry. Results are from one representative experiment of three. (Scale bars, 50 µm.) (B) Quantification of dermal thickness using ImageJ software. Quantification of MMP1. Quantification of pan-collagen fiber detected by Masson’s trichrome staining. Quantification of Ki-67–positive cells. Quantification of epidermal thickness using ImageJ software. Quantification of keratin 10. Quantification of filaggrin. Quantification of COL17A1. Quantification of laminin5. Quantification of type I procollagen protein secreted into culture media quantified by ELISA. Data in B are shown as mean ± SD of three independent fields obtained from three independent samples. ***P < 0.001 compared to DMSO control senescent cells by one-way analysis of variance.
We also assessed the extent to which PDK inhibition affected the release of key SASP components into the medium of the skin equivalents. Similar to our observations with the 2D senescent NHDFs (SI Appendix, Fig. S18), conditioned medium from skin equivalents prepared from either type of senescent NHDFs treated with BX795 contained less GRO, GROα, IL-6, IL-8, MCP-1∼3, and RANTES than was present in conditioned medium from vehicle-treated skin equivalents (SI Appendix, Fig. S19). Altogether, these findings indicated that PDK1 inhibition converts senescent dermal fibroblasts to quiescent fibroblasts, which enhances renewal of the epidermis and preserves skin thickness to limit skin deterioration associated with aging.
Discussion
Cellular senescence contributes to multiple biological processes, including development, tissue repair, and aging (3). Recent studies showed that cellular senescence also plays crucial roles in age-related pathologies by expanding senescent cell population that result in physiological dysfunction, and these cells also promote malignant transformation, cancer stemness, and cancer relapse through their secretory phenotypes (6, 11, 20, 24, 52, 53). As the important roles of cellular senescence in diseases have been revealed, several therapeutic strategies to selectively target senescent cells have been proposed, including the clearance of senescent cells and modulation of its secretory phenotypes (53). In animal models, clearance of senescent cells alleviates age-related diseases, such as osteoarthritis, cognitive impairment, muscle degeneration, and heart disease (4, 5, 7). However, these strategies have potential drawbacks. For instance, elimination of senescent cells can impair tissue regeneration, because senescent cells recruit immune cells to the wound region and provide surrounding cells with proliferation cues during tissue repair and remodeling (24). Suppression of the SASP can also impede immune surveillance of pathogens and cancer cells (24).
To circumvent these limitations, other strategies that aim to reactivate cellular proliferation of senescent cells were attempted. For instance, partial reprogramming by transient expression of Oct4, Sox2, Klf4, and c-Myc not only improves regenerative capacity of multiple organs, including the liver, heart, and skeletal muscle, but also extends lifespan in mouse model of premature aging (54). Although this study showed that cellular senescence can be reversed, it did not rule out the possibility of malignant transformation. Another study showed that premature termination of reprogramming leads to epigenetic dysregulation and induces dedifferentiated dysplastic cells, with the animals developing renal cancer (55). To develop a therapeutic strategy that circumvents the current limitations of senescence therapies, including a risk of tumorigenesis, we explored strategies to convert senescent cells to quiescent cells based on the current molecular understanding of cellular senescence mechanisms.
In this study, we constructed a prior knowledge network by integrating key parts of signaling pathways involved in the DNA damage response, cell-cycle arrest, cell growth, energy metabolism, and the SASP, all of which play major roles in cellular senescence. To simulate the dynamics of the network resulting from the complex interplay among these signaling pathways, we developed Boolean network models by inferring the regulatory logic optimized to fit phosphoprotein array data from NHDFs. To find senescence-to-quiescence control targets, we simulated the ensemble of optimized Boolean network models and performed integrative analysis of the simulation results.
