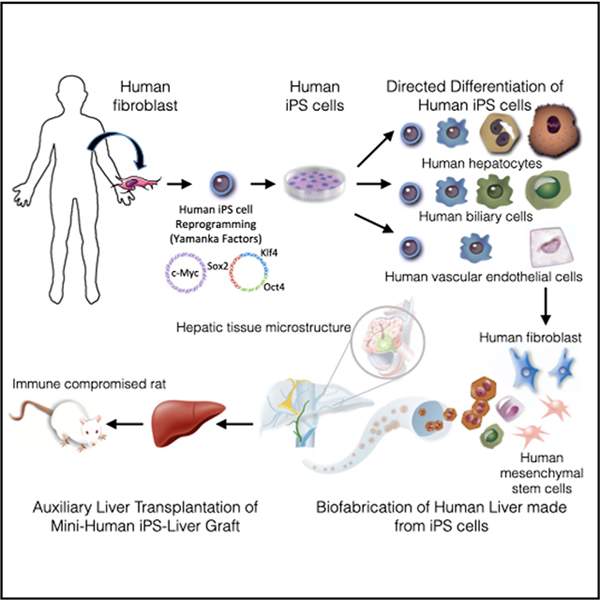
SUMMARY
The availability of an autologous transplantable auxiliary liver would dramatically affect the treatment of liver disease. Assembly and function in vivo of a bioengineered human liver derived from induced pluripotent stem cells (iPSCs) has not been previously described. By improving methods for liver decellularization, recellularization, and differentiation of different liver cellular lineages of human iPSCs in an organ-like environment, we generated functional engineered human mini livers and performed transplantation in a rat model. Whereas previous studies recellularized liver scaffolds largely with rodent hepatocytes, we repopulated not only the parenchyma with human iPSC-hepatocytes but also the vascular system with human iPS-endothelial cells, and the bile duct network with human iPSC-biliary epithelial cells. The regenerated human iPSC-derived mini liver containing multiple cell types was tested in vivo and remained functional for 4 days after auxiliary liver transplantation in immunocompromised, engineered (IL2rg−/−) rats.
In Brief
Takeishi et al. biofabricate human livers for transplantation using human hepatocytes, biliary epithelial cells, and vascular endothelial cells. All originate from induced pluripotent stem cells, human mesenchymal cells, and fibroblasts. The organ-like microenvironment further matures some liver functions and produces tissue structures similar to those found in human livers.
Graphical Abstract
INTRODUCTION
Approximately 30 million people in the USA have liver disorders, and about 40,000 of them will progress to end-stage liver disease, which is responsible for >30,000 deaths annually in the USA (HHS HRSA, 2014; Habka et al., 2015) The only curative treatment for patients with terminal liver failure is liver transplantation. The shortage of donor livers, the high cost of the procedure, and the requirement for lifelong immunosuppression are limits to its application (Ammori et al., 2008). Autologous bioengineered livers derived from the patient’s own cells could change this equation by providing unlimited availability of grafts whose use would not require the need for immunosuppression. To this end, induced pluripotent stem cells (iPSCs) are a valuable autologous cell source that can establish various types of tissue lineages (Takahashi et al., 2007). We (Collin de l’Hortet et al., 2019; Soto-Gutiérrez et al., 2011b; Uygun et al., 2010; Yagi et al., 2013) and others (Baptista et al., 2011; Butter et al., 2018; Hassanein et al., 2017; Kojima et al., 2018; Zhou et al., 2016) have engineered liver grafts by infusing hepatocytes and endothelial cells into the liver parenchymal and vascular compartments using rat liver cells, human cell lines, and human fetal liver cells. Several liver decellularization and recellularization strategies have been described in the literature (Mazza et al., 2015; Ko et al., 2015; Kojima et al., 2018; Zhou et al., 2016), but only limited graft function has been reported using primary cell sources. Recently, we reported the generation of liver grafts using genetically engineered human iPSCs differentiated into liver cells, together with supporting primary human cells to mimic many aspects of human fatty liver disease (Collin de l’Hortet et al., 2019). However, bioengineering of an entire liver graft using human iPSC-derived cells for transplantation has not been described.
It is important to note that complete reestablishment of the liver microarchitecture would require efficient repopulation of the vasculature with endothelial cells. Long-term engraftment of any engineered organ will require a functioning vascular network to provide oxygen and nutrients. The main limitation of bioengineered liver constructs to date is that sparse, or no endothelial cell repopulation of the vasculature, makes them highly susceptible to thrombosis (Bao et al., 2011; Ko et al., 2015; Uygun et al., 2010). Moreover, the incorporation of additional cell types in the bioengineered liver, such as biliary epithelial cells, which would drain bile and remove waste-metabolized products (Beath, 2003), has not been reported, to our knowledge.
In this study, we developed protocols for hepatocyte-, cholangiocyte-, and endothelial-cell differentiation of human iPSCs (Chen et al., 2018). Hepatocyte differentiation was achieved in a low-glucose environment by delivering metabolic and energy maturation cues that included hepatocyte growth factor (HGF), epidermal growth factor (EGF), dexamethasone, hydrocortisone, free fatty acids, cholesterol, bile acids, and rifampicin. Human iPSC-derived hepatocytes (iPSC-Heps) expressed liver-enriched transcription factors and liver-specific microRNAs (miRNAs), and contained mitochondria at levels found in freshly isolated primary human hepatocytes.
Human iPSCs were also differentiated into cholangiocytes that expressed markers found in mature bile ducts, such as cytokeratin 7 (CK7), CK19, SRY-BOX 9 (SOX9), hepatic nuclear factor 1 beta (HNF1β), and cystic fibrosis transmembrane conductance regulator (CFTR). Human iPSC-derived vascular endothelial cells (hiPSC-VECs) engrafted themselves in a decellularized rat liver vascular structure and showed an enhanced expression of angiogenesis and anticoagulation-related genes and functions in the organ-like environment. Finally, we seeded liver scaffolds with human iPSC-derived hepatocytes, endothelial cells, and cholangiocytes, and human primary-liver-derived fibroblast and mesenchymal stem cells, to mimic the liver microstructure. We achieved liver vasculature coverage of 75% and bile-duct coverage of 66% of that observed in normal liver using human iPSC-derived cells. The liver parenchymal cells in engineered iPSC liver grafts expressed cell-cell and cell-extracellular matrix (ECM) molecules and function at levels found in human adult and fetal livers or engineered liver grafts assembled with primary liver cells or in 3D cultures containing freshly isolated primary human fetal and adult hepatocytes. After auxiliary transplantation of human iPSC-derived bioengineered livers in engineered immunocompromised (interleukin [IL] 2rg−/−) rats conditioned to induce liver regeneration, the grafts functioned for four days.
RESULTS
Differentiation of Human iPSCs into Hepatocytes
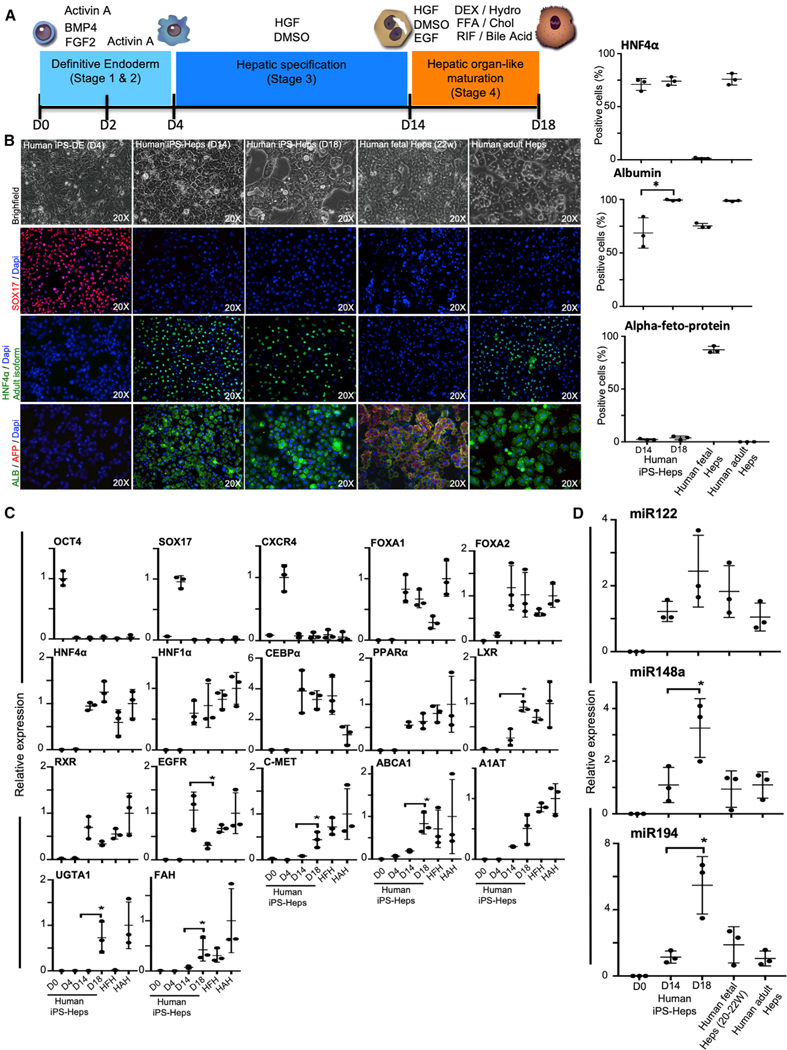
To induce differentiation of human iPSCs toward a hepatocyte phenotype (iPSC-Heps), we used our previously published protocol (Basma et al., 2009; Soto-Gutiérrez et al., 2006, 2007b) with modification of the endoderm induction steps (stages 1 and 2) so that cells were cultured in monolayers for four days rather than as three-dimensional (3D) embryoid bodies (Figure 1A). More than 90% of the resulting cells expressed the definitive endoderm markers SOX17 (Figure 1B) and CXC chemokine receptor 4 (CXCR4; Figure 1C) at day 4 using activin A, bone morphogenetic protein 4 (BMP4), and fibroblast growth factor (FGF)-2. For hepatic maturation, cells were cultured for 14 days in the presence of dimethyl sulfoxide (DMSO) and HGF and, for the last four days, cells were cultured in low glucose in the presence of EGF, dexamethasone, hydrocortisone, free fatty acids, bile acids, cholesterol, and rifampicin (Figure 1A). By day 14 of differentiation (stage 3), after treatment with HGF and DMSO, approximately 70% of cells expressed the adult isoform of HNF4α and albumin, but cells did not express alpha-fetoprotein (AFP; an immature hepatocyte marker; Figure 1B). mRNA analysis of HGF and DMSO-treated cells (stage 3) showed upregulation of liver-enriched transcription factors FOXA1, FOXA2, HNF4α, HNF1α, CCAAT enhancer binding protein alpha (CEBPα), peroxisome proliferator-activated receptor alpha (PPARα), liver X receptor (LXR), and retinoid X receptor (RXR; Figure 1C), and liver specific-microRNAs miR122 (Bandiera et al., 2015), miR148a (Heo et al., 2018), and miR194 (Morimoto et al., 2017; essential for hepatocyte function and homeostasis) to levels approximating those for human fetal and adult hepatocytes.
Figure 1. Generation of Hepatocytes from Human-Induced Pluripotent Stem Cells (iPSCs).
(A) Schematic representation of the protocol used to differentiate human iPSCs to hepatocytes. BMP, bone morphogenetic protein; FGF, fibroblast growth factor; HGF, hepatocyte growth factor; DMSO, dimethyl sulfoxide; EGF, epidermal growth factor; DEX, dexamethasone; FFA, free fatty acids; RIF, rifampicin; Hydro, hydrocortisone; Chol, cholesterol.
(B) Light microscopy images (top) of human iPSC-derived cells at day 4, day 14, and day 18 of hepatic differentiation. Immunofluorescence analyses demonstrating the expression of key definitive endoderm and hepatocyte markers, as indicated, in day 4, day 14, and day 18 using antibodies that recognized SOX17, adult isoform of hepatocyte nuclear factor 4α (HNF4α), alpha-fetoprotein (AFP), and albumin (ALB). Bar graphs showing the levels of positive cell percentage are also shown. iPSCs-Heps, iPSC-derived hepatocytes; iPSC-DE, iPSC-derived definitive endoderm; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
(C) Liver-specific gene expression profile.
(D) MicroRNA-122 (miRNA122), −148a, and −194 of human iPSC-derived hepatocytes (iPSC-Heps) compared to human adult hepatocytes (HAHs) and human fetal hepatocytes (HFHs; gestational age: weeks 20–22). Data are expressed as the fold change relative to HAHs, which is set as 1. HAHs and HFH were used as controls in all experiments. Results are representative of three independent differentiation experiments. ANOVA with Wilcoxon test compared between iPSC-Heps at days 14 and 28: *p < 0.05. Error bars represent mean ± SD of three independent experiments.
Full human liver maturation takes as long as two years from the time of birth (Chen et al., 2018), and is affected by changes in circulation, microbiome, and nutrition (Chen et al., 2018). The liver initially processes nutrients received from maternal blood in utero, and later absorbs maternal milk in the intestines (Beath, 2003; Chen et al., 2018). In preliminary experiments, we analyzed the alterations in the gene-expression patterns using microarrays to determine the effects of the maturation process in human liver and the differences between fetal hepatic state and a fully matured adult human liver (Mendeley data: https://doi.org/10.17632/ncxxgwdwty.1). Clustering and a heatmap revealed that fetal liver samples had a similar expressing pattern to each other, but was significantly different from that of adult samples (Figure S2A). Genes involved in cell-cycle and cell-proliferation pathways showed significantly higher expression in fetal liver when compared to adult liver (Figure S2B). In contrast, genes significantly overexpressed in adult liver were metabolic-related genes, including three major functional groups; fatty acid metabolism-, xenobiotic metabolism-, and glucose metabolism-related pathways (Figures S2B and S2C).
To mimic these metabolic changes, we examine the effect of using media with a low glucose concentration, and adding free fatty acids (palmitic and oleic acid), bile acids (ursodeoxycholic acid), cholesterol, corticoids (dexamethasone and hydrocortisone), EGF, and rifampicin to the culture to promote further hepatocyte maturation of human iPSCs. By day 18 of differentiation (stage 4), approximately 75% of cells expressed the adult isoform of HNF4α and nearly 100% expressed albumin with no expression of AFP expression (Figure 1B). Maturation in the above components increased the expression of genes encoding insulin-signaling/lipogenesis and mature, clinically relevant enzymes such as LXR, C-MET, ATP-binding cassette transporter A1 (ABCA1), alpha-1-antitrypsin (A1AT), uridine diphosphate glucuronosyltransferase 1 A1 (UGTA1), and FAH. We also found a significant increase in the expression of miR148a and miR194 (Figure 1C).
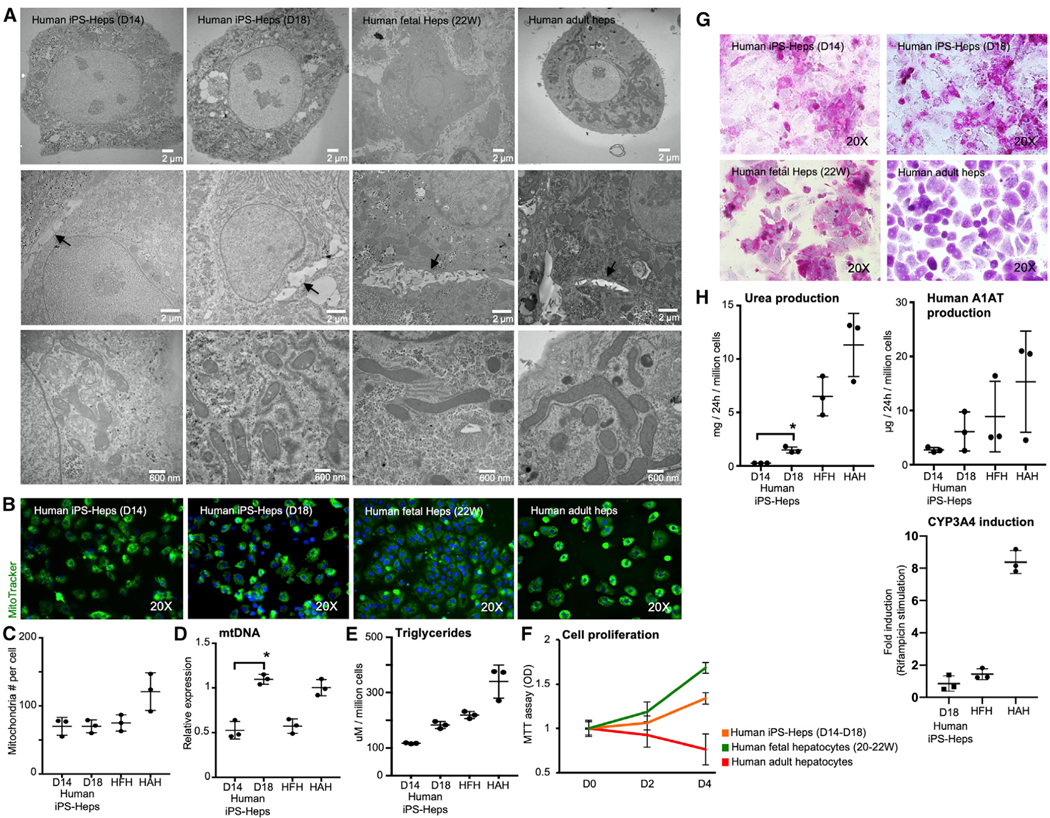
There was a dramatic change in morphology during the course of the differentiation process. Transmission electron microscopy revealed cells with mature round nuclei, evenly distributed chromatin, Golgi complexes, abundant mitochondria with clear membranes, and well-developed bile canaliculi with apical microvilli and tight junctions (Figure 2A). These findings were confirmed using three independent human iPSC lines (Figures S4A–S4D; Figures 2A–2C). As mitochondria are the main energy producer in hepatocytes and play a central role in oxidative metabolism, function, and replication (Yamashina et al., 2009), we examined the mitochondria using the stain MitoTracker and found that there was no quantitative difference in mitochondrial numbers between human iPSC-Heps and control human fetal and adult hepatocytes (Figures 2A–2C), and that human iPSC-Heps (day 18) expressed levels of mitochondrial DNA similar to the levels found in human adult hepatocytes (Figure 2D).
