Abstract
Inflammatory bowel disease (IBD) encompasses a spectrum of gastrointestinal disorders driven by dysregulated immune responses against gut microbiota. We integrated single-cell RNA and antigen receptor sequencing to elucidate key components, cellular states, and clonal relationships of the peripheral and gastrointestinal mucosal immune systems in health and ulcerative colitis (UC). UC was associated with an increase in IgG1+ plasma cells in colonic tissue, increased colonic regulatory T cells characterized by elevated expression of the transcription factor ZEB2, and an enrichment of a γδ T cell subset in the peripheral blood. Moreover, we observed heterogeneity in CD8+ tissue-resident memory T (TRM) cells in colonic tissue, with 4 transcriptionally distinct states of differentiation observed across health and disease. In the setting of UC, there was a marked shift of clonally related CD8+ TRM cells towards an inflammatory state, mediated, in part, by increased expression of the T-box transcription factor Eomesodermin. Taken together, these results provide a detailed atlas of transcriptional changes occurring in adaptive immune cells in the context of ulcerative colitis and suggest a role for CD8+ TRM cells in IBD.
One Sentence Summary:
Single-cell sequencing analyses reveal changes in B and T cell transcriptional states in the context of ulcerative colitis.
Introduction
Inflammatory bowel disease (IBD) encompasses a spectrum of complex intestinal disorders characterized by dysregulated innate and adaptive immune responses to gut microbiota in genetically susceptible hosts (1). IBD is typically categorized as Crohn’s disease (CD) or ulcerative colitis (UC) based on anatomic, clinical, and histopathologic criteria (2). A number of studies aimed at uncovering the cellular and molecular basis of IBD have been undertaken, thereby implicating a number of diverse immune cell types in its pathogenesis, such as macrophages (3, 4), innate lymphoid cells (5–7), and subsets of CD4+ T cells (8–10). Notably, the vast majority of gene expression analyses in IBD have used whole intestinal tissue or bulk populations of cells fluorescence-activated cell sorting (FACS)-purified from peripheral blood or intestinal tissue on the basis of phenotypic cell surface markers. However, intestinal tissue is heterogeneous, comprised of diverse epithelial, stromal, and immune cells, and it has been increasingly appreciated that substantial heterogeneity can exist even within the same immune cell type (11, 12). Thus, gene expression measurements of whole tissue samples likely detect the most highly expressed mRNA transcripts in the most abundant cells, thereby masking many potentially important cell type-specific transcriptional signatures.
Emerging data from a number of laboratories across many fields have demonstrated the necessity and power of an unbiased, marker-agnostic approach in investigating known cell types and discovering new cell subsets and states (11–16). In particular, single-cell RNA-sequencing (scRNA-seq) has been employed to investigate the heterogeneity of intestinal cells in the context of inflammatory bowel disease (17–20). In addition to transcriptomic analyses, the ability to delineate T cell (scTCR-seq) and B cell (scBCR-seq) receptor sequences at the single-cell level has enabled characterization of T and B cell receptor repertoire diversity and identification of clonal relationships (21, 22). Together, these studies have generated exciting insights that could only have been revealed by analyses performed at the single-cell level.
Here we integrated scRNA-seq, scTCR-seq, and scBCR-seq approaches to elucidate key components, cellular states, and clonal relationships of the gastrointestinal mucosal and peripheral immune systems in health and ulcerative colitis. Substantial heterogeneity among T and B cell subsets was observed, including within plasma B cells, γδ T cells, regulatory T (Treg) cells, and CD8+ resident memory T (TRM) cells. It is well established that TRM cells mediate protective responses to microbial infection (23), but a potentially pathogenic role for these cells in autoimmune and inflammatory diseases has been increasingly appreciated (24, 25). We observed clonally related CD8+ TRM cells in four putative differentiation states across health and disease. In ulcerative colitis, CD8+ TRM cells exhibited a marked shift towards an inflammatory differentiation state associated with increased expression of the T-box transcription factor Eomesodermin (Eomes). By virtue of binding to gene targets encoding molecules with inflammatory and effector properties, such as cytokines, cytolytic granules, and killer cell lectin receptors, Eomes may be a critical molecular regulator of a pathogenic CD8+ TRM cell differentiation state in ulcerative colitis.
Results
Single-cell profiling of human immune cells from peripheral blood and colon
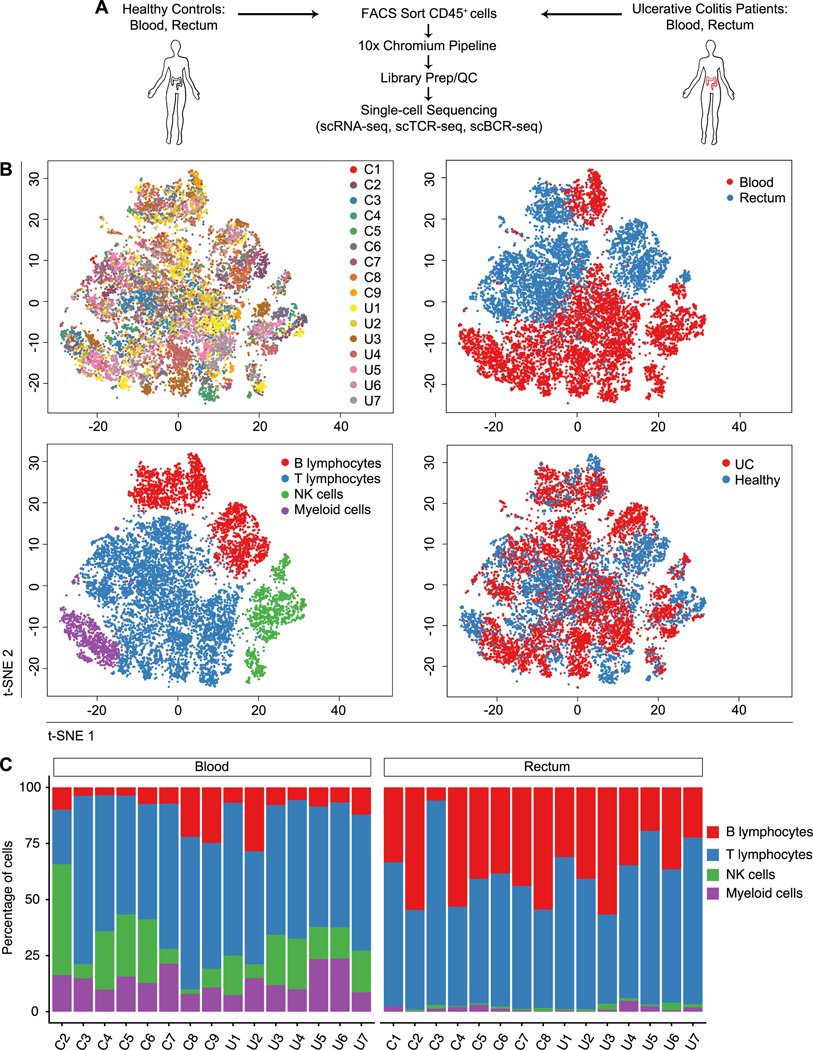
Ulcerative colitis affects the rectum in the vast majority of cases and inflammation extends proximally in a contiguous manner. This uniformity enabled us to sample this same region in every subject and minimize potential non-biologic sources of variability, such as regional differences along the gastrointestinal tract and variable exposures to luminal contents. In order to probe for transcriptional signatures that might be specific to health or disease, we obtained rectal mucosal biopsy and peripheral blood samples from 9 healthy individuals and 7 patients with active ulcerative colitis (Fig. 1A and table S2). Cells from mucosal biopsies and peripheral blood were processed into single-cell suspensions, FACS-purified on the basis of CD45, a pan-immune cell marker (fig. S1A), and subjected to scRNA-seq, scTCR-seq, and scBCR-seq using the 10x Genomics Chromium platform.
Fig. 1. Single-cell analyses reveals cellular composition of the human immune system in health and ulcerative colitis.
(A) Overview of the experimental design and analysis. (B) t-SNE plots of cells from all subjects across all anatomic sites (upper left; “C” labels indicate healthy controls, “U” labels indicate UC patients); colored by anatomic location from which cells were derived (upper right); major immune cell groups (lower left); or by health status (lower right; UC patients vs. healthy individuals). (C) Proportion of each major immune cell group in healthy individuals and UC patients, across both anatomic sites for each subject, as a percentage of all cells.
2116 genes with a mean expression of at least 1 UPM (unique molecular identifier per million reads) were detected, with a total of 10,160 genes detected across the dataset (fig. S1, B and C, and table S3). Data from all subjects across both anatomic sites were merged and unsupervised t-distributed stochastic neighborhood embedding (t-SNE) analysis was performed to visualize the clustering of single cells from all subjects (Fig. 1B). We used expression of canonical genes (see Methods) to annotate the clusters into four broad groups of immune cell types: T lymphocytes, B lymphocytes, NK cells, and myeloid cells. No significant differences in the proportions of each major immune cell group were observed between healthy individuals and UC patients (Fig. 1C), nor was the level of overall BCR or TCR repertoire diversity significantly different between healthy individuals and UC patients (fig. S1, D and E).
