ABSTRACT
Clostridioides difficile is an enteric bacterial pathogen that can a cause nosocomial infection leading to debilitating colitis. The development of a murine model of C. difficile infection has led to fundamental discoveries in disease pathogenesis and the host immune response to infection. Recently, C. difficile endogenously present in the microbiota of mice has been reported and was found to complicate interpretation of mouse studies. Here, we report a novel C. difficile strain, named NTCD-035, isolated from the microbiota of our mouse colony. The presence of NTCD-035 in mice prior to challenge with a highly pathogenic C. difficile strain (VPI10463) led to significantly reduced disease severity. Phylogenetic characterization derived from whole genome sequencing and PCR ribotyping identified the isolate as a novel clade 1, ribotype 035 strain that lacks the pathogenicity locus required to produce toxins. Deficiency in toxin production along with sporulation capacity and secondary bile acid sensitivity was confirmed using in vitro assays. Inoculation of germ-free mice with NTCD-035 did not cause morbidity despite the strain readily colonizing the large intestine. Implementation of a culture-based screening procedure enabled the identification of mice harboring C. difficile in their microbiota, the establishment of a C. difficile-free mouse colony, and a monitoring system to prevent future contamination. Taken together, these data provide a framework for screening mice for endogenously harbored C. difficile and support clinical findings that demonstrate the therapeutic potential of non-toxigenic strains in preventing C. difficile associated disease.
Abbreviations: PaLoc - Pathogenicity locus, CFUs - Colony forming units, TcdA - toxin-A, TcdB - toxin-B, CdtA - binary toxin A, CdtB - binary toxin B, CdtR - binary toxin R, NTCD - non-toxigenic C. difficile
Introduction
Clostridioides difficile, a Gram-positive, spore-forming, obligate anaerobe, is the most common nosocomial infection in the United States, totaling approximately 365,000 infections and 13,000 deaths per year.1–3 A healthy microbiota provides colonization resistance against C. difficile. However, perturbation of the microbiota, such as that observed following antibiotic treatment, impairs colonization resistance to create a biological niche in the lower gastrointestinal tract for C. difficile to grow.4,5 Upon establishment of infection, C. difficile produces cytotoxins that are the main virulence factors that drive the majority of pathologic consequences of infection, including intestinal epithelial damage and diarrhea.6,7 Two toxin proteins, toxin-A (TcdA) and toxin-B (TcdB), as well as regulatory proteins are encoded by a ~ 20kb region termed the pathogenicity locus (PaLoc). Numerous C. difficile strains have been identified that lack the PaLoc. These non-toxigenic strains of C. difficile have the PaLoc replaced with a conserved non-coding 115 bp region and are unable to produce toxin.8–10
Epidemiological surveys of healthy volunteers have shown 4–8% of humans are carriers of C. difficile and about 50% of them carry non-toxigenic C. difficile.11 Non-toxigenic strains of C. difficile are associated with asymptomatic colonization in humans12,13 and correlate with lower rates of C. difficile associated disease.14 This correlation provokes the hypothesis that non-toxigenic C. difficile strains can be used therapeutically to prevent pathogenic strains from colonizing the intestine. Several studies support this hypothesis. For example, prior administration of non-toxigenic C. difficile protects hamsters from pathogenic strains.15–17 In human patients, a phase II clinical trial reported by Gerding et al. showed a significant reduction in recurrence of disease in patients administered non-toxigenic spores.18 Combined, these data support the therapeutic potential of non-toxigenic C. difficile.
The murine model of C. difficile infection has been developed over the past decade as a tool to better understand the immunologic mechanisms that limit or promote pathology. However, the model presents challenges that must be resolved to conduct well-controlled studies. Gewirtz and colleagues described a latent C. difficile strain present in the intestinal microbiota of commercially available mice.19 Further, Lawley et al. demonstrated that mice can latently carry C. difficile, which blooms following antibiotic treatment to induce a “super spreader” state.20 These observations raise the prospect of complications in using the mouse model for C. difficile studies. Identification and screening for contaminating C. difficile in animal colonies is critical to ensure controlled studies in C. difficile research and is a focus of this report.
In this study, the presence of a contaminating endogenous C. difficile strain is identified and isolated from the gastrointestinal tract of naïve mice housed in our animal colony. We demonstrate that colonization with this endogenous C. difficile changes the outcome of infection with pathogenic C. difficile. Through in vitro studies, PCR ribotyping, and whole genome sequencing, we characterize this strain as a novel non-toxigenic F035 ribotype that is a clade 1 member of the C. difficile species. These data demonstrate the importance of thorough animal colony screening for C. difficile strains and identifies a novel probiotic candidate.
Results
Endogenous C. difficile attenuates severe infection from a highly virulent strain of C. difficile
The murine model of C. difficile infection mimics human exposure via administration of broad-spectrum antibiotics to predispose mice to infection followed by oral inoculation with the oxygen tolerant spore form of C. difficile (Figure 1(a)).21 Spore Inoculation with the virulent VPI10463 strain of C. difficile leads to germination into vegetative cells that produce toxins in the large intestine resulting in diarrhea, weight loss, and mortality within the first 72 hours following infection.21,22 Over a series of mouse experiments, we observed a cage-dependent bifurcation of disease severity in mice infected with the VPI10463 strain of C. difficile as determined by weight loss (Figure 1(b)) and survival (Figure 1(c)). These observations raised the possibility that variation in the endogenous microbiota from cage to cage could be driving phenotypic disease differences. Next, the microbiota of uninfected antibiotic treated control mice were screened. Unexpectedly, fecal pellets from uninfected control mice pretreated with two distinct antibiotic regimens grew bacterial colony forming units (CFU) on C. difficile selective agar plates (Figure 1(d)), despite the mice not exhibiting any overt disease symptoms (data not shown).
Figure 1.
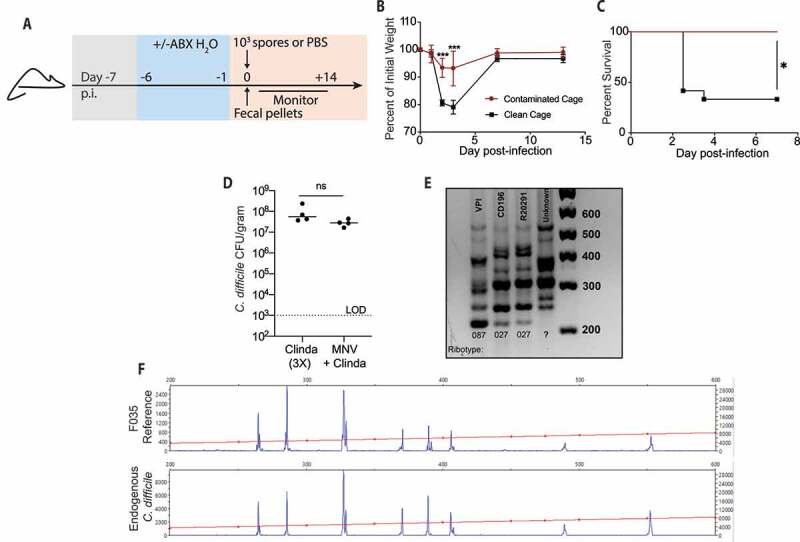
Endogenous C. difficile contamination attenuates severe disease in mice infected with pathogenic VPI
(a) Schematic of antibiotic (ABX) treatment and C. difficile infection. (b-c) Mice were infected with VPI 10463 spores. (b) Percent of initial weight from day of infection. (c) Survival of VPI infected mice contaminated with endogenous C. difficile and non-contaminated mice. (d) C. difficile burden in fecal pellets of C57BL/6 mice following a PBS mock infection. (e) PCR ribotyping of CD196, VPI10463, R20291 C. difficile strains and the unknown C. difficile strain isolated from gastrointestinal track of PBS mock infected C. difficile culture positive mice. (f) PCR ribotype of unknown C. difficile strain and reference F035 ribotype as determined by fluorescent PCR ribotyping. n = 6–8 mice/group. LOD – Limit of Detection. * = p < .05, *** = p < .001.