Figure 2. Characterization of Human iPSC-Heps.
(A) Transmission electron microscopy (TEM) images for cell organelles (upper), bile canaliculi with apical microvilli and tight junctions (middle), and mitochondria (lower).
(B–D) Immunofluorescence analyses demonstrating the mitochondria staining using the stain MitoTracker. Graphs showing (C) the number of mitochondria determined by counting the mitochondria described in TEM, (D) the amount of mitochondria DNA (mtDNA) by PCR, and (E) the amount of triglyceride in the cells.
(F) Cell-number curve of human iPSC-Heps, HFHs (gestational age; weeks 22–23), and HAHs during in vitro culture determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay.
(G) Human iPSC-Heps, as well as HFH and HAHs showed glycogen storage by PAS (periodic acid-Schiff) staining.
(H) Urea and human alpha1 antitrypsin (A1AT) production by human iPSC-Heps was analyzed after in the culture medium by enzyme-linked immunosorbent assay (ELISA). Human-adult CYP450 activity is also shown expressed in fold induction. Results are representative of three independent differentiation experiments. ANOVA with Wilcoxon test compared between human iPSC-Heps at day 14 and day 18: *p < 0.05. Error bars represent mean ± SD of three independent experiments. HFH and HAHs were used as the control in all experiments.
There was also no difference in intracellular triglyceride content (Figure 2E) between the groups, and human iPSC-Heps propagated in culture at a rate similar to that of human fetal hepatocytes during the maturation phase (stage 4; Figure 2F). Human iPSC-Heps at day 18 (stage 4) accumulated glycogen (Figure 2G) and produced significantly higher amounts of urea than did human iPSC-Heps at day 14 (stage 3), and 15%–20% of that produced by freshly isolated human fetal and adult hepatocytes (Figure 2H). The amount of human A1AT secreted into the medium was similar to that generated by human primary hepatocyte controls. As expected, the human-hepatocyte-specific cytochrome P450 activity (CYP3A4) in human iPSC-Heps at day 18 (D18) after exposure to rifampicin was at the level of human fetal hepatocytes (0.85-fold and 1.44-fold induction) and inferior to that observed in adult human hepatocytes (8.38-fold induction; Figure 2H).
Differentiation of Human iPSCs into Cholangiocytes and Assembly of Whole-Liver Human Bile Ducts
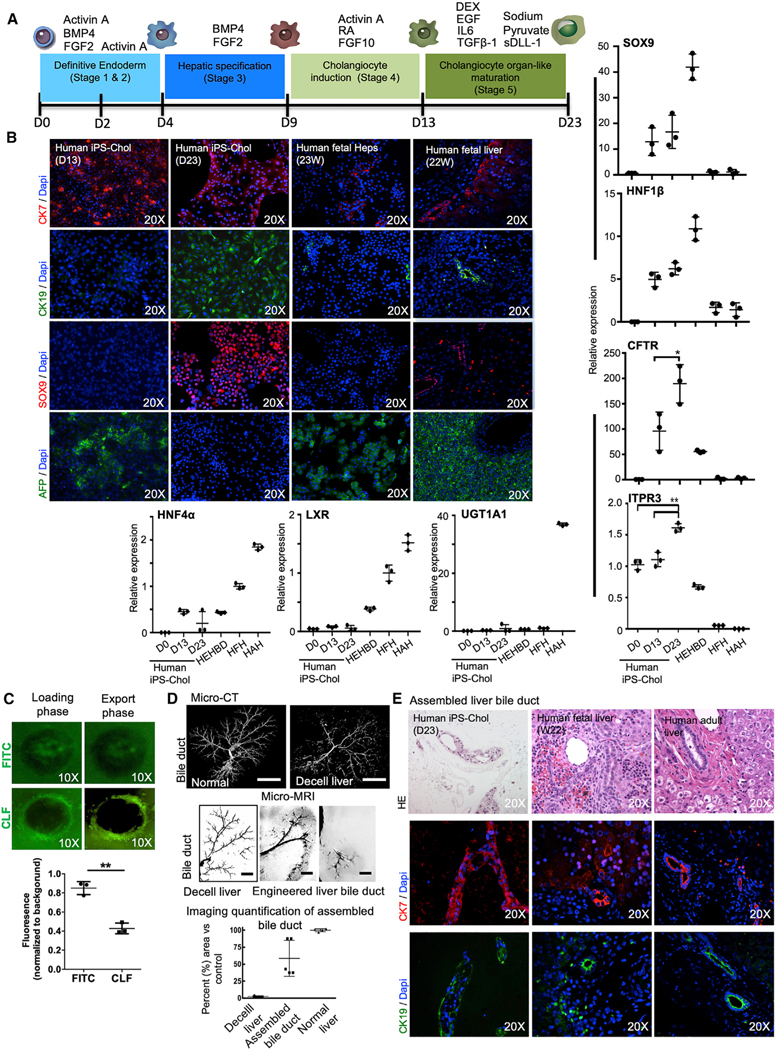
To generate hepatoblasts, day-4 (stages 1 and 2) definitive endoderm cells were exposed to BMP4 and FGF2 (stage 3) for five days (Dianat et al., 2014; Gouon-Evans et al., 2006; Ogawa et al., 2013). To further differentiate hepatoblasts into cholangiocyte progenitors, cells were exposed to Activin A, FGF-10, and retinoic acid for four more days (Sampaziotis et al., 2015; stage 4). mRNA expression for markers of early biliary specification, SOX9, HNF1β, and CFTR, was upregulated (Figure 3B), and CK7 and AFP were detected (Figure 3B), indicating a transition into cholangiocyte progenitors. As Notch and transforming-growth-factor beta (TGFβ) signaling pathways are considered regulators of biliary commitment of hepatoblasts (Flynn et al., 2004; Ogawa et al., 2015; Sampaziotis et al., 2015; Clotman et al., 2005; Schaub et al., 2018), we added (stage 5) EGF, Interleukin-6 (IL6), dexamethasone, sodium pyruvate, TGFβ1, and sDLL-1 (ligand for Notch receptors) to the differentiation program. Following this treatment, the cells expressed CK7 (88%), CK19 (94%), and SOX9 (74%), and AFP expression was lost. mRNA expression of CFTR (p = 0.037, n = 3) and inositol 1,4,5-trisphosphate receptor, type 3 (ITPR3; p < 0.001, n = 3) significantly increased to levels comparable to those in human extrahepatic bile duct (HEHBD) controls, and hepatocyte markers HNF4, LXR, and UGT1A1 became absent or were severely reduced (Figure 3B), indicating differentiation into cells resembling biliary epithelial cells. Next, to characterize the functionality of the generated human iPSC-cholangiocytes, we used three-dimensional (3D) culture conditions through organoid formation for the last maturation stage 5, and evaluated the capacity of human iPSC-cholangiocyte-derived organoids for active export of bile acids (Figure 3C); it showed that human iPSC-chol-angiocyte-derived organoids actively export fluorescent-bile-acid cholyl-lysyl-fluorescein (CLF) from the lumen of CLF-loaded organoids compared to controls loaded with fluorescein isothiocyanate (FITC; Figure 3C).
Figure 3. Generation and Characteristics of Cholangiocytes from Human-iPSCs.
(A) Schematic representation of the protocol generated to differentiate human iPSCs into cholangiocytes (iPSC-Chol). BMP, bone morphogenetic protein; FGF, fibroblast growth factor; RA, retinoic acid; DEX, dexamethasone; EGF, epidermal growth factor; IL, interleukin; TGF, transforming growth factor; sDLL-1, Deltalike protein 1, Delta-1.
(B) Immunostaining analysis showing the proportion of CK7, CK19, SRY-BOX 9 (SOX9), and AFP in the differentiating population at day 13 and day 23 of culture. HFHs (gestational age; week 23) and human fetal liver tissue (gestational age: week 22) were used as control. Also shown: qRT-PCR analyses of the expression of cholangiocyte-specific genes, SOX9; HNF1β; cystic fibrosis transmembrane conductance regulator (CTFR); inositol 1,4,5-trisphosphate receptor, type 3 (ITPR3); and hepatocyte-specific genes HNF4α, LXR, and UGT1A1 in populations at different stages generated from human iPSCs. Values are determined relative to β-actin and presented as fold change relative to the expression in HFHs, which is set as 1. HEHBD, human extrahepatic bile duct; HAHs, human adult hepatocytes; HNF, hepatocyte nuclear factor; CFTR, cystic fibrosis transmembrane conductance regulator; LXR, liver X receptor; UGT, uridine diphosphate glucuronosyltransferase. ANOVA with Wilcoxon test compared between iPS-Chol at day 13 and day 23 for CFTR: *p < 0.05 and for ITPR3: **p < 0.001. Error bars represent mean ± SD of three independent experiments.
(C) Representative images demonstrating active export of the fluorescent bile acid CLF from the lumen of human iPSC-cholangiocyte organoids compared to controls loaded with fluorescein isothiocyanate (FITC). Also shown is fluorescence intensity in the center of organoids. Mean intraluminal fluorescence intensity normalized to background, **p = 0.0001 (two-tailed t test). Results are representative of three independent differentiation experiments.
(D) Assembly of whole organ bile duct in decellularized rat livers was achieved by seeding cholangiocytes directly to the main bile duct. To optimize initially cell-seeding protocols, a human cholangiocyte cell line (MMNK-1) was used and imaging evaluations were performed and then iPSC-Chol were used for all studies. 3D micro-CT angiography of normal and decellularized liver bile duct is shown (n = 5). Scale bars, (micro-CT) 1 cm, (micro-MRI) 4 mm. Representative micro-MRI images of micron-sized iron oxide particle-labeled MMNK-1 seeded into the bile duct of decellularized livers at different depth levels. Quantification of the liver bile duct repopulation is also shown compared to control paired micro-CT image (n = 5).
(E) Hematoxylin and eosin staining and immunostaining of the recellularized liver with iPSC-Chol, which were seeded through the biliary system of the decellularized liver. Human fetal liver tissue (middle, gestational age; week 22) and Human adult liver tissue were used as control.
We next decellularized rat livers, as described in Liver Procurement and Decellularization. We previously showed that decellularized livers preserved vascular and biliary structure (Soto-Gutiérrez et al., 2011b; Uygun et al., 2010; Yagi et al., 2013). We studied and optimized recellularization of the bile duct by perfusing the labeled human-cholangiocyte cell line (MMNK-1) through the biliary structure. Repopulation was assessed at the level of the whole liver using micro-computed tomography (CT) and micro-magnetic resonance imaging (MRI; Figure 3D). Using micro-MRI, each liver lobe was evaluated at different depths and we compared the results seen to a normal biliary system (Masyuk et al., 2001) using micro-CT images, since all cholangiocytes in a normal liver cannot be visualized by micro-MRI (Figures S5A and S5B). The best seeding protocol repopulated 59% ± 24% (n = 5) of the bile ducts relative to those of the native rat liver, as evaluated by micro-imaging (Figure 3D; Masyuk et al., 2001), and confocal microscopy demonstrated a branched, tree-like bile canalicular network in all segments of the liver (Figure S5D). Quantitative histological scoring analysis of individual bile ducts showed bile-duct epithelial cell lining on the interior of the bile-duct walls at 70% ± 18% (n= 7) as compared to that found in the nativerat liver (Figure S5D).
We therefore seeded human iPSC-derived cholangiocytes through the biliary structure of the decellularized liver using the best protocol, consisting of three 2-million cell infusions at 15-min intervals and resulting in the highest level of bile-duct repopulation, with human iPSC-derived cholangiocytes lining 65% ± 8% (n = 3) of the interior bile duct wall (Figure 3E). Furthermore, after two days of bile-duct repopulation process, iPSC-derived cholangiocytes in regenerated bile-duct grafts continued to express cholangiocyte-specific markers, such as CK7 and CK19 (Figure 3E).
Characterization of hiPSC-VECs and Assembly of Whole-Liver Human Vasculature
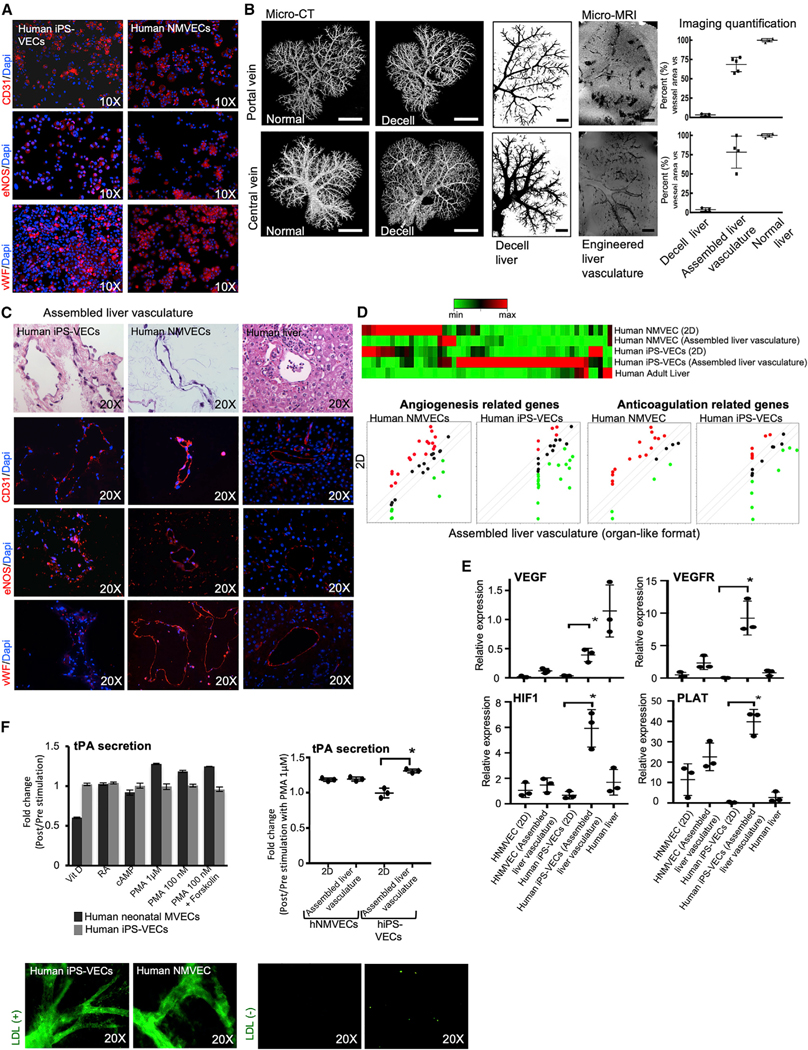
We then studied the changes that hiPSC-VECs undergo as a result of changing from 2D culture to an organ-like culture in a perfused vascular lumen (Jang et al., 2019; Ren et al., 2015). hiPSC-VECs expressed CD31 (83% ± 2%), eNOS (endothelial nitric oxide synthase) (76% ± 3%), and von Willebrand factor (vWF; 34% ± 2%), compared to the expression of CD31 (95% ± 1%), eNOS (83% ± 2%), and vWF (93% ± 1%) in control human neonatal microvascular endothelial cells (hNMVECs; Figure 4A) when cultured for five passages in a 2D format.
Figure 4. Characterization of Human iPSC-Derived Vascular Endothelial Cells (iPSC-VECs).
(A) Characterization of human iPSC-VECs showing homogeneous expression of endothelial markers, CD31, eNOS, and von Willebrand factor (vWF). Human neonatal microvascular endothelial cells (hNMVECs) were used as control.
(B) Endothelial-cell seeding experiments into decellularized rat livers were performed through inferior vena cava (IVC) and portal vein (PV) with a 6-h static culture interval. To optimize initially cell-seeding protocols, a human liver endothelial cell line (TMNK-1) was used and imaging evaluations were performed and then human iPSC-VECs or hNMVECs were used for all studies. Shown is 3D micro-CT angiography of normal and decellularized rat liver vascular compartments (portal and central veins; left). Next, representative micro-MRI images of iron oxide microparticle–labeled TMNK-1 seeded into the portal and central vein of decellularized livers. Quantification of the liver vasculature repopulation is also shown compared to control-paired micro-CT images (n = 5). Scale bars: (micro-CT) 1 cm, (micro-MRI) 4 mm.
(C) Hematoxylin and eosin staining and immunostaining of the recellularized liver tissue with human iPSC-VECs (left) and hNMVECs (middle), which were seeded through vena cava and PV of the decellularized liver. Human adult liver tissue (right) was used as control.
(D) Euclidean hierarchical clustering analysis focusing on the key genes related to angiogenesis and anticoagulation of human iPSC-VECs and hNMVECs (genes differentially expressed in 2D culture versus assembled liver vasculature after recellularization with human iPSC-VECs or human neonatal MVEC, and compared to human adult liver tissue). Green dots represent the genes expressed in the assembled liver vasculature at a higher level when compared to 2D culture, whereas red dots represent genes expressed at a higher level in 2D culture format when compared to assembled liver vasculature.
