Abstract
HLA-DP mismatched allogeneic hematopoietic stem cell transplantation (allo-HCT) is associated with increased risk of aGVHD and decreased risk of relapse with no effects on overall survival (OS). It has been proposed that CMV-reactivation induces expression of HLA-DP molecules on GVHD target tissues by releasing inflammatory cytokines. We hypothesized that the increased GVHD incidence in HLA-DP mismatched allo-SCTs correlates with recipient CMV serostatus or CMV reactivation. In addition, CMV reactivation is associated with increased risk of GVHD with an unknown mechanism. Here, we analyzed the association between HLA-DPB1 and CMV reactivation on cumulative incidence of aGVHD and relapse as well as OS in 613 patients with AML and MDS who underwent matched related or unrelated allo-HCT at MD Anderson Cancer Center from 2005 to 2011. In multivariable analysis, HLA-DPB1 mismatching was associated with increased risk of aGVHD (hazard ratio (HR): 1.53, P<0.001) independent of CMV serostatus and CMV reactivation. Additionally, HLA-DPB1 mismatching was associated with decreased risk of relapse and no effect on OS. CMV reactivation increased risks of aGVHD (HR: 5.82, P<0.001) independent of HLA-DP mismatching with no effect on relapse or OS. In conclusion, our data suggests that HLA-DPB1 mismatching and CMV reactivation increase risk of aGVHD independently.
Keywords: Allogeneic hematopoietic stem cell transplantation, CMV, HLA-DP
INTRODUCTION
Allogeneic hematopoietic stem cell transplantation (allo-HCT) is a potentially curative treatment for a broad spectrum of hematological malignancies. The current standard for a fully matched allo-HCT is transplant from a donor matched at HLA-A, HLA-B, HLA-C, and HLA-DRB1. HLA-DP mismatching is associated with higher rate of grade II-IV acute graft versus host disease (aGVHD) and lower relapse rate resulting in no impact on overall survival (OS).1, 2 For this reason, many centers do not take matching for HLA-DP into consideration when selecting an unrelated donor. HLA-DP antigens are αβ heterodimers encoded by the genes of two loci: (1) DPA1 locus, which has limited polymorphism, and (2) DPB1 locus which is highly polymorphic, with 520 alleles coding for 424 different proteins.3 HLA-DPB1 is a low expression locus (LEL) with both constitutive and inducible expression. Its constitutive expression is restricted to only thymic epithelial cells, antigen-presenting cells such as dendritic cells and mononuclear phagocytes as well as activated T cells and B cells. HLA-DPB1 expression can be induced in other tissues after exposure to interferon gamma and other cytokines.4, 5
Stevanov’c et al have previously demonstrated a link between CMV reactivation and HLA-DPB1 directed aGVHD. Their study suggested that CMV reactivation induces HLA-DPB1 expression resulting in HLA-DPB1 directed GVHD after HLA-DPB1-mismatched CD4+ donor lymphocyte infusion (DLI).4, 6 No study has so far tested whether the increased aGVHD risk in HLA-DPB1 mismatched allo-HCTs correlates with recipient CMV serostatus, or more importantly, CMV reactivation. In addition, CMV reactivation is associated with increased risk of aGVHD with an unknown mechanism.6–8 No study has analyzed whether the increased risk of aGVHD in patients with CMV reactivations is restricted to HLA-DPB1 mismatched allo-HCTs. Here in this retrospective study, we demonstrate that increased risk of aGVHD in HLA-DPB1 mismatched allo-HCT is independent of CMV reactivation. We also demonstrate that CMV reactivation increases risk of aGVHD independently of HLA-DPB1 matching status.