From the ensemble model simulation we identified PDK1 as a promising target of inhibitors to convert senescent cells into quiescent cells while avoiding proliferation. Detailed analysis of the network structure and dynamics revealed that PDK1 is not only located at a structurally important position in the network but also plays a key role in a positive feedback loop composed of PDK1, AKT, IKBKB, and PTEN. In a dynamical system, a positive feedback loop can induce all-or-none bistable responses and stabilize one cellular state instead of others (56). Our simulation results predicted that components of the feedback loop (AKT and IKBKB) and upstream regulators of PDK1 (PIK3CA, IRS1, and IGF1R) are also high-scoring potential inhibitor targets for promoting the transition from senescence to quiescence. Dual inhibition of both mTOR and NF-κB was the most effective combination of all double-node inhibition simulations. Because both mTOR and NF-κB are downstream targets of the positive feedback loop, the modeling analyses suggested that the positive feedback loop composed of PDK1, AKT, IKBKB, and PTEN was a key network motif that sustains the senescent state.
We validated the predicted simulation results using NHDFs. We showed that PDK1 inhibition disrupts the positive feedback loop and suppresses the activity of its downstream targets, mTOR and NF-κB. Markers of cellular senescence, such as SA-β-gal activity and abundant 53BP1, disappeared from senescent cells after transient PDK1 inhibition, and the cells reentered the cell cycle. We demonstrated that the recovery of proliferative potential upon PDK1 inhibition restores skin regeneration capacity using a 2D culture model and a 3D skin equivalent model.
Although we thoroughly examined the multiple cellular senescence markers such as SA-β-gal activity, 53BP1, p21, and p16, we cannot disregard the possibility that a small presenescent cell population might have escaped from cell-cycle arrest. Whether cellular senescence induced by either different cellular stresses or different cell types can also be reversed still remains to be investigated. Moreover, the conversion of senescence to quiescence by PDK1 inhibition should be examined at the organ and organism levels. Because PDK1 is an oncogene that is frequently overexpressed in various cancers, we expect that its inhibition has both an antiaging effect and a tumor-suppressing effect (42).
In summary, our findings provide insight into the complex mechanism of cellular senescence and present a potential therapeutic strategy for reducing age-related diseases associated with the accumulation of senescent cells.
Materials and Methods
Cell Culture.
Three NHDFs derived from skin of different adult donors (ages 21, 33, and 58 y, Caucasian females) were purchased from Lonza and were cultured in DMEM (DMEM, 4.5 g/L glucose) (Lonza) containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin in a 5% CO2 incubator at 37 °C. Population doubling levels (PDL) were estimated with the following equation: PDL = (log10Y − log10X)/log102, where Y indicates the number of cells counted at the end of the passage and X is the number of cells that were seeded. All experiments were performed starting from presenescent NHDFs (PDL 12 to 14, young NHDFs). Representative data were obtained using NHDFs derived from a 33-y-old donor. The results were repeated with two other NHDFs derived from different donors.
Senescence Induction and Chemical Compound Treatment.
NHDFs (PDL 12) were induced to senescence by treatment with 100 ng/mL doxorubicin plus 100 ng/mL IGF1 (senescence-induced drugs) for 7 d. The following day, dimethyl sulfoxide (DMSO), rapamycin (100 nM), rapamycin plus JSH (2.5 µM), or BX795 (100 nM) were added simultaneously with doxorubicin plus IGF1 and cells were further incubated for 7 d. In all cases, chemicals were readded every 48 h. Fifteen days after initial cell plating, cells were subcultured and incubated for various analysis.
Conditioned Medium.
NHDFs (PDL 12) were induced to senescence by treatment with 100 ng/mL doxorubicin plus 100 ng/mL IGF1 for 7 d. After induction of senescence, cells (2 × 106) were seeded in a 10-cm dish and incubated for 24 h with 0.5% FBS. After 24 h, the conditioned medium (CM) was collected, centrifuged at 5,000 × g, and filtered through a 0.2-µm pore filter. For SASP-CM treatment, cells grown in the presence of doxorubicin plus IGF1 for 7 d, followed by 7 d in the additional presence of doxorubicin, IGF1, and BX795, were plated in six-well plates at a seeding density of 2 × 105 cells per well and incubated with SASP-CM for 3 d.
SA-β-Gal Activity Assay.