(E) qRT-PCR-based analyses of the mRNA expression of VEGF, VEGFR, HIF1, and PLAT in cell populations cultured in 2D format and assembled liver vasculature after recellularization with human iPSC-VECs or hNMVECs. Values shown are relative to b-actin and presented as fold change relative to expression in adult liver tissue (human liver), which is set as 1. ANOVA with Wilcoxon test compared between 2D culture and assembled liver vasculature: *p < 0.05 (n = 3). Bars in all graphs represent the mean ± SD of three independent experiments.
(F) Left panel: Fold change of tissue plasminogen activator (tPA) secretion in 2D culture between post- and prestimulation by indicated factors and concentration. Vit D, vitamin D; RA, retinoic acid; PMA, phorbol 12-myristate 13- acetate. Right panel: fold change of tPA secretion in 2D culture and assembled liver vasculature with hNMVECs and iPSC-VECs pre- and post-stimulation with PMA 1 μM. ANOVA with Wilcoxon test compared between 2D culture and assembled liver vasculature: *p < 0.05 (n = 3). Bars in all graphs represent the mean ± SD of three independent experiments. Acetyl LDL Uptake assay at 24 h in assembled liver tissue after recellularization with human iPSC-VECs and hNMVECs with LDL (left) and without LDL (right).
To assemble the liver vasculature, we repopulated decellularized livers through the portal and central vein structures with a recirculation system. To optimize endothelial coverage, we tested different cell concentrations and seeding flow rates. Initially a labeled human liver endothelial cell line (TMNK-1; Soto-Gutiérrez et al., 2006) was used for optimization studies and then hiPSC-VECs or hNMVECs were used for all studies (Figure S5A). We found that the best endothelial coverage of the vascular system was accomplished when cell seeding was performed through both the vena cava and then the portal vein, with an interval of 6 h with no perfusion within the procedures to allow cell attachment. Micro-imaging showed that 68% ± 8% (n = 5) of the portal vein and 78.3% ± 18% (n = 4) of the central vein were repopulated as compared to data from control rat livers (Figure 4B). Histologic analysis of individual vessels showed microvascular endothelial cell coverage on the interior of the portal vein at 86% ± 5% (n = 7) and of the central vein at 81% ± 9% (n = 7) as compared to data from control rat livers (Figure S5C). Next, using the best protocol consisting of seeding 40 million hiPSC-VECs or hNMVECs through both the vena cava and then the portal vein of decellularized rat livers, we found that vascular-endothelial-cell coverage was 75% ± 10% (n = 3) using hiPSC-VECs and 85% ± 9% using control hNMVECs.
Two days after, assembled liver vasculature, populated with either hiPSC-VECs or control hNMVECs, expressed CD31, eNOS, and vWF (Figure 4C). In addition, two days after engraftment, expression of genes important in vascular biology was increased when assembled in the liver vasculature, especially when hiPSC-VECs were used (Figure 4D; Table S1). The gene expression array analysis was confirmed by mRNA qPCR for vascular endothelial growth factor (VEGF), vascular endothelial growth factor receptor (VEGFR), hypoxia-inducible factor 1 alpha (HIF1α), and tissue-type plasminogen activator (PLAT), where expression in assembled liver vasculature engrafted with hiPSC-VECs was significantly higher when compared to those measured in static culture controls (VEGF, p = 0.008, n = 3; VEGFR, p = 0.0005, n = 3; HIF1α, p = 0.002, n = 3; and PLAT, p = 0.0004, n = 3; Figure 4E). The fibrinolytic function of the engineered liver vasculature was further evaluated by assessing the secretion of tissue plasminogen activator (tPA; Kruithof and Dunoyer-Geindre, 2014; Medh et al., 1992) in 2D fibronectin static culture (Figure 4F) and we found that the highest stimulation was achieved with PMA (phorbol 12-myristate 13-acetate) in control hNMVECs. PMA-stimulated-tPA secretion by hiPSC-VECs in assembled liver vasculature was significantly superior to 2D fibronectin static culture (p = 0.002, n = 3). PMA-stimulated-tPA secretion in control hNMVECs was not significantly different. Next, we documented metabolic function of the newly constructed liver vasculature composed of hiPSC-VECs and hNMVECs by evaluating the uptake and metabolism of a fluorescent acetylated low-density lipoprotein (Ac-LDL; Voyta et al., 1984) using confocal microscopy (Figure 4F). This analysis documented the three-dimensional structure of the portal and central venous system (Figure 4F). Thus, hiPSC-VECs were functional and reactive in the engineered liver (Figures 4E and 4F) when compared to standard 2D static culture.
Assembly of Human iPSC-Derived Liver Grafts
After developing protocols for the differentiation and maturation of human iPSCs into hepatocytes and cholangiocytes and testing the capacity of hiPSC-VECs following recellularization of decellularized livers, we repopulated all of the compartments of liver grafts to study lineage interactions and function in a liver architecture. Using a specially designed perfusion and organ culture chamber, the biliary system was repopulated first, followed by repopulation of the parenchymal space, and then vascular system (Figure 5A). Although, hepatocytes constitute most of the parenchymal space, supportive non-parenchymal cells (mesenchymal stromal cells, endothelial cells, and fibroblasts) enhance structural polarity and cellular stabilization (Bhatia et al., 2014; Cohen et al., 2015; Müsch, 2014; Takebe et al., 2013). Based on the normal cellular content of the hepatic lobule (Soto-Gutiérrez et al., 2011a), the parenchymal space was therefore seeded with iPSC-Heps (25 million cells representing 71%), hiPSC-VECs (5 million cells representing 15%), human mesenchymal stromal cells (hMSCs, 2.5 million cells representing 7%), and human fibroblasts (2.5 million cells representing 7%; Figure 5B; Figure S5E). Following repopulation of the compartments (Figure 5B), we found that 28% ± 3% (n = 3) of the cells were positive for the replication marker Ki67 and human albumin (hALB), whereas 19% ± 2% (n = 3) of cells in human fetal livers (16W [16 weeks]) and 2% ± 1% (n = 3) of cells in human adult livers were positive for both Ki67/hALB (Figure 5C), indicating constant regenerative capacity. Moreover, only about 1% (n = 3) cells were double positive for Ki67 and the endothelial marker (CD31) across all groups. We also found that 2.2% ± 1% (n = 3) were double positive for Ki67 and the cholangiocyte marker CK7 in bioengineered human iPSC-livers; and 1% ± 1% (n = 3) in fetal liver and 4.4% ± 1% in human adult livers were Ki67/CK7 positive (Figure 5C). As tissue organization and cell polarization are critical for function and regeneration (Müsch, 2014), we examined tight junctions and cell-cell adhesion in the repopulated grafts. To compare the liver-tissue formation and function abilities of assembled human iPSC-livers to primary cells, we used control-assembled liver grafts that were bioengineered from primary rat liver cells and normal liver tissue. We showed, by immunostaining, that cell-adhesion molecule integrin-beta-1 (ITGB1) was present within two days in the assembled human iPSC-liver (Figure 5D) at levels similar to those found in control-assembled liver grafts bioengineered using rat primary liver cells, adult rat liver, and human fetal and adult livers. ZO-1 proteins, a component of tight junctions, were present in assembled human iPSC-livers at levels similar to that in rat and human adult livers, at higher levels than grafts bioengineered with rat primary livers cells (Figure 5D), while they are not expressed in human fetal livers (24W; n = 3). Finally, Conexin-32 (Cx32), a hepatic enriched gap-junction protein that is expressed in rat and human adult liver, was not expressed in assembled human iPSC-livers or human fetal livers (24W), although it was detected in assembled grafts repopulated with primary rat cells (Figure 5D). These results indicate that key structural proteins important for cell-cell and cell-ECM are present in bioengineered human iPSC-derived liver tissue, although the hepatocyte-hepatocyte transmembrane communication protein Cx32 was not induced in these studies.
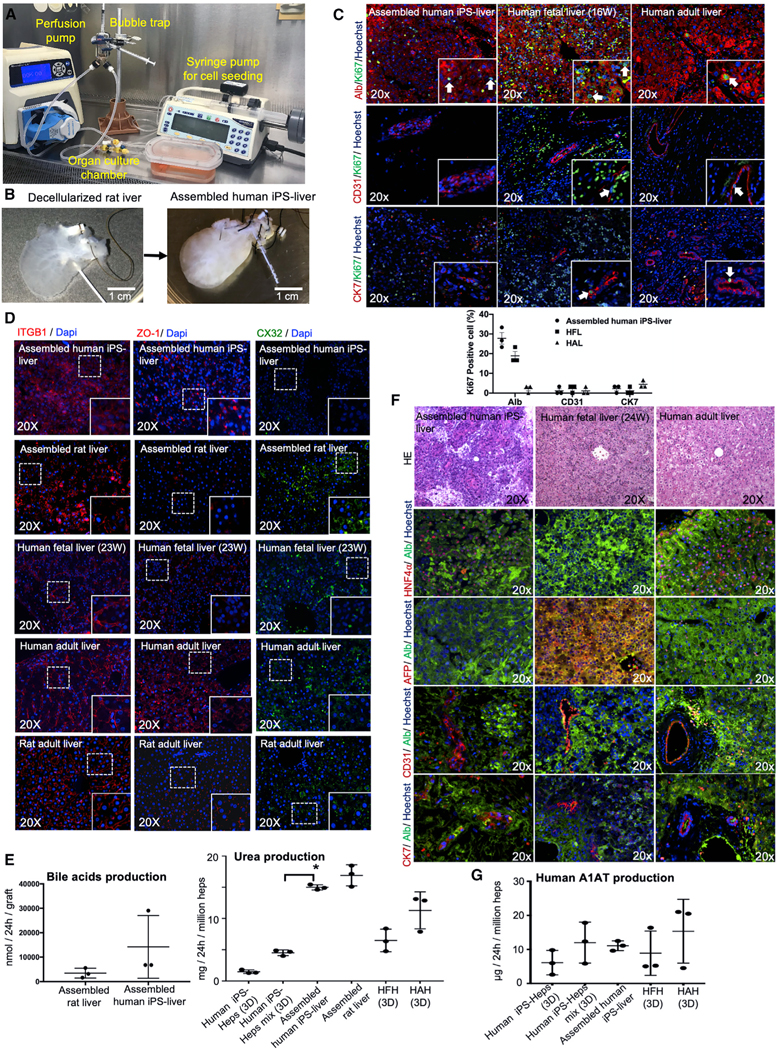
Figure 5. Hepatic Function and Characterization of Engineered Human iPSC-Derived Liver Graft.
(A) Photograph illustrating the organ perfusion and culture chamber used to recellularized rat livers with human iPSC-Heps, iPSC-VECs, human fibroblasts, mesenchymal stem cells, and iPSC-Chol.
(B) Decellularized whole liver matrix (left) and liver after recellularization (right).
(C) Double-immunofluorescence staining for Ki67 and ALB, CD31, and cytokeratin 7 (CK7) of regenerated liver grafts four days after recellularization. Human fetal liver (gestational age; week 16) and adult liver tissues were used as the controls. Bar graphs showing the levels of Ki67 positive cell percentage are also indicated for each cell type.
(D) Immunofluorescence staining for the key markers of cell adhesions and tight junctions, integrin beta-1 (ITGB1), ZO-1, and Conexin32 (CX32), of the regenerated human liver grafts. Bioengineered livers with primary rat liver cells, human fetal liver, human adult liver and rat adult liver were used as controls.
(E) Left: the comparison of bile-acid production between assembled liver grafts derived from rat hepatocytes and assembled liver grafts derived from human iPSCs. Right: regenerated liver tissue assembled with human iPSC-human derived cells (assembled human iPSC-liver) in comparison to that from iPSC-Heps cultured alone in static sandwich (iPSC-Heps [3D]) and human iPSC-Heps cultured with iPSC-VECs, iPS-derived cholangiocytes, mesenchymal stem cells, and fibroblasts (iPSC-Heps mix [3D]). HFHs and HAHs cultured in static sandwich were used as the control in all experiments. To compare between 2D culture and assembled liver, ANOVA with Wilcoxon test was used: *p < 0.05 (n = 3). Bars in all graphs represent the mean ± SD of three independent experiments. Error bars represent mean ± SD of three experimental experiments.
(F) Characterization of human liver graft entirely regenerated from iPSC-derived cells four days after recellularization, showing homogeneous expression of HNF4α and ALB, but no expression of AFP and ALB was detected. Also, double-immunofluorescence staining for CD31 and CK7 are shown. H&E, hematoxylin and eosin. Hoechst (blue stain) was used as counterstaining.
(G) A1AT production from regenerated liver tissue assembled with human iPSC-human derived cells (assembled human iPSC-liver) in comparison to that from iPSC-Heps cultured alone in static sandwich (iPSC-Heps [3D]) and human iPSC-Heps cultured with iPSC-VECs, iPSC-derived cholangiocytes, mesenchymal stem cells, and fibroblasts (iPSC-Heps mix [3D]). HFHs and HAHs cultured in static sandwich were used as the control in all experiments. Error bars represent mean ± SD of three experimental experiments.
There was no significant difference between assembled human iPSC-livers and assembled livers using rat primary liver cells in their ability to secrete bile acids (Figure 5E). Urea production in 3D static cultures was improved when iPSC-Heps were mixed with human iPSC-VECs, human MSCs, and human fibroblasts, compared to iPSC-Heps alone (p = 0.0006, n = 3), and urea production in assembled human iPSC-liver was not different from that observed in assembled primary rat cell liver grafts, but was significantly higher than that in 3D static cultures (p < 0.0001, n = 3; Figure 5E). Assembled human iPSC-liver expressed human albumin and the mature isoform of the hepatic transcription factor HNF4α at similar levels to those seen in human adult livers (Figure 5F), and did not express AFP. Expression of CD31 for endothelial cells and CK7 for bile-duct epithelial cells were expressed in assembled human iPSC-livers at levels similar to that seen in human fetal and adult livers. Finally, A1AT production by assembled human iPSC-livers during culture-perfusion was not statistically different to that produced in static collagen-sandwich (3D) culture (Figure 5G).
Auxiliary Transplantation of Human iPSC-Liver Grafts
To examine the engraftment and function of bioengineered human iPSC-liver grafts in vivo, we performed auxiliary liver transplantation (Matsubara et al., 2015) into immunocompromised (IL2rg−/−; Mashimo et al., 2010), retrorsine-hepatectomy-preconditioned rats (Laconi et al., 1999; Matsubara et al., 2015). To assemble and transplant human iPSC-liver grafts, human iPSCs were differentiated into iPSC-Heps and iPSC-cholangiocytes coordinately. Then, human liver grafts were assembled (bile duct, parenchyma, and vascular compartment) as described above during a two-day period, then regenerated liver grafts were cultured for two days more before transplantation (Figure S5E). Decellularized liver grafts were regenerated by first seeding human iPSC-derived cholangiocytes into the bile duct. A mix of iPSC-Heps (71%), hiPSC-VECs (15%), hMSCs (7%), and human fibroblast (7%) was then seeded into the parenchymal space, and hiPSC-VECs were used to regenerate the liver-graft vascular system. Adequate perfused flow was corroborated in the bioengineered human iPSC-liver grafts (Figure S6A). In an effort to improve any thrombogenesis caused by platelet adhesion to the liver matrix, 1–2 h before auxiliary liver transplantation all bioengineered human iPSC-liver grafts were treated with N-hydroxysuccinimide-polyethylene glycol (NHS-PEG; Deglau et al., 2007), which has proved to block platelet deposition in injured vascular surfaces (Deglau et al., 2007; Figures S6B and S6C). We modified the decellularized liver ECM surface with N-hydroxysuccinimide-polyethylene glycol (NHS-PEG) conjugated to biotin for the purpose of detection, and found that about 46% ± 5.4% of the free space of a decellularized liver was coated (Figure S6B). Moreover, coated livers were perfused with blood in vitro through the portal vein for ~15 min. Thrombus formation was notably reduced in coated-versus-uncoated decellularized livers as indicated by the platelet marker CD41 (Figure S6C). Moreover, heparin was added to the perfusion medium just before the transplant procedure. A right nephrectomy was performed to create space for the donor liver (Figures 6A and 6A1), and the portal vein and inferior vena cava (IVC) were dissected (Figures 6A and 6A2). End-to-side anastomosis was then performed between the donor and recipient IVC, and subsequently between the donor and recipient portal vein (Figures 6A, 6A3, and 6A4). The donor-liver bile duct was stented and inserted into the recipient duodenum. Homogeneous blood perfusion throughout the regenerated grafts was observed immediately after reperfusion (Figures 6A, 6A5, and 6A6). Finally, in an effort to induce regeneration, the left lateral lobe of the recipient rat was removed.