MATERIALS AND METHODS
We retrospectively evaluated all adult patients with AML or MDS who received matched related or unrelated allo-HCT at MD Anderson Cancer Center (MDACC) from January 2005 to December 2011 (total: 613 patients; HLA-DPB1 matched: 363 [59%], and HLA-DPB1 mismatched: 250 [41%]). All patients were matched at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 (HLA level matching of 10/10). HLA typing was based on allelic typing. Patients receiving umbilical cord or haploidentical stem cell transplants, patients who died within 30 days of allo-HCT, and those less than 18 years of age were excluded from the analysis. All patients underwent weekly surveillance by CMV pp65 antigen testing from the time of engraftment and at least until day 100 post allo-HCT. CMV reactivation was defined as presence of > 1pp65 Ag cells/million WBC’s. Preemptive therapy was initiated for patients with > 3 pp65 Ag cells/million WBC’s. Clinical outcomes of interest included OS as well as cumulative incidences (CI) of GVHD, non-relapse mortality (NRM) and relapse. Severity of aGVHD was defined according to Glucksberg criteria.9 OS was estimated using the Kaplan-Meier method and the association between OS and HLA-DPB1 mismatching was determined using a Cox proportional hazards model. The CI of GVHD, NRM, and relapse were determined using the competing risks method (i.e., competing risks for GVHD: relapse and death; NRM: relapse and death) and associations with HLA-DPB1 mismatching were evaluated by proportional subdistribution hazards models.10 Additional factors considered were age in years (>50 vs. ≤ 50), race (others vs. Caucasian), gender (male vs. female), HLA-DPB1 mismatching direction (host versus graft vs. graft versus host), CMV donor (D)/recipient (R) group (D+/R-, D-/R+, D+/R+ vs. D-/R-), transplantation year (2005–2008 vs. 2009–2011), conditioning regimens (myeloablative vs. non-myeloablative), ATG use (yes vs. no), and disease status at transplant (no CR vs. CR). Since CMV reactivation (yes vs. no) occurred after transplantation, it was included in the hazards models as a time-dependent covariate. Statistically significant factors in univariate analyses that were associated with the outcome at P ≤ 0.05 were included in the final multivariable models. The effect of HLA-DPB1 mismatching on transplant outcomes was evaluated in the whole cohort. To analyze the effect of recipient CMV serostatus on HLA-DPB1 related aGVHD, we compared the CI of aGVHD in HLA-DPB1 mismatch/CMV seropositive recipient with HLA-DPB1 mismatch/CMV seronegative recipients. Additionally to analyze the effect of CMV reactivation on HLA-DPB1 mismatch related aGVHD, we compared the CI of aGVHD in HLA-DPB1 mismatch/CMV reactivated recipients with HLA-DPB1 mismatch/no CMV reactivated recipients. For all analysis CMV reactivation was defined as presence of > 1pp65 Ag cells/million WBC’s except for a single analysis testing if CMV reactivation increases the chance of aGVHD in which CMV reactivation was defined as positive when the first day of CMV antigenemia occurred before or up to 7 days after the first day of aGVHD. For the purpose of this analysis, of the 270 with CMV reactivation, 178 (66%) were considered not CMV reactivated because it was discovered more than 7 days after the first day of aGVHD. Lastly, a landmark analysis was produced at day 100 to compare differences in OS, relapse, and NRM between patients who experienced CMV reactivation before 100 days and those who did not. For OS, differences between groups were assessed using the log-rank test while differences between groups for relapse and NRM were assessed using Gray’s test.11 All statistical analyses were performed using SAS 9.3 (SAS Institute Inc., Cary, NC) and StataCorp 2013 (College Station, TX: StataCorp LP).
RESULTS
Characteristics of the patients and transplantation
Total of 613 patients with AML and MDS who underwent allo-HCT at MDACC from 2005–2011 were included. Table 1 shows patient and clinical characteristics. From the whole group 529 patients were CMV seropositive and 84 patients were CMV seronegative. Among CMV seropositive recipients 220 patients (42%) were mismatched at HLA-DPB1 loci and in CMV seronegative recipients 30 patients (36%) were found to be mismatched at HLA-DPB1 loci. The rates of CMV seropositivity among HLA-DPB1 mismatched and HLA-DPB1 matched recipients were 88%, and 86%, respectively.
Table 1.
