Abstract
Causal inference is one of the challenges in epidemiologic studies. Gynecologic diseases have been reported to have association with obesity, however the causality remained controversial except for uterine endometrial cancer. We conducted two‐sample Mendelian randomization (MR) analysis using the large‐scale genome‐wide association study (GWAS) results of gynecologic diseases and body mass index (BMI) in the Japanese population to assess causal effect of BMI on gynecologic diseases. We first conducted GWAS of ovarian cancer, uterine endometrial cancer, uterine cervical cancer, endometriosis, and uterine fibroid (n = 647, 909, 538, 5236, and 645 cases, respectively, and 39 556 shared female controls), and BMI (81 610 males and non‐overlapping 23 924 females). We then applied two‐sample MR using 74 BMI‐associated variants as instrumental variables. We observed significant causal effect of increased BMI on uterine endometrial cancer (β = 0.735, P = .0010 in inverse variance‐weighted analysis), which is concordant with results of European studies. Causal effect of obesity was not apparent in the other gynecologic diseases tested. Our MR analyses provided strong evidence of the causal role of obesity in gynecologic diseases etiology, and suggested a possible preventive effect of intervention for obesity.
Keywords: BMI, GWAS, gynecologic diseases, Mendelian randomization, obesity
We assessed whether obesity is causally associated with gynecologic diseases or not. We performed a Mendelian randomization study and revealed that obesity assessed by measurement of body mass index is causally associated with uterine endometrial cancer in Japanese. Intervention to reduce body weight may prevent uterine endometrial cancer.

1. INTRODUCTION
Ovarian cancer (OC), uterine endometrial cancer (UEC), uterine cervical cancer (UCC), uterine fibroma (UF), and endometriosis are all common proliferative diseases arising from gynecologic organs. They are complex heterogeneous diseases, and both genetic and environmental factors contribute to their development. 1 Recent advancement in the field of genome‐wide association studies (GWAS) has identified population‐specific significantly associated loci associated with gynecologic diseases. 2 As implicated in epidemiological studies, obesity may play an important role in the development of gynecologic diseases, especially in UEC. 1 , 3 , 4 Obesity is thought to increase the risk of UCC by decreased detection of precancerous lesions. 5 Obesity is thought to affect UF through hormonal activity, proinflammatory effect, and hyperinsulinemia. 6 Obesity inversely affects endometriosis, which is thought to be due to hormonal differences between heavy and lean women. 7 However, their relationships remain largely controversial due to difficulty in epidemiologic studies in inferring causal associations.
Recently, novel approaches using genomic data to infer causality have been developed. 8 , 9 Mendelian randomization (MR) is an approach in which genetic variants serve as instrumental variables to estimate the causal effect of exposure phenotypes on outcome phenotypes of interest. The genetic variants are allocated randomly at conception in nature, similar to random allocation in randomized controlled trials, and generate variation in exposure phenotypes that should be unaffected by confounding. 8 Recent achievement in large‐scale GWAS 2 , 10 , 11 , 12 and development of advanced analytical methods of MR 8 enabled a robust estimation of causal effect, therefore MR is widely used for inferring causality and has an impact on clinical medicine. 8 , 9
Here, we conducted an MR analysis to estimate causality of obesity, assessed by measurement of body mass index (BMI), on the 5 gynecologic diseases in the Japanese population. Previous reports in Europeans have identified causal effect of obesity on UEC and several histological subtypes of OC, 13 , 14 , 15 , 16 however other gynecologic diseases have not been widely studied. Also, there has been no study performed in the Japanese population. Therefore, we performed MR analyses using all available gynecologic disease GWAS data in the Japanese biobank, and aimed to provide stronger evidence.