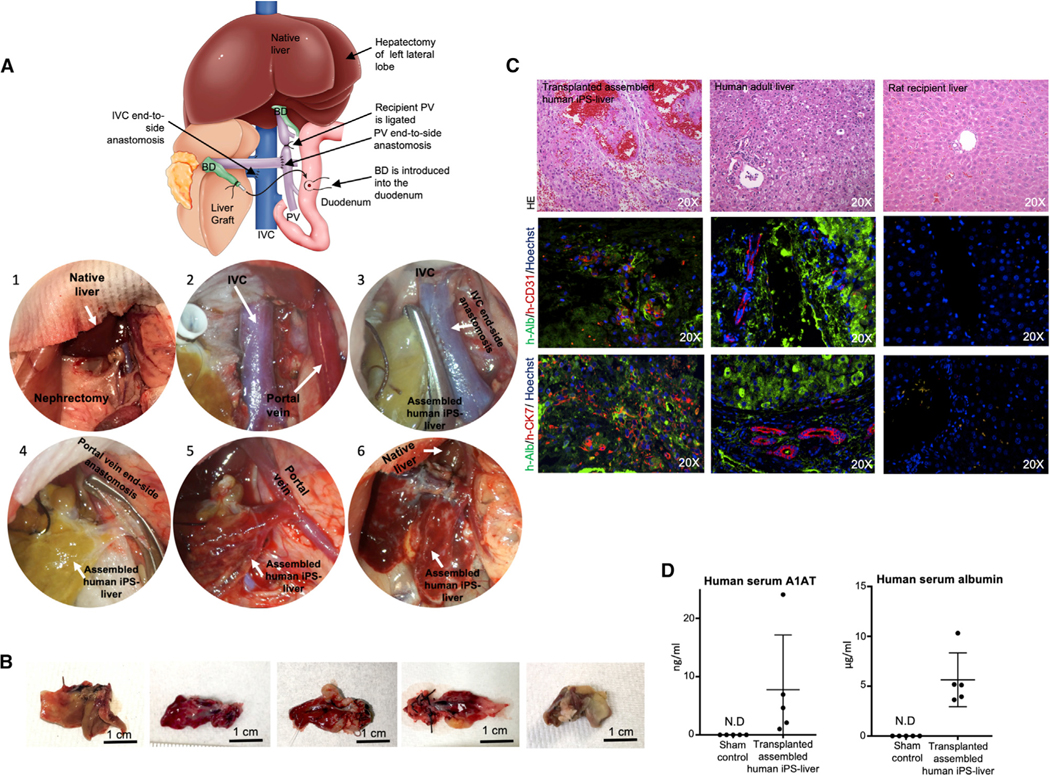
Figure 6. Auxiliary Liver Transplantation of the Engineered Human Liver Graft Derived from iPSCs.
(A) Schematic representation of the auxiliary liver graft transplantation surgical technique for transplantation of human engineered liver grafts. Representative images of graft transplantation: (1) after right nephrectomy, (2) PV) and IVC were exposed. (3) IVC anastomosis (end to side). (4) PV anastomosis (end to side). (5) After reperfusion. (6) Before closing abdomen.
(B) Microscopic finding of the iPSC-liver graft three to four days after transplantation.
(C) Double-immunofluorescent staining of recellularized auxiliary graft after transplantation (left), compared to human adult liver tissue (middle), and rat recipient liver (right). H&E, hematoxylin and eosin; h-ALB, human-specific albumin; h-CD31, human-specific CD31; h-CK7, human-specific cytokeratin 7. Sections were counterstained with Hoechst (blue stain).
(D) The serum concentration of human specific A1AT and human-specific ALB was identified at four days after transplantation of assembled human iPSC-liver by ELISA (n = 5). Bars represent the mean ± SD of five independent experiments.
Transplanted animals (n = 5) were sacrificed at four days after auxiliary liver transplantation. The causes of mortality observed were ileus with poor blood flow into the graft (n = 2), portal vein thrombosis (n = 1), and intestinal ischemia (n = 2). Harvested human iPSC-liver grafts measure 2.5–3 cm and showed liver-like tissue texture (Figure 6B). Histological staining indicated that the hepatocytes retained their morphology and their parenchymal positions (Figure 6C). Immunohistochemical staining for human-specific albumin confirmed that hepatic function was also retained in the transplanted grafts and was localized in proximity to vessels as the human-specific CD31 staining indicated endothelial cell function (Figure 6C). Grafts presented scattered bile-duct-like structures that stained positive for human-specific CK7 in proximity to human-specific albumin-positive tissue. However, there was no anatomical evidence of bile-duct-duodenal-functional reconnection. Human iPSC-liver graft function was evaluated by detection of human serum A1AT and albumin (Figure 6D). Human serum A1AT in transplanted rats was 7.7 ± 9.4 ng/mL and the levels of human serum albumin were 5.6 ± 2.7 μg/mL at the endpoint of the study (Figure 6D). In comparison, to investigate the efficiency of transplanting primary cell-bioengineered liver grafts in retrorsine-conditioned and hepatectomized rats, engineered livers were assembled using the same approach described above, but using primary rat cells and an independent study was performed (n = 8). The recipient survival rate over one week was 50% (n = 4) and at two weeks was 25% (n = 2). Early death-related complications (<3 days) were due to portal vein thrombosis (n = 2) or technical errors (n = 2), and the causes of death at one week were infection and ileus with poor blood flow into the graft (Figure S6D).
DISCUSSION
Generating implantable bioengineered autologous liver grafts using patient-derived cells would provide an alternative strategy for the treatment of patients with terminal liver failure. In this study, we show that an organ-like microenvironment can improve the differentiation of specialized liver lineages in the parenchymal compartments (hepatocyte, cholangiocyte, and vascular endothelial cells) and demonstrate that acellular liver scaffolds repopulated with specialized cells derived from human iPSCs produced human grafts that showed significant hepatic function in vitro and in vivo after transplantation into immunodeficient rats as determined by human serum albumin, A1AT, and postmortem histology.
The goal of decellularization is to remove cells while maintaining the structural, mechanical, and biochemical properties of the ECM scaffold. Numerous whole-organ decellularization protocols have been reported, each with different effects on the ECM (Soto-Gutiérrez et al., 2012). Before in vivo studies, it is difficult to determine which factors are important to success. Here we incorporated DSC monitoring of ECM proteins to more carefully evaluate the micromechanical and biological properties of the decellularized ECM. Liver decellularization required a concentration of Triton X-100 30 times lower than in our previous protocol (Soto-Gutiérrez et al., 2011b) and generated a larger collagen content and improved ECM protein structure and thermal stability. DNA content, a commonly used marker of decellularization, was 3–10 times higher than in previous studies (Baptista et al., 2011; Soto-Gutiérrez et al., 2011b; Uygun et al., 2010), which may lead to an adverse immune response if animal-derived scaffolds are to be used in humans, however, this remains to be tested.
In initial engineering studies, including our own, liver scaffolds were seeded with primary animal or fetal cells (Baptista et al., 2011; Kojima et al., 2018; Soto-Gutiérrez et al., 2011b; Uygun et al., 2010; Yagi et al., 2013). Very recently, we have bio-fabricated human fatty mini livers using genetically engineered human iPSCs differentiated into hepatocyte-like cells and grafted them, together with supporting cells, into an empty liver scaffold to mimic many aspects of human fatty liver disease (Collin de l’Hortet et al., 2019). However, the entire liver vasculature and the bile-duct system were not reconstituted. Here we developed a strategy for producing hepatocytes and cholangiocytes from human iPSCs and demonstrate improvements in gene expression and function when the human iPSC-derived endothelial cells were seeded into a liver architecture where different cell lineages were allowed to interact. We present techniques for repopulation of the biliary, parenchymal, and vascular compartments of a decellularized liver using five different cell types that include human iPSC-derived hepatocytes, cholangiocytes, and vascular endothelial cells, as well as mesenchymal stromal cells and fibroblasts, to rebuild the liver microarchitecture. Finally, we demonstrate the feasibility of transplanting these functional human bioengineered liver grafts.
Human iPSCs are generated by forced expression of specific pluripotency transcription factors (Takahashi et al., 2007). Great progress has been made investigating the hepatocyte differentiation capacity of iPSCs. Although specific culture conditions can vary, most protocols share a three-step strategy based on liver ontology (Basma et al., 2009; Carpentier et al., 2014; Si-Tayeb et al., 2010; Song et al., 2009; Soto-Gutiérrez et al., 2007a, 2007b) and follow the natural stages of liver development (Zaret and Grompe, 2008). Despite progress in advancing the differentiation of human stem cells into hepatocytes in vitro, cells that replicate the ability of human primary adult hepatocytes to metabolize drugs in vitro or can proliferate and replace livers for clinical applications has not been achieved. Most cells usually have a fetal phenotype and immature function (Chen et al., 2018; Rezvani et al., 2016). We have reported a guide to help standardize the analysis of the maturation degree of hepatic-like cells across laboratories by providing a dataset of the expression of 62 genes expressed in 17 fetal and 25 mature human livers (Zabulica et al., 2019) and showed that the main similarities of human iPSC-Heps and human fetal hepatocytes are related to the expression and function of drug metabolism. Clinical application of bioengineered, autologous liver grafts will require the generation of high numbers of iPSC-Heps with functionality equal to that of primary human hepatocytes. Building on our own previous work (Basma et al., 2009; Soto-Gutiérrez et al., 2006, 2007b), we developed a chemically defined system to generate functional hepatocytes with some characteristics of mature phenotype, especially related to expression of clinically relevant liver-specific enzymes (UGTA1 and FAH), mitochondria content, and secretion of liver specific proteins (A1AT and ALB; Basma et al., 2009; Carpentier et al., 2014; Si-Tayeb et al., 2010; Song et al., 2009). Based on nutritional components that promote hepatocyte maturation occurring after birth (Chen et al., 2018), we added a maturation stage that incorporates the low-glucose, cholesterol, and fatty acids present in maternal milk (Ballard and Morrow, 2013), steroids and bile acids (Chen et al., 2018) that stimulate nuclear receptors, and drugs and growth factors important for hepatocyte function (Michalopoulos, 2007). Using this strategy, we were able to extinguish AFP expression (a sign of hepatocyte maturation), maintain expression of hepatocyte-enriched transcription factors important for hepatocyte function, induce expression of clinically relevant enzymes and hepatocyte-specific miRNAs, and induce mitochondria levels equal to or higher than those observed in freshly isolated primary human adult hepatocytes.
We also present a protocol for generation of cholangiocytes from human iPSCs that allowed for early biliary specification, as previously published (Sampaziotis et al., 2015), but also effectively transformed human iPSC-derived hepatoblasts into cholangiocyte progenitors. To enhance natural bile duct maturation, we added a differentiation stage based on the role of Notch and TGFβ signaling (Clotman et al., 2005; Flynn et al., 2004) as well as signaling through IL6 and EGF. These factors induced expression of the mature cholangiocyte markers SOX9, CK19, and CK7, and in most cells eliminated AFP expression. Importantly, CFTR and ITPR3 membrane transporters known to be expressed on mature bile ducts were induced (Sampaziotis et al., 2015; Turner et al., 2011).
Several protocols have been reported on the production of functional vascular endothelial cells from human iPSCs (Olmer et al., 2018; Prasain et al., 2014; Ren et al., 2015). We used commercially available human iPS-VECs for our studies. They functioned and expressed primary human neonatal vascular endothelial cell markers (CD31, eNOS, and vWF) and had comparable ability to reconstitute vascular lumens of decellularized livers, reaching nearly 75% of that found in the native liver. Importantly, human iPS-VECs show expression of genes important for angiogenesis and anticoagulation when assembled in the vascular space.
Two-dimensional differentiation fails to recapitulate cell interactions important for organogenesis and maturation. The three-dimensional assembled liver grafts generated complex organ-like tissues, and we found that assembled human iPSC-liver and assembled rat livers were functionally comparable. However, hepatocyte-specific CX32 was not induced in the assembled human iPSC-livers, indicating that the bioengineered human iPSC-derived liver tissue was still immature. Nevertheless, some functions–namely urea production–were significantly improved in the assembled human iPSC-liver, indicating that the presence of non-parenchymal cells (human iPSC-VECs, MSCs, and fibroblasts) could play a role in hepatocyte functions, which is consistent with previous studies (Soto-Gutiérrez et al., 2006; Takebe et al., 2013).
Since the biliary tree is independent from the vascular system and the parenchymal space, we seeded the bile duct directly in a multistep fashion that led to coverage approaching 65% of that found in the native liver. Whether these cells functionally integrated into the graft is not known. Because hepatocytes are normally surrounded by ECM and many of their functions are regulated by neighboring nonparenchymal cells (Chen et al., 2018; Soto-Gutiérrez et al., 2006), we added human iPSC-VECs, MSCs, and fibroblasts to iPSC-Heps to repopulate the lobular space. Seeding into the parenchymal compartment is highly efficient (Soto-Gutiérrez et al., 2011b; Uygun et al., 2010) because of the lack of an endothelial barrier. Finally, we used a previously reported technique that achieved high efficiency vascular coverage of decellularized lungs (Ren et al., 2015) for our studies.
Our study provided the opportunity to examine the role of nutritional and organ-structure-like microenvironment in each human iPSC-derived hepatic cellular type (hepatocytes, cholangiocytes, and vascular endothelial cells). Recent reports highlight the role of the multicellular components (Camp et al., 2017) and nutritional changes (Avior et al., 2015; Schaub et al., 2018), which impact liver differentiation and maturation. While the auxiliary transplantation studies focus on the survival of the human iPSC-derived mini livers, more work will be required to test their efficacy to function and regenerate in animal models of end-stage liver failure. This study was limited by the small fraction of liver-cell mass used and the vascular complications after transplantation. Future studies should concentrate on procedures to allow continued vascular development using, for instance, nanoparticles and growth-factor-hydrogel modification of acellular scaffolds that have proved to be a successful approach to extend the life of bioengineered lungs after transplantation (Nichols et al., 2018). Moreover, future studies should investigate further maturation of the human iPSC-derived liver grafts in vivo and establishment of bile canaliculus and hepatic artery flow. The strategy shown here represents a significant advance toward our understanding of the production of bioengineered autologous human-liver grafts for transplantation.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alejandro Soto-Gutierrez, MD, PhD (als208@pitt.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The microarray data is available at the Mendeley data repository, https://data.mendeley.com/datasets/ncxxgwdwty/1: Gene expression changes during human liver development: fetal toward adult. The microarray data can be accessed directly via its project DOI (https://doi.org/10.17632/ncxxgwdwty.1) (Fraunhoffer and Soto-Gutiérrez, 2018).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Sprague-Dawley male rats (200–250 g) (Charles River Laboratories, San Diego, CA) were used for liver harvest for whole-organ decellularization, and auxiliary liver transplantation studies. Homozygous F344-Il2rgttm1Kyo rats were used as recipients and were established by T.M. and obtained from the National Bio Resource Project for the Rat in Japan. The animals were cared for in accordance with the guidelines set by the Committee on Laboratory Resources, National Institutes of Health, and Institutional Animal Care Committee of University of Pittsburgh.
Primary human fetal and adult hepatocytes
De-identified tissues were obtained from Magee Women’s Hospital (Pittsburgh, PA) and the University of Washington Department of Pediatrics, Division of Genetic Medicine, Laboratory of Developmental Biology (Seattle, WA) after obtaining a written informed consent by a protocol approved by the Human Research Review Committee of the University of Pittsburgh (Honest broker approval number HB015 and HB000836). Human fetal hepatocytes were isolated and culture from fetal livers as previously described (Tobita et al., 2016). The de-identified normal human liver cells were obtained through the Liver Tissue Cell Distribution System (Pittsburgh, PA) after obtaining a written informed consent by a protocol approved by the Human Research Review Committee of the University of Pittsburgh, which was funded by NIH Contract # HSN276201200017C. Adult human hepatocytes were also obtained from Ira J Fox Laboratory at Children’s Hospital of UPMC, after obtaining a written informed consent by a protocol approved by the Human Research Review Committee and the Institutional Review Board (IRB#: PRO12090466) of the University of Pittsburgh. For human fetal or adult liver tissue and hepatocytes used in this study both genders were used (XX and XY) and the ages for fetal livers ranged from 16 to 24 weeks of gestational age and for adult livers ages ranged from 32yo to 52yo. Specific information on age, gender and cell viability of human liver tissue and hepatocytes used in this study is described in Table S2.
Generation and culture of human iPSC lines
The lines used here were generated from human amnion cells and human fetal fibroblast. Reprogramming was done using two different techniques. Human amnion iPSCs were obtained from Stephen C. Strom laboratory. For both methods iPSCs colonies were isolated around 20–25 days after induction based on morphology. Reprograming of amnion cells was initiated using the viPS lentiviral gene transfer kit (Thermo Fisher Scientific, Waltham, MA), following the manufacturer’s instructions, to ectopically express octamer-binding transcription factor 3/4 (OCT3/4), NANOG, SRY (sex determining region Y)-box 2 (SOX2), LIN28, Kruppel-like factor 4 (KLF4), and C-Myc. Reprograming of fetal fibroblasts was performed using episomal plasmids vectors adapted from a previously described protocol (Okita et al., 2011). Briefly, for each nucleofection, 1 million cells were resuspended in 100 uL of the Amaxa™ NHDF Nucleofector kit (Lonza, Walkersville, MD), containing 3 ug of each of the four episomal plasmids vectors encoding OCT3/4 and p53 shRNA, SOX2 and KLF4, L-MYC and LIN28, and enhanced green fluorescent protein (eGFP) (Addgene, Boston, MA). Cells were nucleofected using the Amaxa 4D-Nucleofector (Lonza, Walkersville, MD) and plated in mTeSR1™ on human embryonic stem cell–qualified Matrigel (Corning, New York, NY)-coated plates. The lines were karyotyped and pluripotency validated by expression of NANOG, OCT4 and membrane markers SSEA and TRA160 at different passages and regularly tested negative for mycoplasma contamination. Both genders (XX and XY) iPSC lines had equal ability to differentiate into liver cells (Figures S4A–S4C).
