Abstract
Centronuclear myopathies (CNMs) are a subtype of congenital myopathies characterized by skeletal muscle weakness and an increase in the number of central myonuclei. SPEG (striated preferentially expressed protein kinase) has been identified as the sixth gene associated with CNM, and it has been shown that striated muscle-specific Speg-knockout (KO) mice have defective triad formation, abnormal excitation-contraction coupling, and calcium mishandling. The impact of SPEG deficiency on the survival and function of myogenic cells remains to be deciphered. In this study, the authors examined the overall population, proliferation, and differentiation of myogenic cells obtained from striated muscle–specific Speg-KO mice and compared them with wild-type (WT) controls. SPEG-deficient skeletal muscles contained fewer myogenic cells, which on further study demonstrated reduced proliferation and delayed differentiation compared with those from WT muscles. Regenerative response to skeletal muscle injury in Speg-KO mice was compared with that of WT mice, leading to the identification of similar abnormalities including fewer satellite cells, fewer dividing cells, and an increase in apoptotic cells in KO mice. Overall, these results reveal specific abnormalities in myogenic cell number and behavior associated with SPEG deficiency. Similar satellite cell defects have been reported in mouse models of MTM1- and DNM2-associated CNM, suggestive of shared underlying pathways.
Centronuclear myopathies (CNMs), a subtype of congenital myopathies, are a heterogeneous group of inherited neuromuscular disorders characterized by muscle weakness, predominant type I fibers, and increased central nuclei.1, 2, 3 Over 60% of CNM cases have been attributed to mutations in the MTM1 [MIM 300415; encoding myotubularin (MTM1)], DNM2 (MIM 602378; encoding dynamin 2), BIN1 (MIM 601248; encoding bridging integrator 1), RYR1 (MIM 180901; encoding ryanodine receptor 1), and TTN (MIM 188840; encoding titin) genes. The authors have identified SPEG (striated preferentially expressed gene) as the sixth gene associated with CNM.4 Patients with SPEG mutations typically present with congenital myopathy–related features with or without dilated cardiomyopathy.4,5
The SPEG protein belongs to the myosin light chain kinase (MLCK) family, and it has four isoforms derived from two transcription start sites and alternative splicing,6 including Spegβ (350 kDa), Spegα (250 kDa), Apeg1 (19 kDa), and Bpeg (94 kDa). Spegα and Spegβ are both expressed in striated muscle (cardiac and skeletal), Apeg1 in vascular smooth muscle, and Bpeg in brain and aorta.6, 7, 8 Spegα is a sensitive marker for striated muscle differentiation,6 its expression being up-regulated during C2C12 myoblast to myotube differentiation in vitro. By comparison, the expression of Spegβ remains uniformly expressed at a minimal level.6
In mice, Spegα expression in the cardiomyocytes of developing heart starts at 11.5 days post coitum.9 Constitutive Speg knockout (KO) mice exhibited significant embryonic or neonatal mortality due to cardiac dysfunction.9 To overcome the embryonic lethal effects, the authors created a conditional Speg-KO mouse and bred them with Mck-cre–expressing mice to excise SPEG in striated muscles postnatally (Speg-KO). The skeletal muscle fibers of Speg-KO mice were noted to be significantly smaller and showed marked variability in fiber size.10 SPEG deficiency led to an abnormal triad structure and defective calcium handling in skeletal muscles,10 indicating a critical role of SPEG during skeletal muscle development and function. SPEG has been identified as an important regulator of cardiac function and development.6,9,11, 12, 13 The underlying mechanism of cardiomyopathy in Speg-KO mice is likely due to defective cardiac progenitor cell function.14 Cardiac progenitor cells harvested from SPEG-deficient mice display defects in clone formation, proliferation, and differentiation in vitro.14 It is unclear whether SPEG plays a similar role in the development of muscle stem cells in skeletal muscles.
Satellite cells (SCs), also known as muscle stem cells, are involved in skeletal muscle growth during fetal and postnatal development. SCs play a critical role in muscle repair and regeneration following injury or disease.15,16 Upon muscle injury, SCs can be activated from a quiescent state to proliferate for self-renewal. Proliferating SCs, also known as myoblasts, then exit cell proliferation, differentiate into myocytes,17,18 providing a source for new myonuclei in muscle growth,19 regeneration,20 or hypertrophy.21 SC dysfunction has been reported in MTM1-deficient22 and DNM2-mutant23,24 mice. However, the impact of SPEG deficiency on SC function has not been studied.
To investigate the role of SPEG in SCs, a striated muscle-specific Speg-KO mouse model (Speg-KO) was used as described earlier.10 The SC numbers were significantly reduced in Speg-KO mice as indicated by fluorescence-activated cell sorting (FACS) and Pax7 immunostaining. Speg-KO myoblasts displayed reduced proliferation and delayed differentiation in comparison to littermate-matched wild-type (WT) myoblasts. In response to injury, skeletal muscles in Speg-KO mice demonstrated a slower healing process and an increase in apoptosis. These results demonstrate that SPEG plays an essential role in skeletal muscle growth and repair by maintaining the SC pool and regulating myogenic cell function.
Materials and Methods
Animals
All studies were approved by the Institutional Animal Care and Use Committee at Children's Hospital Boston (Boston, MA). Speg-KO mice have been generated as previously described.10 In brief, the homozygous Speg-conditional KO mice (Spegfl/fl) were bred with male transgenic mice (MCK-Cre+) who have the Cre recombinase driven by muscle creatine kinase promoter, with Cre activity observed in skeletal and cardiac muscle. Spegfl/fl: MCK-cre+ mice were the Speg-KO mice that were utilized for further experiments whereas littermates with Spegfl/fl: MCK-cre−, Spegfl/wt: MCK-cre+, and Spegfl/wt: MCK-cre− genotypes were used as WT controls.
Myoblast Isolation and FACS
Myogenic cells were extracted from the upper and lower limbs of Speg-KO and WT littermate mice (each genotype n = 6) between 25 and 30 days of life following established methods.25 Briefly, all collected muscles of a mouse were transferred to a fresh Petri dish and minced for approximately 10 minutes using sterile scalpels. Minced muscles were digested at 37°C for 45 minutes using a final concentration of 1.2 U of dispase II (Roche Applied Science, Indianapolis, IN) and 5 mg of collagenase D (Roche Applied Science) per gram of tissue with gentle trituration every 15 minutes. Digested samples were filtered through a 40-μm cell strainer (Thermo Fisher Scientific, Waltham, MA), spun down and treated with Red Blood Cell Lysis Buffer (Qiagen, Valencia, CA). Cells were spun down and resuspended in ice-cold 1× Hank's buffered salt solution (Thermo Fisher Scientific) containing 0.5% bovine serum albumin at a concentration of 1 × 107 cells/mL. Cells were incubated on ice with the antibodies of associated cell surface markers for 20 minutes, including anti-mouse CD45− Alexa Fluor 488 (marker for hematopoietic cells, 0.25 μg/million cells; catalog number 103122; BioLegend, Dedham, MA), anti-mouse Sca-1− Alexa Fluor 488 (Ly-6A/E, marker for mesenchymal stem cells, 0.25 μg/million cells; catalog number 122516; BioLegend), anti-mouse CD31− Alexa Fluor 488 (marker for endothelial cells, 2 μg/million cells; catalog number 102514; BioLegend), and APC anti-mouse CD106 (VCAM-1, marker for SCs, 0.25 μg/million cells; catalog number 105717; BioLegend).
