SUMMARY
ARID1A is frequently mutated in ovarian clear-cell carcinoma (OCCC) and often co-exists with activating mutations of PIK3CA. Although induction of pro-inflammatory cytokines has been observed in this cancer, the mechanism by which the two mutations synergistically activate cytokine genes remains elusive. Here we established an in vitro model of OCCC by introducing ARID1A knock-down and mutant PIK3CA in a normal human ovarian epithelial cell line, which resulted in cell transformation and cytokine gene induction. We demonstrate that loss of ARID1A impairs the recruitment of the Sin3A-HDAC complex, while PIK3CA mutation releases RelA from IkB, leading to cytokine gene activation. We show that an NF-kB inhibitor partly attenuates proliferation of OCCC and improves the efficacy of carboplatin both in cell culture and a mouse model. Our study thus reveals the mechanistic link between ARID1A/PIK3CA mutations and cytokine gene induction in OCCC, and suggests NF-kB inhibition can be a potential therapeutic option.
Keywords: Ovarian clear-cell carcinoma, Pro-inflammatory cytokine, RelA, ARID1A, PIK3CA
INTRODUCTION
Ovarian clear-cell carcinoma (OCCC) consists of about 10-15% of all ovarian cancers (Jones et al., 2010). OCCC is regarded as one of the most aggressive cancers because it is generally refractory to conventional chemotherapies such as taxol or cisplatin (Bast et al., 2009). Genome sequencing of OCCC patient samples has revealed highly recurrent loss-of-function mutations in ARID1A (Jones et al., 2010; Wiegand et al., 2010), a core subunit of the SWI/SNF ATP-dependent chromatin remodeling complex (Wilson and Roberts, 2011). Interestingly, ARID1A mutations often co-exist with activating mutations of PIK3CA, suggesting that they may cooperate in driving OCCC development (Samartzis et al., 2013; Yamamoto et al., 2012). Indeed, Arid1a knock-out/Pik3ca H1047R transgenic mice develop spontaneous OCCC, indicating that the two mutations are sufficient in initiating OCCC (Chandler et al., 2015). Previous studies have revealed that OCCC is associated with increased expression of pro-inflammatory cytokines both in human and mouse models (Chandler et al., 2015; Yamaguchi et al., 2010). Consistently, inhibition of IL-6 in cell culture reduced proliferation of OCCC cells (Chandler et al., 2015; Kumar and Ward, 2014), suggesting induction of pro-inflammatory cytokine might contribute to OCCC tumorigenesis. However, how the two mutations cooperate to induce pro-inflammatory cytokines is not clear. Understanding the molecular mechanism underlying this process can help develop strategies overcoming dysregulation of cytokines, which can serve as a potential treatment for OCCC.
Studying tumorigenesis mechanism in existing cancer cell lines is confounded by the different mutations acquired during their derivation and maintenance. To circumvent this problem, we utilized hTERT-immortalized normal human ovarian epithelial cell lines (Liu et al., 2004). By introducing ARID1A shRNA and PIK3CA mutations to normal human ovarian epithelial cells, we created an in vitro OCCC model. Using this model, we identified RelA NF-kB transcription factor to be a major factor driving cytokine induction. Mechanistically, we demonstrate that PIK3CA acts through AKT-IKK2 pathway to release RelA from IkB. In addition, disruption of ARID1A impairs the recruitment of the Sin3A-histone deacetylase (HDAC) repressor complex to cytokine genes, leading to their derepression. Importantly, inhibition of NF-kB by a chemical inhibitor attenuated the in vitro and in vivo growth of double mutant cells and improved the efficacy of carboplatin, a clinically used cisplatin-derivative. Our study thus not only reveals how mutations of a signaling molecule and an epigenetic factor can cooperate to drive tumorigenesis, but also raises the possibility of inhibiting NF-kB pathway as a potential treatment for OCCC.
RESULTS
ARID1A depletion and PIK3CA mutations transform normal human ovarian epithelial cells
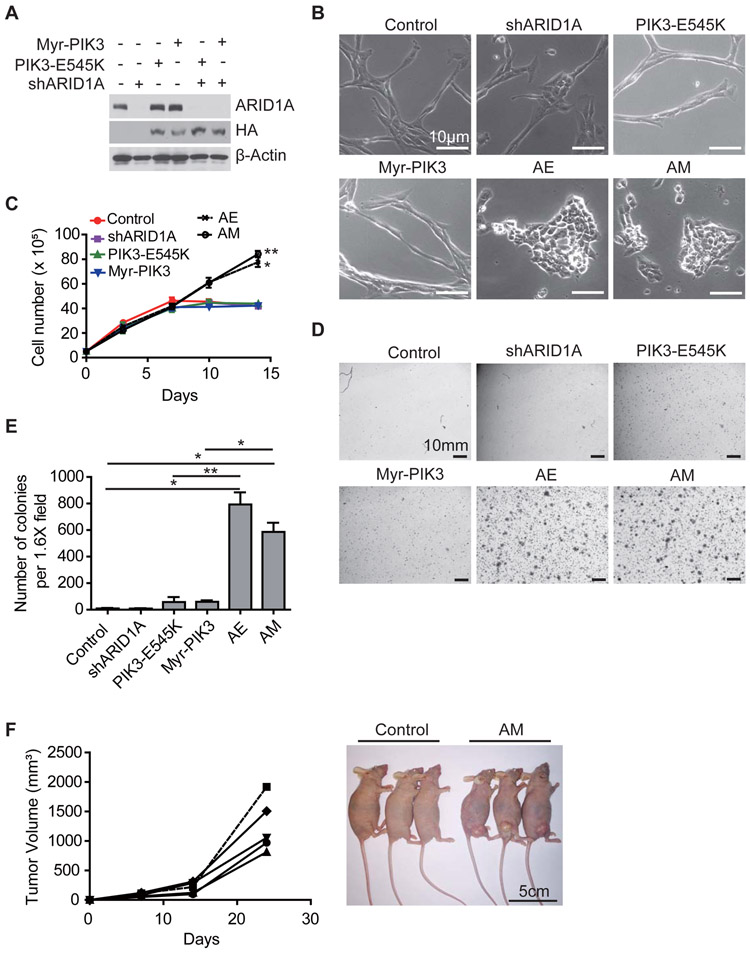
To generate an in vitro model for the study of OCCC containing loss of ARID1A function and gain of PIK3CA function mutations, we introduced single or double mutations into the immortalized normal human ovarian epithelial cell line T80 (Liu et al., 2004) by depleting ARID1A using a shRNA and/or expressing active PIK3CA, respectively (Figure 1A). For PIK3CA, we used either PIK3CA E545K or Myristoylation signal attached PIK3CA (Myr-PIK3CA). PIK3CA E545K is a naturally occurring mutant PIK3CA found in cancer patients that has increased enzymatic activity (Samuels et al., 2005), while Myr-PIK3CA is forcibly recruited to membrane and is constitutively active (Bitler et al., 2015). Although cells expressing ARID1A shRNA or active PIK3CA alone exhibit cell morphology similar to that of the control, cells with both (hereafter AE for ARID1A shRNA and E545K, and AM for ARID1A shRNA and Myr-PIK3CA) have strikingly different morphology with smaller size and disrupted organization (Figure 1B). Although the T80 (AE) and T80 (AM) cells exhibited a similar initial growth rate as that of the control T80 cells or cells with a single mutation, they appeared to have lost cell contact inhibition growth as they kept growing by piling-up on each other (Figure 1C). To ascertain that these observations are not due to a peculiar property of the T80 cell line, we performed similar experiments in T29, another independently established normal human ovarian epithelial cell line (Liu et al., 2004). Results shown in Figure S1 indicate that introduction of the double mutations also caused a morphological change as well as loss of contact inhibition of growth (Figures S1B and S1C).
Figure 1. ARID1A depletion and PIK3CA activation transform normal human ovarian epithelial cell.
(A) Introduction of double mutations into the T80 cell line. T80 cells were transduced with retroviruses expressing an ARID1A shRNA and the indicated PIK3CA alleles. After selection, the transduced cells were analyzed by Western blotting.
