Abstract
Zoonotic spillover, i.e. pathogen transmission from animal to human, has repeatedly introduced RNA viruses into the human population. In some cases, where these viruses were then efficiently transmitted between humans, they caused large disease outbreaks such as the 1918 flu pandemic or, more recently, outbreaks of Ebola and Coronavirus disease. These examples demonstrate that RNA viruses pose an immense burden on individual and public health with outbreaks threatening the economy and social cohesion within and across borders. And while emerging RNA viruses are introduced more frequently as human activities increasingly disrupt wild-life eco-systems, therapeutic or preventative medicines satisfying the “one drug-multiple bugs”-aim are unavailable. As one central aspect of preparedness efforts, this review digs into the development of broadly acting antivirals via targeting viral genome synthesis with host- or virus-directed drugs centering around nucleotides, the genomes’ universal building blocks. Following the first strategy, selected examples of host de novo nucleotide synthesis inhibitors are presented that ultimately interfere with viral nucleic acid synthesis, with ribavirin being the most prominent and widely used example. For directly targeting the viral polymerase, nucleoside and nucleotide analogues (NNAs) have long been at the core of antiviral drug development and this review illustrates different molecular strategies by which NNAs inhibit viral infection. Highlighting well-known as well as recent, clinically promising compounds, structural features and mechanistic details that may confer broad-spectrum activity are discussed. The final part addresses limitations of NNAs for clinical development such as low efficacy or mitochondrial toxicity and illustrates strategies to overcome these.
Keywords: Nucleoside analogues, RNA polymerase, inhibitors, replication
Introduction
When a novel virus acquires the ability to efficiently transmit between humans, it poses an imminent pandemic risk.1,2 Among (re-)emerging RNA viruses, the influenza A virus takes on a prominent role: New strains can and have evolved via reassortment of gene segments within co-infected hosts (antigenic shift)3 and continual mutations in the viral genome (antigenic drift) can lead to vaccine mismatches4 and resistance to existing drugs.5,6 Influenza A virus strains are classified by their surface glycoproteins hemagglutinin (H), which is required for virus attachment, and neuraminidase (N), required for virus budding. Of these proteins, 18 (H) and 11 (N) different subtypes have been identified and they determine the virus’ tropism and pathogenicity. Since the beginning of the 20th century, five major flu pandemics have occurred starting with the 1918 flu pandemic (“Spanish flu”), caused by an H1N1 virus, that led to devastating disease with global death tolls of an estimated 50 million people, accompanied by massive socio-economic impact.7,8 This was followed by the emergence of a novel, H2N2-strain which caused 1–4 million deaths in 1957 (“Asian flu”) and only 11 years later, the emerging H3N2 virus brought about the 1968 flu pandemic (“Hong Kong flu”) that had a similar impact. In 1977, there was an outbreak (“Russian flu”) of what is believed to be a re-emerged H1N1 virus and in 2009, a reassortant H1N1 influenza A virus caused the first flu pandemic of the 21st century (“Swine flu”), only 90 years after the devastating 1918 pandemic.9 This virus (H1N1pdm09), as well as the H3N2 strain are still circulating in the human population causing seasonal outbreaks. Aquatic birds harbor a vast reservoir of avian influenza A viruses (AIVs), which can evolve to highly pathogenic avian influenza A viruses (HPAIVs) and cause severe outbreaks with huge economic cost in farmed poultry.2,10 Occasionally, humans are infected with avian strains by zoonotic spillover, e.g. in the context of poultry farms,1,9,11and numerous events have been reported where people were infected with HPAIV strains H5N1 and H7N9 through close contact with infected animals or via contaminated environments which led to severe disease with high fatality rates.9 Although thus far, sustained human-to-human transmission has not been observed, HPAIVs might acquire this ability via antigenic drift and/or shift.9 The introduction of a novel influenza virus with the ability to efficiently spread among humans into the immuno-naïve human population would naturally lead to massive spread and severe disease,2 potentially cause high fatality rates and have drastic socio-economic consequences. Accordingly, HPAIVs are attributed a high pandemic risk2 and in addition to surveillance, other preparedness efforts are warranted.
Many other RNA viruses continue to repeatedly cross the species barrier from animal to human, and once spillover has occurred, some achieve efficient human-to-human transmission and can thus cause outbreaks of different size. For instance, among the numerous hemorrhagic fever viruses, Filoviridae have repeatedly been introduced from wildlife reservoirs causing severe disease and high mortality in humans.12 The 2014 Western Africa epidemic was the largest Ebola virus outbreak yet, recording almost 30,000 infections with more than 11,000 resulting in death, according to WHO statistics.13 Further, a large variety of negative strand RNA ((–)ssRNA) viruses belonging to the order of Bunyavirales, such as the Andes (hantavirus),14 Lassa (arenavirus),15 and Crimean-Congo hemorrhagic fever virus (nairovirus),16 cause frequent disease outbreaks of different size via cross-species transfer in many parts of the world. Another example of a viral zoonotic disease gaining attention is caused by Nipah virus, a member of the Paramyxoviridae. It causes severe disease in humans with high case fatality rates of up to >70% and with its broad host range, Nipah virus is able to infect and spread among different animals as well as humans, hence its classification as a public health concern.17,18 Among the positive strand RNA ((+)ssRNA) viruses, different coronaviruses have recently emerged from animal reservoirs and caused severe outbreaks of respiratory disease, such as SARS, MERS, and, importantly, the COVID-19 pandemic, the largest pandemic since the 1918 flu.19 The causative agent, SARS-Coronavirus-2, was only recently introduced into the human population and is continuing to spread while this review is being written. It spreads via the respiratory route and has already had most devastating effects on the lives of people around the world with recorded case numbers in the double-digit million range (August 2020); in consequence, it crashes economies and threatens international piece and cooperation. While vaccine candidates are being developed and public health measures are implemented, rapid pandemic response is needed to limit the spread and impact of the virus. Treatment for those suffering from the disease is hampered by the lack of effective antiviral medication; thus, WHO’s solidarity20 as well as a multitude of other recently launched clinical trials are aimed at identifying efficient treatments among available drugs that had previously demonstrated in vitro- or in vivo-potency against related viruses.21
These examples dramatically highlight the urgent need for better and more effective preparedness measures and with it, research to develop broad-spectrum antiviral drugs is gaining momentum. This review examines strategies that ultimately target viral nucleic acid synthesis to stop viral infection. Generally, viral RNA polymerases are considered error-prone and most RNA viruses do not possess any error-correcting function, hence they acquire a comparably high number of mutations during genome replication.22 As a consequence, these viruses evolve rapidly and therefore, they’re considered a high risk for zoonotic disease transmission. Adding to this risk, they can quickly become resistant to treatment or escape vaccine-induced immunity by the same mechanism.23,24 However, this trait can also be exploited to defeat a virus and hence, the viral polymerase sometimes is referred to as its “Achilles heel”.24 Following this approach, viral infection can be treated with compounds that either indirectly or directly block viral nucleic acid synthesis, or those that drive the virus’ mutation rate over a threshold and thereby stop the production of infectious virus (often referred to as lethal mutagenesis).22,23 Virus-targeted examples for such compounds are nucleobase, nucleoside or nucleotide analogues (NNAs) that – after being activated to their corresponding non-natural nucleoside 5’-triphosphate (NTP) form via host cell pathways – are “wrongly” incorporated into the viral genome by the viral polymerase. Host-targeted strategies include compounds that block cellular pathways for the production of natural NTPs, thereby depriving the polymerases of their natural substrates – an established approach which is used in anticancer chemotherapy as well. The following sections will examine both these strategies in more detail.
Targeting viral RNA synthesis via inhibition of the host de novo nucleotide pathways
Upon infection, viruses release their cargo into the host cell and use some of the cell’s own machinery and metabolites to replicate their genome, produce viral progeny and infect new cells. Thus, in a host-targeted approach, the inhibition of certain enzymes or factors of the infected cell can stop the virus from replicating. And since different viruses rely on the same host cell processes, broad-spectrum activity may be achieved. Further, host-targeted compounds benefit from a high barrier to resistance since they aren’t under the control of the error-prone viral replication machinery that often leads to drug-evasion of fast-evolving viruses.25,26 Especially RNA-viruses, owing to their short generation times,22,27 require high amounts of NTPs from the host’s cellular pool to sustain the rapid replication of their genome in acute infection. Hence, by creating an imbalance in the cells’ own NTP pool, the number of mutations that the viral RNA genome acquires during replication can be increased or, simply by depriving the enzyme of its substrates, viral genome synthesis can be stopped altogether. As with all host-directed strategies, however, a balanced inhibition of the cellular pathway is required to maintain cell viability while efficiently blocking the infection.26 With regard to nucleotides, mammalian cells not only rely on de novo biosynthesis but also harbor salvage pathways that recycle purine or pyrimidine nucleotides from e.g. nucleic acid degradation products. While the salvage pathways are able to sustain cell viability, they may not supply sufficient amounts of NTPs to allow fast proliferation of e.g. malignant cells or support viral replication; thus, in addition to antiproliferative therapies, the inhibition of the de novo pathways for nucleotide biosynthesis constitutes a broad-spectrum antiviral strategy.28,29
Blocking de novo pyrimidine nucleotide biosynthesis
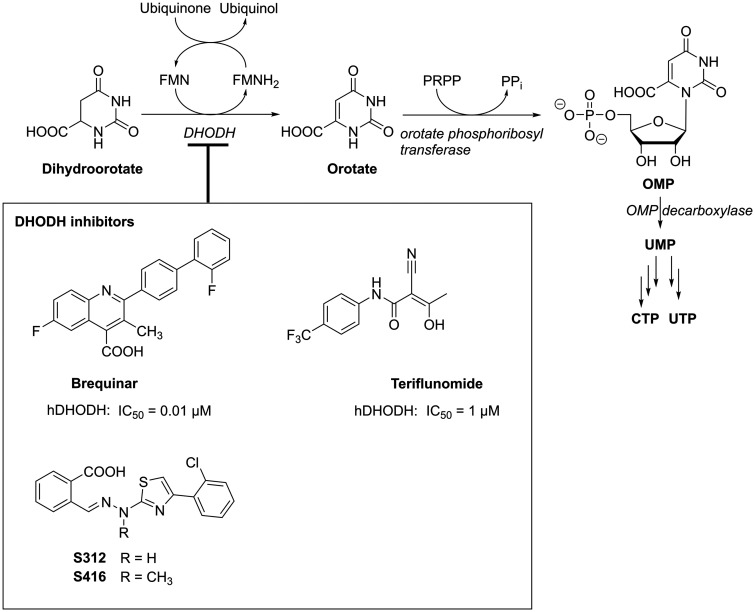
In the de novo pyrimidine nucleotide biosynthetic pathway, dihydroorotate dehydrogenase (DHODH) takes on a central role and its inhibition reduces cellular UTP and CTP,30 and consequently also dCTP and TTP concentrations (Figure 1).32 A very potent inhibitor of DHODH, Brequinar (Figure 1), has been investigated as potential anti-cancer as well as immunosuppressant drug; however, data from clinical trials suggest that this drug is unable to efficiently inhibit DHODH in solid tumors while causing severe side effects systemically.33 These unfavorable therapeutic properties so far excluded Brequinar from gaining approval. One example of a less potent yet more balanced DHODH inhibitor in clinical use is Teriflunomide (Figure 1), an immunosuppressive drug that is applied in the treatment of Multiple Sclerosis. Similarly, its prodrug Leflunomide is applied in Rheumatoid Arthritis28 and both compounds, along with other DHODH inhibitors, have gained attention as potential broad-spectrum antivirals.34 Because they’re targeting a cellular enzyme that is essential in covering the high (d)NTP demand of replicating viruses, these compounds are able to inhibit not only diverse RNA, but also retro- and DNA viruses.26,35 However, so far none of them have successfully completed clinical development as anti-infectives. One limiting factor to their in vivo efficacy may lie in the high plasma levels of uridine, which can enter cells via specific transporters and thus quickly compensate for DHODH inhibition, thereby counteracting the drugs’ effect.28 Still, efforts to develop broadly acting antivirals via targeting DHODH continue and a recent study reported the discovery of two promising DHODH inhibitors S312 and S416 (Figure 1) with low in vivo toxicity starting from a virtual screening.34 Both compounds were able to inhibit the replication of different influenza A viruses, Zika- and Ebola virus, as well as, importantly, SARS-CoV-2.34
Figure 1.
The central role of DHODH in the de novo pyrimidine nucleotide synthesis, two of its known inhibitors, brequinar (not currently approved mainly due to its narrow therapeutic window) and teriflunomide (also used in its prodrug form leflunomide for treating autoimmune disorders), and two recently reported inhibitors, S312 and S416. IC50 values are half-maximal inhibitory compound concentrations reported in Knecht and Loffler.31 PRPP – 5-phosphoribosyl pyrophosphate; FMN – Flavin mononucleotide; OMP – Orotidine 5’-monophosphate.
Blocking de novo purine nucleotide biosynthesis
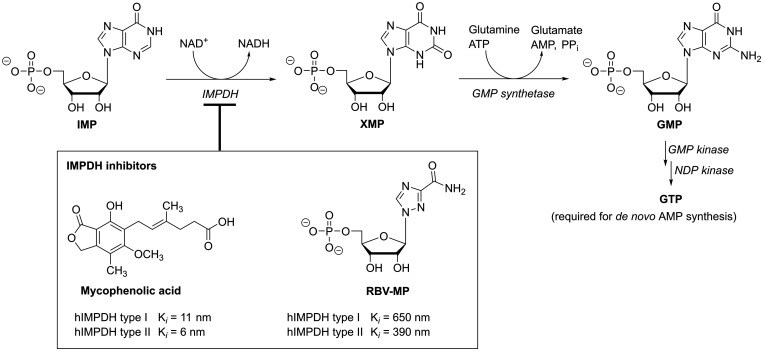
Because of its crucial role in the de novo biosynthesis of guanosine nucleotides, inosine monophosphate dehydrogenase (IMPDH) represents another well-known host-directed target that is clinically exploited in not only anticancer and immunosuppressive, but importantly also in antiviral therapy (Figure 2).37 Similar to the above discussed DHODH inhibitors, compounds that block IMPDH deplete cellular purine nucleotides and thus show antiproliferative and immunosuppressive activities, such as the highly potent mycophenolic acid (MPA).38–40 Yet, even though MPA and its analogues also potently inhibit the replication of diverse viruses via this mechanism,41–43 thus far, their clinical use in antiviral therapy appears limited due to toxicity and unfavorable metabolism leading to rapid inactivation of MPA.44 Another compound that reduces intracellular GTP levels builds on a different molecular scaffold and is widely applied as antiviral in the clinic: Ribavirin (RBV) is a nucleoside analogue that, once converted by host adenosine kinase to the 5’-monophosphate (RBV-MP), mimics IMP and competitively inhibits IMPDH; its binding to the enzyme, however, is much weaker when compared to the uncompetitive inhibitor MPA (Figure 2).36 RBV is approved for the treatment of respiratory syncytial virus (RSV) infections and, in combination with pegylated interferon-α, RBV has been standard treatment for chronic hepatitis C virus (HCV) infections before the introduction of direct-acting antivirals. While RBV has shown activity against various other and, importantly, highly different viruses in cell culture or animal models,45 its therapeutic use remains limited. In addition to uncertain or low efficacy in the clinic,46 RBV can cause severe side effects, such as hemolytic anemia, and teratogenic effects have been observed in animal models.47,48 Still, as currently neither specific nor broad-spectrum drugs are available for many RNA virus infections, RBV often is the only available option in cases of severe disease or epidemic scenarios. For instance, RBV is used for treating viral hemorrhagic fevers caused by Lassa49–51 or Crimean-Congo hemorrhagic fever virus52; all the while its efficacy remains uncertain.46 Further, RBV’s mechanism of action is still extensively discussed53–55 and appears to differ for different viruses and under different experimental settings.56–61 Targets include the here discussed host pathway of de novo purine biosynthesis,56,62 leading to a reduced purine nucleotide pool, as well as downstream effects such as polyamine depletion via the induction of spermidine/spermine acetyltransferase-1 SAT1.61 Additionally, RBV-5’-triphosphate (RBV-TP) can directly target the viral polymerase and its incorporation into the genome causes lethal mutagenesis63,64 (discussed in more detail in a later section), or it can inhibit RNA capping.65,66 While it is difficult to discern these different mechanisms of action, the overall observed in vivo antiviral effect may often be a combination. In fact, different mechanisms may even be synergistic, as GTP depletion, for instance, may reduce competition and thus favor RBV-incorporation by viral polymerase.53
Figure 2.