After incubation, cells were washed with cold 1× Hank's buffered salt solution containing 0.5% bovine serum albumin buffer. To select for live cells by flow cytometry, propidium iodide (PI; marker for dead cells; 1 μg/mL; BMS500PI; Thermo Fisher Scientific) was added at a final concentration of 1 μg/mL. Samples were then analyzed and sorted using a BD FACSAria cell sorter (BD Bioscience, San Jose, CA) to isolate quadruple negative prospective myogenic cells (QNMCs; CD31−CD45− Sca1−PI−) and SCs (CD31−CD45− Sca1−PI− VCAM-1+).
Histology and Immunofluorescence
Cross sections (8-μm thick) of isopentane-frozen tibialis anterior (TA) or quadriceps muscle were stained with hematoxylin and eosin using standard techniques. Immunofluorescence was performed on TA muscles of cardiotoxin-injected mice by standard immunofluorescent staining using mouse anti-Pax7 (marker for SCs; AB 528428, dilution 1:50; Developmental Studies Hybridoma Bank, Iowa City, IA), rabbit anti–Ki-67 (marker for proliferating cells; ab15580, dilution 1:200; Abcam, Cambridge, MA), rabbit anti-SPEG (PA5-53875, dilution 1:100; Thermo Fisher Scientific), and rabbit anti-Caspase 3 primary antibody (marker for apoptotic cells; ab13847, dilution 1:100; Abcam). Goat anti-rabbit IgG (H+L) secondary antibody-Alexa Fluor 594 (A-11012, dilution 1:1000; Thermo Fisher Scientific) or goat anti-mouse IgG (H+L) secondary antibody-Alexa Fluor 488 Phalloidin (A28175, dilution 1:1000; Thermo Fisher Scientific) was used for staining. Slides were coverslipped using Vectashield Mounting Medium with DAPI (H-1200; Vector Laboratories, Burlingame, CA). Images were quantified by counting the number of positive cells within a field and dividing by the total number of fibers (for Pax7 and Caspase 3) or nuclei (for Ki-67) within that field using a Nikon Eclipse 90i microscope in conjunction with NIS-Elements AR software version 4.50 (Nikon Instruments Inc., Melville, NY).
Immunoblot Analysis
Skeletal muscles from Speg-KO and WT littermate mice were dissected, snap frozen in isopentane, and stored at −80°C until analysis. Protein isolation and Western blot procedures were performed as described previously.26 Protein concentration was determined using a colorimetric BCA assay (23225; Thermo Fisher Scientific). Transferred proteins were probed with antibody against mouse anti-Pax7 (AB 528428, dilution 1:100; Developmental Studies Hybridoma Bank), and mouse anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH; MA5-15738, dilution 1:1000; Thermo Fisher Scientific). Secondary horseradish peroxidase–conjugated antibody against mouse (ab205719, dilution 1:3000; Abcam) was detected using enhanced chemiluminescence. Quantification of protein levels normalized to GAPDH was performed using Quantity One software version 4.2.1 (Bio-Rad Laboratories, Inc., Hercules, CA) on an Image Station 440 (Kodak DS; Eastman Kodak Co., Rochester, NY).
Proliferation Assays
QNMCs from 25- to 30-day–old Speg-KO and WT mice (n = 6 of each genotype) were plated onto 96-well plates at a density of either 1000 or 5000 cells per well in skeletal muscle growth medium (Ham's F-10 medium, SH3002501; Thermo Fisher Scientific) supplemented with 20% fetal bovine serum (SH30396; Thermo Fisher Scientific) and 5 ng/mL basic fibroblast growth factor (bFGF; 100-18B; PeproTech, Rocky Hill, NJ). Proliferation was measured at 0, 2, 4, 7, and 10 days after isolation using a CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (TB169; Promega, Madison, WI), as per the manufacturer's instructions. Absorbance was read at 490 nm using a Synergy 2 plate reader (Biotek Instruments, Winooski, VT). The average of three wells for each condition was recorded for each time point, and the experiment was repeated three times. Statistical differences between corrected values were compared using a one-way analysis of variance.
Fusion Assays
For fusion assays, 5000 cells per well were plated in 8-well Permanox slides coated with Matrigel. Cells adhered overnight in high serum medium, then were switched to differentiation medium [Dulbecco's modified Eagle's medium low glucose (MT10013CV; Thermo Fisher Scientific) supplemented with 4% fetal bovine serum]. Differentiation medium was changed daily, and fusion was assessed at days 0, 2, and 5 after the induction of differentiation. Cells were fixed and stained with a rabbit monoclonal anti-desmin antibody (marker for myogenic differentiation; ab216616, dilution 1:50; Abcam) and goat anti-rabbit IgG (H+L) secondary antibody-Alexa Fluor 594 (A-11012, dilution 1:1000; Thermo Fisher Scientific). Slides were coverslipped in Vectashield Hard Set mounting medium containing DAPI and evaluated using a Nikon TE-2000S microscope (Nikon Instruments Inc.). The fusion index was calculated as the ratio of fused nuclei within myotubes to the total number of nuclei. Fusion indices were compared between control and mutant cultures using the t-test.
Apoptosis Assays
QNMCs from Speg-KO and WT mice were plated onto 96-well plates at a density of either 1000 or 5000 cells per well. Apoptosis was measured at 0, 2, 4, 7, and 10 days after isolation using a Dual Apoptosis Assay with NucView 488 Caspase-3 Substrate & Annexin V (#30067; Biotium, Fremont, CA), per the manufacturer's instructions, and read on a single-tube luminometer (Berthold Instruments, Oak Ridge, TN). The average of three wells for each condition was recorded for each time point. Statistical differences between corrected values were compared using a one-way analysis of variance.
Cardiotoxin Injection
The right TA muscle of four isoflurane-anesthetized WT and Speg-KO mice at 30 days of life were injected with 15 to 20 μL of cardiotoxin (11061-96-4; Sigma-Aldrich, St. Louis, MO) diluted to a final concentration of 0.5 μg/μL in sterile PBS. At 5 to 6 days post-injection, the animals were euthanized, and the TA muscles were extracted and snap frozen in isopentane.
Data Analysis and Statistics
Results were analyzed with GraphPad Prism software version 8.0 (GraphPad Software, San Diego, CA), and expressed as means ± SD. The t-test was used to determine statistically significant differences between the WT and Speg-KO groups.