To confirm this outgrowth was C. difficile, bacterial DNA was isolated from a pure culture derived from a colony picked from the agar plate. The 16S rDNA gene region of the bacteria was sequenced by Sanger sequencing methodology and subsequently cross-referenced against the NCBI BLAST database. The search confirmed the bacterium to belong to the C. difficile species, though strain level specificity could not be determined (Suppl. Table 1). To identify the strain of C. difficile isolated from our animal colony, we employed PCR ribotyping, a well-characterized method of classifying C. difficile strains.23 This technique is generally used to track outbreaks of C. difficile in hospitals,24–26 and was adapted to identify and track our novel strain. The unknown C. difficile strain exhibited a banding pattern distinct from the three other strains of C. difficile used in our lab (CD196, VPI10463 and R20291), suggesting the strain’s origins came from an external source (Figure 1(e)). To determine the specific ribotype, fluorescent PCR ribotyping was performed and compared to a database of known C. difficile ribotypes as described by Martinson et al.27 and was determined to be the F035 ribotype (figure 1(f)). The F035 ribotype has been reported to be non-toxigenic24,28 and has been found on the floors and common equipment of hospitals.25 Taken together, these data reveal the presence of a non-toxigenic strain of C. difficile in our animal colony that was capable of attenuating disease severity in our murine C. difficile infection model.
Identification of NTCD-035 as a novel strain of C. difficile that lacks the pathogenicity gene locus by whole genome sequencing
To determine the identity of the C. difficile strain isolated from our mouse colony, whole genome sequencing was performed on this strain, termed NTCD-035 (non-toxigenic C. difficile ribotype 035), along with the CD196 strain as a comparative control. The complete genomes of the two strains were annotated and compared by their core genes (genes present in all genomes) using an in-house pipeline, coreSNPs (https://github.com/chrgu/coreSNPs). Circular representations of the genomes were generated using the Circos software.29 The sequence homology between the core genes for the two strains was 99.3%. Analysis of the complete NTCD-035 genome readily identified genes known to flank the toxin genes. However, NTCD-035 lacks toxin A, toxin B, and toxin regulatory genes that are encoded within the pathogenicity locus as well as all binary toxin gene elements (cdtA, cdtB, cdtR) (Figure 2(a)). These analyses demonstrate this strain lacks the genetic code to produce toxin. Next, NTCD-035 and the reference CD196 strains were compared to publicly available high-quality C. difficile whole genome sequences.30 A phylogenetic tree comparing core genes from various C. difficile strains was generated (Figure 2(b)). While the sequenced CD196 strain phylogenetically matched the publicly deposited CD196 genome, NTCD-035 was distinct from any publicly deposited C. difficile genomes. Phylogenetic characterization based on single nucleotide variants in core genes (Figure 2(b)) and hierarchical clustering based on the presence or absence of accessory genes (Figure 2(c)) determined NTCD-035 clustered with clade 1 isolates, specifically clustering closely to CD105KS strains isolated from soil samples31,32 (Figure 2(b,c)). Due to the high degree of similarity between NTCD-035 and the cohort of CD105KS strains, the genomes were compared to determine if our isolated strain was novel. Hamming distance calculated between the CD105KSE5 and NTCD-035 core genomes revealed 213 single nucleotide variants between strains, suggesting that NTCD-035 is closely related but distinct from CD105KSE5 (Figure 2(d)). In comparison, our lab’s CD196 reference genome had 1 nucleotide variant difference from the publicly deposited CD196 genome, demonstrating the robustness of our whole genome sequencing methods. Presence of toxin genes outside of the pathogenicity locus had previously been described.9,33 Therefore, the NTCD-035 genome was searched via blast for toxin genes (tcdA, tcdB, tcdC, tcdE, tcdR), which came up negative. Instead, a conserved non-coding 115 bp region was identified in the PaLoc region similar to other non-toxigenic C. difficile isolates.8–10 This confirmed the lack of toxin genes in the NTCD-035 strain, a pattern that was repeated in the closely related CD105KS strains (Figure 2(e)). Taken together, these data demonstrate that NTCD-035 is a novel, non-toxigenic C. difficile strain that lacks a pathogenicity locus and binary toxin genes and clusters most closely with strains isolated from soil samples.
Figure 2.
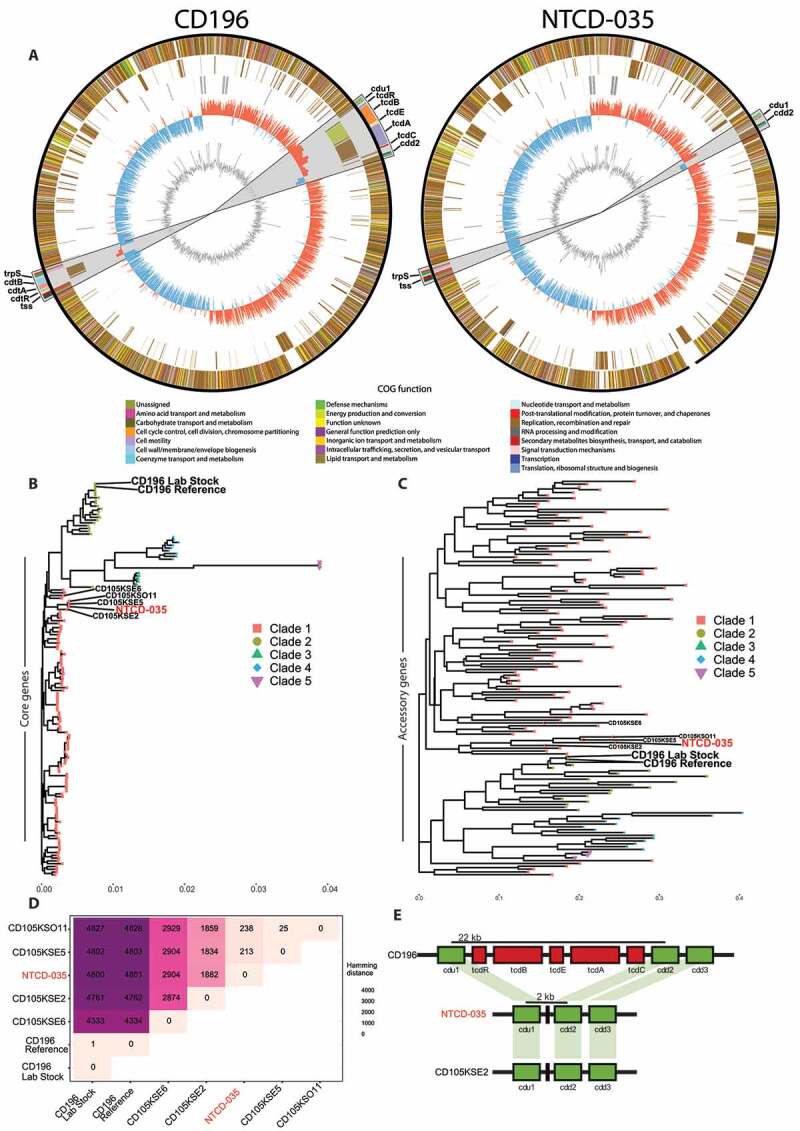
Whole genome sequencing identifies endogenous C. difficile as a novel, non-toxigenic clade 1 member
(a) Circos plots of CD196 and NTCD-035 chromosome contigs. From outside to inside, circles represent shared genes between isolates, unique genes to each isolate, GC skew, and GC content. Color of genes represents annotated gene COG function by Prokka. Grey region 10x zoomed in portion highlighting the region of pathogenicity locus and binary toxin genes. Genomes were rotated with respect to the origin of replication. (b) Phylogenetic analysis of core genes (c) and hierarchical clustering of the presence or absence of accessory genes of C. difficile isolates. (d) Hamming distance of the core genomes between selected C. difficile isolates. (e) Annotated segment of pathogenicity locus comparing distances between cdu1 and cdd2 genes in CD196, NTCD-035, and a similar phylogenetic isolate, CD105KSE2.