The central role of IMPDH in the de novo purine nucleotide synthesis and two of its clinically used inhibitors, MPA (its prodrug MPA-mofetil is used as immunosuppressive drug) and RBV (shown in the active 5’-monophosphate form). Inhibitory constants (Ki) are reported in Hager et al.36 XMP – Xanthosine 5’-monophosphate, NDP – nucleoside 5’-diphosphate.
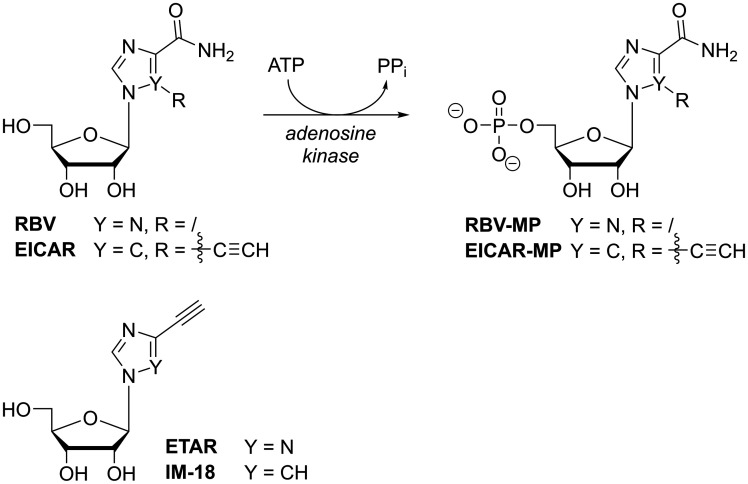
While RBV is the most prominent broad-spectrum antiviral nucleoside analogue, many derivatives have been developed and tested over the past decades. EICAR-MP, the 5’-ethynyl-derivative of RBV-MP (Figure 3), is also an IMP-analogue and inhibitor of IMPDH,67 with even greater potency than RBV in its cytostatic as well as broad-spectrum antiviral effects.68 In contrast to RBV and reminiscent of 6-chloropurine ribonucleotide,69 EICAR irreversibly inhibits IMPDH by covalently alkylating an active-site cysteine residue in a Michael-type addition.70 And with RBV’s clinical impact as anti-HCV treatment, more analogues based on this molecular scaffold continue to emerge, such as ETAR or IM-18,71,72 aiming for the development of pan-flavivirus inhibitors with enhanced potency or reduced cytotoxicity.73
Figure 3.
Metabolic activation of RBV and EICAR to their 5’-monophosphates by host adenosine kinase and chemical structures of ETAR and IM-18.
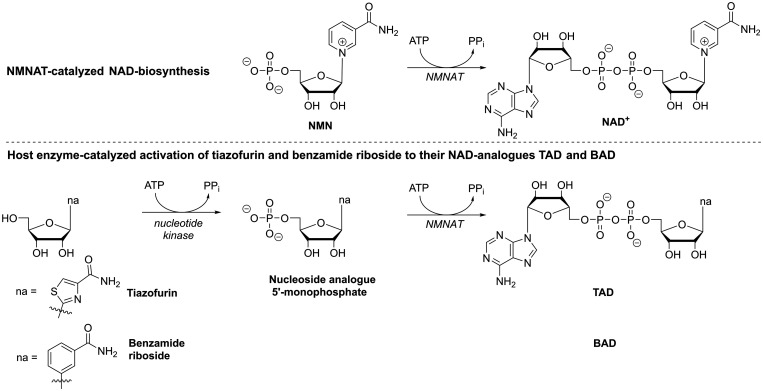
In RBV and some of its close analogues, the carboxamide-moiety in the nucleobase-part (Figure 3) with its rotational flexibility enables the analogues to mimic not only inosine and guanosine but also adenosine nucleotides; hence, these compounds are recognized by different enzymes of host purine metabolism (in Figures 2 and 3, note that adenosine kinase catalyzes RBV’s monophosphorylation while RBV-MP inhibits IMPDH as IMP analogue; this feature also results in what was termed ‘ambiguous base pairing’ which will be discussed in the following section). Moreover, as can be seen from the examples of tiazofurin and benzamide riboside (Figure 4), the carboxamide-moiety can also lead to compounds being recognized as analogues of the natural nicotinamide nucleotide (NMN, Figure 4): Tiazofurin and benzamide riboside monophosphate are converted to the NAD+ analogues TAD and BAD, respectively, by host nicotinamide/nicotinic acid adenylyltransferase (Figure 4). As such they bind to IMPDH in place of the natural cofactor NAD+ and thus inhibit the dehydrogenation reaction.74,75
Figure 4.
Host enzyme-catalyzed metabolic activation of tiazofurin and benzamide ribonucleoside to their NAD+-analogues TAD and BAD by first 5’-monophosphorylation and subsequent dinucleotide formation, catalyzed by nicotinamide/nicotinic acid adenylyltransferase (NMNAT). NMN – nicotinamide ribonucleotide, na – nucleobase analogue.
However, so far, RBV remains the major drug being widely applied in the clinic against various RNA virus infections. For host-directed drugs it is crucial to balance the desired antiviral effect and the inevitable toxicity for the host; thus, the strongest inhibitor might not be the best antiviral candidate. Still, since many RNA virus infections are non-chronic and require only short-term treatment, some degree of toxicity might be tolerable – and especially in face of newly emerging RNA viruses with pandemic potential, host nucleotide synthesis remains a valuable target for developing drugs that can exert a general antiviral effect.
Targeting viral RNA synthesis with direct-acting antiviral nucleobase, nucleoside and nucleotide analogues
RNA viruses universally encode an enzyme that is responsible for synthesizing RNA, the RNA-dependent RNA polymerase (RdRp).76 This enzyme catalyzes the phosphodiester-formation between the 3’-hydroxyl group of the priming (oligo)ribonucleotide and the α-phosphate of the incoming ribonucleoside 5’-triphosphate using RNA as template during genome replication as well as transcription (Figure 5). Viral RdRps work fast and are generally considered low fidelity enzymes: during genome synthesis, they incorporate the wrong nucleotide with a frequency of around 1×10−4 mutations per site for every round of replication (i.e. one mistake every 10,000 nucleotides).24,27,77 Without additional proof-reading capacity, this results in progeny virus that each differ by 1–2 nucleotides from their parent.24 In contrast, some larger DNA viruses as well as the cellular replication machinery harbor proof-reading and DNA repair enzymes and thus, mutation rates are much lower.24 In RNA viruses, the overall low fidelity during genome replication leads to a relatively diverse virus population (‘quasi-species’), and hence, RNA viruses possess the ability to quickly adapt to environmental changes. Consequences include the viruses’ ability to overcome bottlenecks in transmission,78 to evade the immune response, become resistant to drugs,24 and to uphold or even increase pathogenicity when compared to higher-fidelity mutants.79 On the other hand, this trait offers a possible weakness that can directly be addressed by drugs, analogues of the natural NTP substrates, to block the virus from replicating and stop the disease it causes.22
Figure 5.
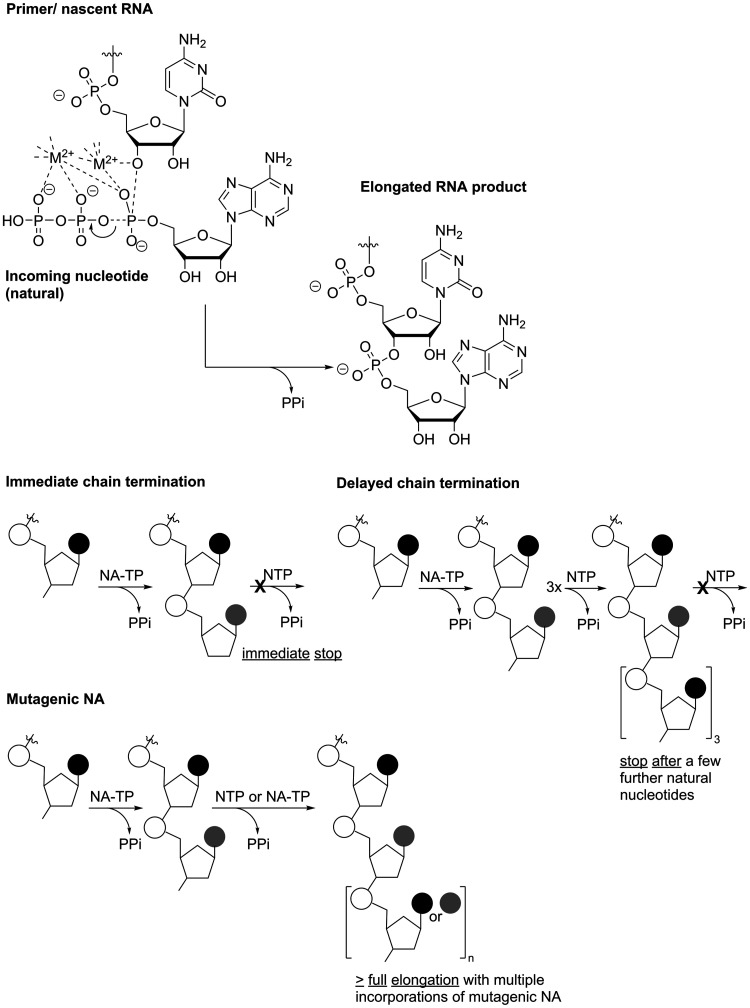
Polymerase-catalyzed RNA elongation. Top: The 3’-end of the nascent RNA attacks the α-phosphate of the incoming NTP, which is activated by M2+-coordination, and forms the phosphate diester after release of pyrophosphate (PPi). Bottom: Schematic of different mechanisms of action of NNAs. NA-TP – nucleoside analogue triphosphate.
The structural features of different viral RdRps have been described and while this is a very active field of research, reviews providing extensive information on this topic are available.80,81 Briefly, like all nucleic acid polymerases, the 3D-structure of RdRp enzymes (or enzymatic domains, as in some viruses, larger proteins harbor several enzymatic activities in distinct sites) resembles a right hand with palm, finger and thumb subdomains. Although overall RdRp sequences can differ extensively, the folding of seven large protein segments (including motifs A-G) that make up about 75% of the total enzyme are highly similar for different viral RNA polymerases and have thus been termed ‘homomorph’ segments.82 What’s more, domains that are directly involved in nucleotide selection and catalysis even show high sequence conservation across different RNA virus families. Hence, the core of the enzyme, the active site that is formed by motifs A-E and that resides in the palm subdomain, seems to maintain high similarity among RNA viruses.83,84 This may be attributed to an early emergence of RdRp from a common ancestor.85 Consequently, since RdRp activity is essential for viral replication and the polymerase’s structure is highly conserved across viral families, its active site represents an attractive target to develop virus-directed drugs that inhibit not only one specific but multiple RNA viruses and can thus serve as broad-spectrum antiviral treatments.86 Moreover, in terms of resistance development, several studies indicate that mutations in the RdRp enzymatic core domain concomitantly lead to attenuated phenotypes in vivo.87–89 Still, so far only few NNAs have been developed that show moderate to good antiviral activities across multiple virus families. This underlines the intricacy of this mission that may be caused by the influence that RdRp amino acids remote from the active site have on nucleotide incorporation fidelity.77 Nonetheless, RdRp is the only enzyme that RNA viruses universally encode. Examples of broadly-active NNAs that act at the RdRp active site have been identified and will be introduced in this chapter alongside those NNAs that have shown to act on a rather narrow group of viruses. Their structural features as well as mechanisms of action will be discussed.
Targeting the viral polymerase, non-natural nucleosides and nucleotides have long been at the core of antiviral therapies. These drugs were among the first to treat HIV-infection and AIDS, where they remain the backbone of combination antiretroviral therapy and are even used as pre-exposure prophylaxis (PrEP), attesting to their efficient and safe profile.90 To exert their effect on viral nucleic acid synthesis, nucleobase, nucleoside or nucleotide analogues (NNAs) have to be processed by intracellular host enzymes to their corresponding 5’-triphosphates. These function as analogues of the natural (d)NTP substrates and can thus be incorporated into the nascent RNA or DNA by the viral polymerase. Depending on their chemical structure as well as the context in which the analogues are incorporated, nucleic acid synthesis may be terminated directly after (immediate chain termination) or once a few additional (natural) nucleotides were added (delayed chain termination; Figure 5). Furthermore, nucleic acid synthesis may continue and even reach completion despite incorporation of an analogue but the analogue may cause an increase in mutational frequency such that no more infectious virus can be produced – a process that was termed lethal mutagenesis (for an insightful review on the different mechanisms by which antiviral NNAs inhibit nucleic acid synthesis, see Deval91) The following sections will discuss and compare some examples of antiviral NNAs that exert their antiviral effect via one of these mechanisms within the context of potential broad-spectrum activity. As, historically, major efforts in the development of anti-RNA virus NNAs have centered on finding new and effective treatments for HCV infection, many of the discussed compounds were originally sourced from anti-HCV-programs.
