Abstract
Marek's disease (MD) is an alphaherpesvirus (Marek's disease virus, MDV)-induced pathology of chickens associated with paralysis, immunosuppression, neurological signs, and T-cell lymphomas. MD is controlled in poultry production via live attenuated vaccines. The purpose of the current study was to compare methods for precipitating exosomes from vaccinated and protected chicken sera (VEX) and tumor-bearing chicken sera (TEX) for biomarker analysis of vaccine-induced protection and MD lymphomas respectively. A standard polyethylene glycol (PEG, 8%) method was compared to a commercial reagent (total exosome isolation reagent, TEI) for exosome yield and RNA content. Although exosomes purified by PEG or TEI were comparable in size and morphology, TEI-reagent yielded 3-4-fold greater concentration. Relative expression of 8 out of 10 G. gallus- and MDV1-encoded miRNAs examined displayed significant difference depending upon the precipitation method used. Standard PEG yields comparable, albeit lower amounts of exosomes than the TEI-reagent and a distinctive miRNA composition.
Keywords: Immunology, Virology, Animal science, Transcriptomics, Veterinary medicine, Hematological system, miRNA, Marek's disease virus, Extracellular vesicle, Exosome precipitation
Immunology; Virology; Animal science; Transcriptomics; Veterinary medicine; Hematological system; miRNA; Marek's disease virus; Extracellular vesicle; Exosome precipitation.
1. Introduction
Cells produce various types of membrane enclosed vesicles into the extracellular environment known as “extracellular vesicles (EVs)” that differ in size, origin, and content (Neerukonda et al., 2017). Exosomes (30–150 nm) are a subset of EVs that originate in late endosomes and carry biologically-functional coding and non-coding RNA species (mRNA, miRNA, rRNA, tRNAs, etc.), proteins, and lipids. Exosomes have been purified from a range of biological fluids including serum, urine, cerebrospinal fluid (CSF), breast milk and saliva (Admyre et al., 2007; Alvarez et al., 2012; Andreu et al., 2016; Medapati et al., 2017). Exosomal miRNA and protein content reflect the physiological or pathological state of the originating cell and tissue, making them potentially useful as non-invasive biomarkers for disease diagnosis and response to prophylaxis or therapy.
In a previous study, we compared the efficiency of Total Exosome Isolation (TEI) reagent to “classical” ultracentrifugation (UC) procedure for its ability to recover exosomes from the serum of Marek's Disease Virus (MDV)-infected chickens (Nath Neerukonda et al., 2019). Particles obtained using TEI reagent and UC purification procedures were evaluated for their size, concentration, morphology, and miRNA content by Nanoparticle tracking analysis (NTA), Transmission electron microscopy (TEM) and quantitative real time PCR (qRT-PCR) respectively. Although the particles purified by either procedure conformed to exosome size range and morphology, TEI reagent yielded greater particle concentration with higher miRNA content compared to UC (Nath Neerukonda et al., 2019).
Commercial exosome precipitation reagents such as ExoQuick™ (Systems Biosciences) and total exosome isolation reagent from InVitrogen™ (TEI) are popular because they are fast, easy to perform, require small sample volumes, and do not involve long periods of ultracentrifugation or specialized equipment (Rekker et al., 2014; Weng et al., 2016). Commercial precipitation reagents employ volume-excluding polymers, for instance, polyethylene glycol (PEG), dextrans, or polyvinyls, and are expensive, especially for high sample number and/or volume (Rider et al., 2016; Weng et al., 2016).
As a less-expensive alternative using commonly-available reagents, we characterized the efficiency of PEG and NaCl precipitation for the isolation of exosomes from chicken serum. PEG/NaCl solution-based methods have been in use for over five decades to precipitate and purify retrovirus particles, macro-analytes, and immunoglobulin (Ig) from a variety of starting samples, including environmental water samples to complex biological fluids such as culture supernatants and serum (Albertsson and Frick, 1960; Atha and Ingham, 1981; Kohno et al., 2002; Lewis and Metcalf, 1988; Polson et al., 1964). Since retroviruses and exosomes share similar biophysical, chemical and morphological properties, the PEG solution-based precipitation strategy was recently adopted and modified to purify exosomes from large volumes of culture supernatants, mouse plasma (C57BL/6 and C3H genetic background), human urine, human CSF and saliva (Rider et al., 2016). Exosome preparations of the highest purity were obtained upon overnight precipitation of HEK293T culture supernatant medium with 8% PEG +0.5 M NaCl followed by an ultracentrifugation wash (100,000 x g) in PBS (Rider et al., 2016). This simple solution containing PEG (Mn: 6000) costs less than 0.01 USD per mL of starting sample compared to a commercial precipitation solution that costs 11.1 USD per mL of starting sample (Rider et al., 2016). Furthermore, PEG is inexpensive and widely available in routine diagnostic laboratories and can be used to collect exosomes together into aggregates which are then precipitated by a low or high speed centrifugation step (Fahie-Wilson and Halsall, 2008; Weng et al., 2016).
In the current study, we compared the efficiency of 8% PEG +0.5 M NaCl (8% PEG) solution to the TEI reagent for its ability to precipitate exosomes from the serum of MDV-infected and tumor-bearing chickens (referred to as TEX) and from MD vaccinated and protected chickens (referred to as VEX). Marek's disease (MD) in commercial poultry is caused by virulent MDV-1 field strains and is controlled by application of live attenuated vaccines. Gross lesions of MD affected poultry include uni- or bilateral paralysis, profound immune suppression, and the rapid formation of visceral or peripheral T-cell lymphomas. The transforming ability of virulent MDV-1 strains is due to direct or indirect actions of viral primary oncoprotein Meq and other gene products (Meq spliced products, vTR, RLORF4, MDV-1 miRNAs) implicated in T-cell transformation (Jarosinski et al., 2005, 2007; Trapp et al., 2006; Yao and Nair, 2014). In an effort to advance serum exosome-based biomarker analysis and discovery in MDV-vaccinated and MD-affected chickens, we tested the efficiency of 8% PEG as a less expensive alternative to TEI reagent for serum exosome purification. We characterized the exosomes purified from both sources using the two methods for their particle size, concentration, morphology and miRNA content by nanoparticle tracking analysis (NTA), transmission electron microscopy (TEM) and qRT-PCR, respectively.