SA-β-gal staining was performed using the Senescence Cells Histochemical Staining Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, cells were washed with phosphate-buffered saline (PBS) and fixed at room temperature for 5 min then incubated in Staining Working Solution at 37 °C for 16 h. For quantification of senescent cells, a minimum of 200 cells were counted to determine the number of SA-β-gal–positive cells using MetaMorph image-analysis software (Molecular Devices).
Immunofluorescence Staining.
NHDF proliferation was evaluated using the Click-iT EdU Imaging Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, cells were treated with 10 µM EdU for 16 h and then fixed, permeabilized, and stained with EdU. After washing three times in PBS for 5 min each, cells were incubated for 60 min with 5% BSA in PBST (blocking buffer); thereafter, the cells were exposed to primary antibodies against 53BP1 (Abcam) in blocking solution overnight at 4 °C. After washing three times in PBS for 5 min each, the cells were incubated for 1 h at room temperature with a 1:1,000 dilution of AlexaFluor 594-labeled secondary antibodies (Invitrogen) in blocking solution. Cell nuclei were stained for 2 min with 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI; Invitrogen). Confocal microscopy images were then acquired using a Zeiss LSM700 confocal microscope and analyzed with Imaris software (Bitplane) for quantification. EdU- or 53BP1-positive cells were scored under a fluorescence microscope and are presented as the percentage of EdU- or 53BP1-positive nuclei relative to the total number of nuclei. At least 200 nuclei were counted.
qRT-PCR Analysis.
Total RNA from cells was prepared with the RNeasy Mini kit (Qiagen), and 1 μg was used to synthesize complementary DNA with the SuperScript II Reverse Transcriptase kit (Life Technologies). The predesigned primCXCL1ers and probe sets for IL-6, IL-8, CXCL1, MCP-1, and ribosomal protein lateral stalk subunit P0 (RPLP0) were obtained from Applied Biosystems (assay IDs: Hs00174131_m1, Hs00174103_m1, Hs00236937_m1, Hs00234140_m1, and Hs00420895_gH; Thermo Fisher Scientific). qRT-PCR was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems). The relative expression level of target genes was normalized to that of RPLP0, and the fold difference was calculated with the 2 − ΔΔCt method.
Immunoblotting.
Cells were lysed in radioimmunoprecipitation buffer composed of PBS (pH 7.4) containing 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate with protease inhibitor mixture (Sigma-Aldrich). Equal amounts of protein (40 μg) were resolved on 4 to 12% NuPAGE gels (Invitrogen) and transferred to a polyvinylidene difluoride membrane. Proteins were detected by enhanced chemiluminescence using ECL Plus (GE Healthcare). Antibodies against PDK1 and phospho-IGF-1R (Tyr1161/Tyr1165/Tyr1166) were purchased from Sigma-Aldrich; antibody against p21 was from Santa Cruz Biotechnology; antibodies against p16, phospho-p53 (Ser15), phospho-IκBα (Ser32/36), phospho-p70S6Kinase (Thr389), phospho-Akt (Thr-308), phospho-Akt (Ser473), IGF-I receptor β (D23H3), E2F-1, p53, p70S6Kinase, Akt and β-actin were from Cell Signaling Technology; and the antibodies against IκBα, NF-κB p65, and phospho NF-κB p65 (Ser536) alpha were from Abcam. Blots were visualized with a LAS-3000 imaging system.
Wound-Healing Assay.
For the in vitro scratch assay, cells (1 × 105 cells per well) were plated in 24-well plate with CytoSelect Wound Healing Inserts (Cell Biolabs, Inc.) and incubated at 37 °C until a monolayer formed. Thereafter, inserts were removed from the plate well and the cells were thoroughly washed with PBS and incubated with fresh medium. Wound closure was monitored by collecting digital images at 0- and 96-h intervals, and digital images were captured by using an inverted microscope (Leica). The images were obtained at the same position before and after incubation. The experiment was repeated three times. The level of wound closure was assessed by the ratio of closure area to initial wound (0 h) as follows:
where Rn represents the percentage of wound closure, An represents the residual area of wound at the metering point (nh), and A0 represents the area of initial wound (0 h).