Endogenous C. difficile strain NTCD-035 sporulates but does not produce toxin
Next, aspects of C. difficile biology were characterized for NTCD-035 and compared to three other pathogenic C. difficile strains (VPI10463, CD196, and R20291) as reference standards. First, the in vitro growth rate of NTCD-035 was found to be comparable to the CD196 and R20291 strains (Figure 3(a)). Second, the absence of the PaLoc in the genome and the inability of NTCD-035 to produce toxin was functionally confirmed. Toxin production of the endogenous strain was quantified and compared to a well-characterized high toxin producing strain (VPI10463) using a cell-based cytotoxicity assay. VPI10463 culture supernatant had high concentrations of toxin that induced cell rounding after a 10,000-fold dilution of the culture supernatant. In contrast, undiluted culture supernatant from NTCD-035 did not cause morphological changes to treated Vero cells, functionally confirming the strain’s inability to produce toxin proteins (Figure 3(b)). Third, the capacity and rate of sporulation was assessed. NTCD-035 generated spores at a similar rate to CD196 and R20291, while VPI10463 generated spores at a slower rate than other strains, consistent with Akeurlund et. al.34 (Figure 3(c)). Fourth, susceptibility of each strain to secondary bile acids was assessed by culturing vegetative C. difficile in the presence of deoxycholic acid or lithocholic acid. Secondary bile acids are essential for colonization resistance to C. difficile infection, as they have been shown to inhibit C. difficile growth,35–38 as well as inhibit C. difficile germination.39–41 NTCD-035 exhibited a secondary bile acid sensitivity profile similar to the CD196 and R20291 strains (Figure 3(d,e)). Finally, antibiotic sensitivity of NTCD-035 to the specific antibiotic cocktail used in our murine infection system (metronidazole, neomycin, vancomycin, or clindamycin) was tested and compared to lab strains. No difference in the antibiotic sensitivity profile was observed in the four C. difficile strains tested. NTCD-035 did not grow in the presence of metronidazole, vancomycin, or clindamycin, while maintaining resistance against neomycin (figure 3(f)). Taken together, our in vitro characterization studies suggest that the isolated strain is physiologically similar to the CD196 and R20291 strains in terms of growth rate, sporulation rate, secondary bile acid and antibiotic sensitivity, but it lacks the capacity to produce functional toxin.
Figure 3.
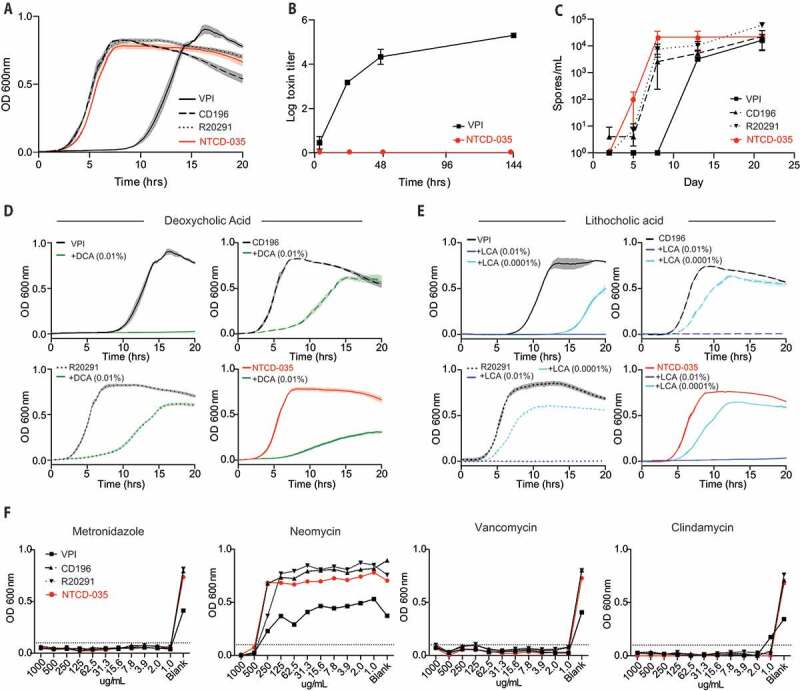
Physiological characteristics of NTCD-035 C. difficile is similar to those of CD196 and R20291 except for the capacity to produce toxin
(a) Growth curves of C. difficile strains as assessed by optical density at 600 nm. (b) Toxin production as assessed by Vero cell assay. C. difficile was cultured anaerobically in BHIS broth at 37°C. Aliquots were taken from liquid culture at various time points and assayed for toxin activity. (c) Sporulation rates of C. difficile strains. C. difficile was cultured anaerobically in BHIS broth at 37°C. Aliquots were taken at various time points, heat killed at 65°C for 60 minutes and anaerobically plated on BHISTA plates to quantify the number of spores. (d-e) Growth curves of C. difficile strains as assessed by optical density at 600 nm (d) in the presence of deoxycholic acid, (e) or lithocholic acid. (f) Antibiotic sensitivity assay assessing C. difficile sensitivity to metronidazole, neomycin, vancomycin, and clindamycin
NTCD-035 lacks pathogenicity in acutely susceptible germ-free mice
Gnotobiotic (germ-free) mice are acutely susceptible to C. difficile infection and present with high morbidity and mortality.42 Therefore, germ-free mice were used to test whether NTCD-035 caused any pathology in mice. Germ-free mice were infected with spores from either NTCD-035 or VPI10463 as a positive control and were subsequently monitored for colonization, weight loss, and mortality. VPI10463 infection resulted in rapid weight loss (Figure 4(a)) and a high mortality rate (Figure 4(b)) within 72 hours post infection. In contrast, mice inoculated with NTCD-035 exhibited no weight loss (Figure 4(a)) and no mortality following infection (Figure 4(b)) despite exhibiting high, persistent C. difficile burden in fecal pellets following inoculation (Figure 4(c)). These data demonstrate that NTCD-035 is avirulent even in acutely susceptible germ-free mice.