2. Materials and methods
2.1. Serum samples
The serum samples used in this study were obtained from a trial comparing the MD efficacy of commercial vaccines that has been described, previously (Nath Neerukonda et al., 2019). Briefly, serum was obtained at the end of vaccine trial that included commercial broiler chickens that were either unvaccinated (n = 3) or vaccinated at one day-of-age with a 1X commercial dose (~2,500 PFU) of CVI988 (Rispens) vaccine. These treatment groups were placed in contact with two-week old, MDV-1-inoculated (vv + MDV, strain, TK2a-inoculated) “shedder” chickens (Neerukonda et al., 2016, 2018). Data for the chicken serum sources were given in Table 1.
Table 1.
Source of seruma exosome samples.
| Bird Tag | Treatment Group | MD Status & sex | miRNA | NTA | TEM |
|---|---|---|---|---|---|
| BL4250 | CVI988-vaccinate | Neg. (M) | + | - | - |
| BL4343 | CVI988-vaccinate | Neg. (F) | + | - | - |
| BL4254 | CVI988-vaccinate | Neg. (M) | + | + | + |
| FU5744 | Unvaccinated contact | + (F, spleen tumor) | + | - | - |
| FU5759 | Unvaccinated contact | + (F, spleen tumor) | + | - | - |
| FU5676 | Unvaccinated contact | + (M, heart tumor) | + | + | + |
Serum was obtained from commercial broiler chickens, provided as embryonated eggs by Mountaire Farms, Inc., Millsboro, DE used in a vaccine study comparing CVI988 vaccines; NTA: Nanoparticle tracking analysis; TEM: transmission electron microscopy.
The performance of the vaccine trial from which serum samples were obtained was in accordance with University of Delaware Institutional Animal Care and Use Committee (IACUC) protocol #64R-2016-0, SOP #1 of MSP.
2.2. Exosome purification
Exosome precipitation was carried out via 8% PEG and TEI reagent procedures. To avoid discrepancies due to inter-bird variations, serum from each bird was subjected to both precipitation procedures (see Table 1), simultaneously. Exosomes precipitated from the sera of CVI-988-vaccinated and protected, and unvaccinated tumor-bearing chickens are designated as Vaccinate Exosomes (VEX) and Tumor Exosomes (TEX), respectively.
Particle precipitation with 8% PEG solution was performed as described previously by Rider et al. (2016). Serum (200 μl) was subjected to centrifugation at 500 ×g for 5 min to pellet cells followed by centrifugation at 2000 ×g for 30 min to pellet cellular debris. Next, a centrifugation step was performed at 12,000 ×g for 45 min to pellet microvesicles and large apoptotic bodies, followed by the transfer of supernatants to an equal volume of a 2X PEG solution (16% PEG +1 M NaCl) at 4 °C, to achieve the desired final concentration (8% PEG +0.5 M NaCl). PEG precipitation was performed by inversion, and incubation at 4 °C overnight (at least 12 h). The next day, samples were centrifuged in a microcentrifuge at maximum speed, 15,000 rpm (Eppendorf, model 5424) for 1 h. The PEG supernatant solution was discarded and the exosome-containing pellets were resuspended in exosome-free PBS. All centrifugation steps were performed at 4 °C.
For the TEI method, exosomes were purified from 200 μl of serum using 1/5th volume of TEI reagent (Invitrogen) according to manufacturer's recommendations, as described previously (Nath Neerukonda et al., 2019).
2.3. Nanoparticle tracking analysis (NTA)
The concentration and size distribution profile of particles precipitated via 8% PEG or TEI reagent were evaluated by tracking their Brownian motion using a Nanosight nanoparticle tracking instrument (NS300, Malvern, Worcestershire, UK) and analyzed with NTA 3.2 Dev Build 3.2.16 software. The following post-acquisition analysis settings were selected: minimum detection threshold 4, automatic blur, and automatic minimum expected particle size. Samples were diluted 1:20 (VEX) or 1:100 (TEX) in PBS to obtain concentration profiles within the detection range of the instrument. This dilution strategy allowed us to achieve measured mean particle concentration 1.6–5.6 × 109 particles per ml. For each sample, three 1 min videos were recorded and analyzed in batch processing mode. Videos were recorded at camera level 12 with minimum expected particle size, track length, and blur setting, all set to default.
2.4. Transmission electron microscopy (TEM)
TEM was performed on negatively stained exosomes as previously described (Nath Neerukonda et al., 2019). Essentially, for TEM analysis, 400 mesh, carbon-coated copper grids were glow-discharged with a PELCO easiGlow™ system to render the surface of the grids hydrophilic. Grids were briefly floated on drops of purified exosome-PBS suspensions, washed by sequential placing on drops of water, and negatively-stained with 2% uranyl acetate. Under the condition where silk-like PEG films and vesicle overcrowding was observed upon negative staining of particles precipitated by either procedure, samples were further diluted (1:10) and resuspended in particle free PBS to improve the resolution.
Air-dried grids were examined using a Zeiss Libra 120 transmission electron microscope at 120 kV. Images were acquired with a Gatan Ultrascan 1000 2k x 2k CCD camera in the bioimaging core at the Delaware Biotechnology Institute at the University of Delaware. For each set of analyses, at least 10 fields were imaged of exosomes, at two magnifications, and representative images are shown.
2.5. Exosomal RNA isolation, reverse transcription and qRT-PCR analysis
Total RNA isolation, reverse transcription (RT) and qRT-PCR analyses of selected Gallus gallus (gga-miR-) and MDV-1 (MDV1-miR-) miRNAs were performed as described (Nath Neerukonda et al., 2019). No RT and no template controls were included. The gga- and MDV1-miRs under study were selected based on their significant and differential expression in VEX relative to TEX as obtained upon Illumina high-throughput sequencing of exosomal small RNAs from a previous independent MD challenge study.
2.6. Data analysis
For statistical analyses, MS Excel and GraphPad Prism 5 (GraphPad Software, CA, USA) were used. miRNA Ct values were presented as Ct means ± standard deviation (SD). To compare significant differences in miRNA Ct values between VEX and TEX, comparative delta Ct method and an unpaired t-test were used. For correlation analyses between precipitation methods, the Pearson's correlation coefficient was calculated. The significance threshold was set to a fold change ≥2 (~1 threshold cycle difference) with a p value ≤ 0.05.
