ABSTRACT
Mycoplasma hyopneumoniae: is the etiological agent of porcine enzootic pneumonia (EP), a disease that impacts the swine industry worldwide. Pathogen-induced damage, as well as the elicited host-response, contribute to disease. Here, we provide an overview of EP epidemiology, control and prevention, and a more in-depth review of M. hyopneumoniae pathogenicity determinants, highlighting some molecular mechanisms of pathogen-host interactions relevant for pathogenesis. Based on recent functional, immunological, and comparative “omics” results, we discuss the roles of many known or putative M. hyopneumoniae virulence factors, along with host molecules involved in EP. Moreover, the known molecular bases of pathogenicity mechanisms, including M. hyopneumoniae adhesion to host respiratory epithelium, protein secretion, cell damage, host microbicidal response and its modulation, and maintenance of M. hyopneumoniae homeostasis during infection are described. Recent findings regarding M. hyopneumoniae pathogenicity determinants also contribute to the development of novel diagnostic tests, vaccines, and treatments for EP.
KEYWORDS: Mycoplasma hyopneumoniae, porcine enzootic pneumonia, pathogenicity, virulence factors, host response
Introduction
Mycoplasma hyopneumoniae is the etiological agent of porcine enzootic pneumonia (EP), a chronic respiratory disease that affects pigs [1]. Infections with M. hyopneumoniae are highly prevalent worldwide, causing major economic losses to the swine industry [2] due to costs of treatment and vaccination, decreased feed conversion rate, and increased mortality resulting from secondary infections [3]. Immunodiagnosis of M. hyopneumoniae infections and vaccination against EP are also challenging due to the still limited repertoire of well-characterized antigens. Some diagnostic and/or vaccinal antigens have been characterized immunologically, i.e., with an assessment of their antigenicity and immunogenicity [4–8], and functionally, with an assessment of their roles in M. hyopneumoniae physiology and pathogen-host interactions [9–15].
In the context of the damage-response framework of microbial pathogenesis [16], EP pathology is mainly determined by the damage caused by interactions between M. hyopneumoniae and the swine host. It is known that M. hyopneumoniae infection leads to epithelial damage of the swine respiratory tract, either directly, considering possible bacterial cytotoxicity [14], or indirectly, by causing a strong and damaging inflammatory response. The damage is usually restricted to the bacterium-caused loss of cilia and cell death, and those caused by the host inflammatory response against the pathogen [2]. The exact mechanisms underlying the immunopathogenesis of M. hyopneumoniae, however, are not clear. It is reasonable to assume that the onset of EP in swine depends on M. hyopneumoniae pathogenicity determinants, classically described as virulence factors, as well as on the triggered host responses, immunological or otherwise. M. hyopneumoniae pathogenicity determinants are considered the bacterial factors that allow the pathogen to override the host defense mechanisms and comprehend molecules that mediate processes such as cell adhesion to the host, response to host environment stress, and immunomodulation. On the other hand, host factors contributing to EP include molecules and processes that mediate innate and adaptive defenses against the pathogen, but also cause damage host tissues, such as lung lesions resulting from exacerbated inflammatory responses.
In the last decade, comparative analyses between pathogenic and nonpathogenic M. hyopneumoniae strains, and between M. hyopneumoniae and the closely related species Mycoplasma flocculare, have considerably improved the knowledge of M. hyopneumoniae pathogenesis. M. hyopneumoniae strains may differ in pathogenicity or virulence, ranging from the nonpathogenic type strain J (ATCC 25,934) to virulent strains isolated from EP outbreaks, such as the Brazilian strain 7448 or the American strain 232 [17–20]. M. flocculare, in turn, is genetically closely related to M. hyopneumoniae but is a commensal species, with its presence in the host being virtually asymptomatic. The “omics” prospective and comparative studies with these strains and species, discussed in a later section, have improved our knowledge on the factors that determine M. hyopneumoniae pathogenicity.
Host EP determinants, on the other hand, have been less assessed, aside from some punctual studies regarding innate and adaptive immunological response and cell death [21–24]. Nevertheless, differences between pathogenic and nonpathogenic strains or closely related species may also depend on the triggered host responses. Recently, comparative studies regarding the interactions between M. hyopneumoniae or M. flocculare and the swine host demonstrated differences in intracellular and secreted protein repertoires of swine cells infected by these mycoplasma species [25,26].
This review includes recent data regarding EP epidemiology, prevention, and control of this disease, and a comprehensive overview of the main M. hyopneumoniae and swine molecular mechanisms and cellular processes underlying the disease. Moreover, it also surveys recent advances toward the identification of both the bacterial and host repertoires of EP determinants, including the most recent efforts involving functional and comparative “omics” approaches to elucidate why some M. hyopneumoniae strains are more virulent than others and why M. flocculare is nonpathogenic despite of sharing most of the virulence factors with M. hyopneumoniae. Interactions between M. hyopneumoniae and swine cells are discussed in the context of their intimate contact during infection, emphasizing molecular/cellular mechanisms related to cell adhesion, biofilm formation, host cell invasion, secretion and signaling, cytotoxicity and apoptosis, immunomodulation and stress response.
Epidemiology, prevention and control of EP
Epidemiology
As far as is known, M. hyopneumoniae is a specific pathogen of domestic pigs (Sus scrofa domesticus) and wild boars (Sus scrofa scrofa), and has a worldwide geographical distribution [27]. Specific data on M. hyopneumoniae prevalence by country are scarce in the literature, as EP does not require mandatory notification in many countries and does not limit commercial trade [2]. An average M. hyopneumoniae prevalence of 30–80% has been reported in domestic pig herds worldwide [28]. In South America, the estimated M. hyopneumoniae prevalence was 48% for pigs in the Mendoza province, Argentina, based on molecular diagnosis [29], and prevalence varying from 52% (based on serology of non-vaccinated animals) to more than 90% (based on molecular diagnosis of slaughtered animals) was reported for Southeastern and Southern Brazil [30,31]. A lower M. hyopneumoniae prevalence, near 10%, was reported for pigs in Africa (Uganda) [32]. For wild boars, the most recent studies carried out in European countries, such as Sweden and Italy, have shown seroprevalences of M. hyopneumoniae of 24.8% and 21.12%, respectively [33,34]. In the last decade, the wild boar population has increased in Europe, which increases the likelihood of its potential contact with domestic pigs, and, thus, the risk of transmission of M. hyopneumoniae and other pathogens [34].
Mycoplasma hyopneumoniae transmission dynamics within swine herds depend on the intensity of the production system used, as recently reviewed by Maes et al. [2]. The first exposure events occur during the lactation period, when piglets are in contact with dams shedding the microorganism [35,36]. At weaning age, colonization with M. hyopneumoniae is important in segregated production systems, since pigs are transferred to clean facilities for the growing and finishing phases [37,38]. Subsequent transmission of M. hyopneumoniae among pen-mates is slow, and a clear understanding of M. hyopneumoniae transmission in the field is still needed to improve infection models used in experimental researches [39–41]. A critical aspect of the epidemiology of M. hyopneumoniae is its long pathogen persistence [42,43], but the factors that determine such persistence are still poorly understood.
An additional complicating factor regarding M. hyopneumoniae epidemiology is the occurrence of distinct strains, with different degrees of virulence, circulating in the field [2]. Therefore, the identification and characterization of the M. hyopneumoniae strains circulating within a herd or geographical region are of utmost importance. Discrimination among M. hyopneumoniae strains usually rely on partial sequencing of the P146 gene [45], multilocus sequence typing (MLST) [46], and multiple-locus variable number tandem repeat analysis (MLVA) [47–49]. More recently, Betlach et al. [49] reviewed published information on M. hyopneumoniae variability in pathogenicity and at the antigenic, proteomic, transcriptomic, and genomic levels, and proposed the variable number of tandem repeats (VNTR)-based common terminology and classification. This VNTR-based system is expected to avoid discrepancies and allow to make inferences across the literature.
Prevention and control
Enzootic pneumonia control and prevention are based on the optimization of management conditions, vaccination and treatment with antibiotics. Factors such as management, biosecurity practices and housing conditions should be optimized within the herd [2]. Decrease of infection levels and/or improvement of the clinical outcome of M. hyopneumoniae infections can be achieved with strategies involving management practices such as all-in/all-out pig flows, medicated and segregated early weaning, and multisite operations [27]. Preventing strategies to avoid the introduction of the pathogen in farms free of M. hyopneumoniae are also important. Garza-Moreno et al. [50] presented different M. hyopneumoniae monitoring strategies of incoming gilts and recipient herds and proposed a farm classification based on their health status. According to clinical signs, lung lesions, and ELISA and PCR results, farms and incoming replacements can then be classified into negative, provisional negative and positive.
Vaccination is today still regarded as the most effective way to control M. hyopneumoniae infections. Gilt replacement acclimation procedures against M. hyopneumoniae in positive farms in Europe and North America showed that vaccination is the main strategy to avoid EP [50]. Anti-M. hyopneumoniae vaccines are used worldwide and consist mainly of inactivated, adjuvanted whole-cell preparations that are administered intramuscularly [27]. Presently, there are at least 26 vaccines approved and commercially available worldwide to prevent M. hyopneumoniae infection [51].
Vaccinated animals present reduced clinical signs and lung lesions, improved performance and reduced number of microorganisms in the respiratory tract [2,38,52,53]. Although vaccination confers overall beneficial effects in most infected herds, the results are often variable [54–56]. These variations in the outcomes of vaccination may be due to many factors, including different infection levels, diversity of the circulating M. hyopneumoniae strains, and unknown aspects of the induced immune responses, along with technical issues, such as improper vaccine storage conditions and administration, and lack of vaccination compliance [44,57]. Furthermore, thus far, commercially available anti-M. hyopneumoniae vaccines have conferred only a limited reduction in the transmission ratios [41,58].
The exact mechanisms of protection needed to avoid M. hyopneumoniae infection are not yet fully understood, but constant efforts are invested in the development of new vaccines that may confer better protection. The latest efforts toward the development of more efficient vaccines against EP have been recently reviewed by Tao et al. [51]. These efforts focus on genetically engineered vaccines and some novel combined vaccines. The most recent genetically engineered vaccines are based on adhesins, such as P97, P95, P46, P42, and P36 delivered as recombinant vectors or recombinant subunits [8,59–62]. However, of a total of 24 genetically engineered vaccines studied over the years, only eleven were tested for their efficacies in pigs.
As M. hyopneumoniae infection can predispose swine to secondary infections, combined vaccines have gained attention, as they prevent multiple diseases at the same time. Combined vaccines developed thus far for EP and other swine respiratory infections consist of a mixture of M. hyopneumoniae bacterin and live attenuated viruses, such as porcine reproductive and respiratory syndrome virus and porcine circovirus 2 (PCV2) [63–65], or genetically engineered antigens from M. hyopneumoniae and other porcine pathogens [66]. There have been studies demonstrating the efficacy of a combined vaccine against M. hyopneumoniae and PCV2 [61,67], and their promising results suggest that bivalent or multivalent vaccines may present advantages over monovalent vaccines.
When EP control by the improvement of management and biosecurity and the implementation of vaccination fails, the clinical disease occurs. Then, the treatment of affected animals with antibiotics is required to maintain animal health and welfare. Treatment of M. hyopneumoniae infections can be accomplished using medication with antibiotics against M. hyopneumoniae and major secondary invading bacteria [68]. Potentially active antibiotics against M. hyopneumoniae include tetracyclines, macrolides, lincosamides, pleuromutilins, amphenicols, aminoglycosides, aminocyclitols and fluoroquinolones [27,69]. M. hyopneumoniae strains with high minimal inhibitory concentration (MIC) values for some antibiotics have been reported, along with a description of resistance mechanisms [70,71]. Therefore, even with the use of antibiotics, improvements in management and/or housing conditions are still necessary to ensure long-lasting effects during and after antimicrobial treatment.
M. hyopneumoniae “omics” studies
The “omics” era started for M. hyopneumoniae in 2004, when the first strains (the pathogenic strains 232 and 7448, and the nonpathogenic type strain J) had their whole genomes sequenced [18,19]. Since then, the genomes of several other strains have been sequenced, and, presently, 21 whole sequenced M. hyopneumoniae genomes are available, 10 already assembled and annotated, and 11 still not fully assembled (https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/190/). Apart that of M. hyopneumoniae J strain, the available sequenced genomes are from American, Brazilian, European, Korean, and Chinese pathogenic strains. These genomes are 0.86–0.96 Mb in size, and in each of them there are 528 to 691 protein-encoding genes. Interestingly, despite their small sizes, up to 30% of their gene contents are still of unknown function [72]. Moreover, 20 to 30% of the M. hyopneumoniae genes code for surface proteins (many of them also of unknown function) [72], therefore pointing out to a complex and still poorly characterized scenario of pathogen-host interactions.
More recently, functional “omics” studies, including transcriptomic, proteomic, and metabolomic, have followed the pioneering descriptive and comparative genomics research. These studies were enriched by data generated for the close relative species M. flocculare and have provided insights into M. hyopneumoniae pathogenicity determinants. M. hyopneumoniae strains and M. flocculare share most (>85%) of the genes that code for known (or predicted as such) pathogenicity determinants, including most adhesins, proteases and antioxidant proteins [19,72]. Moreover, they also share at least 90% of the genes coding for surface proteins. These findings have raised questions regarding the gene products and mechanisms that effectively underlie the differences in pathogenicity or virulence between M. hyopneumoniae strains, and between them and M. flocculare. Therefore, comparative studies between M. hyopneumoniae and M. flocculare have focused on possible differences in gene expression and protein abundances. Transcriptomics, proteomics, secretomics, and metabolomics approaches have been carried out as attempts to correlate differential M. hyopneumoniae transcripts, proteins or metabolites to pathogenicity or virulence [73–76]. In the following sections the main findings of these functional, comparative “omics” studies are integrated with those from several other complementary studies and discussed, to provide an overview of M. hyopneumoniae virulence factors and both bacterial and host EP determinants.
M. hyopneumoniae adhesion to swine epithelial cells
In EP, M. hyopneumoniae adhesion to ciliated epithelial cells of the respiratory tract is the initial event in host colonization. This adhesion process is complex, dynamic, and not fully understood, but it is known to involve several surface-displayed molecules of both the pathogen and the host cells [77,78]. The available information on M. hyopneumoniae and host cell adhesion determinants are discussed in the next sub-sections and are schematically represented in Figure 1.
Figure 1.