Figure 4.
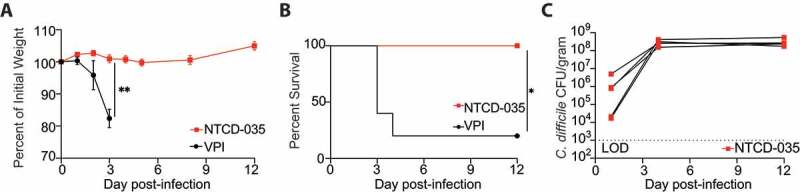
NTCD-035 lacks pathogenicity in germ free mice
(a-c) Germfree C57BL/6 mice were mono-colonized with either the VPI10463 or NTCD-035 spores by oral gavage. (a) Percent of initial weight of infected groups from day of infection. (B) Survival of mice infected with different strains. (c) C. difficile burden in fecal pellets collected from infected mice. n = 5 mice/group. LOD – Limit of Detection. * = p < .05, ** = p < .01.”
Mouse contact tracing determined C. difficile penetrance in the mouse colony and enabled subsequent elimination
To determine the extent of penetrance of the NTCD-035 strain in the resident microbiota of our mouse colony, an epidemiological survey of the university’s animal housing facilities was conducted. First, a screening protocol was established for identification of mice contaminated with C. difficile (Figure 5(a)). Fecal pellets from mice were incubated overnight in C. difficile selective media and plated the following day on C. difficile selective agar plates. Eighty mouse cages were screened, and 10% of the cages tested positive for C. difficile (Figure 5(b)). No trend in mouse genotype or origin of mouse vendor was observed (data not shown). This culture-based enrichment method was capable of detecting C. difficile in fecal pellets that went undetected by direct plating methods or PCR amplification of fecal bacterial DNA. These observations suggest that NTCD-035 is a low abundance member of the mouse’s intestinal microbial community and blooms following antibiotic perturbation. In addition to collection of fecal pellets, the animal changing hood, vivarium floor, water spigots, and cage rack surfaces were swabbed with a sterile cotton swab, brought into anaerobic conditions, and streaked on C. difficile selective plates. After consecutive weeks of positive tests, a thorough reorganization and decontamination of the facility room was initiated, which successfully removed the spores from the environment (Figure 5(c)).
Figure 5.
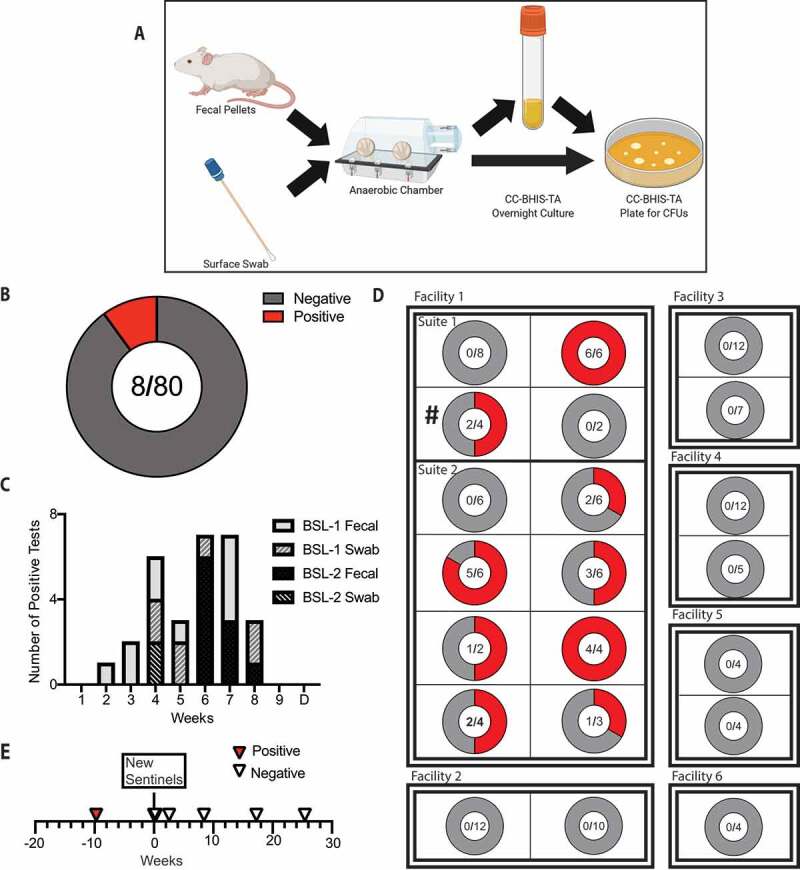
Mouse contact tracing protocol identifies and eliminates the outgrowth of NTCD-035 in the mouse colony
(a) Schematic of screening protocol established. (b) Total cages tested for NTCD-035 in the lab’s mouse breeding room. (c) Number of positive tests per week. ‘D’ denotes decontamination of rodent rooms. (d) C. difficile screen of sentinel mice from representative rooms and suites across campus. ‘#’ denotes the original rodent room identified with NTCD-035 positive mice. (e) C. difficile screen of sentinel mice over time. ‘New sentinels’ indicates the timepoint when naïve sentinel mice were introduced in the room without soiled bedding transfer from previous sentinel cage
To determine the breadth of C. difficile penetrance, sentinel mice were screened from selected rodent rooms within our lab’s mouse facility and from six other mouse facilities across the institution. Sentinel mice are housed in the soiled bedding from stock cages of that particular room and therefore will be exposed to any C. difficile spores harbored by mice in that room due to the coprophagic nature of mice. The feces of approximately half of the sentinel mice tested within our facility were positive for C. difficile. However, the feces of sentinel mice from all other animal facilities tested negative (Figure 5(d)). This data indicated that the C. difficile presence in the mouse colony was localized, and that the origin of C. difficile was not from an outside mouse vendor as the other facilities with negative sentinel mice shared common vendors. Following euthanasia of cages that tested positive for C. difficile and decontamination of the mouse room, new C. difficile negative sentinel mice were established in our lab’s mouse colony. These sentinels remained negative over several months of consistent monitoring (Figure 5(e)). Combined, these epidemiologic methods provide a working template for detecting latent C. difficile in a mouse colony and steps for subsequent elimination.
Discussion
Establishment of murine models of C. difficile infection have led to mechanistic insights into the host response to infection. Examples of such insights include identifying inflammasome signaling43 and innate lymphoid cells22,44 as critical mediators coordinating the early host response following toxin-mediated epithelial damage. Such advances are dependent on maintaining a consistent murine model of C. difficile infection. This report demonstrates that the presence of non-toxigenic C. difficile in the microbiota of mice prior to challenge by toxigenic strains mitigates disease, which skews results of murine C. difficile studies. This report also highlights the importance of screening protocols in the vivarium and serves as a guide for identification and control of an outgrowth in mouse colonies.