METHOD DETAILS
Liver procurement and decellularization
The abdominal cavity was opened with longitudinal incision. Left phrenic vein, right renal artery and vein, right adrenal vein, lumber vein, portal-esophageal shunt and pyloric vein were ligated in that order. A cannula (BD, Franklin Lakes, NJ) was inserted into common bile duct. After heparinization (200 U), cold phosphate buffered saline (PBS) solution was infused from the aorta. After ligation of splenic vein, left renal vein and supra hepatic vena cava, liver was taken out from the abdominal cavity. On the back table, cuffs were put into the portal vein and infra-hepatic inferior vena. Then the liver was frozen at −80°C completely immersed in saline solution. Frozen livers were thawed at 4°C overnight washed with PBS 1 h through perfusion via portal vein at 2 ml/min. Livers were perfused with 0.02% trypsin/0.05% EGTA (Sigma-Aldrich, St Louis, MO) for 2 hours at 37°C followed by 3% Triton X-100/0.05% EGTA or 0.1% Triton X-100/0.05% EGTA (Sigma-Aldrich, St Louis, MO) for 24–36 h. Finally decellularized liver was sterilized with 0.1% paracetic acid solution (Sigma-Aldrich, St Louis, MO) for 2 hours. Next, decellularized livers were washed with sterilized PBS solution with 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen, Carlsbad, CA) and 100 μg/ml gentamicin (Thermo Fisher Scientific, Waltham, MA) and preserved at 4°C up to 7 days.
Optimization of whole-liver decellularization
To generate and optimize a natural liver scaffold to recreate the liver microenvironment for hepatic maturation and tissue assembly using different cell lineages from human iPSCs, we implemented decellularized rat livers. First, we measured collagen content and found that 0.02% trypsin and 0.1% Triton X-100 retained 94% of the fibrillar collagen of the native liver, compared with 53% using our previously published protocol of 0.02% trypsin and 3% Triton X-100 (Soto-Gutiérrez et al., 2011b) (n = 4) (Figure S1A). The capacity of the ECM to support cell ingrowth and tissue remodeling depends on its structural and chemical characteristics (Sun et al., 2013). Differential scanning calorimetry (DSC) (Sun et al., 2013) to measure ECM structural protein stability after decellularization with 0.1% Triton X-100 solution showed at least four transitional events during calorimetric scans between 2°C and ~125°C, with onset of denaturation occurring at ~40°C, 58°C, 70°C and 80°C (Figure S1B). With the 3% Triton X-100 solution, the transitional events at 70°C and 80°C were absent and the thermogram shifted to a lower temperature, indicating less protein stability (57.5 ± 0.3°C versus 54.9 ± 0.9°C, 0.1% versus 3%, p value = 0.002 by Student’s t test, n = 4). As expected, fresh liver showed very small thermal transitions due to the high cellular content. Residual DNA content after either decellularization protocols was < 10% that of native liver but 3–10-fold higher compared with some previous protocols (Baptista et al., 2011; Soto-Gutiérrez et al., 2011b; Uygun et al., 2010) (Figure S1C) (0.1% versus 3%, p = 0.999 by one-way ANOVA, Tukey-Kramer, n = 3). Both decellularization protocols demonstrated complete cellular removal as evidenced by the DNA-binding stain Hoechst (Figure S1C). Scanning electron microscopy (SEM) confirmed that collagen density was better preserved with the 0.1% Triton X-100 solution (Figures S1D and S1E). As the collagen content, DSC data and imaging analysis suggested that the 0.1% Triton X-100 solution better preserves collagens and other ECM elements, this protocol was selected for further study. Fibronectin and laminin components of the basement membrane were preserved (Figure S1F). Furthermore, a qualitative proteomics analysis detected protein in the decellularized liver corresponding to 15% that in the native liver. Gene Ontology analysis indicated the presence of important proteins involved in structural, molecular and biological functions in the decellularized liver (Figure S2; Table S3). However, some of the ECM proteins present might have been missed in the proteomics analysis because they were not dissolved and trypsin digested.
DNA quantification
DNA content was measured using methods described previously (Gilbert et al., 2009; Soto-Gutiérrez et al., 2011b). In brief, the decellularized liver was cut into small strips, weighted and digested using Proteinase K (Invitrogen) for 24–48 h at 50°C until no visible material remained. Phenol-chloroform-isoamyl alcohol (25:24:1; Acros) was then added in equal amounts to the decellularized liver digest and centrifuged for 10 min at 10,000 g. The aqueous top layer containing the DNA was then removed and added to 200 μL of 3M sodium acetate solution to reduce RNA content. Ethanol was then added and the solution frozen at 80C for at least 12 h. Ethanol was then removed, and samples were allowed to fully dry, at which point 1 × TE buffer (Invitrogen) was added. The total amount of DNA was quantified using the NanoDrop (Thermo) using the manufacturer’s instructions. The results were normalized to the weight of liver tissue.
Collagen content
10mg of normal and decellularized 3% Triton and 0.1% Triton liver samples were hydrolyzed with 6N HCL at 120C for 3h. Hydroxyproline content was then measured using a hydroxyproline assay (Sigma, St-Louis, MO) according to the manufacturer’s instructions. The collagen content was calculated indirectly through measurement of Hydroxyproline content according to Jamall et al. (1981). The results were normalized to the wet weight of normal liver.
Differential scanning calorimetry (DSC)
Fresh liver tissue and decellularized extracellular matrices in PBS solution were blot-dried with Kimwipe paper to remove excessive surface water. Samples were hermetically sealed in Tzero crucibles, loaded onto Q2000 differential scanning calorimeter (DSC) (TA Instruments, New Castle, DE) between 20°C and 30°C, and pre-cooled at 3°C/min to 2°C before samples were scanned at 3°C/min up to 125°C for thermo-physical analysis under a pure nitrogen purge flow rate of 50 mL per min. After DSC scan, small holes were punched on the lids of crucibles, and samples were dried in a vacuum oven at 105°C for at least 4 h to determine dry mass. Data were analyzed with the Universal Analysis software (Version 4.5A).
Proteomics assay
Fresh rat liver (n = 1) and decellularized rat liver (n = 1) samples were reduced, alkylated and trypsin digested according to the iTRAQ protocol (Applied Biosystems). Peptides were analyzed by liquid chromatography-matrix assisted laser desorption ionization (LCMALDI) analysis using the Ultimate 3000 nano high pressure liquid chromatography (HPLC) system (Dionex) equipped with a Probot (LC Packings) coupled to a 5800 MALDI time-of-flight/time-of-flight (TOF/TOF) Analyzer. Peptides were loaded onto a C18 PepMap100, 3mm column (LC-Packings) and separated with a gradient of 10%–45% acetonitrile (0.1% trifluoroacetic acid) over 165 minutes. Spectral data were analyzed using Mascot sequence matching software (Matrix Science) against the Ludwig NR database with taxonomy set to Rattus norvegicus. The database contained 51,167 sequences. A list of the proteins analyzed can be found in Table S3. Protein classification and categorization of biological process, cellular component and molecular function of the identified proteins according to Gene Ontology (GO) information obtained from European Bioinformatics Institute (EBI) at http://www.ebi.ac.uk/GOA and http://ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/UNIPROT/gene_%20association.goa_uniprot.gz.
Biological process, cellular component and molecular function categorization of the detected proteins were based upon GO identification numbers (ID #s) with relatively broad categories to better represent the data. Cellular component refers to the place in the cell where a gene product is active. The molecular function is the elemental activities of a protein product at the molecular level. Biological process is defined as a biological objective to which the protein or protein product contributes (Ashburner et al., 2000). After assigning GO ID#s, every GO term was categorized as lower or upper categories and were assigned in three different levels. The third level categories with more than three or four GO proteins were represented in the bar graph for each of the three large categories (Table S3).
Microarray analysis
Liver tissue from fetal and adult human samples was pooled for RNA isolation. Total RNA extraction and purification was performed using QIAGEN RNeasy kit (QIAGEN, San Diego, CA). The first strand of cDNA was synthesized using the primer, while the second strand was made at 16°C by adding E. coli DNA ligase, E. coli DNA polymerase I and RnaseH to the reaction. To blunt the ends of the cDNA synthesized, T4 DNA polymerase was added. cDNA was then purified by phenol/chloroform and ethanol precipitation. Incubation of purified cDNA at 37°C for 4 hours in an in vitro transcription reaction resulted in cRNA (labeled with biotin using MEGAscript system). Then, for Chip hybridization; about 20 μg of cRNA were incubated in a buffer containing 200mM Tris-acetate (pH 8.1), 500mM KOAc and 150mM MgOAc at 95°C for 35 minutes to fragment it. A pre-equilibrated Affymetrix chip was hybridized with the fragmented cRNA at 45°C for 14–16 hours before washing with a low stringency buffer (6X SSPE, 0.1% Tween 20, 0.005% antifoam) for 10 cycles and a stringent buffer (100mM MES, 0.1 M NaCl, 0.1% Tween 20) for 4 cycles. This was followed by staining with SAPE (Strepto-avidin Phycoerythrin) and incubation with biotinylated mouse anti-avidin antibody and restained with SAPE. An HP ChipScanner (Affymetrix Inc, Santa Clara, CA) was used to scan the chip to detect hybridization signals. To perform the microarray analysis; the processed intensity values (from SAM) were imported into R, and the oligo package was used to perform RMA (Robust MultiArray Analysis) normalization, which applies a background correction and log transformation to the data as well. Next, low expressed genes were filtered out using genefilter. This workflow produced a processed ExpressionSet object with expression values for Affymetrix probe sets, to which gene annotation information was added using the annotate and hgu133a.db packages. Principle component analysis (PCA) was performed on the samples. Finally, the limma package was used to apply an empirical Bayes statistical model to calculate a moderated t-statistic and p value for each gene comparing its log-fold expression in adult samples relative to all the fetal samples. 1204 differential expressed genes were identified, using an FDR of 0.05 to improve the stringency of further analysis. Next, we performed GO term enrichment analysis using the topGO package in R to select “Molecular Function” GO terms that were statistically enriched (based on a hypergeometric p value calculation with an FDR cutoff of 0.05) in the pool of differentially expressed genes. 234 GO terms were enriched with a p value of < 0.05. IPA (Ingenuity Pathway Analysis) was run to obtain the canonical pathways differentially expressed in fetal and adults, with the overlapping pathways being identified as well. Network analysis and Upstream regulator analysis was done using IPA as well. The microarray data is available at (https://doi.org/10.17632/ncxxgwdwty.1) (Fraunhoffer and Soto-Gutiérrez, 2018).
Differentiation of human iPSCs into hepatocytes (iPS-Heps)
Our hepatocyte differentiation protocol is summarized in Figure 1A. Human iPSCs were passaged with Accutase (StemCell Technologies, Vancouver, Canada) and re-plated at a density of 1 to 2×105/cm2 growth factor reduced Matrigel (Corning Incorporated, Corning, NY) coated plates in mTeSR. The day after, cells were exposed to a defined differentiation medium containing RPMI (Invitrogen, Carlsbad, CA), 1× B-27 w/o insulin supplement (Invitrogen, Carlsbad, CA), 0.5% Penicillin/Streptomycin (Millipore, Billerica, MA), 0.5% of Non-Essential Amino Acids (Millipore, Billerica, MA), 100 ng/ml Activin A (R&D Systems, Minneapolis, MN), 10 ng/ml BMP4 (R&D Systems, Minneapolis, MN) and 20 ng/ml FGF2 (BD, Franklin Lakes, NJ) for two days and placed in a normal O2 incubator (Stage 1, endoderm induction). Cells were subsequently maintained in a similar medium without FGF2 and BMP4 for two days in ambient O2/5% CO2 incubator (Stage 2, definitive endoderm). Cells were then grown for 10 days in a defined medium containing 45% DMEM low glucose 1g/l (Thermo-Fisher Scientific, Waltham, MA), 45% F-12 (ThermoFisher Scientific, Waltham, MA), 10% CTS KnockOut SR XenoFree Medium (ThermoFisher Scientific, Waltham, MA), 0.5% Non-Essential Amino Acids (Thermo-Fisher Scientific, Waltham, MA), 0.5% L-glutamine (ThermoFisher Scientific, Waltham, MA), 50 ng/ml HGF (Kindly provided by George Michalopoulos) and 1% DMSO (Sigma-Aldrich, Saint Louis, MO), medium was changed every other day (Stage 3, hepatic specification). At the end of Stage 3, cells were detached and either re-plated at a 30%–40% confluence in 3D sandwich culture or seeded into decellularized liver matrix for further maturation. Cells were grown for 4 days in a defined medium containing 45% DMEM low glucose 1g/l (ThermoFisher Scientific, Waltham, MA), 45% F-12 (ThermoFisher Scientific, Waltham, MA), 10% CTS KnockOut SR XenoFree Medium, 0.5% Non-Essential Amino Acids (ThermoFisher Scientific, Waltham, MA), 0.5% L-glutamine (ThermoFisher Scientific, Waltham, MA), 0.1% of Gentamicin/Amphotericin-B (ThermoFisher Scientific, Waltham, MA), 1% of Pennicillin/Streptomycin (ThermoFisher Scientific, Waltham, MA), 50 ng/ml HGF (Kindly provided by George Michalopoulos), 1% DMSO, 0.5uM Dexamethasone (Sigma-Aldrich, Saint Louis, MO), 0.1% of Ascorbic Acid (Sigma-Aldrich, Saint Louis, MO), 0.1% of Bovine Serum Albumin Free of Fatty Acids, 0.1% of Hydrocortisone, 0.1% of Transferrin, 0.1% of Insulin (HCM Bullet Kit, ThermoFisher Scientific, Waltham, MA), 100uM of Urso deoxycolic acid (Sigma-Aldrich, Saint Louis, MO), 20uM of Palmitic Acid (Sigma-Aldrich, Saint Louis, MO), 30 uM of Oleic Acid (Sigma-Aldrich, Saint Louis, MO), 20 uM of Rifampicin (Sigma-Aldrich, Saint Louis, Missouri) and 1x of Cholesterol (ThermoFisher Scientific, Waltham, MA) (Stage 4, hepatic maturation).
Differentiation of human iPSCs into cholangiocytes
Our cholangiocyte differentiation protocol is summarized in Figure 3A. Human iPSCs were passaged with Accutase (StemCell Technologies, Vancouver, Canada) and re-plated at a density of 1 to 2×105/cm2 growth factor reduced Matrigel (Corning Incorporated, Corning, NY) coated plates in mTeSR. The day after, cells were exposed to a defined differentiation medium containing RPMI (Invitrogen, Carlsbad, CA), 1× B-27 w/o insulin supplement (Invitrogen, Carlsbad, CA), 0.5% Penicillin/Streptomycin (Millipore, Billerica, MA), 0.5% of Non-Essential Amino Acids (Millipore, Billerica, MA), 100 ng/ml Activin A (R&D Systems, Minneapolis, MN), 20 ng/ml BMP4 (R&D Systems, Minneapolis, MN) and 10 ng/ml FGF2 (BD, Franklin Lakes, NJ) for two days and placed in ambient O2/5% CO2 incubator (Stage 1, endoderm induction). Cells were subsequently maintained in a similar medium without FGF2 and BMP4 for two days in ambient O2/5% CO2 incubator (Stage 2, definitive endoderm). Cells were then grown for 5 days in a defined medium containing RPMI (Invitrogen, Carlsbad, CA), 1× B-27 with insulin supplement (Invitrogen, Carlsbad, CA), 0.5% Penicillin/Streptomycin (Millipore, Billerica, MA), 0.5% of Non-Essential Amino Acids (Millipore, Billerica, MA), 20 ng/ml BMP4 (R&D Systems, Minneapolis, MN) and 10 ng/ml FGF2 (BD, Franklin Lakes, NJ) at 4% O2/5% CO2 incubator (Stage 3, hepatic specification). To induce biliary specification, cells were culture for another 4d in the presence of RPMI (Invitrogen, Carlsbad, CA), 1X B-27 w/o vitamin A (Invitrogen, Carlsbad, CA), 0.5% Penicillin/Streptomycin (Millipore, Billerica, MA), 0.5% of Non-Essential Amino Acids (Millipore, Billerica, MA), 50 ng/ml FGF10 (R&D Systems, Minneapolis, MN), 50 ng/ml Activin A (R&D Systems, Minneapolis, MN) and 3 μM retinoic acid (Sigma-Aldrich, Saint Louis, MO), medium was changed everyday (Stage 4, cholangiocyte induction). Finally, to induce cholangiocyte maturation, cells were culture in the presence of William’s E Medium (Invitrogen, Carlsbad, CA), 10mM Nicotinamide (Sigma-Aldrich, Saint-Louis, MO), 17mM Sodium bicarbonate (Sigma-Aldrich, Saint-Louis, MO), 0.2mM 2-phospho-L-ascorbic acid trisodium salt (Sigma-Aldrich, Saint-Louis, MO), 14mM Glucose (Sigma-Aldrich, Saint-Louis, MO), 20mM HEPES (Sigma-Aldrich, Saint-Louis, MO), 1X ITS Supplement (BD Biosciences), 6.3mM Sodium pyruvate (Invitrogen), 0.1 μM Dexamethasone (Sigma-Aldrich, Saint Louis, MO), 2mM Glutamax (ThermoFisher Scientific, Waltham, MA), 0.5% of Pennicillin/Streptomycin (ThermoFisher Scientific, Waltham, MA), 20 ng/ml EGF (R&D Systems, Minneapolis, MN), 10 ng/ml human sDLL-1 (PeproTech, Rocky Hill, NJ), 5ng/ml TGFβ1 (Millipore, Billerica, MA), 10ng/ml IL-6 (Miltenyi Biotec, San Diego, CA). The medium was changed every other day and the cells were cultured for a total of 10 d (Stage 5, Cholangiocyte maturation). At the end of the differentiation protocol cells were detached with and used for experiments.