Chain terminators
2’-Ribose modified analogues
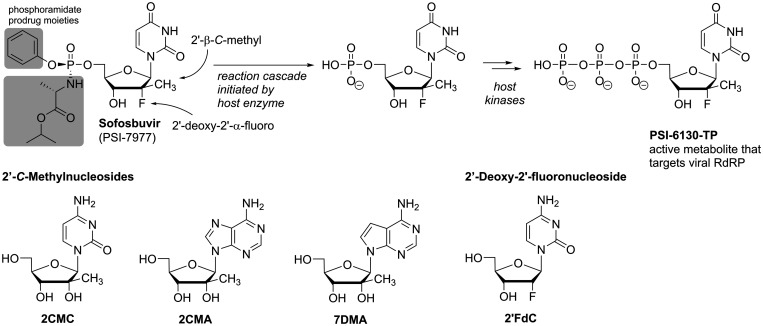
In terms of hepatitis C virus infection, the backbone of treatment has long relied on a combination of pegylated interferon-α (PEG-IFNα) and ribavirin (RBV), a nucleoside analogue already discussed in the previous section. RBV can address multiple targets including the viral RNA polymerase, but suffers from relatively low efficacy (sustained virologic response rate of approx. 30%) paired with severe toxicity,48 which is especially grave in the long-term treatment that chronic HCV infection requires. Consequently, considerable efforts into the development of more potent and safer, direct-acting nucleoside analogues resulted in the approval of sofosbuvir,92 achieving sustained virologic response rates of up to 90% after a 12-week treatment course in combination with PEG-IFNα and RBV.48 Higher response rates of up to 100% were even seen with combinations of sofosbuvir and another direct acting antiviral, thus making it possible to fully dispense RBV from HCV-treatment.48 Sofosbuvir’s chemical structure resembles the natural nucleotide uridine 5’-monophosphate differing only at the 2’-position of the ribose-moiety and additionally containing the phosphoramidate prodrug unit (Figure 6). The prodrug form protects the nucleotide from phosphatases and masks the negative charge of the phosphate so that it can diffuse through cell membranes (pronucleotides, prodrugs of nucleotides, are discussed in more detail in the final section of this review). Ultimately, the prodrug groups are cleaved in a host enzyme-induced cascade reaction to release the ‘naked’ sofosbuvir monophosphate (Figure 6). Although, theoretically, incorporation of the sofosbuvir nucleotide would allow further elongation of the nascent RNA, once incorporated, the 2’-deoxy-2’-α-fluoro-2’-β-C-methylribose motif seems to distort the structure of the RNA and thus prevents elongation (non-obligate chain termination).86,93 While this particular motif is unique among clinically advanced antiviral NNAs and preferred only by HCV polymerase,93,94 the 2’-β-C-methylribose modification has been recognized to equip NNAs with antiviral activity against not only HCV but a multitude of pathogens from various (+)ssRNA virus families.95,96 These include flaviviruses, such as yellow fewer, dengue, west nile,97 zika,98 and tick-borne encephalitis virus,99 that are mainly transmitted by mosquitos and repeatedly cause outbreaks of disease that can have highly debilitating outcomes.100–103 Further, 2’-C-methylcytidine, 2’-C-methyladenosine and 7-deaza-2’-C-methyladenosine (2CMC, 2CMA, 7DMA; Figure 6) have demonstrated potency as broad-spectrum inhibitors of noro-, rota- and sapovirus infection (belonging to Calici- and Reoviridae, the latter are double stranded RNA viruses), which are the leading etiological agents causing viral diarrhea in children.104 The spectrum of activity further includes members of the Picornaviridae,105,106 another (+)ssRNA virus family.
Figure 6.
Chemical structures of 2’-C-methyl- and/or 2’-deoxy-2’-fluororibose-modified nucleoside analogues including anti-HCV pronucleotide sofosbuvir (phosphoramidate prodrug moieties highlighted in grey) and schematic of host cell metabolism to sofosbuvir’s active triphosphate form PSI-6130-TP.
As is reasonable to assume, the RdRps within the group of (+)ssRNA viruses are phylogenetically closer and thus more conserved than when comparing those from (+) and (−)ssRNA viruses.107 In fact, it appears that 2’-C-methylribose-modified NNAs don’t achieve comparable potency against (−)ssRNA viruses. Hence, this motif may contribute to the development of an antiviral nucleoside or nucleotide analogue with a broad antiviral spectrum among (+)ssRNA viruses, some of them, importantly, being pathogens with major significance to public health around the world such as members of the Flavi-, Picorna-, and Coronaviridae. The same strategy may, however, not expand to (-)ssRNA viruses.105
In this regard, a recent RdRp-based phylogenomic analysis reconstructed the evolutionary relationships among RNA viruses and concluded a phylogenetic tree of five major branches. (+)ssRNA viruses are spread about three of these branches, in part overlapping with double strand RNA viruses. In fact, it appears that dsRNA viruses have evolved from (+)ssRNA viruses on two different occasions at least while, in contrast, (−)ssRNA viruses may have evolved from one distinct group of dsRNA viruses. Importantly, (−)ssRNA viruses are limited to only one branch.76 Being based on RdRp-sequences, this analysis again underlines that this viral enzyme may be a promising target to also inhibit various (−)ssRNA viruses in a “one drug-multiple-bugs”-approach. Still, in contrast to RdRp-targeted antiviral candidates against members of the (+)ssRNA viruses, some of which are in advanced clinical development or have even been approved for treatment (greatly driven by efforts to treat and cure HCV infection), similar progress has lagged behind for (−)ssRNA viruses. Within this group, intensive efforts have been put into drug development against influenza virus infections; yet, approved drugs so far predominantly target entry and release of the virus, are thus highly specific and prone to resistance development.108 (Recent advancements include the approval of baloxavir marboxil (Xofluza), an inhibitor of the influenza virus polymerase PA-subunit that catalyzes the cap-snatching reaction to initiate translation;109,110 a structurally and functionally similar viral enzyme activity is used by other (−)ssRNA viruses such as Bunyaviridae,111 hence, baloxavir or similar compounds targeting viral mRNA synthesis may have some broader spectrum of activity.) Importantly, with favipiravir (T-705) a compound that ultimately targets the influenza virus RdRp core enzymatic activity has been approved in Japan as treatment during outbreaks of new or re-emerging influenza virus where other drugs are not effective (Favipiravir is discussed in more detail in the following subsection). Briefly, it has broad antiviral activity but also suffers from low in vivo efficacy and potential teratogenicity. While chain termination has been observed in biochemical assays, favipiravir treatment predominantly seems to lead to the accumulation of mutations in the viral genome (Figure 5).
Still, in terms of non-mutagenic, direct-acting NNAs to broadly target (-)ssRNA virus replication, focused tests as well as screening campaigns have identified some potent inhibitors. Among these were 2’-deoxy-2’-fluororibonucleosides (Figure 6) that, in comparison to the nucleoside analogues discussed above, lack the 2’-C-methyl motif. However, they contain a 2’-fluorine substituent that is also present in Sofosbuvir and that sustains the 3’-endo-conformation typical for ribonucleotides, thus ensuring that these analogues can be recognized by RNA polymerases. While these compounds proved highly active against a panel of (−)ssRNA viruses (including influenza112 and various hemorrhagic fever viruses113,114) with especially 2’FdC demonstrating broad and potent inhibition, clinical advancement is hampered by concerns regarding toxicity, probably because these compounds interfere with host nucleic acid synthesis (mitochondrial toxicity is a common limitation for antiviral drug development with NNAs and is discussed in the final section of this review).115 Biochemical assays with the CCHFV L-protein (harboring the RdRp active site) have further shown that not only the 5’-triphosphate of 2’FdC but also that of 2’-amino-2’-deoxycytidine is a potent, CTP-competitive inhibitor of viral RNA synthesis116; yet, whether this applies to other (−)ssRNA viruses and whether it translates into antiviral activity in infectious assays remains to be determined.
C-Nucleosides
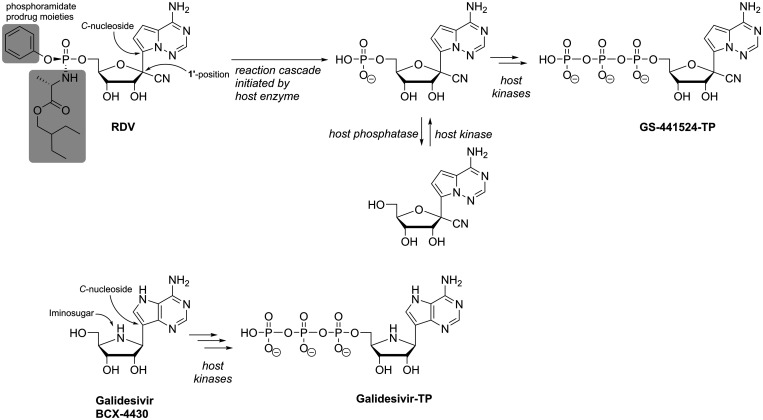
Remdesivir (RDV, GS-5734) is another highly promising example for nucleotide analogues with potential broad-spectrum antiviral activity, importantly spanning (+)ssRNA and (−)ssRNA viruses, that has recently attracted much attention. RDV, similarly to Sofosbuvir a McGuigan-type phosphoramidate pronucleotide, contains the non-natural nucleoside GS-441524 (Figure 7).117,118 This compound originated from a screening campaign of an NNA library against, among others, filoviruses and gained attention after the large West Africa Ebola virus outbreak due to its highly promising inhibitory potency in cell-based and animal studies. In fact, RDV then became part of a clinical trial during the following outbreak of Ebola virus in the Democratic Republic of Congo that started in 2018. However, the RDV treatment arm was discontinued after interim analysis due to superior performance of monoclonal antibodies regarding mortality outcomes.119 In addition to anti-filovirus activity, RDV, or its underlying nucleoside analogue GS-441524, are potent inhibitors of various members of the Flavi-,117 Corona-,120 Paramyxo- and Pneumoviridae,121 hence showing a broad spectrum of antiviral activity including both (+) and (−)ssRNA viruses. With the ongoing and devastating COVID-19-pandemic, RDV has once more moved into the spotlight: It has demonstrated activity against various coronaviruses including SARS-CoV-2 in simple Vero- and primary cell-based models as well as in animal experiments120,122,123 and has been granted approval by the FDA in October 2020 for the treatment of COVID-19 in patients 12 years of age and older. In terms of chemical structure, the nucleoside core of RDV resembles adenosine and includes modifications both at the nucleobase and the ribose-moiety (Figure 7). The ribose 1’-cyano modification is important for target-specificity of the compound, as the simple, 1’-H C-nucleoside suffers from significant cytotoxicity.118 RDV’s broad antiviral spectrum is based on its nucleoside 5’-triphosphate metabolite (i.e. GS-441524-TP) being recognized by diverse viral RdRps. Biochemical assays with polymerase complexes from MERS-CoV, SARS-CoV and SARS-CoV-2 have shown that its incorporation into viral RNA is even favored over the incorporation of the natural adenosine nucleotide.124 In agreement with experimental data, modeling revealed that after RdRp-catalyzed incorporation of the RDV nucleotide into nascent viral RNA, where the 1’-cyano modification is easily accommodated, three subsequent (canonical) nucleotides can be added. Then, however, a steric clash between the protein and the 1’-cyano group leads to deterioration of the RNA structure, which precludes further nucleotide addition and consequently causes delayed chain termination after four translocation events (Figure 5).125 However, to analyze the mechanism of action of nucleotide analogues via their effect on elongation kinetics, SARS-CoV-2 RdRp-catalyzed RNA-synthesis was studied recently on a >1000 nt long RNA template and in the presence of saturating NTP concentration using magnetic tweezers.126 The study compared the kinetic signatures of the enzyme in the presence of GS-441524-TP with those in the presence of the immediate chain terminator 3’-deoxy-ATP and surprisingly, the results disagreed with chain termination as RDV’s mechanism of action. Rather, they revealed that final product length was largely unaffected in the presence of GS-441524-TP, yet, the median replication time was increased while nucleotide incorporation rates were unchanged. Hence, using stochastic models, the authors concluded that instead of prematurely terminating RNA synthesis, the incorporation of RDV-nucleotide transiently stalls SARS-CoV-2 replicase (the authors noted that polymerase stalling may be interpreted as termination events in gel-based assays).126 Importantly, having a comparably large genome (30k nt), coronaviruses are distinct from most other RNA viruses in that they harbor exonuclease activity in their nsp14 protein (ExoN). ExoN performs proof-reading by removing wrongfully incorporated nucleotides (or analogues) and thus restricts many NNAs from being active against the wild-type coronaviruses.127,128 In the case of RDV, while the drug’s efficacy is somewhat sensitive to proof-reading by ExoN (i.e. the antiviral potency is higher in ExoN--CoV mutants compared to the wild-type virus),129 this is to a much lesser extent than for other NNAs.125,130 As modelling has shown, ExoN readily accommodates the RDV nucleotide in the 3’-terminal position of the RNA strand; yet, the 1’-cyano moiety precludes its proper positioning for hydrolysis.125 In combination with the delayed chain termination or stalling mechanism, this may be the reason for RDV’s sustained antiviral potency against wild-type CoVs.
Figure 7.
Chemical structures of broad-spectrum antiviral NNAs Remdesivir (phosphoramidate prodrug moieties highlighted in grey) and Galidesivir. Host cell metabolic activation yields the active TP-forms that the RdRp incorporates into nascent viral RNA, which leads to delayed chain termination (or stalling).
With the imino-C-nucleoside galidesivir (BCX-4430; Figure 7), yet another adenosine analogue has shown some promise for future treatment of various RNA virus infections. Galidesivir is known as a potent transition state analogue inhibitor of the purine nucleoside phosphorylase (PNP) from the protozoan parasite Trichomonas vaginalis131 and it further inhibits Leishmania parasites via blocking their nucleoside hydrolase, an important enzyme in the purine salvage pathway that these purine auxotrophic pathogens rely on.132 In chemical structure, it resembles the antiparasitic immucillins ImmH and ImmG and has thus also been termed immucillin-A (ImmA). Importantly, antiviral screening campaigns identified galidesivir as an inhibitor of diverse members of (+) as well as (−)ssRNA virus families, such as Flavi-, Picorna-, and Coronaviridae, Filo-, Bunya-, Arena-, Ortho- and Paramyxoviridae.133 In terms of antiviral mechanism of action, galidesivir does not inhibit mammalian PNP,131 thus contrasting the antiparasitic properties. Instead, it targets viral RNA polymerases, which incorporate galidesivir nucleotide via its triphosphate form into nascent viral RNA.133 Similar to what is observed with RDV in gel-based assays, nucleic acid synthesis then stops after incorporation of another two natural nucleotides (delayed chain termination, Figure 5).133 While in primary hepatocytes galidesivir is readily converted into galidesivir-5’-triphosphate (Galidesivir-TP; Figure 7) by host kinases, in immortalized cell lines, especially Vero cells, Galidesivir-TP attains only low levels.133 It was suggested that this may limit the observed antiviral potency of the compound in typical screening campaigns and thus lead to underestimation of galidesivir’s potential regarding potency and spectrum of antiviral activity – a limitation that is often observed with NNAs and that can be addressed employing prodrug strategies (which will be discussed in the final section of this review). Importantly, the imino-C-nucleoside galidesivir seems to be devoid of significant cell toxicity up to concentrations that largely exceed antiviral IC50’s.133
The examples of remdesivir and galidesivir impressively show that broad antiviral activity covering not only different viruses of the same family but even spanning diverse members of (+) and (−)ssRNA virus families (including even Coronaviridae which harbor proof-reading activity) can be achieved with chain terminating NNAs (although recent analyses challenge this mechanism of action for RDV).126 Importantly, at the same time these compounds exhibit favorable toxicity profiles, hence optimally positioning them for clinical development. Still, metabolic activation to the ultimately active nucleoside analogue triphosphate may be limited in certain cells and tissues. Thus, to ultimately being successful as broad-spectrum antiviral candidates, technologies to bypass inefficient metabolic activation may become essential.