Results
Speg-KO Mice Have a Smaller SC Population
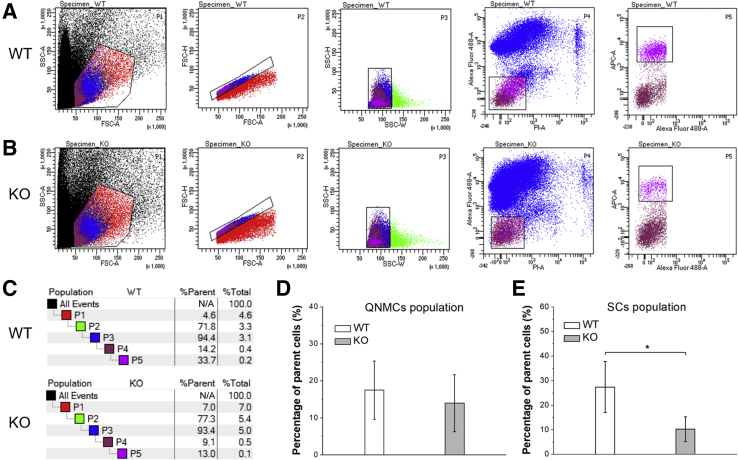
To determine whether the lack of SPEG causes defects in myogenic cells, a FACS strategy was applied to isolate myogenic cells from skeletal muscles of Speg-KO and littermate-matched WT control mice by negative selection for CD45, CD31, Sca1, and PI (corresponding to QNMCs), and positive selection for CD106 (corresponding to SCs). One million cells were loaded in each FACS experiment, and dead cells and doublets of cells were excluded by selected sorting gates. The relative percentage of QNMCs was slightly reduced in Speg-KO tissue (KO: 14.0% ± 7.7%; WT: 17.5% ± 7.9%) (Figure 1), although the difference was not statistically significant. By comparison, the relative percentage of SCs within the QNMC population was severely reduced in Speg-KO tissue (KO: 10.3% ± 5.0%; WT: 27.4% ± 10.4%; P < 0.05). Supplemental Table S1 lists the findings of each FACS experiment in terms of the relative percentage of QNMCs and SCs observed in Speg-KO mice compared with WT.
Figure 1.
FACS profiles of quadruple negative prospective myogenic cell (QNMC) and satellite cell (SC) populations in the skeletal muscles of wild-type (WT) and Speg knockout (KO) mice. The digested tissue from limb muscles of 1-month–old animals were stained with propidium iodide (PI), Alexa Fluor 488 anti-mouse CD31, Alexa Fluor 488 anti-mouse CD45, Alexa Fluor 488 anti-mouse Ly-6A/E (Sca1), and APC anti-mouse CD106 (VCAM1). A: WT profiles. B: KO profiles. The boxed areas in the P1 plots excludes cell debris; the boxed areas in both the P2 and P3 plots exclude the multiplets and doublets to collect single cells; cells in the boxed areas of the P4 and P5 plots are QNMCs and SCs, respectively.C: The population hierarchy of cells from WT and KO mice. D: The percentage of QNMCs in KO mice was comparable to that of WT. E: The percentage of SCs was markedly reduced in KO mice compared with WT n = 6 mice per genotype. ∗P < 0.05. FSC-A, forward scatter area; FSC-H, forward scatter height; SSC-A, side scatter area; SSC-H, side scatter height; SSC-W, side scatter width.
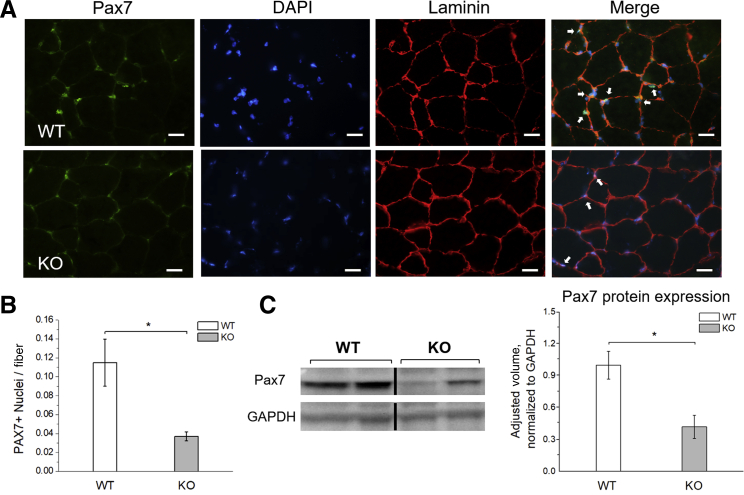
To further evaluate the abundance of SCs in Speg-KO mice, Pax7 immunostaining and immunoblotting were performed on quadriceps muscles from Speg-KO mice and littermate-matched WT at postnatal day 30 (Figure 2). Fewer SCs per fiber (KO: 3.7% ± 0.5%; WT: 11.5% ± 2.5%; P < 0.05) were identified by immunofluorescence and lower Pax7 amounts (KO: 42.0% ± 11.0% of WT values; P < 0.05) was detected by immunoblotting in Speg-KO mice compared with WT. Pax7 findings indicated a marked reduction in the SC population of Speg-KO mice, consistent with FACS experiments.
Figure 2.
Pax7 expression in wild-type (WT) and Speg knockout (KO) skeletal muscle. A: Immunofluorescence results for Pax7 staining in both WT and KO quadricep muscles at 30 days of life. White arrows indicate Pax7-positive nuclei in WT and KO animals. B: Quantification of Pax7-positive nuclei per fiber shows a reduction in the satellite cell population in the KO mice. C: Western blot analyses performed on tissue lysates from quadriceps muscle reveal decreased levels of Pax7 in KO animals. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a control. n = 4 mice per genotype. ∗P ˂ 0.05. Scale bars = 20 μm.