3. Results
3.1. Size and yield of particles precipitated by 8% PEG and TEI reagent kit
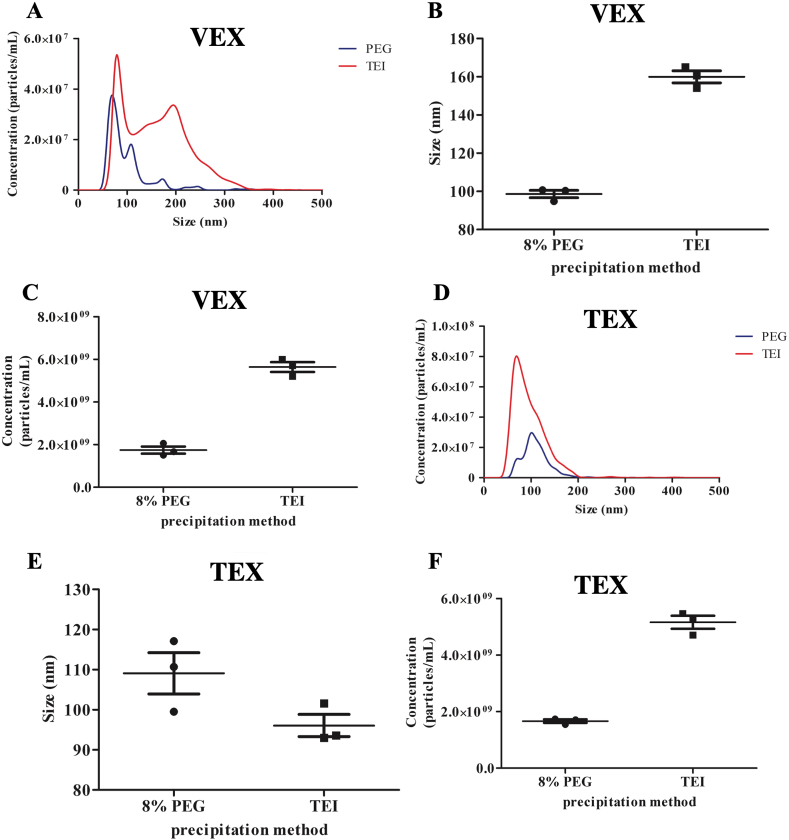
The size distribution profile of 8% PEG-precipitated VEX particles displayed two narrow peaks with a mean particle diameter of 98.7 ± 1.9 nm, and the maximum peak number of detected particles had a diameter of 66.1 ± 3.2 nm (Figures 1A and 1B). The size distribution profile of TEI reagent-precipitated VEX particles displayed one narrow peak and one broader peak with an overall mean particle diameter of 159.9 ± 3.2 nm (Fig. 1A and B). A majority of TEI reagent-precipitated VEX particles sized 79.1 ± 1.0 nm in diameter. The particle subset corresponding to the broader peak comprised a majority of particles sized 195 nm in diameter.
Figure 1.
Size and Concentration of Exosomes Purified by 8% Polyethylene glycol (PEG) and Total Exosome Isolation precipitation (TEI) reagent. Size distribution and concentration profiles of vaccinate exosome (VEX) particles (A, C and E) and tumor exosome (TEX) particles (B, D, and F) purified by PEG and TEI reagent are shown. Error bars denote SEM.
The diameter of particles precipitated by either procedure is within the size range of exosomes (30–150 nm), although the TEI reagent co-precipitated slightly larger particles. Particle concentrations in 8% PEG- and TEI reagent-precipitated VEX fractions were 1.74 and 5.64 billion per mL of serum, respectively, with a 3-fold higher yield by the TEI-reagent (Figure 1C).
TEX fractions precipitated by either procedure harbored particle concentrations well above the detection threshold of the instrument, and had to be further diluted to reach the instrument detection limit (a 5-fold higher dilution compared to VEX fractions; see methods). The size distribution profile of 8% PEG-precipitated TEX particles displayed one narrow peak with a mean particle diameter of 109.1 ± 5.1 nm (Fig. 1D and E). A major proportion of particles had a diameter of 104.7 ± 4.0 nm. Similarly, the size distribution profile of TEI reagent-precipitated TEX particles displayed one narrow peak with a mean particle diameter of 96.1 ± 2.8 nm and a majority of particles having a diameter of 69.0 ± 1.1 nm. With respect to particle concentrations, TEI reagent again yielded 3-fold higher number of particles per ml compared to 8% PEG, with an actual concentration of 5.17 and 1.66 billion particles per ml respectively (Figure 1F).
3.2. Particle visualization by TEM
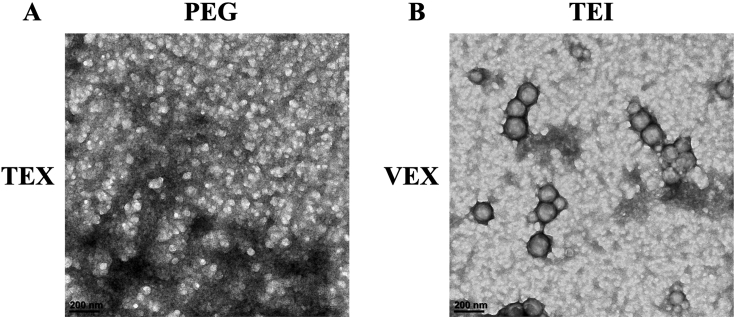
Upon confirming that the size range of particles precipitated by both the procedures conformed to that of exosomes, we performed TEM to visualize the size and morphology of precipitated particles. Notably, when the particles were negatively stained in their original PBS resuspensions, VEX and TEX particles precipitated by either procedure had a spherical shape and appeared coated or held together by a silk-like PEG-film, as previously described (Fig. 2A and B (Weng et al., 2016),). Particles had a diameter of ca. 30–120 nm, confirming them as exosomes.
Figure 2.
TEM of Concentrated TEX and VEX. The micrographs above show exosome aggregates precipitated by 8% PEG (left) and TEI reagent (right). Note multiple exosomes wrapped in silk-like PEG films (A) and as exosome aggregates (B). Bars in the lower left show TEM scale (200 nm).