The data presented here demonstrates that the NTCD-035 strain can readily colonize mice but does not cause pathology even in monocolonized germ-free mice, which are highly susceptible to C. difficile infection.42 Further, NTCD-035 was able to protect mice from subsequent pathogenic C. difficile infection. Previous findings provide insight into potential mechanisms of protection by NTCD-035 via colonization resistance (e.g. niche competition) or immunologic mechanisms. Protection through colonization resistance can be explained by the “founder effect” phenomenon, where the first colonizing strain of one bacterial species metabolically outcompetes subsequent strains for space and resources.45,46 Thus, pre-colonization of a host by non-toxigenic C. difficile could prevent the outgrowth of subsequent pathogenic strains by consuming shared nutrients and occupying the same niche required for the growth and colonization of C. difficile. This concept is supported by data from Wilson and Sheagren who found that non-toxigenic C. difficile had to be administered prior to toxigenic C. difficile challenge to be protective15 as well as by data that demonstrated that non-toxigenic C. difficile needs to be viable at the time of toxigenic C. difficile infection to provide protection.47 Additionally, Ethienne-Mesmin et al. showed their C. difficile strain (LEM1) could outcompete VPI10463 when co-colonized19 providing evidence of protection through metabolic competition. Alternatively, the NTCD-035 strain could indirectly prevent infection with a toxigenic strain by educating the immune system’s response. NTCD-035 does not encode toxin genes, and thus an immune response to toxin proteins could not be elicited. However, shared C. difficile surface proteins have been tested for immunogenic properties and have been found to elicit robust antibody responses that can protect mice against subsequent infections.48 However, in hamster models, previous exposure to non-toxigenic C. difficile was not sufficient to protect hamsters from secondary toxigenic C. difficile infection suggesting the immune response elicited to non-toxigenic C. difficile was not protective.47 The magnitude and protective capacity of the immune response elicited in humans in response to non-toxigenic C. difficile remains to be determined. Future studies investigating the mechanism of NTCD-035 mediated protection will be needed to delineate between these two possible direct and indirect pathways.
This report identified a novel, non-toxigenic ribotype 035 strain of C. difficile, termed NTCD-035, in our mouse colony’s microbiota. Two potential sources of this strain are external mouse vendors or exposure to environmental contamination from the surrounding community. Etienne-Mesmin et. al identified a novel strain of C. difficile that was a minor member of the mouse microbiota from mouse vendors and bloomed following antibiotic treatment.19 Subsequent observations from several laboratories support the concept of commercial mouse vendors as potential reservoirs for endogenous C. difficile (personal communications). In this report, however, commercial mouse vendors were likely not the source because sentinel mice from six different facilities that share common mouse vendors all tested negative for C. difficile. Alternatively, environmental sources are a reservoir for C. difficile. For instance, shoe soles commonly test positive for C. difficile spores, and the spores can be carried from outdoors to the floors of households and hospitals.49,50 This is of particular interest, given that our isolated strain of C. difficile phylogenetically clustered closely with soil isolates based on whole genome sequence analysis. Further, mouse rooms within our lab’s vivarium that share common personnel and foot traffic patterns also had sentinel mice test positive. Last, NTCD-035 is distinct from all strains used in our laboratory. These pieces of evidence lead us to hypothesize that NTCD-035 is an external environmental isolate introduced into the animal colony though the precise source could not be confirmed in this study.
NTCD-035 in the microbiota of our animal colony was identified and successfully eliminated by modifying our mouse handling, cleaning and screening protocol. After initial screening of feces, C. difficile negative mice were transferred to a decontaminated clean mouse room (verified by negative swab test) to establish an endogenous C. difficile free mouse colony. In addition to conventional infection control measures in place throughout UPenn vivaria, such as sterile bedding, food, and use of personal protective equipment (PPE) as have been described by Best et al.,51 supplementary measures were established to prevent future contamination. These measure include use of a rapid-acting sporicidal cleaner, individual autoclaved water bottles for each cage, and disposable surface cover sheets when handling mice in a biosafety cabinet. These increased infection control measures collectively reduced the risk of contamination from C. difficile as evidenced by sentinel mice consistently testing negative following vivarium decontamination.
The use of non-toxigenic C. difficile spores to prevent recurrent C. difficile infection has shown promising results in the laboratory and clinical settings.52 Hamster studies support the idea that administration of non-toxigenic spores to patients most at risk (i.e. patients on antibiotics) will yield the greatest therapeutic benefit.53 In a phase II clinical trial, patients that experience either an initial episode or first recurrence of C. difficile associated disease were administered non-toxigenic C. difficile M3 spores and had a lower relapse rate than placebo administered patients.18 Furthermore, administration of non-toxigenic C. difficile had no major adverse effects when given to healthy subjects with or without antibiotics.54 Collectively, the data presented in this report support these clinical findings for the use of non-toxigenic C. difficile in prevention of pathogenic C. difficile infection and provides evidence for a probiotic based therapeutic approach that has the potential to prevent C. difficile associated disease in high-risk patient populations.
Methods
Mice
C57BL/6 mice were purchased from the Jackson Laboratory and maintained in cages of five mice per cage. All mouse strains used were derived from a C57BL/6 background. All mice were bred and maintained in sterile autoclaved cages, bedding and food under specific pathogen-free conditions and were kept on a grain-based diet (Labdiet 5010, 0001326) at the University of Pennsylvania. Technicians and researchers donned a gown, surgical mask, hairnet, gloves, and shoe covers prior to entry into the animal room and mice were exclusively handled under a laminar flow hood. For gnotobiotic experiments, germfree C57BL/6 mice bred and maintained in a sterile isolator were orally gavaged with C. difficile spores and were transferred to a sterile cage and kept in a biosafety cabinet for the duration of experiment. Mice were provided with autoclaved water ad libitum from water bottles. Sex and age-matched controls were used in all experiments according to institutional guidelines for animal care. All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Antibiotic pretreatment and C. difficile infection
Two to four month old mice were administered drinking water supplemented with neomycin (MilliporeSigma, N1142) (0.25 g/L), metronidazole (MilliporeSigma, M1547) (0.25 g/L), and vancomycin (Novaplus, NDC 0409–6510-01) (0.25 g/L) for seventy-two hours, and then replaced with normal water for the duration of the experiment. Forty-eight hours following cessation of antibiotic water, mice received 200 μg of clindamycin (Sigma, PHR1159) by intraperitoneal injection. Twenty-four hours later, mice received approximately 1,000 C. difficile spores (VPI10463 strain ATCC #43255, or NTCD-035) via oral gavage. After infection, mice were monitored for fecal C. difficile burden and for weight loss. For experiments with clindamycin only, 200ug of clindamycin was administered i.p. every twenty-four hours for three days and fecal pellets were taken twenty-four hours after the last dose of clindamycin.