Skin Equivalents Model.
After induction of senescence for 7 d, followed by an additional 7 d of cotreatment with senescence-induced drugs and DMSO or BX795, skin equivalents were prepared with these NHDFs as previously described (57). Briefly, the dermal layer was constructed by mixing a type I collagen (Advanced Biomatrix) with NHDFs (6.0 × 104 cells per well) in media mixture (DMEM, F12, and NaHCO3, neutralized with NaOH). The mixture was added to each insert of a six-well plate (Snapwell; Corning Inc.) and incubated for 2 h at 37 °C for polymerization. The dermal layer was then cultured in 106 media supplemented with 100 µg/mL ascorbic acid and allowed to contract for 7 d at 37 °C and 5% CO2. To examine the effect of BX795 on replicative senescence, the dermal layer was constructed with NHDFs at early passages (PDL 12 to 14, young cells) or late passages (PDL 57 to 60, replicative senescence cells). The dermal layer was then treated with DMSO or BX795 for 7 d. Human epidermal neonatal keratinocytes (2.0 × 105 cells per well) were seeded on the dermal layer to prepare the skin equivalents. The skin equivalents were cultured for 1 d in EpiLife media and 1 d in the 3D culture media, CnT-3D-PR (CELLnTEC); thereafter, the skin equivalents were fed strictly from the bottom of the 3D culture media, and the surface was exposed to air to promote epidermal differentiation for 8 d. For analysis, the skin equivalents were fixed in 10% neutral buffered formalin and then processed into wax blocks before further analysis.
Immunohistochemistry and Immunofluorescence on Skin Equivalent Sections.
Paraffin samples were dewaxed in xylene and rehydrated through a descending ethanol to water series (3 min per solution). Epitope retrieval was performed at 121 °C for 10 min using Citrate Buffer pH 6.0 Antigen Retriever (64142; Electron Microscopy Sciences), and the slides were cooled prior to three washes in PBS/0.05% Tween 20. The sections were then incubated with water/1% H2O2 for 30 min to block endogenous peroxidase, rinsed with water and PBS/0.05% Tween 20 for 5 min, and incubated with PBS/10% goat serum (Dako) for 20 min before exposure to the following primary antibodies overnight at 4 °C: anti-KRT 10 (Biolegend), anti-FLG (Abcam), anti-MMP-1 (Abcam), and anti-Ki67 (Abcam). Samples were thoroughly washed with water for 10 min and PBS/0.05% Tween 20 for 5 min followed by 30-min incubation with EnVision+ System horseradish peroxidase-labeled polymer anti-mouse antibody (K4001; Dako). Then, the samples were washed in water and PBS before incubation with Liquid DAB+ Substrate Chromogen System (K3468; Dako), for 3 to 5 min. The reaction was stopped by rinsing in tap water, and sections were counterstained in Mayer’s hematoxylin for 3 to 5 min and rinsed in tap water for 1 min and in Scott’s bluing water for 2 min. Finally, the sections were dehydrated in graded ethanol, cleared in xylene, and mounted in Cytoseal 60 (Thermo Scientific). Masson’s trichrome staining was performed using commercially available kits (BBC Biochemical). For immunofluorescence staining, antigen retrieval was performed by boiling the slides for 20 min in Dako target retrieval solution (Dako). Nonspecific staining was blocked by incubation with PBS containing 10% donkey serum and 0.05% Triton X-100 for 30 min. Tissue sections were incubated with COL17A1 (Abcam) and Laminin 5 (Santa Cruz Biotechnology) in Dako Fluorescence Mounting Medium (Dako) at 4 °C overnight and subsequently incubated with appropriate secondary antibodies conjugated with Alexa Fluor 488 (Invitrogen-Molecular Probes) for 2 h at room temperature. After washing in PBS, DAPI (Invitrogen-Molecular Probes) was added for nuclear counterstaining. Coverslips were mounted on glass slides with fluorescent mounting medium (Thermo Electron).
ELISA for Type I Collagen Expression.
