Abstract
Artificial Intelligence (AI) has surpassed dermatologists in skin cancer detection, but dermatology still lags behind radiology in its broader adoption. Building and using AI applications are becoming increasingly accessible. However, complex use cases may still require specialized expertise for design and deployment. AI has many applications in dermatology ranging from fundamental research, diagnostics, therapeutics, and cosmetic dermatology. The lack of standardization of images and privacy concerns are the foremost challenges stifling AI adoption. Dermatologists have a significant role to play in standardized data collection, curating data for machine learning, clinically validating AI solutions, and ultimately adopting this paradigm shift that is changing the way we practice.
Keywords: Artificial intelligence, machine learning, neural networks
Introduction
Artificial Intelligence (AI)—the intelligence possessed by machines—is having a profound impact on every aspect of the healthcare ecosystem, and dermatology is no exception.[1] AI introduces a paradigm shift—a fundamental change—in the way we practice making it necessary for every dermatologist to have a broad understanding of AI. The common question that crops up is how can dermatologists with no technical background understand and contribute to the evolution of AI, thereby helping our patients receive better care and outcomes. In this review, we attempt to answer these questions, simplify the AI jargon, deconstruct some common myths, and lay out some emerging directions in this field.
AI in dermatology for many is almost synonymous with melanoma classifiers.[2] The mole scanning mobile apps have been largely a success story, but the wider impact of AI on dermatology compared to other visually-oriented specialties like radiology has been disappointing.[3] AI can potentially find applications in areas other than skin cancer, including general diagnostic dermatology, cosmetic dermatology, fundamental research, and even practice optimization. We systematically analyze the reasons for this lack of adoption and offer some pragmatic recommendations.
Another common question is—Can AI replace dermatologists? The popular narrative is that AI can never replace us as it learns from us—from the data we collect and labels we assign. Cognitive computing and the other emerging techniques like reinforcement learning (RL) break this barrier; learning beyond available data. However, we do not attempt to change this narrative but offer an alternative perspective.
We adopt a narrow definition of AI, treating it as a synonym for machine learning (ML)—the study of algorithms and statistical models that computers use to perform a task.[4] The rest of the article is structured as follows. First, we introduce readers to the emergence of AI and the potential hype. Then, we describe the commonly used ML methods and the overarching taxonomy. Next, we summarize some existing applications and the typical steps in building and using AI. This is followed by a brief description of the testing and evaluation of AI, and the deployment of machine learning models. Next, we explain the integration of AI into clinical workflows and some common AI concepts such as deep learning, transfer learning, explainable AI, and cognitive computing. We will debunk some myths associated with AI, discuss some challenges in the widespread adoption of AI, and offer some pragmatic recommendations.
The Emergence of AI
AI tools, techniques, and algorithms have been in use for more than a decade. We reported the use of neural networks (NN) for the classification of epidermolysis bullosa way back in 2005.[5] The reemergence of AI is due to an unrelated discovery—the graphical programming unit (GPU) commonly used in gaming consoles that can speed up some of the calculations, several-fold. This led to the democratization of AI techniques, making it accessible to many research teams including those in healthcare.
Healthcare traditionally relies on inferential statistics, attempting to prove or disprove hypotheses that originate from the astute observation of patients. AI extends this approach by finding patterns that are not immediately discernible to the human observer. Pattern finding is easier on images than text because of the richness of data. Hence, AI's initial applications in healthcare were in image intensive specialties such as radiology, pathology, and dermatology.
It is likely that AI, especially in the context of dermatology, is currently in the phase of inflated expectations.[6] We may all experience a trough of disillusionment before we see practical applications of AI in widespread clinical use. However, when it matures, AI can potentially change the way we practice.
Taxonomy
There are several taxonomies and an ever-increasing list of AI algorithms and methods with specific uses. This granular taxonomy of methods and a detailed review of algorithms may not be of interest or utility to the clinicians. We introduce a simple taxonomy and a brief description of some of the AI methods at the risk of oversimplifications. We recommend some good references to those who wish to explore the methods further.[7,8,9]
Supervised methods
Simply put, supervised methods learn from examples. Supervised methods can differentiate melanoma from nonmelanoma if it is trained on a sufficient number of images of both, labelled by experts. Supervised methods are limited by the accuracy of the training set. If the experts make a systematic error in labelling the training images, the AI will make the same mistakes. Classification—assigning cases into predefined groups—is the quintessential supervised learning method.[10] Decision tree is another popular supervised method where a flow-chart like decision pathway is created with each node representing a classification problem.[11,12] Neural networks and deep learning are mostly classification methods that will be discussed in detail later.
Unsupervised methods
Unsupervised methods as the name suggest, find groups or patterns that are not predefined or known. Clustering is the most commonly used unsupervised method.[13] In simple terms, the clustering method identifies hidden groups based on all available data attributes. For example, you can use clustering to determine potential clusters among psoriasis patients resembling each other than those not in the cluster. The clustering algorithm can process all attributes such as the demographic factors, family, and treatment history. Once the algorithm defines the clusters, you can “profile” them into clusters that are likely to respond to PUVA and clusters that are not likely to respond, based on the clinical observations. Then, you can use classification (see the previous section) to assign your future psoriasis patients to one of these clusters for predicting treatment response and subsequent optimization of the treatment protocol. However, the clusters identified by the algorithm may have no clinical significance, and it is essential to use clinical judgement while interpreting the results. Clustering can also be used to find high-value clients in cosmetic dermatology practice for targeted marketing.