(B) Phase contrast images of established cell lines. Scale bar, 50μm.
(C) Growth curve of the established T80 cell lines expressing single or double mutant. 5x105 of the indicated cells were plated on 6-well plates, and counted every two days (n=3). Error bars, S.E.M. Statistical analysis comparing AM or AE cells with control cells was performed on day 14.
(D) Soft agar assay of the established T80 cell lines expressing single or double mutant. Representative images of soft agar colonies. Scale bar, 10mm.
(E) Colony numbers per 1.6x field were counted (n=3). Error bars, S.E.M.
(F) T80 cell line harboring the double mutant can form tumors in nude mice. Six-weeks old female nu/nu mice were subcutaneously injected with 3x105 control T80 or T80 (AM) cells (two independent experiments with 5 mice total in each group). Tumor volume was measured every week for each mouse (left growth curve). Mice were imaged at the time of sacrifice (right panel). Scale bar, 5cm.
*, p-value < 0.05; **, p-value < 0.01. See also Figure S1.
Next, we examined the ability of the T80 cells harboring the double mutations to grow in an anchorage-independent manner. To this end, soft agar assay was performed using cells with single or double mutations. Results shown in Figures 1D and 1E indicated that cells with active PIK3CA displayed a weak ability to grow in soft agar, which was drastically enhanced when combined with ARID1A depletion. Importantly, both PIK3CA mutations exhibited a similar result. To evaluate the tumorigenecity of the cells harboring the two mutations, the T80 (AM) cells were subcutaneously injected into nude mice. Two-weeks after the injection, palpable tumors were observed in all mice tested (5/5), which is in contrast with the mice injected with the same number of control T80 cells (Figure 1F). Collectively, the above results suggest that ARID1A depletion in combination with PIK3CA activation is sufficient to transform normal human ovarian epithelial cells.
ARID1A depletion and PIK3CA activation induce pro-inflammatory cytokine expression
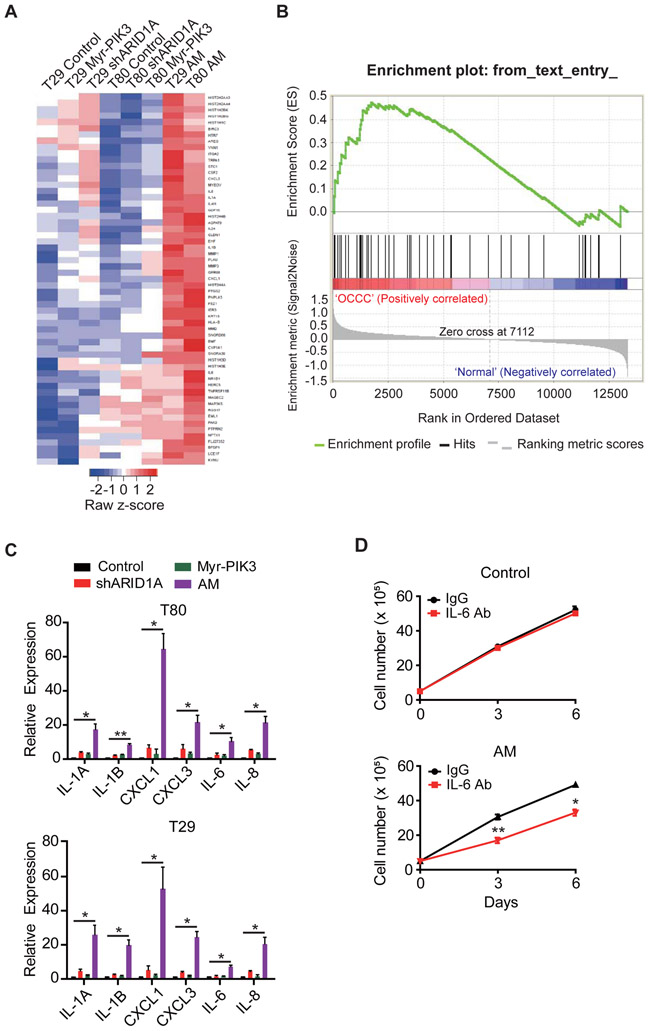
To gain insight into the molecular mechanism of how combined loss of ARID1A and gain of PIK3CA function contributes to transformation of human ovarian epithelial cells, we performed transcriptome analyses by RNA-Seq. To this end, T80 and T29 cells, as well as their derivatives harboring single or double mutations were used for RNA-seq analysis. Comparison of their transcriptomes revealed 59 genes uniquely up-regulated in the cells harboring double mutations [FC>=2 in both T29 (AM) and T80 (AM) cells] (Figure 2A and Supplementary Table S1). To examine if these up-regulated genes are relevant to human OCCC, we performed a Gene Set Enrichment Analysis using the identified 59 genes. By analyzing an available OCCC patient gene expression dataset GSE6008 (Wu et al., 2007), we confirmed that the 59 genes were indeed enriched in the up-regulated gene groups in OCCC patient samples (Figure 2B), supporting the relevance of the upregulation of these genes to human OCCC. Gene Ontology (GO) analysis revealed cytokine/chemokine activity as the most enriched GO term (Figure S2A). KEGG pathway analysis also revealed cytokine signaling as the most enriched pathway (Figure S2B). Among the 59 genes identified, 14 belong to the pro-inflammatory cytokines including IL-1A, IL-1B, IL-6, IL-8, CXCL1 and CXCL3 (Supplementary Table S1). To confirm the RNA-Seq result, we performed qRT-PCR on a subset of cytokine genes. Results shown in Figure 2C indicated that either ARID1A depletion or PIK3CA activation alone only slightly up-regulated cytokine gene expression. However, the combined mutations resulted in a robust activation. Similar results were also obtained in cells harboring the AE mutations (Figure S2C). In contrast to the up-regulated genes, no enrichment of any GO term or pathway was identified in the down-regulated genes, suggesting that up-regulation of these cytokines may underlie the molecular function of the double mutations.
Figure 2. Cytokine genes are aberrantly up-regulated in ARID1A/PIK3CA mutant cells.
(A) Heat-map of the genes commonly up-regulated in T29 (AM) and T80 (AM) cells compared to all other cell lines (FC>=2).
(B) Genes identified in panel (A) are enriched in the up-regulated genes in OCCC. Genes up-regulated in patient OCCC samples relative to normal tissues (from GSE6008) were ranked from left to right, and gene set enrichment on the up-regulated genes can be seen on the top and middle panels.
(C) qRT-PCR verification of the RNA-Seq result. mRNA level of selected cytokines genes were validated by qRT-PCR, normalized by β-actin transcripts (n=3 for all experiments except CXCL1 in T29, of which n=4). Error bars, S.E.M.
(D) IL-6 antibody can partly block T80 (AM), but not T80, cell growth. Control T80 or T80 (AM) cells were incubated with anti-mouse IgG or IL-6 neutralizing antibody (2 ng/ml) (n=3) and the cell proliferation rate was measured. Error bars, S.E.M.
*, p-value < 0.05; **, p-value < 0.01. See also Figure S2 and Supplemental Table S1.
The finding that double mutations result in up-regulation of pro-inflammatory cytokines is intriguing. First, up-regulation of certain cytokines has been previously observed in mouse OCCC model and human OCCC patients (Chandler et al., 2015; Yamaguchi et al., 2010), suggesting its clinical relevance. Second, several of these cytokines, including IL-6, IL-8 and CXCL1, have been previously implicated in promoting cell proliferation, survival, chemoresistance, metastasis and cancer stem cell expansion (Acharyya et al., 2012; Grivennikov et al., 2009; Iliopoulos et al., 2009; Schafer and Brugge, 2007; Waugh and Wilson, 2008). Thus, induction of these oncogenic cytokines may contribute to tumorigenesis. Consistent with a previous report that blocking IL-6 function can reduce proliferation of cancer cells harboring ARID1A/PIK3CA mutations (Chandler et al., 2015), neutralizing antibody against IL-6 was able to slow-down proliferation of the T80 (AM) cells, but not the control T80 cells (Figure 2D). Collectively, the T80 (AM) cells offer an in vitro experimental model for understanding the molecular basis of ARID1A/PIK3CA mutation-induced OCCC.