Transcriptionally distinct IgG1+ plasma cell cluster enriched in ulcerative colitis
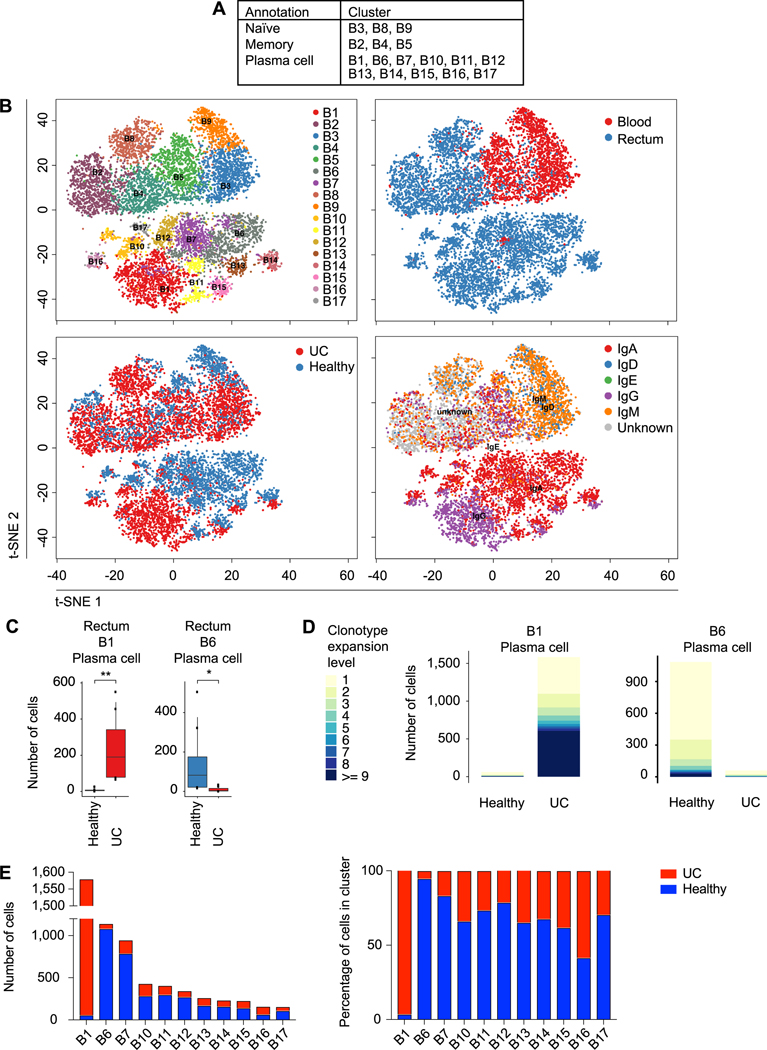
Additional t-SNE and Uniform Manifold Approximation and Projection (UMAP) clustering analyses were performed on B lymphocytes, yielding 17 sub-clusters, all of which included cells from at least two subjects (Fig. 2, A and B, fig. S2, A to E, and table S3). B cell clusters were broadly categorized into naïve, memory, or plasma cell clusters on the basis of differential expression of canonical genes. For example, naïve and memory cells expressed high levels of genes such as CD19 and could be further annotated as naïve (clusters B3, B8, and B9) or memory (clusters B2, B4, and B5) on the basis of high or low expression of IGHD, respectively. Eleven plasma cell clusters were identified on the basis of high expression of PRDM1 and XBP1, and low expression of CD19. Many clusters exhibited strong anatomic associations; only 3 clusters, the B3 and B9 naive clusters and the B5 memory cluster, were found predominantly in the peripheral blood, whereas the rest of the B cell clusters were found preferentially in the intestinal tissue (Fig. 2B, and fig. S2, F and G). We next compared the absolute and relative numbers of each cluster in the healthy and disease states. Strikingly, the B1 plasma cell cluster was almost exclusively derived from UC patients and cells from this cluster were observed to have undergone increased clonal expansion in patients with ulcerative colitis, whereas the B6 plasma cell cluster was highly enriched in healthy individuals and cells from this cluster were observed to have undergone clonal expansion preferentially in these subjects (Fig. 2, C-E, and fig. S2G).
Fig. 2. Enrichment and clonal expansion of an intestinal plasma B cell cluster in ulcerative colitis.
(A) Phenotypic annotations of B lymphocyte clusters. (B) t-SNE plots of B lymphocyte clusters, colored by cluster identity (upper left); anatomic location from which cells were derived (upper right); health status (lower left; “C” labels indicate healthy controls, “U” labels indicate UC patients); and immunoglobulin heavy chain expression, determined using scBCR-seq data (lower right). (C) Quantitation of selected B lymphocyte clusters enriched or depleted in health vs. disease, expressed as absolute numbers. (D) Comparison of clonotypic expansion exhibited by cells from indicated plasma cell clusters, quantitated separately in healthy individuals vs. UC patients. (E) Absolute number (left) and percentage of cells (right) from each cluster derived from healthy individuals (blue) or UC patients (red). Two-sided Wilcoxon rank sum test (C). * p < 0.05, ** p < 0.01.
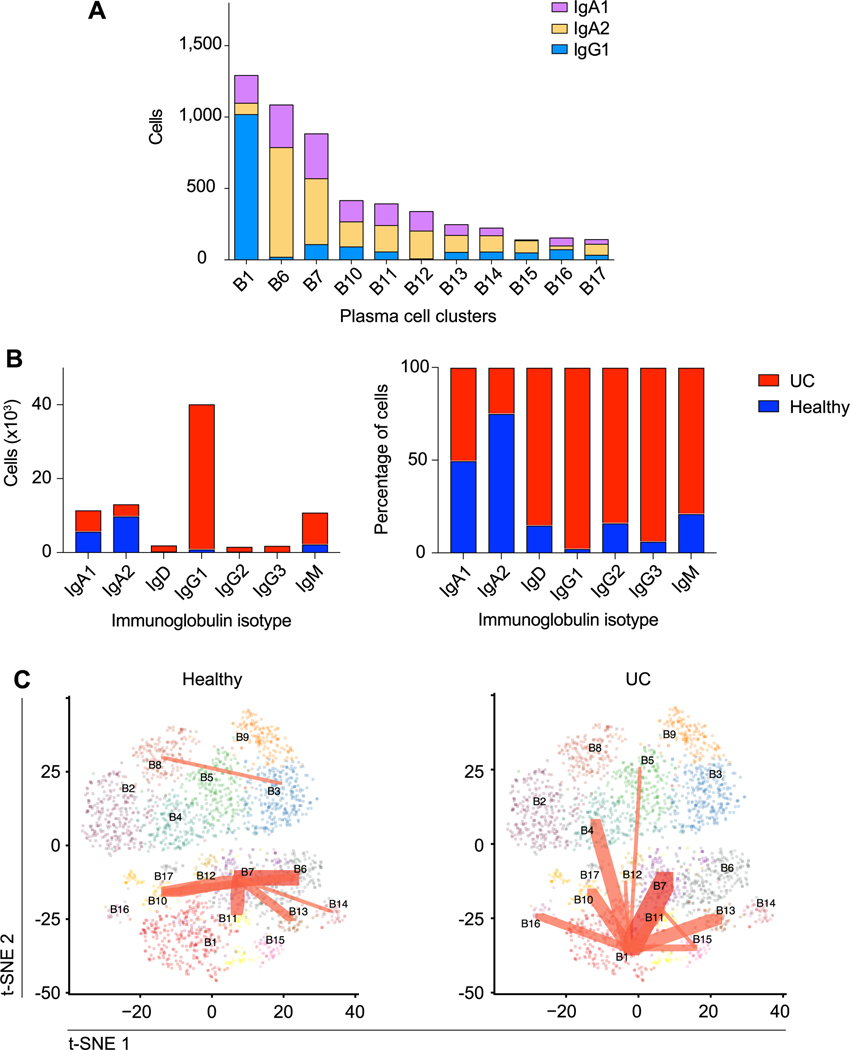
To begin to explore the molecular basis of the heterogeneity among the plasma cell clusters, we performed differential expression analyses among the 11 plasma cell clusters. The UC-enriched B1 cluster exhibited 165 differentially expressed genes compared to all other plasma cell clusters (table S4). Moreover, we observed that cells from the UC-enriched B1 cluster were predominantly IgG1+ (Fig. 3A); additional analyses indicated that the number and proportion of IgG1+ plasma cells were markedly increased in UC patients, whereas the number and proportion of IgA2+ plasma cells were increased in healthy individuals (Fig. 3B). Lastly, we looked for evidence of BCR clonotypes shared among the B cell clusters we annotated. In the healthy setting, clonotypes were shared across multiple plasma cell clusters, but not the B1 plasma cell cluster (Fig. 3C, left panel, fig. S3A). By contrast, in the setting of ulcerative colitis, clonotypes were shared between cells from the B1 plasma cell cluster and most other plasma cell clusters. Taken together, these results suggest the possibility that plasma cells transit between distinct states, the relative proportions of which differ in the setting of health vs. disease.
Fig. 3. Clonal relationships of intestinal plasma B cell clusters in health and ulcerative colitis.
(A) Quantitation of IgA1+, IgA2+, and IgG1+ cells within each plasma cell cluster, determined using scBCR-seq data. (B) Absolute number and percentage of cells from UC patients or healthy individuals are shown for each immunoglobulin isotype (IgA1+, IgA2+, IgD+, IgG1+, IgG2+, and IgM+ only; very few IgG3+, IgG4+, or IgE+ cells were detected). (C) t-SNE plots of plasma cell clusters, colored by cluster identity, with red lines indicating BCR clonotypes shared among clusters and line weight representing number of shared clonotypes, for healthy individuals (left) vs. UC patients (right).
Transcriptionally distinct regulatory T cells in health vs. ulcerative colitis
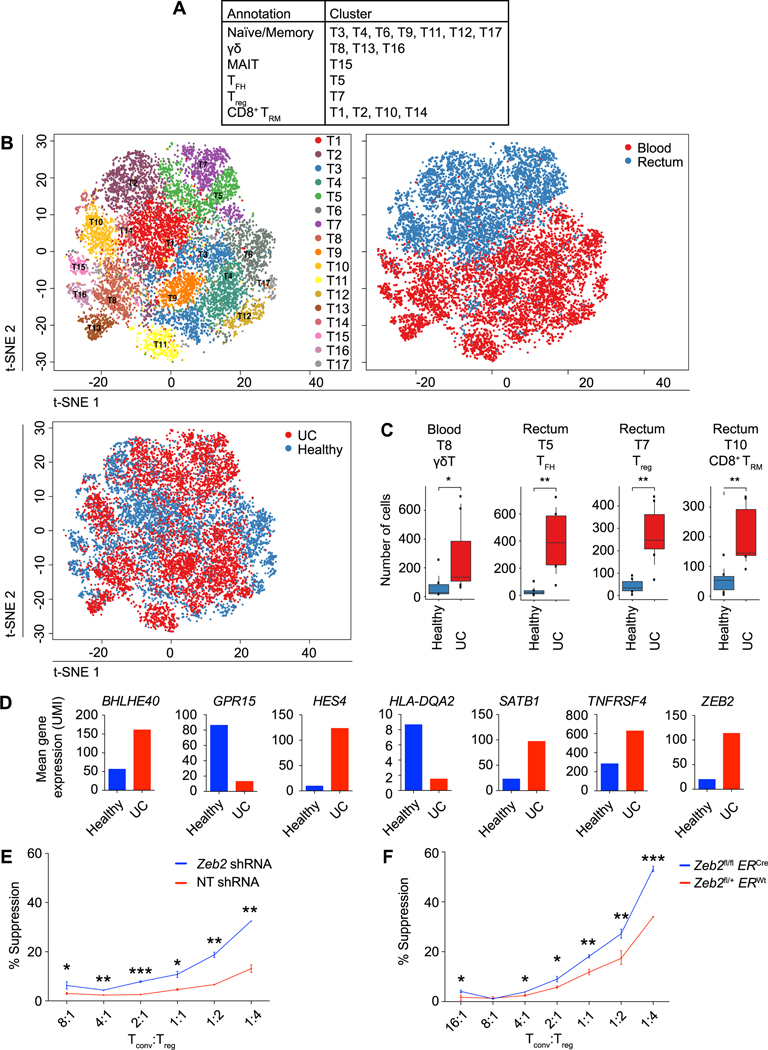
Analogous to our B lymphocyte analyses, additional t-SNE and UMAP analyses were performed on T lymphocytes, yielding 17 sub-clusters, all of which included cells from at least two subjects (Fig. 4, A and B, fig. S4, A to F, and table S3). These T cell clusters were annotated into subsets on the basis of differential expression of canonical genes. Cells from three clusters (T8, T13, and T16) exhibited high expression of genes encoding components of the gamma delta (γδ) T cell receptor, although it should be noted that the T8 cluster also contained TCR alpha beta (αβ) T cells. Seven clusters (T3, T4, T6, T9, T11, T12, and T17) with high expression of SELL (CD62L), CCR7, LEF1, and TCF7 expression were annotated as naïve/memory. Four CD8+ T cell clusters (T1, T2, T10, and T14) expressed transcripts suggestive of a resident memory T (TRM) cell phenotype (23), with high expression of CD69, ITGAE (CD103), CD101, CCR6, and ITGA1 (CD49a) along with low expression of KLF2 and S1PR1. Cells from the T7 cluster expressed transcripts indicative of regulatory T (Treg) cells (26), including high FOXP3 and ILR2RA (CD25) expression along with low IL7R expression. Cells from the T5 cluster expressed transcripts suggestive of T follicular helper (TFH) cells (27), including high CXCR5, PDCD1 (PD1), ICOS, and BCL6 expression. Cells from the T15 cluster expressed transcripts indicative of mucosal-associated invariant T (MAIT) cells (28), including ZBTB16 and TRAV1–2. We next compared the numbers of cells from each of the T lymphocyte clusters derived from healthy individuals and UC patients (Fig. 4C). In the peripheral blood, cluster T8 cells were enriched in UC patients, raising the possibility of previously unrecognized γδ T cell states or subsets that may be more abundant in the setting of disease. In the intestine, the T7 (Treg), T5 (TFH), and T10 (CD8+ TRM) cell clusters were enriched in patients with UC.