Patient and Clinical Characteristics – All Patients and by Recipient CMV Serostatus
| Measure | All (N=613) | R+ (N=529) | R− (N=84) |
|---|---|---|---|
| Diagnosis | |||
| AML | 464 (76) | 413 (78) | 51 (61) |
| MDS | 149 (24) | 116 (22) | 33 (39) |
| HLA DPB1 matching status, n (%) | |||
| Yes | 363 (59) | 309 (58) | 54 (64) |
| No | 250 (41) | 220 (42) | 30 (36) |
| HLA DPB1 mismatching direction, n (%) | |||
| GvH | 37 (15) | 29 (13) | 8 (27) |
| HvG | 17 (7) | 13 (6) | 4 (13) |
| Both | 196 (78) | 178 (81) | 18 (60) |
| Gender, n (%) | |||
| Male | 346 (56) | 293 (55) | 53 (63) |
| Female | 267 (44) | 236 (45) | 31 (37) |
| Age at allo-SCT (years) | |||
| Mean | 53.0 | 52.6 | 55.4 |
| Standard deviation | 12.4 | 12.4 | 11.8 |
| Median | 55.6 | 55.2 | 57.2 |
| Minimum, Maximum | 19.6, 77.0 | 19.6, 74.4 | 20.5, 77.0 |
| Race/Ethnicity, n (%) | |||
| White | 495 (81) | 415 (78) | 80 (95) |
| Hispanic | 69 (11) | 67 (13) | 2 (2) |
| Black | 21 (3) | 20 (4) | 1 (1) |
| Asian | 12 (2) | 12 (2) | 0 |
| Other | 1 (0.2) | 1 (0.2) | 0 |
| Unknown | 15 (2) | 14 (3) | 1 (1) |
| Transplant type, n (%) | |||
| MRD | 297 (48) | 255 (48) | 42 (50) |
| MUD | 316 (52) | 274 (52) | 42 (50) |
| Conditioning regimen, n (%) | |||
| Myeloablative | 497 (81) | 430 (81) | 67 (80) |
| Non myeloablative | 116 (19) | 99 (19) | 17 (20) |
| Conditioning regimen type, n (%) | |||
| Fludarabine+Busulfan+/−ATG | 376 (61) | 321 (61) | 55 (65) |
| Fludarabine+Melphalan+/−ATG | 83 (14) | 71 (13) | 12 (14) |
| Other | 154 (25) | 137 (26) | 17 (20) |
| In vivo T-cell depletion (ATG) | |||
| Yes | 313 (51) | 273 (52) | 40 (48) |
| No | 300 (49) | 256 (48) | 44 (52) |
| CMV risk groups, n (%) | |||
| D+/R+ | 269 (44) | 269 (51) | 0 |
| D−/R− | 50 (8) | 0 | 50 (60) |
| D+/R− | 34 (6) | 0 | 34 (40) |
| D−/R+ | 260 (42) | 260 (49) | 0 |
| Transplant cell source, n (%) | |||
| PBMC | 432 (70) | 369 (70) | 63 (75) |
| Marrow | 181 (30) | 160 (30) | 21 (25) |
| Transplant year, n (%) | |||
| 2005–2008 | 285 (46) | 242 (46) | 43 (51) |
| 2009–2011 | 328 (54) | 287 (54) | 41 (49) |
| Disease risk, n (%) | |||
| Low | 34 (6) | 29 (6) | 5 (6) |
| Int | 175 (29) | 155 (29) | 20 (24) |
| Int-1 | 45 (7) | 33 (6) | 12 (14) |
| Int-2 | 52 (9) | 41 (8) | 11 (13) |
| High | 304 (50) | 268 (51) | 36 (43) |
| Disease status at transplant, n (%) | |||
| Complete response | 333 (54) | 294 (56) | 39 (46) |
| Refractory | 205 (33) | 178 (34) | 27 (32) |
| Untreated | 73 (12) | 56 (11) | 17 (20) |
| Not evaluated | 1 (0.2) | 0 | 1 (1) |
| Unknown | 1 (0.2) | 1 (0.2) | 0 |
| GVHD prophylaxis, n (%) | |||
| Methotrexate+Tacrolimus | 562 (92) | 490 (93) | 72 (86) |
| Methotrexate+Tacrolimus+Others | 0 | 0 | 0 |
| MMF+Tacrolimus | 3 (0.5) | 2 (0.4) | 1 (1) |
| Other | 47 (8) | 37 (7) | 10 (12) |
| None | 1 (0.2) | 0 | 1 (1) |
Abbreviations: R+ = CMV seropositive Recipient; R- = CMV seronegative Recipient; AML = Acute Myelogenous Leukemia; MDS = Myelodysplastic Syndrome; GvH = Graft vs. Host; HvG = Host vs. Graft; MRD = Matched Related Donor; MUD = Matched Unrelated Donor; D = Donor; R = Recipient; PBMC = Peripheral Blood Mononuclear Cells.
