Abstract
An increasing number of studies have clarified the functional roles of RNA-binding proteins (RBPs) in driving post-transcriptional mechanisms of cancer progression. In this study, we integrated data from the RBP database and Gene Expression Omnibus (GEO) data with RNA sequencing (RNA-seq) data from 10 ovarian cancer tissues and 8 normal ovarian tissues and identified an RBP, CUGBP- and ETR-3-like family 2 (CELF2). We found that CELF2 expression was downregulated in ovarian cancer and positively correlated with the overall survival (OS) and progression-free survival (PFS) of patients with ovarian cancer. Altered CELF2 expression led to changes in the proliferation, migration, and invasion of ovarian cancer cells in vitro and in vivo. CELF2 expression increased the stability of its target, FAM198B, by binding to AU/U-rich elements (AREs) in the 3′ untranslated region (3′ UTR). FAM198B knockdown restored the CELF2-mediated suppression of proliferation and migration. We also found that CELF2/FAM198B may repress ovarian cancer progression by inhibiting the mitogen-activated protein kinase/extracellular-regulated protein kinase (MAPK/ERK) signaling pathway. Finally, a curcumin-induced increase in CELF2 expression resulted in increased ovarian cancer cell sensitivity to cisplatin. Our study elucidated a novel mechanism by which the CELF2/FAM198B axis regulates proliferation and metastasis in ovarian cancer, providing novel, potential therapeutic targets for ovarian cancer.
Graphical Abstract
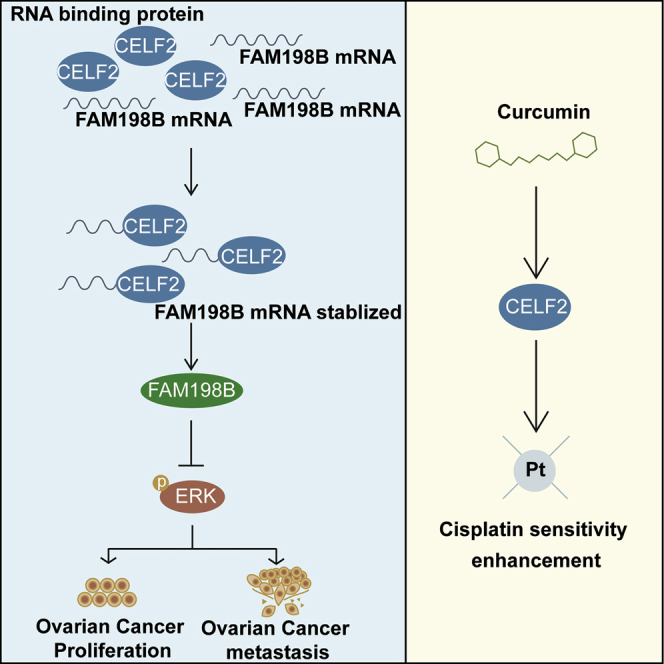
This study identified a key prognosis-related RBP, named CELF2, in ovarian cancer, which functions as a tumor suppressor. CELF2 stabilizes the FAM198B mRNA by binding to AU/U-rich elements within the 3′ untranslated region. Moreover, CELF2/FAM198B may suppress ovarian cancer progression via the MAPK/ERK signaling pathway.
Introduction
Over the past few decades, the incidence of ovarian cancer has increased rapidly, and this disease poses a major threat to women’s health worldwide.1 Patients are usually diagnosed with ovarian cancer at a late stage, and the development of chemoresistant forms will occur in over 70% of patients after initial treatment with cisplatin-based chemotherapy for an extended period of time, resulting in the highest mortality rate among gynecological tumors.2 Therefore, studies aiming to explore the underlying mechanisms involved in the tumorigenesis of ovarian cancer and therapeutic targets to overcome chemoresistance are essential. Post-transcriptional regulatory processes mediated by trans-acting factors, such as RNA-binding proteins (RBPs), microRNAs (miRNAs), and long noncoding RNAs (lncRNAs) play important roles in these oncogenic fitness traits,3, 4, 5 among which the functional roles of RBPs are less well defined.
RBPs are proteins that regulate the abundance and functions of RNA transcripts at multiple post-transcriptional levels, including RNA splicing, localization, polyadenylation, modification, stability, and translation.6,7 Because of these critical roles, functional defects in these proteins have been increasingly identified as major regulators of cancer. For ovarian cancer, several studies have investigated the regulation of several cellular processes by pivotal RBPs. For instance, a recent report illustrated a novel post-transcriptional network that links ovarian cancer progression and immunomodulation within the tumor microenvironment through RBP SORBS2-mediated transcript stabilization.8 More recently, studies from other groups have suggested that the RBP LIN28A, a key factor that maintains the pluripotency of stem cells, regulates the biological behaviors of ovarian cancer cells through ROCK2.9 However, only a small proportion of them have been well elucidated.
In the present study, we performed an integrated transcriptomic analysis to identify key prognosis-related RBPs in ovarian cancer and discovered an RBP named CUGBP- and ETR-3-like family 2 (CELF2), which can function as a tumor suppressor. CELF2 is associated with the proliferation, migration, and metastasis of ovarian cancer cells in vivo and in vitro. More interestingly, we found that CELF2 stabilizes FAM198B mRNAs by binding to its AU/U-rich elements (AREs) within the 3′ untranslated region (3′ UTR). Moreover, CELF2/FAM198B may suppress ovarian cancer progression via the mitogen-activated protein kinase/extracellular-regulated protein kinase (MAPK/ERK) signaling pathway. Finally, a traditional Chinese medicine, named curcumin, may increase the sensitivity of ovarian cancer cells to cisplatin by upregulating CELF2.
Results
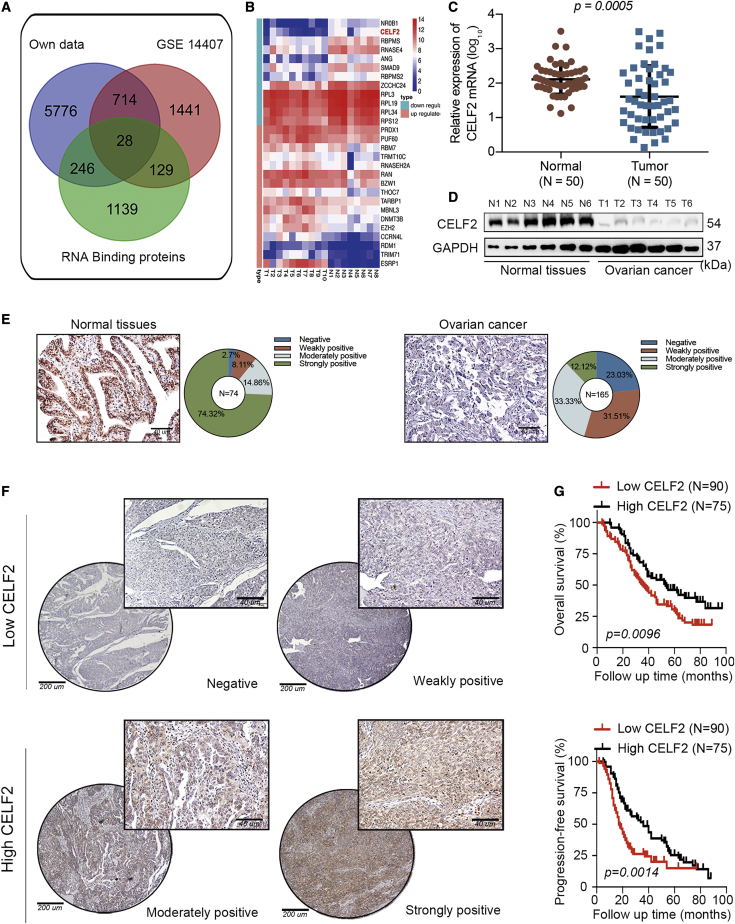
Identification of the RBP CELF2 as a Key Prognostic Tumor Suppressor in Ovarian Cancer Using an Integrated Transcriptomic Analysis
We first conducted RNA sequencing (RNA-seq) in 10 ovarian cancer tissues and 8 normal ovarian tissues to explore the differentially expressed genes in ovarian cancer; we used p <0.05, a fold-change value ≥2, and false discovery rate (FDR) <0.05 as the screening criteria. A total of 6,764 differentially expressed genes were identified, among which 4,656 were upregulated, and 2,108 were downregulated. Next, we profiled the expression of 1,542 RBP genes annotated in the RBP database10 and downloaded previously published RNA-seq data obtained from gene microarray profiles (Gene Expression Omnibus [GEO]: GSE14407) from the GEO database, which consists of 12 ovarian cancer specimens and 12 normal ovarian surface epithelial samples.11 Furthermore, Venn diagrams revealed 28 key dysregulated RBPs in these three categories (Figure 1A). A heatmap was created for these 28 RBPs, of which 16 were upregulated, and 12 were downregulated. Among the 12 RBPs that were downregulated in ovarian cancer, CELF2, which ranked in the top 2 RBPs, according to the fold-change value, attracted our attention, since its expression was positively correlated with the overall survival (OS; p = 0.023) and progression-free survival (PFS; p < 0.001) of patients with ovarian cancer, according to bioinformatics analyses (n = 1,287; http://kmplot.com/analysis/index.php?p=service). However, NR0B1 (ranked top 1) expression did not display a prognostic predictive value for the OS (p = 0.1) or PFS (p = 0.29) of patients with ovarian cancer (Figures 1B and S1).