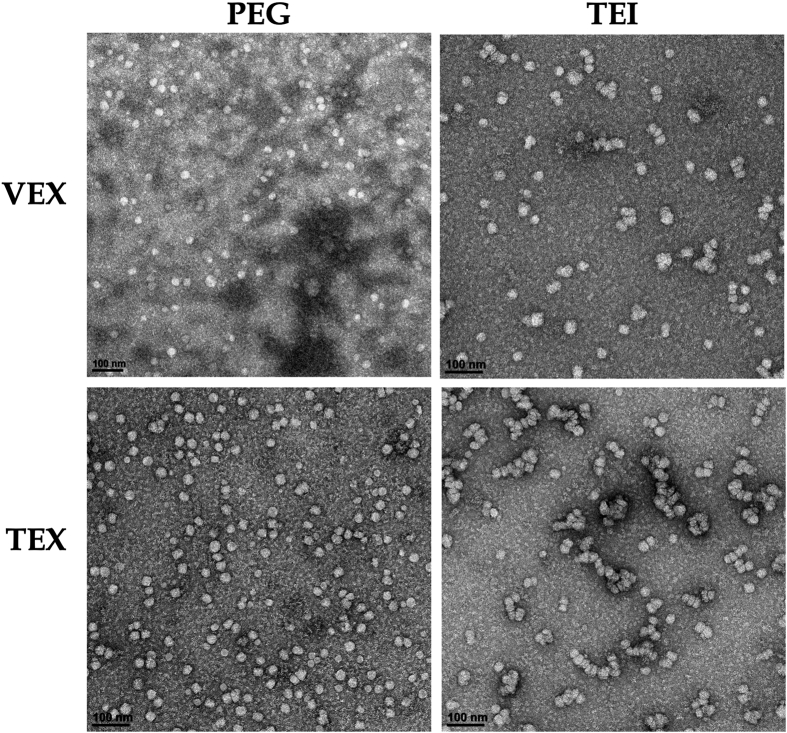
To free exosome aggregates from the PEG polymeric network and improve the resolution, exosome suspensions were further diluted (see methods) in PBS prior to negative staining. Particles precipitated by each procedure displayed typical spherical morphology, although an aggregation of dozens of exosomes was still observed (Figure 3). Collectively, these findings indicate that both precipitation approaches allowed exosome purification in the form of exosome aggregates.
Figure 3.
Visualization of VEX and TEX by TEM. The micrographs above show representative fields of exosomes purified by 8% PEG (left) and TEI reagent (right). The scale bar (lower left in each panel) shows 100 nm.
3.3. Exosomal miRNA analysis by qRT-PCR
Differences in miRNA content in exosomes precipitated by both the procedures were assessed by qRT-PCR. Both precipitation procedures allowed the detection of every investigated miRNA above the detection limit (Ct < 35) (Table 2). Upon examining the raw Ct values, the miRNA threshold detection levels were on average 2.21 cycles lower in TEI reagent-precipitated VEX particles with an overall higher miRNA content compared to 8% PEG-precipitated VEX particles (Table 2). On the other hand, the average Ct value of examined miRNAs in 8% PEG-precipitated TEX particles was 0.76 cycles higher than that of TEI reagent-precipitated TEX particles (Table 2).
Table 2.
Exosomal miRNA Ct values (±SD) as Detected by qRT-PCR.
| miRNA ID | VEX |
TEX |
||
|---|---|---|---|---|
| 8% PEG | TEI | 8% PEG | TEI | |
| gga-miR-146b-5p | 21.84 ± 0.51 | 20.74 ± 0.52 | 20.36 ± 0.89 | 20.8 ± 1.07 |
| MDV1-miR-M6-5p | 26.17 ± 0.58 | 25.31 ± 0.01 | 24.82 ± 0.71 | 25.53 ± 0.94 |
| gga-miR-99a-5p | 26.24 ± 1.19 | 25.23 ± 0.11 | 26.13 ± 0.04 | 25.38 ± 0.64 |
| gga-miR-21-5pa | 26.85 ± 1.04 | 24.18 ± 0.46 | 22.26 ± 0.01 | 24.40 ± 1.47 |
| MDV1-miR-M4-5p | 27.40 ± 0.16 | 25.13 ± 0.36 | 25.21 ± 0.08 | 25.53 ± 0.96 |
| MDV1-miR-M8-5pa | 28.83 ± 1.1 | 26.04 ± 0.38 | 24.13 ± 0.12 | 26.04 ± 0.57 |
| MDV1-miR-M12-3pa | 29.37 ± 0.73 | 26.43 ± 0.59 | 25.40 ± 0.33 | 26.01 ± 0.81 |
| gga-miR-2188-5p | 30.41 ± 0.50 | 28.20 ± 0.54 | 28.90 ± 0.22 | 28.89 ± 1.23 |
| gga-miR-27b-3pa | 30.47 ± 0.70 | 28.15 ± 0.31 | 29.38 ± 0.09 | 28.20 ± 0.46 |
| gga-miR-10b-5pa |
33.54 ± 0.85 |
28.83 ± 0.58 |
29.34 ± 0.44 |
29.95 ± 0.44 |
| Average Ct | 28.11 ± 3.02 | 25.9 ± 2.24 | 25.32 ± 2.85 | 26.08 ± 2.43 |
miRNA abundance differences in VEX were significant different between the purification methods.
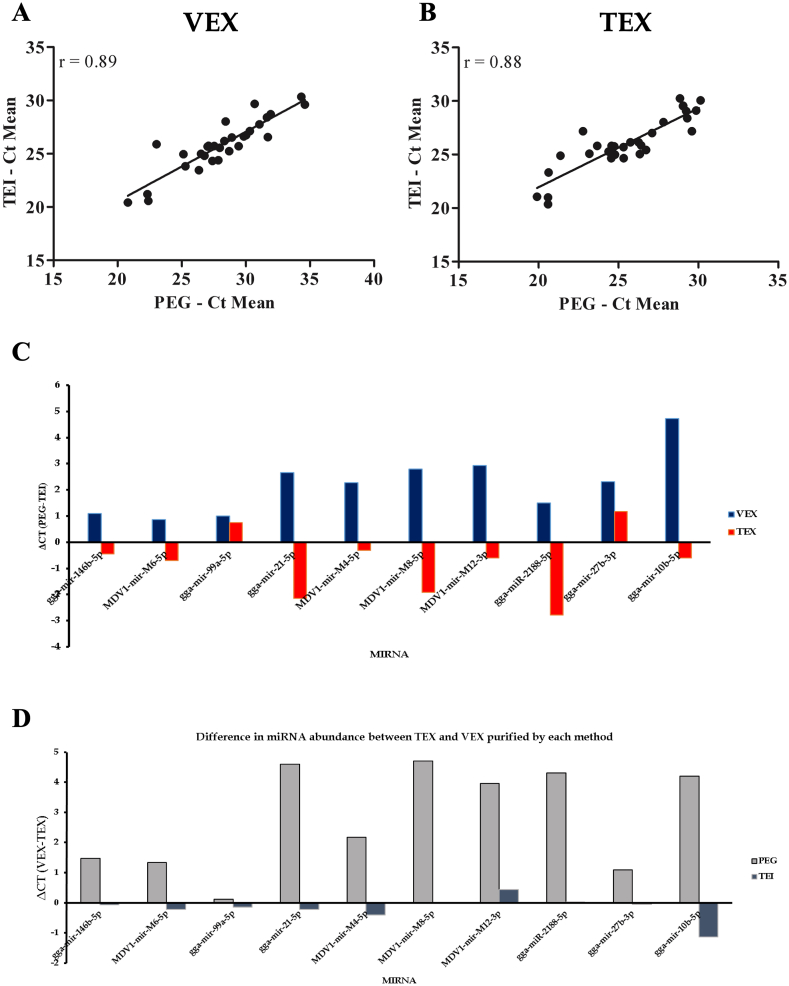
The most abundant miRNA detected in VEX and TEX precipitated by either procedure was gga-miR-146b. While the least abundant miRNA detected in VEX precipitated by 8% PEG was gga-miR-10b, in VEX precipitated by TEI reagent, miR-2188 appeared to be the least abundant and miR-10b remained the second least abundant miRNA (Table 2). Overall, VEX miRNA expression trends displayed a strong and significant correlation between the precipitated procedures (Figure 4A, Pearson r = 0.89, p < 0.0001). The least and second least abundant miRNAs in 8% PEG precipitated TEX were gga-miR-27b and -10b, respectively. On the other hand, whereas miR-27b remained to be the second least abundant miRNA in TEI reagent precipitated TEX, gga-miR-10b appeared as the least abundant. The miRNA expression trends in TEX precipitated by either procedure demonstrated a strong correlation (Figure 4B, Pearson r = 0.88, p < 0.0001).