Reinforcement learning
The above methods are constrained by the amount of data available for the system to learn. The research on problems such as self-driving cars led to the emergence of the concept of cognitive computing—models that simulate human thought processes; and artificial general intelligence (AGI)—a hypothetical future state with the capacity to learn any intellectual task. For simplicity, we shall consider both these related but distinct concepts under the restricted category of reinforcement learning (RL).
In RL, instead of learning from data, an agent learns from the environment by taking actions that maximize some predefined reward.[14] The utility of RL depends on the accuracy with which the environment is modelled. One of our early attempts at modelling laser hair removal is an example of defining “an environment” though it does not extend it to RL.[15]
Neural Network (NN)
NN is not a distinct paradigm as the previous three. NN is a technique that can be classified under supervised methods. However, we discuss it, and the two popular subtypes of NN called deep learning (DL)[16] and convolutional neural networks (CNN)[17] as a separate category here, because of their relative importance in AI especially in relation to dermatology.
NNs are undoubtedly the most popular machine learning technique. NN is modelled based on neurons—with the nodes (synapses) firing when the input exceeds a certain threshold.[5] This simple process is scaled to a very high dimension and the firing thresholds are adjusted (learned) based on data. This probably is similar to how our brains function, though we do not know that for sure.
Neural networks have input, hidden, and output layers, with many nodes in each layer, represented by circles in Figure 1. The nodes are mathematical functions and during the training phase, data is fed through the input layer with each node taking the value of one of the features. If we consider the classification of bullous disorders as an example, one node could represent epidermal vs. dermal blister and another node could represent erythematous vs. nonerythematous base. The hidden layers fire based on their thresholds and finally the output layer [See Figure 1] indicates the predicted class, pemphigus vs. pemphigoid in our example (The absence of both could represent a third class). Typically, only one of the circles in the output layer will light up (if they were metaphorical light bulbs). During the initial training, the “bulbs” light up randomly. As the right class in the training data is known, any error in the classification is fed back to the mathematical functions using a process called backpropagation. This enables tweaking of the threshold for firing in hidden layers to reduce prediction errors (learning). The available data is fed through the network several times (epochs) until the error rate reduces to acceptable levels. This, “trained” network can then be used to predict the class (pemphigus vs. pemphigoid vs. other) for unknown data.
Figure 1.
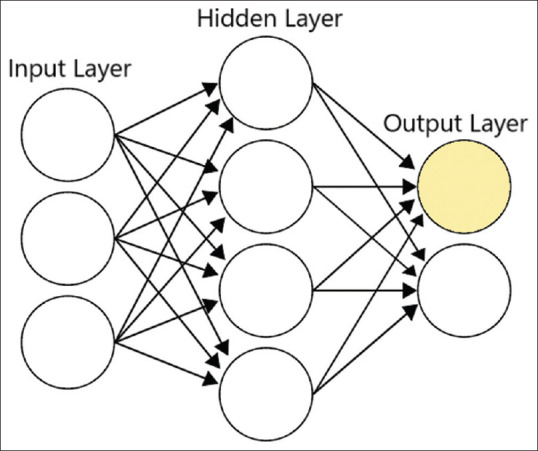
A symbolic representation of NN model with input, hidden, and output layers
In a simple NN, there are only one or a few intermediate layers of nodes between the input and the output. With the growing computational power, it is now possible to increase the number of intermediate layers of nodes, making the model more sensitive at the cost of increased computational complexity. Each layer progressively extracts higher-level features, but it is difficult for humans to interpret or comprehend—hence, described as “black boxes.” The process of estimating the associated weights (learning process) in a multilayered NN is called deep learning.
If the input consists of images, a method is needed to reduce it to binary input features such as “erythema vs. no erythema” as in the previous example. CNN is an innovative method for achieving this by combining small segments of images into a single entity as shown in Figure 2. A clinical image may contain many useful clinical features along with background noise such as clothing and furniture. The CNNs cannot distinguish clinical features from background noise initially, but with sufficient training CNNs may begin to recognize some clinical features. The numerical representation of features and the calculations are complex, but the fundamental principles remain the same as in all NNs—optimization of the firing threshold of the nodes.
Figure 2.
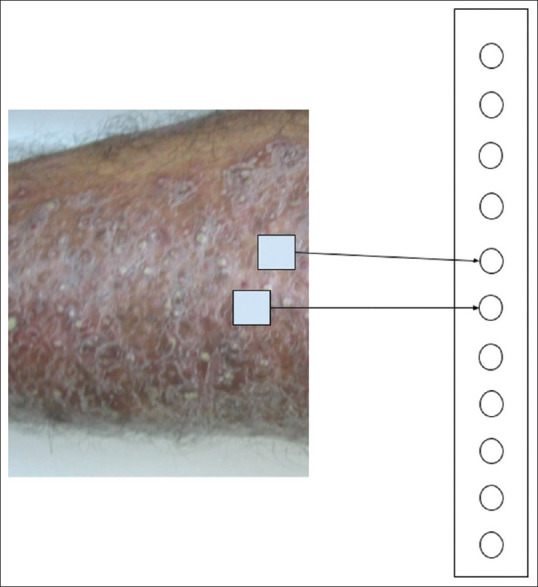
A symbolic representation of CNN converting an image to a vector. The intermediary layers are omitted for clarity
Table 1 summarizes the popular methods in machine learning. Next, we briefly describe some common applications of AI in dermatology.