RelA NF-kB transcription factor is a major contributing factor for pro-inflammatory cytokine activation
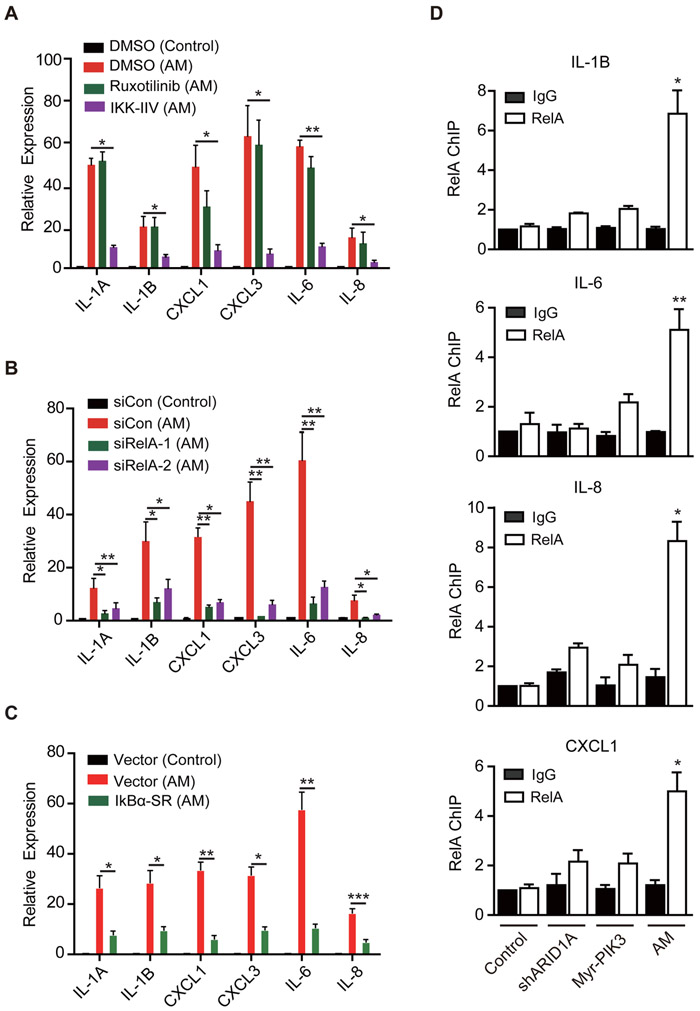
Previous studies have established that the STAT and NF-kB transcription pathways play an important role in cytokine gene induction (Grivennikov and Karin, 2010). To examine whether the two pathways are involved in ARID1A/PIK3CA mutation-caused cytokine induction, we treated the T80 (AM) cells with either a STAT inhibitor, Ruxotinilib (1μM), or an NF-kB inhibitor, IKK-IIV (1μM), and quantified cytokine expression by qRT-PCR. While Ruxotinilib had no significant effect, IKK-IIV markedly reduced the expression of cytokine genes (Figure 3A), indicating that cytokine gene induction in the double mutant cell is mainly mediated through the NF-kB pathway. Given that STAT activation was observed in both mouse OCCC model and human OCCC patients (Chandler et al., 2015; Yamaguchi et al., 2010), it was surprising that STAT inhibition did not result in cytokine gene suppression. Because STAT can be activated by cytokines (Grivennikov and Karin, 2010), our results suggest that STAT activation observed in OCCC is likely a secondary effect of NF-kB-mediated cytokine induction.
Figure 3. RelA NF-kB transcription factor is required for cytokine gene activation in ARID1A/PIK3CA mutant cells.
(A) The NF-kB inhibitor IKK-IIV, but not a STAT inhibitor Ruxotinilib, inhibited cytokine expression in the T80 (AM) cells. T80 (AM) cells were treated with DMSO, Ruxotinilib (1μM) or IKK-IIV (1μM) for three days. Control cells were treated with DMSO. Expression of selected cytokines genes were measured by qRT-PCR, normalized by β-actin (n=3). Error bars, S.E.M.
(B) RelA knock-down reduced cytokine gene expression. Control and T80 (AM) cells were transfected as indicated and the expression of selected cytokines genes were analyzed by qRT-PCR three days after transfection (n=4). Error bars, S.E.M.
(C) RelA inhibition by a dominant-negative IkB partly reduced cytokine gene expression. T80 (AM) cells were infected with empty or IkBα-SR expression retrovirus vector. After selecting with 100 ng/ml Zeocin for two days, surviving cells were analyzed by qRT-PCR (n=4). Error bars, S.E.M.
(D) Cytokine genes are direct RelA targets. ChIP analysis was performed in the indicated cells using antibodies against rabbit IgG (control) or RelA. Immunoprecipitated samples were PCR amplified using primers that amplify the RelA elements of the indicated genes. The signals were normalized with input (n=3). IgG of control cells was set as 1. Error bars, S.E.M. Statistical analysis comparing RelA ChIP in AM and control cells is presented.
*, p-value < 0.05; **, p-value < 0.01; ***, p-value < 0.001. See also Figure S3.
To substantiate the role of NF-kB in cytokine gene activation in the context of ARID1A/PIK3CA double mutations, we performed genetic manipulation. The NF-kB family transcription factors include both RelA and RelB that target overlapping as well as specific genes (Napetschnig and Wu, 2013; Sun, 2011). To determine whether RelA or RelB is involved in cytokine gene activation, we obtained siRNAs that deplete RelA or RelB, respectively (Figure S3A and S3B). While RelA knock-down efficiently decreased cytokine gene expression in T80 (AM) cells, knockdown of RelB had no significant effect (Figures 3B and S3C). RelA and RelB are differentially regulated by the IKK signalosome, which consists of three core proteins: IKK1 (IKKα), IKK2 (IKKβ), and IKKγ with IKK1 and IKK2 functioning as kinases, and IKKγ serving as a regulatory subunit. In response to signals that activate IKK2, IKK2 phosphorylates IkB, leading to its degradation, which then frees RelA from IkB-mediated cytoplasmic sequestration (Mercurio et al., 1997; Xia et al., 2014). To confirm RelA-mediated cytokine induction is part of the IKK signaling, we used a dominant-negative IkBα, IkBα-SR, which carries two serine-to-alanine mutations in IKK2 target sites and thus can block IKK2 signaling when over-expressed (Brown et al., 1995). Results shown in Figure 3C indicate that expression of IkBα-SR in the T80 (AM) cells successfully reduced cytokine gene expression. We note that neither IKK-IIV, IkBα-SR nor RelA depletion completely suppressed cytokine gene expression to the level of control cells. This is likely due to the existence of additional factors contributing to cytokine expression. Nevertheless, our results using multiple approaches indicate that RelA does play a major role in driving cytokine gene expression in the ARID1A/PIK3CA double mutant T80 (AM) cells. Importantly, RelA appears to directly contribute to cytokine gene up-regulation as ChIP-qPCR analysis demonstrates that RelA’s occupancy at the NF-kB binding elements of IL-6, IL-8, IL-1B and CXCL1 genes are all significantly increased in the T80 (AM) cells compared to other control cells (Figure 3D).