C. difficile quantification
Fresh fecal pellets were collected from experimental mice, placed on ice, and transferred into an anaerobic chamber. Feces were resuspended in deoxygenated PBS, and ten-fold serial dilutions were plated on C. difficile selective plates consisting of BHI supplemented with yeast extract, taurocholate (Sigma, T4009), L-cysteine (Sigma, C7352), D-cycloserine (MilliporeSigma, C6880) and cefoxitin (MilliporeSigma, C4786) and grown at 37°C in an anaerobic chamber (Coylabs) overnight.55 Colony forming units were enumerated and data presented as CFUs per gram of feces.
Sanger sequencing
To determine the identity of the bacteria colonies, fecal pellets from contaminated mice were plated out on C. difficile selective plates. Individual colonies were restreaked, picked and grown in C. difficile selective broth. DNA was isolated after overnight broth culture using Qiagen DNeasy PowerSoil kit (Qiagen, 12888) according to manufacturer’s instructions. PCR amplification of the 16S rDNA gene was performed on purified bacterial DNA (primer sequences listed in supplemental table 2), and Sanger sequencing was performed on PCR amplified gene products. Sequence readouts were trimmed using SnapGene Viewer software with a nucleotide base quality score cutoff of 50. Trimmed sequence reads were queried using the NCBI BLAST database.
PCR ribotyping
PCR ribotyping was performed as previously described.56 DNA was isolated from either fecal pellets or from liquid culture using Qiagen DNeasy PowerSoil kit. Following DNA isolation, PCR ribotyping of the 16S gene region was performed (primer sequences listed in supplemental table 2), and PCR products were run on an agarose gel. Fluorescent PCR ribotyping was performed using a protocol as previously described.27,57
Whole genome sequencing
High molecular weight DNA was extracted from vegetative cells grown overnight in BHIS broth. DNA was isolated by phenol-chloroform extraction as previously described.58 Long-read libraries were prepared by using the SQK-RBK004 version of the Rapid Barcoding Kit (Oxford Nanopore Technologies, Oxford, UK) and sequenced on the Oxford Nanopore MinION using a FLO-MIN106 flow cell. Short-read libraries were prepared using the Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA) and were sequenced on the HiSeq 2500 using 2 × 125 bp chemistry. The short reads were quality controlled using the Sunbeam59 while the long reads were processed with our in-house pipeline, Nanoflow (https://github.com/zhaoc1/nanoflow). Nanoflow uses the Albacore command line tool to basecall and demultiplex the fast5 files to fastq files. Porechop and Filtlong were used to trim the adapter sequences and subsample to down to 500 Mbps of the highest quality reads (https://github.com/rrwick/Porechop, https://github.com/rrwick/filtlong). Hybrid assembly was performed via Unicycler in Nanoflow. Unicycler uses Spades to generate short read assembly and scaffolds them with a long read assembly generated from Canu and Nanopolish60,61 (https://github.com/jts/nanopolish). CheckM was used to check the quality of draft genomes.62 Phylogenetic analysis of C. difficile strains was generated with an in-house pipeline, coreSNPs (https://github.com/chrgu/coreSNPs). CoreSNPs uses Prokka to annotate the draft genomes and Roary to analyze the pangenome.63,64 Sequences of the core genes were extracted and concatenated to determine the number of single nucleotide variations (SNVs) between genomes via Hamming distances using an in-house R script. Phylogenetic trees and hierarchical clustering trees were generated by FastTree2.65 111 references genomes were gathered from the RefSeq database and 29 clinical isolates from Dr. Fredric Bushman lab’s database.
Toxin assay
In vitro toxin production was determined using a cell-based cytotoxicity assay modified from previously described protocols.66 C. difficile strains were incubated in deoxygenated BHIS broth and aliquots were taken at various time points after inoculation. Culture supernatant was spun down and filter sterilized (22um) prior to the assay. Vero cells were seeded in a 96 well plate and were incubated at 37°C for 24 hours. Culture supernatant or culture supernatant plus anti-toxin neutralizing antibody (TechLab, T1000) was added at tenfold serial dilutions and incubated for another 24 hours. Cells were then observed for cell rounding.
Sporulation assay
C. difficile sporulation was assessed by growing strains in CC-BHIS broth. Aliquots were taken at various timepoints post inoculation and incubated at 65°C for an hour to heat kill vegetative cells. Heat killed culture was then plated by tenfold serial dilutions on CC-BHIS-TA plates to measure spores by colony forming units.
Growth curves and secondary bile acid inhibition
C. difficile growth and secondary bile acid inhibition was assessed as previously described.37 Broth culture of each strain were grown overnight in BHI media supplemented with yeast extract and L-cystine. The following day, 1% v/v overnight starter culture was added to fresh BHIS media. For analysis of secondary bile acid inhibition, lithocholic acid (Sigma, L6250) or deoxycholic acid (Sigma, D6750) was added to the BHIS media. Deoxycholic acid was dissolved directly into BHIS media while lithocholic acid was first dissolved into sterile DMSO before being diluted 1:1000 into BHIS media. Culture was added to a 96 well plate in a one to one ratio with either BHIS media or BHIS containing secondary bile acids. The plate was incubated at 37°C under anaerobic conditions for 20 hours and growth was assessed by measuring the optical density at 600 nm (OD 600).
Antibiotic sensitivity assay
Assay was modified from previously described protocol.67 An individual colony of each C. difficile strain was grown overnight in BHIS. The culture was diluted 1:1000 and allowed to grow to mid-log phase. This culture was then diluted 1:1000 and plated 1:1 in test BHIS broth plus or minus antibiotics ranging in concentration from 1 mg/mL to 1 μg/mL desired concentration. Plate was incubated overnight at 37°C for 24 hours. Growth was measured on a spectrometer plate reader (OD600).
Facility and mouse screening for C. difficile
For facility screening, the animal changing hood, floor, automated cage water system spigots, and cage racks surfaces were swabbed with a sterile cotton swab, brought into anaerobic conditions, and streaked on C. difficile selective plates. Plates were observed after 24 hours to identify growth of C. difficile. For mouse screening, fresh fecal pellets were collected, placed on ice, and transferred into an anaerobic chamber. Fecal pellets were resuspended in deoxygenated C. difficile selective broth (CCBHISTA) to enrich for C. difficile. After overnight culture, tenfold dilutions were plated on C. difficile selective agar plates to identify outgrowth of C. difficile.
Decontamination of the vivarium
After identification of a contaminating C. difficile strain, a decontamination protocol of the mouse room was implemented. First, mice were screened for endogenous C. difficile in their feces, as described above. Following a negative result, cages were transferred into a separate empty animal room. All surfaces in the room were disinfected with 10% sodium hypochlorite. The cage racks in the new room were autoclaved and the automated water system spigots removed and replaced with individually autoclaved water bottles for each cage. Use of sporicidal cleaner (Clorox Fuzion Cleaner) was implemented for cleaning the biosafety cabinet used to change mouse cages and cart surfaces and wheels brought into the room. Finally, new mouse handling protocols included placing disposable absorbent sheets (Thomas Scientific, 1157M64) on the metal surface of the biosafety cabinet when opening the cages in the hood to reduce the chance of cross contamination between users.