Table 1.
Common machine learning methods
| Method | Prominent Types | Class | Explanation |
|---|---|---|---|
| Regression | Linear and Logistic[18] | Supervised | Estimates the relationship between independent and dependent variables. Logistic regression can also be used for classification. |
| Classification | Naive Bayes,[19] | Supervised | Assigns items into predefined groups. |
| Decision Tree,[11] | |||
| Support Vector Machine,[20] | |||
| Random Forest[21] | |||
| Mixed (Regression and classification) | Neural Networks,[5] | Supervised | Can be used to estimate the outcome or assign into outcome groups. |
| K-Nearest neighbor[22] | |||
| Clustering | K-Means,[23] | Unsupervised | Assign items into previously unknown groups. |
| Hierarchical[24] | |||
| Association | Apriori algorithm[25] | Unsupervised | Finds relationships between various attributes. |
| Dimensionality Reduction | Principal Component Analysis (PCA)[26] | Unsupervised | Reduces the number of attributes. |
| Gradient boosting | XGBoost[27] | Mixed | An ensemble of many learning algorithms. |
| Reinforcement | Q-Learning[28] | Reinforcement | An agent learns from the environment by trial and error. |
| Learning | SARSA[29] | Learning |
Dermatological Applications
As mentioned earlier, melanoma detection remains the most successful and impressive application[1] with various studies showing sensitivity and specificity similar to or in some cases surpassing human dermatologists.[30] AI methods have found to be useful in the segmentation of psoriasis lesions and their risk stratification.[31] Other innovative uses include the application of CNNs in automated acne vulgaris grading,[32] diagnosis of onychomycosis,[33] and in estimating the minimal phototoxic dose from skin color.[34] Recently the use of NNs has been extended beyond melanoma to other pigmented lesions and nonmelanoma skin cancers.[35]
Dermoscopy images are relatively more standardized than conventional digital images of the skin and, therefore, more amenable to machine learning.[36] Pigment networks, an indicator of melanoma, can be identified and image segmentation for detecting areas of interest can be performed with reasonable accuracy on dermoscopy images.[37] HAM10000 is a large dataset of the dermoscopic images of common pigmented skin lesions used for benchmarking AI algorithms in image analytics.[38]
AI finds applications in prognosis and disease monitoring.[39] It has been successfully used for automatic border detection of ulcers and area estimation.[40] The successful stratification of the sensitizing risk of therapeutic agents based on their chemical structure has been reported.[39] Predictive models based on the genetic features for various malignant and nonmalignant conditions have found to be useful. Models for predicting leishmaniasis,[41] foot ulcer progression,[42] and automated facial measurement in facial palsy have shown promising results.[43]
AI techniques can be used for natural language processing (NLP) of free text in documented patient history. NLP has been used on social media posts to evaluate the patient experiences for dermatology patients with various therapeutic interventions.[44] We have proposed a standardization protocol and a computerized tool (LesionMap) for documenting dermatological lesions in an innovative way that can be processed by both NLP techniques and image analytics.[45]
In cosmetic dermatology, AI can be used for skin type and skin tone classifiers, to create ageing and antiaging transformations to the face, and to render the anticipated results of a cosmetic procedure. AI can be a better and therapeutically useful alternative for skin type estimation than a questionnaire-based evaluation. Generative adversarial networks (GAN) is an emerging technology with many applications in rendering treatment effects on facial images.[46]
AI has applications in the realm of dermatopathology,[47] dermatological therapeutics, drug discovery, and fundamental research.[48] A detailed description of these domains is beyond the scope of this review.
Building and Using AI
As AI becomes increasingly ubiquitous, it is important for clinicians to have a high-level understanding of the processes involved in the development and deployment of AI applications. Developing an AI application consists of the following steps:
Identifying a problem that needs to be solved.
Collecting data in the form of images and/or text.
Labelling the data to locate areas of interest.
Building a machine learning model (In simple terms, the model is the representation of the “weights” attributed to each factor, required for prediction).
Testing and evaluating the model accuracy.
Ascertaining the clinical validity and ensuring patient safety.
Deploying the model on the cloud or in a web/mobile app.
Making it available for the users that can be patients or clinicians. Clinical applications are typically integrated into an electronic medical record (EMR).
Maintaining the accuracy of the model as more data become available.
Maintaining and troubleshooting the end-user application (mobile App or EMR).
The role of clinicians and data scientists
The clinicians are vital in the first three steps as described above. They also play a crucial role in the clinical validation and ensuring that the AI application is safe to use on patients. Every AI application may not be clinically useful and embarking on clinically irrelevant models may be a loss of time and resources. The images and clinical history collected electronically during the course of clinical practice are the treasure trove of data for model building. Concerted efforts to collate, organize, and label such data from multiple practitioners in a crowd-sourced manner may be very useful in developing useful AI systems.