PIK3CA-AKT-IKK2 pathway releases RelA from IkB
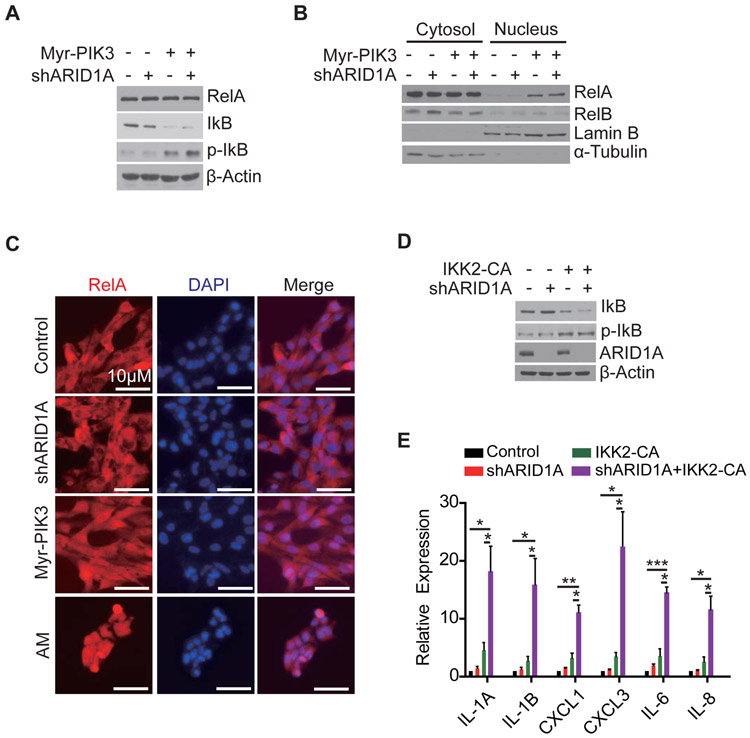
We next attempted to establish a link between PIK3CA activation and RelA-mediated cytokine gene activation. Previous studies in Jurkat cells or human fibroblasts have indicated that AKT, the main downstream target of PIK3CA, can free RelA from IkB by regulating IKK2 (Kane et al., 1999; Romashkova and Makarov, 1999). To examine whether a similar relationship exists in human ovarian epithelial cells, we introduced PIK3CA activation (Myr-) and ARID1A knockdown in the T80 cells. While Myr-PIK3CA expression reduced IkB and increased the phospho-IkB levels, ARID1A knock-down had no effect on IkB status (Figure 4A). Consistently, Myr-PIK3CA expression increased nuclear amount of RelA, but not RelB, as judged by Western blotting (Figure 4B), which was further supported by immuno-staining (Figure 4C).
Figure 4. Active PIK3CA releases RelA from IkB through AKT-IKK2 pathway.
(A) PIK3CA leads to IkB phosphorylation. Western blot analysis of the total and phosphorylated IkB in T80 cells expressing shARID1A, or Myr-PIK3CA, or combination. β-actin serves as a loading control.
(B) PIK3CA leads to nuclear localization of RelA. Indicated cells were fractionated to cytoplasm and nucleus, and analyzed by Western blotting for RelA and RelB. Lamin B and α-tubulin were used as nuclear and cytoplasmic markers, respectively.
(C) Nuclear localization of RelA in PIK3CA cells. Immune-staining of RelA in the indicated cells co-stained with DAPI. Scale bar, 10 μm.
(D) IKK2-CA can replace PIK3CA in mediating IkB phosphorylation. T80 cells were transduced with retroviruses expressing ARID1A shRNA or IKK2-CA or in combination. The total and phosphorylated IkB levels were analyzed by Western blotting.
(E) IKK2-CA causes cytokine gene activation. Cells from (D) were analyzed by qRT-PCR for the indicated cytokine genes, normalized by β-actin transcripts (n=4). Error bars, S.E.M. *, p-value < 0.05; **, p-value < 0.01; ***, p-value < 0.001. See also Figure S4.
To test whether AKT is a downstream effector of ARID1A/PIK3CA double mutation-mediated cytokine induction, we treated T80 (AM) cells with MK-2206, an AKT-specific inhibitor. MK-2206 treatment not only restored the IkB level (Figure S4A), but also suppressed the cytokine induction (Figure S4B), indicating AKT is an integral component of the ARID1A/PIK3CA-mediated cytokine induction pathway. If PIK3CA-AKT pathway releases RelA from IkB through IKK2, then constitutively active IKK2 (IKK2-CA) should be able to replace PIK3CA. To test this hypothesis, we established cell lines expressing IKK2-CA with or without ARID1A shRNA (Figure 4D). We found that cells with both IKK2-CA and ARID1A shRNA robustly induced cytokine gene expression (Figure 4E). This result together with the fact that IKK inhibitor abolishes cytokine expression (Figure 3A) allowed us to conclude that the functional effect of PIK3CA mutation in activation of cytokines is to release RelA from IkB through AKT-IKK2 pathway.
ARID1A recruits Sin3A-HDAC complex to cytokine genes
We next attempted to understand how loss of ARID1A function contributes to RelA-mediated cytokine gene induction. Since ARID1A is a component of the SWI/SNF ATP-dependent remodeling factor, we first considered the possibility that loss of ARID1A might affect chromatin accessibility by RelA, thus changing RelA-mediated cytokine gene expression. To address this possibility, we performed DNaseI hypersensitivity site (DHS) analysis by DNaseI hypersensitive sites sequencing (DNase-seq) (Neph et al., 2012). Consistent with the known function of the SWI/SNF complex in facilitating chromatin access, depletion of ARID1A resulted in loss of 153 (control vs ARID1A shRNA, FC>=3) and 557 (Myr-PIK3CA vs AM, FC>=3) DHSs, respectively (Figure S5A). However, neither ARID1A shRNA alone nor ARID1A/PIK3CA double mutations generated new or increased DHS signal (FC>=3), including the genomic regions of the altered cytokine genes (Figure S5B). Thus, increased cytokine expression in the T80 (AM) cells is not likely due to increased RelA accessibility to the cytokine genes. The lack of a significant effect of loss ARID1 function on the cytokine gene accessibility by RelA is likely due to the fact that the RelA binding sites are already accessible in the control cells (red bars in Figure S5B).
After ruling out the possibility that ARID1A depletion contributes to cytokine induction through regulating chromatin accessibility, we explored alternative mechanisms. Although the SWI/SNF complex is mostly involved in gene activation, it can also repress transcription by recruiting histone deacetylase complexes (Harikrishnan et al., 2005; Zhang et al., 2000). It is possible that ARID1A depletion may affect HDAC complex recruitment, leading to derepression of cytokine gene expression. Before testing if ARID1A recruits the Sin3A-HDAC complex to the cytokine genes, we first demonstrated that ARID1A does bind to the promoters of IL-6 and IL-8 genes in the T80 cells by ChIP assays (Figures 5A and 5B). We then asked whether the histone acetylation status at the ARID1A binding sites is affected by ARID1A depletion. ChIP analysis using a pan-H4 acetylation antibody revealed that knockdown of ARID1A in the T80 (AM) cells resulted in an increase in histone acetylation at the IL-6 and IL-8 promoters (Figure 5C). Consistently, HDAC1 binding to these regions was lost in response to ARID1A knockdown, but not to PIK3CA activation (Figure 5D), supporting a role of ARID1A in the recruitment of HDAC1 to these gene promoters.
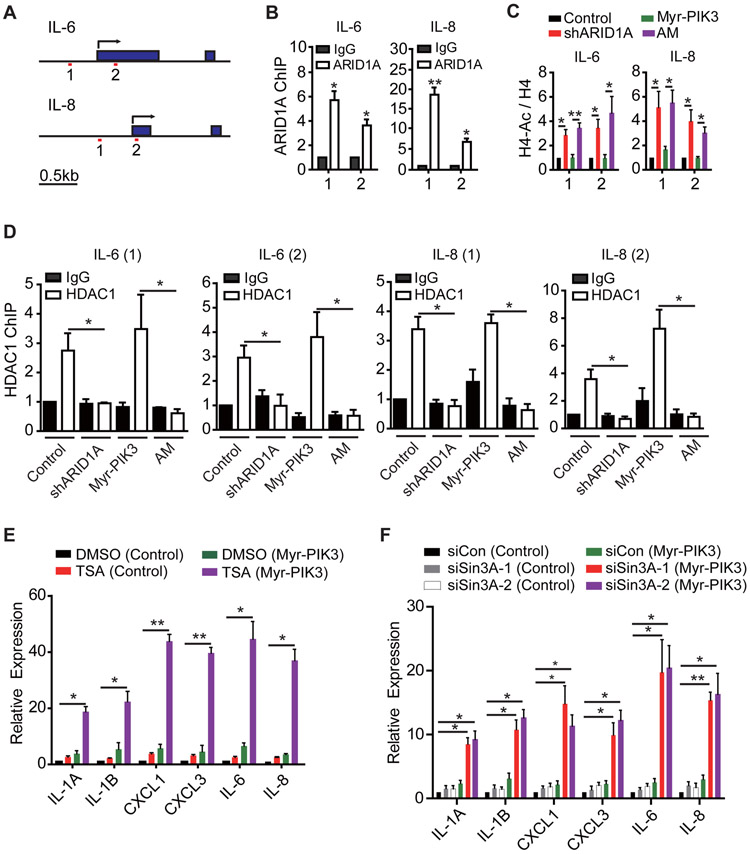
Figure 5. ARID1A suppresses cytokine gene expression by recruiting the Sin3A-HDAC complex.