Figure 1.
CELF2 Expression Is Downregulated in Ovarian Cancer and Predicts Poor Prognosis
(A) Method used to screen differentially expressed RBPs in ovarian cancer through a comprehensive analysis of three datasets. (B) The expression of 28 key RBPs in our microarray data from 10 ovarian cancer tissues and 8 normal ovarian tissues is presented in a heatmap. (C) Expression of the CELF2 mRNA in 50 ovarian cancer specimens and 50 normal ovarian surface epithelial specimens analyzed using qRT-PCR (p = 0.0005). (D) Western blot showing CELF2 levels in six ovarian cancer tissues and six normal ovarian tissues. (E) Representative images and proportions of negative, weakly positive, moderately positive, and strongly positive CELF2 immunohistochemical (IHC) staining in normal ovarian tissues (left panel) and ovarian cancer tissues (right panel) (scale bars, 40 μm). (F) Representative images of negative, weakly positive, moderately positive, and strongly positive IHC staining for CELF2 in ovarian cancer tissues (scale bars, 200 μm and 40 μm). (G) A Kaplan-Meier analysis was performed to assess the associations between CELF2 expression and the OS (p = 0.0096) and PFS (p = 0.0014) of patients with ovarian cancer. Data are presented as mean ± SEM.
Downregulation of CELF2 Is Associated with a Shorter Survival of Patients with Ovarian Cancer
After selecting CELF2 as the core RBP in ovarian cancer, we then examined the expression of the CELF2 mRNA in ovarian cancer samples from the web server Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) and found that CELF2 expression was significantly downregulated in ovarian cancer tissues (N = 426; data from The Cancer Genome Atlas [TCGA]) compared with normal ovarian tissues (N = 88; data from Genotype-Tissue Expression [GTEx]; Figure S2A). Moreover, in four other publicly available datasets of ovarian cancer, CELF2 expression was uniformly downregulated in ovarian cancer tissues compared with normal tissues (Figure S2B). Next, we compared the expression of the CELF2 mRNA in a cohort of 50 fresh-frozen ovarian cancer specimens and 50 normal ovarian surface epithelial specimens collected at our center using qRT-PCR. Consistent with the findings reported in these datasets, the expression of the CELF2 mRNA was downregulated in ovarian cancer tissues (p = 0.0005; Figure 1C). Western blot analyses of six randomly selected ovarian cancer tissues and six normal ovarian tissues revealed reduced levels of the CELF2 protein in tumor tissues (Figure 1D).
CELF2 expression was further validated by immunohistochemistry (IHC) through tissue microarrays (TMAs) from a larger cohort of 74 normal tissues (ovarian surface epithelial tissues or fallopian tube specimens) and 165 ovarian cancer specimens. According to the immunoreactivity scoring system (IRS), CELF2 expression was divided into four grades: negative, weakly positive, moderately positive, and strongly positive, respectively. A total of 74.32% of normal tissues showed strong positive results, whereas 2.70%, 8.11%, and 14.86% were negative, weakly positive, and moderately positive, respectively. However, among patients with ovarian cancer, the proportion of patients classified as strongly positive was 12.12%, whereas 23.03%, 31.51%, and 33.33% of patients were classified as negative, weakly positive, and moderately positive, respectively (Figure 1E). The statistical analysis of the data showed a significant difference between the normal tissues and ovarian cancer tissues (p < 0.01). Representative images of negative, weakly positive, moderately positive, and strongly positive CELF2 expression in ovarian cancer tissues are shown in Figure 1F. We classified negative and weakly positive expression as low expression groups (90 cases) and classified moderately positive and strongly positive expression as high expression groups (75 cases). The analysis of the clinical characteristics of patients with ovarian cancer revealed that CELF2 expression was associated with the International Federation of Gynecology and Obstetrics (FIGO) stage (p = 0.013) and diaphragmatic metastasis (p = 0.035; Table S1). Moreover, Kaplan-Meier survival curves revealed shorter OS (p = 0.0096) and PFS (p = 0.0014; Figure 1G) for patients whose tumor tissues showed CELF2 downregulation, and CELF2 was also an independent prognostic factor for ovarian cancer, according to the Cox regression analysis (Tables S2 and S3). Taken together, CELF2 may represent a prognostic marker for ovarian cancer.
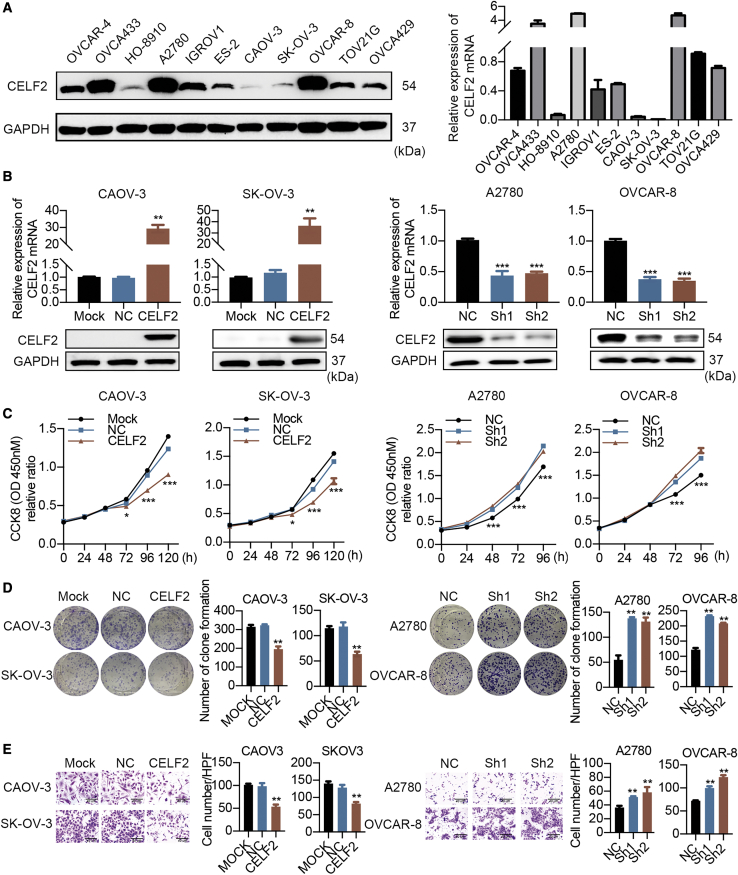
CELF2 Suppresses the Proliferation and Migration of Ovarian Cancer Cells In Vitro
To validate whether CELF2 was previously described as a tumor suppressor, we first examined the levels of CELF2 in 11 ovarian cancer cell lines by western blot and qRT-PCR. CELF2 was expressed at relatively low levels in CAOV-3 and SK-OV-3 cells and high levels in A2780 and OVCAR-8 cells (Figure 2A). Next, CAOV-3 and SK-OV-3 cells were transfected with lentiviruses for stable CELF2 overexpression, and two short hairpin RNAs (shRNAs; Sh1 and Sh2) were used to knock down CELF2 in A2780 and OVCAR-8 cells. The efficiency of overexpression and knockdown was verified (Figure 2B). Cell Counting Kit 8 (CCK-8) assays showed that CELF2 overexpression reduced cell proliferation, whereas CELF2 knockdown exerted the opposite effect (Figure 2C). These results were also confirmed by colony-formation assays (Figure 2D). Transwell assays were employed to assess the alterations in migration caused by CELF2. CELF2 overexpression impeded migration, whereas CELF2 knockdown exhibited a significant increase in migration (Figure 2E). The wound-healing assay produced similar results (Figures S3A–S3D). In vitro results indicate the negative regulation of CELF2 in ovarian cancer cell proliferation and migration.
Figure 2.