Quantitative real time PCR
Total RNA was isolated from human cells using RNeasy Mini kits (QIAGEN, Hilden, Germany) and reverse transcribed using SuperScript III (Invitrogen, Carlsbad, CA) following the manufacturers’ instructions. We performed qPCR with a StepOnePlus system (Applied Biosystems, Foster City, CA) using TaqMan Fast Advanced Master Mix (Life Technologies, Waltham, MA). The probes used are listed in Key Resources Table. Relative gene expression was normalized to β-actin (ACTB) mRNA. Relative expression was calculated using ΔΔCT method. Mature miRNAs were amplified and quantified using Taqman miRNA probes (Life Technologies) (Key Resources Table) and Taqman Universal Master Mix II (Life Technologies). Quantitative PCR reactions were performed using a StepOnePlus System (Life Technologies). Expression values were normalized with Rnu6b, which is standard for miRNA profiling. Relative expression was calculated using DDCT method. Quantitative real time PCR was performed also using Human Endothelial Cell Biology PCR array (See Key Resources Table) following manufacturer’s recommendations. A list of the genes analyzed can be found in Table S1. Genomic DNA was prepared by using a genomic DNA purification kit (Promega, Madison, WI). mtDNA content was analyzed by Sybr green quantitative PCR using primers amplifying NADH-ubiquinone oxidoreductase chain 1 (ND1)(Forward, 5′cacccaagaacagggtttgt-3′, Reverse, 5′-tggccatgggatagttgttaa-3′) normalized to ACTB (forward, 5′-ggcatcctcaccctgaagta-3′, reverse, 5′-gaaggtgtggtgccagattt-3′).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-α-fetoprotein | Invitrogen | Cat#180003; RRID:AB_86566 |
| Rabbit anti-CD31 | Abcam | Cat#ab32457; RRID:AB_726369 |
| Mouse anti-CD31 | DAKO | Cat#M0823; RRID:AB_2114471 |
| Goat anti-CD41 | Santa Cruz Biotechnology | Cat#sc-6602; RRID:AB_2249417 |
| Mouse anti-Connexin 32 | Thermo Fisher Scientific | Cat#13-8200; RRID:AB_2533037 |
| Rabbit anti-Cytokeratin 7 | Abcam | Cat#ab90083; RRID:AB_2041981 |
| Mouse anti-Cytokeratin 19 | Leica Biosystems | Cat#NCL-L-CK19; RRID:AB_563799 |
| Rabbit anti-Fibronectin | Abcam | Cat#ab23751; RRID:AB_447656 |
| Mouse anti-Hepatocyte Nuclear Factor 4 α | Abcam | Cat#ab41898; RRID:AB_732976 |
| Goat anti-Human Albumin | Bethyl | Cat#A80-229A; RRID:AB_67018 |
| Rabbit anti-Integrin Subunit Beta 1 | Abcam | Cat#ab179471; RRID:AB_2773020 |
| Mouse anti-Ki67 | BD PharMingen | Cat#550609; RRID:AB_393778 |
| Goat anti-Laminin | Santa Cruz Biotechnology | Cat#sc-6018; RRID:AB_2134182 |
| Mouse anti-Nanog | Cell Signaling | Cat#4893; RRID:AB_10548762 |
| Rabbit anti-Nitric Oxide Synthase 3 | Santa Cruz Biotechnology | Cat#sc-654; RRID:AB_631423 |
| Rabbit anti-Octamer-binding transcription factor 3/4 | Santa Cruz Biotechnology | Cat#sc-9081; RRID:AB_2167703 |
| Rabbit anti-SRY-Box (SOX) 9 | Millipore | Cat#AB5535; RRID:AB_2239761 |
| Goat anti-SRY-Box (SOX)17 | R & D Systems | Cat#NL1924R; RRID:AB_2195645 |
| Mouse anti-Stage-specific embryonic antigen-4 | BD PharMingen | Cat#560218; RRID:AB_1645389 |
| Mouse anti-TRA-1-60 | BD PharMingen | Cat#560173; RRID:AB_1645379 |
| Rabbit anti-von Willebrand Factor | Abcam | Cat#ab6994; RRID:AB_305689 |
| Mouse anti-ZO-1 | Thermo Fisher | Cat#33-9100; RRID:AB_2533147 |
| Rabbit anti-ZO-1 | Thermo Fisher | Cat#40-2300; RRID:AB_2533457 |
| Biological Samples | ||
| Human adult Hepatocytes | This paper | N/A |
| Human fetal hepatocytes | This paper | N/A |
| Human fibroblast | This paper | N/A |
| Rat hepatocyte | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Accutase | STEMCELL | 7920 |
| acLDL | Thermo Fisher Scientific | L23380 |
| Activin A | R&D Systems | 338-AC |
| 1a,25-dihydroxyvitamin D3 | Sigma-Aldrich | D1530-10UG |
| Amphotericin-B | Thermo Fisher Scientific | 1520018 |
| Ascorbic Acid | Sigma-Aldrich | A1300000 |
| 1 X B27 without insulin supplement | Thermo Fisher Scientific | A1895601 |
| 1 X B27 with insulin supplement | Thermo Fisher Scientific | 17504001 |
| Bismuth chloride | MP biomedicals | 224839-25G |
| BMP4 | R&D Systems | 314-BP |
| CELLMAXX BOVINE ALBUMIN STEM Cell Grade, Low Free Fatty Acid | Thermo Fisher Scientific | 219989980 |
| Chloroform | Sigma Aldrich | C2432-500ML |
| Cholesterol | Sigma Aldrich | 12531-018 |
| Cholyl-lysyl fluorescein | Corning | 451041 |
| Citrate Buffer | Sigma Aldrich | S4641 |
| Collagen I, High Concentration, Rat Tail | Thermo Fisher Scientific | 354249 |
| Collagenase | Sigma Aldrich | C7657 |
| CTS KnockOut SR XenoFree Medium | Thermo Fisher Scientific | 12618-012 |
| Defined Lipid concentrate | Thermo Fisher Scientific | 11905-031 |
| Dexamethasone | Sigma Aldrich | D2915 |
| EGM-2MV SingleQUots Kit | Lonza | CC-3202 |
| Epidermal Growth Factor | R&D Systems | 236-EG-200 |
| FGF 2 Human | BD Biosciences | 354060 |
| FITC | Sigma Aldrich | 124546 |
| FK506 | Astellas pharma | NA |
| Forskolin | Sigma Aldrich | 66675-29-9 |
| GABA | Sigma Aldrich | A2129 |
| Gentamicin | Thermo Fisher Scientific | 15750060 |
| Glutamax | Thermo Fisher Scientific | 35050061 |
| Glutathione | Sigma Aldrich | G6013 |
| Goat Serum | Abcam | ab-7481 |
| HCM Bullet Kit | Lonza | CC-3198 |
| Heparin | Lifeline Cell Technologies | LS-1017 |
| Heparin/Dextrose Catheter Loc Solution | Braintree Scientific | HDS-5 |
| Hepatocyte Growth Factor | Provided from Dr. Michalopoulos | N/A |
| Hoechst 33342 | Sigma Aldrich | B2261 |
| Holo Transferin | CalBiochem | 616424 |
| HyClone Fetal Bovine Serum (U.S.) | Thermo Fisher Scientific | SH3007103 |
| Hydrogen Peroxide | Thermo Fisher Scientific | BP2633-500 |
| iCell Endothelial Cells Medium Supplement | Cellular Dynamics, Fujifilm | #R1112 |
| Insulin | Sigma Aldrich | I9278 |
| Insulin | Sigma Aldrich | 91077C |
| Interleukin 6 | Miltenyi Biotec | 130-095-365 |
| 1X ITS | BD Bioscience | 354351 |
| L-ascorbic acid 2-phosphate magnesium | Sigma Aldrich | A8960 |
| 2-phospho-L-ascorbic acid trisodium salt | Sigma Aldrich | 49752-10G |
| L-Glutamine | Sigma Aldrich | G8540 |
| Linoleic Acid | Sigma Aldrich | L1376-500MG |
| Lithium Chloride | Sigma Aldrich | L4408 |
| L-pipecolic acid | Sigma Aldrich | P2519 |
| Matrigel hESC-Qualified Matrix, LDEV-Free | Thermo Fisher Scientific | 354277 |
| Matrigel Membrane Matrix- Growth Factor Reduced | Thermo Fisher Scientific | CB-40230A |
| MEM Non-Essential Amino Acids Solution (100x) | Thermo Fisher Scientific | 11140-050 |
| MEM Non-Essential Amino Acids Solution (100x) | Millipore | TMS-001-C |
| NHS-PEG, MW5000 | NANOCS | PEG1-0001 |
| Nicotinamide | Sigma-Aldrich | 72340-100G |
| Normal Donkey Serum | Abcam | ab-7475 |
| 10-0 nylon | AROSurgical Instruments Corporation | TK-101038 |
| Oleic Acid | Sigma Aldrich | O1008 |
| Palmitic Acid | Sigma Aldrich | P0500 |
| Peracetic Acid solution | Sigma Aldrich | 269336 |
| Penicillin-Streptomycin (10,000 U/ml) | Thermo Fisher Scientific | 15140-122 |
| Penicillin-Streptomycin 100x | Millipore | TMS-AB2-C |
| Phorbol myristate acetate | Sigma Aldrich | P8139-1MG |
| Proteinase K | Thermo Fisher Scientific | 25530049 |
| Recombinant Human Activin A | Thermo Fisher Scientific | 88518 |
| Recombinant Human Basic Fibroblast Growth Factors (bFGF) | Thermo Fisher Scientific | 354060 |
| Recombinant human basic Fibroblast Growth Factor | Lifeline Cell Technologies | 61977 |
| Recombinant Human BMP-4 | Thermo Fisher Scientific | 314-BP-010 |
| Recombinant Human Epidermal Growth Factor (rhEGF) | Lifeline Cell Technologies | LS-1046 |
| Recombinant human Insulin Growth Factor 1 (rhIGF-1) | Lifeline Cell Technologies | LS-1014 |
| Retinoic acid | Sigma Aldrich | R2625-50MG |
| Retrorsine | Sigma Aldrich | R0382 |
| Rifampicin | Sigma Aldrich | R3501 |
| RT2 SYBR Green ROX qPCR Mastermix | QIAGEN | 330500 |
| sDLL-1 | PeproTech | 140-08 |
| Sp-8-Br-cAMPs | Life Science Institute | B029 |
| Strepto-avidin Phycoerythrin | Thermo Fisher Scientific | S866 |
| TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | 4444557 |
| 1x TE buffer | Thermo Fisher Scientific | 12090015 |
| Thiamine | Sigma Aldrich | T1270 |
| Trace Element B | Corning | 25-022-Cl |
| Trace Element C | Corning | 25-023-Cl |
| Transforming Growth Factor-β1 | Millipore | GF111 |
| Trypsin-EDTA (0.05%), phenol red | Sigma Aldrich | 25300054 |
| Urso deoxycolic acid | Sigma Aldrich | U5127 |
| Critical Commercial Assays | ||
| ABC Goat kit | Vecastain | Cat#PK-6105 |
| ABC Mouse kit | Vectastain | Cat#PK-4000 |
| ABC Rabbit kit | Vectastain | Cat#PK-6101 |
| Bile acid assay kit | Sigma Aldrich | Cat#MAK309 |
| 3-(4,5-Dimethylthiazol-2-yl)-2,5- Diphenyltetrazolium Bromide (MTT) assay | R&D Systems | Cat#4890-025-K |
| Genomic DNA purification kit | Promega | Cat#A1120 |
| RT2 Profiler PCR Array Human Endothelial Cell Biology (PAHS-015Z) | QIAGEN | Cat#330231 |
| MEGAscript system | Thermo Fisher Scientific | Cat#AM1338 |
| Human Alpha-1-Antitrypsin ELISA Kit | Bethyl | Cat#E88-122 |
| Human NHDF Nucleofector kit | Lonza | Cat#VPD-1001 |
| Human tissue plasminogen activator ELISA Kit | Abcam | Cat#ab108914 |
| Hydroxyproline Assay Kit | Sigma Adrich | Cat#MAK008 |
| P450-GloTM CYP3A4 Assay (Luciferin-IPA) | Promega Corporation | Cat#V9001 |
| RNeasy mini kit | QIAGEN | Cat#74104 |
| SuperScript III First-Strand Synthesis System | Thermo Fisher Scientific | Cat#18080-051 |
| Triglyceride Colorimetric Assay | Cayman Chemical Company | Cat#10010303 |
| viPS lentiviral gene transfer kit | Thermo Fisher Scientific | https://www.thermofisher.com/ |
| Experimental Models: Cell Lines | ||
| Adipose-Derived mesenchymal Stem Cells; Normal, Human | American Type Culture Collection (ATCC) | PCS-500-011 |
| Human neonatal microvascular endothelial cells | Lonza | CC-2543 |
| iCell Endothelial Cells, 11713 | Cellular Dynamics, Fujifilm | C1114 |
| Rat microvascular endothelial cells | VEC Technologies, Inc |
https://www.vectechnologies.com/ endothelial-cells/rat/ |
| Cryopreserved rat dermal fibroblast | Cell Applications, Inc | R106-05a |
| Oligonucleotides | ||
| Alpha-1 antitrypsin | Life Technologies | Hs01097800_m1 |
| ATP-binding cassette, sub-familyA(ABCI), member 1 | Life Technologies | Hs01059118_m1 |
| Beta Actin | Life Technologies | Hs01060665_g1 |
| CCAAT-enhancer binding protein alpha | Life Technologies | Hs00269972_s1 |
| C-X-C chemokine receptor type 4 | Life Technologies | Hs00607978_s1 |
| Cystic fibrosis transmembrane conductance regulator | Life Technologies | Hs00357011_m1 |
| Cytochrome P4502C8 | Life Technologies | Hs02383390_s1 |
| Epidermal growth factor receptor | Life Technologies | Hs01076078_m1 |
| Factor five | Life Technologies | Hs00914120_m1 |
| Other Oligonucleotides | See Table S4 | N/A |
| Recombinant DNA | ||
| pCXLE-EGFP | Okita et al., 2011 | Addgene, Cat#27G82 |
| pCXLE-hOCT3/4-shp53-F | Okita et al., 2011 | Addgene, Cat#27G77 |
| pCXLE-hSK | Okita et al., 2011 | Addgene, Cat#27G78 |
| pCXLE-hUL | Okita et al., 2011 | Addgene, Cat#27G8G |
| Software and Algorithms | ||
| Amaxa 4D-Nucleofector | Lonza | N/A |
| HP ChipScnner | Affymetrix Inc | https://www.affymetrix.com |
| ImageJ areas quantification and Gel analysis tool | NIH | https://imagej.nih.gov/ij/ |
| Image Lab | Bio-Rad | https://iwww.bio-rad.com/ |
| Inveon micro-CT system | Siemens Medical Solutions | https://www.siemens-healthineers.com |
| iTRAQ | Applied Biosystems | https://www.thermofisher.com/ |
| JEM-1011 transmission electron microscope | Jeol | https://www.jeol.com |
| JEM-1400Plus transmission electron microscope | Jeol | https://www.jeol.com |
| JMP 10J | SAS Institute | https://www.jmp.com/ |
| 5800 MALDI time-of-flight/time-of-flight system | SCIEX | https://sciex.com/ |
| NanoDrop | Thermo Scientific | https://www.thermofisher.com/ |
| NIS-Elements AR | Nikon | https://www.nikoninstruments.com/ |
| OsiriX | NEWTON GRAPHICS | https://www.newton-graphics.co.jp |
| Prism 7 | GraphPad | https://www.grapgrap.com/ |
| StepOnePlus system | Applied Biosystems | https://www.thermofisher.com/ |
| 7 Tesla micro MRI system | Bruker Biospin | https://www.brunker.com |
| Tzero crucibles, loaded onto Q2000 differential scanning calorimeter | TA Instruments | https://www.tainstruments.com/ |
| Ultimate 3000 nano high pressure liquid chromatography (HPLC) system | DIONEX | https://www.dionex.com |
| Other | ||
| mmu-miR-122-5p | Life Technologies | Cat#2245 |
| mmu-miR-148a-3p | Life Technologies | Cat#470 |
| mmu-miR-194-5p | Life Technologies | Cat#493 |
| Rnu6b | Life Technologies | Cat#1093 |
Transmission electron microscopy
For human fetal and adult hepatocytes and iPS-Heps, cell monolayers were briefly washed with PBS solution. Samples were then fixed with 2.5% glutaraldehyde for 1 hour at room temperature and washed with PBS. Samples were submitted to the University of Pittsburgh Center for Biologic Imaging for post-fixation with 1% osmium tetroxide and 1% potassium ferricyanide for 1 hour at room temperature. Samples were washed with PBS and dehydrated in a graded series of ethanol solution (30%, 50%, 70%, and 90% – 10 minutes each) and three 15-minute changes in fresh 100% ethanol. Infiltration was done with three 1-hour changes of epon. The last change of epon was removed and beam capsules filled with resin were inverted over relevant areas of the monolayers. The resin was allowed to polymerize overnight at 37°C and then for 48 hours at 60°C. Beam capsules and underlying cells were detached from the bottom of the 6-well plate and sectioned. Image acquisition was done using either the JEM-1011 or the JEM-1400Plus transmission electron microscopes (Jeol, Peabody, MA) at 80kV fitted with a side mount AMT 2k digital camera (Advanced Microscopy Techniques, Danvers, MA).
Scanning electron microscopy
Normal fresh liver, decellularized 3% Triton and 0.1% Triton and recellularized (assembled vasculature) liver samples were fixed with glutaraldehyde in preparation for scanning electron microscopy studies. Tissues for immunolabeling and SEM were prepared and imaged as previously described (Soto-Gutiérrez et al., 2006, 2011b). Tissue samples were sectioned into small blocks (8mm3), and were then fixed in 1% Osmium tetroxide for 60 minutes. Samples were dehydrated using graded series of alcohol for 15 minutes each (30% ethanol, 50% ethanol, 70% ethanol, 90% ethanol, 100% ethanol). The samples were then dried at critical point for two hours in absolute alcohol and mounted on an aluminum stub and sputter-coated with gold. Samples were imaged using JEOL 9335 Field Emission Gun.