Fig. 4. CD4+ Treg cells from UC patients and healthy individuals exhibit distinct transcriptional signatures.
(A) Phenotypic annotations of T lymphocyte clusters. (B) t-SNE plots of T lymphocyte clusters, colored by cluster identity (upper left); anatomic location from which cells were derived (upper right); and health status (lower left; “C” labels indicate healthy controls, “U” labels indicate UC patients). (C) Quantitation of T lymphocyte clusters that were enriched or depleted in health vs. disease, expressed as absolute numbers. (D) Mean expression of selected genes that were differentially expressed between T7 cluster CD4+ Treg cells derived from healthy individuals (blue) vs. UC patients (red); see also table S5. (E) In vitro suppression assay using induced Treg cells transduced with non-targeting shRNA (n=3) or Zeb2 shRNA (n=3) or (F) induced Treg cells from tamoxifen-treated ERWtZeb2fl/+ (n=3) and ERCreZeb2fl/fl (n= 3) mice. Putative Treg cells were sorted on the basis of high CD4 and CD25 expression and these cells expressed high levels of Foxp3; see also fig. S5. Error bars indicate s.e.m. Two-sided Wilcoxon rank sum test (B); unpaired Student’s t-test for each Tconv:Treg ratio (D, E). * p < 0.05, ** p < 0.01, *** p < 0.001.
As Treg cells are known to prevent inflammation and autoimmunity (26), it might have been predicted that Treg cells would be numerically reduced in UC patients. However, we observed an enrichment of the T7 Treg cell cluster in patients with UC (Fig. 4C), consistent with previously published reports (8, 9), suggesting that inflammation was not due to insufficient numbers of Treg cells. Rather, we hypothesized that this finding might reflect an expansion of Treg cells in order to control ongoing inflammation. We further reasoned that Treg cells from UC patients might express transcripts leading to defective Treg cell function that could contribute to disease development in spite of increased numbers of these cells. Indeed, comparison of the gene expression profiles of Treg cells from healthy individuals and UC patients revealed 288 differentially expressed genes (Fig. 4D and table S5), some of which have previously been reported to play a role in Treg cell function, suggesting that the current approach was capable of identifying functionally important genes. For example, expression of SATB1, repression of which is necessary for Treg cell suppressive function (29, 30), was increased in Treg cells from UC patients; conversely, KLF2, MYC, and ITGB1 expression were reduced in Treg cells from UC patients, consistent with previously published work demonstrating a role of these factors in Treg cell function (31–33). However, the majority of the genes differentially expressed between Treg cells from healthy individuals vs. UC patients had not been previously linked to Treg cell function. As a first step towards demonstrating that genes identified by these analyses might indeed represent previously unrecognized regulators of Treg cell function, we selected one such gene, ZEB2, which has been previously shown to promote effector function in murine CD8+ T cells (34, 35), for further studies. The observation that ZEB2 was more highly expressed in Treg cells derived from UC patients raised the possibility that downregulation of ZEB2 may be required for optimal Treg cell function. To test this possibility, we transduced in vitro-induced murine Treg cells with a shRNA construct targeting Zeb2. Treg cells transduced with a Zeb2 shRNA construct exhibited enhanced suppressive function compared to control Treg cells transduced with a nontargeting shRNA construct (Fig. 4E). To confirm these results, we treated ERWtZeb2fl/+ and ERCreZeb2fl/fl mice with tamoxifen to induce Zeb2 deletion, FACS-purified naïve CD4+CD25− T cells, and cultured these cells in Treg cell-inducing cytokine conditions. Control and Zeb2-deficient Treg cells were then FACS-purified and tested in an in vitro suppression assay; indeed, we observed that Zeb2-deficient Treg cells exhibited enhanced suppressive function compared to control Treg cells (Fig. 4F and fig. S5).
Differential enrichment of heterogeneous γδ T cell clusters in health vs. ulcerative colitis
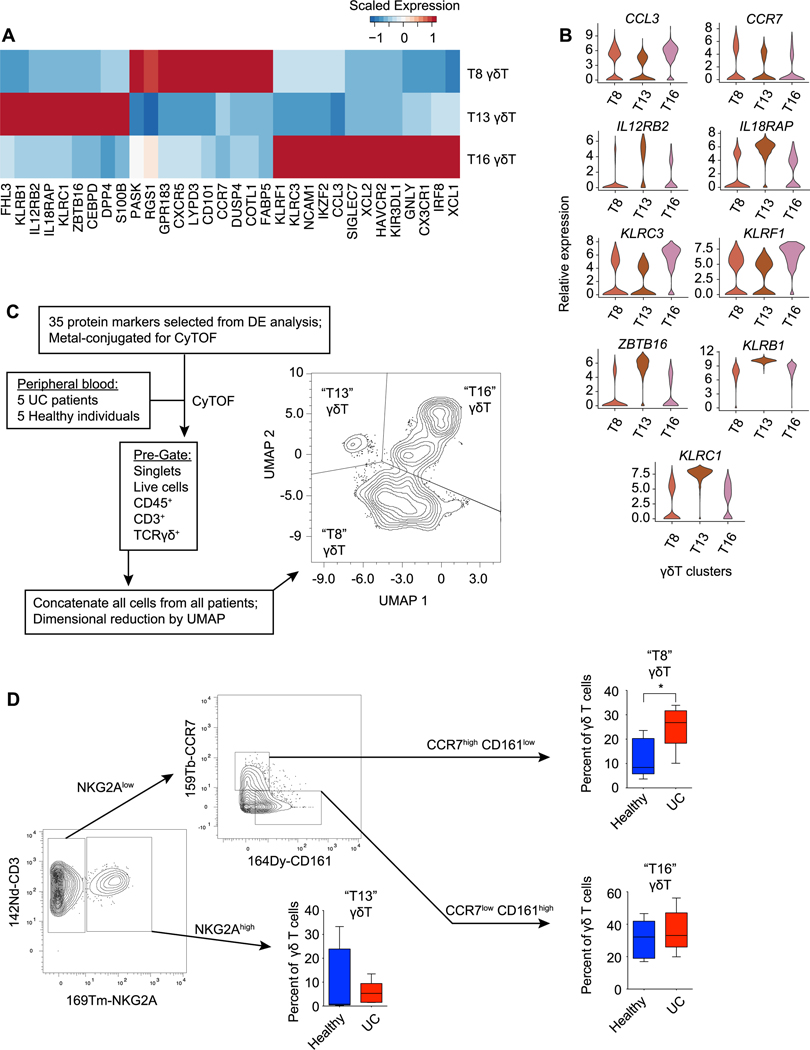
The aforementioned analyses (Fig. 4C) raised the possibility that distinct γδ T cell states or subsets might be differentially enriched in the setting of health vs. ulcerative colitis. To begin to understand the differences among these putative γδ T cell states, we performed differential gene expression analyses of the three γδ T cell clusters (T8, T13, and T16) we defined. These analyses revealed a number of genes that were differentially expressed among the three γδ T cell clusters (Fig. 5, A and B, and table S6). For example, T8 cluster cells expressed high levels of CCR7; T13 cells expressed high levels of KLRB1 (CD161) and S100B; and T16 cells expressed high levels of KLRC1 (NKG2A), GNLY, and XCL1. To confirm whether these γδ T cell states could be discerned in human peripheral blood, we constructed a panel of 35 protein markers based on differentially expressed genes among T8, T13, and T16 cluster cells derived from our scRNA-seq analyses. We then performed mass cytometry using peripheral blood from an independent cohort of healthy individuals and UC patients (fig. S6 and table S7). In addition to unbiased analysis of the dataset using Phenograph (fig. S6), targeted analysis of γδ T cells was performed. γδ T cells were electronically gated on the basis of CD45+CD3+γδTCR+ positivity and then subjected to UMAP analysis, revealing three clusters of γδ T cells (Fig. 5C).
Fig. 5. Differential enrichment of γδ T cell clusters in health vs. ulcerative colitis.
(A) Heatmap of mean expression of selected genes differentially expressed between γδ T cell clusters (T8, T13, and T16). (B) Violin plots of selected genes differentially expressed by γδ T cell clusters (T8, T13, and T16). (C) Overview of the design for mass cytometry (CyTOF) analysis of peripheral blood from healthy individuals (n= 5) and UC patients (n=5), with UMAP analysis of γδ T cells from all patients. (D) Gating strategy to identify CCR7highCD161lowNKG2Alow (“T8”), NKG2Ahigh (“T13”), and CCR7lowCD161high NKG2Alow (“T16”) clusters, with proportion of each cell cluster represented among all γδ T cells for each subject, calculated separately for healthy individuals vs. UC patients. Error bars indicate s.e.m. Unpaired Student’s t-test (D). * p < 0.05.