Effects of CELF2 on the Proliferation and Migration of Ovarian Cancer Cells In Vitro
(A) Western blot and qRT-PCR analyses of CELF2 expression in 11 ovarian cancer cell lines. (B) qRT-PCR and western blot analyses of CELF2 levels following CELF2 overexpression and knockdown. (C–E) CCK-8 (C), colony-formation (D), and Transwell assays (E) (scale bars, 40 μm) were performed to assess the changes in the proliferation and migration induced by CELF2. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
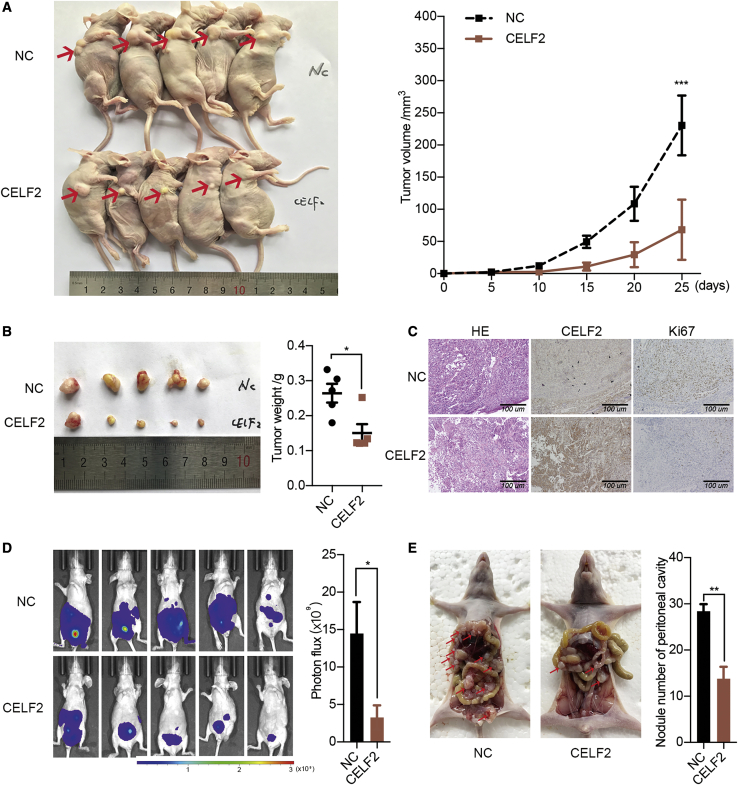
CELF2 Inhibits Ovarian Cancer Tumorigenesis and Metastasis In Vivo
To further verify the effects of CELF2 in vivo, SK-OV-3 cells stably overexpressing CELF2 or a vector were injected subcutaneously into BALB/c nude mice. The volumes of the tumors that developed from the CELF2-overexpressing SK-OV-3 cells were smaller than the negative control (NC) group (Figure 3A). The average tumor weight was decreased in the CELF2 group (Figure 3B). Additionally, staining for the proliferation marker Ki-67 was reduced in tumor tissues overexpressing CELF2 (Figure 3C). Next, SK-OV-3 cells stably expressing CELF2 or a vector were intraperitoneally injected into BALB/c nude mice. The extent of peritoneal metastasis was examined by bioluminescence imaging at 4 weeks postinoculation. CELF2 overexpression reduced tumor metastasis (Figure 3D). After sacrifice, the number of detectable metastatic nodules was reduced in the CELF2 group (Figure 3E). Collectively, these results demonstrate the antitumor effects of CELF2 in vivo.
Figure 3.
Overexpression of CELF2 Inhibited Proliferation and Metastasis In Vivo
(A) Xenografts were established in BALB/c nude mice by subcutaneously injecting CELF2-overexpressing SK-OV-3 cells (CELF2) or vector-expressing cells (NC) and xenograft tumor growth curves of the CELF2 and NC groups (right panel). (B) Final tumor weights of xenograft tumors at sacrifice. (C) Representative images of H&E staining and IHC staining for CELF2 and the proliferation marker Ki-67 in fixed and embedded xenograft tumors (scale bars, 100 μm). (D) Representative images of luciferase signals (left panel) and quantification of photon flux in abdominal cavity metastatic luciferase foci (right panel) after the intraperitoneal injection of CELF2-overexpressing SK-OV-3 cells (CELF2) or vector-expressing cells (NC) in BALB/c nude mice. (E) Representative images of abdominal cavity metastases derived from two groups after sacrifice (left panel) and quantification of the number of metastatic nodules of tumors in the abdominal cavities (right panel). Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
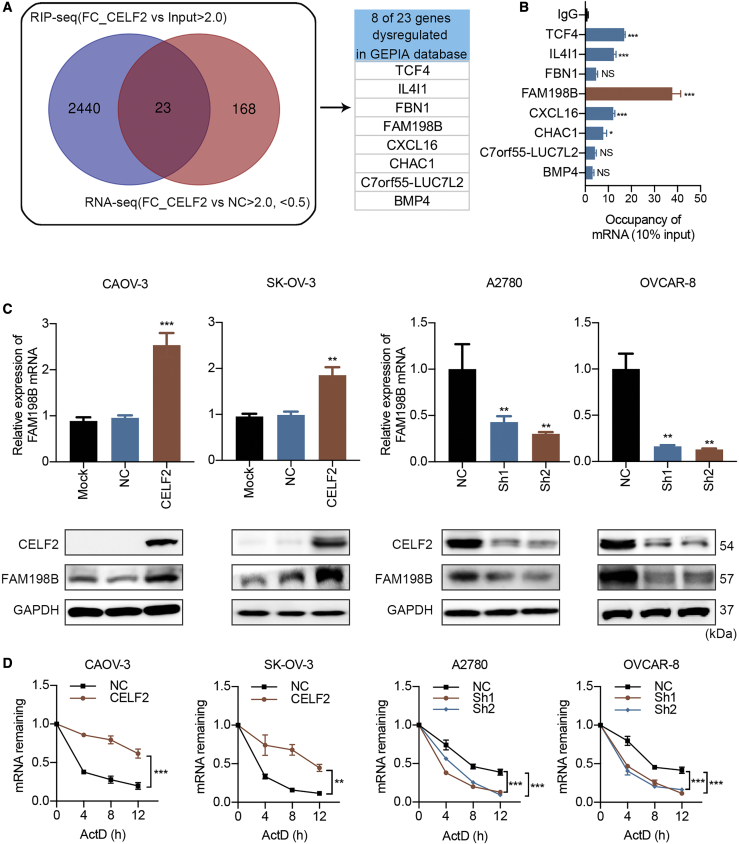
CELF2 Increases the Stability of the FAM198B mRNA in Ovarian Cancer
Given that RBPs mediate post-transcriptional regulation of target genes via a variety of mechanisms, their target genes include mRNA, lncRNA, circular RNA (circRNA), and miRNA,6 among which the most important is mRNA. Although CELF2 had been previously identified as an RBP, its mRNA targets are largely unknown. We performed high-throughput RNA immunoprecipitation (RIP) sequencing (RIP-seq) in A2780 cells to identify potential mRNAs that were directly bound by the CELF2 protein and to identify additional mRNAs regulated by CELF2 in ovarian cancer. We identified 2,464 mRNA targets of the CELF2 protein using the criteria of p < 0.05, a fold-change value >2, and FDR < 0.05. Then, we performed RNA-seq and identified 191 genes that were differentially expressed above the threshold level in CELF2-overexpressing SK-OV-3 cells compared with the expression in NC cells; these genes included 61 upregulated genes and 130 downregulated genes (Figure S4A). Merged with RIP-seq data, we identified 23 genes as potential regulators of ovarian cancer downstream of CELF2. Among the 23 novel putative mRNA targets that were potentially bound and dysregulated by CELF2, eight transcripts were also dysregulated in the GEPIA database; this gene set comprised TCF4, IL4I1, FBN1, FAM198B, CXCL16, CHAC1, C7orf55-LUC7L2, and BMP4 (Figures 4A and S4B). FAM198B was a particularly interesting target, because it displayed the greatest enrichment in the RIP-qPCR data (Figure 4B). We further confirmed that the FAM198B transcript was present in the CELF2 immunocomplex and input but not in the control immunoglobulin G (IgG) immunoprecipitates of the RIP-qPCR product (Figure S4C). Moreover, CELF2 overexpression increased, and CELF2 knockdown decreased the levels of the FAM198B mRNA and protein (Figure 4C). As CELF2 has been identified as a regulator of mRNA stability, we hypothesized that CELF2 might change the transcript abundance of FAM198B through an effect on its stability. We found that FAM198B transcripts exhibited longer half-lives in CELF2-overexpressing CAOV-3 and SK-OV-3 cells following actinomycin-D (Act D; 5 μg/mL) treatment. The opposite result was observed after the knockdown of CELF2 in A2780 and OVCAR-8 cells (Figure 4D), indicating that CELF2 increased the stability of the FAM198B mRNA through a post-transcriptional mechanism.
Figure 4.
CELF2 Stabilized the FAM198B Transcript
(A) Venn diagram illustrating the overlap of 23 mRNA targets of CELF2 identified using the RIP-seq and RNA-seq analysis (left panel); eight transcripts were also dysregulated in the GEPIA database (right panel). (B) qRT-PCR analysis of eight transcripts obtained from RIP with anti-CELF2 and IgG control antibodies in A2780 cells. (C) Levels of the FAM198B mRNA and protein in CELF2-overexpressing CAOV-3 and SK-OV-3 cells and CELF2 knockdown A2780 and OVCAR-8 cells were analyzed using qRT-PCR and western blotting, respectively. (D) After treatment with 5 μg/mL Act D, total RNA was extracted at 0, 4, 8, and 12 h. The half-lives of FAM198B mRNA in CELF2-overexpressing CAOV-3 and SK-OV-3 cells and CELF2-knockdown A2780 and OVCAR-8 cells were measured. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; NS, not significant.