A lot of computing resources are required for developing ML models from the collected data. The hyperparameter tuning—optimizing the model by adjusting its various parameters—is a highly specialized task that is best delegated to specialized labs or trained data scientists. It is a common mistake to entrust this task to software developers who may not have expertise in this domain. Software developers use the models built by data scientists. The bottom line is, any AI venture needs data scientists to maintain their models and software engineers to deploy them. For those technically inclined, we have summarized some of the tools and libraries in Table 2.
Table 2.
Popular tools for AI
| Tool | Category | Details |
|---|---|---|
| Tensorflow[49] | Python library | Free and open-source programming library for machine learning by Google. |
| Popular among machine learning experts. | ||
| PyTorch[50] | Python library | Maintained by Facebook. Known for dynamic graphs, commonly used by researchers. |
| Scikit-learn[51] | Python library | Another open-source python library that was in vogue before Tensorflow and PyTorch |
| Weka[52] | Java Application | Developed at the University of Waikato. |
| Popular in the research community. | ||
| Easy to use with a user-interface. | ||
| KNIME[53] | Java Application | An easy to use interface with drag-and-drop design for common machine learning workflows |
Python, Julia, RProgramming languagesGeneral-purpose programming languages commonly used in machine learning
Testing and evaluating models
Testing and evaluating ML models is a complex undertaking. Typically, some of the data is kept separate for testing. For example, while building a melanoma classifier from a large collection of images, the model is built from 80% of data, and the rest 20% is used to test how accurately the model can predict melanoma in these pictures. As these images were not used for building the model, the accuracy assessment on separate data unseen by the model during the building stage is likely to be more relevant. The assessment of clinical relevance is more challenging and a detailed description of various methods is beyond the scope, but we will try to debunk some common misconceptions below.
The accuracy of a model can be misleading clinically in some cases. It is not really useful if a model can predict a common disease like psoriasis with very high accuracy, but misses SCC in most cases. Accuracy can be misleading in predicting rare conditions. If a diagnosis has only a 0.1% chance, a fake model that always predicts its absence will be 99.9% accurate, just by chance. Hence, the accuracy of models and the sensitivity—the ability to reduce false negatives and specificity—the ability to reduce false positives—should be interpreted cautiously. Clinicians should also be aware of overfitting[54]—a model very accurate on training data, but less so when exposed to new data. Too much tuning can lead to overfitting, but there are statistical methods to reduce overfitting.
Model deployment
AI needs to be useful to clinicians at the point of care. The developed models need to be deployed on suitable infrastructure so that clinicians can get feedback about patients based on specific patient characteristics. Model deployment needs different hardware and software from that used for model building. Most models are deployed on the cloud and shared by many users. Cloud-based model deployments are easy to maintain as new models can be replaced and scaled easily to support increasing or decreasing user demand. Models can also be deployed locally in the user's computer or mobile phones. Though local deployment (sometimes called edge computing[55]) has some advantages such as availability in areas of low or no internet connectivity, the complexity of the models is limited because of the lower system resources and are difficult to maintain.
Integration
Imagine a situation where you have to log in to multiple systems during the course of a patient encounter to get feedback from various AI systems. The clinicians are known to be resistant to such a scenario and the possibility of information fatigue from too much data is a challenge. Hence, it is important to integrate AI applications with existing systems that clinicians use every day such as the EMR. Consumer AI applications for patients are usually delivered through mobile phone Apps. AI designers should be aware of the problem of proliferating the number of apps that patients need to install. Many of these mobile health (mHealth) apps may not be accurate or useful. It is also important to deliver useful information and alerts in a nonintrusive manner. Drishti is a software platform that we designed for such integration.[56]
Challenges in Widespread Adoption
There are several challenges in facilitating the widespread adoption of AI in dermatology. The clinical images in dermatology are often identifiable and raise grave privacy concerns.[57] Hence, it is difficult to obtain consent for collecting clinical images for machine learning. There is no universally accepted standard for capturing, processing, transmitting, and storing images in dermatology. In contrast, other domains such as radiology have standards such as Digital Imaging and Communications in Medicine (DICOM)[58] and systems such as picture archiving and communication system (PACS)[59] for standardizing the imaging workflow. DICOM standard embeds metadata such as the demographic details of the patient in the image. Though there was an effort at extending DICOM to dermatology, it still lacks widespread acceptance.[60] The imaging characteristics such as lighting, positioning, and resolution still remain nonstandard making it difficult to compare images from one source to another,[61] limiting the value for machine learning applications. Dermoscopic images are better in this regard with less variability.
The inherent bias in AI models due to the over-representation of certain skin types and skin color is another major limitation. Most AI models that are available today may not be useful in all parts of the world. This might exacerbate the already prevalent healthcare disparities in dermatology.[48]
Dermatologists should be cognizant of patient privacy whenever an image is captured. Try to restrict the field of capture to the lesion. Be extra careful with facial images, because of the risk of malicious facial recognition bots and algorithms.[62] A concerted effort at standardizing dermatological images or in adapting existing standards such as DICOM for dermatology is needed.[60] It is also important to reduce healthcare disparities and skin color bias in AI models, by developing high-quality image databases including dermatoscopic images.