The UMAP analyses suggested that the combination of 35 protein markers together could distinguish these putative γδ T cell states. We next asked whether these putative γδ T cell states could be discerned using only a few protein markers selected from our differential gene expression analyses. γδ T cells were electronically gated on the basis of CD45+CD3+γδTCR+ positivity and expression of CCR7, CD161, and NKG2A was analyzed. Putative T13 cluster cells could be distinguished from T8 and T16 cluster cells on the basis of high NKG2A expression (Fig. 5D). Putative T8 and T16 cluster cells, both of which expressed low levels of NKG2A, could be distinguished on the basis of CCR7 and CD161 protein expression, with T8 cluster cells tending to express higher levels of CCR7 and T16 cluster cells expressing higher levels of CD161. Lastly, we observed that, as predicted by our scRNA-seq analyses (Fig. 4C), putative T8 cluster cells represented a higher proportion of γδ T cells in the peripheral blood of UC patients compared to healthy controls (Fig. 5D).
Clonal expansion of a UC-associated CD8+ TRM cell cluster with enhanced inflammatory properties
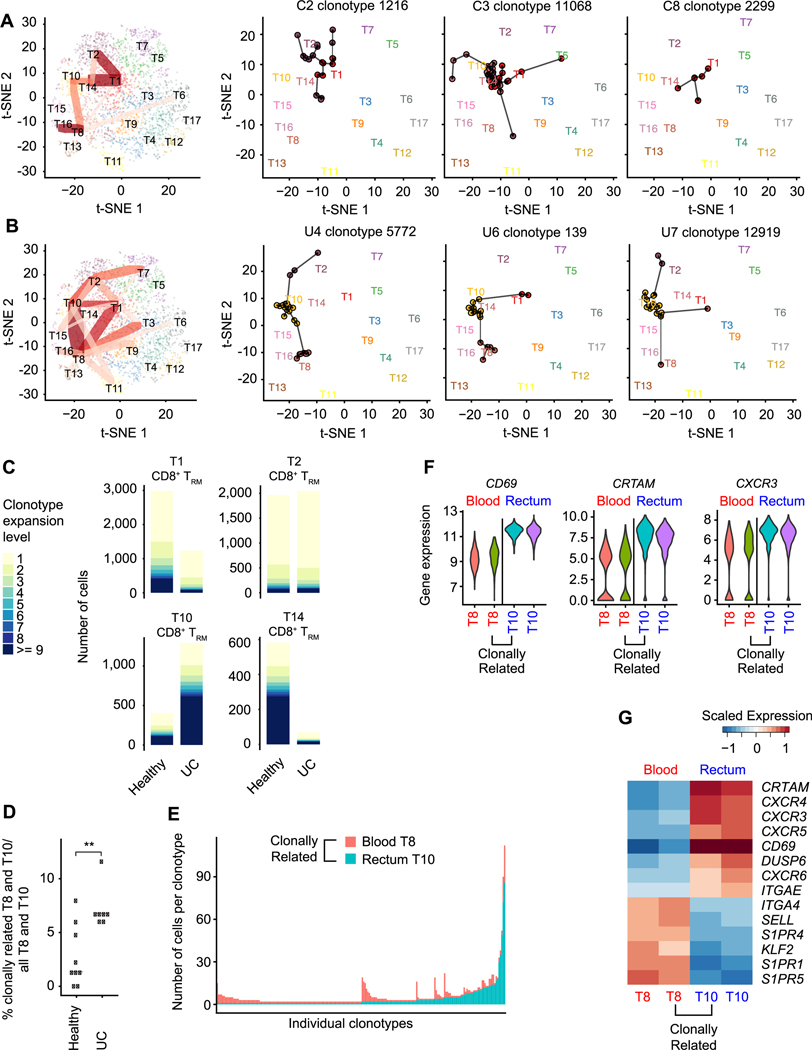
As described above, we annotated four putative CD8+ TRM-like cell clusters (T1, T2, T10, and T14) and hypothesized that these clusters might represent states between which TRM cells transit differentially during health and disease. To investigate this possibility, we looked for evidence of TCR clonotypes shared among the T cell clusters we previously defined. In the healthy setting, clonotypes were shared across cells from the CD8+ TRM T1, T2, and T14 clusters (Fig. 6A and fig. S3B); by contrast, in the setting of ulcerative colitis, clonotypes were shared among the CD8+ TRM T1, T2, and T10 cell clusters (Fig. 6B and fig. S3B). In line with these observations, there was increased clonal expansion of cells from the T1 and T14 clusters preferentially in healthy individuals; conversely, there was increased clonal expansion of cells from the T10 cluster in UC patients (Fig. 6C). Moreover, we observed that in the setting of ulcerative colitis, there was a marked increase in clonotypes shared between the T10 and T8 clusters; because T10 cluster cells were derived predominantly from intestinal tissue whereas T8 cluster cells were almost exclusively derived from peripheral blood (fig. S4D), these results indicated trafficking of clonally related cells between the two compartments. Strikingly, we detected a significantly greater proportion of clonally related T10 and T8 cluster cells in UC patients compared to healthy individuals (Fig. 6D). Moreover, for certain clonotypes, the number of cells in the tissue (T10) was greater than that in the blood (T8), whereas for other clonotypes, the number of cells in the blood was greater than that for tissue (Fig. 6E). Differential expression analyses revealed changes in genes encoding trafficking molecules (23), including decreased expression of CD69 and CRTAM and increased expression of SELL, KLF2, and S1PR1 in blood T8 cells compared to clonally related tissue T10 cells (Fig. 6, F and G). Taken together, these results indicate that in the setting of ulcerative colitis, clonally related CD8+ T cells may modulate their gene expression to enable trafficking between blood and intestinal tissue.
Fig. 6. Clonally expanded cells from a CD8+ TRM cluster enriched in UC patients.
t-SNE plots of T cell clusters of healthy individuals (A) and UC patients (B), colored by cluster identity. For summary plots (left plots), red lines indicate TCR clonotypes shared among clusters and line weight represents number of shared clonotypes; three selected clonotypes from 3 representative healthy individuals (A, right 3 plots) or UC patients (B, right 3 plots) are shown as examples. (C) Comparison of clonotypic expansion exhibited by cells from CD8+ TRM cell clusters (T1, T2, T10, T14) from healthy individuals and UC patients. (D) Quantitation of clonally related T8 and T10 cells in healthy controls and UC patients, represented as proportion of clonally related T8 and T10 cells among all T8 and T10 cells in each individual. (E) Anatomic origin of clonally related blood T8 and rectal T10 cells. Each column represents a single clonotype and is colored based on anatomic location (rectum, blue; peripheral blood, red); numbers of clonally related cells derived from each location are indicated on the y axis. (F, G) Violin plots (F) and heatmap (G) of selected differentially expressed genes in clonally related and clonally unrelated T8 and T10 cells (peripheral blood, red; rectum, blue). Two-sided Wilcoxon rank sum test (D). ** p < 0.01.
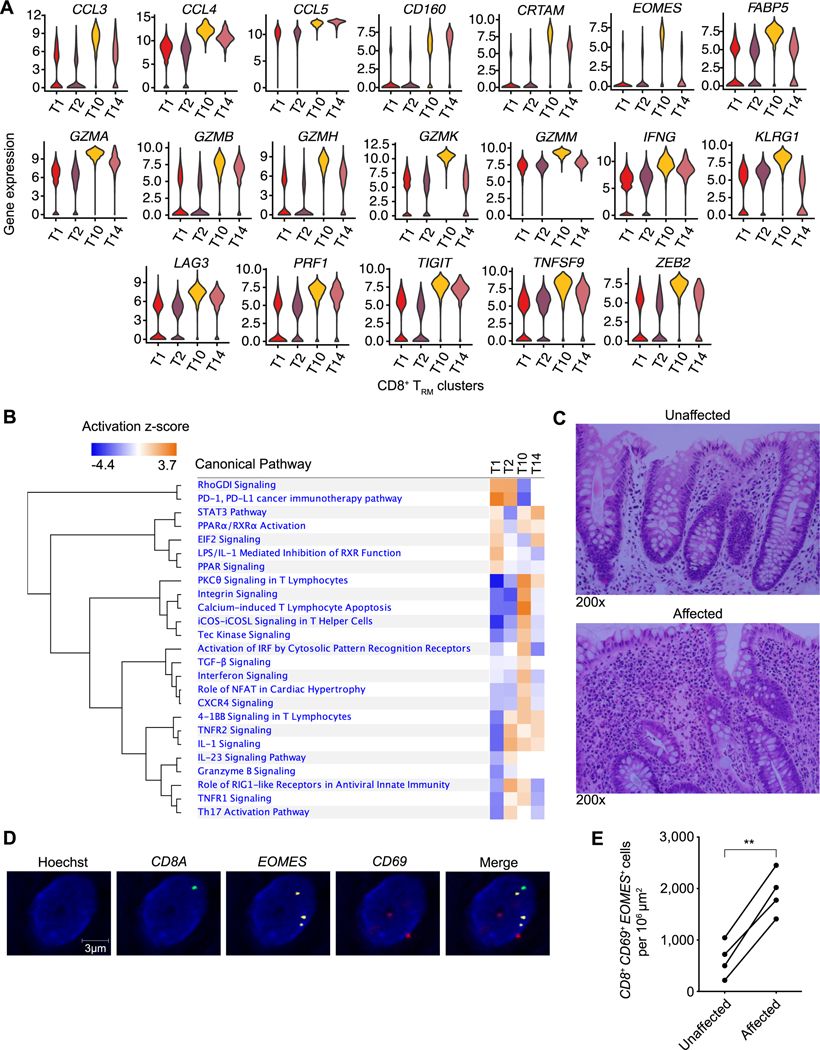
These findings also suggested a spectrum of states between which clonally related CD8+ TRM cells transit differentially in health vs. ulcerative colitis. To begin to understand the differences between these putative CD8+ TRM cell states, we performed differential gene expression and pathway analyses of the four TRM cell clusters (T1, T2, T10, and T14) (Fig. 7, A and B, and table S8). These analyses revealed 481 genes differentially expressed between T10 cluster cells and cells from the other three CD8+ TRM clusters. These included increased expression of transcripts encoding inflammatory molecules and cytolytic granules, such as perforin and Granzymes A, B, H, K, and M; metabolic regulators such as FABP5, which has been shown to support TRM cell differentiation (36); and a number of key transcription factors, including ZEB2 (34, 35) and Eomes (37), both of which have previously been implicated in circulating effector and memory CD8+ T cell differentiation (38). As Eomes has been previously shown to promote its own expression (39), we hypothesized that Eomes might be a critical regulator of the T10 CD8+ TRM transcriptional program.
Fig. 7. Cells from a CD8+ TRM cell cluster with enhanced inflammatory properties are increased in affected colonic tissue from UC patients.