Lethal mutagens
Apart from chain termination, NNAs can exert their antiviral effect by extensively causing mutations during replication, for instance when they behave ambiguous in their base-pairing properties. Since RNA viruses already exhibit low fidelity during genome synthesis, additional mutations rapidly render progeny virus unfit and may thus stop the infection.22,24,27 This strategy has first been described as lethal mutagenesis in the context of HIV.134 In terms of chemical structure, such mutagenic nucleotides require only minor modifications to the nucleobase and none to the ribose-moiety; thus, it is conceivable that such compounds promise to be very broadly active. Nonetheless, analogues certainly need to achieve some specificity for viral over cellular targets to not cause extensive toxicity, which may require additional modifications (one major problem may be mitochondrial toxicity which is discussed further below). Furthermore, mutagenic NNAs appear favorable over many chain terminating NNAs in terms of resistance development.
Carboxamide-substituted nucleobases
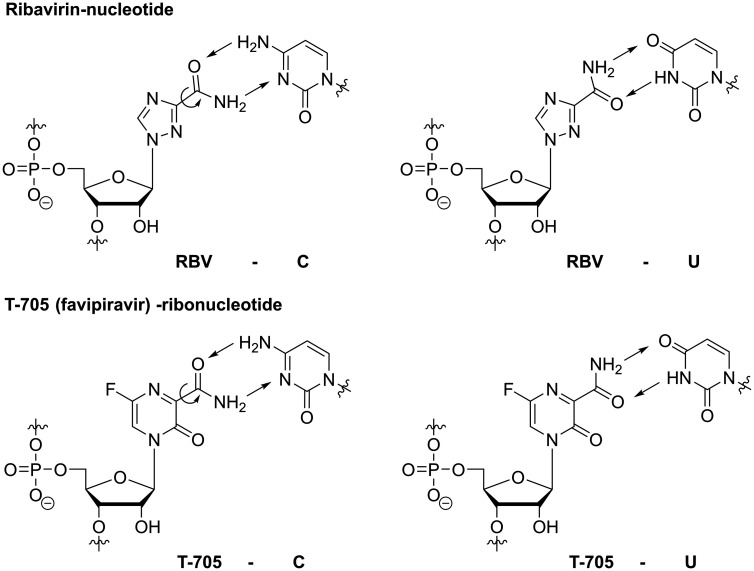
With its diverse mechanisms of action, RBV is also a prominent and extensively studied example of a nucleoside analogue, that – once converted to RBV-triphosphate and then incorporated into viral RNA – induces lethal mutagenesis.63 Its mutagenic potential is explained by the flexibility of the carboxamide substituent at the nucleobase: Rotation around the C(3)-C(=O)-bond allows RBV to mimic guanosine (and inosine, see previous section) or adenosine, thus templating ambiguously for C or U (Figure 8).53 Still, as discussed above, the predominant mechanism by which RBV exerts its antiviral effect remains highly debated and probably differs for different viruses and model systems. Nonetheless, to induce lethal mutagenesis, multiple incorporations of the analogue during viral nucleic acid synthesis are required (Figure 5) and it is conceivable that GTP depletion via IMPDH inhibition (as discussed above) enhances RBV’s chances of incorporation. Hence, these two distinct mechanisms may actually work synergistically. RBV’s efficacy in the clinic, however, remains suboptimal and RBV treatment can cause severe side-effects, e.g. hemolytic anemia via accumulation of its phosphorylated form in erythrocytes,135 and teratogenic effects have been observed in different animal models.136–138
Figure 8.
By rotation of the carboxamide moiety, RBV-nucleotide and T-705-ribonucleotide can base-pair with and thus template for both C and U. Hence, they can exert their antiviral effect through inducing mutations after being incorporated multiple times during viral RNA synthesis. Circled arrows indicate rotation of C-C-bond; straight arrows represent H-bonds.
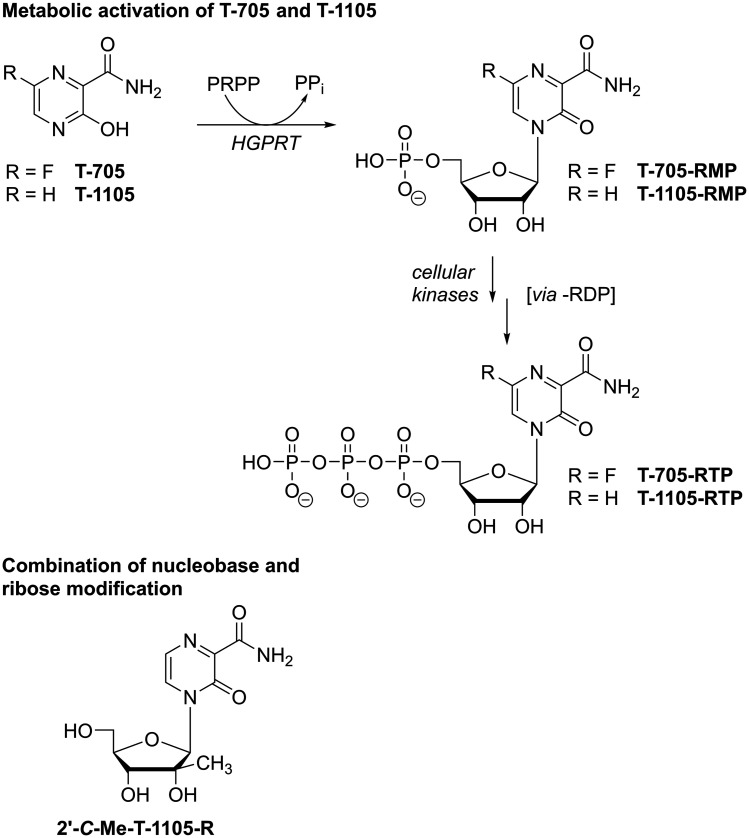
T-705 (favipiravir) is a nucleobase analogue that, just like RBV, contains a carboxamide substituent (Figure 8). Accordingly, once incorporated into viral RNA, the carboxamide’s rotational flexibility allows T-705 to template for both C and U and thus induce mutations, which has been demonstrated both in vitro and in vivo.139–143 Therefore, T-705 must first be converted by host enzymes to the triphosphoribosylated form (T-705-RTP; Figure 9), which is then recognized as a substrate by various viral polymerases.144–146 In addition to acting as a mutagen, biochemical assays also suggested that the incorporation of T-705-ribonucleotide into nascent viral RNA may lead to chain termination or polymerase back-tracking.144,147,148 Consequently, similar to RBV, T-705 has been attributed different mechanisms of action based on in vitro work, yet there’s strong evidence from animal experiments that this compound in fact induces lethal mutagenesis.139–144,147,148 Additionally, in contrast to RBV, no or only very low inhibition of cellular IMPDH has been observed with T-705.141,149 In line with this, cells treated with high concentrations of T-705 appear to experience virtually no toxicity. This, however, is contrasted by the finding that T-705-RTP is an efficient substrate for human mitochondrial RNA polymerase (POLRMT), which typically causes toxic effects of antiviral NNAs.150 So far, a comprehensive explanation for these contradicting data has not been presented; however, in animal experiments, embryotoxic effects have been described for T-705108 and warrant caution regarding its clinical use. Still, T-705 has shown great promise as a broad-spectrum, orally bioavailable antiviral: After it was reported as an inhibitor of influenza virus151 it was found active in vitro and in vivo against a vast number of emerging viruses including diverse hemorrhagic fever viruses.152–155 Favipiravir has subsequently been trialed during outbreaks of Ebola virus156 as well as SARS-CoV-2 (for an extensive overview of T-705’s antiviral activities see recent reviews157–159) From these studies, it appears that T-705 is most active against influenza viruses, which are members of the (−)ssRNA Orthomyxoviridae, and antiviral effects against (+)ssRNA viruses appear to require higher doses.160,161 Yet, due to the different model systems, these comparisons may in some cases be difficult to interpret. In this regard, we and others have shown that the efficiency of T-705’s metabolic activation, which is required to generate the active -RTP form (Figure 9), may differ strongly in different cells and consequently, inhibitory concentrations not only depend on the virus that studied but also on the cell type that is targeted.162–165 Additionally, T-705 suffers from chemical instability once it has been incorporated into a ribonucleoside: we found that in buffered aqueous solution of physiologic pH, T-705-ribonucleoside readily decomposes and the rate of decomposition increases with increasing pH.166 In fact, this may not only limit observed potency but also represent yet another antiviral mechanism with decitabine or AzaC being prominent examples (discussed below). In contrast to T-705, its non-fluorinated analogue T-1105 (Figure 9) proved stable under aqueous conditions. Moreover, T-1105 has demonstrated greater potency to inhibit influenza virus in MDCK cell-based assays and the corresponding ribonucleoside 5’-triphosphate (T-1105-RTP) competes with the purines ATP or GTP during viral RNA synthesis more efficiently than T-705-RTP, which has been shown in the context of human norovirus,145 influenza virus167 as well as SARS-CoV146 polymerase. As mentioned above, however, inefficient metabolic activation to its triphosphate form also limits T-1105’s antiviral potency and hence, this compound suffers from drastic differences in efficacy when studied in different cell types,165 which may somewhat conceal potential broad-spectrum antiviral potency. As mentioned above, both RBV as well as the active forms of T-705 and T-1105 contain an unmodified ribo-glycon. Hence, in terms of countering toxic effects, one study reports the combination of the 2’-C-methylribose motif with the nucleobase analogue T-1105 to prevent POLRMT-targeting of the ribose-unmodified T-1105-RTP (Figure 9).145 Although, compared to the ribose-unmodified version, the 5’triphosphate form of this analogue proved less potent in inhibiting norovirus polymerase, its recognition by POLRMT was also much less efficient,145 demonstrating that small chemical modifications can greatly reduce off-target effects.
Figure 9.
Top: Metabolic activation of T-705 and its non-fluorinated analogue T-1105 to the active ribonucleoside 5’-triphosphate (-RTP) metabolite. PRPP – 5-phosphoribosylpyrophosphate; PPi – pyrophosphate; HGPRT – hypoxanthine guanine phosphoribosyl transferase. Bottom: Chemical structure of 2’-C-methyl-modified ribonucleoside of nucleobase analogue T-1105.
Other features of nucleoside analogues that induce a mutagenic effect
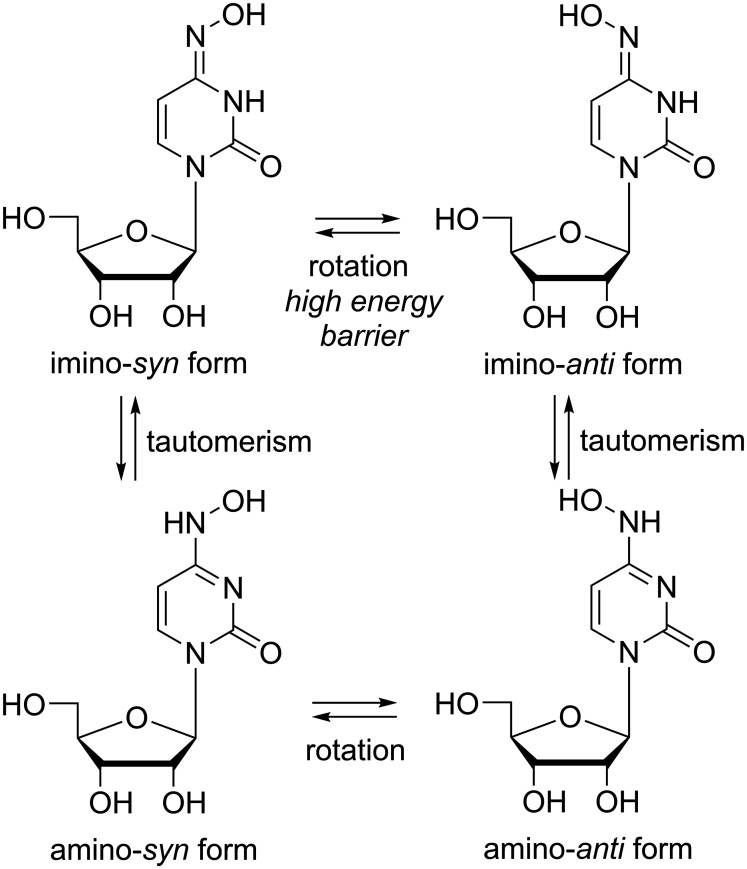
Another ribonucleoside analogue that has demonstrated antiviral potency against influenza and diverse other RNA viruses via inducing mutations is β-d-N4-hydroxycytidine (NHC; Figure 10).168–173 Importantly, this compound is also active against a broad panel of coronaviruses including SARS-CoV-2174,175 and its 5’-isopropylester EIDD-2801, an orally bioavailable prodrug, has advanced to clinical development during the COVID-19 pandemic.176–178 Through its tautomerism and rotational isomerism (Figure 10),179 NHC can replace both pyrimidine nucleosides uracil and cytosine, i.e. it templates for A and G and thus induces lethal mutagenesis.168 Similar to remdesivir and galidesivir - yet contrasting other mutagenic NNAs128 –NHC’s potency is only minorly affected by coronaviral ExoN activity despite its natural ribo-glycon; the mechanism behind NHC’s ability to evade CoV proof-reading has not been elucidated so far. Importantly, in terms of drug evasion, NHC behaves similar to other mutagenic NNAs: passaging experiments did not generate robust resistance in influenza-,173 corona-174 or other RNA viruses.168,172
Figure 10.
Chemical structures of the imino- and amino-forms of the mutagenic nucleoside analogue β-d-N4-hydroxycytidine (NHC).