Figure 4.
Comparison of miRNA species abundance in VEX and TEX purified by 8% PEG and TEI reagent. Panels A and B show correlation analysis of miRNA species abundance in VEX (A), TEX (B). Panel C shows the correlation of the normalized VEX to TEX ratio by species (C) between the exosome precipitation procedures. Pearson correlation coefficient (r) is indicated for the comparison. Panel D shows the relative MDV-1 and chicken (G.gallus) miRNA expression in VEX compared to TEX in 8% PEG-precipitated exosomes (black bars) and TEI-precipitated exosomes (grey bars). The names of cellular and MDV miRNAs and Ct values are given in Table 2 qRT-PCR primer sequences are indicated in Nath Neerukonda et al. (2018). Error bars denote SEM. Asterisk (∗) denotes significant difference in relative expression level (p < 0.05).
In terms of differences in expression of MDV1 and G. gallus miRNAs between 8% PEG- and TEI-precipitated fractions, TEI-precipitated VEX fractions displayed greater abundance of all the miRNAs in the study with their minimum thresholds of detection 0.85–4.72 cycles lower than that of 8% PEG-precipitated fractions (Figure 4C). On the contrary, with the exception of miR-99a and -27b, all the remaining miRNAs displayed greater abundance in 8% PEG-precipitated TEX fractions and their cycle thresholds were 0.44–2.79 cycles lower than that of TEI-precipitated TEX fractions. These findings indicate that disease status may have an impact on the efficiency of the method of choice. However, when we examined miRNA abundance differences between VEX and TEX fractions precipitated through 8% PEG, all miRNAs except miR-99a displayed greater abundance in TEX with at least one threshold cycle difference that corresponds to a 2-fold difference in expression according to 2−ΔCt expression formula (Figure 4C). Among those greater in abundance in TEX, miRs, -10b, -2188, -21, -M8, -M4 and –M12 displayed more than two threshold cycles difference and the latter four miRNAs were also identified to be upregulated in TEX in a previous deep sequencing study of VEX and TEX miRNAs.
On the other hand, differences in miRNA abundance between VEX and TEX fractions obtained by TEI precipitation method were minimal except miR-10b, which displayed greater abundance in VEX with one threshold cycle difference (Figure 4C). All the remaining miRNAs displayed threshold cycle differences ranging between 0-0.43 and none of expression trends correlated with those seen among 8% PEG-precipitated fractions (Figure 4D). Altogether, our findings point to an overall greater abundance of miRNAs in 8% PEG-precipitated fractions or in other words, 8% PEG precipitation methodology yielded an appreciable difference in the miRNA abundance with a higher abundance of oncogenic G. gallus (miR-21, -10b) and MDV-1 (-M4, -M8 and –M12) miRNAs (Figure 4D).
4. Discussion
Current methods of exosome purification include most widely used “classical” ultracentrifugation and commercial polymeric precipitation reagents available through System Biosciences (ExoQuick™), Invitrogen (TEI), and 101Bio (PureExo®) (Thery et al., 2006). Other methods based on size [i.e. size-exclusion chromatography (Boing et al., 2014; de Menezes-Neto et al., 2015; Lai et al., 2010)], filtration [ultrafiltration, microfiltration (Heinemann et al., 2014; Jo et al., 2014; Merchant et al., 2010) and dialysis (Musante et al., 2014)], affinity [i.e. immune-magnetic beads (Clayton et al., 2001), substrate affinity chromatography (He et al., 2014) and microfluidic devices (He et al., 2014)] have also been introduced for exosome purification. We previously reported lower recovery of exosomes from the serum of MDV-infected chickens upon ultracentrifugation compared to TEI reagent procedure (Nath Neerukonda et al., 2019). Moreover, other exosome purification methods based on size and affinity although currently implemented, are limited to low throughput applications, require special equipment, and have been demonstrated to have varying efficiency in terms of exosome recovery (Alvarez et al., 2012; Andreu et al., 2016; Baranyai et al., 2015; Caradec et al., 2014; Momen-Heravi et al., 2013; Royo et al., 2016a, 2016b; Tauro et al., 2012).
In the present study, we described a facile, inexpensive and effective precipitation of exosomes from the serum of MDV-infected chickens using the classic PEG/NaCl method. This precipitation procedure was adapted from the procedure used to concentrate retroviruses and is similar to the recently described “ExtraPEG” procedure used to purify exosomes (Kohno et al., 2002; Rider et al., 2016). Aqueous PEG could wrap hundreds of exosomes together, causing them to aggregate, so that they can easily recovered by a single low speed centrifugation step (Weng et al., 2016). Accordingly, upon negative staining and TEM visualization, exosome aggregates and exosomes held together by silk-like PEG films were observed with 8% PEG precipitated exosomes. This phenomenon was also observed with TEI reagent-precipitated exosomes indicating a similar mechanism of exosome wrapping to be the driving force for exosome isolation.