The expression level of procollagen in the media of skin equivalents was measured using Procollagen Type I CPeptide Kit (TaKaRa) as described by the manufacturer.
Cytokine Antibody Array.
Culture media of skin equivalents were collected and incubated overnight on a RayBio C-Series human inflammation antibody array (RayBiotech) according to the manufacturer’s protocol. Arrays were imaged with an LAS-4000 imager (Fujifilm) and image analysis was performed using Image Studio Lite (LI-COR). The intensities were normalized to internal positive controls and analyzed by Excel-based analysis software tool (RayBiotech). The average density values for each cytokine in each condition were then averaged for each array in order to calculate the fold change.
Statistical Analysis.
Data were evaluated with the unpaired two-sided Student’s t test or one-way analysis of variance followed by Dunnett’s multiple comparisons tests to determine statistical significance after confirming that the data met appropriate assumptions (normality, homogeneity of variance, and independent sampling). Data analysis was performed with SPSS v.20 (SPSS Inc.) or Prism 6 (GraphPad Inc.) software. Statistical significance was determined at a value of P < 0.05.
Prior Knowledge Network Curation.
We curated a comprehensive senescence regulatory network from literature and network databases. The resulting network (Fig. 1A) includes 41 nodes and 77 links (annotations on each link can be found from SI Appendix, Table S2). Each literature source was abstracted into directional activation/inhibition links. These directional signed networks served as the backbone network structure of the Boolean network representation.
Phosphoprotein Array Experiment.
For quiescent cultures, NHDFs (PDL 12) were incubated with low-glucose (1 g/mL) DMEM supplemented with 1% FBS and 100 ng/mL doxorubicin for 1, 6, 24, and 96 h. For senescent cultures, NHDFs (PDL 12) were incubated with high-glucose (4.5 g/mL) DMEM supplemented with 10% FBS, 100 ng/mL doxorubicin, and 100 ng/mL IGF1 for 1, 6, 24, and 96 h. For proliferation cultures, NHDFs (PDL 12) were incubated with high-glucose (4.5 g/mL) DMEM supplemented with 10% FBS and 100 ng/mL IGF1 for 1, 6, and 24 h. At the indicated time, cells were harvested and applied to a Phospho Explorer Antibody Microarray (PEX100; Full Moon Biosystems, Inc.) according to the manufacturer’s protocol. The array contained 1,318 antibodies. Each of the antibodies has four replicates that are printed on coated glass microscope slides, along with multiple positive and negative controls. Slide scanning was performed using the GenePix 4100A scanner (Molecular Devices). The scanned images were graded and quantified using GenePix 7.0 Software (Molecular Devices). The numerical data were analyzed using Genowiz 4.0TM (Ocimum Biosolutions). All sample intensities were normalized with quantile normalization. After these analyses, the protein information was annotated using the UniProt database. Fold change between control and treated sample was calculated using the following formula:
Activation Level Inference from Phosphoprotein Array Data.
From the results of phosphoprotein array analysis, specific antibodies were selected to represent the activation level of each respective node (antibody reference for each node can be found in SI Appendix, Table S4). The activation level was inferred by the AUC of the interpolated mean fold changes on a normalized time scale (SI Appendix, Fig. S4). These AUC data were normalized (z-score normalization) and binarized to represent Boolean on/off states by referring to previous studies (SI Appendix, Table S4).
Boolean Network Modeling of the Cellular Senescence Network.
We modeled the network dynamics using deterministic weighted-sum Boolean state transition logic. In a Boolean network model, the state of any given node on the network can either be on (1) or off (0). Logical state of each node was determined by weighted sum of on (1) nodes directly upstream of the given node and the basal activity of the given node. If the weighted sum exceeds a given threshold, the given node is turned on at the next time point and is turned off otherwise. Network dynamics were modeled by reevaluating the network state at each time point from the initial to final state, synchronously. If the network state reaches an equilibrium, it is considered to have reached an attractor, a network state reflecting a biological steady state, and the simulation is terminated. Multiple sets of weights and basal activities can produce the same weighted-sum transition logic.