Emerging Concepts
AI is constantly evolving and newer techniques are constantly being introduced. We briefly describe some terms that appear in AI articles related to dermatology.
Transfer learning
Building and tuning an image from scratch may need a large amount of data as well as computing power. As mentioned earlier, neural networks have several layers, each composed of multiple nodes. Weights of some of these layers can be drawn from other pretrained models, thereby transferring its knowledge to a different dataset.[63] As a practical example, a melanoma classifier can be built on top of a preexisting skin identifier model. In simple terms, the melanoma model draws from a general skin model. As the model is not built from scratch, less data is required. The number and configurations of the various NN layers are tricky and the performance of the model depends on them. There are architectural patterns that are known to perform well in a given situation such as GoogLeNet,[64] AlexNet,[64] VGGNet,[65] and ResNet.[66] In short, by choosing the appropriate architecture and weights, a good model can be created from a relatively small amount of data. The common perception that AI needs a huge amount of data is not true anymore.
Explainable AI
One of the criticisms of AI is that most of its methods and techniques cannot be understood by humans—most models are black boxes. This is true for NNs as it is difficult to explain why a model predicted a certain outcome. Inexplicability is a limitation in clinical applications. However, some of the other methods that we described such as a decision tree are intuitive and easily interpretable. There are emerging complex statistical techniques to make NN explainable as well.[67] Ongoing research in this field may lead to more transparent models in the future.
Big data and data mining
Big data is conventionally described in terms of 5Vs—high volume, variety, velocity, veracity, and value.[68] To a clinician, a more pragmatic description of big data would be data bigger than what can be handled by the ordinary computers available in the clinic. Data mining is the process of extracting knowledge from such large volumes of data. Data mining—generally performed by a data scientist—can give you insights that are not otherwise apparent, as opposed to working with a hypothesis in conventional data analysis. Data mining[69] is the initial step in machine learning and data mining methods are the same as those mentioned in Table 1.
Multimodal machine learning
Most machine-learning applications consume a single data type. For example, most melanoma classifiers use digital images, whereas NLP-based symptom checkers use text input. Clinicians combine information from various modalities—what they see and what they hear from the patient—in decision making. AI applications having a similar approach, such as combining clinical images with patient history, are called multimodal applications.[70] Multimodal algorithms are closer to the clinical workflow—confirming what we see with what we hear or elicit. Multimodal algorithms are generally more complex than the typical single modality applications.
Precision dermatology
Precision dermatology is an extension of precision medicine in which disease management is personalized taking into account factors such as the genetic, environmental, and behavioral characteristics of an individual. AI, aided by advances in sensors and genomic sequencing, may be a vital step in realizing the promise of precision dermatology.[71]
Digital pathology
Digital pathology is a well-established specialty with a substantial adoption of ML techniques.[72] Various computer vision and image analytics algorithms from machine learning have found application in digital pathology. Digitized pathology slides and collaboration tools built around the digitized images have emerged as a successful model applicable to dermatopathology and dermoscopy as well.[47]
Where Do We Go from Here?
We believe that accuracy cannot measure the utility of an AI application in clinical practice. More collaborative research on factors that make AI applications useful in clinical practice may promote its adoption—a subject we seek to investigate in our research. Emerging concepts such as transfer learning and multimodal approach will advance the sensitivity and specificity of existing algorithms.
Instead of predicting the future, we lay out some emerging directions in this field. We see AI emerging as a triaging tool for clinicians, especially in places where dermatology services are not readily available. Melanoma screening tools may further improve putting it at par with trained dermatologists and available to the masses. We also anticipate skincare AI becoming democratized with services that are easy to use from existing social media platforms. Though AI is unlikely to replace dermatologists, it may completely alter the way we practice this specialty. Some questions that are considered important such as—can AI make the right diagnosis—may lose relevance, as there may be more diagnostic entities in the machine world than we can comprehend.
Dermatologists can play a big role in the evolution of AI. Collecting, standardizing, and tagging data in a privacy-preserving manner is the need of the hour. Leadership by professional bodies such as the Indian Association of Dermatologists, Venereologists and Leprologists (IADVL), gamification (integrating game mechanics to motivate participation),[73] and incorporating AI into the post-graduate training curriculum can improve adoption. It is important to realize that AI has a technical component and needs a multidisciplinary team to build and effectively deploy it for widespread use. Collaboration with technical colleagues should be facilitated along with encouraging the technically inclined E-Health hybrids[74] among dermatologists.