CMV reactivation
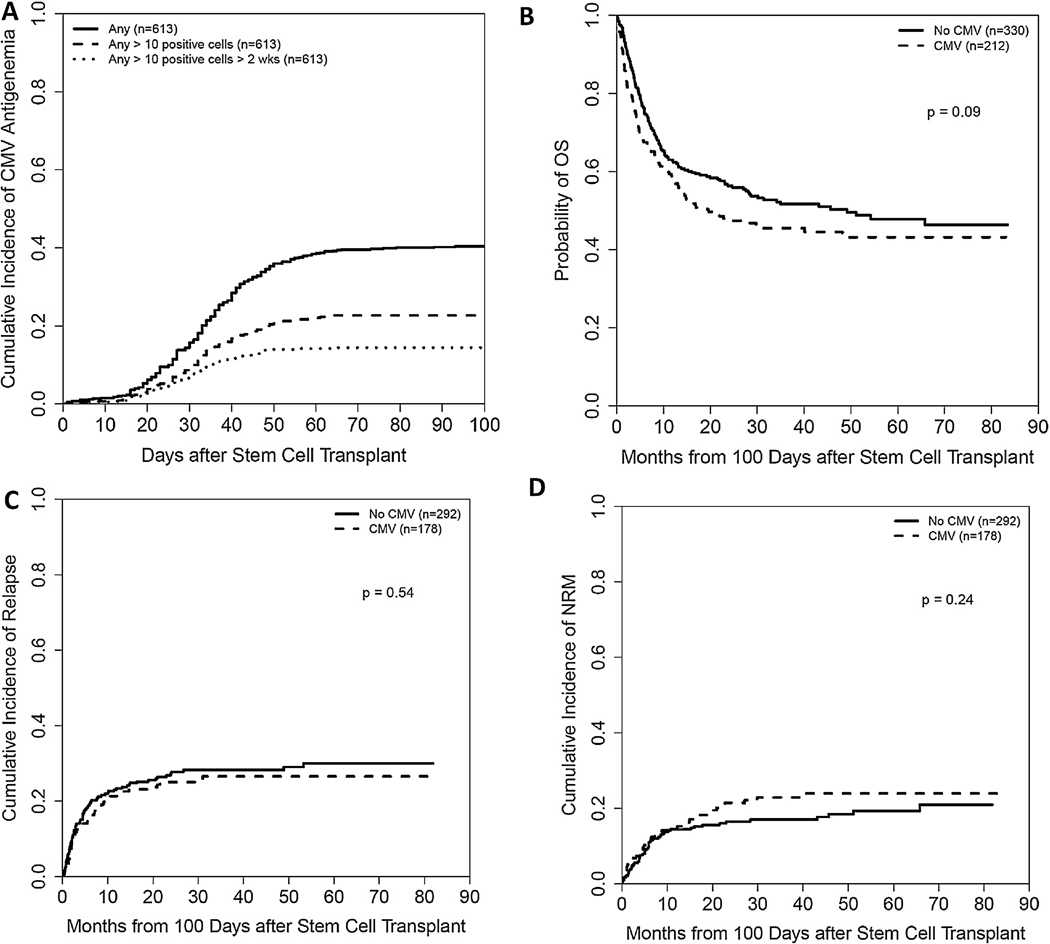
The rates of CMV reactivation in CMV seropositive and CMV seronegative recipients were 49% and 12%, respectively. Figure 1A shows CI of any CMV antigenemia, CMV antigenemia >10 positive cells of any duration and CMV antigenemia >10 positive cells more than 2 weeks. Sixteen out of 613 patients developed CMV disease (3%).
Figure 1.
(A) Cumulative incidence of CMV Antigenemia. (B, C, and D) Landmark analysis at day 100 comparing differences in OS, CI of relapse and NRM between patients who experienced CMV reactivation before day 100 and those who did not.