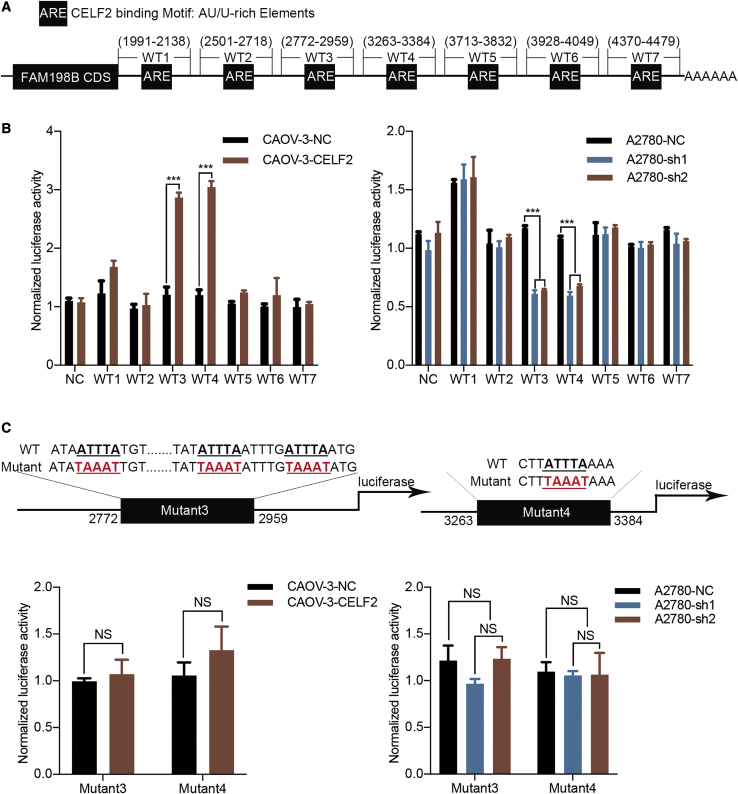
CELF2 Binds to AREs within the FAM198B 3′ UTR in Ovarian Cancer Cells
It is well recognized that CELF2 could predominantly bind to AREs in the 3′ UTR of their target mRNAs,12,13 and we investigated whether CELF2 could bind to the 3′ UTR of FAM198B. To test this idea, the University of California, Santa Cruz (UCSC), Genome Browser (http://genome.ucsc.edu/) was used to analyze FAM198B mRNA and provided a series of AREs in its 3′ UTR. As shown in the schematic, the 3′ UTR of FAM198B mRNA was divided into seven sections, WT1, -2, -3, -4, -5, -6, and -7. Additionally, NC, which does not contain a binding site, was chosen as the NC (Figures 5A and S5). We performed a dual-luciferase assay in CAOV-3 cells and A2780 cells to functionally confirm the ARE regions that were required for CELF2 binding to FAM198B mRNA. The luciferase activity of the reporters carrying WT3 and WT4 was increased upon CELF2 overexpression and decreased when CELF2 was knocked down. In contrast, no alterations in luciferase activity were observed following cotransfection with WT1, -2, -5, -6, or -7 (Figure 5B) or with site-directed mutant 3 and mutant 4 (Figure 5C). Generally, these data indicated that CELF2 binds to AREs in the 3′ UTR-WT3 and 3′ UTR-WT4 sites of FAM198B mRNA to enhance FAM198B expression.
Figure 5.
CELF2 Binding to the 3′ UTR of FAM198B mRNA
(A) Schematic of various regions in the 3′ UTR of the FAM198B mRNA. (B and C) The relative luciferase activity of the reporter containing each region of the FAM198B 3′ UTR (B) or a mutated sequence of CELF2 target binding sites (C) was measured as a ratio of luciferase activity induced by CELF2 overexpression or knockdown to the luciferase activity of a control vector with no additional 3′ UTR sequence. Data are presented as mean ± SEM. ∗∗∗p < 0.001.
FAM198B Is a New Player with a Tumor-Suppressive Role in Ovarian Cancer
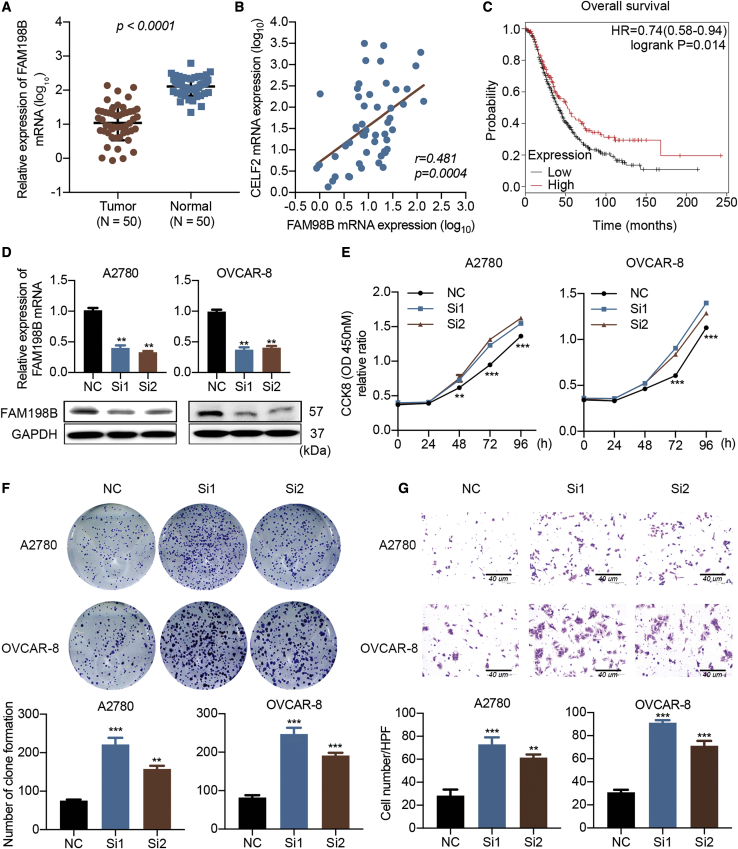
Although CELF2 stabilizes FAM198B by binding to its 3′ UTR in ovarian cancer, the function of FAM198B has rarely been discussed. FAM198B expression was significantly decreased, and a positive correlation was identified between the expression of CELF2 and FAM198B in the GEPIA dataset of ovarian cancer (Figures S6A and S6B). Next, we detected the FAM198B mRNA levels in the aforementioned cohort of 50 ovarian cancer tissues and 50 normal tissues using qRT-PCR and protein levels through TMAs using IHC analyses. Levels of the FAM198B mRNA and protein were decreased in ovarian cancer tissues (Figures 6A and S6C), and FAM198B mRNA expression levels exhibited a positive correlation with the expression of the CELF2 mRNA (p = 0.0004; Figure 6B). Moreover, FAM198B expression correlated with a shorter OS of patients with ovarian cancer (p = 0.014; Figure 6C). We used small interfering RNA (siRNA) to knock down FAM198B expression in A2780 and OVCAR-8 cells and to examine the effect of FAM198B on the phenotype of ovarian cancer cells in vitro (Figure 6D). Based on the results of the CCK-8 proliferation assay and the colony-formation assay, downregulation of FAM198B increased the viability of A2780 and OVCAR-8 cells (Figures 6E and 6F). As shown in Figure 6G, cell migration was increased when FAM198B activity was silenced in A2780 and OVCAR-8 cells. In summary, FAM198B functions as a tumor suppressor in ovarian cancer and suppresses ovarian cancer cell proliferation and migration.
Figure 6.
FAM198B Is a Tumor Suppressor in Ovarian Cancer
(A) Relative expression of the FAM198B mRNA in a cohort of 50 ovarian cancer specimens and 50 normal ovarian surface epithelial specimens determined using qRT-PCR. (B) Correlation of the expression of the CELF2 and FAM198B mRNAs in ovarian tumors (n = 50) from our cohort. (C) Association between FAM198B expression and the OS (p = 0.014) of patients with ovarian cancer in an online database. (D) FAM198B was knocked down by two siRNAs. (E–G) CCK-8 (E), colony-formation (F), and Transwell assays (G) (scale bars, 40 μm) were performed to assess the changes in the proliferation and migration following FAM198B modulation. Data are presented as mean ± SEM. ∗∗p < 0.01 and ∗∗∗p < 0.001.