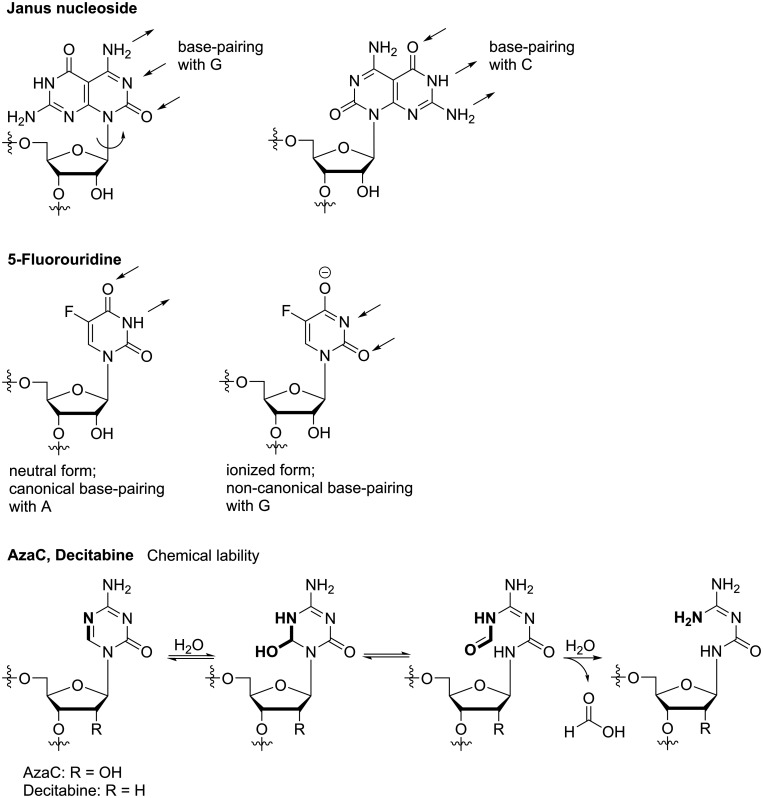
In addition to rotation of a small substituent at the nucleobase, also syn-/anti-isomerization at the gylcosidic bond can result in ambiguous base-pairing. Janus type nucleosides exhibit different hydrogen bond donor/acceptor patterns at the two different sides of their unnatural nucleobase and thus, by rotating at the glycosidic bond, they’re able to mimic different natural nucleotides (Figure 11).180,181 However, so far this concept has not produced any promising antiviral candidate with broad-spectrum potency against RNA viruses. The concept of rotational flexibility has further been extended to rotation within the nucleobase core: fleximers are nucleoside analogues that bear a split purine base, thus, the imidazole and pyrimidine part are no longer fused but connected via a single C-C bond.182 Originally, this approach was followed to study how increased flexibility in the nucleobase would affect enzyme recognition and binding affinity compared to rigid analogues. It was then found that fleximers can bind to atypical enzymes and that they can overcome point mutations, which usually confer resistance to the rigid nucleoside analogue.183 Further, broad-spectrum antiviral activity may be achieved through the fleximers’ ability to take part in “mutually induced fit”.182 Combining the split purine base with an acyclic glycon (as found in the anti-herpesvirus drug acyclovir) has led to promising activity against members of different RNA virus families, including Filo-, Flavi-, and Coronaviridae.184–186 Still, the mechanism of action remains subject of studies and although it has been shown that fleximer-triphosphates can be substrates for nucleic acid polymerases,187 it is still unclear whether the antiviral activity of acyclic fleximers is achieved via this mechanism. Mutagenic properties similar to ribavirin etc. have so far not been reported for fleximers, but it is conceivable that this concept – having flexibility at its core – may in the future extent to inducing lethal mutagenesis. Next to rota- or tautomeric properties, ionization can cause ambiguous base-pairing. In 5-fluorouridine (the ribonucleoside of 5-fluorouracil, 5-FU, the most prominent example following this mechanism) the 5-fluoro substituent strongly decreases the pKa of the NH-group (or, the OH-group of its tautomeric imino-form, respectively; Figure 11). Hence, in addition to 5-FU forming a natural-like base-pair with A, the ionized form of 5-FU is able to hybridize with G.188 5-FU ribonucleotide further inhibits cellular thymidylate synthase, leading to cell death, and is therefore used in anti-tumor therapy.189 Finally, mutagenic properties in NNAs can also be the result of chemical instability of the un-natural nucleobase. For instance, in 5-azacytidine (azaC) and its 2’-deoxy-congener decitabine, the triazine is prone to nucleophilic attack and ring-opening by water, followed by deformylation (Figure 11).190–192 Since phosphorylated metabolites of azaC and decitabine also interfere with cellular processes, these compounds are used in the treatment of myelodysplastic syndrome and leukemia.193,194 The antiviral effect of azaC through mutagenesis has been demonstrated in the context of HIV,195 foot-and-mouth disease virus,196 lymphocytic choriomeningitis virus197 and coronavirus that lacks ExoN-activity.128 However, since these compounds inhibit crucial cellular processes they’re so far not being used in the clinic to treat viral infection.
Figure 11.
Chemical structures and molecular basis for mutagenic properties of Janus-GC nucleoside, 5-FU and AzaC/Decitabine.
Nonetheless, especially the examples of T-705/T-1105 and NHC give rise to hope that the development of broad-spectrum antiviral drugs based on mutagenic NNAs may be attainable. To achieve this goal, cell-based screening campaigns need to integrate cell models that better represent the various target tissues that are clinically relevant for different RNA virus infections, thus factoring in potential cell type-dependent antiviral potency of the compounds. On the other hand, development efforts will require innovative medicinal chemistry strategies to a) enhance efficacy by overcoming poor intracellular activation and b) limit toxicity by enhancing target selectivity. The following section will briefly introduce examples of such strategies.
Problems of virus-targeted nucleoside analogues in antiviral drug development
Though NNAs play essential roles in the treatment of viral diseases such as hepatitis C virus infections, the development of novel members of this drug class is cumbersome and the danger of failure in advanced clinical stages poses high financial risk. One problem is that in vitro antiviral activity may not translate well into in vivo efficacy, as, for instance, NNAs have to overcome metabolic limitations in the activation to their nucleoside analogue triphosphate form within the target tissues. Second, in vivo toxicity of NNAs that only came to light in advanced clinical phases has in the past led to spectacular failures and high attrition rates. Often, this is due to insufficient selectivity of nucleoside analogue triphosphates that interfere with cellular nucleic acid synthesis such as mitochondrial RNA synthesis.
Low selectivity causing toxicity
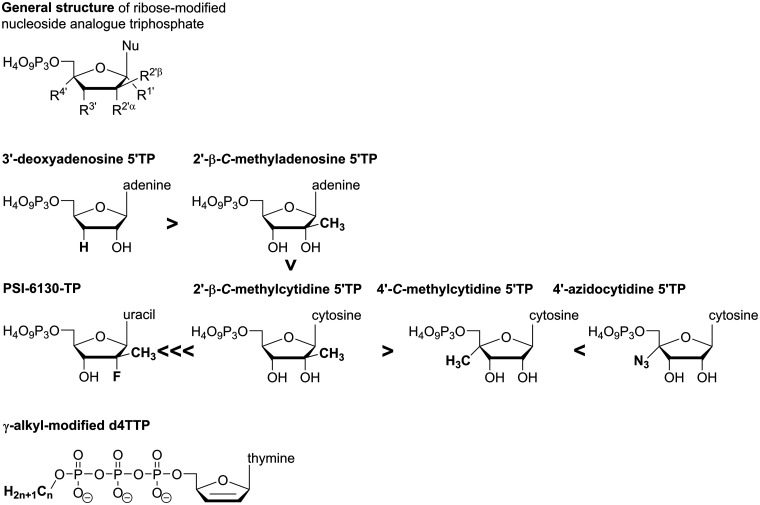
Since cells use nucleotides in diverse processes and harbor (DNA and) RNA polymerases that share their catalytic mechanism with viral polymerases, NNAs can cause severe toxicity when they don’t selectively target the viral enzymes. Consequently, to become successful antiviral drugs, the active metabolites of NNAs must show sufficient selectivity for viral over host enzymes. Examples from the development of antiviral NNAs against DNA viruses, such as herpesviruses and hepatitis B virus, or the retrovirus HIV have shown that for an NNA to be successful, its NTP-form must have sufficiently low substrate properties for host polymerases when compared to the viral polymerase.198 Hence, when an NNA-triphosphate inadvertently targets a host polymerase, consequences are severe toxicity and (late) failure of the candidate in clinical development, as exemplified by the anti-HBV nucleoside analogue fialuridine which causes mitochondrial toxicity via inhibition of DNA polymerase γ.199,200 The problem of mitochondrial toxicity has also led to the attrition of diverse NNAs in development for the treatment of RNA virus infections.115 Ribonucleoside analogue 5’-triphosphates are at risk of being recognized as substrates by host mitochondrial RNA polymerase (POLRMT) and consequently, when analogues disrupt normal mitochondrial RNA synthesis, protein expression is reduced which leads to mitochondrial dysfunction.115,201 In consequence, it was suggested that target selectivity of nucleoside analogue triphosphates should best be studied at an early in vitro stage to avoid failure at more advanced stages in clinical development; available biochemical as well as cell-based assays have been reviewed recently.202 In a joint effort with early testing, the chemical structure of antiviral NNAs must be optimized so that their triphosphate metabolites are disfavored by cellular enzymes, such as POLRMT, while being good substrates for various viral polymerases. However, biochemical studies have shown that POLRMT is quite promiscuous in terms of NTP substrate specificity.115 Lately, following spectacular failures in clinical development, POLRMT substrate properties towards non-natural ribonucleoside triphosphates have been studied with regard to compounds from anti-HCV campaigns, thus including nucleoside analogues modified at the 2’- or 4’-ribose-position.115,150,201 Data for ATP derivatives show that the obligate chain terminator 3’-deoxy ATP is a better substrate for POLRMT than the non-obligate chain terminator 2’-C-methyl ATP.115 Among 2’-modified NTPs, the inhibitory potency of 2’-C-methyl purines towards POLRMT-catalyzed RNA synthesis appears to be approx. 5-fold higher compared to pyrimidine derivatives.201 Interestingly, 2’-deoxy-2’-fluoro-2’-C-methyluridine, the nucleoside analogue contained in the highly successful anti-HCV drug sofosbuvir (Figure 6), clearly stood out with its 5’-triphosphate (PSI-6130-TP) showing very low incorporation efficiency and virtually no inhibition of POLRMT-catalyzed RNA synthesis (IC50 > 500 µM).115,201 Regarding modifications in 2’- versus 4’-ribose position, 2’-C-methyl CTP (the active form of valopicitabine) was a better substrate for POLRMT than 4’-C-methyl CTP; however, changing the 4’-substituent to an azido group (as in balapiravir’s active form) again resulted in enhanced substrate efficiency and pronounced toxicity in the clinic.115 NNAs that elicit their antiviral effect via inducing mutations are generally weaker chain terminators, and for T-705 in vitro studies have shown no strong effect on mitochondrial protein expression even though the compound’s active form, T-705 ribonucleoside 5’-triphosphate, was an efficient substrate for POLRMT.150
These analyses suggest that POLRMT Figure 12 does not discriminate well against non-natural NTPs containing single ribose-modifications, such as the 4’-azido- or 2’-C-methyl motifs. Analogues are incorporated into and hence prematurely terminate the synthesis of mitochondrial RNA transcripts, thus reducing mitochondrial protein levels and causing toxicity. Combining multiple ribose-modifications, as seen for example in sofosbuvir, may decrease the triphosphate’s substrate efficiency towards POLRMT and consequently lead to a potential drug with a much better safety profile. Importantly, however, its distinct pattern of ribose-modifications has precluded sofosbuvir from being broadly active. Here, mutagenic NNAs seem the better option, at least judging from the data available so far. Still, these analogues may cause teratogenic effects and since the problem of ribonucleoside analogue triphosphates targeting POLRMT has only recently moved into the spotlight, more studies are clearly needed to establish general conclusions and to be able to anticipate mitochondrial toxicity from the chemical structure of NNAs.
Figure 12.
Comparison of chemical structures of 3’-deoxy ATP, 2’-C-methyl ATP, PSI-6130-TP, 2’-C-methyl CTP, 4’-C-methyl CTP and 4’-azido CTP; γ-alkyl-modified d4TTP. Bold arrowheads indicate relative substrate properties towards POLRMT.
In terms of countering mitochondrial toxicity by chemical strategies that leave the nucleoside analogue unchanged, an innovative and promising approach has been reported recently in the context of the anti-HIV drug d4T. Here, an alkyl-modification attached to the γ-phosphate group of d4T triphosphate greatly decreased its substrate properties towards host DNA polymerases while having a much smaller effect on its substrate properties towards HIV-RT.203 Thus, the γ-alkyl-modification was able to enhance the selectivity index of the active compounds. If these findings translate also to POLRMT, this strategy holds great potential to generally counter mitochondrial toxicity of any ribonucleotide analogue that is to be developed as broad-spectrum drugs against RNA viruses.
Low efficacy due to inefficient metabolic activation
Nucleobase, nucleoside or nucleotide analogues can only become active on the target viral polymerase once they’ve been converted into the nucleoside analogue 5’-triphosphate. This intracellular metabolic activation is catalyzed by host enzymes (Figure 13) and can thus suffer from being inefficient or even blocked when analogues are not recognized well as substrates by these enzymes. As mentioned above, these metabolic bottlenecks may not be general but depend on specific cell types and hence, varying antiviral potency depending on the cell type has been reported for various NNAs.162,163,165 As this cell type-dependency limits the compounds’ antiviral spectrum, prodrug systems that deliver the active form not only contribute an important factor to attaining highly potent but also broadly active antivirals.
Figure 13.
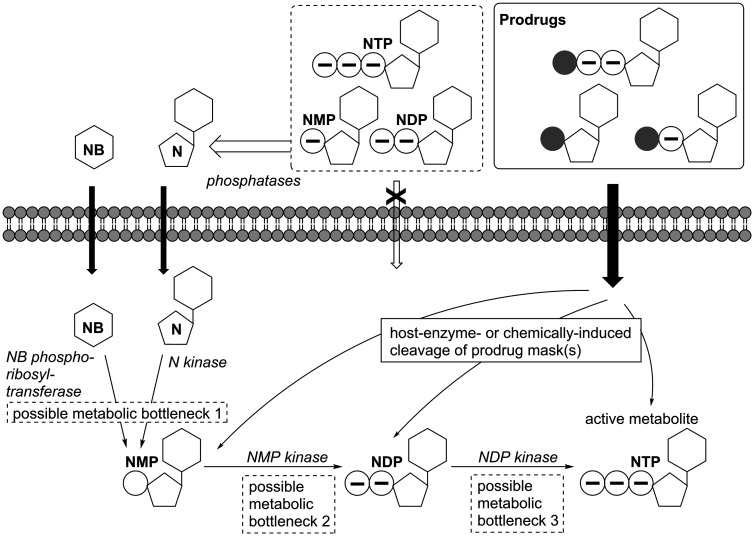
Intracellular activation pathway for nucleobase (NB) or nucleoside (N) analogues and nucleoside 5’-mono- (NMP), -di- (NDP), or -triphosphate (NTP) prodrugs. NB and N can traverse the cell membrane by passive or facilitated diffusion while nucleotides are held back due to their negative charges. Host phosphatases dephosphorylate NMPs, NDPs and NTPs. In the form of prodrugs, these nucleotides are protected from dephosphorylation and the charges are masked; therefore, the prodrugs can pass the cell membrane. Inside the cell, N or NB are metabolized by host enzymes via NMP and NDP to the antivirally active NTP form. Due to insufficient substrate properties of the analogues towards the host enzymes, one or more of these steps may be blocked. Prodrugs are activated intracellularly by ubiquitous host enzymes or chemical reactions and can thus overcome metabolic bottlenecks and deliver the active metabolite.