HLA-DPB1 and transplant outcomes
In multivariable analysis (Table 2) of the whole cohort, HLA-DPB1 mismatching was associated with increased risk of aGVHD (hazard ratio [95% confidence interval]) (1.53 [1.24, 1.90]; P<0.001), decreased risk of relapse (0.73 [0.55, 0.98]; P=0.034), and no effect on OS (1.01 [0.81, 1.26]; P=0.91). This multivariable analysis was adjusted for baseline covariates that were significant in the univariate models in addition to CMV reactivation and HLA-DPB1 mismatching direction (host versus graft vs. graft versus host). These results suggest that increased rate of aGVHD in HLA-DPB1 mismatch group was independent of CMV reactivation. Interaction analysis including CMV reactivation (yes/no) and HLA-DPB1 mismatching (yes/no) showed no statistically significant (p=0.68) differences in CI of aGVHD in four groups, confirming no apparent interaction between HLA-DPB1 and CMV reactivation. Among HLA-DPB1 mismatched group, the CI of grade II-IV aGVHD was 41% in CMV seropositive and 40% in CMV seronegative recipients (P=0.95) suggesting that CMV seropositivity of recipients had no additional effect on the risk of grade II-IV aGVHD in HLA-DPB1 mismatch transplant.
Table 2.
Multivariable analysis of associations between Outcomes and HLA-DPB1 matching status (mismatched vs. matched) and CMV reactivation (yes vs. no).
| Whole cohort (613 patients) | HR (95% CI) | p-value |
|---|---|---|
| HLA-DPB1 matching status (mismatched vs. matched) | ||
| aGVHD | 1.53 (1.24, 1.90) | <0.001 |
| Grade II-IV aGVHD | 1.74 (1.32, 2.31) | <0.001 |
| cGVHD | 0.65 (0.50, 0.86) | 0.002 |
| NRM | 1.25 (0.90, 1.74) | 0.19 |
| CIR | 0.73 (0.55, 0.98) | 0.034 |
| OS | 1.01 (0.81, 1.26) | 0.91 |
| OS | 1.05 (0.83, 1.33) | 0.68 |
| CMV reactivation (yes vs. no) | ||
| aGVHD | 5.88 (4.62, 7.49) | <0.001 |
| Grade II-IV aGVHD | 4.25 (3.04, 5.93) | <0.001 |
| cGVHD | 1.17 (0.81, 1.69) | 0.40 |
| NRM | 0.82 (0.50, 1.34) | 0.43 |
| CIR | 1.26 (0.87, 1.81) | 0.22 |
| OS | 1.06 (0.79, 1.43) | 0.69 |
Abbreviations: HR = hazard ratio; aGVHD = acute graft versus host disease; cGVHD = chronic graft versus host disease; NRM = non-relapse mortality; CIR = cumulative incidence of relapse; OS = overall survival.
CMV reactivation effects on transplant outcomes
In multivariable analysis including ATG use and HLA-DPB1 matching status in the model, CMV reactivation was associated with increased risk of all grade aGVHD (5.88 [4.62, 7.49]; P<0.001] and grade II-IV aGVHD (4.25 [3.04, 5.93]; P<0.001) but had no statistically significant effects on chronic GVHD, relapse, NRM, and OS (Table 2). In the day 100 landmark analysis, CMV reactivation had no statistically significant effects on relapse, NRM, and OS (Figure 1B, 1C, and 1D). Additionally, CMV reactivation was associated with increased incidence of grade II-IV aGVHD in both HLA-DPB1 matched (3.33 [1.91, 5.80]; P<0.001) and HLA-DPB1 mismatched (4.38 [2.90, 6.63]; P<0.001) group suggesting that increased risk of aGVHD in CMV reactivated patients was independent of HLA-DPB1 matching status.
DISCUSSION
This study assesses impacts of HLA-DPB1 and CMV reactivation on GVHD, relapse, and OS after allo-HSCT. Here we demonstrate that HLA-DPB1 and CMV reactivation both increase aGVHD risk independently. Consistent with reported data, we show that HLA-DPB1 mismatching is associated with increased risk of aGVHD and lower relapse resulting in no effects on OS. In contrast to recently published data,12, 13 our study showed that CMV reactivation has no impact on relapse or OS after allo-HSCT.