Our TEM finding is in concordance with a recent study finding that described one- or two-step PEG based precipitation of exosomes from the HeLa supernatants achieved through a final 10% PEG solution without any ionic salts (Weng et al., 2016). In that study, the authors found 10% PEG or higher final concentration (Mn:10 kDa) to be more efficient in exosome recovery compared to lower concentrations tested (5% or 7%). Furthermore, field emission scanning electron microscopy and high resolution TEM of 10% PEG precipitated fractions displayed exosome aggregates coated with silk-like PEG films (Weng et al., 2016). In our study we used slightly lower molecular weight PEG and a lower final PEG concentration to successfully recover exosomes. The choice of critical concentration of PEG required to effectively precipitate exosomes, so they can be acquired by a centrifugation step, depends upon the nature of starting solution (Weng et al., 2016).
In our study, we tested slightly lower final PEG concentration (8%), as our starting point due to the viscosity of serum, as opposed less viscous culture supernatant in other studies that employed 10% final PEG concentration (Weng et al., 2016). On the other hand, 20% final PEG (3350 Da) solution-based precipitation of exosomes from HEK293 supernatants followed by lectin affinity chromatography was also demonstrated to effectively precipitate EVs, including exosomes and ectosomes (Jackson et al., 2017). In addition, 12% final PEG (6–8 kDa) solution was also demonstrated to enrich for EVs, possibly larger ectosomes, from G-protein coupled receptor expressing stable cell lines (Medapati et al., 2017). Together, these findings underscore that, in addition to exosomes, PEG-based precipitation also enriches for other EVs.
The relative abundance of MDV-1- and gga-miRNAs in VEX compared to TEX revealed clear differences depending on precipitation procedure used. MDV-1-miR-M4, -M12 and –M8 belonging to the meq and LAT cluster miRNAs, respectively, and displayed 6-8-fold higher expression in TEX compared to VEX obtained by 8% PEG, as described previously (Nath Neerukonda et al., 2019). MDV-1-miR-M4 is an ortholog of cellular miR-155, and MDV-1 mutants lacking miR-M4 or its seed sequence mutated were severely attenuated in their ability to cause T-cell lymphomas in infected chickens, even though lytic replication per se was largely unaffected (Yu et al., 2014; Zhao et al., 2011). Both MDV-1-miR-M4 and –M12 located antisense to RLORF8 coding region are highly expressed in MD lymphoblastoid cell lines and target RLORF8 3’ UTR (Parnas et al., 2014). Similarly, well known oncomiRs, gga-miR-21 and -10b, displayed lower abundance in 8% PEG-precipitated VEX fractions compared to TEX fractions. MicroRNA 21 (miR-21) is a ubiquitous oncomiR overexpressed in a myriad of carcinomas including lung, prostate, breast, pancreas, colon, head and neck, stomach, esophagus, liver, etc., compared to normal adjacent tissues, and it is the only miRNA that is consistently upregulated in all types of carcinomas (Asangani et al., 2008; Frezzetti et al., 2011; Ma et al., 2011; Si et al., 2007). In addition, it is also upregulated in liquid cancers such as leukemia (Fulci et al., 2007; Volinia et al., 2010), lymphomas (Lawrie et al., 2007; Stik et al., 2013) and multiple myeloma (Pichiorri et al., 2008). In MDA-MB-231 metastatic breast cancer cell line and SUM149 cell line transplanted tumors, miR-10b facilitated migration and metastasis in vitro and in vivo respectively by targeting HoxD10 (Wang et al., 2016). In the context of MD, a previous study investigating H4K4me3 and H3K27me3 chromatin marks in the bursas of 14-day old MD susceptible (L72) and resistant (L63) line of chickens with and without infection with a partially attenuated vv + MDV strain (648A) has reported higher levels of H3K27me3 repressive mark around miR-10b in the L63 line compared to L72, either with or without infection. At 10 dpi, only infected birds from L63 line displayed higher levels of H3K27me3 at miR-10b locus indicative of potential oncogenic functions of miR-10b (Mitra et al., 2015). Finally, gga-miR-99a existed in relatively equivalent proportions in 8% PEG- or TEI-precipitated VEX and TEX fractions. Based on greater expression of MDV-1 meq (-M4, -12) and LAT (-M6, -M8) cluster miRNAs and gga-miR-21 in PEG-precipitated TEX, we would like to propose them to be serum exosomal biomarkers of MD lymphomas in affected chickens.
With regards to miR-21 and -99a, one previous study indicated existence of these circulating miRNAs to a major proportion in non-membrane bound fractions compared to vesicle fractions of healthy human plasma, as Ago2-bound ribonucleoprotein complexes (RNPs) (Arroyo et al., 2011). While we anticipate that 8% PEG may have co-precipitated serum protein complexes, the potential existence of Ago2-bound miRNAs in chicken serum remains to be investigated.
Although the TEI reagent appeared to have yielded 3-fold higher number of particles compared to 8% PEG, this increase in yield did not correlate with increased miRNA abundance levels, suggesting that TEI reagent may have co-precipitated non-vesicle fractions that lacked miRNAs to certain extent.
Taken together, our results indicate, while TEI reagent yielded a greater number of particles compared to 8% PEG that correspond to exosome size range, 8% PEG-precipitated exosome fractions contained greater level of miRNAs. Whether these fractions comprise miRNAs in vesicle or non-vesicle forms requires further investigation. Follow-up studies will focus on Proteinase K treatment of serum samples prior to exosome precipitation with 8% PEG and TEI reagent to evaluate the existence and contribution of Ago2-bound miRNAs to observed miRNA expression.
Declarations
Author contribution statement
Sabari Nath Neerukonda: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Nicholas A. Egan, Joseph Patria, Imane Assakhi, Phaedra Tavlarides-Hontz: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Shannon Modla, Eric R. Muñoz, Matthew B. Hudson: Contributed reagents, materials, analysis tools or data.
Mark S. Parcells: Conceived and designed the experiments; Performed the experiments; Wrote the paper.