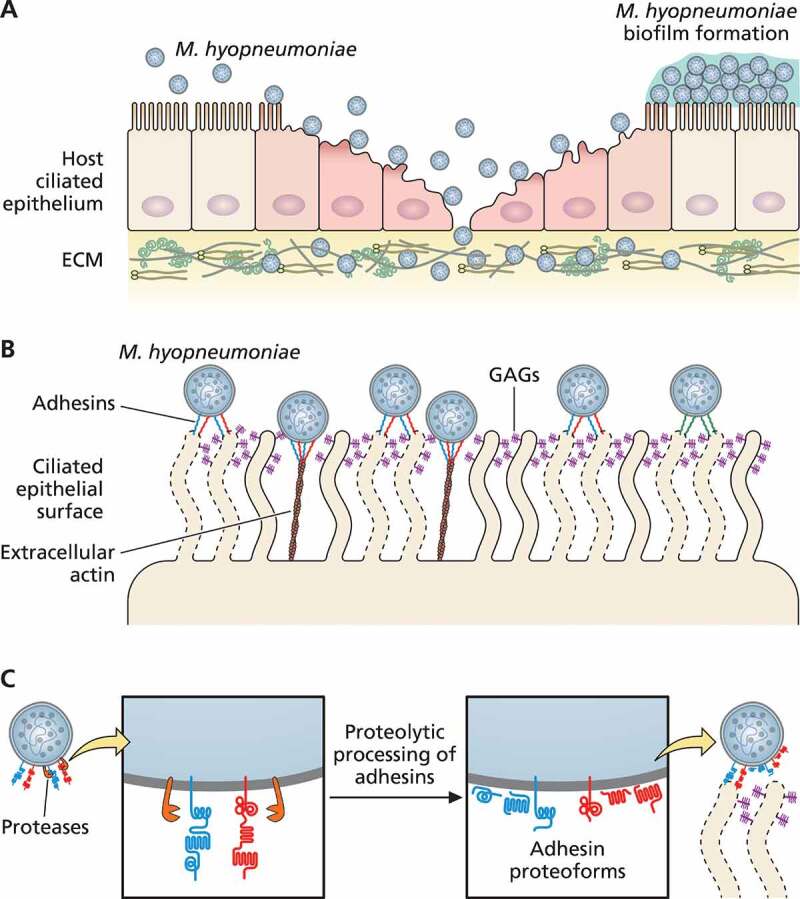
Schematic representation of M. hyopneumoniae adhesion to swine ciliated respiratory epithelium. (a) M. hyopneumoniae cells attach to the cilia of respiratory epithelium, causing ciliostasis, cilium loss, and subsequent epithelial cell death. M. hyopneumoniae cells may form biofilms on the ciliated epithelial surface. M. hyopneumoniae also interacts with molecules from the ECM, such as fibronectin and plasminogen. (b) M. hyopneumoniae adhesion to ciliated cells is mediated by bacterial adhesins that interact with host ligands, as GAGs (displayed on cilia surface), and extracellular actin. (c) Adhesins of M. hyopneumoniae are endoproteolytically processed by surface-displayed proteases, generating a combinatorial library of adhesin proteoforms exposed on the bacterial surface. Dashed lines represent damaged ciliated epithelial cells
M. hyopneumoniae adhesion determinants
The interaction between pathogenic bacteria and host cells during colonization is a critical process for pathogen survival and disease development [79]. Upon inhalation, M. hyopneumoniae must cope with the mucociliary apparatus in the swine respiratory tract, as the so-called lung mucociliary clearance (MCC) is the primary defense mechanism against respiratory pathogens [80]. It is the result of the coordinated interaction of the mucus and a low viscosity periciliary layer with ciliated epithelial cells. The mucus entraps inhaled pathogens and the low viscosity periciliary layer lubricates the airway surface, facilitating the ciliary beating that propels pathogens and particles out of host airways.
Mycoplasma hyopneumoniae attachment to the cilia of the respiratory epithelium allows the pathogen to overcome MCC. It has been long known that the pathogen adhesion causes ciliostasis and subsequent cilium loss and epithelial cell death [81,82]. More recently, in vitro infection assays provided evidence that M. hyopneumoniae infection disrupts the mucociliary function of respiratory epithelial cells by transiently reducing the amounts of mucin 5B secreted in the respiratory epithelium [83]. This reduction causes an uneven distribution of this mucin and might lead to the damage to the epithelial structure, including loss of cilia, observed upon M. hyopneumoniae adhesion to ciliated respiratory cells.
A repertoire of at least 35 M. hyopneumoniae proteins have been previously associated with cell adhesion, including several related to the P97/P102 paralog families and other surface proteins that moonlight as adhesins [78,84,85] (Table 1). However, the number of M. hyopneumoniae adhesins can be higher, considering that its surfaceome includes more than 290 proteins [72,75,86] and that many uncharacterized surface-displayed proteins may bear adhesion properties. Moreover, M. hyopneumoniae pathogenic and nonpathogenic strains, and the nonpathogenic M. flocculare, which differ in pathogenicity and were classically described as having differential adhesion capacities [87], share almost their entire repertoires of known adhesins, according to more recent comparative genomics and proteomics studies [72,75,87], also pointing out to differences beyond the mere sets of known adhesins.
Table 1.
M. hyopneumoniae surface adhesins and adhesion-related proteins and their host ligands
| Protein name |
M. hyopneumoniae strains a |
Host ligands b | Endoproteolytic processing c | References | ||
|---|---|---|---|---|---|---|
| 232 | 7448 | J | ||||
| 46 kDa surface antigen (p46) | mhp511 | MHP7448_0513 | MHJ_0511 | Fibronectin, heparin | Y | [78,94] |
| ABC transporter xylose-binding lipoprotein | mhp623 | MHP7448_0604 | MHJ_0606 | Fibronectin, heparin | Y | [78,94] |
| Acetate kinase | mhp505 | MHP7448_0508 | MHJ_0505 | Heparin | Y | [78] |
| Adenine phosphoribosyltransferase | mhp266 | MHP7448_0114 | MHJ_0110 | Fibronectin, heparin | Y | [78] |
| Adhesin like-protein P146 | mhp684 | MHP7448_0663 | MHJ_0663 | Fibronectin, plasminogen, heparin, porcine epithelial cilia | Y | [78,170] |
| ATP-dependent zinc metalloprotease FtsH | mhp175 | MHP7448_0206 | MHJ_0202 | Fibronectin, heparin | Y | [78] |
| Chaperone protein DnaK (HSP70) | mhp072 | MHP7448_0067 | MHJ_0063 | Fibronectin, heparin | Y | [78,94] |
| Dihydrolipoamide dehydrogenase | mhp504 | MHP7448_0507 | MHJ_0504 | Fibronectin, heparin | Y | [78] |
| Elongation factor Tu (EfTu) | mhp540 | MHP7448_0523 | MHJ_0524 | Fibronectin, heparin | Y | [78,171] |
| Glyceraldehyde 3-phosphate dehydrogenase | mhp036 | MHP7448_0035 | MHJ_0031 | Fibronectin | Y | [78] |
| Hexulose-6-phosphate synthase | mhp441 | MHP7448_0438 | MHJ_0436 | Heparin | Y | [78] |
| L-lactate dehydrogenase (LDH) | mhp245 | MHP7448_0137 | MHJ_0133 | Fibronectin, heparin | Y | [78] |
| Leucyl aminopeptidase | mhp462 | MHP7448_0464 | MHJ_0461 | Plasminogen, heparin | Y | [10] |
| Lipoprotein | mhp390 | MHP7448_0378 | MHJ_0374 | Porcine epithelial cilia | NR | [139] |
| Lppt protein | mhp384 | MHP7448_0372 | MHJ_0368 | Heparin, porcine epithelial cilia | Y | [78,90] |
| M42 glutamyl aminopeptidase | mhp252 | MHP7448_0129 | MHJ_0125 | Plasminogen, heparin | Y | [95] |
| Oligoendopeptidase F | mhp520 | MHP7448_0521 | MHJ_0522 | Heparin | Y | [78] |
| P97-copy 1 | mhp183 | MHP7448_0198 | MHJ_0194 | Fibronectin, plasminogen, heparin, porcine epithelial cilia | Y | [78,91,93,94] |
| P102-copy 1 | mhp182 | MHP7448_0199 | MHJ_0195 | Fibronectin, plasminogen, porcine epithelial cilia | Y | [100] |
| P97-copy 2 | mhp271 | MHP7448_0108 | MHJ_0105 | Fibronectin, heparin, porcine epithelial cilia | Y | [98] |
| P102-copy 2 d | mhp272 | MHP7448_0107 | MHJ_0104 | ND | NR | [18,19] |
| P97-like protein | mhp107 | MHP7448_0272 | MHJ_0264 | Fibronectin, plasminogen, heparin, porcine epithelial cilia | Y | [99] |
| P102-like protein | mhp108 | MHP7448_0271 | MHJ_0263 | Fibronectin, plasminogen, porcine epithelial cilia | Y | [110] |
| Periplasmic sugar-binding protein | mhp145 | MHP7448_0234 | MHJ_0227 | Fibronectin, heparin | Y | [78] |
| Putative MgpA like-protein d | mhp005 | MHP7448_0005 | MHJ_0005 | ND | NR | [19,172] |
| Putative P76 membrane protein (P159) | mhp494 | MHP7448_0497 | MHJ_0494 | Fibronectin, heparin, porcine epithelial cilia | Y | [78,173] |
| Putative P216 surface protein | mhp493 | MHP7448_0496 | MHJ_0493 | Heparin, porcine epithelial cilia | Y | [15,94,102] |
| Putative prolipoprotein P65 | mhp677 | MHP7448_0656 | MHJ_0656 | Fibronectin, heparin | Y | [78] |
| Pyruvate dehydrogenase | mhp264 | MHP7448_0116 | MHJ_0112 | Fibronectin, heparin | Y | [78] |
| Pyruvate dehydrogenase E1-alpha subunit | mhp265 | MHP7448_0115 | MHJ_0111 | Fibronectin, heparin | Y | [78] |
| Uncharacterized protein | mhp009 | MHP7448_0009 | MHJ_0009 | Heparin | Y | [78] |
| Uncharacterized protein | mhp165 | MHP7448_0216 | MHJ_0212 | Heparin | Y | [78] |
| Uncharacterized protein | mhp347 | MHP7448_0335 | MHJ_0326 | Fibronectin, heparin | Y | [78] |
| Uncharacterized protein | mhp385 | MHP7448_0373 | MHJ_0369 | Heparin, porcine epithelial cilia | Y | [78,90] |
| Uncharacterized protein | mhp683 | MHP7448_0662 | MHJ_0662 | Heparin, porcine epithelial cilia | Y | [78,89] |
aNCBI accession numbers corresponding to the genes annotated in the M. hyopneumoniae 232, 7448 and J (RefSeq NC_006360.1, NC_007332.1 and NC_007295.1, respectively).
bHost ligands according to published data available for at least one M. hyopneumoniae strain; ND, not determined.
cEndoproteolytic processing with published experimental evidence available for at least one M. hyopneumoniae strain. Y, yes; NR, not reported.
dP102 copy-2 and putative MgpA-like protein adhesins were predicted as such based on their paralogy/orthology with M. hyopneumoniae P102 copy-1 and Mycoplasma genitalium MgPa adhesins, respectively.
One of the possible explanations for the differences in adhesion capacity between M. hyopneumoniae strains and their nonpathogenic counterpart M. flocculare would be differences in the amount of adhesins presented at the cell surface. Different adhesins may vary in abundance at the cell surface between mycoplasma strains or species, due to differential transcriptional rates of the respective genes, and/or to differential translational rates of the corresponding mRNAs. However, few differences have been found in the expression of ortholog adhesins from M. hyopneumoniae and M. flocculare at transcriptional and proteomic levels [75,76]. Taken together, the available comparative transcriptomic and proteomic results suggest that there are no major overall differences in the repertoires of known adhesins between M. hyopneumoniae pathogenic and nonpathogenic strains and M. flocculare due to gene expression regulation at the transcriptional and translational levels.
Alternative explanations for the observed differences in the adhesion capacity of different M. hyopneumoniae strains and M. flocculare can be found in post-translational events, including their export to the cell membrane and proteolytic processing. Regarding protein export, the most typical M. hyopneumoniae and M. flocculare adhesins, such as members of the P97 and P102 adhesin families, have predicted signal-peptides and are likely exported by the general secretory Sec pathway [74]. Other less typical adhesins, such as some enzymes that moonlight as adhesion proteins at the cell surface, lack a signal-peptide and would be exported by non-classic pathways. However, differential export efficiency apparently is not a major determinant of the differences in the abundance of adhesins in the cell surface, as both M. hyopneumoniae and M. flocculare surfaceomes are similarly enriched with adhesins [75].
Regarding proteolytic processing, it has been described that many adhesins may be targets of endoproteolytic post-translational processing [15,78,88–93] (Table 1). These proteolytic processing events can shape the bacterial surface architecture [78,94], generating several adhesin proteoforms, that may be aimed to different locations and exert alternative functions. At least some of them are displayed at the cell surface, while others may stay in the cytoplasm or be released from the cell membrane to the extracellular milieu. Apart from those adhesin proteoforms with bona fide transmembrane domains, it is not known how processed adhesin proteoforms are anchored in the mycoplasma cell membrane. While some of these proteoforms may interact with and be retained by glycosaminoglycans (GAGs) [15,89,90], others may stay only transiently in the cell surface and then be released as soluble secretion products [74].
A recent comparative analysis of endoproteolytic processing between M. hyopneumoniae and M. flocculare adhesins [94] demonstrated that the five most abundant of them in the M. hyopneumoniae pathogenic strain 7448 are differentially processed compared to their corresponding orthologs in the nonpathogenic M. hyopneumoniae J strain and M. flocculare. Most of the analyzed surface-displayed adhesins from the pathogenic strain were more proteolytically processed (i.e., cleaved at more sites) than the orthologs from the nonpathogenic counterparts, which is consistent with the observed enrichment of several aminopeptidases and endoproteases at the surface of this pathogenic M. hyopneumoniae strain [75]. There is also evidence that adhesins can be differentially proteolytically processed in the cytoplasm, during or after their translation, and/or in the cell membrane, during or after their translocation to the cell surface [94]. Moreover, some adhesin proteoforms generated by proteolytic cleavage were observed in both cytoplasmic and surface compartments, indicating that at least some of the involved proteolytic events occur primarily in the cytoplasm, prior to the translocation of the resulting proteoforms to the cell surface.
The differential adhesin proteoforms generated by proteolytic processing may play differential roles. Those retaining adhesive domains, may retain adhesion properties (i.e., their capacity to bind to host cell or extracellular matrix ligand molecules) and contribute to the overall mycoplasma adhesion capacity. Alternatively, proteoforms devoid of adhesive domains may exert moonlight functions. As many of the generated proteoforms are potentially antigenic [94], and may be presented in the cell surface or even secreted as soluble antigens [74], it is likely that they contribute, at least to some degree, to the mycoplasma strategies of immunomodulation or immunoevasion (discussed in Host immune response and immunomodulation during M. hyopneumoniae infection). Therefore, the mechanism of post-translational proteolytic processing of adhesins (including proteases and their target adhesins) would be a main M. hyopneumoniae pathogenicity determinant, of upmost relevance for EP determination. It would lead to differential presentations of adhesin proteoforms at the cell surface of M. hyopneumoniae strains (and M. flocculare), resulting in, and explaining, at least in part, the observed differential adhesion capacities of these strains and species.
Besides the canonical adhesins and derived proteoforms, other proteins, with different primary function, may moonlight as adhesins. For instance, some typical cytosolic proteins that are also consistently displayed at the cell surface likely exert alternative functions in this ectopic compartment. They may exert moonlight functions by interacting with host components and contributing to host colonization. Among these surface-displayed cytosolic proteins, there are glycolytic enzymes, proteases, chaperones and translation factors that have been characterized as adhesins in different mycoplasma species, including M. hyopneumoniae [10,84,85,95,96]. Interestingly, several of these known moonlighting proteins are overrepresented at the surface of the pathogenic M. hyopneumoniae strain 7448 in comparison to the nonpathogenic J strain and M. flocculare [75]. This overrepresentation also likely contributes to the differences in adhesion capacity observed among these strains and species.