Stevanov’c et al. had suggested that CMV reactivation induces HLA-DPB1 expression in GVHD targets as a result of inflammatory conditions created by immune response to CMV.4, 6 Our data suggest that increased risk of aGVHD in HLA-DPB1 mismatched allo-HCT cohorts is independent of CMV serostatus or CMV reactivation. One explanation for this discrepancy is that CMV infection is not the only factor that can provide an inflammatory milieu after allo-HCT. Inflammation post allo-HCT can be created by myeloablative conditioning, sepsis, and many types of viral infections.14 It is well known that HLA-DPB1 mismatching is one but by far not the only potential target of alloreactivity causing GVHD. It is thus likely that up-regulation of these targets themselves, and/or of the relevant HLA restriction elements presenting them, could be at the basis of increased GVHD risk in the HLA-DPB1 matched setting. Additionally, Petersdorf et al recently reported that rs9277534G and rs9277534A alleles are associated with high and low expression of HLA-DPB1 respectively.15 One limitation of our study is that we did not assess the frequency of these alleles nor evaluate permissive versus non permissive status of HLA-DPB1 mismatching. The clinical association between HLA-DPB1 mismatches and transplant outcomes has been shown to be different in the T cell epitope group permissive vs non-permissive setting. 16–19 Another limitation of our study is that we included both matched related and unrelated allo-allo-HCTs. There are significant immunogenetic differences other than HLA-DPB1 matching status between a genotypically HLA-identical sibling transplant and a phenotypically HLA-identical matched unrelated donor transplant. In our study, matched related donor allo-HCT comprised 79% and 4% of HLA-DPB1 matched and HLA-DPB1 mismatched groups. In the other word majority of HLA-DPB1 matched group were matched related allo-HCTs while majority of HLA-DPB1 mismatched groups were matched unrelated allo-HCTs. These two measures were highly correlated (Pearson correlation=0.7), and as such, were not included together in the multivariable analysis. Surprisingly in our study patients with HLA-DP mismatch group had lower risk of chronic GVHD (Table 2). To our knowledge this has not been reported in previous studies.1, 2, 20
There has been a notion of “CMV vs. leukemia effect” since the 1980s and recent studies by Elmaagacli et al and Green et al suggested a beneficial effect of CMV reactivation on leukemia relapse in patients with AML.12, 13 In our study, CMV reactivation had no effect on relapse after allo-HSCT. This is consistent with a recent CIBMTR study by Teira et al showing no CMV versus leukemia effect in a large multi-institutional study.21 We assessed CMV reactivation by CMV pp65 antigen assay that is less sensitive and reproducible than CMV PCR assay22 and may have resulted in underestimation of CMV reactivation in this study. CMV disease was rare in our study (3%) that is similar to other studies reporting CMV disease in patients receiving preemptive CMV treatment.23
In conclusion, our study suggests that excess aGVHD risks noted in HLA-DPB1 mismatched transplants is independent of recipient’s CMV serostatus or CMV reactivation. These results need to be further evaluated in a larger multi-institutional study preferably including only matched unrelated donor transplants.
ACKNOWLEDGEMENTS
A.G. was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number KL2 TR002346 (principle investigator: Victoria J. Fraser).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Petersdorf EW. Optimal HLA matching in hematopoietic cell transplantation. Current opinion in immunology 2008. October; 20(5): 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw BE, Gooley TA, Malkki M, Madrigal JA, Begovich AB, Horowitz MM, et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood 2007. December 15; 110(13): 4560–4566. [DOI] [PubMed] [Google Scholar]
- 3. http://hla.alleles.org/nomenclature/stats.html.