Speg Is Expressed in SCs
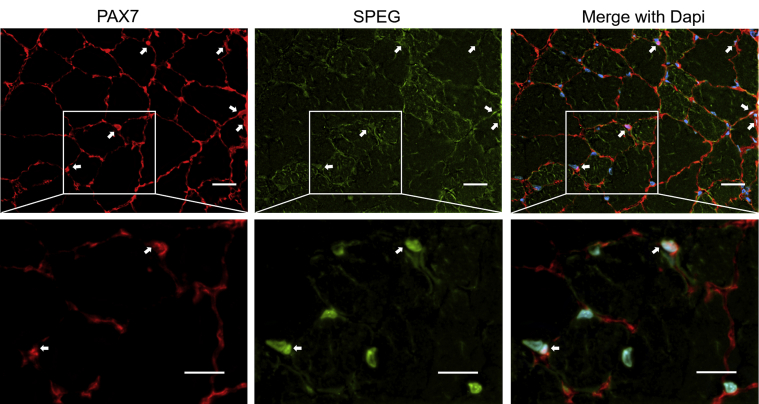
The expression of Speg in single cells from mouse muscle tissue excluding myotubes was evaluated. A publicly available single-cell RNA-seq (scRNA-seq) data set (Tabula Muris database27) classified 1090 cells from 3-month–old mouse skeletal muscles into six cell clusters: B and T lymphocytes, macrophages, endothelial cells, mesenchymal stem cells, and SCs (Supplemental Figure S1A). Multinucleated myotubes, which are typically longer than 100 μm, were excluded because of the restrictions on cell size that can be captured for further analysis (30 μm). Of the total 1090 cells, 540 (49.5%) revealed a gene expression pattern that could be assigned to SCs. Within this cluster, 22 cells (4.1%) were observed to express variable levels of Speg (Supplemental Figure S1, B and C). In the mesenchymal stem cell cluster, 8 of 258 cells (3.1%) were observed to express variable levels of the Speg. Speg was undetectable in B- and T-lymphocytes, macrophages, or endothelial cells. To test this further, the authors coimmunostained SPEG with Pax7 in WT muscle sections and confirmed the expression of SPEG in SCs (Supplemental Figure S2). Overall, scRNA-seq data obtained from dissociated hindlimb skeletal muscles, and the authors’ Pax7/SPEG coimmunostaining results indicate that SPEG is expressed in SCs.
SPEG-Deficient Myogenic Cells Display Slower Proliferation and Delayed Differentiation
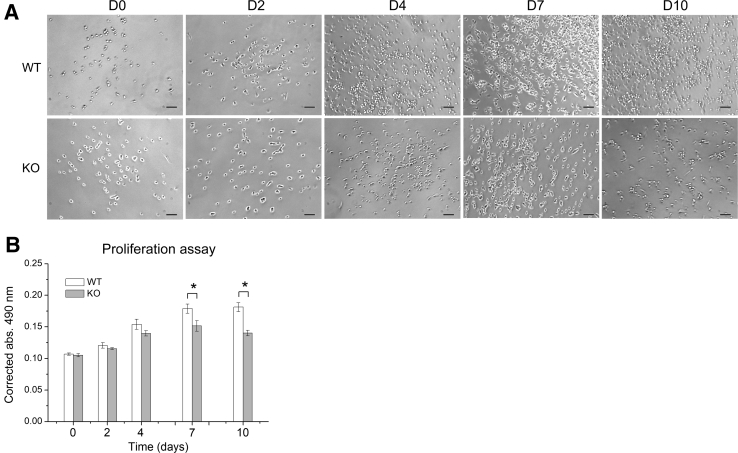
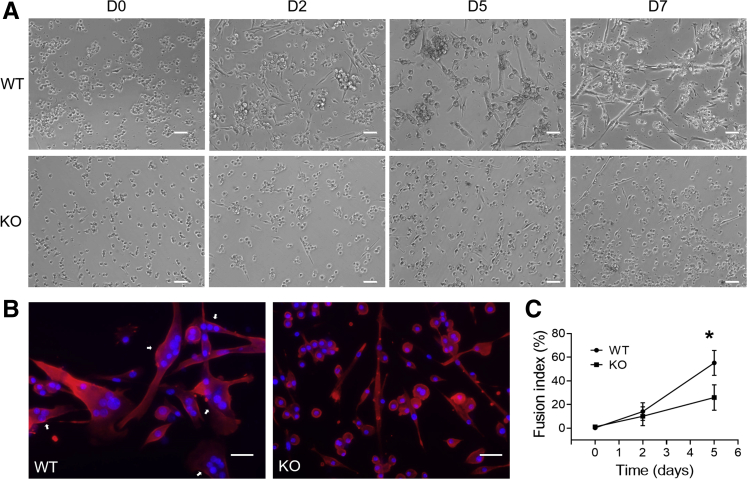
To investigate the properties of SPEG-deficient myogenic cells, isolated QNMCs from FACS experiments were used for cell proliferation, differentiation, and apoptosis tests in vitro. SPEG-deficient and WT myogenic cells were cultured in growth medium over 10 days, and their proliferation was measured using a cell proliferation assay. The number of SPEG-deficient myogenic cells was significantly decreased from WT after 7 days of cell culture (Figure 3). Next, the differentiation of SPEG-deficient myogenic cells and WT into myotubes was induced using differentiation medium and assessed over a 7-day period. The numbers of myotubes formed from Speg-KO cells were fewer than those from WT as time passed (Figure 4A). Fusion assays revealed fewer desmin-expressing myotubes from KO myogenic cells compared with WT cells by immunostaining, and the number of fused nuclei within KO myotubes was significantly less than that of WT (Figure 4B). The majority of WT myogenic cells (55.2% ± 10.6%) were able to differentiate into myotubes after 5-day differentiation medium culture, whereas markedly fewer Speg-KO myogenic cells (26.0% ± 10.8%) were able to differentiate into myotubes (Figure 4C).
Figure 3.
Reduced proliferation of SPEG-deficient quadruple negative prospective myogenic cells (QNMCs) in vitro. A: Phase contrast images of representative clones of wild-type (WT) (top row) and Speg knockout (KO) QNMCs (bottom row) in growth medium over a 10-day period. B: Proliferation rate is significantly lower in freshly isolated KO QNMCs compared with cells from WT littermates after 7 days in culture. n = 6 mice per genotype. ∗P < 0.05. Scale bars = 100 μm. abs, absorption; D, day.
Figure 4.
Delayed differentiation of SPEG-deficient quadruple negative prospective myogenic cells (QNMCs) in vitro. A: Representative phase contrast images of wild-type (WT) and Speg knockout (KO) QNMC clones in differentiation medium over a 7-day period. B: Representative images of immunofluorescence staining for desmin (red) after 5 days of differentiation. DAPI (blue) was used for nuclear staining. White arrows indicate fused nuclei within myotubes. C: Fusion assay reveals a lower fusion index for Speg-KO QNMCs after 5 days in culture. n = 6 mice per genotype. ∗P < 0.05 versus KO. Scale bars: 100 μm (A); 20 μm (B). D, day.
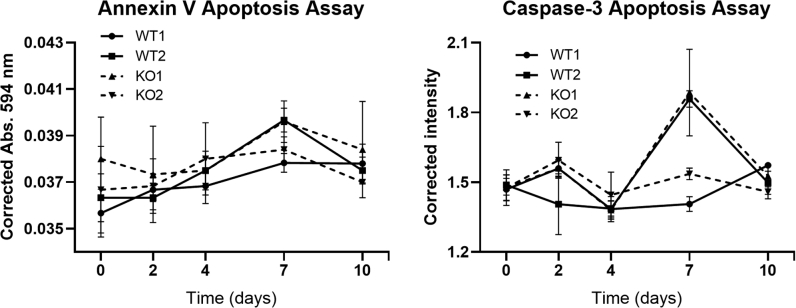
The difference in apoptosis between Speg-KO and WT myogenic cells was evaluated on days 0, 2, 4, 7, and 10 of culture using annexin V and caspase-3 apoptosis assays; and there was no significant difference between them (Supplemental Figure S3). Overall, the growth and differentiation potentials of myogenic cells were significantly reduced in SPEG-deficient cells relative to WT, but not apoptosis.