The active forms, the non-natural NTPs, can’t be applied as such as they 1) would quickly be dephosphorylated by host enzymes before reaching their intracellular viral target and 2) would not be able to pass through the cell membrane due to their highly polar, negatively charged properties. Hence, to overcome limiting steps in the metabolic activation of nucleoside analogues, prodrug strategies have been developed, often in the context of HIV or HCV as highlighted in a recent review.204 In sofosbuvir, for example, the 5’-monophosphate form of the nucleoside analogue is masked as an aryloxy phosphoramidate triester (Figure 6) and those masking groups are cleaved intracellularly via a cascade mechanism that is initiated by host carboxypeptidase or -esterase.205–207 This type of phosphoramidate pronucleotide (‘ProTide’), pioneered by Chris McGuigan and colleagues (for a review see Mehellou et al.208), was designed to release (non-natural) nucleoside monophosphates inside cells. The same technology is also used in remdesivir and while sofosbuvir specifically targets HCV, remdesivir has gained attention as potential anti-Ebola virus and, importantly, anti-coronavirus treatment. Since cleavage of the prodrug moiety still requires host enzyme-catalyzed steps, the prodrug’s efficacy may still be limited by different enzyme content (isoforms and concentrations) in different target tissues.209 Moreover, in terms of general applicability, it has been shown for other NNAs that the bottleneck can also lie in the later steps of metabolic activation, i.e. the formation of the nucleoside analogue di- or even triphosphate.210,211 And although many different prodrug systems have been developed for the intracellular delivery of nucleoside monophosphates (a topic which has been reviewed periodically, see literature212–214) similar strategies for di- and triphosphates were long believed impossible due to their high charges and the inherent chemical instability of the phosphoanhydride bond(s). However, in recent years, the development of the DiPPro- and TriPPPro-technologies demonstrated that chemical delivery systems for modified nucleoside di- and triphosphates are in fact attainable and successful in bypassing late or even all possible metabolic bottlenecks.215,216 This was impressively demonstrated by “switching” former inactive nucleoside analogues into active antivirals by applying the TriPPPro-technology.216 Still, once delivered into target cells, the active nucleoside analogue triphosphate may undergo rapid dephosphorylation by host phosphatases and this may again limit the efficacy of the drug. In this context, the combination of the γ-alkylphosphate modification (mentioned above in the context of lowering toxicity) and a prodrug-moiety, as recently reported for anti-HIV nucleoside analogue d4T triphosphate, appears promising.203 While the prodrug moiety enables the modified NTP to reach the intracellular space and is cleaved there, the γ-alkylphosphate modification was not only shown to increase the target selectivity but to also preclude dephosphorylation and thus stabilize the active nucleoside analogue triphosphate, thereby enhancing its half-life. Altogether, this strategy may increase the antiviral efficacy of NNAs and widen their antiviral spectra; a promising approach that will hopefully facilitate the clinical development of broad-spectrum NNAs as RNA virus therapeutics in the future.
Conclusion
RNA viruses have been responsible for causing devastating epi- and pandemics and the immediate threat of another global health crisis posed by a newly emerging RNA virus has become dramatic reality once more just recently with the emergence of SARS-CoV-2. Still, effective treatments remain scarce. Clinical development for drugs against diseases that are not yet in the large population is often hampered by limited testing opportunities and by being economically unviable. For newly emerging pathogens, the development of specific drugs or vaccines can only start once the pathogen is identified and model systems are available. In this context, the current SARS-CoV-2 pandemic has dramatically put the spotlight on the urgent need for antiviral drugs that are broadly active. Such drugs that inhibit diverse RNA viruses, either by targeting a common cellular process or a highly conserved viral enzyme, would be most promising as immediately available treatments against newly emerging viruses.
RNA viruses rely on a high supply of NTPs from the host metabolism to efficiently replicate. Drugs that inhibit some of the cellular NTP-producing processes can consequently also block viral replication in general and thus possess broad-spectrum antiviral activity. However, when cellular targets are addressed, toxicity may arise and a balance between efficient antiviral action and tolerable host toxicity must be found. In this context, enzymes of the purine or pyrimidine de novo pathways have been identified as potential host targets, as these pathways are required only for fast-dividing cells, such as cells of the immune system or in malignancies, and fast-replicating viruses but not to sustain viability of cells of the normal tissue. Regarding virus-targeted drugs, nucleoside and nucleotide analogues have been in use for several decades and vast antiviral data for many different NNAs is available to date; however, so far, no general pattern for structure-activity relationships emerges. Reasons for this include cell line-dependent metabolic limitations as NNAs need to be converted to their active triphosphate form in the relevant tissue. Consequently, the path towards developing broad-spectrum antiviral NNAs remains misty. Nonetheless, recent progress in the structural biology and biochemistry of viral polymerases, including the achievements in high-throughput as well as mechanistic assays, will help guide future efforts. Together with medicinal chemistry strategies to decrease toxic effects and innovative technologies to bypass metabolic limitations and overcome cell type-dependent potency, these will hopefully yield promising candidates. To counter high attrition rates, especially in advanced clinical development, efforts must further include early pre-clinical test systems for mitochondrial and other toxicity. Consequently, extensive collaboration among scientists from different fields and sectors must be fostered and the efficient translation of basic scientific findings to clinical settings must be supported to ultimately develop broadly-acting antiviral drugs and close this gaping hole in global preparedness.
Acknowledgement
The author wishes to express her gratitude to Prof. Chris Meier (University of Hamburg) and Prof. Lieve Naesens (Rega Institute for Medical Research of KU Leuven) for their wisdom and empowering mentorship, and to all colleagues and collaborators for being outstanding scientists and inspiring teammates. Previous own work in the context of broad-spectrum antiviral research was funded by the Chu Family Foundation, DAAD (Deutscher Akademischer Austauschdienst, German Academic Exchange Service), DFG (Deutsche Forschungsgemeinschaft, German Research Foundation), and DZIF (Deutsches Zentrum für Infektionsforschung, German Center for Infection Research).
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Johanna Huchting https://orcid.org/0000-0001-6368-3273
References
- 1.www.who.int/en/news-room/fact-sheets/detail/influenza-. (avian-and-other-zoonotic) (accessed 18 August 2020).
- 2.Sutton TC. The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses 2018; 10: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steel J, Lowen AC. Influenza a virus reassortment. Curr Top Microbiol Immunol 2014; 385: 377–401. [DOI] [PubMed] [Google Scholar]
- 4.Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine 2007; 25: 6852–6862. [DOI] [PubMed] [Google Scholar]
- 5.Hurt AC, Holien JK, Parker MW, et al. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs 2009; 69: 2523–2531. [DOI] [PubMed] [Google Scholar]
- 6.van der Vries E, Schutten M, Fraaij P, et al. Influenza virus resistance to antiviral therapy. Adv Pharmacol 2013; 67: 217–246. [DOI] [PubMed] [Google Scholar]
- 7.Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull Hist Med 2002; 76: 105–115. [DOI] [PubMed] [Google Scholar]
- 8.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis 2006; 12: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartore S, Bonfanti L, Lorenzetto M, et al. The effects of control measures on the economic burden associated with epidemics of avian influenza in Italy. Poult Sci 2010; 89: 1115–1121. [DOI] [PubMed] [Google Scholar]
- 10.Mostafa A, Abdelwhab EM, Mettenleiter TC, et al. Zoonotic potential of influenza A viruses: a comprehensive overview. Viruses 2018; 10: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013; 368: 1888–1897. [DOI] [PubMed] [Google Scholar]
- 12.Feldmann H, Sprecher A, Geisbert TW. Ebola. N Engl J Med 2020; 382: 1832–1842. [DOI] [PubMed] [Google Scholar]
- 13.WHO, Ebola Virus Disease Fact Sheet, www.who.int/news-room/fact-sheets/detail/ebola-virus-disease (accessed 18 August 2020); WHO, Case Situation Report; https://www.who.int/csr/disease/ebola/en/ (accessed 18 August 2020).
- 14.Cdc. Fact sheet about Andes virus, www.cdc.gov/hantavirus/resources/andes-virus.html (accessed 18 August 2020).
- 15.WHO. Lassa Fever Fact Sheet, www.who.int/news-room/fact-sheets/detail/lassa-fever (accessed 18 August 2020).
- 16.WHO, Crimean-Congo haemorrhagic fever fact sheet, www.who.int/news-room/fact-sheets/detail/crimean-congo-haemorrhagic-fever (accessed 18 August 2020).
- 17.Luby SP, Hossain MJ, Gurley ES, et al. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis 2009; 15: 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WHO. Nipah Virus Fact Sheet, www.who.int/news-room/fact-sheets/detail/nipah-virus (accessed 18 August 2020).
- 19.Guarner J. Three emerging coronaviruses in two decades. Am J Clin Pathol 2020; 153: 420–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO. Solidarity clinical trial for the COVID-19 treatments, www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments (accessed 18 August 2020).
- 21.Stahlmann R, Lode H. Medication for COVID-19 – an overview of approaches currently under study. Dtsch Arztebl Int 2020; 117: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol 1997; 51: 151–178. [DOI] [PubMed] [Google Scholar]
- 23.Carrasco-Hernandez R, Jacome R, Lopez Vidal Y, et al. Are RNA viruses candidate agents for the next global pandemic? A review. ILAR J 2017; 58: 343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol 2018; 16: e3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji X, Li Z. Medicinal chemistry strategies toward host targeting antiviral agents. Med Res Rev 2020; 40: 1519–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann HH, Kunz A, Simon VA, et al. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci USA 2011; 108: 5777–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 2008; 9: 267–276. [DOI] [PubMed] [Google Scholar]
- 28.Okesli A, Khosla C, Bassik MC. Human pyrimidine nucleotide biosynthesis as a target for antiviral chemotherapy. Curr Opin Biotechnol 2017; 48: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei-Jiao G, Shi-Fang L, Yan-Yan C, et al. Antiviral effects of selected IMPDH and DHODH inhibitors against foot and mouth disease virus. Biomed Pharmacother 2019; 118: 109305. [DOI] [PubMed] [Google Scholar]
- 30.Peters GJ. Antipyrimidine effects of five different pyrimidine de novo synthesis inhibitors in three head and neck cancer cell lines. Nucleosides Nucleotides Nucleic Acids 2018; 37: 329–339. [DOI] [PubMed] [Google Scholar]
- 31.Knecht W, Loffler M. Species-related inhibition of human and rat dihydroorotate dehydrogenase by immunosuppressive isoxazol and cinchoninic acid derivatives. Biochem Pharmacol 1998; 56: 1259–1264. [DOI] [PubMed] [Google Scholar]
- 32.Cao L, Weetall M, Trotta C, et al. Targeting of hematologic malignancies with PTC299, a novel potent inhibitor of dihydroorotate dehydrogenase with favorable pharmaceutical properties. Mol Cancer Ther 2019; 18: 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters GJ. Re-evaluation of brequinar sodium, a dihydroorotate dehydrogenase inhibitor. Nucleosides Nucleotides Nucleic Acids 2018; 37: 666–678. [DOI] [PubMed] [Google Scholar]
- 34.Xiong R, Zhang L, Li S, et al. Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2. Protein Cell 2020; 11: 723–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qing M, Zou G, Wang QY, et al. Characterization of dengue virus resistance to brequinar in cell culture. Antimicrob Agents Chemother 2010; 54: 3686–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hager PW, Collart FR, Huberman E, et al. Recombinant human inosine monophosphate dehydrogenase type I and type II proteins. Purification and characterization of inhibitor binding. Biochem Pharmacol 1995; 49: 1323–1329. [DOI] [PubMed] [Google Scholar]
- 37.Jackson RC, Weber G, Morris HP. IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature 1975; 256: 331–333. [DOI] [PubMed] [Google Scholar]
- 38.Naffouje R, Grover P, Yu H, et al. Anti-Tumor potential of IMP dehydrogenase inhibitors: a century-long story. Cancers (Basel) 2019; 11: 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobashigawa J, Miller L, Renlund D, et al. A randomized active-controlled trial of mycophenolate mofetil in heart transplant recipients1. Transplantation 1998; 66: 507–515. [DOI] [PubMed] [Google Scholar]
- 40.Broen JCA, van Laar JM. Mycophenolate mofetil, azathioprine and tacrolimus: mechanisms in rheumatology. Nat Rev Rheumatol 2020; 16: 167–178. [DOI] [PubMed] [Google Scholar]
- 41.Ichimura H, Levy JA. Polymerase substrate depletion: a novel strategy for inhibiting the replication of the human immunodeficiency virus. Virology 1995; 211: 554–560. [DOI] [PubMed] [Google Scholar]
- 42.Smee DF, Bray M, Huggins JW. Antiviral activity and mode of action studies of ribavirin and mycophenolic acid against orthopoxviruses in vitro. Antivir Chem Chemother 2001; 12: 327–335. [DOI] [PubMed] [Google Scholar]
- 43.Takhampunya R, Ubol S, Houng HS, et al. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. J Gen Virol 2006; 87: 1947–1952. [DOI] [PubMed] [Google Scholar]
- 44.Franklin TJ, Jacobs V, Bruneau P, et al. Glucuronidation by human colorectal adenocarcinoma cells as a mechanism of resistance to mycophenolic acid. Adv Enzyme Regul 1995; 35: 91–100. [DOI] [PubMed] [Google Scholar]
- 45.Sidwell RW, Huffman JH, Khare GP, et al. Broad-spectrum antiviral activity of virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 1972; 177: 705–706. [DOI] [PubMed] [Google Scholar]
- 46.Eberhardt KA, Mischlinger J, Jordan S, et al. Ribavirin for the treatment of lassa fever: a systematic review and meta-analysis. Int J Infect Dis 2019; 87: 15–20. [DOI] [PubMed] [Google Scholar]
- 47.Russmann S, Grattagliano I, Portincasa P, et al. Ribavirin-induced anemia: mechanisms, risk factors and related targets for future research. Curr Med Chem 2006; 13: 3351–3357. [DOI] [PubMed] [Google Scholar]
- 48.Mathur P, Kottilil S, Wilson E. Use of ribavirin for hepatitis C treatment in the modern direct-acting antiviral era. J Clin Transl Hepatol 2018; 6: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asogun DA, Gunther S, Akpede GO, et al. Lassa fever: epidemiology, clinical features, diagnosis, management and prevention. Infect Dis Clin North Am 2019; 33: 933–951. [DOI] [PubMed] [Google Scholar]
- 50.World Health Organization. Clinical management of patients with viral haemorrhagic fever: a pocket guide for front-line health workers: interim emergency guidance for country adaptation, https://apps.who.int/iris/. bitstream/handle/10665/205570/9789241549608_eng.pdf (2016, accessed 16 June 2020).