FAM198B Knockdown Ameliorates the CELF2-Mediated Suppression of Growth and Migration
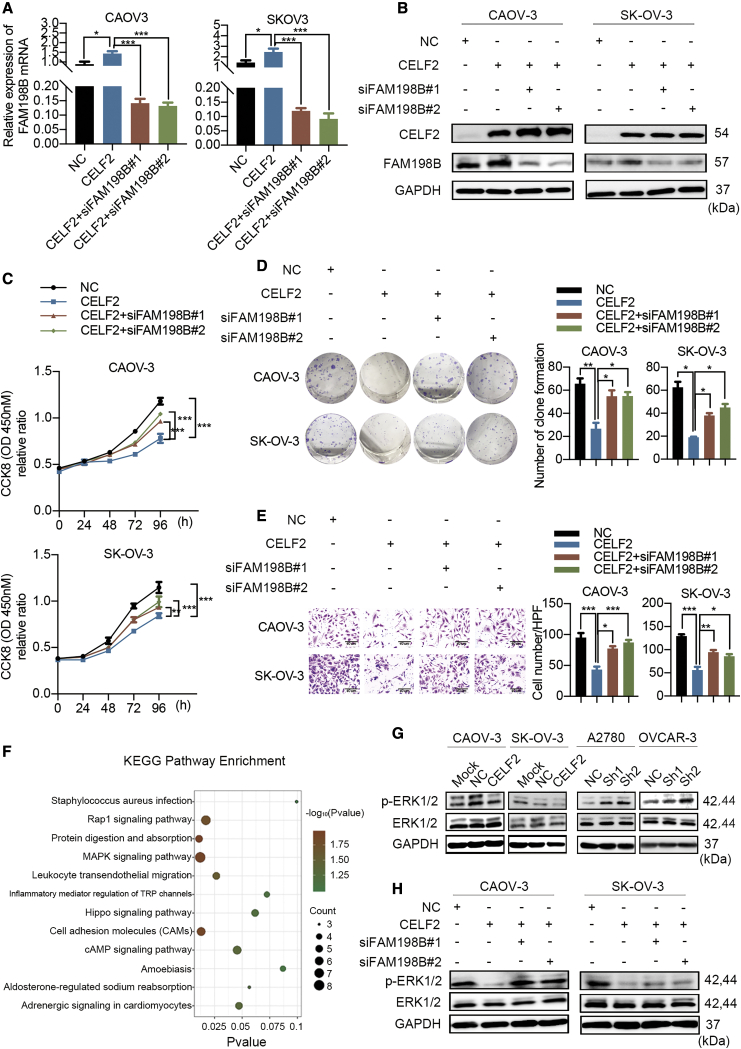
As we found that CELF2 directly binds to FAM198B, we next investigated the coregulation of ovarian cancer cell proliferation and migration by CELF2 and FAM198B. CELF2-overexpressing CAOV-3 and SK-OV-3 cells were transfected with siRNAs targeting FAM198B (siFAM198B#1 and siFAM198B#2), and then qRT-PCR and western blots were performed to assess the knockdown efficiency (Figures 7A and 7B). The CCK-8 and colony-formation assays showed that overexpression of CELF2 restrained cell growth, whereas knockdown of FAM198B reversed this effect, indicating that a reduction in FAM198B expression ameliorated the CELF2-mediated suppression of cell growth (Figures 7C and 7D). Consistent with these findings, the suppression of cell migration by CELF2 overexpression was reversed after FAM198 was downregulated (Figure 7E). Together, these results suggest that FAM198B is a critical downstream target of CELF2.
Figure 7.
CELF2 Inhibited Ovarian Cancer Cell Progression in a FAM198B-Mediated Manner
(A and B) CELF2-overexpressing CAOV-3 and SK-OV-3 cells were transfected with siRNAs targeting FAM198B. (C–E) CCK-8 (C), colony-formation (D), and Transwell assays (E) (scale bars, 40 μm) were performed to assess changes in proliferation and migration. (F) KEGG analysis showing the main signaling pathways associated with these differentially expressed genes targeted by CELF2. (G) Western blot analysis of ERK and p-ERK levels after the overexpression or knockdown of CELF2. (H) Western blot analysis of ERK and p-ERK levels after the knockdown of FAM198B in CELF2-overexpressing CAOV-3 and SK-OV-3 cells. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
CELF2/FAM198B Suppresses the Activity of the MAPK/ERK Pathway in Ovarian Cancer Cells
To explore the putative downstream signaling pathways of the CELF2/FAM198B axis that may mediate tumor-suppressive capacities in ovarian cancer cells, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted for these differentially expressed genes from the RNA-seq results mentioned above in SK-OV-3-CELF2 and SK-OV-3-NC cells (Figure 7F). Among the enriched pathways, the MAPK signaling pathway attracted our attention since a previous report showed that FAM198B inhibits the invasion and metastasis of lung cancer by inhibiting the MAPK/ERK signaling pathway,14 and the MAPK/ERK signaling pathway has a well-established role in promoting the proliferation and metastasis of ovarian cancer cells.15,16 We analyzed the phosphorylation status of ERK (p-ERK) as a surrogate marker of activation of this pathway. As shown in Figure 7G, p-ERK protein expression was decreased when CELF2 was overexpressed in CAOV-3 and SK-OV-3 cells, whereas downregulation of CELF2 in A2780 and OVCAR-8 cells had the opposite effect. However, total ERK expression did not change substantially in all groups. Additionally, FAM198B knockdown reversed the CELF2 overexpression-induced decrease in p-ERK levels (Figure 7H). These data suggest that CELF2/FAM198B may repress ovarian cancer development partially through inhibition of the MAPK/ERK signaling pathway.
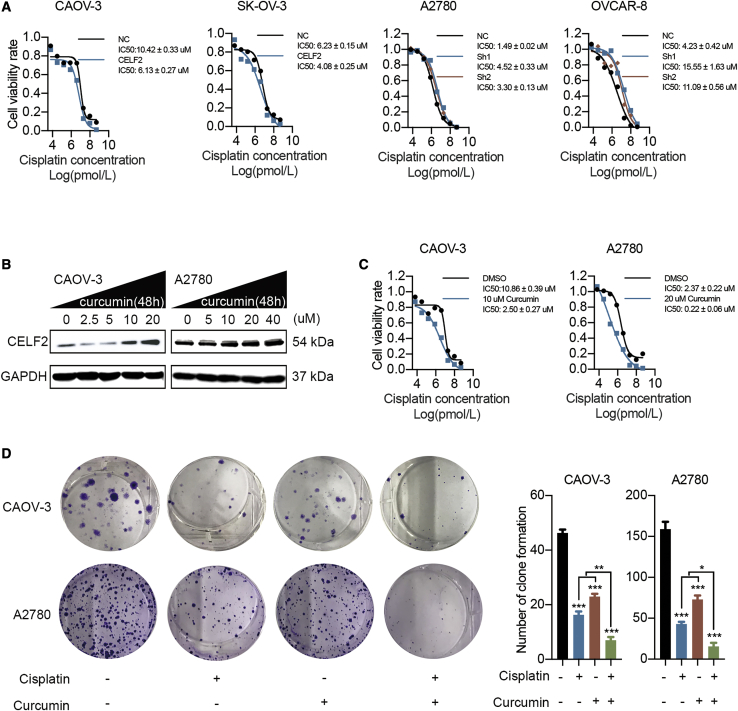
CELF2 Mediates Curcumin-Induced Improvements in the Efficacy of Cisplatin in Ovarian Cancer
Based on previous findings, RBP-, miRNA-, and lncRNA-mediated post-transcriptional regulatory processes also play important roles in regulating the sensitivity to chemotherapeutic agents.17, 18, 19 For the ovarian cancer cells displayed in Figure 8A, the 50% inhibitory concentration (IC50) of cisplatin was measured. Compared with the IC50 value of the NC group, the IC50 values of CELF2-overexpressing CAOV-3 and SK-OV-3 cells were decreased, and the values of CELF2-knockdown A2780 and OVCAR-8 cells increased after treatment with different concentrations of cisplatin, suggesting that the upregulation of CELF2 may be a promising strategy to increase the sensitivity of ovarian cancer cells to cisplatin. Previous reports found that curcumin may increase the expression of CELF2,20 but the real effect needs to be verified in ovarian cancer. Western blot analyses revealed a dose-dependent increase in CELF2 levels in CAOV-3 and A2780 cells after treatment with curcumin (Figure 8B). We then tested whether curcumin mimicked the effect of CELF2 upregulation on increasing the sensitivity of ovarian cancer to cisplatin. Based on the IC50 values (Figure S7), concentrations of curcumin below the IC50 values (10 μM and 20 μM) were administered to CAOV-3 and A2780 cells, respectively, and the cells were treated with cisplatin and curcumin in subsequent experiments. Notably, the IC50 values of cisplatin in CAOV-3 and A2780 cells decreased after the addition of curcumin (Figure 8C). Moreover, based on the results of the colony-formation assay, the combination of cisplatin and curcumin produced a greater inhibition of proliferation than either drug alone in CAOV-3 and A2780 cells (Figure 8D). Taken together, curcumin may increase the sensitivity of ovarian cancer cells to cisplatin by targeting CELF2.
Figure 8.