Organ and cellular imaging
To compare the central and portal vasculature and bile duct between intact and decellularized rat liver, ex vivo liver micro-computed tomography (-CT) was performed. Liver casting was performed with 10 mL 20% Bismuth chloride (MP biomedicals, Solon, OH) with 5% gelatin in PBS. 4% paraformaldehyde fixed (control) or decellularized liver was prepared as described above. The casting solution was injected through the portal vein, inferior vena cava and bile duct and the perfusion was stopped when back pressure was felt. The portal vein and inferior vena cava were ligated above and below the liver and the liver was incubated overnight at room temperature to allow the bismuth-containing gelatin to solidify. Liver vasculature was quantified using a micro-CT system (Inveon micro-CT system, Siemens Medical Solutions, Knoxville, TN). The image resolution and the field of view were set as 30 μm/voxel and 50mm × 50mm × 50mm, respectively with X-ray beam setting at 80kVe and 100mA with a 1000msec exposure time. Acquired pictures were reconstructed and analyzed using OsiriX image processing software program.
To measure and optimize cell seeding within decellularized liver scaffold using micro- magnetic resonance imaging (-MRI), we labeled human liver endothelial cells (TMNK-1) (Matsumura et al., 2004; Soto-Gutiérrez et al., 2006) or human cholangyocites (MMNK-1) (Maruyama et al., 2004) with micron-sized iron oxide particles (Bangs Laboratories, Inc.). Iron oxide micro-particles (1.63 μm) are magnetite cores encapsulated with styrene/divinyl benzene with dragon green fluorescent dye (480-nm excitation, 520-nm emission) soaked in (Kindly provided by Dr. Erik M. Shapiro; Shapiro et al., 2004). For labeling, cells were plated at a density of 1 million cells/cm2 on plastic culture flasks and allowed to attach. Shortly after, 30 iron oxide microspheres/cell were added to the growth medium, and incubated with the cells over night. To remove free particles after labeling, the cells were washed extensively to remove loosely bound free particles. Normally, about 70% of the cells will contain 4–5 iron oxide microspheres (Figure S8A). The cells were then released from the dish by incubation with trypsin and used for cell seeding and imaging studies. The cell assembled liver scaffold was then fixed with 4% paraformaldehyde for 24 hours and then rinsed with neutral PBS twice, and the scaffold was placed in a 60mm Petri dish (IWR) wrapped with polyvinylchloride (Saran Wrap, Dow Chemicals) film to prevent sample drying. The sample was placed in a 72mm-internal diameter volume radiofrequency coil (Bruker Biospin Corporation, Billerica, MA) and the sample was scanned using modified T1-weighted image sequence (TE2.5mse, TR1300msec, Flip angle 30 degrees) with image resolution at 79 μm/voxel and 512×512×256 image size using a 7 Tesla micro MRI system (Bruker BioSpec 70/30 USR, Bruker BioSpin) with ParaVision Acquisition 5.0 software (Bruker, Biospin). For the analysis of cell distribution, different liver lobes were isolated and used for micro MRI. Liver lobes were reconstructed with planar images. As a control of distribution analysis, images of native liver acquired by micro CT were reconstructed to planar slices and compared. For pairing images, planar (2D) images (approximate slice thickness 1 mm) were obtained for each liver lobe. To allow accurate definition in image pairing the major branch of the portal vein, central vein or bile duct was selected for each lobe and manually traced and divided into interbranch segments for anatomical image pairing. Segments were traced beginning at the largest portal vein, central vein or bile duct and moving along the major branch to the smallest. The quantitative analysis of the obtained images of the structures of the liver lobes was performed using the OsiriX image processing software program.
Liver assembly and organ culture
The liver assembly system consisted of a peristaltic pump, bubble trap and syringe infusion pump (Figure 5A). Freshly decellularized rat liver graft (median, superior and inferior right lobes) was connected to the liver assembly system through a portal vein cannula. The inferior vena cava was left open for outlet and the supra hepatic vena cava was ligated. Freshly decellularized rat liver was kept in continuous perfusion through the portal vein. 30–50 mL of culture medium was used in the liver assembly system. Cuffs located in the portal vein and infra-hepatic inferior vena cava allowed easy manipulation of the decellularized rat liver. The main bile duct was also cannulated using a tubing adaptor (BD, Franklin Lakes, NJ). Cell delivery was achieved using a controlled syringe infusion pump (Figure 5A) connected directly to the main bile duct or the perfusion tubing. After a 30–60 min perfusion with medium, first a total of 6 million human iPSC-derived cholangiocytes were infused into the bile duct in four steps at 15-min intervals, each step 1.5 million cells suspended in 0.5mL of medium were perfused over 5 min. Then, approximately 2 h later a mix of iPS-Heps (25 million cells representing 71%), hiPS-VECs (5 million cells representing 15%), human mesenchymal stromal cells (hMSCs) (ATCC, Manassas, VA) (2.5 million cells representing 7%) and human fibroblast (2.5 million cells representing 7%) were infused into the circuit in seven steps at 10-min intervals and were recirculated in the system at room temperature. For the assembly of livers using rat primary cells; we isolated rat hepatocytes using the two-step perfusion protocol as described by Dunn et al., (1991). Viability was assessed by trypan blue exclusion test and was routinely > 90%. Primary rat microvascular endothelial cells (P4-P7) (VEC Technologies, Inc. Rensselaer, NY) and primary rat fibroblasts (Cell Applications, Inc. San Diego, CA) were used as well for histological, bile acid and urea production studies. The engineered liver grafts were transferred to the liver culture system for 24 h with continuous flow at a rate of 15 ml/min for 3h, at 10 ml/min for the next 3h and finally at a rate of 2 ml/min for the remaining 18h.
Next, to assemble the liver vasculature, a total of 40 million human iPS-VECs or human NMVECs were infused into the circuit in two steps at 6 h intervals of no flow (to allow cell attachment) via the inferior vena cava (20 million cells diluted in 10mL of medium) and the portal vein (20 million cells diluted in 10mL of medium) each step was perfused over 5 min at a flow rate of 2 ml/min. The completely engineered human liver graft was cultured for 36 h before functional or transplantation studies at a flow rate of 2 ml/min. The liver culture system consisted of a peristaltic pump, bubble trap and oxygenator. The system was placed in an incubator for temperature control, and the oxygenator was connected to atmospheric gas mixture. The graft was continuously perfused through the portal vein at 2 ml/min with continuous oxygenation that delivered an inflow partial oxygen tension of ~300 mm Hg. Liver assembly was performed using 37% DMEM low glucose 1g/l (ThermoFisher Scientific, Waltham, MA), 37% F-12 (ThermoFisher Scientific, Waltham, MA), 10% CTS KnockOut SR XenoFree Medium (Thermo Fisher Scientific, Waltham, MA), 0.5% Non-Essential Amino Acids (ThermoFisher Scientific, Waltham, MA), 1mM L-glutamine (ThermoFisher Scientific, Waltham, MA), 1% of Penicillin/Streptomycin (ThermoFisher Scientific, Waltham, MA), 50 ng/ml HGF (Kindly provided by George Michalopoulos), 1% DMSO (Sigma-Aldrich, Saint Louis, MO), 0.5 μM Dexamethasone (Sigma-Aldrich, Saint Louis, MO), 0.1% of Ascorbic Acid (Sigma-Aldrich, Saint Louis, MO), 1% of Bovine Serum Albumin Free of Fatty Acids, 0.1% of Hydrocortisone, 0.1% of Transferrin, 0.1% of Insulin, 0.1% Gentamicin (GA1000) (HCM Bullet Kit, ThermoFisher Scientific, Waltham, MA), 100uM of Urso deoxycolic acid (Sigma-Aldrich, Saint Louis, MO), 1× of Cholesterol (ThermoFisher Scientific, Waltham, MA), 20uM of Palmitic Acid (Sigma-Aldrich, Saint Louis, MO), 30 uM of Oleic Acid (Sigma-Aldrich, Saint Louis, MO), 20 uM of Rifampicin (Sigma-Aldrich, Saint Louis, Missouri), 5ng/ml recombinant human Vascular Endothelial Growth Factor (rhVEGF) (Lifeline Cell Technologies, Frederick, MD), 5ng/ml recombinant human Endothelial Growth Factor (rhEGF) (Lifeline Cell Technologies, Frederick, MD), 5ng/ml recombinant human basic Fibroblast Growth Factor (rhbFGF) (Lifeline Cell Technologies, Frederick, MD), 15ng/ml recombinant human Insulin Growth Factor 1 (rhIGF-1) (Lifeline Cell Technologies, Frederick, MD), 0.75 U/ml Heparin (Lifeline Cell Technologies, Frederick, MD), 10% iCell Endothelial Cells Medium Supplement (Cellular Dynamics, Fujifilm, Madison, WI). Additionally, to prevent platelet activation in transplanted human liver grafts; poly-ethylene glycol-carboxymethyl succinimidyl ester (NHS-PEG, MW5000) (NANOCS) was diluted in above described medium at a concentration of 50 mg/ml and the graft was perfused through the portal vein until the engineered liver was filled and soaked (approximately 30ml). The engineered livers were incubated at 37°C for 1 h before auxiliary liver graft transplantation was performed.
Functional analysis
For analysis of hepatocyte function in iPS-Heps and assembled liver grafts, culture media samples or blood serum were collected and analyzed for human alpha-1-antitrypsin (A1AT) content using enzyme linked immunosorbent assay (ELISA) as described by the manufacturer (Bethyl laboratories Inc, Montgomery, TX). Human serum albumin was analyzed by ELISA according to the manufacturer’s instructions (Bethyl laboratories Inc, Montgomery, TX). Urea content was measured using a commercially available kit (Abnova, Walnut, CA.). Triglycerides (Cayman Chemical Company, Ann Arbor, MI) and cell proliferation using the 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) assay (R&D Systems, Minneapolis, MN) were quantified using a colorimetric enzymatic method with commercially available kits. For measurement of bile acids, medium was collected and reaction with 3 alphahydroxysteroid dehydrogenase was measured by fluorescence intensity using the Bile Acid Assay Kit (Sigma-Aldrich, St Louis, MO) according to the manufacturer’s instructions. We measured CYP3A4 activity using P450-Glo CYP3A4 Assay (Luciferin-IPA) (Promega Corporation, Madison, WI, USA, Cat# V9001) according to the manufacturer’s instructions. Cells were exposed to 20 uM of Rifampicin (Sigma-Aldrich, Saint Louis, Missouri) for 48 hours to induce CYP3A4. Cryopreserved human fetal hepatocytes (gestational age: week 24/21) and adult hepatocytes were used as controls.
To test cholangiocytes bile acid transport, differentiating human iPS-Chol were harvest and cultured as 3D culture format, shortly 2×104 cells (the end of Stage 4 of cholangiocytes differentiation), were re-plated in a thick layer of Matrigel Basement Membrane Matrix (Corning, New York, NY). Cells were then further differentiated into mature cholangiocytes using maturation medium (Stage 5). The functional analysis of human iPS-Chol was performed by measuring Cholyl-lysyl-fluorescein (CLF) transports, as described by Sampaziotis et al. (2015). Briefly, the organoids were incubated with 5 mM of CLF (Corning Incorporated) for 30 min at 37°C and then washed with Leibovitz’s medium (Life Technologies). As a control, the same procedure was done using 5 μM of unconjugated fluorescein isothiocyanate (FITC, Sigma-Aldrich). The imagens were collected after the incubation (loading phase) and 10 minutes after (export phase). The measurements were made normalizing the fluorescence by the background and using the loading phase as 1.
To test endothelial cell function, human neonatal microvascular endothelial cells (Lonza, Walkersville, MD) or human iPSC-VECs (iCell Endothelial Cells) (Cellular Dynamics, Fujifilm, Madison, WI) in culture (2D) and assembled vasculature of liver scaffolds were incubated in EBM PLUS Basal Medium supplemented with EGM-2 SingleQuots Kit (Lonza, Walkersville, MD) for human neonatal microvascular endothelial cells and VascuLife VEGF Medium (Cell Technologies, Fremont, CA) supplemented with iCell Endothelial Cells Medium Supplement (Cellular Dynamics, Fujifilm, Madison, WI) for human iPS-derived endothelial cells containing either 1pM of 1a,25-dihydroxyvitamin D3 (Sigma-Aldrich, Saint Louis, MO) or 1 μM retinoic acid (Sigma-Aldrich, Saint Louis, MO) or 1mM Sp-8-BrcAMPs (BIOLOG, Life Science Institute, Bremen, Germany), 1 μM or 100nM Phorbol myristate acetate (PMA) (Sigma-Aldrich, Saint Louis, MO) and a combination of 100nM PMA/10 mM Forskolin (Sigma-Aldrich, Saint Louis, MO) for 24 h. Human tissue plasminogen activator was measured using an ELISA, according to the manufacturer’s instructions (Abcam, Cambridge, MA). Subsequently, assembled vasculature of liver scaffolds was cultured in the liver culture system for 24 h. Next, 10 μg/ml FITC labeled acLDL (Invitrogen, Carlsbad, CA) was added to the perfused culture medium and recirculated for 24 h. Assembled liver scaffolds were washed with fresh medium and liver lobes were dissected and prepared for imaging. Images were captured with a Nikon Eclipse Ti microscope. All experiments were performed using either human neonatal microvascular endothelial cells (Lonza, Walkersville, MD) or human iPSC-VECs (iCell Endothelial Cells) (Cellular Dynamics, Fujifilm, Madison, WI) at passage 1–5.
Immunofluorescence
Human adult or fetal livers, engineered liver grafts or transplanted engineered liver grafts were fixed in 4% paraformaldehyde for 12 h and 70% ethanol overnight at 4°C, and then embedded in paraffin, cut sections (5–7 μm) were mounted on glass slides, for fluorescence staining. Antibodies specific for antigens were acquired for immunofluorescence and list of the antibodies used can be found in Key Resources Table. Slides were deparaffinized and hydrated, washed by PBS and blocked 1h with 10% donkey/goat serum. Antigen retrieval was used, sections were then incubated with primary antibody overnight 4 C and secondary antibody 1h room temperature, sections were covered with mounting media with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). For immunohistochemistry staining, sections, were deparaffinized and processed for staining with hematoxylin and eosin. All procedures followed the kit instructions. Images were captured with a Nikon Eclipse Ti microscope.
Auxiliary liver graft transplantation
All transplanted animals were pre-conditioned with retrorsine, animals were given two injections of retrorsine (Sigma-Aldrich, Saint Louis, MO), 30 mg/kg each, intraperitoneally, 2 weeks apart. Four weeks after the second injection, auxiliary liver transplantation was performed. The auxiliary partial liver transplantation in the rat has been described in detail previously (Matsubara et al., 2015). Briefly, after laparotomy and preparation of the infra-hepatic inferior vena cava (IHIVC), the right renal artery and vein were ligated and right nephrectomy was performed to create the space of heterotopic liver transplantation. After proximal and distal clamping of the IHIVC, the anterior wall of the IHIVC was cut as close to the ligated right renal vein as possible, for the anastomosis with the liver graft by an end-to-side anastomosis using a continuous suture with 10–0 nylon (AROSurgical Instruments corporation, Newport Beach, CA). Next, after clamping the recipient’s portal vein (PV), the splenic vein and the superior mesenteric vein (SMV), the PV was cut and end-to-side anastomosis was performed between the graft PV and recipient’s PV. After finishing both anastomoses, PV was declamped and the graft was re-perfused. Following reperfusion, PV was partially ligated between the pyloric vein and the PV anastomosis. The stented bile duct of the graft was inserted into the duodenum after applying the purse-string suture and fixed to the duodenal wall by tightening the suture. Finally, left lateral lobe of the rat liver was resected for induction of liver regenerative stimulus. All transplanted animals were administrated FK506 (Astellas pharma, Tokyo, Japan) at 1 mg/kg/day to control any rejection episode for the duration of the experiments.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data from at least three sets of samples were used for statistical analysis. The statistical software JMP 10J version (SAS Institute, Cary, NC, USA) was used for all analyses. All experiments were independently performed three times in triplicate. Due to the relatively small sample size, normality testing was not feasible. Data are expressed as mean ± standard deviation (SD) and were compared using an analysis of the Wilcoxon test. Values of p < 0.05 were considered statistically significant.
Supplementary Material
Highlights.
Organ-like microenvironment further matures human iPSC mini livers
Human vascular and biliary network can be engineered in decellularized liver scaffolds
Human iPSC-mini-liver microstructure has similarities to human liver
Human iPSC-derived mini livers can be transplanted in immunodeficient rats
ACKNOWLEDGMENTS
We thank Stephen C. Strom for sharing human iPSCs that have been used in part of this report. We also thank the Center for Biologic Imaging at the University of Pittsburgh for technical support on the transmission electro microscopy study. This project used the UPMC Hillman Cancer Center and Tissue and Research Pathology/Pitt Biospecimen Core shared resource, which is supported in part by award P30CA047904. This project was also partially supported by the Pilot and Feasibility program under the NIH/NIDDK Digestive Disease Research Core Center grant P30DK120531. This work was supported by NIH grants DK099257, DK117881, DK119973, and TR002383 to A.S.-G., P01DK096990 to I.J.F. and A.S.G., DK107697 to E.M.S. and A.S.G., and DK103645 to A.W.D., and by the American Liver Foundation to K.T.