(A) Schematic diagrams of the IL-6 and IL-8 gene loci for ChIP analysis. Transcription start sites (TSSs) are indicated by arrows. Filled boxes indicate exons. The two amplicons at promoter regions are indicated. Scale bar, 0.5kb.
(B) ARID1A binds to IL-6 and IL-8 promoters. T80 cells were subjected to chromatin immunoprecipitation (ChIP) using antibody against IgG or ARID1A. The amplicons are indicated in (A), and the signals are normalized by input (n=3). Error bars, S.E.M.
(C) ARID1A knockdown results in increased histone acetylation. The indicated T80 cells were subjected to ChIP analysis using an H4 or a pan-acetylated H4 antibody. ChIP samples were PCR amplified using the indicated primers. Acetylated H4 was normalized by H4. For each amplicon, control cell was set to 1 (n=4). Error bars, S.E.M.
(D) Loss of HDAC1 recruitment in ARID1A knockdown T80 cell lines. Indicated T80 cells were subjected to ChIP analysis using antibodies against IgG or HDAC1. ChIP samples were PCR amplified using indicated primers, normalized by input (n=3 for all experiments except TSS in IL-8, of which n=4). IgG in control cell was set as 1. Error bars, S.E.M.
(E) Histone deacetylase activity is required for suppressing cytokine gene expression. The indicated cells were treated with DMSO or TSA (100nM) for two days and analyzed by qRT-PCR of selected cytokines, normalized by β-actin (n=3). Error bars indicate S.E.M.
(F) Sin3A depletion resulted in derepression of cytokine genes. Control or Myr-PIK3CA T80 cells were transfected with the indicated siRNAs. Three days after transfection, cells were analyzed by qRT-PCR (n=4). Error bars, S.E.M.
*, p-value < 0.05; **, p-value < 0.01. See also Figure S6.
To examine whether the enzymatic activity of the recruited HDAC is involved in suppressing cytokine gene expression, we tested the effect of the HDAC inhibitor, Trichostatin-A (TSA). Remarkably, TSA treatment greatly induced cytokine gene expression in cells expressing Myr-PIK3CA (Figure 5E), but only modestly in the control cells, indicating a synergistic effect between HDAC inhibition and PIK3CA activation in cytokine induction. We further demonstrated that the HDAC1 involved is part of the Sin3A complex, as depletion of Sin3A by siRNAs, similar to that of HDAC inhibition, was able to synergize with Myr-PIK3CA to induce cytokine expression (Figures 5F and S6). Interestingly, a recent study indicated that ARID1A can recruit the Sin3A-HDAC complex to the telomerase gene in ovarian cancer (Suryo Rahmanto et al., 2016). Collectively, these results support the notion that ARID1A recruits the Sin3A-HDAC complex to suppress cytokine gene expression. Thus, loss of ARID1A can synergize with RelA activated by PIK3CA.
The mechanism of cytokine gene activation is conserved in human OCCC cells
Having established the mechanism of cytokine gene activation in the T80 cell line model, we next asked whether this mechanism is also used for cytokine gene activation in patient-derived OCCC cell lines. To this end, two OCCC cell lines; TOV-21G, which carries both ARID1A and PIK3CA mutations, and ES-2, which does not harbor mutation on either ARID1A or PIK3CA, were used to address the question. Similar to the results obtained in T80, TOV-21G cells had decreased IkB and increased phospho-IkB compared to ES-2 cells (Figure 6A). Consistently, nuclear RelA was higher in TOV-21G cells compared to that in ES-2 cells as judged by both cell fractionation and immune-staining (Figures 6B and 6C). Additionally, IKK-IIV effectively decreased cytokine gene expression in TOV-21G, but not in ES-2 cells (Figure 6D). Furthermore, siRNA-mediated depletion of RelA decreased cytokine gene expression in TOV-21G, but not in ES-2 cells (Figure 6E). ChIP-qPCR analysis confirmed that RelA binds to cytokine genes in TOV-21G, but not in ES-2 cells (Figure 6F). Finally, we analyzed histone acetylation status and HDAC1 recruitment and found that histone acetylation is increased and HDAC1 recruitment is decreased in the TOV-21G cells relative to that in the ES-2 cells, respectively (Figures 6G and 6H). Collectively, these results demonstrate that the mechanism used for cytokine induction in the T80 (AM) cells is conserved in the human OCCC TOV-21G cell line.
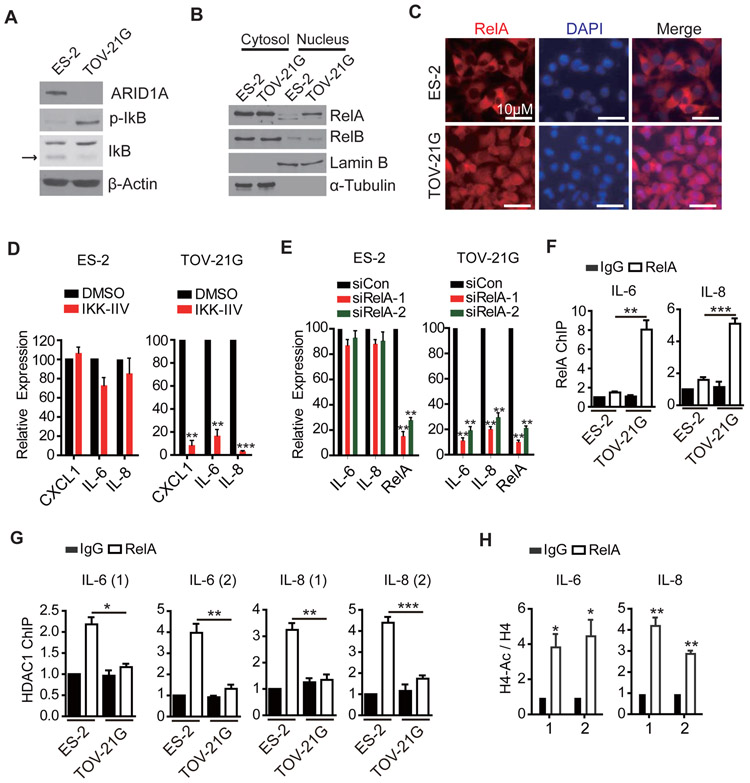
Figure 6. Mechanism of cytokine gene activation is conserved in human OCCC cell line.
(A) The phosphorylation state of IkB in ARID1A/PIK3CA mutant and wild-type OCCC cell lines. ES-2 (wild-type) and TOV-21G (double mutant) cell lysates were analyzed by Western blotting using the indicated antibodies. Arrow points to IkB.
(B) RelA is localized in nucleus in TOV-21G cell. ES-2 and TOV-21G cells were fractionated to cytoplasm and nucleus, and analyzed by Western blot with RelA and RelB antibodies.
(C) Nuclear localization of RelA in TOV-21G cell. Immune-staining of RelA in the indicated cells co-stained with DAPI. Scale bar, 10 μm.