Speg-KO Skeletal Muscles Have Slower Healing and Increased Apoptosis in Response to Injury
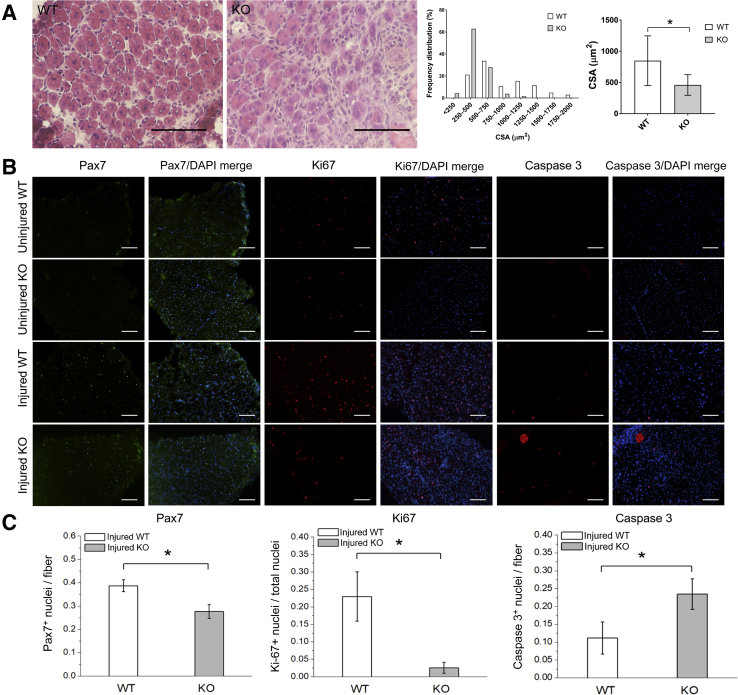
SCs play an important role in the repair and regeneration of skeletal muscles in response to injury, and SPEG-deficient SCs have defects in proliferation and differentiation in addition to being fewer in number. To assess whether SPEG deficiency would have an effect in vivo on skeletal muscle regeneration, cardiotoxin, a myonecrotic agent, was injected into the TA muscles. This produced a focal muscle injury, thereby stimulating the activation, proliferation, and differentiation of quiescent SC stores. Regenerating areas in injured TA muscles were readily detected 5 days after muscle injury by the presence of numerous centrally nucleated fibers in both Speg-KO and WT TA muscles, but the cross-sectional area of Speg-KO TA muscles was significantly less than that of WT muscles (Figure 5A). In addition, Speg-KO mice exhibited fewer SCs (Pax7+ nuclei) and proliferating cells (Ki-67+ nuclei), and more apoptotic cells (caspase3+ nuclei) on immunofluorescence analysis (Figure 5, B and C). Although not all proliferating and apoptotic cells in skeletal muscle regeneration may correspond to SCs, these results provided in vivo evidence of abnormal SC activity in Speg-KO mice, including abnormal levels of both proliferation and apoptosis.
Figure 5.
Reduced proliferation and increased apoptosis in SPEG-deficient cells after muscle injury in vivo. A: At 5 to 6 days after muscle injury by cardiotoxin injections, hematoxylin and eosin staining of the injured transverse abdominal (TA) muscles show signs of muscle regeneration, including numerous centrally nucleated myofibers. The cross-sectional area (CSA) of Speg-KO TA muscles is significant less than that of wild-type (WT) (over 100 fibers were analyzed from each group). B: Immunofluorescence staining of myofibers using antibodies against Pax7 (green) for satellite cells, Ki-67 (red) for proliferating cells, and caspase 3 (red) for apoptotic cells, show evidence of reduced proliferation and increased apoptosis in knockout (KO) mice. C: This was further confirmed by quantification. n = 3 mice per genotype. ∗P < 0.05. Scale bars = 100 μm (A and B).
Speg-KO Skeletal Muscles Have Fewer Activated SCs in Both Uninjured and Injured States
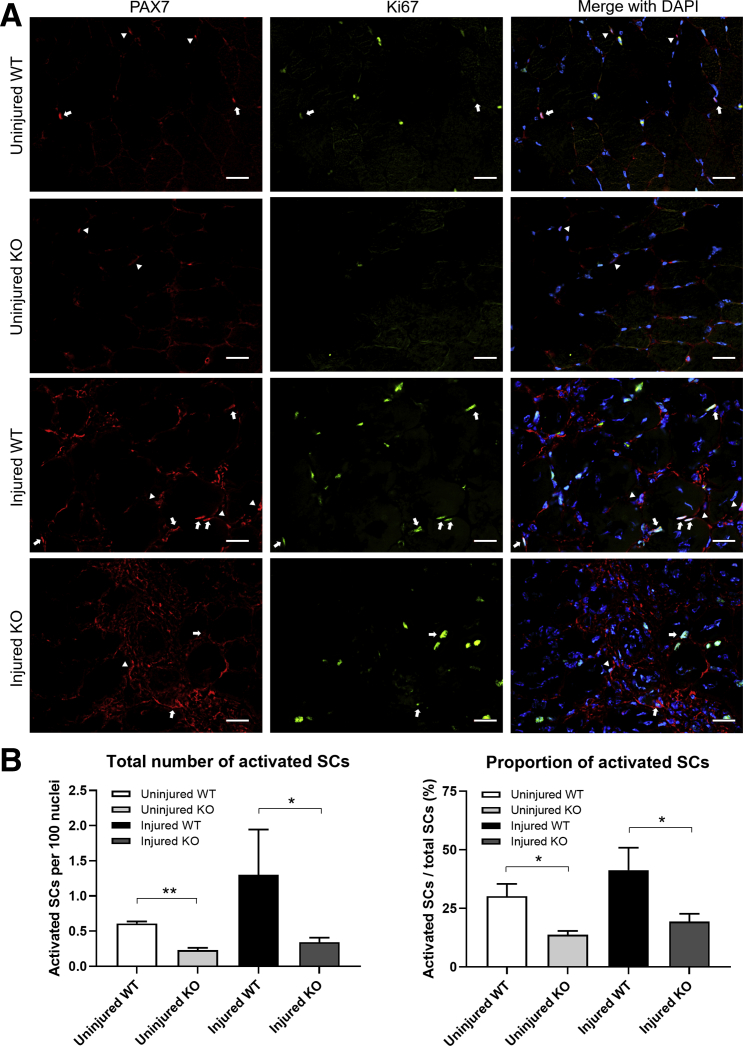
SCs can be activated in response to muscle injury, and activated SCs either are capable of self-renewing or can enter the myogenic pathway and differentiate to restore the damaged muscle.28,29 The authors have shown that Speg-KO mice have fewer SCs and exhibit slower healing when a muscle injury occurs. To evaluate the properties of activated SCs in SPEG-deficient muscles, the number of Pax7- and Ki-67–positive nuclei was evaluated before and after injury (Figure 6). A lower number of activated SC per 100 nuclei was seen in both uninjured (KO: 0.23 ± 0.03; WT: 0.61 ± 0.03; P < 0.01) and injured (KO: 0.34 ± 0.06; WT: 1.3 ± 0.64; P < 0.05) Speg-KO muscle. In addition, a smaller proportion of SCs were activated in both uninjured (KO: 13.7% ± 1.7%; WT: 30.2% ± 5.2%; P < 0.05) and injured (KO: 19.4% ± 3.4%; WT: 41.3% ± 9.6%; P < 0.05) Speg-KO compared with WT muscle. Overall, Pax7 and Ki-67 costaining findings demonstrated a significant decrease in the activated SCs of Speg-KO mice, which may explain the slower healing seen in Speg-KO mice.