(A) Violin plots of selected genes differentially expressed by the four CD8+ TRM cell clusters (T1, T2, T10, T14). (B) Pathway analysis of genes differentially expressed by the T10 CD8+ TRM cluster compared to all other CD8+ TRM clusters. (C) Representative H&E-stained images of unaffected vs. affected colonic tissue from a UC patient used for RNA in situ hybridization (ISH) analyses shown in D, E. Representative ISH images of affected colonic tissue (D) and quantitation of CD8+CD69+EOMES+ cells from unaffected vs. affected regions of colonic tissue from UC patients (n=5) (E). Paired Student’s t-test (E). ** p < 0.01.
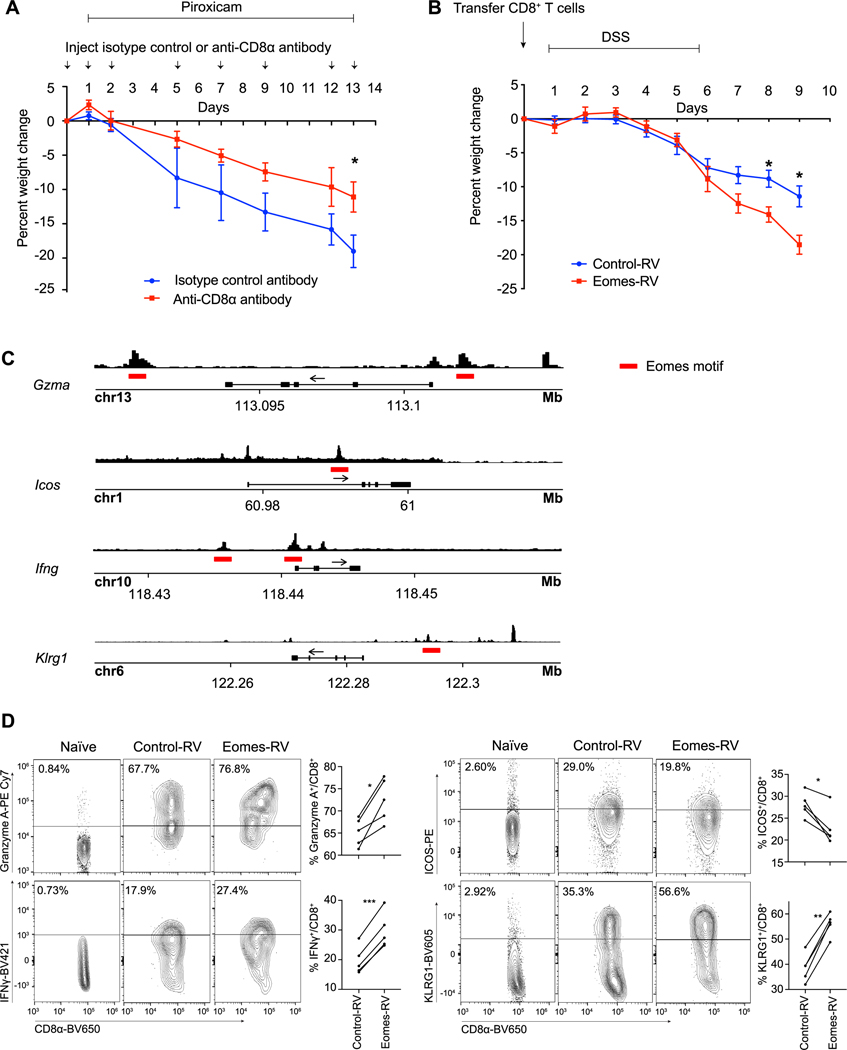
Applying in situ RNA hybridization using EOMES as a marker of T10 CD8+ TRM cells, we first asked whether these cells could be detected in intestinal tissue specimens from an independent cohort of UC patients with active disease. We performed pairwise comparisons between affected vs. unaffected tissue derived from the same UC patients and observed that T10-like cells were increased in affected compared to unaffected tissue in this cohort (Fig. 7, C to E). Next, we investigated whether CD8+ T cells might play a role in mediating intestinal inflammation using an established piroxicam-induced IL-10-deficient mouse model (40). We observed a significant increase in colonic CD8 + T cells in IL-10-deficient mice that were fed piroxicam-containing chow compared to control mice that were fed control chow (fig. S7A). To test whether depletion of CD8+ T cells might ameliorate disease, IL-10-deficient mice fed piroxicam-containing chow were treated with either depleting anti-CD8α or isotype control antibodies and their weight monitored daily over two weeks. Treatment with anti-CD8α antibodies resulted in a substantial depletion of CD8+ T cells in the peripheral blood and colonic tissue (fig. S7B). Moreover, compared to treatment with isotype control antibodies, treatment with anti-CD8α antibodies resulted in a reduction of weight loss and colonic pathology induced by piroxicam (Fig. 8A and fig. S7C), suggesting a role for CD8+ T cells in the piroxicam-induced IL-10-deficient mouse model, consistent with prior reports (40, 41), although it remains possible that CD8α+ dendritic cells and/or CD8αα+ γδ T cells may also play a contributing role in this model. To determine whether ectopic expression of Eomes in CD8+ T cells could influence disease severity in a model of intestinal inflammation, we adoptively transferred CD8+ T cells transduced with control or Eomes retroviral constructs into RAG1-deficient mice prior to challenging them with dextran sulfate sodium (DSS). We observed that mice receiving CD8+ T cells transduced with the Eomes construct lost significantly more weight and exhibited more colonic pathology than mice that received CD8+ T cells transduced with the control construct (Fig. 8B and fig. S7D), suggesting that ectopic Eomes expression is sufficient to confer CD8+ T cells with enhanced pathogenic properties.
Fig. 8. Eomes may regulate the T10 CD8+ TRM cluster transcriptional program.
(A) Percent weight change observed with CD8α depletion (n=5) vs. isotype control (n=5) in a piroxicam-induced IL-10-deficient mouse model, expressed as percent of weight at the start of the experiment. Error bars indicate s.e.m. Data are representative of 2 independent experiments. (B) Percent weight change observed in RAG1-deficient mice receiving 5×105 FACS-sorted, GFP+ control-retrovirus (RV) (n = 13) or Eomes-RV (n= 8) CD8+ T cells and treated with DSS, expressed as percent of weight at the start of the experiment. Error bars indicate s.e.m. Data are representative of 2 independent experiments. (C) ATAC-seq tracks with putative Eomes motifs (indicated with red lines) near accessible promoter regions for selected genes are shown. P14 CD8+ T cells were adoptively transferred into congenic recipients subsequently infected with LCMV-Armstrong; cells were FACS-sorted at days 7 and 30 post-infection (2 technical replicates per time point) and subjected to ATAC-seq. Representative day 7 post-infection ATAC-seq tracks are shown. (D) P14 CD8+ T cells were transduced with control-RV (CD45.1) or Eomes-RV (CD45.1.2) constructs and adoptively transferred into congenic recipients (CD45.1.2) subsequently infected with LCMV-Armstrong (n=5). Expression of selected proteins by control-RV- vs. Eomes-RV-expressing CD8+ T cells was analyzed by FACS at 7 days after infection; staining of naïve CD8+ T cells from an uninfected mouse are shown as a control. Data are representative of 2 independent experiments. Unpaired Student’s t-test (A, B) or paired Student’s t-test (D). * p < 0.05, ** p < 0.01, *** p < 0.001.
We next investigated what gene targets Eomes might be acting upon to mediate these effects. Putative gene targets of Eomes have been previously identified in thymic innate memory (TIM) CD8+ T cells (42) and in vitro-differentiated CD8+ T cells (39) using chromatin immunoprecipitation sequencing approaches, but such approaches are not feasible in intestinal CD8+ TRM cells due to technical challenges with cell numbers. Moreover, the low numbers of intestinal CD8+ T cells that can be recovered in commonly used intestinal inflammation models precluded their use as a model system with which to identify Eomes gene targets in intestinal CD8+ TRM cells. Thus, to identify putative gene targets of Eomes specifically in intestinal CD8+ TRM cells, we applied the Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) in the context of the lymphocytic choriomeningitis virus (LCMV) model system in which intestinal CD8+ TRM cells have been widely studied (43). P14 CD8+CD45.1+ T cells, which have transgenic expression of a T cell receptor that recognizes an immunodominant epitope of LCMV, were adoptively transferred into congenic CD45.2+ wild-type recipient mice subsequently infected with LCMV one day later. Donor CD45.1+ P14 T cells were FACS-sorted from the small intestine epithelial lymphocyte compartment of recipient mice at 7 and 30 days after infection and processed for ATAC-seq. We searched for predicted Eomes binding motifs in accessible enhancer and promoter regions and looked for overlap of these genes with the T10 CD8+ TRM cluster transcriptional signature. These analyses confirmed known Eomes gene targets such as Ifng and Gzma (37), but identified other molecules, such as Klrg1, a killer lectin receptor, and Icos, a costimulatory molecule, as putative Eomes gene targets (Fig. 8C and table S9). To test whether ectopic expression of Eomes resulted in increased expression of putative gene targets we identified by ATAC-seq analysis, we transduced congenically distinct CD8+ T cells with control (CD45.1) or Eomes (CD45.1.2) retroviral constructs prior to adoptive transfer into recipient mice (CD45.2) subsequently infected with LCMV, and used flow cytometry to examine protein expression of several putative targets in intestinal CD8+ T cells at 7 days after infection. Indeed, compared to intestinal CD8+ T cells expressing control constructs, intestinal CD8+ T cells expressing Eomes constructs expressed higher levels of IFNγ, Granzyme A, and KLRG1 protein, and lower levels of ICOS protein (Fig. 8D). Taken together, these findings suggest the possibility that in ulcerative colitis, upregulation of key factors such as Eomes in intestinal CD8+ TRM cells may promote their differentiation into a pathogenic state and confer enhanced inflammatory and cytolytic properties.
Discussion
Recent studies have begun to apply single-cell transcriptomic approaches to investigate the mechanisms underlying the complex dysregulation of the immune system in inflammatory bowel disease (18, 19). Our integrated single-cell transcriptomic and antigen-receptor sequencing analyses have resulted in several insights into the immunobiology of ulcerative colitis. First, we annotated multiple clusters of plasma cells in intestinal tissue and observed that BCR clonotypes were shared among cells from many of these clusters, raising the possibility that plasma cells may transit among a spectrum of states. Plasma cells from UC patients exhibited a marked shift toward a specific IgG1+ cluster (B1), in contrast to plasma cells from healthy individuals, which were predominantly IgA+, in accordance with early immunohistochemical findings first reported in the 1970s (44) and confirmed in subsequent studies (45–47). It has been shown that colitogenic intestinal bacteria can be coated by high levels of IgA (48), suggesting that the ability of healthy individuals to produce IgA in the gut microenvironment may enable them to control specific inflammatory commensals that might otherwise initiate intestinal inflammation. Alternatively or in addition, IgG antibodies may themselves be pathogenic, as it was previously proposed that an increase in anti-commensal IgG antibodies in patients with ulcerative colitis may lead to inflammation through IgG-mediated FcγR receptor activation and type 17 immunity (45).