(D) The NF-kB inhibitor IKK-IIV suppresses cytokine gene expression in TOV-21G, but not in ES-2, OCCC cell line. Cells were treated with DMSO or IKK-IIV (1μM) for three days. Their effect on cytokine gene expression was analyzed by qRT-PCR (n=3). Error bars, S.E.M.
(E) RelA knock-down suppresses cytokine gene expression in TOV-21G, but not in ES-2, OCCC cell line. Cells were transfected as indicated and the expression of selected cytokine genes were analyzed by qRT-PCR three days after transfection (n=4). Error bars, S.E.M.
(F) RelA binds at IL-6 and IL-8 promoters in TOV-21G cells. ChIP analysis was performed using antibodies against rabbit IgG (control) or RelA. The signals were normalized to input (n=4). IgG of control cells was set as 1. Error bars, S.E.M.
(G) HDAC1 binding at IL-6 and IL-8 is decreased in TOV-21G cells. ChIP analysis was performed using antibodies against IgG or HDAC1, and processed as (F). Error bars, S.E.M.
(H) Histone acetylation at IL-6 and IL-8 promoters is decreased in TOV-21G cells. ChIP analysis was performed using an H4 (for normalization) or a pan-acetylated H4 antibody. For each amplicon, control cell was set to 1 (n=4). Error bars, S.E.M.
*, p-value < 0.05; **, p-value < 0.01
NF-kB inhibitor attenuates growth of ARID1A and PIK3CA mutant cells and improves the efficacy of carboplatin
The fact that PIK3CA activating mutation caused cytokine activation through RelA raises the possibility that NF-kB inhibitors could be used as a novel therapy for OCCC with ARID1A and PIK3CA mutations. Pro-inflammatory cytokines have been previously implicated in several aspects of tumorigenesis including cell proliferation, survival, and in conferring resistance to chemotherapy, especially cisplatin (Acharyya et al., 2012; Grivennikov et al., 2009; Iliopoulos et al., 2009; Schafer and Brugge, 2007). Therefore, we thought that NF-kB inhibitor may attenuate cell proliferation and improve the efficacy of cisplatin through suppression of cytokine gene activation. The latter effect is particularly relevant given that OCCC is known to be cisplatin resistant (Bast et al., 2009). To this end, we first tested the effect of a NF-kB inhibitor on cell proliferation and found that while the proliferation rate of control cells or cells with a single mutation were not affected, growth of T80 (AM) cells was partly attenuated by the NF-kB inhibitor IKK-IIV (Figure 7A, black and purple curves). The partial attenuation might be due to either a partial rescue of cytokine gene expression by IKK-IIV or due to a limited contribution of cytokines to cell proliferation. Interestingly, when cells were treated with carboplatin, a clinically used derivative of cisplatin, only T80 (AM) cells showed persistent growth while the growth of other cells was effectively arrested (Figure 7A, green curves). Importantly, when carboplatin was combined with IKK-IIV, growth of the T80 (AM) cells was also arrested (Figure 7A, red curves). Of note, recombinant IL-6 and IL-8 was able to rescue this effect (Figure S7), indicating that IL-6 and IL-8 activation is mainly responsible for the chemoresistant effect. Consistent with its effect on the T80 (AM) cells, IKK-IIV partially attenuated proliferation of TOV-21G, but not that of the ES-2 cells (Figure 7B, black and purple curves). Importantly, when combined with carboplatin, IKK-IIV completely arrested TOV-21G cell proliferation (Figure 7B, red curves).
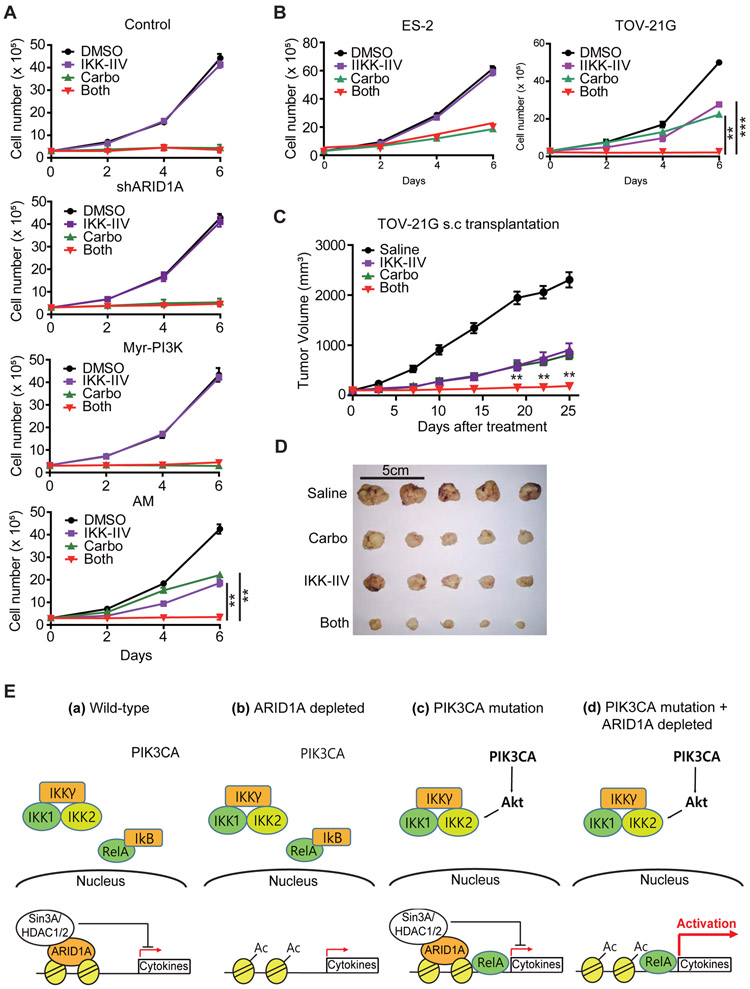
Figure 7. NF-kB inhibitor suppresses proliferation of ARID1A/PIK3CA mutant cells and synergizes with carboplatin.
(A) NF-kB inhibitor synergizes with carboplatin in suppressing T80 (AM) cell proliferation. 3x105 of indicated cells were plated on 6-well plates. One day after seeding, cells were treated with the indicated drugs and counted every two days (n=3). Error bars, S.E.M.
(B) Synergistic effect of IKK-IIV and carboplatin in suppressing TOV-21G, but not ES-2, cell proliferation. Cells were assayed as in (A) (n=3). Error bars, S.E.M.
(C) Combined treatment of IKK-IIV and carboplatin inhibits TOV-21G tumor growth in a mouse xenograft model. 3x106 TOV-21G cells were subcutaneously transplanted to 6-weeks old female nude mice and housed until tumors reached palpable size of about 100mm3. Mice were randomized into four groups for the indicated treatment and tumor sizes were measured at the indicated days (n=5 per group). Statistical analysis comparing combined treatment and single treatments is performed (either carboplatin only or IKK-IIV only, which resulted in the same range of p-values). Error bars, S.E.M.
(D) Image of dissected tumors at the time of mouse sacrifice shown in (C). Scale bar, 5cm.
(E) Working model of how ARID1A/PIK3CA mutation induces cytokine expression. (a) In wild-type cells, cytokine genes are repressed by the Sin3A-HDAC complex recruited by ARID1A. At the same time, RelA is sequestered by IkB. (b) In ARID1A mutant cells, the recruitment of Sin3-HDAC complex is abolished leading to histone acetylation. However, RelA is still sequestered by IkB. Thus, cytokine induction is limited. (c) In PIK3CA active cells, the AKT-IKK2 pathway releases RelA from IkB. However, the presence of Sin3A-HDAC complex limits cytokine induction. (d) In presence of both mutations, cytokine genes are robustly induced, which contributes to cell proliferation and drug resistance.