Host cell adhesion determinants
The interaction between M. hyopneumoniae adhesins (or adhesin proteoforms) and host cells also depends on the corresponding host ligands on the cell surface or in the extracellular matrix (ECM). Preliminary studies demonstrated that some swine ciliary glycolipids could act as receptors for M. hyopneumoniae attachment [97], but the bacterial adhesins involved in these pathogen-host interactions remain unknown. In the last decade, it has been demonstrated that adhesive proteoforms from the M. hyopneumoniae P97/P102 adhesin family can bind to GAGs from proteoglycans exposed on the cilia surface, which thereby could act as receptors [89,90]. Indeed, adhesins and adhesin proteoforms of the P97/P102 family display short linear motifs enriched in positively charged amino acids, which promote their binding to anionic molecules, such as GAGs and heparin [78,90,93,98–100]
Apart from the cilia-exposed glycans, some swine ECM molecules, such as fibronectin and plasminogen, also provide binding sites for surface adhesins of M. hyopneumoniae, contributing to host colonization [78,93,98,101,102]. Moreover, interactions between ECM molecules and bacterial surface proteins have been described for several mycoplasma species, including Mycoplasma pneumoniae, Mycoplasma gallisepticum and Mycoplasma bovis, among others [103–107]. Interestingly, the fibronectin-binding ability of M. hyopneumoniae may mediate the adherence to swine respiratory cilia and can provide a mechanism for host cytoskeleton rearrangements, which may facilitate bacterial internalization [108]. Additionally, the plasminogen-binding ability of M. hyopneumoniae may facilitate its traffic via the circulatory system and penetration into host organs, such as liver, kidneys and spleen, from which it has already been isolated [109,110].
More recently, it was also demonstrated that extracellular actin is used as a surface receptor by different proteoforms of M. hyopneumoniae P97 adhesin and another 143 proteins, including lipoproteins, glycolytic enzymes, chaperones and translation factors, among others [77]. Interestingly, anti-actin antibodies inhibit 90% of the ability of M. hyopneumoniae to adhere and colonize PK-15 swine cells, indicating that extracellular actin is an important receptor for M. hyopneumoniae infection. Surface proteins of M. hyopneumoniae also interact with other cytoskeletal proteins besides extracellular actin, such as vimentin, keratin, tubulin, myosin, and tropomyosin [77,111].
M. hyopneumoniae biofilm formation, host cell invasion and systemic trafficking across the porcine respiratory epithelium barrier
Direct contact of M. hyopneumoniae cells with each other and with host cells or ECM molecules may render the pathogen capable of forming biofilms. Biofilm formation is a strategy used by many bacteria, including other mycoplasma species [112–114], to cope with host immune response or with antimicrobial effects, thereby rendering bacteria extremely adaptive and thus contributing to virulence. Recent studies have shown that at least some M. hyopneumoniae strains are indeed capable of forming biofilms, in experimental in vitro conditions, on abiotic surfaces or on host cell monolayers, and within the respiratory tract of experimentally infected swine [115,116]. Biofilm formation makes M. hyopneumoniae more resistant to antibiotics, at least in vitro [116], providing evidence of the importance of biofilm formation for pathogen survival. The molecular interactions and cellular processes underlying M. hyopneumoniae biofilm formation are thus far mostly unknown. However, at least in vitro, biofilm formation involves the generation of a subpopulation of unstable large cell variants that may contribute to the release of extracellular DNA, essential for forming biofilms on abiotic surfaces [115].
Along with biofilm formation capacity, another valuable strategy for mycoplasma survival and pathogenesis is the ability to invade host cells, which is well described for Mycoplasma penetrans and M. pneumoniae, for example [117,118]. M. hyopneumoniae is usually regarded as an extracellular mycoplasma. However, the demonstration that it binds to fibronectin and plasmin at the site of infection [100,110,119,120], an interaction classically associated to host cell invasion by bacteria [121,122], suggested that it could also penetrate porcine cells. Indeed, it was recently demonstrated that M. hyopneumoniae could invade in vitro-infected host-derived epithelial cells [108]. Host cell invasion is mediated by endocytic pathways, which are initiated by interactions between mycoplasma surface proteins and host fibronectin and integrin β1. Remarkably, within porcine cells, at least some bacterial cells can survive phagolysosomal fusion and escape into the cytosol, providing evidence of an alternative intracellular form for M. hyopneumoniae. Moreover, infected host porcine cells could function as a source of pathogen cells for re-infection, as internalized, dormant M. hyopneumoniae could eventually leave to the extracellular environment. In line with that, evidence that M. hyopneumoniae may influence the endosomal trafficking and modulate the maturation of early endosomes was provided by proteomic analyses of swine epithelial cells infected with this pathogen [25]. In such a scenario, infected host cells could also function as a reservoir for the traffic of M. hyopneumoniae within the respiratory tract, thereby contributing to the chronic infection status.
As it is mostly recovered from trachea and lung lesions from infected pigs, M. hyopneumoniae has been considered an exclusive respiratory pathogen [109]. However, at least in experimentally infected pigs, M. hyopneumoniae cells have been re-isolated from inner organs, such as liver, spleen, brain, kidneys, and lymph nodes, although at frequencies lower that of the respiratory tract [109,123,124]. These findings demonstrate that M. hyopneumoniae is able to disseminate to extrapulmonary sites within the swine host, but the mechanisms that allow the systemic trafficking of the bacterium across the respiratory epithelial barrier remain elusive. One possible mechanism mediating this M. hyopneumoniae trafficking might involve its cell invasion capacity, discussed above. As it has been recently demonstrated that M. hyopneumoniae could also, at least in vitro, invade porcine macrophages and avoid phagocytosis [125], it can be speculated that, by invading these and possibly other immune cells, the bacterium could be spread to other organs.
Evidence of another possible mechanism that could mediate M. hyopneumoniae trafficking to alternative host sites was revelead more recently by experiments using an in vitro air-liquid culture system of porcine bronchial epithelial cells [83]. With that, it was shown that M. hyopneumoniae cells could migrate across the epithelial barrier by the paracellular route, but not by the transcellular route. In this in vitro model, M. hyopneumoniae reversibly disrupts the tight junctions between epithelial cells, increasing the permeability and damaging the integrity of the epithelial barrier. In line with that, several studies have demonstrated that M. hyopneumoniae infection facilitates the activation of plasminogen to plasmin, which contributes to the degradation of several ECM and cellular junction components [10,95,100,110,120]. Such disruption of the porcine respiratory epithelial barrier would contribute to the extrapulmonary dissemination of M. hyopneumoniae and to the persistence of infection. Overall, further studies are needed to elucidate how and to what extant M. hyopneumoniae disseminates to different extrapulmonary niches in natural infections.
Protein secretion of M. hyopneumoniae and its impact on pathogen-host interactions
Protein secretion is a vital process for all organisms and has a particular role in the pathogenesis of bacterial infections. Gram-negative and Gram-positive bacteria have at least 6 reported secretion/translocation systems [126]. The general secretory (Sec) pathway comprises an essential, ubiquitous and universal export machinery for most proteins that translocate through (soluble secretome) or integrate into (insoluble secretome) the cell membrane [127]. In mycoplasmas, however, the Sec secretory pathway is apparently incomplete, as some genes coding for components of this pathway were not found in their genomes. In the M. hyopneumoniae genome, only the genes coding for SecY, SecG, SecD/F, YidC and SecA proteins were identified, and the absence of the SecE protein may indicate an incomplete SecYEG transmembrane channel [18,19,72]. However, the secretome analysis of M. hyopneumoniae provided evidences that the Sec secretory pathway is functional, as several detected proteins in the soluble secretome fraction were predicted to be secreted by this secretion pathway [74]. This evidence suggests that other more divergent and still unknown proteins may be part of the Sec pathway, which is consistent with the fact that ~35% of the M. hyopneumoniae genome has thus far no functional annotation.
In contrast, most of the detected proteins in the M. hyopneumoniae soluble secretome were predicted as secreted by Sec-independent secretion pathways [74]. This finding suggests that M. hyopneumoniae may present alternative secretion mechanisms not yet identified due to their lack of conservation with other well-characterized secretion pathways. In line with that, protein secretion in extracellular vesicles was observed in several mycoplasma species [128,129], suggesting that M. hyopneumoniae may alternatively use a vesicle system to secrete proteins.
The soluble secretome of M. hyopneumoniae can be considered a reservoir of virulence factors (Table 2), being composed of several adhesins, lipoproteins, and nucleases [26,74]. The detection of adhesins in the soluble secretome fraction may result from the extensive proteolytic processing of these proteins on the M. hyopneumoniae cell surface (discussed in M. hyopneumoniae adhesion determinants), eventually resulting in the release of some adhesin proteoforms in the extracellular milieu. These adhesin proteoforms may act as extracellular antigens that could trigger the host immunological defenses (discussed in Host immune response and immunomodulation during M. hyopneumoniae infection). The potential relevance of the M. hyopneumoniae secretome for pathogenesis is evident by comparative analyses with M. flocculare, the secretome of which is less complex and presents few orthologs of known M. hyopneumoniae virulence factors.
Table 2.
Putative virulence factors found in the M. hyopneumoniae secretome and/or surfaceome
| Protein name |
M. hyopneumoniae strain a |
Function associated to pathogenicity b | Subcellular localization c | References | ||
|---|---|---|---|---|---|---|
| 232 | 7448 | J | ||||
| 46 kDa surface antigen (p46) | mhp511 | MHP7448_0513 | MHJ_0511 | Adhesion | Se/Su | [26,78] |
| Adhesin like-protein P146 | mhp684 | MHP7448_0663 | MHJ_0663 | Adhesion | Se/Su | [26,170] |
| Aminopeptidase | mhp252 | MHP7448_0129 | MHJ_0125 | Proteolytic processing, immunomodulation, adhesion | Su | [75,95,151] |
| ATP-dependent protease binding protein | mhp278 | MHP7448_0101 | MHJ_0098 | Heat shock protein, proteolytic processing | Su | [75,78] |
| ATP-dependent zinc metalloprotease FtsH | mhp175 | MHP7448_0206 | MHJ_0202 | Proteolytic processing, adhesion | Su | [75,78] |
| Chaperone protein DnaJ | mhp073 | MHP7448_0068 | MHJ_0064 | Chaperone, post-translational processing | Su | [75,169] |
| Elongation factor Tu (EfTu) | mhp540 | MHP7448_0523 | MHJ_0524 | Immunomodulation, adhesion | Su | [75,156] |
| Hemolysin C | mhp663 | MHP7448_0643 | MHJ_0643 | Cytotoxicity | Su | [75,174] |
| Leucyl aminopeptidase | mhp462 | MHP7448_0464 | MHJ_0461 | Proteolytic processing, adhesion | Su | [10,75] |
| Lipoprotein | mhp164 | MHP7448_0217 | MHJ_0213 | Cytotoxicity | Se/Su | [21,26,75,138] |
| Lipoprotein | mhp502 | MHP7448_0505 | MHJ_0502 | Cytotoxicity | Se/Su | [21,26,75,138] |
| Lipoprotein | mhp378 | MHP7448_0367 | MHJ_0363 | Cytotoxicity | Se/Su | [21,74,75,138] |
| Lipoprotein | mhp345 | MHP7448_0333 | MHJ_0324 | Cytotoxicity | Su | [21,75,138] |
| Lipoprotein | mhp377 | MHP7448_0366 | MHJ_0362 | Cytotoxicity | Su | [21,75,138] |
| Lipoprotein | mhp379 | MHP7448_0368 | MHJ_0364 | Cytotoxicity | Su | [21,75,138] |
| L-lactate dehydrogenase (LDH) | mhp245 | MHP7448_0137 | MHJ_0133 | Adhesion | Se/Su | [26,78] |
| Lon protease (ATP-dependent protease La) | mhp541 | MHP7448_0524 | MHJ_0525 | Proteolytic processing | Su | [75] |
| Lppt protein | mhp384 | MHP7448_0372 | MHJ_0368 | Adhesion | Se/Su | [26,74,90] |
| Membrane nuclease, lipoprotein | mhp597 | MHP7448_0580 | MHJ_0581 | Surface nuclease, cytotoxicity, imunomodulation | Se/Su | [11,26,74] |
| Oligoendopeptidase F | mhp520 | MHP7448_0521 | MHJ_0522 | Proteolytic processing, immunomodulation, adhesion | Su | [9,75,151] |
| Outer membrane protein-P95 | mhp280 | MHP7448_0099 | MHJ_0096 | Surface antigen | Se/Su | [169; 26, 75] |
| P102-copy 1 | mhp182 | MHP7448_0199 | MHJ_0195 | Adhesion | Se/Su | [26,74,100] |
| P102-copy 2 d | mhp272 | MHP7448_0107 | MHJ_0104 | Adhesion | Se/Su | [26,74] |
| p37-like ABC transporter substrate-binding lipoprotein | mhp371 | MHP7448_0360 | MHJ_0356 | Cytotoxicity | Su | [21,75,138] |
| P60-like lipoprotein | mhp364 | MHP7448_0353 | MHJ_0348 | Cytotoxicity | Se/Su | [21,74,138] |
| P97-copy 1 | mhp183 | MHP7448_0198 | MHJ_0194 | Adhesion | Se/Su | [26,74,93] |
| P97-copy 2 | mhp271 | MHP7448_0108 | MHJ_0105 | Adhesion | Se/Su | [26,74,98] |
| Phosphopentomutase | mhp221 | MHP7448_0161 | MHJ_0157 | DNA damage response | Su | [175] |
| Protein GrpE (HSP-70 cofactor) | mhp011 | MHP7448_0011 | MHJ_0011 | Chaperone, post-translational processing | Se/Su | [169; 26, 75] |
| Putative lipoprotein | mhp390 | MHP7448_0378 | MHJ_0374 | Cytotoxicity | Se/Su | [21,26,138] |
| Putative lipoprotein | mhp640 | MHP7448_0621 | MHJ_0622 | Cytotoxicity | Su | [21,75,138] |
| Putative MgpA like-protein d | mhp005 | MHP7448_0005 | MHJ_0005 | Adhesion | Se/Su | [74] |
| Putative P216 surface protein | mhp493 | MHP7448_0496 | MHJ_0493 | Adhesion | Se/Su | [15,26,102] |
| Putative P76 membrane protein (P159) | mhp494 | MHP7448_0497 | MHJ_0494 | Adhesion | Se/Su | [26,173] |
| Putative prolipoprotein P65 | mhp677 | MHP7448_0656 | MHJ_0656 | Adhesion | Se/Su | [26,78] |
| Signal-peptidase I | mhp028 | MHP7448_0026 | MHJ_0022 | Proteolytic processing, citotoxicity | Su | [13,14] |
| Thiol peroxidase | mhp283 | MHP7448_0096 | MHJ_0093 | Antioxidant protection | Se | [12,26] |
| Thioredoxin | mhp396 | MHP7448_0384 | MHJ_0380 | Antioxidant protection | Se | [26] |
| Trigger factor | mhp233 | MHP7448_0149 | MHJ_0145 | Chaperone, post-translational processing | Se/Su | [169; 26, 75] |
| XAA-PRO aminopeptidase | mhp680 | MHP7448_0659 | MHJ_0659 | Proteolytic processing, immunomodulation | Se | [9,75] |
aNCBI accession numbers corresponding to the genes annotated in the M. hyopneumoniae 232, 7448 and J sequenced genomes (RefSeq NC_006360.1, NC_007332.1 and NC_007295.1, respectively).
bFunction(s) associated to pathogenicity predicted in silico or according to functional data available for at least one M. hyopneumoniae strain.
cSubcellular localization according to published proteomic data available for at least one M. hyopneumoniae strain. Se, secreted; Su, surface-displayed.