- 51.Nigeria centre for disease control. Viral haemorrhagic fevers preparedness and response plan, https://ncdc.gov.ng/themes/common/docs/ protocols/24_1502192155.pdf (2017, accessed 16 June 2020)
- 52.Fillatre P, Revest M, Tattevin P. Crimean-Congo hemorrhagic fever: an update. Med Mal Infect 2019; 49: 574–585. [DOI] [PubMed] [Google Scholar]
- 53.Graci JD, Cameron CE. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol 2006; 16: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Te HS, Randall G, Jensen DM. Mechanism of action of ribavirin in the treatment of chronic hepatitis C. Gastroenterol Hepatol (N Y) 2007; 3: 218–225. [PMC free article] [PubMed] [Google Scholar]
- 55.Nyström K, Waldenström J, Tang K-W, et al. Ribavirin: pharmacology, multiple modes of action and possible future perspectives. Future Virology 2019; 14: 153–160. [Google Scholar]
- 56.Wray SK, Gilbert BE, Noall MW, et al. Mode of action of ribavirin: effect of nucleotide Pool alterations on influenza virus ribonucleoprotein synthesis. Antiviral Res 1985; 5: 29–37. [DOI] [PubMed] [Google Scholar]
- 57.Leyssen P, Balzarini J, De Clercq E, et al. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J Virol 2005; 79: 1943–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreno H, Gallego I, Sevilla N, et al. Ribavirin can be mutagenic for arenaviruses. J Virol 2011; 85: 7246–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olschlager S, Neyts J, Gunther S. Depletion of GTP Pool is not the predominant mechanism by which ribavirin exerts its antiviral effect on Lassa virus. Antiviral Res 2011; 91: 89–93. [DOI] [PubMed] [Google Scholar]
- 60.Julian TR, Baugher JD, Rippinger CM, et al. Murine norovirus (MNV-1) exposure in vitro to the purine nucleoside analog ribavirin increases quasispecies diversity. Virus Res 2016; 211: 165–173. [DOI] [PubMed] [Google Scholar]
- 61.Tate PM, Mastrodomenico V, Mounce BC. Ribavirin induces polyamine depletion via nucleotide depletion to limit virus replication. Cell Rep 2019; 28: 2620–2633. [DOI] [PubMed] [Google Scholar]
- 62.Streeter DG, Witkowski JT, Khare GP, et al. Mechanism of action of 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (virazole), a new broad-spectrum antiviral agent. Proc Natl Acad Sci USA 1973; 70: 1174–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crotty S, Maag D, Arnold JJ, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med 2000; 6: 1375–1379. [DOI] [PubMed] [Google Scholar]
- 64.Cameron CE, Castro C. The mechanism of action of ribavirin: lethal mutagenesis of RNA virus genomes mediated by the viral RNA-dependent RNA polymerase. Curr Opin Infect Dis 2001; 14: 757–764. [DOI] [PubMed] [Google Scholar]
- 65.Scheidel LM, Stollar V. Mutations that confer resistance to mycophenolic acid and ribavirin on sindbis virus map to the nonstructural protein nsP1. Virology 1991; 181: 490–499. [DOI] [PubMed] [Google Scholar]
- 66.Bougie I, Bisaillon M. The broad spectrum antiviral nucleoside ribavirin as a substrate for a viral RNA capping enzyme. J Biol Chem 2004; 279: 22124–22130. [DOI] [PubMed] [Google Scholar]
- 67.Balzarini J, Karlsson A, Wang L, et al. Ethynyl-1-beta-D-ribofuranosylimidazole-4-carboxamide). a novel potent inhibitor of inosinate dehydrogenase activity and guanylate biosynthesis. J Biol Chem 1993; 268: 24591–24598. [PubMed] [Google Scholar]
- 68.De Clercq E, Cools M, Balzarini J, et al. Antiviral activities of 5-ethynyl-1-beta-D-ribofuranosylimidazole-4-carboxamide and related compounds. Antimicrob Agents Chemother 1991; 35: 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gilbert HJ, Drabble WT. Active-site modification of native and mutant forms of inosine 5'-monophosphate dehydrogenase from Escherichia coli K12. Biochem J 1980; 191: 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang W, Papov VV, Minakawa N, et al. Inactivation of inosine 5'-monophosphate dehydrogenase by the antiviral agent 5-ethynyl-1-beta-D-ribofuranosylimidazole-4-carboxamide 5'-monophosphate. Biochemistry 1996; 35: 95–101. [DOI] [PubMed] [Google Scholar]
- 71.McDowell M, Gonzales SR, Kumarapperuma SC, et al. A novel nucleoside analog, 1-beta-d-ribofuranosyl-3-ethynyl-[1,2,4]triazole (ETAR), exhibits efficacy against a broad range of flaviviruses in vitro. Antiviral Res 2010; 87: 78–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okano Y, Saito-Tarashima N, Kurosawa M, et al. Synthesis and biological evaluation of novel imidazole nucleosides as potential anti-dengue virus agents. Bioorg Med Chem 2019; 27: 2181–2186. [DOI] [PubMed] [Google Scholar]
- 73.Eyer L, Nencka R, de Clercq E, et al. Nucleoside analogs as a rich source of antiviral agents active against arthropod-borne flaviviruses. Antivir Chem Chemother 2018; 26: 2040206618761299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou T, Kurnasov O, Tomchick DR, et al. Structure of human nicotinamide/nicotinic acid mononucleotide adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin. J Biol Chem 2002; 277: 13148–13154. [DOI] [PubMed] [Google Scholar]
- 75.Jager W, Salamon A, Szekeres T. Metabolism of the novel IMP dehydrogenase inhibitor benzamide riboside. Curr Med Chem 2002; 9: 781–786. [DOI] [PubMed] [Google Scholar]
- 76.Wolf YI, Kazlauskas D, Iranzo J, et al. Origins and evolution of the global RNA virome. mBio 2018; 9: e02329–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castro C, Arnold JJ, Cameron CE. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res 2005; 107: 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Warmbrod KL, Patterson EI, Kautz TF, et al. Viral RNA-dependent RNA polymerase mutants display an altered mutation spectrum resulting in attenuation in both mosquito and vertebrate hosts. PLoS Pathog 2019; 15: e1007610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vignuzzi M, Stone JK, Arnold JJ, et al. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 2006; 439: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferrero D, Ferrer-Orta C, Verdaguer N. Viral RNA-Dependent RNA polymerases: a structural overview. Subcell Biochem 2018; 88: 39–71. [DOI] [PubMed] [Google Scholar]
- 81.Te Velthuis AJ. Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci 2014; 71: 4403–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lang DM, Zemla AT, Zhou CL. Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucleic Acids Res 2013; 41: 1464–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Das K, Arnold E. Negative-Strand RNA virus L proteins: one machine, many activities. Cell 2015; 162: 239–241. [DOI] [PubMed] [Google Scholar]
- 84.Amroun A, Priet S, de Lamballerie X, et al. Bunyaviridae RdRps: structure, motifs, and RNA synthesis machinery. Crit Rev Microbiol 2017; 43: 753–778. [DOI] [PubMed] [Google Scholar]
- 85.de Farias ST, Dos Santos Junior AP, Rego TG, et al. Origin and evolution of RNA-dependent RNA polymerase. Front Genet 2017; 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boehr AK, Arnold JJ, Oh HS, et al. 2'-C-methylated nucleotides terminate virus RNA synthesis by preventing active site closure of the viral RNA-dependent RNA polymerase. J Biol Chem 2019; 294: 16897–16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eyer L, Kondo H, Zouharova D, et al. Escape of Tick-Borne flavivirus from 2'-C-methylated nucleoside antivirals is mediated by a single conservative mutation in NS5 that has a dramatic effect on viral fitness. J Virol 2017; 91: e01028–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delang L, Yen PS, Vallet T, et al. Differential transmission of antiviral drug-resistant Chikungunya viruses by aedes mosquitoes. mSphere 2018; 3: e00230–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eyer L, Nougairède A, Uhlířová M, et al. An E460D substitution in the NS5 protein of tick-borne encephalitis virus confers resistance to the inhibitor galidesivir (BCX4430) and also attenuates the virus for mice. J Virol 2019; 93: e00367–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.WHO. Topics, HIV/AIDS, Pre-exposure prophylaxis, www.who.int/hiv/topics/prep/en/ (accessed 18 August 2020).
- 91.Deval J. Antimicrobial strategies: inhibition of viral polymerases by 3'-hydroxyl nucleosides. Drugs 2009; 69: 151–166. [DOI] [PubMed] [Google Scholar]
- 92.Sofia MJ, Bao D, Chang W, et al. Discovery of a beta-d-2'-deoxy-2'-alpha-fluoro-2'-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem 2010; 53: 7202–7218. [DOI] [PubMed] [Google Scholar]
- 93.Potisopon S, Ferron F, Fattorini V, et al. Substrate selectivity of dengue and zika virus NS5 polymerase towards 2'-modified nucleotide analogues. Antiviral Res 2017; 140: 25–36. [DOI] [PubMed] [Google Scholar]
- 94.Good SS, Moussa A, Zhou XJ, et al. Preclinical evaluation of at-527, a novel guanosine nucleotide prodrug with potent, pan-genotypic activity against hepatitis C virus. PLoS One 2020; 15: e0227104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carroll SS, Tomassini JE, Bosserman M, et al. Inhibition of hepatitis C virus RNA replication by 2'-modified nucleoside analogs. J Biol Chem 2003; 278: 11979–11984. [DOI] [PubMed] [Google Scholar]
- 96.Deval J, Symons JA, Beigelman L. Inhibition of viral RNA polymerases by nucleoside and nucleotide analogs: therapeutic applications against positive-strand RNA viruses beyond hepatitis C virus. Curr Opin Virol 2014; 9: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Benzaria S, Bardiot D, Bouisset T, et al. 2'-C-Methyl branched pyrimidine ribonucleoside analogues: potent inhibitors of RNA virus replication. Antivir Chem Chemother 2007; 18: 225–242. [DOI] [PubMed] [Google Scholar]
- 98.Zmurko J, Marques RE, Schols D, et al. The viral polymerase inhibitor 7-Deaza-2'-C-methyladenosine is a potent inhibitor of in vitro zika virus replication and delays disease progression in a robust mouse infection model. PLoS Negl Trop Dis 2016; 10: e0004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eyer L, Valdes JJ, Gil VA, et al. Nucleoside inhibitors of tick-borne encephalitis virus. Antimicrob Agents Chemother 2015; 59: 5483–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.WHO, Dengue Fact Sheet, www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed 5 June 2020).
- 101.WHO, West Nile virus fact sheet, www.who.int/news-room/fact-sheets/detail/west-nile-virus (accessed 5 June 2020).
- 102.WHO, Yellow Fever Fact Sheet, www.who.int/news-room/fact-sheets/detail/yellow-fever (accessed 5 June 2020).
- 103.WHO, Zika Virus Fact Sheet, www.who.int/news-room/fact-sheets/detail/zika-virus (accessed 5 June 2020).
- 104.Van Dycke J, Arnoldi F, Papa G, et al. A single nucleoside viral polymerase inhibitor against norovirus, rotavirus, and Sapovirus-induced diarrhea. J Infect Dis 2018; 218: 1753–1758. [DOI] [PubMed] [Google Scholar]
- 105.Migliaccio G, Tomassini JE, Carroll SS, et al. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem 2003; 278: 49164–49170. [DOI] [PubMed] [Google Scholar]
- 106.Lefebvre DJ, De Vleeschauwer AR, Goris N, et al. Proof of concept for the inhibition of foot-and-mouth disease virus replication by the anti-viral drug 2'-C-methylcytidine in severe combined immunodeficient mice. Transbound Emerg Dis 2014; 61: e89-91–e91. [DOI] [PubMed] [Google Scholar]
- 107.Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol 1993; 28: 375–430. [DOI] [PubMed] [Google Scholar]
- 108.Mifsud EJ, Hayden FG, Hurt AC. Antivirals targeting the polymerase complex of influenza viruses. Antiviral Res 2019; 169: 104545. [DOI] [PubMed] [Google Scholar]
- 109.Dias A, Bouvier D, Crepin T, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009; 458: 914–918. [DOI] [PubMed] [Google Scholar]
- 110.Omoto S, Speranzini V, Hashimoto T, et al. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci Rep 2018; 8: 9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Olschewski S, Cusack S, Rosenthal M. The cap-snatching mechanism of bunyaviruses. Trends Microbiol 2020; 28: 293–303. [DOI] [PubMed] [Google Scholar]
- 112.Kumaki Y, Day CW, Smee DF, et al. In vitro and in vivo efficacy of fluorodeoxycytidine analogs against highly pathogenic avian influenza H5N1, seasonal, and pandemic H1N1 virus infections. Antiviral Res 2011; 92: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Welch SR, Guerrero LW, Chakrabarti AK, et al. Lassa and ebola virus inhibitors identified using minigenome and recombinant virus reporter systems. Antiviral Res 2016; 136: 9–18. [DOI] [PubMed] [Google Scholar]
- 114.Welch SR, Scholte FEM, Flint M, et al. Identification of 2'-deoxy-2'-fluorocytidine as a potent inhibitor of Crimean-Congo hemorrhagic fever virus replication using a recombinant fluorescent reporter virus. Antiviral Res 2017; 147: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arnold JJ, Sharma SD, Feng JY, et al. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog 2012; 8: e1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tchesnokov EP, Bailey-Elkin BA, Mark BL, et al. Independent inhibition of the polymerase and deubiquitinase activities of the Crimean-Congo hemorrhagic fever virus full-length L-protein. PLoS Negl Trop Dis 2020; 14: e0008283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cho A, Saunders OL, Butler T, et al. Synthesis and antiviral activity of a series of 1'-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg Med Chem Lett 2012; 22: 2705–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Siegel D, Hui HC, Doerffler E, et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-Nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J Med Chem 2017; 60: 1648–1661. [DOI] [PubMed] [Google Scholar]
- 119.Mulangu S, Dodd LE, Davey RT, Jr, et al. ; PALM Consortium Study Team. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med 2019; 381: 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 2017; 9: eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lo MK, Jordan R, Arvey A, et al. GS-5734 and its parent nucleoside analog inhibit filo-, pneumo-, and paramyxoviruses. Sci Rep 2017; 7: 43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brown AJ, Won JJ, Graham RL, et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res 2019; 169: 104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pruijssers AJ, George AS, Schafer A, et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep 2020; 32: 107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gordon CJ, Tchesnokov EP, Woolner E, et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 2020; 295: 6785–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shannon A, Le NT, Selisko B, et al. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res 2020; 178: 104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Seifert M, Bera SC, van Nies P, et al. Signatures and mechanisms of efficacious therapeutic ribonucleotides against SARS-CoV-2 revealed by analysis of its replicase using magnetic tweezers. bioRxiv 2020: 2020.2008.2006.240325. DOI: 10.1101/2020.08.06.240325. [Google Scholar]
- 127.Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA 2018; 115: E162–E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Smith EC, Blanc H, Surdel MC, et al. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog 2013; 9: e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Agostini ML, Andres EL, Sims AC, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 2018; 9: e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pruijssers AJ, Denison MR. Nucleoside analogues for the treatment of coronavirus infections. Curr Opin Virol 2019; 35: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Evans GB, Tyler PC, Schramm VL. Immucillins in infectious diseases. ACS Infect Dis 2018; 4: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Freitas EO, Nico D, Alves-Silva MV, et al. Immucillins ImmA and ImmH are effective and non-toxic in the treatment of experimental visceral leishmaniasis. PLoS Negl Trop Dis 2015; 9: e0004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Warren TK, Wells J, Panchal RG, et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 2014; 508: 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Perales C, Gallego I, de Avila AI, et al. The increasing impact of lethal mutagenesis of viruses. Future Med Chem 2019; 11: 1645–1657. [DOI] [PubMed] [Google Scholar]
- 135.Page T, Connor JD. The metabolism of ribavirin in erythrocytes and nucleated cells. Int J Biochem 1990; 22: 379–383. [DOI] [PubMed] [Google Scholar]