The capabilities of AI may be over-hyped at present. There is a growing tendency to portray AI as the panacea for all the woes. It is important for us to make sure that AI applications are realistic and we do not expect it to do everything. After the trough of disillusionment, AI in some form will stay on and change the way we practice dermatology. One of the challenges in the adoption of any information system is the resistance in embracing change and AI is no exception.[74] It is vital for dermatologists to accept AI as a disruptive change but at the same time realize that AI can never be the “human” behind the white coat.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Du-Harpur X, Watt FM, Luscombe NM, Lynch MD. What is AI Applications of artificial intelligence to dermatology. Br J Dermatol. 2020 doi: 10.1111/bjd.18880. doi: 10.1111/bjd.18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sboner A, Eccher C, Blanzieri E, Bauer P, Cristofolini M, Zumiani G, et al. A multiple classifier systems for early melanoma diagnosis. Artif Intell Med. 2003;27:29–44. doi: 10.1016/s0933-3657(02)00087-8. [DOI] [PubMed] [Google Scholar]
- 3.Li CX, Shen CB, Xue K, Shen X, Jing Y, Wang ZY, Xu F, et al. Artificial intelligence in dermatology: Past, present, and future. Chin Med J. 2019;132:2017–20. doi: 10.1097/CM9.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop CM. Pattern Recognition and Machine Learning. New York: Springer; 2016. [Google Scholar]
- 5.Eapen BR. ‘Neural network’ algorithm to predict severity in epidermolysis bullosa simplex. Indian J Dermatol Venereol Leprol. 2005;71:106–8. doi: 10.4103/0378-6323.13995. [DOI] [PubMed] [Google Scholar]
- 6.O'Leary DE. Gartner's hype cycle and information system research issues. Int J Account Inf Syst. 2008;9:240–52. [Google Scholar]
- 7.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–30. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alanazi HO, Abdullah AH, Qureshi KN. A critical review for developing accurate and dynamic predictive models using machine learning methods in medicine and health care. J Med Syst. 2017;41:69. doi: 10.1007/s10916-017-0715-6. [DOI] [PubMed] [Google Scholar]
- 9.Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: A practical introduction. BMC Med Res Methodol. 2019;19:64. doi: 10.1186/s12874-019-0681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arasi MA, El-Horbaty E-SM, El-Sayed A. 1st AiCIS. IEEE; 2018. Classification of dermoscopy images using naive bayesian and decision tree techniques; pp. 7–12. [Google Scholar]
- 12.Eapen BR. Malignancy in dermatomyositis: A Bayesian Belief Network approach. Indian J Dermatol Venereol Leprol. 2007;73:445. [Google Scholar]
- 13.Fidan U, Ozkan N, Calikusu I. TIPTEKNO. IEEE; 2016. Clustering and classification of dermatologic data with Self Organization Map (SOM) method; pp. 1–4. [Google Scholar]
- 14.Akrout M, Farahmand A-M, Jarmain T, Abid L. International Conference on Med Image Comput Comput Assist Interv. Springer; 2019. Improving skin condition classification with a visual symptom checker trained using reinforcement learning; pp. 549–57. [Google Scholar]
- 15.Eapen BR. Agent-based model of laser hair removal: A treatment optimization and patient education tool. Indian J Dermatol Venereol Leprol. 2009;75:383–7. doi: 10.4103/0378-6323.53135. [DOI] [PubMed] [Google Scholar]
- 16.Yap J, Yolland W, Tschandl P. Multimodal skin lesion classification using deep learning. Exp Dermatol. 2018;27:1261–7. doi: 10.1111/exd.13777. [DOI] [PubMed] [Google Scholar]
- 17.Brinker TJ, Hekler A, Enk AH, Klode J, Hauschild A, Berking C, et al. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur J Cancer. 2019;111:148–54. doi: 10.1016/j.ejca.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Tyrrell J, Paterson C, Curnow A. Regression analysis of protoporphyrin IX measurements obtained during dermatological photodynamic therapy. Cancers (Basel) 2019;11:72. doi: 10.3390/cancers11010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Priya V, Sathya N. Classification and prediction of dermatitis dataset using naive bayes and value weighted naive bayes algorithms. Int Res J Eng Technol. 2019;6:1077–81. [Google Scholar]
- 20.Jiang B, Cui L, Zi Y, Jia Y, He C. Skin surface lipid differences in sensitive skin caused by psychological stress and distinguished by support vector machine. J Cosmet Dermatol. 2019;18:1121–7. doi: 10.1111/jocd.12793. [DOI] [PubMed] [Google Scholar]
- 21.Kaur P, Kumar R, Kumar M. A healthcare monitoring system using random forest and internet of things (IoT) Multimed Tools Appl. 2019;78:19905–16. [Google Scholar]
- 22.Datta S, Misra D, Das S. A feature weighted penalty based dissimilarity measure for k-nearest neighbour classification with missing features. Pattern Recognit Lett. 2016;80:231–7. [Google Scholar]
- 23.Anas M, Gupta K, Ahmad S. Skin cancer classification using k-means clustering. Int J Tech Res Appl. 2017;5:62–5. [Google Scholar]
- 24.Jung YS, Bae JM, Kim BJ, Kang J-S, Cho SB. Periorbital melasma: Hierarchical cluster analysis of clinical features in Asian patients. Skin Res Technol. 2017;23:552–7. doi: 10.1111/srt.12370. [DOI] [PubMed] [Google Scholar]