Footnotes
DECLARATION OF INTERESTS
H.Y. and A.S.-Gare inventors on a patent application that involves some of the perfusion technology used in this work (WO/2011/002926); K.H., K.M., J.G.-L., H.Y., and A.S.-G. have an international patent related to this work that describes methods of preparing artificial organs and related compositions for transplantation and regeneration (WO/2015/168254). K.T., A.C.H., J.G.-L., Y.W., T.M., and A.S.-G. have a provisional international patent application that describes hepatic differentiation of human pluripotent stem cells and liver repopulation (PCT/US2018/018032). A.S.-G., J.G.-L., K.T., A.C.H., Y.W., T.M., and I.J.F. are co-founders and have a financial interest in Von Baer Wolff, Inc. a company focused on biofabrication of autologous human hepatocytes from stem cells technology and programming liver failure and their interests are managed by the Conflict of Interest Office at the University of Pittsburgh in accordance with their policies.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107711.
REFERENCES
- Ammori JB, Pelletier SJ, Lynch R, Cohn J, Ads Y, Campbell DA, and Englesbe MJ (2008). Incremental costs of post-liver transplantation complications. J. Am. Coll. Surg 206, 89–95. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. ; The Gene Ontology Consortium (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avior Y, Levy G, Zimerman M, Kitsberg D, Schwartz R, Sadeh R, Moussaieff A, Cohen M, Itskovitz-Eldor J, and Nahmias Y. (2015). Microbial-derived lithocholic acid and vitamin K2 drive the metabolic maturation of pluripotent stem cells-derived and fetal hepatocytes. Hepatology 62, 265–278. [DOI] [PubMed] [Google Scholar]
- Ballard O, and Morrow AL (2013). Human milk composition: nutrients and bioactive factors. Pediatr. Clin. North Am 60, 49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandiera S, Pfeffer S, Baumert TF, and Zeisel MB (2015). miR-122–a key factor and therapeutic target in liver disease. J. Hepatol 62, 448–457. [DOI] [PubMed] [Google Scholar]
- Bao J, Shi Y, Sun H, Yin X, Yang R, Li L, Chen X, and Bu H. (2011). Construction of a portal implantable functional tissue-engineered liver using perfusion-decellularized matrix and hepatocytes in rats. Cell Transplant. 20, 753–766. [DOI] [PubMed] [Google Scholar]
- Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, and Soker S. (2011). The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology 53, 604–617. [DOI] [PubMed] [Google Scholar]
- Basma H, Soto-Gutiérrez A, Yannam GR, Liu L, Ito R, Yamamoto T, Ellis E, Carson SD, Sato S, Chen Y, et al. (2009). Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology 136, 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beath SV (2003). Hepatic function and physiology in the newborn. Semin. Neonatol 8, 337–346. [DOI] [PubMed] [Google Scholar]
- Bhatia SN, Underhill GH, Zaret KS, and Fox IJ (2014). Cell and tissue engineering for liver disease. Sci. Transl. Med 6, 245sr2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butter A, Aliyev K, Hillebrandt KH, Raschzok N, Kluge M, Seiffert N, Tang P, Napierala H, Muhamma AI, Reutzel-Selke A, et al. (2018). Evolution of graft morphology and function after recellularization of decellularized rat livers. J. Tissue Eng. Regen. Med 12, e807–e816. [DOI] [PubMed] [Google Scholar]
- Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, Kanton S, Kageyama J, Damm G, Seehofer D, et al. (2017). Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538. [DOI] [PubMed] [Google Scholar]
- Carpentier A, Tesfaye A, Chu V, Nimgaonkar I, Zhang F, Lee SB, Thorgeirsson SS, Feinstone SM, and Liang TJ (2014). Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. J. Clin. Invest 124, 4953–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Soto-Gutierrez A, Baptista PM, and Spee B. (2018). Biotechnology challenges to in vitro maturation of hepatic stem cells. Gastroenterology 154, 1258–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, and Lemaigre FP (2005). Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 19, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Levy G, and Nahmias Y. (2015). Coculture and long-term maintenance of hepatocytes. Methods Mol. Biol 1250, 161–173. [DOI] [PubMed] [Google Scholar]
- Collin de l’Hortet A, Takeishi K, Guzman-Lepe J, Morita K, Achreja A, Popovic B, Wang Y, Handa K, Mittal A, Meurs N, et al. (2019). Generation of human fatty livers using custom-engineered induced pluripotent stem cells with modifiable SIRT1 metabolism. Cell Metab. 30, 385–401 e389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglau TE, Johnson JD, Villanueva FS, and Wagner WR (2007). Targeting microspheres and cells to polyethylene glycol-modified biological surfaces. J. Biomed. Mater. Res. A 81, 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianat N, Dubois-Pot-Schneider H, Steichen C, Desterke C, Leclerc P, Raveux A, Combettes L, Weber A, Corlu A, and Dubart-Kupperschmitt A. (2014). Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology 60, 700–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JC, Tompkins RG, and Yarmush ML (1991). Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol. Prog 7, 237–245. [DOI] [PubMed] [Google Scholar]
- Flynn DM, Nijjar S, Hubscher SG, de Goyet J.d.V., Kelly DA, Strain AJ, and Crosby HA (2004). The role of Notch receptor expression in bile duct development and disease. J. Pathol 204, 55–64. [DOI] [PubMed] [Google Scholar]
- Fraunhoffer N, and Soto-Gutié rrez A. (2018). Gene expression changes during human liver development: fetal towards adult. Mendeley Data. Published online July 30, 2018. 10.17632/ncxxgwdwty.1. [DOI] [Google Scholar]
- Gilbert TW, Freund JM, and Badylak SF (2009). Quantification of DNA in biologic scaffold materials. J. Surg. Res 152, 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V, Boussemart L, Gadue P, Nierhoff D, Koehler CI, Kubo A, Shafritz DA, and Keller G. (2006). BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat. Biotechnol 24, 1402–1411. [DOI] [PubMed] [Google Scholar]
- Habka D, Mann D, Landes R, and Soto-Gutierrez A. (2015). Future economics of liver transplantation: A 20-year cost modeling forecast and the prospect of bioengineering autologous liver grafts. PLoS ONE 10, e0131764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanein W, Uluer MC, Langford J, Woodall JD, Cimeno A, Dhru U, Werdesheim A, Harrison J, Rivera-Pratt C, Klepfer S, et al. (2017). Recellularization via the bile duct supports functional allogenic and xenogenic cell growth on a decellularized rat liver scaffold. Organogenesis 13, 16–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo MJ, Kim TH, You JS, Blaya D, Sancho-Bru P, and Kim SG (2018). Alcohol dysregulates miR-148a in hepatocytes through FoxO1, facilitating pyroptosis via TXNIP overexpression. Gut. 68, gutjnl-2017–315123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HHS HRSA (2014). OPTN/SRTR 2012 Annual Data Report (Department of Health and Human Services, Health Resources and Services Administration; ), pp. 1–185. [Google Scholar]
- Jamall IS, Finelli VN, and Que Hee SS (1981). A simple method to determine nanogram levels of 4-hydroxyproline in biological tissues. Anal. Biochem 112, 70–75. [DOI] [PubMed] [Google Scholar]
- Jang S, Collin de l’Hortet A, and Soto-Gutierrez A. (2019). Induced pluripotent stem cell-derived endothelial cells: overview, current advances, applications, and future directions. Am. J. Pathol 189, 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko IK, Peng L, Peloso A, Smith CJ, Dhal A, Deegan DB, Zimmerman C, Clouse C, Zhao W, Shupe TD, et al. (2015). Bioengineered transplantable porcine livers with re-endothelialized vasculature. Biomaterials 40, 72–79. [DOI] [PubMed] [Google Scholar]
- Kojima H, Yasuchika K, Fukumitsu K, Ishii T, Ogiso S, Miyauchi Y, Yamaoka R, Kawai T, Katayama H, Yoshitoshi-Uebayashi EY, et al. (2018). Establishment of practical recellularized liver graft for blood perfusion using primary rat hepatocytes and liver sinusoidal endothelial cells. Am. J. Transplant. 18, 1351–1359. [DOI] [PubMed] [Google Scholar]
- Kruithof EK, and Dunoyer-Geindre S. (2014). Human tissue-type plasminogen activator. Thromb. Haemost. 112, 243–254. [DOI] [PubMed] [Google Scholar]
- Laconi S, Curreli F, Diana S, Pasciu D, De Filippo G, Sarma DS, Pani P, and Laconi E. (1999). Liver regeneration in response to partial hepatectomy in rats treated with retrorsine: a kinetic study. J. Hepatol 31, 1069–1074. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Kobayashi N, Westerman KA, Sakaguchi M, Allain JE, Totsugawa T, Okitsu T, Fukazawa T, Weber A, Stolz DB, et al. (2004). Establishment of a highly differentiated immortalized human cholangiocyte cell line with SV40T and hTERT. Transplantation 77, 446–451. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, and Serikawa T. (2010). Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS ONE 5, e8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuk TV, Ritman EL, and LaRusso NF (2001). Quantitative assessment of the rat intrahepatic biliary system by three-dimensional reconstruction. Am. J. Pathol 158, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Yokota S, Ono Y, Fukumitsu K, Handa K, Guzman-Lepe J, Perez-Gutierrez AR, Yagi H, Fox IJ, Murase N, et al. (2015). Surgical model of auxiliary partial liver transplantation in the rat. Protoc. Exch [Google Scholar]
- Matsumura T, Takesue M, Westerman KA, Okitsu T, Sakaguchi M, Fukazawa T, Totsugawa T, Noguchi H, Yamamoto S, Stolz DB, et al. (2004). Establishment of an immortalized human-liver endothelial cell line with SV40T and hTERT. Transplantation 77, 1357–1365. [DOI] [PubMed] [Google Scholar]
- Mazza G, Rombouts K, Rennie Hall A, Urbani L, Vinh Luong T, Al-Akkad W, Longato L, Brown D, Maghsoudlou P, Dhillon AP, et al. (2015). Decellularized human liver as a natural 3D-scaffold for liver bioengineering and transplantation. Sci. Rep 5, 13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medh RD, Santell L, and Levin EG (1992). Stimulation of tissue plasminogen activator production by retinoic acid: synergistic effect on protein kinase C-mediated activation. Blood 80, 981–987. [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK (2007). Liver regeneration. J. Cell. Physiol 213, 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto A, Kannari M, Tsuchida Y, Sasaki S, Saito C, Matsuta T, Maeda T, Akiyama M, Nakamura T, Sakaguchi M, et al. (2017). An HNF4a-microRNA-194/192 signaling axis maintains hepatic cell function. J. Biol. Chem 292, 10574–10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müsch A. (2014). The unique polarity phenotype of hepatocytes. Exp. Cell Res 328, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JE, La Francesca S, Niles JA, Vega SP, Argueta LB, Frank L, Christiani DC, Pyles RB, Himes BE, Zhang R, et al. (2018). Production and transplantation of bioengineered lung into a large-animal model. Sci. Transl. Med 10, eaao3926. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Surapisitchat J, Virtanen C, Ogawa M, Niapour M, Sugamori KS, Wang S, Tamblyn L, Guillemette C, Hoffmann E, et al. (2013). Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development 140, 3285–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Ogawa S, Bear CE, Ahmadi S, Chin S, Li B, Grompe M, Keller G, Kamath BM, and Ghanekar A. (2015). Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat. Biotechnol 33, 853–861. [DOI] [PubMed] [Google Scholar]
- Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, et al. (2011). A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412. [DOI] [PubMed] [Google Scholar]
- Olmer R, Engels L, Usman A, Menke S, Malik MNH, Pessler F, Göhring G, Bornhorst D, Bolten S, Abdelilah-Seyfried S, et al. (2018). Differentiation of human pluripotent stem cells into functional endothelial cells in scalable suspension culture. Stem Cell Reports 10, 1657–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasain N, Lee MR, Vemula S, Meador JL, Yoshimoto M, Ferkowicz MJ, Fett A, Gupta M, Rapp BM, Saadatzadeh MR, et al. (2014). Differentiation of human pluripotent stem cells to cells similar to cord-blood endothelial colony-forming cells. Nat. Biotechnol 32, 1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Moser PT, Gilpin SE, Okamoto T, Wu T, Tapias LF, Mercier FE, Xiong L, Ghawi R, Scadden DT, et al. (2015). Engineering pulmonary vasculature in decellularized rat and human lungs. Nat. Biotechnol 33, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Rezvani M, Grimm AA, and Willenbring H. (2016). Assessing the therapeutic potential of lab-made hepatocytes. Hepatology 64, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaziotis F, de Brito MC, Madrigal P, Bertero A, Saeb-Parsy K, Soares FAC, Schrumpf E, Melum E, Karlsen TH, Bradley JA, et al. (2015). Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol 33, 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub JR, Huppert KA, Kurial SNT, Hsu BY, Cast AE, Donnelly B, Karns RA, Chen F, Rezvani M, Luu HY, et al. (2018). De novo formation of the biliary system by TGFβ-mediated hepatocyte transdifferentiation. Nature 557, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, and Koretsky AP (2004). MRI detection of single particles for cellular imaging. Proc. Natl. Acad. Sci. USA 101, 10901–10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, and Duncan SA (2010). Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, Song X, Guo Y, Zhao Y, Qin H, et al. (2009). Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 19, 1233–1242. [DOI] [PubMed] [Google Scholar]
- Soto-Gutiérrez A, Kobayashi N, Rivas-Carrillo JD, Navarro-Alvarez N, Zhao D, Okitsu T, Noguchi H, Basma H, Tabata Y, Chen Y, et al. (2006). Reversal of mouse hepatic failure using an implanted liver-assist device containing ES cell-derived hepatocytes. Nat. Biotechnol 24, 1412–1419. [DOI] [PubMed] [Google Scholar]
- Soto-Gutiérrez A, Navarro-Alvarez N, Rivas-Carrillo JD, Tanaka K, Chen Y, Misawa H, Okitsu T, Noguchi H, Tanaka N, and Kobayashi N. (2007a). Construction and transplantation of an engineered hepatic tissue using a polyaminourethane-coated nonwoven polytetrafluoroethylene fabric. Transplantation 83, 129–137. [DOI] [PubMed] [Google Scholar]
- Soto-Gutiérrez A, Navarro-Alvarez N, Zhao D, Rivas-Carrillo JD, Lebkowski J, Tanaka N, Fox IJ, and Kobayashi N. (2007b). Differentiation of mouse embryonic stem cells to hepatocyte-like cells by co-culture with human liver nonparenchymal cell lines. Nat. Protoc 2, 347–356. [DOI] [PubMed] [Google Scholar]
- Soto-Gutiérrez A, Navarro-Alvarez N, and Kobayashi N. (2011a). Hepatocytes. In Molecular Pathology of Liver Diseases, Monga SPS, ed. (Springer; ), pp. 17–26. [Google Scholar]
- Soto-Gutiérrez A, Zhang L, Medberry C, Fukumitsu K, Faulk D, Jiang H, Reing J, Gramignoli R, Komori J, Ross M, et al. (2011b). A whole-organ regenerative medicine approach for liver replacement. Tissue Eng. Part C Methods 17, 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Gutiérrez A, Wertheim JA, Ott HC, and Gilbert TW (2012). Perspectives on whole-organ assembly: moving toward transplantation on demand. J. Clin. Invest 122, 3817–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WQ, Xu H, Sandor M, and Lombardi J. (2013). Process-induced extracellular matrix alterations affect the mechanisms of soft tissue repair and regeneration. J. Tissue Eng 4, 2041731413505305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. [DOI] [PubMed] [Google Scholar]
- Tobita T, Guzman-Lepe J, Takeishi K, Nakao T, Wang Y, Meng F, Deng CX, Collin de l’Hortet A, and Soto-Gutié rrez A. (2016). SIRT1 disruption in human fetal hepatocytes leads to increased accumulation of glucose and lipids. PLoS ONE 11, e0149344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R, Lozoya O, Wang Y, Cardinale V, Gaudio E, Alpini G, Mendel G, Wauthier E, Barbier C, Alvaro D, and Reid LM (2011). Human hepatic stem cell and maturational liver lineage biology. Hepatology 53, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uygun BE, Soto-Gutiérrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, Milwid J, Kobayashi N, Tilles A, Berthiaume F, et al. (2010). Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat. Med 16, 814–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyta JC, Via DP, Butterfield CE, and Zetter BR (1984). Identification and isolation of endothelial cells based on their increased uptake of acetylated-low density lipoprotein. J. Cell Biol 99, 2034–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Fukumitsu K, Fukuda K, Kitago M, Shinoda M, Obara H, Itano O, Kawachi S, Tanabe M, Coudriet GM, et al. (2013). Human-scale whole-organ bioengineering for liver transplantation: a regenerative medicine approach. Cell Transplant. 22, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashina S, Sato N, Kon K, Ikejima K, and Watanabe S. (2009). Role of mitochondria in liver pathophysiology. Drug Discov. Today Dis. Mech 6, e25–e30. [Google Scholar]
- Zabulica M, Srinivasan RC, Vosough M, Hammarstedt C, Wu T, Gramignoli R, Ellis E, Kannisto K, Collin de l’Hortet A, Takeishi K, et al. (2019). Guide to the assessment of mature liver gene expression in stem cell-derived hepatocytes. Stem Cells Dev. 28, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, and Grompe M. (2008). Generation and regeneration of cells of the liver and pancreas. Science 322, 1490–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Huang Y, Guo Y, Wang L, Ling C, Guo Q, Wang Y, Zhu S, Fan X, Zhu M, et al. (2016). Decellularization and recellularization of rat livers with hepatocytes and endothelial progenitor cells. Artif. Organs 40, E25–E38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data is available at the Mendeley data repository, https://data.mendeley.com/datasets/ncxxgwdwty/1: Gene expression changes during human liver development: fetal toward adult. The microarray data can be accessed directly via its project DOI (https://doi.org/10.17632/ncxxgwdwty.1) (Fraunhoffer and Soto-Gutiérrez, 2018).