Boolean Logic Inference Using Evolutionary Algorithm.
The weights and basal activities of the curated prior knowledge network (PKN) were inferred using the pairs of input node states and the binarized phosphoproteomic AUC values of 14 key proteins (SI Appendix, Table S4) as input–output solution pairs. Evolutionary algorithm framework from the python package Pygmo (58) was used to infer the weights and basal activities that fit all of the input–output solution pairs of quiescence, senescence, and proliferation. For this purpose, self-adaptive differential evolution (59) was used where the cost function was as follows:
where n is the total number of conditions: quiescence, senescence, and proliferation; m is the total number of phenotypic nodes represented by the expected output; yij is the binarized value of phosphoprotein array data at jth node and ith condition; and oij is the Boolean state of the attractor from the network model simulation at jth node and ith condition. Only the networks with zero cost, the Boolean networks with outputs that match the expected outputs perfectly, were selected for the ensemble model. Because a number of Boolean logic combinations may satisfy all of the required the input–output solution pairs, 5,000 models that satisfy the input–output solution pairs were generated.
Perturbation Simulation for Targets That Can Reverse Senescence to Quiescence.
The senescence-reversing targets that can convert senescence state, a network equilibrium state under senescence input, to quiescence state while avoiding the proliferation state were determined through brute-force inhibition simulations of all nodes. A targeted inhibition of a specific node was simulated in the network model by assigning the corresponding node to constant off (0). To quantify senescence to quiescence conversion rate, we defined a measure, as the ratio of network models where the [node] inhibition converts the senescence attractor to the quiescence attractor. Similarly, we defined as the ratio of network models where the [node] inhibition converts the senescence attractor to the proliferation attractor. The optimal senescence-reversing target was screened by the score where . When inhibited, nodes with high are more likely to drive the network into quiescence from senescence equilibrium and less likely to drive the network into proliferation.
Analysis of PDK1 Inhibition Dynamics.
Only the 2,401 Boolean network models for which PDK1 inhibition produced the quiescence phenotype from the senescence equilibrium were collected. The state transition trajectory from the senescence state to the quiescence state caused by PDK1 inhibition were monitored for the 2,401 Boolean network models. The Boolean network models reached the quiescence attractor within 15 time steps (SI Appendix, Fig. S12A) and the cost function decreased as time steps increased and converged to zero when the Boolean network models reached the quiescence attractor (SI Appendix, Fig. S12B). Because each network has a different length of transition trajectory, we unified the transition time steps into 10 by interpolating between node values. For each network model, we calculated the difference between node state of the senescence attractor and interpolated node value for 10 time steps and averaged the differences for each time step (SI Appendix, Table S8). The total effect of PDK1 inhibition on a specific node, , is defined as the difference between the average node state after the network reached its attractor from senescence equilibrium with PDK1 inhibition, , and the average node state after the network reached its attractor under senescence input without PDK inhibition, . Thus, , is equal to the node values in the 10th time step shown in SI Appendix, Table S8.
Signal Flow Visualization.
A signal flow strength from node A to node B is defined as the product of average activation level of the outgoing node and average link weight as follows:
where SSAB is the signal flow strength from node A to node B, n is the number of individual network models in the ensemble simulation (n = 2,401), NAi is the adjusted activation level of node A in the ith network model, and WABj is the weight between node A and node B in the jth network model. The adjusted activation level of a node is defined as a Boolean state linearly adjusted from a range of [0, 1] to [−1, 1].
Supplementary Material
Acknowledgments
This work was supported by National Research Foundation of Korea grants funded by the Korea Government, the Ministry of Science and Information & Communication Technology (ICT) (2020R1A2B5B03094920) and an Electronics and Telecommunications Research Institute grant funded by the Korean government (20ZS1100, Core Technology Research for Self-Improving Integrated Artificial Intelligence System). It was also partially supported by Korea Advanced Institute of Science and Technology Grand Challenge 30 Project and a research grant from Amorepacific R&D Center. We thank Sea Choi for her critical reading and valuable comments.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920338117/-/DCSupplemental.
Data Availability.