Curcumin Enhanced the Efficacy of Cisplatin in Ovarian Cancer by Upregulating CELF2
(A) The IC50 value of cisplatin was determined in CELF2-overexpressing CAOV-3 and SK-OV-3 cells and CELF2 knockdown A2780 and OVCAR-8 cells. (B) CAOV-3 and A2780 cells were treated with increasing curcumin concentrations and then subjected to western blotting. (C) The IC50 value of cisplatin was measured after the addition of curcumin to CAOV-3 and A2780 cells. (D) A colony-formation assay was performed to assess the proliferation of CAOV-3 and A2780 cells after the addition of cisplatin and curcumin alone and in combination. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Discussion
As the role of RBPs in cancer emerges, the ability of RBPs to interact with thousands of RNAs makes it an appropriate group of proteins to be selectively dysregulated in cancer.21 An interesting hypothesis is that dysregulation of members of the RBP family may collectively contribute to the transcriptomic imbalance in tumor cells, thus driving oncogenicity.22 The transcriptomes of ovarian cancer and normal ovarian tissues were sequenced, and the data were combined with the analysis of the existing public database to test this hypothesis; 28 RBPs exhibited concordant differential expression in ovarian cancer, providing evidence supporting the hypothesis that altered RBPs may be involved in the coordinated networks and contribute to ovarian cancer tumorigenicity. Furthermore, combined with the fold change and the survival analysis of genes in ovarian cancer, we identified CELF2 as a key prognostic RBP for further study.
The CELF2 gene is located on chromosome 10p, a region frequently lost in human cancers,23,24 suggesting that the region encoding CELF2 potentially encodes tumor suppressors. The role of CELF2 as a tumor suppressor has been studied in multiple types of cancer, including colon cancer,25 pancreatic cancer,20 lung cancer,26 gastric cancer,27 and breast cancer.28 However, the functional role of CELF2 in ovarian cancer has not yet been identified. The present study was the first to report the decreased expression of CELF2 in ovarian cancer. We further demonstrated that ovarian cancer patients with high levels of CELF2 had better outcomes than those with low levels, indicating that CELF2 might serve as a potential biomarker for prognostic evaluation in ovarian cancer.
As an RBP, CELF2 should have the ability to modulate various post-transcriptional events. We investigated additional downstream target genes that bind to and are regulated by CELF2, and the combination of RIP-seq and RNA-seq analyses further identified FAM198B as a selective target gene, which is a newly discovered gene. Hsu et al.14 described FAM198B as a prognostic marker in lung adenocarcinoma that inhibits the metastasis of lung adenocarcinoma by blocking ERK-mediated matrix metalloproteinase-1 (MMP-1) expression. Our study found that FAM198B also acted as a tumor-suppressor gene, which was downregulated in ovarian cancer, and inhibition of FAM198B improved the proliferation and migration capacity of ovarian cancer cells. Moreover, we found that CELF2 could regulate FAM198B expression by stabilizing its mRNA. Previous studies indicated that CELF2 could regulate target gene expression by strongly binding to AREs in the 3′ UTR of its target mRNAs.12,13,29 Consistent with this, we also demonstrated that the 3′ UTR of FAM198B mRNA contains two functional CELF2 binding sites, as confirmed by mutational analysis, which contributed to the stability of FAM198B mRNA. To date, most studies have investigated the mechanism by which CELF2 regulates target genes by alternative splicing.30, 31, 32 However, the regulation of mRNA stability by CELF2 is less well characterized. In the present study, we revealed a novel mechanism in which CELF2 functions as an important post-transcriptional regulator to stabilize the FAM198B transcript by binding directly to target AREs in the 3′ UTR of the FAM198B mRNA. Moreover, FAM198B knockdown reversed the tumor growth inhibition mediated by CELF2 overexpression, further confirming that the mechanism by which CELF2 suppresses tumor growth, at least partially, depends on its ability to induce FAM198B expression in ovarian cancer.
The MAPK/ERK cascade generally participates in cell proliferation, metastasis, and survival in most malignant tumors.15,33 Mounting evidence has indicated the pivotal role of MAPK/ERK signaling during tumor development and progression.34,35 In the present study, we provide evidence that CELF2 inhibited p-ERK, and FAM198B knockdown restored p-ERK, indicating that the CELF2/FAM198B axis may play a role in regulating p-ERK. Nevertheless, the detailed mechanism remains to be determined. CELF2 and FAM198B may be used to target the ERK pathway in the future.
In line with these findings, the present study also proved that CELF2 could be used as a therapeutic target in ovarian cancer. Recurrent and metastatic ovarian cancer is often resistant to standard cisplatin-based chemotherapy,36,37 and strategies to increase the sensitivity of cisplatin are very important to improve the prognosis of patients with ovarian cancer. Our results indicated that overexpression of CELF2 could sensitize cells to cisplatin, whereas knockdown of CELF2 had the opposite effect. On the other hand, it has been found that curcumin, a common Chinese herbal medicine, increases the expression of CELF2 in pancreatic cancer.20 Curcumin is an active ingredient in the spice turmeric, which has been demonstrated to have therapeutic efficacy for many cancer types.38, 39, 40 In ovarian cancer, owing to its effective anticancer effect, curcumin and its derivatives have been put into several clinical trials.41 Notably, recent studies have demonstrated that curcumin acts synergistically with conventional chemotherapeutic drugs to eradicate resistant cancer cell lines.42,43 As shown in the study by Zhang et al.,44 curcumin restores MEG3 levels and decreases the extracellular vesicle-mediated transfer of miR-214 in ovarian cancer cells, thereby reducing cisplatin resistance, suggesting that curcumin is a strong chemosensitizer that may improve the therapeutic potential of cisplatin. Similar findings were also found in vitro in this study. Combination therapy represents a potentially promising strategy to treat resistant ovarian cancer. Curcumin was found to dysregulate the expression of several drug-resistance proteins, such as ATP-binding cassette drug transporters,45 P-glycoproteins,46 and multi-drug-resistant (MDR) proteins,47 which resulted in the sensitivity of tumor cells to chemotherapy. In this study, we identified a novel mechanism by which a therapy combining curcumin and cisplatin improves the response of ovarian cancer cells to cisplatin treatment via CELF2 upregulation. Since cisplatin is currently one of the most effective chemotherapeutic drugs used for treating ovarian cancer, new treatment strategies based on curcumin and drugs targeting CELF2 or FAM198B may be an enticing candidate for improving the rate of response to cisplatin in ovarian cancer patients.
Conclusions
In the present study, we used a combination of computational, biochemical, and functional approaches to identify key RBPs in ovarian cancer and characterized a novel mechanism by which CELF2 increased the stability of the FAM198B mRNA to subsequently restrain ovarian cancer progression by inhibiting p-ERK, providing new insights into the theoretical basis of tumorigenesis and elucidating the importance of CELF2 and FAM198B in ovarian cancer progression and gene-targeted precision therapy for individuals with ovarian cancer. Notably, whereas this study focuses on CELF2, the RNA-seq results described here identified multiple dysregulated RBPs in ovarian cancer, which may serve as a tractable platform for the identification of new roles of RBPs in ovarian cancer.
Materials and Methods
Please see a complete description of Materials and Methods in the Supplemental Materials and Methods.
Cell Culture
The human ovarian cancer cell lines OVCAR-4, OVCA433, HO-8910, A2780, IGROV1, ES-2, CAOV-3, SK-OV-3, OVCAR-8, TOV21G, and OVCA429 were obtained from Shanghai Cell Bank Type Culture Collection (Shanghai, China), and all cell lines were authenticated by short tandem repeat (STR) profiling and tested for mycoplasma contamination. Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, CA, USA), supplemented with 10% fetal bovine serum (FBS; Gibco, CA, USA) and a 1% penicillin-streptomycin (Gibco, CA, USA) solution. The cells were maintained at a specific density and seeded into 6-, 24-, or 96-well culture plates at the appropriate cell density 1 day before treatment.
Tissue Samples
The use and collection of the samples were reviewed and approved by the Institutional Ethics Committee of the Fudan University Shanghai Cancer Center, and informed consent was obtained from each patient. Formalin-fixed, paraffin-embedded samples and frozen specimens of ovarian cancer and normal ovarian tissues were obtained from Fudan University Shanghai Cancer Center. All ovarian cancer specimens analyzed in this study were high-grade serous ovarian carcinoma (HGSOC) specimens, the most common and deadly form of ovarian epithelial cancer. No chemotherapy or radiation therapy was administered prior to tumor excision. All samples were examined by experienced pathologists who confirmed the disease diagnosis.
Plasmids and siRNA
The CELF2 overexpression plasmid was constructed by cloning the full-length CELF2 cDNA into the pCDH-CMV-MCS-EF1-puro vector (System Biosciences, CA, USA). Plasmids carrying shRNAs targeting CELF2 were generated using the U6-MCS-CMV-ZsGreen1-PGK-Puro vector (LncBio, Shanghai, China). The above plasmids, along with FAM198B-targeted siRNA and matched empty vector controls, were all purchased from Lncbio (Shanghai, China). All fragments were confirmed by sequencing. The target sequences of the CELF2 shRNAs and FAM198B siRNAs are listed in Table S4.
RNA Extraction and qRT-PCR
Total RNA was extracted from cells and samples using the TRIzol reagent (Life Technologies, CA, USA), according to the manufacturer’s protocol. qRT-PCR was conducted using TB Green PCR Master Mix (TaKaRa, Dalian, China) in an ABI 7900HT Real-Time PCR system (Applied Biosystems, USA). The relative quantification was normalized to β-actin with the 2−ΔΔCT formula. The primers used for qRT-PCR are listed in Table S5.