Figure 6.
Fewer activated satellite cells in Speg knockout (KO) skeletal muscle before and after muscle injury in vivo. A: Immunofluorescence staining of myofibers using antibodies against Pax7 (red) and Ki-67 (green) shows fewer activated satellite cells in KO mice in both uninjured and injured states. Pax7 stains all satellite cells. Pax7 and Ki-67 together stain only the activated ones. Arrows indicate Pax7-positive nuclei; arrowheads indicate both Pax7- and Ki-67–positive nuclei. B: This was further confirmed by quantification. n = 3 mice per genotype. ∗P < 0.05, ∗∗P < 0.01. Scale bars = 20 μm. SC, satellite cell; WT, wild type.
Discussion
In this study, the impact of SPEG deficiency on myogenic stem cells and their function was evaluated. Previously, SPEG deficiency has been seen to lead to an abnormal triad structure, defective excitation–contraction coupling, and calcium mishandling,10 but its effects at the cellular level are unknown. To study this, the number of myogenic cells in SPEG-deficient skeletal muscle were first quantified using both FACS and Pax7 immunostaining and immunoblotting, and were seen to be markedly reduced. Proliferation and differentiation assays on myogenic cells revealed that SPEG-deficient myogenic cells displayed slow proliferation and delayed differentiation in vitro, indicating abnormal myogenic cell behavior in Speg-KO mice. To further evaluate these effects in vivo, a Speg-KO mouse model was utilized to analyze the effects of SPEG deficiency on muscle regeneration after injury. Speg-KO skeletal muscles healed more slowly after injury, with evidence of fewer SCs and dividing cells using Pax7 and Ki-67 staining.
A mouse model with postnatal striated muscle–specific excision of Speg using Mck-cre (Speg-KO) was utilized because constitutive Speg-KO mice die in utero.9 By delaying Speg excision using Mck-cre, the Speg-KO mice can survive up to 3 months of life.10 Mck-cre is normally detected in post-differentiation stages of myogenesis and identified in post-fusion myotubes starting from day 3 after induction of myogenic differentiation.30 It is therefore interesting to observe the reduction in SC number in Speg-KO mice, although QNMCs were not statistically different. The authors hypothesize that SPEG interacts with the extrinsic cues including extracellular matrix and basal lamina, which may affect SC survival or proliferation in the skeletal muscle.31,32 In addition, SPEG-deficient myogenic cells exhibited slower proliferation at day 7 of culture and not prior (Figure 3). By this time, the myoblasts became confluent, and started to show early differentiation. Analyzing the scRNA-seq data from in vitro human pluripotent stem cell–derived muscle cells shows that there is a small amount of MCK present in the myoblast stage that goes up with differentiation to myocytes (Supplemental Figure S4). The authors suggest that the MCK increase during late myoblast stage may explain the reduced proliferation in SPEG-deficient myogenic cells in vitro.
SPEG, MTM1, and DNM2 mutations are associated with CNM in humans, and a depletion of SCs has been previously described with MTM1-22,33,34 and DNM2-23,24 related mouse models. The defects seen in Speg-KO mice were similar to those in Mtm1 and Dnm2-mutant mouse models, with all three models displaying fewer SCs and delayed muscle regeneration in response to injury (Table 1).22, 23, 24 Furthermore, the proliferation rate in both SPEG- and MTM1-deficient myoblasts was reduced, but not described in Dnm2-mutant myoblasts. In addition, apoptosis was increased in both Speg- and Mtm1-KO mice following TA muscle injury in vivo. Apoptosis was not evaluated in Dnm2-mutant mice. Finally, the rate of differentiation in SPEG-deficient myoblasts was reduced, whereas it was unchanged in both Mtm1 and Dnm2-mutant myoblasts. Although the rate of differentiation in Mtm1 mutant myoblasts was not affected, it has been reported that MTM1 is involved with the final stage of myotube maturation.35 The reduced differentiation of SPEG-deficient myoblasts is likely due to a key role of SPEG with Spegα expression up-regulated throughout the muscle differentiation. Overall, SPEG, MTM1, and DNM2 may be critical for pathways involved in SC survival, maintenance, and function.
Table 1.
Summary of SC Defects in SPEG, MTM1, and DNM2-Mutant Mouse Models
| Mouse model | SC number | Myoblast (in vitro) |
Muscle regeneration (in vivo) |
Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Proliferation | Differentiation | Apoptosis | SC number | Proliferation | Apoptosis | |||
| Spegfl/fl: MCK-cre+ | ↓ | ↓ | ↓ | No change | ↓ | ↓ | ↑ | The current study |
| Mtm1−/y | ↓ | ↓ | No change | ↑ | ↓ | ↓ | ↑ | Lawlor et al22 |
| Dnm2R465W/+ | ↓ | Unknown | No change | Unknown | ↓ | Unknown | Unknown | Fongy et al,23 Almeida et al24 |
SC, satellite cell; ↓, decrease; ↑, increase.
The mechanisms by which SPEG deficiency reduces SC survival and disrupts their function is yet to be deciphered. SPEG is expressed in both SCs and mesenchymal stem cells,27 and Spegα is up-regulated during C2C12 cell differentiation,6 which suggests its role in myogenic cell differentiation. SPEG may influence SC activity through pathways mediating intracellular calcium homeostasis. The authors’ previous study suggests SPEG's critical role in regulating calcium handling during muscle contraction.10 Interestingly, calcium channel disruption has been reported to prevent rat SC activation and proliferation triggered by the mechanical stimulus in vitro.36 Further, perturbation of calcium release from internal stores impairs the skeletal muscle regeneration of the mouse injured TA muscle in vivo.37,38 Thus, SPEG's role in calcium handling may be critical for SC function and skeletal muscle regeneration.