2. MATERIALS AND METHODS
2.1. Gynecologic and BMI GWAS in Japanese
Gynecologic disease GWAS was conducted as previously described. 2 Briefly, we enrolled OC, UEC, UCC, UF, and endometriosis cases, and shared female controls without gynecologic, malignant, and MHC‐related diseases from the BioBank Japan Project (BBJ), then performed association analyses under the generalized linear model. 2 BMI GWAS data were adopted from a previous report, 17 and we re‐conducted the analysis using genetic data of non‐overlapping BBJ individuals without gynecologic diseases under the generalized linear model. These studies were established, published, and was the largest GWAS among Japanese. In the original BMI GWAS, associations of the lead variants were replicated in multiple biobank cohorts, suggesting their robustness. BMI was measured by weight (kg), divided by squared height (m2), and not adjusted by waist‐hip ratio. Both males and females were included in the BMI GWAS to ensure a valid estimation of BMI‐associated variants by increasing the sample size, which then avoids bias in MR analysis. Among the lead variants previously reported in the Japanese BMI GWAS, 17 we selected ones available in both BMI and gynecologic GWAS. We confirmed that the association of the BMI‐related variants was strong enough (P‐value < 5.0 × 10−8) to be utilized as instrumental variables in MR analysis. When the lead variants were not available in gynecologic GWAS, proxy variants in linkage disequilibrium (LD) (r 2 ≥ 0.5 in east Asian subjects of 1000 Genome Project phase 3v5) were selected. This study was approved by the ethical committee of Osaka University Graduate School of Medicine. Genotype data are deposited at National Bioscience Database Center, with the accession number of hum0014 at https://ddbj.nig.ac.jp/jga/viewer/view/study/JGAS00000000114.
2.2. MR analysis
We adopted the two‐sample MR method implemented in the package twosampleMR (http://www.mrbase.org/) of R statistical software (v.3.5.1). We applied 2 types of MR analyses, inverse variance‐weighted (IVW) and MR‐Egger methods. We estimated the effect of BMI as an exposure on gynecologic diseases as outcomes, by adopting the variants significantly associated (P < 5.0 × 10−8) with BMI as instrumental variables. We applied Bonferroni correction to assess the significance of association, both in the discovery and replication analyses; significance threshold was calculated as a type 1 error rate α = .05 divided by the number of methods and phenotypes tested in the analysis. As the sensitivity test, we applied a heterogeneity test, leave‐one‐out analysis, and funnel plots implemented in the twosamplemr package. We also applied the Mendelian randomization pleiotropy residual sum and outlier (MR‐PRESSO) method implemented in the R package mr‐presso and assessed pleiotropic effect.
2.3. Replication study in UK Biobank Europeans
We applied the same methods to the publicly available summary statistics of the European population. We obtained the gynecologic disease GWAS summary statistics from the UK Biobank (UKBB) (http://www.nealelab.is/uk‐biobank, Phenotype Codes: C56, C54, C53, D25, and N80). The summary statistics of the BMI GWAS conducted among the subjects not overlapping with those in UKBB were obtained from the GIANT consortium (https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files). 18
3. RESULTS
3.1. Gynecologic and BMI GWAS in Japanese
The numbers of the subjects for each gynecologic disease are listed in Table 1. We enrolled 647, 909, 538, 5236, and 645 cases of OC, UEC, UCC, UF, and endometriosis, respectively, and 39 556 shared female controls. 2 Whole genome sequencing‐based genotype imputation yielded more than 7 000 000 variants available for MR analysis. 19 For BMI, independent 105 534 subjects were included in the analysis. These GWAS were the largest Japanese studies available to date, which would provide the highest statistical power to estimate causal effect. Several variants were missing in the GWAS due to extremely low allele frequency among the case subjects, we thus removed these variants from the instrumental variables. As a result, we incorporated the 73 lead variants for OC and endometriosis, and 74 lead variants for UEC, UCC and UF, including proxy variants in LD.
TABLE 1.
Effect size estimates by each method
| Disease | No. case | No. control | IVW | MR‐Egger | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | OR (95% CI) | P | Beta | SE | OR (95% CI) | P | Intercept (95% CI) | P | |||
| Ovarian cancer | 647 | 39 556 | 0.409 | 0.265 | 1.12 (0.97 to 1.29) | .12 | 1.093 | 0.702 | 1.35 (0.93 to 1.96) | .12 | −0.021 (−0.059 to 0.018) | .30 |
| Uterine endometrial cancer | 909 | 39 556 | 0.735 | 0.223 | 1.22 (1.08 to 1.38) | .0010 | 1.625 | 0.583 | 1.56 (1.14 to 2.12) | .0068 | −0.027 (−0.059 to 0.0050) | .10 |
| Uterine cervical cancer | 538 | 39 556 | 0.062 | 0.290 | 1.02 (0.87 to 1.19) | .83 | 0.157 | 0.759 | 1.04 (0.70 to 1.56) | .84 | −0.0029 (−0.045 to 0.039) | .89 |
| Uterine fibroid | 5236 | 39 556 | 0.081 | 0.131 | 1.02 (0.95 to 1.10) | .54 | 0.054 | 0.345 | 1.01 (0.84 to 1.22) | .88 | 0.00080 (−0.018 to 0.020) | .93 |
| Endometriosis | 645 | 39 556 | −0.062 | 0.286 | 0.98 (0.84 to 1.15) | .83 | −0.262 | 0.767 | 0.93 (0.62 to 1.40) | .73 | 0.0060 (−0.036 to 0.048) | .78 |
Sample sizes for each GWAS and causal effect estimates of BMI on each gynecologic disease are shown.