Funding statement
This work was supported by the College of Agriculture and Natural Resources Seed Grant to MSP.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Admyre C., Johansson S.M., Qazi K.R., Filen J.J., Lahesmaa R., Norman M., Neve E.P., Scheynius A., Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- Albertsson P.A., Frick G. Partition of virus particles in a liquid two-phase system. Biochim. Biophys. Acta. 1960;37:230–237. doi: 10.1016/0006-3002(60)90228-6. [DOI] [PubMed] [Google Scholar]
- Alvarez M.L., Khosroheidari M., Kanchi Ravi R., DiStefano J.K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- Andreu Z., Rivas E., Sanguino-Pascual A., Lamana A., Marazuela M., Gonzalez-Alvaro I., Sanchez-Madrid F., de la Fuente H., Yanez-Mo M. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracell. Vesicles. 2016;5:31655. doi: 10.3402/jev.v5.31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., Tait J.F., Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Nat. Acad. Sci. U. S. A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani I.A., Rasheed S.A., Nikolova D.A., Leupold J.H., Colburn N.H., Post S., Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Atha D.H., Ingham K.C. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J. Biol. Chem. 1981;256:12108–12117. [PubMed] [Google Scholar]
- Baranyai T., Herczeg K., Onodi Z., Voszka I., Modos K., Marton N., Nagy G., Mager I., Wood M.J., El Andaloussi S., Palinkas Z., Kumar V., Nagy P., Kittel A., Buzas E.I., Ferdinandy P., Giricz Z. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One. 2015;10 doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boing A.N., van der Pol E., Grootemaat A.E., Coumans F.A., Sturk A., Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caradec J., Kharmate G., Hosseini-Beheshti E., Adomat H., Gleave M., Guns E. Reproducibility and efficiency of serum-derived exosome extraction methods. Clin. Biochem. 2014;47:1286–1292. doi: 10.1016/j.clinbiochem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Clayton A., Court J., Navabi H., Adams M., Mason M.D., Hobot J.A., Newman G.R., Jasani B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods. 2001;247:163–174. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- de Menezes-Neto A., Saez M.J., Lozano-Ramos I., Segui-Barber J., Martin-Jaular L., Ullate J.M., Fernandez-Becerra C., Borras F.E., Del Portillo H.A. Size-exclusion chromatography as a stand-alone methodology identifies novel markers in mass spectrometry analyses of plasma-derived vesicles from healthy individuals. J. Extracell. Vesicles. 2015;4:27378. doi: 10.3402/jev.v4.27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahie-Wilson M., Halsall D. Polyethylene glycol precipitation: proceed with care. Ann. Clin. Biochem. 2008;45:233–235. doi: 10.1258/acb.2008.007262. [DOI] [PubMed] [Google Scholar]
- Frezzetti D., De Menna M., Zoppoli P., Guerra C., Ferraro A., Bello A.M., De Luca P., Calabrese C., Fusco A., Ceccarelli M., Zollo M., Barbacid M., Di Lauro R., De Vita G. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene. 2011;30:275–286. doi: 10.1038/onc.2010.416. [DOI] [PubMed] [Google Scholar]
- Fulci V., Chiaretti S., Goldoni M., Azzalin G., Carucci N., Tavolaro S., Castellano L., Magrelli A., Citarella F., Messina M., Maggio R., Peragine N., Santangelo S., Mauro F.R., Landgraf P., Tuschl T., Weir D.B., Chien M., Russo J.J., Ju J., Sheridan R., Sander C., Zavolan M., Guarini A., Foa R., Macino G. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- He M., Crow J., Roth M., Zeng Y., Godwin A.K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14:3773–3780. doi: 10.1039/c4lc00662c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann M.L., Ilmer M., Silva L.P., Hawke D.H., Recio A., Vorontsova M.A., Alt E., Vykoukal J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A. 2014;1371:125–135. doi: 10.1016/j.chroma.2014.10.026. [DOI] [PubMed] [Google Scholar]
- Jackson C.E., Scruggs B.S., Schaffer J.E., Hanson P.I. Effects of inhibiting VPS4 support a general role for ESCRTs in extracellular vesicle biogenesis. Biophys. J. 2017;113:1342–1352. doi: 10.1016/j.bpj.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosinski K., Kattenhorn L., Kaufer B., Ploegh H., Osterrieder N. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Nat. Acad. Sci. 2007;104:20025–20030. doi: 10.1073/pnas.0706295104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosinski K.W., Osterrieder N., Nair V.K., Schat K.A. Attenuation of Marek's disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J. Virol. 2005;79:11647–11659. doi: 10.1128/JVI.79.18.11647-11659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo W., Kim J., Yoon J., Jeong D., Cho S., Jeong H., Yoon Y.J., Kim S.C., Gho Y.S., Park J. Large-scale generation of cell-derived nanovesicles. Nanoscale. 2014;6:12056–12064. doi: 10.1039/c4nr02391a. [DOI] [PubMed] [Google Scholar]
- Kohno T., Mohan S., Goto T., Morita C., Nakano T., Hong W., Sangco J.C., Morimatsu S., Sano K. A new improved method for the concentration of HIV-1 infective particles. J. Virol. Methods. 2002;106:167–173. doi: 10.1016/s0166-0934(02)00162-3. [DOI] [PubMed] [Google Scholar]
- Lai R.C., Arslan F., Lee M.M., Sze N.S., Choo A., Chen T.S., Salto-Tellez M., Timmers L., Lee C.N., El Oakley R.M., Pasterkamp G., de Kleijn D.P., Lim S.K. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Lawrie C.H., Soneji S., Marafioti T., Cooper C.D., Palazzo S., Paterson J.C., Cattan H., Enver T., Mager R., Boultwood J., Wainscoat J.S., Hatton C.S. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54:1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Kumar M., Choudhury S.N., Becker Buscaglia L.E., Barker J.R., Kanakamedala K., Liu M.F., Li Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Nat. Acad. Sci. U. S. A. 2011;108:10144–10149. doi: 10.1073/pnas.1103735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medapati M.R., Singh A., Korupally R.R., Henderson D., Klonisch T., Manda S.V., Chelikani P. Characterization of GPCRs in extracellular vesicle (EV) Methods Cell Biol. 2017;142:119–132. doi: 10.1016/bs.mcb.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Merchant M.L., Powell D.W., Wilkey D.W., Cummins T.D., Deegens J.K., Rood I.M., McAfee K.J., Fleischer C., Klein E., Klein J.B. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin. Appl. 2010;4:84–96. doi: 10.1002/prca.200800093. [DOI] [PubMed] [Google Scholar]