- 4.Stevanovic S, van Bergen CA, van Luxemburg-Heijs SA, van der Zouwen B, Jordanova ES, Kruisselbrink AB, et al. HLA class II upregulation during viral infection leads to HLA-DP-directed graft-versus-host disease after CD4+ donor lymphocyte infusion. Blood 2013. September 12; 122(11): 1963–1973. [DOI] [PubMed] [Google Scholar]
- 5.Ciurea SO, Thall PF, Wang X, Wang SA, Hu Y, Cano P, et al. Donor-specific anti-HLA Abs and graft failure in matched unrelated donor hematopoietic stem cell transplantation. Blood 2011. November 24; 118(22): 5957–5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peggs KS. Attack of the T-cell clones. Blood 2013. September 12; 122(11): 1847–1848. [DOI] [PubMed] [Google Scholar]
- 7.Cantoni N, Hirsch HH, Khanna N, Gerull S, Buser A, Bucher C, et al. Evidence for a Bidirectional Relationship between Cytomegalovirus Replication and acute Graft-versus-Host Disease. Biol Blood Marrow Tr 2010. September; 16(9): 1309–1314. [DOI] [PubMed] [Google Scholar]
- 8.Nachbaur D, Bonatti H, Oberaigner W, Eibl B, Kropshofer G, Gast G, et al. Survival after bone marrow transplantation from cytomegalovirus seropositive sibling donors. Lancet 2001. October 6; 358(9288): 1157–1159. [DOI] [PubMed] [Google Scholar]
- 9.Glucksbe H, Storb R, Fefer A, Buckner CD, Neiman PE, Clift RA, et al. Clinical Manifestations of Graft Versus Host Disease in Human Recipients of Marrow from Hl-a-Matched Sibling Donors. Transplantation 1974; 18(4): 295–304. [DOI] [PubMed] [Google Scholar]
- 10.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 1999. June; 94(446): 496–509. [Google Scholar]
- 11.Gray RJ. A Class of K-Sample Tests for Comparing the Cumulative Incidence of a Competing Risk. Ann Stat 1988. September; 16(3): 1141–1154. [Google Scholar]
- 12.Elmaagacli AH, Koldehoff M. Cytomegalovirus replication reduces the relapse incidence in patients with acute myeloid leukemia. Blood 2016. July 21; 128(3): 456–459. [DOI] [PubMed] [Google Scholar]
- 13.Green ML, Leisenring WM, Xie H, Walter RB, Mielcarek M, Sandmaier BM, et al. CMV reactivation after allogeneic HCT and relapse risk: evidence for early protection in acute myeloid leukemia. Blood 2013. August 15; 122(7): 1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohty M, Blaise D, Faucher C, Vey N, Bouabdallah R, Stoppa AM, et al. Inflammatory cytokines and acute graft-versus-host disease after reduced-intensity conditioning allogeneic stem cell transplantation. Blood 2005. December 15; 106(13): 4407–4411. [DOI] [PubMed] [Google Scholar]
- 15.Petersdorf EW, Malkki M, O’HUigin C, Carrington M, Gooley T, Haagenson MD, et al. High HLA-DP Expression and Graft-versus-Host Disease. The New England journal of medicine 2015. August 13; 373(7): 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pidala J, Lee SJ, Ahn KW, Spellman S, Wang HL, Aljurf M, et al. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood 2014. October 16; 124(16): 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischhauer K, Shaw BE, Gooley T, Malkki M, Bardy P, Bignon JD, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol 2012. April; 13(4): 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zino E, Frumento G, Marktel S, Sormani MP, Ficara F, Di Terlizzi S, et al. A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood 2004. February 15; 103(4): 1417–1424. [DOI] [PubMed] [Google Scholar]
- 19.Crocchiolo R, Zino E, Vago L, Oneto R, Bruno B, Pollichieni S, et al. Nonpermissive HLA-DPB1 disparity is a significant independent risk factor for mortality after unrelated hematopoietic stem cell transplantation. Blood 2009. August 13; 114(7): 1437–1444. [DOI] [PubMed] [Google Scholar]
- 20.Morishima Y, Kashiwase K, Matsuo K, Azuma F, Morishima S, Onizuka M, et al. Biological significance of HLA locus matching in unrelated donor bone marrow transplantation. Blood 2015. February 12; 125(7): 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teira P, Battiwalla M, Ramanathan M, Barrett AJ, Ahn KW, Chen M, et al. Early cytomegalovirus reactivation remains associated with increased transplant-related mortality in the current era: a CIBMTR analysis. Blood 2016. May 19; 127(20): 2427–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allice T, Cerutti F, Pittaluga F, Varetto S, Franchello A, Salizzoni M, et al. Evaluation of a novel real-time PCR system for cytomegalovirus DNA quantitation on whole blood and correlation with pp65-antigen test in guiding pre-emptive antiviral treatment. J Virol Methods 2008. March; 148(1–2): 9–16. [DOI] [PubMed] [Google Scholar]
- 23.Hakimi Z, Ferchichi S, Aballea S, Odeyemi I, Toumi M, English M, et al. Burden of cytomegalovirus disease in allogeneic hematopoietic cell transplant recipients: a national, matched cohort study in an inpatient setting. Curr Res Transl Med 2018. November; 66(4): 95–101. [DOI] [PubMed] [Google Scholar]