Beta and standard error (SE) is based on per standard deviation (SD) increase of BMI, and OR is based on per 1 kg/m2 increase of BMI.
BMI GWAS was conducted among the independent 105 534 subjects with standardized phenotypic values.
BMI, body mass index; CI, confidence interval; GWAS, genome‐wide association study; IVW, inverse variance‐weighted; OR, odds ratio.
3.2. MR analysis
MR analysis results are shown in Table 1, Figure 1, and Figure S1. Estimated causal effect size (β) per standard deviation (SD) unit increase of BMI was 0.409, 0.735, 0.062, 0.081, and −0.062 respectively for OC, UEC, UCC, UF, and endometriosis in IVW (P = .12, .0010, .83, .54, and .83, respectively), and 1.093, 1.625, 0.157, 0.054, and −0.262, respectively in MR‐Egger (P = .12, .0068, .84, .88, and .73, respectively). While the MR‐Egger analysis result was marginally significant, the BMI effect on UEC in IVW analysis remained significant (P = .0010) after Bonferroni's correction considering the number of phenotypes and methods (P < .05/10 = .005). The results of heterogeneity tests, including leave‐one‐out analysis, funnel plots, and mr‐presso analysis, were not significant (results not shown), which led us to the conclusion that the potential bias conferred by the pleiotropic variants was negligible in our analysis. We did not inversely conduct MR analysis assessing UEC effect on obesity, as no variants were identified in the UEC GWAS.
FIGURE 1.
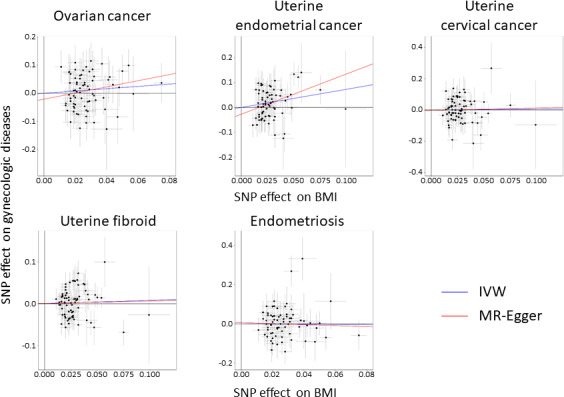
Scatter plots of Mendelian randomization (MR) tests assessing the effect of body mass index (BMI) on each gynecologic disease. Each dot represents effect sizes of each single nucleotide polymorphism (SNP) on BMI (x‐axis) and gynecologic diseases (y‐axis), and regression slopes show the estimated causal effect of BMI on gynecologic diseases. For all plots, inverse variance‐weighted (IVW) results are shown in blue and MR‐Egger regression results are shown in red
3.3. Replication study in UKBB Europeans
The number of cases and controls in the gynecologic GWAS was not specified, but 194 174 subjects in total were analyzed. The number of subjects in BMI GWAS was 449 889. In total, 131 lead variants associated with BMI were also available in gynecologic GWAS. MR analysis results are shown in Table S1, Figure S2, and Figure S3. BMI effect on UEC was nominally significant with directional concordance with that in Japanese (β = 4.04 × 10−3, P = .010 in IVW analysis).
4. DISCUSSION
In this study, we utilized the large‐scale GWAS results of gynecologic diseases in the Japanese population and demonstrated that obesity had a causal effect on UEC that was replicated with the same directional effect in UKBB Europeans.