Supplementary Material
Acknowledgments
We thank members of the Abt lab for helpful discussion and critical reading of the manuscript. We thank the PennCHOP Microbiome Program for whole genome sequencing. We thank Dymtro Kobuley of the Penn Gnotobiotic Core Facility with assistance in procuring germfree mice. We thank Dr. Fredric Bushman for thoughtful comments on the manuscript and for generously sharing his whole genome sequence database of C. difficile clinical isolates.
Funding Statement
This work was supported by the NIH-NIAID under Grant R00 AI125786 and a PennCHOP Microbiome Pilot Grant
Disclosure of Potential Conflicts of Interest
The authors report no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, et al. Burden of clostridium difficile infection in the United States. N Engl J Med. 2015;372(9):825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (U.S.) . Antibiotic Resistance Threats in the United States, 2019. Centers for Disease Control and Prevention (U.S.); 2019. doi: 10.15620/cdc:82532 [DOI]
- 3.Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, Wilson LE, Holzbauer SM, Phipps EC, Dumyati GK, et al. Trends in U.S. burden of clostridioides difficile infection and outcomes. N Engl J Med. 2020;382(14):1320–1330. doi: 10.1056/NEJMoa1910215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seekatz AM, Young VB.. Clostridium difficile and the microbiota. J Clin Invest. 2014;124(10):4182–4189. doi: 10.1172/JCI72336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7(7):526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 6.Chandrasekaran R, Lacy DB. The role of toxins in clostridium difficile infection. FEMS Microbiol Rev. 2017;41(6):723–750. doi: 10.1093/femsre/fux048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter GP, Chakravorty A, Nguyen TAP, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, et al. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during clostridium difficile infections. mBio. 2015;6(3):3. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-streiber C. Definition of the single integration site of the pathogenicity locus in clostridium difficile. Gene. 1996;181(1–2):29–38. doi: 10.1016/s0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 9.Monot M, Eckert C, Lemire A, Hamiot A, Dubois T, Tessier C, Dumoulard B, Hamel B, Petit A, Lalande V, et al. Clostridium difficile: new insights into the evolution of the pathogenicity locus. Sci Rep. 2015;5(1):15023. doi: 10.1038/srep15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH, et al. Evolutionary history of the clostridium difficile Pathogenicity Locus. Genome Biol Evol. 2014;6(1):36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natarajan M, Walk ST, Young VB, Aronoff DM. A clinical and epidemiological review of non-toxigenic Clostridium difficile. Anaerobe. 2013;22:1–5. doi: 10.1016/j.anaerobe.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato H, Kita H, Karasawa T, Maegawa T, Koino Y, Takakuwa H, Saikai T, Kobayashi K, Yamagishi T, Nakamura S, et al. Colonisation and transmission of clostridium difficile in healthy individuals examined by PCR ribotyping and pulsed-field gel electrophoresis. J Med Microbiol. 2001;50(8):720–727. doi: 10.1099/0022-1317-50-8-720. [DOI] [PubMed] [Google Scholar]
- 13.Miyajima F, Roberts P, Swale A, Price V, Jones M, Horan M, Beeching N, Brazier J, Parry C, Pendleton N, et al. Characterisation and carriage ratio of clostridium difficile strains isolated from a community-dwelling elderly population in the United Kingdom. Plos One. 2011;6(8):e22804. doi: 10.1371/journal.pone.0022804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. Primary symptomless colonisation by clostridium difficile and decreased risk of subsequent diarrhoea. Lancet Lond Engl. 1998;351(9103):633–636. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 15.Wilson KH, Sheagren JN. Antagonism of toxigenic clostridium difficile by nontoxigenic C. difficile. J Infect Dis. 1983;147(4):733–736. doi: 10.1093/infdis/147.4.733. [DOI] [PubMed] [Google Scholar]
- 16.Sambol SP, Merrigan MM, Tang JK, Johnson S, Gerding DN. Colonization for the prevention of clostridium difficile disease in hamsters. J Infect Dis. 2002;186(12):1781–1789. doi: 10.1086/345676. [DOI] [PubMed] [Google Scholar]
- 17.Nagaro KJ, Phillips ST, Cheknis AK, Sambol SP, Zukowski WE, Johnson S, Gerding DN. Nontoxigenic clostridium difficile protects hamsters against challenge with historic and epidemic strains of toxigenic BI/NAP1/027 C. difficile. Antimicrob Agents Chemother. 2013;57(11):5266–5270. doi: 10.1128/AAC.00580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerding DN, Meyer T, Lee C, et al. Administration of spores of nontoxigenic clostridium difficile strain M3 for prevention of recurrent C difficile infection: a randomized clinical trial. JAMA. 2015;313(17):1719–1727. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 19.Etienne-Mesmin L, Chassaing B, Adekunle O, Mattei LM, Bushman FD, Gewirtz AT. Toxin-positive clostridium difficile latently infect mouse colonies and protect against highly pathogenic C. difficile. Gut. 2018;67(5):860–871. doi: 10.1136/gutjnl-2016-313510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, et al. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77(9):3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, Kelly CP. A mouse model of clostridium difficile-associated disease. Gastroenterology. 2008;135(6):1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Abt MC, Lewis BB, Caballero S, Xiong H, Carter R, Sušac B, Ling L, Leiner I, Pamer E. Innate immune defenses mediated by two ILC subsets are critical for protection against acute clostridium difficile infection. Cell Host Microbe. 2015;18(1):27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit J-C. Development of a new PCR-ribotyping method for clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175(2):261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- 24.Rotimi VO, Jamal WY, Mokaddas EM, Brazier JS, Johny M, Duerden BI. Prevalent PCR ribotypes of clinical and environmental strains of Clostridium difficile isolated from intensive-therapy unit patients in Kuwait. J Med Microbiol. 2003;52(Pt 8):705–709. doi: 10.1099/jmm.0.05207-0. [DOI] [PubMed] [Google Scholar]
- 25.Best EL, Parnell P, Thirkell G, Verity P, Copland M, Else P, Denton M, Hobson RP, Wilcox MH. Effectiveness of deep cleaning followed by hydrogen peroxide decontamination during high clostridium difficile infection incidence. J Hosp Infect. 2014;87(1):25–33. doi: 10.1016/j.jhin.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Al-Thani AA, Hamdi WS, Al-Ansari NA, Doiphode SH. Polymerase chain reaction ribotyping of clostridium difficileisolates in Qatar: a hospital-based study. BMC Infect Dis. 2014;14(1):502. doi: 10.1186/1471-2334-14-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinson JNV, Broadaway S, Lohman E, Johnson C, Alam MJ, Khaleduzzaman M, Garey KW, Schlackman J, Young VB, Santhosh K, et al. Evaluation of portability and cost of a fluorescent PCR ribotyping protocol for clostridium difficile epidemiology. J Clin Microbiol. 2015;53(4):1192–1197. doi: 10.1128/JCM.03591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terhes G. Distribution of Clostridium difficile PCR ribotypes in regions of Hungary. J Med Microbiol. 2006;55(3):279–282. doi: 10.1099/jmm.0.46141-0. [DOI] [PubMed] [Google Scholar]