Western Blot and Antibodies
Western blotting was performed as described in our previous study.48 The following antibodies were used in the present study: anti-CELF2 (1:2,000; Abcam, MA, USA), anti-FAM198B (1:2,000; Sigma-Aldrich, MO, USA), anti-ERK1/2 (1:3,000; Proteintech, IL, USA), anti-p-ERK1/2 (1:3,000; Proteintech, IL, USA), and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:5,000; Proteintech, IL, USA).
RIP Assay
The RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA, USA), according to the manufacturer’s protocol. Briefly, A2780 cells were lysed with RIP lysis buffer (Millipore, MA, USA) and then incubated with 5 μg of rabbit polyclonal anti-CELF2 (Abcam, MA, USA) or rabbit IgG isotype control (Sigma-Aldrich, MO, USA) at 4°C overnight. The RNA-protein immunocomplexes were brought down by protein A/G magnetic beads, followed by RNA purification. cDNA generation and qRT-PCR were performed as described earlier. The fold enrichment for each target was measured by comparing the CT values of the CELF2-immunoprecipitated fraction to the IgG isotype fraction and normalized using the ΔCt formula. For sequencing, rRNAs were depleted using the TruSeq Stranded Total RNA Library Prep Kit (Illumina, CA, USA). The cDNA libraries were quantified using an Agilent Bioanalyzer 2100 (Agilent Technologies, USA) and sequenced on an Illumina HiSeq sequencer according to the manufacturer’s instructions at CloudSeq (Shanghai, China).
Tumor Xenograft Models
All animal studies were approved by the Institutional Animal Care and Use Committee of Fudan University. 5- to 6-week-old female nude mice (BALB/c) were purchased from Shanghai Slack Laboratory (Shanghai, China) and housed under specific pathogen-free (SPF) conditions at the animal care facility of the Experimental Animal Center of School of Pharmacy. For tumor formation, 8 × 106 CELF2-overexpressing and vector-transfected SK-OV-3 cells were suspended in 0.1 mL of sterile PBS, and cells were subsequently injected subcutaneously into the flanks of randomly selected mice (n = 5 per group). Tumor volumes were measured every 5 days after the appearance of tumors, and the tumor volume was calculated as (length × width2) × 0.5. After 4 weeks, the mice were sacrificed, and the tumors were harvested, weighed, and analyzed using IHC analyses. An orthotopic model was generated by intrabursal injection of ovarian cancer cell lines to assess the peritoneal metastasis of ovarian cancer. After 4 weeks, BALB/c nude mice were intraperitoneally injected with 10 μL of D-luciferin (15 μg/μL)/g of body weight for the in vivo imaging analysis, and the mice were anesthetized and imaged using an In Vivo Imaging System (IVIS) Lumina system (Xenogen, MA, USA). The number of metastatic nodules was counted at sacrifice.
Statistics
All data are presented as the mean ± SEM from at least three independent experiments. Statistical analyses were performed using IBM SPSS 23.0 and GraphPad Prism 7 software. Student’s t tests or one-way ANOVA were applied to assess differences between or among different groups. The chi-square test was used to analyze the relationship between the level of the CELF2 protein and clinicopathological parameters. Spearman correlation analysis was performed to assess the relationship between different factors. OS and PFS curves were plotted using the Kaplan-Meier method and analyzed using the log-rank test. Univariate and multivariate regression analyses, based on the Cox proportional hazards model, were used to analyze the risk factors for patient prognosis. The results of all statistical analyses were reported as p values from two-tailed tests, and p <0.05 was considered statistically significant (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
Accession Number
RNA-seq and RIP-seq data were deposited into figshare.49
Author Contributions
Q.G. and Y.W. contributed cell and molecular experiments, analyzed the data, and wrote the paper. X.G. and L.C. contributed animal experiments. F.X. and H.Z. collected the human tissue samples. J.Z., H.W., X.J., and X.W. designed the study and participated in the data analysis. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported, in part, by grants from the National Natural Science Foundation of China (numbers 81972431, 81902641, and 81902640); Science and Technology Commission of Shanghai Municipality (number KW1711); Shanghai Sailing Program (number 20YF1408000); and Shanghai Anticancer Association EYAS PROJECT (number SACA-CY19A07).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.011.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Jayson G.C., Kohn E.C., Kitchener H.C., Ledermann J.A. Ovarian cancer. Lancet. 2014;384:1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 3.Staicu C.E., Predescu D.V., Rusu C.M., Radu B.M., Cretoiu D., Suciu N., Crețoiu S.M., Voinea S.C. Role of microRNAs as Clinical Cancer Biomarkers for Ovarian Cancer: A Short Overview. Cells. 2020;9:169. doi: 10.3390/cells9010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhan L., Li J., Wei B. Long non-coding RNAs in ovarian cancer. J. Exp. Clin. Cancer Res. 2018;37:120. doi: 10.1186/s13046-018-0793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blagden S., Abdel Mouti M., Chettle J. Ancient and modern: hints of a core post-transcriptional network driving chemotherapy resistance in ovarian cancer. Wiley Interdiscip. Rev. RNA. 2018;9:e1432. doi: 10.1002/wrna.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hentze M.W., Castello A., Schwarzl T., Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 7.Chatterji P., Rustgi A.K. RNA Binding Proteins in Intestinal Epithelial Biology and Colorectal Cancer. Trends Mol. Med. 2018;24:490–506. doi: 10.1016/j.molmed.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L., Wang W., Huang S., Yang Z., Xu L., Yang Q., Zhou X., Wang J., Shen Q., Wang C. The RNA binding protein SORBS2 suppresses metastatic colonization of ovarian cancer by stabilizing tumor-suppressive immunomodulatory transcripts. Genome Biol. 2018;19:35. doi: 10.1186/s13059-018-1412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong Y., Yang S., Wang W., Wei P., He S., Ma H., Yang J., Wang Q., Cao L., Xiong W. The interaction of Lin28A/Rho associated coiled-coil containing protein kinase2 accelerates the malignancy of ovarian cancer. Oncogene. 2019;38:1381–1397. doi: 10.1038/s41388-018-0512-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerstberger S., Hafner M., Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowen N.J., Walker L.D., Matyunina L.V., Logani S., Totten K.A., Benigno B.B., McDonald J.F. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med. Genomics. 2009;2:71. doi: 10.1186/1755-8794-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukhopadhyay D., Houchen C.W., Kennedy S., Dieckgraefe B.K., Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 13.Sureban S.M., Murmu N., Rodriguez P., May R., Maheshwari R., Dieckgraefe B.K., Houchen C.W., Anant S. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132:1055–1065. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Hsu C.Y., Chang G.C., Chen Y.J., Hsu Y.C., Hsiao Y.J., Su K.Y., Chen H.Y., Lin C.Y., Chen J.S., Chen Y.J. FAM198B Is Associated with Prolonged Survival and Inhibits Metastasis in Lung Adenocarcinoma via Blockage of ERK-Mediated MMP-1 Expression. Clin. Cancer Res. 2018;24:916–926. doi: 10.1158/1078-0432.CCR-17-1347. [DOI] [PubMed] [Google Scholar]
- 15.Chen K., Liu M.X., Mak C.S.-L., Yung M.M.-H., Leung T.H.-Y., Xu D., Ngu S.-F., Chan K.K.-L., Yang H., Ngan H.-Y.S., Chan D.W. Methylation-associated silencing of miR-193a-3p promotes ovarian cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways. Theranostics. 2018;8:423–436. doi: 10.7150/thno.22377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burotto M., Chiou V.L., Lee J.M., Kohn E.C. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120:3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi J., Areecheewakul S., Li Y., Yang S., Zhang Y., Ebeid K., Li L., Thiel K.W., Zhang J., Dai D. MTDH/AEG-1 downregulation using pristimerin-loaded nanoparticles inhibits Fanconi anemia proteins and increases sensitivity to platinum-based chemotherapy. Gynecol. Oncol. 2019;155:349–358. doi: 10.1016/j.ygyno.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin F., Wang Y., Li M., Zhu Y., Liang H., Wang C., Wang F., Zhang C.Y., Zen K., Li L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540. doi: 10.1038/cddis.2016.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phatak P., Byrnes K.A., Mansour D., Liu L., Cao S., Li R., Rao J.N., Turner D.J., Wang J.Y., Donahue J.M. Overexpression of miR-214-3p in esophageal squamous cancer cells enhances sensitivity to cisplatin by targeting survivin directly and indirectly through CUG-BP1. Oncogene. 2016;35:2087–2097. doi: 10.1038/onc.2015.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakstaite A., Maziukiene A., Silkuniene G., Kmieliute K., Dauksa A., Paskauskas S., Gulbinas A., Dambrauskas Z. Upregulation of cugbp2 increases response of pancreatic cancer cells to chemotherapy. Langenbecks Arch. Surg. 2016;401:99–111. doi: 10.1007/s00423-015-1364-1. [DOI] [PubMed] [Google Scholar]