- 25.Lee DE, Yoon T. Analysis of high-risk human papillomavirus using decision tree and apriori algorithm. Proceedings of the 2nd ICCBB. 2018:22–6. [Google Scholar]
- 26.He YF, Wang RZ, Meng H, Li L, Wu Z, Dong YM. Establishment of a PCA model for skin health evaluation. Biotechnol Biotechnol Equip. 2018;32:1060–4. [Google Scholar]
- 27.Pham HN, Koay CY, Chakraborty T, Gupta S, Tan BL, Wu H, et al. ICSSE. IEEE; 2019. Lesion segmentation and automated melanoma detection using deep convolutional neural networks and XGBoost; pp. 142–7. [Google Scholar]
- 28.Krakow EF, Hemmer M, Wang T, Logan B, Arora M, Spellman S, et al. Tools for the precision medicine era: How to develop highly personalized treatment recommendations from cohort and registry data using Q-learning. Am J Epidemiol. 2017;186:160–72. doi: 10.1093/aje/kwx027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebrahimi MS, Pourghassem H, Ashourian M. 17th ICBME. IEEE; 2010. Lesion detection in dermoscopy images using SARSA reinforcement algorithm; pp. 1–4. [Google Scholar]
- 30.Fujisawa Y, Otomo Y, Ogata Y, Nakamura Y, Fujita R, Ishitsuka Y, et al. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br J Dermatol. 2019;180:373–81. doi: 10.1111/bjd.16924. [DOI] [PubMed] [Google Scholar]
- 31.Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput Methods Programs Biomed. 2017;150:9–22. doi: 10.1016/j.cmpb.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Shen X, Zhang J, Yan C, Zhou H. An automatic diagnosis method of facial acne vulgaris based on convolutional neural network. Sci Rep. 2018;8:1–10. doi: 10.1038/s41598-018-24204-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han SS, Park GH, Lim W, Kim MS, Na JI, Park I, et al. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: Automatic construction of onychomycosis datasets by region-based convolutional deep neural network. PLoS One. 2018;13:e0191493. doi: 10.1371/journal.pone.0191493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubegni P, Cevenini G, Flori ML, Barbini P, Andreassi L. Relationship between minimal phototoxic dose and skin colour plus sun exposure history: A neural network approach. Photodermatol Photoimmunol Photomed. 1998;14:26–30. doi: 10.1111/j.1600-0781.1998.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 35.Tschandl P, Codella N, Akay BN, Argenziano G, Braun RP, Cabo H, et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: An open, web-based, international, diagnostic study. Lancet Oncol. 2019;20:938–47. doi: 10.1016/S1470-2045(19)30333-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez AR, Giro-i-Nieto X, Burdick J, Marques O. 13th IASTED. IEEE; 2017. Skin lesion classification from dermoscopic images using deep learning techniques; pp. 49–54. [Google Scholar]
- 37.García Arroyo JL, García Zapirain B. Detection of pigment network in dermoscopy images using supervised machine learning and structural analysis. Comput Biol Med. 2014;44:144–57. doi: 10.1016/j.compbiomed.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Tschandl P, Rosendahl C, Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data. 2018;5:180161. doi: 10.1038/sdata.2018.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomsen K, Iversen L, Titlestad TL, Winther O. Systematic review of machine learning for diagnosis and prognosis in dermatology. J Dermatolog Treat. 2020;31:496–510. doi: 10.1080/09546634.2019.1682500. [DOI] [PubMed] [Google Scholar]
- 40.Veredas FJ, Luque-Baena RM, Martín-Santos FJ, Morilla-Herrera JC, Morente L. Wound image evaluation with machine learning. Neurocomputing. 2015;164:112–22. [Google Scholar]
- 41.Mollalo A, Sadeghian A, Israel GD, Rashidi P, Sofizadeh A, Glass GE. Machine learning approaches in GIS-based ecological modeling of the sand-fly Phlebotomus Papatasi, a vector of zoonotic cutaneous leishmaniasis in Golestan province, Iran. Acta Trop. 2018;188:187–94. doi: 10.1016/j.actatropica.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Goyal M, Reeves ND, Davison AK, Rajbhandari S, Spragg J, Yap MH. TETCI Conference. IEEE; 2018. Dfunet: Convolutional neural networks for diabetic foot ulcer classification; pp. 1–12. [Google Scholar]
- 43.Guarin DL, Dusseldorp J, Hadlock TA, Jowett N. A machine learning approach for automated facial measurements in facial palsy. JAMA Facial Plast Surg. 2018;20:335–7. doi: 10.1001/jamafacial.2018.0030. [DOI] [PubMed] [Google Scholar]
- 44.Okon E, Rachakonda V, Hong HJ, Callison-Burch C, Lipoff JB. Natural language processing of Reddit data to evaluate dermatology patient experiences and therapeutics? J Am Acad Dermatol. 2019:S0190. doi: 10.1016/j.jaad.2019.07.014. doi: 10.1016/j.jaad. 2019.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Eapen BR, Archer N, Sartipi K. LesionMap: A method and tool for the semantic annotation of dermatological lesions for documentation and machine learning. JMIR Dermatol. 2020;3:e18149. [Google Scholar]