In addition to the differences in the virulence factor content, the overall protein content and abundance in M. hyopneumoniae and M. flocculare secretomes are differential. Both species possess the genes coding for the Sec secretory pathway components, although the partial Sec secretory machinery may be supplemented by unknown proteins not shared by these mycoplasma species, leading to differential secretion efficiencies. Moreover, M. hyopneumoniae and M. flocculare may use differential secretion signals and secretion pathways, which resulted in the observed differences in secretome contents. More studies are needed to elucidate the secretion mechanisms used by these and other mycoplasma species.
The insoluble secretome (or surfaceome) of M. hyopneumoniae also acts as a reservoir of virulence factors (Table 2), which strongly contribute to EP establishment. Proteomic studies focused on surface proteins have demonstrated that this cell fraction is enriched with adhesion proteins (discussed in section 4.1), lipoproteins, and proteases, among others [75,86]. The roles of these proteins in immunomodulation and cell damage are discussed in the following sections.
Host immune response and immunomodulation during M. hyopneumoniae infection
Upon M. hyopneumoniae adherence in the swine respiratory tract (discussed in section 4.1), the host immune response starts to adjust in order to cope with infection. Histopathological changes during M. hyopneumoniae infection are frequently observed in the host respiratory tract [130]. They are characterized by a prominent accumulation of mononuclear cells and infiltration of lymphocytes, plasma cells, and neutrophils in the alveolar lumina and septa. Infected lungs present bronchoalveolar exudate, enlargement of alveolar septa and lymphoreticular hyperplasia of the bronchus-associated lymphoid tissue (BALT). Moreover, several studies have demonstrated that the accumulation of immune cells increases the production and secretion of different pro-inflammatory cytokines, including interleukin (IL)-1β, tumor necrosis factor (TNF)-α, IL-6, IL-8, and IL-18 [120,131,132]. The resultant peribronchiolar and perivascular infiltration of mononuclear leukocytes, along with the increase of cytokine production, are, therefore, associated with immunopathological EP lesions. Altogether, these observations support the idea that the host immune cell response against M. hyopneumoniae can be considered one of the main causes of the lung lesions observed in EP (Figure 2).
Figure 2.
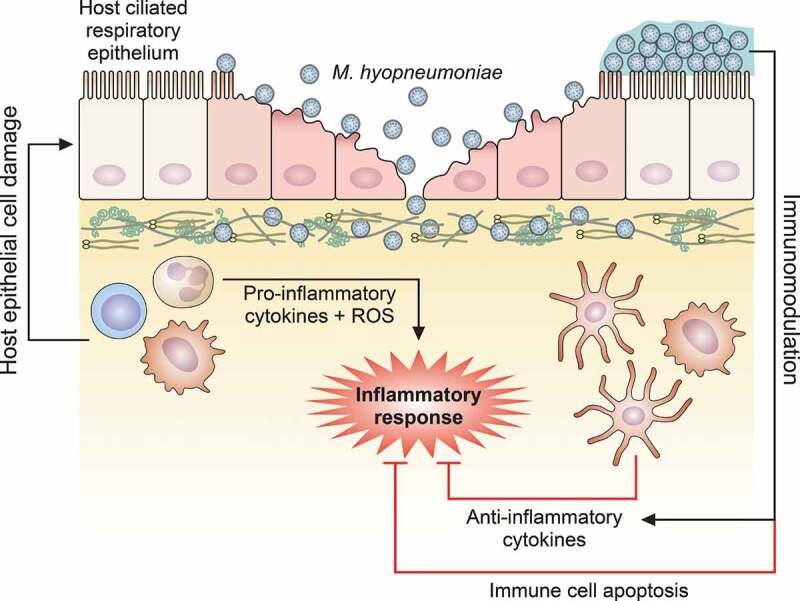
Schematic representation of host immune response modulation mediated by M. hyopneumoniae. M. hyopneumoniae infection elicits an acute inflammatory response in swine lungs, represented by a prominent infiltration and accumulation of immune cells that secrete pro-inflammatory cytokines and release ROS, triggering a microbicidal response. However, this pro-inflammatory microbicidal response causes damage to the host respiratory epithelium. Moreover, M. hyopneumoniae cells can persist in the respiratory tract modulating the host immune response, eliciting the secretion of anti-inflammatory cytokines by dendritic cells and macrophages, and inducing apoptosis on immune cells accumulated in the respiratory tract. The representation of M. hyopneumoniae cells attached to the ciliated respiratory epithelium is described in Figure 1
Despite of the acute host inflammatory immune response elicited by M. hyopneumoniae infection, EP is considered a chronic disease. The persistence of this pathogen in infected pigs is associated with its capacity to modulate and/or evade of host defenses (Figure 2). This is supported by comparative studies on the porcine immune responses elicited by M. hyopneumoniae, M. flocculare, and M. hyorhinis (which also colonizes the porcine respiratory tract) revealing that M. flocculare and M. hyorhinis induce the secretion of higher levels of TNF-α and IL-12, respectively, than M. hyopneumoniae [131]. Moreover, M. hyopneumoniae infection also induces the expression of high levels of IL-10 but decreases the levels of IL-12 and interferon-γ (IFN-γ) expression in dendritic cells [133]. In line with that, it was demonstrated that a surface protein from M. hyopneumoniae elicited the secretion of high levels of IL-10 in murine splenocytes [134]. These findings suggest that M. hyopneumoniae infection modulates the host defenses to favor the Th2 immune response, which was also reported in dendritic cells against mycoplasma infection [133], promoting the development of a chronic disease. On the other hand, one study reported a specific systemic humoral immune response found to be predominately involving the IgG2 subclass, suggesting a dominant Th1-mediated immune response to M. hyopneumoniae [135]. In fact, there are several inconsistencies regarding the type of predominant immune response induced by M. hyopneumoniae [133,135–137]. Such inconsistencies observed in the published literature suggest that M. hyopneumoniae can induce mixed Th1/Th2 responses, but more studies are necessary to confirm this dual response and to understand how it is elicited.
The functional modulation of porcine antigen-presenting cells, such as dendritic cells, was also reported in M. hyopneumoniae infections [133]. It was demonstrated that M. hyopneumoniae down-regulates the expression of the CD1a protein in porcine dendritic cells. Since this protein is responsible for displaying antigenic lipids to T cell receptors, its down-regulation by the pathogen (by a still unknown mechanism), decreases the host dendritic cell capacity for antigen presentation. Moreover, M. hyopneumoniae also reduces the overall populations of dendritic and T cells in the porcine nasal cavity in long-term infections, weakening the immune function in the upper respiratory tract [133].
The ability of M. hyopneumoniae to reduce the populations of immune cells may be associated with the cytotoxic potential of some bacterial proteins displayed at its surface (Figure 2). M. hyopneumoniae cytotoxicity has been considered a virulence mechanism, as the bacterial infection induces apoptosis of immune cells, weakening the immune system, and apoptosis of respiratory epithelial cells, resulting in physical damage to the host tissue. Lipid-associated membrane proteins (LAMPs) from M. hyopneumoniae are known to be crucial for M. hyopneumoniae cytotoxicity to host cells, as they induce cell death by apoptosis or necrosis and modulate the inflammatory response [21,22,138]. It was demonstrated that these LAMPs induce apoptosis in porcine peripheral blood mononuclear cells, alveolar macrophages and lung epithelial cells in vitro by increasing the levels of nitric oxide (NO) and reactive oxygen species (ROS) and by caspase-3 activation. Besides LAMPs, the M. hyopneumoniae P68 surface lipoprotein was also identified as an inflammatory and pro-apoptotic mediator to swine immune cells [139]. In this context, apoptosis induction in host immune cells may elicit an immunosuppressive effect, which likely plays an important role in immunomodulation and evasion.
Nitric oxide and ROS are important players in host immune response, contributing to inflammation and microbicidal response. NO is used as a signaling molecule involved in the regulation of vascular hemodynamics and mediates interaction and recruitment of immune cells during infection [140–142]. ROS are key toxic metabolites to kill bacterial pathogens [143]. These molecules are produced intracellularly by phagocytic cells, such as neutrophils and macrophages, eliminating phagocytized bacteria in a process called respiratory burst response [144]. However, bacterial pathogens can subvert these microbicidal responses and induce apoptosis in host cells by increasing the host production and release of NO and ROS [142,145]. Indeed, NO and ROS are also crucial signal molecules for host cell apoptosis, being implicated in the apoptotic cascade and in the activation of initiator and effector caspases [146]. Porcine cells treated with M. hyopneumoniae LAMPs presented high levels of NO and superoxide anion radicals, which form peroxynitrite, and lead to oxidative stress and activation of the apoptotic cascade [21,22,138]. This cytopathogenic mechanism was also observed in other mycoplasma species, including M. hyorhinis, Mycoplasma synoviae, and M. pneumoniae [147–149].
The success of host colonization by several mycoplasma species depends on their ability to rapidly alter the antigenic repertoire of their surface, using different genetic systems, such as those of phase- or antigenic variation caused by DNA slippage [150]. However, for M. hyopneumoniae, no evidence of such a mechanism has been found. Instead, the differential proteolytic processing resulting in the presence of differential proteoforms on the M. hyopneumoniae surface (discussed in M. hyopneumoniae adhesion determinants) may contribute to antigenic variation, which can be associated with immune response modulation and/or evasion. In this sense, M. hyopneumoniae surface-associated proteases would be indirectly involved in the modulation of the host immune response, by generating proteoforms of adhesins (and possibly of other surface proteins) presenting different epitope sets.
Furthermore, M. hyopneumoniae surface proteases have also been associated with immunomodulation through the proteolytic degradation of pro-inflammatory peptides, such as bradykinin, kininogen, substance P, neurokinin A, and neuropeptide Y [9,151]. It is known that bradykinin and kininogen are involved in the host innate immune response, being associated with bronchoconstriction, MCC and cough induction [152]. Moreover, substance P, neurokinin A and neuropeptide Y are known as inducers of the inflammatory response, eliciting the secretion of high levels of pro-inflammatory cytokines and chemokines [153,154]. Therefore, the proteolytic degradation of these peptides can be considered a virulence mechanism associated with ciliostasis, impairing MCC, and modulation of host immune response.
Mycoplasma hyopneumoniae also displays the ability to evade cellular immune responses. Recently, it was demonstrated that M. hyopneumoniae evades the phagocytic uptake by porcine alveolar macrophages in vitro, although the mechanism underlying this resistance to phagocytosis is still unclear [125]. The presence of convalescent sera, as a source of specific antibodies and complement components for opsonization, did not improve M. hyopneumoniae phagocytosis by cell. This result indicates that, although M. hyopneumoniae can induce the production of antibodies in the host [155], they are not protective. Another mechanism of host immune evasion by M. hyopneumoniae may be the inhibition of the complement pathway. Indeed, it has recently shown that several M. hyopneumoniae surface-displayed proteins bind complement factor H, namely elongation factor thermo unstable (EF-Tu), P146, pyruvate dehydrogenase (acetyl-transferring) E1 component subunit alpha (PdhA), P46, pyruvate dehydrogenase E1 component subunit beta (PdhB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and three different hypothetical proteins [156]. Factor H binding to the M. hyopneumoniae cell surface can confer to the bacterium the ability to inactivate the C3b eventually deposited on the M. hyopneumoniae surface, thereby avoiding the consequences of complement activation [157]. The binding of factor H by EF-Tu also contributed to decrease C3 deposition on the M. hyopneumoniae surface and increased M. hyopneumoniae adhesion to epithelial cells [156]. By counteracting complement activation effects on M. hyopneumoniae, these mechanisms contribute to immune evasion and allow more efficient colonization of the respiratory epithelium by the pathogen.
Moreover, it has been shown that M. hyopneumoniae is also able to evade neutrophil/macrophage extracellular traps (NETs or METs, respectively), at least in vitro [11,158]. In line with that, it has also been demonstrated that the surface nuclease MnuA is responsible for the degradation of NETs, allowing M. hyopneumoniae to escape the host immune defense [11].
Overall, despite fostering a strong immune response, M. hyopneumoniae can persist for several months in the swine host [42,43]. In this context, the ability of M. hyopneumoniae to modulate the host immune responses is an important feature that impacts virulence and disease progression. The entire set of immunomodulation mechanisms used by M. hyopneumoniae, however, remains elusive and is worthy of future investigation.
Cell damage induced by M. hyopneumoniae infection
The mechanisms underlying M. hyopneumoniae pathogenesis described thus far are bacterium-induced loss of cilia and cell death, which can also be correlated to the damage caused by the intense host microbicidal response against this pathogen [2]. On the other hand, there were additional studies that described the presentation of cytotoxic proteins at the bacterial cell surface and the release of toxic bacterial metabolites as a M. hyopneumoniae virulence mechanism causing damage in swine tissues as will be discussed below.
As discussed in Host immune response and immunomodulation during M. hyopneumoniae infection, the cytotoxicity of proteins displayed at the M. hyopneumoniae cell surface has been associated with immunosuppression because, at least in some cases, these proteins induce apoptosis in host immune cells. In addition, they have also been associated with epithelial cell death, causing physical damage to the swine respiratory epithelium (Figure 3), which may contribute to the lung lesions observed in swine with EP. For instance, M. hyopneumoniae LAMPs induce apoptosis in porcine lung epithelial cell line by activating caspase 3, caspase 8, cytochrome C, Bax, and p38 MAPK pathways [22]. Besides LAMPs, the MnuA nuclease and a putative type I signal peptidase (SPase I) from M. hyopneumoniae also showed pro-apoptotic effects on porcine epithelial cells [11,14]. MnuA is also secreted and, extracellularly, contributes to both NETs degradation and to host immunomodulation (as discussed in Host immune response and immunomodulation during M. hyopneumoniae infection). The putative SPase I, in turn, is likely involved in the secretion of many proteins, including at least some adhesins, to the cell surface or extracellular milieu (as discussed in Protein secretion of M. hyopneumoniae and its impact on pathogen-host interactions). The additional roles of M. hyopneumoniae MnuA and SPase I in M. hyopneumoniae biology may be the result of the specific evolutionary history of Mollicutes, which might have led to the acquisition of additional functions by some proteins to compensate in part for the observed genome reduction events [159].
Figure 3.