- 136.Kilham L, Ferm VH. Congenital anomalies induced in hamster embryos with ribavirin. Science 1977; 195: 413–414. [DOI] [PubMed] [Google Scholar]
- 137.Ferm VH, Willhite C, Kilham L. Teratogenic effects of ribavirin on hamster and rat embryos. Teratology 1978; 17: 93–101. [DOI] [PubMed] [Google Scholar]
- 138.Kochhar DM, Penner JD, Knudsen TB. Embryotoxic, teratogenic, and metabolic effects of ribavirin in mice. Toxicol Appl Pharmacol 1980; 52: 99–112. [DOI] [PubMed] [Google Scholar]
- 139.Baranovich T, Wong SS, Armstrong J, et al. T-705 (favipiravir) induces lethal mutagenesis in influenza a H1N1 viruses in vitro. J Virol 2013; 87: 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Arias A, Thorne L, Goodfellow I. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. Elife 2014; 3: e03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vanderlinden E, Vrancken B, Van Houdt J, et al. Distinct effects of T-705 (favipiravir) and ribavirin on influenza virus replication and viral RNA synthesis. Antimicrob Agents Chemother 2016; 60: 6679–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Escribano-Romero E, Jimenez de Oya N, Domingo E, et al. Extinction of West Nile virus by favipiravir through lethal mutagenesis. Antimicrob Agents Chemother 2017; 61: e01400–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Goldhill DH, Langat P, Xie H, et al. Determining the mutation bias of favipiravir in influenza virus using next-generation sequencing. J Virol 2019; 93: e01217–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jin Z, Smith LK, Rajwanshi VK, et al. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5'-triphosphate towards influenza a virus polymerase. PLoS One 2013; 8: e68347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jin Z, Tucker K, Lin X, et al. Biochemical evaluation of the inhibition properties of favipiravir and 2'-C-methyl-cytidine triphosphates against human and mouse norovirus RNA polymerases. Antimicrob Agents Chemother 2015; 59: 7504–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shannon A, Selisko B, Le NT, et al. Rapid incorporation of favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat Commun 2020; 11: 4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sangawa H, Komeno T, Nishikawa H, et al. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob Agents Chemother 2013; 57: 5202–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dulin D, Arnold JJ, van Laar T, et al. Signatures of nucleotide analog incorporation by an RNA-dependent RNA polymerase revealed using high-throughput magnetic tweezers. Cell Rep 2017; 21: 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Furuta Y, Takahashi K, Kuno-Maekawa M, et al. Mechanism of action of T-705 against influenza virus. Antimicrob Agents Chemother 2005; 49: 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jin Z, Kinkade A, Behera I, et al. Structure-activity relationship analysis of mitochondrial toxicity caused by antiviral ribonucleoside analogs. Antiviral Res 2017; 143: 151–161. [DOI] [PubMed] [Google Scholar]
- 151.Furuta Y, Takahashi K, Fukuda Y, et al. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother 2002; 46: 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gowen BB, Wong MH, Jung KH, et al. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrob Agents Chemother 2007; 51: 3168–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Julander JG, Smee DF, Morrey JD, et al. Effect of T-705 treatment on Western equine encephalitis in a mouse model. Antiviral Res 2009; 82: 169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Oestereich L, Ludtke A, Wurr S, et al. Successful treatment of advanced ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res 2014; 105: 17–21. [DOI] [PubMed] [Google Scholar]
- 155.Bixler SL, Bocan TM, Wells J, et al. Efficacy of favipiravir (T-705) in nonhuman primates infected with Ebola virus or Marburg virus. Antiviral Res 2018; 151: 97–104. [DOI] [PubMed] [Google Scholar]
- 156.Sissoko D, Laouenan C, Folkesson E, et al. Experimental treatment with favipiravir for ebola virus disease (the JIKI trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med 2016; 13: e1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Furuta Y, Gowen BB, Takahashi K, et al. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res 2013; 100: 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Furuta Y, Komeno T, Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci 2017; 93: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Delang L, Abdelnabi R, Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res 2018; 153: 85–94. [DOI] [PubMed] [Google Scholar]
- 160.Julander JG, Shafer K, Smee DF, et al. Activity of T-705 in a hamster model of yellow fever virus infection in comparison with that of a chemically related compound, T-1106. Antimicrob Agents Chemother 2009; 53: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rocha-Pereira J, Jochmans D, Dallmeier K, et al. Favipiravir (T-705) inhibits in vitro norovirus replication. Biochem Biophys Res Commun 2012; 424: 777–780. [DOI] [PubMed] [Google Scholar]
- 162.Lanko K, Eggermont K, Patel A, et al. Replication of the zika virus in different iPSC-derived neuronal cells and implications to assess efficacy of antivirals. Antiviral Res 2017; 145: 82–86. [DOI] [PubMed] [Google Scholar]
- 163.Franco EJ, Rodriquez JL, Pomeroy JJ, et al. The effectiveness of antiviral agents with broad-spectrum activity against chikungunya virus varies between host cell lines. Antivir Chem Chemother 2018; 26: 2040206618807580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bixler SL, Bocan TM, Wells J, et al. Intracellular conversion and in vivo dose response of favipiravir (T-705) in rodents infected with Ebola virus. Antiviral Res 2018; 151: 50–54. [DOI] [PubMed] [Google Scholar]
- 165.Huchting J, Vanderlinden E, Van Berwaer R, et al. Cell line-dependent activation and antiviral activity of T-1105, the non-fluorinated analogue of T-705 (favipiravir). Antiviral Res 2019; 167: 1–5. [DOI] [PubMed] [Google Scholar]
- 166.Huchting J, Winkler M, Nasser H, et al. Synthesis of T-705-Ribonucleoside and T-705-Ribonucleotide and studies of chemical stability. ChemMedChem 2017; 12: 652–659. [DOI] [PubMed] [Google Scholar]
- 167.Huchting J, Vanderlinden E, Winkler M, et al. Prodrugs of the phosphoribosylated forms of hydroxypyrazinecarboxamide pseudobase T-705 and its De-Fluoro analogue T-1105 as potent influenza virus inhibitors. J Med Chem 2018; 61: 6193–6210. [DOI] [PubMed] [Google Scholar]
- 168.Stuyver LJ, Whitaker T, McBrayer TR, et al. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob Agents Chemother 2003; 47: 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Costantini VP, Whitaker T, Barclay L, et al. Antiviral activity of nucleoside analogues against norovirus. Antivir Ther 2012; 17: 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Reynard O, Nguyen XN, Alazard-Dany N, et al. Identification of a new ribonucleoside inhibitor of ebola virus replication. Viruses 2015; 7: 6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ehteshami M, Tao S, Zandi K, et al. Characterization of beta-d-N(4)-hydroxycytidine as a novel inhibitor of chikungunya virus. Antimicrob Agents Chemother 2017; 61: e02395–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Urakova N, Kuznetsova V, Crossman DK, et al. Beta-d-N (4)-hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J Virol 2018; 92: e01965–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yoon JJ, Toots M, Lee S, et al. Orally efficacious Broad-Spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob Agents Chemother 2018; 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Agostini ML, Pruijssers AJ, Chappell JD, et al. Small-Molecule antiviral beta-d-N (4)-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J Virol 2019; 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sheahan TP, Sims AC, Zhou S, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 2020; 12: eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.COVID-19 first in human study to evaluate safety, tolerability, and pharmacokinetics of EIDD-2801 in healthy volunteers phase 1 clinical trial, www.ClinicalTrials.gov (identifier NCT04392219) (accessed 20 July 2020).
- 177.A safety tolerability and efficacy of EIDD-2801 to eliminate infectious virus detection in persons with COVID-19. Phase 2 clinical trial, retrieved from www.ClinicalTrials.gov (identifier NCT04405570) (accessed 20 July 2020).
- 178.The Safety of EIDD-2801 and Its Effect on Viral Shedding of SARS-CoV-2 (The END-COVID Study). Phase 2 clinical trial, www.ClinicalTrials.gov (identifier NCT04405739) (accessed 20 July 2020).
- 179.Leś A, Adamowicz L, Rode W. Structure and conformation of N4-hydroxycytosine and N4-hydroxy-5-fluorocytosine. A theoretical ab initio study. Biochim Biophys Acta 1993; 1173: 39–48. [DOI] [PubMed] [Google Scholar]
- 180.Branda N, Kurz G, Lehn J-M. JANUS WEDGES: a new approach towards nucleobase-pair recognition. Chem Commun 1996; 2443–2444. [Google Scholar]
- 181.Yang H-Z, Pan M-Y, Jiang D-W, et al. Synthesis of janus type nucleoside analogues and their preliminary bioactivity. Org Biomol Chem 2011; 9: 1516–1522. [DOI] [PubMed] [Google Scholar]
- 182.Seley-Radtke K. Flexibility-Not just for yoga anymore! Antivir Chem Chemother 2018; 26: 2040206618756788.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Peters HL, Ku TC, Seley-Radtke KL. Flexibility as a strategy in nucleoside antiviral drug design. Curr Med Chem 2015; 22: 3910–3921. [DOI] [PubMed] [Google Scholar]
- 184.Peters HL, Jochmans D, de Wilde AH, et al. Design, synthesis and evaluation of a series of acyclic fleximer nucleoside analogues with anti-coronavirus activity. Bioorg Med Chem Lett 2015; 25: 2923–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yates MK, Raje MR, Chatterjee P, et al. Flex-nucleoside analogues – novel therapeutics against filoviruses. Bioorg Med Chem Lett 2017; 27: 2800–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yates MK, Chatterjee P, Flint M, et al. Probing the effects of pyrimidine functional group switches on acyclic fleximer analogues for antiviral activity. Molecules 2019; 24: 3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Vichier-Guerre S, Dugue L, Pochet S. 2'-Deoxyribonucleoside 5'-triphosphates bearing 4-phenyl and 4-pyrimidinyl imidazoles as DNA polymerase substrates. Org Biomol Chem 2019; 17: 290–301. [DOI] [PubMed] [Google Scholar]
- 188.Yu H, Eritja R, Bloom LB, et al. Ionization of bromouracil and fluorouracil stimulates base mispairing frequencies with guanine. J Biol Chem 1993; 268: 15935–15943. [PubMed] [Google Scholar]
- 189.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003; 3: 330–338. [DOI] [PubMed] [Google Scholar]
- 190.Notari RE, DeYoung JL. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J Pharm Sci 1975; 64: 1148–1157. [DOI] [PubMed] [Google Scholar]
- 191.Zielinski WS, Sprinzl M. Chemical synthesis of 5-azacytidine nucleotides and preparation of tRNAs containing 5-azacytidine in its 3'-terminus. Nucleic Acids Res 1984; 12: 5025–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rogstad DK, Herring JL, Theruvathu JA, et al. Chemical decomposition of 5-aza-2'-deoxycytidine (decitabine): kinetic analyses and identification of products by NMR, HPLC, and mass spectrometry. Chem Res Toxicol 2009; 22: 1194–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Aimiuwu J, Wang H, Chen P, et al. RNA-dependent inhibition of ribonucleotide reductase is a major pathway for 5-azacytidine activity in acute myeloid leukemia. Blood 2012; 119: 5229–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Daskalakis M, Blagitko-Dorfs N, Hackanson B. Decitabine. Recent Results Cancer Res 2010; 184: 131–157. [DOI] [PubMed] [Google Scholar]
- 195.Dapp MJ, Clouser CL, Patterson S, et al. 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1. J Virol 2009; 83: 11950–11958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sierra S, Davila M, Lowenstein PR, et al. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J Virol 2000; 74: 8316–8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ruiz-Jarabo CM, Ly C, Domingo E, et al. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 2003; 308: 37–47. [DOI] [PubMed] [Google Scholar]
- 198.St Clair MH, Furman PA, Lubbers CM, et al. Inhibition of cellular alpha and virally induced deoxyribonucleic acid polymerases by the triphosphate of acyclovir. Antimicrob Agents Chemother 1980; 18: 741–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lewis W, Griniuviene B, Tankersley KO, et al. Depletion of mitochondrial DNA, destruction of mitochondria, and accumulation of lipid droplets result from fialuridine treatment in woodchucks (marmota monax). Lab Invest 1997; 76: 77–87. [PubMed] [Google Scholar]
- 200.Semino-Mora C, Leon-Monzon M, Dalakas MC. Mitochondrial and cellular toxicity induced by fialuridine in human muscle in vitro. Lab Invest 1997; 76: 487–495. [PubMed] [Google Scholar]
- 201.Feng JY, Xu Y, Barauskas O, et al. Role of mitochondrial RNA polymerase in the toxicity of nucleotide inhibitors of hepatitis C virus. Antimicrob Agents Chemother 2016; 60: 806–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Feng JY. Addressing the selectivity and toxicity of antiviral nucleosides. Antivir Chem Chemother 2018; 26: 2040206618758524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Meier C, Zhao C, Weber S, et al. Prodrugs of gamma-Alkyl-modified nucleoside triphosphates - improved inhibition of HIV reverse transcriptase. Angew Chem Int Ed Engl 2020; 59: 22063-22071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dousson CB. Current and future use of nucleo(s)tide prodrugs in the treatment of hepatitis C virus infection. Antivir Chem Chemother 2018; 26: 2040206618756430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bourdin C, McGuigan C, Brancale A, et al. Synthesis and evaluation against hepatitis C virus of 7-deaza analogues of 2'-C-methyl-6-O-methyl guanosine nucleoside and L-Alanine ester phosphoramidates. Bioorg Med Chem Lett 2013; 23: 2260–2264. [DOI] [PubMed] [Google Scholar]
- 206.Birkus G, Wang R, Liu X, et al. Cathepsin a is the major hydrolase catalyzing the intracellular hydrolysis of the antiretroviral nucleotide phosphonoamidate prodrugs GS-7340 and GS-9131. Antimicrob Agents Chemother 2007; 51: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Saboulard D, Naesens L, Cahard D, et al. Characterization of the activation pathway of phosphoramidate triester prodrugs of stavudine and zidovudine. Mol Pharmacol 1999; 56: 693–704. [PubMed] [Google Scholar]
- 208.Mehellou Y, Balzarini J, McGuigan C. Aryloxy phosphoramidate triesters: a technology for delivering monophosphorylated nucleosides and sugars into cells. ChemMedChem 2009; 4: 1779–1791. [DOI] [PubMed] [Google Scholar]
- 209.Sun D. Remdesivir for treatment of COVID-19: Combination of pulmonary and IV administration may offer aditional benefit. Aaps J 2020; 22: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Jordan PC, Stevens SK, Tam Y, et al. Activation pathway of a nucleoside analog inhibiting respiratory syncytial virus polymerase. ACS Chem Biol 2017; 12: 83–91. [DOI] [PubMed] [Google Scholar]
- 211.Pertenbreiter F, Balzarini J, Meier C. Nucleoside mono- and diphosphate prodrugs of 2',3'-dideoxyuridine and 2',3'-dideoxy-2',3'-didehydrouridine. ChemMedChem 2015; 10: 94–106. [DOI] [PubMed] [Google Scholar]
- 212.Ray AS, Hostetler KY. Application of kinase bypass strategies to nucleoside antivirals. Antiviral Res 2011; 92: 277–291. [DOI] [PubMed] [Google Scholar]
- 213.Wagner CR, Iyer VV, McIntee EJ. Pronucleotides: toward the in vivo delivery of antiviral and anticancer nucleotides. Med Res Rev 2000; 20: 417–451. [DOI] [PubMed] [Google Scholar]
- 214.Mackman RL, Cihlar T. Prodrug strategies in the design of nucleoside and nucleotide antiviral therapeutics. Annual Reports in Medicinal Chemistry. Academic Press, 2004, pp. 305–321.
- 215.Gollnest T, de Oliveira TD, Schols D, et al. Lipophilic prodrugs of nucleoside triphosphates as biochemical probes and potential antivirals. Nat Commun 2015; 6: 8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gollnest T, Dinis de Oliveira T, Rath A, et al. Membrane-permeable triphosphate prodrugs of nucleoside analogues. Angew Chem Int Ed Engl 2016; 55: 5255–5258. [DOI] [PubMed] [Google Scholar]