In summary, the authors have shown that SPEG deficiency causes abnormalities in myogenic cell number and function, characterized by fewer SCs with reduced proliferation and delayed differentiation, and poor muscle regeneration after injury in vivo. These findings provide evidence that SPEG is critical for skeletal muscle development and regeneration. Future studies are needed to elucidate the underlying mechanisms of impaired SC survival and function due to SPEG deficiency.
Footnotes
Supported by NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant R01 AR068429 (P.B.A.).
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2020.08.012.
Supplemental Data
Supplemental Figure S1.
Publicly available single-cell RNA-seq (scRNA-seq) data from mouse skeletal muscles (Tabula Muris, https://tabula-muris.ds.czbiohub.org, last accessed June 6, 2020). A: Graph-based clustering of fluorescence-activated cell sorting (FACS)-isolated single cells identifies distinct clusters corresponding to different cell populations. B: Speg expression in distinct cell clusters. C: Variable levels of Speg expression among different clusters. CPM, counts per million mapped reads.
Supplemental Figure S2.
SPEG (green) colocalizes with Pax7 (red) in wild-type mouse skeletal muscles. White arrows indicate both SPEG- and Pax7-positive nuclei. Boxed areas are shown at higher magnification in the bottom row. Scale bars = 20 μm.
Supplemental Figure S3.
Apoptosis assay of wild-type (WT) and Speg knockout (KO) quadruple negative prospective myogenic cells at 0, 2, 4, 7, and 10 days in culture. n = 4 per genotype, each sample was tested in triplicate. Abs, absorption.
Supplemental Figure S4.
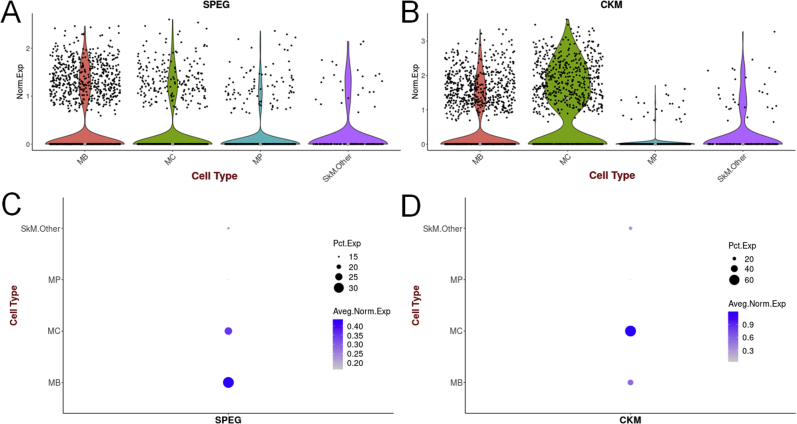
Publicly available single-cell RNA-seq (scRNA-seq) data from in vitro hPSC-derived muscle cells (UCSC Cell Browser; https://skeletal-muscle.cells.ucsc.edu/; last accessed August 6, 2020). A: SPEG expression in myogenic subset. B: CKM expression in myogenic subset. C: Variable levels of SPEG expression among different cell muscle groups. D: Variable levels of CKM expression among different cell muscle groups. CKM, creatine kinase, muscle; hPSC, human pluripotent stem cell; MB, myoblasts; MC, myocytes; MP, myogenic progenitors; SkM, skeletal muscle.
References
- 1.Jungbluth H., Gautel M. Pathogenic mechanisms in centronuclear myopathies. Front Aging Neurosci. 2014;6:339. doi: 10.3389/fnagi.2014.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phadke R. Myopathology of congenital myopathies: bridging the old and the new. Semin Pediatr Neurol. 2019;29:55–70. doi: 10.1016/j.spen.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Tasfaout H., Cowling B.S., Laporte J. Centronuclear myopathies under attack: a plethora of therapeutic targets. J Neuromuscul Dis. 2018;5:387–406. doi: 10.3233/JND-180309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal P.B., Pierson C.R., Joshi M., Liu X., Ravenscroft G., Moghadaszadeh B., Talabere T., Viola M., Swanson L.C., Haliloglu G., Talim B., Yau K.S., Allcock R.J., Laing N.G., Perrella M.A., Beggs A.H. SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy. Am J Hum Genet. 2014;95:218–226. doi: 10.1016/j.ajhg.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qualls A.E., Donkervoort S., Herkert J.C., D'Gama A.M., Bharucha-Goebel D., Collins J., Chao K.R., Foley A.R., Schoots M.H., Jongbloed J.D.H., Bonnemann C.G., Agrawal P.B. Novel SPEG mutations in congenital myopathies: genotype-phenotype correlations. Muscle Nerve. 2019;59:357–362. doi: 10.1002/mus.26378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh C.M., Fukumoto S., Layne M.D., Maemura K., Charles H., Patel A., Perrella M.A., Lee M.E. Striated muscle preferentially expressed genes alpha and beta are two serine/threonine protein kinases derived from the same gene as the aortic preferentially expressed gene-1. J Biol Chem. 2000;275:36966–36973. doi: 10.1074/jbc.M006028200. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh C.M., Yet S.F., Layne M.D., Watanabe M., Hong A.M., Perrella M.A., Lee M.E. Genomic cloning and promoter analysis of aortic preferentially expressed gene-1. Identification of a vascular smooth muscle-specific promoter mediated by an E box motif. J Biol Chem. 1999;274:14344–14351. doi: 10.1074/jbc.274.20.14344. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh C.M., Yoshizumi M., Endege W.O., Kho C.J., Jain M.K., Kashiki S., de los Santos R., Lee W.S., Perrella M.A., Lee M.E. APEG-1, a novel gene preferentially expressed in aortic smooth muscle cells, is down-regulated by vascular injury. J Biol Chem. 1996;271:17354–17359. doi: 10.1074/jbc.271.29.17354. [DOI] [PubMed] [Google Scholar]
- 9.Liu X., Ramjiganesh T., Chen Y.-H., Chung S.W., Hall S.R., Schissel S.L., Padera R.F., Jr., Liao R., Ackerman K.G., Kajstura J., Leri A., Anversa P., Yet S.-F., Layne M.D., Perrella M.A. Disruption of striated preferentially expressed gene locus leads to dilated cardiomyopathy in mice. Circulation. 2009;119:261–268. doi: 10.1161/CIRCULATIONAHA.108.799536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huntoon V., Widrick J.J., Sanchez C., Rosen S.M., Kutchukian C., Cao S., Pierson C.R., Liu X., Perrella M.A., Beggs A.H., Jacquemond V., Agrawal P.B. SPEG-deficient skeletal muscles exhibit abnormal triad and defective calcium handling. Hum Mol Genet. 2018;27:1608–1617. doi: 10.1093/hmg/ddy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quan C., Du Q., Li M., Wang R., Ouyang Q., Su S., Zhu S., Chen Q., Sheng Y., Chen L., Wang H., Campbell D.G., MacKintosh C., Yang Z., Ouyang K., Wang H.Y., Chen S. A PKB-SPEG signaling nexus links insulin resistance with diabetic cardiomyopathy by regulating calcium homeostasis. Nat Commun. 2020;11:2186. doi: 10.1038/s41467-020-16116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quan C., Li M., Du Q., Chen Q., Wang H., Campbell D., Fang L., Xue B., MacKintosh C., Gao X. SPEG controls calcium reuptake into the sarcoplasmic reticulum through regulating SERCA2a by its second kinase-domain. Circ Res. 2019;124:712–726. doi: 10.1161/CIRCRESAHA.118.313916. [DOI] [PubMed] [Google Scholar]