**, p-value < 0.01; ***, p-value < 0.001. See also Figure S7.
To examine whether the combined therapy is also effective in vivo, we transplanted TOV-21G cells into nude mice by subcutaneous injection. When the tumors reached palpable size (~100mm3), we divided the mice into four groups randomly and subject them to the following treatment: saline control, carboplatin only (30mg/kg, once per week), IKK-IIV only (25mg/kg, twice per week), and carboplatin plus IKK-IIV, respectively. While single treatments showed partial attenuation of tumor growth, the combined treatment completely arrested tumor growth (Figures 7C and 7D). These results demonstrate that the NF-kB inhibitor IKK-IIV is able to selectively suppress proliferation of ARID1A/PIK3CA double mutant cells, as well as to have the potential to improve the efficacy of carboplatin in OCCC treatment.
DISCUSSION
Understanding the molecular mechanisms underlying how cancer initiating mutations cause uncontrolled cellular proliferation is critical for developing effective treatment. Despite the identification of large number of epigenetic factor mutations in various cancers, how these mutations contribute to tumorigenesis is poorly understood. Here, we report how a loss-of-function mutation in an epigenetic factor (ARID1A) synergizes with a gain-of-function mutation in a signaling factor (PIK3CA) to contribute to tumorigenesis in OCCC through inducing pro-inflammatory cytokine gene expression (Figure 7E). Mechanistically, loss of ARID1A releases cytokine genes from Sin3A-HDAC-mediated repression, while activating mutation of PIK3CA frees RelA from IkB so that RelA can enter the nucleus to activate cytokine genes. These mechanistic insights prompted us to test the therapeutic potential of a NF-kB inhibitor in combination with carboplatin, which is supported by our data presented in Figure 7. Our results thus provide the missing mechanistic link between ARID1A/PIK3CA mutations and cytokine gene induction.
Loss of Sin3A-HDAC repression underlies cytokine gene induction in OCCC
Components of SWI/SNF complex, including ARID1A, are frequently mutated in different types of cancers (Kadoch and Crabtree, 2015). Understanding how these mutations contribute to oncogenesis is of great importance. Although the DNaseI accessibility to the cytokine genes is not altered by loss of ARID1A, the recruitment of the Sin3A-HDAC complex to the cytokine genes is impaired by loss of ARID1A. HDAC1 and HDAC2 exist in at least two distinctive protein complexes that include NuRD, and Sin3A (Zhang et al., 1997; Zhang et al., 1998). The NuRD complex has established function in cancer (Lai and Wade, 2011; Wang et al., 2009). Therefore, HDAC inhibitors have been widely used as an anti-cancer drug with promising outcomes (Minucci and Pelicci, 2006). In contrast, the role of Sin3A-HDAC complex in cancer has been unclear. Interestingly, our results indicate that the Sin3A-HDAC complex serves as a tumor suppressor in OCCC. Consistent with the fact that an HDAC inhibitor failed to show beneficial effects in cisplatin-resistant ovarian cancers in a phase II clinical trial (Mackay et al., 2010), our results suggest that HDAC inhibitors would not be effective for OCCC, and could even cause unfavorable outcomes.
An unanswered, but important remaining question is whether chromatin-remodeling function of ARID1A contributes to OCCC development. It is hard to imagine that recruitment of Sin3A-HDAC complex is the sole function of ARID1A contributing to OCCC. Future studies analyzing the relationship between transcriptional changes and DNase I sensitivity changes may reveal additional ARID1A functions. It is likely that the combined effect of chromatin remodeling-dependent and -independent function of ARID1A contribute to oncogenesis in OCCC.
Gain of RelA activation drives aberrant cytokine gene induction in OCCC
The NF-kB pathway plays a central role in cancer biology through activating multiple genes involved in cell proliferation, survival, drug resistance, angiogenesis and metastasis (Baud and Karin, 2009; Karin et al., 2004). Thus, NF-kB inhibition has been reported to increase the efficacy of several anticancer reagents (Nakanishi and Toi, 2005). However, the role of NF-kB in OCCC has been poorly characterized. Our results suggest that the RelA NF-kB transcription factor has an important role in OCCC development and progression. Constitutive activation of NF-kB has been observed in many types of cancers, especially in hematological cancers (Horie, 2013). In these tumors, NF-kB is usually activated by mutations of genes that directly regulate NF-kB. Constitutive NF-kB activity is also found in some solid tumors where NF-kB is usually activated by inflammatory cues (e.g. smoking or intestinal inflammation) (Grivennikov et al., 2009; Schwitalla et al., 2013; Takahashi et al., 2010). Unlike these known NF-kB activation mechanisms, OCCC uses a unique strategy to activate NF-kB. In OCCC, a combination of two mutations that respectively derepress and activate cytokine gene transcription. It is important to note that up-regulation of pro-inflammatory cytokines in OCCC is achieved cell autonomously by the two mutations without the involvement of any environmental factors.
Potential histone acetyltransferases that activate cytokines in OCCC
Our current study did not identify the responsible histone acetyltransferases (HATs) cooperating with RelA in OCCC. So far, two HATs have been implicated in NF-kB mediated cytokine transcription. The first are the p300/CBP, which have been implicated in cytokine induction in TNF-α stimulated HUVEC and leukemia cells (Gerritsen et al., 1997; Perkins et al., 1997). The other is Tip60, which has also been implicated in cytokine induction in TNF-α stimulated HepG2 cells (Kim et al., 2012). To explore whether the two HATs are involved in cytokine gene induction in OCCC, we treated both T80 (AM) and TOV-21G cells with inhibitor of p300 (C636) or Tip60 (Nu9056) or even their combination. However, none of these treatments reduced cytokine gene expression (M.K. Unpublished data). In addition, despite multiple attempts, we failed to detect binding of p300 or Tip60 at cytokine gene promoters in T80 (AM) cells. Thus, it is likely that HATs other than p300 or Tip60 may be involved in cytokine gene induction in OCCC. Identification of the responsible HAT may provide a target for OCCC treatment.
NF-kB inhibitor as a potential therapeutic option for ARID1A/PIK3CA mutant cancers
Our results show that NF-kB inhibition could selectively inhibit the growth of ARID1A/PIK3CA mutant cells, while leaving normal cells or OCCC without these mutations unaffected. In addition, we demonstrate both in cell culture and mouse model that NF-kB inhibition potentiates the effect of carboplatin, which is currently the first line of treatment for ovarian cancer but is not effective for OCCC. Furthermore, cytokines up-regulated in our system are implicated in cancer-stromal interaction, which can drive processes involved in cancer progression such as angiogenesis (e.g. IL-8) (Waugh and Wilson, 2008) and metastasis (e.g. CXCL1) (Acharyya et al., 2012). Therefore, we expect that NF-kB inhibition could also benefit OCCC treatment by blocking cancer progression. It would be informative to test this hypothesis in a genetically engineered mouse model of OCCC in the future. Additionally, the use of an NF-kB inhibitor would be a better strategy than suppressing expression of a specific cytokine. It is worth noting that a recent study revealed combined mutations of ARID1A/PIK3CA in gastric adenocarcinoma (Zang et al., 2012). It would be interesting to test if our findings in OCCC can also be applied to this type of gastric cancer.
EXPERIMENTAL PROCEDURES
Mouse xenograft assay
All animal studies were performed in accordance with guidelines of the Institutional Animal Care and Use Committee at Harvard Medical School. Six-week old female nude mice were purchased from Jackson Laboratory (stock No. 002019, homozygous for Foxn). 3x106 cells (mixed with 50% Matrigel, Corning) were subcutaneously injected into the right part of back skin. Tumor sizes were measured using an electronic caliper. For treatment experiments, tumors were grown until they reached palpable size of ~100 mm3. Then, mice were randomized into four groups and treated with saline, carboplatin only (30mg/kg, once per week), IKK-IIV only (25mg/kg, twice per week), and a combination by intraperitoneal injection.