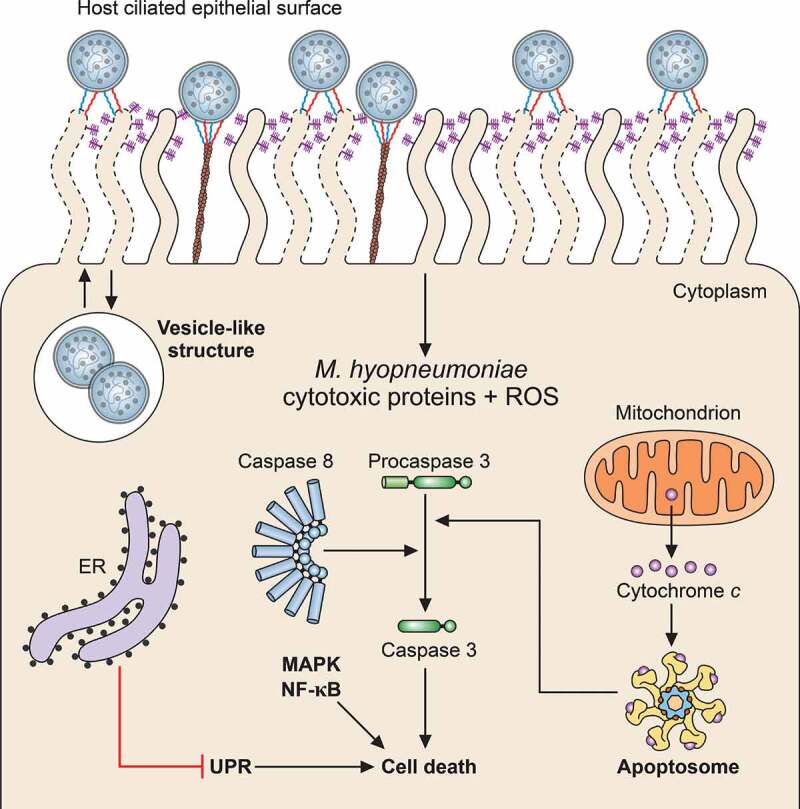
Schematic representation of host cell damage pathways induced by M. hyopneumoniae. M. hyopneumoniae cells interact with host ciliated epithelium causing cell damage through surface-displayed cytotoxic proteins and ROS release. These cytotoxic molecules can induce host cell death by triggering different pathways, including the apoptosis cascades associated with cytochrome C, caspase-3 and −8 (apoptosome), UPR, MAPK and NF-κB. Internalization of M. hyopneumoniae into a vesicle-like structure in the host cell cytosol and trafficking between the intra- and extracellular milieus are also represented. The representation of M. hyopneumoniae cells attached to the ciliated respiratory epithelium is described in Figure 1. ER, endoplasmic reticulum
Mycoplasmas can also produce ROS, by which they can induce cell death and tissue damage [160,161]. In line with that, it was demonstrated that M. hyopneumoniae can produce hydrogen peroxide (H2O2) when glycerol is available as a carbon source, while M. flocculare is unable to produce detectable amounts of this toxic molecule [162]. Notably, the gene encoding glycerol-3-phosphate oxidase (glpO), one of the enzymes responsible for H2O2 production in other mycoplasmas, is present in the genomes of multiple M. hyopneumoniae strains, but is absent in the M. flocculare genome [72]. However, the exact mechanism of H2O2 production in M. hyopneumoniae strains is still unknown. Moreover, the production of H2O2 by M. hyopneumoniae has the potential to cause oxidative stress to host cells and, at least in vitro, was associated with the antioxidant response elicited in swine respiratory epithelial cells [163].
In vitro experimental infections of a swine respiratory epithelial cell line with M. hyopneumoniae have also shown that several genes related to ciliary motility, ciliogenesis and ciliary polarization were down-regulated in infected host cells [163]. This evidence suggests a scenario in which M. hyopneumoniae infection could modulate the expression of genes related to the mucociliary apparatus, explaining, at least in part, the ciliostasis and loss of cilia observed in infected host tissues.
Besides the cilia damage, M. hyopneumoniae infection causes apoptosis-related cell events involving endoplasmic reticulum (ER) stress (Figure 3). Proteomic analyses of a tracheal swine cell line infected with M. hyopneumoniae revealed that infection induces dysregulation of Ca2+ homeostasis and ER stress [25]. It was also shown that ER stress leads to activation of the cytoprotective unfolded protein response (UPR), which, despite triggering signal transduction events associated with host defenses, is also associated with host cell apoptosis and, consequently, with damage to the ciliated respiratory epithelium. On the other hand, it was demonstrated that M. hyopneumoniae infection could also suppress one of the UPR pathways (the NF-κB pathway), counteracting at least in part the pro-apoptotic effects and favoring the survival of infected cells [164]. These apparently antagonistic effects of M. hyopneumoniae infection on UPR may represent alternative mechanisms favorable to the pathogen at different times of infection. An anti-apoptotic effect may be interesting in the initial steps of infection, to allow bacterial adherence and colonization of the host respiratory tract. The pro-apoptotic effect would be more prominent later on, contributing to tissue damage.
Additional evidences of swine epithelial cell death triggered by M. hyopneumoniae infection were also provided by secretome analyses of cells infected in vitro with pathogenic and nonpathogenic strains and with M. flocculare [26]. Cell death-related proteins were detected only in culture supernatants of swine cells infected with a M. hyopneumoniae pathogenic strain, suggesting a specific host response to pathogenic M. hyopneumoniae. Among these secreted cell death-related proteins, there were danger-associated molecule patterns (DAMPs), known to be secreted by host cells undergoing apoptosis during pathogen infections to alert the immune system and trigger a pro-inflammatory immune response.
M. hyopneumoniae homeostasis maintenance during host colonization
The persistence of M. hyopneumoniae infection relies not only on its ability to modulate and evade the host immune response, but also on its arsenal of protective mechanisms to cope with different stress conditions imposed by the swine host. Moreover, M. hyopneumoniae survival also depends on the uptake of nutrients provided by host cells. In this final section, these aspects, which are essential for M. hyopneumoniae cell homeostasis, will be presented.
As discussed in Host immune response and immunomodulation during M. hyopneumoniae infection and Cell damage induced by M. hyopneumoniae infection, M. hyopneumoniae infection is marked by the production of ROS by both host and mycoplasma cells. However, it is not yet clear how this pathogen can protect itself from endogenously and host produced toxic metabolites. The M. hyopneumoniae anti-toxic metabolites arsenal is limited, as its genome lacks genes encoding important antioxidant proteins [19,72]. Thus far, four antioxidant enzymes have been identified, namely thioredoxin, thioredoxin reductase, NADH-oxidase and peroxiredoxin, and all of them were overrepresented in the M. hyopneumoniae 7448 pathogenic strain in comparison to the nonpathogenic strain J and M. flocculare [75]. This evidence provided a clear link between protection against oxidative stress and M. hyopneumoniae pathogenicity. Furthermore, the M. hyopneumoniae peroxiredoxin (MhPrx) was functionally characterized and shown to protect DNA from ROS-mediated damage in vitro [12,165]. Therefore, MhPrx may play an essential role in pathogen survival.
In functional “omics” studies, virtually no changes in transcript or protein levels were observed for M. hyopneumoniae genes/proteins classically involved in oxidative stress between stress and control conditions [166–168]. At the proteome level, it was observed that three of the four known M. hyopneumoniae proteins involved in the oxidative stress response (thioredoxin, NADH-oxidase and MhPrx) were among the top 20 most abundant proteins, even in the absence of oxidative stress conditions [167]. Altogether, these transcriptomic and proteomic results suggest that the anti-oxidative stress arsenal of M. hyopneumoniae is constitutively expressed, enabling the pathogen being always prompt to respond to this kind of stress. Interestingly, pathogenic and nonpathogenic strains of M. hyopneumoniae presented similar abundances of antioxidant proteins under oxidative stress [167]. On the other hand, the protein repertoire of M. hyopneumoniae pathogenic strain 7448 was enriched with potential virulence factors, as adhesins, nucleases and lipoproteins, upon exposure to oxidative stress in comparison to control conditions (absence of oxidative stress) and to the M. hyopneumoniae nonpathogenic J strain.
Mycoplasma hyopneumoniae must also cope with temperature shifts in the host environment due to the release of pyrogenic cytokines as a mechanism of the immune response [48,132]. A transcriptomic study regarding the expression of heat stress-related genes demonstrated that several genes, including those coding for the heat shock proteins DnaK, DnaJ, Lon proteases, and ATP-dependent serine proteinase were affected by temperature shifts [166]. However, quantitative proteomic analyses did not reveal significant differences in the abundance of these proteins between heat stress and control conditions [167]. As discussed above for the response to oxidative stress, in temperature stress conditions, the heat stress-related proteins were found to be among the most abundant, while some potential virulence factors were more abundant in the M. hyopneumoniae pathogenic 7448 strain under heat stress.
Taken together, the transcriptomic and the proteomic findings suggest that M. hyopneumoniae may require the constitutive presence of proteins involved with stress protection. These findings also suggest that the synthesis of at least some virulence factors, including adhesins, nucleases, and lipoproteins, can be triggered by oxidative and temperature stresses, which would imply increased virulence in infection (stress) conditions. Further studies will be required to improve our understanding of this relationship between stress response and virulence, and also to identify, among the non-annotated genes of M. hyopneumoniae, those possibly encoding so far unknown stress-response proteins.
Along with the M. hyopneumoniae capacity to respond to stress conditions, its metabolic capacity is essential for survival in the host environment and may contribute to virulence. The M. hyopneumoniae transcription unit coding for myo-inositol catabolism proteins provides a good example of that (and thus far the only one characterized regarding this aspect). In silico metabolic reconstructions for swine respiratory mycoplasmas demonstrated that the metabolic capacities of M. hyopneumoniae, M. hyorhinis and M. flocculare are similar [73]. However, among mycoplasma species, M. hyopneumoniae is the only one with a transcription unit coding for myo-inositol catabolism proteins in its genome [19,73], suggesting that it can use inositol as an alternative carbon source [169]. Indeed, it was demonstrated that M. hyopneumoniae strains uptake myo-inositol from culture medium, while M. hyorhinis and M. flocculare are unable to do so [162]. This capacity to uptake and metabolize the myo-inositol from the host system would allow M. hyopneumoniae to persist longer than M. hyorhinis and M. flocculare in the host respiratory tract [176,177].
Conclusion
Regardless of its inherent simplicity, as a small, wall-less bacterium with reduced genome, M. hyopneumoniae utilizes several pathogenicity mechanisms, some of which are not fully understood. These mechanisms include the adherence to host ciliated epithelial cells, modulation of immune response and other host defenses, and induction of cell damage by both pathogen and host components. Moreover, comparative “omics” studies between M. hyopneumoniae and a nonpathogenic closely related species, M. flocculare, have provided evidence of proteins and functions associated with pathogenicity, as the production of differential adhesin proteoforms presented in the cell surface, the secretion of differential repertoires of proteins, and the overrepresentation of proteases, antioxidant proteins, and adhesion-related proteins in pathogenic M. hyopneumoniae strains. A schematic representation of the M. hyopneumoniae pathogenicity determinants discussed in this review is presented in Figure 4. In this complex scenario of pathogen-host interplay, more studies are needed to corroborate the available evidence and to elucidate further aspects of M. hyopneumoniae pathogenicity, especially those related to the in vivo infection process. Overall, the identification of M. hyopneumoniae pathogenicity determinants and their mechanisms of action are of utmost relevance to discover new and efficient targets for the development of novel diagnostic methods, therapeutic drugs, and preventive vaccines against EP.
Figure 4.
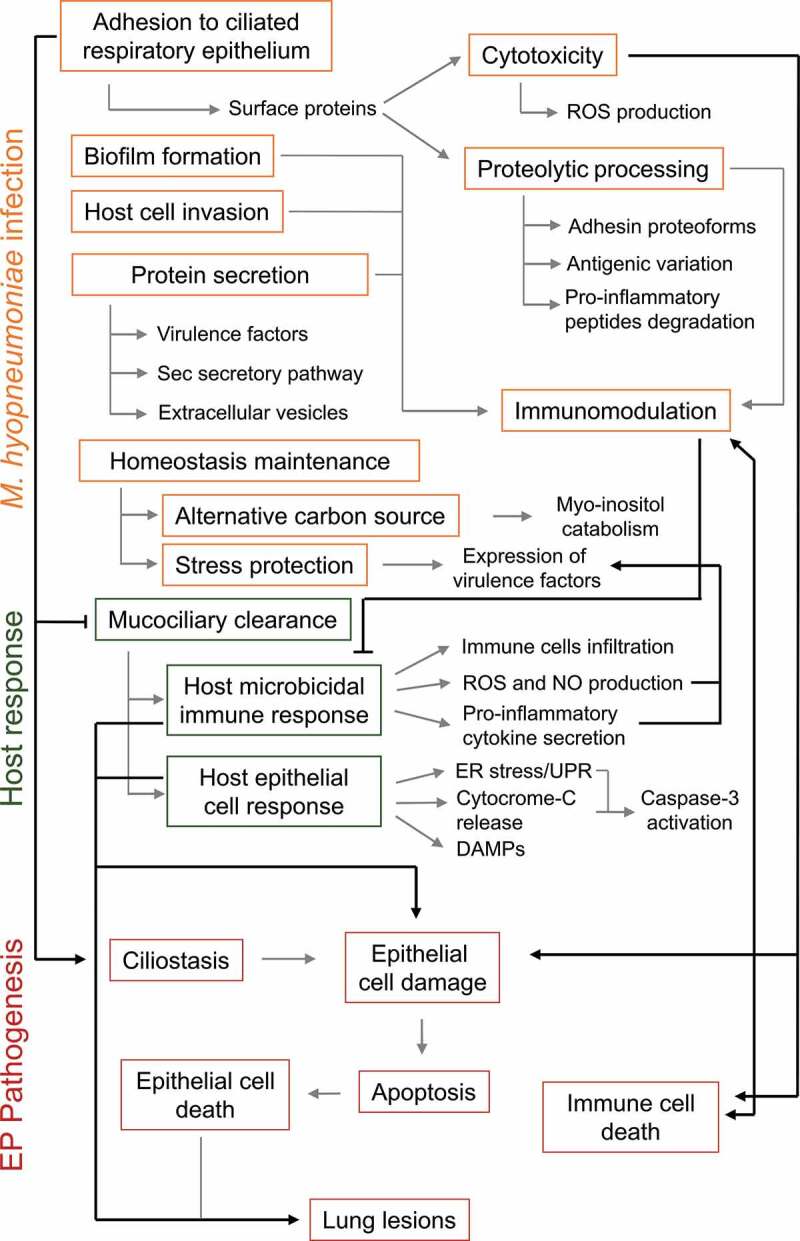
Schematic representation of M. hyopneumoniae pathogenicity determinants and host responses involved in EP establishment. Infection success depends on M. hyopneumoniae capacity to escape from the host mucociliary clearance and to adhere to ciliated cells of the porcine respiratory epithelium. Upon adhesion, M. hyopneumoniae causes ciliostasis and may form biofilms or invade host epithelial cells. M. hyopneumoniae also has a cytotoxic effect on host cells, causing cell damage and epithelial and immune cell death by apoptosis. This activity contributes to the modulation of the induced host immunological responses and may lead to lung lesions. Cell damage and epithelial cell death is also an outcome of host microbicidal and epithelial response to M. hyopneumoniae infection. Immunomodulation may also be associated with bacterial antigenic variation generated by proteolytic processing of surface adhesins and other surface proteins, and with the secretion of several virulence factors, as well as host pro-inflammatory peptides. During infection, M. hyopneumoniae may maintain its homeostasis using myo-inositol from lung tissue as an alternative carbon source. M. hyopneumoniae must also deal with stressing conditions, and its exposure to the host environment triggers the expression of several virulence factors. M. hyopneumoniae pathogenicity mechanisms, host response mechanisms and EP pathogenesis outcomes are shown in orange, green and red rectangles, respectively. Molecular processes and molecules involved in each mechanism or outcome are pointed out by gray arrows. Associations among M. hyopneumoniae pathogenicity mechanisms, host responses and EP pathogenesis outcomes are pointed out by black arrows
Acknowledgements
FMALZ was a recipient of a Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil) Ph.D. fellowship (140742/2015-8) and a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil) PDSE fellowship (PDSE 19/2016 - 88881.133246/2016-01). JAP received a CAPES post-doctoral fellowship (CAPES-Biologia Computacional - Process Number: 611 23038.010043/2013-02), and is a recipient of a CAPES postdoctoral fellowship (PNPD – 88887.464453/2019-00).