All study data are included in the paper, SI Appendix, and Datasets S1 and S2.
References
- 1.Campisi J., et al. , From discoveries in ageing research to therapeutics for healthy ageing. Nature 571, 183–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McHugh D., Gil J., Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 217, 65–77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodier F., Campisi J., Four faces of cellular senescence. J. Cell Biol. 192, 547–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baar M. P., et al. , Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169, 132–147.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang J., et al. , Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childs B. G., et al. , Senescent cells: An emerging target for diseases of ageing. Nat. Rev. Drug Discov. 16, 718–735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeon O. H., et al. , Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 23, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blagosklonny M. V., Cell cycle arrest is not senescence. Aging (Albany NY) 3, 94–101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blagosklonny M. V., Geroconversion: Irreversible step to cellular senescence. Cell Cycle 13, 3628–3635 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Zamudio R. I., Robinson L., Roux P.-F., Bischof O. J. C., SnapShot: Cellular senescence pathways. Cell 170, 816–816.e1 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Campisi J., di Fagagna F. d. A., Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Kim H., et al. , Expression profiles of p53-, p16INK4a-, and telomere-regulating genes in replicative senescent primary human, mouse, and chicken fibroblast cells. Exp. Cell Res. 272, 199–208 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Iwasa H., Han J., Ishikawa F., Mitogen activated protein kinase p38 defines the common senescence‐signalling pathway. Genes Cells 8, 131–144 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Wu D., Prives C., Relevance of the p53–MDM2 axis to aging. Cell Death Differ. 25, 169–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salminen A., Kaarniranta K., AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res. Rev. 11, 230–241 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Xu Y., Li N., Xiang R., Sun P., Emerging roles of the p38 MAPK and PI3K/AKT/mTOR pathways in oncogene-induced senescence. Trends Biochem. Sci. 39, 268–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demidenko Z. N., et al. , Rapamycin decelerates cellular senescence. Cell Cycle 8, 1888–1895 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Liang Y., Vanhoutte P. M., SIRT1 and AMPK in regulating mammalian senescence: A critical review and a working model. FEBS Lett. 585, 986–994 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Blagosklonny M. V., Rapamycin, proliferation and geroconversion to senescence. Cell Cycle 17, 2655–2665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coppé J.-P., Desprez P.-Y., Krtolica A., Campisi J., The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young A. R., Narita M., SASP reflects senescence. EMBO Rep. 10, 228–230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien Y., et al. , Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tilstra J. S., et al. , NF-κB inhibition delays DNA damage–induced senescence and aging in mice. J. Clin. Invest. 122, 2601–2612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S., Schmitt C. A., The dynamic nature of senescence in cancer. Nat. Cell Biol. 21, 94–101 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Herranz N., Gil J., Mechanisms and functions of cellular senescence. J. Clin. Invest. 128, 1238–1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beauséjour C. M., et al. , Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J. 22, 4212–4222 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milanovic M., et al. , Senescence-associated reprogramming promotes cancer stemness. Nature 553, 96–100 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Sage J., Miller A. L., Pérez-Mancera P. A., Wysocki J. M., Jacks T. J. N., Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424, 223–228 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Yu Y., et al. , Targeting the senescence-overriding cooperative activity of structurally unrelated H3K9 demethylases in melanoma. Cancer Cell 33, 322–336.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D., et al. , The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Türei D., Korcsmáros T., Saez-Rodriguez J. J. N. m., OmniPath: Guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 13, 966–967 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Abdian N., et al. , Comparison of human dermal fibroblasts (HDFs) growth rate in culture media supplemented with or without basic fibroblast growth factor (bFGF). Cell Tissue Bank. 16, 487–495 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Helikar T., Konvalina J., Heidel J., Rogers J. A., Emergent decision-making in biological signal transduction networks. Proc. Natl. Acad. Sci. U.S.A. 105, 1913–1918 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H.