Second, we envision that the current dataset can be utilized as a starting point to identify genes previously unknown to be dysregulated in IBD in an immune cell type-specific manner for further investigation. As an example, we observed an enrichment of Treg cells (contained within cluster T7) in patients with UC, raising the possibility that these cells might be functionally impaired despite being present in adequate numbers. Further analyses revealed a number of transcripts that were differentially expressed between Treg cells derived from healthy individuals compared to those from patients with UC, many of which were not previously known to have a role in Treg cells. We selected ZEB2 for further study, which, owing to its observed upregulation in Treg cells from UC patients, was hypothesized to impair Treg cell function; indeed, knockdown or deletion of Zeb2 resulted in enhanced murine Treg cell suppressive activity. This finding, together with the observations that the expression of several previously known regulators of murine Treg cells were also altered in Treg cells from UC patients, suggests the potential value of the dataset in selecting putative regulators of healthy vs. disease Treg cell states for further study.
A third insight that derives from these analyses is the finding of heterogeneity among CD8+ TRM cells in the human intestine. TRM cells are a subset of memory T lymphocytes that reside within tissues and provide essential protection at body surfaces (23), but to date there has been only limited evidence for heterogeneity among murine (49, 50) and human TRM cells (51). TRM cells have been implicated in human autoimmune diseases such as vitiligo and psoriasis (52, 53), and recent studies have suggested a role for CD4+ TRM cells (24, 25) in inflammatory bowel diseasee. We detected four transcriptionally distinct clusters of CD8+ TRM cells, one of which (T10) contained cells that had undergone significant clonal expansion predominantly in patients with UC. The finding that TCR clonotypes were shared among cells from the four CD8+ TRM cell clusters supports the hypothesis that these clusters represent states between which CD8+ TRM cells transit; in the setting of ulcerative colitis, we observed a marked shift of cells towards the putative T10 differentiation state. Moreover, the observation that increased numbers of cells in the peripheral blood that were clonally related to T10 CD8+ TRM cells in the intestine were increased in UC is intriguing in light of recent reports that murine and human TRM cells can exit tissue and recirculate (54, 55). Taken together, our data suggest a role for CD8+ TRM cells in ulcerative colitis and raise the possibility that during IBD exacerbations, CD8+ TRM cells may exit intestinal tissue and recirculate, providing a potential explanation for the tendency for IBD to affect multiple organ systems outside of the gastrointestinal tract.
Compared to cells in the other TRM states, cells in the T10 differentiation state expressed higher levels of genes encoding molecules that confer inflammatory and effector properties, such as cytokines, cytolytic granules, and killer lectin receptors. Our analyses nominate the T-box transcription factor Eomes as a regulator of the putative T10 CD8+ TRM cell transcriptional state. Eomes and T-bet are highly homologous members of the T-box family of transcription factors and are highly expressed by activated CD8+ T cells and resting and activated NK cells (37). Eomes and T-bet have cooperative functions (56) in inducing effector functions and enhanced expression of CD122, the receptor controlling IL-15 responsiveness, which underlies proliferative renewal following clearance of microbial pathogen (57, 58). Eomes also has functions that are distinct from those of T-bet, such as promoting self-renewal of long-lived memory cells (59), and has been shown to be upregulated in CD8+ T cells during chronic infection (60). In the context of murine skin TRM cell differentiation in response to microbial infection, both Eomes and T-bet undergo initial upregulation, but are subsequently downregulated in order to enable responsiveness to TGFβ signaling and continued TRM cell differentiation (61). Eomes is extinguished by 2–4 weeks after infection, at least in skin TRM cells, but low levels of T-bet are necessary for the maintenance of CD122 and survival of TRM cells (61–63); it is therefore intriguing that cells in the putative T10 CD8+ TRM transcriptional state expressed high levels of Eomes. Notably, this T10 CD8+ TRM cluster, which exhibited high expression of transcription factors such as EOMES, appears to be transcriptionally distinct from a previously described CD4+CD8+IL17A+ T cell cluster (19), which expressed high levels of RORA and RORC, identified by scRNA-seq; moreover, it remains unknown how the T10 CD8+ TRM cluster described here relates to a CD3+CD4−CD8−IL-17A+ T cell cluster recently identified using mass cytometry (9). Future work will further investigate the degree of heterogeneity among intestinal CD8+ T cells with respect to function and plasticity in health and inflammatory bowel disease.
Our data suggest a model in which TRM cells exist in equilibrium across several differentiation states in the healthy condition. In the setting of ulcerative colitis, TRM cells may upregulate Eomes, which binds to a number of downstream gene targets. Based on our ATAC-seq analyses in murine intestinal CD8+ T cells, putative gene targets may include inflammatory cytokines (Ifng), cytolytic granules (Gzma), chemokines (Ccl3, Ccl4, Ccl5); molecules that promote survival (Bach2, Cd27, Il2rb), killer cell lectin receptors (Klrb1, Klrc1, Klrd1, Klrg1, Krk1); co-stimulatory molecules (Tnfrsf18 (GITR), Tnfrsf4 (OX40R), Icos); and trafficking molecules such as Crtam (64). A caveat of the current study is the use of an infection system, owing to technical limitations with experimental colitis models, with which to identify putative Eomes gene targets in intestinal CD8+ TRM cells. Nonetheless, it appears that upregulation of Eomes in CD8+ TRM cells, through its actions on a diverse set of gene targets, may promote the acquisition of an inflammatory and pathogenic tissue-resident memory T cell transcriptional program.
Overall, our work has resulted in an integrated single-cell transcriptomic and antigen-receptor sequencing dataset that expands the single-cell data available in human inflammatory bowel disease. The study identifies alterations in immune cell types and clonal relationships that occur in the context of disease, including plasma cells, regulatory T cells, γδ T cells, and CD8+ TRM cells, and will enable other investigators to identify additional ulcerative colitis-associated changes in a cell type- and tissue-specific manner for further study. This is likely to accelerate mechanistic and functional investigations into the role of specific genes in relevant immune cell types and states in ulcerative colitis.
Study Design
The purpose of this study was to gain a broader understanding of the heterogeneity and clonal relationships of adaptive immune cells in the context of human ulcerative colitis. To this end, we generated and analyzed single-cell RNA and antigen-receptor sequencing data generated from human peripheral blood and gastrointestinal mucosal tissue samples.
Materials and Methods
Human subjects
The Human Research Protection Programs at the University of California, San Diego and the San Diego VA Healthcare System approved the study. Intestinal biopsies and peripheral blood were obtained from patients undergoing colonoscopy at the University of California, San Diego and the San Diego VA Healthcare System after obtaining informed consent. Healthy individuals were undergoing colonoscopy as part of routine clinical care for colorectal cancer screening/surveillance or non-inflammatory gastrointestinal symptoms that included constipation or rectal bleeding. Inclusion criteria included age over 18 years old and absence of significant comorbidities or colorectal cancer. Ulcerative colitis patients with active endoscopic disease were selected. Details of the study subjects are provided in table S2.
Human peripheral blood mononuclear cell isolation
Blood was collected in BD Vacutainer CPT tube and centrifuged at 400 x g for 25 minutes. The buffy coat layer was removed, washed, and counted. Cells were resuspended in freezing buffer (10%(v/v) Dimethylsulfoxide (DMSO, Sigma Aldrich), 40%(v/v) complete RPMI [RPMI (Corning) +10%(v/v) fetal bovine serum (FBS, Life Technologies) +100 U/mL penicillin/100 μg/mL streptomycin (Life Technologies)], 50%(v/v) FBS), placed into a freezing container (Mr. Frosty), and stored at −80°C. Cells were recovered, washed, and filtered, and utilized for mass cytometry (CyTOF) as described below or labeled with anti-human CD45 (2D1) (Biolegend) for sorting. CD45+ immune cells were sorted on a FACSAria2 (BD Biosciences) utilizing gating strategy shown in fig. S1A.
Human intestinal cell isolation
Four intestinal biopsies were obtained with endoscopic biopsy forceps from the rectum and collected in a conical tube with Hank’s buffered saline solution (HBSS, Corning). Intestinal biopsies were transferred into freezing buffer (10%(v/v) DMSO, 40%(v/v) complete RPMI, 50%(v/v) FBS) and stored at −80°C. Biopsies were recovered, incubated in HBSS on a shaker, then incubated twice in HBSS + 5 mM dithiothreitol (DTT, Thermo Fisher Scientific) with shaking, then washed in HBSS. Intestinal biopsies were mechanically dissociated, then placed into 10 mL of digestion mixture [complete RPMI + 1.5 mg/mL Collagenase type VIII (Sigma-Aldrich) + 50 μg/mL DNase I (Roche)] on a rocker at 37°C for 20 minutes, filtered and stained with anti-human CD45. CD45+ immune cells were sorted on a FACSAria2 utilizing gating strategy shown in fig. S1A.
10x Genomics library preparation and sequencing
Cells were washed and resuspended in phosphate buffered saline (PBS) + 0.04%(w/v) bovine serum albumin per the manufacturer’s guidelines. Single cell libraries were prepared according to the protocol for 10x Genomics for Single Cell V(D)J + 5’ Gene Expression. Approximately 20,000 sorted CD45+ cells were loaded and partitioned into Gel Bead In-Emulsions (GEM). scRNA libraries were sequenced on a HiSeq4000 (Illumina). The B and T cell receptor libraries were amplified according to the manufacturer’s protocol and sequenced on a NovaSeqS4 (Illumina).
Mice
All mice were housed under specific pathogen-free conditions in an American Association of Laboratory Animal Care-approved facility at the University of California San Diego (UCSD), and all procedures were approved by the UCSD Institutional Animal Care and Use Committee. C57BLJ/6 CD45.1, CD45.2, CD45.1.2, P14 TCR transgenic (CD45.1 or CD45.1.2, both maintained on a C57BL6/J background), RAG1-deficient, and IL-10-deficient mice were bred at UCSD or purchased from Jackson Laboratories. Mice with a loxP-flanked Zeb2 allele (35, 65) were bred with Rosa26Cre-ERT2 (ERCre) mice (66) and were maintained on a C57BL/J6 background. Rosa26Cre-ERT2-mediated deletion of the floxed Zeb2 gene was induced by oral gavage of 1 mg tamoxifen (Cayman Chemical Company) emulsified in 100 μl sunflower seed oil (Sigma-Aldrich) for 5 consecutive days, then rested for 5 days. Cells for regulatory T cell suppression assays were obtained from male mice that were 12–28 weeks old.