Funding Statement
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico [140742/2015-8]; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [PNPD – 88887.464453/2019-00]; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [PDSE 19/2016 - 88881.133246/2016-01]; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES Biologia Computacional - Process Number: 611 23038.010043/2013-02).
Disclosure statement
The authors declare no conflict of interest.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- [1].Thacker EL, Minion CF.. Mycoplasmosis. In: Jeffrey J. Zimmerman, Locke A. Karriker, Alejandro Ramirez, Ken J. Schwartz and Gregory W. Stevenson, editors. Diseases of swine. Iowa state: university press; 2010. p. 779–797. [Google Scholar]
- [2].Maes D, Sibila M, Kuhnert P, et al. Update on mycoplasma hyopneumoniae infections in pigs: knowledge gaps for improved disease control. Transbound Emerg Dis. 2018;65(1):110-124. DOI: 10.1111/tbed.12677 [DOI] [PubMed] [Google Scholar]
- [3].Holst S, Yeske P, Pieters M. Elimination of mycoplasma hyopneumoniae from breed-to-wean farms: a review of current protocols with emphasis on herd closure and medication. J Swine Health Prod. 2015;23(6):321–330. [Google Scholar]
- [4].Bai Y, Gan Y, Hua LZ, et al. Application of a sIgA-ELISA method for differentiation of Mycoplasma hyopneumoniae infected from vaccinated pigs. Vet Microbiol. 2018;223:86–92. [DOI] [PubMed] [Google Scholar]
- [5].Feng ZX, Bai Y, Yao JT, et al. Use of serological and mucosal immune responses to Mycoplasma hyopneumoniae antigens P97R1, P46 and P36 in the diagnosis of infection. Vet J. 2014;202:128–133. [DOI] [PubMed] [Google Scholar]
- [6].Galli V, Simionatto S, Marchioro SB, et al. Immunisation of mice with Mycoplasma hyopneumoniae antigens P37, P42, P46 and P95 delivered as recombinant subunit or DNA vaccines. Vaccine. 2012;31(1):135–140. [DOI] [PubMed] [Google Scholar]
- [7].Marchioro SB, Fisch A, Gomes CK, et al. Local and systemic immune responses induced by a recombinant chimeric protein containing Mycoplasma hyopneumoniae antigens fused to the B subunit of Escherichia coli heat-labile enterotoxin LTB. Vet Microbiol. 2014;173(1–2):166–171. [DOI] [PubMed] [Google Scholar]
- [8].Virginio VG, Gonchoroski T, Paes JA, et al. Immune responses elicited by Mycoplasma hyopneumoniae recombinant antigens and DNA constructs with potential for use in vaccination against porcine enzootic pneumonia. Vaccine. 2014;32(44):5832–5838. [DOI] [PubMed] [Google Scholar]
- [9].Jarocki VM, Raymond BBA, Tacchi JL, et al. Mycoplasma hyopneumoniae surface-associated proteases cleave bradykinin, substance P, neurokinin A and neuropeptide Y. Sci Rep. 2019;9:14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jarocki VM, Santos J, Tacchi JL, et al. MHJ_0461 is a multifunctional leucine aminopeptidase on the surface of Mycoplasma hyopneumoniae. Open Biol. 2015;5:140175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li P, Zhang Y, Li X, et al. Mycoplasma hyopneumoniae Mhp597 is a cytotoxicity, inflammation and immunosuppression associated nuclease. Vet Microbiol. 2019;235:53–62. [DOI] [PubMed] [Google Scholar]
- [12].Machado C, Pinto P, Zaha A, et al. A peroxiredoxin from Mycoplasma hyopneumoniae with a possible role in H2O2 detoxification. Microbiology. 2009;155(10):3411–3419. [DOI] [PubMed] [Google Scholar]
- [13].Moitinho-Silva L, Heineck BL, Reolon LA, et al. Mycoplasma hyopneumoniae type I signal peptidase: expression and evaluation of its diagnostic potential. Vet Microbiol. 2012;154(3–4):282–291. [DOI] [PubMed] [Google Scholar]
- [14].Paes JA, Virginio VG, Cancela M, et al. Pro-apoptotic effect of a Mycoplasma hyopneumoniae putative type I signal peptidase on PK(15) swine cells. Vet Microbiol. 2017b;201:170–176. [DOI] [PubMed] [Google Scholar]
- [15].Tacchi JL, Raymond BB, Jarocki VM, et al. Cilium adhesin P216 (MHJ_0493) is a target of ectodomain shedding and aminopeptidase activity on the surface of Mycoplasma hyopneumoniae. J Proteome Res. 2014;13:2920–2930. [DOI] [PubMed] [Google Scholar]
- [16].Pirofski LA, Casadevall A. The damage-response framework as a tool for the physician-scientist to understand the pathogenesis of infectious diseases. J Infect Dis. 2018;218:S7–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mare CJ, Switzer WP. New species: mycoplasma hyopneumoniae; a causative agent of virus pig pneumonia. Vet Med Small Anim Clin. 1965;60:841–846. [PubMed] [Google Scholar]
- [18].Minion F, Lefkowitz E, Madsen M, et al. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J Bacteriol. 2004;186(21):7123–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vasconcelos AT, Ferreira HB, Bizarro CV, et al. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J Bacteriol. 2005;187:5568–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zielinski GC, Ross RF. Effect of growth in cell cultures and strain on virulence of mycoplasma hyopneumoniae for swine. Am J Vet Res. 1990;51:344–348. [PubMed] [Google Scholar]
- [21].Bai F, Ni B, Liu M, et al. Mycoplasma hyopneumoniae-derived lipid-associated membrane proteins induce apoptosis in porcine alveolar macrophage via increasing nitric oxide production, oxidative stress, and caspase-3 activation. Vet Immunol Immunopathol. 2013;155(3):155–161. [DOI] [PubMed] [Google Scholar]
- [22].Ni B, Bai FF, Wei Y, et al. Apoptosis induced by lipid-associated membrane proteins from Mycoplasma hyopneumoniae in a porcine lung epithelial cell line with the involvement of caspase 3 and the MAPK pathway. Genet Mol Res. 2015;14(3):11429–11443. [DOI] [PubMed] [Google Scholar]
- [23].Ni L, Song C, Wu X, et al. RNA-seq transcriptome profiling of porcine lung from two pig breeds in response to Mycoplasma hyopneumoniae infection. PeerJ. 2019;7:e7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Trueeb BS, Braun RO, Auray G, et al. Differential innate immune responses induced by Mycoplasma hyopneumoniae and Mycoplasma hyorhinis in various types of antigen presenting cells. Vet Microbiol. 2020;240:108541. [DOI] [PubMed] [Google Scholar]
- [25].Leal Zimmer FMA, Moura H, Barr JR, et al. Intracellular changes of a swine tracheal cell line infected with a Mycoplasma hyopneumoniae pathogenic strain. Microb Pathog. 2019a;137:103717. [DOI] [PubMed] [Google Scholar]
- [26].Leal Zimmer FMDA, Paludo GP, Moura H, et al. Differential secretome profiling of a swine tracheal cell line infected with mycoplasmas of the swine respiratory tract. J Proteomics. 2019b;192:147–159. [DOI] [PubMed] [Google Scholar]
- [27].Maes D, Segales J, Meyns T, et al. Control of Mycoplasma hyopneumoniae infections in pigs. Vet Microbiol. 2008;126(4):297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].He Y, Xu MJ, Zhou DH, et al. Seroprevalence of Mycoplasma hyopneumoniae in pigs in subtropical southern China. Trop Anim Health Prod. 2011;43(3):695–698. [DOI] [PubMed] [Google Scholar]
- [29].Sosa C, Blois A, Ibáñez F, et al. Genetic diversity of mycoplasma hyopneumoniae in Mmendoza province. Rev Argent Microbiol. 2019;51:229–233. [DOI] [PubMed] [Google Scholar]
- [30].Pacce VD, Oliveira NRD, Jorge S, et al. Occurrence of mycoplasma hyopneumoniae in slaughter pigs from Southern Brazil. BrazilJ Veterinary Res Animal Sci. 2019;5:6. [Google Scholar]
- [31].Vicente AF, Catto D, Allendorf SD, et al. Seropositivity for mycoplasma hyopneumoniae in pigs at a slaughterhouse in the central region of São Paulo. Arquivo Brasileiro De Medicina Veterinária E Zootecnia. 2013;65:5. [Google Scholar]
- [32].Oba P, Wieland B, Mwiine FN, et al. Status and gaps of research on respiratory disease pathogens of swine in Africa. Porcine Health Manag. 2020;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bertelloni F, Mazzei M, Cilia G, et al. Serological Survey on Bacterial and Viral Pathogens in Wild Boars Hunted in Tuscany. Ecohealth. 2020;17(1):85–93. [DOI] [PubMed] [Google Scholar]
- [34].Malmsten A, Magnusson U, Ruiz-Fons F, et al. A serologic survey of pathogens in wild boar (sus scrofa) in sweden. J Wildl Dis. 2018;54:229–237. [DOI] [PubMed] [Google Scholar]
- [35].Calsamiglia M, Pijoan C. Colonisation state and colostral immunity to Mycoplasma hyopneumoniae of different parity sows. Vet Rec. 2000;146(18):530–532. [DOI] [PubMed] [Google Scholar]
- [36].Nathues H, Doehring S, Woeste H, et al. Individual risk factors for Mycoplasma hyopneumoniae infections in suckling pigs at the age of weaning. Acta Vet Scand. 2013;55:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fano E, Pijoan C, Dee S, et al. Effect of mycoplasma hyopneumoniae colonization at weaning on disease severity in growing pigs. Can J Vet Res. 2007;71:195–200. [PMC free article] [PubMed] [Google Scholar]
- [38].Sibila M, Nofrarías M, López-Soria S, et al. Chronological study of Mycoplasma hyopneumoniae infection, seroconversion and associated lung lesions in vaccinated and non-vaccinated pigs. Vet Microbiol. 2007;122:97–107. [DOI] [PubMed] [Google Scholar]
- [39].Meyns T, Maes D, Dewulf J, et al. Quantification of the spread of Mycoplasma hyopneumoniae in nursery pigs using transmission experiments. Prev Vet Med. 2004;66(1–4):265–275. [DOI] [PubMed] [Google Scholar]
- [40].Roos LR, Fano E, Homwong N, et al. A model to investigate the optimal seeder-to-naïve ratio for successful natural Mycoplasma hyopneumoniae gilt exposure prior to entering the breeding herd. Vet Microbiol. 2016;184:51–58. [DOI] [PubMed] [Google Scholar]
- [41].Villarreal I, Meyns T, Dewulf J, et al. The effect of vaccination on the transmission of Mycoplasma hyopneumoniae in pigs under field conditions. Vet J. 2011;188(1):48–52. [DOI] [PubMed] [Google Scholar]
- [42].Pieters M, Pijoan C, Fano E, et al. An assessment of the duration of Mycoplasma hyopneumoniae infection in an experimentally infected population of pigs. Vet Microbiol. 2009;134(3–4):261–266. [DOI] [PubMed] [Google Scholar]
- [43].Takeuti KL, de Barcellos DESN, de Lara AC, et al. Detection of Mycoplasma hyopneumoniae in naturally infected gilts over time. Vet Microbiol. 2017;203:215–220. [DOI] [PubMed] [Google Scholar]
- [44].Mayor D, Zeeh F, Frey J, et al. Diversity of Mycoplasma hyopneumoniae in pig farms revealed by direct molecular typing of clinical material. Vet Res. 2007;38(3):391–398. [DOI] [PubMed] [Google Scholar]
- [45].Mayor D, Jores J, Korczak BM, et al. Multilocus sequence typing (MLST) of Mycoplasma hyopneumoniae: a diverse pathogen with limited clonality. Vet Microbiol. 2008;127(1–2):63–72. [DOI] [PubMed] [Google Scholar]
- [46].de Castro LA, Rodrigues Pedroso T, Kuchiishi SS, et al. Variable number of tandem aminoacid repeats in adhesion-related CDS products in Mycoplasma hyopneumoniae strains. Vet Microbiol. 2006;116:258–269. [DOI] [PubMed] [Google Scholar]
- [47].Dos Santos LF, Sreevatsan S, Torremorell M, et al. Genotype distribution of Mycoplasma hyopneumoniae in swine herds from different geographical regions. Vet Microbiol. 2015;175:374–381. [DOI] [PubMed] [Google Scholar]
- [48].Hwang, M.H., Damte, D., Lee, J.S., Gebru, E., Chang, Z.Q., Cheng, H., Jung, B.Y., Rhee, M.H., Park, S.C . 2011. Mycoplasma hyopneumoniae induces proinflammatory cytokine and nitric oxide production through NFκB and MAPK pathways in RAW264.7 cells. Vet Res Commun 35, 21–34. [DOI] [PubMed] [Google Scholar]
- [49].Garza-Moreno L, Segalés J, Pieters M, et al. Acclimation strategies in gilts to control Mycoplasma hyopneumoniae infection. Vet Microbiol. 2018;219:23–29. [DOI] [PubMed] [Google Scholar]
- [50].Betlach AM, Maes D, Garza-Moreno L, et al. Mycoplasma hyopneumoniae variability: current trends and proposed terminology for genomic classification. Transbound Emerg Dis. 2019. doi: 10.1111/tbed.13233 [DOI] [PubMed] [Google Scholar]
- [51].Vranckx K, Maes D, Sacristán REP, et al. A longitudinal study of the diversity and dynamics of Mycoplasma hyopneumoniae infections in pig herds. Vet Microbiol. 2012;156:315–321. [DOI] [PubMed] [Google Scholar]
- [52].Tao Y, Shu J, Chen J, et al. A concise review of vaccines against Mycoplasma hyopneumoniae. Res Vet Sci. 2019;123:144–152. [DOI] [PubMed] [Google Scholar]
- [53].Meyns T, Dewulf J, de Kruif A, et al. Comparison of transmission of Mycoplasma hyopneumoniae in vaccinated and non-vaccinated populations. Vaccine. 2006;24(49–50):7081–7086. [DOI] [PubMed] [Google Scholar]
- [54].Villarreal I, Vranckx K, Calus D, et al. Effect of challenge of pigs previously immunised with inactivated vaccines containing homologous and heterologous Mycoplasma hyopneumoniae strains. BMC Vet Res. 2012;8(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Beffort L, Weiß C, Fiebig K, et al. Field study on the safety and efficacy of intradermal versus intramuscular vaccination against Mycoplasma hyopneumoniae. Vet Rec. 2017;181(13):348. [DOI] [PubMed] [Google Scholar]
- [56].Maes D, Sibila M, Kuhnert P, et al. Update on Mycoplasma hyopneumoniae infections in pigs: knowledge gaps for improved disease control. Transbound Emerg Dis. 2018;65(Suppl 1):110–124. [DOI] [PubMed] [Google Scholar]
- [57].Chae C. Porcine respiratory disease complex:iInteraction of vaccination and porcine circovirus type 2, porcine reproductive and respiratory syndrome virus, and Mycoplasma hyopneumoniae. Vet J. 2016;212:1–6. [DOI] [PubMed] [Google Scholar]
- [58].Feng ZX, Wei YN, Li GL, et al. Development and validation of an attenuated Mycoplasma hyopneumoniae aerosol vaccine. Vet Microbiol. 2013;167:417–424. [DOI] [PubMed] [Google Scholar]