Epidemiologic studies have suggested that obesity affects the likelihood of gynecologic diseases. 1 , 3 , 4 MR‐Egger analysis in European population did not reach statistical significance, possibly due to the conservative nature of the method. However, observations that IVW results both in Japanese and European populations were concordant with conventional epidemiological studies should support previous epidemiological findings. 1 , 3 , 4 Direct comparison of the effect size between current study and the previous MR and epidemiological studies is difficult because the studies were designed differently and the effect sizes were described in different scales and units. 3 , 15 , 20 , 21 The direction of effects was concordant among multiple MR and epidemiological studies, suggesting that our MR analysis provided further evidence of a causal effect of obesity on UEC. These observations suggested that intervention for obesity itself could be a potential prevention, leading to a better management strategy for this malignancy, as well as identifying novel drug targets.
Although the direction of BMI effect on OC was concordant with that in the previous European studies, 14 , 16 our results failed to reach statistical significance (P = .12 in IVW analysis). Furthermore, the results of our analysis among the European population were directionally discordant between IVW and MR‐Egger methods and also failed to reach statistical significance. The discrepant results were probably caused by pleiotropic effects, 1 variant with outstanding effect, and/or the difference in the proportion of histologic subtypes in the cohorts. However, all of the heterogeneity test, leave‐one‐out analysis, funnel plots, and MR‐PRESSO failed to show a significant pleiotropic effect (data not shown), leading us to conclude that the pleiotropic effect might exist but be negligible in our analysis. Since MR‐Egger uses two‐step least squares, the result might have been strongly affected by outstanding effect. Also, it is well known that high‐grade serous subtype is prevalent in Europeans, but clear cell carcinoma is also common in Japanese. Either way, it is difficult to obtain a definitive conclusion from the current study. The association of obesity and OC still remains controversial, and further studies are thus warranted.
Conversely, we reported the first MR analysis of other gynecologic diseases (UCC, UF, and endometriosis), which suggested a non‐causal association of BMI with them. The discrepant results shown in MR‐Egger analysis among Europeans might suggest population specificity and/or pleiotropic effect. We considered that from our analysis alone it would be difficult to make conclusions. These associations are also controversial in epidemiologic studies, however, our observation suggested causal effect of obesity on these diseases is limited. Further application of MR approaches on a wider range of risk factors, as well as validation studies in different cohort, should be warranted.
It is well known that age at menopause is related to gynecologic malignancies and obesity. We thought that it is possible that menopausal status might affect our results. Further study is needed in the future including, for example, stratified analysis based on menopausal status, multivariable MR analysis, and/or interaction‐based MR analysis.
Also, discrepant results among different gynecologic diseases might be attributed to menopausal status, obesity, estrogenic hormonal effect, and the immune system. Further investigation is warranted.
Although our study utilized the largest GWAS in the Japanese population, possible limitations of our study include the relatively small number of case subjects in the gynecologic GWAS, lack of replication and validation studies especially for non‐UEC gynecologic diseases, and consideration on age at menopause and hormonal effect. However, we observed a causal relationship of obesity with UEC. To explore reverse causality, as well as to assess the robustness by replication and validation, and to incorporate considerations for other variables, further investigation on different cohorts with larger sample sizes and with detailed phenotypic information is warranted.
5. CONCLUSIONS
In summary, our MR analysis identified that obesity had a significant causal effect on UEC, which was observed both in Japanese and European populations. Our results provided strong evidence of the causal association and suggested that intervention for obesity could be a potential management strategy against UEC.
DISCLOSURE
Authors declare that there is no conflict of interest.
Supporting information
Supinfo
ACKNOWLEDGMENTS
BBJ was supported by the Tailor‐Made Medical Treatment Program (the BioBank Japan Project) of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and the Japan Agency for Medical Research and Development (AMED). We thank Dr. Yukihide Momozawa at the RIKEN Center for Integrative Medical Sciences for supporting the study. This study was supported by the Bioinformatics Initiative of Osaka University Graduate School of Medicine. YO was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (19H01021, 20K21834), and AMED (JP20km0405211, JP20ek0109413, JP20ek0410075, JP20gm4010006, and 20km0405217).