RNA-seq
RNA-seq libraries were made from poly(A)+ mRNA using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs). Briefly, mRNA was purified from 1 μg total RNA using NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) and fragmented by heating at 94 °C for 15 minutes. The fragmented RNA was reverse transcribed with random primers. After second strand DNA synthesis, the cDNA fragments were dA tailed and adaptor ligated. PCR amplification was carried out to get sufficient amount of libraries for sequencing. The libraries were sequenced on an Illumina Hiseq2500 in single-end mode (Illumina). Sequencing reads were mapped to human genome hg19 using Tophat 2.0.13 (Trapnell et al., 2009). Expression level of each gene was quantified with normalized FPKM (fragments per kilobase of exon per million mapped fragments) using Cufflinks 2.2.1 (Trapnell et al., 2010). Gene Set Enrichment Analysis was carried out using GSEA v2.2.2 application from Broad Institute with expression data downloaded from GSE6008 (Wu et al., 2007) and the set of upregulated genes in double mutant cells as gene set input. The default enrichment cutoff was used in the analysis.
DNase I hypersensitive sites sequencing
DNase-seq libraries were prepared from about 1 million cells following a published protocol with minor modifications (He et al., 2014). The nuclei were digested with DNase I (Roche) at 60 U/ml final concentration at 37°C for 5 minutes. The reactions were stopped by adding stop buffer containing EDTA and proteinase K. DNA was purified by phenyl-chloroform extraction and ethanol precipitation. Small fragments around 150 bp were selected with SPRIselect (Beckman Coulter). Sequencing libraries were prepared with NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs). The libraries were sequenced on an Illumina Hiseq2500 in single-end mode (Illumina). Sequencing reads were mapped to human genome hg19 using Bowtie2 2.1.0 (Langmead and Salzberg, 2012). Differential DHSs with 3-fold difference were called using ChIPDiff (Xu et al., 2008).
Statistical analysis
All graphs were drawn using GraphPad Prism. Statistical analyses were performed by paired student’s t-test (two-tailed) comparing indicated pairs in each figure.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Jin-Sung Liu for the T80 and T29 cell lines; Luis Tuesta for critical reading of the manuscript. M.K. is supported by a NIH F32 Postdoctoral Training Fellowship. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBER
The accession number for the sequencing data generated in this study is GEO: GSE86004.
REFERENCES
- Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, et al. (2012). A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 150, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bast RC Jr., Hennessy B, and Mills GB (2009). The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer 9, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud V, and Karin M (2009). Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov 8, 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih Ie M, Conejo-Garcia JR, et al. (2015). Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 21, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Gerstberger S, Carlson L, Franzoso G, and Siebenlist U (1995). Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science 267, 1485–1488. [DOI] [PubMed] [Google Scholar]
- Chandler RL, Damrauer JS, Raab JR, Schisler JC, Wilkerson MD, Didion JP, Starmer J, Serber D, Yee D, Xiong J, et al. (2015). Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat Commun 6, 6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, and Collins T (1997). CREB-binding protein/p300 are transcriptional coactivators of p65. Proceedings of the National Academy of Sciences of the United States of America 94, 2927–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al. (2009). IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, and Karin M (2010). Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev 21, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, et al. (2005). Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet 37, 254–264. [DOI] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, et al. (2014). Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods 11, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie R (2013). Molecularly-targeted Strategy and NF-kappaB in lymphoid malignancies. J Clin Exp Hematop 53, 185–195. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, and Struhl K (2009). An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr., Vogelstein B, et al. (2010). Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, and Crabtree GR (2015). Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv 1, e1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LP, Shapiro VS, Stokoe D, and Weiss A (1999). Induction of NF-kappaB by the Akt/PKB kinase. Curr Biol 9, 601–604. [DOI] [PubMed] [Google Scholar]
- Karin M, Yamamoto Y, and Wang QM (2004). The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov 3, 17–26. [DOI] [PubMed] [Google Scholar]
- Kim JW, Jang SM, Kim CH, An JH, Kang EJ, and Choi KH (2012). New molecular bridge between RelA/p65 and NF-kappaB target genes via histone acetyltransferase TIP60 cofactor. The Journal of biological chemistry 287, 7780–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, and Ward AC (2014). Role of the interleukin 6 receptor family in epithelial ovarian cancer and its clinical implications. Biochim Biophys Acta 1845, 117–125. [DOI] [PubMed] [Google Scholar]
- Lai AY, and Wade PA (2011). Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 11, 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, Patterson A, Marquez RT, Auersperg N, Yu Y, Hahn WC, et al. (2004). A genetically defined model for human ovarian cancer. Cancer Res 64, 1655–1663. [DOI] [PubMed] [Google Scholar]
- Mackay HJ, Hirte H, Colgan T, Covens A, MacAlpine K, Grenci P, Wang L, Mason J, Pham PA, Tsao MS, et al. (2010). Phase II trial of the histone deacetylase inhibitor belinostat in women with platinum resistant epithelial ovarian cancer and micropapillary (LMP) ovarian tumours. Eur J Cancer 46, 1573–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, et al. (1997). IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278, 860–866. [DOI] [PubMed] [Google Scholar]
- Minucci S, and Pelicci PG (2006). Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6, 38–51. [DOI] [PubMed] [Google Scholar]
- Nakanishi C, and Toi M (2005). Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer 5, 297–309. [DOI] [PubMed] [Google Scholar]
- Napetschnig J, and Wu H (2013). Molecular basis of NF-kappaB signaling. Annu Rev Biophys 42, 443–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. (2012). An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, and Nabel GJ (1997). Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science 275, 523–527. [DOI] [PubMed] [Google Scholar]
- Romashkova JA, and Makarov SS (1999). NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature 401, 86–90. [DOI] [PubMed] [Google Scholar]
- Samartzis EP, Noske A, Dedes KJ, Fink D, and Imesch P (2013). ARID1A mutations and PI3K/AKT pathway alterations in endometriosis and endometriosis-associated ovarian carcinomas. Int J Mol Sci 14, 18824–18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. (2005). Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7, 561–573. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, and Brugge JS (2007). IL-6 involvement in epithelial cancers. J Clin Invest 117, 3660–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38. [DOI] [PubMed] [Google Scholar]
- Sun SC (2011). Non-canonical NF-kappaB signaling pathway. Cell Res 21, 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryo Rahmanto Y, Jung JG, Wu RC, Kobayashi Y, Heaphy CM, Meeker AK, Wang TL, and Shih IM (2016). Inactivating ARID1A Tumor Suppressor Enhances TERT Transcription and Maintains Telomere Length in Cancer Cells. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Ogata H, Nishigaki R, Broide DH, and Karin M (2010). Tobacco smoke promotes lung tumorigenesis by triggering IKKbeta- and JNK1-dependent inflammation. Cancer Cell 17, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al. (2009). LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672. [DOI] [PubMed] [Google Scholar]
- Waugh DJ, and Wilson C (2008). The interleukin-8 pathway in cancer. Clin Cancer Res 14, 6735–6741. [DOI] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. (2010). ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 363, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, and Roberts CW (2011). SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11, 481–492. [DOI] [PubMed] [Google Scholar]
- Wu R, Hendrix-Lucas N, Kuick R, Zhai Y, Schwartz DR, Akyol A, Hanash S, Misek DE, Katabuchi H, Williams BO, et al. (2007). Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signaling pathways. Cancer cell 11, 321–333. [DOI] [PubMed] [Google Scholar]
- Xia Y, Shen S, and Verma IM (2014). NF-kappaB, an active player in human cancers. Cancer Immunol Res 2, 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wei CL, Lin F, and Sung WK (2008). An HMM approach to genome-wide identification of differential histone modification sites from ChIP-seq data. Bioinformatics 24, 2344–2349. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Mandai M, Oura T, Matsumura N, Hamanishi J, Baba T, Matsui S, Murphy SK, and Konishi I (2010). Identification of an ovarian clear cell carcinoma gene signature that reflects inherent disease biology and the carcinogenic processes. Oncogene 29, 1741–1752. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Tsuda H, Takano M, Tamai S, and Matsubara O (2012). Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod Pathol 25, 615–624. [DOI] [PubMed] [Google Scholar]
- Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al. (2012). Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet 44, 570–574. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, and Dean DC (2000). Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101, 79–89. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, and Reinberg D (1997). Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89, 357–364. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, and Reinberg D (1998). The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.