- 46.Diamant N, Zadok D, Baskin C, Schwartz E, Bronstein AM. ICIP. IEEE; 2019. Beholder-Gan: Generation and Beautification of Facial Images with Conditioning on Their Beauty Level; pp. 739–43. [Google Scholar]
- 47.Olsen TG, Jackson BH, Feeser TA, Kent MN, Moad JC, Krishnamurthy S, et al. Diagnostic performance of deep learning algorithms applied to three common diagnoses in dermatopathology. J Pathol Inform. 2018;9:32–3. doi: 10.4103/jpi.jpi_31_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adamson AS, Smith A. Machine learning and health care disparities in dermatology. JAMA Dermatol. 2018;154:1247–8. doi: 10.1001/jamadermatol.2018.2348. [DOI] [PubMed] [Google Scholar]
- 49.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. Tensorflow: A system for large-scale machine learning. 12th USENIX Symposium. OSDI – 16. 2016:265–83. [Google Scholar]
- 50.Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. Adv Neural Inf Process Syst. MIT Press; 2019. Pytorch: An imperative style, high-performance deep learning library; pp. 8024–35. [Google Scholar]
- 51.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine learning in python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 52.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: An update. SIGKDD Explor. 2009;11:10–8. [Google Scholar]
- 53.Berthold MR, Cebron N, Dill F, Gabriel TR, Kotter T, Meinl T, et al. KNIME-the Konstanz Information Miner: Version 2.0 and beyond. SIGKDD Explor. 2009;11:26–31. [Google Scholar]
- 54.Dietterich T. Overfitting and under computing in machine learning. ACM Comput Surv. 1995;27:326–7. [Google Scholar]
- 55.Shi W, Cao J, Zhang Q, Li Y, Xu L. Edge computing: Vision and challenges. IEEE Internet Things J. 2016;3:637–46. [Google Scholar]
- 56.Eapen BR, Archer N, Sartipi K, Yuan Y. IEEE/ ACM 1st International Workshop (SEH) Vol. 2019. IEEE; 2019. Drishti: A sense-plan-act extension to open mHealth framework using FHIR; pp. 49–52. [Google Scholar]
- 57.Kunde L, McMeniman E, Parker M. Clinical photography in dermatology: Ethical and medico-legal considerations in the age of digital and smartphone technology. Aust J Dermatol. 2013;54:192–7. [Google Scholar]
- 58.Mildenberger P, Eichelberg M, Martin E. Introduction to the DICOM standard. Eur Radiol. 2002;12:920–7. doi: 10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- 59.Huang HK. PACS-Based Multimedia Imaging Informatics: Basic Principles and Applications. New Jersey: Wiley; 2019. [Google Scholar]
- 60.Chartier A. DICOM (Digital Imaging and Communications in Medicine) in dermatology Teledermatology. London: The Royal Society of Medicine Press Limited; 2002. [Google Scholar]
- 61.Scheinfeld N. Photographic images, digital imaging, dermatology, and the law. Arch Dermatol. 2004;140:473–6. doi: 10.1001/archderm.140.4.473. [DOI] [PubMed] [Google Scholar]
- 62.Brundage M, Avin S, Clark J, Toner H, Eckersley P, Garfinkel B, et al. The malicious use of artificial intelligence: Forecasting, prevention, and mitigation. arXiv preprint. 2018 arXiv: 1802.07228. [Google Scholar]
- 63.Torrey L, Shavlik J. Transfer learning In Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques Pennsylvania IGI Global. 2010:242–64. [Google Scholar]
- 64.Ballester P, Araujo RM. Thirtieth AAAI Conference on Artificial Intelligence. AAAI; 2016. On the performance of GoogLeNet and AlexNet applied to sketches; pp. 1124–8. [Google Scholar]
- 65.Guo S, Luo Y, Song Y. Random forests and VGG-NET: An algorithm for the ISIC 2017 skin lesion classification challenge. arXiv preprint. 2017 arXiv: 1703.05148. [Google Scholar]
- 66.Budhiman A, Suyanto S, Arifianto A. 2019 – ISRITI. IEEE; 2019. Melanoma cancer classification using ResNet with data augmentation; pp. 17–20. [Google Scholar]
- 67.Holzinger A. World Symposium on DISA. IEEE; 2018. From machine learning to explainable AI; pp. 55–66. [Google Scholar]
- 68.Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA. 2013;309:1351–2. doi: 10.1001/jama.2013.393. [DOI] [PubMed] [Google Scholar]
- 69.Barati E, Saraee MH, Mohammadi A, Adibi N, Ahmadzadeh MR. A survey on utilization of data mining approaches for dermatological (skin) diseases prediction. JSHI. 2011;2:1–11. [Google Scholar]
- 70.Baltrusaitis T, Ahuja C, Morency L-P. Multimodal machine learning: A survey and taxonomy. IEEE Trans. Pattern Anal Mach Intell. 2018;41:423–43. doi: 10.1109/TPAMI.2018.2798607. [DOI] [PubMed] [Google Scholar]
- 71.Wongvibulsin S, Ho BK-T, Kwatra SG. Embracing machine learning and digital health technology for precision dermatology. J Dermatolog Treat. 2020;31:491–5. doi: 10.1080/09546634.2019.1623373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madabhushi A, Lee G. Image Analysis and Machine Learning in Digital Pathology: Challenges and Opportunities. London: Elsevier; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balducci F, Buono P. Building a qualified annotation dataset for skin lesion analysis trough gamification. Proceedings of the ICAVI. 2018:1–5. [Google Scholar]
- 74.Heeks R. Health information systems: Failure, success and improvisation. Int J Med Inform. 2006;75:125–37. doi: 10.1016/j.ijmedinf.2005.07.024. [DOI] [PubMed] [Google Scholar]