Supplementary Material
Table S3. Quality control metrics for single-cell sequencing data
Table S4. Differential gene expression analyses of plasma cell clusters
Table S5. Differential gene expression analyses between γδ T cell clusters T8, T13, and T16
Table S6. Differential gene expression analyses between T7 Cluster cells from UC patients vs. healthy individuals
Table S7. Metal conjugated antibodies for mass cytometry
Table S8. Differential gene expression analyses of CD8+ tissue resident memory cell clusters
Table S9. Putative Eomes gene targets in intestinal CD8+ T cells
Table S1. Raw data file
Fig. S1. Overview of experimental design, quality control metrics of single-cell sequencing data, and assessment of overall antigen receptor diversity in healthy individuals and UC patients
Fig. S2. Single-cell RNA-sequencing reveals heterogeneous B lymphocyte clusters
Fig. S3. Analyses of clonal relationships in health and ulcerative colitis
Fig. S4. Single-cell RNA-sequencing reveals heterogeneous T lymphocyte clusters
Fig. S5. Analysis of Zeb2 knockdown or Zeb2-deficient Treg cells
Fig. S6. Mass cytometry analysis of peripheral blood
Fig. S7. CD8+ T cell analysis and histopathology in mouse models of intestinal inflammation
Table S2. Demographics for patient samples
Acknowledgments:
We thank Lindsay Arambula Tsai for assistance with figures. We thank members of the Chang, Goldrath, and Yeo laboratories for technical advice, helpful discussion, and critical reading of the manuscript. Single-cell RNA-, TCR-, and BCR-sequencing using the 10x Genomic platform were performed at the UCSD IGM Genomics Center and supported by NIH grants P30KC063491, P30CA023100, and S10OD026929. Microscopy was performed at the UCSD Microscopy Core and supported by NIH grant NS047101. Mass cytometry was performed at the La Jolla Institute for Immunology Flow Cytometry Core and supported by NIH grant S10OD018499. Current address for N.S.K. is Cancer Immunology Discovery, Pfizer Oncology Research and Development, La Jolla, CA 92121.
Funding: This work was supported by the NIDDK-funded San Diego Digestive Diseases Research Center (P30DK120515) and the CCSG grant (P30CA23100), and funded by grants from the Kenneth Rainin Foundation (W.J.S., G.W.Y., J.T.C.) and the NIH: TR001444 and DK123406 (B.S.B); DK007202 (M.S.T.); AI082850 and AI00880 (Z.H., A.B.); AI123202, AI129973, and BX003424 (J.T.C.); AI132122 (A.W.G., G.W.Y., J.T.C.); and MH107367 (G.W.Y.).
Competing interests: B.S.B. was a consultant for Abbvie, Prometheus Laboratories, and Pfizer in the past 3 years. P.S.D. is on the steering committee for Takeda; is a consultant for Takeda and Janssen; has received honoraria for speaker events from Takeda, travel support from Takeda and Janssen, and grant support from Takeda and Pfizer. G.W.Y. is co-founder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Locana and Eclipse BioInnovations. A.W.G. is on the SAB of Pandion Therapeutics and ArsenalBio. W.J.S reports research grants from Atlantic Healthcare Limited, Amgen, Genentech, Gilead Sciences, Abbvie, Janssen, Takeda, Lilly, Celgene/Receptos, Pfizer, Prometheus Laboratories (now Prometheus Biosciences); consulting fees from Abbvie, Allergan, Amgen, Arena Pharmaceuticals, Avexegen Therapeutics, BeiGene, Boehringer Ingelheim, Celgene, Celltrion, Conatus, Cosmo, Escalier Biosciences, Ferring, Forbion, Genentech, Gilead Sciences, Gossamer Bio, Incyte, Janssen, Kyowa Kirin Pharmaceutical Research, Landos Biopharma, Lilly, Oppilan Pharma, Otsuka, Pfizer, Progenity, Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories), Reistone, Ritter Pharmaceuticals, Robarts Clinical Trials (owned by Health Academic Research Trust, HART), Series Therapeutics, Shire, Sienna Biopharmaceuticals, Sigmoid Biotechnologies, Sterna Biologicals, Sublimity Therapeutics, Takeda, Theravance Biopharma, Tigenix, Tillotts Pharma, UCB Pharma, Ventyx Biosciences, Vimalan Biosciences, Vivelix Pharmaceuticals; and stock or stock options from BeiGene, Escalier Biosciences, Gossamer Bio, Oppilan Pharma, Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories), Progenity, Ritter Pharmaceuticals, Shoreline Biosciences, Ventyx Biosciences, Vimalan Biosciences. Spouse: Iveric Bio - consultant, stock options; Progenity - consultant, stock; Oppilan Pharma - consultant, stock options; Escalier Biosciences – prior employee, stock options; Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories) - employee, stock options; Shoreline Biosciences – stock options; Ventyx Biosciences – stock options; Vimalan Biosciences – stock options.
Footnotes
Data and materials availability: The data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE125527.
References
- 1.Khor B, Gardet A, Xavier RJ, Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham C, Cho JH, Inflammatory bowel disease. N Engl J Med 361, 2066–2078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, Sakuraba A, Kitazume MT, Sugita A, Koganei K, Akagawa KS, Hibi T, Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest 118, 2269–2280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Na YR, Stakenborg M, Seok SH, Matteoli G, Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nature reviews. Gastroenterology & hepatology 16, 531–543 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Bal SM, Golebski K, Spits H, Plasticity of innate lymphoid cell subsets. Nature reviews. Immunology, (2020). [DOI] [PubMed] [Google Scholar]
- 6.Lim AI, Menegatti S, Bustamante J, Le Bourhis L, Allez M, Rogge L, Casanova JL, Yssel H, Di Santo JP, IL-12 drives functional plasticity of human group 2 innate lymphoid cells. The Journal of experimental medicine 213, 569–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou L, Chu C, Teng F, Bessman NJ, Goc J, Santosa EK, Putzel GG, Kabata H, Kelsen JR, Baldassano RN, Shah MA, Sockolow RE, Vivier E, Eberl G, Smith KA, Sonnenberg GF, Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 568, 405–409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maul J, Loddenkemper C, Mundt P, Berg E, Giese T, Stallmach A, Zeitz M, Duchmann R, Peripheral and intestinal regulatory CD4+ CD25(high) T cells in inflammatory bowel disease. Gastroenterology 128, 1868–1878 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Mitsialis V, Wall S, Liu P, Ordovas-Montanes J, Parmet T, Vukovic M, Spencer D, Field M, McCourt C, Toothaker J, Bousvaros A, Center BI, Crohn’s BWH, Colitis C, Shalek AK, Kean L, Horwitz B, Goldsmith J, Tseng G, Snapper SB, Konnikova L, Single-Cell Analyses of Colon and Blood Reveal Distinct Immune Cell Signatures of Ulcerative Colitis and Crohn’s Disease. Gastroenterology, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogino T, Nishimura J, Barman S, Kayama H, Uematsu S, Okuzaki D, Osawa H, Haraguchi N, Uemura M, Hata T, Takemasa I, Mizushima T, Yamamoto H, Takeda K, Doki Y, Mori M, Increased Th17-inducing activity of CD14+ CD163 low myeloid cells in intestinal lamina propria of patients with Crohn’s disease. Gastroenterology 145, 1380–1391 e1381 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Arsenio J, Kakaradov B, Metz PJ, Kim SH, Yeo GW, Chang JT, Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nature immunology 15, 365–372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, Yu B, Wehrens EJ, Lopez J, Kim SH, Zuniga EI, Goldrath AW, Chang JT, Yeo GW, Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing. Nature immunology 18, 422–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahar Halpern K, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Stokar-Avihail A, David E, Amit I, Itzkovitz S, Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR, Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SA, Sim S, Neff NF, Skotheim JM, Wernig M, Quake SR, Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner A, Regev A, Yosef N, Revealing the vectors of cellular identity with single-cell genomics. Nature biotechnology 34, 1145–1160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, Sharma E, Wills Q, Bowden R, Richter FC, Ahern D, Puri KD, Henault J, Gervais F, Koohy H, Simmons A, Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 175, 372–386 e317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang LS, Nayar S, Greenstein AJ, Dubinsky M, Walker L, Leader A, Fine JS, Whitehurst CE, Mbow ML, Kugathasan S, Denson LA, Hyams JS, Friedman JR, Desai PT, Ko HM, Laface I, Akturk G, Schadt EE, Salmon H, Gnjatic S, Rahman AH, Merad M, Cho JH, Kenigsberg E, Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 178, 1493–1508 e1420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, Sud M, Andrews E, Velonias G, Haber AL, Jagadeesh K, Vickovic S, Yao J, Stevens C, Dionne D, Nguyen LT, Villani AC, Hofree M, Creasey EA, Huang H, Rozenblatt-Rosen O, Garber JJ, Khalili H, Desch AN, Daly MJ, Ananthakrishnan AN, Shalek AK, Xavier RJ, Regev A, Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730 e722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, Ashley N, Johnson E, Hublitz P, Bao L, Lukomska J, Andev RS, Bjorklund E, Kessler BM, Fischer R, Goldin R, Koohy H, Simmons A, Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, Konno H, Guo X, Ye Y, Gao S, Wang S, Hu X, Ren X, Shen Z, Ouyang W, Zhang Z, Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Goldstein LD, Chen YJ, Wu J, Chaudhuri S, Hsiao YC, Schneider K, Hoi KH, Lin Z, Guerrero S, Jaiswal BS, Stinson J, Antony A, Pahuja KB, Seshasayee D, Modrusan Z, Hotzel I, Seshagiri S, Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun Biol 2, 304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackay LK, Kallies A, Transcriptional Regulation of Tissue-Resident Lymphocytes. Trends in immunology 38, 94–103 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, This S, Danne C, Campion S, Duncan SH, Owens BMJ, Uhlig HH, McMichael A, Oxford IBDCI, Bergthaler A, Teichmann SA, Keshav S, Powrie F, Circulating and Tissue-Resident CD4(+) T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology 153, 1320–1337 e1316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zundler S, Becker E, Spocinska M, Slawik M, Parga-Vidal L, Stark R, Wiendl M, Atreya R, Rath T, Leppkes M, Hildner K, Lopez-Posadas R, Lukassen S, Ekici AB, Neufert C, Atreya I, van Gisbergen K, Neurath MF, Hobit- and Blimp-1-driven CD4(+) tissue-resident memory T cells control chronic intestinal inflammation. Nature immunology 20, 288–300 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Josefowicz SZ, Lu LF, Rudensky AY, Regulatory T cells: mechanisms of differentiation and function. Annual review of immunology 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crotty S, T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 50, 1132–1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller AN, Corbett AJ, Wubben JM, McCluskey J, Rossjohn J, MAIT cells and MR1-antigen recognition. Curr Opin Immunol 46, 66–74 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Beyer M, Thabet Y, Muller RU, Sadlon T, Classen S, Lahl K, Basu S, Zhou X, Bailey-Bucktrout SL, Krebs W, Schonfeld EA, Bottcher J, Golovina T, Mayer CT, Hofmann A, Sommer D, Debey-Pascher S, Endl E, Limmer A, Hippen KL, Blazar BR, Balderas R, Quast T, Waha A, Mayer G, Famulok M, Knolle PA, Wickenhauser C, Kolanus W, Schermer B, Bluestone JA, Barry SC, Sparwasser T, Riley JL, Schultze