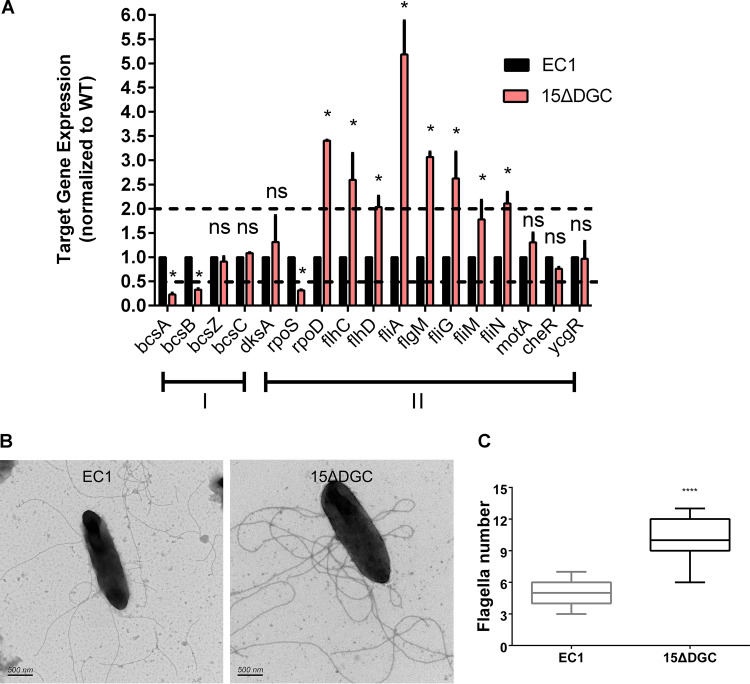
FIG 3.
Quantitative reverse transcription-PCR (qRT-PCR) and transmission electron microscopy (TEM) analyses of wild-type EC1 and mutant 15ΔDGC. (A) Quantification of expression of the genes related to biofilm (I) and flagellar transcription (II) hierarchies by qRT-PCR. The following genes were quantified in this experiment: bcsABZC, exopolysaccharide cellulose genes (34); dksA and rpoS, encoding FlhDC transcriptional inhibition regulators (50, 62); rpoD, encoding a general sigma factor (σ70) that works in cooperation with FlhDC to direct RNA polymerase to transcribe class II genes (63); flhC and flhD, encoding a class I master regulator of the flagellar transcription hierarchy and a flagellar transcriptional activator (36); fliA, encoding a class II RNA polymerase sigma factor for a flagellar operon, which directly transcribes class III genes encoding flagellin, chemotactic signaling (Che), and motor (Mot) proteins (64); flgM, encoding the fliA-specific anti-sigma factor (65); motA, encoding a flagellar energy transduction protein (36); cheR, encoding a chemotaxis protein methyltransferase (66); and ycgR, encoding a flagellar brake protein (40). The dotted lines represent 2-fold and 50% gene expression levels compared to the wild type, which was set to a value of 1. *, P < 0.05; ns, P > 0.05 (by two-way ANOVA with multiple comparisons) (n ≥ 3 independent experiments). (B) Representative TEM of wild-type and 15ΔDGC cells after negative staining with 2% (wt/vol) phosphotungstic acid for 2 min. (C) Numbers of flagella of EC1 and 15ΔDGC. ****, P < 0.0001 (by Student’s unpaired t test).