- 13.Quick A.P., Wang Q., Philippen L.E., Barreto-Torres G., Chiang D.Y., Beavers D., Wang G., Khalid M., Reynolds J.O., Campbell H.M. SPEG (striated muscle preferentially expressed protein kinase) is essential for cardiac function by regulating junctional membrane complex activity. Circ Res. 2017;120:110–119. doi: 10.1161/CIRCRESAHA.116.309977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., Hall S.R.R., Wang Z., Huang H., Ghanta S., Di Sante M., Leri A., Anversa P., Perrella M.A. Rescue of neonatal cardiac dysfunction in mice by administration of cardiac progenitor cells in utero. Nat Commun. 2015;6:8825. doi: 10.1038/ncomms9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacco A., Doyonnas R., Kraft P., Vitorovic S., Blau H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang N.C., Rudnicki M.A. Satellite cells: the architects of skeletal muscle. Curr Top Dev Biol. 2014;107:161–181. doi: 10.1016/B978-0-12-416022-4.00006-8. [DOI] [PubMed] [Google Scholar]
- 17.Kadi F., Charifi N., Denis C., Lexell J., Andersen J.L., Schjerling P., Olsen S., Kjaer M. The behaviour of satellite cells in response to exercise: what have we learned from human studies? Pflugers Arch. 2005;451:319–327. doi: 10.1007/s00424-005-1406-6. [DOI] [PubMed] [Google Scholar]
- 18.Relaix F., Zammit P.S. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139:2845–2856. doi: 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- 19.Moss F., Leblond C. Satellite cells as the source of nuclei in muscles of growing rats. Anat Rec. 1971;170:421–435. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- 20.Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 21.Serrano A.L., Baeza-Raja B., Perdiguero E., Jardí M., Muñoz-Cánoves P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008;7:33–44. doi: 10.1016/j.cmet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Lawlor M.W., Alexander M.S., Viola M.G., Meng H., Joubert R., Gupta V., Motohashi N., Manfready R.A., Hsu C.P., Huang P., Buj-Bello A., Kunkel L.M., Beggs A.H., Gussoni E. Myotubularin-deficient myoblasts display increased apoptosis, delayed proliferation, and poor cell engraftment. Am J Pathol. 2012;181:961–968. doi: 10.1016/j.ajpath.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fongy A., Falcone S., Laine J., Prudhon B., Martins-Bach A., Bitoun M. Nuclear defects in skeletal muscle from a Dynamin 2-linked centronuclear myopathy mouse model. Sci Rep. 2019;9:1580. doi: 10.1038/s41598-018-38184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almeida C., Bitoun M., Vainzof M. Satellite cell alteration in DNM2-related centronuclear myopathy. Neuromuscul Disord. 2017;27:S174. [Google Scholar]
- 25.Liu L., Cheung T.H., Charville G.W., Rando T.A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agrawal P.B., Joshi M., Savic T., Chen Z., Beggs A.H. Normal myofibrillar development followed by progressive sarcomeric disruption with actin accumulations in a mouse Cfl2 knockout demonstrates requirement of cofilin-2 for muscle maintenance. Hum Mol Genet. 2012;21:2341–2356. doi: 10.1093/hmg/dds053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabula Muris Consortium Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung T.H., Rando T.A. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuang S., Rudnicki M.A. The emerging biology of satellite cells and their therapeutic potential. Trends Mol Med. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Bi P., Yue F., Sato Y., Wirbisky S., Liu W., Shan T., Wen Y., Zhou D., Freeman J., Kuang S. Stage-specific effects of Notch activation during skeletal myogenesis. Elife. 2016;5:e17355. doi: 10.7554/eLife.17355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pannerec A., Marazzi G., Sassoon D. Stem cells in the hood: the skeletal muscle niche. Trends Mol Med. 2012;18:599–606. doi: 10.1016/j.molmed.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shichiji M., Biancalana V., Fardeau M., Hogrel J.Y., Osawa M., Laporte J., Romero N.B. Extensive morphological and immunohistochemical characterization in myotubular myopathy. Brain Behav. 2013;3:476–486. doi: 10.1002/brb3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawlor M.W., Viola M.G., Meng H., Edelstein R.V., Liu F., Yan K., Luna E.J., Lerch-Gaggl A., Hoffmann R.G., Pierson C.R., Buj-Bello A., Lachey J.L., Pearsall S., Yang L., Hillard C.J., Beggs A.H. Differential muscle hypertrophy is associated with satellite cell numbers and Akt pathway activation following activin type IIB receptor inhibition in Mtm1 p. R69C mice. Am J Pathol. 2014;184:1831–1842. doi: 10.1016/j.ajpath.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavriilidis C., Laredj L., Solinhac R., Messaddeq N., Viaud J., Laporte J., Sumara I., Hnia K. The MTM1-UBQLN2-HSP complex mediates degradation of misfolded intermediate filaments in skeletal muscle. Nat Cell Biol. 2018;20:198–210. doi: 10.1038/s41556-017-0024-9. [DOI] [PubMed] [Google Scholar]
- 36.Hara M., Tabata K., Suzuki T., Do M.-K.Q., Mizunoya W., Nakamura M., Nishimura S., Tabata S., Ikeuchi Y., Sunagawa K., Anderson J.E., Allen R.E., Tatsumi R. Calcium influx through a possible coupling of cation channels impacts skeletal muscle satellite cell activation in response to mechanical stretch. Am J Physiol Cell Physiol. 2012;302:C1741–C1750. doi: 10.1152/ajpcell.00068.2012. [DOI] [PubMed] [Google Scholar]
- 37.Sakuma K., Nishikawa J., Nakao R., Watanabe K., Totsuka T., Nakano H., Sano M., Yasuhara M. Calcineurin is a potent regulator for skeletal muscle regeneration by association with NFATc1 and GATA-2. Acta Neuropathol. 2003;105:271–280. doi: 10.1007/s00401-002-0647-0. [DOI] [PubMed] [Google Scholar]
- 38.Sakuma K., Nakao R., Aoi W., Inashima S., Fujikawa T., Hirata M., Sano M., Yasuhara M. Cyclosporin A treatment upregulates Id1 and Smad3 expression and delays skeletal muscle regeneration. Acta Neuropathol. 2005;110:269–280. doi: 10.1007/s00401-005-1049-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.