- 29.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bushman FD, Conrad M, Ren Y, et al. Multi-omic analysis of the interaction between clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe. 2020;doi: 10.1016/j.chom.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rashid SRJ, Clokie MRJ, Millard AD. Draft genome sequences of three novel clostridium isolates from Northern Iraq. Genome Announc. 2016;4(1):1. doi: 10.1128/genomeA.00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashid SJ, Barylski J, Hargreaves KR, Millard AA, Vinner GK, Clokie MRJ. Two novel myoviruses from the north of iraq reveal insights into clostridium difficile phage diversity and biology. Viruses. 2016;8(11):11. doi: 10.3390/v8110310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramírez-Vargas G, López-Ureña D, Badilla A, Orozco-Aguilar J, Murillo T, Rojas P, Riedel T, Overmann J, González G, Chaves-Olarte E, et al. Novel clade C-I clostridium difficile strains escape diagnostic tests, differ in pathogenicity potential and carry toxins on extrachromosomal elements. Sci Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-32390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Åkerlund T, Persson I, Unemo M, et al. Increased sporulation rate of epidemic clostridium difficile type 027/NAP1. J Clin Microbiol. 2008;46(4):1530–1533. doi: 10.1128/JCM.01964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winston JA, Theriot CM. Impact of microbial derived secondary bile acids on colonization resistance against clostridium difficile in the gastrointestinal tract. Anaerobe. 2016;41:44–50. doi: 10.1016/j.anaerobe.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theriot CM, Koenigsknecht MJ, Carlson PE, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to clostridium difficile infection. Nat Commun. 2014;5(1):1–10. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis BB, Carter RA, Pamer EG. Bile acid sensitivity and in vivo virulence of clinical clostridium difficile isolates. Anaerobe. 2016;41:32–36. doi: 10.1016/j.anaerobe.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for clostridium difficile spores. J Bacteriol. 2008;190(7):2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francis MB, Allen CA, Sorg JA. Muricholic acids inhibit clostridium difficile spore germination and growth. Plos One. 2013;8(9):e73653. doi: 10.1371/journal.pone.0073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu D, Sorg JA, Sun X. Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front Cell Infect Microbiol. 2018;8:29. doi: 10.3389/fcimb.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thanissery R, Winston JA, Theriot CM. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe. 2017;45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family lachnospiraceae. Infect Immun. 2012;80(11):3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasegawa M, Kamada N, Jiao Y, Liu MZ, Núñez G, Inohara N. Protective role of commensals against clostridium difficile infection via an IL-1β–mediated positive-feedback loop. J Immunol. 2012;189(6):3085–3091. doi: 10.4049/jimmunol.1200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frisbee AL, Saleh MM, Young MK, et al. IL-33 drives group 2 innate lymphoid cell-mediated protection during clostridium difficile infection. Nat Commun. 2019;10(1):1–13. doi: 10.1038/s41467-019-10733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501(7467):426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Litvak Y, Bäumler AJ. The founder hypothesis: A basis for microbiota resistance, diversity in taxa carriage, and colonization resistance against pathogens. PLoS Pathog. 2019;15:2. doi: 10.1371/journal.ppat.1007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borriello SP, Barclay FE. Protection of hamsters against clostridium difficile ileocaecitis by prior colonisation with non-pathogenic strains. J Med Microbiol. 1985;19(3):339–350. doi: 10.1099/00222615-19-3-339. [DOI] [PubMed] [Google Scholar]
- 48.Péchiné S, Bruxelle JF, Janoir C, Collignon A. Targeting clostridium difficile surface components to develop immunotherapeutic strategies against clostridium difficile infection. Front Microbiol. 2018:9. doi: 10.3389/fmicb.2018.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alam MJ, Anu A, Walk ST, Garey KW. Investigation of potentially pathogenic clostridium difficile contamination in household environs. Anaerobe. 2014;27:31–33. doi: 10.1016/j.anaerobe.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Alam MJ, Walk ST, Endres BT, Basseres E, Khaleduzzaman M, Amadio J, Musick WL, Christensen JL, Kuo J, Atmar RL, et al. Community environmental contamination of toxigenic clostridium difficile. Open Forum Infect Dis. 2017;4(1):1. doi: 10.1093/ofid/ofx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Best EL, Freeman J, Wilcox MH. Models for the study of Clostridium difficile infection. Gut Microbes. 2012;3(2):145–167. doi: 10.4161/gmic.19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerding DN, Sambol SP, Johnson S. Non-toxigenic clostridioides (formerly clostridium) difficile for prevention of c. difficile infection: from bench to bedside back to bench and back to bedside. Front Microbiol. 2018:9. doi: 10.3389/fmicb.2018.01700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merrigan MM, Sambol SP, Johnson S, Gerding DN. New approach to the management of clostridium difficile infection: colonisation with non-toxigenic C. difficile during daily ampicillin or ceftriaxone administration. Int J Antimicrob Agents. 2009;33:S46–S50. doi: 10.1016/S0924-8579(09)70017-2. [DOI] [PubMed] [Google Scholar]
- 54.Villano SA, Seiberling M, Tatarowicz W, Monnot-Chase E, Gerding DN. Evaluation of an oral suspension of VP20621, spores of nontoxigenic clostridium difficile strain M3, in healthy subjects. Antimicrob Agents Chemother. 2012;56(10):5224–5229. doi: 10.1128/AAC.00913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorg JA, Dineen SS. Laboratory maintenance of clostridium difficile. Curr Protoc Microbiol. 2009;12(1):9A.1.1–9A.1.10. doi: 10.1002/9780471729259.mc09a01s12. [DOI] [PubMed] [Google Scholar]
- 56.Janezic S. Direct PCR-ribotyping of clostridium difficile. Methods Mol Biol Clifton NJ. 2016;1476:15–21. doi: 10.1007/978-1-4939-6361-4_2. [DOI] [PubMed] [Google Scholar]
- 57.Walk ST, Micic D, Jain R, Lo ES, Trivedi I, Liu EW, Almassalha LM, Ewing SA, Ring C, Galecki AT, et al. Clostridium difficile ribotype does not predict severe infection. Clin Infect Dis Off Publ Infect Dis Soc Am. 2012;55(12):1661–1668. doi: 10.1093/cid/cis786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bouillaut L, McBride SM, Sorg JA. Genetic Manipulation of Clostridium difficile. Curr Protoc Microbiol. 2011;0 9:Unit–9A.2. doi: 10.1002/9780471729259.mc09a02s20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke EL, Taylor LJ, Zhao C, Connell A, Lee -J-J, Fett B, Bushman FD, Bittinger K. Sunbeam: an extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome. 2019;7(1):46. doi: 10.1186/s40168-019-0658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prokka TS. Rapid Prokaryotic Genome Annotation. Bioinf (Oxford, England). doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 64.Aj P, Ca C, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinf (Oxford, England). doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mn P, Ps D, Ap A. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jarchum I, Liu M, Lipuma L, Pamer EG. Toll-like receptor 5 stimulation protects mice from acute clostridium difficile colitis. Infect Immun. 2011;79(4):1498–1503. doi: 10.1128/IAI.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abt MC, Buffie CG, Sušac B, Becattini S, Carter RA, Leiner I, Keith JW, Artis D, Osborne LC, Pamer EG, et al. TLR-7 activation enhances IL-22–mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med. 2016;8(327):327ra25. doi: 10.1126/scitranslmed.aad6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