- [59].Martelli P, Terreni M, Guazzetti S, et al. Antibody response to Mycoplasma hyopneumoniae infection in vaccinated pigs with or without maternal antibodies induced by sow vaccination. J Vet Med B Infect Dis Vet Public Health. 2006;53(5):229–233. [DOI] [PubMed] [Google Scholar]
- [60].Pieters M, Fano E, Pijoan C, et al. An experimental model to evaluate mycoplasma hyopneumoniae transmission from asymptomatic carriers to unvaccinated and vaccinated sentinel pigs. Can J Vet Res. 2010;74:157–160. [PMC free article] [PubMed] [Google Scholar]
- [61].de Oliveira NR, Jorge S, Gomes CK, et al. A novel chimeric protein composed of recombinant Mycoplasma hyopneumoniae antigens as a vaccine candidate evaluated in mice. Vet Microbiol. 2017;201:146–153. [DOI] [PubMed] [Google Scholar]
- [62].Jorge S, de Oliveira NR, Marchioro SB, et al. The Mycoplasma hyopneumoniae recombinant heat shock protein P42 induces an immune response in pigs under field conditions. Comp Immunol Microbiol Infect Dis. 2014;37(4):229–236. [DOI] [PubMed] [Google Scholar]
- [63].Tassis PD, Tsakmakidis I, Papatsiros VG, et al. A randomized controlled study on the efficacy of a novel combination vaccine against enzootic pneumonia (Mycoplasma hyopneumoniae) and porcine Circovirus type 2 (PCV2) in the presence of strong maternally derived PCV2 immunity in pigs. BMC Vet Res. 2017;13(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Virginio VG, Bandeira NC, Leal FM, et al. Assessment of the adjuvant activity of mesoporous silica nanoparticles in recombinant. Heliyon. 2017;3:e00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Drexler CS, Witvliet MH, Raes M, et al. Efficacy of combined porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae vaccination in piglets. Vet Rec. 2010;166(3):70–74. [DOI] [PubMed] [Google Scholar]
- [66].Jeong J, Park C, Choi K, et al. A new single-dose bivalent vaccine of porcine circovirus type 2 and Mycoplasma hyopneumoniae elicits protective immunity and improves growth performance under field conditions. Vet Microbiol. 2016;182:178–186. [DOI] [PubMed] [Google Scholar]
- [67].Park C, Jeong J, Choi K, et al. Efficacy of a new bivalent vaccine of porcine circovirus type 2 and Mycoplasma hyopneumoniae (Fostera™ PCV MH) under experimental conditions. Vaccine. 2016;34(2):270–275. [DOI] [PubMed] [Google Scholar]
- [68].Roques E, Girard A, Gagnon CA, et al. Antibody responses induced in mice immunized with recombinant adenovectors expressing chimeric proteins of various porcine pathogens. Vaccine. 2013;31(24):2698–2704. [DOI] [PubMed] [Google Scholar]
- [69].Duivon D, Corrégé I, Hémonic A, et al. Field evaluation of piglet vaccination with a. Porcine Health Manag. 2018;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Maes D, Boyen F, Haesebrouck F, et al. Antimicrobial treatment of Mycoplasma hyopneumoniae infections. Vet J. 2020;259-260:105474. [DOI] [PubMed] [Google Scholar]
- [71].Vicca J, Stakenborg T, Maes D, et al. In vitro susceptibilities of Mycoplasma hyopneumoniae field isolates. Antimicrob Agents Chemother. 2004;48(11):4470–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Felde O, Kreizinger Z, Sulyok KM, et al. Antibiotic susceptibility testing of Mycoplasma hyopneumoniae field isolates from Central Europe for fifteen antibiotics by microbroth dilution method. PLoS One. 2018;13(12):e0209030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Qiu G, Rui Y, Zhang J, et al. Macrolide-resistance selection in Tibetan pigs with a high load of Mycoplasma hyopneumoniae. Microb Drug Resist. 2018;24(7):1043–1049. [DOI] [PubMed] [Google Scholar]
- [74].Siqueira FM, Thompson CE, Virginio VG, et al. New insights on the biology of swine respiratory tract mycoplasmas from a comparative genome analysis. BMC Genomics. 2013;14:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ferrarini MG, Siqueira FM, Mucha SG, et al. Insights on the virulence of swine respiratory tract mycoplasmas through genome-scale metabolic modeling. BMC Genomics. 2016;17:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Paes JA, Lorenzatto KR, de Moraes SN, et al. Secretomes of Mycoplasma hyopneumoniae and Mycoplasma flocculare reveal differences associated to pathogenesis. J Proteomics. 2017a;154:69–77. [DOI] [PubMed] [Google Scholar]
- [77].Paes JA, Machado LDPN, Dos Anjos Leal FM, et al. Comparative proteomics of two Mycoplasma hyopneumoniae strains and Mycoplasma flocculare identified potential porcine enzootic pneumonia determinants. Virulence. 2018;9:1230–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Siqueira FM, Gerber AL, Guedes RL, et al. Unravelling the transcriptome profile of the Swine respiratory tract mycoplasmas. PLoS One. 2014;9:e110327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Raymond BBA, Madhkoor R, Schleicher I, et al. Extracellular Actin Is a Receptor for. Front Cell Infect Microbiol. 2018b;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tacchi JL, Raymond BB, Haynes PA, et al. Post-translational processing targets functionally diverse proteins in mycoplasma hyopneumoniae. Open Biol. 2016;. 6(2):150210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hammerschmidt S, Rohde M, Preer KT. Extracellular matrix interactions with gram-positive pathogens. Microbiol Spectr. 2019;7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bustamante-Marin XM, Ostrowski LE. Cilia and mucociliary clearance. Cold Spring Harb Perspect Biol. 2017;9(4):a028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Blanchard B, Vena M, Cavalier A, et al. Electron microscopic observation of the respiratory tract of SPF piglets inoculated with Mycoplasma hyopneumoniae. Vet Microbiol. 1992;30(4):329–341. [DOI] [PubMed] [Google Scholar]
- [84].DeBey MC, Ross RF. Ciliostasis and loss of cilia induced by Mycoplasma hyopneumoniae in porcine tracheal organ cultures. Infect Immun. 1994;62:5312–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wang H, Zhang Z, Xie X, et al. Paracellular pathway-mediated Mycoplasma hyopneumoniae Migration across Porcine Airway Epithelial Barrier under Air-Liquid Interface Conditions. Infect Immun. 2020;88(10):e00470-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chen R, Yu Y, Feng Z, et al. Featured species-specific loops are found in the crystal structure of Mhp Eno, a Cell Surface Adhesin From Mycoplasma hyopneumoniae. Front Cell Infect Microbiol. 2019;9:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Yu Y, Liu M, Hua L, et al. Fructose-1,6-bisphosphate aldolase encoded by a core gene of Mycoplasma hyopneumoniae contributes to host cell adhesion. Vet Res. 2018a;49(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Reolon LA, Martello CL, Schrank IS, et al. Survey of surface proteins from the pathogenic Mycoplasma hyopneumoniae strain 7448 using a biotin cell surface labeling approach. PLoS One. 2014;9(11):e112596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Young TF, Thacker EL, Erickson BZ, et al. A tissue culture system to study respiratory ciliary epithelial adherence of selected swine mycoplasmas. Vet Microbiol. 2000;71(3–4):269–279. [DOI] [PubMed] [Google Scholar]
- [90].Berry IJ, Jarocki VM, Tacchi JL, et al. N-terminomics identifies widespread endoproteolysis and novel methionine excision in a genome-reduced bacterial pathogen. Sci Rep. 2017;7:11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bogema DR, Scott NE, Padula MP, et al. Sequence TTKF ↓ QE defines the site of proteolytic cleavage in Mhp683 protein, a novel glycosaminoglycan and cilium adhesin of Mycoplasma hyopneumoniae. J Biol Chem. 2011;286:41217–41229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Deutscher AT, Tacchi JL, Minion FC, et al. Mycoplasma hyopneumoniae surface proteins Mhp385 and Mhp384 bind host cilia and glycosaminoglycans and are endoproteolytically processed by proteases that recognize different cleavage motifs. J Proteome Res. 2012;11(3):1924–1936. [DOI] [PubMed] [Google Scholar]
- [93].Djordjevic SP, Cordwell SJ, Djordjevic MA, et al. Proteolytic processing of the Mycoplasma hyopneumoniae cilium adhesin. Infect Immun. 2004;72(5):2791–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Machado LDPN, Paes JA, Souza Dos Santos P, et al. Evidences of differential endoproteolytic processing on the surfaces of mycoplasma hyopneumoniae and mycoplasma flocculare. Microb Pathog. 2019;140:103958. [DOI] [PubMed] [Google Scholar]
- [95].Raymond BB, Jenkins C, Seymour LM, et al. Proteolytic processing of the cilium adhesin MHJ_0194 (P123J) in Mycoplasma hyopneumoniae generates a functionally diverse array of cleavage fragments that bind multiple host molecules. Cell Microbiol. 2015;17:425–444. [DOI] [PubMed] [Google Scholar]
- [96].Machado LDPN, Paes JA, Souza Dos Santos P, et al. Evidences of differential endoproteolytic processing on the surfaces of Mycoplasma hyopneumoniae and Mycoplasma flocculare. Microb Pathog. 2020;140:103958. [DOI] [PubMed] [Google Scholar]
- [97].Robinson MW, Buchtmann KA, Jenkins C, et al. MHJ_0125 is an M42 glutamyl aminopeptidase that moonlights as a multifunctional adhesin on the surface of Mycoplasma hyopneumoniae. Open Biol. 2013;3:130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Widjaja M, Harvey KL, Hagemann L, et al. Elongation factor Tu is a multifunctional and processed moonlighting protein. Sci Rep. 2017;7:11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhang Q, Young TF, Ross RF. Glycolipid receptors for attachment of Mycoplasma hyopneumoniae to porcine respiratory ciliated cells. Infect Immun. 1994;62:4367–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Deutscher AT, Jenkins C, Minion FC, et al. Repeat regions R1 and R2 in the P97 paralogue Mhp271 of Mycoplasma hyopneumoniae bind heparin, fibronectin and porcine cilia. Mol Microbiol. 2010;78(2):444–458. [DOI] [PubMed] [Google Scholar]
- [101].Seymour LM, Falconer L, Deutscher AT, et al. Mhp107 is a member of the multifunctional adhesin family of Mycoplasma hyopneumoniae. J Biol Chem. 2011;286(12):10097–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Seymour LM, Jenkins C, Deutscher AT, et al. Mhp182 (P102) binds fibronectin and contributes to the recruitment of plasmin(ogen) to the Mycoplasma hyopneumoniae cell surface. Cell Microbiol. 2012;14:81–94. [DOI] [PubMed] [Google Scholar]
- [103].Burnett TA, Dinkla K, Rohde M, et al. P159 is a proteolytically processed, surface adhesin of Mycoplasma hyopneumoniae: defined domains of P159 bind heparin and promote adherence to eukaryote cells. Mol Microbiol. 2006;60(3):669–686. [DOI] [PubMed] [Google Scholar]
- [104].Wilton J, Jenkins C, Cordwell SJ, et al. Mhp493 (P216) is a proteolytically processed, cilium and heparin binding protein of Mycoplasma hyopneumoniae. Mol Microbiol. 2009;71(3):566–582. [DOI] [PubMed] [Google Scholar]
- [105].Grimmer J, Dumke R. Organization of multi-binding to host proteins: the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of Mycoplasma pneumoniae. Microbiol Res. 2019;218:22–31. [DOI] [PubMed] [Google Scholar]
- [106].Gründel A, Jacobs E, Dumke R. Interactions of surface-displayed glycolytic enzymes of Mycoplasma pneumoniae with components of the human extracellular matrix. Int J Med Microbiol. 2016;306(8):675–685. [DOI] [PubMed] [Google Scholar]
- [107].Hagemann L, Gründel A, Jacobs E, et al. The surface-displayed chaperones GroEL and DnaK of mycoplasma pneumoniae interact with human plasminogen and components of the extracellular matrix. Pathog Dis. 2017;75(3). [DOI] [PubMed] [Google Scholar]
- [108].Huang J, Zhu H, Wang J, et al. Fructose-1,6-bisphosphate aldolase is involved in Mycoplasma bovis colonization as a fibronectin-binding adhesin. Res Vet Sci. 2019;124:70–78. [DOI] [PubMed] [Google Scholar]
- [109].Qi J, Zhang F, Wang Y, et al. Characterization of Mycoplasma gallisepticum pyruvate dehydrogenase alpha and beta subunits and their roles in cytoadherence. PLoS One. 2018;13(12):e0208745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Raymond BBA, Turnbull L, Jenkins C, et al. Mycoplasma hyopneumoniae resides intracellularly within porcine epithelial cells. Sci Rep. 2018c;8(1):17697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Marois C, Le Carrou J, Kobisch M, et al. Isolation of Mycoplasma hyopneumoniae from different sampling sites in experimentally infected and contact SPF piglets. Vet Microbiol. 2007;120(1–2):96–104. [DOI] [PubMed] [Google Scholar]
- [112].Seymour LM, Deutscher AT, Jenkins C, et al. A processed multidomain Mycoplasma hyopneumoniae adhesin binds fibronectin, plasminogen, and swine respiratory cilia. J Biol Chem. 2010;285(44):33971–33978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Widjaja M, Berry IJ, Jarocki VM, et al. Cell surface processing of the P1 adhesin of mycoplasma pneumoniae identifies novel domains that bind host molecules. Sci Rep. 2020;10:6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Bürki S, Frey J, Pilo P. Virulence, persistence and dissemination of Mycoplasma bovis. Vet Microbiol. 2015;179(1–2):15–22. [DOI] [PubMed] [Google Scholar]
- [115].Chen H, Yu S, Hu M, et al. Identification of biofilm formation by Mycoplasma gallisepticum. Vet Microbiol. 2012;161(1–2):96–103. [DOI] [PubMed] [Google Scholar]
- [116].Simmons WL, Daubenspeck JM, Osborne JD, et al. Type 1 and type 2 strains of Mycoplasma pneumoniae form different biofilms. Microbiology. 2013;159(Pt_4):737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Raymond BBA, Jenkins C, Turnbull L, et al. Extracellular DNA release from the genome-reduced pathogen Mycoplasma hyopneumoniae is essential for biofilm formation on abiotic surfaces. Sci Rep. 2018a;8:10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tassew DD, Mechesso AF, Park NH, et al. Biofilm formation and determination of minimum biofilm eradication concentration of antibiotics in Mycoplasma hyopneumoniae. J Vet Med Sci. 2017;79:1716–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Dallo SF, Baseman JB. Intracellular DNA replication and long-term survival of pathogenic mycoplasmas. Microb Pathog. 2000;29(5):301–309. [DOI] [PubMed] [Google Scholar]