- 21.Neelamraju Y., Hashemikhabir S., Janga S.C. The human RBPome: from genes and proteins to human disease. J. Proteomics. 2015;127(Pt A):61–70. doi: 10.1016/j.jprot.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 22.Dang H., Takai A., Forgues M., Pomyen Y., Mou H., Xue W., Ray D., Ha K.C.H., Morris Q.D., Hughes T.R., Wang X.W. Oncogenic Activation of the RNA Binding Protein NELFE and MYC Signaling in Hepatocellular Carcinoma. Cancer Cell. 2017;32:101–114.e8. doi: 10.1016/j.ccell.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Ibraheemi A., Martinez A., Weiss S.W., Kozakewich H.P., Perez-Atayde A.R., Tran H., Parham D.M., Sukov W.R., Fritchie K.J., Folpe A.L. Fibrous hamartoma of infancy: a clinicopathologic study of 145 cases, including 2 with sarcomatous features. Mod. Pathol. 2017;30:474–485. doi: 10.1038/modpathol.2016.215. [DOI] [PubMed] [Google Scholar]
- 24.Gao C., Su Y., Koeman J., Haak E., Dykema K., Essenberg C., Hudson E., Petillo D., Khoo S.K., Vande Woude G.F. Chromosome instability drives phenotypic switching to metastasis. Proc. Natl. Acad. Sci. USA. 2016;113:14793–14798. doi: 10.1073/pnas.1618215113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natarajan G., Ramalingam S., Ramachandran I., May R., Queimado L., Houchen C.W., Anant S. CUGBP2 downregulation by prostaglandin E2 protects colon cancer cells from radiation-induced mitotic catastrophe. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;294:G1235–G1244. doi: 10.1152/ajpgi.00037.2008. [DOI] [PubMed] [Google Scholar]
- 26.Yeung Y.T., Fan S., Lu B., Yin S., Yang S., Nie W., Wang M., Zhou L., Li T., Li X. CELF2 suppresses non-small cell lung carcinoma growth by inhibiting the PREX2-PTEN interaction. Carcinogenesis. 2020;41:377–389. doi: 10.1093/carcin/bgz113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Liu L., Sun Y., Xue Y., Qu J., Pan S., Li H., Qu H., Wang J., Zhang J. miR-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed. Pharmacother. 2018;101:406–413. doi: 10.1016/j.biopha.2018.02.104. [DOI] [PubMed] [Google Scholar]
- 28.Piqué L., Martinez de Paz A., Piñeyro D., Martínez-Cardús A., Castro de Moura M., Llinàs-Arias P., Setien F., Gomez-Miragaya J., Gonzalez-Suarez E., Sigurdsson S. Epigenetic inactivation of the splicing RNA-binding protein CELF2 in human breast cancer. Oncogene. 2019;38:7106–7112. doi: 10.1038/s41388-019-0936-x. [DOI] [PubMed] [Google Scholar]
- 29.Dasgupta T., Ladd A.N. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip. Rev. RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallory M.J., Allon S.J., Qiu J., Gazzara M.R., Tapescu I., Martinez N.M., Fu X.D., Lynch K.W. Induced transcription and stability of CELF2 mRNA drives widespread alternative splicing during T-cell signaling. Proc. Natl. Acad. Sci. USA. 2015;112:E2139–E2148. doi: 10.1073/pnas.1423695112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang E.T., Ward A.J., Cherone J.M., Giudice J., Wang T.T., Treacy D.J., Lambert N.J., Freese P., Saxena T., Cooper T.A., Burge C.B. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015;25:858–871. doi: 10.1101/gr.184390.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz A.S., Preussner M., Bunse M., Karni R., Heyd F. Activation-Dependent TRAF3 Exon 8 Alternative Splicing Is Controlled by CELF2 and hnRNP C Binding to an Upstream Intronic Element. Mol. Cell. Biol. 2017;37 doi: 10.1128/MCB.00488-16. e00488–e00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apps J.R., Carreno G., Gonzalez-Meljem J.M., Haston S., Guiho R., Cooper J.E., Manshaei S., Jani N., Hölsken A., Pettorini B. Tumour compartment transcriptomics demonstrates the activation of inflammatory and odontogenic programmes in human adamantinomatous craniopharyngioma and identifies the MAPK/ERK pathway as a novel therapeutic target. Acta Neuropathol. 2018;135:757–777. doi: 10.1007/s00401-018-1830-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei C.H., Wu G., Cai Q., Gao X.C., Tong F., Zhou R., Zhang R.G., Dong J.H., Hu Y., Dong X.R. MicroRNA-330-3p promotes cell invasion and metastasis in non-small cell lung cancer through GRIA3 by activating MAPK/ERK signaling pathway. J. Hematol. Oncol. 2017;10:125. doi: 10.1186/s13045-017-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Yen J.H., Lin C.Y., Chuang C.H., Chin H.K., Wu M.J., Chen P.Y. Nobiletin Promotes Megakaryocytic Differentiation through the MAPK/ERK-Dependent EGR1 Expression and Exerts Anti-Leukemic Effects in Human Chronic Myeloid Leukemia (CML) K562 Cells. Cells. 2020;9:877. doi: 10.3390/cells9040877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christie E.L., Bowtell D.D.L. Acquired chemotherapy resistance in ovarian cancer. Ann. Oncol. 2017;28:viii13–viii15. doi: 10.1093/annonc/mdx446. [DOI] [PubMed] [Google Scholar]
- 37.Bookman M.A. Optimal primary therapy of ovarian cancer. Ann. Oncol. 2016;27(Suppl 1):i58–i62. doi: 10.1093/annonc/mdw088. [DOI] [PubMed] [Google Scholar]
- 38.Willenbacher E., Khan S.Z., Mujica S.C.A., Trapani D., Hussain S., Wolf D., Willenbacher W., Spizzo G., Seeber A. Curcumin: New Insights into an Ancient Ingredient against Cancer. Int. J. Mol. Sci. 2019;20:1808. doi: 10.3390/ijms20081808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giordano A., Tommonaro G. Curcumin and Cancer. Nutrients. 2019;11:2376. doi: 10.3390/nu11102376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao S., Pi C., Ye Y., Zhao L., Wei Y. Recent advances of analogues of curcumin for treatment of cancer. Eur. J. Med. Chem. 2019;180:524–535. doi: 10.1016/j.ejmech.2019.07.034. [DOI] [PubMed] [Google Scholar]
- 41.Terlikowska K.M., Witkowska A.M., Zujko M.E., Dobrzycka B., Terlikowski S.J. Potential application of curcumin and its analogues in the treatment strategy of patients with primary epithelial ovarian cancer. Int. J. Mol. Sci. 2014;15:21703–21722. doi: 10.3390/ijms151221703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan B.L., Norhaizan M.E. Curcumin Combination Chemotherapy: The Implication and Efficacy in Cancer. Molecules. 2019;24:2527. doi: 10.3390/molecules24142527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen P., Huang H.P., Wang Y., Jin J., Long W.G., Chen K., Zhao X.H., Chen C.G., Li J. Curcumin overcome primary gefitinib resistance in non-small-cell lung cancer cells through inducing autophagy-related cell death. J. Exp. Clin. Cancer Res. 2019;38:254. doi: 10.1186/s13046-019-1234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J., Liu J., Xu X., Li L. Curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and miR-214 in ovarian cancer. Cancer Chemother. Pharmacol. 2017;79:479–487. doi: 10.1007/s00280-017-3238-4. [DOI] [PubMed] [Google Scholar]
- 45.Murakami M., Ohnuma S., Fukuda M., Chufan E.E., Kudoh K., Kanehara K., Sugisawa N., Ishida M., Naitoh T., Shibata H. Synthetic Analogs of Curcumin Modulate the Function of Multidrug Resistance-Linked ATP-Binding Cassette Transporter ABCG2. Drug Metab. Dispos. 2017;45:1166–1177. doi: 10.1124/dmd.117.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao M.D., Li J.Q., Chen F.Y., Dong W., Wen L.J., Fei W.D., Zhang X., Yang P.L., Zhang X.M., Zheng C.H. Co-Delivery of Curcumin and Paclitaxel by “Core-Shell” Targeting Amphiphilic Copolymer to Reverse Resistance in the Treatment of Ovarian Cancer. Int. J. Nanomedicine. 2019;14:9453–9467. doi: 10.2147/IJN.S224579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah K., Mirza S., Desai U., Jain N., Rawal R. Synergism of Curcumin and Cytarabine in the Down Regulation of Multi-Drug Resistance Genes in Acute Myeloid Leukemia. Anticancer. Agents Med. Chem. 2016;16:128–135. doi: 10.2174/1871520615666150817115718. [DOI] [PubMed] [Google Scholar]
- 48.Wang S., Li J., Xie J., Liu F., Duan Y., Wu Y., Huang S., He X., Wang Z., Wu X. Programmed death ligand 1 promotes lymph node metastasis and glucose metabolism in cervical cancer by activating integrin β4/SNAI1/SIRT3 signaling pathway. Oncogene. 2018;37:4164–4180. doi: 10.1038/s41388-018-0252-x. [DOI] [PubMed] [Google Scholar]
- 49.Guo, Q. (2020). CELF2 stabilize FAM198B in ovarian cancer. figshare, 10.6084/m9.figshare.12451820.v1. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.