-S., et al. , A systems-biological study on the identification of safe and effective molecular targets for the reduction of ultraviolet B-induced skin pigmentation. Sci. Rep. 5, 10305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi M., Shi J., Jung S. H., Chen X., Cho K.-H., Attractor landscape analysis reveals feedback loops in the p53 network that control the cellular response to DNA damage. Sci. Signal. 5, ra83 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Wu Z., Jiang D., Kang L., “Evolutionary algorithm for identifying discontinuous parameters of inverse problems” in International Conference on Computational Science, Shi Y., van Albada G. D., Dongarra J., Sloot P. M. A., Eds. (Springer, 2007), vol. 4490, pp. 1131–1138. [Google Scholar]
- 38.Michalewicz Z., Evolutionary computation techniques for nonlinear programming problems. Int. Trans. Oper. Res. 1, 223–240 (1994). [Google Scholar]
- 39.Huang S., Eichler G., Bar-Yam Y., Ingber D. E., Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys. Rev. Lett. 94, 128701 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Bonzanni N., et al. , Hard-wired heterogeneity in blood stem cells revealed using a dynamic regulatory network model. Bioinformatics 29, i80–i88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polikar R., Ensemble based systems in decision making. IEEE Circuit Syst. Mag. 6, 21–45 (2006). [Google Scholar]
- 42.Raimondi C., Falasca M., Targeting PDK1 in cancer. Curr. Med. Chem. 18, 2763–2769 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Salminen A., Kauppinen A., Kaarniranta K., Emerging role of NF-κB signaling in the induction of senescence-associated secretory phenotype (SASP). Cell Signal. 24, 835–845 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Gates A. J., Rocha L. M., Control of complex networks requires both structure and dynamics. Sci. Rep. 6, 24456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bafna V., Berman P., Fujito T., A 2-approximation algorithm for the undirected feedback vertex set problem. SIAM J. Discrete Math. 12, 289–297 (1999). [Google Scholar]
- 46.Zañudo J. G. T., Yang G., Albert R., Structure-based control of complex networks with nonlinear dynamics. Proc. Natl. Acad. Sci. U.S.A. 114, 7234–7239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiedler B., Mochizuki A., Kurosawa G., Saito D., Dynamics and control at feedback vertex sets. I: Informative and determining nodes in regulatory networks. J. Theor. Biol. 25, 563–604 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Dimri G. P., et al. , A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Figueroa R., Lindenmaier H., Hergenhahn M., Nielsen K. V., Boukamp P., Telomere erosion varies during in vitro aging of normal human fibroblasts from young and adult donors. Cancer Res. 60, 2770–2774 (2000). [PubMed] [Google Scholar]
- 50.Giangreco A., Goldie S. J., Failla V., Saintigny G., Watt F. M., Human skin aging is associated with reduced expression of the stem cell markers beta1 integrin and MCSP. J. Invest. Dermatol. 130, 604–608 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Werner S., Smola H., Paracrine regulation of keratinocyte proliferation and differentiation. Trends Cell Biol. 11, 143–146 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Demaria M., et al. , Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7, 165–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niedernhofer L. J., Robbins P. D., Senotherapeutics for healthy ageing. Nat. Rev. Drug Discov. 17, 377 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Ocampo A., et al. , In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167, 1719–1733.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alderton G. K., Pluripotency: Partial reprogramming induces cancer. Nat. Rev. Cancer 14, 216–217 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Xiong W., Ferrell J. E. Jr, A positive-feedback-based bistable ‘memory module’that governs a cell fate decision. Nature 426, 460–465 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Park S., et al. , TIMP3 is a CLOCK‐dependent diurnal gene that inhibits the expression of UVB‐induced inflammatory cytokines in human keratinocytes. The FASEB Journal 32, 1510–1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biscani F., Izzo D., Yam C. H., A global optimisation toolbox for massively parallel engineering optimisation. arXiv:1004.3824 (22 April 2010).
- 59.Brest J., Zumer V., Maucec M. S., “Self-adaptive differential evolution algorithm in constrained real-parameter optimization” in 2006 IEEE International Conference on Evolutionary Computation (IEEE, 2006), pp. 215–222. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the paper, SI Appendix, and Datasets S1 and S2.