- [120].Yavlovich A, Tarshis M, Rottem S. Internalization and intracellular survival of Mycoplasma pneumoniae by non-phagocytic cells. FEMS Microbiol Lett. 2004;233(2):241–246. [DOI] [PubMed] [Google Scholar]
- [121].Raymond BB, Djordjevic S. Exploitation of plasmin(ogen) by bacterial pathogens of veterinary significance. Vet Microbiol. 2015;178:1–13. [DOI] [PubMed] [Google Scholar]
- [122].Woolley LK, Fell SA, Djordjevic SP, et al. Plasmin activity in the porcine airways is enhanced during experimental infection with Mycoplasma hyopneumoniae, is positively correlated with proinflammatory cytokine levels and is ameliorated by vaccination. Vet Microbiol. 2013;164(1–2):60–66. [DOI] [PubMed] [Google Scholar]
- [123].Bhattacharya S, Ploplis VA, Castellino FJ. Bacterial plasminogen receptors utilize host plasminogen system for effective invasion and dissemination. J Biomed Biotechnol. 2012;2012:482096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Sinha B, François PP, Nüsse O, et al. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cell Microbiol. 1999;1:101–117. [DOI] [PubMed] [Google Scholar]
- [125].Friis NF. Mycoplasm suipneumoniae and mycoplasma flocculare in comparative pathogenicity studies. Acta Vet Scand. 1974;15:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Le Carrou J, Laurentie M, Kobisch M, et al. Persistence of Mycoplasma hyopneumoniae in experimentally infected pigs after marbofloxacin treatment and detection of mutations in the parC gene. Antimicrob Agents Chemother. 2006;50(6):1959–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Deeney AS, Maglennon GA, Chapat L, et al. Mycoplasma hyopneumoniae evades phagocytic uptake by porcine alveolar macrophages in vitro. Vet Res. 2019;50(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Green ER, Mecsas J. Bacterial secretion systems: an overview. Microbiol Spectr. 2016;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Smets D, Loos MS, Karamanou S, et al. Protein transport across the bacterial plasma membrane by the sec pathway. Protein J. 2019;38(3):262–273. [DOI] [PubMed] [Google Scholar]
- [130].Chernov VM, Mouzykantov AA, Baranova NB, et al. Extracellular membrane vesicles secreted by mycoplasma Acholeplasma laidlawii PG8 are enriched in virulence proteins. J Proteomics. 2014;110C:117–128. [DOI] [PubMed] [Google Scholar]
- [131].Gaurivaud P, Ganter S, Villard A, et al. Mycoplasmas are no exception to extracellular vesicles release: revisiting old concepts. PLoS One. 2018;13(11):e0208160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Rodríguez F, Batista M, Hernández JN, et al. Relationship between expression of interleukin-5 and Interleukin-13 by epithelial cells and bronchiolar changes in pigs infected with mycoplasma hyopneumoniae. J Comp Pathol. 2016;154(2–3):165–168. [DOI] [PubMed] [Google Scholar]
- [133].Fourour S, Marois-Créhan C, Martelet L, et al. Intra-species and inter-species differences in cytokine production by porcine antigen-presenting cells stimulated by Mycoplasma hyopneumoniae, M. hyorhinis, and M. flocculare. Pathogens. 2019;8(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Woolley LK, Fell S, Gonsalves JR, et al. Evaluation of clinical, histological and immunological changes and qPCR detection of Mycoplasma hyopneumoniae in tissues during the early stages of mycoplasmal pneumonia in pigs after experimental challenge with two field isolates. Vet Microbiol. 2012;161(1–2):186–195. [DOI] [PubMed] [Google Scholar]
- [135].Shen Y, Hu W, Wei Y, et al. Effects of Mycoplasma hyopneumoniae on porcine nasal cavity dendritic cells. Vet Microbiol. 2017;198:1–8. [DOI] [PubMed] [Google Scholar]
- [136].Leal FMDA, Virginio VG, Martello CL, et al. Mycoplasma hyopneumoniae and Mycoplasma flocculare differential domains from orthologous surface proteins induce distinct cellular immune responses in mice. Vet Microbiol. 2016;50–57. DOI: 10.1016/j.vetmic.2016.05.008 [DOI] [PubMed] [Google Scholar]
- [137].Garcia-Morante B, Segalés J, Fraile L, et al. Potential use of local and systemic humoral immune response parameters to forecast Mycoplasma hyopneumoniae associated lung lesions. PLoS One. 2017;12(4):e0175034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Muneta Y, Minagawa Y, Shimoji Y, et al. Immune response of gnotobiotic piglets against Mycoplasma hyopneumoniae. J Vet Med Sci. 2008;70(10):1065–1070. [DOI] [PubMed] [Google Scholar]
- [139].Thanawongnuwech R, Thacker EL. Interleukin-10, interleukin-12, and interferon-gamma levels in the respiratory tract following mycoplasma hyopneumoniae and PRRSV infection in pigs. Viral Immunol. 2003;16:357–367. [DOI] [PubMed] [Google Scholar]
- [140].Bai F, Ni B, Liu M, et al. Mycoplasma hyopneumoniae-derived lipid-associated membrane proteins induce inflammation and apoptosis in porcine peripheral blood mononuclear cells in vitro. Vet Microbiol. 2015;175(1):58–67. [DOI] [PubMed] [Google Scholar]
- [141].Liu W, Zhou D, Yuan F, et al. Surface proteins mhp390 (P68) contributes to cilium adherence and mediates inflammation and apoptosis in Mycoplasma hyopneumoniae. Microb Pathog. 2019;126:92–100. [DOI] [PubMed] [Google Scholar]
- [142].Chandrasekar BS, Yadav S, Victor ES, et al. Interferon-gamma and nitric oxide synthase 2 mediate the aggregation of resident adherent peritoneal exudate cells: implications for the host response to pathogens. PLoS One. 2015;10(6):e0128301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Thom SR, Bhopale VM, Milovanova TN, et al. Nitric-oxide synthase-2 linkage to focal adhesion kinase in neutrophils influences enzyme activity and β2 integrin function. J Biol Chem. 2013;288:4810–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Yadav S, Pathak S, Sarikhani M, et al. Nitric oxide synthase 2 enhances the survival of mice during Salmonella Typhimurium infection-induced sepsis by increasing reactive oxygen species, inflammatory cytokines and recruitment of neutrophils to the peritoneal cavity. Free Radic Biol Med. 2018;116:73–87. [DOI] [PubMed] [Google Scholar]
- [145].Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57(1):395–418. [DOI] [PubMed] [Google Scholar]
- [146].El-Benna J, Hurtado-Nedelec M, Marzaioli V, et al. Priming of the neutrophil respiratory burst: role in host defense and inflammation. Immunol Rev. 2016;273:180–193. [DOI] [PubMed] [Google Scholar]
- [147].Rose SJ, Bermudez LE, Flynn JL. Mycobacterium avium biofilm attenuates mononuclear phagocyte function by triggering hyperstimulation and apoptosis during early infection. Infect Immun. 2014;82(1):405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Poderoso JJ, Helfenberger K, Poderoso C. The effect of nitric oxide on mitochondrial respiration. Nitric Oxide. 2019;88:61–72. [DOI] [PubMed] [Google Scholar]
- [149].Choi SY, Lim JW, Shimizu T, et al. Reactive oxygen species mediate Jak2/Stat3 activation and IL-8 expression in pulmonary epithelial cells stimulated with lipid-associated membrane proteins from Mycoplasma pneumoniae. Inflamm Res. 2012;61(5):493–501. [DOI] [PubMed] [Google Scholar]
- [150].Dusanic D, Bencina D, Oven I, et al. Mycoplasma synoviae induces upregulation of apoptotic genes, secretion of nitric oxide and appearance of an apoptotic phenotype in infected chicken chondrocytes. Vet Res. 2012;43(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Obara H, Harasawa R. Nitric oxide causes anoikis through attenuation of E-cadherin and activation of caspase-3 in human gastric carcinoma AZ-521 cells infected with Mycoplasma hyorhinis. J Vet Med Sci. 2010;72(7):869–874. [DOI] [PubMed] [Google Scholar]
- [152].Citti C, Nouvel LX, Baranowski E. Phase and antigenic variation in mycoplasmas. Future Microbiol. 2010;5:1073–1085. [DOI] [PubMed] [Google Scholar]
- [153].Moitinho-Silva L, Kondo MY, Oliveira LC, et al. Mycoplasma hyopneumoniae in vitro peptidase activities: identification and cleavage of kallikrein-kinin system-like substrates. Vet Microbiol. 2013;163:264–273. [DOI] [PubMed] [Google Scholar]
- [154].Polosa R, Hasani A, Pavia D, et al. Acute effect of inhaled bradykinin on tracheobronchial clearance in normal humans. Thorax. 1992;47:952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Onaga T. Tachykinin: recent developments and novel roles in health and disease. Biomol Concepts. 2014;5(3):225–243. [DOI] [PubMed] [Google Scholar]
- [156].Prod’homme T, Weber MS, Steinman L, et al. A neuropeptide in immune-mediated inflammation, Y? Trends Immunol. 2006;27(4):164–167. [DOI] [PubMed] [Google Scholar]
- [157].Neto JC, Strait EL, Raymond M, et al. Antibody responses of swine following infection with Mycoplasma hyopneumoniae, M. hyorhinis, M. hyosynoviae and M. flocculare. Vet Microbiol. 2014. DOI: 10.1016/j.vetmic.2014.08.008 [DOI] [PubMed] [Google Scholar]
- [158].Yu Y, Wang J, Han R, et al. Evades complement activation by binding to factor H via elongation factor thermo unstable (EF-Tu). Virulence. 2020;11:1059–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Weiler JM, Daha MR, Austen KF, et al. Control of the amplification convertase of complement by the plasma protein beta1H. Proc Natl Acad Sci U S A. 1976;73:3268–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Henthorn CR, Chris Minion F, Sahin O. Utilization of macrophage extracellular trap nucleotides by Mycoplasma hyopneumoniae. Microbiology. 2018;164(11):1394–1404. [DOI] [PubMed] [Google Scholar]
- [161].Sirand-Pugnet P, Citti C, Barré A, et al. Evolution of mollicutes: down a bumpy road with twists and turns. Res Microbiol. 2007;158(10):754–766. [DOI] [PubMed] [Google Scholar]
- [162].Hames C, Halbedel S, Hoppert M, et al. Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J Bacteriol. 2009;191:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Vilei EM, Frey J. Genetic and biochemical characterization of glycerol uptake in mycoplasma mycoides subsp. mycoides SC: its impact on H(2)O(2) production and virulence. Clin Diagn Lab Immunol. 2001;8:85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Ferrarini MG, Mucha SG, Parrot D, et al. Hydrogen peroxide production and myo-inositol metabolism as important traits for virulence of Mycoplasma hyopneumoniae. Mol Microbiol. 2018. DOI: 10.1111/mmi.13957 [DOI] [PubMed] [Google Scholar]
- [165].Mucha SG, Ferrarini MG, Moraga C, et al. Mycoplasma hyopneumoniae J elicits an antioxidant response and decreases the expression of ciliary genes in infected swine epithelial cells. Sci Rep. 2020;10:13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Pan Q, Wang X, Liu T, et al. Mycoplasma hyopneumoniae inhibits porcine beta-defensin 2 production by blocking the unfolded protein response to facilitate epithelial adhesion and infection. Infect Immun. 2020;88(7). DOI: 10.1128/IAI.00164-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Gonchoroski T, Virginio VG, Thompson CE, et al. Evolution and function of the Mycoplasma hyopneumoniae peroxiredoxin, a 2-Cys-like enzyme with a single Cys residue. Mol Genet Genomics. 2017;292(2):297–305. [DOI] [PubMed] [Google Scholar]
- [168].Madsen ML, Nettleton D, Thacker EL, et al. Transcriptional profiling of Mycoplasma hyopneumoniae during heat shock using microarrays. Infect Immun. 2006;74(1):160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Paes JA, Leal Zimmer FMA, Moura H, et al. Differential responses to stress of two Mycoplasma hyopneumoniae strains. J Proteomics. 2019;199:67–76. [DOI] [PubMed] [Google Scholar]
- [170].Schafer ER, Oneal MJ, Madsen ML, et al. Global transcriptional analysis of Mycoplasma hyopneumoniae following exposure to hydrogen peroxide. Microbiology. 2007;153(11):3785–3790. [DOI] [PubMed] [Google Scholar]
- [171].Ferreira HB, Castro LA. A preliminary survey of M. hyopneumoniae virulence factors based on comparative genomic analysis. São Paulo Gene Mol Microbiol. 2007;30(1):245–255. [Google Scholar]
- [172].Bogema DR, Deutscher AT, Woolley LK, et al. Characterization of cleavage events in the multifunctional cilium adhesin Mhp684 (P146) reveals a mechanism by which Mycoplasma hyopneumoniae regulates surface topography. MBio. 2012;3(2): e00282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Yu Y, Wang H, Wang J, et al. Elongation factor thermo unstable (EF-Tu) moonlights as an adhesin on the surface of mycoplasma hyopneumoniae by binding to fibronectin. Front Microbiol. 2018b;9:974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Deng X, Zhu Y, Dai P, et al. Three polypeptides screened from phage display random peptide library may be the receptor polypeptide of Mycoplasma genitalium adhesion protein. Microb Pathog. 2018;120:140–146. [DOI] [PubMed] [Google Scholar]
- [175].Raymond BB, Tacchi JL, Jarocki VM, et al. P159 from Mycoplasma hyopneumoniae binds porcine cilia and heparin and is cleaved in a manner akin to ectodomain shedding. J Proteome Res. 2013;12:5891–5903. [DOI] [PubMed] [Google Scholar]
- [176].Minion FC, Jarvill-Taylor K. Membrane-associated hemolysin activities in mycoplasmas. FEMS Microbiol Lett. 1994;116(1):101–106. [DOI] [PubMed] [Google Scholar]
- [177].Khil PP, Camerini-Otero RD. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol Microbiol. 2002;44(1):89–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