Masuda T, Ogawa K, Kamatani Y, Murakami Y, Kimura T, Okada Y. A Mendelian randomization study identified obesity as a causal risk factor of uterine endometrial cancer in Japanese. Cancer Sci. 2020;111:4646–4651. 10.1111/cas.14667
This work was carried out at Osaka University Graduate School of Medicine, 2‐2 Yamadaoka, Suita, Osaka 565‐0871, Japan.
REFERENCES
- 1. Nagai K, Hayashi K, Yasui T, et al. Disease history and risk of comorbidity in women's life course: a comprehensive analysis of the Japan Nurses' Health Study baseline survey. BMJ Open. 2015;5(3):e006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Masuda T, Low SK, Akiyama M, et al. GWAS of five gynecologic diseases and cross‐trait analysis in Japanese. Eur J Hum Genet. 2019;28(1):95‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhaskaran K, Douglas I, Forbes H, dos‐Santos‐Silva I, Leon DA, Smeeth L. Body‐mass index and risk of 22 specific cancers: a population‐based cohort study of 5·24 million UK adults. Lancet. 2014;384(9945):755‐765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body‐mass index and incidence of cancer: a systematic review and meta‐analysis of prospective observational studies. Lancet. 2008;371(9612):569‐578. [DOI] [PubMed] [Google Scholar]
- 5. Clarke MA, Fetterman B, Cheung LC, et al. Epidemiologic evidence that excess body weight increases risk of cervical cancer by decreased detection of precancer. J Clin Oncol. 2018;36(12):1184‐1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pavone D, Clemenza S, Sorbi F, Fambrini M, Petraglia F. Epidemiology and risk factors of uterine fibroids. Best Pract Res Clin Obstet Gynaecol. 2018;46:3‐11. [DOI] [PubMed] [Google Scholar]
- 7. Parveen P, Pinar O, Endometriosis KLT. Epidemiology, diagnosis and clinical management. Physiol Behav. 2017;6(1):34‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bowden J, Smith GD, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pingault JB, O'Reilly PF, Schoeler T, Ploubidis GB, Rijsdijk F, Dudbridge F. Using genetic data to strengthen causal inference in observational research. Nat Rev Genet. 2018;19(9):566‐580. [DOI] [PubMed] [Google Scholar]
- 10. Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Okada Y, Momozawa Y, Sakaue S, et al. Deep whole‐genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat Commun. 2018;9(1):1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirata J, Hosomichi K, Sakaue S, et al. Genetic and phenotypic landscape of the major histocompatibilty complex region in the Japanese population. Nat Genet. 2019;51(3):470–480. [DOI] [PubMed] [Google Scholar]
- 13. O'Mara TA, Glubb DM, Amant F, et al. Identification of nine new susceptibility loci for endometrial cancer. Nat Commun. 2018;9(1):3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao C, Patel CJ, Michailidou K, et al. Mendelian randomization study of adiposity‐related traits and risk of breast, ovarian, prostate, lung and colorectal cancer. Int J Epidemiol. 2016;45(3):896‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Painter JN, O'Mara TA, Marquart L, et al. Genetic risk score mendelian randomization shows that obesity measured as body mass index, but not waist:hip ratio, is causal for endometrial cancer. Cancer Epidemiol Biomarkers Prev. 2016;25(11):1503‐1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dixon SC, Nagle CM, Thrift AP, et al. Adult body mass index and risk of ovarian cancer by subtype: a Mendelian randomization study. Int J Epidemiol. 2016;45(3):884‐895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Akiyama M, Okada Y, Kanai M, et al. Genome‐wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet. 2017;49(10):1458‐1467. [DOI] [PubMed] [Google Scholar]
- 18. Turcot V, Lu Y, Highland HM, et al. Protein‐altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50(1):26‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akiyama M, Ishigaki K, Sakaue S, et al. Characterizing rare and low‐frequency height‐associated variants in the Japanese population. Nat Commun. 2019;10(1):4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delahanty RJ, Beeghly‐Fadiel A, Xiang YB, et al. Association of obesity‐related genetic variants with endometrial cancer risk: a report from the shanghai endometrial cancer genetics study. Am J Epidemiol. 2011;174(10):1115‐1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lauby‐Secretan B, Ph D, Scoccianti C, Ph D, Loomis D, Ph D. Body fatness and cancer — viewpoint of the IARC Working Group. N Engl J Med. 2016;375(8):794‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supinfo