- Mitra A., Luo J., He Y., Gu Y., Zhang H., Zhao K., Cui K., Song J. Histone modifications induced by MDV infection at early cytolytic and latency phases. BMC Genomics. 2015;16:311. doi: 10.1186/s12864-015-1492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F., Balaj L., Alian S., Mantel P.Y., Halleck A.E., Trachtenberg A.J., Soria C.E., Oquin S., Bonebreak C.M., Saracoglu E., Skog J., Kuo W.P. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013;394:1253–1262. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L., Tataruch D., Gu D., Benito-Martin A., Calzaferri G., Aherne S., Holthofer H. A simplified method to recover urinary vesicles for clinical applications, and sample banking. Sci. Rep. 2014;4:7532. doi: 10.1038/srep07532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath Neerukonda S., Egan N.A., Patria J., Assakhi I., Tavlarides-Hontz P., Modla S., Muñoz E.R., Hudson M.B., Parcells M.S. Comparison of exosomes purified via ultracentrifugation (UC) and Total Exosome Isolation (TEI) reagent from the serum of Marek’s disease virus (MDV)-vaccinated and tumor-bearing chickens. J. Virol. Methods. 2019;263:1–9. doi: 10.1016/j.jviromet.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Neerukonda S.N., Egan N.A., Parcells M.S. Exosomal communication during infection, inflammation and virus-associated pathology. J. Cancer Ther. Sci. 2017;1(1):1–13. [Google Scholar]
- Neerukonda S.N., Katneni U.K., Bott M., Golovan S.P., Parcells M.S. Induction of the unfolded protein response (UPR) during Marek's disease virus (MDV) infection. Virology. 2018;522:1–12. doi: 10.1016/j.virol.2018.06.016. [DOI] [PubMed] [Google Scholar]
- Neerukonda S.N., Katneni U.K., Golovan S., Parcells M.S. Evaluation and validation of reference gene stability during Marek’s disease virus (MDV) infection. J. Virol. Methods. 2016;236:111–116. doi: 10.1016/j.jviromet.2016.07.017. [DOI] [PubMed] [Google Scholar]
- Parnas O., Corcoran D.L., Cullen B.R. Analysis of the mRNA targetome of microRNAs expressed by Marek's disease virus. mBio. 2014;5 doi: 10.1128/mBio.01060-13. e01060-01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichiorri F., Suh S.S., Ladetto M., Kuehl M., Palumbo T., Drandi D., Taccioli C., Zanesi N., Alder H., Hagan J.P., Munker R., Volinia S., Boccadoro M., Garzon R., Palumbo A., Aqeilan R.I., Croce C.M. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Nat. Acad. Sci. U. S. A. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson A., Potgieter G.M., Largier J.F., Mears G.E., Joubert F.J. The fractionation of protein mixtures by linear polymers of high molecular weight. Biochim. Biophys. Acta. 1964;82:463–475. doi: 10.1016/0304-4165(64)90438-6. [DOI] [PubMed] [Google Scholar]
- Rekker K., Saare M., Roost A.M., Kubo A.L., Zarovni N., Chiesi A., Salumets A., Peters M. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014;47:135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- Rider M.A., Hurwitz S.N., Meckes D.G., Jr. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Scientific Rep. 2016;6:23978. doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo F., Diwan I., Tackett M.R., Zuniga P., Sanchez-Mosquera P., Loizaga-Iriarte A., Ugalde-Olano A., Lacasa I., Perez A., Unda M., Carracedo A., Falcon-Perez J.M. Comparative miRNA analysis of urine extracellular vesicles isolated through five different methods. Cancers (Basel) 2016;8 doi: 10.3390/cancers8120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo F., Zuniga-Garcia P., Sanchez-Mosquera P., Egia A., Perez A., Loizaga A., Arceo R., Lacasa I., Rabade A., Arrieta E., Bilbao R., Unda M., Carracedo A., Falcon-Perez J.M. Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. J. Extracell Vesicles. 2016;5:29497. doi: 10.3402/jev.v5.29497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M.L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.Y. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Stik G., Dambrine G., Pfeffer S., Rasschaert D. The oncogenic MicroRNA OncomiR-21 overexpressed during Marek's disease lymphomagenesis is transactivated by the viral oncoprotein meq. J. Virol. 2013;87:80–93. doi: 10.1128/JVI.02449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro B.J., Greening D.W., Mathias R.A., Ji H., Mathivanan S., Scott A.M., Simpson R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3, Unit 3 22. [DOI] [PubMed] [Google Scholar]
- Trapp S., Parcells M.S., Kamil J.P., Schumacher D., Tischer B.K., Kumar P.M., Nair V.K., Osterrieder N. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 2006;203:1307–1317. doi: 10.1084/jem.20052240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Galasso M., Costinean S., Tagliavini L., Gamberoni G., Drusco A., Marchesini J., Mascellani N., Sana M.E., Abu Jarour R., Desponts C., Teitell M., Baffa R., Aqeilan R., Iorio M.V., Taccioli C., Garzon R., Di Leva G., Fabbri M., Catozzi M., Previati M., Ambs S., Palumbo T., Garofalo M., Veronese A., Bottoni A., Gasparini P., Harris C.C., Visone R., Pekarsky Y., de la Chapelle A., Bloomston M., Dillhoff M., Rassenti L.Z., Kipps T.J., Huebner K., Pichiorri F., Lenze D., Cairo S., Buendia M.A., Pineau P., Dejean A., Zanesi N., Rossi S., Calin G.A., Liu C.G., Palatini J., Negrini M., Vecchione A., Rosenberg A., Croce C.M. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Z., Zhao X., Zuo X., Peng Z. miR-10b promotes invasion by targeting HOXD10 in colorectal cancer. Oncol. Lett. 2016;12:488–494. doi: 10.3892/ol.2016.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y., Sui Z., Shan Y., Hu Y., Chen Y., Zhang L., Zhang Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst. 2016;141:4640–4646. doi: 10.1039/c6an00892e. [DOI] [PubMed] [Google Scholar]
- Yao Y., Nair V. Role of virus-encoded microRNAs in Avian viral diseases. Viruses. 2014;6:1379–1394. doi: 10.3390/v6031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.H., Teng M., Sun A.J., Yu L.L., Hu B., Qu L.H., Ding K., Cheng X.C., Liu J.X., Cui Z.Z., Zhang G.P., Luo J. Virus-encoded miR-155 ortholog is an important potential regulator but not essential for the development of lymphomas induced by very virulent Marek's disease virus. Virology. 2014;448:55–64. doi: 10.1016/j.virol.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Xu H., Yao Y., Smith L.P., Kgosana L., Green J., Petherbridge L., Baigent S.J., Nair V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]