JL, Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nature immunology 12, 898–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, Yasuda K, Motooka D, Nakamura S, Kondo M, Taniuchi I, Kohwi-Shigematsu T, Sakaguchi S, Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nature immunology 18, 173–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klann JE, Kim SH, Remedios KA, He Z, Metz PJ, Lopez J, Tysl T, Olvera JG, Ablack JN, Cantor JM, Boland BS, Yeo G, Zheng Y, Lu LF, Bui JD, Ginsberg MH, Petrich BG, Chang JT, Integrin Activation Controls Regulatory T Cell-Mediated Peripheral Tolerance. J Immunol 200, 4012–4023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pabbisetty SK, Rabacal W, Maseda D, Cendron D, Collins PL, Hoek KL, Parekh VV, Aune TM, Sebzda E, KLF2 is a rate-limiting transcription factor that can be targeted to enhance regulatory T-cell production. Proceedings of the National Academy of Sciences of the United States of America 111, 9579–9584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saravia J, Zeng H, Dhungana Y, Bastardo Blanco D, Nguyen TM, Chapman NM, Wang Y, Kanneganti A, Liu S, Raynor JL, Vogel P, Neale G, Carmeliet P, Chi H, Homeostasis and transitional activation of regulatory T cells require c-Myc. Sci Adv 6, eaaw6443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, Kaech SM, The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. The Journal of experimental medicine 212, 2041–2056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, Seuntjens E, Stryjewska A, Zweier C, Roychoudhuri R, Gattinoni L, Bird LM, Higashi Y, Kondoh H, Huylebroeck D, Haigh J, Goldrath AW, Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. The Journal of experimental medicine 212, 2027–2039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE, Divito SJ, Fuhlbrigge R, Puigserver P, Krueger JG, Hotamisligil GS, Clark RA, Kupper TS, Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL, Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302, 1041–1043 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Chang JT, Wherry EJ, Goldrath AW, Molecular regulation of effector and memory T cell differentiation. Nature immunology 15, 1104–1115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, He Y, Hao J, Ni L, Dong C, High Levels of Eomes Promote Exhaustion of Anti-tumor CD8(+) T Cells. Frontiers in immunology 9, 2981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holgersen K, Kvist PH, Markholst H, Hansen AK, Holm TL, Characterisation of enterocolitis in the piroxicam-accelerated interleukin-10 knock out mouse--a model mimicking inflammatory bowel disease. J Crohns Colitis 8, 147–160 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Punit S, Dube PE, Liu CY, Girish N, Washington MK, Polk DB, Tumor Necrosis Factor Receptor 2 Restricts the Pathogenicity of CD8(+) T Cells in Mice With Colitis. Gastroenterology 149, 993–1005 e1002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Istaces N, Splittgerber M, Lima Silva V, Nguyen M, Thomas S, Le A, Achouri Y, Calonne E, Defrance M, Fuks F, Goriely S, Azouz A, EOMES interacts with RUNX3 and BRG1 to promote innate memory cell formation through epigenetic reprogramming. Nat Commun 10, 3306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, Wang W, Pipkin ME, Goldrath AW, Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandtzaeg P, Baklien K, Fausa O, Hoel PS, Immunohistochemical characterization of local immunoglobulin formation in ulcerative colitis. Gastroenterology 66, 1123–1136 (1974). [PubMed] [Google Scholar]
- 45.Castro-Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D, Stewart BJ, Jing C, Strongili K, Labzin LI, Monk EJM, Saeb-Parsy K, Bryant CE, Clare S, Parkes M, Clatworthy MR, Anti-commensal IgG Drives Intestinal Inflammation and Type 17 Immunity in Ulcerative Colitis. Immunity 50, 1099–1114 e1010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macpherson A, Khoo UY, Forgacs I, Philpott-Howard J, Bjarnason I, Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut 38, 365–375 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uo M, Hisamatsu T, Miyoshi J, Kaito D, Yoneno K, Kitazume MT, Mori M, Sugita A, Koganei K, Matsuoka K, Kanai T, Hibi T, Mucosal CXCR4+ IgG plasma cells contribute to the pathogenesis of human ulcerative colitis through FcgammaR-mediated CD14 macrophage activation. Gut 62, 1734–1744 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA, Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurd NS, He Z, Louis TL, Milner JJ, Omilusik KD, Jin W, Tsai MS, Widjaja CE, Kanbar JN, Olvera JG, Tysl T, Quezada LK, Boland BS, Huang WJ, Murre C, Goldrath AW, Yeo GW, Chang JT, Early precursors and molecular determinants of tissue-resident memory CD8(+) T lymphocytes revealed by single-cell RNA sequencing. Sci Immunol 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milner JJ, Toma C, He Z, Kurd NS, Nguyen QP, McDonald B, Quezada L, Widjaja CE, Witherden DA, Crowl JT, Shaw LA, Yeo GW, Chang JT, Omilusik KD, Goldrath AW, Heterogenous Populations of Tissue-Resident CD8(+) T Cells Are Generated in Response to Infection and Malignancy. Immunity 52, 808–824 e807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar BV, Kratchmarov R, Miron M, Carpenter DJ, Senda T, Lerner H, Friedman A, Reiner SL, Farber DL, Functional heterogeneity of human tissue-resident memory T cells based on dye efflux capacities. JCI Insight 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheuk S, Schlums H, Gallais Serezal I, Martini E, Chiang SC, Marquardt N, Gibbs A, Detlofsson E, Introini A, Forkel M, Hoog C, Tjernlund A, Michaelsson J, Folkersen L, Mjosberg J, Blomqvist L, Ehrstrom M, Stahle M, Bryceson YT, Eidsmo L, CD49a Expression Defines Tissue-Resident CD8(+) T Cells Poised for Cytotoxic Function in Human Skin. Immunity 46, 287–300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clark RA, Resident memory T cells in human health and disease. Science translational medicine 7, 269rv261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fonseca R, Beura LK, Quarnstrom CF, Ghoneim HE, Fan Y, Zebley CC, Scott MC, Fares-Frederickson NJ, Wijeyesinghe S, Thompson EA, Borges da Silva H, Vezys V, Youngblood B, Masopust D, Developmental plasticity allows outside-in immune responses by resident memory T cells. Nature immunology 21, 412–421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klicznik MM, Morawski PA, Hollbacher B, Varkhande SR, Motley SJ, Kuri-Cervantes L, Goodwin E, Rosenblum MD, Long SA, Brachtl G, Duhen T, Betts MR, Campbell DJ, Gratz IK, Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL, Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nature immunology 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, Ma A, Ahmed R, Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. The Journal of experimental medicine 195, 1541–1548 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldrath AW, Sivakumar PV, Glaccum M, Kennedy MK, Bevan MJ, Benoist C, Mathis D, Butz EA, Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells. The Journal of experimental medicine 195, 1515–1522 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL, Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 185, 4988–4992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, Wherry EJ, Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, Tscharke DC, Heath WR, Inouye M, Carbone FR, Gebhardt T, The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nature immunology 14, 1294–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, Cauley LS, Craft J, Kaech SM, CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 41, 633–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, Bevan MJ, The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J Immunol 189, 3462–3471 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boles KS, Barchet W, Diacovo T, Cella M, Colonna M, The tumor suppressor TSLC1/NECL-2 triggers NK-cell and CD8+ T-cell responses through the cell-surface receptor CRTAM. Blood 106, 779–786 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Higashi Y, Maruhashi M, Nelles L, Van de Putte T, Verschueren K, Miyoshi T, Yoshimoto A, Kondoh H, Huylebroeck D, Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis 32, 82–84 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM, Differentiation of CD8 memory T cells depends on Foxo1. The Journal of experimental medicine 210, 1189–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen YJ, Chitre AS, Chiang EY, Iftikhar H, O’Gorman WE, Au-Yeung A, Takahashi C, Goldstein LD, Poon C, Keerthivasan S, de Almeida Nagata DE, Du X, Lee HM, Banta KL, Mariathasan S, Das Thakur M, Huseni MA, Ballinger M, Estay I, Caplazi P, Modrusan Z, Delamarre L, Mellman I, Bourgon R, Grogan JL, Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579, 274–278 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Troutaud D, Drouet M, Decourt C, Le Morvan C, Cogne M, Age-related alterations of somatic hypermutation and CDR3 lengths in human Vkappa4-expressing B lymphocytes. Immunology 97, 197–203 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koning D, Costa AI, Hoof I, Miles JJ, Nanlohy NM, Ladell K, Matthews KK, Venturi V, Schellens IM, Borghans JA, Kesmir C, Price DA, van Baarle D, CD8+ TCR repertoire formation is guided primarily by the peptide component of the antigenic complex. J Immunol 190, 931–939 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Risso D, Ngai J, Speed TP, Dudoit S, Normalization of RNA-seq data using factor analysis of control genes or samples. Nature biotechnology 32, 896–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eisenberg E, Levanon EY, Human housekeeping genes, revisited. Trends in genetics : TIG 29, 569–574 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luecken MD, Theis FJ, Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 15, e8746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Risso D, Perraudeau F, Gribkova S, Dudoit S, Vert JP, A general and flexible method for signal extraction from single-cell RNA-seq data. Nat Commun 9, 284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Quality control metrics for single-cell sequencing data
Table S4. Differential gene expression analyses of plasma cell clusters
Table S5. Differential gene expression analyses between γδ T cell clusters T8, T13, and T16
Table S6. Differential gene expression analyses between T7 Cluster cells from UC patients vs. healthy individuals
Table S7. Metal conjugated antibodies for mass cytometry
Table S8. Differential gene expression analyses of CD8+ tissue resident memory cell clusters
Table S9. Putative Eomes gene targets in intestinal CD8+ T cells
Table S1. Raw data file
Fig. S1. Overview of experimental design, quality control metrics of single-cell sequencing data, and assessment of overall antigen receptor diversity in healthy individuals and UC patients
Fig. S2. Single-cell RNA-sequencing reveals heterogeneous B lymphocyte clusters
Fig. S3. Analyses of clonal relationships in health and ulcerative colitis
Fig. S4. Single-cell RNA-sequencing reveals heterogeneous T lymphocyte clusters
Fig. S5. Analysis of Zeb2 knockdown or Zeb2-deficient Treg cells
Fig. S6. Mass cytometry analysis of peripheral blood
Fig. S7. CD8+ T cell analysis and histopathology in mouse models of intestinal inflammation
Table S2. Demographics for patient samples